Abstract
In male heterogametic systems, the X Chromosome is epigenetically differentiated between males and females, to facilitate dosage compensation. For example, the X Chromosome in female mammals is largely inactivated. Relative to well-studied male heterogametic systems, the extent of epigenetic differentiation between male and female Z Chromosomes in female heterogametic species, which often lack complete dosage compensation, is poorly understood. Here, we examined the chromosomal DNA methylation landscapes of male and female Z Chromosomes in two distantly related avian species, namely chicken and white-throated sparrow. We show that, in contrast to the pattern in mammals, male and female Z Chromosomes in these species exhibit highly similar patterns of DNA methylation, which is consistent with weak or absent dosage compensation. We further demonstrate that the epigenetic differences between male and female chicken Z Chromosomes are localized to a few regions, including a previously identified male hypermethylated region 1 (MHM1; CGNC: 80601). We discovered a novel region with elevated male-to-female methylation ratios on the chicken Z Chromosome (male hypermethylated region 2 [MHM2]; CGNC: 80602). The MHM1 and MHM2, despite little sequence similarity between them, bear similar molecular features that are likely associated with their functions. We present evidence consistent with female hypomethylation of MHMs and up-regulation of nearby genes. Therefore, despite little methylation differentiation between sexes, extremely localized DNA methylation differences between male and female chicken Z Chromosomes have evolved and affect expression of nearby regions. Our findings offer new insights into epigenetic regulation of gene expression between sexes in female heterogametic systems.
Chromosomal sex determination is widespread in animals, having originated multiple times in different lineages (for reviews, see Bachtrog et al. 2014; Graves 2016). For example, birds have a female-heterogametic sex chromosome system in which males are homozygous for the Z Chromosomes and females are heterozygous for the Z and W Chromosomes. In comparison, eutherian mammals have evolved a male heterogametic system in which females are homozygous for the X Chromosomes and males are heterozygous for the X and the Y Chromosome.
Notwithstanding diversity, several common characteristics are shared among sex chromosome systems, indicating similar underlying evolutionary principles. A critical step during sex chromosome evolution is the cessation of recombination between the proto-sex chromosomes (Charlesworth 1978; Lahn and Page 1999), which can occur in a step-wise fashion, leading to the distinct evolutionary strata of the nonrecombining chromosome (Lahn and Page 1999; Nam and Ellegren 2008; Zhou et al. 2014). As recombination diminishes between the sex chromosomes, the effective population size of the nonrecombining chromosome declines, leading to its genetic degeneration (Charlesworth and Charlesworth 2000; Yi and Charlesworth 2000; Bachtrog 2013). In addition to the degeneration of the nonrecombining chromosome, dosage compensation between homogametic and heterogametic individuals is often observed in sex chromosome systems. In the well-studied male heterogametic system, dosage compensation tends to be achieved in a chromosomal-wide fashion. In fruit flies, transcription of the single male X Chromosome is up-regulated (Vicoso and Bachtrog 2009; Conrad and Akhtar 2012). In mammals, the X Chromosome is largely inactivated in females. For instance, in humans, only ∼15%–25% of genes escape the inactivation and another 10% exhibit levels of inactivation that vary among individuals (Carrel and Willard 2005; Cotton et al. 2015; Graves 2016; Tukiainen et al. 2017). Inactivation is generally achieved via epigenetic mechanisms such as DNA methylation and histone modifications (Hellman and Chess 2007; Sharp et al. 2011; Brockdorff and Turner 2015; Lucchesi and Kuroda 2015; Marin et al. 2017).
Analyses of female heterogametic systems show a picture very different from the XY systems (Mank 2009; Vicoso and Bachtrog 2011; Vicoso et al. 2013). For example, dosage compensation is incomplete in birds (Ellegren et al. 2007; Itoh et al. 2007; Adolfsson and Ellegren 2013; Wang et al. 2014; Mullon et al. 2015; Graves 2016; Marin et al. 2017). Z-linked genes are generally more highly expressed in males (with two Z Chromosomes) than in females (with one Z Chromosome), and only a subset of genes exhibit expression doses that are similar between sexes (Itoh et al. 2007, 2010; Uebbing et al. 2013, 2015). Some female heterogametic species, on the other hand, exhibit global dosage compensation (Smith et al. 2014; Huylmans et al. 2017), indicating greater diversity in the level of dosage compensation in female heterogamety compared to male heterogamety. Consequently, epigenetic differentiation of the Z Chromosomes between sexes in female heterogametic systems is likely regulated by mechanisms different from that of the X Chromosomes in male heterogametic systems. However, data on epigenetic differences between males and females of female heterogametic taxa are scarce.
Nevertheless, several previous studies have suggested a potential role for DNA methylation in avian sex chromosome regulation. For example, the distribution of CpG islands has been shown to have a significant impact on Z-linked gene dosage compensation in chickens (Melamed and Arnold 2009). In addition, earlier targeted studies have identified a region of the chicken Z Chromosome that is differentially methylated between males and females. DNA methylation in this region, previously referred to as the “male hypermethylated region (MHM),” is higher in males than in females (Teranishi et al. 2001). Genes near the MHM locus display pronounced reduction in male-to-female expression ratios in Galloanserae (landfowl and waterfowl) (Melamed and Arnold 2007; Mank and Ellegren 2009; Melamed et al. 2009; Wright et al. 2015). However, in the absence of chromosome-wide epigenetic profiling, it was unknown whether this region represents the only epigenetic locus of differentiation between the sexes or whether it originated by male hypermethylation (as assumed) or female hypomethylation.
To fill this gap of knowledge, here, we analyzed whole-genome methylation and chromatin accessibility maps from chickens (Gallus gallus) and white-throated sparrows (Zonotrichia albicollis), two avian species that have diverged approximately 90 million years ago (Jarvis et al. 2014; Zhou et al. 2014). We also investigated the evolutionary origins of epigenetic divergence between sexes and its potential impacts on gene expression, by examining transcriptome data from 130 chicken samples and 16 samples of outgroup species. The results from these analyses provide novel insights into how gene dosage on the Z and W Chromosomes of female heterogametic systems are epigenetically regulated.
Results
DNA methylation and transcription in the chicken genome
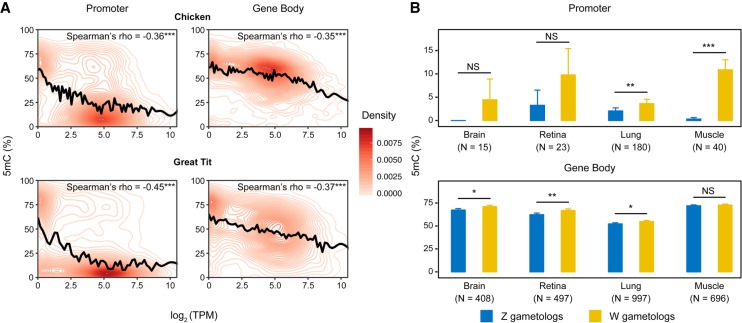
Studies in mammalian systems have contributed to a model of DNA methylation in which methylation of cis-regulatory regions (promoters and enhancers) reduces transcription of associated genes (Schübeler 2015). This model predicts a negative correlation between promoter methylation and gene expression. This prediction has been supported by research in mammals (Schübeler 2015), but it has yet to be fully explored in avian systems. To test the relationship between DNA methylation and expression in birds, we compared brain whole-genome DNA methylation maps with transcriptome data from the same developmental stage in chicken (embryonic day 18 [E18]) (Uebbing et al. 2015; Lee et al. 2017); we also compared methylation with expression in an adult great tit (Laine et al. 2016). CpG methylation and gene expression were negatively correlated not only at promoters but also at gene bodies (Fig. 1A; Supplemental Fig. S1). This observation is consistent with the model of transcriptional silencing by DNA methylation (Schübeler 2015), which can be applied to most loci in the chicken genome.
Figure 1.
DNA methylation and transcription in avian genomes. (A) Density of data points as a function of CpG methylation (5mC [%]) and gene expression (log2 [TPM]) in female chickens and a male great tit (for male chickens, see Supplemental Fig. S1). The relationship between methylation and gene expression was smoothed with cubic splines (black lines). Number of genes with CpG methylation data: chicken: N = 11,662 for promoters and N = 11,723 for gene bodies; great tit: N = 14,694 for promoters and N = 14,721 for gene bodies. (B) Comparison of DNA methylation between aligned CpGs of Z and W gametologs. N depicts the number of Z-W-aligned CpGs with at least three mapped reads in each sample. Statistical significance was evaluated using paired Mann–Whitney U tests. For A and B, the promoter of a gene was defined as upstream 1.5 kb to downstream 500 bp of its transcription start site. (***) P < 0.001, (**) P < 0.01, (*) P < 0.05, (NS) not significant.
We found that differential DNA methylation of the Z and W Chromosomes was associated with differential expression of the Z and W gametologs (homologous genes on the sex chromosomes). In the 26 pairs of Z/W gametologs (see Methods), both promoters and gene bodies of the W Chromosome were significantly hypermethylated relative to the Z Chromosome, concordant with lower expression of W relative to Z gametologs (Supplemental Fig. S2). This pattern was consistent across tissue types and developmental stages (Fig. 1B), although the differences in methylation between the W and Z gametologs were not significant in some samples (brain and retina E18), likely due to low sequencing depths. This result indicates that DNA methylation may epigenetically regulate expression of Z and W gametologs.
Novel Z Chromosome locus with significant sex differences in DNA methylation and chromatin accessibility
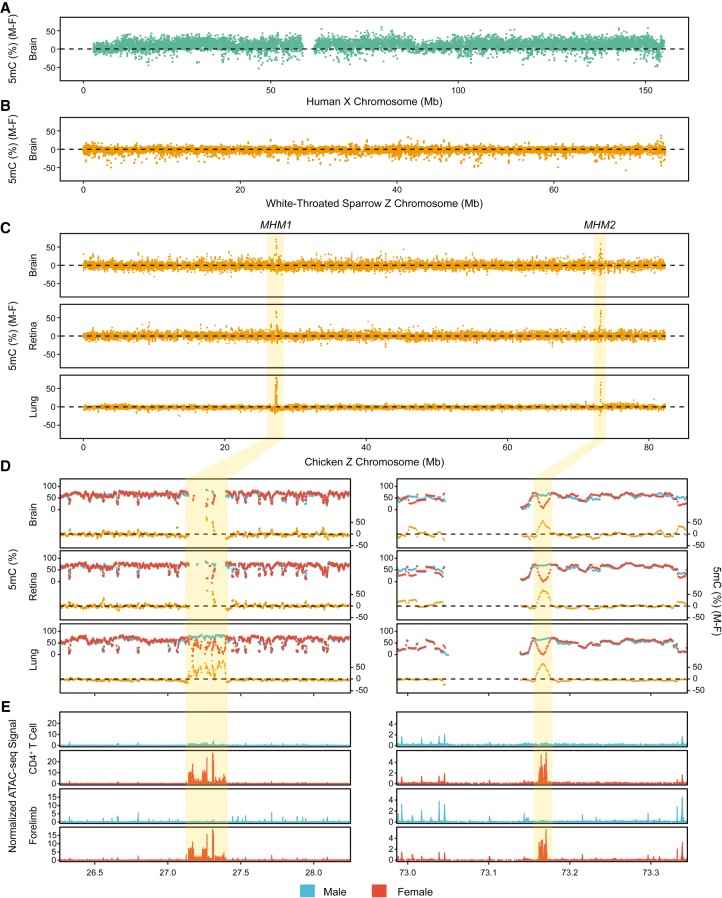
We investigated how sex differences in DNA methylation vary across avian Z Chromosomes using whole-genome bisulfite sequencing (WGBS) data from chicken (Galloanserae) and white-throated sparrow (Neoaves). The human X Chromosome exhibits substantial differences in DNA methylation between the sexes (Fig. 2A). The male X Chromosome is more hypermethylated over the majority of the chromosome compared to the female X Chromosome, which is consistent with the finding that the active X Chromosome is hypermethylated in gene bodies (Hellman and Chess 2007) and intergenic regions (Keown et al. 2017).
Figure 2.
Sex differences in DNA methylation on sex chromosomes and chromatin accessibility patterns of MHMs. (A) Differences in DNA methylation between male and female X Chromosomes in human brains (5mC [%] [M-F]). (B) Differences in DNA methylation between male and female Z Chromosomes in the white-throated sparrow. (C) Differences in DNA methylation between male and female Z Chromosomes in chicken. Two regions with extreme sex differences in DNA methylation (MHM1 and MHM2) are highlighted. For A–C, methylation values were plotted using a 10-kb window size with a 2-kb step size. (D) A zoomed-in view of the MHM loci. Methylation levels (5mC [%]) for males (blue) and females (red) are shown in the upper lines. Lower lines (orange) depict sex differences in DNA methylation. (E) Both MHMs (shaded areas) display increased chromatin accessibility in females compared with males. ATAC-seq reads were merged per sex and normalized to fragment pileup per million reads for direct comparison between sexes. For either CD4+ T cells (Foissac et al. 2019) or forelimb (E4.5) (Sackton et al. 2019), the two loci contained significantly female-biased peaks, tested using bdgdiff from the MACS2 program (Zhang et al. 2008). The vast majority of tissue/cell types with available data show similar patterns (Supplemental Fig. S5).
In contrast to the pattern observed for the human X Chromosome (Fig. 2A), methylation of the Z Chromosome in two divergent avian species is overall highly similar between males and females (Fig. 2B,C; Supplemental Fig. S3). In chicken, but not in white-throated sparrow, we found two regions on the Z Chromosome within which DNA methylation is extremely male-biased. This bias was consistent across tissues (Fig. 2C,D; Methods). The first region localized to 27.140–27.398 Mb of the chicken Z Chromosome (Fig. 2C,D). This region, which was previously identified using cDNA clones and methylation-sensitive restriction digest assays, has been referred to as the “male hypermethylated region (MHM)” (Teranishi et al. 2001; Itoh et al. 2010). Here, we report another region on the Z Chromosome with substantial sex differences in DNA methylation across all tissue types examined (Fig. 2C,D). This novel region is located at 73.160–73.173 Mb of the Z Chromosome. The Chicken Gene Nomenclature Consortium (CGNC) has assigned male hypermethylated region 1 (MHM1; CGNC: 80601) to this previously identified region, and male hypermethylated region 2 (MHM2; CGNC: 80602) to this newly identified region. In contrast, autosomes show highly similar DNA methylation patterns between males and females in both species, supporting that extreme regional methylation differences are restricted to the chicken Z Chromosome (Supplemental Fig. S4; Methods).
Additionally, we examined recent ATAC-seq data (Foissac et al. 2019; Sackton et al. 2019) and found that chromatin accessibility is elevated in females, relative to males, at both MHM1 and MHM2 (Fig. 2E; Supplemental Fig. S5). Another region (5,974,978–5,975,264 base pairs [bp]), composed of tandem repeats (repeat unit: CCTTT) on the Z Chromosome, also exhibited increased chromatin accessibility in females across tissues/cell types (Supplemental Fig. S6). However, this region was much shorter than the other two regions (∼300 bp compared to ∼250 and ∼10 kb for MHM1 and MHM2, respectively) and does not harbor any CpGs. Due to these features, we could not examine the correspondence with DNA methylation. As genes within 100 kb of the female-biased peak at ∼5.9 Mb have undetectable or very low expression, in the remainder of the manuscript we present in-depth analyses of the two MHM loci.
Unusual molecular characteristics of MHM2 that parallel MHM1
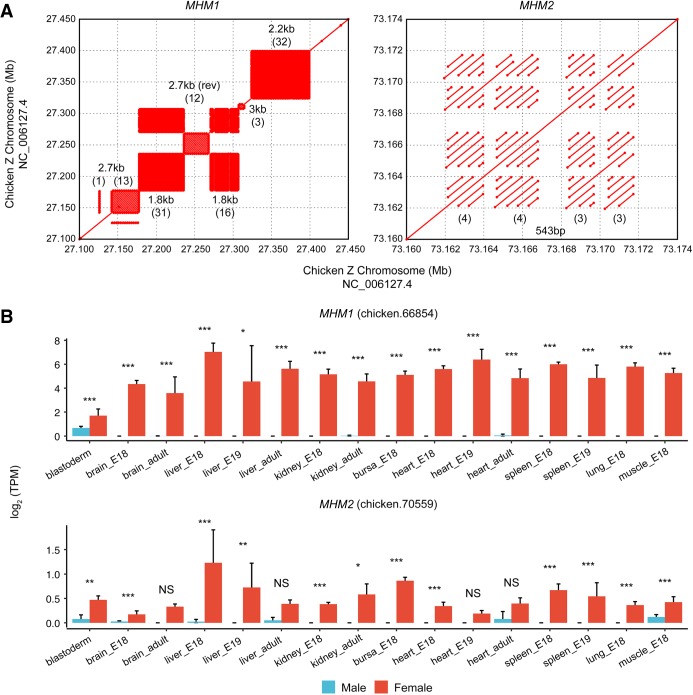
The alignment between the MHM1 and MHM2 loci revealed that the two regions do not share any homology, implying that these two loci originated independently. Nevertheless, MHM2 exhibits distinct characteristics that parallel those of the MHM1. First, similar to the MHM1 locus, the novel MHM2 locus is highly repetitive (Methods). Specifically, the MHM2 locus comprises four repetitive blocks of tandem repeats, with three to four iterations of a 542-bp repeat unit per block (Fig. 3A). Second, by annotating transcripts using RNA-seq data from 130 chicken samples across tissues and developmental stages (see Methods), we found that, like MHM1, MHM2 encodes long noncoding RNAs (lncRNAs) (Supplemental Fig. S7). Specifically, three poly(A)-transcripts are located within the second repetitive block of MHM2. As expected from the patterns of epigenetic differences, the expression of MHM1 and MHM2 lncRNAs are both female-biased (Fig. 3B).
Figure 3.
Molecular characteristics of MHM1 and MHM2 loci. (A) Both loci are highly repetitive. Each rectangular area filled with matches in dot plots depicts a block of tandem repeats. (B) Expression differences between males and females for lncRNAs transcribed from MHMs. For each locus, one example lncRNA gene with the highest average expression is shown. Significant expression differences between males and females were detected using DESeq2 with raw counts generated from StringTie. (***) Q < 0.001, (**) Q < 0.01, (*) Q < 0.05, (NS) not significant.
Although MHM1 is located on the oldest stratum (S0) shared by all three avian lineages, MHM2 is located on a younger stratum (S1), which is shared only by the Galloanserae and Neoaves (Wang et al. 2014; Zhou et al. 2014). The repetitive units of MHM1 and MHM2 exhibit distinct phylogenetic distributions. Whereas the repeat units of MHM1 are observed only in Galloanserae, the repeat unit of MHM2 exists as a single copy in some Neoaves species (Supplemental Table S1; Supplemental Fig. S8). The absence of the MHM2 repeat unit in Palaeognathae or other Neoaves lineages (e.g., passerines) (Supplemental Fig. S8) suggests that this sequence may have originated in the ancestor of Neognathae birds but was subsequently lost in some Neoaves.
Genes near MHM1 and MHM2 show similarly reduced male-to-female expression ratios
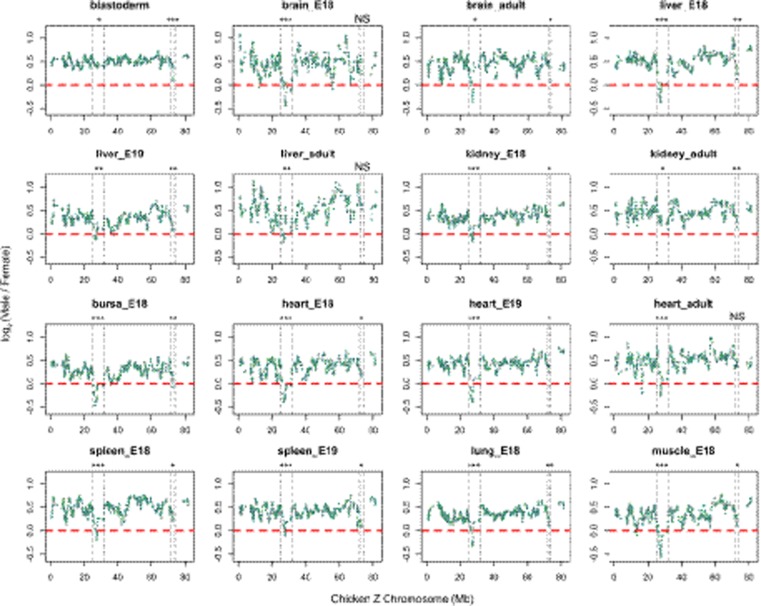
Male-to-female expression ratios across the avian Z Chromosomes are generally greater than 1, as dosage compensation is incomplete (Ellegren et al. 2007; Itoh et al. 2007; Graves 2016). However, microarray data has shown that genes located near MHM1 display pronounced reduction in male-to-female expression ratios in the chicken (Melamed and Arnold 2007; Mank and Ellegren 2009). Using RNA-seq data, we investigated whether genes adjacent to MHM1 displayed reduced male-to-female ratio consistent with the previous microarray studies. In addition, we looked for similar patterns near the novel MHM2 locus. We found that genes neighboring MHM1 or MHM2 exhibited reduced male-to-female ratios compared to the rest of the Z Chromosome in most somatic tissues across developmental stages in chicken (Fig. 4; Supplemental Figs. S9, S10). By performing a changepoint analysis to detect abrupt changes in ratios across the Z Chromosome (see Methods), we found that the boundary for MHM1-affected genes is 25–32 Mb, which is narrower than previously reported (25–35 Mb) (Melamed and Arnold 2007). The boundary for MHM2-affected genes was 72.5–73.5 Mb.
Figure 4.
Sex differences in gene expression across the Z Chromosome. The log2 (Male/Female) values are plotted using a 10-gene window. Genes with average log2(TPM) lower than 1 in either males or females were filtered out. The dashed lines depict the borders of potential MHM1- or MHM2-affected protein-coding genes identified by a changepoint analysis (Methods). For MHM1, the border is 25–32 Mb; and for MHM2, the border is 72.5–73.5 Mb. One-tailed Mann–Whitney U tests were used to test whether the MHM1- or MHM2-neighboring regions consist of genes with lower Male/Female ratios than the Z background. (***) P < 0.001, (**) P < 0.01, (*) P < 0.05, (NS) not significant.
How did the sex difference in DNA methylation and expression of genes at and near MHMs originate?
The difference in DNA methylation between male and female Z Chromosomes could have been caused by an increase of DNA methylation in the male Z Chromosome (male hypermethylation) or reduction of DNA methylation in the female Z Chromosome (female hypomethylation). Likewise, the reduced male-to-female ratio of gene expression could have arisen via down-regulation of the Z-linked genes in males or their up-regulation in females (Wright et al. 2015). We investigated available data on DNA methylation and gene expression from chickens and other avian species to gain insight into the evolutionary origin of these sex differences.
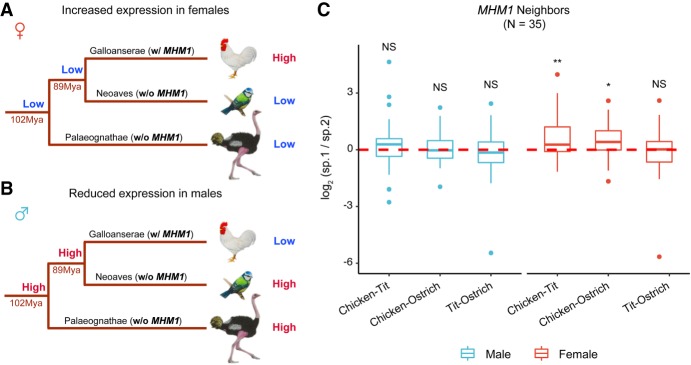
Comparison of expression levels of MHM-neighboring genes with the rest of the Z-linked genes yielded inconclusive results (Supplemental Fig. S11). An important assumption in this analysis is that the expression levels observed for genes on the current Z Chromosome are similar to the ancestral expression levels, which may not necessarily be the case. Taking an alternative approach similar to the methods used in previous studies (Brawand et al. 2011; Julien et al. 2012; Mank 2013; Cortez et al. 2014; Marin et al. 2017), we used data from outgroups to infer ancestral expression of MHM-neighboring genes. Specifically, we used available brain RNA-seq data from two divergent outgroups—blue tit (a passerine in Neoaves) and ostrich (in Palaeognathae) (Adolfsson and Ellegren 2013; Mueller et al. 2015), both lacking MHM loci (Fig. 5A,B). Comparing the expression of MHM1-neighboring genes among the three species—chicken, blue tit, and ostrich—we show that their expression is higher in female chicken than in female ostrich or blue tit. In contrast, there were no differences among the species for the male samples (Fig. 5C). This result is consistent with the scenario that sex differences in expression of these genes originated due to up-regulation in females (Fig. 5A). On the other hand, MHM2 is located in a younger stratum (S1) which is shared only by chicken and blue tit (in ostrich, this region is located in the pseudoautosomal region) (Wang et al. 2014; Zhou et al. 2014), and the comparison between chicken and blue tit was inconclusive (Supplemental Fig. S12). Therefore, we do not have enough evidence to fully support either scenario (up-regulation in females or down-regulation in males) for the evolution of MHM2.
Figure 5.
Alternative models for the evolution of MHM1 region in chicken. Reduced male-to-female expression ratios near MHM1 in chicken could be due to female up-regulation (A) or male down-regulation (B). In scenario A, low expression in females is the ancestral state, and up-regulation in females has evolved recently in chicken. An expression pattern similar to that shown in A, in other words, higher expression in female chickens than in female tit and ostrich, would suggest that the reduced male-to-female expression ratios seen in chicken evolved due to up-regulation in females. In scenario B, high expression in males is ancestral and the depicted pattern of expression would suggest recently evolved down-regulation in males. For A and B, the time for the split of avian lineages is from Jarvis et al. (2014) and Zhou et al. (2014). (C) Pairwise species differences in expression of the MHM1-neighboring genes (adult brain). Expression was averaged across samples per species, and the pairwise expression difference (the ratio of expression of species 1 to the expression of species 2) was log2-scaled. Only orthologs present in all species were used (numbers of genes compared are in parentheses). The statistical significance was evaluated using paired Mann–Whitney U tests. (**) P < 0.01, (*) P < 0.05, (NS) not significant.
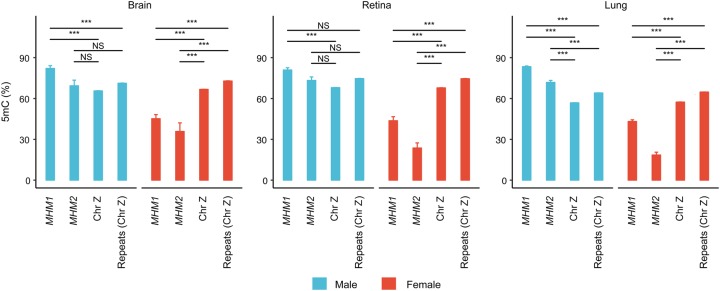
Next, we compared the DNA methylation levels of MHM1 and MHM2 with the rest of the Z Chromosome. Since both MHM loci are highly repetitive, we also compared methylation of MHM with that of other repetitive sequences on the Z Chromosome. Both MHM loci were significantly hypomethylated relative to either the entire Z Chromosome or repetitive sequences on the Z Chromosome in females, and this pattern was consistent across all tissues examined (Fig. 6). In males, although the MHM loci generally exhibited hypermethylation relative to the background, several comparisons are not significant. Thus, we found robust female-hypomethylation, and potentially weak male-hypermethylation of the MHM loci. Female-hypomethylation of MHM loci is consistent with the up-regulation of nearby genes (Fig. 5A).
Figure 6.
Hypomethylation of MHM loci relative to the entire Z Chromosome or repeats on the Z Chromosome in females. In the graphs, DNA methylation of MHM1 and MHM2 is compared with that of the Z Chromosome or repeats on the Z Chromosome for males and females. Coordinates of repetitive sequences were obtained from RepeatMasker track (last update: 2016-04-14) of the UCSC Table Browser. Statistical significance was evaluated using Mann–Whitney U tests. (***) P < 0.001, (NS) not significant.
Discussion
Much of our current knowledge about the epigenetic regulation of gene expression and dosage compensation comes from studies of male heterogametic mammalian systems. By comparison, information about these processes in female heterogametic systems is scarce. With the increasing wealth of genomic data, birds are emerging as a rich source of models in which to investigate the evolution of sex chromosomes. The extent of genetic degeneration of the nonrecombining W Chromosome is highly variable across birds (Zhou et al. 2014). Avian systems also offer new perspectives on the evolution of dosage compensation. In mammals, the nonrecombining Y Chromosomes are nearly completely degenerated, and dosage compensation is achieved by global inactivation of one of the X Chromosomes in females. In contrast, birds exhibit incomplete dosage compensation even in the chicken, in which the nonrecombining W Chromosome is largely degenerated. In addition, data on a recently recombination-suppressed avian chromosome indicate that partial dosage compensation can evolve prior to substantial genetic degeneration (Sun et al. 2018). Thus, avian systems offer an excellent opportunity to investigate the evolution of sex chromosomes and the genomic impact of reduced recombination in general.
We integrated recently generated nucleotide-resolution DNA methylation maps with comprehensive transcriptome data in the chicken. Similar to the genomes of other vertebrates, the chicken genome is heavily methylated (Elango and Yi 2008; Suzuki and Bird 2008). Genic DNA methylation is negatively correlated with gene expression, as expected when considering the currently accepted model of transcriptional suppression by DNA methylation (Schübeler 2015). A recently published study of the great tit also led to a similar conclusion (Laine et al. 2016). At the chromosomal level, male and female Z Chromosomes show little difference in DNA methylation (Fig. 2C), which is consistent with the lack of global dosage compensation.
Despite this global similarity, there are two regions within which DNA methylation of the chicken Z Chromosomes is highly differentiated between males and females. The first region corresponds to a previously described locus identified via a targeted approach (Teranishi et al. 2001). Since its discovery nearly two decades ago, this region (referred to as MHM1 in our study) has been considered the only region exhibiting differential DNA methylation between the male and female chicken Z Chromosomes. Here, we identified an additional novel region on the chicken Z Chromosome with highly differentiated DNA methylation, which we refer to as MHM2. There is little sequence similarity between the two MHM loci, suggesting independent evolutionary origins. MHM1 sequences are present in Galloanserae but absent from the other two avian lineages we examined, while MHM2 sequences are present in Galloanserae and several Neoaves lineages (Supplemental Table S1; Supplemental Fig. S8). There are several similarities between MHM1 and MHM2: (1) MHMs exhibit higher chromatin accessibility in females than males as confirmed by recently generated ATAC-seq data (Fig. 2E; Supplemental Fig. S5), which is consistent with previous studies of MHM1 that identified the enrichment of H4K16ac (Bisoni et al. 2005; Itoh et al. 2010, 2011); (2) MHM-derived lncRNAs are transcribed in a female-biased manner for both loci in the tissues examined so far (Fig. 3B); (3) MHM-neighboring genes display reduced male-to-female expression ratios (Fig. 4); and (4) both loci are highly repetitive (Fig. 3A). These convergent features between the two MHMs indicate that epigenetic regulation of these regions may have evolved via similar molecular mechanisms.
The reduced male-to-female expression ratios of genes near both MHM1 and MHM2 (Fig. 4) suggest that these regions may influence the expression of these genes. According to current models in chicken, long noncoding RNAs from the MHM1 bind to neighboring genes and increase their expression in females (Melamed and Arnold 2007; Mank and Ellegren 2009; Wright et al. 2015). Indeed, the repetitive structure of both MHMs parallels the tandem repeat blocks in the placental XIST and marsupial Rsx lncRNAs, which are independently evolved master regulators of X inactivation (Grant et al. 2012; Brockdorff 2018). For example, tandem repeat blocks within XIST can bind RNA-binding proteins and mediate XIST localization (Chu et al. 2015; Ridings-Figueroa et al. 2017). MHM2 has fewer repeat units compared to the MHM1 (Fig. 3A), and correspondingly, the region exhibiting reduced male-to-female expression ratios near MHM2 (∼1 Mb) is narrower than the one near MHM1 (∼7 Mb) (Fig. 4). These observations are consistent with the significance of repetitive structures of MHMs and their lncRNAs affecting the expression of nearby genes.
We tested whether the reduced male-to-female expression ratios of MHM-nearby genes are due to a reduction of expression in males or to an increase of expression in females. Wright et al. (2015) compared the expression of MHM1-nearby genes to the Z chromosomal average and reported that four out of six species displayed significantly lower expression of MHM1-nearby genes in males compared to females. Using RNA-seq data from a larger number of samples, we did not find consistent differences between male and female expression of MHM-nearby genes using a similar approach (Supplemental Fig. S11). However, the disparity between the results of Wright et al. (2015) and current results may be due to different developmental stages of samples used in the two studies. For example, Wright et al. (2015) used adult spleen, whereas we used spleen samples from embryonic stages.
We took an alternative approach to compare expression of current Z-linked genes with expression of these genes on proto-sex chromosomes (Brawand et al. 2011; Julien et al. 2012; Mank 2013; Cortez et al. 2014; Marin et al. 2017). The results were consistent with up-regulation of MHM1-nearby genes in females (Fig. 5) but inconclusive for MHM2-nearby genes (Supplemental Fig. S12). Comparing DNA methylation inside MHMs versus the entire Z Chromosome (with or without repetitive sequences) indicated significant hypomethylation of MHMs in females for all tissues and hypermethylation in males for some tissues (Fig. 6). Female-hypomethylation is also consistent with the ATAC-seq peaks, which show highly accessible chromatin in the MHM loci only in females (Fig. 2E; Supplemental Fig. S5). In conclusion, we find evidence supporting female-hypomethylation of MHM1 and up-regulation of MHM1-neighboring genes, while MHM2 regions require more data and further analysis. In globally heavily methylated genomes such as mammals and birds (Elango and Yi 2008; Suzuki and Bird 2008), hypomethylated genomic regions often harbor regulatory signals and are used as promoters or enhancers (Roadmap Epigenomics Consortium et al. 2015; Mendizabal and Yi 2016). Therefore, we hypothesize that MHM1 hypomethylation in females is associated with female-specific regulatory function.
One hypothesized function of MHM1 is that it might drive sexual differentiation by silencing the candidate avian sex-determining gene DMRT1 in females (Teranishi et al. 2001). However, similar to other genes near MHM1, DMRT1 is expressed at similar levels between sexes in the majority of somatic tissues and is even significantly female-biased in samples from brain (E18) (Supplemental Fig. S13). This result indicates limited effects of MHM1 on DMRT1 transcription in the majority of samples analyzed. Indeed, a previous study found that misexpression of MHM1 in gonad affected DMRT1 not at the mRNA level but at the posttranscriptional level (Roeszler et al. 2012). The role of DMRT1 in sex determination in chicken, which may depend on developmental stage and tissue, needs to be further evaluated. To this end, we examined a sample from chicken blastoderm at 12 h, which represents the earliest developmental stage at which chicken embryos are accessible postlaying (Ayers et al. 2013), much earlier than the embryonic stage examined by Teranishi et al. (2001). Compared to other tissues and developmental stages, the blastoderm shows relatively weak effects of sex on expression of genes near MHM1 (Fig. 4). The male-to-female expression ratio of MHM2-nearby genes, on the other hand, was strongly reduced in the blastoderm stage (Fig. 4).
Other researchers have hypothesized that in chicken, epigenetic differentiation between male and female Z Chromosomes is implicated in the regulation of genes with functions critical for females (Mank and Ellegren 2009; Wright et al. 2015). We and others (e.g., Wright et al. 2015) have not identified such functional enrichment. However, at least one of the genes near MHM1 is known to play a vital role in female reproduction. The gene VLDLR encodes a very low-density lipoprotein receptor, which is expressed in the cell surfaces of oocytes and regulates vitellogenesis (yolk deposition). In chickens with a naturally occurring mutation in this gene, vitellogenesis is disrupted and the hens do not lay eggs (Ho et al. 1974; Nimpf et al. 1989). Males heterozygous for the mutation are phenotypically normal carriers with unaffected fertility (Ho et al. 1974). Thus, the role of VLDLR is profoundly female-specific. Two other genes, RLN3 near MHM1 and CARTPT near MHM2 (encoding relaxin 3 and CART prepropeptide, respectively) show female-biased expression. Both genes encode neuropeptides that are thought to regulate food intake in chickens (Tachibana et al. 2003; Higgins et al. 2010). In rodents, the effects of these peptides on food intake have been found to be somewhat larger in females than males (Asnicar et al. 2001; Lenglos et al. 2015); however, such differences have not been reported for birds. The exact functions of MHM loci in chickens need experimental validation, for example, via knockout or knockdown of MHMs in females. Our study offers a picture, with chromosome-wide resolution, of epigenetic differentiation between male and female sex chromosomes in chicken. In addition, the work provides new candidate regions for future studies of how gene expression doses are regulated and evolve in a female heterogametic system.
Methods
Enhanced chicken genome assembly
We used an enhanced Gallus_gallus-5.0 (GCA_000002315.3) assembly for our analyses. Specifically, the W Chromosome was replaced by the one from the latest GRCg6a (GCA_000002315.5) assembly (Bellott et al. 2017) because of its improved quality (Gallus_gallus-5.0: 5.16 Mb vs. GRCg6a: 6.81 Mb).
Whole-genome bisulfite sequencing data and DNA methylation calling
To compare DNA methylation between Z and W gametologs and to compare DNA methylation between males and females, we collected previously published chicken WGBS data sets across four tissues (brain, retina, lung, and muscle) (Supplemental Table S2; Li et al. 2015; Lee et al. 2017; Zhang et al. 2017), which comprised either samples from both sexes (brain, retina, and lung) or from females alone (muscle). We also collected brain WGBS data from an adult male great tit to determine the relationship between gene expression and DNA methylation (Laine et al. 2016).
To compare the sex differences in DNA methylation between chicken and the white-throated sparrow, we performed brain WGBS of the white-throated sparrow (tan morph) for a male adult and a female adult. WGBS libraries were prepared using a custom protocol. First, DNA was extracted from the brain (hypothalamus region) using a QIAGEN DNeasy Blood and Tissue DNA kit. The DNA samples were then sheared on a Covaris ultrasonicator to 200–600 bp. The DNA fragment ends were repaired, and A-overhangs were added before bisulfite compatible adaptors were ligated to the DNA fragments overnight. Then, the DNA fragments were bisulfite-converted and PCR-amplified to increase concentration and enrich for adaptor-ligated DNA fragments. WGBS libraries were then sequenced using Illumina HiSeq X Ten or HiSeq 2500 at Macrogen Clinical Laboratory. Additionally, to compare sex differences in DNA methylation between birds and humans, we obtained fractional methylation data of the human brain from Zeng et al. (2012) (accession number in Supplemental Table S2).
WGBS reads were trimmed with Trim Galore! (version 0.4.5) (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) using a quality cutoff of 30 and then aligned to the enhanced assembly for chicken samples (or to the “Parus_major1.1” assembly for the great tit sample) using Bismark v0.18.1 in Bowtie 2 mode (Krueger and Andrews 2011). Subsequently, mapped reads were deduplicated using deduplicate_bismark, non-bisulfite-converted reads were filtered using filter_non_conversion (with the --percentage_cutoff 20 option), and fractional methylation was extracted using bismark_methylation_extractor. Methylation data were then merged across samples for each sex. Only CpGs with at least three mapped reads were retained for further analysis.
Comparison of DNA methylation between Z and W gametologs
In total, 26 Z-W pairs were annotated in NCBI (the W sequence was from the latest GRCg6a version) (Supplemental Table S3). Since the transcription start site (TSS) of W gametologs may be poorly annotated, promoters of Z gametologs (defined as upstream 1.5 kb to downstream 500 bp of TSS) were blasted against the W sequence to identify homologous promoters on W in BLASTN 2.7.1+ (-task blastn). Gene bodies or promoters of Z-W pairs were then aligned using MAFFT v7.407 (Katoh and Standley 2013). Only aligned CpGs with more than three reads covered for both Z and W gametologs were retained for further analysis.
Identification of regions with extreme sex differences in DNA methylation
To identify regions harboring extreme sex differences in DNA methylation, we computed DNA methylation levels averaged across CpGs, across the whole chicken genome, using a 10-kb window size and a 1-kb step size (a threshold required that at least 20 CpGs with methylation data be present in a window). The differences in methylation between males and females were then calculated. We identified regions with extreme sex differences in DNA methylation, defined as those outside the range of the first quartile minus three times the interquartile range and the third quartile plus three times the interquartile range (i.e., <Q1 – 3 × IQR or >Q3 + 3 × IQR). To identify potentially non-tissue-specific outlier regions, we retained outlier windows common to the brain, retina, and lung (Supplemental Code). Except for MHM1 and MHM2 on the chicken Z Chromosome, we did not detect extreme sex differences in DNA methylation anywhere in the genome, including on any autosomes (macrochromosomes are shown as examples in Supplemental Fig. S4).
ATAC-seq data, signal normalization, and differential peak identification
We collected recently published ATAC-seq data from liver, CD4+ T cells (Foissac et al. 2019), and flight muscles/bones (Supplemental Table S2; Sackton et al. 2019) in chicken. ATAC-seq reads were trimmed with Trim Galore! (version 0.4.5) using a quality cutoff of 30, and the processed reads were aligned to the enhanced chicken genome using Bowtie 2 2.3.4.2 (Langmead and Salzberg 2012) with -X 2000 --no-mixed --no-discordant parameters. Aligned reads were then merged across samples for each sex. Normalized ATAC-seq signals were obtained by running callpeak from the MACS2 2.1.1.20160309 program (Zhang et al. 2008) with the -p 0.01 -B --nomodel --SPMR --shift -100 --extsize 200 options. The signals (fragments pileup per million reads) for each sex at the two MHM loci were plotted with ggbio 1.28.5 (Yin et al. 2012). To find statistically significant ATAC-seq peaks, we reran callpeak without --SPMR and identified differential peaks using bdgdiff.
RNA-seq data, transcriptome assembly, and lncRNA identification
RNA-seq data from 130 chicken samples across multiple somatic tissues (blastoma, brain, bursa of Fabricius, heart, kidney, liver, lung, muscle, spleen) and developmental stages (12 h, E18, E19, and adults) were collected from multiple sources (Supplemental Table S2; Brawand et al. 2011; Julien et al. 2012; Ayers et al. 2013; Uebbing et al. 2015; Zimmer et al. 2016; Marin et al. 2017; Chickspress: http://geneatlas.arl.arizona.edu/). Raw reads were trimmed with Trim Galore! (version 0.4.5) and then aligned to the enhanced chicken genome using HISAT2 (Kim et al. 2015). We filtered out secondary alignments using SAMtools 1.7 (Li et al. 2009) to ensure that only primary alignments were retained for further estimation of gene expression.
To identify lncRNA genes within MHM loci and quantify the transcripts, we used StringTie 1.3.4d (Pertea et al. 2015) to assemble aligned reads into transcripts. The GTF annotation of transcripts is available at Figshare (http://dx.doi.org/10.6084/m9.figshare.8066612) and as Supplemental Material. Then, the coding potential of these transcripts was predicted using FEELnc v.0.1.1 (Wucher et al. 2017) under the shuffle mode, and transcripts with the coding potential score smaller than 0.4214 (a cutoff chosen by the program) were defined as lncRNAs. It should be noted that since our collected RNA-seq data sets are poly(A)-selected, we could capture only polyadenylated lncRNAs.
Quantification of gene expression and detection of differential expression
We measured the expression of lncRNAs by rerunning StringTie against the annotation of the new transcript assembly with the -e -b –A <gene_abund.tab> options. Meanwhile, we quantified protein-coding genes on the Z Chromosome by running StringTie against the chicken Ensembl annotation (Gallus_gallus-5.0.92 release) with the same options. The transcripts per kilobase million (TPM) values were extracted to represent gene expression levels for further analysis (Supplemental Code) and were log2-scaled (specifically, log2[TPM + 1]).
We averaged expression across samples within each sex, and genes with average expression above 1 in either sex were retained. The running average (window size of 10 genes with a slide size of one gene) of the male-to-female ratios on the Z Chromosome was computed with gtools 3.8.1 (Supplemental Code; https://cran.r-project.org/package=gtools). Expression valleys (dips in male/female ratios) near MHM1 and MHM2 were tested using a one-tailed Mann–Whitney U test.
Additionally, we tested for differential expression between males and females using DESeq2 1.18.1 (Supplemental Code; Love et al. 2014), using raw counts generated with prepDE.py from the StringTie package.
Identification of repeat units in MHM loci
We detected tandem repeat units of MHM1 and MHM2 using XSTREAM (Newman and Cooper 2007). We identified five different repeat units of MHM1: 2.7-kb, 1.8-kb, 2.7-kb (rev) (reverse complement of the 2.7-kb unit), 3-kb, and 2.2-kb (Fig. 3A). Among them, the 2.7-kb (rev) (JF692775) and 1.8-kb units (JF692776, named “sMHM-BamHI”) were previously identified by Itoh et al. (2011), and the 2.2-kb unit was discovered by Teranishi et al. (2001) (AB046698). As described by Itoh et al. (2011), the five repeat units share a core sequence (JF692776: 957–1219) which is potentially important for their function. For MHM2, a 543-bp unit is iterated 3–4 times per repeat block (Fig. 3A). All repeat units of MHM loci are relatively GC-rich (GC content: 55%–60%), while the GC content for the chicken Z Chromosome (the Gallus_gallus-5.0 assembly) is 40.74%. The sequences of all repeat units are available at Figshare (http://dx.doi.org/10.6084/m9.figshare.8066621) and as Supplemental Material.
Detection of the boundaries of MHM-affected genes
To obtain accurate boundaries of MHM-affected genes (dosage valleys), we performed a changepoint analysis using the R package “changepoint.np” (Supplemental Code; https://cran.r-project.org/package=changepoint.np). This package identifies multiple changepoints in timeseries-like data, without any assumptions regarding the distribution. The detected changepoints are reported in Supplemental Table S4. The changepoints across tissues/developmental stages were merged to obtain all potentially affected genes near MHM1 or MHM2.
Cross-species expression comparison
We obtained RNA-seq data from brain samples of great tit (Laine et al. 2016), blue tit (Mueller et al. 2015), and ostrich (Supplemental Table S2; Adolfsson and Ellegren 2013). The reads were preprocessed, and expression levels (in log2[TPM + 1]) were measured using the same pipeline as for chicken, except that reads were aligned to the “Parus_major1.1” assembly for the great tit sample, to the “cyaCae2” assembly for blue tit samples, and to the “ASM69896v1” assembly for ostrich samples.
To identify cross-species orthologs (chicken - blue tit - ostrich), we ran Proteinortho V5.16b (Lechner et al. 2011) on the protein sequences of three species (NCBI annotations for the blue tit and ostrich and Ensembl annotation for the chicken), with -synteny -identity = 50 -cov = 50 -selfblast options. Using this method, we identified 9991 orthologs present in chicken, blue tit, and ostrich.
To ensure comparability of expression across samples from different species, we used a median scaling method (Brawand et al. 2011; Julien et al. 2012; Marin et al. 2017). For genes with expression that was consistently within the inner quartile range (2111 genes across all 23 samples), the median expression per sample was computed. We then scaled the medians to a common value (mean of all medians) by scaling factors, and the scaling factor was used to calculate expression values for all genes in each sample (Supplemental Code).
Sample sexing
Since the sexes of several WGBS (brain and retina) and ATAC-seq (flight muscles/bones) samples were unknown, we sexed the samples based on the percentage of reads that could be mapped to the W Chromosome out of all aligned reads (W%). The distribution of percent W reads revealed two clear groups corresponding to the two sexes (Supplemental Fig. S14), and we determined the sex based on this stratification (Supplemental Table S5).
Identification of MHM repeat units in other avian species
We blasted all MHM repeat units against all representative genomes of birds (taxid: 8782) in NCBI (last access: 4/28/2019) using BLASTN in “blastn” mode. This mode is optimized for cross-species searches. We set the threshold of E-value to 10−5. To confirm the presence or absence of MHM2 in other avian species, we aligned the Z Chromosome of these species to the chicken Z Chromosome using LASTZ 1.04.00 (Harris 2007) with the --notransition --nogapped --step=20 --rdotplot options. The output dot plots were zoomed in to MHM2-neighboring regions and visualized in R (R Core Team 2019).
Self-alignment of MHM loci
We self-aligned each MHM loci to test for repetitive sequences. Specifically, NUCmer from the MUMmer 4.0.0beta2 (Marçais et al. 2018) was run to align each MHM locus with itself with --maxmatch --nosimplify options. We then used mummerplot to draw dot plots for these self-alignments. The two MHM loci were also aligned against each other using the same approach, but no alignment was generated.
Gene Ontology (GO) analysis
The PANTHER Overrepresentation Test (Released 20171205) from the PANTHER Classification System version 11 (Mi et al. 2017) was used to test for enrichment of GO terms in MHM- or MHM2-neighboring genes. Ensembl gene IDs were used for these genes, and the reference list used was the Gallus gallus annotation (PANTHER annotation version 13.1). We found that no GO terms from the biological process or molecular function were enriched in these sets of genes.
Data access
The WGBS data from two white-throated sparrows generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA540850. The GTF annotation of transcripts using 130 RNA-seq chicken samples have been submitted to Figshare (DOI: http://dx.doi.org/10.6084/m9.figshare.8066612) and is available as Supplemental Material. The Chicken Gene Nomenclature Consortium (CGNC) has assigned records to the male hypermethylated region 1 (MHM1; CGNC: 80601) and male hypermethylated region 2 (MHM2; CGNC: 80602). The National Center for Biotechnology Information (NCBI) will make the corresponding Entrez Gene IDs upon the next update, and when the Entrez Gene IDs are available, the two identifiers will be linked at both NCBI and CGNC. The sequences of all MHM repeat units have been submitted to Figshare (DOI: http://dx.doi.org/10.6084/m9.figshare.8066621) and are available as Supplemental Material. Custom scripts generated in this study are available as Supplemental Code.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Science Foundation (IOS 1656247) and the National Institutes of Health (R01MH082833) to D.L.M. and S.V.Y., as well as the Elizabeth Smithgall Watts endowment and the Georgia Tech School of Biological Sciences to S.V.Y. We thank the Yi laboratory for comments on the manuscript and Xiulan Pan for help with the illustration.
Author contributions: D.S. and S.V.Y. conceived the study and analyzed the data. D.S. and S.V.Y. wrote the manuscript. S.V.Y. supervised the work. D.L.M. provided samples of white-throated sparrow DNA and edited the manuscript. T.S.L. and P.C. performed WGBS of the white-throated sparrows.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.248641.119.
References
- Adolfsson S, Ellegren H. 2013. Lack of dosage compensation accompanies the arrested stage of sex chromosome evolution in ostriches. Mol Biol Evol 30: 806–810. 10.1093/molbev/mst009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asnicar MA, Smith DP, Yang DD, Heiman ML, Fox N, Chen Y-F, Hsiung HM, Köster A. 2001. Absence of cocaine- and amphetamine-regulated transcript results in obesity in mice fed a high caloric diet. Endocrinology 142: 4394–4400. 10.1210/endo.142.10.8416 [DOI] [PubMed] [Google Scholar]
- Ayers KL, Davidson NM, Demiyah D, Roeszler KN, Grützner F, Sinclair AH, Oshlack A, Smith CA. 2013. RNA sequencing reveals sexually dimorphic gene expression before gonadal differentiation in chicken and allows comprehensive annotation of the W-chromosome. Genome Biol 14: R26 10.1186/gb-2013-14-3-r26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. 2013. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet 14: 113–124. 10.1038/nrg3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, Mank JE, Peichel CL, Kirkpatrick M, Otto SP, Ashman T-L, Hahn MW, Kitano J, Mayrose I, Ming R, et al. 2014. Sex determination: why so many ways of doing it? PLoS Biol 12: e1001899 10.1371/journal.pbio.1001899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellott DW, Skaletsky H, Cho TJ, Brown L, Locke D, Chen N, Galkina S, Pyntikova T, Koutseva N, Graves T, et al. 2017. Avian W and mammalian Y chromosomes convergently retained dosage-sensitive regulators. Nat Genet 49: 387–394. 10.1038/ng.3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisoni L, Batlle-Morera L, Bird AP, Suzuki M, McQueen HA. 2005. Female-specific hyperacetylation of histone H4 in the chicken Z chromosome. Chromosome Res 13: 205–214. 10.1007/s10577-005-1505-4 [DOI] [PubMed] [Google Scholar]
- Brawand D, Soumillon M, Necsulea A, Julien P, Csárdi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, et al. 2011. The evolution of gene expression levels in mammalian organs. Nature 478: 343–348. 10.1038/nature10532 [DOI] [PubMed] [Google Scholar]
- Brockdorff N. 2018. Local tandem repeat expansion in Xist RNA as a model for the functionalisation of ncRNA. Noncoding RNA 4: E28 10.3390/ncrna4040028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N, Turner BM. 2015. Dosage compensation in mammals. Cold Spring Harb Perspect Biol 7: a019406 10.1101/cshperspect.a019406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel L, Willard HF. 2005. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434: 400–404. 10.1038/nature03479 [DOI] [PubMed] [Google Scholar]
- Charlesworth B. 1978. Model for evolution of Y-chromosomes and dosage compensation. Proc Natl Acad Sci 75: 5618–5622. 10.1073/pnas.75.11.5618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. 2000. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci 355: 1563–1572. 10.1098/rstb.2000.0717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Zhang QFC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. 2015. Systematic discovery of Xist RNA binding proteins. Cell 161: 404–416. 10.1016/j.cell.2015.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad T, Akhtar A. 2012. Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nat Rev Genet 13: 123–134. 10.1038/nrg3124 [DOI] [PubMed] [Google Scholar]
- Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, Waters PD, Grützner F, Kaessmann H. 2014. Origins and functional evolution of Y chromosomes across mammals. Nature 508: 488–493. 10.1038/nature13151 [DOI] [PubMed] [Google Scholar]
- Cotton AM, Price EM, Jones MJ, Balaton BP, Kobor MS, Brown CJ. 2015. Landscape of DNA methylation on the X chromosome reflects CpG density, functional chromatin state and X-chromosome inactivation. Hum Mol Genet 24: 1528–1539. 10.1093/hmg/ddu564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elango N, Yi SV. 2008. DNA methylation and structural and functional bimodality of vertebrate promoters. Mol Biol Evol 25: 1602–1608. 10.1093/molbev/msn110 [DOI] [PubMed] [Google Scholar]
- Ellegren H, Hultin-Rosenberg L, Brunström B, Dencker L, Kultima K, Scholz B. 2007. Faced with inequality: Chicken do not have a general dosage compensation of sex-linked genes. BMC Biol 5: 40 10.1186/1741-7007-5-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foissac S, Djebali S, Munyard K, Vialaneix N, Rau A, Muret K, Esquerré D, Zytnicki M, Derrien T, Bardou P, et al. 2019. Transcriptome and chromatin structure annotation of liver, CD4+ and CD8+ T cells from four livestock species. bioRxiv 10.1101/316091 [DOI] [PMC free article] [PubMed]
- Grant J, Mahadevaiah SK, Khil P, Sangrithi MN, Royo H, Duckworth J, McCarrey JR, VandeBerg JL, Renfree MB, Taylor W, et al. 2012. Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature 487: 254–258. 10.1038/nature11171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JAM. 2016. Evolution of vertebrate sex chromosomes and dosage compensation. Nat Rev Genet 17: 33–46. 10.1038/nrg.2015.2 [DOI] [PubMed] [Google Scholar]
- Harris RS. 2007. “Improved pairwise alignment of genomic DNA.” PhD thesis, The Pennsylvania State University. [Google Scholar]
- Hellman A, Chess A. 2007. Gene body–specific methylation on the active X chromosome. Science 315: 1141–1143. 10.1126/science.1136352 [DOI] [PubMed] [Google Scholar]
- Higgins SE, Ellestad LE, Trakooljul N, McCarthy F, Saliba J, Cogburn LA, Porter TE. 2010. Transcriptional and pathway analysis in the hypothalamus of newly hatched chicks during fasting and delayed feeding. BMC Genomics 11: 162 10.1186/1471-2164-11-162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho KJ, Lawrence WD, Lewis LA, Liu LB, Taylor CB. 1974. Hereditary hyperlipidemia in nonlaying chickens. Arch Pathol 98: 161–172. [PubMed] [Google Scholar]
- Huylmans AK, Macon A, Vicoso B. 2017. Global dosage compensation is ubiquitous in Lepidoptera, but counteracted by the masculinization of the Z Chromosome. Mol Biol Evol 34: 2637–2649. 10.1093/molbev/msx190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Melamed E, Yang X, Kampf K, Wang S, Yehya N, Van Nas A, Replogle K, Band MR, Clayton DF, et al. 2007. Dosage compensation is less effective in birds than in mammals. J Biol 6: 2 10.1186/jbiol53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Replogle K, Kim YH, Wade J, Clayton DF, Arnold AP. 2010. Sex bias and dosage compensation in the zebra finch versus chicken genomes: general and specialized patterns among birds. Genome Res 20: 512–518. 10.1101/gr.102343.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Kampf K, Arnold AP. 2011. Possible differences in the two Z chromosomes in male chickens and evolution of MHM sequences in Galliformes. Chromosoma 120: 587–598. 10.1007/s00412-011-0333-x [DOI] [PubMed] [Google Scholar]
- Jarvis ED, Mirarab S, Aberer AJ, Li B, Houde P, Li C, Ho SYW, Faircloth BC, Nabholz B, Howard JT, et al. 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346: 1320–1331. 10.1126/science.1253451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien P, Brawand D, Soumillon M, Necsulea A, Liechti A, Schütz F, Daish T, Grützner F, Kaessmann H. 2012. Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol 10: e1001328 10.1371/journal.pbio.1001328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keown CL, Berletch JB, Castanon R, Nery JR, Disteche CM, Ecker JR, Mukamel EA. 2017. Allele-specific non-CG DNA methylation marks domains of active chromatin in female mouse brain. Proc Natl Acad Sci 114: E2882–E2890. 10.1073/pnas.1611905114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. 2011. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572. 10.1093/bioinformatics/btr167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn BT, Page DC. 1999. Four evolutionary strata on the human X chromosome. Science 286: 964–967. 10.1126/science.286.5441.964 [DOI] [PubMed] [Google Scholar]
- Laine VN, Gossmann TI, Schachtschneider KM, Garroway CJ, Madsen O, Verhoeven KJF, de Jager V, Megens HJ, Warren WC, Minx P, et al. 2016. Evolutionary signals of selection on cognition from the great tit genome and methylome. Nat Commun 7: 10474 10.1038/ncomms10474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner M, Findeiss S, Steiner L, Marz M, Stadler PF, Prohaska SJ. 2011. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics 12: 124 10.1186/1471-2105-12-124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Rasoul BA, Holub AS, Lejeune A, Enke RA, Timp W. 2017. Whole genome DNA methylation sequencing of the chicken retina, cornea and brain. Sci Data 4: 170148 10.1038/sdata.2017.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenglos C, Calvez J, Timofeeva E. 2015. Sex-specific effects of relaxin-3 on food intake and brain expression of corticotropin-releasing factor in rats. Endocrinology 156: 523–533. 10.1210/en.2014-1743 [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li R, Wang Y, Hu X, Zhao Y, Li L, Feng C, Gu X, Liang F, Lamont SJ, et al. 2015. Genome-wide DNA methylome variation in two genetically distinct chicken lines using MethylC-seq. BMC Genomics 16: 851 10.1186/s12864-015-2098-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi JC, Kuroda MI. 2015. Dosage compensation in Drosophila. Cold Spring Harb Perspect Biol 7: a019398 10.1101/cshperspect.a019398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE. 2009. The W, X, Y and Z of sex-chromosome dosage compensation. Trends Genet 25: 226–233. 10.1016/j.tig.2009.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE. 2013. Sex chromosome dosage compensation: definitely not for everyone. Trends Genet 29: 677–683. 10.1016/j.tig.2013.07.005 [DOI] [PubMed] [Google Scholar]
- Mank JE, Ellegren H. 2009. All dosage compensation is local: gene-by-gene regulation of sex-biased expression on the chicken Z chromosome. Heredity 102: 312–320. 10.1038/hdy.2008.116 [DOI] [PubMed] [Google Scholar]
- Marçais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A. 2018. MUMmer4: a fast and versatile genome alignment system. PLoS Comput Biol 14: e1005944 10.1371/journal.pcbi.1005944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin R, Cortez D, Lamanna F, Pradeepa MM, Leushkin E, Julien P, Liechti A, Halbert J, Brüning T, Möossinger K, et al. 2017. Convergent origination of a Drosophila-like dosage compensation mechanism in a reptile lineage. Genome Res 27: 1974–1987. 10.1101/gr.223727.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed E, Arnold AP. 2007. Regional differences in dosage compensation on the chicken Z chromosome. Genome Biol 8: R202 10.1186/gb-2007-8-9-r202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed E, Arnold AP. 2009. The role of LINEs and CpG islands in dosage compensation on the chicken Z chromosome. Chromosome Res 17: 727–736. 10.1007/s10577-009-9068-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed E, Elashoff D, Arnold AP. 2009. Evaluating dosage compensation on the chicken Z chromosome: Should effective dosage compensation eliminate sexual bias? Heredity 103: 357–359. 10.1038/hdy.2009.91 [DOI] [PubMed] [Google Scholar]
- Mendizabal I, Yi SV. 2016. Whole-genome bisulfite sequencing maps from multiple human tissues reveal novel CpG islands associated with tissue-specific regulation. Hum Mol Genet 25: 69–82. 10.1093/hmg/ddv449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD. 2017. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45: D183–D189. 10.1093/nar/gkw1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller JC, Kuhl H, Timmermann B, Kempenaers B. 2015. Characterization of the genome and transcriptome of the blue tit Cyanistes caeruleus: polymorphisms, sex-biased expression and selection signals. Mol Ecol Resour 16: 549–561. 10.1111/1755-0998.12450 [DOI] [PubMed] [Google Scholar]
- Mullon C, Wright AE, Reuter M, Pomiankowski A, Mank JE. 2015. Evolution of dosage compensation under sexual selection differs between X and Z chromosomes. Nat Commun 6: 7720 10.1038/ncomms8720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K, Ellegren H. 2008. The chicken (Gallus gallus) Z chromosome contains at least three nonlinear evolutionary strata. Genetics 180: 1131–1136. 10.1534/genetics.108.090324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AM, Cooper JB. 2007. XSTREAM: a practical algorithm for identification and architecture modeling of tandem repeats in protein sequences. BMC Bioinformatics 8: 382 10.1186/1471-2105-8-382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimpf J, Radosavljevic MJ, Schneider WJ. 1989. Oocytes from the mutant restricted ovulator hen lack receptor for very low-density lipoprotein. J Biol Chem 264: 1393–1398. [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33: 290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Ridings-Figueroa R, Stewart ER, Nesterova TB, Coker H, Pintacuda G, Godwin J, Wilson R, Haslam A, Lilley F, Ruigrok R, et al. 2017. The nuclear matrix protein CIZ1 facilitates localization of Xist RNA to the inactive X-chromosome territory. Genes Dev 31: 876–888. 10.1101/gad.295907.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeszler KN, Itman C, Sinclair AH, Smith CA. 2012. The long non-coding RNA, MHM, plays a role in chicken embryonic development, including gonadogenesis. Dev Biol 366: 317–326. 10.1016/j.ydbio.2012.03.025 [DOI] [PubMed] [Google Scholar]
- Sackton TB, Grayson P, Cloutier A, Hu Z, Liu JS, Wheeler NE, Gardner PP, Clarke JA, Baker AJ, Clamp M, et al. 2019. Convergent regulatory evolution and loss of flight in paleognathous birds. Science 364: 74–78. 10.1126/science.aat7244 [DOI] [PubMed] [Google Scholar]
- Schübeler D. 2015. Function and information content of DNA methylation. Nature 517: 321–326. 10.1038/nature14192 [DOI] [PubMed] [Google Scholar]
- Sharp AJ, Stathaki E, Migliavacca E, Brahmachary M, Montgomery SB, Dupre Y, Antonarakis SE. 2011. DNA methylation profiles of human active and inactive X chromosomes. Genome Res 21: 1592–1600. 10.1101/gr.112680.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G, Chen YR, Blissard GW, Briscoe AD. 2014. Complete dosage compensation and sex-biased gene expression in the moth Manduca sexta. Genome Biol Evol 6: 526–537. 10.1093/gbe/evu035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Huh I, Zinzow-Kramer WM, Maney DL, Yi SV. 2018. Rapid regulatory evolution of a nonrecombining autosome linked to divergent behavioral phenotypes. Proc Natl Acad Sci 115: 2794–2799. 10.1073/pnas.1717721115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. 2008. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 9: 465–476. 10.1038/nrg2341 [DOI] [PubMed] [Google Scholar]
- Tachibana T, Takagi T, Tomonaga S, Ohgushi A, Ando R, Denbow DM, Furuse M. 2003. Central administration of cocaine- and amphetamine-regulated transcript inhibits food intake in chicks. Neurosci Lett 337: 131–134. 10.1016/S0304-3940(02)01321-6 [DOI] [PubMed] [Google Scholar]
- Teranishi M, Shimada Y, Hori T, Nakabayashi O, Kikuchi T, Macleod T, Pym R, Sheldon B, Solovei I, Macgregor H, et al. 2001. Transcripts of the MHM region on the chicken Z chromosome accumulate as non-coding RNA in the nucleus of female cells adjacent to the DMRT1 locus. Chromosome Res 9: 147–165. 10.1023/A:1009235120741 [DOI] [PubMed] [Google Scholar]
- Tukiainen T, Villani A-C, Yen A, Rivas MA, Marshall JL, Satija R, Aguirre M, Gauthier L, Fleharty M, Kirby A, et al. 2017. Landscape of X chromosome inactivation across human tissues. Nature 550: 244 10.1038/nature24265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uebbing S, Künstner A, Mäkinen H, Ellegren H. 2013. Transcriptome sequencing reveals the character of incomplete dosage compensation across multiple tissues in flycatchers. Genome Biol Evol 5: 1555–1566. 10.1093/gbe/evt114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uebbing S, Konzer A, Xu L, Backström N, Brunström B, Bergquist J, Ellegren H. 2015. Quantitative mass spectrometry reveals partial translational regulation for dosage compensation in chicken. Mol Biol Evol 32: 2716–2725. 10.1093/molbev/msv147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Bachtrog D. 2009. Progress and prospects toward our understanding of the evolution of dosage compensation. Chromosome Res 17: 585–602. 10.1007/s10577-009-9053-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Bachtrog D. 2011. Lack of global dosage compensation in Schistosoma mansoni, a female-heterogametic parasite. Genome Biol Evol 3: 230–235. 10.1093/gbe/evr010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Emerson JJ, Zektser Y, Mahajan S, Bachtrog D. 2013. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol 11: e1001643 10.1371/journal.pbio.1001643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhang J, Yang W, An N, Zhang P, Zhang G, Zhou Q. 2014. Temporal genomic evolution of bird sex chromosomes. BMC Evol Biol 14: 250 10.1186/s12862-014-0250-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AE, Zimmer F, Harrison PW, Mank JE. 2015. Conservation of regional variation in sex-specific sex chromosome regulation. Genetics 201: 587–598. 10.1534/genetics.115.179234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wucher V, Legeai F, Hédan B, Rizk G, Lagoutte L, Leeb T, Jagannathan V, Cadieu E, David A, Lohi H, et al. 2017. FEELnc: a tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic Acids Res 45: e57 10.1093/nar/gkw1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi SJ, Charlesworth B. 2000. Contrasting patterns of molecular evolution of the genes on the new and old sex chromosomes of Drosophila miranda. Mol Biol Evol 17: 703–717. 10.1093/oxfordjournals.molbev.a026349 [DOI] [PubMed] [Google Scholar]
- Yin T, Cook D, Lawrence M. 2012. ggbio: an R package for extending the grammar of graphics for genomic data. Genome Biol 13: R77 10.1186/gb-2012-13-8-r77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Konopka G, Hunt BG, Preuss TM, Geschwind D, Yi SV. 2012. Divergent whole-genome methylation maps of human and chimpanzee brains reveal epigenetic basis of human regulatory evolution. Am J Hum Genet 91: 455–465. 10.1016/j.ajhg.2012.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Yan FB, Li F, Jiang KR, Li DH, Han RL, Li ZJ, Jiang RR, Liu XJ, Kang XT, et al. 2017. Genome-wide DNA methylation profiles reveal novel candidate genes associated with meat quality at different age stages in hens. Sci Rep 7: 45564 10.1038/srep45564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Zhang JL, Bachtrog D, An N, Huang QF, Jarvis ED, Gilbert MTP, Zhang GJ. 2014. Complex evolutionary trajectories of sex chromosomes across bird taxa. Science 346: 1246338 10.1126/science.1246338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer F, Harrison PW, Dessimoz C, Mank JE. 2016. Compensation of dosage-sensitive genes on the chicken Z chromosome. Genome Biol Evol 8: 1233–1242. 10.1093/gbe/evw075 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.