Abstract
CD4+ T follicular helper (TFH) cells provide help to B cells and promote antibody-mediated immune responses. Increasing evidence supports the existence of TFH populations that secrete cytokines typically associated with the effector functions of other CD4+ T cell subsets. These include T helper 1 (TH1)-biased TFH (TFH1) cells that have recognized roles in both immune responses to pathogens and also the pathogenesis of autoimmune disease. Given their apparent importance to human health, there is interest in understanding the mechanisms that regulate TFH1 cell formation and function. However, their origin and the molecular requirements for their differentiation are unclear. Here, we describe a population of murine TH1-derived, TFH1-like cells that express the chemokine receptor Cxcr3 and produce both the TH1 cytokine interferon-γ and the TFH-associated cytokine interleukin-21 (IL-21). Furthermore, these TFH1-like cells promote B cell activation and antibody production at levels indistinguishable from conventional IL-6-derived TFH-like cells. Regarding their regulatory requirements, we find that IL-12 signaling is necessary for the differentiation and function of this TFH1-like cell population. Specifically, IL-12-dependent activation of STAT4, and unexpectedly STAT3, promotes increased expression of IL-21 and the TFH lineage-defining transcription factor Bcl-6 in TFH1-like cells. Taken together, these findings provide insight into the potential origin and differentiation requirements of TFH1 cells.
Subject terms: Gene regulation in immune cells, Transcription
Introduction
Naïve T helper cells differentiate into a number of effector subsets that coordinate pathogen-specific immune responses including T helper 1 (TH1) and T follicular helper (TFH) cell populations. TH1 cells mediate immune responses to intracellular pathogens in large part through the production of the pro-inflammatory cytokine interferon-γ (IFN-γ), while TFH cells aid in antibody-mediated immunity by activating B cells via cognate cell-cell interactions and secretion of the cytokine IL-211–3. As with other T helper cell subsets, the differentiation of TH1 and TFH cells is coordinated by the interplay between cell-extrinsic cytokine signals and cell-intrinsic transcription factor networks. For TH1 cells, IL-12-dependent activation of Signal Transducer and Activator of Transcription 4 (STAT4) drives the expression of the transcriptional regulator T-bet and expression of the TH1 gene program, while IL-6- or IL-21-dependent activation of STAT3 and up-regulation of the transcriptional repressor Bcl-6 play important roles in TFH cell differentiation4–11.
Early work regarding TH1 and TFH cells suggested that static populations of these cells developed in parallel during immune responses. However, it is now generally accepted that many T helper cell subsets are subject to phenotypic plasticity driven by signals from complex cytokine milieus during the course of an immune response12–18. With regard to TFH populations, recent reports from both murine and human studies describe ‘hybrid’ TFH cells that exhibit both B cell helper activity and secretion of effector cytokines normally expressed by other T helper subsets19. These include TH1-biased ‘TFH1’ cells, which are capable of producing the TH1 cytokine IFN-γ in addition to IL-21. These cells have been identified as important responders in murine infection models, and have also been described in humans infected with HIV, mycobacterium tuberculosis, and plasmodium, among other pathogens20–27. Additionally, cell populations phenotypically similar to TFH1 cells have been implicated in the onset of autoimmune disease, including systemic lupus erythematosus28. To date, however, the origin of these cells, including the regulatory mechanisms that direct both their differentiation and function, remains enigmatic.
Here, we describe the step-by-step in vitro differentiation of a murine TH1-derived, TFH1-like cell population that exhibits phenotypic and functional characteristics similar to TFH1 cells observed in vivo, in both murine and human settings. Specifically, these cells express elevated levels of Cxcr3 and are capable of producing both IFN-γ and IL-21. Interestingly, we find that TFH1-like cells provide B cell help similar to conventional TFH-like cells generated in the presence of IL-6. Mechanistically, we find that the differentiation and function of TFH1-like cells requires IL-12-dependent activation of both STAT4 and STAT3, which cooperatively drive the expression of both Bcl-6 and IL-21. Finally, and somewhat surprisingly, we found that while STAT3 activation required signals from IL-12, it was independent of autocrine IL-21 signaling. Taken together, the findings presented here provide insight into the potential origin and differentiation requirements of recently described TFH1 cell populations that have increasingly recognized roles in host immune responses and autoimmune disease.
Results
TH1-derived TFH1-like cells express both T-bet and Bcl-6
Increasing evidence suggests that TFH cells exhibit considerable heterogeneity, and that phenotypically distinct TFH subsets arise in response to diverse immune challenges. These include TH1-biased TFH1 cells, which secrete the cytokines IFN-γ and IL-21 and express the chemokine receptor Cxcr325,29. While TFH1 cells have been observed in a number of clinical and experimental settings in vivo, the regulatory mechanisms underlying their development remain unclear20,22,27,29. A previous study from our laboratory demonstrated that TH1 cells upregulate a TFH-like gene program in response to decreased signals from environmental IL-2 (Supplementary Fig. 1A and17). These findings were in agreement with several other studies demonstrating that IL-2 signaling is a potent repressor of TFH cell differentiation30–32. Given the TH1 origin of this TFH-like population, we sought to determine whether these cells were phenotypically and functionally similar to TFH1 cells described in vivo.
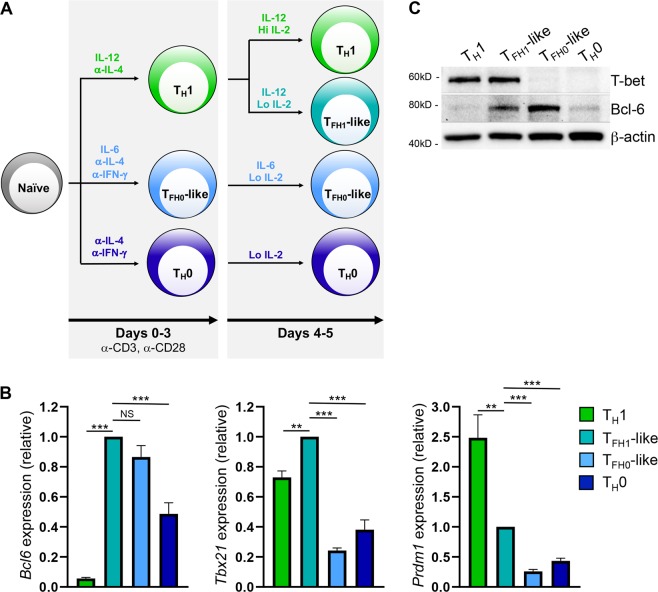
A hallmark feature of TFH cell populations is their elevated expression of the transcriptional repressor Bcl-633–35. As such, we began by comparing Bcl-6 expression in in vitro-differentiated murine TH1 cells, TH1-derived TFH-like (‘TFH1-like’) cells, previously described conventional TFH-like cells differentiated in the presence of IL-6 (‘TFH0-like’), and non-polarized TH0 cells36 (Fig. 1A). Relative to TH1 and TH0 cells, both TFH1-like and TFH0-like populations displayed elevated expression of the TFH lineage-defining transcription factor Bcl-6 at both the transcript and protein level (Fig. 1B,C). In contrast, the Bcl-6 antagonist Blimp-1 was highly expressed only in the TH1 cell population (Fig. 1B). In addition to Bcl-6, TFH1 cells have also been shown to express the TH1 lineage-defining transcription factor T-bet18,37. As such, we next assessed T-bet expression levels across the above cell populations. Indeed, both transcript and protein analyses revealed that only the TFH1-like population expressed both T-bet and Bcl-6 (Fig. 1B,C). Collectively, these data demonstrate that TH1-derived TFH1-like cells express both Bcl-6 and T-bet similar to findings from TFH1 cells observed in vivo18,37.
Figure 1.
TFH1-like cells derived from TH1 cells uniquely co-express Bcl-6 and T-bet. (A) Schematic depicting culturing conditions utilized for the differentiation of the indicated cell populations. Briefly, naïve CD4+ T cells were cultured on plate-bound anti-CD3 and anti-CD28 as follows: TH1 (5 ng/mL IL-12, 5 μg/mL anti-IL-4), TFH0-like (50 ng/mL IL-6, 10 μg/mL anti-IL-4, 10 μg/mL anti-IFN-γ), or TH0 (10 μg/mL anti-IL-4, 10 μg/mL anti-IFN-γ). After 3 days, cells were removed from stimulation and plated under the following conditions: TH1 (5 ng/mL rmIL-12, 2.5 μg/mL anti-IL-4, 500 U/mL rhIL-2), TFH1-like (expanded from TH1 population; 5 ng/mL rmIL-12, 2.5 μg/mL anti-IL-4, 10 U/mL rhIL-2), TFH0-like (10 μg/mL anti-IL-4, 10 μg/mL anti-IFN-γ, 50 ng/mL rmIL-6, 10 U/mL rhIL-2), or TH0 (10 μg/mL anti-IL-4, 10 μg/mL anti-IFN-γ, 10 U/mL IL-2) for an additional 2 days. (B) qRT-PCR was used to assess expression of the indicated genes. The data were normalized to Rps18 and presented as fold change relative to the TFH1-like sample (mean of n = 4–7 ± s.e.m.). **P < 0.01, ***P < 0.001; one-way ANOVA with Tukey multiple-comparison test. (C) Immunoblot analysis of Bcl-6 and T-bet protein expression in the indicated T helper cell populations. β-actin serves as a loading control. Shown is a representative blot of four independent experiments.
TFH1-like cells express elevated levels of Cxcr3, ICOS, and CD40 ligand
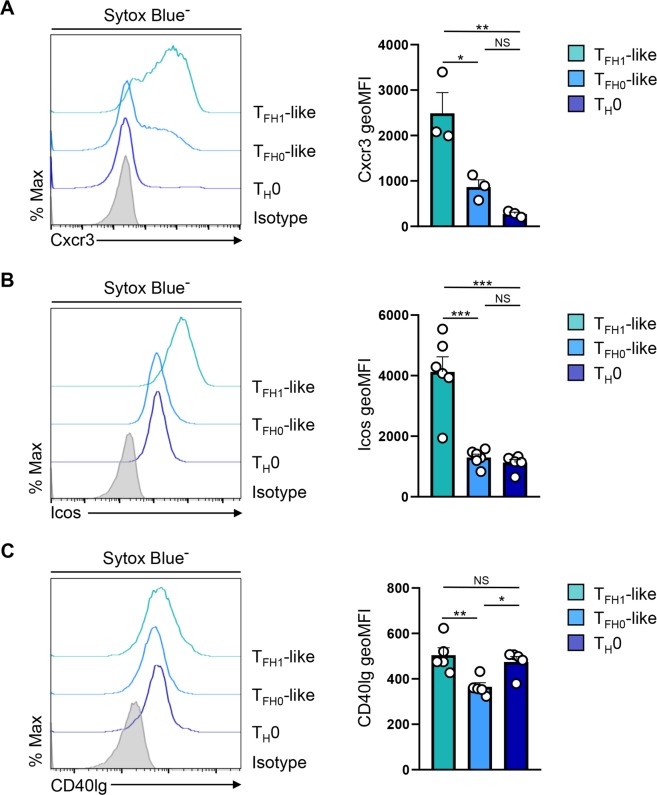
A second distinguishing feature of TFH1 cells found in vivo is elevated surface expression of the chemokine receptor Cxcr324,25. Indeed, gene expression analysis demonstrated that TFH1-like cells had elevated levels of Cxcr3 expression compared to their TH1 cell counterparts (Supplementary Fig. 1B). Therefore, we next used flow cytometric analysis to determine the relative expression of Cxcr3 on the surface of TFH1-like and TFH0-like populations. Indeed, TFH1-like cells exhibited significantly more surface expression of Cxcr3 compared to the TFH0 population (Fig. 2A). Interestingly, two cell surface proteins that are critical for the B cell helper activity of TFH cells, ICOS and CD40 ligand, were also more highly expressed on TFH1-like cells (Fig. 2B,C). To determine whether there were further differences in expression of the TFH gene program between the TFH1- and TFH0-like populations, we performed additional transcript analyses and found that, while there was no significant difference in the expression of Bcl6 or Btla, a number of other TFH-associated genes, including the chemokine receptor Cxcr5, were more highly expressed in the TFH0-like population (Fig. 1A and Supplementary Fig. 2A,B). Conversely, as with ICOS and CD40 ligand, TFH1-like cells expressed significantly higher levels of TFH genes known to be critical for effective B cell helper activity (Supplementary Fig. 2C). Taken together, these findings demonstrate that TFH1-like cells preferentially express Cxcr3 alongside a number of proteins required to provide T cell help to B cells.
Figure 2.
TFH1-like cells express elevated levels of Cxcr3, ICOS, and CD40 ligand. (A) Flow cytometry analysis of Cxcr3 surface expression on the indicated T helper cell populations. Mean fluorescence intensity (MFI) is also shown (mean of n = 3 ± s.e.m.). (B) Flow cytometry analysis of ICOS and CD40 ligand surface expression by the indicated T helper cell populations. Mean fluorescence intensity (MFI) is also shown (mean of n = 5–6 ± s.e.m.). *P < 0.05, **P < 0.01, ***P < 0.001; one-way ANOVA with Tukey multiple-comparison test.
TFH1-like cells produce both IFN-γ and IL-21
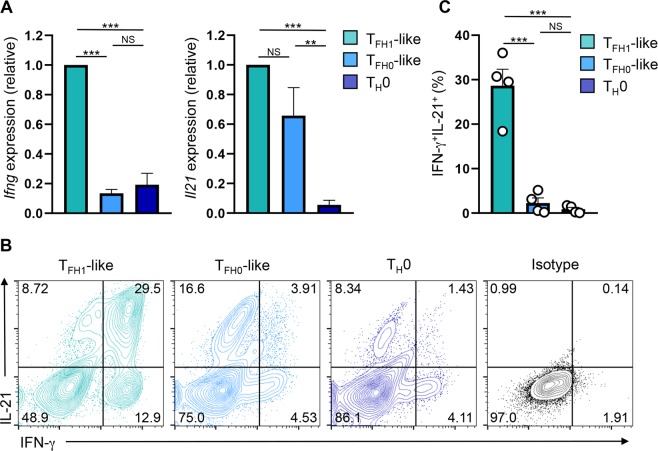
A functional characteristic of TFH1 cell populations is their expression of both IL-21 and IFN-γ29,38. To determine whether TFH1-like cells similarly exhibit this function, we evaluated their ability to simultaneously produce IL-21 and IFN-γ via flow cytometry. While both TFH1-like and TFH0-like populations expressed IL-21, TFH1-like cells were superior producers of IFN-γ (Fig. 3A,B). Importantly, a significantly higher percentage of TFH1-like cells were IFN-γ+IL-21+, as compared to the TFH0-like population (Fig. 3C). Interestingly, we did not observe significant differences in the expression of either IFN-γ or IL-21 between TH1 and TFH1-like cells (Supplementary Fig. 1C). This was interesting, as it has been reported that IFN-γ expression is subject to Bcl-6-dependent repression during the differentiation of conventional TFH cell populations35. Collectively, these data demonstrate that in addition to exhibiting a TFH1 phenotype, TH1-derived TFH1-like cells also exhibit functional characteristics associated with TFH1 cells in the form of dual production of IFN-γ and IL-2118,25.
Figure 3.
TFH1-like cells express both IFN-γ and IL-21. (A) qRT-PCR analysis of Ifng and Il21 expression in the indicated cell populations following stimulation with PMA/Ionomycin for 2.5 hrs. The data were normalized to Rps18 and presented as fold change relative to the TFH1-like sample (mean of n = 5 ± s.e.m.). (B) Flow cytometry analysis of intracellular expression of IL-21 and IFN-γ in the indicated cell populations. Shown are representative data from four independent experiments. (C) Percentage of IFN-γ+IL-21+ cells as assessed by flow cytometry analysis (mean of n = 4 ± s.e.m.). **P < 0.01, ***P < 0.001; one-way ANOVA with Tukey multiple-comparison test.
TFH1-like cells are capable of activating B cells and inducing antibody production
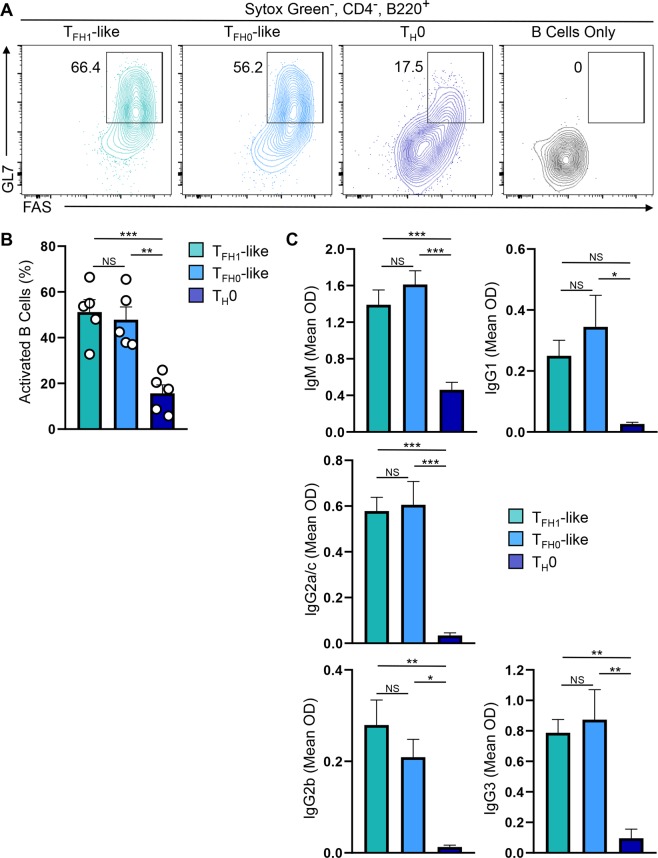
To extend our functional analyses, we next compared B cell helper activity between the two TFH-like populations. Consistent with their increased IL-21 production and expression of TFH cell markers, both TFH-like cell populations were more effective at promoting B cell activation than non-polarized TH0 cells (Fig. 4A,B). Furthermore, co-culture experiments demonstrated that both TFH-like populations were capable of inducing antibody production by B cells, while TH0 cells were poor providers of B cell help (Fig. 4C). Interestingly, we did not observe differences in the ability of TFH0-like and TFH1-like cells to preferentially induce isotype switching. This may be due to the presence of multiple cytokines in the culture media, including both IL-21 and IFN-γ, as well as others that were not analyzed (e.g. IL-10). Regardless, our data support a functional role for TFH1-like cells, similar to that of TFH0-like cells, in supporting B cell antibody production (Fig. 4C). Together, these data demonstrate that TH1-derived TFH1-like cells are capable of performing functions attributed to bona fide TFH cells and, interestingly, are functionally similar to more conventional IL-6-derived TFH cells19,25,27.
Figure 4.
TFH1-like cells activate B cells and induce antibody production. (A) B cells were cultured with the indicated cell population (3:1 B/T ratio) and activation status was assessed by flow cytometry analysis of GL7 and FAS expression. Shown is representative data from five independent experiments. (B) The percentage of activated B cells (FAS+GL7+ cells) as assessed by flow cytometry in ‘A’ (mean of n = 5 ± s.e.m.). (C) ELISA analysis of antibody production by B cells cultured with the indicated cell population (3:1 B/T ratio) for 5 days (mean OD of n = 5 ± s.e.m.). **P < 0.01, ***P < 0.001; one-way ANOVA with Tukey multiple-comparison test.
TFH gene expression patterns and B cell helper activity are dependent upon IL-12
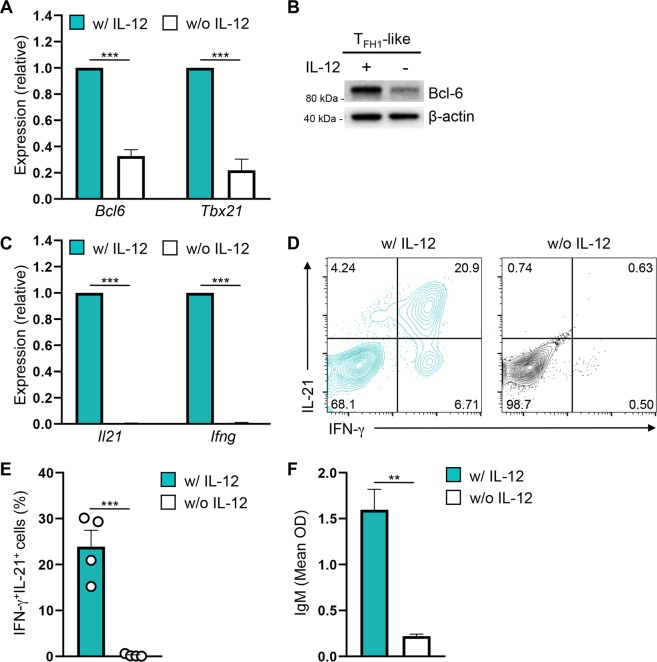
We next sought to determine specific cytokine signals and transcription factors responsible for driving the TFH1-like phenotype. A notable difference between the TFH1-like and TFH0-like cell populations is that the TH1-derived TFH1-like cells are cultured in the presence of IL-12, rather than IL-6. While IL-12 has been reported to be an important factor in the in vitro and in vivo differentiation of human TFH cell populations, the role of IL-12 in promoting murine TFH cell differentiation is less clear39–42. In order to assess the role of IL-12 in TFH1-like cell differentiation, we cultured TFH1-like cells with and without IL-12 and assessed their expression of notable TFH1 cell transcription factors and cell surface receptors. Strikingly, expression of both T-bet and Bcl-6 was significantly reduced in the absence of IL-12 (Fig. 5A,B). Additional analyses revealed that while many TFH genes were unaffected by the loss of IL-12, the expression of the key TFH-associated gene Icos was significantly reduced at both the transcript and protein level (Supplementary Fig. 3).
Figure 5.
IL-12 signaling promotes Bcl-6, IL-21, and ICOS expression in TFH1-like cells. (A) qRT-PCR to assess expression of the indicated genes in TFH1-like cells cultured with (teal bars) or without (white bars) IL-12. The data were normalized to Rps18 and presented as fold change relative to TFH1-like cells cultured with IL-12 (mean of n = 3 ± s.e.m.). (B) Immunoblot analysis of Bcl-6 protein expression in TFH1-like cells cultured with or without IL-12. Shown is a representative blot of three independent experiments. β-actin was used as a loading control. (C) qRT-PCR to assess expression of the indicated genes in TFH1-like cells cultured with (blue bars) or without (white bars) IL-12. The data were normalized to Rps18 and presented as fold change relative to TFH1-like cells cultured with IL-12 (mean of n = 3 ± s.e.m.). (D) Flow cytometry analysis of intracellular expression of IL-21 and IFN-γ in TFH1-like cells cultured with or without IL-12. Shown is representative data from four independent experiments. (E) The percent of IFN-γ+IL-21+ cells as assessed by flow cytometry analysis in ‘D’ (mean of n = 4 ± s.e.m.). (F) ELISA analysis of IgM production by B cells co-cultured with the indicated TFH1-like population at a 3:1 B/T cell ratio for 5 days (mean OD of n = 3 ± s.e.m.). **P < 0.01, ***P < 0.001; unpaired Student’s t-test.
We next wanted to assess the importance of IL-12 signaling to TFH1-like cell function. As such, we assessed the ability of TFH1-like cells cultured in the presence and absence of IL-12 to produce cytokines and induce B cell-mediated antibody production. Importantly, production of IL-21 and IFN-γ, as well as the percentage of IFN-γ+IL-21+ cells, was reduced in the absence of signals from IL-12 (Fig. 5C–E). Furthermore, TFH1-like cells cultured without IL-12 were poor inducers of B cell antibody production as compared to IL-12-cultured controls (Fig. 5F). Taken together, these data demonstrate that IL-12 signaling is a potent inducer of the phenotypic and functional properties of TFH1-like cells.
STAT4 and STAT3 associate with the Bcl6 and Il21 loci downstream of IL-12 signaling
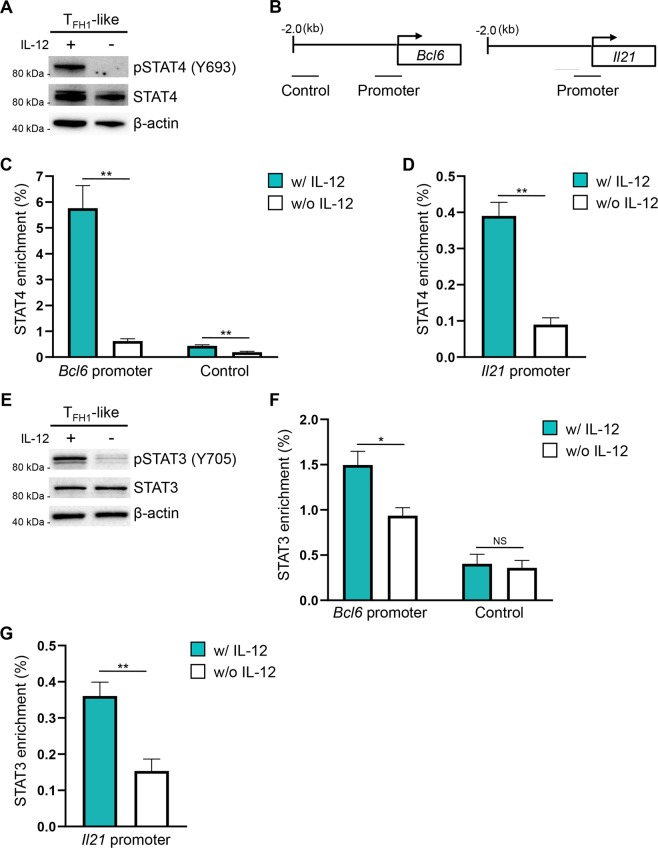
We next sought to identify transcription factors downstream of IL-12 signaling that may regulate expression of key TFH genes in TFH1-like cells. We began by focusing on STAT4, which is activated (phosphorylated) downstream of signals from IL-12 (Fig. 6A). A previous study described STAT4 association with the Bcl6 and Il21 loci42. Indeed, we observed increased STAT4 enrichment at both the Bcl6 and Il21 promoters in TFH1-like cells cultured in the presence of IL-12, as compared to cells cultured without IL-12 (Fig. 6B–D). These findings suggest that STAT4 is a positive regulator of both Bcl-6 and IL-21 expression in TFH1-like cells.
Figure 6.
IL-12 signaling results in increased association of STAT4 and STAT3 with the Bcl6 and Il21 loci. (A) Immunoblot analysis to assess STAT4 activation (pSTAT4 Y693) in TFH1-like cells cultured with or without IL-12. STAT4 and β-actin serve as controls for total STAT4 and equal protein loading, respectively. Shown is a representative blot of three independent experiments. (B) Schematic depicting the location of amplicons within the Bcl6 and Il21 gene loci utilized for ChIP analyses. (C,D) ChIP assays to evaluate STAT4 enrichment at the Bcl6 and Il21 loci in TFH1-like cells cultured with or without IL-12. Data are presented as percent enrichment relative to a “total” input sample (mean of n = 4 ± s.e.m.). (E) Immunoblot analysis of activated STAT3 (pSTAT3 Y705) in TFH1-like cells cultured with or without IL-12. STAT3 and β-actin are shown as controls for total STAT3 and equal protein loading, respectively. Shown is a representative blot from three independent experiments. (F,G) ChIP assays to quantify STAT3 enrichment at the Bcl6 and Il21 loci in TFH1-like cells cultured with or without IL-12. Data are presented as percent enrichment relative to a “total” input sample (mean of n = 4 ± s.e.m.). *P < 0.05, **P < 0.01; unpaired Student’s t-test.
In addition to STAT4, STAT3 has also been identified as an important positive regulator of TFH gene expression5,17,43,44. STAT3 is activated in response to signaling from a number of cytokines including IL-21. Our previous findings demonstrated that IL-12 signaling was a potent driver of IL-21 expression by the TFH1-like population. As such, we considered the possibility that STAT3 activation may also require upstream signals from IL-12 in TFH1-like cells. Indeed, we found that STAT3 activation was reduced in the absence of IL-12 (Fig. 6E). Consistent with these data, we observed decreased STAT3 enrichment at the Bcl6 and Il21 promoters in TFH1-like cells cultured without IL-12 (Fig. 6F,G). We next sought to determine whether the observed STAT3 activation was due to IL-12-dependent, autocrine IL-21 signaling. However, we observed no difference in STAT3 activation or in the expression of Bcl-6 when IL-21R-deficient cells were differentiated under TFH1-like polarizing conditions, suggesting that STAT3 activation downstream of IL-12 signals was independent of IL-21 (Supplementary Figs. 4A–C). Collectively, these data implicate cooperative, IL-12-dependent activities of both STAT4 and STAT3 in the differentiation and functional regulation of TFH1-like cells.
Discussion
Recent work has established that TH1-biased TFH cell populations exist in vivo and that these cells play roles in both healthy immune responses and autoimmune disease20,24,25,28,29. To date, the cytokine signals and transcriptional mechanisms underlying their formation and function are unclear. Here, we demonstrate that in vitro-generated TH1 cells are capable of differentiating into a TFH1-like cell population that exhibits phenotypic and functional characteristics associated with TFH1 cells. Similar to TFH1 cells described in vivo, TFH1-like cells express both the TFH lineage-defining factor Bcl-6 and the TH1 lineage-defining factor T-bet, in addition to the chemokine receptor Cxcr3. Functionally, our findings demonstrate that the TFH1-like cell population is capable of producing both IFN-γ and IL-21 and providing help to B cells that results in B cell activation and antibody production.
While there is a general consensus on the phenotypic and functional properties of TFH1 cells, there is still a debate as to their cellular origin. The findings presented here support a developmental pathway wherein TH1 cells give rise to a hybrid TFH1-like cell state by inducing the expression of Bcl-6 and other aspects of the TFH gene program. In agreement with our findings, a recent report demonstrated that IFN-γ-producing TFH cells generated in vivo require prior expression of T-bet37. Alternatively, it has been suggested that TFH1 cells are conventional TFH cells that gain T-bet expression and the ability to produce IFN-γ29. While our work does not necessarily support this model, it is important to note that our findings also do not exclude this as a potential mechanism. Future studies will be necessary to determine whether a dominant differentiation pathway for TFH1 cells exists or whether there may be multiple origins from which these specialized TFH cell populations arise.
Our current findings also provide mechanistic insight into the potential cytokine signals and downstream transcriptional mechanisms required for the differentiation of TFH1 cells. In this regard, we find that the expression of a number of genes that promote induction of the TFH1-like cell state is dependent upon signals from the cytokine IL-12. Specifically, we find that IL-12 is required for the elevated expression of Bcl-6 and ICOS that is observed in the TFH1-like population, as well as the production of IFN-γ and IL-21. These data are in agreement with previous work in human cells demonstrating that IL-12 is required for the development of a CD4+ T cell population that co-expresses IFN-γ and IL-2126. Furthermore, our findings are also in agreement with work identifying a role for IL-12 in positively regulating the development and function of human TFH cells in both in vitro and in vivo settings40,41,43,45.
Interestingly, while the role of IL-12 in human TFH cell development is relatively well defined, the requirement for IL-12 signals in murine TFH differentiation is less clear16,18,42. As a potential explanation for this discrepancy, we find that while IL-12 signaling in the form of STAT4 and STAT3 activation is required to drive the expression of a subset of TFH genes, including Bcl6, Icos, and Il21, signals from IL-12 appear to play a complementary but secondary role to those derived from IL-217. Thus, our findings support a model wherein combined IL-12 and IL-2 signals drive TH1 cell differentiation, while IL-12 signals in the absence of strong IL-2 signaling promote the alternative TFH1-like phenotype. Indeed, our previous work, and that of others, has demonstrated that the IL-2/STAT5 signaling axis functions as a potent negative regulator of TFH cell development17,30–32.
Intriguingly, our work implicates an IL-12/STAT4/STAT3 signaling axis in the positive regulation of TFH1 cell development, as IL-12 signaling is required for both STAT4 and STAT3 activation in the TFH1-like population. In agreement with these findings, STAT4 activation has been previously implicated in the generation of IFNγ-producing T-bet+Bcl-6+ TFH cells generated in response to viral infection18. Though our findings suggest that IL-12-dependent activation of STAT3 promotes the expression of the TFH1 phenotype, the identity of the signals that drive the phosphorylation of STAT3 remain unclear. Classically, IL-12 signaling has been associated with STAT4 activation. However, previous work has indicated that IL-12 signals are also capable of inducing STAT3 activation46. We also considered the possibility that STAT3 activation may arise from autocrine IL-21 signaling. However, experiments with IL-21R−/− T cells demonstrated that STAT3 activation is independent of IL-21. Thus, whether STAT3 activation is occurring directly downstream of IL-12 signaling, or downstream of signals from an IL-12-dependent cytokine, is as yet unclear.
Collectively, the work presented here supports a possible TH1 origin for the TFH1 cells observed in vivo, and also provides insights into the regulatory requirements that govern their development. Given their role in infection and autoimmune disease, a better understanding of such regulatory requirements may identify potential therapeutic targets, which will allow for more selective manipulation of TFH1 cell populations in efforts to treat human disease.
Methods
Primary cells and cell culture
All mouse strains [C57BL/6J, C57BL/6NJ, IL-21R−/− (C57BL/6NJ background)] were obtained from the Jackson Laboratory. Naïve CD4+ T cells were purified from the spleens and lymph nodes of 5–8 week old mice via negative selection using the BioLegend Mojosort kit according to the manufacturer’s instructions. For all experiments, cells were cultured in complete IMDM (“cIMDM”: IMDM [Life Technologies], 10% FBS [Life Technologies], 1% Penicillin-Streptomycin [Life Technologies], and 50 μM β-mercaptoethanol [Sigma-Aldrich]). Following isolation, cells were plated at a density of 3–5 × 105 cells per well and stimulated using plate-bound anti-CD3ε (5 µg/ml; BD Biosciences) and anti-CD28 (10 μg/ml; BD Biosciences) under the following polarizing conditions: TH1 (5 ng/mL rmIL-12 [R&D Systems], 5 μg/mL anti-IL-4), TFH0-like (10 μg/mL anti-IFN-γ, 10 μg/mL anti-IL-4, 50 ng/mL rmIL-6 [R&D Systems]), TH0 (10 μg/mL anti-IFN-γ [XMG1.2; BioLegend], 10 μg/mL anti-IL-4 [11B11; BioLegend]). After 3 days, cells were removed from stimulation and expanded to plate at 5–7 × 105 cells/well in fresh media under the following conditions: TH1 (5 ng/mL rmIL-12, 2.5 μg/mL anti-IL-4, 500 U/mL rhIL-2 [NIH]), TFH1-like (expanded from TH1 population; 5 ng/mL rmIL-12, 2.5 μg/mL anti-IL-4, 10 U/mL rhIL-2), TFH0-like (10 μg/mL anti-IFN-γ, 10 μg/mL anti-IL-4, 50 ng/mL rmIL-6, 10 U/mL rhIL-2), TH0 (10 μg/mL anti-IFN-γ, 10 μg/mL anti-IL-4, 10 U/mL IL-2) for an additional 48 h. Where indicated, IL-12 was omitted from the TFH1-like culturing conditions. The Institutional Animal Care and Use Committee of Virginia Tech approved all experiments involving the use of mice. All methods were performed in accordance with the approved guidelines.
T and B cell co-culture and analysis of helper activity
B cells were purified from the spleens and lymph nodes of age- and sex-matched 5–8 week old C57BL/6J mice using the MojoSort Mouse Pan B cell isolation kit (BioLegend), according to the manufacturer’s instructions. For each indicated population, 1 × 105 T cells were mixed with B cells at a 1:3 T cell:B cell ratio and stimulated using plate-bound anti-CD3ε (5 μg/mL) under TFH1-like or TFH0-like conditions. Where indicated, IL-12 was omitted from the TFH1-like conditions. T and B cells were co-cultured for either 2 days, at which point B cell activation was analyzed by flow cytometry, or 4 days, where supernatant was collected for ELISA analysis of antibody production.
Antibody production was measured using the BD Pharmingen Mouse Immunoglobulin Isotyping ELISA kit according to the manufacturer’s instructions. OD450 values were calculated by subtracting OD450 readings taken from supernatants from B cells cultured alone in the indicated polarizing conditions from the OD450 values of co-cultured samples.
RNA isolation and qRT-PCR
RNA was purified using the NucleoSpin RNA Kit (Macherey-Nagel). Complementary DNA was generated using the Superscript IV First Strand Synthesis System (Thermo Fisher). qRT-PCR reactions were performed with 10–20 ng cDNA per reaction, gene-specific primers (Rps18 forward: 5′-GGAGAACTCACGGAGGATGAG-3′, Rps18 reverse: 5′-CGCAGCTTGTTGTCTAGACCG-3′; Bcl6 forward: 5′-CCAACCTGAAGACCCACACTC-3′, Bcl6 reverse: 5′-GCGCAGATGGCTCTTCAGAGTC-3′; Tbx21 forward: 5′-GTGACTGCCTACCAGAACGC-3′, Tbx21 reverse: 5′-AGGGGACACTCGTATCAACAG-3′; Il21 forward: 5′-TGGATCCTGAACTTCTATCAGCTCC-3′, Prdm1 forward: 5′-CTTGTGTGGTATTGTCGGGAC-3′, Prdm1 reverse: 5′-CACGCTGTACTCTCTCTTGG-3′; Il21 reverse: 5′-AGGCAGCCTCCTCCTGAGC-3′; Ifng forward: 5′-CTACCTTCTTCAGCAACAGC-3′, Ifng reverse: 5′-GCTCATTGAATGCTTGGCGC-3′; Il21 forward: 5′- TGGATCCTGAACTTCTATCAGCTCC, Il21 reverse: 5′- AGGCAGCCTCCTCCTGAGC; Cxcr5 forward: 5′-GTACCTAGCCATCGTCCATGC-3′, Cxcr5 reverse: 5′-GTGCACTGTGGTAAGGAGTCG-3′; Btla forward: 5′-CATCCCAGATGCCACCAATGC-3′, Btla reverse: 5′-CAGAAAGCAGAGCAGGCAGAC-3′; Icos forward: 5′-CTCACCAAGACCAAGGGAAGC-3′, Icos reverse: 5′-CCACAACGAAAGCTGCACACC-3′; Cd40l forward: 5′-AGCCAACAGTAATGCAGCATCCG-3′, Cd40l reverse: 5′-AGCCAGAGGCCGACGATGAATG-3′; Sh2d1a forward: 5′-CTGGATGGAAGCTATCTGCTG-3′, Sh2d1a reverse: 5′-CAGGTGCTGTCTCGGCACTCC-3′; Il6ra forward: 5′-CCACATAGTGTCACTGTGCG-3′, Il6ra reverse: 5′-GGTATCGAAGCTGGAACTGC-3′; Tnfsf8 forward: 5′-GCAGCTACTTCTACCTCAGCAC-3′, Tnfsf8 reverse: 5′-GTGCCATCTTCGTTCCATGACAG-3′; Pdcd1 forward: 5′-CGTCCCTCAGTCAAGAGGAG-3′, Pdcd1 reverse: 5′-GTCCCTAGAAGTGCCCAACA-3′; Cxcr3 forward: 5′- CCTTGAGGTTAGTGAACGTC-3′, Cxcr3 reverse: 5′-GCTGGCAGGAAGGTTCTGTC-3′) and SYBR Select Mastermix (Life Technologies). All samples were normalized to Rps18 as a control and are presented either as mRNA relative to Rps18, or as fold change relative to the control sample, as indicated. To measure Il21 and Ifng transcript expression, the indicated T helper populations were re-stimulated with PMA and Ionomycin for 2.5–4 h.
Immunoblot analyses
Immunoblot analyses were performed as described previously44. Antibodies used to detect chosen proteins were as follows: Bcl-6 (1:500, BD Pharmingen), T-bet (1:1,000, Santa Cruz), pSTAT3 Y705 (1:20,000, Abcam), STAT3 (1:5000, Santa Cruz), pSTAT4 Y693 (1:1000, Cell Signaling), and STAT4 (1:2500, BioLegend). For all experiments, β-actin (1:15,000, GenScript) expression was monitored to ensure equivalent protein loading. Original and uncropped images of immunoblots can be found in Supplementary Fig. 5.
Flow cytometry
For extracelluar epitopes, cells were harvested and washed 1x with FACS buffer (PBS w/2% FBS, 1% BSA) prior to staining with flurochrome-conjugated anti-ICOS PE (eBioscience), anti-CD40lg PE (Biolegend), anti-Cxcr3 PE (Biolegend), anti-FAS BV421 (BD Bioscience), anti-GL7 AF488 (BD Bioscience), anti-CD4 AF488 (R&D), anti-B220 PECy7 (BD Bioscience), anti-IL21R PE (Biolegend), or the appropriate isotype control. Cells were incubated for 1 hour at room temperature and subsequently washed 2x with FACS buffer. Cell viability was determined by incubating the cells with either Sytox Blue (ThermoFisher) or Sytox Green (ThermoFisher). Samples containing B cells were subjected to Fc block anti-CD16/CD32 (Invitrogen). For cytokine staining, cells were treated with Golgi stop (BD Bioscience) and restimulated with PMA and Ionomycin for 2 h. Cells were fixed and permeablized using the BD Cytofix/Cytoperm kit as detailed by the manufacturer. Cells were incubated with IL-21R subunit/Fc chimera (R&D Systems), washed with Perm/Wash buffer (BD), stained with F(ab’)2 anti-Human IgG Fc R-PE (Life Technologies), washed, and finally stained with anti-IFN-γ AF700 (R&D Systems). All incubations were carried out for 30 minutes at 4 °C. Goat F(ab’)2 IgG R-PE (Life Technologies) and Rat IgG2a AF700 (R&D Systems) were used as isotype controls. Samples were analyzed on the BD Accuri C6 or the Sony SH800 flow cytometers and data evaluated using Flowjo software.
ChIP
Chromatin was prepared from the indicated T helper cell population as previously described17. The resulting chromatin was incubated with either anti-STAT4 (Santa Cruz), anti-STAT3 (Santa Cruz), or control antibody (Abcam) and immunoprecipitated using Protein G Dynabeads (Life Technologies). Precipitated DNA was analyzed via quantitative PCR using SYBR Select Mastermix (Life Technologies) and gene-specific primers: (Bcl6 promoter forward: 5′-GCGGAGCAATGGTAA AGCCC-3′, and reverse: 5′-CTGGTGTCCGGCCTTTCCTAG-3′; Bcl6 control forward: 5′-GTACTCCAACAACAGCACAGC-3′, and reverse: 5′-GTGGCTCGTTAAATCACAGAGG-3′; Il21 promoter forward: 5′-CAC ACACCTTGGTGAATGCTG-3′, and reverse: 5′-CCATTGGCTAGGTGTACGTGTG-3′). Samples were normalized to total input followed by the subtraction of the isotype control to account for unspecific binding.
Statistical analyses
All data represent at least three independent experiments. Error bars represent the standard error of the mean. For statistical analysis, unpaired t tests or one-way ANOVA with Tukey multiple comparison tests were performed to assess statistical significance, as appropriate for a given experiment. P values < 0.05 were considered statistically significant.
Supplementary information
Acknowledgements
This work was supported by grants from The National Institutes of Health grants (AI134972 and AI127800) to K.J.O.
Author Contributions
M.D.P. and K.A.R. performed experiments, analyzed data, and wrote the manuscript. B.K.S. and D.M.J. performed experiments and analyzed data. K.J.O. supervised the research, designed the study, analyzed data, and wrote the manuscript.
Data Availability
The datasets produced in this study will be made available upon reasonable request. Requests should be sent to the corresponding author.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Michael D. Powell and Kaitlin A. Read contributed equally.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-50614-1.
References
- 1.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lazarevic V, Glimcher LH. T-bet in disease. Nat Immunol. 2011;12:597–606. doi: 10.1038/ni.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu J, Paul WE. CD4 T cells: fates, functions, and faults. Blood. 2008;112:1557–1569. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eto D, et al. IL-21 and IL-6 are critical for different aspects of B cell immunity and redundantly induce optimal follicular helper CD4 T cell (Tfh) differentiation. PloS one. 2011;6:e17739. doi: 10.1371/journal.pone.0017739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nurieva RI, et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29:138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poholek AC, et al. In vivo regulation of Bcl6 and T follicular helper cell development. J Immunol. 2010;185:313–326. doi: 10.4049/jimmunol.0904023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsieh CS, et al. Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science. 1993;260:547–549. doi: 10.1126/science.8097338. [DOI] [PubMed] [Google Scholar]
- 8.Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996;382:174–177. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- 9.Manetti R, et al. Natural killer cell stimulatory factor (interleukin 12 [IL-12]) induces T helper type 1 (Th1)-specific immune responses and inhibits the development of IL-4-producing Th cells. J Exp Med. 1993;177:1199–1204. doi: 10.1084/jem.177.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seder RA, Gazzinelli R, Sher A, Paul WE. Interleukin 12 acts directly on CD4+ T cells to enhance priming for interferon gamma production and diminishes interleukin 4 inhibition of such priming. Proc Natl Acad Sci USA. 1993;90:10188–10192. doi: 10.1073/pnas.90.21.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thierfelder WE, et al. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature. 1996;382:171–174. doi: 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- 12.Basu R, Hatton RD, Weaver CT. The Th17 family: flexibility follows function. Immunol Rev. 2013;252:89–103. doi: 10.1111/imr.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geginat J, et al. Plasticity of human CD4 T cell subsets. Frontiers in immunology. 2014;5:630. doi: 10.3389/fimmu.2014.00630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakayamada S, Takahashi H, Kanno Y, O’Shea JJ. Helper T cell diversity and plasticity. Curr Opin Immunol. 2012;24:297–302. doi: 10.1016/j.coi.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinmann AS. Regulatory mechanisms that control T-follicular helper and T-helper 1 cell flexibility. Immunol Cell Biol. 2014;92:34–39. doi: 10.1038/icb.2013.49. [DOI] [PubMed] [Google Scholar]
- 16.Lu KT, et al. Functional and epigenetic studies reveal multistep differentiation and plasticity of in vitro-generated and in vivo-derived follicular T helper cells. Immunity. 2011;35:622–632. doi: 10.1016/j.immuni.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oestreich KJ, Mohn SE, Weinmann AS. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat Immunol. 2012;13:405–411. doi: 10.1038/ni.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weinstein JS, et al. STAT4 and T-bet control follicular helper T cell development in viral infections. J Exp Med. 2018;215:337–355. doi: 10.1084/jem.20170457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin L, et al. Insights Into the Molecular Mechanisms of T Follicular Helper-Mediated Immunity and Pathology. Frontiers in immunology. 2018;9:1884. doi: 10.3389/fimmu.2018.01884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baiyegunhi, O. et al. Frequencies of Circulating Th1-Biased T Follicular Helper Cells in Acute HIV-1 Infection Correlate with the Development of HIV-Specific Antibody Responses and Lower Set Point Viral Load. J Virol92 (2018). [DOI] [PMC free article] [PubMed]
- 21.Carpio VH, et al. IFN-gamma and IL-21 Double Producing T Cells Are Bcl6-Independent and Survive into the Memory Phase in Plasmodium chabaudi Infection. PloS one. 2015;10:e0144654. doi: 10.1371/journal.pone.0144654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, et al. Mycobacterium tuberculosis-Specific IL-21+ IFN-gamma+ CD4+T Cells Are Regulated by IL-12. PloS one. 2016;11:e0147356. doi: 10.1371/journal.pone.0147356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moon, H. et al. Early Development in the Peritoneal Cavity of CD49dhigh Th1 Memory Phenotype CD4+ T Cells with Enhanced B Cell Helper Activity. J Immunol (2015). [DOI] [PubMed]
- 24.Obeng-Adjei N, et al. Circulating Th1-Cell-type Tfh Cells that Exhibit Impaired B Cell Help Are Preferentially Activated during Acute Malaria in Children. Cell reports. 2015;13:425–439. doi: 10.1016/j.celrep.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Velu V, et al. Induction of Th1-Biased T Follicular Helper (Tfh) Cells in Lymphoid Tissues during Chronic Simian Immunodeficiency Virus Infection Defines Functionally Distinct Germinal Center Tfh Cells. J Immunol. 2016;197:1832–1842. doi: 10.4049/jimmunol.1600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu S, et al. IL-12 induced the generation of IL-21- and IFN-gamma-co-expressing poly-functional CD4+ T cells from human naive CD4+ T cells. Cell Cycle. 2015;14:3362–3372. doi: 10.1080/15384101.2015.1093703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zander RA, et al. Th1-like Plasmodium-Specific Memory CD4(+) T Cells Support Humoral Immunity. Cell reports. 2017;21:1839–1852. doi: 10.1016/j.celrep.2017.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jain S, Stock A, Macian F, Putterman C. A Distinct T Follicular Helper Cell Subset Infiltrates the Brain in Murine Neuropsychiatric Lupus. Frontiers in immunology. 2018;9:487. doi: 10.3389/fimmu.2018.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Velu V, Mylvaganam G, Ibegbu C, Amara RR. Tfh1 Cells in Germinal Centers During Chronic HIV/SIV Infection. Frontiers in immunology. 2018;9:1272. doi: 10.3389/fimmu.2018.01272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ballesteros-Tato A, et al. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity. 2012;36:847–856. doi: 10.1016/j.immuni.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. J Exp Med. 2012;209:243–250. doi: 10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nurieva RI, et al. STAT5 protein negatively regulates T follicular helper (Tfh) cell generation and function. J Biol Chem. 2012;287:11234–11239. doi: 10.1074/jbc.M111.324046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnston RJ, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nurieva RI, et al. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu D, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Xin G, et al. Single-cell RNA sequencing unveils an IL-10-producing helper subset that sustains humoral immunity during persistent infection. Nature communications. 2018;9:5037. doi: 10.1038/s41467-018-07492-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fang D, et al. Transient T-bet expression functionally specifies a distinct T follicular helper subset. J Exp Med. 2018;215:2705–2714. doi: 10.1084/jem.20180927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yap XZ, Hustin LSP, Sauerwein RW. TH1-Polarized TFH Cells Delay Naturally-Acquired Immunity to Malaria. Frontiers in immunology. 2019;10:1096. doi: 10.3389/fimmu.2019.01096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma CS, et al. Early commitment of naive human CD4(+) T cells to the T follicular helper (T(FH)) cell lineage is induced by IL-12. Immunol Cell Biol. 2009;87:590–600. doi: 10.1038/icb.2009.64. [DOI] [PubMed] [Google Scholar]
- 40.Schmitt N, et al. IL-12 receptor beta1 deficiency alters in vivo T follicular helper cell response in humans. Blood. 2013;121:3375–3385. doi: 10.1182/blood-2012-08-448902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmitt N, Liu Y, Bentebibel SE, Ueno H. Molecular Mechanisms Regulating T Helper 1 versus T Follicular Helper Cell Differentiation in Humans. Cell reports. 2016;16:1082–1095. doi: 10.1016/j.celrep.2016.06.063. [DOI] [PubMed] [Google Scholar]
- 42.Nakayamada S, et al. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity. 2011;35:919–931. doi: 10.1016/j.immuni.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma CS, et al. Functional STAT3 deficiency compromises the generation of human T follicular helper cells. Blood. 2012;119:3997–4008. doi: 10.1182/blood-2011-11-392985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Read KA, et al. Integrated STAT3 and Ikaros Zinc Finger Transcription Factor Activities Regulate Bcl-6 Expression in CD4+ Th Cells. J Immunol. 2017;199:2377–2387. doi: 10.4049/jimmunol.1700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitt N, et al. The cytokine TGF-beta co-opts signaling via STAT3-STAT4 to promote the differentiation of human TFH cells. Nat Immunol. 2014;15:856–865. doi: 10.1038/ni.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jacobson NG, et al. Interleukin 12 signaling in T helper type 1 (Th1) cells involves tyrosine phosphorylation of signal transducer and activator of transcription (Stat)3 and Stat4. J Exp Med. 1995;181:1755–1762. doi: 10.1084/jem.181.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets produced in this study will be made available upon reasonable request. Requests should be sent to the corresponding author.