Summary
Members of the Arabidopsis thaliana TCP transcription factor (TF) family affect plant growth and development. We systematically quantified the effect of mutagenizing single or multiple TCP TFs and how altered vegetative growth or branching influences final seed yield. We monitored rosette growth over time and branching patterns and seed yield characteristics at the end of the lifecycle. Subsequently, an approach was developed to disentangle vegetative growth and to determine possible effects on seed yield. Analysis of growth parameters showed all investigated tcp mutants to be affected in certain growth aspects compared with wild‐type plants, highlighting the importance of TCP TFs in plant development. Furthermore, we found evidence that all class II TCPs are involved in axillary branch outgrowth, either as inhibitors (BRANCHED‐like genes) or enhancers (JAW‐ and TCP5‐like genes). Comprehensive phenotyping of plants mutant for single or multiple TCP TFs reveals that the proposed opposite functions of class I and class II TCPs in plant growth needs revision and shows complex interactions between closely related TCP genes instead of full genetic redundancy. In various instances, the alterations in vegetative growth or in branching patterns result into negative trade‐off effects on seed yield that were missed in previous studies, showing the importance of comprehensive and quantitative phenotyping.
Keywords: TCP transcription factor, plant growth, yield, phenotyping, Arabidopsis thaliana, plant architecture
Significance Statement
This study shows the importance of the TCP transcription factor family during Arabidopsis growth and development, coupled to yield and plant architecture. It was found that all class II TCPs are involved in axillary bud outgrowth and that the proposed opposite functioning of class I and class II TCPs requires revision.
Introduction
The major challenge of modern agriculture is to produce increasing amounts of high quality biomass for food, feed, and bio‐based products, with a minimal ecological footprint. Final yield, in the form of seeds, fruits, or leaves (e.g. leafy vegetables), depends strongly on plant architecture, organ size, and tissue longevity (Busov et al., 2008; Cai et al., 2016). Therefore, these traits have been the subject of breeding since the dawn of agriculture.
Our research aims at identifying genes involved in the control of plant development and architecture and finding possible correlations between their functioning and yield characteristics. We focus on key regulatory factors belonging to the TCP TF family in the plant model species Arabidopsis thaliana. Genes of this family orchestrate numerous growth‐related processes during Arabidopsis development (for review see Uberti Manassero et al., 2013). The TCP family consists of two classes with supposed opposite function; TCPs of class I are thought to be activators of growth and genes in class II to act as growth suppressors (Li et al., 2005). Examples in Arabidopsis include the role of BRANCHED1 (BRC1) and BRC2 in the outgrowth of axillary branches (Aguilar‐Martínez et al., 2007), determination of leaf shape and curvature by JAW‐TCPs (Nath et al., 2003; Palatnik et al., 2003) and effects on leaf size by TCP5‐like genes (Efroni et al., 2008). Nevertheless, for one of the founding members of TCPs, CYCLOIDIA (CYC) in Antirrhinum majus, both growth activating and repressing functions were found, depending on the developmental stage and cellular context (Luo et al., 1995), revealing lack of complete linkage between TCP classification and function.
TCP gene functions appear to be conserved between species and a mutation in the maize TEOSINTE BRANCHED1 (TB1) locus, the orthologue of Arabidopsis BRANCHED‐like genes, resulted in an higher expression and increased apical dominance, which has been instrumental in the domestication of maize from its ancestor teosinte (Doebley et al., 1995, 1997). Branching phenotypes have been observed as well in rice plants with a mutation in the TB1‐like gene (Takeda et al., 2003). The JAW‐TCPs (TCP2, ‐3, ‐4, ‐10, and ‐24) are simultaneously targeted by a microRNA (miR319) in Arabidopsis. Overexpression of miR319 leads to the jaw‐D mutant, which has a striking crinkled leaf phenotype due to an overproduction of cells in the leaf margins (Nath et al., 2003; Palatnik et al., 2003; Efroni et al., 2008). Similarly, tomato miR319 is required for normal leaf development by regulating the tomato JAW‐TCP homologue LANCEOLATE (Ori et al., 2007).
The above‐mentioned examples indicate the importance of the TCP TF family for the regulation of growth and plant architecture in the model species Arabidopsis and various crops. The more detailed our knowledge on the specific function of these TCPs and their potential combined effects on plant development, the more we will be able to precisely direct breeding for yield optimization by altering for example leaf growth and branching patterns (tillering).
Even though many reports about individual members of the TCP family of TFs are available (reviewed by Danisman, 2016), a comprehensive and detailed phenotyping of the various single and combined tcp gene mutants is lacking. This analysis is essential to understanding the role and importance of each individual TCP gene, the genetic interactions between the different TCP genes, and how effects on one trait influence other traits and/or final yield parameters. Most phenotypes observed so far consist of snapshots at a given time point, specific stage or on specific tissues or organs. This inevitably limits the documentation of phenotypes, which occur in a dynamic manner. Furthermore, it does not allow for an analysis on how phenotypes, such as leaf size and final seed yield, might be linked.
One of the difficulties of studying genes affecting growth and growth speed is that phenotypes are often only noticeable when careful measurements are performed. For example, an increase in leaf surface area of five percent is very difficult to identify with the naked eye, while it could give rise to a substantial increase in biomass production. Furthermore, it is evident that mutations resulting in altered developmental progression, such as tcp mutants affecting leaf growth rate, can be recorded only if a temporal component is included in the analysis (Boyes et al., 2001). Plants mutated for TCP20 for instance, show an increase in leaf pavement cell size in young developing leaves, whereas no difference in final leaf size could be detected (Danisman et al., 2012).
Functional redundancy has been shown and suggested for various members of the TCP family (Cubas et al., 1999; Danisman et al., 2013). For example, a single knockout of a gene of the Arabidopsis CIN‐TCP clade produces only mild phenotypes, whereas knocking‐down the whole clade shows dramatic changes in leaf development (Palatnik et al., 2003; Schommer et al., 2008). In this study, we analysed a well defined set of single and combined tcp mutants for several growth and yield parameters. Analysing various combinations of mutants (e.g. single, double, and triple) enables further analyses of the proposed functional redundancy within the TCP family and to decipher the contribution of individual TCPs to specific growth and development related functions.
In this study, temporal phenotyping analyses were performed using the ‘Phenovator’ platform (Flood et al., 2016). Using this phenotyping platform, all the above‐mentioned traits and characteristics were monitored in various tcp mutant lines and compared with wild‐type plants. The plants were imaged during the vegetative developmental stage at several moments of the day, allowing the measurement of rosette size, growth rate, and photosynthetic efficiency. Phenotyping of the branching traits, yield, and seed characteristics was performed on the matured and full‐grown plants. We converted the data on growth and development into an equation representing growth over time and coupled these results to other phenotypic parameters. This provided insights into the function of several members of the TCP TF family, as well as on the presumed redundancy of several TCPs. For all the genotypes studied, a difference in at least one of the growth equation parameters was observed, seemingly unlinked to which class (I or II) of TCP TFs the respective mutant belongs. The latter questions the validity of the historical functional division of the TCP family of TFs based on sequence similarity. Our phenotypic analysis revealed that there is no example showing absolute redundancy among related genes from a subclass of TCPs and their growth phenotypes. Furthermore, we showed that, under our growing conditions, an alteration in branching pattern (regardless of an increase or decrease in number of branches), leads to a decrease in final yield. Lastly, we hypothesised that all class II TCPs influence branching architecture of a plant grown with a higher plant density, possibly through involvement in light‐sensing.
Results
Mutant selection
The full set of tcp mutants that was analysed for developmental, architectural, and yield parameters can be found in Table S1(a). This set was further classified into subgroups using defined sequence and function‐associated characteristics. The family of TCP transcription factors has been divided into two classes based on differences in their TCP domains (Cubas et al., 1999). The second class of TCPs (Class II) can be subdivided into the groups CYC/TB1 (BRANCHED‐likes) and CIN based on sequence homology. For all these class II TCP genes biological functions have been assigned based on their respective mutant phenotypes (Alonso et al., 2003; Palatnik et al., 2003; Aguilar‐Martínez et al., 2007; Efroni et al., 2008; van Es et al., 2018). Sequence‐based subdivision of class I TCPs is less profound and slightly different parameters in phylogeny studies yield rather different phylogenetic trees (Martín‐Trillo and Cubas, 2010; Aguilar‐Martínez and Sinha, 2013; Danisman et al., 2013; Danisman, 2016). It is therefore difficult to determine subgroups and to predict potential functional redundancy among class I TCPs, solely based on sequence information. An interesting approach was taken by Danisman et al. (2013), who combined sequence similarity, gene expression patterns, and protein−protein interaction capacity to predict functional redundancy and to classify members of the Arabidopsis TCP family. With that knowledge in mind, we created two subgroups of class I TCPs: ‘TCP20‐likes’ and ‘TCP15‐likes’. In addition, we created the group ‘TCP20‐associated’, for which the selection criteria included information on downstream targets of TCP20 (Danisman et al., 2012), besides the redundancy prediction (Danisman et al., 2013).
Several of the selected tcp mutants have strong developmental phenotypes and were used for validation of the approach and critical evaluation of the followed methodology. An example is the jaw‐D mutant, which is known to exhibit slowly developing leaves that eventually grow larger than leaves of wild‐type plants (Efroni et al., 2008). Obvious candidates to study the relation between branching and yield are the mutants in the BRANCHED‐like TCPs (Aguilar‐Martínez et al., 2007). Additionally, we focused on the analysis of potential functional redundancy within the TCP TF family, and included among others the double mutant tcp14tcp15 (Kieffer et al., 2011), tcp8tcp15, as well as the single tcp8 and tcp15 mutants (group ‘TCP15‐likes’).
Experimental set‐up and technical constrains of the ‘Phenovator’ platform
The Phenovator platform (Flood et al., 2016) allows the growth of 1440 plants simultaneously in a grid of 24 by 60 plants (Figure S1a). This grid was divided into 28 plots in which all 24 selected genotypes (Table S1a) were randomly positioned to allow for the correction of a possible positional bias. A regression analysis was performed to check for a possible effect of the plant's position in the growth chamber. The average size of all plants over time, as well as the maximum size was plotted on the X‐ and Y‐coordinates corresponding to their position in the growth chamber (Figure S1a). We found a correlation between the X‐coordinate and both maximum and average growth for our plants (Figure S1b). No significant correlation between Y‐coordinate and projected leaf area (PLA) was detected (Figure S1c). The R‐package SpATS (Rodríguez‐Álvarez et al., 2016; Velazco et al., 2017) was used to correct for the observed spatial effect in the X‐position. Although the growth chamber is expected to have near identical conditions regardless of the position within the chamber, the observed positional effect in the X‐direction might be due to small differences in for example air flow, humidity or nutrient availability. The latter mentioned effects could be introduced due to the inflow of water with nutrients from one side of the chamber (X ≈ 0; Figure S1a).
Arabidopsis rosette development follows an S‐curve that includes an oscillatory component
Plant growth consists of two opposing factors: the intrinsic tendency toward unlimited increase (exponential growth phase) and restraints imposed by environmental resistance and ageing (resulting in an asymptotic maximum area) (Zeide, 1993). These two factors result in an S‐curve growth function (Tessmer et al., 2013). Furthermore, plant leaves move rhythmically up and down in a circadian fashion (Engelmann et al., 1992). Singular spectrum analysis (SPA) on the PLA confirmed that the progress of Arabidopsis rosette growth contains two parts, an S‐curve and an oscillatory function (Figure S2a).
An essential part in analysing plant size in a time series is to convert the data into an equation representing growth over time. Creating a growth‐curve equation fitting the observed data enables turning different sizes and growth speeds into easily distinguishable parameters. The near‐infrared (NIR) data were used to determine PLA as the latter provides a good estimate of above‐ground biomass (Leister et al., 1999) and can therefore be used to determine plant growth. This provides insight into growth dynamics of all mutant lines over time and enables a step beyond investigating average final plant sizes only.
The S‐curve (Figure S2b) contains parameters that account for the distance between the two asymptotes (β1), that describe the magnitude of the derivative (β2), and the point at which the derivative of the function reaches its maximum (β3). Next, the function to describe the circadian rhythm is an oscillatory function (Figure S2c) with certain amplitude (β4) and phase (β5). These two functions combined allowed us to determine the actual growth function (1). With the described combination of starting values and bounds for the parameters (see Experimental procedures), the algorithm converges for all the time series and the values for β1–β5 could be determined. An example of a fitted function is shown in Figure S2(d). The obtained β‐values allow us to look at the growth function in more detail and to extract the most likely biological meaning of the different β‐parameters. For instance, the parameter β1 could represent the maximum possible growth; β2 a measure for the rate of growth in the exponential phase and β3 a measure for the time until the rosette of a plant starts its rapid growth phase. β4 is the change in measured rosette area relative to the real rosette area at that moment as a result of oscillatory leaf movement and β5 is the starting point of this oscillation related to the circadian rhythm.
Growth parameter analyses
The nature of the measurements enables measurement of the rosette surface area, but not distinguishing individual leaves and their shape and size. Although the β‐values in the growth equation described above cannot be directly linked to a particular growth parameter, we used a careful interpretation to disassemble the dynamic growth at plant level. Average β‐values and their standard deviation per plant line were plotted (Figure S3a) and the parameters β2 and β3 were scaled to the lowest value to enhance readability of the graph. Statistical tests show differences in β‐values comparing the mutant lines with wild‐type control plants (Figure S3b). In short, all tcp mutants studied show alterations in developmental progress compared with wild‐type control plants. Subsequently, a principal component analysis (PCA) was performed to visualise potential differences for the various tcp mutants (Figure 1).
Figure 1.
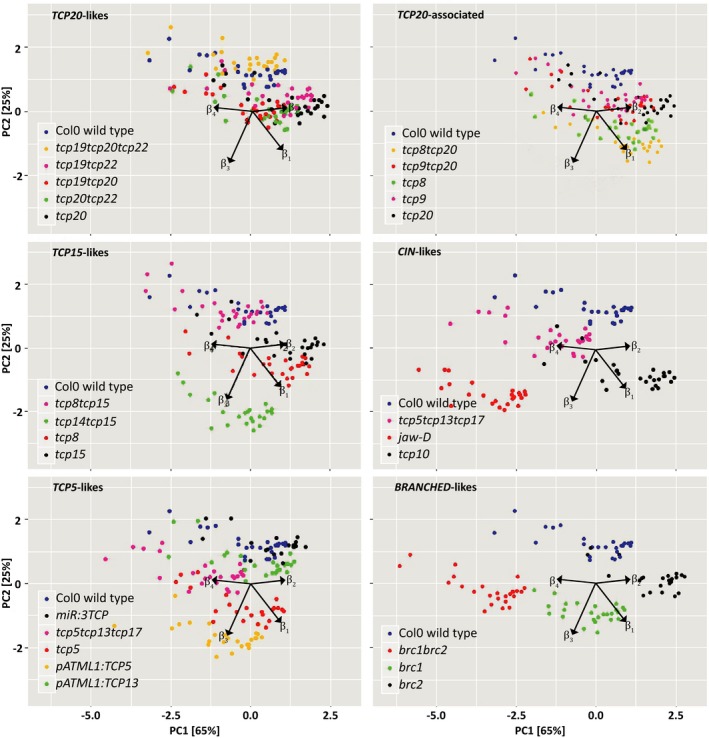
Principal component analysis (PCA) plots for individual β‐values of all lines in this study. Results of a PCA for all the βs to examine a possible difference in mutants versus wild‐type. For visual clarity the plots are made up for selected subgroups of tcp mutants, always including the Col0 wild‐type as the control. Each data point in the PCA plot represents an individual plant. The β1, β2, β3 and β4 variable loadings are depicted as arrows. Combined, principal components 1 and 2 explain 90% of all variance (PC1 explains 65% of the variation, PC2 explains 25%).
A group of class I TCPs more closely studied on a cellular level and for possible redundancy are the TCP20‐likes and TCP20‐associated (Danisman et al., 2012, 2013; Table S1a). Cells in leaves of several of these mutants are shown to enter the elongation phase earlier than leaf cells of wild‐type control plants. In line with this, we found that tcp19tcp20tcp22, tcp9, and tcp20 exhibit a lower β3 than wild‐type control. Contrastingly, the double tcp9tcp20 mutant does not show a difference in β3 compared with wild‐type.
The other class I TCPs studied here, the TCP15‐likes, are characterised by a higher β1 and β3, with tcp8tcp15 as notable exception, having only a lower β1 compared with wild‐type control plants. The latter could indicate that the combination of tcp8 and tcp15 single mutations attenuates their individual growth and development effects.
A clear example of a class II mutant with altered development is the jaw‐D mutant, shown to have a significantly lower β2 but higher β3. This represents the plants’ characteristic and well known slow developmental progress (Palatnik et al., 2003) through slow growth rate (β2) and longer time to reach the rapid growth phase (β3). Together with the tcp5tcp13tcp17 triple mutant, jaw‐D is the only mutant that has a lower β2 compared with the wild‐type, as visualized by the PCA plots in Figure 1. By contrast, tcp10 has a higher β1 than wild‐type, i.e. a larger final size, fitting earlier research (Schommer et al., 2008). At a cellular level, β2 might be linked to the speed of cell elongation, fitting with the observed growth phenotypes of the jaw‐D and the tcp5tcp13tcp17 triple mutant.
The tcp5 single mutant shows a phenotype opposite of that of the triple tcp5tcp13tcp17 mutant as it has a significant higher β1 and β3. The mutant overexpressing TCP5 in the epidermis only (pATML1:TCP5; van Es et al., 2018) shows a higher β2 and β3. Overexpressing TCP13 in a similar manner results in a higher β1 and a lower β3.
Members of the BRANCHED‐like group of TCPs are best known for their role in branching. Next to altered branching patterns, plants mutated for these genes show an altered developmental progression of plant growth. Plants of the brc1brc2 double mutant reach their maximum growth faster (β3) and have a significantly smaller final size (β1) than the wild‐type control plants. Interestingly, the single mutants brc1 and brc2 show opposite behavior compared with the double brc1brc2 mutant concerning β1 and deviate from the double in that brc1 has a higher β3 value, whereas brc2 appears to have a higher growth speed (β2).
It must be noted that our interpretation of the final size of a plant is influenced by the duration of the measurements, i.e. some plants might not have reached full size by the time the PLA measurements had stopped. Another way to visualise growth is by plotting the PLA over time for the raw data versus the model (Figure 2a), showing their resemblance. This was followed by plotting the raw data for the brc mutants and Col‐0 control in Figure 2(b), revealing the lower β1 for the brc1brc2 mutant for example. Figure 2(b) shows that the higher growth speed (β2) might have led to the overall bigger brc2 plant, whereas brc1 only ‘catches up’ later, having reached its maximum growth speed (β3) faster.
Figure 2.
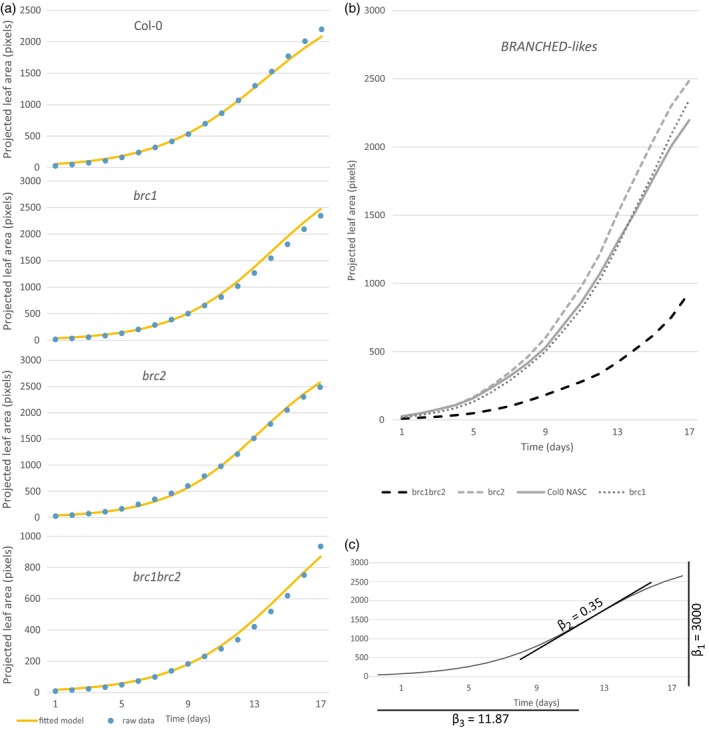
Visualisation of model versus raw data of BRANCHED TCPs.
(a) Shows the fitted model (yellow line) and the raw data (blue data points) for four plant lines during the 17 days of measurement. A comparison of the fitted model of the three branched mutants and a wild‐type control (b) showing the different growth curves. An example of a fitted S‐curve for three β‐values shown in (c). Shown in (a) and (b) is the average projected leaf area per plant for each line, with measurements from one time point during the day, omitting the circadian rhythm for visual clarity.
Photosynthetic capacity of plants
Next to size measurements based on PLA, the ‘Phenovator’ camera is monitoring photosynthetic efficiency of the plants under study. For this purpose, the system uses pulse amplitude modulated (PAM) chlorophyll fluorescence imaging to measure the light‐use efficiency of PSII electron transport (ΦPSII) (Genty et al., 1989; Baker, 2008; Flood et al., 2016). Based on the average output of the ΦPSII measurement for all wild‐type and tcp mutant lines, no significant differences were found (Figure S4), revealing that none of the analysed TCP transcription factors was having a direct effect on photosynthetic efficiency during the investigated timespan of plant development.
Seed yield characteristics
Several characteristics of seeds were determined. Final yield was measured by weighing all the seeds for eight plants per genotype. Seed characteristics were explored in more detail by measuring seed size and seed number per silique on four siliques of five plants per genotype. We found that average seed size was not significantly affected in any of the analysed mutants (Figure 3a), whereas several lines showed a reduction in the average number of seeds per silique and total seed weight (Figure 3b,c). Only the seeds of the double mutant tcp8tcp15 were significantly lighter compared with wild‐type seeds (Figure 3d). Correlation analyses of the different seed characteristics revealed that the number of seeds is negatively correlated with both seed area and seed weight and that seed weight is positively correlated with seed area (Figure S5a–c). Several lines showed a decrease in total seed weight (i.e. total yield), such as the brc1brc2 double mutant and jaw‐d, but also two tcp15‐related double mutants: tcp8tcp15 and tcp14tcp15.
Figure 3.
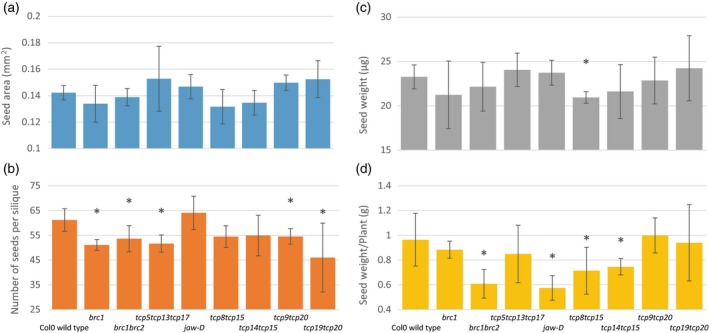
Genotypes showing differences in yield characteristics.
Quantification of yield characteristics such as average seed area (a), average seed number per silique (b), total seed weight per plant and average weight of the individual seed in (c) and (d) respectively. Only genotypes are shown for which statistically significant differences were found compared with wild‐type control for at least one of the investigated parameters. Average seed number and seed area were determined on four siliques on five plants per genotype. Statistically significant differences with wild‐type (Student's t‐test, P < 0.05) are indicated by an asterisk.
TCPs affecting branching parameters; old acquaintances and new friends
To obtain insight in the effects of mutating or modifying expression levels of particular TCPs on plant architecture, the number and type of branches were quantified for all lines studied. In Figure 4(b), data on all the lines that showed a significant difference to Col‐0 are presented. We found, as expected, that brc1 and brc1brc2 double mutants showed an increase in secondary branches (Aguilar‐Martínez et al., 2007). Conversely, several other lines showed a reduction in some aspects of branching, most notably the jaw‐D (class II TCPs) and tcp14tcp15 (class I TCPs) mutant lines, having a significant reduction in the number of secondary branches.
Figure 4.
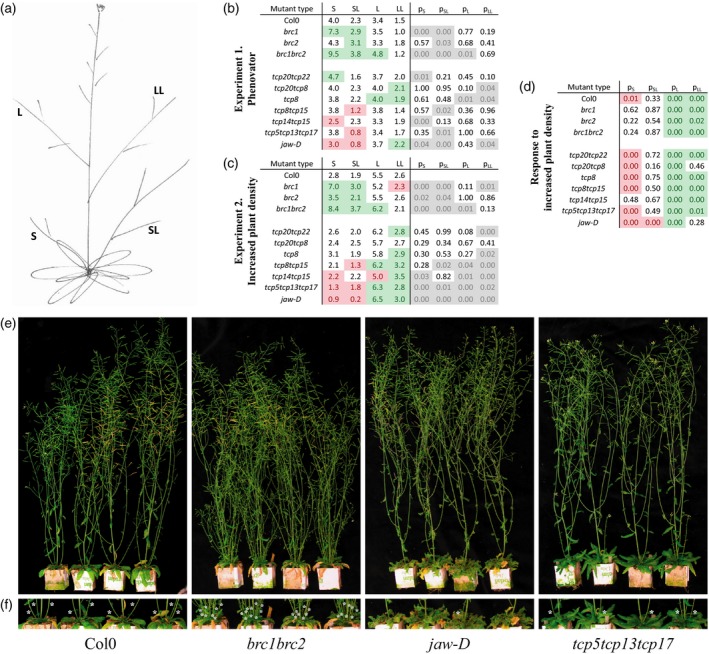
Branching phenotypes.
(a) Schematic visualization of the position of secondary branches (S), lateral branches (L), branches on secondary branches (SL) and branches on lateral branches (LL).
(b) Branching parameters obtained from plants grown in the ‘Phenovator’ phenotyping system.
(c) Branching parameters for the same mutants as shown in (b) but grown under higher plant density in a separate ‘Increased plant density’ experiment. In both (b) and (c) red and green indicate a significant decrease or increase in number of branches respectively, compared with wild‐type control, followed by a table with the outcome of Student's t‐tests (P < 0.05, n = 25; grey shaded) per branching parameter. Only genotypes are shown for which statistically significant differences were found compared to wild‐type control.
(d) Shows the response to the increased planting density per line, red indicates a statistically significant decrease of branching, green a significant increase (Student's t‐tests, P < 0.05) in the ‘Increased plant density’ experiment compared to the ‘Phenovator’ experiment.
(e) Pictures of representative plants of the various lines in ‘Experiment 2. Increased plant density’, showing the observed differences in branching phenotype compared with wild‐type control for the brc1brc2, jaw‐D and tcp5tcp13tcp17 class II TCP mutants.
(f) Zoom‐in of the pictures in (e), highlighting the secondary branching phenotypes.
Surprisingly, both an increase as well as a decrease in the number of branches has a negative effect on the total seed weight per plant grown under our conditions (brc1brc2, jaw‐D, and tcp14tcp15 in Figure 3d). We cannot exclude that these genes are also involved in seed characteristics independently from their function in the development of branches. However, this possibility seems unlikely for the BRANCHED‐like genes as they are reported to be expressed in axillary buds only. Interestingly, the number of secondary branches is not correlated to the number of lateral branches (Figure S5d).
Plants produce fewer branches as a result of a low red to far‐red (R:FR) ratio, known as the shade‐avoidance response, which can be achieved by high plant density (Casal, 2012). Taking this knowledge into account, we investigated branching under slightly higher plant density in a separate experiment. We grew the plants 6 cm apart, compared with the 9 × 7.5 cm in the initial ‘Phenovator’ experiment. In general, the number of secondary branches was reduced when the plants were grown at higher density, as nicely exemplified for the Col‐0 wild‐type plants (Figure 4d–f). As expected, we also found insensitivity of brc1 for different R:FR ratios (Aguilar‐Martínez et al., 2007), validating the experiment's effectiveness (Figure 4d). Interestingly, two other clades of class II TCPs (tcp5‐likes and jaw‐D mutants) showed a strong reduction in number of secondary branches grown under higher planting density (Figure 4d–f), a phenotype unnoticed in previous experiments. Additionally and surprisingly, we observed more lateral branches under higher planting density for all investigated lines. Overall, observed phenotypic alterations in the Phenovator experiment could be confirmed and observed effects on secondary branching were enhanced due to the higher plant density, as expected.
Discussion
The aim of this research was to provide a comprehensive overview of differences in developmental, vegetative growth, and seed yield characteristics between a carefully chosen set of tcp mutants and wild‐type plants. We have developed an approach that describes the growth curve during the vegetative stage of development, using four different biologically meaningful parameters. Variation in these parameters among different tcp mutant lines could be clearly represented by a PCA (Figure 1). Interestingly, for all of the genotypes studied, a difference in at least one of the growth equation parameters was observed, which is not surprising as TCPs are involved in different aspects of plant development (reviewed by Martín‐Trillo and Cubas, 2010; Uberti Manassero et al., 2013; Nicolas and Cubas, 2015, 2016; Danisman, 2016). Next to this, characterising branching parameters and seed yield characteristics on fully mature plants revealed that several tcp mutant lines behaved differently compared with wild‐type with respect to number and weight of seeds (Figure 3) and the number of branches (Figure 4).
The role of TCP transcription factors in growth regulation during the vegetative stage of plant development
Even though our study does not venture into the cellular level of development, we can speculate on the underlying cellular mechanism causal for the observed differences in the individual growth equation parameters (β‐values) based on whole plant imaging. β3 for example is supposed to represent the moment a plant enters the rapid growth phase. This moment is typically marked, at the cellular level, by a switch from cell proliferation into cell elongation (Andriankaja et al., 2012). Some well studied class II tcp mutants are thought to reside longer in the cell proliferation phase producing more, but often smaller, cells for example jaw‐D and related cin mutants (Nath et al., 2003; Crawford et al., 2004; Challa et al., 2019). In the jaw‐D mutant this is represented by a higher β3 compared with wild‐type as it simply takes this mutant longer to reach the phase of rapid and exponential rosette growth. The jaw‐D mutant shares its relatively slow progress of growth with a line mutated for the closely related TCP genes TCP5, TCP13, and TCP17 (tcp5tcp13tcp17 mutant; Efroni et al., 2008). A delay in leaf and rosette development can also be caused by a reduced speed of cell elongation, a lower β2, which was indeed observed in the jaw‐D and the tcp5tcp13tcp17 triple mutant.
Several genotypes such as tcp19tcp20tcp22, tcp9, and tcp20 exhibit a lower β3 compared with wild‐type, suggesting that they enter the cell elongation phase earlier, as was previously shown for tcp20 (Danisman et al., 2012). The single tcp9 and the double tcp9tcp20 mutants showed a cellular phenotype similar to that of tcp20 (Danisman et al., 2012). In our experiment however, the double tcp9tcp20 does not show difference in β3, suggesting that TCP9 has unique functions and in this is epistatic over TCP20. Additionally, several class I mutants show a similar effect on β3 as the class II jaw‐D mutant, for example the double mutants tcp14tcp15 and tcp8tcp20.
Focussing on β1, which represents the maximum of growth, only brc1brc2 and tcp8tcp15 have a lower β1 than wild‐type. Several other mutants show a higher β1, such as the single branched mutants as well as tcp15 and the tcp14tcp15 double mutant. Molecularly, β1 is hard to interpret as the final size is influenced by numerous factors other than altered growth speed and timing of rapid growth; for instance, the maximum growth is also dependent on the flowering time, a trait that has not been measured in this study. Additionally, some lines might not have reached their final size during the measurement period.
Next to the growth parameters, we also studied branching patterns. Best studied in this respect are the branched mutants that produce more branches than wild‐type, most notably the brc1brc2 double and the brc1 single mutants (Aguilar‐Martínez et al., 2007). In addition, we found that the jaw‐D and tcp5tcp13tcp17 (class II), and tcp14tcp15 (class I) mutant lines showed a significant reduction in the number of secondary branches (Figure 4b). This suggests that next to the BRANCHED TCPs, more TCPs are involved in the control of axillary bud outgrowth, either directly by acting in the meristem, or indirectly, for example through changes in sink‐source relationships, hormone levels or availability of carbon resources. Promoter GUS fusions of several class II TCPs revealed TCP3 (a JAW‐like TCP) and TCP5 expression in axillary buds (Koyama et al., 2007), enabling cell‐autonomous control over axillary bud outgrowth. When grown under higher plant density, significantly fewer branches are observed in jaw‐D, tcp5tcp13tcp17 and tcp14tcp15 mutants and significantly more secondary branches were found in the various brc mutants and tcp20tcp22 (Figure 4c). These effects are most likely related to the altered R:FR light ratio. In Sorghum bicolor, the photoreceptor phyB is thought to negatively regulate the BRC1 homologue SbTB1 (Kebrom et al., 2010) and a recent study showed that TCP5, TCP13, and TCP17 directly upregulate several PHYTOCHROME INTERACTING FACTOR's (PIFs) (Zhou et al., 2017), resembling known interactors of phyB. Additionally, TCP15 activity was shown to be modulated by high light intensities (Viola et al., 2015). Altogether, this leads to the hypothesis that these specific TCPs are involved in light‐sensing or signaling and thereby influencing branching architecture of a plant under different light conditions.
TCPs do not show expected redundant behavior for phenotypes tested
The BRANCHED subgroup of TCPs are believed to act redundantly; however, they show striking growth phenotypes. Visualising their rosette growth pattern reveals that both the brc1 and brc2 single mutants eventually outgrow Col‐0 control plants, although brc1 grows slower in the first few days of measuring. The brc1brc2 double mutant conversely grows slower and remains smaller throughout the duration of the measurements (Figure 2b). It must be noted that this observed rosette phenotype could be due to the duration of the measurement and that the brc1brc2 double mutant might eventually catch up or even outgrow wild‐type plants. These observed growth phenotypes suggest that the two Arabidopsis BRC genes control each other, possibly through a negative feed‐back regulation. This has been shown for BRC2, which was upregulated in the brc1 mutant, possibly compensating for the loss of BRC1 function (Aguilar‐Martínez et al., 2007). Similarly, BRC1 could be strongly upregulated in the brc2 mutant, explaining the opposite growth behavior when comparing with the double mutant.
A closer look into the rosette growth of the ‘TCP15‐likes’ (Figure 1) shows another interesting pattern of single and double mutants. The double mutant tcp8tcp15 seems almost identical to Col‐0 wild‐type, whereas both tcp8 and tcp15 look vastly different indicating that the combination of tcp8 and tcp15 attenuates their individual growth and development effects.
Overall, full redundancy would imply that single mutants show behavior identical to wild‐type and only double or higher order mutants would exhibit mutant phenotypes. Based on our data, there are no examples showing true redundancy among the investigated groups of related TCPs and their growth phenotypes. The single mutant behavior we have observed would therefore imply only partial redundancy among the analysed members of the TCP family of TFs. An example of known partial redundancy is the miR319 regulation of the JAW‐D TCPs; a single knockout of a gene in this clade produces only mild phenotypes, whereas knocking out the whole clade shows dramatic changes in leaf development (Schommer et al., 2008). This effect can be seen in the higher β1 for the tcp10 single mutant, which corresponds to an increase in leaf area as previously observed (Schommer et al., 2008).
Previous research ranked pairs of a number of TCP TFs on their likeliness to be functionally redundant, based on protein sequence, gene expression and Y2H analysis (Danisman et al., 2013). Interestingly, this list showed that the closely related TCP19 and TCP20 ranked high in potential functional overlap, which was confirmed by a detached leaf senescence experiment. Although TCP8 is as closely related to TCP20 as is TCP19 based on sequence similarity, no shared function of TCP8 with TCP20 was found (Danisman et al., 2013). The data presented here show that during rosette development, TCP19 and TCP20 do not seem to share a function in growth (both tcp20 and tcp19tcp20 are similarly different from wild‐type, independent of the presence or absence of a functional TCP19 gene), whereas the double mutant tcp8tcp20 seems to enhance the tcp20 single mutant phenotype (Figure 1). Further research into the exact molecular function of each TCP protein is necessary to be conclusive on the potential partial functional redundancy.
Two classes of TCPs, showing antagonistic behavior?
Historically, the TCP family of TFs has been divided into two classes based on sequence characteristics (Cubas et al., 1999). Members of these two classes are believed to act antagonistically, either by promoting cell growth and proliferation (class I) or through the repression of these processes (class II) (Martín‐Trillo and Cubas, 2010). Whether looking at different β‐values in Figure S3(b) or at the distribution of the data points in the PCA plot in Figure 1, one cannot easily identify differences between class I and class II TCPs. This could be due to the nature of our measurements, looking at whole rosette growth as opposed to the cellular level in which the opposite function is observed (Nath et al., 2003; Crawford et al., 2004; Danisman et al., 2012). This notwithstanding, our results question this strict division into two antagonistically functioning classes. Mutants from particular members of class I and class II TCP genes show similar changes in specific β‐parameters. More comparative research will be necessary to elude either antagonistic or similar behavior. Supporting our data, the CYCLOIDEA gene in Antirrhinum is known to both suppress and promote growth of the dorsal petals depending on the developmental stage (Luo et al., 1995).
Next to the growth phenotypes we describe, branching patterns also lack a strict opposite behavior for plant mutagenized in either class I or II TCPs. The increase in branching of brc1brc2 double mutants would imply that a class I mutant should exhibit fewer branches. This is the case for tcp14tcp15, but also the class II mutants jaw‐D and tcp5tcp13tcp17 show a decrease in some of the investigated branching parameters. Growing plants with an increase in plant density seemed to increase this effect (Figure 4). This similarity in phenotypes could indicate overlapping functions, or at least similar final phenotypic effects for proteins of both classes, and therefore questions the functional division of the TCP family of TFs solely based on sequence variation.
The link between plant development and final yield
Our study has provided an excellent body of data allowing for a closer look at the effect of unique plant developmental alterations on plant yield under defined environmental conditions. We have collected data on the architecture of the plants, their developmental progress as well as yield characteristics in terms of total seed production. We have confirmed the link between several seed characteristics by correlation analyses (Figure S5a–c) and showed that an increase in number of seeds is negatively correlated with both seed area and seed weight. This negative correlation has been found before (Gnan et al., 2014) and is in line with an earlier proposed model describing a fixed amount of resources allocated to reproduction (Paul‐Victor and Turnbull, 2009).
Strikingly, plants that appear to have several significantly different growth parameters (β's) compared with wild‐type, such as jaw‐D, tcp14tcp15, and brc1brc2, show altered branching patterns or yield characteristics as well. However, a different progression of growth and rosette development (the β parameters) does not necessarily result in differences in yield or branching patterns. Telling examples are the tcp8 and tcp15 single mutants, both show a higher β1, β2, and β3 compared with the wild‐type, but no differences in seed yield characteristics have been observed. It has previously been reported that inflorescence architecture is of great influence on final seed yield characteristics. For example, prolonged life span of the Arabidopsis inflorescence in a fruitful mutant resulted in more seeds (Balanzà et al., 2018). Furthermore, pruning in Arabidopsis led to the development of longer and larger siliques that contained fewer, but bigger seeds (Bennett et al., 2012). This was attributed again to a reallocation of resources which could also be applicable in the brc1 and brc1brc2 mutants. The production of more branches in these mutants reduces the amount of resources left for seed production, resulting in a reduction in seed yield. This does not explain however the reduction in seed production in the jaw‐D, tcp5tcp13tcp17, and tcp14tcp15 mutants for instance, whose lack of branching could provide additional resources for seed development.
Altogether, this study makes clear that it is indeed challenging to increase and optimize final crop yield by changing the activity of key developmental regulatory genes, such as TCP transcriptions factors. Previous simple classification into activators and repressors of growth appear not to hold and functions were shown to be context and developmental‐stage dependent. Furthermore, despite conservation of overall functions for TCP transcription factors, there are various species‐specific differences, among others due to species or lineage‐specific gene duplications that directly affect possibilities for knowledge transfer from model species to crops.
Experimental procedures
Plant materials
Information about all lines and genotypes used in this study is shown in Table S1(a). Col‐0 wild‐type (NASC) was used as control for the T‐DNA mutant lines. The Col‐0 wild‐type used as background for the ATML1 pro :TCP5/13‐GFP transformations served as the control for those particular lines (Col‐0 ‘WUR’ in Table S1a). ATML1 pro :TCP5‐GFP and ATML1 pro :TCP13‐GFP lines were previously generated (van Es et al., 2018).
System set‐up and plant growing conditions
All lines mentioned in Table S1(a) were grown under long‐day conditions (16/8 light/dark cycle at 21°C) on Rockwool and their seeds were harvested simultaneously for use in the large‐scale phenotyping experiments. The seeds were sown on wet filter paper and stratified for 2 days at 4°C to ensure uniform germination. After stratification, the seeds were placed on wet Rockwool (www.grodan.com), that was pre‐soaked in a nutrient solution designed for Arabidopsis (van Rooijen et al., 2015; Flood et al., 2016). The seeds were placed in the phenotyping system (Phenovator, Flood et al., 2016) at day zero. The Phenovator allows to grow 1440 plants simultaneously in a grid of 24 by 60, placed upon an ebb and flood hydroponic system. The analysis software is unable to distinguish between overlapping plants and therefore only half of the available positions were used, resulting in a planting grid of 9 × 7.5 cm. The grid was divided into 28 plots, in which all 24 genotypes were randomly positioned to prevent a possible positional bias. The plants were grown in the Phenovator under a 16/8 day/night rhythm at a constant temperature of 20°C, relative humidity of 70%, and constant irradiance of 200 μm m−2 sec−1.
Measurement schematics
The Phenovator system (Flood et al., 2016) was used to measure two different parameters: PSII operating efficiency (ΦPSII) and near‐infrared reflection at 790 nm (NIR). The raw data are provided in Table S2. ΦPSII was measured daily at Zeitgeber (ZT) one, five, eight, 11, and 13. Data obtained through the NIR reflection were used to determine PLA. The wavelength (790 nm) was chosen so that plants could be measured both day and night without disturbing their circadian rhythm. The plants were imaged from day 8 until day 25, until overlapping leaves of neighbouring plants made distinguishing between individual plants impossible. For a detailed overview of the light and measurement regime described above, see Table S1(b).
Subsequently, full‐grown plants were phenotyped on their level of branching, total seed set as well as several yield characteristics such as number of seeds per silique, size of seeds, and the seed weight. Phenotyping of branching was done by manually counting the number of lateral shoots and branches on the main inflorescence. Total yield was determined by weighing all the seeds for eight plants per genotype. Seeds size and number per silique was measured for four siliques on five plants per genotype. The seeds were pictured using a set‐up designed for imaging seeds and their germination: the ‘germinator’ (Joosen et al., 2010). We used this software for seed imaging, after which the pictures were analysed by the ‘Analyse particles’ function in ImageJ, obtaining surface area.
Data handling and pre‐processing of growth experiment
Raw data generated by the Phenovator (Table S2) are converted into .csv files with data on the physiological parameters (e.g. ΦPSII or PLA through the NIR images). Non‐germinated seeds were identified by a lack of signal in the PAM dataset the 25th day of the experiment. Coordinates in which no signal was observed were removed from both NIR and PAM datasets. The SpATS package (Rodríguez‐Álvarez et al., 2016) was used to correct for the observed spatial bias in the PLA dataset.
Creation of a formula to describe the progress of growth for all plants
The PLA time series consisted of 110 data points per coordinate (i.e. individual plant) and was analysed by singular spectral analysis (SSA) using the R‐package Rssa (Golyandina et al., 2013). Elementary components (ECs) were retrieved by ‘residuals()’. Based on visual assessment of the ECs growth function (1) was constructed, which consists of two parts: one describing an S‐curve, accounting for the actual growth of the plants (Zeide, 1993; Tessmer et al., 2013) and one accounting for the circadian rhythm (Engelmann et al., 1992):
| (1) |
The parameters of function (1) were then estimated by non‐linear regression, using the ‘nls’ function in R, for each individual coordinate (i.e. individual plant) in the time series. The specific algorithm used was originally introduced as the NL2Sol algorithm (Dennis et al., 1977). Lower and upper bounds that are implemented in the port function, and starting values are required. For the parameters in the first part of f(t) in between curly brackets referred, to as g(t), a set of starting values for each of the time series was determined using their basic properties:
| (2a) |
| (2b) |
| (2c) |
Estimates for limt→∞ g(t), g(0), and g’(0) were used to determine starting values of the βs. As the first EC resulting from the SSA is used as an approximation for g(t) as it contributes to g(t) for 99% in terms of covariance. For each of the time series this component is a vector of length 110; EC = {EC1,EC2,…,EC110}, with the corresponding time values as t = {t 1,t 2,…,t 110}.The required approximations are:
| (3) |
| (4) |
| (5) |
The necessary bounds were assessed using the available biological information, such as growth stages of Arabidopsis (Boyes et al., 2001). Note that the exact values of these bounds are not extremely important, as long as they are biologically meaningful and the optimal parameter value lays within the bounds this will ensure that the algorithm converges. For β1, the natural lower bound is β1,lower = 0 and the upper bound was set to β1,upper = 3 * max{EC} as the plants were measured until 25 days after sowing, which approaches the time a wild‐type Arabidopsis plant needs to reach maximum rosette size (Boyes et al., 2001). The measurements started 8 days after sowing and continued for 17 days, so it is unlikely that a plant has already reached the point of most rapid growth (β3) at time 0 and conversely, 30 days is a safe upper bound, assuming a plant has reached its maximum size almost 40 days after sowing. The parameter β2 is closely related to β3 and was inferred by:
| (6) |
Here t tang,0 is the time coordinate for which the tangent of g(t) at the inflection point intersects the t‐axis. The part of the function g(t) left of the inflection point is convex. Therefore, every tangent in that part lies below the function. This, combined with the expectation that g(0) is very small, as the plants just started growing at time 0, implies that t tang,0 ≤ 0. Most of the growth of the plant happens between β3 − t tang,0 and β3 + (β3 − t tang,0), that is within a time span of 2(β3 − t tang,0). We assumed 2(β3 − ttang,0) ≥ 4 as it is highly unlikely that most of the growth of the plants happens within 4 days. Combining these bounds with equation (6) yields which allowed us to formulate the following estimations of bounds:
| (7a) |
| (7b) |
| (7c) |
The second part of f(t), shown in function (1) in between square brackets as h(t), is an oscillatory function with a period of 1 day, describing leaf movement represented by β4. We assumed a starting value of β4,start = 0.05. As it is unlikely that the circadian movement of the rosette leaves will result in more than 50% change in area, a reasonable upper bound is therefore β4,upper = 0.5. The moment the circadian rhythm starts is represented by β5, i.e. the phase‐shift of the oscillation. The range of β5 is naturally bound between 0 ≤ β5 ≤ 2*π. The average value of β5,start = π was chosen. Finally, identification and visualisation of differences between average β‐values (Per analysed line; see Table S1) was performed by PCA using prcomp() in R, scale set to ‘true’. To test whether the differences in the measured variables between the wild‐type and mutants are significant, some two‐sided t‐tests were performed at a significance level of 0.2.
Data handling yield characteristics and branching parameters
Yield characteristics were described as mentioned above and several PCA were performed on all measured variables. Again the prcomp() command was used to perform this analysis. A single parameter is introduced to capture most of the variance of the four branching parameters, defined as the linear combination given by the first principal component, resulting from the PCA between the four branching parameters. Regression analysis using the lm() command was performed to test whether correlations between the variables number, area, and weight exist.
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Figure S1. Regression analysis of positional effects in the experimental set‐up.
Figure S2. Designing a growth function based on rosette size measurements over time.
Figure S3. Average values for all four beta parameters.
Figure S4. Comparison of photosynthetic efficiency for the various tcp mutants.
Figure S5. Correlation analysis of the seed and branching parameters.
Table S1. Mutant lines and measurement regime used in this study.
Table S2. Raw ΦPSII and NIR data generated by the Phenovator platform.
Acknowledgements
We would like to thank J. Harbinson and A.E. Prinzenberg for their help in the experimental set‐up, initial data analyses and fruitful discussions. We greatly appreciate the members of the PDS research group for their help in setting up the ‘Phenovator’ experiment and J. Busscher‐Lange for her assistance with quantifying the branching phenotypes. W.T. Kruijer recommended the use of the SpATS package. Our work is supported by grants from the Dutch Scientific Organization (NWO); (NWO‐JSTP grant 833.13.008), CAPES/NUFFIC (no. 010/07) and CAPES/NUFFIC (no. 033/2012). This research was partly financed through an ‘Enabling Technologies Hotel’ (ZonMw 435002014).
References
- Aguilar‐Martínez, J.A. , Poza‐Carrión, C. and Cubas, P. (2007) Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell, 19, 458–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M. , Stepanova, A.N. , Leisse, T.J. , et al. (2003) Genome‐Wide Insertional Mutagenesis of Arabidopsis thaliana . Science, 301, 653‐657. [DOI] [PubMed] [Google Scholar]
- Aguilar‐Martínez, J.A. and Sinha, N. (2013) Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Frontiers in plant science, 4, 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriankaja, M. , Dhondt, S. , De Bodt, S. et al. (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: a not‐so‐gradual process. Dev. Cell, 22, 64–78. [DOI] [PubMed] [Google Scholar]
- Baker, N.R. (2008) Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 59, 89–113. [DOI] [PubMed] [Google Scholar]
- Balanzà, V. , Martínez‐Fernández, I. , Sato, S. , Yanofsky, M.F. , Kaufmann, K. , Angenent, G.C. , Bemer, M. and Ferrándiz, C. (2018) Genetic control of meristem arrest and life span in Arabidopsis by a FRUITFULL‐APETALA2 pathway. Nat. Commun. 9, 564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, E. , Roberts, J.A. and Wagstaff, C. (2012) Manipulating resource allocation in plants. J. Exp. Bot. 63, 3391–3400. [DOI] [PubMed] [Google Scholar]
- Boyes, D.C. , Zayed, A.M. , Ascenzi, R. , McCaskill, A.J. , Hoffman, N.E. , Davis, K.R. and Görlach, J. (2001) Growth stage‐based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell, 13, 1499–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busov, V.B. , Brunner, A.M. and Strauss, S.H. (2008) Genes for control of plant stature and form. New Phytol. 177, 19. [DOI] [PubMed] [Google Scholar]
- Cai, G. , Yang, Q. , Chen, H. , Yang, Q. , Zhang, C. , Fan, C. and Zhou, Y. (2016) Genetic dissection of plant architecture and yield‐related traits in Brassica napus . Sci. Rep. 6, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal, J.J. (2012) Shade avoidance. Arabidopsis Book, 10, e0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challa, K.R. , Rath, M. and Nath, U. (2019) The CIN‐TCP transcription factors promote commitment to differentiation in Arabidopsis leaf pavement cells via both auxin‐dependent and independent pathways. PLoS Genet. 15, e1007988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, B.C.W. , Nath, U. , Carpenter, R. and Coen, E.S. (2004) Cincinnata controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum . Plant Physiol. 135, 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas, P. , Lauter, N. , Doebley, J. and Coen, E. (1999) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222. [DOI] [PubMed] [Google Scholar]
- Danisman, S. (2016) TCP transcription factors at the interface between environmental challenges and the plant's growth responses. Front. Plant Sci. 7, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman, S. , van der Wal, F. , Dhondt, S. et al. (2012) Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 159, 1511–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman, S. , Van Dijk, A.D.J. , Bimbo, A. , Van Der Wal, F. , Hennig, L. , De Folter, S. , Angenent, G.C. and Immink, R.G.H. (2013) Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. J. Exp. Bot. 64, 5673–5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis, J.E. , Gay, D.M. and Welsch, R.E. (1977) An adaptive nonlinear least square algorithm. ACM Trans. Math. Softw. 7, 348–368. [Google Scholar]
- Doebley, J. , Stec, A. and Gustus, C. (1995) teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics, 141, 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J. , Stec, A. and Hubbard, L. (1997) The evolution of apical dominance in maize. Nature, 386, 485–488. [DOI] [PubMed] [Google Scholar]
- Efroni, I. , Blum, E. , Goldshmidt, A. and Eshed, Y. (2008) A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell, 20, 2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmann, W. , Simon, K. and Phen, C.J. (1992) Leaf movement rhythm in A. thaliana . Z. Naturforsch., 47, 925–928. [Google Scholar]
- van Es, S.W. , Silveira, S.R. , Rocha, D.I. , Bimbo, A. , Martinelli, A.P. , Dornelas, M.C. , Angenent, G.C. and Immink, R.G. (2018) Novel functions of the Arabidopsis transcription factor TCP5 in petal development and ethylene biosynthesis. Plant J. 94, 867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flood, P.J. , Kruijer, W. , Schnabel, S.K. , van der Schoor, R. , Jalink, H. , Snel, J.F.H. , Harbinson, J. and Aarts, M.G.M. (2016) Phenomics for photosynthesis, growth and reflectance in Arabidopsis thaliana reveals circadian and long‐term fluctuations in heritability. Plant Methods, 12, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genty, B. , Briantais, J.M. and Baker, N.R. (1989) The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta Gen. Subj. 990, 87–92. [Google Scholar]
- Gnan, S. , Priest, A. and Kover, P.X. (2014) The genetic basis of natural variation in seed size and seed number and their trade‐off using Arabidopsis thaliana magic lines. Genetics, 198, 1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golyandina, N. , Korobeynikov, A. , Shlemov, A. and Usevich, K. (2013) Multivariate and 2D extensions of singular spectrum analysis with the Rssa package. J. Stat. Softw. 67, 1–78. [Google Scholar]
- Joosen, R.V.L. , Kodde, J. , Willems, L.A.J. , Ligterink, W. , Van Der Plas, L.H.W. and Hilhorst, H.W.M. (2010) Germinator: a software package for high‐throughput scoring and curve fitting of Arabidopsis seed germination. Plant J. 62, 148–159. [DOI] [PubMed] [Google Scholar]
- Kebrom, T.H. , Brutnell, T.P. and Finlayson, S.A. (2010) Suppression of Sorghum axillary bud outgrowth by shade, phyB and defoliation signalling pathways. Plant Cell Environ. 33, 48–58. [DOI] [PubMed] [Google Scholar]
- Kieffer, M. , Master, V. , Waites, R. and Davies, B. (2011) TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 68, 147‐158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama, T. , Furutani, M. , Tasaka, M. and Ohme‐Takagi, M. (2007) TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary‐specific genes in Arabidopsis. Plant Cell, 19, 473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister, D. , Varotto, C. , Pesaresi, P. , Niwergall, A. and Salamini, F. (1999) Large‐scale evaluation of plant growth in Arabidopsis thaliana by non‐invasive image analysis. Plant Physiol. Biochem. 37, 671–678. [Google Scholar]
- Li, C. , Potuschak, T. , Colón‐Carmona, A. , Gutiérrez, R.A. and Doerner, P. (2005) Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl Acad. Sci. USA, 102, 12978–12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, D. , Carpenter, R. , Vincent, C. , Copsey, L. and Coen, E. (1995) Origin of floral asymmetry in Antirrhinum. Nature, 383, 794–799. [DOI] [PubMed] [Google Scholar]
- Martín‐Trillo, M. and Cubas, P. (2010) TCP genes: a family snapshot ten years later. Trends Plant Sci. 15, 31–39. [DOI] [PubMed] [Google Scholar]
- Nath, U. , Crawford, B.C.W. , Carpenter, R. and Coen, E. (2003) Genetic control of surface curvature. Science, 299, 1404–1407. [DOI] [PubMed] [Google Scholar]
- Nicolas, M. and Cubas, P. (2015) Plant Transcription Factors In The Role of TCP Transcription Factors in Shaping Flower Structure, Leaf Morphology, and Plant Architecture. (González D., ed). Amsterdam, The Netherlands: Elsevier Inc. [Google Scholar]
- Nicolas, M. and Cubas, P. (2016) TCP factors: new kids on the signaling block. Curr. Opin. Plant Biol. 33, 33–41. [DOI] [PubMed] [Google Scholar]
- Ori, N. , Cohen, A.R. , Etzioni, A. , et al. (2007) Regulation of LANCEOLATE by miR319 is required for compound‐leaf development in tomato. Nat. Genet. 39, 787–791. [DOI] [PubMed] [Google Scholar]
- Palatnik, J.F. , Allen, E. , Wu, X.L. , Schommer, C. , Schwab, R. , Carrington, J.C. and Weigel, D. (2003) Control of leaf morphogenesis by microRNAs. Nature, 425, 257–263. [DOI] [PubMed] [Google Scholar]
- Paul‐Victor, C. and Turnbull, L.A. (2009) The effect of growth conditions on the seed size/number trade‐off. PLoS ONE, 4, e6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Álvarez, M.X. , Boer, M.P. , , van Eeuwijk, F.A. and Eilers, P.H.C. (2016) Spatial Models for Field Trials. 1–39. http://arxiv.org/abs/1607.08255
- van Rooijen, R. , Aarts, M.G.M. and Harbinson, J. (2015) Natural genetic variation for acclimation of photosynthetic light use efficiency to growth irradiance in Arabidopsis thaliana . Plant Physiol. 167, 1412–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer, C. , Palatnik, J.F. , Aggarwal, P. , Chételat, A. , Cubas, P. , Farmer, E.E. , Nath, U. and Weigel, D. (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6, 1991–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, T. , Suwa, Y. , Suzuki, M. , Kitano, H. , Ueguchi‐Tanaka, M. , Ashikari, M. , Matsuoka, M. and Ueguchi, C. (2003) The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 33, 513–520. [DOI] [PubMed] [Google Scholar]
- Tessmer, O.L. , Jiao, Y. , Cruz, J.A. , Kramer, D.M. and Chen, J. (2013) Functional approach to high‐throughput plant growth analysis. BMC Syst. Biol. 7, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uberti Manassero, N.G. , Viola, I.L. , Welchen, E. and Gonzalez, D.H. (2013) TCP transcription factors: architectures of plant form. Biomol. Concepts, 4, 111–127. [DOI] [PubMed] [Google Scholar]
- Velazco, J.G. , Rodríguez‐Álvarez, M.X. , Boer, M.P. , Jordan, D.R. , Eilers, P.H.C. , Malosetti, M. and van Eeuwijk, F.A. (2017) Modelling spatial trends in Sorghum breeding field trials using a two‐dimensional P‐spline mixed model. Theor. Appl. Genet. 130, 1375–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola, I.L. , Camoirano, A. and Gonzalez, D.H. (2015) Redox‐Dependent Modulation of Anthocyanin Biosynthesis by the TCP Transcription Factor TCP15 during Exposure to High Light Intensity Conditions in Arabidopsis. Plant Physiol. 170, 74‐85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeide, B. (1993) Analysis of growth equations. For. Sci. 39, 594–616. [Google Scholar]
- Zhou, Y. , Zhang, D. , An, J. , Yin, H. , Fang, S. , Chu, J. , Zhao, Y. and Li, J. (2017) TCP transcription factors regulate shade avoidance via directly mediating the expression of both PHYTOCHROME INTERACTING FACTORs and auxin biosynthetic genes. Plant Physiol. 176, 1850–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Regression analysis of positional effects in the experimental set‐up.
Figure S2. Designing a growth function based on rosette size measurements over time.
Figure S3. Average values for all four beta parameters.
Figure S4. Comparison of photosynthetic efficiency for the various tcp mutants.
Figure S5. Correlation analysis of the seed and branching parameters.
Table S1. Mutant lines and measurement regime used in this study.
Table S2. Raw ΦPSII and NIR data generated by the Phenovator platform.


