Abstract
Brain mineralocorticoid receptors (MRs) and glucocorticoid receptors (GRs) respond to the same glucocorticoid hormones but can have differential effects on cellular function. Several lines of evidence suggest that MR‐specific target genes must exist and might underlie the distinct effects of the receptors. The present study aimed to identify MR‐specific target genes in the hippocampus, a brain region where MR and GR are co‐localised and play a role in the stress response. Using genome‐wide binding of both receptor types, we previously identified MR‐specific, MR‐GR overlapping and GR‐specific putative target genes. We now report altered gene expression levels of such genes in the hippocampus of forebrain MR knockout (fbMRKO) mice, killed at the time of their endogenous corticosterone peak. Of those genes associated with MR‐specific binding, the most robust effect was a 50% reduction in Jun dimerization protein 2 (Jdp2) mRNA levels in fbMRKO mice. Down‐regulation was also observed for the MR‐specific Nitric oxide synthase 1 adaptor protein (Nos1ap) and Suv3 like RNA helicase (Supv3 l1). Interestingly, the classical glucocorticoid target gene FK506 binding protein 5 (Fkbp5), which is associated with MR and GR chromatin binding, was expressed at substantially lower levels in fbMRKO mice. Subsequently, hippocampal Jdp2 was confirmed to be up‐regulated in a restraint stress model, posing Jdp2 as a bona fide MR target that is also responsive in an acute stress condition. Thus, we show that MR‐selective DNA binding can reveal functional regulation of genes and further identify distinct MR‐specific effector pathways.
Keywords: glucocorticoids, Jdp2, mineralocorticoid receptor knockout, restraint stress, transcription
1. INTRODUCTION
Endogenous glucocorticoid hormones affect brain function via two closely‐related nuclear receptors: the mineralocorticoid receptor (MR) and the glucocorticoid receptor (GR). The ligand concentration in part determines the specific MR/GR responses. High affinity MRs are occupied by endogenous corticosteroids under basal conditions and have been found to be more relevant in the initial phase of a stress response.1, 2 By contrast, the lower affinity GRs are activated only at higher glucocorticoid levels, around the peak of the circadian rhythm and during a stress response. Although GRs are expressed widely throughout the central nervous system, brain glucocorticoid binding MRs are mainly restricted to limbic areas.3
In the hippocampus, MR and GR are crucial for spatial memory and the modulation of cognition, mood and behaviour.3 Within the CA1 hippocampal subregion, MR and GR mediate opposite glucocorticoid effects on pyramidal neurone excitability4 via transcriptional mechanisms.5 Also, spatial learning in rodents is differentially affected by MR and GR signalling, with MR modulating response selection and GR being essential for memory consolidation.6, 7 Because of intrinsic MR‐mediated effects that oppose those of GR, it has long been argued that MR‐specific target genes must exist.8 The existence of MR‐specific transcriptional coregulators9, 10 also supports this idea. However, many of the effects that can be attributed specifically to MR function so far comprise rapid non‐genomic effects, mediated by the membrane variant of the receptor.11, 12
Several classical genomic MR‐targets have been described in various tissues over the past two decades, such as FK506 binding protein 5 (Fkbp5),13 glucocorticoid‐induced leucine zipper (Gilz),14 period circadian clock 1 (Per1)15 and serum/glucocorticoid regulated kinase 1 (Sgk1).16 However, these genes are all known to be GR responsive as well.17, 18, 19, 20 Of note, not only can the two receptors bind their target DNA as homodimers, but also heterodimerisation of MR/GR has been described.15 Although MR‐selective transrepression and transactivation may occur,21, 22 to date, no hippocampal genomic targets have been reported that are strictly MR‐dependent. Transcriptional changes have been attributed to MR function,23 although they were not formally proven to be direct targets of the receptor and might thus be affected by MR activity in an indirect manner. However, although the presence of glucocorticoid response element (GRE) appears to be crucial for both MR and GR DNA binding in the hippocampus, binding sites for NeuroD transcription factors were found selectively at MR‐bound loci.24 NeuroD factors could coactivate glucocorticoid‐induced transactivation and were indeed present near MR‐specific binding sites, suggesting that specific GRE‐dependent MR target genes do exist.
The present study investigated whether direct MR binding to the hippocampal DNA led to expression regulation of the nearby gene. Based on our recent work that defined MR‐specific, MR‐GR overlapping and GR‐specific chromatin binding sites and the corresponding putative target genes within the rat hippocampus,24 we examined mRNA levels of several genes in each of these categories. Forebrain MR knockout (fbMRKO) mice showed altered expression for a subset of genes, including down‐regulation of the mixed MR/GR target Fkbp5, as well as MR‐specific Jun dimerization protein 2 (Jdp2), Nitric oxide synthase 1 adaptor protein (Nos1ap) and Suv3 like RNA helicase (Supv3 l1) mRNA levels. Subsequently, corticosterone responsiveness of Jdp2, which is one of the genes with an MR‐bound promoter, was validated in mice that were exposed to different durations of restraint stress.
2. MATERIALS AND METHODS
2.1. Animals
Male homozygous fbMRKO and control c57bl/6 mice (n = 7) aged 8‐9 weeks were housed under a 12:12 hour light/dark reversed photocycle (lights off 9.00 am). The fbMRKO mice were generated using MRflox mice, with MR exon 3 flanked by loxP sites, and mice expressing Cre recombinase controlled by the CAMKIIα gene.25 Male MRflox/floxCamKCreCre/wt mice were crossed with female MRflox/flox mice to generate fbMRKO (MRflox/flox_Cre) and control (MRflox/flox_wt) offspring. Because the breeding unexpectedly generated more fbMRKO than control mice, only part of the control animals were littermates. No differences were found in expression levels between littermate and non‐littermate controls for any of the genes measured. Mice were transferred to a novel cage 20 minutes before harvesting the tissue and then killed around the time of their endogenous corticosterone peak, between 9.30 am and 12.00 pm. We assessed the expression of MR, overlapping and GR putative target genes in this condition because both receptor types are activated at peak of the diurnal corticosterone rhythm. The novel cage was included in the protocol to ensure MR and GR binding for chromatin immunoprecipitation (ChIP) analysis in the same animals, under the assumption that mRNA levels will not be affected in this short time span. Trunk blood was collected from all mice and hippocampal hemispheres were freshly dissected and snap‐frozen in liquid nitrogen for later analysis.
For validation of Jdp2 down‐regulation, male fbMRKO (n = 14) and littermate controls (n = 10) aged 8‐12 weeks were housed under a 12:12 hour light/dark photocycle (lights on 7.00 am). Mice were bred as described above. Mice were killed under baseline conditions, between 9.30 am and 10.30 am. Brains were collected and snap‐frozen in liquid nitrogen for later analysis.
For MR binding site validation in the mouse brain, male c57bl/6 mice (n = 5) aged 16‐19 weeks were housed under a 12:12 hour light/dark photocycle (lights on 8.00 am) and were killed in the afternoon 60 minutes after an i.p. injection of 3.0 mg kg‐1 corticosterone (Sigma, St Louis, MO, USA) dissolved in 5% ethanol in saline, ensuring MR binding. Hippocampal hemispheres were freshly dissected and snap‐frozen in liquid nitrogen for later analysis.
Male Balb/c mice (n = 3‐6) aged 8‐15 weeks were housed under a 12:12 hour light/dark photocycle (lights on 6.00 am) and were exposed to various periods of restraint stress (0, 30, 60, 120 and 240 minutes) and killed directly afterwards, between 9.30 am and 2.00 pm. At this time of the diurnal corticosterone trough, both MR and GR DNA binding can be enhanced in response to stress15 and consequential gene expression changes compared to non‐stressed control mice could be revealed. Trunk blood was collected from all mice, and hippocampal hemispheres were freshly dissected and snap‐frozen in liquid nitrogen for later analysis.
All experiments were performed in accordance with the European Commission Council Directive 2010/63/EU and the Dutch law on animal experiments and approved by the animal ethical committee from Utrecht University, University of Amsterdam, or the German Regierungspräsidium Tübingen.
2.2. Plasma measurements
Trunk blood was centrifuged for 10 minutes at 7000 g, after which plasma was transferred to new tubes. Corticosterone levels of the fbMRKO experiment were determined using an enzyme immunoassay (Immunodiagnostic Systems, East Boldon, UK) and adrenocorticotrophic hormone (ACTH) and corticosterone levels of the restraint stress mice were determined using an enzyme‐linked immunosorbent assay (IBL International, Hamburg, Germany) in accordance with the manufacturers’ instructions.
2.3. Target gene selection
MR‐specific, MR‐GR overlapping and GR‐specific binding sites were annotated to the nearest gene.24 To increase the chances of correct annotation and identifying functional target genes, we focused on binding sites located intragenic or in the proximal promoter (up to‐5 kb). Furthermore, hippocampal expression26 of the putative target genes, the degree of coexpression with NeuroD factors (Neurod1/2/6) and face validity of ChiP‐sequencing (ChIP‐seq) peaks were assessed. The total numbers of putative target genes measured for MR‐specific, overlapping (including classical targets) and GR‐specific subset were 12, 10 and 9, respectively.
2.4. ChIP‐quantitative polymerase chain reaction (PCR)
For MR binding validation in the mouse, we performed ChIP‐quantitative PCR on hippocampal tissue of wild‐type (WT) mice (n = 5) as described previously.27 Hippocampal hemispheres were cryosectioned at 30 μm before cross‐linking with 2 mmol L‐1 disuccinimidyl glutarate, followed by 1% formaldehyde. Fixated tissue was suspended, nuclei were isolated and sonicated for 10 rounds (30 seconds ON/30 seconds OFF) using a Bioruptor Pico (Diagenode, Seraing, Belgium). Chromatin of two hemispheres of the same animal was pooled and used for a single ChIP sample (500 μL) to measure MR binding with 5 μg of anti‐MR antibody (21854‐1‐AP; ProteinTech, Rosemont, IL, USA). Immunoprecipitation was performed with 50 μL of magnetic Protein A beads (Dynabeads; Invitrogen, Carlsbad, CA, USA). The background signal was detected for each sample with a sequential ChIP using 5 μg of control immunoglobulin (Ig)G antibody (ab37415; Abcam, Cambridge, MA, USA). Pellets were dissolved in 50 μL 10 mmol L‐1 Tris‐HCl (pH 8). Subsequently, a quantitative PCR was performed on 5 × diluted ChIP samples, with primers that were designed to span the GRE of the MR binding sites (Table 1).
Table 1.
Primer sequences used for a quantitative polymerase chain reaction in mouse hippocampal chromatin immunoprecipitation samples (for binding site details, see Table 3)
| Binding site | Nearest gene | Forward and reverse (5′‐ to 3′) | Product length (bp) |
|---|---|---|---|
| GR3000_1726 | Acsl6 | CCTGCCAGGAGAGCAGATGTGTGCAGGAAGGCAAGTTCT | 178 |
| MR3000_740GR3000_34 | Fkbp5 | TGCCAGCCACATTCAGAACATCAAGTGAGTCTGGTCACTGC | 122 |
| MR3000_1054 | Jdp2 | AAGTAAGACCGCGACCTACAAAATACCCAGTGCAGAGACGAA | 192 |
| MR300_473GR3000_599 | Kif1c | GCTGGGGTGTACACAGATGGTGACTAGCCAGAGCAGTATGTC | 156 |
| GR3000_106 | Mrpl48 | AGCTGTGCTTTGGAAGCCTACATAAGGTGGGCCACACTCC | 170 |
| MR300_196 | Nos1ap | CCTCCGATGCTGCTTGGATACAGACCGAGCCAGCGATAAG | 197 |
| MR3000_738GR3000_12 | Per1 | GGAGGCGCCAAGGCTGAGTGCGGCCAGCGCACTAGGGAAC | 73 |
| MR300_503 | Rilpl1 | CAGGCAGATGCCAGGCTCCCATGCCTGTTCCTCTAGT | 106 |
| MR3000_359 | Supv3 l1 | TGCAGGGATTCGATGGACAGCTCTGAGCCACCTCTCAAGC | 165 |
| MR3000_641GR3000_1603 | Zfp219 | AGTCCATCACATTCTGTTGCTTTCTAGTCAGCTATGACCATGCAGT | 131 |
2.5. Real‐time quantitative PCR
Mouse hippocampal hemispheres were homogenised in TriPure (Roche, Basel, Switzerland) by shaking the tissue with 1.0‐mm diameter glass beads for 20 seconds at 6.5 m s‐1 in a FastPrep‐24 5G instrument (MP Biomedicals, Santa Ana, CA, USA). Total RNA was isolated, cDNA was generated and quantitative real‐time PCR was performed as described previously.24 Because Actb (β‐actin) expression was regulated between fbMRKO and control mice, genes of interest were normalised against the in both experiments stably expressed housekeeping gene Rplp0, encoding a ribosomal protein. Primer sequences are listed in Table 2.
Table 2.
Primer sequences used for a quantitative real‐time polymerase chain reaction in the mouse hippocampus
| Gene | Full name | Forward and reverse (5′‐ to 3′) | Product length (bp) |
|---|---|---|---|
| Acsl6 | Acyl‐CoA synthetase long‐chain family member 6 | TCTCAGGGAATGGACCCTGTCCTCTTGGTAGGACAGCCAC | 135 |
| Bhlhb9 | Basic helix‐loop‐helix domain containing, class B9 | AACTCACCTGGCCAGCAATCCTCTGGCTGCCTTGGGATTT | 187 |
| C4ST1 (Chst11) | Chondroitin 4‐sulfotransferase 1 | GAATTTGCCGGATGGTGCTGAGCAGATGTCCACACCGAAG | 117 |
| Camk1d | Calcium/calmodulin‐dependent protein kinase ID | GCATCGAGAACGAGATTGCCCCAGACACAAGTTGCATGACC | 114 |
| Camkk2 | Calcium/calmodulin‐dependent protein kinase kinase 2 | AGAACTGCACACTGGTCGAGACCAGGATCACAGTTGCCAG | 85 |
| Fkbp5 | FK506 binding protein 5 | TCCTGGGAGATGGACACCAATTCCCGTACTGAATCACGGC | 113 |
| Gilz (Tsc22d3) | Glucocorticoid‐induced leucine zipper | TGGCCCTAGACAACAAGATTGAGCCCACCTCCTCTCTCACAGCAT | 78 |
| Hsd17b11 | Hydroxysteroid (17‐β) dehydrogenase 11 | CGCAGGACCCTCAGATTGAAGGAGCAGTAAGCCAGCAAGA | 167 |
| Jdp2 | Jun dimerization protein 2 | TACGCTGACATCCGCAACATCGTCTAGCTCACTCTTCACGG | 100 |
| Kif1c | Kinesin family member 1C | TTAATGCCCGTGAGACCAGCAAGCTTTTGGGGGCATCCTT | 106 |
| Mrpl48 | Mitochondrial ribosomal protein L48 | CAGTATGTCCACCGCCTCTGCTCGCTCATGGGTGGTAAGG | 145 |
| Nos1ap | Nitric oxide synthase 1 adaptor protein | TGGAATTCAGCCGAGGTGTGGGAAGGGAGCAGCATTCGAG | 131 |
| Nr3c1 (GR) | Nuclear receptor subfamily 3, group C, member 1 | CCCTCCCATCTAACCATCCTT ACATAAGCGCCACCTTTCTG | 89 |
| Nr3c2 (MR) | Nuclear receptor subfamily 3, group C, member 2 | TCCAAGATCTGCTTGGTGTGCCCAGCTTCTTTGACTTTCG | 239 |
| Per1 | Period circadian clock 1 | ACGGCCAGGTGTCGTGATTACCCTTCTAGGGGACCACTCA | 162 |
| Rilpl1 | Rab interacting lysosomal protein‐like 1 | ACGAGCTCAAGTCCAAGGTGAGTCGCTTGATCCCCGATTC | 148 |
| Rplp0 | Ribosomal protein, large, P0 | GGACCCGAGAAGACCTCCTTGCACATCACTCAGAATTTCAATGG | 85 |
| Sgk1 | Serum/glucocorticoid regulated kinase 1 | AGAGGCTGGGTGCCAAGGATCACTGGGCCCGCTCACATTT | 129 |
| Supv3 l1 | Suv3 like RNA helicase | CTCACTCGGCCTCTAGACAAGTCCACGTCCAGAGAATGGGA | 170 |
| Zfp219 | Zinc finger protein 219 | GATCTGCAGCGCTACTCCAATGCACGAGTCTCAGACCAAC | 96 |
2.6. In situ hybridisation
Frozen brains were sectioned at 18 μm in a cryostat microtome, collected on SuperFrost Plus slides (Thermo Scientific, Walthaqm, MA, USA) and stored at −80°C until further use. In situ hybridisation using 35S UTP‐labelled ribonucleotide probes for Jdp2 was performed as described previously.28
2.7. Statistical analysis
In the fbMRKO experiment, independent t‐tests were used and P < 0.01 was considered as a statistically significance cut‐off to correct for multiple gene testing. For the ChIP‐quantitative PCR validation, we performed one‐tailed paired t tests. The predictable directionality (ie, the MR signal is higher than the background IgG signal) justifies the use of a one‐tailed test. Because a decrease in signal might also be relevant, we note that all statisitically significant results were at P < 0.025 and therefore would also be significant using a two‐tailed test. We considered a paired test appropriate because MR and IgG are measured on the same chromatin sample and this allows correction for the corresponding background levels. Again, one‐tailed unpaired t tests gave essentially the same results. For the gene Nos1ap, one of the samples was excluded from analysis because of a missing value as a result of non‐detectable IgG levels. For the time course of restraint stress, a one‐way ANOVA was performed with Holm–Sidak's multiple comparison post‐hoc tests. In the in situ measurements of the fbMRKO animals, unpaired t tests were performed. P < 0.05 was considered statistically significantly significant unless stated otherwise. prism, version 7 (GraphPad Software Inc., San Diego, CA, USA) was used to analyse the data. All graphs show individual values and data are further depicted as the mean ± SEM.
3. RESULTS
To explore the functional effects of previously detected MR/GR DNA binding (ie, transcription regulation), binding sites were associated with their nearest gene. This resulted in lists of MR‐specific, MR‐GR overlapping and GR‐specific putative target genes.24 Gene expression levels, for a subset of each category (Table 3), were measured in fbMRKO mice at the time of their diurnal corticosterone peak. MR mRNA expression was indeed abolished and GR mRNA was slightly up‐regulated in the hippocampus of fbMRKO mice (Figure 1A), confirming earlier reports.25 MR protein levels also showed efficient knockdown Bonapersona V., Damsteegt R., Adams M.L., van Weert L., Meijer O.C., Joëls M., Sarabdjitsingh R.A.(unpublished). Furthermore, no differences were found in plasma corticosterone levels of these animals at the time of death (Figure 1B). Because the studied target loci were originally detected in the rat brain,24 we validated MR binding in mice. ChIP‐quantitative PCR confirmed hippocampal MR binding at the Jdp2 (P = 0.0124), Kif1c (P = 0.0087), Nos1ap (P = 0.0172), Rilpl1 (P = 0.0098) and Zfp219 (P = 0.0049) loci in WT mice, although this signal did not exceed background IgG levels at the GR‐specific sites near Acsl6 (P = 0.4410) and Mrpl48 (P = 0.2142) (Figure 1C). Only for Supv3 l1 (P = 0.1784) were we unable to detect the expected MR binding. Also, for classical target genes Fkbp5 (P = 0.0246) and Per1 (P = 0.0066), MR enrichment was demonstrated.
Table 3.
Selected putative target genes to validate
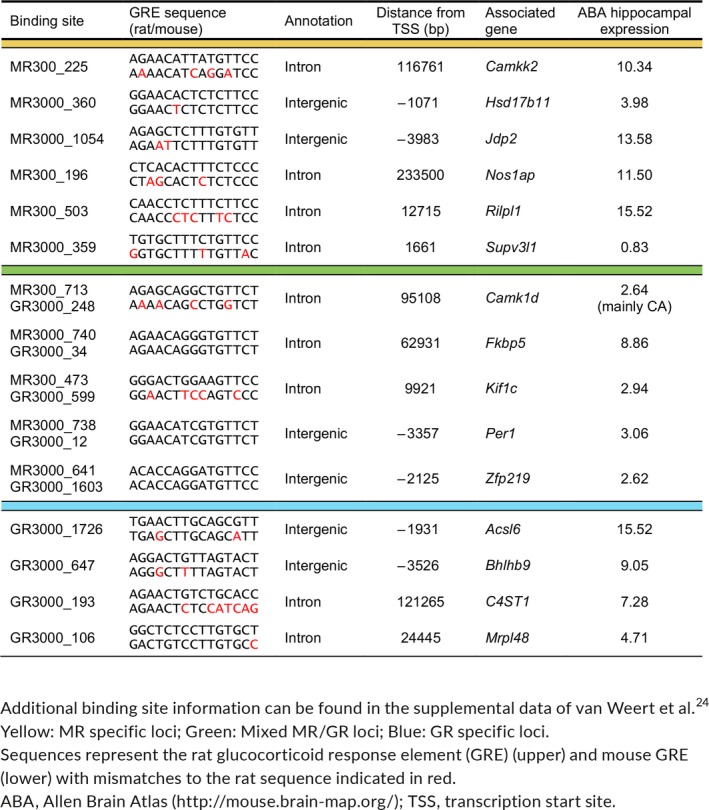
Figure 1.
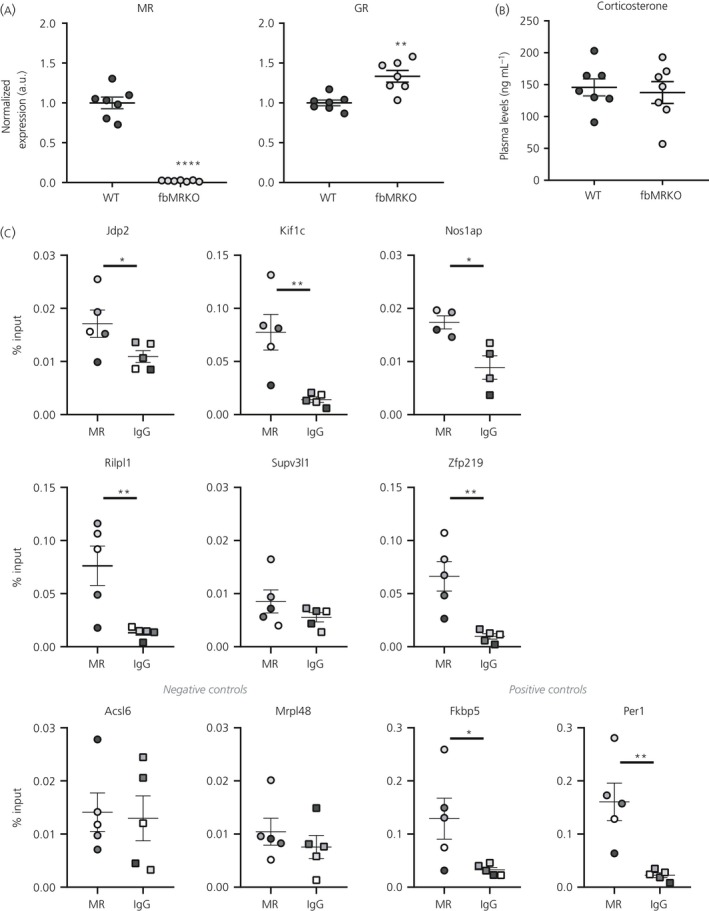
Validation of mineralocorticoid receptor (MR) detection in wild‐type (WT) mice and absence of MR in forebrain MR knockout (fbMRKO) mice. (A) Hippocampal mRNA levels showing MR down‐regulation and slight GR up‐regulation, as well as (B) unaltered plasma corticosterone levels in fbMRKO vs WT mice, as assessed by independent t‐tests. (C) MR binding assessed by a chromatin immunoprecipitation‐quantitative polymerase chain reaction in the hippocampus of WT mice, along with an immunoglobulin (Ig)G background signal per sample, as assessed by one‐tailed paired t‐tests. Corresponding measurements are depicted in the same colour. GR‐specific targets Acsl6 and Mrpl48 served as negative controls; classical glucocorticoid targets Fkbp5 and Per1 served as positive controls. a.u., arbitrary unit. *P < 0.05, **P < 0.01, ****P < 0.0001
Several MR‐specific putative targets showed lower expression levels in the fbMRKO compared to WT mice (Figure 2A). The most robust effect was found in the Jdp2 mRNA levels, which were reduced by 50% (P < 0.0001). Other differentially expressed genes were MR‐specific Nos1ap (P = 0.0005) and Supv3 l1 (P = 0.0061), as well as MR‐GR overlapping Camk1d (P = 0.0016) and Kif1c (P = 0.0022), which were also all down‐regulated in the fbMRKO compared to WT mice (Figure 2A,B). Moreover, two of the GR‐specific genes, Acsl6 (P = 0.0002) and Mrpl48 (P = 0.0065), were expressed at lower levels, and C4ST1 showed a trend for lowered expression (P = 0.0138) (Figure 2C).
Figure 2.
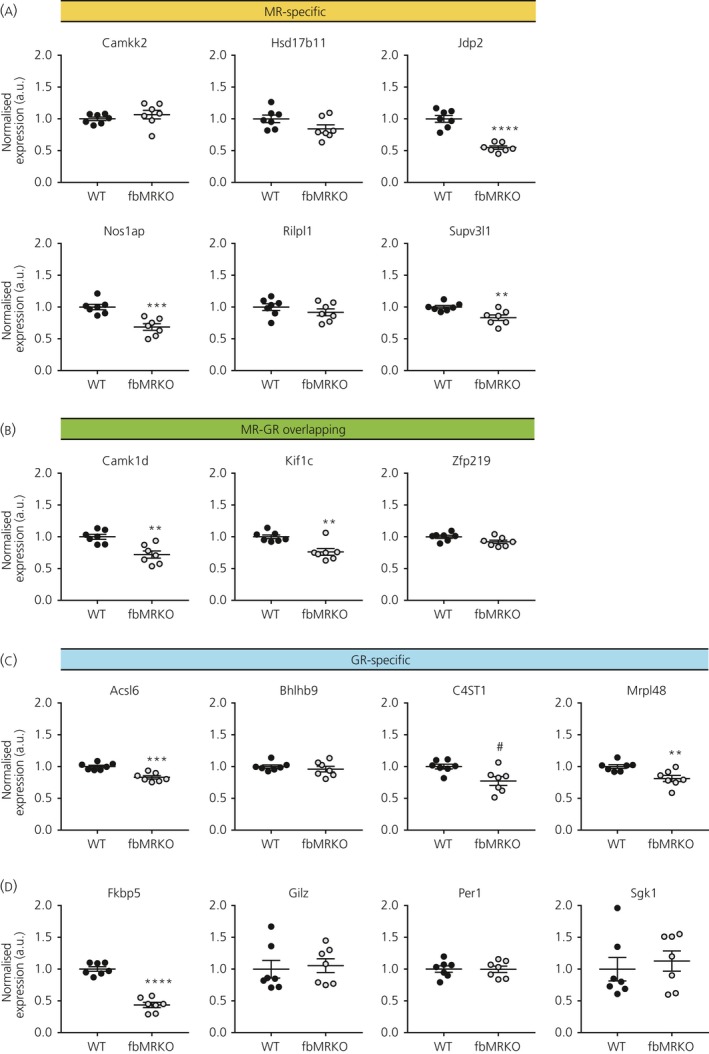
Hippocampal mRNA levels of glucocorticoid target genes assessed in wild‐type (WT) and forebrain mineralocorticoid receptor (MR) knockout (fbMRKO) mice. Gene expression of (A) MR‐specific, (B) overlapping and (C) glucocorticoid receptor (GR)‐specific targets and (D) classical glucocorticoid targets in fbMRKO versus WT mice, as assessed by independent t‐tests with P < 0.01 as the statistically significant cut‐off. Other genes measured but not differentially expressed between WT and fbMRKO mice were: Adam23, Arl8b, Dgkb, Els1, Myo16 and Nob1 as MR‐specific targets; Grb2, Luzp1 and Map1lc3b as overlapping targets; Arntl, B3galt1, Map2k5, Pglyrp1 and Slc3a2 as GR‐specific targets. a.u., arbitrary unit. #P < 0.05 (considered a trend), **P < 0.01, ***P < 0.001, ****P < 0.0001
Besides the brain‐related putative MR/GR target genes, we measured the expression of the classical target genes Fkbp5, Gilz, Per1 and Sgk1 (Figure 2D). These genes are all known to be bound and/or regulated by both MR and GR, comprising our identified MR‐GR overlapping target subset contained Fkbp5 and Per1 (Table 3). Of the four classical targets, only Fkbp5 was down‐regulated in fbMRKO mice, reaching 44% of the levels observed in WT animals (P < 0.0001).
Next, we aimed to show regulation of the target genes in an acute stress context. Even though MR is substantially occupied by ligand under basal glucocorticoid levels, MR (and GR) DNA binding and subsequent transcriptional effects can be enhanced by a rise of corticosterone.15 Hippocampal gene expression was assessed in mice that were exposed to restraint stress of different durations (0, 30, 60, 120 and 240 minutes). Plasma corticosterone levels were increased after all durations of restraint stress, although they tended to return to baseline at 120 and 240 minutes, in line with the fact that ACTH levels were normalised at these time points (Figure 3A).
Figure 3.
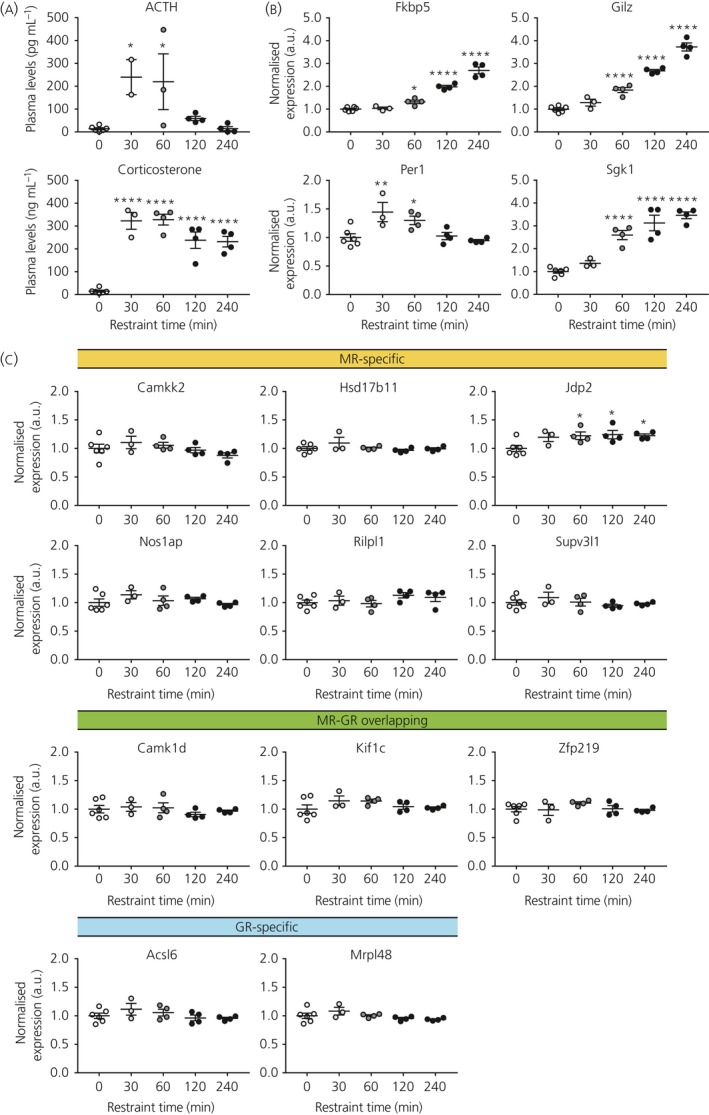
Hippocampal mRNA levels of glucocorticoid target genes assessed in a restraint stress model. A, Plasma adrenocorticotrophic hormone (ACTH) and corticosterone levels after different durations of restraint stress. B, Validation of time‐dependent classical glucocorticoid target gene activation upon restraint stress. C, Gene expression of MR‐specific, overlapping and GR‐specific targets after different durations of restraint stress. All assessed by one‐way ANOVA with Holm‐Sidak's post‐hoc tests. a.u., arbitrary unit. *P < 0.05, **P < 0.01, ****P < 0.0001
Of the classical glucocorticoid target genes, Fkbp5, Gilz and Sgk1 were up‐regulated after 60, 120 and 240 minutes of restraint (Figure 3B). Per1 showed a transient increase, with elevated levels at 30 and 60 minutes, which had declined again from 120 minutes of restraint stress. Interestingly, the MR‐exclusive target gene Jdp2 that was mostly affected in fbMRKO mice showed an increase in response to stress (Figure 3C) in those animals exposed to restraint for 60‐240 minutes. Other genes associated with MR and/or GR binding loci that we had selected for validation did not show transcriptional effects upon restraint stress (Figure 3C).
Finally, we confirmed Jdp2 down‐regulation measured by in situ hybridisation in an independent experiment in fbMRKO (Figure 4). In the absence of MR, Jdp2 mRNA levels were decreased in the principal neurones of the dorsal hippocampus, as was apparent from a significantly lower expression in the CA2 (P = 0.0001), CA3 (P = 0.0357) and dentate gyrus (P = 0.0005) subregions. For the CA1, this occurred at the trend level (P = 0.0901).
Figure 4.
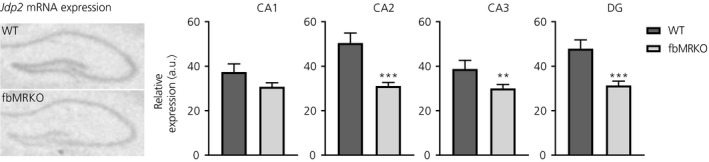
Validation of hippocampal Jdp2 down‐regulation in forebrain mineralocorticoid receptor (MR) knockout (fbMRKO) mice compared to wild‐type (WT) mice, detected by in situ hybridisation, as assessed by unpaired t‐tests. Left: representative scanned autoradiograph film per genotype. Gene expression is quantified per subregion of the hippocampus: cornu ammonis (CA)1, CA2, CA3 and the dentate gyrus (DG). a.u., arbitrary unit. **P < 0.01, ***P < 0.001
4. DISCUSSION
Based on non‐overlapping MR‐GR binding sites, we defined putative MR‐specific and GR‐specific hippocampal target genes. We identified Jdp2 as a likely MR‐specific transcriptional target that is both down‐regulated in fbMRKO mice and up‐regulated in response to restraint stress. Also, Nos1ap and Supv3 l1, two other genes linked to MR‐specific binding sites, were expressed at a lower levels in fbMRKO mice but did not change upon restraint stress. Classical glucocorticoid target genes Fkbp5, Gilz, Per1 and Sgk1 all responded to restraint stress by increased transcription. Of these targets, only Fkbp5 showed a substantially lower hippocampal expression in the absence of MR.
Both technical and biological factors could explain the limited success with respect to validating MR‐specific genomic targets. The annotation of binding sites to the nearest gene is not without error because it is possible that another neighbouring gene is affected by the binding locus assessed. We do not have data on spatial chromatin organisation or RNA polymerase activity in the same experimental set‐up, which could enable the proper linking of binding loci to the actual site of transcriptional activity.29 To lower the chance of false positive annotations, we focused on binding sites that were located within genes or (proximal) promoter regions. However, even in cases where the putative target is legitimate, we might still have false negative results regarding gene expression changes. Because the hippocampus consists of several subregions and various cell types, we might not be able to detect MR‐dependent regulation that is constrained to a subset of hippocampal cells. Although the ChIP‐seq signal can be sufficiently strong to withstand dilution, gene regulation might be diluted when the average gene expression over the whole hippocampus is assessed because the fold change in hippocampal mRNA expression tends to be modest in response to steroids.30 Despite possible false negative results, we were able to find robust changes in several MR‐specific and classical glucocorticoid target genes.
It is of note that gene regulation by MR knockout and restraint stress was validated in a mouse model, whereas the MR/GR binding loci were obtained from experiments in rats. We were able to show MR binding in the mouse hippocampus at five out of six MR targets originally detected in the rat brain. Evolutionary conservation can increase the predictive value of functional GREs.31, 32 Moreover, because brain MR/GR‐mediated regulation is considered to be part of a general adaptive response, the genes regulated in the rat would also be expected to be affected in mice. However, the species difference is an additional potential cause for absence of mRNA regulation.
The fbMRKO animals become MR deficient during embryonic development and loss of MR protein is completed after birth.25 In the present study, down‐regulated MR expression was validated and slight up‐regulation of GR expression in the hippocampus was observed as described previously.25 It is possible that MR‐dependent gene expression is normalised as a result of compensation by GR or other factors. We cannot exclude that such compensatory mechanisms might also affect expression of Jdp2, Nos1ap and Supv3 l1 in fbMRKO mice. Also, redundancy in gene regulation is not uncommon and, although complete dependence of target genes to a single transcription factor can occur,33 it is rare in case of MR and GR signalling. In addition, the binding of nuclear receptors such as MR can have permissive effects on chromatin and could be necessary but not sufficient for transcription. Indeed, as little as 13% of GR binding sites can be linked to transcriptional activity.34 Thus, the lack of transcriptional effects might reflect a context dependency.
To begin looking at MR regulation in a relevant context, we chose a restraint stress paradigm in WT mice as a more physiological setting. Mice were stressed in the morning to ensure that basal corticosterone levels were low, and MR activation not necessarily fully maximal.35 The classical glucocorticoid target genes all responded in this acute stress situation and, of the MR‐specific targets identified in fbMRKO mice, only Jdp2 expression was affected. Non‐regulated genes in the restraint stress experiment might still be MR‐dependent but at a lower EC50,36 or in different contexts, including behavioural paradigms in which fbMRKO animals show changed phenotypes, such as working memory in a radial maze.25
For the genes associated with GR‐specific chromatin binding, Acsl6 and Mrpl48 showed lower expression levels in fbMRKO mice. In general, the effect size on specifically GR‐associated target gene expression was less pronounced. The fact that these GR targets are down‐regulated and the expression of GR itself is slightly up‐regulated in fbMRKO mice appears to be contradictory. However, this could be a result of indirect effects of MR deficiency. Another explanation is that GR binding takes place at a negative GRE, where GR leads to repression (instead of activation) of the nearby gene.37, 38
More interestingly, several overlapping targets were down‐regulated in fbMRKO mice: the newly identified Camk1d and Kif1c, and the classical target Fkbp5. This suggests that MR is needed for expression of these genes in the hippocampus. The GR compensatory up‐regulation does not appear to prevent dysregulation of these combined target genes in the absence of MR. It is likely that heterodimerisation of MR and GR is involved in the regulation of overlapping binding sites. Fkbp5 expression was recently shown to be modulated by MR‐GR heterodimers.15 The observation that Fkbp5 expression is lowered in fbMRKO mice can represent functional consequences of the absence of one of the heterodimerisation partners. Fkbp5 is part of an ultra‐short feedback loop in which it is induced by glucocorticoids, whereas, in turn, Fkbp5 prevents GR activation.39 Besides the observed up‐regulation of GR expression itself, the lowered Fkbp5 levels could contribute to a compensatory mechanism by relieving the repression of GR function to overcome the lack of MR signalling.
Overall, the Jdp2 gene was the most robust MR target identified in the present study. Initially, Jdp2 was discovered as a negative regulator of activator protein‐1 (AP‐1) function, by dimerising to c‐Jun and preventing transcriptional effects.40 Later, it was found that Jdp2 can also act in a stimulating fashion as a coactivator for the progesterone receptor.41 In this latter study, Jdp2 was also shown to have a coactivating effect on transactivation by GR, as confirmed by Garza et al.42 We found Jdp2 to be a bona fide MR target. A feedforward mechanism could be speculated in which MR can increase Jdp2 levels, which, in turn, could enhance GR activity. A recent ChIP‐seq study in mouse neuroblastoma cells found the Jdp2 binding motif near both MR‐ and GR‐bound sites.43 Besides the differential affinity of MR and GR for their hormone, temporal responses to glucocorticoids could be accounted for by such a feedforward loop. Feedforward models have been described previously for GR44 and other nuclear receptors.45, 46 It is worth noting that Jdp2 has been implicated in AP‐1 modulation during fear extinction,47 and polymorphisms in the Nos1ap gene have been linked to post‐traumatic stress disorder and depression,48 also demonstrating a functional role of these genes in the stress system.
In conclusion, we found three novel hippocampal MR‐specific target genes, comprising Jdp2, Nos1ap and Supv3 l1, of which Jdp2 is also responsive in an acute stress situation. Dissecting the glucocorticoid response in MR‐specific, common and GR‐specific pathways will enable us to better understand the stress physiology and pathophysiology of stress‐related disorders.
CONFLICT OF INTERESTS
The authors declare that they have no conflicts of interest.
ACKNOWLEDGEMENTS
We thank Trea Streefland, Ute Burret, Sylvie Lesuis and Karianne Schuurman for technical assistance, as well as Marian Joëls for critically reading the manuscript. This research was supported by NWO ALW grant 823.02.002, COST Action ADMIRE BM1301 and the Deutsche Forschungsgemeinschaft CRC1149/C02 INST 40/492‐1.
van Weert LTCM, Buurstede JC, Sips HCM, et al. Identification of mineralocorticoid receptor target genes in the mouse hippocampus. J Neuroendocrinol. 2019;31:e12735 10.1111/jne.12735
van Weert and Buurstede contributed equally.
REFERENCES
- 1. Reul JM, de Kloet ER. Two receptor systems for corticosterone in rat brain: microdistribution and differential occupation. Endocrinology. 1985;117:2505‐2511. [DOI] [PubMed] [Google Scholar]
- 2. Joels M, Karst H, DeRijk R, de Kloet ER. The coming out of the brain mineralocorticoid receptor. Trends Neurosci. 2008;31:1‐7. [DOI] [PubMed] [Google Scholar]
- 3. de Kloet ER, Joels M, Holsboer F. Stress and the brain: from adaptation to disease. Nat Rev Neurosci. 2005;6:463‐475. [DOI] [PubMed] [Google Scholar]
- 4. Joels M, de Kloet ER. Mineralocorticoid receptor‐mediated changes in membrane properties of rat CA1 pyramidal neurons in vitro. Proc Natl Acad Sci USA. 1990;87:4495‐4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Karst H, Karten YJ, Reichardt HM, de Kloet ER, Schutz G, Joels M. Corticosteroid actions in hippocampus require DNA binding of glucocorticoid receptor homodimers. Nat Neurosci. 2000;3:977‐978. [DOI] [PubMed] [Google Scholar]
- 6. Oitzl MS, de Kloet ER. Selective corticosteroid antagonists modulate specific aspects of spatial orientation learning. Behav Neurosci. 1992;106:62‐71. [DOI] [PubMed] [Google Scholar]
- 7. Oitzl MS, Reichardt HM, Joels M, de Kloet ER. Point mutation in the mouse glucocorticoid receptor preventing DNA binding impairs spatial memory. Proc Natl Acad Sci USA. 2001;98:12790‐12795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Datson NA, van der Perk J, de Kloet ER, Vreugdenhil E. Identification of corticosteroid‐responsive genes in rat hippocampus using serial analysis of gene expression. Eur J Neuorsci. 2001;14:675‐689. [DOI] [PubMed] [Google Scholar]
- 9. Pascual‐Le Tallec L, Lombes M. The mineralocorticoid receptor: a journey exploring its diversity and specificity of action. Mol Endocrinol. 2005;19:2211‐2221. [DOI] [PubMed] [Google Scholar]
- 10. Yang J, Fuller PJ, Morgan J, et al. Use of phage display to identify novel mineralocorticoid receptor‐interacting proteins. Mol Endocrinol. 2014;28:1571‐1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Joels M, Sarabdjitsingh RA, Karst H. Unraveling the time domains of corticosteroid hormone influences on brain activity: rapid, slow, and chronic modes. Pharmacol Rev. 2012;64:901‐938. [DOI] [PubMed] [Google Scholar]
- 12. Karst H, Berger S, Turiault M, Tronche F, Schutz G, Joels M. Mineralocorticoid receptors are indispensable for nongenomic modulation of hippocampal glutamate transmission by corticosterone. Proc Natl Acad Sci USA. 2005;102:19204‐19207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Latouche C, Sainte‐Marie Y, Steenman M, et al. Molecular signature of mineralocorticoid receptor signaling in cardiomyocytes: from cultured cells to mouse heart. Endocrinology. 2010;151:4467‐4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Soundararajan R, Zhang TT, Wang J, Vandewalle A, Pearce D. A novel role for glucocorticoid‐induced leucine zipper protein in epithelial sodium channel‐mediated sodium transport. J Biol Chem. 2005;280:39970‐39981. [DOI] [PubMed] [Google Scholar]
- 15. Mifsud KR, Reul JM. Acute stress enhances heterodimerization and binding of corticosteroid receptors at glucocorticoid target genes in the hippocampus. Proc Natl Acad Sci USA. 2016;113:11336‐11341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen SY, Bhargava A, Mastroberardino L, et al. Epithelial sodium channel regulated by aldosterone‐induced protein sgk. Proc Natl Acad Sci USA. 1999;96:2514‐2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baughman G, Wiederrecht GJ, Chang F, Martin MM, Bourgeois S. Tissue distribution and abundance of human FKBP51, and FK506‐binding protein that can mediate calcineurin inhibition. Biochem Biophys Res Comm. 1997;232:437‐443. [DOI] [PubMed] [Google Scholar]
- 18. D'Adamio F, Zollo O, Moraca R, et al. A new dexamethasone‐induced gene of the leucine zipper family protects T lymphocytes from TCR/CD3‐activated cell death. Immunity. 1997;7:803‐812. [DOI] [PubMed] [Google Scholar]
- 19. Conway‐Campbell BL, Sarabdjitsingh RA, McKenna MA, et al. Glucocorticoid ultradian rhythmicity directs cyclical gene pulsing of the clock gene period 1 in rat hippocampus. J Neuroendocrinol. 2010;22:1093‐1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Webster MK, Goya L, Ge Y, Maiyar AC, Firestone GL. Characterization of sgk, a novel member of the serine/threonine protein kinase gene family which is transcriptionally induced by glucocorticoids and serum. Mol Cell Biol. 1993;13:2031‐2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meijer OC, de Kloet ER. Corticosterone and serotonergic neurotransmission in the hippocampus: functional implications of central corticosteroid receptor diversity. Crit Rev Neurobiol. 1998;12:1‐20. [PubMed] [Google Scholar]
- 22. Meinel S, Ruhs S, Schumann K, et al. Mineralocorticoid receptor interaction with SP1 generates a new response element for pathophysiologically relevant gene expression. Nucleic Acids Res. 2013;41:8045‐8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meijer OC, de Kloet ER. A role for the mineralocorticoid receptor in a rapid and transient suppression of hippocampal 5‐HT1A receptor mRNA by corticosterone. J Neuroendocrinol. 1995;7:653‐657. [DOI] [PubMed] [Google Scholar]
- 24. van Weert LTCM, Buurstede JC, Mahfouz A, et al. NeuroD factors discriminate mineralocorticoid from glucocorticoid receptor DNA Binding in the male rat brain. Endocrinology. 2017;158:1511‐1522. [DOI] [PubMed] [Google Scholar]
- 25. Berger S, Wolfer DP, Selbach O, et al. Loss of the limbic mineralocorticoid receptor impairs behavioral plasticity. Proc Natl Acad Sci USA. 2006;103:195‐200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lein ES, Hawrylycz MJ, Ao N, et al. Genome‐wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168‐176. [DOI] [PubMed] [Google Scholar]
- 27. Singh AA, Schuurman K, Nevedomskaya E, et al. Optimized ChIP‐seq method facilitates transcription factor profiling in human tumors. Life Sci Alliance. 2019;2:e201800115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Santarelli S, Zimmermann C, Kalideris G, et al. An adverse early life environment can enhance stress resilience in adulthood. Psychoneuroendocrinology. 2017;78:213‐221. [DOI] [PubMed] [Google Scholar]
- 29. Davies JO, Oudelaar AM, Higgs DR, Hughes JR. How best to identify chromosomal interactions: a comparison of approaches. Nat Methods. 2017;14:125‐134. [DOI] [PubMed] [Google Scholar]
- 30. Datson NA, van den Oever JM, Korobko OB, Magarinos AM, de Kloet ER, McEwen BS. Previous history of chronic stress changes the transcriptional response to glucocorticoid challenge in the dentate gyrus region of the male rat hippocampus. Endocrinology. 2013;154:3261‐3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. So AY, Cooper SB, Feldman BJ, Manuchehri M, Yamamoto KR. Conservation analysis predicts in vivo occupancy of glucocorticoid receptor‐binding sequences at glucocorticoid‐induced genes. Proc Natl Acad Sci USA. 2008;105:5745‐5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Datson NA, Polman JA, de Jonge RT, et al. Specific regulatory motifs predict glucocorticoid responsiveness of hippocampal gene expression. Endocrinology. 2011;152:3749‐3757. [DOI] [PubMed] [Google Scholar]
- 33. Finotto S, Krieglstein K, Schober A, et al. Analysis of mice carrying targeted mutations of the glucocorticoid receptor gene argues against an essential role of glucocorticoid signalling for generating adrenal chromaffin cells. Development. 1999;126:2935‐2944. [DOI] [PubMed] [Google Scholar]
- 34. Vockley CM, D'Ippolito AM, McDowell IC, et al. Direct GR binding sites potentiate clusters of TF binding across the human genome. Cell. 2016;166:pp. 1269–81 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meijer OC, Van Oosten RV, De Kloet ER. Elevated basal trough levels of corticosterone suppress hippocampal 5‐hydroxytryptamine(1A) receptor expression in adrenally intact rats: implication for the pathogenesis of depression. Neuroscience. 1997;80:419‐426. [DOI] [PubMed] [Google Scholar]
- 36. Reddy TE, Pauli F, Sprouse RO, et al. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19:2163‐2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sharma D, Bhave S, Gregg E, Uht R. Dexamethasone induces a putative repressor complex and chromatin modifications in the CRH promoter. Mol Endocrinol. 2013;27:1142‐1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Surjit M, Ganti KP, Mukherji A, et al. Widespread negative response elements mediate direct repression by agonist‐liganded glucocorticoid receptor. Cell. 2011;145:224‐241. [DOI] [PubMed] [Google Scholar]
- 39. Binder EB. The role of FKBP5, a co‐chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psychoneuroendocrinology. 2009;34(Suppl 1):S186‐S195. [DOI] [PubMed] [Google Scholar]
- 40. Aronheim A, Zandi E, Hennemann H, Elledge SJ, Karin M. Isolation of an AP‐1 repressor by a novel method for detecting protein‐protein interactions. Mol Cell Biol. 1997;17:3094‐3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wardell SE, Boonyaratanakornkit V, Adelman JS, Aronheim A, Edwards DP. Jun dimerization protein 2 functions as a progesterone receptor N‐terminal domain coactivator. Mol Cell Biol. 2002;22:5451‐5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Garza AS, Khan SH, Moure CM, Edwards DP, Kumar R. Binding‐folding induced regulation of AF1 transactivation domain of the glucocorticoid receptor by a cofactor that binds to its DNA binding domain. PLoS One. 2011;6:e25875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rivers CA, Rogers MF, Stubbs FE, Conway‐Campbell BL, Lightman SL, Pooley JR. Glucocorticoid receptor tethered mineralocorticoid receptors increase glucocorticoid‐induced transcriptional responses. Endocrinology. 2019;160:1044‐1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sasse SK, Zuo Z, Kadiyala V, et al. Response element composition governs correlations between binding site affinity and transcription in glucocorticoid receptor feed‐forward loops. J Biol Chem. 2015;290:19756‐19769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pabona JM, Simmen FA, Nikiforov MA, et al. Kruppel‐like factor 9 and progesterone receptor coregulation of decidualizing endometrial stromal cells: implications for the pathogenesis of endometriosis. J Clin Endocrinol Metab. 2012;97:E376‐E392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Villanueva CJ, Vergnes L, Wang J, et al. Adipose subtype‐selective recruitment of TLE3 or Prdm16 by PPARgamma specifies lipid storage versus thermogenic gene programs. Cell Metab. 2013;17:423‐435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guedea AL, Schrick C, Guzman YF, et al. ERK‐associated changes of AP‐1 proteins during fear extinction. Mol Cell Neurosci. 2011;47:137‐144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bruenig D, Morris CP, Mehta D, et al. Nitric oxide pathway genes (NOS1AP and NOS1) are involved in PTSD severity, depression, anxiety, stress and resilience. Gene. 2017;625:42‐48. [DOI] [PubMed] [Google Scholar]


