Significance
The clinical effectiveness of FDA-approved anticancer rapalogs is limited because resistance is observed in multiple-tumor settings. In order to improve the efficacy of rapalogs, there is a need to improve our ability to predict and overcome rapamycin resistance. Here we report that high FRAT1/2 expression confers rapamycin resistance by excluding GSK3 from the nucleus in multiple-cancer cell lines. FRAT1 expression is frequently high in multiple cancers, and our observations provide evidence for using FRAT1/2 expression as a biomarker for rapamycin sensitivity. Moreover, our findings reveal a major mechanism for how mTORC1 regulates nuclear protein phosphorylation and signaling, and uncover a potential therapeutic approach for targeting mTORC1-driven cancers.
Keywords: mTOR, GSK3, FRAT, Resistance
Abstract
The mTORC1 pathway regulates cell growth and proliferation by properly coupling critical processes such as gene expression, protein translation, and metabolism to the availability of growth factors and hormones, nutrients, cellular energetics, oxygen status, and cell stress. Although multiple cytoplasmic substrates of mTORC1 have been identified, how mTORC1 signals within the nucleus remains incompletely understood. Here, we report a mechanism by which mTORC1 modulates the phosphorylation of multiple nuclear events. We observed a significant nuclear enrichment of GSK3 when mTORC1 was suppressed, which promotes phosphorylation of several proteins such as GTF2F1 and FOXK1. Importantly, nuclear localization of GSK3 is sufficient to suppress cell proliferation. Additionally, expression of a nuclear exporter of GSK3, FRAT, restricts the nuclear localization of GSK3, represses nuclear protein phosphorylation, and prevents rapamycin-induced cytostasis. Finally, we observe a correlation between rapamycin resistance and FRAT expression in multiple-cancer cell lines. Resistance to Food and Drug Administration (FDA)-approved rapamycin analogs (rapalogs) is observed in many tumor settings, but the underling mechanisms remain incompletely understood. Given that FRAT expression levels are frequently elevated in various cancers, our observations provide a potential biomarker and strategy for overcoming rapamycin resistance.
The protein kinase complex mechanistic target of rapamycin complex 1 (mTORC1) is a central regulator of mammalian cell growth and proliferation that is dysregulated in various cancer types, including colorectal, breast, lung, renal cell carcinoma, glioblastoma, and prostate cancers (1). Several rapalogs (rapamycin and its derivatives) have been developed and Food and Drug Administration (FDA)-approved for cancer treatment (2). However, the impact of rapalogs in the clinic has been limited, as resistance to rapalogs is frequently observed upon long-term treatment. In order to improve anticancer efficacy of rapalogs, there is a need to improve our understanding of these resistance mechanisms. As a central signaling cascade, once activated, mTORC1 promotes multiple anabolic processes by phosphorylating a number of downstream substrates, including S6K1, 4EBPs, Grb10, Lipin1, and others (1). Changes in mTORC1 signaling are also understood to influence several biological processes that are spatially restricted to the nucleus, including gene transcription, messenger RNA (mRNA) splicing, and chromosome remodeling. However, despite the significant advances made in understanding the mTORC1 pathway in the past 2 decades, how mTORC1 signaling controls nuclear events is still not fully defined, and whether nuclear signaling contributes to rapamycin resistance observed during chronic treatment is unknown.
GSK3 is known to phosphorylate a large number of substrates important in regulating cell growth and metabolism (3). Inappropriate regulation of GSK3 is linked to several disorders, including cancer (3, 4). In contrast to mTORC1, GSK3α/β have relatively high activity in resting cells, which is suppressed upon stimulation by multiple growth factors (3). This suppression is predominantly achieved via phosphorylation at Ser21 or Ser9, respectively, on GSK3α/β by multiple kinases (5). For example, inhibitory phosphorylation of GSK3 was shown to be regulated by mTORC1/S6K1 and contribute to mTORC1-mediated suppression of GSK3 activity (6). Interestingly, high levels of inhibitory GSK3 phosphorylation or GSK3 inhibitors were reported to confer rapamycin resistance in certain cell types (7, 8). GSK3 also phosphorylates and inhibits multiple nuclear proteins upon pharmacological mTORC1 suppression, such as TIP60, c-Myc, and FOXK1 (6, 9, 10). Therefore, one prediction is that, upon rapamycin treatment, GSK3 inhibitory phosphorylation is lost, resulting in inhibition of proliferation. However, it has been reported that GSK3 phosphorylation was not significantly suppressed by rapamycin treatment in multiple cell lines (8). Additionally, we have observed that GSK3-dependent regulation of FOXK1 phosphorylation occurred under conditions where GSK3 phosphorylation was not significantly affected upon mTORC1 suppression, suggesting a possible phosphorylation-independent mechanism for regulation of GSK3 by mTORC1 signaling (9). Thus, the relationship between mTORC1, GSK3, and rapamycin resistance remains unclear.
Here we report that the nuclear−cytoplasmic localization of GSK3 is regulated by mTORC1 signaling, which subsequently contributes to multiple nuclear protein phosphorylation events. The mTORC1 suppression leads to accumulation of GSK3 in the nucleus, thereby promoting the phosphorylation of GSK3 targets such as FOXK1 and GTF2F1. Critically, we found the GSK3 interacting proteins, frequently rearranged in advanced T lymphomas 1 and 2 (FRAT1 and 2), which promote GSK3 nuclear export, prevent rapamycin-induced nuclear accumulation of GSK3 and phosphorylation of its targets. Given these observations, we asked whether mTORC1-GSK3 nuclear signaling was a major contributor to cell proliferation and whether failure to promote GSK3 nuclear signaling might contribute to rapamycin resistance. In support of this possibility, we observed that ectopic expression of GSK3 that is forced to locate in the nucleus (NLS-GSK3) promotes phosphorylation of proteins such as FOXK1 and GTF2F1, and, importantly, suppresses cancer cell proliferation. We also observed that overexpression FRAT1/2 in rapamycin-sensitive cancer cells promotes resistance to the cytostatic effects of rapamycin, consistent with the acquisition of drug resistance. Consistently, cells with high FRAT1/2 expression are relatively insensitive to rapamycin treatment, and down-regulation of FRAT1/2 with RNA interference (RNAi) resensitizes cells to rapamycin. Our observations in xenograft models further support the conclusion that elevated expression of FRAT1/2, and suppression of GSK3 nuclear flux, are potential mechanisms by which cancer cells acquire rapamycin resistance. Collectively, our findings demonstrate that monitoring FRAT1/2 expression levels and/or GSK3 localization may be important biomarkers for rapamycin sensitivity, and that targeting FRAT1/2 expression or its ability to interact with GSK3 may represent therapeutic approaches to overcoming rapamycin resistance.
Results
Nuclear Enrichment of GSK3 Promotes Protein Phosphorylation upon mTORC1 Suppression.
The mTORC1 and downstream effectors such as ULK1/2, S6 protein kinases (S6K1/2), and SRPK2 regulate the phosphorylation of a variety of proteins that affect multiple cellular events associated with metabolism, mRNA processing and translation, survival signaling, autophagy, and cell growth (1, 11). An important observation made upon analysis of the mTORC1 phosphoproteome revealed that, in addition to the promotion of phosphorylation via mTORC1 signaling, the phosphorylation of ∼100 proteins appeared to be suppressed by mTORC1 (12). To investigate this unexpected role of mTORC1 in suppressing protein phosphorylation, we characterized phosphorylation of a transcription factor FOXK1, and demonstrated that mTORC1 suppresses FOXK1 phosphorylation in a GSK3-dependent manner (9). Furthermore, many of the phosphoproteins identified in this analysis are also nuclear resident proteins with potential, conserved GSK3 sites (SI Appendix, Table S1). To determine whether there are additional proteins whose phosphorylation is suppressed by mTORC1, we utilized available reagents to monitor the phosphorylation of another nuclear protein upon mTORC1 inhibition, the general transcription factor GTF2F1. Upon mTORC1 inhibition, GTF2F1 displayed decreased migration on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE) analysis as observed with FOXK1, indicating that GTF2F1 phosphorylation is suppressed by mTORC1 signaling (Fig. 1A). The appearance of slower-migrating forms of GTF2F1 following mTORC1 inhibition was prevented by the small-molecule GSK3 inhibitor, CHIR99021, as well as by phosphatase treatment, demonstrating that the mobility shifts were indeed due to phosphorylation (SI Appendix, Fig. S1 A and B). Furthermore, suppression and reactivation of mTORC1 by serum starvation and restimulation promoted the phosphorylation and dephosphorylation, respectively, of GTF2F1 (SI Appendix, Fig. S1C). Thus, GTF2F1 is another target under the control of mTORC1-dependent suppression of GSK3-regulated nuclear protein phosphorylation.
Fig. 1.
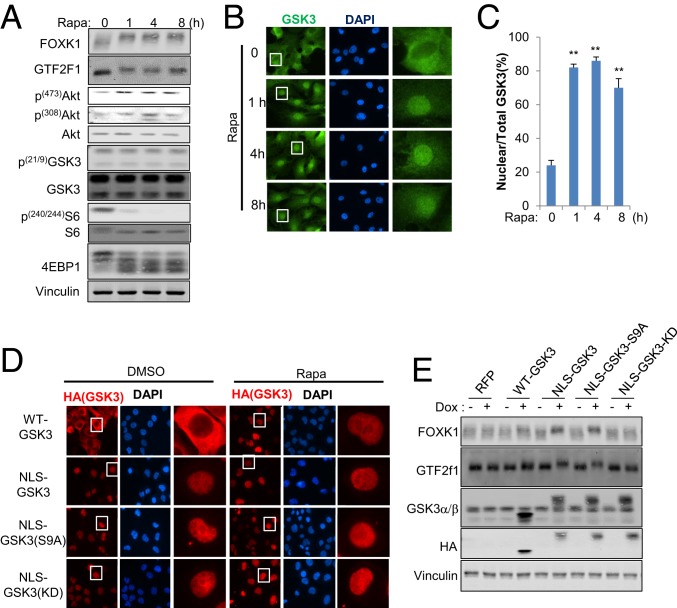
Cytoplasmic/nuclear translocation of GSK3 mediates nuclear protein phosphorylation upon mTORC1 suppression. (A–C) HCC 4006 cells were exposed to 20 nM rapamycin for the indicated times, and (A) whole-cell lysate was extracted and subjected to Western blot (WB) analysis with the indicated antibodies. (B) Immunostaining (IF) was performed with GSK3 antibody and observed with a fluorescence microscope at 600× magnification. The white boxes represent single cells selected for further visualization (Right). (C) Nuclear translocation was quantified and assessed as the ratios between nuclear and total fluorescence in at least 10 cells, and values are expressed as mean ± SEM. One-way ANOVA with Dunnett’s posttest, **P < 0.001, n = 3. (D and E) Cell lines were established with pTRIPZ-RFP, pTRIPZ-HA-GSK3-(WT), pTRIPZ-HA-NLS-GSK3β, pTRIPZ-HA-NLS-GSK3β-(S9A), or pTRIPZ-HA-NLS-GSK3β-(KD) in HCC4006 cells. (D) Cells were exposed to 1 μM Doxycycline for 24 h and further exposed to 20 nM rapamycin for 4 h where indicated. Immunostaining (IF) was performed with HA antibody and observed with a fluorescence microscope at 600× magnification. The white boxes represent single cells selected for further visualization (Right). (E) Cells were exposed to 1 μM Doxycyclin for 24 h where indicated, and whole-cell lysate was subjected to WB analysis with the indicated antibodies. Data are representative of at least 3 independent experiments.
Importantly, we did not observe significant changes in GSK3 phosphorylation, an established mechanism for GSK3 regulation, for up to 8 h of rapamycin treatment (Fig. 1A). Therefore, we next investigated additional potential mechanisms for regulation of GSK3 by mTORC1. FOXK1 and GTF2F1 are both nuclear resident proteins; thus we assessed the cytoplasmic/nuclear distribution of GSK3 in rapamycin-treated or nontreated cells, using an antibody we validated in wild-type (WT) and GSK3α/β-deficient mouse embryonic fibroblasts (MEFs) (SI Appendix, Fig. S1D). In a nonsmall cell lung cancer (NSCLC) cell line, HCC4006 cells, we found that GSK3 was predominantly distributed in the cytoplasm of growing cells, and a significant nuclear accumulation of GSK3 was observed upon rapamycin treatment that was sustained for at least 8 h (Fig. 1 B and C). Similarly, suppression of mTORC1 by serum starvation induced a significant nuclear enrichment of GSK3 which was rapidly reversed by serum restimulation for 30 min (SI Appendix, Fig. S1 E and F). The regulation of GSK3 nuclear localization mirrored FOXK1 and GTF2F1 phosphorylation under these conditions (SI Appendix, Fig. S1C). To further investigate and validate these findings, we established inducible systems to regulate the expression of WT-GSK3, NLS (nuclear localization signal)-tagged GSK3β, NLS-tagged GSK3β-S9A (nonphosphorylated Ser9 and active), and NLS-tagged GSK3β-KD (kinase-dead) in HCC4006 cells. Consistently, WT-GSK3β was predominantly localized in the cytoplasm under full serum growth conditions and became enriched in the nucleus following rapamycin treatment. However, NLS-GSK3β, NLS-GSK3β-S9A, and NLS-GSK3β-KD remained in the nucleus in the presence or absence of rapamycin (Fig. 1D and SI Appendix, Fig. S1G). Using these cell systems, we observed significant phosphorylation of FOXK1 and GTF2F1 in response to NLS-GSK3β or NLS-GSK3β-S9A expression in contrast to WT-GSK3β and NLS-GSK3β-KD (Fig. 1E). Further supporting this observation, FOXK1 and GTF2F1 phosphorylation by NLS-GSK3β was significantly suppressed by GSK3 inhibitor CHIR99021 (SI Appendix, Fig. S1H). These data are consistent with a role for nuclear translocation of GSK3β in promoting nuclear protein phosphorylation upon mTORC1 suppression.
Constitutive Expression of Nuclear GSK3β Suppresses Cell Proliferation.
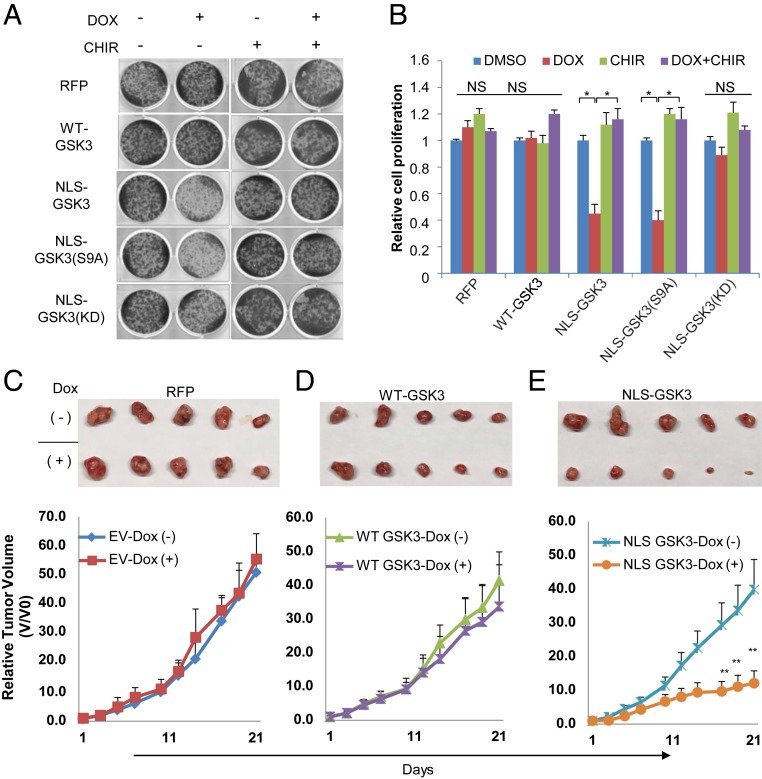
GSK3 has been reported to control cell proliferation in various cancer cell experimental settings (13, 14). Since enrichment of GSK3 in the nucleus has the potential to regulate the phosphorylation of multiple proteins involved in chromatin remodeling, transcription, RNA processing, and other nuclear processes, we next asked whether the cytoplasmic/nuclear translocation of GSK3 contributes to the regulation of cell proliferation. Strikingly, with the inducible system described above, we observed a significant suppression of cell proliferation by expressing NLS-GSK3β or NLS-GSK3β-(S9A), which was significantly prevented by CHIR99021 treatment, whereas WT-GSK3β or NLS-GSK3β-(KD) had little effect (Fig. 2 A and B). These results are consistent with the observation that nuclear GSK3 signals to regulate FOXK1 and GTF2F1 phosphorylation (Fig. 1E), and our previous observation that phosphorylated FOXK1 is suppressed in its ability to stimulate several growth-associated metabolic processes (9). Critically, this is further supported by mouse xenograft experiments with RFP, WT-GSK3β, and NLS-GSK3β cells (Fig. 2 C–E and SI Appendix, Fig. S2), where we observed a significant suppression of tumor growth using HCC4006 cells expressing NLS-GSK3β compared to RFP or WT-GSK3β. This is consistent with the fact that GSK3 promotes phosphorylation and inactivates many nuclear proteins that promote cell proliferation, which include Myc, JunB, and FOXK1 (3, 9).
Fig. 2.
Constitutive expression of nuclear GSK3β suppresses cell proliferation. Cell lines were established as in Fig. 1D. (A) Clonogenic assay was performed with or without 1 μM Doxycycline, in the presence or absence of GSK3 inhibitor 0.5 μM CHIR in the media and (B) cell proliferation was estimated by quantification of clonogenic cell growth as described in Experimental Procedures. Values are expressed as mean ± SEM. Student t test, *P < 0.01, **P < 0.001, n = 3. NS, not statistically significant. (C–E) Indicated cells (2 × 106) were injected into mice through s.c. injection as described in Experimental Procedures. Doxycycline (625 mg/kg) was included in the food where indicated, 5 mice were used in each group, and tumor growth was monitored. Values are expressed as mean ± SD. Student t test, **P < 0.01.
Restriction of Nuclear GSK3 and Protein Phosphorylation by FRAT.
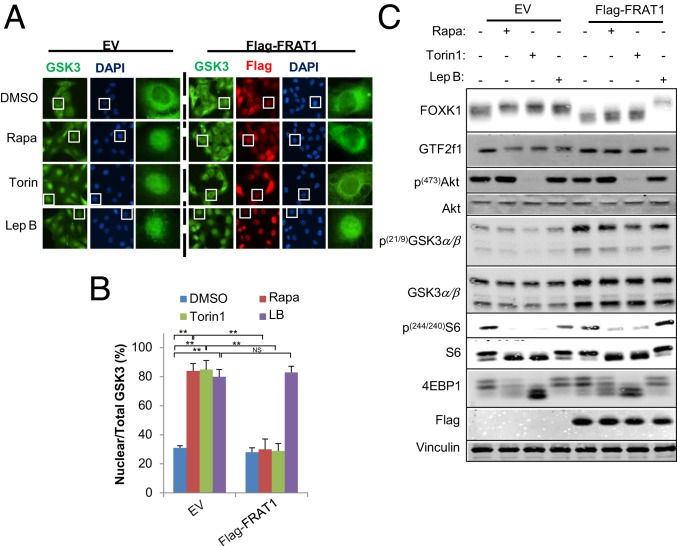
To investigate potential mechanisms for regulating GSK3 cell distribution, we first investigated the possible involvement of FRAT1/2. FRAT1/2 are known GSK3-interacting proteins that function as exporters of GSK3 from the nucleus (15). Critically, we observed that nuclear enrichment of GSK3 when mTORC1 was inhibited was efficiently prevented by transient expression of FRAT1 (Fig. 3 A and B). Additionally, the exportin 1 inhibitor leptomycin B significantly increased nuclear GSK3 either in the absence or presence of FRAT1 overexpression, consistent with the dynamics of the FRAT1−GSK3 interaction and the role of FRAT as a GSK3 nuclear exporter (Fig. 3 A and B). Similar results were observed with FRAT2 overexpression in HCC4006 cells (SI Appendix, Fig. S3). Strikingly, rapamycin- or torin1-induced, GSK3-mediated, FOXK1 and GTF2F1 phosphorylation were efficiently prevented by FRAT1 overexpression, mirroring the cytoplasmic enrichment of GSK3. Further supporting the idea that nuclear GSK3 regulates protein phosphorylation, leptomycin B, which traps GSK3 in the nucleus, promoted the phosphorylation of FOXK1 and GTF2F1 in the absence or presence of FRAT1 overexpression without affecting mTORC1 activity (Fig. 3C). This is consistent with a role for nuclear translocation of GSK3β in promoting nuclear protein phosphorylation upon mTORC1 suppression.
Fig. 3.
Restriction of nuclear GSK3 and protein phosphorylation by FRAT. Cell lines were established with ectopic expression of FRAT1 and exposed to indicated inhibitors (rapamycin 20 nM, Torin1 200 nM, leptomycin B 100 nM) for 4 h. (A) Cells were subsequently subjected to immunostaining with indicated antibodies and observed with a fluorescence microscope at 600× magnification. The white boxes represent single cells selected for further visualization (Right). (B) Nuclear translocation was quantified and assessed as the ratios between nuclear and total fluorescence in at least 10 cells, and values are expressed as mean ± SEM. One-way ANOVA with Dunnett’s posttest, **P < 0.001, n = 3. (C) Whole-cell lysate was extracted and subsequently subjected for WB analysis with the indicated antibodies. Data are representative of at least 3 independent experiments.
Acquisition of Rapamycin Resistance by FRAT.
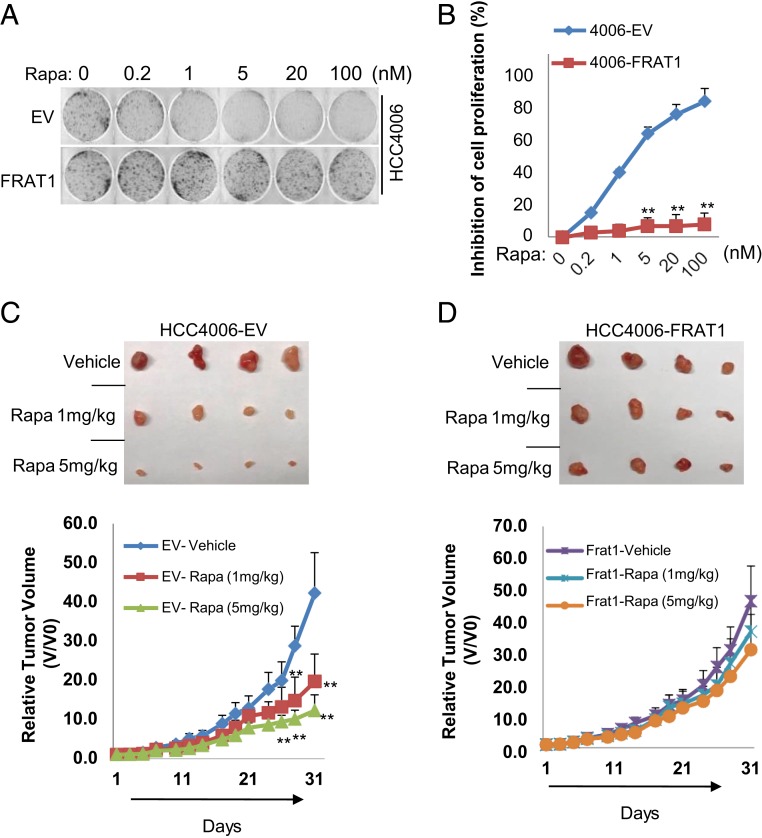
We have observed a nuclear enrichment of GSK3 upon mTORC1 suppression (Fig. 1) and shown that nuclear GSK3 significantly suppresses cell proliferation (Fig. 2). Thus, we next asked whether nuclear flux of GSK3 mediates rapamycin-induced cytostasis. Using HCC4006 cells stably expressing empty vector (HCC4006-EV) or FRAT1 (HCC4006-FRAT1), we found that the inhibition concentration 50% (IC50) for rapamycin was about 4.5 nM in the control cells but increased to over 100 nM upon FRAT1 overexpression (Fig. 4 A and B). Similar results were observed with FRAT2 expression in an additional NSCLC cell line A549 (SI Appendix, Fig. S4). This observation was further supported by in vivo xenograft analysis with the HCC4006 cells. Rapamycin significantly suppressed tumor growth of HCC4006-EV cells, while having relatively little effect on HCC4006-FRAT1 tumors (Fig. 4 C and D). These results implicate a potential role of FRAT expression in acquisition of rapamycin resistance in cancer cells by preventing nuclear accumulation of GSK3, which is required for growth inhibition.
Fig. 4.
Acquisition of rapamycin resistance by FRAT. FRAT1 expressing cell lines were established as in Fig. 3. (A) Clonogenic assay was performed with the indicated concentrations of rapamycin in the media, and (B) inhibition of cell growth was estimated as described in Experimental Procedures. Values are expressed as mean ± SEM. Two-tailed Student t test, **P < 0.001, n = 3. (C and D) Indicated cells were injected (5 × 105) into mice through s.c. inoculation as described in Experimental Procedures. Four mice were used in each group and treated with vehicle or rapamycin (1 mg/kg or 5 mg/kg) 3 times a week, and tumor growth was monitored. Values are expressed as mean ± SD. Two-tailed Student t test, **P < 0.01.
Negative Correlation of FRAT Expression Levels and Rapamycin Sensitivity.
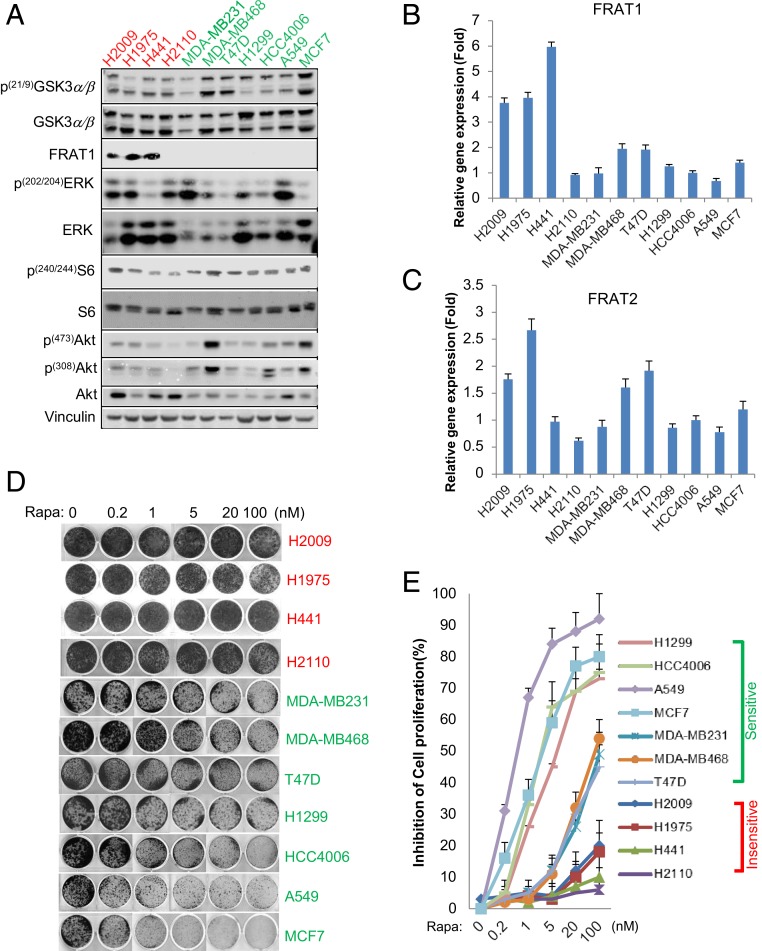
To determine whether there is a general correlation between FRAT expression and rapamycin responsiveness, we assessed multiple non-small cell lung and breast cancer cell lines. We found that 3 out of 11 cancer cells tested (NCI-H2009, NCI-H1975, and NCI-H441) have relatively high expression of FRAT1, FRAT2, or both (Fig. 5 A–C). Importantly, these cell lines are relatively insensitive to rapamycin treatment, with IC50 higher than 100 nM (Fig. 5 D and E), which is consistent with our previous observations with ectopic expression of FRAT1/2 (Fig. 4 A and B). In contrast, most cells with low FRAT1/2 expression are relatively sensitive (IC50 less than 100 nM) to rapamycin, including MDA-MB231, MDA-MB468, T47D, H1299, HCC4006, A549, and MCF7 cells (Fig. 5 D and E). H2110 cells, which express low levels of FRAT1/2, represented one exception which remained relatively insensitive to rapamycin treatment. Additional mechanisms for rapamycin resistance have been described which may explain the insensitivity of this cell line. However, rapamycin sensitivity of most of the cancer cell lines tested shows good correlations with FRAT1/2 expression. Further supporting the critical role of GSK3 nuclear accumulation in rapamycin-induced cytostasis, we also observed that either pharmacologic inhibition of GSK3 or suppression of GSK3 expression diminished rapamycin-induced cytostasis in multiple cell lines expressing low FRAT1/2, including HCC4006, A549, H1299, MDA-MB231, and T47D cells (SI Appendix, Fig. S5 A–E).
Fig. 5.
Negative correlation of FRAT expression levels and rapamycin sensitivity. (A) Whole-cell lysate from various cell lines was subjected to WB analysis with indicated antibodies. (B and C) Total RNA was extracted from the indicated cell lines and subjected to RT-qPCR analysis with primers against FRAT1/2. (D) Clonogenic assays with multiple cell lines under the indicated concentrations of rapamycin and (E) inhibition of cell growth were estimated by quantification of clonogenic cell growth as described in Experimental Procedures. Data are representative of at least 3 independent experiments.
Suppression of FRAT1/2 Potentiates Rapamycin-Induced Cytostasis.
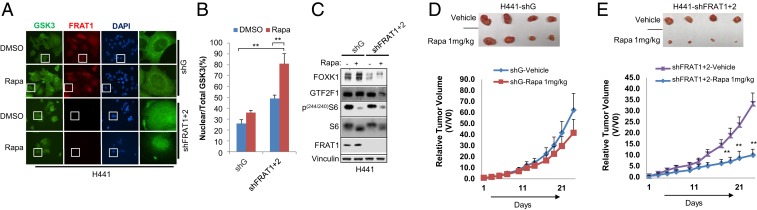
To further confirm the critical role of FRAT1/2 expression and nuclear flux of GSK3 upon mTORC1 suppression in determining rapamycin sensitivity, we used NCI-H441 cells which have high FRAT1/2 expression and are rapamycin-insensitive. Strikingly, nuclear GSK3 accumulation in NCI-H441 cells was relatively modest in response to mTORC1 inhibition. Consistently, insufficient phosphorylation of FOXK1 and GTF2F1 was observed upon rapamycin treatment. However, when both FRAT1 and FRAT2 expression was suppressed, rapamycin-induced nuclear GSK3 localization and FOXK1 and GTF2F1 phosphorylation was significantly increased in NCI-H441 cells (Fig. 6 A–C and SI Appendix, Fig. S6A). Importantly, suppressing FRAT1/2 expression resulted in a gain of rapamycin sensitivity where the IC50 was decreased to ∼16 nM (SI Appendix, Fig. S6 B and C). Similar results were observed in another rapamycin-insensitive cell line, H1975 (SI Appendix, Fig. S6). Consistently, this result was further supported by in vivo xenograft analysis. Rapamycin suppressed tumor growth more significantly when FRAT1/2 was suppressed in NCI-H441 cells (Fig. 6 D and E). These results clearly support a critical role for FRAT expression in determining rapamycin sensitivity in cells in vitro and in mouse tumor models.
Fig. 6.
Suppression of FRAT1/2 potentiates rapamycin-induced cytostasis. Cell lines were established with knockdown of FRAT1/2 in H441 cells. (A) Cells were exposed to 20 nM rapamycin for 4 h where indicated and subsequently subjected to immunostaining with the indicated antibodies and observed with a fluorescence microscope at 600× magnification. The white boxes represent single cells selected for further visualization (Right). (B) GSK3 nuclear translocation was quantified and assessed as the ratios between nuclear and total fluorescence in at least 10 cells, and values are expressed as mean ± SEM. One-way ANOVA with Dunnett’s posttest, **P < 0.001, n = 3. (C) Whole-cell lysate was subjected to WB analysis with the indicated antibodies. Data are representative of at least 3 independent experiments. (D and E) Indicated cells (5 × 105) were injected into mice through s.c. inoculation as described in Experimental Procedures. Four mice were used in each group and treated with vehicle or rapamycin (1 mg/kg or 5 mg/kg) 3 times a week, and tumor growth was monitored. Values are expressed as mean ± SD. Two-tailed Student t test, **P < 0.01.
Discussion
The mTORC1 protein kinase complex integrates mitogenic, nutrient, amino acid, stress, and oxygen status to dictate the strength and duration of downstream signaling to regulate a variety of cellular processes alone or in coordination with other signaling systems. Here we reveal a previously unknown mechanism of GSK3 regulation by mTORC1 which links this critical pathway to multiple nuclear events. Although many cytoplasmic mTORC1 targets have been identified and characterized, less is known regarding how mTORC1 directly or indirectly regulates the phosphorylation and regulation of nuclear targets associated with gene transcription, chromosome remodeling, mRNA splicing, ribosomal DNA synthesis and other critical nuclear processes involved in the regulation of cell growth, metabolism, survival, and proliferation (1). Our results indicate that the molecular basis for this link is at least in part centered around the nuclear accumulation of GSK3 upon suppression of mTORC1 signaling (SI Appendix, Fig. S8). In some conditions, mTORC1/S6K1 suppress GSK3α/β signaling through inhibitory phosphorylation at Ser21/9 (3, 6). It is noteworthy that mTORC1-regulated FOXK1 phosphorylation and GSK3 localization occurred in S6K1/2-deficient MEFs (SI Appendix, Fig. S7 A–D) and independent of GSK3 phosphorylation in our cell systems (Fig. 1A and SI Appendix, Fig. S7 E and F). Nuclear accumulation of WT-GSK3, NLS-GSK3β, or NLS-GSK3β-S9A is associated with significantly promoting FOXK1 and GTF2F1 phosphorylation, in contrast to cytoplasmic localized WT-GSK3β or nuclear-localized GSK3 (NLS-GSK3β-KD) (Fig. 1E). Further supporting the critical role of nuclear GSK3 in promoting nuclear protein phosphorylation, NLS-GSK3-induced FOXK1 and GTF2F1 phosphorylation was significantly suppressed by the GSK3 inhibitor, CHIR99021 (SI Appendix, Fig. S1H). FOXK1 is a major regulator of metabolic reprogramming downstream of mTORC1 (9). The general initiation factor for transcription by RNA polymerase II, GTF2F1, is also required for transcriptional activation by serum response factor (16); however, the role of phosphorylation in regulating its activity is not known. Meanwhile, from analysis of the mTORC1 phosphoproteome datasets, several additional potential GSK3 targets have been revealed, which remain to be characterized (SI Appendix, Table S1) (12). These results implicate an extensive regulation by mTORC1/GSK3 in regulating diverse protein phosphorylation events in the nucleus which potentially links mTORC1 to multiple critical nuclear processes.
Our observations support the hypothesis that nuclear localization of GSK3 promotes rapamycin-induced cytostasis. Expression of NLS-GSK3 significantly suppresses cancer cell progression both in cells and in xenograft models (Fig. 2). This is consistent with the observations that many nuclear proteins that promote cell proliferation, such as c-Myc, JunB, Cyclin D, FOXK1, and others, are phosphorylated by GSK3 and down-regulated by several distinct mechanisms (3, 9). Thus, diminishing nuclear GSK3 signaling upon mTORC1 inhibition may be suppressing rapamycin responsiveness, and, indeed, our data support this model in multiple-cancer cell lines (SI Appendix, Fig. S5). One mechanism that accounts for the regulation of GSK3 cellular distribution and rapamycin resistance is linked to expression levels of FRAT1/2, known nuclear exporters of GSK3 (Fig. 3A) (15). In cells expressing high levels of FRAT, sufficient accumulation of nuclear GSK3 is suppressed following rapamycin treatment, preventing GSK3 from acting as a negative regulator of its nuclear targets. Ectopically expressed FRAT1/2 confer rapamycin resistance in multiple cell lines (Fig. 4 and SI Appendix, Fig. S4), and multiple NSCLC cells with high endogenous FRAT1/2 are relatively resistant to rapamycin treatment (Fig. 5). Furthermore, suppressing FRAT1/2 levels with RNAi potentiates rapamycin-induced cytostasis in most cancer cells with high endogenous FRAT1/2 which are rapamycin-insensitive (Fig. 6 D and E and SI Appendix, Fig. S6). Further supporting this conclusion, upon FRAT knockdown, a small elevation of nuclear GSK3 was observed, and this was potentiated upon mTORC1 suppression (Fig. 6 A and B), which is consistent with the increased sensitivity to rapamycin-induced inhibition of cell proliferation in FRAT knockdown of H441 and H1975 cells (SI Appendix, Fig. S6 B and C). The lack of a more dramatic accumulation of GSK3 in the nucleus may be due to incomplete knockdown of FRAT1/2, or alternative FRAT-independent mechanisms that mediate GSK3 translocation upon mTORC1 inhibition which may also contribute to this regulatory process. These findings provide evidence for a mechanism by which high FRAT1/2 expression promotes rapamycin resistance in multiple-cancer cell lines (Fig. 5). Given the frequently observed, high expression of FRAT1/2 in multiple cancers (17–20), additional studies are needed to determine how expression of these GSK3 interactors is regulated. Our findings also indicate that high FRAT1/2 expression in patients can potentially be used as a biomarker for rapamycin sensitivity. Thus, targeting FRAT1/2 expression may be a valuable strategy for sensitizing cancers to rapamycin therapies.
High mTORC1 pathway activation has been observed in multiple cancers. However, the effectiveness of FDA-approved rapalogs (rapamycin derivatives) is limited, as clinically acquired resistance has been observed upon prolonged exposure and the mechanisms remain unclear (21). Therefore, understanding and overcoming rapamycin resistance in cancer patients is a major objective in cancer therapy. Our observations provide insights toward understanding the role of mTORC1/GSK3 in regulating multiple nuclear events and uncover a mechanism by which FRAT expression confers rapamycin resistance by excluding GSK3 from the nucleus. Thus, targeting FRAT expression may be a valuable strategy in overcoming rapamycin resistance.
Experimental Procedures
Cell Lines and Reagents.
WT and Gsk3α−/− MEFs were provided by Jim Woodgett (University of Toronto, Toronto, ON, Canada). NSCLC cells, breast cancer cells, and other cell lines used in the present study were obtained from ATCC unless otherwise noticed. Cells were cultured in Dulbecco’s Modified Eagle’s medium or Roswell Park Memorial Institute (RPMI) 1640 containing 10% fetal bovine serum. Anti-FOXK1 and anti-GAPDH antibodies were purchased from Abcam. Anti-HA antibody was purchased from Santa Cruz Biotechnology. Leptomycin B, Anti-FLAG-M2 antibody, anti-GTF2F1, and anti-Vinculin antibody were purchased from Sigma. Anti-phospho-S6, anti-S6, anti-phospho-ERK, anti-ERK, anti-phospho Gsk3α/β, anti-Gsk3α/β and anti-phospho Akt, and anti-Akt and anti-S6K1/2 antibodies were purchased from Cell Signaling Technology. CHIR99021 was from Selleck Chem.
RNA Extraction and RT-qPCR Analysis.
Total RNA was isolated using RNeasy mini kit (QIAGEN) and used for complementary DNA synthesis with superscript III first-strand synthesis supermix kit (Life Technologies) according to the manufacturers’ instructions. qPCR was performed using QuantiTect SYBR green qPCR kit on Roche LightCycler 480. Primers were purchased from IDT, and melting curve analysis was performed at the end of PCR.
Immunoblot Analysis.
Whole-cell lysate was extracted with 1× SDS sample buffer (50 mM Tris⋅HCl pH 6.8, 10% glycerol, 2% SDS, 10% 2-mercaptoethanol, 0.1% bromophenol blue). Samples were boiled and electrophoresed by SDS/PAGE. Proteins were then transferred to nitrocellulose membranes. The membranes were blocked for 1 h prior to probe with primary antibodies overnight. Membranes were subsequently incubated with secondary antibodies for 1 h and developed with LI-COR odyssey. All immunoblots in this study are representative of at least 3 independent experiments.
Clonogenic Assay.
Either 1,000 or 2,000 cells were seeded in 12- or 6-well/plates and incubated for 1 to ∼2 wk. After colonies were clearly observed, they were fixed with 4% formaldehyde, stained with crystal violet (0.5% wt/vol). After rinsing 4 times with phosphate-buffered saline (PBS) buffer, the images of the wells were scanned. For quantification, methanol was added to each well plate, and optical density was measured at 570 nm as described (22).
Immunofluorescence Staining.
Cells were plated on cover glass, and, the next day, cells were fixed with 4% paraformaldehyde for 10 min at room temperature. The cells were rinsed with PBS 4 times and incubated with a blocking solution containing 5% bovine serum albumin in PBS for 15 min. The cells were then incubated with indicated antibodies in blocking solution for 3.5 h followed by washing 4 times with PBS. Secondary antibodies conjugated to a fluorochrome (Alexa Fluor; Thermo Fisher Scientific) in blocking buffer were then added to cover glasses and incubated for another 1.5 h at room temperature. Cells were rinsed with PBS 4 times and incubated with Hoechst 33258 solution (DNA staining) for 15 min. After washing with PBS 4 times, cells were mounted with mounting buffer, and images were observed by fluorescence microscopy.
Animal Studies.
For our mouse xenograft studies, we followed Institutional Animal Care and Use Committee (IACUC; Weill Cornell Medical College)-approved protocols and guidelines (IRB protocol no. 2014-0060). Indicated amount of cells (1 × 106 cells) were injected subcutaneously (s.c.) into 5- to 6-wk-old female nude mice (Envigo or Taconic). After s.c. tumors formed, mice were randomly divided into 2 to ∼3 groups for intraperitoneal injection 3 d/wk with vehicle or rapamycin (1 mg/kg or 5 mg/kg body weight) or fed food containing Doxycyclin and grown for additional 3 to ∼4 wk. Rapamycin was dissolved with PEG400:water (50%:50%).
Establishment of Stable Cell Lines.
To generate lentiviruses, short hairpin RNA plasmids or overexpression plasmids were transfected into 293T cells together with the packaging (Δ8.9) and envelope (Glycoprotein G of the vescular stomatitis virus [VSVG]) plasmids, and medium was changed the next day. After 24 h, viral supernatants were harvested, and new medium was added. For infection, cells were infected with viral supernatants in the presence of a serum-containing medium supplemented with 4 μg/mL polybrene. After 16 h, viral-containing medium was removed, and cells were grown in serum-containing medium for 24 h. Cells were treated with puromycin (2 μg/mL) or blasticidin (10 μM) for selection. The knockdown or overexpression of target protein was confirmed by immunoblot analysis.
Statistics.
Data were expressed as average ± SEM of at least 3 independent experiments performed in triplicate. One-way ANOVA or 2-tailed Student’s t test was used to determine differences between each group, followed by the Dunnett’s or Tukey’s posttest or pairwise comparisons as appropriate.
Supplementary Material
Acknowledgments
We thank Yasir Ibrahim, Gina Lee, Sungyun Cho, and other members of the J.B. laboratory for critical discussions and technical assistance. L.H. is a Korea Research Institute of Bioscience and Biotechnology (KRIBB)–World Class Institute (WCI) Foundation Postdoctoral Fellow. J.B. is a Lymphangioleiomyomatosis (LAM) Foundation Established Investigator. The work was supported by National Institutes of Health Grants GM51405 and HL121266 to J.B.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902397116/-/DCSupplemental.
References
- 1.Saxton R. A., Sabatini D. M., mTOR signaling in growth, metabolism, and disease. Cell 169, 361–371 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Kim L. C., Cook R. S., Chen J., mTORC1 and mTORC2 in cancer and the tumor microenvironment. Oncogene 36, 2191–2201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beurel E., Grieco S. F., Jope R. S., Glycogen synthase kinase-3 (GSK3): Regulation, actions, and diseases. Pharmacol. Ther. 148, 114–131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manning B. D., Toker A., AKT/PKB signaling: Navigating the network. Cell 169, 381–405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jope R. S., Johnson G. V., The glamour and gloom of glycogen synthase kinase-3. Trends Biochem. Sci. 29, 95–102 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Zhang H. H., Lipovsky A. I., Dibble C. C., Sahin M., Manning B. D., S6K1 regulates GSK3 under conditions of mTOR-dependent feedback inhibition of Akt. Mol. Cell 24, 185–197 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laks D. R., et al. , A molecular cascade modulates MAP1B and confers resistance to mTOR inhibition in human glioblastoma. Neuro Oncol. 20, 764–775 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koo J., et al. , GSK3 is required for rapalogs to induce degradation of some oncogenic proteins and to suppress cancer cell growth. Oncotarget 6, 8974–8987 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He L., et al. , mTORC1 promotes metabolic reprogramming by the suppression of GSK3-dependent Foxk1 phosphorylation. Mol. Cell 70, 949–960.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin S. Y., et al. , GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science 336, 477–481 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Lee G., et al. , Post-transcriptional regulation of de novo lipogenesis by mTORC1-S6K1-SRPK2 signaling. Cell 171, 1545–1558.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu Y., et al. , Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332, 1322–1326 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dembowy J., Adissu H. A., Liu J. C., Zacksenhaus E., Woodgett J. R., Effect of glycogen synthase kinase-3 inactivation on mouse mammary gland development and oncogenesis. Oncogene 34, 3514–3526 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armanious H., et al. , Clinical and biological significance of GSK-3β inactivation in breast cancer-an immunohistochemical study. Hum. Pathol. 41, 1657–1663 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Franca-Koh J., Yeo M., Fraser E., Young N., Dale T. C., The regulation of glycogen synthase kinase-3 nuclear export by Frat/GBP. J. Biol. Chem. 277, 43844–43848 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Joliot V., Demma M., Prywes R., Interaction with RAP74 subunit of TFIIF is required for transcriptional activation by serum response factor. Nature 373, 632–635 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Guo G., et al. , Overexpression of FRAT1 is associated with malignant phenotype and poor prognosis in human gliomas. Dis. Markers 2015, 289750 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan Y., et al. , The clinical significance of FRAT1 and ABCG2 expression in pancreatic ductal adenocarcinoma. Tumour Biol. 36, 9961–9968 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Guo G., et al. , FRAT1 expression and its correlation with pathologic grade, proliferation, and apoptosis in human astrocytomas. Med. Oncol. 28, 1–6 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y., et al. , Overexpression of FRAT1 correlates with malignant phenotype and advanced stage in human non-small cell lung cancer. Virchows Arch. 459, 255–263 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Martelli A. M., Buontempo F., McCubrey J. A., Drug discovery targeting the mTOR pathway. Clin. Sci. (Lond.) 132, 543–568 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Feoktistova M., Geserick P., Leverkus M., Crystal violet assay for determining viability of cultured cells. Cold Spring Harb. Protoc. 2016, pdb.prot087379 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.