Abstract
Undifferentiated uterine sarcoma is a diagnosis of exclusion with limited molecular genetic data available. Recent recognition of high-grade endometrial stromal sarcomas with diverse genotypes suggests that some tumors classified as undifferentiated uterine sarcomas may represent misdiagnosed high-grade endometrial stromal sarcomas. Archival material from 10 tumors diagnosed as undifferentiated uterine sarcomas in 2009–2017 were collected. BCOR immunohistochemistry and fluorescence in situ hybridization (FISH) using break-apart probes flanking BCOR, ZC3H7B, CCNB3, YWHAE, NUTM2, JAZF1, and BCORL1 were performed. Tumors lacking or harboring gene rearrangement with no known fusion partner by FISH were subjected to targeted RNA sequencing. Morphology was correlated with FISH and sequencing results. BCOR expression was moderate to strong in ≥50% of cells in eight tumors, while weak in <5% cells and negative in two. FISH detected mutually exclusive ZC3H7B-BCOR and YWHAE-NUTM2 fusions in three uniform undifferentiated uterine sarcomas; two pleomorphic tumors harbored YWHAE rearrangement with no known partner. Targeted RNA sequencing of five FISH-negative uniform undifferentiated uterine sarcomas detected BRD8-PHF1 and YWHAE-NUTM2B fusions and BCOR internal tandem duplication in four of them. Tumors with YWHAE-NUTM2 fusions and BCOR genetic abnormalities showed morphology characteristic of high-grade endometrial stromal sarcomas. No fusions were detected by sequencing in the tumor with YWHAE rearrangement only by FISH. Most tumors classified as undifferentiated uterine sarcomas represent misdiagnosed high-grade endometrial stromal sarcomas. BCOR expression in ≥50% of cells may help triage tumors for molecular confirmation of high-grade endometrial stromal sarcoma-related genetic abnormalities. Novel YWHAE rearrangements may define a subset of true undifferentiated pleomorphic sarcomas.
Keywords: endometrial stromal sarcoma, undifferentiated uterine sarcoma, undifferentiated endometrial sarcoma, YWHAE, BCOR, PHF1
INTRODUCTION
Undifferentiated uterine sarcoma is an aggressive uterine sarcoma subtype that lacks morphologic and immunophenotypic evidence of cell differentiation. Previously known as “undifferentiated endometrial sarcomas,” these tumors may arise in the endometrium or myometrium, consist of highly atypical cells arranged in sheets and storiform or herringbone patterns. Rhabdoid morphology and myxoid matrix may be seen. Destructive myometrial invasion, brisk mitotic activity, tumor necrosis, and vascular involvement are frequently present. While clinical data is limited, the prognosis is reportedly dismal (1). The diagnosis of undifferentiated uterine sarcoma is one of exclusion after consideration of more common uterine sarcoma subtypes such as leiomyosarcoma and endometrial stromal sarcoma.
In practice, undifferentiated uterine sarcomas often exhibit a variety of morphologic features. This observation led to a proposed subclassification of undifferentiated uterine sarcomas into pleomorphic and uniform types, with marked nuclear pleomorphism characteristic of the former and relative nuclear isomorphism indicative of the latter (1). By immunohistochemistry, pleomorphic and uniform undifferentiated uterine sarcomas also appear distinct with estrogen and progesterone receptors as well as b-catenin and cyclin D1 more frequently expressed among uniform tumors and aberrant p53 staining patterns limited to pleomorphic ones (1).
Molecular genetic data among undifferentiated uterine sarcomas are limited due to the rarity of this tumor type. Few early reports suggest that undifferentiated uterine sarcomas have complex karyotypes and frequently harbor TP53 mutations (2, 3). Subsequent studies have identified JAZF1-SUZ12 (4), YWHAE-NUTM2 (5, 6), and ZC3H7B-BCOR (7, 8) gene fusions among tumors classified as undifferentiated uterine sarcomas. Those harboring YWHAE and BCOR rearrangements likely represent high-grade endometrial stromal sarcomas that were previously unrecognized due to lack of available novel fusion detection methods at the time of diagnosis.
These recent studies suggest that some tumors morphologically classified as undifferentiated uterine sarcomas may in fact represent misdiagnosed high-grade endometrial stromal sarcomas. We performed a combination of BCOR immunohistochemistry, fluorescence in situ hybridization (FISH), and next-generation targeted RNA sequencing to define the morphologic and genetic landscape of lesions classified as undifferentiated uterine sarcomas.
MATERIAL AND METHODS
Case Selection
Undifferentiated uterine sarcomas diagnosed between 2009 and 2017 were retrospectively identified by searching the pathology database at three institutions (Memorial Sloan Kettering Cancer Center, New York, NY, USA; Massachusetts General Hospital, Boston, MA, USA; and Instituto Português de Oncologia de Lisboa Francisco Gentil, Lisbon, Portugal). Search terms included “undifferentiated uterine sarcoma,” “undifferentiated endometrial sarcoma,” and “undifferentiated sarcoma.” After this initial search, 10 tumors with hematoxylin-and-eosin and immunohistochemical stained slides as well as formalin-fixed paraffin-embedded tumoral material were retrieved. All available slides and pathology reports were reviewed by two pathologists (SC and PC), and morphologic features were recorded. Tumors were also subclassified as pleomorphic or uniform based on definitions previously described by Kurihara and colleagues (1). Available clinical data, including patient age and FIGO stage based on the 2009 FIGO staging system for uterine sarcomas (9), were also extracted from medical records.
Immunohistochemistry
Immunohistochemical staining for BCOR was performed on all tumors. In brief, a commercially available monoclonal antibody, clone C-10 (sc-514576; Santa Cruz, Dallas, TX) at 1:150 dilution (1.7 μg/ml) was applied to five μm formalin-fixed paraffin-embedded whole tissue sections as previously described (10). The staining was performed on the Leica Bond-3 autostaining system (Leica, Buffalo Grove, IL, USA) using heat-based antigen retrieval, a high pH buffer solution (Leica, ER2, 30 min), 30 min primary incubation time, and a polymer detection system (Refine, Leica). A carrier-based multitissue block comprising of normal skin, colon, lung, testis, spleen, placenta, pancreas, liver, and kidney served as negative controls (11). Intensity of the staining (strong, moderate, weak, and negative) and estimated percentage of positive tumor cells (nuclear staining only) were evaluated.
FISH
Break-apart FISH using probes flanking BCOR (8), ZC3H7B (8), CCNB3 (12), YWHAE (13)), NUTM2 (13), JAZF1 (14), and BCORL1 (15) genes was performed on five μm formalin-fixed, paraffin-embedded whole tissue sections of tumors with unknown gene rearrangement status and BCOR nuclear immunoexpression of any intensity in ≥50% of tumor cells as previously described (10). Custom probes were made by bacterial artificial chromosomes (BAC) clones obtained from BAC/PAC Resources (Children’s Hospital Oakland Research Institute, Oakland, CA, USA). BAC clones were labeled with nick translation and validated on normal metaphase chromosomes. Slides were de-paraffinized, pre-treated, and hybridized with denatured probes overnight, followed by post-hybridization washes and counterstaining with DAPI. Slides were examined on a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany) using Isis 5 software (Metasystems). Gene rearrangement was confirmed if break-apart signals were seen in at least 20% of 200 tumor nuclei counted.
Targeted RNA sequencing
Tumor samples lacking or harboring BCOR, ZC3H7B, CCNB3, YWHAE, NUTM2, JAZF1 and BCORL1 rearrangements with no fusion partner by FISH were subjected to the Archer FusionPlex Custom Solid Panel (ArcherDX Inc., Boulder, CO, USA), a next-generation targeted RNA sequencing assay utilizing the Anchored Multiplex PCR (AMP) technology that detects gene fusions and oncogenic isoforms in selected protein-coding exons of 62 genes (16). Tumor RNA was extracted from five μm, formalin-fixed, paraffin-embedded tumoral tissue sections followed by cDNA synthesis and library preparation. Final targeted amplicons were sequenced on an Illumina MiSeq. Data were analyzed using the Archer Software (version 4.0.10; ArcherDX Inc.).
RESULTS
Clinicopathologic features
Patient age ranged from 50 to 71 (median, 54) years, and tumor size ranged from 3.0 to 13.0 (median, 8.4) cm (Table 1). Myometrial invasion was destructive in three tumors, and tongue-like in seven. Lymphovascular invasion was seen in five tumors. Eight patients had FIGO stage I tumors, while the remaining two had FIGO stage IV disease based on the 2009 FIGO staging system for uterine sarcomas (9). Eight tumors were subclassified as undifferentiated uterine sarcoma of uniform type in which three were spindled and five were spindled and rounded, while two tumors were considered undifferentiated pleomorphic sarcomas.
Table 1.
Clinicopathologic and molecular features.
| Case | Age, y | Stage | Year | Morphology | BCOR expression | FISH | RNA sequencing | Revised diagnosis |
|---|---|---|---|---|---|---|---|---|
| 1 | 58 | IB | 2009 | Pleomorphic | M-S, 50% | YWHAE rearrangement only | Failed | UUS |
| 2 | 50 | IA | 2010 | Uniform | S, ≥95% | YWHAE-NUTM2B | Not performed | HGESS |
| 3 | 53 | IB | 2011 | Uniform | W, <5% | ZC3H7B-BCOR | Not performed | HGESS |
| 4 | 55 | IVA | 2013 | Uniform | M-S, 50% | Negative | BRD8-PHF1 | HGESS |
| 5 | 71 | IB | 2014 | Uniform | S, ≥95% | ZC3H7B-BCOR | Not performed | HGESS |
| 6 | 54 | IB | 2016 | Uniform | Negative | Negative | Negative | UUS |
| 7 | 52 | IA | 2016 | Uniform | S, ≥95% | Negative | BCOR ITD | HGESS |
| 8 | 57 | IVA | 2017 | Pleomorphic | S, ≥95% | YWHAE rearrangement only | Negative | UUS |
| 9 | 54 | IB | 2017 | Uniform | S, ≥95% | Negative | YWHAE-NUTM2B | HGESS |
| 10 | 61 | IB | 2017 | Uniform | S, ≥95% | Negative | BCOR ITD | HGESS |
FISH indicates fluorescence in situ hybridization; HGESS, high-grade endometrial stromal sarcoma; M, moderate; S, strong; UUS, undifferentiated uterine sarcoma; W, weak.
Immunohistochemical findings
Nuclear BCOR expression was strong in ≥95% of cells in six tumors, moderate to strong in 50% of cells in two tumors, and weak in <5% of cells in one tumor (Table 1). No BCOR expression was seen in the remaining tumor with appropriate positive control. CD10, ER, PR, and cyclin D1 expression profiles of all tumors are presented in Supplementary Table I.
FISH findings
FISH was successful in all cases. Rearrangements were detected in 5 of 10 (50%) tumors (Table 1). ZC3H7B-BCOR and YWHAE-NUTM2 gene fusions were identified in two (spindled only) and one (spindled and rounded) undifferentiated uterine sarcomas of uniform histology, respectively (Figures 1 and 2). YWHAE rearrangement with no identifiable gene partner was detected in two pleomorphic tumors (Figure 3). No BCOR, ZC3H7B, CCNB3, YWHAE, NUTM2, JAZF1 and BCORL1 gene rearrangements were detected in the remaining five tumors.
Figure 1.
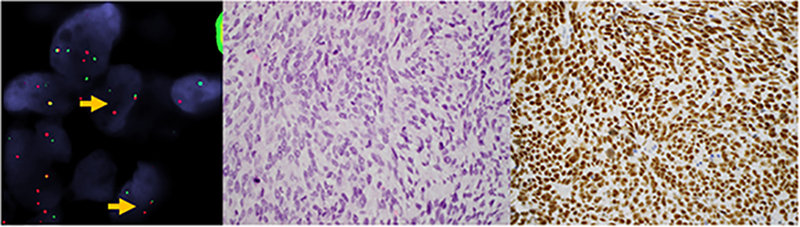
Uterine sarcoma harboring ZC3H7B-BCOR fusion. A) FISH demonstrates separation of red and green probe signals flanking the BCOR gene (arrows) in a tumor composed of B) spindle cells with intermediate grade nuclei, open chromatin pattern, and scant cytoplasm in a myxoid background. C) Strong BCOR expression was seen in ≥95% of cells.
Figure 2.
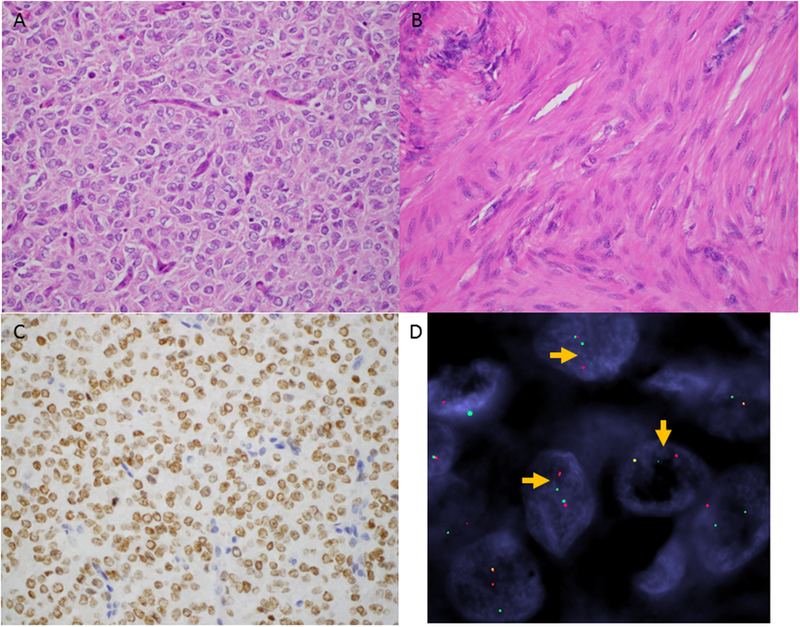
Uterine sarcoma harboring YWHAE-NUTM2 fusion. The tumor is composed of (A) a high-grade round cell and (B) low-grade spindle cell components. C) Strong BCOR expression is seen in ≥95% of cells of the high-grade round cell component. D) YWHAE rearrangement is confirmed by FISH with clear separation of the red and green probe signals flanking the gene (arrows).
Figure 3.
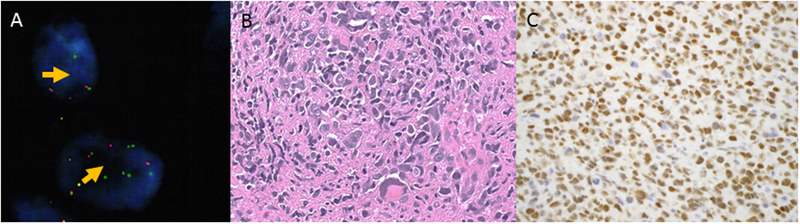
Undifferentiated pleomorphic sarcoma harboring YWHAE gene rearrangement. A) YWHAE rearrangement is confirmed by separation of green and red probe signals flanking the gene (arrows) by FISH. B) Markedly pleomorphic cells (C) exhibit strong BCOR immunoreactivity.
Targeted RNA sequencing findings
Targeted RNA sequencing was performed on seven tumors, including five that were negative for gene rearrangements and two with YWHAE gene rearrangement detected only by FISH. Sequencing was successful in six cases. BCOR exon 15 internal tandem duplication and BRD8-PHF1 fusion (exons 9 and 2) were detected in two and one tumors, respectively (Figures 4 and 5). YWHAE-NUTM2B gene fusion (exons 5 and 2) was detected in another tumor in which rearrangements were not detected by FISH (Figure 6). Sequencing did not identify fusions in one tumor harboring YWHAE rearrangement only and another tumor negative for rearrangements by FISH. Overall, endometrial stromal sarcoma-associated gene abnormalities were detected in four of six (67%) tumors sequenced.
Figure 4.
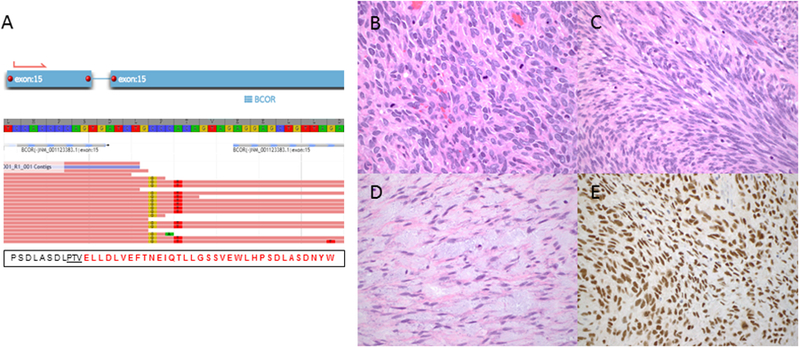
Uterine sarcoma harboring BCOR internal tandem duplication. A) Schematic illustrates BCOR exon 15 internal tandem duplication detected by targeted RNA sequencing. The tumor consists of B) high-grade round cell, C) high-grade spindle cell, and D) low-grade fibromyxoid spindle cell components. E) Strong BCOR expression in ≥95% of tumor cells is seen in all components.
Figure 5.
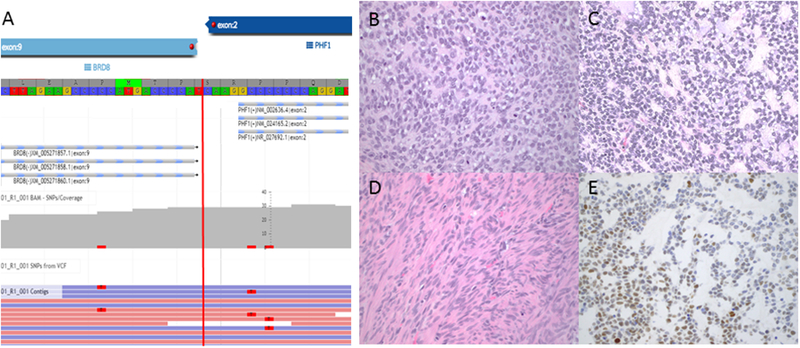
BRD8-PHF1 fusion-positive sarcoma. A) Schematic illustrates the fusion of exon 9 of BRD8 with exon 2 of PHF1. The round cell component is arranged in sheets (B) and small nests and trabeculae embedded in myxoid matrix (C). D) A bland spindle cell component is also present. E) Moderate BCOR expression is seen in 50% of tumor cells.
Figure 6.
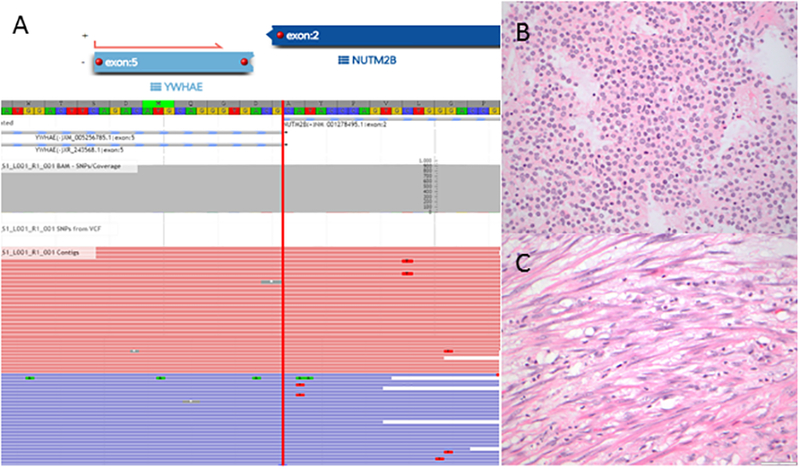
YWHAE-NUTM2 fusion detected in a tumor negative for YWHAE rearrangement by FISH. A) Schematic illustrates fusion of YWHAE exon 5 and NUTM2B exon 2 by targeted RNA sequencing. The tumor consists of a (B) high-grade round cell and (C) low-grade fibromyxoid components.
Genotype-phenotype correlation
YWHAE-NUTM2 fusion-positive tumors
YWHAE-NUTM2 fusions were detected in two tumors. One was confirmed by FISH; the other was negative for YWHAE and NUTM2 rearrangements by FISH, but the YWHAE-NUTM2B fusion was detected by targeted RNA sequencing. Both tumors consisted of high-grade round and low-grade spindle cell components. Sheets of round cells with intermediate size round to ovoid nuclei, open chromatin, and scant to moderate eosinophilic cytoplasm were present adjacent to fascicles of bland spindle cells with elongated nuclei, homogeneous chromatin, and ample eosinophilic cytoplasm resembling fibroblastic low-grade endometrial stromal sarcoma (Figures 2A–B and 6B). Mitotic index was 10 and 15 per 10 high power fields in the round cell components of both cases, respectively. One of the tumors also displayed myxoid matrix in the low-grade spindle cell component (Figure 6C). Both demonstrated tongue-like myometrial invasion, and lymphovascular involvement was present in one tumor. BCOR expression was strong in ≥95% of tumor cells in the round cell components of both tumors (Figures 2C). CD10, ER, PR, and cyclin D1 were performed as part of the diagnostic work up in one of the tumors; cyclin D1 was diffusely positive, while CD10, ER, and PR were negative in the round cell component. The morphology seen in both cases is similar to typical YWHAE-NUTM2 high-grade endometrial stromal sarcomas (17), and thus reclassified as such. One tumor was diagnosed as undifferentiated uterine sarcoma in 2010, before the recognition of the unique histologic features of YWHAE-NUTM2 high-grade endometrial stromal sarcomas. The other tumor was diagnosed as undifferentiated uterine sarcoma in 2017 due to the absence of YWHAE rearrangement by FISH.
YWHAE gene rearrangement-only tumors
Both tumors harboring YWHAE rearrangement with no known partner by FISH demonstrated sheets of highly atypical cells with extensive nuclear pleomorphism (Figure 3A–B). Both showed destructive myometrial invasion. One tumor also demonstrated hyalinized and myxoid stroma, tumor necrosis, and a mitotic index of 56 per 10 high power fields (Figure 3B). No lymphovascular invasion was identified. The tumor was strongly positive for BCOR in ≥95% of cells and weakly positive for cyclin D1 in <50% of cells, while negative for CD10, SMA, desmin, melan A, S100, and ALK (Figure 3C). The other tumor consisted of spindle and round cells with marked nuclear pleomorphism, prominent nucleoli, and moderate eosinophilic cytoplasm. Infarct necrosis, lymphovascular invasion, and 18 mitotic figures per 10 high power fields were seen. BCOR expression was moderate to strong in 50% of tumor cells. Immunohistochemical stains were not performed as a part of the diagnostic work up in this tumor.
ZC3H7B-BCOR fusion-positive tumors
Both tumors were composed of spindle cells with intermediate size, round to ovoid to elongated nuclei, homogeneous chromatin, inconspicuous or one to three pinpoint nucleoli, and scant to moderate eosinophilic cytoplasm (Figure 1B). Mitotic index was 12 and 39 per 10 high power fields, respectively. Abundant myxoid matrix as well as tongue-like myometrial and lymphovascular invasion was seen in both tumors. BCOR expression was strong in ≥95% of cells in one tumor (Figure 1C) while weak in <5% of cells in the other, correlating with previous reports of BCOR expression being informative in only 50% of BCOR-rearranged high-grade endometrial stromal sarcomas (8, 10). Immunohistochemistry performed in one case showed that the tumor was weakly positive for CD10, while negative for ER, PR, desmin, SMA, and h-caldesmon. Both tumors were diagnosed in 2014 and 2011 before the recognition of high-grade endometrial stromal sarcomas with ZC3H7B-BCOR fusion (7, 8). Based on histologic features similar to those described in published reports, these two tumors were re-classified as high-grade endometrial stromal sarcoma.
BCOR internal tandem duplication tumors
Both tumors harboring BCOR exon 15 internal tandem duplication showed similar morphologic features. They were comprised of high-grade round cell, high-grade spindle cell, and low-grade spindle cell components with variable amounts of myxoid matrix (Figure 4B–D). The round cell component consisted of sheets of cells with intermediate size round to ovoid nuclei, coarse chromatin, small nucleoli, and scant pale cytoplasm. The high-grade spindle cell component consisted of haphazard fascicles and bundles of cells with elongated pleomorphic nuclei, coarse chromatin, and ample eosinophilic cytoplasm. The low-grade spindle cell component resembled low-grade fibrous or fibromyxoid endometrial stromal sarcoma and consisted of bundles of bland cells with elongated nuclei, homogeneous chromatin, inconspicuous nucleoli, and abundant eosinophilic cytoplasm. The mitotic index was brisk (21 and 36 per 10 high power fields, respectively) in the high-grade round and spindle cell components, but low (less than one per 10 high power fields) in the low-grade spindle cell component. Tongue-like myometrial and lymphovascular invasion was seen. Strong BCOR expression was seen in ≥95% of tumor cells in both high- and low-grade components (Figure 4E). The tumors were also focally positive for CD10, desmin, and ER, while negative for SMA. Both tumors morphologically resembled published reports of high-grade endometrial stromal sarcoma with BCOR internal tandem duplication (10, 18) and were re-classified as such after re-review of the morphology and molecular results.
BRD8-PHF1 fusion-positive tumor
The tumor consisted of round and spindle cells components (Figure 5B–D). The round cells had intermediate-size, round to ovoid nuclei with coarse chromatin, one to two small nucleoli and scant cytoplasm. They were arranged in cords, trabeculae, small nests and solid sheets embedded in abundant myxoid stroma. In these foci, the mitotic index reached 32 per 10 high power fields. In contrast, the spindle cell component consisted of fascicles of bland spindle cells with elongated nuclei, homogeneous chromatin, and abundant eosinophilic cytoplasm resembling low-grade fibroblastic endometrial stromal sarcoma. Mitotic index in these areas was less than one per 10 high power fields. Delicate arteriolar vasculature was present, but no tumor cell whorling around arterioles was seen. Infarct type necrosis was present throughout. Tongue-like myometrial and vascular invasion was also identified. Moderate to strong BCOR expression was seen in 50% of tumor cells (Figure 5E). The tumor was also diffusely positive for cyclin D1 and focally positive for CD10 and c-kit, while negative for EMA, ER, PR, CK18, inhibin, calretinin, and desmin. Given the recent publication of BRD8-PHF1 fusion in a low-grade endometrial stromal sarcoma, the current tumor was re-classified as high-grade endometrial stromal sarcoma based on the fusion status and presence of high-grade histologic features.
DISCUSSION
Using a combination of BCOR immunohistochemistry, FISH, and next-generation targeted RNA sequencing, we defined the genetic landscape of a series of tumors classified as undifferentiated uterine sarcoma. Known endometrial stromal sarcoma-associated genetic aberrations were detected in 70% of tumors tested and included ZC3H7B-BCOR (n=2), YWHAE-NUTM2 (n=2), and BRD8-PHF1 (n=1) fusions as well as BCOR internal tandem duplications (n=2). Based on re-review of the morphology and identification of characteristic genetic abnormalities, these tumors were re-classified as high-grade endometrial stromal sarcomas. Novel YWHAE gene rearrangements with no known partner were also detected by FISH in two additional undifferentiated uterine sarcomas with marked nuclear pleomorphism and may represent novel fusions in this sarcoma subtype. BCOR expression in ≥50% of cells corresponded to the presence of gene fusion or internal tandem duplication in 89% (eight of nine) of tumors, suggesting its utility in triaging tumors for molecular confirmation of high-grade endometrial stromal sarcoma-associated genetic abnormalities.
Undifferentiated uterine sarcoma has long been regarded as a diagnosis of exclusion after consideration of more common uterine sarcoma entities such as leiomyosarcoma and endometrial stromal sarcoma using a combination of morphologic evaluation and immunohistochemical stains. Three of our tumors were diagnosed as undifferentiated uterine sarcoma before 2012, the year YWHAE-NUTM2 high-grade endometrial stromal sarcoma was first described (17, 19). An additional five tumors were also diagnosed as undifferentiated uterine sarcoma between 2012 and 2016, before endometrial stromal sarcomas with BCOR genetic abnormalities (7, 8, 10, 18) and BRD8-PHF1 fusion (20) were subsequently characterized. Among these eight tumors previously considered undifferentiated uterine sarcoma prior to 2017, five (63%) were now reclassified as high-grade endometrial stromal sarcoma after morphologic evaluation and detection of characteristic endometrial stromal sarcoma-associated genetic abnormalities by molecular genetic assays that were not available for clinical use at the time of initial diagnosis. This suggests that most tumors that are classified as undifferentiated uterine sarcomas in fact represent misdiagnosed high-grade endometrial stromal sarcomas with various genotypes that may now be accurately recognized through a combination of morphologic, immunohistochemical, and molecular tools.
In 2017, when high-grade endometrial stromal sarcomas with YWHAE and BCOR genetic abnormalities should be recognized, one of our tumors was diagnosed as undifferentiated uterine sarcoma based on the absence of the suspected genetic abnormality by FISH. It displayed histologic (high-grade round and low-grade spindle cell components) and immunophenotypic features (cyclin D1, BCOR positive; ER, PR, CD10 negative) characteristic of YWHAE-NUTM2 high-grade endometrial stromal sarcoma. No YWHAE gene rearrangements were detected by FISH clinically at the time of diagnosis and in our study. YWHAE-NUTM2B gene fusion, however, was detected by target RNA sequencing. This case illustrates the utility of target RNA sequencing in its sensitivity in detecting cryptic gene rearrangements in addition to its advantage over FISH and DNA-based fusion assays in novel fusion discovery. Confirmation of the suspected genetic abnormality with an alternative molecular assay is also necessary when it is not initially detected by a single method and there is strong suspicion of the disease entity based on morphology and immunophenotype.
It is not surprising that YWHAE-NUTM2 fusions as well as BCOR gene rearrangements and internal tandem duplications were found among our tumors that demonstrated nuclear isomorphism. Interestingly, however, novel YWHAE gene rearrangements with no known fusion partners were detected by FISH in two undifferentiated uterine sarcomas demonstrating marked nuclear pleomorphism and strong BCOR expression in at least 50% of tumor cells by immunohistochemistry. Targeted RNA sequencing was attempted in both cases, but RNA quality was insufficient to pass the quality control standards in one tumor, no gene fusions or isoforms were detected in the other. Our current RNA sequencing panel uses primers targeting YWHAE exon 5, the most common YWHAE exon described in YWHAE-NUTM2 fusions. It is possible that in at least one of our cases, the rearrangement detected by FISH involves YWHAE exons 1–4. Confirmation of YWHAE rearrangement and an alternative fusion partner among a larger series of undifferentiated pleomorphic sarcomas would be of great interest in identifying yet another molecular subset of uterine sarcomas. For the time being, however, we would still consider these tumors as undifferentiated pleomorphic sarcomas given the lack of nuclear isomorphism typically seen in both high- and low-grade endometrial stromal sarcomas.
A BRD8-PHF1 fusion was detected by targeted RNA sequencing in one of our tumors that was previously considered an undifferentiated uterine sarcoma due to the absence of YWHAE rearrangement. While PHF1 rearrangements have been well recognized among low-grade endometrial stromal sarcomas, fusion with an alternative partner, BRD8, was only recently described (20). This tumor reportedly showed sex cord-like differentiation and dense collagenous matrix deposition along with strong CD10 expression; based on the published histologic images, the tumor appeared morphologically low-grade. In contrast, however, our tumor was composed of monomorphic cells with intermediate size nuclei and prominent nucleoli arranged in strands, cords, and solid sheets. Mitotic index was brisk with 32 per 10 high power fields. The cells were embedded in myxoid stroma and associated with tongue-like myometrial and lymphovascular invasion. Variable cyclin D1, c-kit, and CD10 expression was present; ER and PR were both negative. Of interest, BCOR expression was moderate to strong in 50% of cells, similar to the pattern of cyclin D1 staining. Based on histologic features that exceed those acceptable for low-grade endometrial stromal sarcoma and the presence of an endometrial stromal sarcoma-associated gene fusion, this tumor was ultimately reclassified as a high-grade endometrial stromal sarcoma rather than undifferentiated uterine sarcoma. A larger cohort of BRD8-PHF1 fusion-positive tumors is required to confirm its inclusion among other well-known high-grade endometrial stromal sarcomas.
BCOR expression by immunohistochemistry has previously been shown to aid identification of genetically diverse high-grade endometrial stromal sarcomas, including those with variant morphologies (10). In our current study, BCOR immunohistochemistry was useful in predicting YWHAE and BCOR genetic abnormalities in the majority of tested tumors. BCOR was informative in all tumors harboring YWHAE rearrangement and BCOR internal tandem duplication and in only one of two tumors harboring BCOR rearrangement, confirming previously published observations (10). While the BCOR antibody clone C-10 detects the epitope encoded by exons 1, 2 and 3, and part of exon 4 of BCOR, BCOR immunoexpression does not appear to correlate with BCOR breakpoints in the fusion transcripts (10).
In summary, most tumors classified as undifferentiated uterine sarcomas based on morphologic and immunohistochemical features harbor endometrial stromal sarcoma-associated genetic aberrations and likely represent misdiagnosed high-grade endometrial stromal sarcomas of various genotypes. Entities that must be considered prior to a diagnosis of undifferentiated uterine sarcoma include high-grade endometrial stromal sarcomas harboring YWHAE, BCOR, and PHF1 rearrangements as well as BCOR internal tandem duplication. Novel YWHAE rearrangements may define a subset of bonafide undifferentiated pleomorphic sarcomas. BCOR immunoexpression may be helpful in triaging cases for molecular profiling.
Supplementary Material
REFERENCES
- 1.Kurihara S, Oda Y, Ohishi Y, et al. Endometrial stromal sarcomas and related high-grade sarcomas: immunohistochemical and molecular genetic study of 31 cases. Am J Surg Pathol. 2008;32:1228–1238. [DOI] [PubMed] [Google Scholar]
- 2.Halbwedl I, Ullmann R, Kremser ML, et al. Chromosomal alterations in low-grade endometrial stromal sarcoma and undifferentiated endometrial sarcoma as detected by comparative genomic hybridization. Gynecol Oncol. 2005;97:582–587. [DOI] [PubMed] [Google Scholar]
- 3.Gil-Benso R, Lopez-Gines C, Navarro S, et al. Endometrial stromal sarcomas: immunohistochemical, electron microscopical and cytogenetic findings in two cases. Virchows Arch. 1999;434:307–314. [DOI] [PubMed] [Google Scholar]
- 4.Koontz JI, Soreng AL, Nucci M, et al. Frequent fusion of the JAZF1 and JJAZ1 genes in endometrial stromal tumors. Proc Natl Acad Sci U S A. 2001;98:6348–6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Croce S, Hostein I, Ribeiro A, et al. YWHAE rearrangement identified by FISH and RT-PCR in endometrial stromal sarcomas: genetic and pathological correlations. Mod Pathol. 2013;26:1390–1400. [DOI] [PubMed] [Google Scholar]
- 6.Sciallis AP, Bedroske PP, Schoolmeester JK, et al. High-grade endometrial stromal sarcomas: a clinicopathologic study of a group of tumors with heterogenous morphologic and genetic features. Am J Surg Pathol. 2014;38:1161–1172. [DOI] [PubMed] [Google Scholar]
- 7.Hoang LN, Aneja A, Conlon N, et al. Novel high-grade endometrial stromal sarcoma: a morphologic mimicker of myxoid leiomyosarcoma. Am J Surg Pathol. 2017;41:12–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis N, Soslow RA, Delair DF, et al. ZC3H7B-BCOR high-grade endometrial stromal sarcomas: a report of 17 cases of a newly defined entity. Mod Pathol. 2018;31:674–684. [DOI] [PubMed] [Google Scholar]
- 9.Prat J FIGO staging for uterine sarcomas. Int J Gynaecol Obstet. 2009;104:177–178. [DOI] [PubMed] [Google Scholar]
- 10.Chiang S, Lee CH, Stewart CJR, et al. BCOR is a robust diagnostic immunohistochemical marker of genetically diverse high-grade endometrial stromal sarcoma, including tumors exhibiting variant morphology. Mod Pathol. 2017;30:1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frosina D, Jungbluth AA. A novel technique for the generation of multitissue blocks using a carrier. Appl Immunohistochem Mol Morphol. 2016;24:668–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kao YC, Owosho AA, Sung YS, et al. BCOR-CCNB3 fusion positive sarcomas: a clinicopathologic and molecular analysis of 36 cases with comparison to morphologic spectrum and clinical behavior of other round cell sarcomas. Am J Surg Pathol. 2018;42:604–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kao YC, Sung YS, Zhang L, et al. Recurrent BCOR internal tandem duplication and YWHAE-NUTM2B fusions in soft tissue undifferentiated round cell sarcoma of infancy: overlapping genetic features with clear cell sarcoma of kidney. Am J Surg Pathol. 2016;40:1009–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antonescu CR, Sung YS, Chen CL, et al. Novel ZC3H7B-BCOR, MEAF6-PHF1, and EPC1-PHF1 fusions in ossifying fibromyxoid tumors--molecular characterization shows genetic overlap with endometrial stromal sarcoma. Genes Chromosomes Cancer. 2014;53:183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kao YC, Sung YS, Zhang L, et al. Expanding the molecular signature of ossifying fibromyxoid tumors with two novel gene fusions: CREBBP-BCORL1 and KDM2A-WWTR1. Genes Chromosomes Cancer. 2017;56:42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20:1479–1484. [DOI] [PubMed] [Google Scholar]
- 17.Lee CH, Marino-Enriquez A, Ou W, et al. The clinicopathologic features of YWHAE-FAM22 endometrial stromal sarcomas: a histologically high-grade and clinically aggressive tumor. Am J Surg Pathol. 2012;36:641–653. [DOI] [PubMed] [Google Scholar]
- 18.Marino-Enriquez A, Lauria A, Przybyl J, et al. BCOR internal tandem duplication in high-grade uterine sarcomas. Am J Surg Pathol. 2018;42:335–341. [DOI] [PubMed] [Google Scholar]
- 19.Lee CH, Ou WB, Marino-Enriquez A, et al. 14-3-3 fusion oncogenes in high-grade endometrial stromal sarcoma. Proc Natl Acad Sci U S A. 2012;109:929–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Micci F, Brunetti M, Dal Cin P, et al. Fusion of the genes BRD8 and PHF1 in endometrial stromal sarcoma. Genes Chromosomes Cancer. 2017;56:841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


