 Hydroxyapatite nanoparticles (HAP-NPs) are an inorganic component of natural bone that are mainly used in tissue engineering field due to their bioactivity, osteoconductivity, biocompatibility, non-inflammatory, and non-toxicity properties.
Hydroxyapatite nanoparticles (HAP-NPs) are an inorganic component of natural bone that are mainly used in tissue engineering field due to their bioactivity, osteoconductivity, biocompatibility, non-inflammatory, and non-toxicity properties.
Abstract
Hydroxyapatite nanoparticles (HAP-NPs) are an inorganic component of natural bone and are mainly used in the tissue engineering field due to their bioactivity, osteoconductivity, biocompatibility, non-inflammatory, and non-toxicity properties. However, the current toxicity data for HAP-NPs regarding human health are limited, and only a few results from basic studies have been published. Therefore, the present study was designed to investigate the beneficial role of chitosan nanoparticles (CsNPs) and curcumin nanoparticles (CurNPs) in alleviating nephrotoxicity induced by HAP-NPs in male rats. The results showed that HAP-NPs caused a reduction in antioxidant enzymes and induced lipid peroxidation, nitric oxide production and DNA oxidation. Moreover, HAP-NP administration was associated with intense histologic changes in kidney architecture and immunoreactivity to proliferating cell nuclear antigen (PCNA). However, the presence of CsNPs and/or CurNPs along with HAP-NPs reduced the levels of oxidative stress through improving the activities of antioxidant enzymes. Also, the rats administered the nanoparticles showed a moderate improvement in glomerular damage which matched that of the control group and showed mild positive reactions to PCNA–ir in glomeruli and renal tubules in the cortical and medullary portions. These novel insights confirm that the presence of chitosan and curcumin in nanoforms has powerful biological effects with enhanced bioactivity and bioavailability phenomena compared to their microphase counterparts. Also, they were able to ameliorate the nephrotoxicity induced by HAP-NPs.
1. Introduction
Hydroxyapatite (HAP) is an inorganic component of natural bone1 and is extensively used in the tissue engineering (TE) field due to its excellent bioactivity, biocompatibility, non-toxicity, osteoconductivity, and non-inflammatory properties.2 Hydroxyapatite has been used as a resin for plasmid DNA and protein regulation, since ions on its surface are negatively and positively charged (PO43– and Ca2+), which can bind electrostatically with basic and acidic bio-macromolecules, respectively.3 Due to HAP similarity to the inorganic component of bone matrix, it has been involved in the development of new preparation methods in which it can be biomedically applied to repair hard tissues or as a drug delivery vehicle. Its use as a bone substitute or replacement comprises filling bones and teeth, partial or complete bone augmentation, and coating in dental implants and orthopedics.4
Even though hydroxyapatite on the nanoscale is documented to be biocompatible,5 the safety and toxicity concerns of HAP-NPs are growing regardless of their favorable prospect in various biomedical applications.6 Due to their nanoscale particle size, it is unknown whether they are safe when used in medical applications, as they can be internalized by the cells and interact with biological molecules, affecting cells in a harmful manner through altering the cell response and leading to toxicological response.7 Moreover, acute toxicity studies of HAP-NPs in vivo,8 interaction with different cell sources,9 and in vitro sensitization studies10 have been documented in several references.
Antioxidant activity is supposed to be one of the most known functions of chitosan as many studies have addressed chitosan's ability to prevent lipid oxidation and inhibit reactive oxygen species (ROS) in biological and food systems.11 Chitosan can scavenge free radicals or chelate metal ions through donation of lone pairs of electrons or a hydrogen atom.12 The amino groups (NH2) and hydroxyl group (OH) in chitosan are the main functional groups for its antioxidant activity, and it is difficult to detach due to its strong hydrogen bonding and semi-crystalline structure.13 Chitosan nanoparticles are a natural polymer that is used in nanomedicine production, because they have attractive features for drug delivery and have proven to be very functional when used in the nanoscale form. Properties such as the cationic character and the solubility of chitosan in aqueous medium have been reported as an evidence of this polysaccharide success.14
Also, curcumin is a well-known antioxidant and a highly pleiotropic molecule12 that is capable of exerting an extensive range of pharmacological activities like anti-cancer, antioxidant, antibacterial, anti-inflammatory, hypoglycaemic,15 anti-microbial, anti-atherosclerotic,16 and wound healing properties.17 Moreover, curcumin has been found to instantly interact with various intracellular signaling molecules.18 Its beneficial effects have been specified to be intermediated through the modification of multiple cell signaling molecules.19 Also, it has been reported to have the capability to directly scavenge reactive oxygen species (ROS).20 Curcumin's poor bioavailability is a major disadvantage related to its use as a therapeutic agent. However, curcumin in the nanoform is considered as the improved form of curcumin that shows better bioavailability and solubility. Nanoparticles such as micelles, nanogels, liposomes, and polymeric nanoparticles can be used to deliver therapeutic concentrations of curcumin, which in turn reinforces the therapeutic efficacy of curcumin.21
The goals of this study were (i) to demonstrate hydroxyapatite nanoparticles’ involvement in oxidative stress and DNA fragmentation; (ii) to investigate the histopathological and immunohistochemical alterations induced by HAP-NPs; and (iii) to explore whether the nanoforms of chitosan and curcumin are capable of ameliorating the nephrotoxicity induced by HAP-NPs.
2. Materials and methods
2.1. Tested compounds and doses
Hydroxyapatite nanoparticles (HAP-NPs) were prepared using the following reagents: sodium carbonate, Riedel-de-Haën, Germany, and sodium hydroxide, El-Nasr pharmaceutical chemicals Co., Egypt. Chitosan nanoparticles (CsNPs) and curcumin nanoparticles (CurNPs) were purchased from Nanotech Egypt for Photo Electronics. HAP-NPs were dissolved in distilled water, and the dose (300 mg per kg bw) was chosen according to Sabry.22 Also, CsNPs were dissolved in acetic acid (Mw 310–375 kDa), and the dose (280 mg per kg bw) was chosen according to Tang and Abdel-Wahhab.23–25 CurNPs were dissolved in distilled water, and the dose (15 mg per kg bw) was chosen according to Yadav.26
2.2. Characterization of nanoparticles of hydroxyapatite, chitosan, and curcumin
All samples were examined for morphology and characterized using a high resolution transmission electron microscope (HR-TEM).
2.3.1. Transmission electron microscopy analysis
HAP-NPs, CsNPs, and CurNPs were analyzed using high resolution transmission electron microscopy (HR-TEM), which is an imaging mode of specialized transmission electron microscopes that allows for direct imaging of the atomic structure of the sample. The HR-TEM JEOL JEM 2100Plus is a versatile TEM for both large-scale 2D screening and tomography. For TEM analysis, the samples were placed on carbon-coated copper grids and left to dry for 5 min; the excess solution was removed using blotting paper at room temperature. The HR-TEM runs Serial EM and thus is capable of automated multi-position acquisitions. Depending on the type and thickness of the specimen, the acceleration voltage can be chosen from 80 to 200 kV.27,28
2.3. Animals and experimental design
This study was performed in strict accordance with the guidelines of the Institutional Animal Care and Use Committee (IACUC) and approved by the Animal Care and Use Committee of Alexandria University (ethics approval no. 1468-104, revised 2018), provided that animals will not suffer at any stage of the experiment. The study was conducted on 80 male Wistar rats weighing 170 ± 175 g obtained from the Faculty of Medicine, Alexandria University, Egypt. The animals were housed in a comfortable environment on basal diet and tap water which were provided ad libitum. They were monitored during the period of treatment. Food and water intake, and body weights were weekly recorded throughout the whole experimental period. After acclimation for 2 weeks, they were randomly divided equally into 8 groups, and each cage housed a maximum of 10 rats. Group 1 was used as control; group 2 was orally treated with CsNPs alone (280 mg per kg bw); group 3 was treated orally with CurNPs alone (15 mg per kg bw); group 4 was orally treated with both CsNPs and CurNPs; group 5 was orally treated with HAP-NPs alone (300 mg per kg bw); group 6 was orally treated with both CsNPs and HAP-NPs; group 7 was orally treated with CurNPs and HAP-NPs; and group 8 was the combination group as it received CsNPs, CurNPs and HAP-NPs. The animals were orally treated with respective doses daily for 45 days.
2.4. Blood sample collection and kidney tissue preparation
At the end of the 45th day, all animals were anesthetized and then sacrificed, and blood samples were collected in heparin collection tubes. For plasma separation, blood was centrifuged at 860g for 20 minutes, and plasma was kept at –80 °C until analysis. Kidneys were removed and washed with saline solution (0.9%), and then connective tissues and adhering fats were immediately removed. Kidneys were minced and then homogenized in a Potter–Elvehjem type homogenizer. The homogenates were then centrifuged at 10 000g for 20 minutes at 4 °C to pellet the cell debris, and the supernatant was collected and stored for the analysis of parameters under study.
2.5. Measured parameters
2.5.1. Oxidative stress markers and kidney antioxidant parameters
Thiobarbituric acid-reactive substances (TBARS) were measured by the method of Tappel and Zalkin.29 Total antioxidant capacity (TAC) in kidney homogenates was assayed according to the Koracevic method.30 Nitric oxide (NO) level was assayed according to Montgomery and Dymock.31 Superoxide dismutase (SOD) activity was measured using the method of Misra and Fridovich.32 Glutathione peroxidase (GPx) activity was determined following the Chiu method.33 Glutathione S-transferase (GST) activity was analyzed according to the Habig method.34 Catalase (CAT) activity was assayed following the Luck method.35 Glutathione (GSH) levels were assayed using the Jollow method.36 The abovementioned assays were conducted in compliance with the instruction manual of Biodiagnostic Kit, Egypt.
2.5.2. Assay of DNA fragmentation
8-OH-2-deoxyguanosine (8-OHdG) was measured in the samples of DNA using the 8-OHdG commercial ELISA kit (ab201734, Abcam, Cambridge, UK) following the protocol of the manufacturer.
2.5.3. Histopathological examination
Tissues taken from the rats’ kidneys were cut and immediately fixed in 10% formalin solution and dehydrated using ascending grades of alcohol and xylene. They were then placed in molten wax and xylene for about 10 minutes, embedded in paraffin wax, and then sectioned by a rotary microtome to obtain sections (thickness, 4–6 μm). After that, they were stained with H&E for investigating histopathological changes following the Drury method.37
2.5.4. Proliferating cell nuclear antigen immunoreactivity (PCNA-ir) measurement
Kidney distribution of PCNA receptor subunits was determined in deparaffinized sections (thickness, 5 μm) using an avidin–biotin–peroxidase (IHC) method (Elite–ABC; Vector Laboratories, CA, USA), and the anti-PCNA monoclonal antibody (dilution 1 : 100; DAKO Japan Co, Tokyo, Japan) was used.
2.6. Statistical analysis
Data were reported as mean ± SE. Statistical analysis of the parameters was performed using the general linear model (GLM) produced by Statistical Analysis Systems Institute.38 Duncan's new multiple range test was followed to test the significant differences between means according to the Duncan method.39
3. Results
3.1. Characterization of nanoparticles of hydroxyapatite, chitosan, and curcumin
3.1.1. Transmission electron microscopy analysis
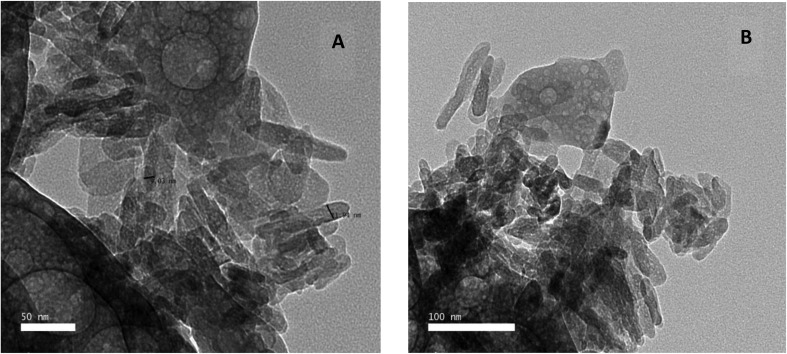
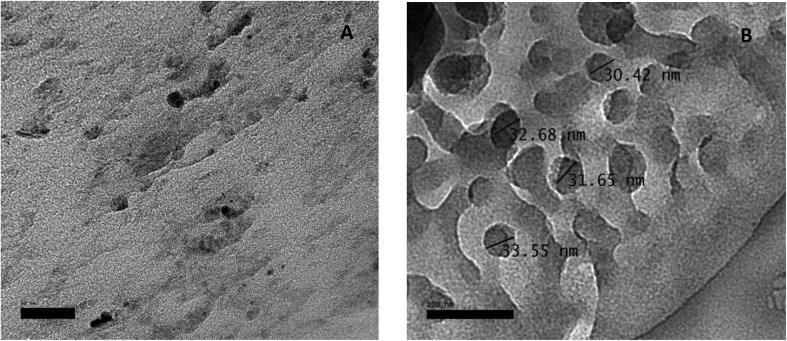
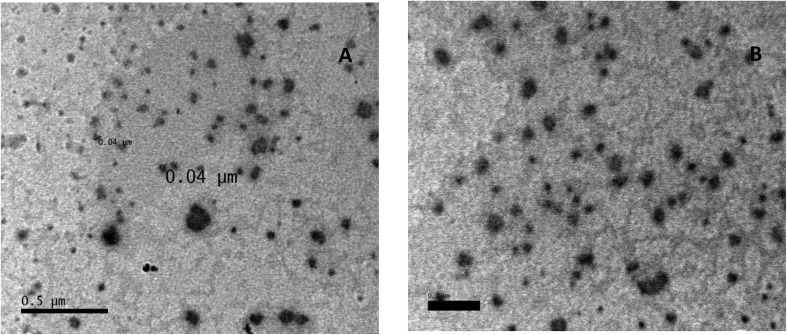
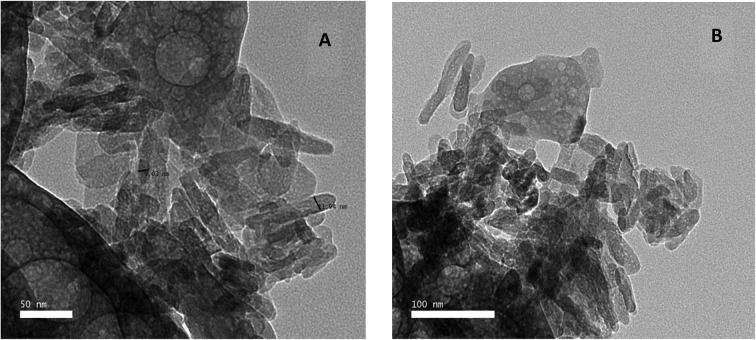
The structure and morphology of the samples were further confirmed using high resolution transmission electron microscopy (HR-TEM) images of HAP-NPs, CsNPs, and CurNPs as shown in Fig. 1–3. The TEM analysis confirmed the presence of needle-like crystal morphology of HAP-NPs at different scale bars 50–100 nm (Fig. 1). TEM images of chitosan nanoparticles (CsNPs) are shown in Fig. 2. These nanoparticles have a solid and consistent structure and are also spherical in shape at different scale bars (Fig. 2(A and B)). TEM images of curcumin nanoparticles shown in Fig. 3 show the well-defined crystalline morphology of nano-curcumin. The average particle size of curcumin nanocrystals is 0.04 μm as shown in Fig. 3(A).
Fig. 1. TEM micrographs demonstrating the appearance of n-HAP needle-like crystals.
Fig. 2. TEM images and size distribution of chitosan nanoparticles.
Fig. 3. TEM images and size distribution of curcumin nanoparticles.
3.2. Oxidative stress parameters and antioxidant enzymes
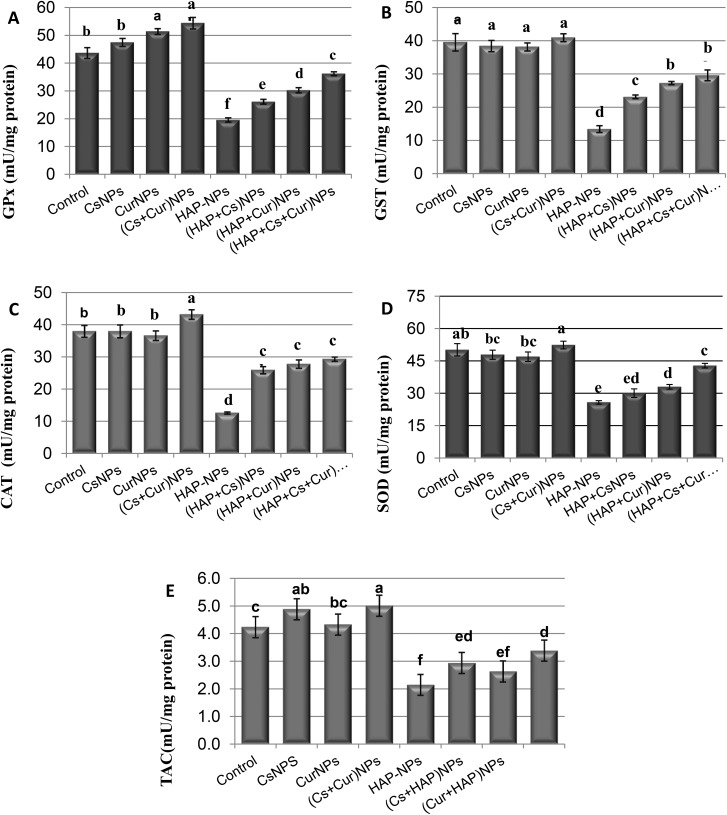
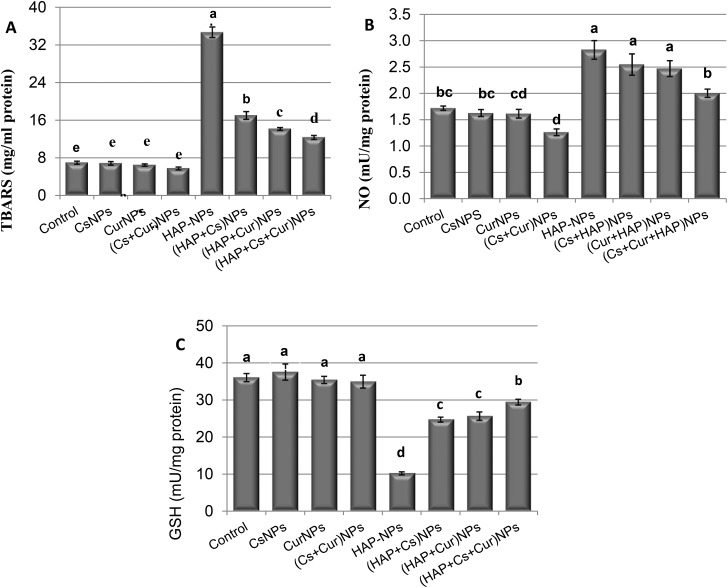
The activities of GPx, GST, CAT, SOD and TAC, and also the levels of TBARS, NO and GSH were measured in the kidneys of the adult male rats orally treated daily with CsNPs, CurNPs, and HAP-NPs alone or in combination, as shown in Tables 1 and 2 and Fig. 4 and 5. The data showed that there were no significant differences between the effect of CsNPs and CurNPs treated alone on GPx, GST, CAT, SOD, TAC, TBARS, NO, and GSH. However, the presence of CsNPs and CurNPs in combination significantly increased the activities of all these enzymes and decreased the levels of TBARS and NO in the kidney compared to the control group. Treatment with HAP-NPs alone showed a significant decrease in GPx, GST, CAT, SOD, GSH and TAC levels, but showed a significant increase in the levels of TBARS and NO compared to the control group. However, the presence of CsNPs and CurNPs alone or in combination with HAP-NPs was capable of increasing the activities of GPx, GST, CAT, SOD, TAC and GSH and decreasing the activities of TBARS and NO compared to the HAP-NP treated group but did not reach the control group values.
Table 1. Kidney levels of glutathione peroxidase, glutathione S-transferase, catalase, superoxide dismutase and total antioxidant capacity of male rats treated orally with nanoparticles of chitosan, curcumin, and hydroxyapatite alone or in combination.
| Experimental groups | Parameter |
||||
| GPx (mU mg–1 protein) | GST (mU mg–1 protein) | CAT (mU mg–1 protein) | SOD (mU mg–1 protein) | TAC (mU mg–1 protein) | |
| Control | 43.7 ± 1.95b | 39.6 ± 2.59a | 38.0 ± 1.84b | 50.2 ± 2.86ab | 4.22 ± 0.20c |
| CsNPs | 47.5 ± 1.43b | 38.5 ± 1.69a | 38.0 ± 1.99b | 47.9 ± 2.12bc | 4.8 ± 0.14ab |
| CurNPs | 51.4 ± 1.03a | 38.2 ± 1.19a | 36.6 ± 1.47b | 47 ± 2.20bc | 4.3 ± 0.14bc |
| (Cs + Cur) NPs | 54.4 ± 2.07a | 41.0 ± 1.19a | 43.2 ± 1.45a | 52.4 ± 1.79a | 5.0 ± 0.28a |
| HAP-NPs | 19.9 ± 0.77f | 13.5 ± 1.02d | 12.6 ± 0.33d | 25.8 ± 0.80e | 2.1 ± 0.12f |
| (HAP + Cs) NPs | 26.2 ± 0.86e | 23.2 ± 0.54c | 25.9 ± 1.10c | 30 ± 1.99ed | 2.9 ± 0.19ed |
| (HAP + Cur) NPs | 30.3 ± 0.92d | 27.3 ± 0.48b | 27.8 ± 1.29c | 33.0 ± 1.08d | 2.6 ± 0.21ef |
| (HAP + Cs + Cur) NPs | 36.2 ± 0.70c | 29.6 ± 1.58b | 29.3 ± 0.66c | 42.8 ± 1.07c | 3.4 ± 0.19d |
Table 2. Kidney levels of thiobarbituric acid-reactive substances, nitric oxide and glutathione of male rats treated orally with nanoparticles of chitosan, curcumin, and hydroxyapatite alone or in combination.
| Experimental groups | Parameter |
||
| TBARS (mg ml–1 protein) | NO (mU mg–1 protein) | GSH (mU mg–1 protein) | |
| Control | 7.0 ± 0.32e | 1.7 ± 0.04bc | 36.1 ± 1.08a |
| CsNPs | 6.9 ± 0.30e | 1.6 ± 0.07bc | 37.6 ± 2.16a |
| CurNPs | 6.5 ± 0.25e | 1.6 ± 0.08cd | 35.5 ± 0.95a |
| (Cs + Cur) NPs | 5.8 ± 0.30e | 1.3 ± 0.06d | 35.0 ± 1.71a |
| HAP-NPs | 24.7 ± 1.11a | 2.8 ± 0.18a | 10.2 ± 0.40d |
| (HAP + Cs) NPs | 17.0 ± 0.82b | 2.5 ± 0.20a | 24.7 ± 0.67c |
| (HAP + Cur) NPs | 14.2 ± 0.30c | 2.5 ± 0.15a | 25.7 ± 1.12c |
| (HAP + Cs + Cur) NPs | 12.4 ± 0.41d | 2.0 ± 0.08b | 29.5 ± 0.77b |
Fig. 4. Levels of GPX, GST, CAT, SOD and TAC in rat kidneys treated orally with CsNPs, CurNPs and HAP-NPs alone or in combination.
Fig. 5. Levels of TBARS, NO and GSH of male rats treated orally with CsNPs, CurNPs, and HAP-NPs alone or in combination.
3.3. Oxidative DNA damage
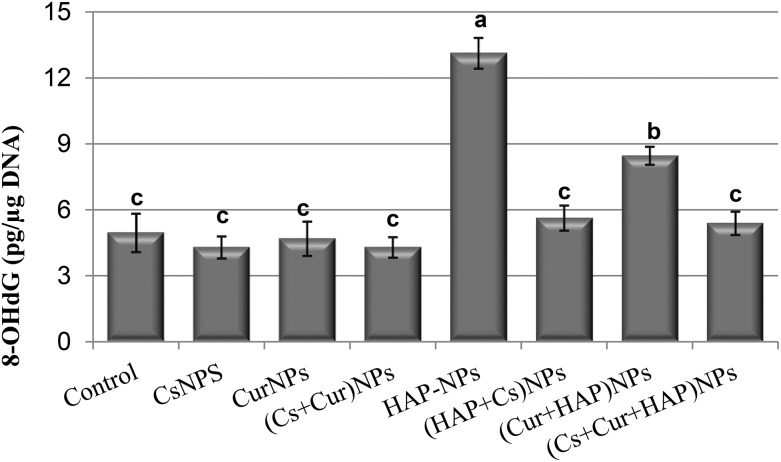
The results of 8-OH-2-deoxyguanosine (8-OHdG) content in the renal tissues of the studied groups are listed in Table 3 and shown in Fig. 6. The rats treated with CsNPs or CurNPs alone or in combination showed no significant effect on the level of 8-OHdG compared to the control group. On the other hand, the group treated with the hydroxyapatite nanoparticles alone showed a significant increase in the level of renal 8-OHdG to about 2.6-fold compared to the control group values, while treatment with CsNPs or CurNPs alone or in combination with HAP-NPs significantly ameliorated the renal content of 8-OHdG; however, the levels were still significantly higher than the control group values. It is apparent that CsNPs showed a better effect than CurNPs, while the treatment with both of them resulted in a significant reduction in and complete normalization of renal 8-OHdG levels, as shown in Table 3 and Fig. 6.
Table 3. Renal tissue content of 8-OH-2-deoxyguanosine of male rats treated orally with nanoparticles of chitosan, curcumin, and hydroxyapatite alone or in combination.
| Experimental groups | 8-OHdG (pg μg–1 DNA) |
| Control | 4.97 ± 1.5c |
| CsNPs | 4.30 ± 0.9c |
| CurNPs | 4.70 ± 1.3c |
| (Cs + Cur) NPs | 4.30 ± 0.79c |
| HAP-NPs | 13.13 ± 1.21a |
| (HAP + Cs) NPs | 5.63 ± 0.99c |
| (HAP + Cur) NPs | 8.47 ± 0.70b |
| (HAP + Cs + Cur) NPs | 5.40 ± 0.92c |
Fig. 6. Renal tissue content of 8-OH-2-deoxyguanosine of male rats treated orally with CsNPs, CurNPs, and HAP-NPs alone or in combination.
3.4. Histopathological examination
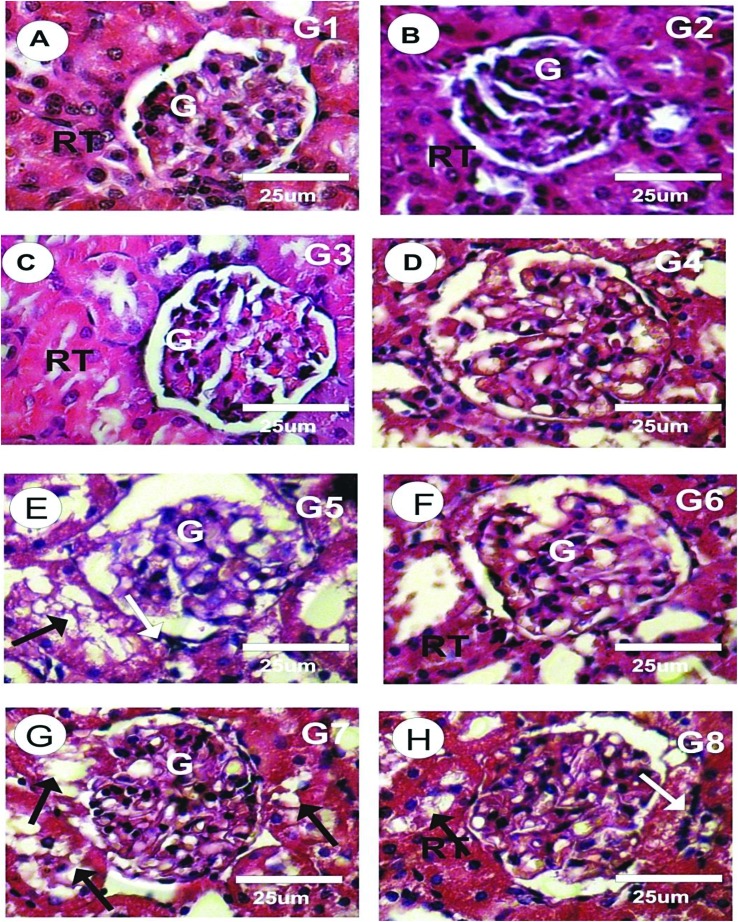
As shown in Fig. 7A, the histological examination of the H&E stained kidney sections in the control rats revealed normal histological structures of the glomeruli and renal tubules. Cells of the Bowman's capsule's outer layer formed a simple squamous epithelium, while cells of the inner layer were very complex in shape and could not be detected by histological staining. The glomeruli were round to oval in shape. No histopathological abnormalities were found in the kidney sections of the rats treated with CsNPs as in G2, or with CurNPs as in G3, or with both CsNPs and CurNPs as in G4, as shown in Fig. 7B–D, respectively. The sections of the kidney treated with HAP-NPs revealed inconstant pathological changes in several parts of the urinary tubules and glomeruli, as shown in Fig. 7E. The most intense changes were in the Malpighian as corpuscles lost their configuration and the renal tubules appeared with a wide lumen, marked medullar tubular epithelial degeneration, focal tubular epithelial necrosis, moderate hemorrhage, mild to moderate atrophic glomerulus with degenerated epithelium and marked congestion in the renal blood vessels, as shown in Fig. 7E. The kidney sections of the group treated with HAP-NPs and CsNPs together revealed a moderate degree of improvement in glomerular damage, which resembled that of the control group, with minimal vacuolization of tubular cells, as shown in Fig. 7F. The sections of the group treated with HAP-NPs and CurNPs together revealed a mild degree of improvement in glomerular damage with a wide lumen in the renal tubules, but some of the renal tubules were still degenerated (Fig. 7G). The group treated with HAP-NPs along with CsNPs and CurNPs revealed a very good improvement with normal structure as in the control group (Fig. 7H).
Fig. 7. Photomicrographs of kidney sections of different experimental groups stained by H&E.
3.5. Proliferating cell nuclear antigen immunoreactivity
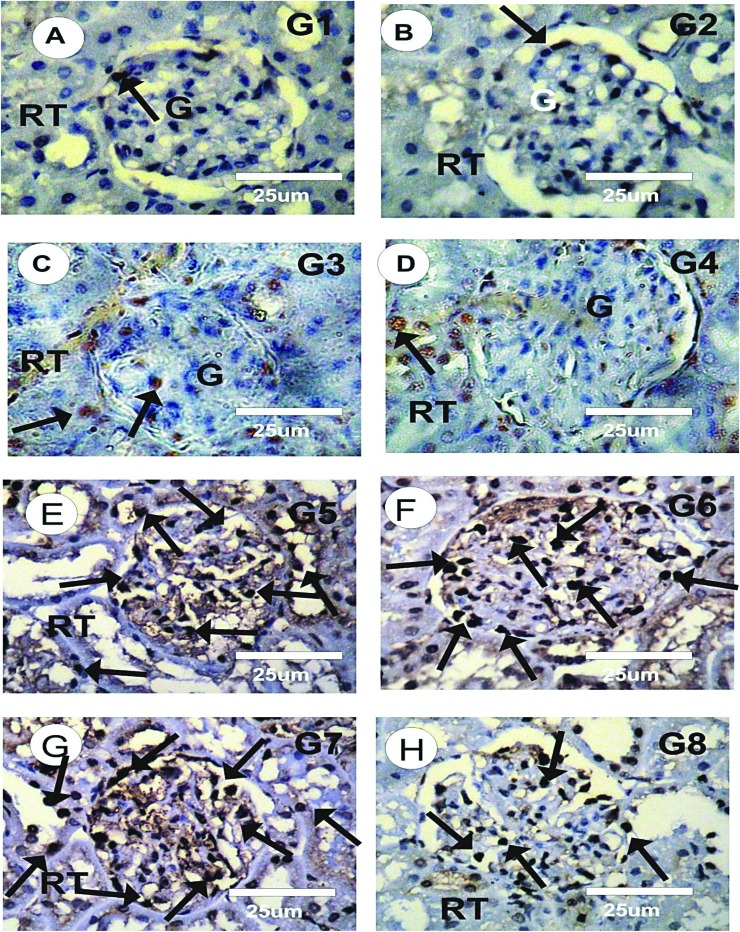
PCNA immunoreactivity (PCNA-ir) detection of kidney sections in different experimental groups was performed as shown in Figures 8A-H. The kidney sections of the control rats showed faint or negative reaction for PCNA-ir (grade 0) in glomeruli and renal tubules, as shown in Fig. 8A. The kidney sections of rats treated with CsNPs (G2), with CurNPs (G3), and with both CsNPs and CurNPs (G4) showed mild reaction for PCNA-ir (grade 1), as shown in Fig. 8B–D. Strong positive reactions for PCNA-ir (grade 5) were detected in the kidney sections of the rats treated with HAP-NPs alone, as shown in Fig. 8E. The intensity of PCNA-ir in the HAP-NP treated group showed a significant increase when compared to the control group. However, moderate positive reactions for PCNA–ir (grade 3) were observed in the kidney sections of the rats treated with HAP-NPs and CsNPs together, while moderate to marked positive reactions for PCNA–ir (grade 4) were observed in the kidney sections of the rats treated with HAP-NPs and CurNPs together, as shown in Fig. 8F–G. The kidney sections of the group treated with HAP-NPs along with CsNPs and CurNPs revealed mild positive reactions for PCNA–ir (grade 1), as shown in Fig. 8H.
Fig. 8. Photomicrographs of kidney sections of different experimental groups stained with PCNA-ir.
4. Discussion
Nano-sized particles have the ability to internalize cells, where they can interact with biological molecules, alter cell response, and affect the cell in deleterious behavior, leading to toxicological response.7 Comprehensive and adequate estimate of toxicological effects induced by exposure to nanoparticles should take into consideration the different points through which these particles enter the body and translocate into organs, tissues, and biological systems after being distributed in the body. Despite the promising potential of hydroxyapatite nanoparticles in various applications, their toxicity is of growing concern. So, in order to indicate the suitable protective intervention against these NPs, it is important to understand its toxicity mechanism.
Kidney is considered as one of the main sites inducing xenobiotic toxicity.40 Its specific toxicity is assigned to the high flow rate of blood, which in turn delivers elevated concentrations of xenobiotic to the kidney. In particular, the proximal tubule epithelium is more vulnerable to nephrotoxicity; these cells express various transporters, which enable active intake and intracellular accumulation of metabolites or toxic compounds. Proximal tubular epithelial cells can bio-activate nontoxic compounds into reactive intermediates which might cause toxicity through damaging cellular macromolecules, and also they are highly metabolically active.40
Nemenqani indicated that oxidative stress induction is the main mechanism of nanoparticles’ toxicity. When nanoparticles enter the cell, they disturb the redox balance between oxidants and antioxidants, which in turn induce the intracellular oxidative stress. Excessive oxidative stress may also modify lipids, proteins, and nucleic acids, which in turn deteriorates the antioxidant defense systems or leads to cell death.41 Meantime, nanoparticles can increase gene expression of the death receptor and cause DNA damage through increased production of ROS.42 Also, it was documented that the increased production of ROS induced by nanoparticles in lysosomes can cause DNA mutations or induce DNA single- or double-strand breaks.43
At the level of redox status of kidney tissues, HAP-NPs caused significant reduction in different antioxidant enzymes, including SOD, GPx, CAT, GST, and GSH systems. This deterioration in the kidney antioxidant defense system is linked to the enhanced production of nitric oxide and ROS, which exacerbates oxidative stress in renal tissues due to exposure to HAP-NPs. The depletion of GSH could explain the inhibited activities of GSH-dependent enzymes (GST and GPx), while the main route to the depletion of GSH levels is the protection against oxidative stress that was confirmed by the elevated level of 8-OHdG due to ROS overproduction induced by HAP-NPs. This ROS overload was faced by the low molecular weight antioxidant, e.g. glutathione, that results in GSH depletion.
The present results are in agreement with those of Chen, who demonstrated that the exposure to HAP-NPs caused oxidative stress with the elevation of the levels of H2O2 and MDA and a decrease in SOD and GSH levels.44 Also, Xu found that HAP-NPs induced ROS and SOD intracellular accumulation in cell lines.45 Turkez demonstrated that exposure to HAP-NPs had dose-dependent effects on inducing genotoxicity, cytotoxicity, and oxidative damage in human blood cells.46 Also, HAP-NP exposure resulted in significant increase of micronuclei, sister chromatid exchange, 8-OH-dG levels, and chromosome aberration rates as compared to untreated culture.
Moreover, Sies and Akerboom (1984) reported that after administration of nanoparticles, GSH can act as a conjugating agent in their metabolism.47 When these nanoparticles induce oxidative stress by generating hydroperoxides or H2O2, GSH can also be oxidized in a reaction catalyzed by GPx. It was also reported that GSH depletion in tissues leads to deterioration of the cellular defense against ROS and may result in peroxidative injury.47 As GSH plays an important role in cell's protection against induced oxidative stress, it also acts as a vital antioxidant and a major cofactor for antioxidant enzymes participating in cellular redox reactions. Likewise, HAP-NPs decreased the total SOD activity in C6 glioma cells.48 Also, an in vivo study documented a dose-dependent decrease in the activity of GPx in dermally HAP-NP exposed rats.49
However, at the molecular level, oxidative stress induced by HAP-NPs in kidney tissues caused significant elevation in the oxidative DNA marker, 8-OHdG. Reactive oxygen species are highly reactive with a set of biological molecules, among which DNA is the most important one.50 8-Oxoguanine (8-oxo-G) is one of the distinguished forms of oxidatively generated DNA base modifications and also a sensitive oxidative DNA damage marker.51 If this base modification (8-OH-dG) was not repaired by the repair mechanism, guanine (G) to thymine (T) substitution may happen upon replication. Alternatively, 8-Oxo-2′-deoxyguanosine-5′-Triphosphate (8-Oxo-dGTP) (in the nucleotide pool) may be mis-integrated opposite adenine (A) producing A–C substitution.52 These mutagenic effects of 8-OH-dG formation may denote genotoxicity hazards induced by HAP-NPs.
The antioxidant effects of CsNPs were confirmed in many previous studies; for example, Wen reported that CsNPs’ protective role against H2O2-induced cell injury was through boosting the endogenous antioxidants’ activities (SOD, GPx and CAT) and enhancing their gene expressions.53 Furthermore, El-Denshary documented that treatment with CsNPs succeeded in ameliorating the antioxidant capacity of the body and reducing the oxidative stress as indicated by the increase in antioxidant enzymes (CAT, SOD, GPx) and decrease of MDA levels.54 Also, the present results compare with those of Jeon, who reported that the treatment with chitosan significantly decreased MDA levels and increased activities of the antioxidant enzymes, CAT and SOD, against carbon tetrachloride chronic (CCl4)-induced toxicity.55 Also, Xie showed that CsNPs inhibited lipid peroxidation by scavenging the hydroxyl radicals.13 It was found that chitosan administration with a dose of 100 mg per kg bw showed a significant increase in antioxidant enzymes (SOD, GPx, GR, CAT) and non-enzymatic antioxidants (GSH, vitamin E and vitamin C) but showed a significant decrease in lipid peroxidation levels in the benzidine induced bladder cancer cell line.56,57 Also, Prashanth reported that chitosan was able to scavenge O2˙– and ˙OH radicals and offered protection against calf thymus DNA damage.58
Flora found that CurNPs at the dose of 15 mg kg–1 reduced free radical generation and also restored the antioxidant enzymes’ activity in rats treated with lead.59 The antioxidant activity of CurNPs along with their enhanced bioavailability makes them more vulnerable to be an effective agent against lead poisoning. Nehra found that treatment with CurNPs significantly decreased redundant leakage of ROS and lipid peroxidation in cardiomyocytes under hypoxia.60 Yadav found that CurNPs ameliorated toxic effects induced by fluoride and arsenic in rat tissues and blood through reducing the levels of ROS and restoring the blood glutathione level.61
These results clearly demonstrated the boosting effects of CsNPs and CurNPs on the antioxidant status of renal tissues through the activation of antioxidant enzymes (SOD, GPx, GST and CAT), increasing total antioxidant capacity (TAC) and correction of glutathione. Also, CsNPs and CurNPs have significantly decreased the production of nitric oxide as indicated by the lower nitric oxide level. These ameliorative effects on antioxidants and free radicals result in correction of the oxidative stress status of renal tissues as evidenced in the present study by the lower levels of MDA; the markers of lipid peroxidation and 8-OH-dG; and the marker of oxidative DNA damage. The obtained results agree with those of Ahmed-Farid, who reported that CurNPs increased the tissue energy rate in addition to a significant decrease in 8-OHdG levels and decreased nitrosative and oxidative stresses.62 CurNPs are also considered as a powerful free radical oxidant scavenger via electron transfer and hydrogen atom donation.63
It is obvious from the previous results that CsNP and/or CurNP administration along with HAP-NPs has significantly corrected the impairments of most studied parameters related to renal toxicity that has been induced by the treatment with HAP-NPs alone. However, at the histological level, HAP-NPs caused severe changes in the Malpighian corpuscles, which lost their characteristic configuration, and the renal tubules appeared with a wide lumen, marked cortical and medullar tubular epithelial degeneration, focal tubular epithelial necrosis, moderate hemorrhage, mild to moderate atrophic glomerulus and degenerated epithelium, and marked congestion in the renal blood vessels. Also, strong positive reactions for PCNA-ir (grade 5) were detected in kidney sections of rats treated with HAP-NPs. These data may indicate the entry of HAP-NPs into the renal tissues, cells and cellular organelles. However, the administration of CsNPs and/or CurNPs showed a moderate improvement in glomerular damage, which matched that of the control group with only minimal vacuolization in tubular cells. Kidney sections of the rats administered HAP-NPs along with either CsNPs or CurNPs or both revealed very good improvements with normal structure as in the control group.
5. Conclusion
Collectively, the present results concluded that oral intake of HAP-NPs induced renal toxicity at different levels, including kidney antioxidant system, oxidative stress, DNA damage and renal tissue content of 8-OHdG, histological structure and immunohistochemical reactivity towards PCNA. Given the substantial use of HAP-NPs in the field of medicine, the search for protective interceptions against its toxicity is of great medical prominence. The presented results obviously indicated that CsNPs and CurNPs have alleviated the nephrotoxicity induced by HAP-NPs with prominent ameliorative effects observed with CsNPs either administered alone or in combination with CurNPs. Our results also confirm that the nanoforms of these two compounds have powerful biological effects with enhanced bioactivity and bioavailability phenomena compared to their microphase counterparts.
Conflicts of interest
The authors gratefully acknowledge that there are no conflicts of interests.
References
- Azzaoui K., Lamhamdi A., Mejdoubi E. M., Berrabah M., Hammouti B., Elidrissi A., Fouda M. M. G., Al-Deyab S. S. Carbohydr. Polym. 2014;111:41–46. doi: 10.1016/j.carbpol.2014.04.058. [DOI] [PubMed] [Google Scholar]
- Murugan R., Ramakrishna S. Biomaterials. 2004;25:3829–3835. doi: 10.1016/j.biomaterials.2003.10.016. [DOI] [PubMed] [Google Scholar]
- Schröder E., Jönsson J., Poole T. L. Anal. Biochem. 2003;313:176–178. doi: 10.1016/s0003-2697(02)00567-5. [DOI] [PubMed] [Google Scholar]
- Zhou H., Lee J. Acta Biomater. 2011;7:2769–2781. doi: 10.1016/j.actbio.2011.03.019. [DOI] [PubMed] [Google Scholar]
- Remya N. S., Syama S., Gayathri V., Varma H. K., Mohanan P. V. Colloids Surf., B. 2011;117:389–397. doi: 10.1016/j.colsurfb.2014.02.004. [DOI] [PubMed] [Google Scholar]
- Xiao X., Wang W., Liu D., Zhang H., Gao P., Geng L., Wang Z. Sci. Rep. 2015;5:9409. doi: 10.1038/srep09409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazi M. A., Herranz B., Santos H. A. Biomatter. 2012;2:296–312. doi: 10.4161/biom.22347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Xiao Z., Xiao Y., Wang Z., Li F., Li M., Peng X. Oncol. Lett. 2014;7:1485–1492. doi: 10.3892/ol.2014.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues S. C., Salgado C. L., Sahu A., Garcia M. P., Fernandes M. H., Monteiro F. J. J. Biomed. Mater. Res., Part A. 2013;101:1080–1094. doi: 10.1002/jbm.a.34394. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Ren C., Zhang B., Yang H., Lang Y., Min L., Mo A. Int. J. Nanomed. 2014;9:485. doi: 10.2147/IJN.S52990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. W., Thomas R. L. Food Chem. 2007;101:308–313. [Google Scholar]
- Lin S. B., Chen S. H., Peng K. C. J. Sci. Food Agric. 2009;89:238–244. [Google Scholar]
- Xie W., Xu P., Liu Q. Bioorg. Med. Chem. Lett. 2001;11:1699–1701. doi: 10.1016/s0960-894x(01)00285-2. [DOI] [PubMed] [Google Scholar]
- Grenha A., Al-Qadi S., Seijo B., Remuñán-Lopez C. J. Drug Delivery Sci. Technol. 2010;20:33–43. [Google Scholar]
- Sharma S., Kulkarni S. K., Chopra K. Clin. Exp. Pharmacol. Physiol. 2006;33:940–945. doi: 10.1111/j.1440-1681.2006.04468.x. [DOI] [PubMed] [Google Scholar]
- Nishiyama T., Mae T., Kishida H., Tsukagawa M., Mimaki Y., Kuroda M., Kitahara M. J. Agric. Food Chem. 2005;53:959–963. doi: 10.1021/jf0483873. [DOI] [PubMed] [Google Scholar]
- Aggarwal B. B., Sung B. Trends Pharmacol. Sci. 2009;30:85–94. doi: 10.1016/j.tips.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Gupta S. C., Prasad S., Kim J. H., Patchva S., Webb L. J., Priyadarsini I. K., Aggarwal B. B. Nat. Prod. Rep. 2011;28:1937–1955. doi: 10.1039/c1np00051a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon N., Aggarwal B. B., Newman R. A., Wolff R. A., Kunnumakkara A. B., Abbruzzese J. L., Kurzrock R. Clin. Cancer Res. 2008;14:4491–4499. doi: 10.1158/1078-0432.CCR-08-0024. [DOI] [PubMed] [Google Scholar]
- Gupta S. C., Patchva S., Aggarwal B. B. AAPS J. 2013;15:195–218. doi: 10.1208/s12248-012-9432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shome S., Talukdar A. D., Choudhury M. D., Bhattacharya M. K., Upadhyaya H. J. Pharm. Pharmacol. 2016;68:1481–1500. doi: 10.1111/jphp.12611. [DOI] [PubMed] [Google Scholar]
- Sabry R., Preparation of hydroxyapatite nanoparticles by using emulsion liquid membrane [PhD Thesis], Al-Azhar University, Egypt, 2012. [Google Scholar]
- Tang Z. X., Qian J. Q., Shi L. E. Appl. Biochem. Biotechnol. 2007a;136:77–96. doi: 10.1007/BF02685940. [DOI] [PubMed] [Google Scholar]
- Tang Z. X., Shi L. E., Qian J. Q. Biochem. Eng. J. 2007b;34:217–223. [Google Scholar]
- Abdel-Wahhab M. A., Aljawish A., El-Nekeety A. A., Abdel-Aiezm S. H., Abdel-Kader H. A., Rihn B. H., Joubert O. Toxicol. Rep. 2015;2:737–747. doi: 10.1016/j.toxrep.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav A., Lomash V., Samim M., Flora S. J. Chem.-Biol. Interact. 2012;199:49–61. doi: 10.1016/j.cbi.2012.05.011. [DOI] [PubMed] [Google Scholar]
- Spence J. C. H., High-Resolution Electron Microscopy, Oxford Univ. Press, Oxford, UK, 2003. [Google Scholar]
- Spence J. C. H., Kolar H. R., Hembree G., Humphreys C. J., Barnard J., Datta R., Justo J. F. Philos. Mag. 2006;86:4781–4796. [Google Scholar]
- Tappel A. L., Zalkin H. Arch. Biochem. Biophys. 1959;80:333–336. [Google Scholar]
- Koracevic D., Koracevic G., Djordjevic V., Andrejevic S., Cosic V. J. Clin. Pathol. 2001;54:356–361. doi: 10.1136/jcp.54.5.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery H. A. C., Dymock J. F. Analyst. 1961;86:414–416. [Google Scholar]
- Misra H. P., Fridovich I. J. Biol. Chem. 1972;247:3170–3175. [PubMed] [Google Scholar]
- Chiu D. T., Stults F. H., Tappel A. L. Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol. 1976;445:558–566. doi: 10.1016/0005-2744(76)90110-8. [DOI] [PubMed] [Google Scholar]
- Habig W. H., Pabst M. J., Jakoby W. B. J. Biol. Chem. 1974;249:7130–7139. [PubMed] [Google Scholar]
- Luck H., Catalase, in Method of Enzymatic Analysis, ed. M. V. Bergmayer, Verlag. Chemic., Academic Press, New, 1974, p. 885. [Google Scholar]
- Jollow D. J., Mitchell J. R., Zampaglione N. A., Gillette J. R. Pharmacology. 1974;11:151–169. doi: 10.1159/000136485. [DOI] [PubMed] [Google Scholar]
- Drury R. A., Wallington E. A. and Carleton S., in Histological Techniques, Oxford University Press, London, New York, Toronto, 5th edn, 1980, pp. 241–242. [Google Scholar]
- SAS, Statistical Analysis System. SAS Procedure Guide. Release 6.03 Edition, SAS Institute Inc., Cary, Nc, U.S.A., 1998. [Google Scholar]
- Duncan D. B. Biometrics. 1955;11:1–42. [Google Scholar]
- Hart S. E. and Kinter L. B., Assessing renal effects of toxicants in vivo, in Toxicology of the Kidney, ed. J. B. Tarloff and L. H. Lash, 2005, pp. 81–147. [Google Scholar]
- Nemenqani D., El-Gharib O., Ahmed A. M., Baiuomy A. R. Int. Res. J. Appl. Basic Sci. 2015;9:502–509. [Google Scholar]
- Wang Y., Aker W. G., Hwang H. M., Yedjou C. G., Yu H., Tchounwou P. B. Sci. Total Environ. 2011;409:4753–4762. doi: 10.1016/j.scitotenv.2011.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Liu C., Yang D., Zhang H., Xi Z. J. Appl. Toxicol. 2009;29:69–78. doi: 10.1002/jat.1385. [DOI] [PubMed] [Google Scholar]
- Chen Q., Xue Y., Sun J. J. Appl. Toxicol. 2014;34:1256–1264. doi: 10.1002/jat.3073. [DOI] [PubMed] [Google Scholar]
- Xu Z., Liu C., Wei J., Sun J. J. Appl. Toxicol. 2012b;32:429–435. doi: 10.1002/jat.1745. [DOI] [PubMed] [Google Scholar]
- Turkez H., Yousef M. I., Sönmez E., Togar B., Bakan F., Sozio P., Stefano A. D. J. Appl. Toxicol. 2014;34:373–379. doi: 10.1002/jat.2958. [DOI] [PubMed] [Google Scholar]
- Sies H., Akerboom T. P. Methods Enzymol. 1984;105:445–451. doi: 10.1016/s0076-6879(84)05062-x. [DOI] [PubMed] [Google Scholar]
- Xu J., Xu P., Li Z., Huang J., Yang Z. J. Biomed. Mater. Res., Part A. 2012a;100:738–745. doi: 10.1002/jbm.a.33270. [DOI] [PubMed] [Google Scholar]
- Parayanthala-Valappil M., Santhakumar S., Arumugam S. Int. J. Biomater. 2014:2014. doi: 10.1155/2014/476942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangerich A., Knutson C. G., Parry N. M., Muthupalani S., Ye W., Prestwich E., Cui L., McFaline J. L., Mobley M., Ge Z., Taghizadeh K. Proc. Natl. Acad. Sci. U. S. A. 2012;109:1820–1829. doi: 10.1073/pnas.1207829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke M. S., Evans M. D., Dizdaroglu M., Lunec J. FASEB J. 2003;17:195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- Cheng K. C., Cahill D. S., Kasai H., Nishimura S., Loeb L. A. J. Biol. Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- Wen Z. S., Liu L. J., Qu Y. L., OuYang X. K., Yang L. Y., Xu Z. R. Mar. Drugs. 2013;11:3582–3600. doi: 10.3390/md11103582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Denshary E. S., Aljawish A., El-Nekeety A. A., Hassan N. S., Saleh R. H., Rihn B. H., Abdel-Wahhab M. A. Soft Nanosci. Lett. 2015;5:36–50. [Google Scholar]
- Jeon T. L., Hwang S. G., Park N. G., Jung Y. R., Shin S. I., Choi S. D., Park D. K. Toxicology. 2003;187:67–73. doi: 10.1016/s0300-483x(03)00003-9. [DOI] [PubMed] [Google Scholar]
- Mohamed N. E. Am. J. Sci. 2011;7:406–417. [Google Scholar]
- Kuppusamy S., Karuppaiah J. Asian Pac. J. Trop. Dis. 2012;2:769–773. [Google Scholar]
- Prashanth K. V. H., Dharmesh S. M., Rao K. S. J., Tharanathan R. N. Carbohydr. Res. 2007;342:190–195. doi: 10.1016/j.carres.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Flora G., Gupta D., Tiwari A. Biol. Trace Elem. Res. 2013;152:31–40. doi: 10.1007/s12011-012-9586-3. [DOI] [PubMed] [Google Scholar]
- Nehra S., Bhardwaj V., Ganju L., Saraswat D. PLoS One. 2015;10:0139121. doi: 10.1371/journal.pone.0139121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav A., Flora S. J. S., Kushwaha P. Def. Life Sci. J. 2016;1:69–77. [Google Scholar]
- Ahmed-Farid O. A., Nasr M., Ahmed R. F., Bakeer R. M. J. Biomed. Sci. 2017;24:66. doi: 10.1186/s12929-017-0373-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzegar A., Moosavi-Movahedi A. A. PLoS One. 2011;6:26012. doi: 10.1371/journal.pone.0026012. [DOI] [PMC free article] [PubMed] [Google Scholar]