Abstract
Burkholderia contaminans is a member of the Burkholderia cepacia complex (Bcc), a pathogen with increasing prevalence among cystic fibrosis (CF) patients and the cause of numerous outbreaks due to the use of contaminated commercial products. The antibiotic resistance determinants, particularly β-lactamases, have been poorly studied in this species. In this work, we explored the whole genome sequence (WGS) of a B. contaminans isolate (FFH 2055) and detected four putative β-lactamase-encoding genes. In general, these genes have more than 93% identity with β-lactamase genes found in other Bcc species. Two β-lactamases, a class A (Pen-like, suggested name PenO) and a class D (OXA-like), were further analyzed and characterized. Amino acid sequence comparison showed that Pen-like has 82% and 67% identity with B. multivorans PenA and B. pseudomallei PenI, respectively, while OXA-like displayed strong homology with class D enzymes within the Bcc, but only 22–44% identity with available structures from the OXA family. PCR reactions designed to study the presence of these two genes revealed a heterogeneous distribution among clinical and industrial B. contaminans isolates. Lastly, blaPenO gene was cloned and expressed into E. coli to investigate the antibiotic resistance profile and confers an extended-spectrum β-lactamase (ESBL) phenotype. These results provide insight into the presence of β-lactamases in B. contaminans, suggesting they play a role in antibiotic resistance of these bacteria.
Introduction
Cystic fibrosis (CF) is a genetic disorder that affects approximately 70,000–100,000 people worldwide, imposing a life-limiting condition to patients harboring this disorder [1]. In persons with CF, Pseudomonas aeruginosa is one of the most important pathogens that can cause life-threatening infections. Regrettably, many other bacterial genera have also emerged as significant pathogens in persons with CF [2]. Moreover, prolonged antibiotic therapy received by these patients is a propitious scenario for the evolution of antibiotic resistant pathogens [3].
The Burkholderia cepacia complex (Bcc) is composed of >20 closely related species that are opportunistic human pathogens that mainly have a significant impact in patients with CF [4]. The Bcc complex is resistant to a wide range of antibiotics, and therefore infections caused by these bacteria are very difficult to eradicate. Strikingly, infections may derive into “cepacia syndrome” with fatal consequences (necrotizing pneumonia, respiratory failure, and sepsis/bacteremia) [5–7]. An additional negative aspect of the Bcc is their ability to contaminate man-made products such as pharmaceuticals, cosmetics, and disinfectants [4, 8, 9]. Nosocomial outbreaks due to the use of Bcc contaminated products are often reported [10].
Since B. cenocepacia and B. multivorans are the most prevalent Bcc among persons with CF in the United States, Canada, France, United Kingdom, and Belgium, these Bcc species are well studied and characterized [11–13]. However, in recent years, B. contaminans, a species defined in 2009, has been widely identified as a nosocomial opportunistic pathogen of rising importance due to outbreaks that took place in different parts of the world [14]. Presently, B. contaminans is reported as the most frequent Bcc species isolated in CF patients from Portugal, Spain, and Argentina, and is associated with infections in immunocompromised and hospitalized patients exposed to contaminated products in Germany and United States [15–20]. Due to its recent speciation, few studies are available on B. contaminans, and little is known about its virulence factors, susceptibility to antibiotics, prognosis of infected patients, and other issues of clinical relevance [21]. Recently, whole genome sequencing was performed on several B. contaminans strains and these sequences are available in the GenBank database [22–24].
As with all Bcc members, B. contaminans possesses innate resistance to a wide range of antimicrobials; as a result, the clinical treatment of infections is problematic. The main antibiotic resistance mechanisms described in Bcc include the unique structure of the lipopolysaccharide (LPS) in the outer membrane, the presence of efflux pumps, the degradation of antibiotics by means of enzymes such as β-lactamases, and the ability to form biofilms [25]. Adding to this, Bcc displays the possibility to acquire even more antibiotic resistant determinants through horizontal gene transfer [26, 27].
Among the antibiotic resistance mechanisms mentioned above, β-lactamases are not well characterized in Bcc. Inducible chromosomal class A β-lactamases (PenA and PenB) were investigated in B. cenocepacia and B. multivorans and are reported in other Bcc members [28–32]. Most studies focused on studying this single class A β-lactamase, as it appears to be the dominant β-lactam resistance mechanism in Bcc. However, the genomes of Bcc harbor other putative β-lactamase (bla) genes. For instance, in the study describing the whole genome of B. cenocepacia J2315, at least four bla genes encoding potential β-lactamases were found though their activity is not yet established [33]. With regard to B. contaminans, studies on β-lactamases have not been conducted. The presence of genes associated with β-lactamases is mentioned in a work describing the genome of B. contaminans MS14, an isolate obtained from soil, but the number of genes and the characterization of these enzymes was not pursued [22].
In this work, we explore the genome of the B. contaminans strain FFH 2055 that was isolated from the sputum of a person with CF in Argentina [34]. We determined the presence of four putative β-lactamase genes in B. contaminans strain FFH 2055 and conducted comparative analyses of the bla genes with other Bcc and non-Bcc species. The presence of select bla genes was further explored within 46 B. contaminans isolates. In addition, molecular models were generated of the class D OXA β-lactamase as well as the class A Pen-like β-lactamase, which based on previous nomenclature in the Bcc field, will be denoted as PenO [35]. As the Pen-like β-lactamases are the dominant β-lactam resistance mechanism in Burkholderia spp., the antibiotic resistance profile of PenO was further evaluated by cloning and expressing blaPenO in Escherichia coli.
Materials and Methods
Bacterial Strain and Genome Sequences
Burkholderia contaminans FFH 2055 was isolated from sputum of a 6-year-old girl with CF in Argentina. The patient was chronically infected with this species until her death at the age of 12. The strain was preserved at − 80 °C in Tryptic Soy Broth (TSB) with 40% glycerol for future studies. The draft genome of FFH 2055 was recently published [34]. Sequences were retrieved from GenBank (accession LASC01000001 to LASC01000008) and analyzed to search for putative bla genes using the RAST server [36, 37]. Also, 46 B. contaminans isolates obtained from persons with CF (n = 21), non-CF patient comparators (n = 13), and industrial contaminated products (n = 12) were employed to investigate the distribution of blaPenO and blaOXA-like genes (Table 1).
Table 1.
Putative β-lactamases in B. contaminans FFH2055 and % identity to non-Bcc and Bcc species
| Putative β-lactamase genes in B. contaminans FFH2055 Locus tag (bp length) |
Amino acid comparison in GenBank Best match % of identity accession product |
Nucleotide comparison in Burkholderia.com % identity Locus tag |
||
|---|---|---|---|---|
| Best match | Identity with B. cenocepacia J2315 | Identity with B. multivorans ATCC 17676 | ||
| Class A β-lactamase WR30_RS27340 (939) |
Jeongeupia sp. USM3 65% WP_070527757.1 Class A β-lactamase |
B. cepacia MSMB1339WGS MSMB1339WGS_28 93.31% WK01_RS09215 |
89.53% BCAM2165 |
82.44% Bmul_3689 |
| Class B β-lactamase WR30_RS21040 (1014) |
Amantichitinum ursilacus 55% WP_053936944.1 MBL fold metallo-hydrolase |
B. vietnamiensis FL-7–710-S2-D0 79.70% WJ18_RS07505 |
72.16% BCAS0557 |
– |
| Class C β-lactamase WR30_RS20060 (1170) |
Vogesella sp. LIG4 63% WP_088965050.1 Class C β-lactamase |
B. lata sp. 383 96.83% BCEP18194_RS04915 |
89.64% BCAS0156 |
81.89% Bmul_6008 |
| Class D β-lactamase WR30_RS31065 (810) |
Klebsiella michiganensis 61% WP_065366333.1 class D β-lactamase |
B. cepacia MSMB2211WGS_28 94.69% WL58_RS12165 |
87.79% BCAM0393 |
– |
Bioinformatics Analysis
The nucleotide and translated amino acid sequences of the putative β-lactamase-encoding genes in B. contaminans FFH 2055 were investigated in genome sequences of other B. contaminans strains as well as other Bcc species by performing BLAST N searches in GenBank and the Burkholderia.com database [38]. A gene encoding a Pen-like class A β-lactamase (WR30_RS27340, named blaPenO) was selected for further studies. Translated amino acid sequences were analyzed using NCBI BLAST X 2.7.0+ [39]. Sequence and structural alignments of amino acid sequences were performed using T-Coffee Expresso and ESPript/ENDscript [40, 41].
Protein Structure Prediction and Molecular Modeling of Hypothetical Pen‑Like and OXA‑Like β‑Lactamases
A prediction of the α-helical content of PenO was performed with AGADIR software [42], and theoretical three-dimensional (3D) models for the enzyme were predicted by using the Swiss-Model tool (http://swissmodel.expasy.org) and Yasara, using a standard homology modeling protocol [43]. All models were visualized with PyMOL 1.6 [44].
General Molecular Biology Techniques
The blaPenO gene was amplified by PCR reaction using primers listed in Table S1 and cloned into the pBC SK(+) vector (CHLR) (Adgene). E. coli TOP10 cells were then transformed with the pBC SK(+) blaPenO (pblapenO) and antibiotic susceptibility tests were performed (see below).
In addition, PCR reactions were used to detect the blaPenO and blaOXA-like genes among a collection of B. contaminans isolates. Primers employed in these reactions were designed using Primer 3 software (v 0.4.0) and are listed in Table S1 [45, 46]. B. contaminans isolates were suspended in sterile distilled water, boiled for 10 min, and centrifuged at 14000 rpm for 5 min. A 5 μl bacterial suspension was used to carry out PCR reactions. Cycling conditions were 94 °C for 2 min; 30 cycles of 94 °C for 1 min, annealing temperature for 1 min, and 72 °C for 1 min; and finally, 72 °C for 10 min.
Antimicrobial Susceptibility Testing
Disk diffusion assays with various β-lactam antibiotics (AMP: ampicillin; AMC: amoxicillin-clavulanate; CTX: cefotaxime; CRO: ceftriaxone; CAZ: ceftazidime; CZ: cefazolin; FEP: cefepime; FOX: cefoxitin; IMP: imipenem; MEM: meropenem) were performed following the Clinical and Laboratory Standards Institute (CLSI) guidelines (Table 2) [47]. The disk diffusion tests served as a screening tool to determine potential substrates for the enzyme. Minimum inhibitory concentrations (MIC) by E test for AMP, CAZ, MEM, and IMP were also performed as recommended.
Table 2.
Origin, source, and distribution of blaPenO and blaOXA-like genes among B. contaminans isolates
| Isolates |
Distribution of blaPenO and blaOXA-like |
||||
|---|---|---|---|---|---|
| Origin (number of isolates) | Source (number of isolates) |
blaPenO+ blaOXA-like+ |
blaPenO− blaOXA-like+ |
blaPenO+ blaOXA-like− |
blaPenO− blaOXA-like− |
| CF patients (21) | Children’s Hospital Juan P. Garrahan (9) | 8 | 0 | 0 | 1 |
| Children’s Hospital Ricardo Gutierrez (12) | 10 | 1 | 0 | 1 | |
| Non-CF patients (13) | University Hospital José de San Martín (5) | 5 | 0 | 0 | 0 |
| Hospital Ramos Mejía (5) | 4 | 0 | 1 | 0 | |
| Children’s Hospital Ricardo Gutierrez (3) | 3 | 0 | 0 | 0 | |
| Industrial (12) | Purified water and facilities (5) | 4 | 0 | 0 | 1 |
| Pharmaceutical products (3) | 1 | 1 | 0 | 1 | |
| Ultrasound gel (1) | 1 | 0 | 0 | 0 | |
| Disinfectants and housecleaners (3) | 1 | 1 | 1 | 0 | |
Results
Putative β‑Lactamases Present in the B. contaminans FFH 2055 Genome
The total number of putative bla genes in B. contaminans FFH 2055 genome was eighteen. However, non-β-lactamases, such as hydrolases and proteases, possess similar structures/amino acid sequences to β-lactamases; thus, proteins are often annotated as β-lactamases, even though they are not actually β-lactamases. As a result, a secondary analysis for the presence of a signal peptide as well as conserved β-lactamase motifs in translated amino acid sequences was conducted. As β-lactamases are periplasmic enzymes, they must possess a signal peptide (analysis tool: SignalP 4.1 Server). Analysis of the signal peptides in the 18 putative β-lactamases reduced the list to 5 β-lactamases (two class A enzymes, one class B, one class C, and one class D). Each β-lactamase class possesses conserved motifs that can be used to further identify potential β-lactamases. The following motifs should be present within each class: class A: SXXK motif, SDN loop, RXEXXXXXXXXXXXXD Ω-loop, and KTG motif; class B: HXHXD motif; class C: SXXK motif, YSN motif, and KTG motif; and class D: SXXK motif and KTG motif. Of the two class A β-lactamases, one lacked the SXXK, SDN, and KTG motifs, and thus was removed from the list.
As a result of this analysis, four putative bla genes were identified in B. contaminans FFH 2055. These genes encode for putative class A Pen-like (WR30_RS27340), class B (WR30_RS21040), class C (WR30_RS20060), and class D OXA (WR30_RS31065) β-lactamases (Table 1). The nucleotide sequences of WR30_RS27340 and WR30_RS20060 were found in all other B. contaminans genomes currently deposited in GenBank (n = 17) with 95–100% identity. Meanwhile, WR30_RS21040 and WR30_RS31065 were detected in one and 12 genomes (100% and 95–100% identity), respectively.
Comparative Analysis of the Four β‑Lactamase Amino Acid and Nucleotide Sequences to Non‑Bcc and Bcc
To further investigate these β-lactamases, a comparison to non-Burkholderial as well as Bcc β-lactamases was conducted using the GenBank and Burkholderia.com databases. According to GenBank, the four B. contaminans FHH 2055 β-lactamases were found to be most similar to bacteria (i.e., Jeongeupia sp. USM3, Amantichitinum ursilacus, Vogesella sp. LIG4, and Klebsiella michiganensis) isolated from environmental sources with amino acid sequence identity ranging from 55 to 65% (Table 1). A BLAST search of the Burkholderia.com database demonstrated that three of bla nucleotide sequences shared more than 93% identity with β-lactamase-encoding genes found in other Bcc species (Table 1). Only the gene coding for a putative metallo-β-lactamase (class B β-lactamase) displayed a lower identity with the closest match to a B. vietnamiensis strain (79.7% identity). The highest identity of individual genes was found within B. cepacia and B. lata strains (Table 1).
Furthermore, a BLAST search at amino acid level using the four putative β-lactamases detected in B. contaminans FFH 2055 revealed that two of them (class A and class C β-lactamase) were also found in all the Bcc species available genomes in the GenBank (only B. arboris does not have sequenced genomes submitted) with more than 75% and 40% of coverage and identity, respectively. The Penlike class A β-lactamase is critical for β-lactam resistance in Bcc, while the class C β-lactamase, which was characterized in B. multivorans, may not actually be a β-lactamase, but a low molecular mass penicillin binding protein; further studies are in progress (References: PMID: 30012762 and 29983287). The class B β-lactamase was not detected in B. multivorans, B. dolosa, B. latens, B. metallica, B. seminalis, B. lata, B. metallica, B. pseudomultivorans, B. stagnalis, and B puraquae. With respect to the class D β-lactamases, none were detected in B. vietnamiensis, B. dolosa, B ambifaria, and B. latens (Figure S1). Of the β-lactamases, the class A β-lactamase and class C β-lactamase are most prevalent in Bcc.
As B. cenocepacia and B. multivorans are the most prevalent species in Europe and North America, the four putative β-lactamase genes were compared to the type strain of these species (B. cenocepacia J2315 and B. multivorans ATCC 17616). The four bla genes found in B. contaminans FFH 2055 shared between 70% and 90% identity with genes of B. cenocepacia, while the blaOXA-like and the class B β-lactamase genes were not found in the B. multivorans strain (Table 1). Interestingly, the B. cenocepacia genes that shared similarity with blaPenO and class B β-lactamase were not annotated as β-lactamases when genome of B. cenocepacia J2315 was described [33].
Distribution of blaPenO and blaOXA‑like Genes Among B. contaminans Isolates
We decided to focus our studies on two selected bla genes, named blaPenO and blaOXA-like, coding for a class A β-lactamase and a class D β-lactamase, respectively. PenO was selected, because it is the dominant β-lactamase in Bcc. As evidence to date suggests that the class C β-lactamase may not function as a β-lactamase (PMID: 30012762), the OXA-like β-lactamase was also selected for analysis as it is next most often detected β-lactamase in Bcc.
By PCR reactions, blaPenO and blaOXA-like genes were detected in most of the B. contaminans isolates evaluated (84.8% blaPenO positive reactions and 87% blaOXA-like positive reactions). Slight differences were observed considering the origin of the isolates. Both PCR reactions yielded an amplification product in 37 out of 46 isolates (18 from 21 CF isolates, 12 from 13 clinical non-CF isolates, 7 from 12 industrial isolates), while in two isolates collected from CF patients and two clinical isolates, both PCR reactions resulted in negative results (Table 2). The highest percentage of isolates lacking one of these genes was obtained among industrial isolates (33.3% blaPenO and 25% blaOXA-like, respectively). On the other hand, the clinical isolates obtained from non-CF patients accounted the highest percentage of positive amplification for blaPenO and blaOXA-like, with only one isolate displaying a negative amplification result for blaOXA-like. We note that sequence heterogeneity within the blaPenO and blaOXA-like genes may have limited the detection of some of these bla genes in the isolates [35].
Genetic Environment of blaPenO and blaOXA‑like
The surrounding of blaPenO gene displayed homologous sequences from the environment of blapenB previously described in B. cenocepacia (Figure S2) [29]. A gene encoding a putative LysR regulator (PenR) (WR30_RS27345) is located upstream and is transcribed in an opposite orientation of blaPenO. The nucleotide sequence of this gene shares more than 94% identity with LysR encoding genes detected in B. lata, B. cepacia, and B. cenocepacia strains among other Bcc species. The 127-bp intergenic sequence between blaPenO and penR gene also revealed 85% similarity with the intergenic region of B. cenocepacia J2315 displaying identical sequences for the two putative − 10 and − 35 promoter boxes and the two potential ribosome-binding sites described (Fig. S2) [29].
Regarding blaOXA-like genetic environment, it is closely related to strains of other Bcc species like B. cepacia, B. cenocepacia, and B. multivorans, flanked by a putative acyltransferase and a putative porin gene (Fig. S3). Although previous described promoters were not detected in the upstream intergenic region, a BLAST comparison of this 327 bp did not reveal any matches with non-Burkholderia species, but shares more than 80% identity with sequences of B. cenocepacia, B. cepacia, and B. lata strains among other Bcc species, located upstream of a putative class D β-lactamase gene (Fig. S3). In addition, insertion sequences, transposons, or other mobile elements were not detected around blaPenO and blaOXA-like (data not shown).
Amino Acid Sequence Analysis and Molecular Modeling of PenO and OXA‑Like β‑Lactamases
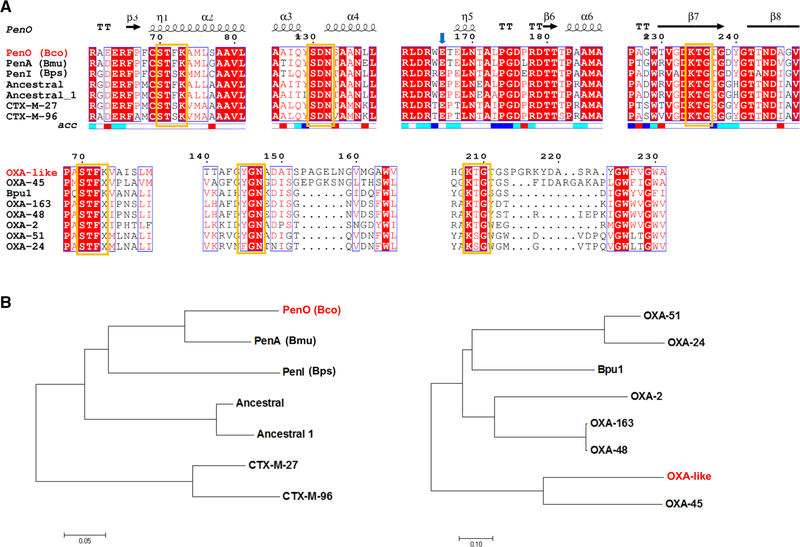
Compared to other class A β-lactamases, B. contaminans PenO enzyme possesses all the conserved residues and motifs that model the active site (Fig. 1a): motif 1 containing the active-site serine (S70) residue (STFK); motif 2 (SDN); motif 3 (KTG); and a Ω-loop including the essential glutamic acid residue (E166). According to the amino acid relationship with other class A enzymes (Fig. 1b), B. contaminans PenO possesses 82% and 67% amino acid identity with B. multivorans PenA and B. pseudomallei PenI enzymes, respectively [30], and whose crystallographic structures were also solved (PDB 3W4Q and 3W4O). Regarding other Pen-like β-lactamases described in Bcc members, amino acid identities ranges from 74% (B. ubonensis PenH) to 88% (B. cenocepacia PenB1). In comparison with non-Bcc Burkholderia species, PenO displays identities from 57% (B. gladioli PenN) to 63% (B. thailandensis PenL). PenO also showed 55–57% amino acid identity with extended-spectrum CTX-M β-lactamases, and shares some common residues with PER enzymes, essential for the active-site coordination (see below).
Fig. 1.
a Multiple sequence alignment of PenO and OXA-like and other related class A and D β-lactamases, respectively, were constructed with ESPript 3.0 (http://espript.ibcp.fr/ESPript/ESPript/) and showed the regions containing the conserved active-site residues. Yellow boxes indicate the putative conserved motifs. Secondary structure is provided only for PenO, which possesses high homology with the reference sequence/model. b Neighbor-joining-based tree showing the amino acid sequence relationship of both enzymes with the closest β-lactamases. Sequences included were selected from the closest enzymes according to the Blast result. For those enzymes which are species-specific, the species name is provided in parenthesis: Bco: B. contaminans; Bmu: B. multivorans; Bps: B. pseudomallei. (Color figure online)
On the other hand, B. contaminans OXA-like β-lactamase showed the class D conserved motifs (Fig. 1a): STFK (motif 1) containing the active-site serine (S70); YGN (motif 2); and KTG (motif 3). OXA-like β-lactamase displayed significant homology with class D enzymes within the Bcc, 52–55% amino acid identity with OXA-57 from B. pseudomallei, although only 22–44% amino acid identity with available structures from the OXA family, being closely related to OXA-45 and related enzymes (Fig. 1b).
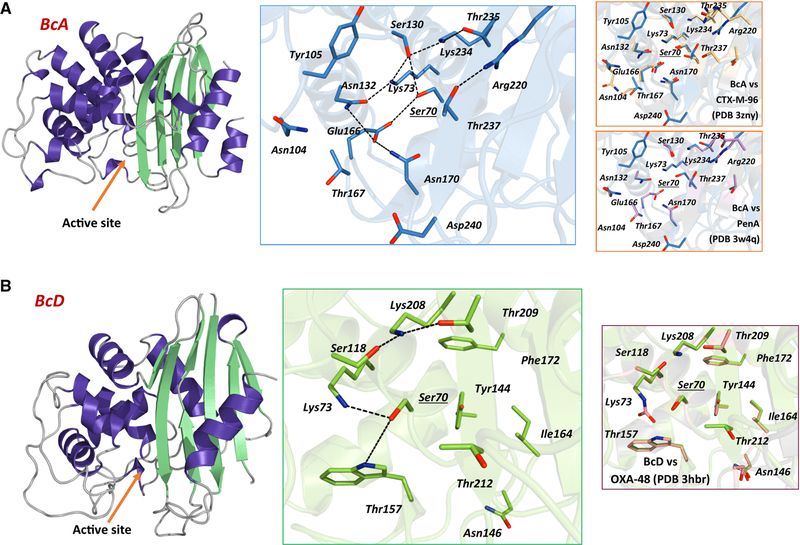
In silico molecular modeling of both PenO and OXA-like showed that these β-lactamases possess the typical overall fold of other class A and D β-lactamases, respectively, being the active-site cavity located between the “all α” and the “α/β” domains (Fig. 2).
Fig. 2.
Left: theoretical molecular model of PenO (up) and OXA-like (down) β-lactamases, indicating the active-site entry (orange arrow); Middle: detail of the active-site coordination, indicating conserved or important residues for the structure and function of class A β-lactamases; right: comparison of PenO vs CTX-M-96 (PDB 3ZNY) and PenA (PDB 3W4Q), and OXA-like vs OXA-48 (PDB 3HBR). (Color figure online)
In PenO, the active site is composed of conserved residues that are also found in other class A β-lactamases (Fig. 2b). Among the most interesting features, the presence of R220 (also found in Pen β-lactamases) and D240 are highlighted. A conserved arginine is found in most clinically important ESBL, i.e., CTX-M (R276), TEM/SHV (R244), and PER (R220) whose guanidinium lateral chain always points towards the active site [48–50]; as in the case of PER β-lactamases, R220 could also create a hydrogen bond network essential for the activity in Pen β-lactamases [51]. An aspartic acid residue at 240 is commonly found in enzymes like the cefotaxime-hydrolyzing CTX-M, which is replaced by a glycine in those variants having a slight “ceftazidimase” behavior [52, 53]. Interestingly, the putative Ω-loop fold seems to create a broader active-site cavity compared to extended-spectrum CTX-M β-lactamases, which together with the residues mentioned above could contribute to an efficient hydrolysis towards some β-lactams. Noteworthy is the presence of a threonine at 167 (also observed in PenA) replacing the conserved proline in most class A β-lactamases, which could influence the folding of the Ω-loop and probably also in the activity.
Remarkably, the OXA-like β-lactamase includes interesting structural features that deserve further analysis. Apart from the conserved residues usually found in class D enzymes (Figs. 1, 2), OXA-like β-lactamase remarkably carries a six-residue insertion after the YGN conserved motif (PAGELN), which is also found in OXA-45 (EPGKSN) [54]. As in other clinically important OXAs, including OXA-48-derived carbapenemases, OXA-like β-lactamase presents several mutations in the β4–β5 loop [55]; as in OXA-45, this “hot spot” region is composed of ten residues compared to five in OXA-48, OXA-24, and OXA-51 carbapenemases, while in OXA-2 and OXA-163 there is only one residue (data not shown).
Antimicrobial Phenotypic Profile of PenO β‑Lactamase
As the Pen-like β-lactamases are present in all Bcc, the antimicrobial resistance profile of PenO β-lactamase was determined. Disk diffusion assays using several β-lactam antibiotics against the parent B. contaminans FHH 2055 and E. coli TOP10 cells containing plasmid, pblaPenO, were determined according to CLSI guidelines [47]. CLSI only provides breakpoints for ceftazidime, meropenem, and ticarcillin-clavulanic acid for Bcc. Thus, β-lactam breakpoints for Enterobacteriaceae were used to assist interpretation of the resistance profiles.
The B. contaminans strain demonstrated resistance towards ampicillin, amoxicillin-clavulanic acid, cefazolin, cefoxitin, cefotaxime, and ceftriaxone (Table 3). Compared to E. coli TOP10 harboring the empty vector, changes in the inhibition zones were observed for ampicillin, amoxicillin-clavulanic acid, cefazolin, cefoxitin, cefotaxime, ceftriaxone, and cefepime, when pblaPenO plasmid was introduced (Table 3). Thus, PenO behaves similarly to an extended-spectrum β-lactamase (ESBL). MIC results were also obtained for the E. coli strains and E. coli with pblaPenO possessed higher MICs towards all the β-lactams tested with the exception of carbapenems (Table 4).
Table 3.
Average inhibition zones (in mm) obtained by disk diffusion tests from E. coli TOP10 derived recombinant cells producing PenO β-lactamase
| AMP | AMC | CTX | CRO | CAZ | CZ | FEP | FOX | IMP | MEM | |
|---|---|---|---|---|---|---|---|---|---|---|
| B. contaminans FFH 2055 | 6 | 6 | 17 | 12 | 26 | 6 | 20 | 6 | 17 | 24 |
| E. coli TOP10 pBC SK+ | 21 | 22 | 36 | 34 | 34 | 25 | 35 | 25 | 30 | 35 |
| E. coli TOP10—pblaPenO | 6 | 12 | 20 | 10 | 26 | 6 | 25 | 21 | 30 | 35 |
Average disk diffusion values were determined from three independent trials
AMP ampicillin, AMC amoxicillin-clavulanate, CTX cefotaxime, CRO ceftriaxone, CAZ ceftazidime, CZ cefazolin, FEP cefepime, FOX cefoxitin, IMP imipenem, MEM meropenem
Table 4.
Minimum inhibitory concentration (MIC) values (μg/ml) for E. coli TOP10 derived recombinant cells producing PenO β-lactamase
| MIC | AMP | AMC | CTX | CRO | CAZ | CZ | FEP | MEM | IMP |
|---|---|---|---|---|---|---|---|---|---|
| E. coli TOP10 pBCSK+ | 4 | 2 | 0.047 | 0.047 | 0.38 | 3 | 0.047 | 0.032 | 0.5 |
| E. coli TOP10—pblaPenO | >256 | 16 | 2 | 6 | 2 | >256 | 0.75 | 0.032 | 0.5 |
Conclusion
Although class A β-lactamases of the Pen family were reported in Bcc species and PenA in B. multivorans was studied and characterized, the knowledge about other β-lactamases in the Bcc is scarce [31, 55]. Particularly, studies are not available about these enzymes in B. contaminans, an emerging opportunistic pathogen that is also frequently recovered from contaminated products. The analysis of B. contaminans FFH 2055 revealed the presence of at least four putative genes coding different classes of these enzymes. A comparative analysis with other Bcc genomes demonstrated the presence, although not homogeneously, of a similar number of putative β-lactamases genes in other Bcc species. A deeper study of two of these genes, blaPenO and blaOXA-like, allowed us to report the relationship with other β-lactamases, the hypothetical structure of its products, and to hypothesize that a heterogeneous distribution of these genes may occur among B. contaminans isolates. According to nomenclature of Pen-like family described in the Burkholderia genus, the name of PenO is suggested for class A β-lactamase detected in B. contaminans. The antibiotic susceptibility testing suggested that PenO is likely an ESBL.
Supplementary Material
Acknowledgements
Research reported in this publication was supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, the Veterans Affairs Merit Review Program BX002872 (KMP-W) and BX001974 (RAB) from the United States (U.S.) Department of Veterans Affairs Biomedical Laboratory Research and Development Service, and the Geriatric Research Education and Clinical Center VISN 10 to RAB. The contents do not represent the views of the U.S. Department of Veterans Affairs or the United States Government. This study was partially supported by National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Numbers R21AI114508, R01AI100560, R01AI063517, and R01AI072219 to RAB. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We want to thank Dr. Ana P. Meo (Hospital Ramos Mejía) and Dr. Carlos Vay (Hospital de Clínicas General San Martín) for providing the non-CF B. contaminans isolates for this study. Special thanks are offered to Dr. German M. Traglia for his technical help in bioinformatics analysis.
Funding This work was funded by grants from Agencia Nacional de Promoción Científica y Tecnológica (PICT 2014–0457 to PP). P. Power is a researcher for the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, Argentina). JF has a SOAR-ELEVAR Scholar Fellowship from Latina/o Graduate Students from the U.S. Department of Education.
Footnotes
Conflict of interest The authors declare that they have no conflict of interest.
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s00284–019-01653–4) contains supplementary material, which is available to authorized users.
Compliance with Ethical Standards
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kelly J (2017) Environmental scan of cystic fibrosis research worldwide. J Cyst Fibros 16(3):367–370. 10.1016/j.jcf.2016.11.002 [DOI] [PubMed] [Google Scholar]
- 2.Gilligan PH (2014) Infections in patients with cystic fibrosis: diagnostic microbiology update. Clin Lab Med 34(2):197–217. 10.1016/j.cll.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sherrard LJ, Tunney MM, Elborn JS (2014) Antimicrobial resistance in the respiratory microbiota of people with cystic fibrosis. Lancet 384(9944):703–713. 10.1016/S0140-S [DOI] [PubMed] [Google Scholar]
- 4.Mahenthiralingam E, Baldwin A, Dowson CG (2008) Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. J Appl Microbiol 104(6):1539–1551. 10.1111/j.1365-2672.2007.03706.x [DOI] [PubMed] [Google Scholar]
- 5.De Smet B, Mayo M, Peeters C, Zlosnik JE, Spilker T, Hird TJ, LiPuma JJ, Kidd TJ, Kaestli M, Ginther JL, Wagner DM, Keim P, Bell SC, Jacobs JA, Currie BJ, Vandamme P (2015) Burkholderia stagnalis sp. nov. and Burkholderia territorii sp. nov., two novel Burkholderia cepacia complex species from environmental and human sources. Int J Syst Evol Microbiol 65(7):2265–2271. 10.1099/ijs.0.000251 [DOI] [PubMed] [Google Scholar]
- 6.Isles A, Maclusky I, Corey M, Gold R, Prober C, Fleming P, Levison H (1984) Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J Pediatr 104(2):206–210 [DOI] [PubMed] [Google Scholar]
- 7.Zhou J, Chen Y, Tabibi S, Alba L, Garber E, Saiman L (2007) Antimicrobial susceptibility and synergy studies of Burkholderia cepacia complex isolated from patients with cystic fibrosis. Antimicrob Agents Chemother 51(3):1085–1088. 10.1128/AAC.00954-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song JE, Kwak YG, Um TH, Cho CR, Kim S, Park IS, Hwang JH, Kim N, Oh GB (2017) Outbreak of Burkholderia cepacia pseudobacteraemia caused by intrinsically contaminated commercial 0.5% chlorhexidine solution in neonatal intensive care units. J Hosp Infect. 10.1016/j.jhin.2017.09.012 [DOI] [PubMed] [Google Scholar]
- 9.Torbeck L, Raccasi D, Guilfoyle DE, Friedman RL, Hussong D (2011) Burkholderia cepacia: this decision is overdue. PDA J Pharm Sci Technol 65(5):535–543. 10.5731/pdajpst.2011.00793 [DOI] [PubMed] [Google Scholar]
- 10.Vonberg RP, Gastmeier P (2007) Hospital-acquired infections related to contaminated substances. J Hosp Infect 65(1):15–23. 10.1016/j.jhin.2006.09.018 [DOI] [PubMed] [Google Scholar]
- 11.Baldwin A, Mahenthiralingam E, Drevinek P, Pope C, Waine DJ, Henry DA, Speert DP, Carter P, Vandamme P, LiPuma JJ, Dowson CG (2008) Elucidating global epidemiology of Burkholderia multivorans in cases of cystic fibrosis by multilocus sequence typing. J Clin Microbiol 46(1):290–295. 10.1128/JCM.01818-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lipuma JJ (2010) The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev 23(2):299–323. 10.1128/CMR.00068-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zlosnik JE, Zhou G, Brant R, Henry DA, Hird TJ, Mahenthiralingam E, Chilvers MA, Wilcox P, Speert DP (2015) Burkholderia species infections in patients with cystic fibrosis in British Columbia, Canada. 30 years’ experience. Ann Am Thorac Soc 12(1):70–78. 10.1513/AnnalsATS.201408-395OC [DOI] [PubMed] [Google Scholar]
- 14.Vanlaere E, Baldwin A, Gevers D, Henry D, De Brandt E, LiPuma JJ, Mahenthiralingam E, Speert DP, Dowson C, Vandamme P (2009) Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int J Syst Evol Microbiol 59(Pt 1):102–111. 10.1099/ijs.0.001123-0 [DOI] [PubMed] [Google Scholar]
- 15.Coutinho CP, Barreto C, Pereira L, Lito L, Melo Cristino J, Sa-Correia I (2015) Incidence of Burkholderia contaminans at a cystic fibrosis centre with an unusually high representation of Burkholderia cepacia during 15 years of epidemiological surveillance. J Med Microbiol 64(8):927–935. 10.1099/jmm.0.000094 [DOI] [PubMed] [Google Scholar]
- 16.Martin M, Christiansen B, Caspari G, Hogardt M, von Thomsen AJ, Ott E, Mattner F (2011) Hospital-wide outbreak of Burkholderia contaminans caused by prefabricated moist washcloths. J Hosp Infect 77(3):267–270. 10.1016/j.jhin.2010.10.004 [DOI] [PubMed] [Google Scholar]
- 17.Martina P, Bettiol M, Vescina C, Montanaro P, Mannino MC, Prieto CI, Vay C, Naumann D, Schmitt J, Yantorno O, Lagares A, Bosch A (2013) Genetic diversity of Burkholderia contaminans isolates from cystic fibrosis patients in Argentina. J Clin Microbiol 51(1):339–344. 10.1128/JCM.02500-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medina-Pascual MJ, Valdezate S, Carrasco G, Villalon P, Garrido N, Saez-Nieto JA (2015) Increase in isolation of Burkholderia contaminans from Spanish patients with cystic fibrosis. Clin Microbiol Infect 21(2):150–156. 10.1016/j.cmi.2014.07.014 [DOI] [PubMed] [Google Scholar]
- 19.Moehring RW, Lewis SS, Isaacs PJ, Schell WA, Thomann WR, Althaus MM, Hazen KC, Dicks KV, Lipuma JJ, Chen LF, Sexton DJ (2014) Outbreak of bacteremia due to Burkholderia contaminans linked to intravenous fentanyl from an institutional compounding pharmacy. JAMA Intern Med 174(4):606–612. 10.1001/jamainternmed.2013.13768 [DOI] [PubMed] [Google Scholar]
- 20.Peterson AE, Chitnis AS, Xiang N, Scaletta JM, Geist R, Schwartz J, Dement J, Lawlor E, Lipuma JJ, O’Connell H, Noble-Wang J, Kallen AJ, Hunt DC (2013) Clonally related Burkholderia contaminans among ventilated patients without cystic fibrosis. Am J Infect Control 41(12):1298–1300. 10.1016/j.ajic.2013.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nunvar J, Kalferstova L, Bloodworth RA, Kolar M, Degrossi J, Lubovich S, Cardona ST, Drevinek P (2016) Understanding the pathogenicity of Burkholderia contaminans, an emerging pathogen in cystic fibrosis. PLoS ONE 11(8):e0160975 10.1371/journal.pone.0160975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng P, Wang X, Baird SM, Showmaker KC, Smith L, Peterson DG, Lu S (2016) Comparative genome-wide analysis reveals that Burkholderia contaminans MS14 possesses multiple antimicrobial biosynthesis genes but not major genetic loci required for pathogenesis. MicrobiologyOpen 5(3):353–369. 10.1002/mbo3.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haim MS, Mollerach M, Van Domselaar G, Teves SA, Degrossi J, Cardona ST (2016) Draft genome sequences of Burkholderia contaminans FFI-28, a strain isolated from a contaminated pharmaceutical solution. Genome Announcements. 10.1128/genomeA.01177-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jung JY, Ahn Y, Kweon O, LiPuma JJ, Hussong D, Marasa BS, Cerniglia CE (2017) Improved high-quality draft genome sequence and annotation of Burkholderia contaminans LMG 23361(T). Genome Announcements. 10.1128/genomeA.00245-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burns J (2006) Antibiotic resistance of Burkholderia spp In: Coenye PVT (ed) Burkholderia: molecular microbiology and genomics. Horizon Bioscience, Norfolk, pp 81–91 [Google Scholar]
- 26.Drevinek P, Mahenthiralingam E (2010) Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clin Microbiol Infect 16(7):821–830. 10.1111/j.1469-0691.2010.03237.x [DOI] [PubMed] [Google Scholar]
- 27.Ramirez MS, Vargas LJ, Cagnoni V, Tokumoto M, Centron D (2005) Class 2 integron with a novel cassette array in a Burkholderia cenocepacia isolate. Antimicrob Agents Chemother 49(10):4418–4420. 10.1128/AAC.49.10.4418-4420.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trepanier S, Prince A, Huletsky A (1997) Characterization of the penA and penR genes of Burkholderia cepacia 249 which encode the chromosomal class A penicillinase and its LysRtype transcriptional regulator. Antimicrob Agents Chemother 41(11):2399–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poirel L, Rodriguez-Martinez JM, Plesiat P, Nordmann P (2009) Naturally occurring Class A ss-lactamases from the Burkholderia cepacia complex. Antimicrob Agents Chemother 53(3):876–882. 10.1128/AAC.00946-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papp-Wallace KM, Taracila MA, Gatta JA, Ohuchi N, Bonomo RA, Nukaga M (2013) Insights into beta-lactamases from Burkholderia species, two phylogenetically related yet distinct resistance determinants. J Biol Chem 288(26):19090–19102. 10.1074/jbc.M113.458315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papp-Wallace KM, Becka SA, Taracila MA, Zeiser ET, Gatta JA, LiPuma JJ, Bonomo RA (2017) Exploring the role of the Omega-loop in the evolution of Ceftazidime resistance in the PenA beta-lactamase from Burkholderia multivorans, an important cystic fibrosis pathogen. Antimicrob Agents Chemother. 10.1128/AAC.01941-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Everaert A, Coenye T (2016) Effect of beta-Lactamase inhibitors on in vitro activity of beta-Lactam antibiotics against Burkholderia cepacia complex species. Antimicrob Resist Infect Control 5:44 10.1186/s13756-016-0142-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holden MT, Seth-Smith HM, Crossman LC, Sebaihia M, Bentley SD, Cerdeno-Tarraga AM, Thomson NR, Bason N, Quail MA, Sharp S, Cherevach I, Churcher C, Goodhead I, Hauser H, Holroyd N, Mungall K, Scott P, Walker D, White B, Rose H, Iversen P, Mil-Homens D, Rocha EP, Fialho AM, Baldwin A, Dowson C, Barrell BG, Govan JR, Vandamme P, Hart CA, Mahenthiralingam E, Parkhill J (2009) The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J Bacteriol 191(1):261–277. 10.1128/JB.01230-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bloodworth RA, Selin C, Lopez De Volder MA, Drevinek P, Galanternik L, Degrossi J, Cardona ST (2015) Draft genome sequences of Burkholderia contaminans, a Burkholderia cepacia complex species that is increasingly recovered from cystic fibrosis patients. Genome Announcements. 10.1128/genomeA.00766-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Becka SA, Zeiser ET, Marshall SH, Gatta JA, Nguyen K, Singh I, Greco C, Sutton GG, Fouts DE, LiPuma JJ, Papp-Wallace KM (2018) Sequence heterogeneity of the PenA carbapenemase in clinical isolates of Burkholderia multivorans. Diagn Microbiol Infect Dis 92(3):253–258. 10.1016/j.diagmicrobio.2018.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R (2014) The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42(Database issue):D206–D214. 10.1093/nar/gkt1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winsor GL, Khaira B, Van Rossum T, Lo R, Whiteside MD, Brinkman FS (2008) The Burkholderia genome database: facilitating flexible queries and comparative analyses. Bioinformatics 24(23):2803–2804. 10.1093/bioinformatics/btn524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gouet P, Robert X, Courcelle E (2003) ESPript/END-script: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res 31(13):3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Notredame C, Higgins DG, Heringa J (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302(1):205–217. 10.1006/jmbi.2000.4042 [DOI] [PubMed] [Google Scholar]
- 42.Muñoz V, Serrano L (1997) Development of the multiple sequence approximation within the AGADIR model of alpha-helix formation: comparison with Zimm-Bragg and Lifson-Roig formalisms. Biopolymers 41(5):495–509 [DOI] [PubMed] [Google Scholar]
- 43.Krieger E, Darden T, Nabuurs SB, Finkelstein A, Vriend G (2004) Making optimal use of empirical energy functions: force-field parameterization in crystal space. Proteins 57(4):678–683. 10.1002/prot.20251 [DOI] [PubMed] [Google Scholar]
- 44.Schrödinger L The PyMOL molecular Graphics System., 1.5.0.4 edn.
- 45.Koressaar T, Remm M (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics 23(10):1289–1291. 10.1093/bioinformatics/btm091 [DOI] [PubMed] [Google Scholar]
- 46.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3–new capabilities and interfaces. Nucleic Acids Res 40(15):e115 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clinical and Laboratory Standards Institute (2017) Performance standards for antimicrobial susceptibility testing; twenty-sixth informational supplement M100-S27, vol 37 vol 1. Clinical and Laboratory Standards Institute, Wayne [Google Scholar]
- 48.Bret L, Chaibi EB, Chanal-Claris C, Sirot D, Labia R, Sirot J (1997) Inhibitor-resistant TEM (IRT) b-lactamases with different substitutions at position 244. Antimicrob Agents Chemother 41(11):2547–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perez-Llarena FJ, Cartelle M, Mallo S, Beceiro A, Perez A, Villanueva R, Romero A, Bonnet R, Bou G (2008) Structure-function studies of arginine at position 276 in CTX-M b-lactamases. J Antimicrob Chemother 61(4):792–797. 10.1093/jac/dkn031 [DOI] [PubMed] [Google Scholar]
- 50.Ruggiero M, Kerff F, Herman R, Sapunaric F, Galleni M, Gutkind G, Charlier P, Sauvage E, Power P (2014) Crystal structure of the extended-spectrum beta-lactamase PER-2 and insights into the role of specific residues in the interaction with beta-lactams and beta-lactamase inhibitors. Antimicrob Agents Chemother. 10.1128/AAC.00089-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruggiero M, Curto L, Brunetti F, Sauvage E, Galleni M, Power P, Gutkind G (2017) Impact of mutations at Arg220 and Thr237 in PER-2 beta-lactamase on conformation, activity, and susceptibility to inhibitors. Antimicrob Agents Chemother. 10.1128/AAC.02193-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y, Delmas J, Sirot J, Shoichet B, Bonnet R (2005) Atomic resolution structures of CTX-M beta-lactamases: extended spectrum activities from increased mobility and decreased stability. J Mol Biol 348(2):349–362. 10.1016/j.jmb.2005.02.010 [DOI] [PubMed] [Google Scholar]
- 53.Delmas J, Chen Y, Prati F, Robin F, Shoichet BK, Bonnet R (2008) Structure and dynamics of CTX-M enzymes reveal insights into substrate accommodation by extended-spectrum beta-lactamases. J Mol Biol 375(1):192–201. 10.1016/j.jmb.2007.10.026 [DOI] [PubMed] [Google Scholar]
- 54.Toleman MA, Rolston K, Jones RN, Walsh TR (2003) Molecular and biochemical characterization of OXA-45, an extendedspectrum class 2d’ beta-lactamase in Pseudomonas aeruginosa. Antimicrob Agents Chemother 47(9):2859–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stojanoski V, Chow DC, Fryszczyn B, Hu L, Nordmann P, Poirel L, Sankaran B, Prasad BV, Palzkill T (2015) Structural basis for different substrate profiles of two closely related class d beta-lactamases and their inhibition by halogens. Biochemistry 54(21):3370–3380. 10.1021/acs.biochem.5b00298 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.