Abstract
Enteric viruses infect the mammalian gastrointestinal tract which is home to a diverse community of intestinal bacteria. Accumulating evidence suggests that certain enteric viruses utilize these bacteria to promote infection. While this is not surprising considering their proximity, multiple viruses from different viral families have been shown to bind directly to bacteria or bacterial components to aid in viral replication, pathogenesis, and transmission. These data suggest that the concept of a single virus infecting a single cell, independent of the environment, needs to be reevaluated. In this review, I will discuss the current knowledge of enteric virus-bacterial interactions and discuss the implications for viral pathogenesis and transmission.
Keywords: Virus, microbiota, bacteria, enteric
Introduction
The microbiota is now widely recognized for being vital to human health (1). Consisting of bacteria, fungus, and viruses, the microbiota inhabit multiple sites on the human body where they have developed an ecological niche that is a benefit to both the host and colonizing organisms. Among these sites, the gastrointestinal tract (GI) is home to the most extensive collection of the microbiota, including a dense community of commensal bacteria. This diverse community is made up of approximately 1013 bacteria from 500-100 different species (2–4). A majority of these bacteria reside in the lower gastrointestinal tract, however, due to pH, oxygen, and nutrient availability, distinct bacterial populations inhabit both the small and large intestine (5). Further, intestinal bacteria vary amongst individuals and can be influenced by genetics, diet, and lifestyle (6–8). Additionally, imbalances in the microbial composition have been linked to many human diseases including inflammatory bowel diseases, type 2 diabetes, and obesity (9–14).
Enteric viruses initiate infection in the mammalian GI tract where they encounter these microbiota. Previous data demonstrate that multiple enteric viruses utilize the microbiota to promote replication, pathogenesis, transmission. Many of these viruses have been shown to interact directly with intestinal bacteria, and in this review, I will highlight our latest understanding of the interaction between enteric viruses and intestinal bacteria and the consequences. Specifically, I will discuss the influence of bacteria on members of the picornavirus, reovirus, and retrovirus families. Additional enteric viruses also exploit the microbiota; however, these viruses will be discussed separately in this special volume. Overall these studies suggest that the conventional view of a single virus infecting a cell is too limited. Other factors, including the microbiota, need to be considered when investigating the outcome of viral infections in vivo.
Intestinal bacteria impact enteric viral infections
It comes to no surprise that, given the proximity of enteric viruses and intestinal bacteria, recent research indicates that bacteria can impact enteric viral infections. While these bacteria could impede viral infection by acting as a physical barrier in the intestine, recent compelling evidence has shown that some enteric viruses utilize bacteria to promote infection. Poliovirus, a non-enveloped enteric virus in the Picornaviridae family that initiates infection in the intestine, can disseminate to the central nervous system to cause paralytic poliomyelitis. In mice depleted of intestinal bacteria by antibiotics, poliovirus replication and lethality is significantly reduced compared to untreated mice (15). These data indicate that bacteria promote poliovirus intestinal replication and pathogenesis. Further, data reveal that poliovirus can bind to bacteria through interactions with bacterial surface components, lipopolysaccharide (LPS) and peptidoglycan, to enhance infectivity (16, 17).
Rotavirus, a non-enveloped RNA virus from the Reoviridae family, is a leading cause of diarrheal disease in children worldwide (18). Depletion of intestinal bacteria by antibiotics reduced diarrhea duration and rotavirus fecal shedding in mice (19). Further, germ-free mice also exhibit a delay in rotavirus fecal shedding, implicating a role for bacteria in rotavirus infection. Reovirus, another member of the Reoviridae family, also had reduced replication and pathogenesis when intestinal bacteria are depleted (15). Similar to poliovirus, both Grampositive and Gram-negative bacteria can bind and enhance reovirus infectivity (20). This interaction is likely facilitated by viral binding to LPS and peptidoglycan, suggesting that reovirus, like poliovirus, may utilize a broad array of different intestinal bacterial isolates to facilitate infection in the intestine.
Finally, mouse mammary tumor virus (MMTV) is an enveloped retrovirus that infects lymphoid cells in Peyer’s patches of the intestine (21). MMTV can establish persistence in mice and is transmitted from infected females to suckling pups in milk. Commensal bacteria are required to maintain viral persistence as the virus can be abolished in germ-free mice (22) Specifically, MMTV can bind to LPS via LPS-binding proteins that become integrated into the viral envelope during egress (23). Overall these data provide compelling evidence that some enteric viruses have evolved to utilize intestinal bacteria to promote infection.
Bacteria enhance viral thermostability
Recent advances have shed light on the mechanistic outcome for these virus-bacterial interactions. Data suggest that enteric viruses bind to bacteria to stabilize the virion to retain infectivity. For example, both Gram-positive and Gram-negative bacteria enhance the thermostability of poliovirus (Fig. 1A) (15). Binding of poliovirus to LPS retain viral particles in their infectious state, limiting inactivation and premature release of viral RNA. (16). Further a mutation in the poliovirus VP1 capsid protein, T99K, reduces LPS binding and environmental fitness, suggesting that bacterial enhancement of thermostability may also play an essential role in viral transmission from host-to-host. These effects are also seen in other members of the Picornaviridae family as well. LPS and peptidoglycan enhance the thermostability of Coxsackievirus A21, Coxsackievirus B5, and Echovirus 30 (24), and bacteria improve the stability of Coxsackievirus B3, Aichi, and mengovirus during bleach treatment (25). Overall, these data suggest that bacterial-mediated enhancement of thermostability may be a shared mechanism for picornaviruses.
Figure 1.
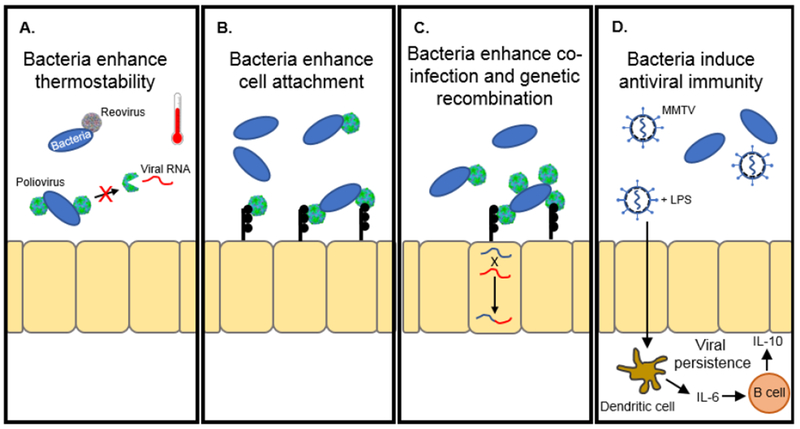
Bacteria promote enteric virus infection. (A) The binding of bacteria to poliovirus and reovirus enhance thermostability. For poliovirus, enhanced thermostability limits premature release of viral RNA. Poliovirus and reovirus adapted from PBD 1HXS and 2CSE respectively. (B) Bacteria increase poliovirus binding to the poliovirus receptor on permissive cells. (C) Poliovirus bound to bacteria increase co-infection and genetic recombination between progeny virions. (D) MMTV bound to LPS induce an antiviral response to allow for viral persistence.
Similar to poliovirus, recent evidence suggests that bacteria also enhance the stability of reovirus. Both Gram-positive and Gram-negative bacteria improve reovirus thermostability in vitro through interactions with LPS and peptidoglycan (20). Furthermore, intermediate infectious subvirion particles (ISVPs) also are stabilized by LPS and peptidoglycan. These ISVPs are formed by proteolytic processing of the virion and required for attachment and entry of reovirus into host cells and contribute to intestinal infection (26–28). These data suggest that the binding site of these bacterial components to reovirus does not involve capsid proteins, μ1 or σ3, since they are either cleaved or absent on ISVPs. Interestingly, individual bacterial components have different effects on specific strains of reovirus. While LPS and peptidoglycan enhance have both the Type 1 Lang (TIL) and Type 3 Dearing (T3D) strains of reovirus, lipoteichoic acid and chitin only enhanced T3D thermostability, but not the TIL strain. Therefore, specific reovirus strains may have different affinities for distinct bacterial components; however, if these affinities affect strain-specific replication and pathogenesis differences remains unknown.
Bacteria enhance viral attachment to host cells
Another mechanism for bacterial-viral interactions is through enhancement of viral attachment to the host cell. In addition to enhancing poliovirus thermostability, incubation of poliovirus with bacteria increase attachment to poliovirus receptor (PVR)-expressing cells (Fig. 1B) (15). Evidence suggests that this enhancement is through direct interaction with the receptor since poliovirus, incubated with LPS, bound to significantly more purified PVR than virus incubated in PBS alone (16). Interestingly, the VP1-T99K poliovirus mutant, which reduces LPS binding, still retained LPS-mediated enhancement of cell attachment. These data suggest that the affinity and concentration of LPS molecules may influence poliovirus cell attachment and thermostability.
Bacteria can facilitate viral co-infection and promote viral genetic recombination
In addition to the virion thermostability and cell attachment, Erickson et al. demonstrated that binding of poliovirus to specific bacterial strains increased viral co-infection of host cells (Fig. 1C) (17). Using DsRed- and GFP-expressing poliovirus, the authors found that poliovirus mixed with certain bacterial isolates increased the number of dual-infected HeLa cells. These data are not surprising since electron microscopy studies revealed that multiple poliovirus virions bound to a single bacterium. Interestingly, the increase in co-infection rates correlated with viral binding to bacterial isolates that had increased adherence to HeLa cells. Further, using drug- and temperature-sensitive poliovirus mutants, the authors found that bacterial enhancement of co-infection also increased the likelihood of genetic recombination amongst poliovirus virions. Overall, these data suggest that poliovirus may utilize bacteria to improve viral fitness in the mammalian GI tract through increased genetic recombination.
Bacteria promote viral immune evasion
Commensal bacteria provide vital stimulation to develop a mature host immune system (29, 30). While these immune signals from bacteria promote colonization resistance to pathogenic organisms, evidence suggests that some enteric viruses have exploited these pathways to evade the immune system. For example, antibiotics enhance the antibody response to a rotavirus infection, suggesting that bacteria can modulate the immune response to rotavirus (19). The mechanism behind this response, however, is unclear. Similarly, MMTV can subvert the host immune response to establish persistence in mice. Immune evasion for MMTV requires functional Toll-like receptor 4 (TLR-4) signaling and the immunosuppressive cytokine IL-10; however, the mechanism was largely unclear (31). Kane et al. shed light on this mechanism when they demonstrated that MMTV uses commensal bacteria to facilitate persistence by binding to bacterial LPS (Fig. 1D) (22). To enable binding to LPS, MMTV incorporates LPS-binding factors, such as MD-2 and CD14, into the viral envelope during egress from infected cells (23). These LPS-binding factors play a critical role in the transfer of the LPS molecule to TLR-4 and can help trigger LPS-induced signaling pathways (32). Upon binding of LPS to the viral membrane, MMTV can stimulate the TLR-4 pathway leading to IL-6 production of the immunosuppressive IL-10 cytokine. This cascade activates an immune evasion pathway, facilitating persistent MMTV infections.
Negative or unclear effects of bacteria on enteric viruses
While this review focuses on the beneficial outcome of bacterial and viral interactions, these effects may not be conserved. Mouse adenovirus 1 does not require intestinal bacteria for intestinal infection (33). Additionally, defensins, host antimicrobial peptides induced in response to intestinal bacteria, can neutralize some adenoviruses types (34). Interestingly, recent data suggest that α-defensin may enhance mouse adenovirus type 2, a mouse enteric pathogen, suggesting that further work is necessary to determine the role of intestinal bacteria on enteric adenoviruses (35). Further, even though some experimental data suggest that intestinal bacteria may promote rotavirus infection, other data contradict this. For example, probiotics have been shown to reduce the duration of viral diarrhea and administration of Lactobacillus rhamnosus GG reduces rotavirus shedding (36–38). Additionally, soluble factors from commensal bacteria can block rotavirus infection in vitro, and bacterial flagellin can eliminate chronic rotavirus infection through induction of IL-22 and IL-18 in vivo (39). Overall, these data suggest that some enteric viruses may utilize intestinal bacteria to promote infection, while others may not.
Conclusions
Traditionally, viral infections have been viewed in the context of a single virus infecting a host cell. This straightforward view has allowed a reductionist approach to understanding the fundamental processes of the viral life cycle. This reductionist approach, however, does not always paint a full picture of the complex interactions that arise during a viral infection in vivo. In light of this potential complexity, recent evidence indicates that additional extrinsic factors, such the microbiota, may play a significant role in establishing a viral infection within a host. Overall, these new factors have established a new field of study in virology where viral infections are understood in the context of the environment for which they initiate infection. Future experiments identifying these complex interactions and their mechanisms need to be elucidated to provide a clear picture of the viral cycle, not only within the cell, but within the entire host.
In light of all these recent findings, there is clearly a need for more studies to understand how intestinal bacteria and the microbiota impact viral infections. Many questions remain as to the precise mechanisms of bacterial-mediated viral enhancement. First, which specific bacteria are required to enhance viral infection? Specific bacteria required to enhance infection for one enteric virus likely not enhance another. Data suggest that poliovirus and reovirus have different binding affinities to particular bacteria and bacterial components, and identifying the mechanism behind these binding affinities may provide potential therapeutic targets. Second, does biological sex influence these interactions? Microbiota differences between males and females exist and have been shown to influence type 1 diabetes in a mouse model (40). Recently, Coxsackievirus B3 replication in the intestine was shown to be sex-dependent (41). Since bacteria interact with Coxsackievirus B3 (25), perhaps sex-dependent microbiota may influence viral replication through direct or indirect mechanisms. Third, do bacteria affect other non-enteric viruses? Data suggest that bacteria can affect the immune response to influenza virus (42–45); however, the impact of bacteria on other viruses is unclear. Since the bacteria colonize multiple sites on the human body, likely other viruses have interactions with bacteria. Additionally, viruses may play a complementary role in promoting bacterial infections (46); however, additional data is required to determine if this impacts infections in the gastrointestinal tract. Finally, what is the role of other components of the microbiota? Much of the current data has focused on the effect of the bacterial component of the microbiota on enteric viruses, but fungal inhabitants of the intestine and other viruses may also influence enteric viral infections as well. Additional studies are needed to investigate these interactions in the intestine to determine their influence on viral infection.
Finally, it is important to note that while current data suggest that antibiotics may be an effective antiviral therapy, the side effects of disruption of commensal bacteria would largely outweigh any potential gain. Antiviral effects in mice require a substantial depletion of intestinal bacteria, with multiple antibiotics given prophylactically, which would likely disrupt the health benefits that these bacteria provide. Future experiments identifying specific interactions between enteric viruses and specific bacteria or bacterial components may offer a better opportunity for therapeutics. For example, the precise modulation of these virus-bacteria interactions may provide benefits in controlling outbreaks by disrupting viral transmission.
In conclusion, the role of the microbiota on enteric viruses represents a promising area of future study. It is imperative that this field moves forward so that we can truly understand the overall environmental factors that underlie a viral infection within a host. While we are only beginning to uncover the mechanisms behind interactions between enteric viruses and bacteria, I anticipate that future studies will continue to shed light on other factors of the microbiota that influence enteric viral infections. This knowledge will be vital as we continue to understand the complex interactions and environments within the human body that influence viral infections.
Acknowledgments
Research in the Robinson laboratory is supported in by funding from the National Institutes of Health, NIDDK (K01 DK110216), and the Indiana Clinical and Translational Sciences Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
• Of special interest
* Of outstanding interest
- 1.Clemente JC, Ursell LK, Parfrey LW, Knight R. 2012. The impact of the gut microbiota on human health: an integrative view. Cell 148:1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hao WL, Lee YK. 2004. Microflora of the gastrointestinal tract: a review. Methods Mol Biol 268:491–502. [DOI] [PubMed] [Google Scholar]
- 3.Sender R, Fuchs S, Milo R. 2016. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol 14:e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sender R, Fuchs S, Milo R. 2016. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell 164:337–340. [DOI] [PubMed] [Google Scholar]
- 5.Tropini C, Earle KA, Huang KC, Sonnenburg JL. 2017. The Gut Microbiome: Connecting Spatial Organization to Function. Cell Host Microbe 21:433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, Kachman SD, Moriyama EN, Walter J, Peterson DA, Pomp D. 2010. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci U S A 107:18933–18938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Annalisa N, Alessio T, Claudette TD, Erald V, Antonino de L, Nicola DD. 2014. Gut microbioma population: an indicator really sensible to any change in age, diet, metabolic syndrome, and life-style. Mediators Inflamm 2014:901308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 104:13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malinen E, Rinttila T, Kajander K, Matto J, Kassinen A, Krogius L, Saarela M, Korpela R, Palva A. 2005. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol 100:373–382. [DOI] [PubMed] [Google Scholar]
- 11.Kerckhoffs AP, Ben-Amor K, Samsom M, van der Rest ME, de Vogel J, Knol J, Akkermans LM. 2011. Molecular analysis of faecal and duodenal samples reveals significantly higher prevalence and numbers of Pseudomonas aeruginosa in irritable bowel syndrome. J Med Microbiol 60:236–245. [DOI] [PubMed] [Google Scholar]
- 12.Gerritsen J, Smidt H, Rijkers GT, de Vos WM. 2011. Intestinal microbiota in human health and disease: the impact of probiotics. Genes Nutr 6:209–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sorensen SJ, Hansen LH, Jakobsen M. 2010. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 5:e9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, Hooper LV, Dermody TS, Pfeiffer JK. 2011. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science 334:249–252. [DOI] [PMC free article] [PubMed] [Google Scholar]; * For the first time demonstrated that bacteria promote replciation and pathogenesis of an enteric virus
- 16.Robinson CM, Jesudhasan PR, Pfeiffer JK. 2014. Bacterial lipopolysaccharide binding enhances virion stability and promotes environmental fitness of an enteric virus. Cell Host Microbe 15:36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erickson AK, Jesudhasan PR, Mayer MJ, Narbad A, Winter SE, Pfeiffer JK. 2018. Bacteria Facilitate Enteric Virus Co-infection of Mammalian Cells and Promote Genetic Recombination. Cell Host Microbe 23:77–88 e75. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This paper highlights that intestinal bacteria can enhance genetic recombination and promote viral fitness in the intestine.
- 18.Greenberg HB, Estes MK. 2009. Rotaviruses: from pathogenesis to vaccination. Gastroenterology 136:1939–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uchiyama R, Chassaing B, Zhang B, Gewirtz AT. 2014. Antibiotic treatment suppresses rotavirus infection and enhances specific humoral immunity. J Infect Dis 210:171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berger AK, Yi H, Kearns DB, Mainou BA. 2017. Bacteria and bacterial envelope components enhance mammalian reovirus thermostability. PLoS Pathog 13:e1006768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golovkina TV, Shlomchik M, Hannum L, Chervonsky A. 1999. Organogenic role of B lymphocytes in mucosal immunity. Science 286:1965–1968. [DOI] [PubMed] [Google Scholar]
- 22.Kane M, Case LK, Kopaskie K, Kozlova A, MacDearmid C, Chervonsky AV, Golovkina TV. 2011. Successful transmission of a retrovirus depends on the commensal microbiota. Science 334:245–249. [DOI] [PMC free article] [PubMed] [Google Scholar]; * A seminal paper demonstrating that commensal bacteria are required for MMTV persistence through modulation of the host immune response.
- 23.Wilks J, Lien E, Jacobson AN, Fischbach MA, Qureshi N, Chervonsky AV, Golovkina TV. 2015. Mammalian Lipopolysaccharide Receptors Incorporated into the Retroviral Envelope Augment Virus Transmission. Cell Host Microbe 18:456–462. [DOI] [PMC free article] [PubMed] [Google Scholar]; • An important finding that MMTV encoporates LPS binding factors into the viral envelop to bind to LPS to influence an antiviral immune response.
- 24.Waldman P, Meseguer A, Lucas F, Moulin L, Wurtzer S. 2017. Interaction of Human Enteric Viruses with Microbial Compounds: Implication for Virus Persistence and Disinfection Treatments. Environ Sci Technol 51:13633–13640. [DOI] [PubMed] [Google Scholar]
- 25.Aguilera ER, Nguyen Y, Sasaki J, Pfeiffer JK. 2019. Bacterial Stabilization of a Panel of Picornaviruses. mSphere 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amerongen HM, Wilson GA, Fields BN, Neutra MR. 1994. Proteolytic processing of reovirus is required for adherence to intestinal M cells. J Virol 68:8428–8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bodkin DK, Nibert ML, Fields BN. 1989. Proteolytic digestion of reovirus in the intestinal lumens of neonatal mice. J Virol 63:4676–4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nibert ML, Chappell JD, Dermody TS. 1995. Infectious subvirion particles of reovirus type 3 Dearing exhibit a loss in infectivity and contain a cleaved sigma 1 protein. J Virol 69:5057–5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abt MC, Artis D. 2013. The dynamic influence of commensal bacteria on the immune response to pathogens. Curr Opin Microbiol 16:4–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belkaid Y, Hand TW. 2014. Role of the microbiota in immunity and inflammation. Cell 157:121–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jude BA, Pobezinskaya Y, Bishop J, Parke S, Medzhitov RM, Chervonsky AV, Golovkina TV. 2003. Subversion of the innate immune system by a retrovirus. Nat Immunol 4:573–578. [DOI] [PubMed] [Google Scholar]
- 32.Park BS, Lee JO. 2013. Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp Mol Med 45:e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gounder AP, Myers ND, Treuting PM, Bromme BA, Wilson SS, Wiens ME, Lu W, Ouellette AJ, Spindler KR, Parks WC, Smith JG. 2016. Defensins Potentiate a Neutralizing Antibody Response to Enteric Viral Infection. PLoS Pathog 12:e1005474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson SS, Wiens ME, Smith JG. 2013. Antiviral mechanisms of human defensins. J Mol Biol 425:4965–4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson SS, Bromme BA, Holly MK, Wiens ME, Gounder AP, Sul Y, Smith JG. 2017. Alpha-defensin-dependent enhancement of enteric viral infection. PLoS Pathog 13:e1006446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaila M, Isolauri E, Saxelin M, Arvilommi H, Vesikari T. 1995. Viable versus inactivated lactobacillus strain GG in acute rotavirus diarrhoea. Arch Dis Child 72:51–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Majamaa H, Isolauri E, Saxelin M, Vesikari T. 1995. Lactic acid bacteria in the treatment of acute rotavirus gastroenteritis. J Pediatr Gastroenterol Nutr 20:333–338. [DOI] [PubMed] [Google Scholar]
- 38.Saavedra J 2000. Probiotics and infectious diarrhea. Am J Gastroenterol 95:S16–18. [DOI] [PubMed] [Google Scholar]
- 39.Zhang B, Chassaing B, Shi Z, Uchiyama R, Zhang Z, Denning TL, Crawford SE, Pruijssers AJ, Iskarpatyoti JA, Estes MK, Dermody TS, Ouyang W, Williams IR, Vijay-Kumar M, Gewirtz AT. 2014. Viral infection. Prevention and cure of rotavirus infection via TLR5/NLRC4-mediated production of IL-22 and IL-18. Science 346:861–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, Antonopoulos D, Umesaki Y, Chervonsky AV. 2013. Gender bias in autoimmunity is influenced by microbiota. Immunity 39:400–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson CM, Wang Y, Pfeiffer JK. 2017. Sex-Dependent Intestinal Replication of an Enteric Virus. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dolowy WC, Muldoon RL. 1964. Studies of Germfree Animals. I. Response of Mice to Infection with Influenza a Virus. Proc Soc Exp Biol Med 116:365–371. [DOI] [PubMed] [Google Scholar]
- 43.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, Iwasaki A. 2011. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A 108:5354–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, Paley MA, Antenus M, Williams KL, Erikson J, Wherry EJ, Artis D. 2012. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 37:158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.David SC, Norton T, Tyllis T, Wilson JJ, Singleton EV, Laan Z, Davies J, Hirst TR, Comerford I, McColl SR, Paton JC, Alsharifi M. 2019. Direct interaction of wholeinactivated influenza A and pneumococcal vaccines enhances influenza-specific immunity. Nat Microbiol doi: 10.1038/s41564-019-0443-4. [DOI] [PubMed] [Google Scholar]
- 46.Rowe HM, Meliopoulos VA, Iverson A, Bomme P, Schultz-Cherry S, Rosch JW. 2019. Direct interactions with influenza promote bacterial adherence during respiratory infections. Nat Microbiol doi: 10.1038/s41564-019-0447-0. [DOI] [PMC free article] [PubMed] [Google Scholar]


