Abstract
Bacterial pathogens assemble adhesive surface structures termed pili or fimbriae to initiate and sustain infection of host tissues. Uropathogenic Escherichia coli, the primary causative agent of urinary tract infections, expresses type 1 and P pili required for colonization of the bladder and kidney, respectively. These pili are assembled by the conserved chaperone–usher (CU) pathway, in which a periplasmic chaperone works together with an outer membrane (OM) usher protein to build and secrete the pilus fiber. Previously, we found that the small molecule and antiparasitic drug nitazoxanide (NTZ) inhibits CU pathway–mediated pilus biogenesis in E. coli by specifically interfering with proper maturation of the usher protein in the OM. The usher is folded and inserted into the OM by the β-barrel assembly machine (BAM) complex, which in E. coli comprises five proteins, BamA–E. Here, we show that sensitivity of the usher to NTZ is modulated by BAM expression levels and requires the BamB and BamE lipoproteins. Furthermore, a genetic screen for NTZ-resistant bacterial mutants isolated a mutation in the essential BamD lipoprotein. These findings suggest that NTZ selectively interferes with an usher-specific arm of the BAM complex, revealing new details of the usher folding pathway and BAM complex function. Evaluation of a set of NTZ derivatives identified compounds with increased potency and disclosed that NTZ's nitrothiazole ring is critical for usher inhibition. In summary, our findings indicate highly specific effects of NTZ on the usher folding pathway and have uncovered NTZ analogs that specifically decrease usher levels in the OM.
Keywords: outer membrane, bacteria, protein folding, small molecule, protein secretion, Bam complex, fimbriae, nitazoxanide, pili, usher protein, bacterial outer membrane, virulence factor
Introduction
Attachment to host tissues is a critical initial step in bacterial pathogenesis, permitting colonization of specific niches within the host. Pili, also known as fimbriae, are organelles expressed by bacteria that perform a variety of adhesive and virulence-associated functions, including binding to host receptors (1, 2). In Gram-negative bacteria, a large family of pili are assembled by the conserved chaperone–usher (CU)5 pathway (3–5). CU pili are hairlike surface structures that mediate bacterial adhesion and invasion into host cells as well as biofilm formation. Pilus-mediated adhesion is particularly important for infection of sites such as the urinary tract, allowing bacteria to resist washout from fluid flow (6). The tissue tropic type 1 (Fim) and P (Pap) pili expressed by uropathogenic Escherichia coli (UPEC) are prototypical examples of pili assembled by the CU pathway. These pili function in the establishment of urinary tract infections by UPEC, with type 1 pili binding to mannosylated proteins in the bladder, leading to cystitis, and P pili binding to kidney glycolipids, leading to pyelonephritis (7).
In the CU pathway, newly synthesized pilus subunits are first translocated from the cytoplasm to the periplasm via the Sec general secretory pathway (Fig. S1) (8). In the periplasm, the subunits form binary complexes with pilus-specific chaperone proteins (FimC or PapD), which facilitate subunit folding and inhibit premature subunit–subunit interactions (9, 10). These chaperone–subunit complexes then interact with the outer membrane (OM) usher (FimD or PapC), which comprises a gated transmembrane β-barrel channel with periplasmic N- and C-terminal domains (Fig. S1) (11–13). The usher catalyzes the exchange of chaperone–subunit for subunit–subunit interactions, promotes ordered polymerization of the pilus fiber, and provides the channel for secretion of the pilus fiber to the cell surface (4, 5). The usher is required for pilus biogenesis; in the absence of the usher, chaperone–subunit complexes accumulate in the periplasm, but pilus assembly and secretion does not occur. The usher is also the rate-limiting factor for pilus biogenesis, with the number of usher molecules determining the number of pili assembled on the bacterial surface (14, 15).
Pili are critical for initiating and sustaining infection and thus represent attractive targets for the development of anti-virulence therapeutics. Anti-virulence therapeutics offer an alternative to traditional antibiotics and a promising approach to combat the ever-increasing rate of antibiotic resistance among pathogenic bacteria (16, 17). By specifically targeting systems required to cause disease within the host, such therapeutics should limit detrimental side effects on the beneficial commensal bacteria and decrease the selective pressures that lead to antibiotic resistance (18, 19). A number of approaches are under investigation for developing anti-virulence therapeutics that target pili, including vaccines against pilus subunits, competitive inhibitors of pilus-mediated adhesion, and small molecules that disrupt pilus biogenesis, termed pilicides (20, 21).
In previous studies, the small molecule nitazoxanide (NTZ) was found to inhibit assembly of aggregative adherence fimbriae (AAF) by the CU pathway on the surface of enteroaggregative E. coli (22). NTZ is a synthetic nitrothiazolyl-salicylamide compound (see Fig. 7) used clinically to treat parasitic diseases such as giardiasis and cryptosporidiosis (23, 24). We subsequently showed that, in addition to AAF, NTZ also inhibits biogenesis of the E. coli type 1 and P pili (25). This suggests that NTZ has broad inhibitory activity against CU pili. Treatment of E. coli with NTZ was found to interfere with proper folding of the usher protein in the bacterial OM, specifically its transmembrane β-barrel domain, leading to degradation of the usher by the periplasmic protease DegP (25). Given that the usher is essential for assembly and secretion of the pilus fiber, loss of functional usher proteins explains the loss of pili upon NTZ treatment. Notably, NTZ appears to selectively interfere with folding of the usher, because levels of other outer membrane proteins (OMPs) remain unaffected by drug treatment (25). This mechanism of action is unique compared with other existing pilicides, which target different aspects of the CU pathway, such as chaperone–subunit interactions or chaperone–subunit binding to the usher (21, 26–28).
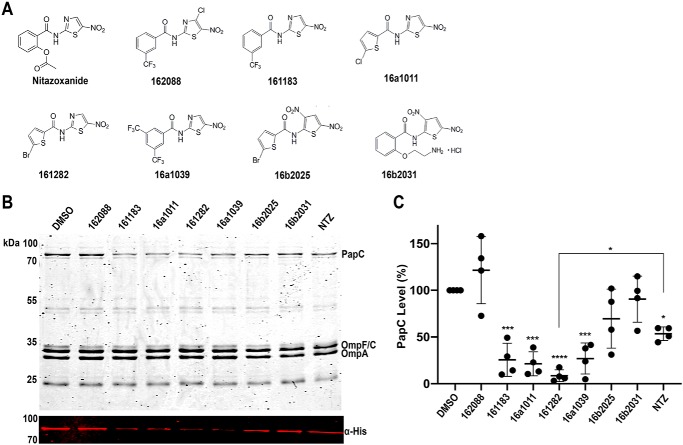
Figure 7.
Effect of NTZ analogs on OM usher assembly. Structures of NTZ-derived analogs (A). Strain BW25113/pMJ3 (PapCHis) was grown in the presence of 32 μm NTZ or the indicated NTZ derivatives (B). OM fractions isolated from normalized bacterial cultures were subjected to SDS-PAGE and Coomassie Blue staining to observe PapC and the overall OMP profile (top). Samples were also probed with anti-His tag antibody to visualize PapC (bottom). PapC levels were measured by densitometry of the anti-His tag blot, and percentage PapC levels were calculated relative to 0 μg/ml NTZ (C). The graph shows individual data points and means ± S.D. (error bars) from four independent experiments. *, p < 0.05; ***, p < 0.001; ****, p < 0.0001 for comparison of each compound with DMSO vehicle control.
Folding of bacterial OMPs occurs via the conserved β-barrel assembly machine (BAM) complex (29–31). In E. coli, the BAM complex comprises the integral OM β-barrel protein BamA and four accessory lipoproteins, BamB–E (Fig. S1). Following translocation into the periplasm by the Sec translocon, nascent unfolded β-barrel OMPs bind chaperones such as SurA and Skp, which transport these substrates to the BAM machinery (29). The BAM complex recruits the nascent OMP–chaperone complexes and catalyzes efficient OMP insertion and assembly in the OM (32). Should a β-barrel substrate fail to be efficiently inserted into the OM, it can be targeted for degradation by DegP (33, 34). Many questions remain regarding the mechanistic contributions of the individual BAM components (BamA–E) to OMP folding. BamA and BamD constitute the core components of the complex and are both essential for bacterial viability (35, 36). BamD is implicated in recruitment of OMP substrates, whereas BamA plays a central role in OMP folding and insertion into the OM. The nonessential components BamB, BamC, and BamE perform accessory roles in enhancing the kinetics of OMP biogenesis (37–40). OMPs vary in size and complexity and are differentially affected by deletion of individual accessory BAM components (30, 41, 42). This suggests that distinct, specialized arms of the BAM complex exist for the folding and membrane insertion of different OMP substrates. Analysis of the type 1 pilus usher FimD showed a strong dependence on SurA and BamB for folding in the OM, whereas deletion of BamC or BamE only minimally affected usher folding (43).
We hypothesized that NTZ inhibits pilus biogenesis by interfering with a specific aspect of BAM complex function required for folding of the usher, but not other OMPs. Consistent with this hypothesis, we show here that sensitivity of the PapC usher to NTZ is modulated by expression levels of the BAM complex and requires the BamB and BamE lipoproteins. Furthermore, using an unbiased genetic screen, we identify a point mutation in the essential BamD lipoprotein that confers reduced sensitivity to NTZ. These results define new aspects of the usher folding pathway and new functionality for the BamD and BamE lipoproteins, supporting a model in which NTZ targets a specialized arm of the BAM complex required for usher folding. We also report the identification of NTZ analogs with increased potency in specifically inhibiting the usher and determine that the nitrothiazole ring of NTZ is critical for the drug's inhibitory activity against the usher.
Results
Sensitivity to NTZ correlates with BAM expression levels
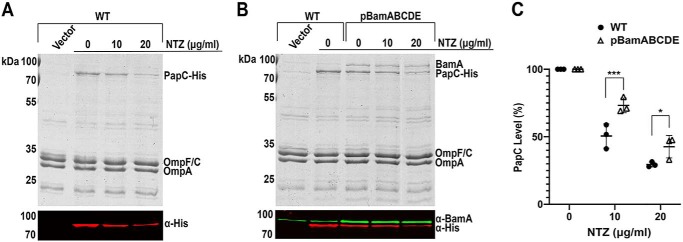
We hypothesized that NTZ interferes with folding of the usher in the OM by altering activity of the BAM complex. If the BAM machinery is the direct target of NTZ, we should observe altered NTZ activity in bacterial strains with increased or decreased levels of the BAM complex; increased BAM expression should confer resistance to NTZ (more folded usher), whereas decreased BAM expression should potentiate NTZ activity (less folded usher). To test this, we quantitated levels of the P pilus usher PapC in OM fractions isolated from E. coli strains that over- or underexpressed the BAM complex, compared with the WT parental strain. For overexpression of the BAM complex, E. coli strain BW25113 expressing His-tagged PapC (pMJ3) was compared with the same strain transformed with a plasmid containing each of the bam genes under control of an inducible promoter (pBamABCDE). OM fractions were isolated from the strains grown in the presence of 0, 10, or 20 μg/ml NTZ; the fractions were blotted with anti-His tag antibody; and PapC levels were quantitated by densitometry. In the WT strain background, treatment with 10 μg/ml NTZ caused a 50% decrease in usher levels, and treatment with 20 μg/ml NTZ caused a 70% decrease in usher levels, compared with vehicle-treated control (0 μg/ml NTZ) (Fig. 1, A and C). In contrast, overexpression of the BAM complex from pBamABCDE resulted in reduced sensitivity to NTZ, with usher levels decreasing by only 27% at 10 μg/ml NTZ and 57% at 20 μg/ml NTZ (Fig. 1, B and C). Examination of the OM fractions with an anti-BamA antibody confirmed overexpression of the BAM complex in the strain containing pBamABCDE, with BamA levels four times higher compared with the parental WT strain (Fig. 1B). The changes in BamA, as well as PapC, expression levels were also apparent in Coomassie Blue–stained gels of the OM fractions (Fig. 1, A and B). Moreover, inspection of the Coomassie-stained gels confirmed our previous findings (25) that treatment of bacteria with NTZ selectively affects levels of the usher, with other OMPs (such as the major porins OmpA, OmpF, and OmpC) remaining unchanged. We also did not observe discernable changes in the overall OMP profile of bacteria overexpressing the BAM complex compared with the parental WT strain.
Figure 1.
Effect of Bam overexpression on sensitivity to NTZ. Strains BW25113/pMJ3 (PapCHis) + pTRYC (Vector) (A) and BW25113/pMJ3 + pBamABCDE (B) were grown in the presence of the indicated concentrations of NTZ. E. coli containing vector pMON6235Δcat served as a negative control for PapC expression. OM fractions isolated from normalized bacterial cultures were subjected to SDS-PAGE and Coomassie Blue staining to observe PapC and the overall OMP profile (top panels). Samples were also probed with anti-His tag and anti-BamA antibodies to visualize PapC and BamA, respectively (bottom panels). PapC levels were measured by densitometry of the anti-His tag blot, and percentage PapC levels were calculated relative to 0 μg/ml NTZ (C). The graph shows individual data points and means ± S.D. (error bars) from three independent experiments. *, p < 0.05; ***, p < 0.001 for comparison of OM PapC levels at each NTZ concentration.
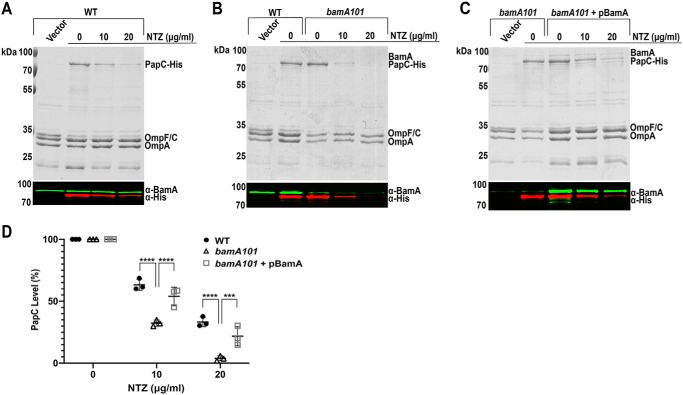
We next examined the effect of BAM complex underexpression on sensitivity to NTZ. Because BAM complex activity is essential for bacterial viability, we used the MC4100bamA101 mutant strain (44) for this purpose. This strain contains a transposon insertion in the upstream promoter region of bamA, resulting in an ∼10-fold decrease in BamA expression (45). BamA is the central component of the BAM complex (36); therefore, reducing BamA levels results in a corresponding decrease in overall levels of functional BAM complex. Parental strain MC4100 and the MC4100bamA101 mutant strain, both transformed with pMJ3 (expressing His-tagged PapC), were grown in the presence of 0, 10, or 20 μg/ml NTZ, and OM fractions were isolated. Quantitation of anti-His tag immunoblots showed that strain MC4100/pMJ3 exhibited a similar dose-dependent decrease in OM usher levels in response to NTZ as observed for BW25113/pMJ3 (Fig. 2, A and D). In comparison, the bamA101 strain exhibited increased sensitivity to NTZ, with usher levels decreasing by 68% at 10 μg/ml NTZ and 96% at 20 μg/ml NTZ (Fig. 2, B and D). Complementation of the bamA101 mutant with a BamA expression plasmid (pBamA) restored WT sensitivity of the user to NTZ (Fig. 2, C and D), confirming the specificity of the mutant phenotype. Examination of the OM fractions with an anti-BamA antibody confirmed decreased expression of BamA in the bamA101 mutant compared with the parental WT strain, as well as restoration of BamA expression in the complemented strain (Fig. 2, B and C). The changes in PapC and BamA levels in the different strain backgrounds and NTZ treatment conditions were also apparent in Coomassie Blue–stained gels of the OM fractions. Taken together, these experiments show an inverse correlation between BAM expression levels and the activity of NTZ; BAM overexpression resulted in increased resistance of the usher to NTZ, and BAM underexpression resulted in increased sensitivity of the usher to NTZ. These findings are consistent with the BAM complex as a relevant target for the mechanism of action of NTZ.
Figure 2.
Effect of Bam underexpression on sensitivity to NTZ. Strains MC4100/pMJ3 (PapCHis) (A), MC4100bamA101/pMJ3 (B), and MC4100bamA101/pMJ3 + pBamA (C) were grown in the presence of the indicated concentrations of NTZ. E. coli containing vector pMON6235Δcat served as a negative control for PapC expression. OM fractions isolated from normalized bacterial cultures were subjected to SDS-PAGE and Coomassie Blue staining to observe PapC and the overall OMP profile (top panels). Samples were also probed with anti-His tag and anti-BamA antibodies to visualize PapC and BamA, respectively (bottom panels). PapC levels were measured by densitometry of the anti-His tag blot, and percentage PapC levels were calculated relative to 0 μg/ml NTZ (D). The graph shows individual data points and means ± S.D. (error bars) from three independent experiments. ***, p < 0.001; ****, p < 0.0001 for comparison of OM PapC levels at each NTZ concentration.
The effect of NTZ on usher levels depends on BamB and BamE
If the BAM complex is the target of NTZ, NTZ binding must alter assembly or function of the complex in a manner that specifically affects folding of the usher protein, but not other OMPs. The BAM proteins form a stable complex in the OM that can be isolated from bacteria by pulldown assays using any individual component (46, 47). To determine whether NTZ affects assembly of the Bam complex, E. coli strain BW25113 expressing the BAM complex with a His-tagged BamE (pBamABCDE-His6) was cultured in the presence of 0 or 20 μg/ml NTZ, OM fractions were isolated, and the detergent-extracted complex was captured by nickel affinity chromatography. Proteins eluted from the column were then analyzed by staining with Coomassie Blue or immunoblotting with anti-His tag and anti-BamA antibodies. We did not observe any changes in BAM protein stoichiometry or loss of co-purifying proteins in response to NTZ treatment (Fig. S2), indicating that NTZ does not interfere with the formation or stability of the BAM complex.
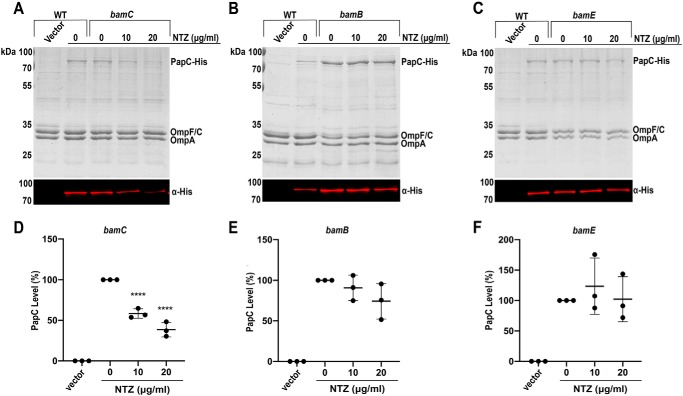
The folding of OMPs is differentially impacted by the loss of individual nonessential BAM lipoproteins (BamB, BamC, and BamE) (30, 41, 42), suggesting that different OMPs require distinct subcomplexes of the BAM machinery. To determine whether NTZ alters the function of the BAM complex in a manner specific to the usher, we first investigated contributions of the nonessential BAM components to folding of PapC and sensitivity to NTZ. The E. coli BW25113 WT strain and ΔbamB, ΔbamC, and ΔbamE mutant derivatives, each expressing His-tagged PapC (plasmid pMJ3), were grown in the presence of 0, 10, or 20 μg/ml NTZ; OM fractions were isolated; and usher levels were analyzed by Coomassie staining and immunoblotting with anti-His tag antibody. For the ΔbamC strain, growth in the presence of NTZ resulted in a similar dose-dependent decrease in PapC levels as observed for the WT strain (Fig. 3, A and D), suggesting that BamC does not play a role in the sensitivity of the usher to NTZ. In addition, in the absence of NTZ, the ΔbamC strain expressed similar levels of PapC compared with the WT strain, and the overall OMP profile was also unchanged (Fig. 3A). This is consistent with previous studies in which deletion of BamC caused only minor defects in general OMP folding (41, 48). In contrast to the ΔbamC mutant, the ΔbamB and ΔbamE strains appeared resistant to NTZ, with growth in the presence of the drug no longer causing a decrease in PapC levels (Fig. 3). This suggests that BamB and BamE are required for NTZ activity against the usher. Interestingly, PapC levels increased in the ΔbamB strain compared with the WT strain, whereas deletion of bamE did not alter PapC levels (Fig. 3, B and C). The behavior of the usher was in contrast to the overall OMP levels, which were reduced in the ΔbamB and ΔbamE strains (Fig. 3, B and C).
Figure 3.
Effect of Bam deletion mutants on sensitivity to NTZ. Strains BW25113ΔbamC/pMJ3 (PapCHis) (A), BW25113ΔbamB/pMJ3 (B), and BW25113ΔbamE/pMJ3 (C) were grown in the presence of the indicated concentrations of NTZ. WT bacteria containing vector only (pMON6235Δcat) or PapC (pMJ3) served as negative and positive controls for PapC expression, respectively. OM fractions isolated from normalized bacterial cultures were subjected to SDS-PAGE and Coomassie Blue staining to observe PapC and the overall OMP profile (top panels). Samples were also probed with anti-His tag antibody to visualize PapC (bottom panel). PapC levels were measured by densitometry of the anti-His tag blot, and percentage PapC levels were calculated relative to 0 μg/ml NTZ for each strain (D–F). Graphs show individual data points and means ± S.D. (error bars) from three independent experiments. ****, p < 0.0001 for comparison with 0 μg/ml NTZ.
BamB was previously shown to be required for folding of the FimD usher in the OM (43), whereas a role for BamE in the usher folding pathway has not been described. To probe the basis for the NTZ phenotypes of the ΔbamB and ΔbamE mutants, we investigated effects of the mutations on folding of the usher in the OM and function of the usher in pilus assembly. To discern effects on usher folding, we exploited the intrinsic resistance of β-barrel proteins such as the usher to denaturation by SDS, which results in heat-modifiable mobility on SDS-PAGE, where a fraction of the protein remains folded and migrates more rapidly than its predicted molecular mass in the absence of heat (49, 50). To determine the function of the usher in pilus biogenesis, we measured assembly of adhesive P pili (from plasmid pFJ29, encoding an inducible pap operon) on the bacterial surface using a hemagglutination (HA) assay (51). The HA titer was measured as the maximum -fold dilution of bacteria able to agglutinate human red blood cells.
For the ΔbamB mutant, analysis of OM samples incubated in SDS sample buffer at 25 °C revealed that mutant was unable to produce any folded PapC usher (Fig. 4A). Complementation of the ΔbamB mutant with a BamB expression plasmid (pBamB) restored usher folding back to WT levels (Fig. 4A). Consistent with a lack of folded usher, the ΔbamB strain was unable to assemble adhesive P pili, with an HA titer of 0 (Fig. 4C). The lack of folded usher in the ΔbamB mutant provides an explanation for its apparent resistance to NTZ, as NTZ specifically interferes with usher folding. Thus, the absence of an NTZ-dependent decrease in usher levels in the ΔbamB mutant represents an accumulation of unfolded usher that is not degraded, rather than a decreased sensitivity to NTZ. In contrast to the ΔbamB mutant, we found that the ΔbamE mutant was still able to produce folded usher, although the amount of SDS-stable, faster-migrating species was lower compared with the WT strain (Fig. 4B). Complementation of the ΔbamE mutant with a BamE expression plasmid (pBamE) restored usher folding back to WT levels (Fig. 4B). Intriguingly, the ΔbamE mutant was fully functional for assembly of adhesive pili, with an HA titer 4 times greater than for the WT strain (Fig. 4C). The increased HA titer is not explained by a change in usher levels, which were comparable between the ΔbamE and WT strains (Fig. 3C). Together, these results implicate BamE in the mechanism of action of NTZ and reveal a previously unknown role for BamE in the usher folding pathway.
Figure 4.
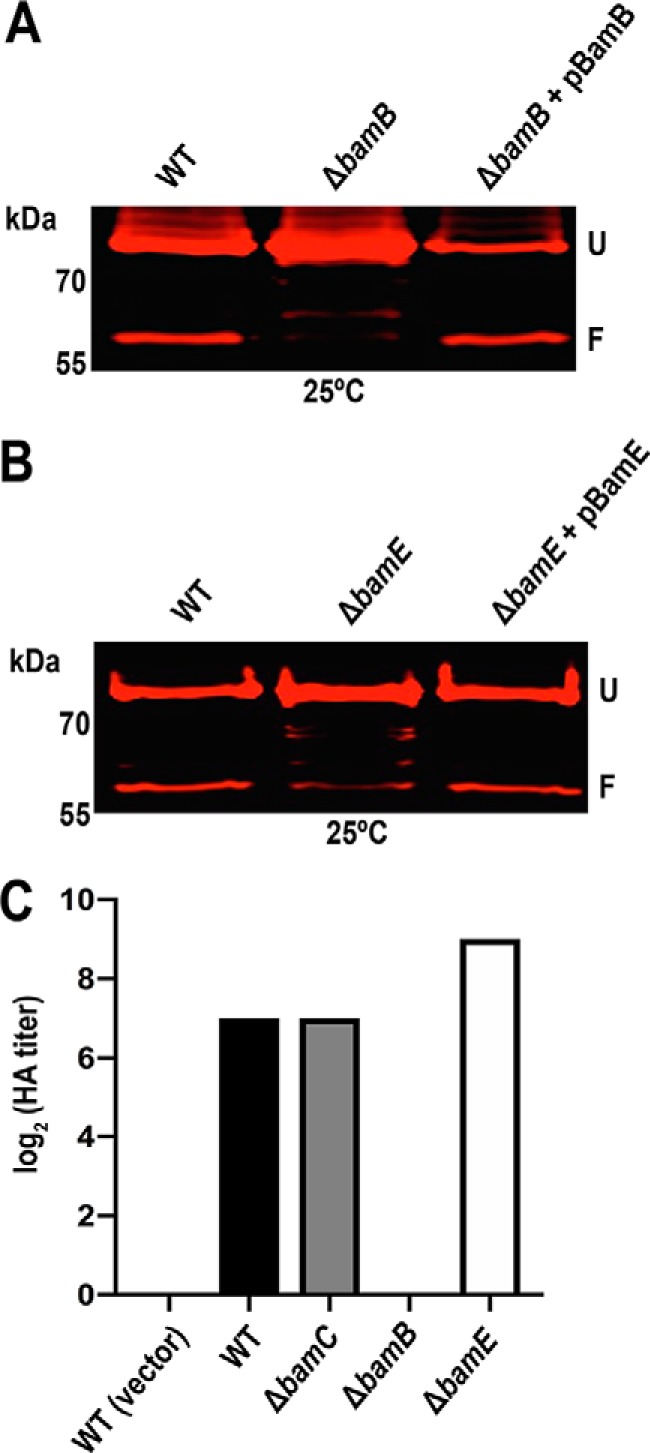
Contributions of BamB and BamE to usher folding and function. OM fractions were isolated from normalized bacterial cultures of WT BW25113, the ΔbamB and ΔbamE deletion mutants, and complemented strains, each expressing His-tagged PapC (pMJ3) (A and B). The OM fractions were incubated in SDS sample buffer at 25 °C, subjected to SDS-PAGE, and probed with anti-His tag antibody to visualize PapC. Positions of the folded (F) and unfolded (U) PapC species are indicated on the right of each gel image. WT BW25113 and the ΔbamB, ΔbamC, and ΔbamE deletion mutants, each expressing P pili (pJF29), were tested for HA activity (C). Serial dilutions of normalized whole bacteria were incubated with human red blood cells, and the HA titer was recorded as the maximum -fold dilution of bacteria able to agglutinate the blood cells. Bars, the log2 transformed titers, which were calculated from three independent experiments of three replicates each; all values for each of the experiments and replicates were identical.
A BamDP100S mutation reduces sensitivity to NTZ
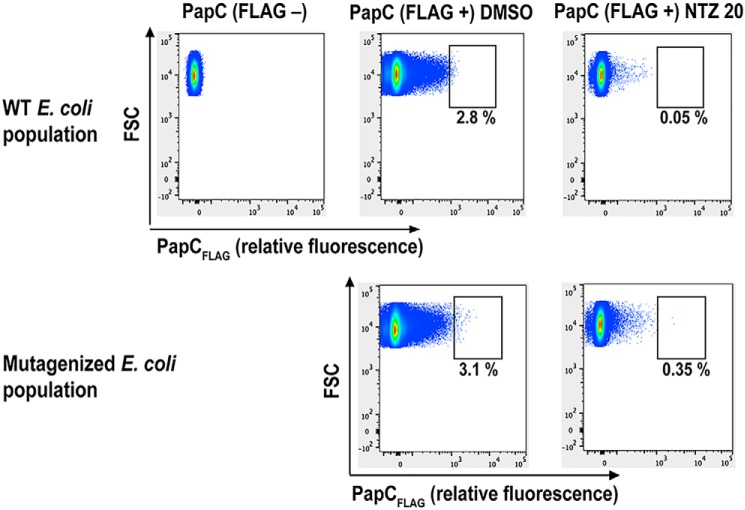
As an unbiased approach to identify the target and mechanism of action of NTZ, we performed a genetic screen to isolate mutants resistant to NTZ (i.e. strains that retain high OM usher levels when grown in the presence of drug). We used nitrosoguanidine mutagenesis (52) to generate a library in E. coli BW25113 expressing a PapC construct (plasmid pJP1) with a 3X-FLAG tag insertion in a surface-exposed loop (Fig. S3A). The FLAG tag allowed detection of the usher in intact bacteria using a phycoerythrin (PE)-conjugated anti-FLAG antibody, as confirmed by fluorescence microscopy (Fig. S3B). Using flow cytometry of intact bacteria, we confirmed that growth in the presence of 20 μg/ml NTZ caused a large decrease in fluorescence of the bacteria, indicating loss of PapCFLAG in the OM (Fig. 5). We then set up a gate that contained almost no highly fluorescent bacteria in the WT population treated with 20 μg/ml NTZ. Analysis of the nitrosoguanidine mutagenized bacteria showed a 7-fold increase in the percentage of bacteria within this gate after NTZ treatment, indicating the emergence of resistant clones (Fig. 5). Using this gate for fluorescence-activated cell sorting (FACS), we isolated ∼200 mutagenized E. coli cells that remained highly fluorescent following NTZ treatment. Viable clones (∼50) were then individually assessed for their sensitivity to NTZ by growth in the presence or absence of 20 μg/ml NTZ. OM fractions were isolated and analyzed by Coomassie Blue staining and immunoblotting using an anti-FLAG antibody to detect PapC levels. Through this analysis, five mutant strains were verified to maintain higher usher levels in the presence of NTZ compared with the WT strain. These five mutant strains, as well as our WT strain, were then subjected to whole-genome sequencing to identify the mutation(s) that conferred resistance to NTZ. Each of the resistant strains had 100–200 nonsynonymous mutations (Data File S1).
Figure 5.
Screen for mutants resistant to NTZ. WT BW25113/pJP1 (PapCFLAG) and a nitrosoguanidine-mutagenized library of BW25113/pJP1 were grown in the presence of vehicle control (DMSO) or 20 μg/ml NTZ. Normalized bacteria were labeled with a PE-conjugated anti-FLAG antibody to detect PapCFLAG levels in the OM, and the bacteria were analyzed by flow cytometry. Representative flow cytometry plots are shown. The gate used for selection of highly fluorescent bacteria (high levels of PapCFLAG) following NTZ treatment is indicated, along with the percentage of total bacteria falling within the gate.
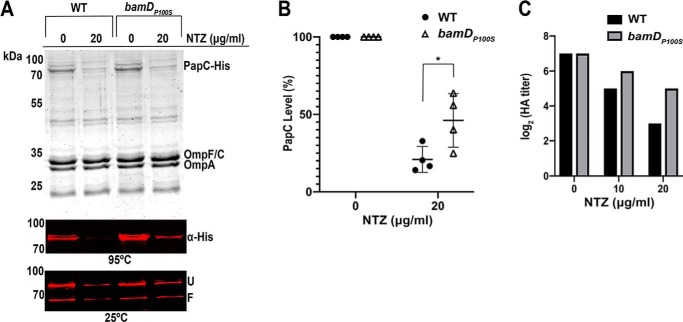
A mutation of particular interest isolated from our screen was a proline-to-serine mutation in residue 100 of the BamD lipoprotein. To confirm that this mutation confers resistance to NTZ, we constructed a bamDP100S mutation in the chromosome of WT E. coli BW25113. The WT and bamDP100S strains, each expressing His-tagged PapC (pMJ3), were grown in the presence of 0 or 20 μg/ml NTZ, OM fractions were isolated, and relative usher levels were analyzed by immunoblotting with anti-His tag antibody and quantitation by densitometry. Growth of the bamDP100S mutant in the presence of 20 μg/ml NTZ caused a 54% decrease in OM usher levels, compared with a 79% decrease for WT BW25113 (Fig. 6, A and B). Thus, the P100S mutation in BamD, by itself, reduces sensitivity to NTZ. In the absence of NTZ, expression levels of the PapC usher in the OM were comparable between WT BW25113 and the bamDP100S mutant (Fig. 6A). Furthermore, inspection of Coomassie Blue–stained gels showed that the general OMP profile was not altered in the bamDP100S mutant compared with the WT strain in the presence or absence of NTZ (Fig. 6A). This indicates that the BamDP100S mutation specifically affects sensitivity of the usher to NTZ. Analysis of heat-modifiable mobility showed that, in the absence of NTZ, the bamDP100S mutant produced folded PapC at similar levels as the WT strain (Fig. 6A). The usher also remained fully functional in the bamDP100S strain, with an HA titer similar to WT in the absence of NTZ (Fig. 6C). However, when grown in the presence of increasing concentrations of NTZ, the HA titer decreased by only 4-fold for the bamDP100S mutant compared with 16-fold for the WT strain (Fig. 6C). Together, these results demonstrate that the BamDP100S mutation enables bacteria to assemble more functional usher in the presence of NTZ.
Figure 6.
Effect of bamDP100S mutation on sensitivity to NTZ and usher folding and function. Strains BW25113/pMJ3 (PapCHis) and BW25113bamDP100S/pMJ3 were grown in the presence of the indicated concentrations of NTZ. OM fractions isolated from normalized bacterial cultures were subjected to SDS-PAGE and Coomassie Blue staining to observe PapC and the overall OMP profile (A, top panel). Samples were also incubated in SDS sample buffer at 25 or 95 °C, subjected to SDS-PAGE, and probed with anti-His tag antibody to visualize PapC (A, middle and bottom panels). Positions of the folded (F) and unfolded (U) PapC species are indicated on the right for samples incubated at 25 °C. PapC levels were measured by densitometry of the anti-His tag blot (95 °C) and percentage PapC levels were calculated relative to 0 μg/ml NTZ (B). The graph shows individual data points and means ± S.D. (error bars) from four independent experiments. *, p < 0.05 for comparison of OM PapC levels at 20 μg/ml NTZ. WT and bamDP100S BW25113, each expressing P pili (pJF29), were tested for HA activity (C). Serial dilutions of normalized whole bacteria were incubated with human red blood cells, and the HA titer was recorded as the maximum -fold dilution of bacteria able to agglutinate the blood cells. Bars, log2 transformed titers, which were calculated from three independent experiments of three replicates each; all values for each of the experiments and replicates were identical.
The inhibitory effect of NTZ on usher folding depends on its nitrothiazolyl ring
NTZ is a broadly active compound with a wide range of reported activities (53–55). At the concentrations used for our experiments, NTZ does not exhibit toxicity toward E. coli. Instead, NTZ specifically affects pilus biogenesis by the CU pathway by interfering with folding of the usher protein. To generate information on the structure–activity relationship of NTZ and its specificity in disrupting usher folding, we screened analogs derived from the NTZ scaffold (Fig. 7A) for effects on levels of the PapC usher in the OM. E. coli strain BW25113 expressing His-tagged PapC (pMJ3) was grown in the presence of vehicle only (DMSO), 32 μm (10 μg/ml) NTZ, or 32 μm NTZ derivative. OM fractions isolated from bacteria were immunoblotted with anti-His tag antibody, and PapC levels were quantified by densitometry (Fig. 7, B and C). Treatment of bacteria with 32 μm NTZ caused a 47% decrease in OM usher levels compared with control. Nitrothiazole analogs of NTZ (161183, 16a1011, 161282, and 16a1039) exhibited greater potency compared with NTZ, whereas dinitrothiophene analogs (16b2025 and 16b2031) had no appreciable effect on OM usher levels (Fig. 7, B and C). Substitution of a chloro group at position 4 on the nitrothiazole ring (162088) abolished the drug's effect as well. This identifies the nitrothiazole ring of NTZ as critical for the inhibitory effect of the drug on the usher and, by extension, CU pilus assembly. The most potent NTZ derivative was 161282, a thiophene analog of 2-amino-5-nitrothiazole (Fig. 7). Treatment of bacteria with 32 μm compound 161282 decreased OM usher levels by 91% compared with vehicle-treated control (Fig. 7B). Analysis of the OM fractions by Coomassie staining revealed no effect of NTZ or the NTZ analogs on levels of other OMPs. This indicates that the increased potency of the nitrothiazole analogs is due to increased efficacy against the usher, rather than a general targeting of OMP biogenesis or other nonspecific effect, consistent with the mechanism of action of NTZ.
Discussion
Pili are critical virulence factors for a wide range of pathogenic bacteria. Their roles in initiating infection, promoting colonization, and dictating tropism for different sites within the host make pili attractive targets for therapeutic intervention (2–4). We have determined that the small molecule NTZ acts as a pilicide to disrupt pilus assembly by the conserved CU pathway (25). In previous work using the type 1 and P pilus systems expressed by UPEC, we found that treatment of bacteria with NTZ causes misfolding and subsequent degradation of the usher protein (25). The usher is an essential and rate-limiting factor for pilus biogenesis, explaining the loss of pili upon NTZ treatment. Here, we investigated the mechanism by which NTZ affects usher folding. We present evidence that NTZ acts on an usher-specific aspect of the BAM complex and assign new roles for BamD and BamE in the usher folding pathway.
Prior analysis of the FimD usher found that proper folding and insertion into the OM is mediated by the BAM complex and requires the SurA periplasmic chaperone and BamB lipoprotein (34, 43, 56). The translocation and assembly module (TAM) proteins TamA and TamB have also been shown to work together with the BAM complex to increase the efficiency of usher folding in the OM (57). The BAM complex mediates the folding of a broad range of OMPs in addition to the usher; however, NTZ selectively disrupts usher folding, leaving other OMPs unaffected (25). A goal of the current study was to define the usher-specific folding pathway that is targeted by NTZ. We found that the sensitivity of ΔtamA or ΔtamB deletion mutant strains to NTZ was similar to the WT strain, suggesting that NTZ is not targeting the function of the TAM complex.6 SurA assists in the delivery of many OMPs to the OM, including the major porins (58). Treatment of bacteria with NTZ does not alter porin levels or the general OMP profile, and therefore NTZ is unlikely to target SurA activity. In contrast, evidence has accumulated for the existence of distinct arms of the BAM complex for folding particular substrates (30, 41, 42), leading us to hypothesize the BAM system as an NTZ target.
Consistent with the BAM system as a target of NTZ, we show here that overexpression of the BAM complex leads to increased resistance of the PapC usher to NTZ. BAM overexpression by itself did not alter PapC levels in the OM; nor did it alter the overall OMP profile. Importantly, this argues that the observed resistance of PapC to NTZ upon Bam overexpression was not simply due to increased production of the usher overcoming the effects of the drug. Instead, this result supports a model in which NTZ disrupts an usher-specific aspect of BAM function, with BAM overexpression providing a larger pool of functional complexes to compensate for complexes rendered incompetent for usher assembly by NTZ. Also consistent with the BAM system as a target of NTZ, we observed increased sensitivity of the usher to NTZ in the bamA101 mutant strain, which has reduced BamA expression and therefore reduced levels of functional BAM complexes (45). Thus, in this case, fewer BAM complexes competent for usher folding should remain following NTZ treatment. The bamA101 mutation did not by itself alter steady-state PapC levels in the OM, although the levels of other OMPs, such as the porins, decreased compared with the WT strain. Therefore, the increased sensitivity of PapC to NTZ in the bamA101 strain cannot be explained by a lower starting level of usher molecules. The different response of PapC compared with other OMPs such as the porins to the bamA101 mutation also supports the existence of a distinct folding pathway on the BAM complex for the usher.
We took advantage of the sensitivity of the usher to NTZ to use the drug as a tool to probe the usher-specific folding pathway on the BAM complex. In agreement with prior analysis of the FimD usher (43), we found that production of properly folded and functional PapC in the OM required BamB but not BamC. An interesting difference between our results and the prior FimD study is that we observed an accumulation of unfolded PapC in the OM in the ΔbamB mutant, whereas FimD levels decreased (43). Our results suggest that, in the absence of BamB, the unfolded PapC is trapped at an intermediate step along its folding pathway, presumably on the BAM complex, where it is inaccessible to degradation by the DegP protease. Alternatively, the absence of BamB prevents handoff of PapC to the BAM complex, and PapC is protected by the chaperone SurA. PapC levels no longer decreased in response to NTZ in the ΔbamB strain. This apparent resistance to NTZ is attributable to the accumulation of unfolded usher in the ΔbamB mutant. Thus, NTZ would have no apparent effect in the ΔbamB strain, as the mutant BAM complex is incompetent for usher folding, and the unfolded usher is trapped at a stage where it is not subject to degradation. Further, this suggests that NTZ targets a step in the usher folding pathway subsequent to BamB function. In contrast to the ΔbamB mutant, PapC remained sensitive to NTZ in the ΔbamC strain, consistent with the lack of a role for BamC in the usher folding pathway.
In addition to BamB, our analysis revealed a previously unrecognized role for BamE in the usher folding pathway and a requirement for BamE in the mechanism of action of NTZ. The ΔbamE mutant was resistant to NTZ, with PapC levels no longer decreasing in response to the drug. Total levels of PapC in the OM were similar between the ΔbamE mutant and WT strain, and thus this resistance was not due to increased usher expression. In contrast to the ΔbamB mutant, PapC remained fully functional for pilus assembly in the ΔbamE strain, with an HA titer even greater than for the WT strain. Given that the usher must be folded to assemble pili, this shows that the NTZ resistance of the ΔbamE strain was not due to an accumulation of unfolded usher. However, loss of BamE did alter usher stability, with less folded species appearing in the heat-modifiable mobility assay. The basis for the NTZ resistance and increased HA titer of the ΔbamE mutant remains to be determined, but our results suggest a role for BamE, either directly or indirectly, in achieving optimal final stability of the usher. To our knowledge, such a phenotype has not been described previously. The function of BamE is poorly understood, and loss of BamE causes only minor defects in general OMP assembly (30, 47). BamE has been shown to modulate the conformation of BamA through its interactions with BamD (46, 59–61). If NTZ targets the BAM complex directly, an altered conformation of the BAM complex in the ΔbamE mutant might affect the drug-binding site. It was recently shown that BamB and BamE have specialized, nonoverlapping roles in coordinating BamA–BamD interactions during the assembly of OMPs in complex with the stress-sensing lipoprotein RcsF (39, 40, 62). Our results suggest that BamB and BamE also play distinct roles in the usher folding pathway. Whereas deletion of BamB completely abolished usher folding, deletion of BamE resulted in a less stable yet functional state of the usher. Therefore, the folding pathway of the usher in the ΔbamE mutant might be changed in such a way that it is no longer susceptible to interference by NTZ.
A further piece of evidence for the BAM complex as a target of NTZ came from our isolation of a BamDP100S mutation in a screen for mutations conferring decreased sensitivity of the usher to the drug. The nitrosoguanidine mutagenesis protocol used for our screen generated multiple nonsynonymous mutations in each of the resistant strains (Data File S1). We focused here on the role of the BAM complex in the mechanism of NTZ and constructed the bamDP100S mutation in a clean genetic background. Other mutations identified in our screen will be the subject of a separate study. In the absence of NTZ, the BamDP100S mutation did not cause changes to usher expression levels, usher folding, or usher function in pilus assembly. The mutation also did not alter the general OMP profile in the presence or absence of drug. Thus, the BamDP100S mutation presumably affects an usher-specific aspect of the BAM complex that is targeted by NTZ, possibly decreasing affinity of NTZ for its binding site or altering the usher folding pathway in a way that bypasses the drug's effect. Studies are under way to determine whether NTZ binds directly to the BAM complex and the impact of the BamDP100S mutation on this binding. An alternate possibility is that NTZ binds to an interface between the unfolded usher and the BAM complex in a way that affects productive folding by the BAM complex. Proline 100 of BamD is located on a flexible loop that is part of an inner periplasmic ring formed by BamD and POTRA domains 1, 2, and 5 of BamA (Fig. S4) (31, 63, 64). This region is located under the BamA β-barrel pore and is thought to serve as the first contact point for interaction of substrates with the BAM complex (31, 63, 65, 66). The resistant phenotypes of the ΔbamE and bamDP100S mutants suggest that BamE and BamD cooperate in the usher folding pathway and that NTZ disrupts a specialized BamE-BamD-BamA arm of the BAM complex.
This study analyzed steady-state protein levels, and further work is needed to understand the contributions of BamE and BamD to the kinetics of usher folding in the OM and whether the specificity of NTZ for the usher might relate to differences in folding kinetics or turnover rates for the usher compared with other OMPs. Whereas the BAM complex is generally conserved in Gram-negative bacteria, there are differences in BAM complex structure and domain composition among species (67–69). The individual contributions of BAM complex components to OMP assembly may thus vary in distantly related organisms. However, the BamE and BamD lipoproteins and the proline 100 residue of BamD are conserved in many different organisms, including Klebsiella pneumoniae, Proteus mirabilis, Enterobacter cloacae, Yersinia pestis, Salmonella enterica and Pseudomonas aeruginosa. The mechanism of NTZ is thus likely to be conserved across multiple Gram-negative pathogens, and NTZ may affect a wide variety of CU pathways.
NTZ is used clinically as an anti-parasitic agent (23, 24). NTZ also exhibits broad-spectrum activity against anaerobic bacteria and has a wide range of other reported activities (53–55). In contrast to this broad activity, we have identified a highly specific effect of NTZ on the usher folding pathway. With the goal of obtaining more potent and narrowly active NTZ analogs that could help define the target and mechanism of action of the drug against the usher, we screened a set of analogs derived from the NTZ scaffold. Our studies identified the nitrothiazole ring of NTZ as critical for inhibition of PapC levels in the OM. Among the most potent compounds were 16a1011 and 161282, which incorporate halide-substituted thiophenes coupled to 2-amino-5- nitrothiazole (Fig. 7A). Consistent with the proposed mechanism of action for NTZ, the NTZ analogs specifically decreased usher levels in the OM, with no effects on the general OMP profile. These results hold promise for the development of novel NTZ analogs that could be used as anti-virulence therapeutics to disrupt pilus biogenesis by the CU pathway in diverse bacterial pathogens.
Experimental procedures
Strains, plasmids, and growth conditions
The E. coli strains and plasmids used in this study are listed in Table S1. Unless otherwise stated, E. coli overnight cultures were diluted 1:100 into fresh LB medium containing appropriate antibiotics (100 μg/ml ampicillin, 15 μg/ml tetracycline) and 0–20 μg/ml NTZ as indicated. For 0 μg/ml NTZ, a final concentration of 0.2% DMSO was added as a vehicle-only control, and 0.2% DMSO final concentration was maintained for all NTZ concentrations. Cultures were grown at 37 °C with aeration. When the cultures reached OD600 of 0.6, the expression of plasmid-encoded genes was induced for 1 h by the addition of the appropriate inducing agent: 50 μm isopropyl-β-d-thiogalactopyranoside (IPTG) or 0.1% arabinose.
The BW25113 ΔbamB, ΔbamC, and ΔbamE deletion mutants were obtained from the Keio collection (70) and verified by PCR. Strain BW25113bamDP100S was generated using the no-SCAR (scarless Cas9-assisted recombineering) system, as described (71, 72). Primers used are listed in Table S2.
Plasmid pBamB was generated by digesting pBAD18::bamB (36) with EcoRI and HindIII and ligating the bamB fragment into pTRYC (73). Plasmid pBamE was generated by amplifying the bamE-His8 fragment from pBamE-His (47) using a forward primer containing an EcoRI site and a reverse primer with a HindIII site. All primers used for cloning are listed in Table S2. The fragment was then ligated into pTRYC. Plasmids pBamABCDE and pBamA were generated by subcloning the desired bam genes from plasmid pJH114 (37) into pTRYC. The bamE gene from pJH114 also contains a C-terminal His tag. For cloning of these genes into pTRYC, a NdeI site was introduced into the multiple cloning site of pTRYC using the QuikChange XL mutagenesis protocol (Stratagene) to generate pTRYC-NdeI. The desired bam genes were then amplified from pJH114 using a forward primer containing a NdeI site and a reverse primer with an XmaI site and then ligated into pTRYC-NdeI. Plasmid pJP1 encoding the PapC usher with a 3X-FLAG tag insertion at residue Asn-197 was generated by site-directed, ligase-independent mutagenesis (SLIM) as described (74, 75) from plasmid pFJ20 (76).
NTZ and NTZ analog preparation
NTZ (Cayman Chemical) and NTZ analogs were obtained in powder form and dissolved in DMSO to give a stock concentration of 10 mg/ml. Synthesis of NTZ analogs has been described previously (77). All structures were confirmed by NMR and were >95% pure by LC-MS.
HA assay
HA assays were conducted as described (51) by serial dilution in microtiter plates. E. coli strain BW25113/pFJ29 was used to test P pilus functionality. Plasmid pACYC184 served as the vector control for pFJ29. Following growth in 0–20 μg/ml NTZ and induction with 50 μm IPTG, bacteria were harvested and washed with PBS before being resuspended and normalized to an OD540 of 1.0. Following serial dilution of the bacteria in microtiter plates, human erythrocytes were added, and HA titers were determined visually as the highest dilution of bacteria able to maintain agglutination. For each assay, three independent experiments were performed, and each experiment contained three replicates.
Isolation and analysis of the OM
OM fractions were isolated from the indicated E. coli strains grown and induced as described. PapC usher was expressed from plasmid pMJ3 or pJP1, with plasmid pMON6235Δcat serving as the vector control for the PapC plasmids. The entire BAM complex was expressed form plasmid pBamABCDE, with plasmid pTRYC serving as the vector control. BamA, BamB, and BamE were expressed from pBamA, pBamB, and pBamE, respectively. Bacteria were harvested, washed, resuspended and normalized to an OD600 of 1.0 in 20 mm Tris-HCl (pH 8.0) plus 1× complete protease inhibitor mixture. The OM fractions were isolated as described (49). The OM pellets were resuspended in 20 mm Tris-HCl and 0.3 m NaCl, followed by the addition of SDS sample buffer. The OM samples were incubated at 25 °C or heated to 95 °C for 10 min. Samples were analyzed by SDS-PAGE and immunoblotted with a mouse monoclonal anti-His6 antibody (1:1,000; Covance) for strains expressing pMJ3 or with a mouse monoclonal anti-FLAG antibody (1:1,000; Sigma-Aldrich) for strains expressing pJP1, followed by a secondary IRDye 680 goat anti-mouse conjugated IR antibody (1:40,000; LI-COR Biosciences). Samples were immunoblotted with anti-BamA antibody (1:30,000) (47), followed by a secondary IRDye 800 goat anti-rabbit–conjugated IR antibody (1:15,000; LI-COR Biosciences). Blots were visualized, and band densitometry was performed using a LI-COR Odyssey CLx imager. The specificity of the anti-FLAG antibody is demonstrated by a lack of signal in bacteria expressing non-FLAG-tagged PapC (Fig. 5). The specificity of the anti-His6 antibody is demonstrated by a lack of signal in vector controls (Figs. 1 and 2). The specificity of the anti-BamA antibody is demonstrated by expected changes in signal strength for the BamA WT, mutant, and complemented strains (Figs. 1 and 2).
Purification of the BAM complex
The BAM complex was purified as described previously (37). E. coli strain BW25113/pBamABCDE-His6 was cultured in the presence of either 0 or 20 μg/ml NTZ and lysed by passage through a French press, and a detergent-solubilized clarified lysate was passed over a nickel affinity column to capture the His-tagged BamE, together with stably associated proteins. Protein fractions eluted from the column with imidazole were then concentrated, normalized, and analyzed by Coomassie staining and immunoblotting with the anti-His tag and anti-BamA antibodies.
Nitrosoguanidine mutagenesis
A mutant library of BW25113 expressing the FLAG-tagged PapC usher (pJP1) was generated using N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) (Bio-Rad), as described (52). BW25113/pJP1 was grown to an OD600 of 1.0. A 2-ml aliquot of cells was incubated with 50 μl of MNNG (2.5 mg/ml in 95% ethanol) for 10 min at 37 °C. Cells were harvested by centrifugation (10 min, 1,500 × g) and washed twice with 2 ml of M63 medium. Cells were then resuspended in 5 ml of LB medium and grown overnight at 37 °C with aeration. The mutagenized BW25113/pJP1 bacteria were then subcultured in the presence of 0–20 μg/ml NTZ and subjected to FACS.
FACS
WT and nitrosoguanidine-mutagenized BW25113/pJP1 were grown in the presence of 0–20 μg/ml NTZ and induced with 0.1% l-arabinose for expression of the FLAG-tagged PapC usher. Bacteria were harvested, washed, resuspended, and normalized to an OD600 of 1.0 in PBS. A 500-μl aliquot was resuspended in PBS containing 1% BSA and rocked at 25 °C for 30 min. Bacteria were then resuspended in 500 μl of 1:1000 PE-conjugated anti-FLAG antibody (BioLegend) and rocked at 25 °C for 1 h. Bacteria were washed three times with PBS and subjected to FACS using a BD FACS ARIA IIIU high-speed cell sorter. Gating on side and forward scatter was used to focus on bacterial cells, and doublets were excluded. Mutagenized bacteria that maintained high levels of fluorescence (high levels of PapCFLAG) in the presence of NTZ were sorted as single cells into 96-well plates containing LB medium. Plates were then incubated at 37 °C, 300 rpm for 24 h and stored at −80 °C after the addition of 5% glycerol.
Whole-genome sequencing
Five mutant FLAG+ strains as well as the parent strain BW25113 were submitted as frozen cell pellets to GENEWIZ for whole-genome sequencing. Genomic DNA was isolated as specified for Gram-negative bacteria and sequenced by MiSeq (Illumina) to generate 150-bp paired-end data. Reads from each isolate were mapped onto the reference genome (E. coli BW25113, accession number NZ_CP009273), and variants were identified.
Statistical analysis
Densitometry analysis of protein bands was performed using a LI-COR Odyssey CLx imager on samples from three independent experiments. Statistical significance was calculated by one-way analysis of variance and by Bonferroni's multiple-comparison post-test using Prism version 8 (GraphPad Software). p values of <0.05 were considered significant.
Author contributions
J. J. P., P. C., N. W. R., P. S. H., and D. G. T. conceptualization; J. J. P., P. C., and D. G. T. formal analysis; J. J. P., P. C., and N. S. H. investigation; J. J. P., P. C., N. S. H., N. W. R., P. S. H., and D. G. T. methodology; J. J. P. writing-original draft; J. J. P., P. C., N. S. H., N. W. R., P. S. H., and D. G. T. writing-review and editing; P. S. H. and D. G. T. resources; D. G. T. supervision; D. G. T. funding acquisition.
Supplementary Material
Acknowledgments
We thank T. J. Silhavy (Princeton University) for the anti-BamA antibody, strains MC4100 and MC4100bamA101, plasmids pBAD18::bamB and pBamE-His, and for advice, inciteful discussions, and critical reading of the manuscript. We thank H. D. Bernstein (National Institutes of Health) for plasmid pJH114. We thank A. W. Karzai and the Karzai laboratory (Stony Brook University) for access to and assistance with the Keio E. coli mutant collection. We thank M. Tong and M. Seeliger (Stony Brook University) for advice and assistance with experiments. We thank the staff at the Flow Cytometry Core Research Facility at the Stony Brook University Hospital for assistance with the cell-sorting experiments.
This work was supported by NIGMS, National Institutes of Health (NIH), Grant R01GM062987 and NIAID, NIH, Grant R21AI121639 (to D. G. T.). This work was also supported by NIH Grant S10OD020155 (to M. Seeliger, Stony Brook University). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Data File S1, Tables S1 and S2, and Figs. S1–S4.
J. J. Psonis and D. G. Thanassi, unpublished results.
- CU
- chaperone–usher
- UPEC
- uropathogenic E. coli
- OM
- outer membrane
- NTZ
- nitazoxanide
- AAF
- aggregative adherence fimbriae
- OMP
- outer membrane protein
- HA
- hemagglutination
- PE
- phycoerythrin
- FACS
- fluorescence-activated cell sorting
- LB
- lysogeny broth
- OD
- optical density
- IPTG
- isopropyl-β-d-thiogalactopyranoside
- n-SCAR
- scarless Cas9-assisted recombineering
- SLIM
- site-directed, ligase-independent mutagenesis
- MNNG
- N-methyl-N′-nitro-N-nitrosoguanidine.
References
- 1. Thanassi D. G., Bliska J. B., and Christie P. J. (2012) Surface organelles assembled by secretion systems of Gram-negative bacteria: diversity in structure and function. FEMS Microbiol. Rev. 36, 1046–1082 10.1111/j.1574-6976.2012.00342.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chahales P., and Thanassi D. G. (2015) Structure, function, and assembly of adhesive organelles by uropathogenic bacteria. Microbiol. Spectr. 3, 10.1128/microbiolspec.UTI-0018-2013 10.1128/microbiolspec.UTI-0018-2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zav'yalov V., Zavialov A., Zav'yalova G., and Korpela T. (2010) Adhesive organelles of Gram-negative pathogens assembled with the classical chaperone/usher machinery: structure and function from a clinical standpoint. FEMS Microbiol. Rev. 34, 317–378 10.1111/j.1574-6976.2009.00201.x [DOI] [PubMed] [Google Scholar]
- 4. Werneburg G. T., and Thanassi D. G. (2018) Pili assembled by the chaperone/usher pathway in Escherichia coli and Salmonella. EcoSal Plus 8, ESP-0007-2017 10.1128/ecosalplus.ESP-0007-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Geibel S., and Waksman G. (2014) The molecular dissection of the chaperone-usher pathway. Biochim. Biophys. Acta 1843, 1559–1567 10.1016/j.bbamcr.2013.09.023 [DOI] [PubMed] [Google Scholar]
- 6. Kline K. A., Fälker S., Dahlberg S., Normark S., and Henriques-Normark B. (2009) Bacterial adhesins in host-microbe interactions. Cell Host Microbe 5, 580–592 10.1016/j.chom.2009.05.011 [DOI] [PubMed] [Google Scholar]
- 7. Flores-Mireles A. L., Walker J. N., Caparon M., and Hultgren S. J. (2015) Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 13, 269–284 10.1038/nrmicro3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lycklama A., Nijeholt J. A., and Driessen A. J. (2012) The bacterial Sec-translocase: structure and mechanism. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1016–1028 10.1098/rstb.2011.0201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sauer F. G., Fütterer K., Pinkner J. S., Dodson K. W., Hultgren S. J., and Waksman G. (1999) Structural basis of chaperone function and pilus biogenesis. Science 285, 1058–1061 10.1126/science.285.5430.1058 [DOI] [PubMed] [Google Scholar]
- 10. Choudhury D., Thompson A., Stojanoff V., Langermann S., Pinkner J., Hultgren S. J., and Knight S. D. (1999) X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 285, 1061–1066 10.1126/science.285.5430.1061 [DOI] [PubMed] [Google Scholar]
- 11. Remaut H., Tang C., Henderson N. S., Pinkner J. S., Wang T., Hultgren S. J., Thanassi D. G., Waksman G., and Li H. (2008) Fiber formation across the bacterial outer membrane by the chaperone/usher pathway. Cell 133, 640–652 10.1016/j.cell.2008.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Phan G., Remaut H., Wang T., Allen W. J., Pirker K. F., Lebedev A., Henderson N. S., Geibel S., Volkan E., Yan J., Kunze M. B., Pinkner J. S., Ford B., Kay C. W., Li H., et al. (2011) Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate. Nature 474, 49–53 10.1038/nature10109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Du M., Yuan Z., Yu H., Henderson N., Sarowar S., Zhao G., Werneburg G. T., Thanassi D. G., and Li H. (2018) Handover mechanism of the growing pilus by the bacterial outer-membrane usher FimD. Nature 562, 444–447 10.1038/s41586-018-0587-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Norgren M., Båga M., Tennent J. M., and Normark S. (1987) Nucleotide sequence, regulation and functional analysis of the papC gene required for cell surface localization of Pap pili of uropathogenic Escherichia coli. Mol. Microbiol. 1, 169–178 10.1111/j.1365-2958.1987.tb00509.x [DOI] [PubMed] [Google Scholar]
- 15. Nishiyama M., Ishikawa T., Rechsteiner H., and Glockshuber R. (2008) Reconstitution of pilus assembly reveals a bacterial outer membrane catalyst. Science 320, 376–379 10.1126/science.1154994 [DOI] [PubMed] [Google Scholar]
- 16. Fauci A. S., and Morens D. M. (2012) The perpetual challenge of infectious diseases. N. Engl. J. Med. 366, 454–461 10.1056/NEJMra1108296 [DOI] [PubMed] [Google Scholar]
- 17. Cegelski L., Marshall G. R., Eldridge G. R., and Hultgren S. J. (2008) The biology and future prospects of antivirulence therapies. Nat. Rev. Microbiol. 6, 17–27 10.1038/nrmicro1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Allen R. C., Popat R., Diggle S. P., and Brown S. P. (2014) Targeting virulence: can we make evolution-proof drugs? Nat. Rev. Microbiol. 12, 300–308 10.1038/nrmicro3232 [DOI] [PubMed] [Google Scholar]
- 19. Paharik A. E., Schreiber H. L. 4th, Spaulding C. N., Dodson K. W., and Hultgren S. J. (2017) Narrowing the spectrum: the new frontier of precision antimicrobials. Genome Med. 9, 110 10.1186/s13073-017-0504-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Psonis J. J., and Thanassi D. G. (2019) Therapeutic approaches targeting the assembly and function of chaperone-usher pili. EcoSal Plus 8, 10.1128/ecosalplus.ESP-0033-2018 10.1128/ecosalplus.ESP-0033-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pinkner J. S., Remaut H., Buelens F., Miller E., Aberg V., Pemberton N., Hedenström M., Larsson A., Seed P., Waksman G., Hultgren S. J., and Almqvist F. (2006) Rationally designed small compounds inhibit pilus biogenesis in uropathogenic bacteria. Proc. Natl. Acad. Sci. U.S.A. 103, 17897–17902 10.1073/pnas.0606795103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shamir E. R., Warthan M., Brown S. P., Nataro J. P., Guerrant R. L., and Hoffman P. S. (2010) Nitazoxanide inhibits biofilm production and hemagglutination by enteroaggregative Escherichia coli strains by blocking assembly of AafA fimbriae. Antimicrob. Agents Chemother. 54, 1526–1533 10.1128/AAC.01279-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Romero Cabello R., Guerrero L. R., Muñóz Garcia M. R., and Geyne Cruz A. (1997) Nitazoxanide for the treatment of intestinal protozoan and helminthic infections in Mexico. Trans. R. Soc. Trop. Med. Hyg. 91, 701–703 10.1016/S0035-9203(97)90531-9 [DOI] [PubMed] [Google Scholar]
- 24. Gilles H. M., and Hoffman P. S. (2002) Treatment of intestinal parasitic infections: a review of nitazoxanide. Trends Parasitol. 18, 95–97 10.1016/S1471-4922(01)02205-X [DOI] [PubMed] [Google Scholar]
- 25. Chahales P., Hoffman P. S., and Thanassi D. G. (2016) Nitazoxanide inhibits pilus biogenesis by interfering with folding of the usher protein in the outer membrane. Antimicrob. Agents Chemother. 60, 2028–2038 10.1128/AAC.02221-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lo A. W., Van de Water K., Gane P. J., Chan A. W., Steadman D., Stevens K., Selwood D. L., Waksman G., and Remaut H. (2014) Suppression of type 1 pilus assembly in uropathogenic Escherichia coli by chemical inhibition of subunit polymerization. J. Antimicrob. Chemother. 69, 1017–1026 10.1093/jac/dkt467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Piatek R., Zalewska-Piatek B., Dzierzbicka K., Makowiec S., Pilipczuk J., Szemiako K., Cyranka-Czaja A., and Wojciechowski M. (2013) Pilicides inhibit the FGL chaperone/usher assisted biogenesis of the Dr fimbrial polyadhesin from uropathogenic Escherichia coli. BMC Microbiol. 13, 131 10.1186/1471-2180-13-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Svensson A., Larsson A., Emtenäs H., Hedenström M., Fex T., Hultgren S. J., Pinkner J. S., Almqvist F., and Kihlberg J. (2001) Design and evaluation of pilicides: potential novel antibacterial agents directed against uropathogenic Escherichia coli. Chembiochem 2, 915–918 [DOI] [PubMed] [Google Scholar]
- 29. Knowles T. J., Scott-Tucker A., Overduin M., and Henderson I. R. (2009) Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat. Rev. Microbiol. 7, 206–214 10.1038/nrmicro2069 [DOI] [PubMed] [Google Scholar]
- 30. Konovalova A., Kahne D. E., and Silhavy T. J. (2017) Outer membrane biogenesis. Annu. Rev. Microbiol. 71, 539–556 10.1146/annurev-micro-090816-093754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Noinaj N., Gumbart J. C., and Buchanan S. K. (2017) The β-barrel assembly machinery in motion. Nat. Rev. Microbiol. 15, 197–204 10.1038/nrmicro.2016.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ricci D. P., and Silhavy T. J. (2019) Outer membrane protein insertion by the β-barrel assembly machine. EcoSal Plus 8, ESP-0035–2018 10.1128/ecosalplus.ESP-0035-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krojer T., Sawa J., Schäfer E., Saibil H. R., Ehrmann M., and Clausen T. (2008) Structural basis for the regulated protease and chaperone function of DegP. Nature 453, 885–890 10.1038/nature07004 [DOI] [PubMed] [Google Scholar]
- 34. Watts K. M., and Hunstad D. A. (2008) Components of SurA required for outer membrane biogenesis in uropathogenic Escherichia coli. PLoS One 3, e3359 10.1371/journal.pone.0003359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Malinverni J. C., Werner J., Kim S., Sklar J. G., Kahne D., Misra R., and Silhavy T. J. (2006) YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol. Microbiol. 61, 151–164 10.1111/j.1365-2958.2006.05211.x [DOI] [PubMed] [Google Scholar]
- 36. Wu T., Malinverni J., Ruiz N., Kim S., Silhavy T. J., and Kahne D. (2005) Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121, 235–245 10.1016/j.cell.2005.02.015 [DOI] [PubMed] [Google Scholar]
- 37. Roman-Hernandez G., Peterson J. H., and Bernstein H. D. (2014) Reconstitution of bacterial autotransporter assembly using purified components. Elife 3, e04234 10.7554/eLife.04234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hagan C. L., Kim S., and Kahne D. (2010) Reconstitution of outer membrane protein assembly from purified components. Science 328, 890–892 10.1126/science.1188919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tata M., and Konovalova A. (2019) Improper coordination of BamA and BamD results in Bam complex jamming by a lipoprotein substrate. MBio 10, e00660–19 10.1128/mBio.00660-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hart E. M., Gupta M., Wühr M., and Silhavy T. J. (2019) The synthetic phenotype of ΔbamB ΔbamE double mutants results from a lethal jamming of the Bam complex by the lipoprotein RcsF. MBio 10, e00662–00619 10.1128/mBio.00662-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Charlson E. S., Werner J. N., and Misra R. (2006) Differential effects of yfgL mutation on Escherichia coli outer membrane proteins and lipopolysaccharide. J. Bacteriol. 188, 7186–7194 10.1128/JB.00571-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mahoney T. F., Ricci D. P., and Silhavy T. J. (2016) Classifying β-barrel assembly substrates by manipulating essential Bam complex members. J. Bacteriol. 198, 1984–1992 10.1128/JB.00263-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Palomino C., Marín E., and Fernández L. A. (2011) The fimbrial usher FimD follows the SurA-BamB pathway for its assembly in the outer membrane of Escherichia coli. J. Bacteriol. 193, 5222–5230 10.1128/JB.05585-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ricci D. P., Hagan C. L., Kahne D., and Silhavy T. J. (2012) Activation of the Escherichia coli β-barrel assembly machine (Bam) is required for essential components to interact properly with substrate. Proc. Natl. Acad. Sci. U.S.A. 109, 3487–3491 10.1073/pnas.1201362109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aoki S. K., Malinverni J. C., Jacoby K., Thomas B., Pamma R., Trinh B. N., Remers S., Webb J., Braaten B. A., Silhavy T. J., and Low D. A. (2008) Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol. Microbiol. 70, 323–340 10.1111/j.1365-2958.2008.06404.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rigel N. W., Schwalm J., Ricci D. P., and Silhavy T. J. (2012) BamE modulates the Escherichia coli β-barrel assembly machine component BamA. J. Bacteriol. 194, 1002–1008 10.1128/JB.06426-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sklar J. G., Wu T., Gronenberg L. S., Malinverni J. C., Kahne D., and Silhavy T. J. (2007) Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 104, 6400–6405 10.1073/pnas.0701579104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Onufryk C., Crouch M. L., Fang F. C., and Gross C. A. (2005) Characterization of six lipoproteins in the σE regulon. J. Bacteriol. 187, 4552–4561 10.1128/JB.187.13.4552-4561.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ng T. W., Akman L., Osisami M., and Thanassi D. G. (2004) The usher N terminus is the initial targeting site for chaperone-subunit complexes and participates in subsequent pilus biogenesis events. J. Bacteriol. 186, 5321–5331 10.1128/JB.186.16.5321-5331.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sugawara E., Steiert M., Rouhani S., and Nikaido H. (1996) Secondary structure of the outer membrane proteins OmpA of Escherichia coli and OprF of Pseudomonas aeruginosa. J. Bacteriol. 178, 6067–6069 10.1128/jb.178.20.6067-6069.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Henderson N. S., Ng T. W., Talukder I., and Thanassi D. G. (2011) Function of the usher N-terminus in catalysing pilus assembly. Mol. Microbiol. 79, 954–967 10.1111/j.1365-2958.2010.07505.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Silhavy T. J., Berman M. L., and Enquist L. W. (eds) (1984) Experiments with Gene Fusions, pp. 129–130, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 53. Dubreuil L., Houcke I., Mouton Y., and Rossignol J. F. (1996) In vitro evaluation of activities of nitazoxanide and tizoxanide against anaerobes and aerobic organisms. Antimicrob. Agents Chemother. 40, 2266–2270 10.1128/AAC.40.10.2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rossignol J. F. (2014) Nitazoxanide: a first-in-class broad-spectrum antiviral agent. Antiviral Res. 110, 94–103 10.1016/j.antiviral.2014.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hoffman P. S., Sisson G., Croxen M. A., Welch K., Harman W. D., Cremades N., and Morash M. G. (2007) Antiparasitic drug nitazoxanide inhibits the pyruvate oxidoreductases of Helicobacter pylori, selected anaerobic bacteria and parasites, and Campylobacter jejuni. Antimicrob. Agents Chemother. 51, 868–876 10.1128/AAC.01159-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Justice S. S., Hunstad D. A., Harper J. R., Duguay A. R., Pinkner J. S., Bann J., Frieden C., Silhavy T. J., and Hultgren S. J. (2005) Periplasmic peptidyl prolyl cis-trans isomerases are not essential for viability, but SurA is required for pilus biogenesis in Escherichia coli. J. Bacteriol. 187, 7680–7686 10.1128/JB.187.22.7680-7686.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stubenrauch C., Belousoff M. J., Hay I. D., Shen H. H., Lillington J., Tuck K. L., Peters K. M., Phan M. D., Lo A. W., Schembri M. A., Strugnell R. A., Waksman G., and Lithgow T. (2016) Effective assembly of fimbriae in Escherichia coli depends on the translocation assembly module nanomachine. Nat. Microbiol. 1, 16064 10.1038/nmicrobiol.2016.64 [DOI] [PubMed] [Google Scholar]
- 58. Sklar J. G., Wu T., Kahne D., and Silhavy T. J. (2007) Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 21, 2473–2484 10.1101/gad.1581007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. McCabe A. L., Ricci D., Adetunji M., and Silhavy T. J. (2017) Conformational changes that coordinate the activity of BamA and BamD allowing β-barrel assembly. J. Bacteriol. 199, e00373–00317 10.1128/JB.00373-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee J., Sutterlin H. A., Wzorek J. S., Mandler M. D., Hagan C. L., Grabowicz M., Tomasek D., May M. D., Hart E. M., Silhavy T. J., and Kahne D. (2018) Substrate binding to BamD triggers a conformational change in BamA to control membrane insertion. Proc. Natl. Acad. Sci. U.S.A. 115, 2359–2364 10.1073/pnas.1711727115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rigel N. W., Ricci D. P., and Silhavy T. J. (2013) Conformation-specific labeling of BamA and suppressor analysis suggest a cyclic mechanism for β-barrel assembly in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 110, 5151–5156 10.1073/pnas.1302662110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Konovalova A., Perlman D. H., Cowles C. E., and Silhavy T. J. (2014) Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of β-barrel proteins. Proc. Natl. Acad. Sci. U.S.A. 111, E4350–E4358 10.1073/pnas.1417138111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gu Y., Li H., Dong H., Zeng Y., Zhang Z., Paterson N. G., Stansfeld P. J., Wang Z., Zhang Y., Wang W., and Dong C. (2016) Structural basis of outer membrane protein insertion by the BAM complex. Nature 531, 64–69 10.1038/nature17199 [DOI] [PubMed] [Google Scholar]
- 64. Iadanza M. G., Higgins A. J., Schiffrin B., Calabrese A. N., Brockwell D. J., Ashcroft A. E., Radford S. E., and Ranson N. A. (2016) Lateral opening in the intact β-barrel assembly machinery captured by cryo-EM. Nat. Commun. 7, 12865 10.1038/ncomms12865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Warner L. R., Gatzeva-Topalova P. Z., Doerner P. A., Pardi A., and Sousa M. C. (2017) Flexibility in the periplasmic domain of BamA is important for function. Structure 25, 94–106 10.1016/j.str.2016.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bergal H. T., Hopkins A. H., Metzner S. I., and Sousa M. C. (2016) The structure of a BamA-BamD fusion illuminates the architecture of the β-barrel assembly machine core. Structure 24, 243–251 10.1016/j.str.2015.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Volokhina E. B., Beckers F., Tommassen J., and Bos M. P. (2009) The β-barrel outer membrane protein assembly complex of Neisseria meningitidis. J. Bacteriol. 191, 7074–7085 10.1128/JB.00737-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Arnold T., Zeth K., and Linke D. (2010) Omp85 from the thermophilic cyanobacterium Thermosynechococcus elongatus differs from proteobacterial Omp85 in structure and domain composition. J. Biol. Chem. 285, 18003–18015 10.1074/jbc.M110.112516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Stubenrauch C., Grinter R., and Lithgow T. (2016) The modular nature of the β-barrel assembly machinery, illustrated in Borrelia burgdorferi. Mol. Microbiol. 102, 753–756 10.1111/mmi.13527 [DOI] [PubMed] [Google Scholar]
- 70. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., and Mori H. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Reisch C. R., and Prather K. L. (2015) The no-SCAR (scarless Cas9 assisted recombineering) system for genome editing in Escherichia coli. Sci. Rep. 5, 15096 10.1038/srep15096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Reisch C. R., and Prather K. L. J. (2017) Scarless Cas9 assisted recombineering (no-SCAR) in Escherichia coli, an easy-to-use system for genome editing. Curr. Protoc. Mol. Biol. 117, 31.38.31–31.38.20 10.1002/cpmb.29 [DOI] [PubMed] [Google Scholar]
- 73. Wright K. J., Seed P. C., and Hultgren S. J. (2007) Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cell Microbiol. 9, 2230–2241 10.1111/j.1462-5822.2007.00952.x [DOI] [PubMed] [Google Scholar]
- 74. Chiu J., Tillett D., Dawes I. W., and March P. E. (2008) Site-directed, ligase-independent mutagenesis (SLIM) for highly efficient mutagenesis of plasmids greater than 8kb. J. Microbiol. Methods 73, 195–198 10.1016/j.mimet.2008.02.013 [DOI] [PubMed] [Google Scholar]
- 75. Chiu J., March P. E., Lee R., and Tillett D. (2004) Site-directed, ligase-independent mutagenesis (SLIM): a single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res. 32, e174 10.1093/nar/gnh172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jacob-Dubuisson F., Striker R., and Hultgren S. J. (1994) Chaperone-assisted self-assembly of pili independent of cellular energy. J. Biol. Chem. 269, 12447–12455 [PubMed] [Google Scholar]
- 77. Ballard T. E., Wang X., Olekhnovich I., Koerner T., Seymour C., Salamoun J., Warthan M., Hoffman P. S., and Macdonald T. L. (2011) Synthesis and antimicrobial evaluation of nitazoxanide-based analogues: identification of selective and broad spectrum activity. ChemMedChem 6, 362–377 10.1002/cmdc.201000475 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.