Abstract
Wnt‐signaling is one of the most abundant pathways involved in processes such as cell‐proliferation, ‐polarity, and ‐differentiation. Altered Wnt‐signaling has been linked with several neurodevelopmental disorders including attention‐deficit/hyperactivity disorder (ADHD) as well as with cognitive functions, learning and memory. Particularly, lipoprotein receptor‐related protein 5 (LRP5) or LRP6 coreceptors, responsible in the activation of the canonical Wnt‐pathway, were associated with cognitive alterations in psychiatric disorders. Following the hypothesis of Wnt involvement in ADHD, we investigated the association of genetic variations in LRP5 and LRP6 genes with three independent child and adolescent ADHD (cADHD) samples (total 2,917 participants), followed by a meta‐analysis including previously published data. As ADHD is more prevalent in males, we stratified the analysis according to sex and compared the results with the recent ADHD Psychiatric Genomic Consortium (PGC) GWAS. Meta‐analyzing our data including previously published cADHD studies, association of LRP5 intronic rs4988319 and rs3736228 (Ala1330Val) with cADHD was observed among girls (OR = 1.80 with 95% CI = 1.07–3.02, p = .0259; and OR = 2.08 with 95% CI = 1.01–4.46, p = .0026, respectively), whereas in boys association between LRP6 rs2302685 (Val1062Ile) and cADHD was present (OR = 1.66, CI = 1.20–2.31, p = .0024). In the PGC‐ADHD dataset (using pooled data of cADHD and adults) tendency of associations were observed only among females with OR = 1.09 (1.02–1.17) for LRP5 rs3736228 and OR = 1.18 (1.09–1.25) for LRP6 rs2302685. Together, our findings suggest a potential sex‐specific link of cADHD with LRP5 and LRP6 gene variants, which could contribute to the differences in brain maturation alterations in ADHD affected boys and girls, and suggest possible therapy targets.
Keywords: attention‐deficit hyperactivity disorder, gender, genetics, polymorphism, SNP
1. INTRODUCTION
Attention‐deficit/hyperactivity disorder (ADHD), a neurodevelopmental disorder, is one of the most common psychiatric and behavioral disorders in children and adolescents, with more than 5% of the pediatric population affected worldwide, often persisting into adulthood (Polanczyk, Willcutt, Salum, Kieling, & Rohde, 2014; Thomas, Sanders, Doust, Beller, & Glasziou, 2015). Although both the cause and pathophysiology of ADHD are largely unknown, there is a growing body of evidence supporting interactions of multiple genetic and environmental factors during early development thus providing a neurobiological susceptibility to the disorder (Curatolo, D'Agati, & Moavero, 2010).
ADHD diagnosis in children and adolescents is more frequent in boys than girls, with males being 2–4 times more likely to meet diagnostic criteria than females (Davies, 2014). Moreover, age‐at‐onset, severity, and comorbidities were found to be sex‐dependent (Davies, 2014). Just recently, a fMRI study reported significant volume reduction in putamen and thalamus of girls with ADHD, whereas a significant subcortical volume reduction was observed in ADHD patients independent of sex (Rosch et al., 2018). Subsequently, the authors discuss the importance to study the trajectories of such neurodevelopmental disorders together with the sex‐dimorphic neuroanatomical development, as maturation timelines between boys and girls are substantially different (Rosch et al., 2018). Differential susceptibility to pre‐ and postnatal stress has been discussed to affect brain development and maturation in a sex‐dimorphic manner, which was hypothesized to add to the genetic load that already leads to the different ADHD frequencies between genders (Van den Bergh et al., 2017). The aforementioned recent findings as examples, point to the importance of studying genetic susceptibility for ADHD not only at the diagnostic level, but also at age‐at‐onset and sex differences.
Wnt‐signaling pathways orchestrate cellular proliferation, polarity and differentiation; processes that are crucial for healthy tissue morphogenesis, especially in the embryonic stage (MacDonald, Tamai, & He, 2009). Two most known Wnt‐pathways exist: a canonical pathway and a noncanonical pathway. In the canonical pathway, the secreted Wnt glycoproteins bind to Frizzled receptors, as well as to either lipoprotein receptor‐related protein 5 (LRP5) or LRP6 coreceptors, to initiate a signaling cascade, in which downstream β‐catenin is harnessed as a cotranscription factor in the nucleus (Tamai et al., 2000; Wehrli et al., 2000). Canonical Wnt‐signaling has a pivotal role both in the developing and mature brain. During development the Wnt pathway regulates the balance between proliferation and differentiation of neuronal progenitor and precursor cells (Noelanders & Vleminckx, 2016). Furthermore, Wnt‐signaling also affects neuronal stem cell proliferation and differentiation in the mature brain (Bengoa‐Vergniory & Kypta, 2015), and it has a supportive role in the maturation of dendrites and spines (Hussaini et al., 2014).
Several neurodevelopmental psychiatric disorders have been shown to overlap not only at the behavioral levels, but also at the genetic levels, as it has been repeatedly shown among ADHD, autism spectrum disorder (ASD), intellectual disability (ID), bipolar disorder, and psychosis (Brainstorm Consortium et al., 2018; Bulik‐Sullivan et al., 2015; Khanzada, Butler, & Manzardo, 2017; Polimanti & Gelernter, 2017; Taurines et al., 2012; van Hulzen et al., 2017; Zhao & Nyholt, 2017). Interestingly, the Wnt‐signaling pathway seems to be one of the overlapping pathways, showing pathological alterations in the above disorders (Kwan, Unda, & Singh, 2016; Mulligan & Cheyette, 2017; Oron & Elliott, 2017; Zhao & Nyholt, 2017). For example, genome‐wide association studies (GWAS) reported the 4p15.31 region to be nominally associated with ADHD (not reaching genome‐wide p‐value < 5 × 10−8), where the KCNIP4 (potassium voltage‐gated channel interacting protein 4) gene is located (Lasky‐Su et al., 2008; Lesch et al., 2008; Neale et al., 2008). KCINP4 is known to play a role in the negative feedback loop of the Wnt/β‐catenin pathway. In a later candidate gene study, six KCNIP4 single markers and one haplotype block were found to associate with adult ADHD (aADHD; Weissflog et al., 2013). Another GWAS analyzed a subpopulation of ADHD cases, which had concomitant oppositional defiant disorder, and found the β‐catenin‐pathway to be highlighted in the enrichment analysis (Aebi et al., 2015). Moreover, Wnt‐signaling has been also associated with learning and memory, especially with deficits in working and spatial memory (Fortress, Schram, Tuscher, & Frick, 2013; Maguschak & Ressler, 2011; Maguschak & Ressler, 2012). Epidemiological studies pointed to Wnt‐involvement in behavioral problems including hyperactivity (Hussaini et al., 2014; Maguschak & Ressler, 2012). In ASD, highly comorbid disorder in ADHD, a wide range of evidence points to the involvement of the Wnt‐pathway (Belinson et al., 2016; Caracci, Avila, & De Ferrari, 2016; Packer, 2016; Zhang, Yuan, Wang, & Li, 2014). Lastly, an indirect evidence of the Wnt‐signaling involvement in ADHD could be demonstrated by our research group, showing that methylphenidate (a psychostimulant and one of the first line treatment in ADHD therapy) influences cell proliferation and differentiation supporting neuronal maturation (Grünblatt, Bartl, & Walitza, 2018). More specifically, we could demonstrate that methylphenidate activates Wnt‐signaling, which was not due to the dopamine transporter inhibition—the main know therapeutic mechanism of this drug—as selective dopamine transporter inhibitor GBR‐12909 treatment demonstrated the opposite effects (Grünblatt et al., 2018).
As the two possible coreceptors LRP5 and LRP6 play major role in Wnt‐pathway activation, we conducted literature search selecting functional gene variants affecting either receptor function, gene expression or epigenetic modulations, such as histone modifications or DNA methylation levels (see Table 1 and Supporting Information Table S1). Published genetic associations with psychiatric, neurological, or metabolic diseases are also listed in Table 1. Based on the hypothesis of the involvement of Wnt‐signaling in ADHD, we carried out genetic association analyses of the selected four LRP5 and LRP6 gene variants with ADHD in three independent European samples (total 2,917 participants), and a meta‐analysis with previously published data. Furthermore, due to sex‐discrepancy in ADHD frequency, we stratified the analysis according to sex and compared our results to the recent Psychiatric Genomic Consortium (PGC) ADHD GWAS results (Demontis et al., 2017; Martin et al., 2018).
Table 1.
Literature summary linking LRP5 and LRP6 gene variants with ADHD and other disorders
| Gene | SNPa | Functional effects in HEK293T cells | Functional effects in neuronal cells or animal models | Association studies |
|---|---|---|---|---|
| LRP5 | Loci/gene | NA | Overexpression of LRP5 & LRP6 in SH‐SY5Y cells protected against oxidative stress and reduced tau phosphorylation (Zhang, Bahety, & Ee, 2015). | BMD (Estrada et al., 2012); schizophrenia and major depressive disorder (Zhao & Nyholt, 2017) |
| LRP5 | rs4988319 | NA | NA | BMD (Tran, Nguyen, Eisman, & Nguyen, 2008); Wagner syndrome (Rothschild et al., 2013) |
| LRP5 | rs3736228 | LRP5‐V1330 demonstrated reduced Wnt3a signaling compared to wild‐type (Urano et al., 2009) | NA | Males low diastolic blood pressure (Suwazono et al., 2006); obesity and BMI (Guo et al., 2006); male bone fractures (van Meurs et al., 2006); BMD (Estrada et al., 2012; Tran et al., 2008); risk of having metabolic syndrome (Yang et al., 2013) |
| LRP6 | Loci/gene | NA | Neural tube defects in humans as well as in mice (Allache et al., 2014; Carter et al., 2005; Kokubu et al., 2004; Lei et al., 2015; Pinson, Brennan, Monkley, Avery, & Skarnes, 2000); Lrp6 mutant mice (insertion mutation) demonstrate suboptimal development of brain regions (e.g., forebrain, midbrain, and hindbrain), and defected neurogenesis of dopaminergic neurons (Castelo‐Branco et al., 2010; Pinson et al., 2000; Zhou, Zhao, & Pleasure, 2004), as well as age‐dependent synaptic loss and memory impairments in these mice (Liu et al., 2014) | AD, diabetes mellitus type 2, osteoporosis (Wang, Luo, Xu, Zhou, & Zhang, 2017) |
| LRP6 | rs1012672 | LRP6‐variant demonstrated reduced Wnt signaling compared to wild‐type (Xu et al., 2014) | NA | AD (miR‐141 miR‐23a miR‐23b; Mallick & Ghosh, 2011); AD (Alarcon et al., 2013; De Ferrari et al., 2007) |
| LRP6 | rs2302685 | LRP6‐variant demonstrated reduced Wnt signaling compared to wild‐type (De Ferrari et al., 2007; Xu et al., 2014) | NA | Risk of ischemic stroke (Harriott et al., 2015); AD (Alarcon et al., 2013; De Ferrari et al., 2007); male bone fractures (van Meurs et al., 2006) |
Abbreviations: AD = Alzheimer's disease; BMD = bone mineral density; BMI = body mass index; NA = not available.
Details on annotation, GTEx and epigenetic findings are presented in the Supporting Information Table S1.
2. METHODS
2.1. Study samples
2.1.1. Zurich child and adolescent ADHD (cADHD) patients and parents including unrelated control‐sample
One hundred and ninety six Caucasian nuclear families (146 families with both parents and 50 families with one parent) were recruited and the index patients (aged 6–21 years) were phenotypically characterized in the outpatient units of the Department of Child and Adolescent Psychiatry and Psychotherapy, University Hospital of Psychiatry Zurich. Families were included if at least the index patient fulfilled the diagnostic criteria for ADHD (F90.0 or F90.1) according to ICD‐10 (Dilling, Freyberger, & Stieglitz, 1996; World Health Organization, 2016). Accordingly, this resulted in 727 individuals (258 probands with ADHD (males = 179, females = 79) and 469 controls (males = 203, females = 266); total of 382 male and 345 female participants). The ADHD diagnoses of the parents and siblings were reported by the parents. The psychiatric diagnostics of the index patient was assessed by a child and adolescent psychiatrist or psychologist under supervision of a senior psychiatrist in the clinic. The index patient was required to be ≥6 years and to have an IQ over 75 as assessed with either the Wechsler Intelligence Scale for Children (WISC; Tewes, Rossmann, & Schallberger, 1999; Wechsler, 1991), the Kaufman Assessment Battery for Children (K‐ABC; Kaufman & Kaufman, 1983; Melchers & Preuss, 1994), the Culture Fair Test (CFT‐20‐R; Weiss, 2006), Snijders‐Oomen Nonverbal Intelligence Test (SON‐R; Tellegen, Winkel, & Laros, 2003) or Intelligence and Development Scales (IDS; Grob, Meyer, & Hagmann‐von Arx, 2009). Exclusion criteria were: (a) potentially confounding and severe psychiatric diagnoses such as psychosis, any pervasive developmental disorder, primary mood or anxiety disorder, and Tourette's disorder, (b) neurological disorders such as epilepsy, (c) a history of any acquired brain damage or evidence of the fetal alcohol syndrome, (d) premature deliveries (delivery before 37th gestational week), and/or (e) maternal reports of severe prenatal, perinatal or postnatal complications.
In the case–control setting, the 196 index patients (males = 148, females = 48) were compared to genetically independent 124 Caucasian healthy controls (males = 72, females = 52, aged between 5 and 18 years) who were recruited at the Departments of Child and Adolescent Psychiatry of the Universities of Würzburg and Zurich. The index patient in the case–control study was part of the family study and the inclusion and exclusion criteria were the same as described above. Informed written consent was obtained in all cases from the participants and their parents. The study was approved by the ethical commissions of all involved universities in accordance with the latest version of the Declaration of Helsinki, including an ethical permission granted by the Ethic Committees from Würzburg, and the Cantonal Ethic Committee of Zurich (Ref. Nr. KFO 140/03 and KEK‐ZH‐Nr. 2016–00101). Demographic characteristics of Zurich ADHD cases and controls are summarized in Supporting Information Table S2a and S2b.
2.1.2. Replication samples (Hungarian and German cADHD)
The SNP association study was replicated in samples from Budapest, Hungary and Würzburg, Germany, which were described in detail previously (Kereszturi et al., 2007; Walitza et al., 2005). The replication sample included child and adolescent patients with ADHD and their parents (Würzburg; Walitza et al., 2005), and ADHD patients, their available parents and unrelated controls (Budapest; Kereszturi et al., 2007) with the following characteristics: There were 171 Caucasian nuclear families (106 families with both parents and 65 families with one parent) with additional cADHD cases and healthy young adult controls from Budapest. In the family‐based setting this yielded 181 probands with ADHD according to ICD‐10 (males = 157, females = 24) and 312 controls (males = 132, females = 180), whereas the case–control study consisted of 206 cADHD patients (males = 180, females = 26, aged between 5 and 17 years) and 262 healthy controls (males = 160, females = 102, aged between 18 and 29 years). The Würzburg‐trios and duos included 387 children and adolescents affected with ADHD also according to ICD‐10 (males = 296, females = 91) and 540 controls (males = 275, females = 265).
The Ethics Committees of the respective universities approved the study and written informed consents were obtained from the participants and their parents after the study have been fully explained (see previous publications; Kereszturi et al., 2007; Walitza et al., 2005).
2.1.3. Genotyping
The study samples were genotyped for rs4988319 and rs3736228 in LRP5, and rs1012672 and rs2302685 in LRP6, which were chosen based on previous genetic findings (described in the introduction) and the availability of validated or functionally tested TaqMan assays. DNA was isolated either from whole blood collected in ethylenediaminetetraacetic acid tubes using QIAamp DNA Blood Maxi Kit (Qiagen), or from saliva collected in the Oragene DNA collection kit (DNA Genotek, Canada) and isolated as per manufacturer's protocol. DNA concentrations, A260/A280, and A260/A230 ratios were measured using a spectrophotometer (NanoVue Plus, GE). The study population was genotyped with DNA (10 ng), TaqMan® Genotyping Master Mix (Applied Biosystems), and LRP5 or LRP6 SNP Genotyping Assays (Applied Biosystems—see Supporting Information Table S3) combined in a 384‐well plate. Real‐time PCR was performed in a C1000™ CFX384™ Thermal cycler (Bio‐Rad) using TaqMan® SNP Genotyping Assay PCR standard protocol. Genotypes were determined by the allelic discrimination program of Bio‐Rad CFX Manager™ Software version 2.1. Samples were run in duplicates to ensure reproducibility. In case of ambiguity in duplicates, genotyping was repeated in a separate run to resolve the discrepancy. No‐template controls were included in every run to exclude impurities.
2.1.4. Statistical analysis
All association studies were run on the PLINK v1.7 (URL: http://pngu.mgh.harvard.edu/purcell/plink/; Purcell et al., 2007). Each study group (case–control study) was tested for Hardy–Weinberg equilibrium (Supporting Information Table S4). For the case–control association study, the Fisher's Exact Test was conducted and significance was set at p < .00417 following multiple testing corrections (four SNPs, three groups = male, female, and all together). For the family association study, Mendel errors test (none were found) followed by the transmission disequilibrium test was conducted as well as a parent‐of‐origin analysis. Power was calculated using the Genetic Association Study (GAS) calculator http://csg.sph.umich.edu/abecasis/cats/gas_power_calculator/index.html.
2.1.5. Meta‐analysis
We conducted literature search to find any publications that described genetic association studies for LRP5 and LRP6 SNPs in connection to ADHD or GWAS in European (Caucasian) ADHD. No previous GAS was available for these two genes in ADHD. However, we could find the GWAS by Hinney et al. (2011), and the GWAS results of the PGC (Demontis et al., 2017; Martin et al., 2018) containing the SNPs analyzed in the current study. A meta‐analysis was conducted with the currently studied populations (Zurich, Budapest, and Würzburg) together with the cADHD results from Hinney et al. (2011) using the MIX 2.0 Pro v.2.0.1.4 (BiostatXL, 2011. http://www.meta-analysis-made-easy.com). The PGC‐ADHD results were not added into the meta‐analysis, as the results from the PGC GWAS would outweigh our three study samples, due to a larger sample size, as well as high heterogeneity since the PGC did not contain solely cADHD patients. Therefore, the results of the PGC‐ADHD were used only as comparison to the meta‐analysis results. Variability, due to between‐study heterogeneity, was estimated by I 2 and funnel plots (Supporting Information Table S5), followed by Begg's and Egger's regression test (Begg & Mazumdar, 1994; Egger, Davey Smith, Schneider, & Minder, 1997; Supporting Information Table S6) to evaluate publication bias due to heterogeneity, and the quality of the studies was assessed based on traditional epidemiological considerations as previously described in Liu et al. (2015) (Supporting Information Table S7 and S8). Following heterogeneity tests, fixed‐effects model was used to conduct the mea‐analysis when I 2 demonstrated no significant heterogeneity in the samples, whereas the random‐effects model meta‐analysis was run if heterogeneity was found (see type of test used for each test in Supporting Information Table S5).
2.1.6. Gene expression patterns and functional findings
To elaborate whether the four SNPs studied may have any potential functional effects on gene expression or epigenetic targets, the SNPnexus (http://www.snp-nexus.org/; Chelala, Khan, & Lemoine, 2009; Dayem Ullah, Lemoine, & Chelala, 2012; Dayem Ullah, Lemoine, & Chelala, 2013) and eQTLs for both genes according to GTEx (https://www.gtexportal.org/home/; Consortium, 2013) were run (see results Supporting Information Table S1). Furthermore, the gene‐expression profiles in various brain regions compared to whole blood and nerve‐tibial were extracted from GTEx data for LRP5 and LRP6 stratified by sex. Welch two‐sided t test was conducted between the two sexes for each of the tissue analyzed. Since ADHD is a neurodevelopmental disorder, and most likely the transcript patterns alter with age, we downloaded the expression profiles of the two genes from the BRAINSPAN consortium (http://www.brainspan.org/) focusing on several brain regions (dorsolateral prefrontal cortex, orbital frontal cortex, hippocampus, amygdala, striatum, and cerebellum) to create age trajectories for the expression of the two genes.
3. RESULTS
3.1. LRP5 and LRP6 genetic associations in cADHD
Linkage disequilibrium (LD) values between the two LRP5 SNPs and the LRP6 SNPs demonstrated weak linkage in all studied cohorts, similar to the NIH LDlink data for European populations (extracted from https://analysistools.nci.nih.gov/LDlink/?tab=home; see Supporting Information Table S9). Therefore, we could conclude that the resulted association was independent of each other.
In the case–control study a nominal significant association between LRP5 rs3736228 and ADHD was observed in the Zurich sample (OR = 2.043, 95% CI 1.209–3.453, p = .0067, power = 0.944; Supporting Information Table S10). This association was significant after stratification by sex only in ADHD females (OR = 3.614, 95% CI 1.519–8.6, p = .0024, power = 1.0). Nominal significant sex‐specific association was also present at this LRP5 SNP in the Budapest case–control sample (OR = 2.549, 95% CI 1.218–5.334, p = .0109, power = 0.995). Moreover, LRP5 rs4988319 was nominally associated with ADHD in the female subgroup from Budapest (OR = 2.12, 95% CI 1.007–4.462, p = .0444, power = 0.923).
Although no significant association between LRP6 rs1012672 or rs2302685 and ADHD was detected in any one of the three European samples, some tendencies could be observed (Supporting Information Table S10). However, following sex stratification, nominal significant association of the LRP6 rs2302685 was observed among males using the case–control design at both the Zurich and Budapest ADHD samples (OR = 1.971, 95% CI 1.13–3.438, p = .0159, power = 0.904; OR = 1.516, 95% CI 1.009–2.277, p = .0441, power = 0.519, respectively). Family based association analyses of the LRP6 SNPs yielded only tendency toward association in the Zurich sample (Supporting Information Table S10).
3.2. Meta‐analysis of LRP5 and LRP6 SNPs in children and adolescents with ADHD
In order to evaluate the associations found with ADHD, we performed a meta‐analysis including both the case–control and family studies (Zurich, Würzburg, and Budapest) with available published data from Hinney et al. (2011). As described in the methods section, the large PGC‐ADHD GWAS (Demontis et al., 2017; Martin et al., 2018) was not added into the current meta‐analysis and was only used as a comparison study, because it consisted of both cADHD and aADHD, as well as due to its weight comparing to the other studies (Supporting Information Table S7‐S8). Following heterogeneity analysis (Supporting Information Table S5 and Figure S1) a fixed‐effect model analysis was conducted since no significant heterogeneity was detected. At the LRP5 SNPs we could not find significant association with cADHD (Figures 1a and 2a). On the other hand, a tendency was observed at LRP6 rs1012672 (total n = 4,712; OR = 1.262, p = .0559, power = 0.884; Figure 3a), and a nominal significant association was detected at LRP6 rs2302685 with ADHD (total n = 2,917; OR = 1.206, 95% CI 1.03–1.413; p = .0197; power = 0.794; Figure 4a).
Figure 1.
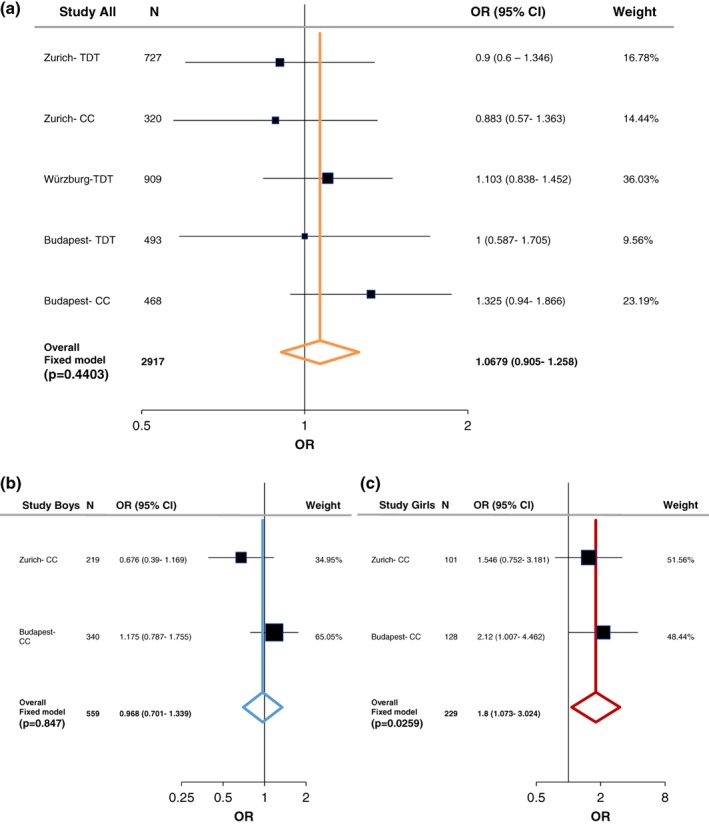
Summary and meta‐analysis of all cohorts and published association analyses of the LRP5 (rs4988319) gene variation with attention‐deficit hyperactivity disorder (ADHD) following sex stratification. (a) Forest plot for rs4988319 in male/female combined cohorts. (b) Forest plot for rs4988319 in males. (c) Forest plot for rs4988319 in females. Black whiskers in the forest plot represent 95% confidence intervals (CI) for odds ratio; the weight of the study is reflected in symbol size. Sample demographics, individual statistics, heterogeneity, literature bias statistics, quality assessments and scores, and type of tests was summarized in Supporting Information Tables S5–S9. Abbreviations: CC = case–control; TDT = transmission disequilibrium test [Color figure can be viewed at wileyonlinelibrary.com]
Figure 2.
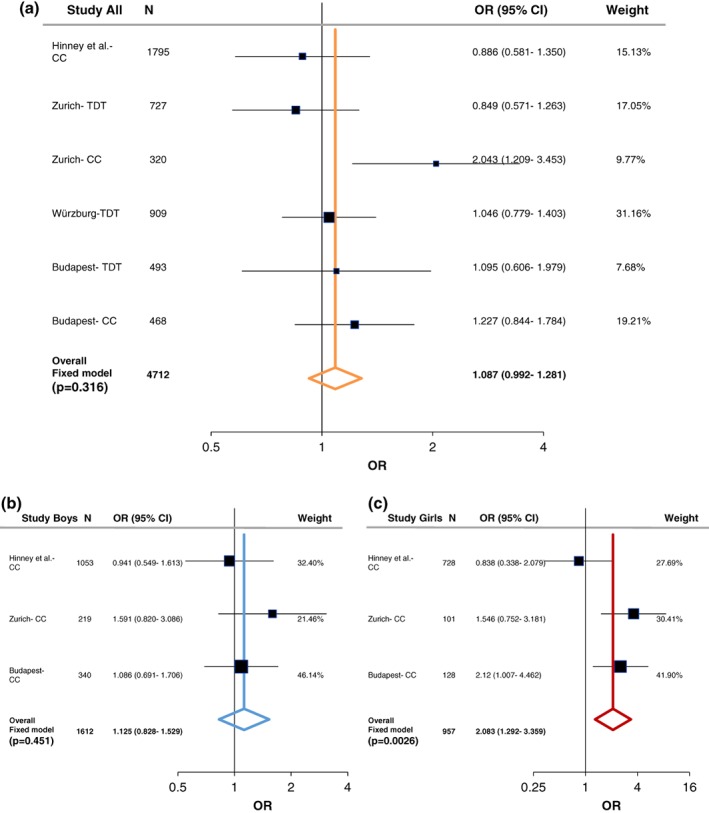
Summary and meta‐analysis of all cohorts and published association analyses of the LRP5 (rs3736228) gene variation with attention‐deficit hyperactivity disorder (ADHD) following sex stratification. (a) Forest plot for rs3736228 in male/female combined cohorts. (b) Forest plot for rs3736228 in males. (c) Forest plot for rs3736228 in females. Black whiskers in the forest plot represent 95% confidence intervals (CI) for odds ratio; the weight of the study is reflected in symbol size. Sample demographics, individual statistics, heterogeneity, literature bias statistics, quality assessments and scores, and type of tests was summarized in Supporting Information Tables S5–S9. Abbreviations: CC = case–control; TDT = transmission disequilibrium test [Color figure can be viewed at wileyonlinelibrary.com]
Figure 3.
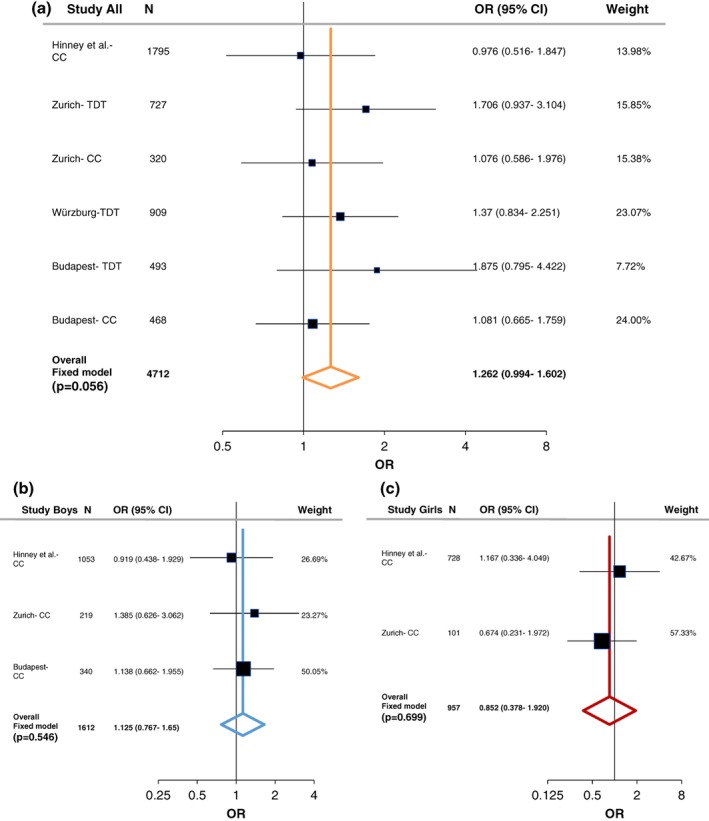
Summary and meta‐analysis of all cohorts and published association analyses of the LRP6 (rs1012672) gene variation with attention‐deficit hyperactivity disorder (ADHD) following sex stratification. (a) Forest plot for rs1012672 in male/female combined cohorts. (b) Forest plot for rs1012672 in males. (c) Forest plot for rs1012672 in females. Black whiskers in the forest plot represent 95% confidence intervals (CI) for odds ratio; the weight of the study is reflected in symbol size. Sample demographics, individual statistics, heterogeneity, literature bias statistics, quality assessments and scores, and type of tests was summarized in Supporting Information Tables S5–S9. Abbreviations: CC = case–control; TDT = transmission disequilibrium test [Color figure can be viewed at wileyonlinelibrary.com]
Figure 4.
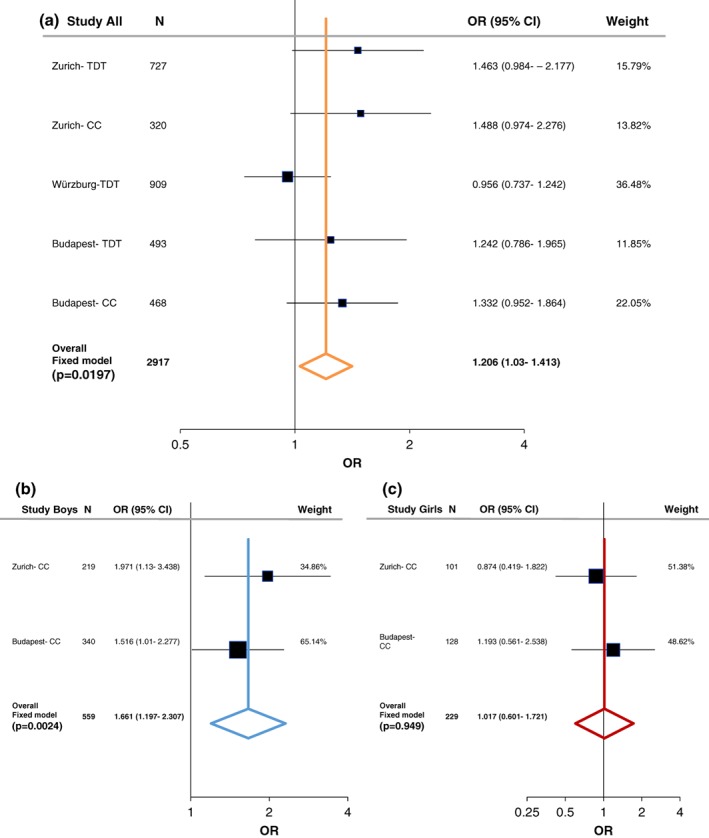
Summary and meta‐analysis of all cohorts and published association analyses of the LRP6 (rs2302685) gene variation with attention‐deficit hyperactivity disorder (ADHD) following sex stratification. (a) Forest plot for rs2302685 in male/female combined cohorts. (b) Forest plot for rs2302685 in males. (c) Forest plot for rs2302685 in females. Black whiskers in the forest plot represent 95% confidence intervals (CI) for odds ratio; the weight of the study is reflected in symbol size. Sample demographics, individual statistics, heterogeneity, literature bias statistics, quality assessments and scores, and type of tests was summarized in Supporting Information Tables S5–S9. Abbreviations: CC = case–control; TDT = transmission disequilibrium test [Color figure can be viewed at wileyonlinelibrary.com]
Following sex stratification, a nominal significant association of LRP5 rs4988319 and a significant association of LRP5 rs3736228 was observed with cADHD among females (total n = 229; OR = 1.8, 95% CI 1.073–3.024, p = .0259, power = 0.731; total n = 957; OR = 2.083, 95% CI 1.292–3.359, p = .0026, power = 0.998; respectively; Figures 1c and 2c). On the other hand, at LRP6 rs2303685 significant association with cADHD was present in males (total n = 559; OR = 1.661, 95% CI 1.196–2.307, p = .0024, power = 0.846; Figure 4b).
To test the sex effect, we ran a regression analysis (generalized linear mixed model) looking at the sex and SNP dosage and their interaction including the sites as a random effect in order to account for possible site‐effects. The regression analysis showed significant association between sex and ADHD but no significant SNP × sex interaction was detected (Supporting Information Table S11). Nevertheless, some suggestive sex effects were found supporting the meta‐analysis results, as the probability to have ADHD increased with the number of risk allele at LRP5 rs4988319 in females (Supporting Information Figure S2a). Similarly, a modest increase for having ADHD could be seen for those with the risk allele of LRP5 rs3736228 (Supporting Information Figure S2b). Whereas at LRP6 rs2302685 the probability to have ADHD increased with the number of risk allele only in males (Supporting Information Figure S2d).
3.3. PGC‐ADHD GWAS results for LRP5 and LRP6 SNPs
In order to compare the current findings that focused on Caucasian cADHD patients, we extracted the summary statistics of the newest (November 2017) available PGC data for European ancestry ADHD patients (child and adolescent as well as adults) and then stratified data by sex (Demontis et al., 2017; Martin et al., 2018). Only a suggestive association was observed between ADHD and LRP5 rs3736228 among females (OR = 1.0942, 95% CI 1.0197–1.1742, p = .2779). While, LRP6 rs2302685 showed association with ADHD only in the female subgroup of the PGC cohort (OR = 1.177, 95% CI 1.0858–1.2503, p = .0208). Therefore, none of the associations found in the meta‐analysis could be confirmed in the PGC‐ADHD data (Supporting Information Table S10). Nevertheless, observing all SNPs' association results of the PGC‐ADHD dataset at the LRP5 gene (Supporting Information Figure S3), a nominally significant signal could be seen in the male/female combined population at rs4988321 (p = .032), which is in moderate LD with rs3736228 (D′ = 1.0, R 2 = 0.227 at CEU; D′ = 0.967, R 2 = 0.266 at EUR) but not with rs4988319 (D′ = 0.219, R 2 = 0.009 at CEU; D′ = 0.514, R 2 = 0.056 at EUR). This association was specifically stronger (p = .0086) at the female population in the sex stratified Manhattan plot (Supporting Information Figure S3c). Similarly, around the studied LRP6 SNPs, some nominally significant signals could be observed in the male/female combined study population (Supporting Information Figure S4a), that were particularly enhanced in the stratified female ADHD population (Supporting Information Figure S4b).
3.4. Sex and age dependent LRP5 and LRP6 gene expression patterns in brain tissue
According to GTEx database, LRP5 transcript expression in various brain regions is rather low compared to the nerve‐tibial, but higher than whole blood samples (Supporting Information Figure S5). Similar pattern was observed for LRP6 transcript with slightly higher expression levels in brain regions (Supporting Information Figure S6). LRP5 transcript was expressed significantly higher in male brain‐spinal cord, whereas slightly higher expression in female could be observed in the cerebellum, nucleus accumbens, putamen, and substantia nigra, however in the last three not reaching significance. LRP6 transcript was expressed significantly higher in male brain‐cortex, and slightly more expressed in male caudate, spinal cord, and nerve‐tibial, however not reaching significance. As the GTEx data represents aged population, age dependent gene‐expression was extracted from the BRAINSPAN database (Supporting Information Figure S7). From the six brain regions extracted, LRP5 and LRP6 expression show a strong age dependency with higher levels at embryonal and early postnatal stages compared to middle age subjects (up to 40 years of age), in which the expression becomes lower starting from around 19 years of age (corresponding to 1,000 weeks).
4. DISCUSSION
The involvement of Wnt‐signaling in neurodevelopmental and neurodegenerative disorders have been widely discussed, particularly in ASD and AD with several studies pointing to its role in cognition and behavior (Kwan et al., 2016; Libro, Bramanti, & Mazzon, 2016; Mulligan & Cheyette, 2017; Rios, Cisternas, Arrese, Barja, & Inestrosa, 2014; Wang et al., 2017; Zhang et al., 2014). In the current study, we tested the hypothesis that LRP5 and LRP6 gene variants, coding for essential receptors for the Wnt‐pathway activation, associate with ADHD among children and adolescents, in a sex‐specific manner. Among the four studied gene variants, our meta‐analysis showed significant association between LRP5 rs3736228 (Ala1330Val) and cADHD in girls, while LRP6 rs2303685 (Val1062Ile) was associated with cADHD in boys. This phenomenon could also be observed in the more heterogeneous ADHD population studied in the PGC‐ADHD, consisting of both cADHD and aADHD samples, in which nominally significant SNP signals on the Manhattan plots were found to be specific for females at LRP5, however no clear‐cut result for LRP6 gene variants emerged. This discrepancy between our results and the PGC‐ADHD might be due to higher heterogeneity observed in the PGC‐ADHD sample, considering the age of onset that may play a major role with these gene variants. Indeed, BRAINSPAN dataset analysis (Supporting Information Figure S7) demonstrated age‐dependent gene expression of both LRP5 and LRP6 with higher transcript levels at brain developmental stages while lower in adulthood. Independent to age effects, in GTEx dataset some hint for higher expression of LRP5 in aged females and of LRP6 in aged males was observed (Supporting Information Figures S5 and S6, respectively), point to sex‐dependent regulation.
The Wnt‐signaling genes were shown to display sex differences in gene expression levels in a study of human placental transcriptomics of male and female embryos (Sedlmeier et al., 2014). In this study, the authors tested the effect of maternal dietary n‐3 polyunsaturated fatty acid supplementation during pregnancy, measuring the offspring's fat mass and development. Interestingly, the authors revealed significant sex‐dependent gene expression difference in control placentae per se, in which Wnt‐signaling genes were down regulated in male compared to female placentae (Sedlmeier et al., 2014). Furthermore, n‐3 unsaturated fatty acid supplementation affected the placental transcriptomics much stronger in females, as demonstrated with decreased LRP6 mRNA levels in the n‐3 supplemented female placentae (Sedlmeier et al., 2014), whereas male placenta transcriptomics did not result in such alterations. In addition, the authors showed significant positive correlation with placental estradiol‐17β/testosterone ratio and LRP6 expression (Sedlmeier et al., 2014). One possible explanation to this sex‐dimorphic effect is that there is at least one estrogen receptor alpha response element within the gene body of the Wnt‐signaling genes, which might affect their transcription regulation via sex hormones (Sedlmeier et al., 2014).
Lastly, some limitations should be mentioned, such as the relatively modest sample size of the sex‐stratified ADHD cohorts at certain gene variants in the meta‐analysis, the heterogeneity of the PGC‐ADHD used as a comparison study, and the limited number of variants studied within the two genes. Therefore, some caution should be taken at the interpretation of our results, especially at those analyses where only two independent study cohorts could be used. Nevertheless, our study's strength lies in the more homogenous, childhood‐onset ADHD samples recruited from European populations with the same diagnostic tools and similar inclusion/exclusion criteria.
In summary, we were able to demonstrate genetic associations between nonsynonymous LRP5 and LRP6 variants with cADHD in a sex‐specific manner. These results could highlight genetic variants with modified receptor functions in the Wnt‐signaling playing important role in brain development and maturation, which is particularly vulnerable in children and adolescents with ADHD. However, this hypothesis should be further investigated at the molecular and cellular levels, for example, using animal models or using induced pluripotent stem cell neuronal modeling techniques. In the light of our previous findings that methylphenidate activates Wnt‐signaling (Grünblatt et al., 2018), we suggest that LRP5/6 coreceptors could be regarded as possible new therapeutic targets in ADHD treatment, especially in childhood when brain maturation is still ongoing.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Table S1: Supplementary Table.
Appendix S1: Supplementary Material.
ACKNOWLEDGMENTS
The authors thank the families, patients, and control volunteers who participated in this research. The authors would like to acknowledge Psychiatric Genomics Consortium (PGC) ADHD for providing us with the newest summary statistics of the GWAS, Ms. Miryame Hofmann and Mrs. Susanne Kunert‐Dümpelmann for their laboratory technical support. The authors acknowledge the national grant that supported the Hungarian study (NAP‐B KTIA_NAP_13‐2014‐0011), the Deutsche Forschungsgemeinschaft (DFG) grant that supported the German study (KFO125; HE1446/9‐1, HI865/2‐1) and the postdoctoral grant support for Anna Maria Werling by the University of Zurich, the “Filling the Gap program”.
Grünblatt E, Nemoda Z, Werling AM, et al. The involvement of the canonical Wnt‐signaling receptor LRP5 and LRP6 gene variants with ADHD and sexual dimorphism: Association study and meta‐analysis. Am J Med Genet Part B. 2019;180B:365–376. 10.1002/ajmg.b.32695
Funding information Hungarian Brain Research Program, Grant/Award Number: NAP‐B KTIA_NAP_13‐2014‐0011; University of Zurich, Filling the Gap, Grant/Award Number: Postdoctoral to Anna Maria Werling; University of Zurich; Deutsche Forschungsgemeinschaft, Grant/Award Number: HE1446/9‐1HI865/2‐1KFO125
REFERENCES
- Aebi, M. , van Donkelaar, M. M. , Poelmans, G. , Buitelaar, J. K. , Sonuga‐Barke, E. J. , Stringaris, A. , … van Hulzen, K. J. (2015). Gene‐set and multivariate genome‐wide association analysis of oppositional defiant behavior subtypes in attention‐deficit/hyperactivity disorder. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics, 171(5), 573–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon, M. A. , Medina, M. A. , Hu, Q. , Avila, M. E. , Bustos, B. I. , Perez‐Palma, E. , … De Ferrari, G. V. (2013). A novel functional low‐density lipoprotein receptor‐related protein 6 gene alternative splice variant is associated with Alzheimer's disease. Neurobiology of Aging, 34(6), e1709–e1718. [DOI] [PubMed] [Google Scholar]
- Allache, R. , Lachance, S. , Guyot, M. C. , De Marco, P. , Merello, E. , Justice, M. J. , … Kibar, Z. (2014). Novel mutations in Lrp6 orthologs in mouse and human neural tube defects affect a highly dosage‐sensitive Wnt non‐canonical planar cell polarity pathway. Human Molecular Genetics, 23(7), 1687–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg, C. B. , & Mazumdar, M. (1994). Operating characteristics of a rank correlation test for publication bias. Biometrics, 50(4), 1088–1101. [PubMed] [Google Scholar]
- Belinson, H. , Nakatani, J. , Babineau, B. A. , Birnbaum, R. Y. , Ellegood, J. , Bershteyn, M. , … Wynshaw‐Boris, A. (2016). Prenatal beta‐catenin/Brn2/Tbr2 transcriptional cascade regulates adult social and stereotypic behaviors. Molecular Psychiatry, 21(10), 1417–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengoa‐Vergniory, N. , & Kypta, R. M. (2015). Canonical and noncanonical Wnt signaling in neural stem/progenitor cells. Cellular and Molecular Life Sciences, 72(21), 4157–4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brainstorm Consortium , Anttila, V. , Bulik‐Sullivan, B. , Finucane, H. K. , Walters, R. K. , et al. (2018). Science, 360(6395), pii: eaap8757. doi: 10.1126/science.aap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik‐Sullivan, B. , Finucane, H. K. , Anttila, V. , Gusev, A. , Day, F. R. , Loh, P. R. , … Neale, B. M. (2015). An atlas of genetic correlations across human diseases and traits. Nature Genetics, 47(11), 1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caracci, M. O. , Avila, M. E. , & De Ferrari, G. V. (2016). Synaptic Wnt/GSK3beta signaling hub in autism. Neural Plasticity, 2016, 9603751. doi: 10.1155/2016/9603751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, M. , Chen, X. , Slowinska, B. , Minnerath, S. , Glickstein, S. , Shi, L. , … Ross, M. E. (2005). Crooked tail (cd) model of human folate‐responsive neural tube defects is mutated in Wnt coreceptor lipoprotein receptor‐related protein 6. Proceedings of the National Academy of Sciences of the United States of America, 102(36), 12843–12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelo‐Branco, G. , Andersson, E. R. , Minina, E. , Sousa, K. M. , Ribeiro, D. , Kokubu, C. , … Arenas, E. (2010). Delayed dopaminergic neuron differentiation in Lrp6 mutant mice. Developmental Dynamics, 239(1), 211–221. [DOI] [PubMed] [Google Scholar]
- Chelala, C. , Khan, A. , & Lemoine, N. R. (2009). SNPnexus: A web database for functional annotation of newly discovered and public domain single nucleotide polymorphisms. Bioinformatics, 25(5), 655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, G. T. (2013). The genotype‐tissue expression (GTEx) project. Nature Genetics, 45(6), 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curatolo, P. , D'Agati, E. , & Moavero, R. (2010). The neurobiological basis of ADHD. Italian Journal of Pediatrics, 36(1), 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, W. (2014). Sex differences in attention deficit hyperactivity disorder: Candidate genetic and endocrine mechanisms. Frontiers in Neuroendocrinology, 35(3), 331–346. [DOI] [PubMed] [Google Scholar]
- Dayem Ullah, A. Z. , Lemoine, N. R. , & Chelala, C. (2012). SNPnexus: A web server for functional annotation of novel and publicly known genetic variants (2012 update). Nucleic Acids Research, 40(Web Server issue), W65–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayem Ullah, A. Z. , Lemoine, N. R. , & Chelala, C. (2013). A practical guide for the functional annotation of genetic variations using SNPnexus. Briefings in Bioinformatics, 14(4), 437–447. [DOI] [PubMed] [Google Scholar]
- De Ferrari, G. V. , Papassotiropoulos, A. , Biechele, T. , Wavrant De‐Vrieze, F. , Avila, M. E. , Major, M. B. , … Moon, R. T. (2007). Common genetic variation within the low‐density lipoprotein receptor‐related protein 6 and late‐onset Alzheimer's disease. Proceedings of the National Academy of Sciences of the United States of America, 104(22), 9434–9439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis, D. , Walters, R. , Martin, J. , Mattheisen, M. , Als, T. , Agerbo, E. , … Neale, B. M. (2017). Discovery of the first genome‐wide significant risk loci for ADHD. June 3, 2017 ed. bioRxiv: 10.1101/145581. [DOI]
- Dilling, H. , Freyberger, H. J. , & Stieglitz, R. D. (1996). ICD‐10 field trial of the diagnostic criteria for research in German‐speaking countries. Introduction. Psychopathology, 29(5), 258–259. [DOI] [PubMed] [Google Scholar]
- Egger, M. , Davey Smith, G. , Schneider, M. , & Minder, C. (1997). Bias in meta‐analysis detected by a simple, graphical test. BMJ, 315(7109), 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada, K. , Styrkarsdottir, U. , Evangelou, E. , Hsu, Y. H. , Duncan, E. L. , Ntzani, E. E. , … Rivadeneira, F. (2012). Genome‐wide meta‐analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nature Genetics, 44(5), 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortress, A. M. , Schram, S. L. , Tuscher, J. J. , & Frick, K. M. (2013). Canonical Wnt signaling is necessary for object recognition memory consolidation. The Journal of Neuroscience, 33(31), 12619–12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob, A. , Meyer, C. , & Hagmann‐von Arx, P. (2009). Intelligence and Development Scales (IDS). Intelligenz‐ und Entwicklungsskalen für Kinder von 5–10 Jahren. Bern: Verlag Hans Huber. [Google Scholar]
- Grünblatt, E. , Bartl, J. , & Walitza, S. (2018). Methylphenidate enhances neuronal differentiation and reduces proliferation concomitant to activation of Wnt signal transduction pathways. Translational Psychiatry, 8(1), 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y. F. , Xiong, D. H. , Shen, H. , Zhao, L. J. , Xiao, P. , Guo, Y. , … Deng, H. W. (2006). Polymorphisms of the low‐density lipoprotein receptor‐related protein 5 (LRP5) gene are associated with obesity phenotypes in a large family‐based association study. Journal of Medical Genetics, 43(10), 798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harriott, A. M. , Heckman, M. G. , Rayaprolu, S. , Soto‐Ortolaza, A. I. , Diehl, N. N. , Kanekiyo, T. , … Ross, O. A. (2015). Low density lipoprotein receptor related protein 1 and 6 gene variants and ischaemic stroke risk. European Journal of Neurology, 22(8), 1235–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinney, A. , Scherag, A. , Jarick, I. , Albayrak, O. , Putter, C. , Pechlivanis, S. , … Psychiatric GCAs . (2011). Genome‐wide association study in German patients with attention deficit/hyperactivity disorder. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics, 156B(8), 888–897. [DOI] [PubMed] [Google Scholar]
- Hussaini, S. M. , Choi, C. I. , Cho, C. H. , Kim, H. J. , Jun, H. , & Jang, M. H. (2014). Wnt signaling in neuropsychiatric disorders: Ties with adult hippocampal neurogenesis and behavior. Neuroscience and Biobehavioral Reviews, 47, 369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman, A. , & Kaufman, N. (1983). K‐ABC: Kaufman‐assessment battery for children. Circle Pines, MN: American Guidance Service. [Google Scholar]
- Kereszturi, E. , Kiraly, O. , Csapo, Z. , Tarnok, Z. , Gadoros, J. , Sasvari‐Szekely, M. , & Nemoda, Z. (2007). Association between the 120‐bp duplication of the dopamine D4 receptor gene and attention deficit hyperactivity disorder: Genetic and molecular analyses. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics, 144B(2), 231–236. [DOI] [PubMed] [Google Scholar]
- Khanzada, N. S. , Butler, M. G. , & Manzardo, A. M. (2017). GeneAnalytics pathway analysis and genetic overlap among autism Spectrum disorder, bipolar disorder and schizophrenia. International Journal of Molecular Sciences, 18(3), 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokubu, C. , Heinzmann, U. , Kokubu, T. , Sakai, N. , Kubota, T. , Kawai, M. , … Imai, K. (2004). Skeletal defects in ringelschwanz mutant mice reveal that Lrp6 is required for proper somitogenesis and osteogenesis. Development, 131(21), 5469–5480. [DOI] [PubMed] [Google Scholar]
- Kwan, V. , Unda, B. K. , & Singh, K. K. (2016). Wnt signaling networks in autism spectrum disorder and intellectual disability. Journal of Neurodevelopmental Disorders, 8, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasky‐Su, J. , Anney, R. J. , Neale, B. M. , Franke, B. , Zhou, K. , Maller, J. B. , … Faraone, S. V. (2008). Genome‐wide association scan of the time to onset of attention deficit hyperactivity disorder. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics, 147B(8), 1355–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, Y. , Fathe, K. , McCartney, D. , Zhu, H. , Yang, W. , Ross, M. E. , … Finnell, R. H. (2015). Rare LRP6 variants identified in spina bifida patients. Human Mutation, 36(3), 342–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch, K. P. , Timmesfeld, N. , Renner, T. J. , Halperin, R. , Roser, C. , Nguyen, T. T. , … Jacob, C. (2008). Molecular genetics of adult ADHD: Converging evidence from genome‐wide association and extended pedigree linkage studies. Journal of Neural Transmission, 115(11), 1573–1585. [DOI] [PubMed] [Google Scholar]
- Libro, R. , Bramanti, P. , & Mazzon, E. (2016). The role of the Wnt canonical signaling in neurodegenerative diseases. Life Sciences, 158, 78–88. [DOI] [PubMed] [Google Scholar]
- Liu, C. C. , Tsai, C. W. , Deak, F. , Rogers, J. , Penuliar, M. , Sung, Y. M. , … Bu, G. (2014). Deficiency in LRP6‐mediated Wnt signaling contributes to synaptic abnormalities and amyloid pathology in Alzheimer's disease. Neuron, 84(1), 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Yang, A. , Zhang, Q. , Yang, G. , Yang, W. , Lei, H. , … Yu, K. (2015). Association between genetic variants in SLC25A12 and risk of autism spectrum disorders: An integrated meta‐analysis. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics, 168B(4), 236–246. [DOI] [PubMed] [Google Scholar]
- MacDonald, B. T. , Tamai, K. , & He, X. (2009). Wnt/beta‐catenin signaling: Components, mechanisms, and diseases. Developmental Cell, 17(1), 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguschak, K. A. , & Ressler, K. J. (2011). Wnt signaling in amygdala‐dependent learning and memory. The Journal of Neuroscience, 31(37), 13057–13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguschak, K. A. , & Ressler, K. J. (2012). A role for WNT/beta‐catenin signaling in the neural mechanisms of behavior. Journal of Neuroimmune Pharmacology, 7(4), 763–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick, B. , & Ghosh, Z. (2011). A complex crosstalk between polymorphic microRNA target sites and AD prognosis. RNA Biology, 8(4), 665–673. [DOI] [PubMed] [Google Scholar]
- Martin, J. , Walters, R. , Demontis, D. , Mattheisen, M. , Lee, S. , Robinson, E. , … Neale, B. M. (2018). A genetic investigation of sex bias in the prevalence of attention deficit hyperactivity disorder. June 15, 2018. Biol. Psychiatry, 83(12), 1044–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchers, P. , & Preuss, U. (1994). K‐ABC: Kaufman‐Assessment battery for children. Deutsche Bearbeitung. Amsterdam: Swets&Zeitglinger. [Google Scholar]
- Mulligan, K. A. , & Cheyette, B. N. (2017). Neurodevelopmental perspectives on Wnt signaling in psychiatry. Molecular Neuropsychiatry, 2(4), 219–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale, B. M. , Lasky‐Su, J. , Anney, R. , Franke, B. , Zhou, K. , Maller, J. B. , … Faraone, S. V. (2008). Genome‐wide association scan of attention deficit hyperactivity disorder. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics, 147B(8), 1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noelanders, R. , & Vleminckx, K. (2016). How Wnt signaling builds the brain: Bridging development and disease. The Neuroscientist, 23(3), 314–329. [DOI] [PubMed] [Google Scholar]
- Oron, O. , & Elliott, E. (2017). Delineating the common biological pathways perturbed by ASD's genetic etiology: Lessons from network‐based studies. International Journal of Molecular Sciences, 18(4), 828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer, A. (2016). Enrichment of factors regulating canonical Wnt signaling among autism risk genes. Molecular Psychiatry, 23(3), 492–493. [DOI] [PubMed] [Google Scholar]
- Pinson, K. I. , Brennan, J. , Monkley, S. , Avery, B. J. , & Skarnes, W. C. (2000). An LDL‐receptor‐related protein mediates Wnt signalling in mice. Nature, 407(6803), 535–538. [DOI] [PubMed] [Google Scholar]
- Polanczyk, G. V. , Willcutt, E. G. , Salum, G. A. , Kieling, C. , & Rohde, L. A. (2014). ADHD prevalence estimates across three decades: An updated systematic review and meta‐regression analysis. International Journal of Epidemiology, 43(2), 434–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polimanti, R. , & Gelernter, J. (2017). Widespread signatures of positive selection in common risk alleles associated to autism spectrum disorder. PLoS Genetics, 13(2), e1006618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell, S. , Neale, B. , Todd‐Brown, K. , Thomas, L. , Ferreira, M. A. , Bender, D. , … Sham, P. C. (2007). PLINK: A tool set for whole‐genome association and population‐based linkage analyses. American Journal of Human Genetics, 81(3), 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios, J. A. , Cisternas, P. , Arrese, M. , Barja, S. , & Inestrosa, N. C. (2014). Is Alzheimer's disease related to metabolic syndrome? A Wnt signaling conundrum. Progress in Neurobiology, 121, 125–146. [DOI] [PubMed] [Google Scholar]
- Rosch, K. S. , Crocetti, D. , Hirabayashi, K. , Denckla, M. B. , Mostofsky, S. H. , & Mahone, E. M. (2018). Reduced subcortical volumes among preschool‐age girls and boys with ADHD. Psychiatry Research Neuroimaging, 271, 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild, P. R. , Brezin, A. P. , Nedelec, B. , Burin, d. , Roziers, C. , Ghiotti, T. , … Valleix, S. (2013). A family with Wagner syndrome with uveitis and a new versican mutation. Molecular Vision, 19, 2040–2049. [PMC free article] [PubMed] [Google Scholar]
- Sedlmeier, E. M. , Brunner, S. , Much, D. , Pagel, P. , Ulbrich, S. E. , Meyer, H. H. , … Bader, B. L. (2014). Human placental transcriptome shows sexually dimorphic gene expression and responsiveness to maternal dietary n‐3 long‐chain polyunsaturated fatty acid intervention during pregnancy. BMC Genomics, 15, 941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwazono, Y. , Kobayashi, E. , Uetani, M. , Miura, K. , Morikawa, Y. , Ishizaki, M. , … Nogawa, K. (2006). Low‐density lipoprotein receptor‐related protein 5 variant A1330V is a determinant of blood pressure in Japanese males. Life Sciences, 78(21), 2475–2479. [DOI] [PubMed] [Google Scholar]
- Tamai, K. , Semenov, M. , Kato, Y. , Spokony, R. , Liu, C. , Katsuyama, Y. , … He, X. (2000). LDL‐receptor‐related proteins in Wnt signal transduction. Nature, 407(6803), 530–535. [DOI] [PubMed] [Google Scholar]
- Taurines, R. , Schwenck, C. , Westerwald, E. , Sachse, M. , Siniatchkin, M. , & Freitag, C. (2012). ADHD and autism: Differential diagnosis or overlapping traits? A selective review. Attention Deficit and Hyperactivity Disorders, 4(3), 115–139. [DOI] [PubMed] [Google Scholar]
- Tellegen, P. , Winkel, M. , & Laros, J. (2003). Snijders Oomen Non‐verbal Intelligence Test Revised (SON‐R). Göttingen, Germany: Hofgrefe‐Verlag. [Google Scholar]
- Tewes, U. , Rossmann, P. , & Schallberger, U. (1999). Der Hamburg‐Wechsler‐Intelligenztest für Kinder (HAWIK‐III)‐ 3. Auflage. Bern: Huber‐Verlag. [Google Scholar]
- Thomas, R. , Sanders, S. , Doust, J. , Beller, E. , & Glasziou, P. (2015). Prevalence of attention‐deficit/hyperactivity disorder: A systematic review and meta‐analysis. Pediatrics, 135(4), e994–e1001. [DOI] [PubMed] [Google Scholar]
- Tran, B. N. , Nguyen, N. D. , Eisman, J. A. , & Nguyen, T. V. (2008). Association between LRP5 polymorphism and bone mineral density: A Bayesian meta‐analysis. BMC Medical Genetics, 9, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano, T. , Shiraki, M. , Usui, T. , Sasaki, N. , Ouchi, Y. , & Inoue, S. (2009). A1330V variant of the low‐density lipoprotein receptor‐related protein 5 (LRP5) gene decreases Wnt signaling and affects the total body bone mineral density in Japanese women. Endocrine Journal, 56(4), 625–631. [DOI] [PubMed] [Google Scholar]
- Van den Bergh, B. R. H. , van den Heuvel, M. I. , Lahti, M. , Braeken, M. , de Rooij, S. R. , Entringer, S. , … Schwab, M. (2017). Prenatal developmental origins of behavior and mental health: The influence of maternal stress in pregnancy. Neuroscience and Biobehavioral Reviews, In press, doi: 10.1016/j.neubiorev.2017.07.003. [DOI] [PubMed] [Google Scholar]
- van Hulzen, K. J. E. , Scholz, C. J. , Franke, B. , Ripke, S. , Klein, M. , McQuillin, A. , … Reif, A. (2017). Genetic overlap between attention‐deficit/hyperactivity disorder and bipolar disorder: Evidence from genome‐wide association study meta‐analysis. Biological Psychiatry, 82(9), 634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meurs, J. B. , Rivadeneira, F. , Jhamai, M. , Hugens, W. , Hofman, A. , van Leeuwen, J. P. , … Uitterlinden, A. G. (2006). Common genetic variation of the low‐density lipoprotein receptor‐related protein 5 and 6 genes determines fracture risk in elderly white men. Journal of Bone and Mineral Research, 21(1), 141–150. [DOI] [PubMed] [Google Scholar]
- Walitza, S. , Renner, T. J. , Dempfle, A. , Konrad, K. , Wewetzer, C. , Halbach, A. , … Lesch, K. P. (2005). Transmission disequilibrium of polymorphic variants in the tryptophan hydroxylase‐2 gene in attention‐deficit/hyperactivity disorder. Molecular Psychiatry, 10(12), 1126–1132. [DOI] [PubMed] [Google Scholar]
- Wang, Z. M. , Luo, J. Q. , Xu, L. Y. , Zhou, H. H. , & Zhang, W. (2017). Harnessing low‐density lipoprotein receptor protein 6 (LRP6) genetic variation and Wnt signaling for innovative diagnostics in complex diseases. The Pharmacogenomics Journal, 18(3), 351–358. [DOI] [PubMed] [Google Scholar]
- Wechsler, D. (1991). Examiner's manual: Wechsler intelligence scale for children—third edition. San Antonio, TX: The Psychological Corporation. [Google Scholar]
- Wehrli, M. , Dougan, S. T. , Caldwell, K. , O'Keefe, L. , Schwartz, S. , Vaizel‐Ohayon, D. , … DiNardo, S. (2000). Arrow encodes an LDL‐receptor‐related protein essential for wingless signalling. Nature, 407(6803), 527–530. [DOI] [PubMed] [Google Scholar]
- Weiss, R. (2006). Grundintelligenztest Skala 2‐Revision (CFT 20‐R) mit Wortschatztest und Zahlenfolgentest—Revision (Ws/ZF‐R). Göttingen, Germany: Hofgrefe‐Verlag. [Google Scholar]
- Weissflog, L. , Scholz, C. J. , Jacob, C. P. , Nguyen, T. T. , Zamzow, K. , Gross‐Lesch, S. , … Reif, A. (2013). KCNIP4 as a candidate gene for personality disorders and adult ADHD. European Neuropsychopharmacology, 23(6), 436–447. [DOI] [PubMed] [Google Scholar]
- World Health Organization . (2016). 10th International Classification of Diseases (ICD‐10). 10th revision, Geneva, Switzerland, World Health Organization. [Google Scholar]
- Xu, Y. , Gong, W. , Peng, J. , Wang, H. , Huang, J. , Ding, H. , & Wang, D. W. (2014). Functional analysis LRP6 novel mutations in patients with coronary artery disease. PLoS One, 9(1), e84345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C. W. , Li, C. I. , Liu, C. S. , Bau, D. T. , Lin, C. H. , Lin, W. Y. , … Lin, C. C. (2013). The joint effect of cigarette smoking and polymorphisms on LRP5, LEPR, near MC4R and SH2B1 genes on metabolic syndrome susceptibility in Taiwan. Molecular Biology Reports, 40(1), 525–533. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Bahety, P. , & Ee, P. L. (2015). Wnt co‐receptor LRP5/6 overexpression confers protection against hydrogen peroxide‐induced neurotoxicity and reduces tau phosphorylation in SH‐SY5Y cells. Neurochemistry International, 87, 13–21. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Yuan, X. , Wang, Z. , & Li, R. (2014). The canonical Wnt signaling pathway in autism. CNS & Neurological Disorders Drug Targets, 13(5), 765–770. [DOI] [PubMed] [Google Scholar]
- Zhao, H. , & Nyholt, D. R. (2017). Gene‐based analyses reveal novel genetic overlap and allelic heterogeneity across five major psychiatric disorders. Human Genetics, 136(2), 263–274. [DOI] [PubMed] [Google Scholar]
- Zhou, C. J. , Zhao, C. , & Pleasure, S. J. (2004). Wnt signaling mutants have decreased dentate granule cell production and radial glial scaffolding abnormalities. The Journal of Neuroscience, 24(1), 121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Supplementary Table.
Appendix S1: Supplementary Material.


