Abstract
Type I and type III interferons (IFNs) share several properties in common, including the induction of signaling pathways, the activation of gene transcripts, and immune responses, against viral infection. Recent advances in the understanding of the molecular basis of innate and adaptive immunity have led to the re-examination of the role of these IFNs in autoimmune diseases. To date, a variety of IFN-regulated genes, termed IFN signature genes, have been identified. The expressions of these genes significantly increase in systemic lupus erythematosus (SLE), highlighting the role of type I and type III IFNs in the pathogenesis of SLE. In this review, we first discussed the signaling pathways and the immunoregulatory roles of type I and type III IFNs. Next, we discussed the roles of these IFNs in the pathogenesis of autoimmune diseases, including SLE. In SLE, IFN-stimulated genes induced by IFN signaling contribute to a positive feedback loop of autoimmunity, resulting in perpetual autoimmune inflammation. Based on this, we discussed the use of several specific IFN blocking strategies using anti-IFN-α antibodies, anti-IFN-α receptor antibodies, and IFN-α-kinoid or downstream small molecules, which intervene in Janus kinase (JAK)-signal transducer and activator of transcription (STAT) pathways, in clinical trials for SLE patients. Hopefully, the development of novel regimens targeting IFN signaling pathways will shed light on promising future therapeutic applications for SLE patients.
Keywords: interferon, interferon receptor signaling, dendritic cell, T cell, systemic lupus erythematosus
1. Introduction
The systemic lupus erythematosus (SLE) is characterized by a wide array of immune tolerance breakdown with systemic inflammation involving the dysregulation of immune responses. Due to the complexity of its clinical manifestations, limitations of laboratory examination, and a current lack of effective medication, SLE is one of the most difficult-to-control autoimmune diseases. The incidence of SLE varies around the world, with the highest incidence reported in North America (23.2/100,000 person-years) [1] and the lowest incidence reported in Africa (0.3/100,000 person-years) [2]. In general, European countries have lower incidences of SLE, while Asia, Australasia, and the Americas have higher incidences [3]. For each age and ethnic group, women are more vulnerable to SLE than men, and most cases in women occur in middle adulthood. Although the differences in epidemiology results remain unclear, the pathogenesis of SLE appears to involve a complex interplay of immunological, genetic, and environmental risk factors. Advancements in genome-wide association studies (GWAS) have at least partially elucidated the complex genetic architecture of SLE and have identified differences in risk variants across different continental populations [4,5]. More than 90 risk loci have been identified and have collectively been used to establish several critical pathways involved in SLE pathogenesis, including innate immune responses, lymphocyte activation, and immune complex clearing [6]. In particular, these pathways result from the inadequate clearance of nuclear debris and immune complexes, the dysregulation of the innate immune system driven by innate receptors, such as Toll-like receptors (TLRs) and interferon (IFN) signaling, and aberrant immune responses of the adaptive immune system mediated by B-cell and T-cell signals [7]. In serial gene expression microarray studies, the upregulation of IFN-inducible genes (IFIGs) contributes to over 50% of pathogenesis in human SLE patients [8,9], suggesting a strong association between IFNs and the development of SLE. As current treatments are usually of limited efficacy, it is critical to exploit novel targets related to SLE pathogenesis. Hopes have strongly rested on biological agents since several biological therapies have shown great efficacy in patients with other autoimmune rheumatic diseases. However, biological therapies for the treatment of SLE appear to be relatively unsuccessful, and newly developed biological agents have failed to meet their primary endpoints in large-scale clinical trials [10]. Given the key role of IFNs in the initiation and perpetuation of autoimmune responses in SLE, many efforts have been made to obtain an in-depth understanding of IFNs and the development of targeted SLE therapies through intervening in IFN signaling pathways.
In this review, we have focused on the signaling pathways of type I and type III IFNs and outlined their immune-regulatory function in the pathogenesis of SLE. We also discussed current therapies, which target the signaling pathways of type I and type III IFNs, and their results in large-scale clinical trials in SLE.
2. IFNs and IFN Signaling Pathways
IFNs are critical cytokines involved in the manipulation of complicated antiviral responses. More than 20 forms of IFN have been identified, and these have been further classified into three families–type I, type II, and type III–based on their distinct structures, receptor ligation, and biological activities. IFNs belong to the class II cytokine family (which also includes interleukin (IL)-10, IL-19, IL-20, IL-22, IL-24, and IL-26) and share a conserved structure comprised of six α-helices. The type II IFN family has only one member, IFN-γ, which mediates pro-inflammatory and immunomodulatory functions, unlike type I and type III IFNs. The role of type II IFNs in autoimmunity has been reviewed elsewhere [11] and has not been discussed herein.
2.1. Type I IFNs
Type I IFNs are the largest family of IFN and comprise five classes in humans, namely IFN-α, IFN-β [12], IFN-ω, IFN-ε, and IFN-κ [13]. Of these, the IFN-α class is the largest and can be further divided into 12 subtypes, which encode 13 highly homologous genes clustered on chromosome 9 [14]. The production of type I IFNs is induced by pathogen-associated molecular patterns (PAMPs), composed of evolutionarily conserved molecules like nucleic acids in viruses and nucleic acids, peptidoglycan, flagellin, or lipopolysaccharides (LPS) in bacteria, which are sensed by corresponding pattern recognition receptors (PRRs). PRRs include TLRs, retinoic acid-inducible gene 1 (RIG-I)-like receptors (RLRs), and nucleotide oligomerization domain (NOD)-like receptors (NLRs) [15] (Figure 1). When activated via PRRs, type I IFNs are mainly produced by plasmacytoid dendritic cells (pDCs), although type I IFNs can be produced by almost all nucleated cells [16]. Mechanistically, through the expression of TLR7 or TLR9 in endosomal membranes, pDCs sense the RNA or DNA of pathogens via receptor-mediated endocytosis and initiate an activation chain involving myeloid differentiation factor 88 (MyD88) and downstream transcription factors, such as interferon regulatory factor (IRF)-3 and IRF-7, finally inducing the expression of type I IFNs [17,18]. Additionally, type I IFN can also be produced through TLR-independent cytosolic recognition systems, which are mediated by RNA-sensing molecules. For example, cytosolic RIG-I and melanoma differentiation-associated protein 5 (MDA5) sense viral RNA and promote downstream signaling cascades involving mitochondrial antiviral-signaling protein (MAVS), tumor necrosis factor receptor-associated factor 2 (TRAF2), TRAF6, Fas-associated protein with the death domain (FADD), and receptor-interacting protein-1 (RIP1) [19,20], which eventually lead to the activation of IRF-3, IRF-7, and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB), which in turn leads to the induction of type I IFN genes [21,22]. The production of type I IFNs is mainly controlled by various serial gene transcriptions, especially the IRF system, in a strictly highly ordered process.
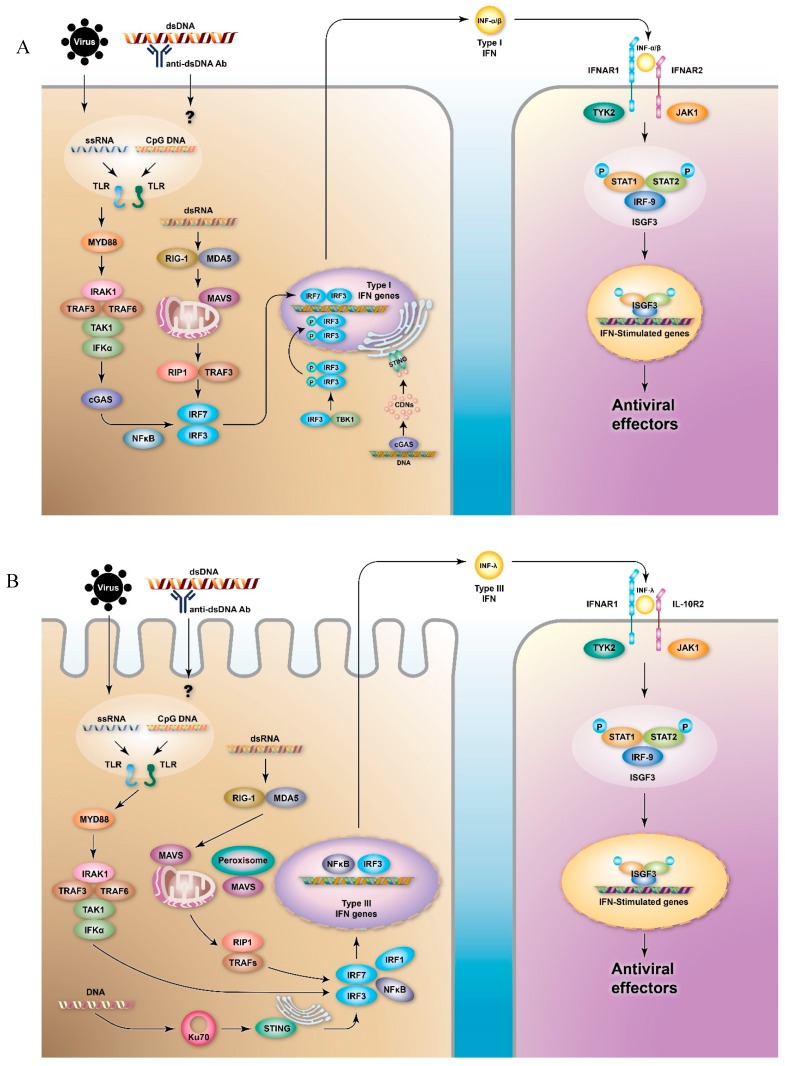
Figure 1.
Signaling pathways of type I and type III interferons (IFNs). The production of (A) type I and (B) type III IFNs can be induced by virus infection or by immune complexes, which are sensed by pattern recognition receptors (PRRs), especially toll-like receptors (TLRs) and retinoic acid-inducible gene 1 (RIG-I)-like receptors (RLRs). Differential signaling molecules lead to the activation of the transcription factors nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) and interferon regulatory factors (IRFs), and eventually to the activation of IFN gene transcripts. The secreted IFNs ligate to the type I or type III IFN receptors (IFN-α/β receptor (IFNAR)1/IFNAR2 or IFN-λ receptor (IFNLR)1/interleukin-10 receptor (IL-10R)2, respectively) of the neighboring cells and stimulate the production of IFN-stimulated genes (ISGs) via Janus kinase (JAK)-signal transducer and activator of transcription (STAT) pathways, which results in the production of several antiviral effectors. Cyclic GMP-AMP synthase (cGAS) transforms DNA into cyclic di-nucleotides (CDNs), which can be recognized by stimulator of interferon genes (STING). STING triggers the activation of tank-binding kinase 1 (TBK1) to phosphorylate IRF-3 and induces the transcription of type I IFNs. In the transcription of type III IFNs, the activation of STING is involved in Ku70.
Another DNA-sensing PRR is involved in the activation of stimulator of interferon genes (STING). STING is a transmembrane protein, which resides in the endoplasmic reticulum (ER) of epithelial and endothelial cells and a variety of hematopoietic cells, such as macrophages and dendritic cells (DCs). When pathogen-related DNA is present in the cytosol of a cell, cyclic GMP-AMP synthase (cGAS) transforms it into cyclic di-nucleotides (CDNs), and the resulting CDNs bind to STING. This triggers the activation of Tank-binding kinase 1 (TBK1), by which TBK1 phosphorylates IRF-3, thus inducing the transcription of type I IFNs [23] (Figure 1A). After the activation of TBK1, STING is phosphorylated by a serine/threonine UNC-51-like kinase, UNC-51-like autophagy activating kinase 1/autophagy-related protein 1 (ULK1/ATG1), and consequently, IRF-3 function is suppressed [24].
All of the produced type I IFNs further exert functions via ligating to a shared heterodimeric receptor, IFN-α/β receptor (IFNAR), which is composed of IFNAR1 and IFNAR2 subunits. The expression of IFNAR is ubiquitous in most nucleated cells; however, the level of expression varies between cell types [25,26]. IFNs can especially interplay with pDC, macrophages, natural killer cells, and T- and B-cells, which increases their survival, maturation, and activity [13] and indicates their pleiotropic effects on the modulation of both the innate and the adaptive immune responses. The engagement of IFNs with IFNAR activates the receptor-associated protein tyrosine kinases Janus kinase 1 (JAK1) and tyrosine kinase 2 (TYK2), which in turn phosphorylates the latent cytoplasmic transcription factors signal transducer and activator of transcription 1 (STAT1) and STAT2. Subsequently, the phosphorylated STAT1 and STAT2 dimerize and translocate to the nucleus, and the STAT1/STAT2 dimers in the nucleus assemble with IRF-9 to form a tri-molecular complex called IFN-stimulated gene factor 3 (ISGF3). Following the activation of the transcription of interferon-stimulated genes (ISGs), ISGF3 further binds to its cognate DNA sequences, IFN-stimulated response elements (ISREs), which results in a cellular antiviral state [27,28] (Figure 1A). Additionally, type I IFN receptors can also activate phosphorylated STAT1 homodimers to bind to gamma-activated sequences (GASs), which induces the activation of pro-inflammatory genes [29] (Figure 2).
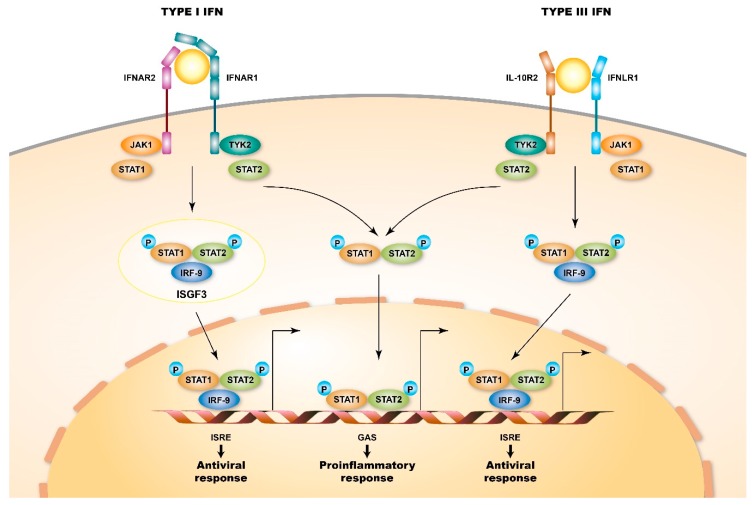
Figure 2.
Signaling pathways of type I and type III IFN receptors. Type I and type III IFN receptors are heterodimers and consist of different receptor chains (IFNAR1 and IFNAR2 for the type I IFN receptor and IFNLR1 and IL10R2 for the type III IFN receptor). Both receptors are associated with two kinases from the JAK family: JAK1 and TYK2 (tyrosine kinase 2). When ligating to their cognate ligand, JAK kinases auto-phosphorylate the IFN receptor, which results in the recruitment of signal transducer and activator of transcription (STAT) proteins, phosphorylation, dimerization, and nuclear translocation. In particular, STAT1, STAT2, and IFN-regulatory factor 9 (IRF9) form interferon-stimulated gene factor 3 (ISGF3) complexes and bind to IFN-stimulated response element (ISRE) sequences, thus activating classical antiviral genes. Additionally, STAT1 homodimers bind to gamma-activated sequences (GASs), thus inducing the activation of pro-inflammatory genes.
2.2. Type III IFNs
Type III IFNs, also called IFN-λs, are the most recently identified members of the IFN family. Type III IFNs are comprised of IFN-λ1 (IL-29), IFN-λ2 (IL-28A), IFN-λ3 (IL-28B) [30], and IFN-λ4 [31]. IFN-λs can be produced by various cell types; however, are mainly produced by pDCs [32,33], as are type I IFNs. However, intestinal epithelial cells (IECs) can preferentially produce IFN-λs [34], and the production of IFN-λs is more abundant at mucosal sites in epithelial and myeloid cells in response to viral infection [35], suggesting a predominant role of IFN-λs in the mucosal immune system response against viral infection. Similar to type I IFNs, IFN-λs are expressed following the detection of PAMPs by PRRs, such as TLRs [36] and RLRs [37]. The sensing of PAMPs by RLRs results in the recruitment of MAVS to mitochondria complemented by peroxisomes, which results in the activation of the transcription factors NF-κB and IRFs and eventually in the induction of the expression of IFN-λs [38] (Figure 1B). Importantly, the cytosolic sensor Ku70 appears to be preferentially involved in the production of IFN-λs rather than type I IFNs [39]. Although the precise mechanism remains unclear, experiments involving the transfection of DNA or herpes simplex virus-2 infection have shown that viral DNA can bind to Ku70, further recruits STING, and subsequently activates downstream IRFs [40] (Figure 1B). Furthermore, IFN-λ can be induced by type I IFNs through ISGs [41]. Besides, the type I and III IFNs that are induced following viral infection can also be temporally regulated. Type I IFNs genes are more rapidly induced and resolved; by contrast, the following IFNL genes are induced in a delayed but sustained fashion [42,43]. The differential expressions of type I and type III IFNs may be involved in the induction of type III IFNs and require the co-activation of both IRF and NF-κB signals [44]. Additionally, the IFN-inducible nature of IFN-λs indicates that these genes can be directly induced by ISGF3 or other IFN-induced transcription factors, such as IRF-1.
Structurally, type III IFN receptors are heterodimeric receptors, unlike type I IFN receptors, and are composed of high-affinity IFN-λ receptor (IFNLR)1 chains (IL-28Rα) and low-affinity IL-10R2 chains. Despite the difference in the structures of type I and type III IFN receptors, the engagement of IFN-λs with type III IFN receptors activates the JAK-STAT signaling cascade, recruits IRF-9, and ultimately activates ISGF3, which drives the transcription of ISGs; this is similar to the engagement of type I IFNs to their cognate type I IFN receptor. Although IFNAR is ubiquitously expressed in most cell types, the expression of IFNLR1 seems to be limited to cells with epithelial origins, such as hepatocytes and intestinal cells [45,46], and to cells with immune origins, such as natural killer (NK) cells, pDCs, and DCs [47,48,49].
3. Immunoregulatory Function of Type I and Type III IFNs
In addition to the primary effects on viral defense, type I IFN signaling is also involved in the activation of innate immune cells and the regulation of adaptive immune responses. Type I IFNs can promote the differentiation of monocytes into mature DCs mediated by granulocyte-macrophage colony-stimulating factor (GM-CSF) [50,51], and further induce the phenotypic maturation of DCs by enhancing the expression of major histocompatibility complex (MHC) class I, MHC class II, and co-stimulatory molecules [52,53], along with cell adhesion molecules to facilitate the migration of DCs into draining lymph nodes [54]. Additionally, IFN-α drives DCs to produce CXC-chemokine ligand 9 (CXCL9) and CXCL10, which chemoattract T cells [55]. Overall, type I IFNs enhance the ability of DCs to prime T cells in secondary lymphoid organs, which facilitates immune responses. Interestingly, when exposed to chronic viral antigens, DCs from persistently infected mice exhibit an immunotolerant phenotype characterized by increased IL-10 production and the increased expression of programmed cell death 1 ligand 1 (PD-L1), which is mediated by persistent type I IFN signaling. When the type I IFN signaling is blocked with anti-IFNAR antibodies, the immunotolerant state is reversed, DCs are further stimulated, and virus-specific CD4+ T cell immunity is enhanced, which consequently improves infection control [56,57]. Altogether, the temporal control of type I IFN signaling affects DC function; short-term IFN exposure promotes the ability of DCs to prime T-cells, while prolonged exposure can induce immune-regulatory DCs to attenuate T cell-mediated immunity against viruses.
IFNAR signaling exerts a dual effect on promoting or inhibiting T cell priming, presumably via a temporal relationship between IFNAR signaling and T cell receptor (TCR) engagement. These opposite effects are involved in different intracellular signaling pathways which are initiated after IFNAR engagement in T cells, whether concomitant with or before TCR signaling [27]. When type I IFN signaling occurs via STAT1, it leads to a pro-inflammatory, anti-proliferative, and pro-apoptotic state; however, when such signaling occurs via STAT3, STAT4, and STAT5, it leads to an anti-inflammatory, pro-proliferative, and anti-apoptotic state, which favors cell survival, proliferation, and differentiation [58,59]. The competition between STAT1 and STAT3/STAT4/STAT5 appears to result from the inability of TCR-activated T cells to phosphorylate STAT1, which leads to type I IFN signals being propagated by other STATs and consequently favors a pro-cell survival and pro-proliferative state [60].
In addition to affecting T cells, type I IFNs also induce DCs to produce B cell stimulatory cytokines, including B-cell activating factor (BAFF) and a proliferation-inducing ligand (APRIL), which cause B cells to undergo immunoglobulin class-switching [61]. Furthermore, the ability of IFN-α to augment the antibody response is greatly reduced by the conditioned deletion of IFNAR1 in B cells since B cells are unable to respond to type I IFNs; this indicates that IFN-α directly acts on B cells in the regulation of antibody responses [62]. In specific conditions, type I IFNs can also regulate the growth and survival of B-cell precursors. Direct IFN-α treatment greatly decreases bone marrow cellularity and splenic cellularity and reduces the number of B lineage cells [63], while anti-IFNAR1 antibody treatment promotes the survival and differentiation of B cells and also neutralizes the production of antibodies against lymphocytic choriomeningitis mammarenavirus (LCMV) infection [64,65]. Over-activated type I IFNs can enhance the survival of IL-6-producing transitional B cells, which has been reported to contribute to the pathogenesis of SLE [66].
Type III IFNs also modulate the responses of T- and B-cells. Studies suggest that type III IFNs can modulate T-cell responses. However, this modulation appears to be indirect and possibly occurs through DCs, since the expression of IFNLR in T cells is minimal and the response of T cells to IFN-λ remains uncertain, although some reports link IFN-λs to the skewing of T cells toward a Th1 phenotype [67]. Moreover, recent studies have shown that CD4 T-cells can express the IFN-λ1-specific receptor IL-28α, which is responsive to IFN-λ1 and further reduces existing Th2 responses by suppressing Th2 cytokines (IL-4 and IL-13) [68,69]. In addition to an increase in the number of Th1-cytokines (e.g., IFN-γ) and suppressed Th2 cytokines (e.g., IL-4, IL-5, IL-13), recombinant IL-λ3 (IL-28B) also inhibits H1N1-stimulated B cell proliferation and IgG production, and the blockade of IL-28Rα augments influenza vaccine responses [70]. In an allergic airway murine model, IL-λ2 (IL-28A) was found to ameliorate disease activity and suppress Th2 and Th17 responses with the induction of IFN-γ. Additionally, in IL-28Rα knockout mice, IL-λ2 was found to exacerbate allergic airway inflammation by augmenting Th2 and Th17 responses and IgE levels [49]. Altogether, these results suggest that IFN-λs have a Th1-skewing property and that specific virus–host and immune cell interactions possibly regulate the type III IFN signaling pathways involved in T cell proliferation and maturation. However, inconsistent results have been found across different models regarding the regulation of B cell responses by type III IFN signaling, and different IFN-λs seem to have different immunoregulatory functions. For example, IFN-λ1 can augment TLR-mediated B cell activation, possibly through the upregulation of TLR7 expression [71]. Furthermore, IFN-λ3 seems to be able to increase the production of antigen-specific anti-HIV antibodies [67]. However, conflicting results suggest that IFN-λ3 can cause an increase in IgG-production and the H1N1-stimulated activation of B cells [70], suggesting that IFN-λs can up- or downregulate B cell responses in specific triggering environments.
4. Type I and Type III IFNs in Autoimmunity
The dysregulated sensing of PAMPs leads to the persistent activation of innate immune signaling and eventually to autoimmune responses. IFN signaling is tightly regulated by several gene transcripts; IFN risk allele variants result in unwanted persistent IFN signaling and autoimmune inflammation, including SLE, rheumatoid arthritis (RA), and vasculitis. Various mechanisms have been proposed to cause sustained type I IFN signaling, and a resultant increase in autoimmunity has been proposed. For example, an impaired RNase H2 complex, which functions as a genome surveillance enzyme, promotes DNA strand breaks and triggers a DNA damage response [72] along with cGAS-dependent type I IFN activation [73]. Interestingly, STING knockout mice aggravate several autoimmune-disease phenotypes of lupus mouse models [74], suggesting an immune-regulatory role of STING in the pathogenesis of SLE. In humans, mutations in TMEM173, which encodes STING, have been linked to familial inflammatory syndrome with lupus-like manifestations [75]. These results suggest genetic contributions of STING to the development of SLE, probably due to aberrancy of IFN activation. Aicardi-Goutières Syndrome (AGS), a severe disease driven by type I IFNs, is characterized by progressive encephalopathy, chilblain-like skin lesions, glaucoma, hypothyroidism, and lupus-like symptoms [76]. Deficiencies in the AGS-associated genes TREX1 and SAMHD1 cause chronic low-level DNA damage and result in the activation of type I IFNs [77,78].
Genes involved in IFN signaling pathways, such as MDA5, TLR7, IRF-5, and IRF-7, are involved in the progression of Sjogren’s syndrome [79,80], a chronic autoimmune inflammatory state that involves focal lymphocytic infiltration into the exocrine glands, leading to dry eyes and mouth. Rheumatoid arthritis (RA) is autoimmune inflammatory arthritis characterized by pathological synovitis (pannus formation) and bone erosion. Genetic variants associated with the type I IFN signaling pathways have been linked with the development and progression of RA, suggesting a role of type I IFNs as a trigger factor for RA development [81]. Furthermore, the level of serum IFN-λ1 is markedly elevated in RA patients [82]. All these studies suggest that an interplay between type I and type III IFNs is involved in the pathogenesis of autoimmune diseases. Interestingly, a study of an animal model of collagen-induced arthritis (a typical mouse model for RA) revealed that treatment with IFN-λ2 reversed the development of arthritis by reducing the number of pro-inflammatory IL-17-producing Th17 and γδ T-cells in the inflamed joints and inguinal lymph nodes as well as by restricting the recruitment of IL-1β-expressing neutrophils [83]. By contrast, in the inflamed intestine of a dextran sodium sulfate (DSS)-induced colitis mouse model, IFN-λ specifically activated a translation-independent signaling pathway that diminished the production of reactive oxygen species and decreased degranulation in neutrophils [84]. Other autoimmune inflammations, such as psoriasis, are characterized by keratinocyte hyperproliferation, loss of skin barrier function, and immune cell infiltration. In psoriatic skin lesions, IFN-λ1 increases the sensitivity of keratinocytes and ISG expression, possibly by mediating Th17 cells [85]. Additionally, elevated IFN-λ expression in psoriatic lesions is associated with the upregulation of the chemokines CXCL10 and CXCL11 compared with non-lesional skin, resulting in epidermal T-cell infiltrations [86].
5. Type I IFNs in SLE
In 1969, it was reported that using an IFN inducer (poly I:C) accelerated autoimmune inflammation and autoantibody production in New Zealand Black and New Zealand White NZB/NZW F1hybrid strains mice, a typical animal model for the study of SLE [87]. Furthermore, in human SLE patients, observational studies have shown that increased serum levels of IFN correlate well to disease activity [88]. Additionally, it has been reported that IFN therapy can induce autoimmunity and cause SLE [89,90]. Furthermore, GWAS of SLE patients have demonstrated that key genetic variants are involved in over-activation or regulatory deficits in the innate immune responses that are closely correlated to type I IFNs, by which IRF-5 and STAT4 risk alleles additively increase the probability of developing SLE [91,92]. Additionally, risk alleles that operate in IFN pathway genes have also been implicated in the pathogenesis of lupus in GWAS [93]. Therefore, dysregulated type I interferon signaling associated with SLE is now recognized as a new family of diseases termed “interferonopathies” [94].
pDCs are the main source of type I IFN production. In SLE, DNA- and RNA-containing immune complexes (ICs) are engulfed by pDCs by ligation to FcγRIIa and are then delivered into the endosomal compartment and sensed by TLR-7 and TLR-9 [95]. Another source of type I IFN production in SLE appears to be neutrophils. One of the defense mechanisms of neutrophils against pathogens is the formation of neutrophil extracellular traps (NETs), which contain primarily nuclear materials, such as dsDNA and histones. Lupus neutrophils can be primed by type I IFNs and release NETs upon exposure to SLE-derived anti-ribonucleoprotein antibodies. In turn, SLE NETs can activate pDCs, thus producing high levels of IFN-α in a TLR9-dependent fashion [96]; this suggests that the interplay between pDC and neutrophils is involved in the production of type I IFNs in SLE (Figure 3).
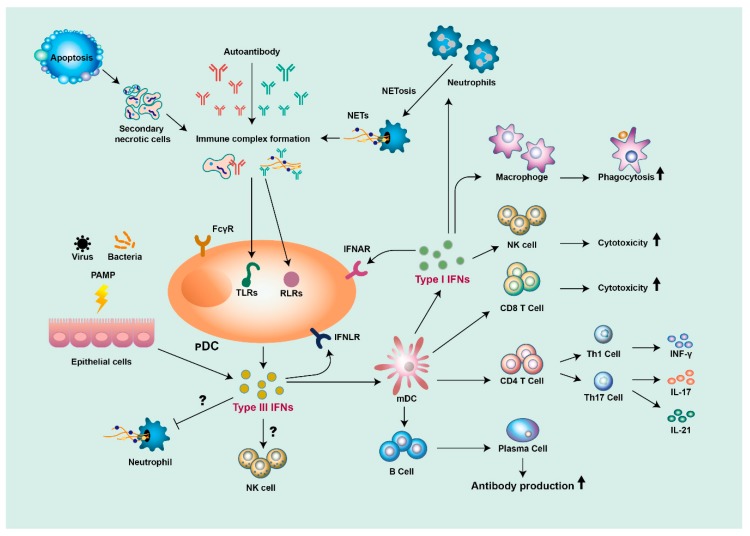
Figure 3.
The roles of type I and type III IFNs in the pathogenesis of SLE (systemic lupus erythematosus). Self-nucleic acid from apoptotic cells, or from neutrophil extracellular traps (NETs) released from neutrophils, are detected by autoantibodies to form immune complexes, stimulating plasmacytoid dendritic cells (pDCs) to produce type I and type III IFNs. Epithelial cells also produce type III IFNs in response to pathogen-associated molecular patterns (PAMPs). Both type I and type III IFNs stimulate myeloid DCs (mDCs) to activate T- and B-cells, which leads to the production of diverse proinflammatory cytokines and autoantibodies. Type I IFNs can also promote cytotoxicity function in macrophages and natural killer (NK) cells. Both type I and type III IFNs can contribute to a positive feedback loop of inflammation.
Although type I IFNs have been regarded as key IFNs in the pathogenesis of SLE, different classes of type I IFNs seem to have different functions in the regulation of SLE. For example, in SLE, IFN-α is a key regulator of autoimmune responses and act on multiple target cells and pathways. For instance, IFN-α stimulates the maturation of myeloid DCs from monocytes and enhances the expression of MHC class I and class II on DCs for antigen presentation [97]. Moreover, IFN-α promotes the development of CD4+ T-cells skewing to Th1 differentiation along with CD8+ T-cell maturation and suppresses the development of Th2, Th17, and regulatory T cells [98,99,100] (Figure 3). Furthermore, IFN-α stimulates the differentiation of B cells and increases the production of autoantibodies through the production of BAFF [79]. In SLE patients, IFN-α can prime monocytes for enhanced NLRP3 inflammasome activity and IL-1β production in an IRF-1-dependent manner [101], and neutrophils for NETosis upon exposure to SLE-derived anti-ribonucleoprotein antibodies [96].
IFN-β is mainly produced by dendritic cells, epithelial cells, and macrophages. In contrast to IFN-α, which is mainly driven by TLR activation, IFN-β is induced through the activation of the STING-IRF-3 pathway driven by the recognition of cytoplasmic nucleic acids by RNA or DNA sensors. Another difference between IFN-α and IFN-β is that IFN-β seems to play an immunoregulatory role in SLE pathogenesis; a study showed that treating patients with multiple sclerosis, an inflammatory demyelinating disease of the central nervous system, with IFN-β reduces the incidence of clinical relapses and brain disease activity [102]. The immunoregulatory function of IFN-β is possibly mediated by increased IL-10 production and the inhibition of inflammasome activation and IL-1 production [103]. Interestingly, circulating B cells from SLE patients show an increased level of IFN-β, especially in SLE patients with renal disease and in those with autoantibodies [104], suggesting the potential involvement of different types of IFN pathways in SLE pathogenesis.
Unlike other type I IFN family members, IFN-κ is selectively expressed in epidermal keratinocytes, and its levels significantly increase upon viral infection [105]. IFN-κ can trigger monocytes and DC to release cytokines and are also particularly effective in inhibiting inducible IL-12 release from monocytes [106]. Although few studies of IFN-κ have focused on SLE, genetic studies have shown that IFN-κ SNPs are associated with increased serum levels of type I IFN in SLE patients [107]. In SLE patients, increased skin production of IFN-κ by lupus keratinocytes drives the overproduction of IL-6 [108], suggesting a specific pathological role of IFN-κ in cutaneous lupus.
6. Type III IFNs in SLE
Although type III IFNs are thought to function as a pathogen defense by maintaining the mucosal immune system, it has also been suggested that type III IFNs are involved in the pathogenesis of SLE. Similar to type I IFNs, serum levels of IFN-λ1 are elevated in SLE patients and, more importantly, are positively correlated with lupus disease activity and anti-dsDNA levels, as well as being negatively correlated with complement level [109], perhaps partially due to an IFN-λ1-mediated increase in the level of Th17-derived cytokines [110] (Figure 3). Notably, elevated serum levels of IFN-λ3 are significantly correlated with higher SLE disease activity and decreased complement level, and genetically, the IFNL3/4 SNP alleles that contain dysfunctional IFNL4 in combination with low production of IFN-λ3 seem to be risk factors for lupus nephritis [111]. Persistently increased levels of IFN-λ have been suggested to be associated with unfavorable histological response in the treatment of lupus nephritis [112]. An SLE observational study found that IFN-λ1 is usually associated with milder disease, while high levels of type I IFNs tend to be associated with severe SLE with nephritis and arthritis [113]. Among the different type III IFNs, IFN-λ3 appears to be associated with the extent of lupus activity and, in particular, with active serosal and cutaneous disease, which may be triggered by anti-Ro/SSA antibodies [114]. Altogether, although the mechanism of the role of IFN-λs in SLE remains unclear, the evidence is accumulating for pathological adjuvant activity in the development and disease progression of SLE.
7. Targeting Type I IFN and IFN Signaling Pathways in SLE
Studies of murine lupus models and serial studies of SLE patients have shown that the IFN pathways are involved in a significant increase in the expression of IFN-regulated genes, the broad enhancement and regulation of immune cells, and autoimmune inflammation in SLE. Targeting IFNs and associated signaling molecules in IFN-mediated pathways has led to important achievements in the development of novel drugs for SLE. Presently, the main development in the search for anti-SLE drugs which target IFN pathways involves targeting type I IFNs, including employing monoclonal antibodies against IFN-α or its receptor IFNAR, or the use of a therapeutic vaccine to induce polyclonal anti-IFN-α neutralizing antibodies. Although therapeutic applications targeting type III IFNs have not yet been developed, trials investigating the use of small molecules to target JAK-STAT pathways are ongoing. IFN-targeting therapies are summarized in Table 1.
Table 1.
Therapies targeting type I interferons (IFNs) and IFN signaling pathways in systemic lupus erythematosus (SLE).
| Therapy | Mechanism of Action | Current Development Stage | Ref. |
|---|---|---|---|
| Rontalizumab | Humanized IgG1 mAb against IFN-α | Phase II, completed | [115] |
| Sifalimumab | Fully human IgG1 against IFN-α | Phase IIb, completed | [116] |
| AGS-009 | Humanized IgG4 mAb against IFN-α | Phase I, completed | [117] |
| Anifrolumab | Fully human IgG1κ mAb against IFNAR1 | Phase II (MUSE trial), completed Phase III (TULIP 1 and 2), completed |
[118] NCT02446899 NCT02446912 |
| IFN-α kinoid (IFN-K) | Therapeutic vaccine of IFN-α2b | Phase I/II, completed | [119] |
| Baricitinib | JAK1/JAK2 inhibitor | Phase II, completed Phase III, recruiting |
[120] NCT03616964 NCT03616912 |
| Tofacitinib | JAK1/JAK2/JAK3 inhibitor | Phase I, completed Phase I/II, recruiting |
NCT02535689 NCT03288324 |
| Ruxolitinib | JAK1/JAK2 inhibitor | Preclinical | [121] |
| CC-930 | JAK1/JAK2/JAK3 inhibitor | Phase II, terminated (discoid lupus) | NCT01466725 |
| GSK2586184 | JAK1 inhibitor | Phase I, completed Phase II, terminated |
NCT01687309 NCT01777256 |
There are three therapeutic monoclonal antibodies, which directly inhibit IFN-α, namely rontalizumab, sifalimumab, and AGS-009. Rontalizumab is a humanized IgG1 monoclonal antibody, which neutralizes IFN-α and has an acceptable safety profile. Rontalizumab was found to cause a sustained decline in the expression of interferon regulated genes (IRGs) in a phase I dose-escalation study of mildly active SLE patients [122]. In the following phase II study (NCT00962832), 238 patients with moderate-to-severe active SLE were recruited and randomized to receive either rontalizumab or placebo [115]. The efficacy response rates between the two groups, which were assessed by the British Isles Lupus Assessment Group (BILAG) index (primary endpoint) and the SLE responder index response (SRI)-4 at week 24 (secondary endpoint), were found to be similar. Therefore, the development of rontalizumab has been discontinued due to failure to meet the primary endpoint. Sifalimumab is a fully human IgG1 monoclonal antibody, which neutralizes IFN-α. It showed both a satisfactory tolerability profile and efficacy profile in a phase I study [123]. In the following phase IIb trial (NCT01283139), 431 patients with moderate-to-severe active SLE were randomized to receive monthly intravenous sifalimumab at a dose of 200, 600, or 1200 mg, or placebo [116]. The results showed that a greater percentage of patients who received sifalimumab (all dosages) met the primary endpoint (percentage of patients achieving an SRI response at week 52) compared to the placebo group. Therefore, sifalimumab may be a promising treatment option for adults with SLE. AGS-009 is a humanized IgG4 monoclonal anti-IFN-α antibody, which has shown satisfactory tolerability profiles in a phase Ia single-dose-escalation study [117].
In addition to directly targeting IFN-α, anifrolumab, a fully human IgG1κ monoclonal antibody directed against IFNAR1, targets the type I IFN receptor. After a successful phase I and early phase II studies, a phase IIb double-blind placebo-controlled randomized control trial of 305 adults with moderate-to-severe SLE (MUSE trial, NCT01438489) showed positive results on the composite primary endpoint SRI-4 [118]. This trial also found that anifrolumab is especially effective for a high baseline IFN gene signature compared with a low baseline IFN gene signature and that anifrolumab showed promising results in patients with cutaneous and arthritic manifestations. Additionally, two phase III trials (TULIP 1 and TULIP 2, NCT02446899 and NCT02446912) assessed the efficacy and safety of anifrolumab in moderately-to-severely active autoantibody-positive SLE patients. In August 2018, the promoter announced that the TULIP1 Phase III trial did not reach its primary endpoint (SRI-4 at week 52); however, the final results are still pending.
Interferon-α-kinoid (IFN-K) is a therapeutic vaccine comprised of IFN-α2b with a carrier protein to induce the production of polyclonal anti-IFN-α neutralizing antibodies. In earlier animal studies, it was found that using IFN-K to immunize human IFN-α transgenic mice could induce the mice to produce antibodies that neutralize IFN-α in sera from SLE patients [124,125]. Moreover, in phase I/II randomized double-blind placebo-controlled dose-escalation study (NCT01058343), IFN-K was found to be well-tolerated and significantly reducing the expression of the IFN signature compared to placebo in 28 patients with mild-to-moderate SLE [119]. However, no differences were observed in disease activity scores (SLEDAI-2K and BILAG) nor were significant changes in baseline levels of C3, C4, or anti-dsDNA levels observed between the groups. Another phase IIb randomized double-blind placebo-controlled study is ongoing to evaluate the effect of IFN-α-kinoid on the neutralization of the IFN gene signature and its clinical efficacy in SLE patients (NCT02665364).
Other candidate targets transduced by type I and type III IFN signaling pathways are involved in JAK-STAT signaling pathways. Type I and type III IFNs signal through JAKs to induce IFN-stimulated gene expression; therefore, several JAK inhibitors used for the treatment of other autoimmune diseases are also under evaluation for their efficacy in treating SLE (Table 1). Additionally, following satisfactory results in phase II trials, two phase III randomized double-blind placebo-controlled parallel-group studies of the use of the JAK1/JAK2 inhibitor baricitinib in SLE patients are in progress (NCT03616964 and NCT03616912) [120].
So far, various agents, which target IFN-α receptors or type I IFN receptors, have failed to meet their primary endpoints in SLE clinical trials. Other than the drugs not being efficacious, several reasons can be proposed for these clinical trial failures, including the clinical heterogeneity of eligible patients, the outcome measurement not being suitable for the clinical trials, the measurement being unable to reflect changes accurately over time, site-investigator bias for the recruitment, or other errors in trial design [126,127]. For rontalizumab, although a phase II study showed safe and well-tolerated profiles in patients with active extrarenal lupus, it did not achieve its primary efficacy endpoint, probably due to a discrepancy in the numbers of SLE patient subgroups (high or low IFN signature metric (ISM)) leading to suboptimal drug efficacy [115]. In a sifalimumab trial, although it demonstrated significant clinical effects across several different validated endpoints and in organ-specific assessments of disease activity [116], further clinical trials were not performed due to the loss of sponsorship. In a phase III study of anifrolumab, TULIP 1 failed to reach its primary goal, with the proportion of patients achieving an SRI-4 at week 52 being similar in the anifrolumab and placebo groups [118]; meanwhile, the final results for TULIP 2 are still pending. It is conceivable that the IFN signatures in the recruited SLE patients restricted the efficacy of anifrolumab on the disease. However, more detailed patient recruitment with genetic background selection may improve trials of the efficacy of anifrolumab to treat SLE.
Other potential future SLE therapies may involve the inhibition of IFN receptor subunits, such as Tyk2. The inhibition of Tyk2 selective inhibitors (BMS-986165) was addressed in early clinical and preclinical investigations [128], and a phase II study is ongoing to characterize the long-term safety and tolerability of Tyk2 selective inhibitors in SLE (NCT03920267). Future SLE therapies may also target the inhibition of other IFN receptor subunits, such as IFNLR1 (IL-28Rα) and IL-10R2; however, to the best of the authors’ knowledge, no clinical trials are ongoing.
8. Concluding Remarks
Type I and type III IFNs were originally investigated for their antiviral activity; however, recent evidence has shown that they also possess an immunomodulatory function in autoimmunity. Type I and type III IFNs share several properties in common; however, there appear to be temporal and kinetic differences in the signaling pathways of the consequent immune pathologies. Despite these differences, both type I and type III IFNs are central mediators in the pathogenesis of SLE. Therefore, novel therapeutic strategies targeting IFNs and IFN signaling pathways are being developed, and these seem promising for modulating disease activity in SLE. In the future, the selection of the most effective drugs for SLE patients may be based on the expression of the IFN signature or an in-depth personalized characterization involving IFN signaling pathways.
Acknowledgments
The authors wish to thank Ingrid Kuo for creating the illustrations used herein.
Author Contributions
Conceptualization, I.-T.C. and J.-Y.C.; writing-original draft preparation, I.-T.C.; writing-review and editing, I.-T.C., H.-T.T., and J.-Y.C.
Funding
This work was supported by grants from the National Science Council, Taiwan (105-2628-B-281-001-MY3) (for Dr. Chyuan) and 107-2314-B-182-059-MY3, CMRPG5F0222, CMRPG5H0022 (for Dr. Chen).
Conflicts of Interest
The authors have no conflicts of interest to declare.
References
- 1.Feldman C.H., Hiraki L.T., Liu J., Fischer M.A., Solomon D.H., Alarcon G.S., Winkelmayer W.C., Costenbader K.H. Epidemiology and sociodemographics of systemic lupus erythematosus and lupus nephritis among US adults with Medicaid coverage, 2000–2004. Arthritis Rheum. 2013;65:753–763. doi: 10.1002/art.37795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor H.G., Stein C.M. Systemic lupus erythematosus in Zimbabwe. Ann. Rheum. Dis. 1986;45:645–648. doi: 10.1136/ard.45.8.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rees F., Doherty M., Grainge M.J., Lanyon P., Zhang W. The worldwide incidence and prevalence of systemic lupus erythematosus: A systematic review of epidemiological studies. Rheumatology (Oxford) 2017;56:1945–1961. doi: 10.1093/rheumatology/kex260. [DOI] [PubMed] [Google Scholar]
- 4.Gorji A.E., Roudbari Z., Alizadeh A., Sadeghi B. Investigation of systemic lupus erythematosus (SLE) with integrating transcriptomics and genome wide association information. Gene. 2019;706:181–187. doi: 10.1016/j.gene.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Deng Y., Tsao B.P. Updates in Lupus Genetics. Curr. Rheumatol. Rep. 2017;19:68. doi: 10.1007/s11926-017-0695-z. [DOI] [PubMed] [Google Scholar]
- 6.Moser K.L., Kelly J.A., Lessard C.J., Harley J.B. Recent insights into the genetic basis of systemic lupus erythematosus. Genes Immun. 2009;10:373–379. doi: 10.1038/gene.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Z., Davidson A. Taming lupus-a new understanding of pathogenesis is leading to clinical advances. Nat. Med. 2012;18:871–882. doi: 10.1038/nm.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Q.Z., Zhou J., Lian Y., Zhang B., Branch V.K., Carr-Johnson F., Karp D.R., Mohan C., Wakeland E.K., Olsen N.J. Interferon signature gene expression is correlated with autoantibody profiles in patients with incomplete lupus syndromes. Clin. Exp. Immunol. 2010;159:281–291. doi: 10.1111/j.1365-2249.2009.04057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baechler E.C., Batliwalla F.M., Karypis G., Gaffney P.M., Ortmann W.A., Espe K.J., Shark K.B., Grande W.J., Hughes K.M., Kapur V., et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc. Natl. Acad. Sci. USA. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Touma Z., Gladman D.D. Current and future therapies for SLE: Obstacles and recommendations for the development of novel treatments. Lupus Sci. Med. 2017;4:e000239. doi: 10.1136/lupus-2017-000239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green D.S., Young H.A., Valencia J.C. Current prospects of type II interferon gamma signaling and autoimmunity. J. Biol. Chem. 2017;292:13925–13933. doi: 10.1074/jbc.R116.774745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taniguchi T., Mantei N., Schwarzstein M., Nagata S., Muramatsu M., Weissmann C. Human leukocyte and fibroblast interferons are structurally related. Nature. 1980;285:547–549. doi: 10.1038/285547a0. [DOI] [PubMed] [Google Scholar]
- 13.Pestka S., Krause C.D., Walter M.R. Interferons, interferon-like cytokines and their receptors. Immunol. Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 14.Honda K., Takaoka A., Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Barrat F.J., Elkon K.B., Fitzgerald K.A. Importance of Nucleic Acid Recognition in Inflammation and Autoimmunity. Annu. Rev. Med. 2016;67:323–336. doi: 10.1146/annurev-med-052814-023338. [DOI] [PubMed] [Google Scholar]
- 16.Mackern-Oberti J.P., Llanos C., Vega F., Salazar-Onfray F., Riedel C.A., Bueno S.M., Kalergis A.M. Role of dendritic cells in the initiation, progress and modulation of systemic autoimmune diseases. Autoimmun. Rev. 2015;14:127–139. doi: 10.1016/j.autrev.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 17.Gilliet M., Cao W., Liu Y.J. Plasmacytoid dendritic cells: Sensing nucleic acids in viral infection and autoimmune diseases. Nat. Rev. Immunol. 2008;8:594–606. doi: 10.1038/nri2358. [DOI] [PubMed] [Google Scholar]
- 18.Smith N., Vidalain P.O., Nisole S., Herbeuval J.P. An efficient method for gene silencing in human primary plasmacytoid dendritic cells: Silencing of the TLR7/IRF-7 pathway as a proof of concept. Sci. Rep. 2016;6:29891. doi: 10.1038/srep29891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K.J., Takeuchi O., Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 20.Xu L.G., Wang Y.Y., Han K.J., Li L.Y., Zhai Z., Shu H.B. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 21.Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 22.Yoneyama M., Kikuchi M., Matsumoto K., Imaizumi T., Miyagishi M., Taira K., Foy E., Loo Y.M., Gale M., Jr., Akira S., et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 23.McCaffary D. STING signalling: An emerging common pathway in autoimmunity and cancer. Immunopharmacol. Immunotoxicol. 2017;39:253–258. doi: 10.1080/08923973.2017.1350704. [DOI] [PubMed] [Google Scholar]
- 24.Konno H., Konno K., Barber G.N. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155:688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Negrotto S.C.J.D.G., Lapponi M.J., Etulain J., Rivadeneyra L., Pozner R.G., Gomez R.M., Schattner M. Expression and functionality of type I interferon receptor in the megakaryocytic lineage. J. Thromb. Haemost. 2011;9:2477–2485. doi: 10.1111/j.1538-7836.2011.04530.x. [DOI] [PubMed] [Google Scholar]
- 26.Katlinski K.V., Gui J., Katlinskaya Y.V., Ortiz A., Chakraborty R., Bhattacharya S., Carbone C.J., Beiting D.P., Girondo M.A., Peck A.R., et al. Inactivation of Interferon Receptor Promotes the Establishment of Immune Privileged Tumor Microenvironment. Cancer Cell. 2017;31:194–207. doi: 10.1016/j.ccell.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Boxel-Dezaire A.H., Rani M.R., Stark G.R. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25:361–372. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 28.Majoros A., Platanitis E., Kernbauer-Holzl E., Rosebrock F., Muller M., Decker T. Canonical and Non-Canonical Aspects of JAK-STAT Signaling: Lessons from Interferons for Cytokine Responses. Front. Immunol. 2017;8:29. doi: 10.3389/fimmu.2017.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stanifer M.L., Pervolaraki K., Boulant S. Differential Regulation of Type I and Type III Interferon Signaling. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20061445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kotenko S.V., Gallagher G., Baurin V.V., Lewis-Antes A., Shen M., Shah N.K., Langer J.A., Sheikh F., Dickensheets H., Donnelly R.P. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 31.Prokunina-Olsson L., Muchmore B., Tang W., Pfeiffer R.M., Park H., Dickensheets H., Hergott D., Porter-Gill P., Mumy A., Kohaar I., et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 2013;45:164–171. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolk K., Witte K., Witte E., Proesch S., Schulze-Tanzil G., Nasilowska K., Thilo J., Asadullah K., Sterry W., Volk H.D., et al. Maturing dendritic cells are an important source of IL-29 and IL-20 that may cooperatively increase the innate immunity of keratinocytes. J. Leukoc. Biol. 2008;83:1181–1193. doi: 10.1189/jlb.0807525. [DOI] [PubMed] [Google Scholar]
- 33.Yin Z., Dai J., Deng J., Sheikh F., Natalia M., Shih T., Lewis-Antes A., Amrute S.B., Garrigues U., Doyle S., et al. Type III IFNs are produced by and stimulate human plasmacytoid dendritic cells. J. Immunol. 2012;189:2735–2745. doi: 10.4049/jimmunol.1102038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pott J., Mahlakoiv T., Mordstein M., Duerr C.U., Michiels T., Stockinger S., Staeheli P., Hornef M.W. IFN-lambda determines the intestinal epithelial antiviral host defense. Proc. Natl. Acad. Sci. USA. 2011;108:7944–7949. doi: 10.1073/pnas.1100552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jewell N.A., Cline T., Mertz S.E., Smirnov S.V., Flano E., Schindler C., Grieves J.L., Durbin R.K., Kotenko S.V., Durbin J.E. Lambda interferon is the predominant interferon induced by influenza A virus infection in vivo. J. Virol. 2010;84:11515–11522. doi: 10.1128/JVI.01703-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ank N., Iversen M.B., Bartholdy C., Staeheli P., Hartmann R., Jensen U.B., Dagnaes-Hansen F., Thomsen A.R., Chen Z., Haugen H., et al. An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. J. Immunol. 2008;180:2474–2485. doi: 10.4049/jimmunol.180.4.2474. [DOI] [PubMed] [Google Scholar]
- 37.Onoguchi K., Yoneyama M., Takemura A., Akira S., Taniguchi T., Namiki H., Fujita T. Viral infections activate types I and III interferon genes through a common mechanism. J. Biol. Chem. 2007;282:7576–7581. doi: 10.1074/jbc.M608618200. [DOI] [PubMed] [Google Scholar]
- 38.Odendall C., Dixit E., Stavru F., Bierne H., Franz K.M., Durbin A.F., Boulant S., Gehrke L., Cossart P., Kagan J.C. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat. Immunol. 2014;15:717–726. doi: 10.1038/ni.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X., Brann T.W., Zhou M., Yang J., Oguariri R.M., Lidie K.B., Imamichi H., Huang D.W., Lempicki R.A., Baseler M.W., et al. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J. Immunol. 2011;186:4541–4545. doi: 10.4049/jimmunol.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sui H., Zhou M., Imamichi H., Jiao X., Sherman B.T., Lane H.C., Imamichi T. STING is an essential mediator of the Ku70-mediated production of IFN-lambda1 in response to exogenous DNA. Sci. Signal. 2017;10 doi: 10.1126/scisignal.aah5054. [DOI] [PubMed] [Google Scholar]
- 41.Ank N., West H., Bartholdy C., Eriksson K., Thomsen A.R., Paludan S.R. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J. Virol. 2006;80:4501–4509. doi: 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolen C.R., Ding S., Robek M.D., Kleinstein S.H. Dynamic expression profiling of type I and type III interferon-stimulated hepatocytes reveals a stable hierarchy of gene expression. Hepatology. 2014;59:1262–1272. doi: 10.1002/hep.26657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin J.D., Feng N., Sen A., Balan M., Tseng H.C., McElrath C., Smirnov S.V., Peng J., Yasukawa L.L., Durbin R.K., et al. Distinct Roles of Type I and Type III Interferons in Intestinal Immunity to Homologous and Heterologous Rotavirus Infections. PLoS Pathog. 2016;12:e1005600. doi: 10.1371/journal.ppat.1005600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomson S.J., Goh F.G., Banks H., Krausgruber T., Kotenko S.V., Foxwell B.M., Udalova I.A. The role of transposable elements in the regulation of IFN-lambda1 gene expression. Proc. Natl. Acad. Sci. USA. 2009;106:11564–11569. doi: 10.1073/pnas.0904477106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sommereyns C., Paul S., Staeheli P., Michiels T. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008;4:e1000017. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mordstein M., Neugebauer E., Ditt V., Jessen B., Rieger T., Falcone V., Sorgeloos F., Ehl S., Mayer D., Kochs G., et al. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J. Virol. 2010;84:5670–5677. doi: 10.1128/JVI.00272-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y., Li T., Chen Y., Wei H., Sun R., Tian Z. Involvement of NK Cells in IL-28B-Mediated Immunity against Influenza Virus Infection. J. Immunol. 2017;199:1012–1020. doi: 10.4049/jimmunol.1601430. [DOI] [PubMed] [Google Scholar]
- 48.Megjugorac N.J., Gallagher G.E., Gallagher G. Modulation of human plasmacytoid DC function by IFN-lambda1 (IL-29) J. Leukoc. Biol. 2009;86:1359–1363. doi: 10.1189/jlb.0509347. [DOI] [PubMed] [Google Scholar]
- 49.Koltsida O., Hausding M., Stavropoulos A., Koch S., Tzelepis G., Ubel C., Kotenko S.V., Sideras P., Lehr H.A., Tepe M., et al. IL-28A (IFN-lambda2) modulates lung DC function to promote Th1 immune skewing and suppress allergic airway disease. EMBO Mol. Med. 2011;3:348–361. doi: 10.1002/emmm.201100142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paquette R.L., Hsu N.C., Kiertscher S.M., Park A.N., Tran L., Roth M.D., Glaspy J.A. Interferon-alpha and granulocyte-macrophage colony-stimulating factor differentiate peripheral blood monocytes into potent antigen-presenting cells. J. Leukoc. Biol. 1998;64:358–367. doi: 10.1002/jlb.64.3.358. [DOI] [PubMed] [Google Scholar]
- 51.Santini S.M., Lapenta C., Logozzi M., Parlato S., Spada M., Di Pucchio T., Belardelli F. Type I interferon as a powerful adjuvant for monocyte-derived dendritic cell development and activity in vitro and in Hu-PBL-SCID mice. J. Exp. Med. 2000;191:1777–1788. doi: 10.1084/jem.191.10.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simmons D.P., Wearsch P.A., Canaday D.H., Meyerson H.J., Liu Y.C., Wang Y., Boom W.H., Harding C.V. Type I IFN drives a distinctive dendritic cell maturation phenotype that allows continued class II MHC synthesis and antigen processing. J. Immunol. 2012;188:3116–3126. doi: 10.4049/jimmunol.1101313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Della Bella S., Nicola S., Riva A., Biasin M., Clerici M., Villa M.L. Functional repertoire of dendritic cells generated in granulocyte macrophage-colony stimulating factor and interferon-alpha. J. Leukoc. Biol. 2004;75:106–116. doi: 10.1189/jlb.0403154. [DOI] [PubMed] [Google Scholar]
- 54.Rouzaut A., Garasa S., Teijeira A., Gonzalez I., Martinez-Forero I., Suarez N., Larrea E., Alfaro C., Palazon A., Dubrot J., et al. Dendritic cells adhere to and transmigrate across lymphatic endothelium in response to IFN-alpha. Eur. J. Immunol. 2010;40:3054–3063. doi: 10.1002/eji.201040523. [DOI] [PubMed] [Google Scholar]
- 55.Padovan E., Spagnoli G.C., Ferrantini M., Heberer M. IFN-alpha2a induces IP-10/CXCL10 and MIG/CXCL9 production in monocyte-derived dendritic cells and enhances their capacity to attract and stimulate CD8+ effector T cells. J. Leukoc. Biol. 2002;71:669–676. [PubMed] [Google Scholar]
- 56.Teijaro J.R., Ng C., Lee A.M., Sullivan B.M., Sheehan K.C., Welch M., Schreiber R.D., de la Torre J.C., Oldstone M.B. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science. 2013;340:207–211. doi: 10.1126/science.1235214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilson E.B., Yamada D.H., Elsaesser H., Herskovitz J., Deng J., Cheng G., Aronow B.J., Karp C.L., Brooks D.G. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science. 2013;340:202–207. doi: 10.1126/science.1235208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanabe Y., Nishibori T., Su L., Arduini R.M., Baker D.P., David M. Cutting edge: Role of STAT1, STAT3, and STAT5 in IFN-alpha beta responses in T lymphocytes. J. Immunol. 2005;174:609–613. doi: 10.4049/jimmunol.174.2.609. [DOI] [PubMed] [Google Scholar]
- 59.Gimeno R., Lee C.K., Schindler C., Levy D.E. Stat1 and Stat2 but not Stat3 arbitrate contradictory growth signals elicited by alpha/beta interferon in T lymphocytes. Mol. Cell Biol. 2005;25:5456–5465. doi: 10.1128/MCB.25.13.5456-5465.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van De Wiele C.J., Marino J.H., Whetsell M.E., Vo S.S., Masengale R.M., Teague T.K. Loss of interferon-induced Stat1 phosphorylation in activated T cells. J. Interferon Cytokine Res. 2004;24:169–178. doi: 10.1089/jir.2004.24.169. [DOI] [PubMed] [Google Scholar]
- 61.Le Bon A., Schiavoni G., D’Agostino G., Gresser I., Belardelli F., Tough D.F. Type interferons potently enhance humoral immunity and can promote isotype switching by stimulating dendritic cells in vivo. Immunity. 2001;14:461–470. doi: 10.1016/S1074-7613(01)00126-1. [DOI] [PubMed] [Google Scholar]
- 62.Le Bon A., Thompson C., Kamphuis E., Durand V., Rossmann C., Kalinke U., Tough D.F. Cutting edge: Enhancement of antibody responses through direct stimulation of B and T cells by type I IFN. J. Immunol. 2006;176:2074–2078. doi: 10.4049/jimmunol.176.4.2074. [DOI] [PubMed] [Google Scholar]
- 63.Lin Q., Dong C., Cooper M.D. Impairment of T and B cell development by treatment with a type I interferon. J. Exp. Med. 1998;187:79–87. doi: 10.1084/jem.187.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fallet B., Narr K., Ertuna Y.I., Remy M., Sommerstein R., Cornille K., Kreutzfeldt M., Page N., Zimmer G., Geier F., et al. Interferon-driven deletion of antiviral B cells at the onset of chronic infection. Sci. Immunol. 2016;1 doi: 10.1126/sciimmunol.aah6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moseman E.A., Wu T., de la Torre J.C., Schwartzberg P.L., McGavern D.B. Type I interferon suppresses virus-specific B cell responses by modulating CD8(+) T cell differentiation. Sci. Immunol. 2016;1 doi: 10.1126/sciimmunol.aah3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu M., Guo Q., Wu C., Sterlin D., Goswami S., Zhang Y., Li T., Bao C., Shen N., Fu Q., et al. Type I interferons promote the survival and proinflammatory properties of transitional B cells in systemic lupus erythematosus patients. Cell Mol. Immunol. 2019;16:367–379. doi: 10.1038/s41423-018-0010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morrow M.P., Pankhong P., Laddy D.J., Schoenly K.A., Yan J., Cisper N., Weiner D.B. Comparative ability of IL-12 and IL-28B to regulate Treg populations and enhance adaptive cellular immunity. Blood. 2009;113:5868–5877. doi: 10.1182/blood-2008-11-190520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dai J., Megjugorac N.J., Gallagher G.E., Yu R.Y., Gallagher G. IFN-lambda1 (IL-29) inhibits GATA3 expression and suppresses Th2 responses in human naive and memory T cells. Blood. 2009;113:5829–5838. doi: 10.1182/blood-2008-09-179507. [DOI] [PubMed] [Google Scholar]
- 69.Jordan W.J., Eskdale J., Srinivas S., Pekarek V., Kelner D., Rodia M., Gallagher G. Human interferon lambda-1 (IFN-lambda1/IL-29) modulates the Th1/Th2 response. Genes Immun. 2007;8:254–261. doi: 10.1038/sj.gene.6364382. [DOI] [PubMed] [Google Scholar]
- 70.Egli A., Santer D.M., O’Shea D., Barakat K., Syedbasha M., Vollmer M., Baluch A., Bhat R., Groenendyk J., Joyce M.A., et al. IL-28B is a key regulator of B- and T-cell vaccine responses against influenza. PLoS Pathog. 2014;10:e1004556. doi: 10.1371/journal.ppat.1004556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.de Groen R.A., Groothuismink Z.M., Liu B.S., Boonstra A. IFN-lambda is able to augment TLR-mediated activation and subsequent function of primary human B cells. J. Leukoc. Biol. 2015;98:623–630. doi: 10.1189/jlb.3A0215-041RR. [DOI] [PubMed] [Google Scholar]
- 72.Gunther C., Kind B., Reijns M.A., Berndt N., Martinez-Bueno M., Wolf C., Tungler V., Chara O., Lee Y.A., Hubner N., et al. Defective removal of ribonucleotides from DNA promotes systemic autoimmunity. J. Clin. Investig. 2015;125:413–424. doi: 10.1172/JCI78001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mackenzie K.J., Carroll P., Lettice L., Tarnauskaite Z., Reddy K., Dix F., Revuelta A., Abbondati E., Rigby R.E., Rabe B., et al. Ribonuclease H2 mutations induce a cGAS/STING-dependent innate immune response. EMBO J. 2016;35:831–844. doi: 10.15252/embj.201593339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sharma S., Campbell A.M., Chan J., Schattgen S.A., Orlowski G.M., Nayar R., Huyler A.H., Nundel K., Mohan C., Berg L.J., et al. Suppression of systemic autoimmunity by the innate immune adaptor STING. Proc. Natl. Acad. Sci. USA. 2015;112:E710–E717. doi: 10.1073/pnas.1420217112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jeremiah N., Neven B., Gentili M., Callebaut I., Maschalidi S., Stolzenberg M.C., Goudin N., Fremond M.L., Nitschke P., Molina T.J., et al. Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J. Clin. Investig. 2014;124:5516–5520. doi: 10.1172/JCI79100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Orcesi S., La Piana R., Fazzi E. Aicardi-Goutieres syndrome. Br. Med. Bull. 2009;89:183–201. doi: 10.1093/bmb/ldn049. [DOI] [PubMed] [Google Scholar]
- 77.Ablasser A., Hemmerling I., Schmid-Burgk J.L., Behrendt R., Roers A., Hornung V. TREX1 deficiency triggers cell-autonomous immunity in a cGAS-dependent manner. J. Immunol. 2014;192:5993–5997. doi: 10.4049/jimmunol.1400737. [DOI] [PubMed] [Google Scholar]
- 78.Maelfait J., Bridgeman A., Benlahrech A., Cursi C., Rehwinkel J. Restriction by SAMHD1 Limits cGAS/STING-Dependent Innate and Adaptive Immune Responses to HIV-1. Cell Rep. 2016;16:1492–1501. doi: 10.1016/j.celrep.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sjostrand M., Johansson A., Aqrawi L., Olsson T., Wahren-Herlenius M., Espinosa A. The Expression of BAFF Is Controlled by IRF Transcription Factors. J. Immunol. 2016;196:91–96. doi: 10.4049/jimmunol.1501061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maria N.I., Steenwijk E.C., AS I.J., van Helden-Meeuwsen C.G., Vogelsang P., Beumer W., Brkic Z., van Daele P.L., van Hagen P.M., van der Spek P.J., et al. Contrasting expression pattern of RNA-sensing receptors TLR7, RIG-I and MDA5 in interferon-positive and interferon-negative patients with primary Sjogren’s syndrome. Ann. Rheum. Dis. 2017;76:721–730. doi: 10.1136/annrheumdis-2016-209589. [DOI] [PubMed] [Google Scholar]
- 81.Rodriguez-Carrio J., Lopez P., Suarez A. Type I IFNs as biomarkers in rheumatoid arthritis: Towards disease profiling and personalized medicine. Clin. Sci. (Lond.) 2015;128:449–464. doi: 10.1042/CS20140554. [DOI] [PubMed] [Google Scholar]
- 82.Wu Q., Yang Q., Sun H., Li M., Zhang Y., La Cava A. Serum IFN-lambda1 is abnormally elevated in rheumatoid arthritis patients. Autoimmunity. 2013;46:40–43. doi: 10.3109/08916934.2012.730587. [DOI] [PubMed] [Google Scholar]
- 83.Blazek K., Eames H.L., Weiss M., Byrne A.J., Perocheau D., Pease J.E., Doyle S., McCann F., Williams R.O., Udalova I.A. IFN-lambda resolves inflammation via suppression of neutrophil infiltration and IL-1beta production. J. Exp. Med. 2015;212:845–853. doi: 10.1084/jem.20140995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Broggi A., Tan Y., Granucci F., Zanoni I. IFN-lambda suppresses intestinal inflammation by non-translational regulation of neutrophil function. Nat. Immunol. 2017;18:1084–1093. doi: 10.1038/ni.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wolk K., Witte K., Witte E., Raftery M., Kokolakis G., Philipp S., Schonrich G., Warszawska K., Kirsch S., Prosch S., et al. IL-29 is produced by T(H)17 cells and mediates the cutaneous antiviral competence in psoriasis. Sci. Transl. Med. 2013;5 doi: 10.1126/scitranslmed.3006245. [DOI] [PubMed] [Google Scholar]
- 86.Witte E., Kokolakis G., Witte K., Warszawska K., Friedrich M., Christou D., Kirsch S., Sterry W., Volk H.D., Sabat R., et al. Interleukin-29 induces epithelial production of CXCR3A ligands and T-cell infiltration. J. Mol. Med. (Berl.) 2016;94:391–400. doi: 10.1007/s00109-015-1367-y. [DOI] [PubMed] [Google Scholar]
- 87.Steinberg A.D., Baron S., Talal N. The pathogenesis of autoimmunity in New Zealand mice, I. Induction of antinucleic acid antibodies by polyinosinic-polycytidylic acid. Proc. Natl. Acad. Sci. USA. 1969;63:1102–1107. doi: 10.1073/pnas.63.4.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hooks J.J., Moutsopoulos H.M., Geis S.A., Stahl N.I., Decker J.L., Notkins A.L. Immune interferon in the circulation of patients with autoimmune disease. N. Engl. J. Med. 1979;301:5–8. doi: 10.1056/NEJM197907053010102. [DOI] [PubMed] [Google Scholar]
- 89.Dumoulin F.L., Leifeld L., Sauerbruch T., Spengler U. Autoimmunity induced by interferon-alpha therapy for chronic viral hepatitis. Biomed. Pharmacother. 1999;53:242–254. doi: 10.1016/S0753-3322(99)80095-X. [DOI] [PubMed] [Google Scholar]
- 90.Ronnblom L.E., Alm G.V., Oberg K.E. Autoimmunity after alpha-interferon therapy for malignant carcinoid tumors. Ann. Intern. Med. 1991;115:178–183. doi: 10.7326/0003-4819-115-3-178. [DOI] [PubMed] [Google Scholar]
- 91.Sigurdsson S., Nordmark G., Garnier S., Grundberg E., Kwan T., Nilsson O., Eloranta M.L., Gunnarsson I., Svenungsson E., Sturfelt G., et al. A risk haplotype of STAT4 for systemic lupus erythematosus is over-expressed, correlates with anti-dsDNA and shows additive effects with two risk alleles of IRF5. Hum. Mol. Genet. 2008;17:2868–2876. doi: 10.1093/hmg/ddn184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Abelson A.K., Delgado-Vega A.M., Kozyrev S.V., Sanchez E., Velazquez-Cruz R., Eriksson N., Wojcik J., Linga Reddy M.V., Lima G., D’Alfonso S., et al. STAT4 associates with systemic lupus erythematosus through two independent effects that correlate with gene expression and act additively with IRF5 to increase risk. Ann. Rheum. Dis. 2009;68:1746–1753. doi: 10.1136/ard.2008.097642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Harley I.T., Kaufman K.M., Langefeld C.D., Harley J.B., Kelly J.A. Genetic susceptibility to SLE: New insights from fine mapping and genome-wide association studies. Nat. Rev. Genet. 2009;10:285–290. doi: 10.1038/nrg2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Crow Y.J. Type I interferonopathies: A novel set of inborn errors of immunity. Ann. N. Y. Acad. Sci. 2011;1238:91–98. doi: 10.1111/j.1749-6632.2011.06220.x. [DOI] [PubMed] [Google Scholar]
- 95.Gonzalez-Navajas J.M., Lee J., David M., Raz E. Immunomodulatory functions of type I interferons. Nat. Rev. Immunol. 2012;12:125–135. doi: 10.1038/nri3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Garcia-Romo G.S., Caielli S., Vega B., Connolly J., Allantaz F., Xu Z., Punaro M., Baisch J., Guiducci C., Coffman R.L., et al. Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci. Transl. Med. 2011;3 doi: 10.1126/scitranslmed.3001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Eloranta M.L., Alm G.V., Ronnblom L. Disease mechanisms in rheumatology--tools and pathways: Plasmacytoid dendritic cells and their role in autoimmune rheumatic diseases. Arthritis Rheum. 2013;65:853–863. doi: 10.1002/art.37821. [DOI] [PubMed] [Google Scholar]
- 98.Curtsinger J.M., Valenzuela J.O., Agarwal P., Lins D., Mescher M.F. Type I IFNs provide a third signal to CD8 T cells to stimulate clonal expansion and differentiation. J. Immunol. 2005;174:4465–4469. doi: 10.4049/jimmunol.174.8.4465. [DOI] [PubMed] [Google Scholar]
- 99.Le Buanec H., Gougeon M.L., Mathian A., Lebon P., Dupont J.M., Peltre G., Hemon P., Schmid M., Bizzini B., Kunding T., et al. IFN-alpha and CD46 stimulation are associated with active lupus and skew natural T regulatory cell differentiation to type 1 regulatory T (Tr1) cells. Proc. Natl. Acad. Sci. USA. 2011;108:18995–19000. doi: 10.1073/pnas.1113301108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Moschen A.R., Geiger S., Krehan I., Kaser A., Tilg H. Interferon-alpha controls IL-17 expression in vitro and in vivo. Immunobiology. 2008;213:779–787. doi: 10.1016/j.imbio.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 101.Liu J., Berthier C.C., Kahlenberg J.M. Enhanced Inflammasome Activity in Systemic Lupus Erythematosus Is Mediated via Type I Interferon-Induced Up-Regulation of Interferon Regulatory Factor 1. Arthritis Rheumatol. 2017;69:1840–1849. doi: 10.1002/art.40166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Interferon beta-1b in the treatment of multiple sclerosis: Final outcome of the randomized controlled trial. The IFNB Multiple Sclerosis Study Group and The University of British Columbia MS/MRI Analysis Group. Neurology. 1995;45:1277–1285. doi: 10.1212/WNL.45.7.1277. [DOI] [PubMed] [Google Scholar]
- 103.Guarda G., Braun M., Staehli F., Tardivel A., Mattmann C., Forster I., Farlik M., Decker T., Du Pasquier R.A., Romero P., et al. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34:213–223. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 104.Hamilton J.A., Wu Q., Yang P., Luo B., Liu S., Li J., A L.M., Sanz I., Chatham W.W., Hsu H.C., et al. Cutting Edge: Intracellular IFN-beta and Distinct Type I IFN Expression Patterns in Circulating Systemic Lupus Erythematosus B Cells. J. Immunol. 2018;201:2203–2208. doi: 10.4049/jimmunol.1800791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.LaFleur D.W., Nardelli B., Tsareva T., Mather D., Feng P., Semenuk M., Taylor K., Buergin M., Chinchilla D., Roshke V., et al. Interferon-kappa, a novel type I interferon expressed in human keratinocytes. J. Biol. Chem. 2001;276:39765–39771. doi: 10.1074/jbc.M102502200. [DOI] [PubMed] [Google Scholar]
- 106.Nardelli B., Zaritskaya L., Semenuk M., Cho Y.H., LaFleur D.W., Shah D., Ullrich S., Girolomoni G., Albanesi C., Moore P.A. Regulatory effect of IFN-kappa, a novel type I IFN, on cytokine production by cells of the innate immune system. J. Immunol. 2002;169:4822–4830. doi: 10.4049/jimmunol.169.9.4822. [DOI] [PubMed] [Google Scholar]
- 107.Harley I.T., Niewold T.B., Stormont R.M., Kaufman K.M., Glenn S.B., Franek B.S., Kelly J.A., Kilpatrick J.R., Hutchings D., Divers J., et al. The role of genetic variation near interferon-kappa in systemic lupus erythematosus. J. Biomed. Biotechnol. 2010;2010 doi: 10.1155/2010/706825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stannard J.N., Reed T.J., Myers E., Lowe L., Sarkar M.K., Xing X., Gudjonsson J.E., Kahlenberg J.M. Lupus Skin Is Primed for IL-6 Inflammatory Responses through a Keratinocyte-Mediated Autocrine Type I Interferon Loop. J. Investig. Dermatol. 2017;137:115–122. doi: 10.1016/j.jid.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu Q., Yang Q., Lourenco E., Sun H., Zhang Y. Interferon-lambda1 induces peripheral blood mononuclear cell-derived chemokines secretion in patients with systemic lupus erythematosus: Its correlation with disease activity. Arthritis Res. Ther. 2011;13:R88. doi: 10.1186/ar3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Oke V., Brauner S., Larsson A., Gustafsson J., Zickert A., Gunnarsson I., Svenungsson E. IFN-lambda1 with Th17 axis cytokines and IFN-alpha define different subsets in systemic lupus erythematosus (SLE) Arthritis Res. Ther. 2017;19:139. doi: 10.1186/s13075-017-1344-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen J.Y., Wang C.M., Chen T.D., Jan Wu Y.J., Lin J.C., Lu L.Y., Wu J. Interferon-lambda3/4 genetic variants and interferon-lambda3 serum levels are biomarkers of lupus nephritis and disease activity in Taiwanese. Arthritis Res. Ther. 2018;20:193. doi: 10.1186/s13075-018-1683-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zickert A., Oke V., Parodis I., Svenungsson E., Sundstrom Y., Gunnarsson I. Interferon (IFN)-lambda is a potential mediator in lupus nephritis. Lupus Sci. Med. 2016;3:e000170. doi: 10.1136/lupus-2016-000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Oke V., Gunnarsson I., Dorschner J., Eketjall S., Zickert A., Niewold T.B., Svenungsson E. High levels of circulating interferons type I, type II and type III associate with distinct clinical features of active systemic lupus erythematosus. Arthritis Res. Ther. 2019;21:107. doi: 10.1186/s13075-019-1878-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Amezcua-Guerra L.M., Marquez-Velasco R., Chavez-Rueda A.K., Castillo-Martinez D., Masso F., Paez A., Colin-Fuentes J., Bojalil R. Type III Interferons in Systemic Lupus Erythematosus: Association Between Interferon lambda3, Disease Activity, and Anti-Ro/SSA Antibodies. J. Clin. Rheumatol. 2017;23:368–375. doi: 10.1097/RHU.0000000000000581. [DOI] [PubMed] [Google Scholar]
- 115.Kalunian K.C., Merrill J.T., Maciuca R., McBride J.M., Townsend M.J., Wei X., Davis J.C., Jr., Kennedy W.P. A Phase II study of the efficacy and safety of rontalizumab (rhuMAb interferon-alpha) in patients with systemic lupus erythematosus (ROSE) Ann. Rheum. Dis. 2016;75:196–202. doi: 10.1136/annrheumdis-2014-206090. [DOI] [PubMed] [Google Scholar]
- 116.Khamashta M., Merrill J.T., Werth V.P., Furie R., Kalunian K., Illei G.G., Drappa J., Wang L., Greth W., CD1067 Study Investigators Sifalimumab, an anti-interferon-alpha monoclonal antibody, in moderate to severe systemic lupus erythematosus: A randomised, double-blind, placebo-controlled study. Ann. Rheum. Dis. 2016;75:1909–1916. doi: 10.1136/annrheumdis-2015-208562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tcherepanova I., Curtis M., Sale M., Miesowicz F., Nicolette C. SAT0193 Results of a randomized placebo controlled phase ia study of AGS-009, a humanized anti-interferon-α monoclonal antibody in subjects with systemic lupus erythematosus. Ann. Rheum. Dis. 2014;71:533–537. doi: 10.1136/annrheumdis-2012-eular.3140. [DOI] [Google Scholar]
- 118.Furie R., Khamashta M., Merrill J.T., Werth V.P., Kalunian K., Brohawn P., Illei G.G., Drappa J., Wang L., Yoo S., et al. Anifrolumab, an Anti-Interferon-alpha Receptor Monoclonal Antibody, in Moderate-to-Severe Systemic Lupus Erythematosus. Arthritis Rheumatol. 2017;69:376–386. doi: 10.1002/art.39962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lauwerys B.R., Hachulla E., Spertini F., Lazaro E., Jorgensen C., Mariette X., Haelterman E., Grouard-Vogel G., Fanget B., Dhellin O., et al. Down-regulation of interferon signature in systemic lupus erythematosus patients by active immunization with interferon alpha-kinoid. Arthritis Rheum. 2013;65:447–456. doi: 10.1002/art.37785. [DOI] [PubMed] [Google Scholar]
- 120.Wallace D.J., Furie R.A., Tanaka Y., Kalunian K.C., Mosca M., Petri M.A., Dorner T., Cardiel M.H., Bruce I.N., Gomez E., et al. Baricitinib for systemic lupus erythematosus: A double-blind, randomised, placebo-controlled, phase 2 trial. Lancet. 2018;392:222–231. doi: 10.1016/S0140-6736(18)31363-1. [DOI] [PubMed] [Google Scholar]
- 121.Chan E.S., Herlitz L.C., Jabbari A. Ruxolitinib Attenuates Cutaneous Lupus Development in a Mouse Lupus Model. J. Investig. Dermatol. 2015;135:1912–1915. doi: 10.1038/jid.2015.107. [DOI] [PubMed] [Google Scholar]
- 122.McBride J.M., Jiang J., Abbas A.R., Morimoto A., Li J., Maciuca R., Townsend M., Wallace D.J., Kennedy W.P., Drappa J. Safety and pharmacodynamics of rontalizumab in patients with systemic lupus erythematosus: Results of a phase I, placebo-controlled, double-blind, dose-escalation study. Arthritis. Rheum. 2012;64:3666–3676. doi: 10.1002/art.34632. [DOI] [PubMed] [Google Scholar]
- 123.Merrill J.T., Wallace D.J., Petri M., Kirou K.A., Yao Y., White W.I., Robbie G., Levin R., Berney S.M., Chindalore V., et al. Safety profile and clinical activity of sifalimumab, a fully human anti-interferon alpha monoclonal antibody, in systemic lupus erythematosus: A phase I, multicentre, double-blind randomised study. Ann. Rheum. Dis. 2011;70:1905–1913. doi: 10.1136/ard.2010.144485. [DOI] [PubMed] [Google Scholar]
- 124.Mathian A., Amoura Z., Adam E., Colaone F., Hoekman M.F., Dhellin O., Vandepapeliere P., Haroche J., Piette J.C., Lebon P., et al. Active immunisation of human interferon alpha transgenic mice with a human interferon alpha Kinoid induces antibodies that neutralise interferon alpha in sera from patients with systemic lupus erythematosus. Ann. Rheum. Dis. 2011;70:1138–1143. doi: 10.1136/ard.2010.141101. [DOI] [PubMed] [Google Scholar]
- 125.Zagury D., Le Buanec H., Mathian A., Larcier P., Burnett R., Amoura Z., Emilie D., Peltre G., Bensussan A., Bizzini B., et al. IFNalpha kinoid vaccine-induced neutralizing antibodies prevent clinical manifestations in a lupus flare murine model. Proc. Natl. Acad. Sci. USA. 2009;106:5294–5299. doi: 10.1073/pnas.0900615106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Medina-Rosas J., Al-Rayes H., Moustafa A.T., Touma Z. Recent advances in the biologic therapy of lupus: The 10 most important areas to look for common pitfalls in clinical trials. Expert Opin. Biol. Ther. 2016;16:1225–1238. doi: 10.1080/14712598.2016.1214263. [DOI] [PubMed] [Google Scholar]
- 127.Rodriguez-Pinto I., Espinosa G., Cervera R. The problems and pitfalls in systemic lupus erythematosus drug discovery. Expert Opin. Drug Discov. 2016;11:525–527. doi: 10.1080/17460441.2016.1181056. [DOI] [PubMed] [Google Scholar]
- 128.Wrobleski S.T., Moslin R., Lin S., Zhang Y., Spergel S., Kempson J., Tokarski J.S., Strnad J., Zupa-Fernandez A., Cheng L., et al. Highly Selective Inhibition of Tyrosine Kinase 2 (TYK2) for the Treatment of Autoimmune Diseases: Discovery of the Allosteric Inhibitor BMS-986165. J. Med. Chem. 2019 doi: 10.1021/acs.jmedchem.9b00444. [DOI] [PubMed] [Google Scholar]