Summary
The Type VI secretion system (T6SS) is a bacterial nanomachine that delivers effector proteins into prokaryotic and eukaryotic preys. This secretion system has emerged as a key player in regulating the microbial diversity in a population. In the plant pathogen Agrobacterium tumefaciens, the signalling cascades regulating the activity of this secretion system are poorly understood. Here, we outline how the universal eubacterial second messenger cyclic di‐GMP impacts the production of T6SS toxins and T6SS structural components. We demonstrate that this has a significant impact on the ability of the phytopathogen to compete with other bacterial species in vitro and in planta. Our results suggest that, as opposed to other bacteria, c‐di‐GMP turns down the T6SS in A. tumefaciens thus impacting its ability to compete with other bacterial species within the rhizosphere. We also demonstrate that elevated levels of c‐di‐GMP within the cell decrease the activity of the Type IV secretion system (T4SS) and subsequently the capacity of A. tumefaciens to transform plant cells. We propose that such peculiar control reflects on c‐di‐GMP being a key second messenger that silences energy‐costing systems during early colonization phase and biofilm formation, while low c‐di‐GMP levels unleash T6SS and T4SS to advance plant colonization.
Introduction
Interspecies bacterial competition plays a key role in shaping microbial populations and determining what bacterial species are dominant in a given niche (Kapitein and Mogk, 2014; Bernal et al., 2017a; McNally et al., 2017; Chassaing and Cascales, 2018). It is indeed increasingly clear that any specific environmental niche, including within various parts of the human body (Rojo et al., 2017), will profile the establishment of specific microbial populations. How bacteria respond to environmental variations or intrusion of ‘foreign organisms’ by triggering an attack/defence war game with other bacteria is not clearly understood. Yet, despite the numerous protein secretion systems encoded within bacterial genomes, one has emerged as being the ‘weapon’ of choice through which bacteria mediate interspecies competition, for example, in the human gut (Russell et al., 2014b; Chatzidaki‐Livanis et al., 2016; Sana et al., 2016; Anderson et al., 2017) or in planta (Ma et al., 2014; Bernal et al., 2017a; 2017b). This system, termed the Type VI Secretion System (T6SS), is found on the genome of a wide variety of Gram‐negative bacteria and can deliver a remarkable array of toxins such as nucleases, amidases and phospholipases (Russell et al., 2011; Russell et al., 2012; Ma et al., 2014; Russell et al., 2014a; Alcoforado Diniz et al., 2015). It is noticeable that in Gram‐positive bacteria the type VII secretion system (T7SS) is now emerging as the antibacterial nano‐weapon (Cao et al., 2016). Whereas many of these toxins are antibacterial (Russell et al., 2014a), some are also designed to target eukaryotic host cells (Hachani et al., 2016) and fungi (Trunk et al., 2018) or have a dual function like phospholipases (Jiang et al., 2014). These toxins can be encoded as part of a large T6SS cluster and thus genetically linked with genes encoding core T6SS components, or be found independently in so‐called vgrG/hcp islands (Hachani et al., 2014; Whitney et al., 2014). It has recently emerged that T6SS toxins can specifically interact with VgrG proteins (Bondage et al., 2016; Cianfanelli et al., 2016a; Flaugnatti et al., 2016), the puncturing device of the T6SS nanomachine, or can be found fused to VgrG thus forming what is referred to as an evolved VgrG (Pukatzki et al., 2007; Durand et al., 2012) or fused to a PAAR domain which loads the toxin onto a cognate VgrG (Whitney et al., 2015; Quentin et al., 2018). Some bacterial species have been shown to encode numerous T6SSs, with experimental evidence highlighting specific target or prey such as the H1‐T6SS in Pseudomonas aeruginosa being associated with interbacterial competition while the H2‐T6SS was proposed to preferentially target eukaryotic cells (Sana et al., 2012; Hachani et al., 2014; Jones et al., 2014). Now, it seems that a clear‐cut system specificity is unlikely to be the case as the H2‐T6SS was recently shown to also be antibacterial (Allsopp et al., 2017).
Agrobacterium tumefaciens is a soil bacterium that causes crown gall disease in a wide range of plants as a result of the delivery of T‐DNA to plant cells via the Type IV Secretion System (T4SS) (Pitzschke and Hirt, 2010; Christie et al., 2014; Hwang et al., 2017). Agrobacterium tumefaciens strain C58 has also been shown to carry a single T6SS cluster composed of two divergently transcribed operons and featuring three toxins, Tae, a putative peptidoglycan amidase, and two DNases, Tde1 and Tde2 (Fig. 1A) (Ma et al., 2014). The Tde effectors have been shown to play a role in interbacterial competition and plant colonization. While ample information is now available consistently describing the structural components of the T6SS nanomachine (Cianfanelli et al., 2016b; Joshi et al., 2017; Nguyen et al., 2018) and its associated arsenal of toxins (Durand et al., 2014; Hachani et al., 2016; Lien and Lai, 2017), the type of regulatory elements that control expression of the system is variable from one species to another and has not always been conclusively determined. Well characterized signalling cascades have been shown to play a role in activating the T6SS such as the Gac/Rsm cascade (Moscoso et al., 2011; Allsopp et al., 2017) and the LasR/MvfR (PqsR) quorum sensing systems in P. aeruginosa (Lesic et al., 2009; Majerczyk et al., 2016; Sana et al., 2012), or, for example, the autoinducer‐2 quorum sensing system and QstR in Vibrio cholerae (Ishikawa et al., 2009; Jaskolska et al., 2018). The environmental stimuli that may trigger T6SS expression are also poorly understood but some modulators have been proposed to be bile salts (Reen et al., 2012; Bachmann et al., 2015), contact dependent and counter‐attack based induction (Russell et al., 2011; Basler et al., 2013), part of the competence regulon in V. cholerae (Borgeaud et al., 2015), or danger signal released from lysing cells in a P. aeruginosa population (LeRoux et al., 2015). To date in A. tumefaciens C58, only the ExoR/ChvG/ChvI signalling cascade has been shown to be capable of regulating the T6SS, with induction of this system at low environmental pH leading to activation of the T6SS (Wu et al., 2012). This is believed to play a key role in allowing A. tumefaciens dominate around the site of a wound in a plant, as low pH is characteristic of this niche (Wu et al., 2012; Heckel et al., 2014).
Figure 1.
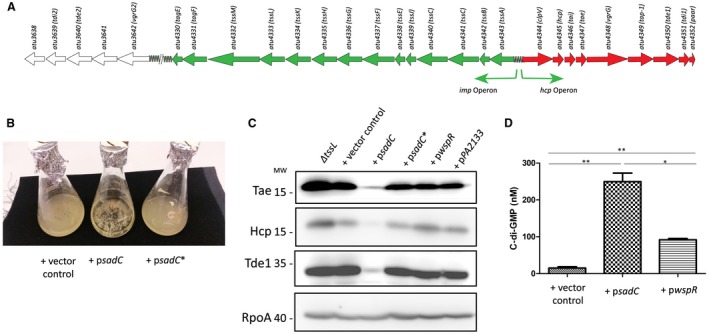
Impact of c‐d‐GMP on Tde1, Tae and Hcp levels. A. The T6SS cluster composed of the divergently transcribed imp (Green) and hcp (Red) operons encoding 14 and 9 genes, respectively, and the remote island starting from atu3642 (vgrG‐2) encoded by A. tumefaciens strain C58. B. Expression of wild‐type SadC and a catalytically inactive SadC (psadC*) in wild‐type A. tumefaciens grown to early exponential phase in 523 medium. C. Western blots using Tde1, Tae, Hcp and RpoA (Loading Control) antibodies on whole lysates of ΔtssL cells (No secretion control) or wild‐type cells transformed with either pBBRMCS4 (vector control), psadC, psadC*, pwspR and pPA2133 grown to early exponential phase in 523 medium. D. C‐di‐GMP levels were quantified via LC‐MS/MS. Transformed cells were grown for 16 h in 523 medium and cells equivalent to an OD600 5 were collected. Samples of interest were compared to a standard curve derived from measurements of known concentrations of pure c‐di‐GMP to determine the concentration (in nM) of c‐di‐GMP in the samples. All experiments are the mean of two independent biological experiments with standard deviation error bars. Statistical significance was determined using students t‐test with p < 0.05*, p < 0.01 **, p < 0.001***.
Response to external stimuli can be further driven by intracellular second messengers and this is the case for the universal eubacterial second messenger molecule cyclic di‐GMP (c‐di‐GMP) (Jenal et al., 2017), which is tightly associated with biofilm development in a number of bacterial species including P. aeruginosa and A. tumefaciens (Heindl et al., 2014; Valentini and Filloux, 2016). The c‐di‐GMP signalling network can impact virulence in both human and plant pathogens as, for example, seen with the DgcP cyclase in controlling infections by P. aeruginosa or Pseudomonas savastanoi pv savastanoi, an olive tree pathogen (Aragon et al., 2015). Remarkably, in P. aeruginosa, c‐di‐GMP signalling has been shown to regulate T6SS, but while high levels correlate with increased levels in biofilm formation and T6SS activity (Moscoso et al., 2011), low levels of this second messenger are associated with increased motility and Type III secretion system (T3SS) activity (Moscoso et al., 2011). The biofilm part of this paradigm has been confirmed within A. tumefaciens. Indeed, one of the pioneering studies on c‐di‐GMP as a signalling molecule has been carried out in this organism, where it was shown that the activity of a cellulose synthase was dependent on the binding of c‐di‐GMP (Amikam and Benziman, 1989). Subsequent studies have demonstrated the ability of high levels of c‐di‐GMP to increase the levels of a number of different A. tumefaciens polysaccharides and as a result biofilm formation (Heindl et al., 2014; Feirer et al., 2015). In the present study, we thus investigate the role of c‐di‐GMP in regulating the T6SS in A. tumefaciens C58 using a non‐native diguanylate cyclase (DGC) from P. aeruginosa as well as native A. tumefaciens DGCs. We discover that contrary to the paradigm established in P. aeruginosa, high levels of c‐di‐GMP represses the T6SS activity in A. tumefaciens. We show that this occurs at the transcriptional level and has a significant impact on the ability of A. tumefaciens to attack and outcompete other bacterial species. We also demonstrate that this is not specific to a single A. tumefaciens DGC but that several of these enzymes can have a significant impact. Furthermore, we show that elevated levels of c‐di‐GMP within the cell also impact the expression of key structural components of the T4SS. This transcriptional influence subsequently impacts the capacity of A. tumefaciens to transform plant cells as determined by a transient transformation assay. This duality is quite remarkable and suggests that at a specific stage during the A. tumefaciens colonization process interaction with the rhizosphere and with the plant cells should both be kept silent.
Results
The P. aeruginosa diguanylate cyclase SadC supresses T6SS activity in A. tumefaciens C58
The universal second messenger c‐di‐GMP regulates a wide variety of bacterial phenotypes including motility, protein secretion, exopolysaccharide production, virulence, or biofilm formation (Romling and Balsalobre, 2012; Romling et al., 2013; Jenal et al., 2017). Across almost all eubacteria, paradigms have emerged that correlate high intracellular levels of c‐di‐GMP with loss of motility, increased polysaccharide production and biofilm formation (Valentini and Filloux, 2016; Conner et al., 2017). The T6SS has also emerged as being subject to c‐di‐GMP control, with high levels of c‐di‐GMP associated with activation and low levels associated with suppression of this secretion system (Moscoso et al., 2011; Romling et al., 2013) as shown clearly with P. aeruginosa and to a lesser extent with Vibrio alginolyticus. In the latter case, it was proposed that the PppA phosphatase has a negative impact on both the activity of the T6SS and the levels of c‐di‐GMP (Sheng et al., 2013). In the case of Pseudomonas fluorescens it was also proposed that c‐di‐GMP could directly bind to the ClpB2 ATPase one of the core component in the T6SS (Trampari et al., 2015). In A. tumefaciens C58, it has been shown that high levels of c‐di‐GMP are associated with increased extracellular polysaccharide (EPS) production and biofilm formation (Heindl et al., 2014; Feirer et al., 2015), but the impact of c‐di‐GMP on T6SS has not been explored. Agrobacterium tumefaciens has one T6SS cluster which is composed of two divergently transcribed operons, the imp operon which primarily encodes the structural components of the secretion system and the hcp operon that encodes two of the known A. tumefaciens T6SS toxins, i.e. Tae and Tde1 (Wu et al., 2008; Lin et al., 2014). There is also an orphan cluster that encodes Tde2, the only known T6SS toxin not directly encoded in the A. tumefaciens primary cluster (Fig. 1A) (Ma et al., 2014). Here, we investigate the impact of c‐di‐GMP on the functionality of this system, and introduced plasmids encoding the well characterized P. aeruginosa diguanylate cyclases (DGC) SadC and WspR (Guvener and Harwood, 2007; Merritt et al., 2007a; Dahlstrom and O'Toole, 2017; McCarthy et al., 2017a), a mutated version of SadC that has the canonical catalytic site GGEEF domain changed to AAAEF (psadC*) and a known functional phosphodiesterase PA2133 (Ueda and Wood, 2010), into a wild‐type strain of A. tumefaciens. The recombinant strains were grown to early exponential phase and the levels of the T6SS DNase effector Tde1, amidase effector Tae and T6SS nanotube building block Hcp were assessed by western blotting (Ma et al., 2014). Firstly, it was striking to observe that the clumping/hyperbiofilm phenotype resulting from SadC overexpression and usually associated with high levels of c‐di‐GMP (Moscoso et al., 2014) was no longer seen when the catalytically inactive sadC* mutant was expressed indicating as expected that this enzyme was no longer capable of generating c‐di‐GMP (Fig. 1B). The reduction in activity of this mutated version of SadC was confirmed by LC‐MS/MS and was shown to be comparable to another mutated version of SadC that had the GGEEF domain changed to GGAAF (Supporting Information Fig. S1). Furthermore, it was readily observed that expression of the sadC‐encoded membrane bound DGC dramatically abrogated Tde1, Tae and Hcp production. Levels seen in the strains containing plasmids encoding the SadC catalytic site mutant were comparable to strains carrying the empty vector (pBBRMCS4) (Fig. 1C) suggesting that the intracellular levels of c‐di‐GMP were effectively responsible for the reduction in the level of these T6SS components and toxins. The expression of the wspR‐encoded cytoplasmic DGC did not alter the levels of Tde1, Tae or Hcp (Fig. 1C). This suggests that the intracellular activity of WspR in A. tumefaciens might not be strong enough to impact the levels of Tde1/Hcp or that membrane association of the DGC is instrumental to the phenotype. Note that the overexpression of the cytoplasmic phosphodiesterase (PDE) PA2133 also did not significantly impact the levels of Tde1, Tae or Hcp (Fig. 1C). The proposed reduced activity of WspR in A. tumefaciens was confirmed by quantifying the levels of c‐di‐GMP using LC‐MS/MS. This analysis confirmed that SadC produced significantly more c‐di‐GMP than WspR when expressed in A. tumefaciens (Fig. 1D). Overall, these findings are opposite to the established P. aeruginosa paradigm of positive regulation of the T6SS by c‐di‐GMP. Such variation in control is likely depending on how and where c‐di‐GMP is acting and this may dramatically differ from one bacterial species to the other. For example, in P. aeruginosa, the control exerted by master regulators such as RetS/LadS is observable on all H1‐T6SS genes (Allsopp et al., 2017) whereas in Pseudomonas syringae it can only be seen on a subset of T6SS genes (e.g. icmF) but not on others (e.g. hcp) (Records and Gross, 2010).
SadC has a significant impact on T6SS‐dependent interbacterial competition in vitro and in planta
The T6SS is known to play a key role in shaping the microbiota and this is particularly true in the plant rhizosphere (Bernal et al., 2017a; Bernal et al., 2017b). To investigate if the impact of c‐di‐GMP on T6SS was biologically significant and capable of influencing the structure of a bacterial population, bacterial competition assays were performed as described in experimental procedures and using as an attacker strain A. tumefaciens which constitutively expresses wild‐type (psadC) or mutant version of sadC (psadC*). The bacterial prey Escherichia coli carries the pRL662 plasmid conferring gentamicin resistance and survivors after A. tumefaciens contact could be recovered as colony forming units (CFU) on gentamicin‐containing plates. Expression of wild‐type sadC in A. tumefaciens significantly reduced the ability of A. tumefaciens to kill the prey bacterium E. coli (Fig. 2A) when compared to a strain harbouring the vector control or expressing the non‐functional version of sadC, sadC*. The low levels of SadC‐dependent killing were comparable to those seen with an A. tumefaciens mutant strain lacking a core gene for the T6SS machinery, tssL, thus resulting in T6SS inactivation and confirming the T6SS downregulation through SadC activity. To fully rule out that the observed reduction in killing was not a consequence of the increased cell to cell aggregation seen when overexpressing psadC, the assay was repeated in P. aeruginosa, where increased expression of sadC is also known to impact cell to cell aggregation (Moscoso et al., 2014). Competition assays between P. aeruginosa PAO1 expressing sadC and E. coli demonstrated increased levels of killing, contrary to what was observed in the A. tumefaciens competition assays (Supporting Information Fig. S2). A similar increase in killing was seen when using a P. aeruginosa ΔrsmA mutant, which is known to have elevated levels of sadC expression (Moscoso et al., 2014). This suggests that the impact seen on cell‐to‐cell aggregation by the expression of sadC does not impact the activity of the T6SS.
Figure 2.
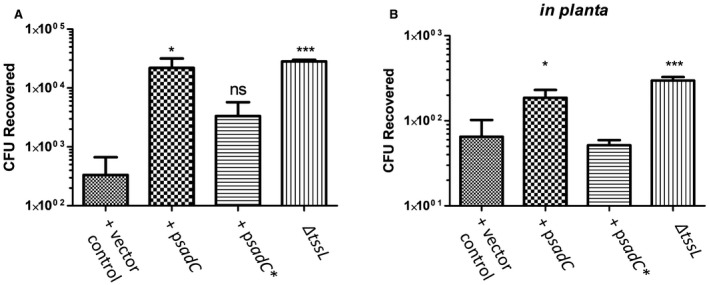
Impact of c‐di‐GMP on the ability of A. tumefaciens to kill other bacteria. A. A. tumefaciens strains with either an empty vector control, a plasmid expressing wild‐type SadC or a plasmid expressing a catalytically inactive SadC (psadC*) were incubated for 14 h on I medium with of E. coli DH10B containing pRL662 which harbours a gentamicin cassette. Bacteria were then resuspended in PBS and plated on LB agar containing gentamicin. B. The bacterial strains described above were resuspended in 1/2 Murashige and Skoog (MS) medium (pH 5.5) and immediately injected into the leaves of 6–8‐day‐old N. benthamiana plants. After 14 h incubation, coupons were cut from these leaves, homogenized in PBS and plated on LB supplemented with gentamicin (50 µg ml−1). All experiments are the mean of three independent biological experiments with standard deviation error bars. Statistical significance was determined using students t‐test comparing each strain to the vector control with p < 0.05 *, p < 0.01 **, p < 0.001***.
To add further biological significance to this observation and partially recreate a physiologically relevant environment, the competition assays were repeated but bacteria were infiltrated in the leaves of the Nicotiana benthamiana plant as previously described (Ma et al., 2014). This assay more or less phenocopies our in vitro findings with expression of sadC leading to significant reduction in prey killing, and thus higher CFU recovery from the plant leaves of gentamicin resistant E. coli cells (Fig. 2B). These findings demonstrate that the intracellular levels of c‐di‐GMP can have a significant impact on A. tumefaciens ability to engage via the T6SS in interspecies competition in planta.
Endogenous A. tumefaciens DGCs suppress the T6SS activity
The use of a well‐characterized P. aeruginosa DGC to increase the intracellular levels of c‐di‐GMP in A. tumefaciens does not guarantee that DGCs encoded within the A. tumefaciens genome can have a similar impact on T6SS. To investigate this, we focused on endogenous A. tumefaciens DGCs, 16 of which are encoded within the genome (Romling et al., 2013), of these we selected 6 that encoded transmembrane domains and carry the canonical GGDEF domain, namely Atu2091, Atu2691, Atu1207 and Atu5372 as well as the two previously described DGCs DgcA (Atu1257) and DgcC (Atu2179) both of which have been shown to impact EPS production (Xu et al., 2013) (Supporting Information Fig. S3). The rationale here was that both SadC and WspR have been shown to be active DGCs, but only the membrane bound SadC was capable of impacting c‐di‐GMP associated phenotypes (Fig. 1C). This could suggest that the localization of the the DGC may have a critical impact on phenotypic outcomes. The genes encoding these DGCs were amplified and cloned under the control of the IPTG‐inducible lac promoter in the pTrc200 vector. Using agar plates‐containing Congo red, it is possible to reveal exopolysaccharide (EPS) production in response of increased intracellular levels of c‐di‐GMP (Howie and Brewer, 2009; Heckel et al., 2014; Heindl et al., 2014; Feirer et al., 2015; Feirer et al., 2017). It was clear that some of the A. tumefaciens transformed with the above recombinant plasmids yielded colonies displaying a wrinkly phenotype (Fig. 3A). This confirmed that as previously described DgcA and DgcC could impact EPS production (Xu et al., 2013) but also that Atu2091 and Atu5372 were increasing the levels of exopolysaccharide production, suggesting they are active cyclases, while Atu1207 and Atu2691 had little or no effect (Fig. 3A).
Figure 3.
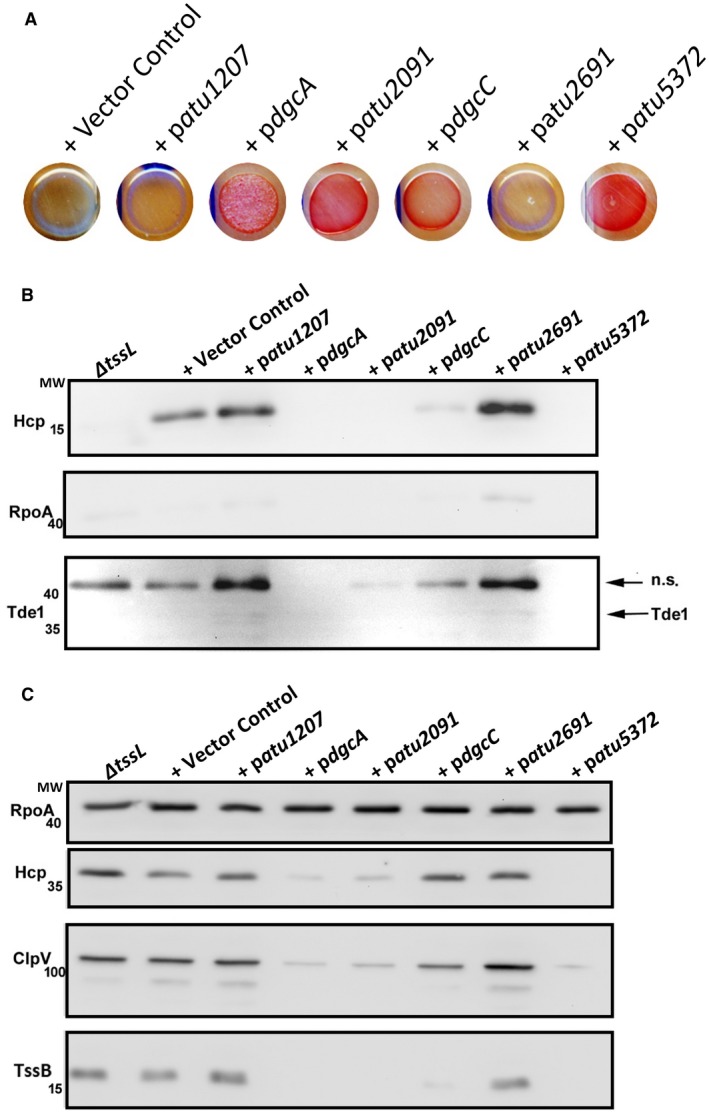
Impact of native DGCs on T6SS. A. Expression of a range of six different DGCs whose genes were cloned into wild‐type A. tumefaciens and spotted onto Congo red agar containing 0.5 mM IPTG and Spectinomycin 200 µg ml−1. B. Western blot using Hcp, Tde1, and RpoA (Loading Control) antibodies on supernatants of cells transformed with either empty vector control, pdgcA, patu2091, pdgcC, patu2691, patu1207 or patu5372 from cells grown to early exponential phase in 523 medium. C. Western blot using Hcp, ClpV, TssB, Tde1 and RpoA (Loading Control) antibodies on whole cell lysates of cells transformed with either empty vector control, pdgcA, patu2091, pdgcC, patu2691, patu1207 or patu5372 from cells grown to early exponential phase in 523 medium.
To assess if anyone of these DGCs was capable of shutting down the T6SS, as SadC did, a secretion assay was performed using a Hcp and a Tde1 specific antibody as described above. This assay revealed that Atu2091, Atu5372 and DgcA were all capable of down‐regulating the secretion of Hcp and Tde1 to non‐detectable levels, and these levels were comparable to those observed with the non‐functional T6SS mutant A. tumefaciens ΔtssL (Fig. 3B). DgcC also had a negative impact on Hcp levels but to a lesser extent. The cellular levels of Hcp, TssB (subunit of the T6SS sheath) and ClpV (T6SS AAA+ ATPase), revealed a similar profile to what was seen with Hcp secretion, with expression of Atu2091, Atu5372 and DgcA leading to reduced levels of these proteins and DgcC having a lesser effect (Fig. 3C). As for the EPS assay, Atu1207 and Atu2691, did not shut off T6SS activity, thus suggesting that they may not be active or are generating levels of c‐di‐GMP too low to trigger observable phenotypic changes.
We then performed in vitro killing assays with each of the recombinant strains. Significant reductions in killing, comparable with what is observed with the tssL mutant, were seen upon expression of dgcA and atu5372 while a reduced but not statistically significant killing was observed for strains expressing atu2091 and dgcC (Fig. 4A). No reduction in killing was observed with strains expressing atu1207 and atu2691. To confirm that the observed phenotypes were a direct result of altered levels of c‐di‐GMP, the levels of c‐di‐GMP were quantified in strains expressing either patu5372 or patu2691. As expected, the strain expressing patu2691 did not display elevated levels of c‐di‐GMP compared to the vector control, while a strain expressing atu5372 had levels of c‐di‐GMP higher than those seen in a strain expressing the known active P. aeruginosa DGC, sadC (Figs 4B and 1D). It was intriguing to observe that some of the DGCs tested were unable to impact the T6SS suggesting that strength in DGC activity might be an issue in signalling and that, such as with a rheostat, once a threshold of intracellular c‐di‐GMP is reached, some phenotypes could be triggered while others might need a higher threshold. This observation might also be in line with the established concept for the need of spatial pools of c‐di‐GMP (Christen et al., 2010), and specific subcellular localization of some DGCs such as within the cytoplasmic membrane, or their direct interaction with the effector generating the output (McCarthy et al., 2017a), would overcome the threshold issue.
Figure 4.
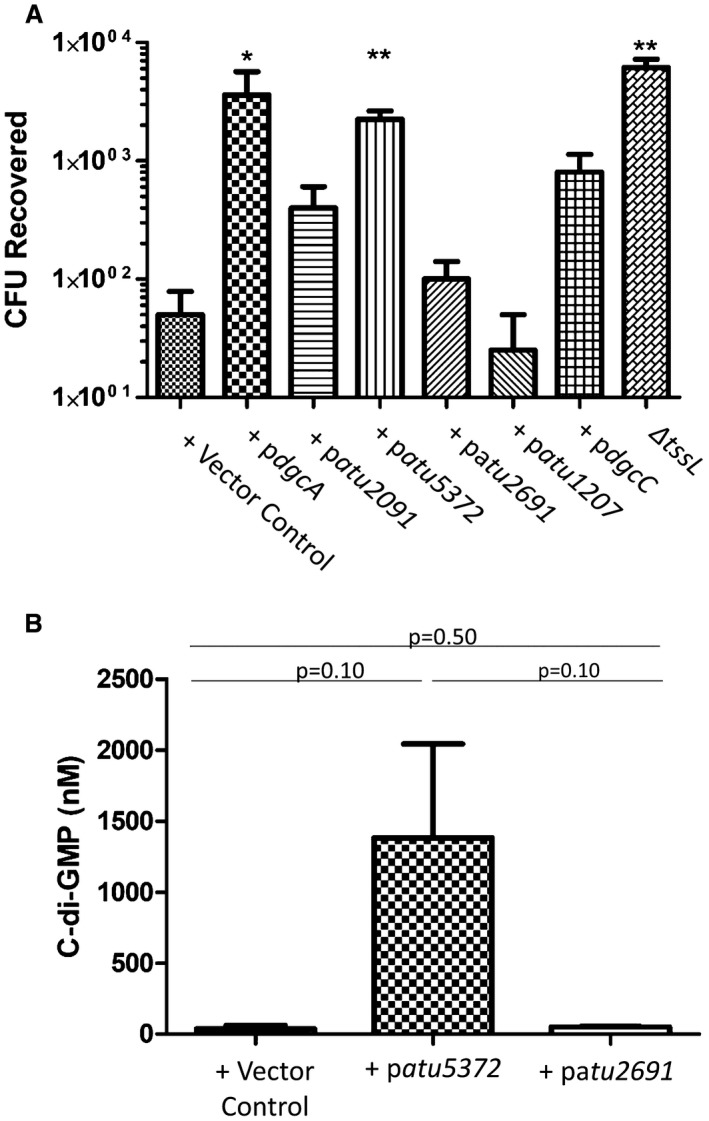
Impact of native DGC on interspecies killing and cdi‐GMP levels. A. A. tumefaciens strains with either an empty vector control, pdgcA, patu2091, pdgcC, patu2691, patu1207 and patu5372 were incubated for 14 h on a I medium (pH5.5) with E. coli containing pRL662 which harbours a gentamicin cassette. Bacteria were then resuspended in PBS and plated on LB agar‐containing gentamicin. All experiments are the mean of three independent biological experiments with standard deviation error bars. Statistical significance was determined using students t‐test comparing each strain with the vector control, p < 0.05 *, p < 0.01 ** B. C‐di‐GMP levels were quantified via LC‐MS/MS in strains expressing either an empty vector control, atu5372 or atu2691. Native DGSc were induced with 0.5 mM IPTG for 16 h in 523 rich medium and cells equivalent to an OD600 5 were collected. Samples of interest were compared to a standard curve derived from measurements of known concentrations of pure cdi‐GMP to determine the concentration (in nM) of c‐di‐GMP in the samples. All experiments are the mean of two independent biological experiments with standard deviation error bars.
C‐di‐GMP regulates both the imp and hcp T6SS clusters at the transcriptional level independently of the ExoR signalling cascade
Although T6SS regulation in A. tumefaciens is relatively poorly understood, one pathway involved is the ExoR/ChvI/ChvG signalling system which is known to impact EPS production, horizontal gene transfer, motility and virulence (Wu et al., 2012; Heckel et al., 2014). The ChvG/ChvI two‐component system positively regulates the T6SS while ExoR acts negatively and is a periplasmic repressor (Wu et al., 2012). In acidic pH conditions such as those found around a plant wound site, ExoR is degraded, this allows the autophosphorylation of ChvG, which can then transfer a phosphoryl group to its cognate response regulator ChvI for activation of T6SS (Wu et al., 2012) (see Discussion). To assess if the impact on T6SS through c‐di‐GMP signalling involves this cascade, psadC was introduced into an A. tumefaciens exoR mutant which is known to have elevated activity of T6SS. If the elevated pool of c‐di‐GMP is acting through the ExoR cascade, then c‐di‐GMP transmission should be interrupted in the exoR mutant and no longer be able to impact on the T6SS. This was assessed in standard 523 medium, but also in minimal media (AB‐MES pH7), a condition where ExoR‐mediated repression of T6SS has previously been demonstrated (Wu et al., 2012). Remarkably, it was observed that in the exoR mutant, expression of sadC and thus high levels of c‐di‐GMP are still capable of down‐regulating the T6SS, as seen by monitoring Hcp, ClpV and TssB production (Fig. 5A) suggesting that c‐di‐GMP is acting independently of the ExoR network. Since all data presented thus far of c‐di‐GMP impacting the T6SS were at the protein level, we also wanted to establish if this regulation was originally exerted at the transcriptional level. To address this, RNA was isolated from A. tumefaciens transformed with either the cloning vector, a vector expressing a nonspecific protein GFP, psadC, psadC*, patu2691, a proposed inactive DGC or patu5372, a native A. tumefaciens DGC that was shown to have a significant impact on clumping, interspecies killing and Hcp secretion. qRT‐PCR was performed on cDNA synthesized using RNA extracted from these strains and primers specific to genes belonging to both divergent operons of the A. tumefaciens T6SS cluster, namely tssB, fha, hcp and clpV (Figs 1A and 5B,C). Remarkably, all of these genes were significantly down‐regulated when atu5372 or sadC was expressed, but not upon expression of an inactive sadC*, a proposed inactive DGC atu2691 or a nonspecific protein GFP, confirming that the impact of c‐di‐GMP on T6SS also occurs at the transcriptional level and that all T6SS genes are impacted.
Figure 5.
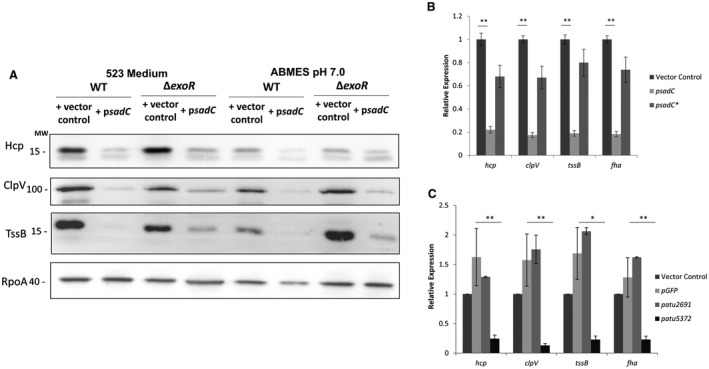
c‐di‐GMP impacts T6SS at the transcriptional level. A. Western blot using Hcp, TssB, ClpV, and RpoA (Loading control) antibodies on whole cell lysate of ΔexoR cells transformed with either Empty Vector control or psadC grown to early exponential phase in 523 medium or AB‐MES pH7.0. B. qRT‐PCR on RNA isolated from cells transformed with either an empty vector control, psadC or psadC* grown to early exponential phase. Expression quantified using primers specific to either tssB, fha, hcp, clpV and 16s rRNA which serves as a normalization control. Statistical significance was determined using students t‐test with p < 0.05 *, p < 0.01 **, p < 0.001*** comparing the vector control to psadC. C. qRT‐PCR on RNA isolated from cells transformed with either an empty vector control, pGFP, patu2691 or patu5372 grown to early exponential phase. Expression quantified using primers specific to either tssB, fha, hcp, clpV or 16s rRNA which serves as a normalization control. Statistical significance was determined using students t‐test with p < 0.05 *, p < 0.01 **, p < 0.001*** comparing each strain to the expression control pGFP. All experiments are the mean of three independent biological experiments with standard deviation error bars.
C‐di‐GMP negatively impacts expression of components of the T4SS and the ability of A. tumefaciens to transform plant cells
The significant impact of c‐di‐GMP on the activity of the T6SS and biofilm formation led us to ask whether other determinants involved in infection progression, such as the T4SS, were also responsive to the intracellular levels of c‐di‐GMP. The rationale here was in line with previous reports where protein secretion systems, e.g. the T6SS and T3SS, are antagonistically controlled (Moscoso et al., 2011; McCarthy et al., 2017b). As described above for the control of EPS production, the regulation of the T4SS is dependent on activation of the VirA/VirG signalling by acidity or phenols such as acetosyringone (AS), in which the transcription of the gene encoding the VirG response regulator is activated via the ExoR/ChvI/ChvG signalling system (Heckel et al., 2014). Activation of this cascade subsequently confers the ability of A. tumefaciens to transfer T‐DNA into the host plant cell. We thus analysed the expression of a number of different T4SS genes (virB1, virB2) and T4SS effector gene virE2 in cells expressing the native DGCs (Fig. 6A). qRT‐PCR was performed on cDNA synthesized using RNA isolated from cells transformed with either a vector control, a vector expressing a nonspecific protein GFP, patu2691 or patu5372. Our results suggested that increasing the levels of c‐di‐GMP within the cell significantly down‐regulates the expression of T4SS machinery and effector genes, while expression of patu2691, a proposed inactive DGC or a non‐specific protein pGFP did not negatively impact expression (Fig. 6B). Similar results were seen when cells were expressing psadC, although only partial but significant restoration of expression was seen when expressing psadC* (Fig. 6C).
Figure 6.
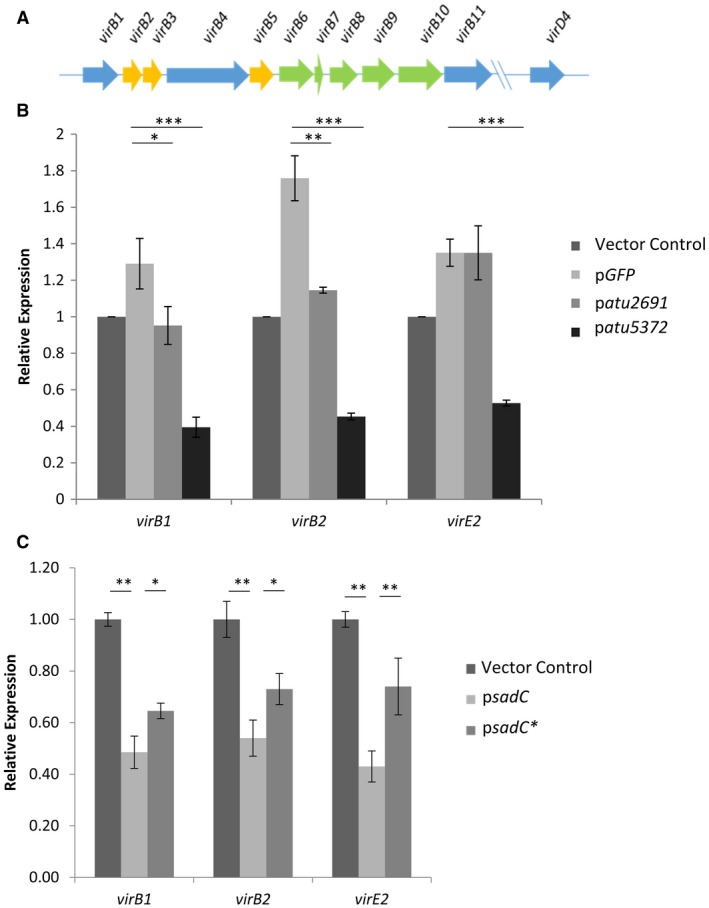
Impact of c‐di‐GMP on T4SS. A. Operon encoding components of the T4SS, virB1‐11 represents the structural components of the T4SS machinery while virD4 is involved in coupling DNA transfer. B. qRT‐PCR on RNA isolated from cells transformed with either an empty vector control, pGFP, patu2691 or patu5372 grown in AB‐MES medium (pH 5.5) containing 200 μM acetorysingone (AS) for virulence gene induction. Expression quantified using primers specific to either virB1, virB2, virE2 or 16s rRNA which serves as a normalization control. All experiments are the mean of three independent biological experiments with standard deviation error bars. Statistical significance was determined using students's t‐test with p < 0.05 *, p < 0.01 **, p < 0.001*** when compared to the protein expression control pGFP. C. qRT‐PCR on RNA isolated from cells transformed with either an empty vector control, psadC, psadC* grown to early exponential phase. Expression quantified using primers specific to virB1, virB2, virE2 and 16s RNA which serves as a normalization control. All experiments are the mean of three independent biological experiments with standard deviation error bars. Statistical significance was determined using students's t‐test with p < 0.05 *, p < 0.01 **, p < 0.001***.
To further interrogate this and determine if this impact on transcription is biologically relevant, an AGROBEST assay was performed (Wu et al., 2014). This is an Agrobacterium‐mediated Arabidopsis transformation assay (Agrobacterium‐mediated enhanced seedling transformation) that utilizes β‐glucuronidase (GUS) as a reporter carried on the T‐DNA to determine the impact of native regulatory elements on Agrobacterium transformation activity (Wu et al., 2014). Four‐day‐old Arabidopsis seedlings were infected with A. tumefaciens strain C58C1 (pTiB6S3ΔT)H carrying either an empty vector control, a vector expressing a nonspecific protein GFP, patu2691 or patu5372 and expressing a GUS reporter. The assay was allowed to proceed for 3 days before seedlings were stained with 5‐bromo‐4‐chloro‐3‐indolyl glucuronide (X‐Gluc) to visualize the GUS staining. Remarkably, only expression of patu5372 leads to a dramatic reduction in GUS staining (Fig. 7A) indicating that, in concordance with the qRT‐PCR data, the activity of the T4SS is significantly impacted by the intracellular levels of c‐di‐GMP. Similar findings were observed when cells were expressing psadC, although expression of psadC* could not completely abrogate this (Fig. 7B).
Figure 7.
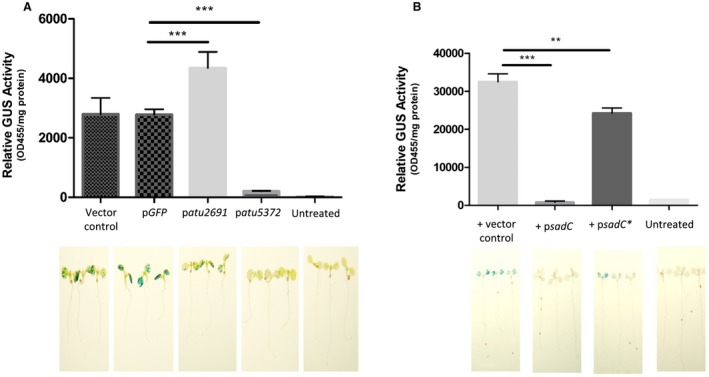
Impact of c‐di‐GMP on plant transformation. A. Four‐day‐old Arabidopsis seedlings were infected with Agrobacterium strain C58C1(pTiB6S3ΔT)H carrying pBISN1 and either an empty vector control, pGFP, patu2691 or patu5372. These cells were pre‐incubated in I medium (pH5.5) supplemented with 200 μM AS to induce expression of the vir genes. Seedlings were stained with 5‐bromo‐4‐chloro‐3‐indolyl glucuronide (X‐Gluc) to visualize the GUS activity. Statistical significance was determined using students's t‐test with p < 0.05 *, p < 0.01 **, p < 0.001*** when compared to the protein expression control pGFP. B. Four‐day‐old Arabidopsis seedlings were infected with Agrobacterium strain C58C1(pTiB6S3ΔT)H carrying pBISN1 and either an empty vector control, psadC or psadC*. These cells were pre‐incubated in I medium (pH5.5) supplemented with 200 μM AS to induce expression of the vir genes. Seedlings were stained with 5‐bromo‐4‐chloro‐3‐indolyl glucuronide (X‐Gluc) to visualize the GUS activity. All experiments are the mean of three independent biological experiments with standard deviation error bars. Statistical significance was determined using students's t‐test with p < 0.05 *, p < 0.01 **, p < 0.001*** when compared to the vector control.
Discussion
Agrobacterium tumefaciens is a phytopathogen capable of causing tumorigenesis in a wide variety of plant species through T‐DNA transfer via the T4SS (Gelvin, 2010). The start of this transfer process is centered on A. tumefaciens sensing characteristic changes in the rhizosphere such as a drop in pH and phytochemical release, indicative of a plant wound. Intriguingly, a drop in pH has also been shown to induce not only the T4SS but also the T6SS. The latter is central to A. tumefaciens outcompeting other bacteria within this niche whereas the former allows the unimpeded transfer of T‐DNA and eventual formation of a crown gall (Wu et al., 2012). Other than low pH, the understanding of what factors regulate the activity of the T6SS in A. tumefaciens is relatively limited. In this study, we investigate the role of c‐di‐GMP in regulating T6SS and T4SS in A. tumefaciens. Previously characterized DGCs and PDEs from P. aeruginosa were transformed into A. tumefaciens. The membrane bound DGC called SadC (Merritt et al., 2007a) was shown to significantly induce a clumping phenotype, characteristic of high levels of c‐di‐GMP (Heindl et al., 2014) (Fig. 1B), and down‐regulate the expression of both the T6SS toxins and T6SS machinery components (Fig. 1C). This is contrary to the established paradigm in P. aeruginosa where high levels of c‐di‐GMP are associated with increased levels of T6SS (Supporting Information Fig. S2) (Moscoso et al., 2011). We demonstrated that the ability to impact T6SS was dependent on SadC being active as a version of SadC that features a mutated GGDEF domain had no impact on bacterial clumping or T6SS activity (Fig. 1B and C). The biological relevance of this down‐regulation is validated by the inability of a strain of A. tumefaciens transformed with a plasmid expressing sadC to kill a prey bacterium in vitro or in the native environment of a plant (Fig. 2A and B).
The use of an active DGC from P. aeruginosa could create an artificial pseudo‐circuit inhibiting the activation of the T6SS. To rule this out and to explore the capacity of native A. tumefaciens DGCs to down‐regulate T6SS, the genes of six different A. tumefaciens DGCs, that have similar features as SadC, e.g. predicted transmembrane domains, were cloned and expressed. Three of these DGCs, DgcA, Atu2091 and Atu5372, were capable of down regulating the secretion of Hcp to non detectable levels, comparable to those observed in an A. tumefaciens T6SS mutant, ΔtssL (Fig. 3B). When further assessed, these DGCs were all capable of inhibiting interbacterial killing by A. tumefaciens both in vitro and in planta to differing degrees. These findings confirm the hypothesis that in A. tumefaciens high levels of c‐di‐GMP down‐regulate T6SS but they also offer an intriguing insight into the specificity exhibited by different DGCs. The differing levels of killing observed in strains expressing different DGCs (Fig. 4A) highlights the importance of c‐di‐GMP threshold levels to trigger specific output, and may also lend support to an emerging phenomenon whereby the spatial and temporal localization of a DGC can significantly influence the resulting phenotypic impact (Romling et al., 2013; Dahlstrom and O'Toole, 2017).
The regulation of the T6SS is shown to occur at the transcriptional level with both divergent transcriptional units in the T6SS cluster being significantly down‐regulated (Fig. 5B and C). The only characterized signalling cascade involved in the regulation of T6SS in A. tumefaciens is the ExoR/ChvI/ChvG system (Wu et al., 2012; Heckel et al., 2014); however, we collected data suggesting that the impact of c‐di‐GMP acts independently of this regulatory network (Fig. 5A). This finding prompted us to explore the influence of intracellular c‐di‐GMP levels on the T4SS, another ExoR/ChvI/ChvG target. Intriguingly, we observed a significant transcriptional down‐regulation in the expression of key components of the T4SS machinery when a DGC was expressed, sadC or atu5372, and this requires c‐di‐GMP synthesis since an inactive SadC, SadC*, displays only traces of inhibition of T‐DNA transfer into Arabidopsis plant cells (Fig. 7AB). Intriguingly, it seems here that c‐di‐GMP signalling is independent of the master regulator for T4SS and T6SS expression, i.e. ChvI/ChvG/ExoR. This is slightly different from what is observed with P. aeruginosa, in which the master network controlling T6SS activity, Gac/Rsm, is entangled with c‐di‐GMP signalling. In this case, the gene encoding the DGC SadC, is directly controlled by the translational repressor RsmA (Moscoso et al., 2014), which in turn modulates c‐di‐GMP levels. Note that the absence of ExoR, relieves the kinase ChvG which can then phosphorylate the response regulator ChvI and therefore activate the T6SS and the T4SS genes. In this case if c‐di‐GMP was binding ChvI or ChvG to inhibit their activity, such inhibition should have been detected in an exoR mutant, which was not the case (Fig. 5A). Alternatively, a plausible explanation is that c‐di‐GMP could modulate the activity of another protein, e.g. a membrane protein interacting with ExoR. This protein could capture the periplasmic ExoR when it is not bound to c‐di‐GMP. Instead, when such an integral membrane protein is bound to c‐di‐GMP, interaction with ExoR does not occur and ExoR is free and available to inhibit the ChvI kinase (Fig. 8). This type of control at the interface cytoplasm/periplasm and across the cytoplasmic membrane was previously shown in the case of the P. fluorescens Lap system (Hinsa and O'Toole, 2006; Newell et al., 2009). In this case, the degenerated LapD cyclase/phosphodiesterase is an integral cytoplasmic protein, which can bind c‐di‐GMP and then interact with the periplasmic LapG protease. When LapD is c‐di‐GMP‐bound, it prevents the LapG protease from cleaving and releasing the surface adhesin LapA, which contributes to biofilm formation only when it remains associated with the cell surface (Navarro et al., 2011; Dahlstrom and O'Toole, 2017). This is in agreement with the dogma‐high c‐di‐GMP/high biofilm. Future work could focus on identifying possible missing links such as the one suggested above. Interestingly the active cyclases are membrane embedded, such as SadC in contrast to WspR, and that may reinforce the idea that the signalling could be a transmembrane inside‐out signalling, where the membrane cyclase or an interacting partner therein plays a key role in the process (Navarro et al., 2011). In the inside‐out signalling mechanism described by the O'Toole laboratory, the transmission of the signal involves HAMP domains so a directed approach could be used to assess known c‐di‐GMP‐related proteins with a HAMP domain (Navarro et al., 2011). Alternatively a non‐biased approach using random transposon mutagenesis in a strain overexpressing a DGC and carrying a reporter fusion for T6SS or T4SS expression could be used. Screens could be aimed at identifying mutants whose T4SS or T6SS activity goes high up even in the context of DGC expression, thus pinpointing any players in the circuitry. In any case, c‐di‐GMP signalling has shown that the combination and sequence of events that lead from synthesis to output is far from a simple and stereotyped trajectory. Examples from within A. tumefaciens have shown that a dual DGC‐PDE, called DcpA, can see its activity balanced from DGC to PDE based on its interaction with another protein called PruA, a pterin reductase. In the presence of PruA, DcpA is a PDE while in its absence it is a DGC. This implies that when DcpA is expressed in a heterologous host, such as E. coli in which PruA is absent, the main activity is PDE, while in Agrobacterium it is mostly a DGC (Feirer et al., 2015). This has also lended support to our rationale to analyse the impact of native Agrobacterium DGC versus heterologous ones from P. aeruginosa to fully validate our phenotypic observations.
Figure 8.
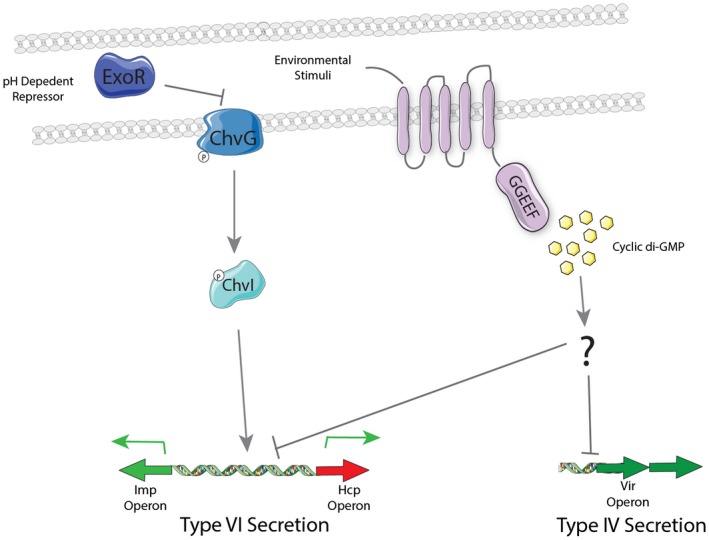
Regulation of T6SS and T4SS in A. tumefaciens. In acidic pH conditions such as those found around a plant wound site, the periplasmic regulator ExoR is degraded, this allows the activation of ChvG via autophosphorylation, ChvG can then activate its cognate response regulator ChvI, which then activates the T6SS. The activation of native DGCs by environmental stimuli leads to an increase in the local levels of cyclic di‐GMP. These increased levels of c‐di‐GMP are capable of repressing the transcription of the T6SS and the T4SS via an as yet uncharacterized regulator(s).
In P. aeruginosa, the switch in lifestyle from biofilm to motile is also tightly linked to a concomittant switch from T6SS to T3SS respectively (Moscoso et al., 2011; Valentini and Filloux, 2016; McCarthy et al., 2017b). This makes sense if considering that the T6SS may help killing unwanted members of a biofilm population and instead prefering the T3SS cytotoxicty when adopting an acute infectious style. In the case of Agrobacterium, simultaneous down‐regulation of the T6SS and T4SS when biofilm formation is induced gives an intriguing insight into the phases of A. tumefaciens infection. The upregulation of motility genes, the T6SS and the T4SS by low pH and phytochemical release associated with a plant wound is the first step in A. tumefaciens targeting the wound site and ensuring a mono‐infection by killing competing species (Merritt et al., 2007b; Wu et al., 2012). Once at the site of the wound it is possible that intracellular levels of c‐di‐GMP are elevated stimulating the production of EPS, attachment and biofilm formation (Heindl et al., 2014). T6SS and T4SS are concurrently downregulated possibly as an economy measure allowing more efficient channelling of resources towards establishing infection at the site of the wound. Once initial sustainable colonization is established, variation in c‐di‐GMP levels can then allow further colonization and again competition with other bacteria and modification of the plant cell environment. The orchestration of c‐di‐GMP levels and its variation in different cells and at a different localization in each cell, would be in accordance with a model, where the spatio‐temporal dynamics of c‐di‐GMP is key (Christen et al., 2010; Abel et al., 2013; Kulasekara et al., 2013; Skotnicka et al., 2016). It will be instrumental to the fate of the community at a population level but may of course be specific to some individuals, offsprings of which will constitute a new sub‐population with dictinct aims, such as, for example, dispersal from the biofilm. Changes in c‐di‐GMP within a cell have been demonstrated on numerous instances to significantly alter the behavior of that cell, examples of this include the temporally oscilating global pools of c‐di‐GMP in Caulobacter crescentus which play a key role in regulating the swarmer to stalked cell transition and in Myxococcus xanthus where flucatuations in c‐di‐GMP levels control the developmental cycle that results in fruiting body formation (Abel et al., 2013; Kulasekara et al., 2013; Ozaki et al., 2014; Lori et al., 2015; Rotem et al., 2015; Skotnicka et al., 2016).
The impact seen in A. tumefaciens is to down‐regulate both T4SS and T6SS, so this duality may have significant impact on shaping future bio‐control efforts in the agri‐sector. Exploiting the knowledge that both the T4SS and the T6SS are down regulated in the presence of elevated levels of c‐di‐GMP (Fig. 8) may also prompt the development of exogenous soil treatments that could influence the accumulation of this signalling molecule within the cell and in turn influence its physiology/pathogenicity. Similar approaches have proven successful in disrupting P. aeruginosa biofilms in the cystic fibrosis lung, whereby exposure of the lung to nitric oxide induces biofilm dispersal through the modulation of the activity of phosphodiesterases, thus manipulating the intracellular levels of c‐di‐GMP (Barraud et al., 2006; Barraud et al., 2009; Howlin et al., 2017). In all, while the hypothesis that c‐di‐GMP is fine tuning the behaviour of A. tumefaciens as it transitions from the rhizosphere to the plant surface remains to be verified, here we have demonstrated that c‐di‐GMP is capable of significantly influencing the ability of A. tumefaciens to compete with other bacterial species and thus may shape the microbial diversity within the soil and also significantly impacts the capacity of A. tumefaciens to carry out one of its most defining features, the ability to transform the plant cell with interkingdom DNA transfer.
Experimental procedures
Bacterial strains and plasmids
Strains, plasmids and primer sequences used in this study are shown in Supporting Information Table [Link], [Link] and [Link], [Link]. Escherichia coli was cultured in Lysogeny Broth, whereas 523 medium (Kado and Heskett, 1970) was routinely used for A. tumefaciens strains unless otherwise indicated. Growth conditions were previously described (Ma et al., 2009). When required, appropriate antibiotics were added to the medium as follows: for E. coli, 50 µg ml−1 ampicillin, 50 µg ml−1 gentamicin (Gm), 50 µg ml−1 kanamycin (Km) and 100 µg ml−1 spectinomycin; and for A. tumefaciens, 200 µg ml−1 spectinomycin. About 1mM Isopropyl‐β‐D‐thiogalactopyranoside (IPTG) was used when necessary. Site directed mutagenesis was performed on pBBRMCS4‐sadC using primers (Supporting Information Table S1) targeted to the catalytic site GGDEF site with specific nucleotide changes corresponding to mutation of the active site to AAAEF. These primers were used to amplify the full plasmid, followed by ligation with T4 ligase and subsequent transformation into E. coli DH5α. Congo red assays were performed as previously described (Moscoso et al., 2011).
Interbacterial competition on agar plates
Interbacterial competition assays were performed as previously described (Ma et al., 2014; Bondage et al., 2016). In brief, overnight cultures of E. coli DH10B containing pRL662 derivative conferring gentamicin resistance were grown in LB at 37°C. The A. tumefaciens cells were adjusted to OD600 0.1, whereas the E. coli DH10B were adjusted to OD600 0.01, mixed at a 10:1 ratio, and 10 μl was spotted on LB (pH7.0) agar and incubated for 16 h at 28°C. Cells were harvested, serially diluted, and plated in triplicates on LB agar with or without gentamicin for colony forming units (CFU) counting. All experiments are the mean of a minimum of three independent biological experiments with standard deviation error bars. Statistical significance was determined using students t‐test with p < 0.05 *, p < 0.01 **. Bacterial competitions with P. aeruginosa were carried out as described before (Pissaridou et al., 2018). Briefly, strains harbouring indicated plasmids were grown over night with appropriate antibiotics and competition assays were performed the next day on LB agar plates without antibiotics using a 1:1 ratio of attacker to prey and incubating at 37°C for 24 h. Attacker strains were P. aeruginosa PAO1 and PAO1 ΔrsmA and prey strain was E. coli Top10 pRL662‐gfp. Competitions were recovered and serially diluted prior to spot plating on LB agar plates with antibiotics for selection and grown overnight at 37°C. Survival was assessed by quantitative colony counts on selective media (using gentamicin for selection of E. coli and ampicillin for P. aeruginosa).
Measurement of c‐di‐GMP levels
For c‐di‐GMP quantification, samples were prepared as described previously (Valentini et al., 2016) and analysed by liquid‐chromatography mass spectrometry (LC‐MS/MS). In brief, A. tumefaciens C58 strains harbouring pBBRMCS4 vectors constitutively expressing DGCs were grown overnight in 20 mL 523 medium and cells equivalent to an OD600 5 were collected by centrifugation. Strains harbouring pTrC200 vector or derivatives were grown overnight, adjusted to an OD600 0.1 and grown to an OD600 0.7. Native DGCs were induced with 0.5 mM IPTG for 16 h and cells equivalent to an OD600 5 were collected. Collected cells were resuspended in extraction solution (Acetonitrile/methanol/water, 2/2/1, v/v/v), incubated on ice for 15 min and heated for 10 min at 95–99°C. Cells were centrifuged for 10 min at 4°C, 20,800 x g and supernatant fluid was collected. Extraction was repeated twice and supernatant fluids of the 3 extraction steps were combined and incubated at −20°C overnight. Extraction fluids were centrifuged again and supernatant fluid was analysed at the BIOLOG Life Science Institute (Biolog, Bremen) via LC‐MS/MS. Samples of interest were compared to a standard curve derived from measurements of known concentrations of pure c‐di‐GMP to determine the concentration (in nM) of c‐di‐GMP in the samples.
Interbacterial competition assay in planta
Interbacterial competition assays in planta were performed as previously described (Ma et al., 2014). The intra‐species A. tumefaciens competition assay was performed with a 10:1 attacker‐to‐target ratio by leaf infiltration of N. benthamiana. Briefly, 523 overnight‐cultured A. tumefaciens cells were sub‐cultured at 28°C in the same medium for further growth to OD600 1.0–1.5. The harvested cells were resuspended in 1/2 Murashige and Skoog (MS) medium (pH 5.5) to an appropriate OD600. The attacker (OD600 5) and target, E. coli DH10B containing pRL662 (OD600 0.5) were mixed equally before infiltration into 2‐month‐old leaves of N. benthamiana with use of a needleless syringe. After 24‐h incubation at room temperature, the infiltrated spot was punched out, ground in 0.9% NaCl, serially diluted, and plated in triplicates on LB agar containing appropriate antibiotic to select for the target cells. All experiments are the mean of three independent biological experiments with standard deviation error bars. Statistical significance was determined using students t‐test with p < 0.05 *, p < 0.01 **.
Agrobacterium infection in Arabidopsis seedlings and GUS activity assay
The procedures for the seedling transient transformation assay were adapted from (Kim et al., 2006; Wu et al., 2014) with modifications. Agrobacterium tumefaciens cells were pre‐induced by addition of 200 μM of acetosyringone at 28°C for 16 h and resuspended in appropriate buffer with cell density OD600 0.02. Four‐day‐old A. thaliana seedlings were then incubated in 1ml of the buffer in a well of a 6‐well plate for another 3 days. The seedlings were then removed from the buffer for GUS staining or GUS activity assay. For GUS staining, seedlings were stained with 5‐bromo‐4‐chloro‐3‐indolyl glucuronide (X‐Gluc) at 37°C for 6 h as described. GUS activity assay was determined by the conversion of 4‐methylumbelliferyl‐β‐D‐glucuronide (4‐MUG) to 4‐methylumbelliferone (4‐MU). 4‐MU fluorescence (excitation. 356 nm, emission. 455 nm) was measured with a 96 microtiter‐plate reader (Bio‐Tek Synergy Mx) and amount of protein in the reaction was determined by a Bradford assay. The relative GUS activity was expressed as OD455 μg−1 protein (Wu et al., 2014).
RNA extraction and quantitative RT‐PCR
Strains were grown to early exponential phase before cells were pelleted. Total RNA was extracted from pellets by Total RNA Extraction Kit (Arrowtech). First‐strand cDNA was synthesized from 4 μg of total RNA with SuperScript III Reverse Transcriptase (Invitrogen) and random primers. Quantitative PCR were performed in QuantStudio 12 K Flex Real‐Time PCR machine (Applied Biosystems) with Power SYBR Green PCR Master Mix (Invitrogen).
Production of T6SS components and T6SS‐dependent secretion assay
The T6SS components and effectors in whole cell and during secretion assays were performed as described previously (Lin et al., 2014). Strains were grown to early exponential phase in both AB‐MES pH7 medium and 523 medium. Whole cell extract and secreted proteins were separated by SDS/PAGE and transferred onto a PVDF or nitrocellulose membrane using a transfer apparatus (Bio‐Rad). The membrane was probed with primary antibody against Tde1 epitope (Ma et al., 2014), Hcp (Wu et al., 2008), Tae (Ma et al., 2014), ClpV (Wu et al., 2012) and RpoA (Lin et al., 2014) was used as an internal control then incubated with horseradish peroxidase‐conjugated anti‐rabbit secondary antibody (1:20,000) and visualized with the ECL system (Perkin‐Elmer).
Author contributions
RMC, AF, MY, EML designed research. RMC, MY, KG, YCW performed research. RMC, AF, MY, EML analysed data. RMC and AF wrote manuscript.
Supporting information
Acknowledgements
We are grateful to Sharon Longford (NTU, Singapore) for the design of the graphical abstract. Work in Alain Filloux's laboratory was supported by the BBSRC grant BB/L007959/1 and funding for Lai lab is supported by the Ministry of Science and Technology of Taiwan grant no. 104‐2311‐B‐001‐025‐MY3. Funding is also supported by the BBSRC Taiwan‐UK International Partnering Award A.F (BB/M02735X/1) and E.M.L. (grant no. 105‐2911‐I‐001‐503).
Ronan R. McCarthy and Manda Yu were contributed equally.
Contributor Information
Ronan R. McCarthy, Email: ronan.mccarthy@brunel.ac.uk.
Alain Filloux, Email: a.filloux@imperial.ac.uk.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Abel, S. , Bucher, T. , Nicollier, M. , Hug, I. , Kaever, V. , Abel zur Wiesch, P. and Jenal, U. (2013) Bi‐modal distribution of the second messenger c‐di‐GMP controls cell fate and asymmetry during the caulobacter cell cycle. PLoS Genetics, 9, e1003744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcoforado Diniz, J. , Liu, Y.C. and Coulthurst, S.J. (2015) Molecular weaponry: diverse effectors delivered by the Type VI secretion system. Cellular Microbiology, 17, 1742–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allsopp, L.P. , Wood, T.E. , Howard, S.A. , Maggiorelli, F. , Nolan, L.M. , Wettstadt, S. , et al (2017) RsmA and AmrZ orchestrate the assembly of all three type VI secretion systems in Pseudomonas aeruginosa . Proceedings of the National Academy of Sciences of the United States of America, 114, 7707–7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amikam, D. and Benziman, M. (1989) Cyclic diguanylic acid and cellulose synthesis in Agrobacterium tumefaciens . Journal of Bacteriology, 171, 6649–6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, M.C. , Vonaesch, P. , Saffarian, A. , Marteyn, B.S. and Sansonetti, P.J. (2017) Shigella sonnei encodes a functional T6SS used for interbacterial competition and niche occupancy. Cell Host & Microbe, 21, 769–776.e3. [DOI] [PubMed] [Google Scholar]
- Aragon, I.M. , Perez‐Mendoza, D. , Moscoso, J.A. , Faure, E. , Guery, B. , Gallegos, M.T. , et al (2015) Diguanylate cyclase DgcP is involved in plant and human Pseudomonas spp. infections. Environmental Microbiology, 17, 4332–4351. [DOI] [PubMed] [Google Scholar]
- Bachmann, V. , Kostiuk, B. , Unterweger, D. , Diaz‐Satizabal, L. , Ogg, S. and Pukatzki, S. (2015) Bile salts modulate the mucin‐activated type VI secretion system of pandemic Vibrio cholerae . PLoS Neglected Tropical Diseases, 9, e0004031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud, N. , Hassett, D.J. , Hwang, S.H. , Rice, S.A. , Kjelleberg, S. and Webb, J.S. (2006) Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa . Journal of Bacteriology, 188, 7344–7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud, N. , Schleheck, D. , Klebensberger, J. , Webb, J.S. , Hassett, D.J. , Rice, S.A. , et al (2009) Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di‐GMP levels, and enhanced dispersal. Journal of Bacteriology, 191, 7333–7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler, M. , Ho, B.T. and Mekalanos, J.J. (2013) Tit‐for‐tat: type VI secretion system counterattack during bacterial cell‐cell interactions. Cell, 152, 884–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal, P. , Allsopp, L.P. , Filloux, A. and Llamas, M.A. (2017a) The Pseudomonas putida T6SS is a plant warden against phytopathogens. The ISME Journal, 11, 972–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal, P. , Llamas, M.A. and Filloux, A. (2017b) Type VI secretion systems in plant‐associated bacteria. Environmental Microbiology, 20(1), 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondage, D.D. , Lin, J.S. , Ma, L.S. , Kuo, C.H. and Lai, E.M. (2016) VgrG C terminus confers the type VI effector transport specificity and is required for binding with PAAR and adaptor‐effector complex. Proceedings of the National Academy of Sciences of the United States of America, 113, E3931–3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgeaud, S. , Metzger, L.C. , Scrignari, T. and Blokesch, M. (2015) The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science, 347, 63–67. [DOI] [PubMed] [Google Scholar]
- Cao, Z. , Casabona, M.G. , Kneuper, H. , Chalmers, J.D. and Palmer, T. (2016) The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nature microbiology, 2, 16183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassaing, B. and Cascales, E. (2018) Antibacterial weapons: targeted destruction in the microbiota. Trends in Microbiology, 26, 329–338. [DOI] [PubMed] [Google Scholar]
- Chatzidaki‐Livanis, M. , Geva‐Zatorsky, N. and Comstock, L.E. (2016) Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proceedings of the National Academy of Sciences of the United States of America, 113, 3627–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen, M. , Kulasekara, H.D. , Christen, B. , Kulasekara, B.R. , Hoffman, L.R. and Miller, S.I. (2010) Asymmetrical distribution of the second messenger c‐di‐GMP upon bacterial cell division. Science, 328, 1295–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie, P.J. , Whitaker, N. and Gonzalez‐Rivera, C. (2014) Mechanism and structure of the bacterial type IV secretion systems. Biochimica et biophysica acta, 1843, 1578–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianfanelli, F.R. , Alcoforado Diniz, J. , Guo, M. , De Cesare, V. , Trost, M. and Coulthurst, S.J. (2016a) VgrG and PAAR proteins define distinct versions of a functional type VI secretion system. PLoS Path, 12, e1005735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianfanelli, F.R. , Monlezun, L. and Coulthurst, S.J. (2016b) Aim, load, fire: the type VI secretion system, a bacterial nanoweapon. Trends in Microbiology, 24, 51–62. [DOI] [PubMed] [Google Scholar]
- Conner, J.G. , Zamorano‐Sanchez, D. , Park, J.H. , Sondermann, H. and Yildiz, F.H. (2017) The ins and outs of cyclic di‐GMP signaling in Vibrio cholerae . Current Opinion in Microbiology, 36, 20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlstrom, K.M. and O'Toole, G.A. (2017) A symphony of cyclases: specificity in diguanylate cyclase signaling. Annual Review of Microbiology, 71, 179–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand, E. , Derrez, E. , Audoly, G. , Spinelli, S. , Ortiz‐Lombardia, M. , Raoult, D. , et al (2012) Crystal structure of the VgrG1 actin cross‐linking domain of the Vibrio cholerae type VI secretion system. The Journal of Biological Chemistry, 287, 38190–38199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand, E. , Cambillau, C. , Cascales, E. and Journet, L. (2014) VgrG, Tae, Tle, and beyond: the versatile arsenal of Type VI secretion effectors. Trends in Microbiology, 22, 498–507. [DOI] [PubMed] [Google Scholar]
- Feirer, N. , Xu, J. , Allen, K.D. , Koestler, B.J. , Bruger, E.L. , Waters, C.M. , et al (2015) A pterin‐dependent signaling pathway regulates a dual‐function diguanylate cyclase‐phosphodiesterase controlling surface attachment in Agrobacterium tumefaciens . mBio, 6, e00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feirer, N. , Kim, D. , Xu, J. , Fernandez, N. , Waters, C.M. and Fuqua, C. (2017) The Agrobacterium tumefaciens CheY‐like protein ClaR regulates biofilm formation. Microbiology, 163(11), 1680–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaugnatti, N. , Le, T.T. , Canaan, S. , Aschtgen, M.S. , Nguyen, V.S. , Blangy, S. , et al (2016) A phospholipase A1 antibacterial Type VI secretion effector interacts directly with the C‐terminal domain of the VgrG spike protein for delivery. Molecular Microbiology, 99, 1099–1118. [DOI] [PubMed] [Google Scholar]
- Gelvin, S.B. (2010) Plant proteins involved in Agrobacterium‐mediated genetic transformation. Annual Review of Phytopathology, 48, 45–68. [DOI] [PubMed] [Google Scholar]
- Guvener, Z.T. and Harwood, C.S. (2007) Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic‐di‐GMP in response to growth on surfaces. Molecular Microbiology, 66, 1459–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachani, A. , Allsopp, L.P. , Oduko, Y. and Filloux, A. (2014) The VgrG proteins are “a la carte” delivery systems for bacterial type VI effectors. The Journal of Biological Chemistry, 289, 17872–17884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachani, A. , Wood, T.E. and Filloux, A. (2016) Type VI secretion and anti‐host effectors. Current Opinion in Microbiology, 29, 81–93. [DOI] [PubMed] [Google Scholar]
- Heckel, B.C. , Tomlinson, A.D. , Morton, E.R. , Choi, J.H. and Fuqua, C. (2014) Agrobacterium tumefaciens exoR controls acid response genes and impacts exopolysaccharide synthesis, horizontal gene transfer, and virulence gene expression. Journal of Bacteriology, 196, 3221–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heindl, J.E. , Wang, Y. , Heckel, B.C. , Mohari, B. , Feirer, N. and Fuqua, C. (2014) Mechanisms and regulation of surface interactions and biofilm formation in Agrobacterium . Frontiers in Plant Science, 5, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsa, S.M. and O'Toole, G.A. (2006) Biofilm formation by Pseudomonas fluorescens WCS365: a role for LapD. Microbiology, 152, 1375–1383. [DOI] [PubMed] [Google Scholar]
- Howie, A.J. and Brewer, D.B. (2009) Optical properties of amyloid stained by Congo red: history and mechanisms. Micron, 40, 285–301. [DOI] [PubMed] [Google Scholar]
- Howlin, R.P. , Cathie, K. , Hall‐Stoodley, L. , Cornelius, V. , Duignan, C. , Allan, R.N. , et al (2017) Low‐dose nitric oxide as targeted anti‐biofilm adjunctive therapy to treat chronic Pseudomonas aeruginosa infection in cystic fibrosis. Molecular Therapy, 25, 2104–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, H.‐H. , Yu, M. and Lai, E.‐M. (2017) Agrobacterium‐Mediated Plant Transformation: Biology and Applications. The Arabidopsis Book, e0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa, T. , Rompikuntal, P.K. , Lindmark, B. , Milton, D.L. and Wai, S.N. (2009) Quorum sensing regulation of the two hcp alleles in Vibrio cholerae O1 strains. PLoS One, 4, e6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskolska, M. , Stutzmann, S. , Stoudmann, C. and Blokesch, M. (2018) QstR‐dependent regulation of natural competence and type VI secretion in Vibrio cholerae . Nucleic Acids Research, 46, 10619–10634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal, U. , Reinders, A. and Lori, C. (2017) Cyclic di‐GMP: second messenger extraordinaire. Nature Reviews Microbiology, 15, 271–284. [DOI] [PubMed] [Google Scholar]
- Jiang, F. , Waterfield, N.R. , Yang, J. , Yang, G. and Jin, Q. (2014) A Pseudomonas aeruginosa type VI secretion phospholipase D effector targets both prokaryotic and eukaryotic cells. Cell Host & Microbe, 15, 600–610. [DOI] [PubMed] [Google Scholar]
- Jones, C. , Hachani, A. , Manoli, E. and Filloux, A. (2014) An rhs gene linked to the second type VI secretion cluster is a feature of the Pseudomonas aeruginosa strain PA14. Journal of Bacteriology, 196, 800–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, A. , Kostiuk, B. , Rogers, A. , Teschler, J. , Pukatzki, S. and Yildiz, F.H. (2017) Rules of engagement: the type VI secretion system in Vibrio cholerae . Trends in Microbiology, 25, 267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kado, C.I. and Heskett, M.G. (1970) Selective media for isolation of Agrobacterium, Corynebacterium, Erwinia, Pseudomonas, and Xanthomonas . Phytopathology, 60, 969–976. [DOI] [PubMed] [Google Scholar]
- Kapitein, N. and Mogk, A. (2014) Type VI secretion system helps find a niche. Cell Host & Microbe, 16, 5–6. [DOI] [PubMed] [Google Scholar]
- Kim, K.W. , Franceschi, V.R. , Davin, L.B. and Lewis, N.G. (2006) Beta‐glucuronidase as reporter gene: advantages and limitations. Methods in Molecular Biology, 323, 263–273. [DOI] [PubMed] [Google Scholar]
- Kulasekara, B.R. , Kamischke, C. , Kulasekara, H.D. , Christen, M. , Wiggins, P.A. and Miller, S.I. (2013) c‐di‐GMP heterogeneity is generated by the chemotaxis machinery to regulate flagellar motility. eLife, 2, e01402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoux, M. , Kirkpatrick, R.L. , Montauti, E.I. , Tran, B.Q. , Peterson, S.B. , Harding, B.N. , et al. (2015). Kin cell lysis is a danger signal that activates antibacterial pathways of Pseudomonas aeruginosa . eLife, 4, e05701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesic, B. , Starkey, M. , He, J. , Hazan, R. and Rahme, L.G. (2009) Quorum sensing differentially regulates Pseudomonas aeruginosa type VI secretion locus I and homologous loci II and III, which are required for pathogenesis. Microbiology, 155, 2845–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien, Y.W. and Lai, E.M. (2017) Type VI secretion effectors: methodologies and biology. Frontiers in Cellular and Infection Microbiology, 7, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J.S. , Wu, H.H. , Hsu, P.H. , Ma, L.S. , Pang, Y.Y. , Tsai, M.D. , et al (2014) Fha interaction with phosphothreonine of TssL activates type VI secretion in Agrobacterium tumefaciens . PLoS Path, 10, e1003991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lori, C. , Ozaki, S. , Steiner, S. , Bohm, R. , Abel, S. , Dubey, B.N. , et al (2015) Cyclic di‐GMP acts as a cell cycle oscillator to drive chromosome replication. Nature, 523, 236–239. [DOI] [PubMed] [Google Scholar]
- Ma, L.S. , Lin, J.S. and Lai, E.M. (2009) An IcmF family protein, ImpLM, is an integral inner membrane protein interacting with ImpKL, and its walker a motif is required for type VI secretion system‐mediated Hcp secretion in Agrobacterium tumefaciens . Journal of Bacteriology, 191, 4316–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L.S. , Hachani, A. , Lin, J.S. , Filloux, A. and Lai, E.M. (2014) Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host & Microbe, 16, 94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerczyk, C. , Schneider, E. and Greenberg, E.P. (2016). Quorum sensing control of Type VI secretion factors restricts the proliferation of quorum‐sensing mutants. eLife, 5, e14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, R.R. , Mazon‐Moya, M.J. , Moscoso, J.A. , Hao, Y. , Lam, J.S. , Bordi, C. , et al (2017a) Cyclic‐di‐GMP regulates lipopolysaccharide modification and contributes to Pseudomonas aeruginosa immune evasion. Nature Microbiology, 2, 17027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, R.R. , Valentini, M. and Filloux, A. (2017b) Contribution of cyclic di‐GMP in the control of type III and type VI secretion in Pseudomonas aeruginosa . Methods in Molecular Biology, 1657, 213–224. [DOI] [PubMed] [Google Scholar]
- McNally, L. , Bernardy, E. , Thomas, J. , Kalziqi, A. , Pentz, J. , Brown, S.P. , et al . (2017) Killing by Type VI secretion drives genetic phase separation and correlates with increased cooperation. Nature Communications, 8, 14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt, J.H. , Brothers, K.M. , Kuchma, S.L. and O'Toole, G.A. (2007a) SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. Journal of Bacteriology, 189, 8154–8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt, P.M. , Danhorn, T. and Fuqua, C. (2007b) Motility and chemotaxis in Agrobacterium tumefaciens surface attachment and biofilm formation. Journal of Bacteriology, 189, 8005–8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscoso, J.A. , Mikkelsen, H. , Heeb, S. , Williams, P. and Filloux, A. (2011) The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c‐di‐GMP signalling. Environmental Microbiology, 13, 3128–3138. [DOI] [PubMed] [Google Scholar]
- Moscoso, J.A. , Jaeger, T. , Valentini, M. , Hui, K. , Jenal, U. and Filloux, A. (2014) The diguanylate cyclase SadC is a central player in Gac/Rsm‐mediated biofilm formation in Pseudomonas aeruginosa . Journal of Bacteriology, 196, 4081–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, M.V. , Newell, P.D. , Krasteva, P.V. , Chatterjee, D. , Madden, D.R. , O'Toole, G.A. , et al (2011) Structural basis for c‐di‐GMP‐mediated inside‐out signaling controlling periplasmic proteolysis. PLoS Biology, 9, e1000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell, P.D. , Monds, R.D. and O'Toole, G.A. (2009) LapD is a bis‐(3',5')‐cyclic dimeric GMP‐binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0‐1. Proceedings of the National Academy of Sciences of the United States of America, 106, 3461–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, V.S. , Douzi, B. , Durand, E. , Roussel, A. , Cascales, E. and Cambillau, C. (2018) Towards a complete structural deciphering of Type VI secretion system. Current Opinion in Structural Biology, 49, 77–84. [DOI] [PubMed] [Google Scholar]
- Ozaki, S. , Schalch‐Moser, A. , Zumthor, L. , Manfredi, P. , Ebbensgaard, A. , Schirmer, T. , et al (2014) Activation and polar sequestration of PopA, a c‐di‐GMP effector protein involved in Caulobacter crescentus cell cycle control. Molecular Microbiology, 94, 580–594. [DOI] [PubMed] [Google Scholar]
- Pissaridou, P. , Allsopp, L.P. , Wettstadt, S. , Howard, S.A. , Mavridou, D.A.I. and Filloux, A. (2018) The Pseudomonas aeruginosa T6SS‐VgrG1b spike is topped by a PAAR protein eliciting DNA damage to bacterial competitors. Proceedings of the National Academy of Sciences of the United States of America, 115, 12519–12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke, A. and Hirt, H. (2010) New insights into an old story: Agrobacterium‐induced tumour formation in plants by plant transformation. The EMBO Journal, 29, 1021–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki, S. , Ma, A.T. , Revel, A.T. , Sturtevant, D. and Mekalanos, J.J. (2007) Type VI secretion system translocates a phage tail spike‐like protein into target cells where it cross‐links actin. Proceedings of the National Academy of Sciences of the United States of America, 104, 15508–15513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin, D. , Ahmad, S. , Shanthamoorthy, P. , Mougous, J.D. , Whitney, J.C. and Raunser, S. (2018) Mechanism of loading and translocation of type VI secretion system effector Tse6. Nature Microbiology, 3, 1142–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Records, A.R. and Gross, D.C. (2010) Sensor kinases RetS and LadS regulate Pseudomonas syringae type VI secretion and virulence factors. Journal of Bacteriology, 192, 3584–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reen, F.J. , Woods, D.F. , Mooij, M.J. , Adams, C. and O'Gara, F. (2012) Respiratory pathogens adopt a chronic lifestyle in response to bile. PLoS One, 7, e45978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo, D. , Mendez‐Garcia, C. , Raczkowska, B.A. , Bargiela, R. , Moya, A. , Ferrer, M. , et al (2017) Exploring the human microbiome from multiple perspectives: factors altering its composition and function. FEMS Microbiology Reviews, 41, 453–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling, U. and Balsalobre, C. (2012) Biofilm infections, their resilience to therapy and innovative treatment strategies. Journal of Internal Medicine, 272, 541–561. [DOI] [PubMed] [Google Scholar]
- Romling, U. , Galperin, M.Y. and Gomelsky, M. (2013) Cyclic di‐GMP: the first 25 years of a universal bacterial second messenger. Microbiology and Molecular Biology Reviews, 77, 1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem, O. , Nesper, J. , Borovok, I. , Gorovits, R. , Kolot, M. , Pasternak, Z. , et al (2015) An extended cyclic di‐GMP network in the predatory bacterium Bdellovibrio bacteriovorus. Journal of Bacteriology, 198, 127–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, A.B. , Hood, R.D. , Bui, N.K. , LeRoux, M. , Vollmer, W. and Mougous, J.D. (2011) Type VI secretion delivers bacteriolytic effectors to target cells. Nature, 475, 343–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, A.B. , Singh, P. , Brittnacher, M. , Bui, N.K. , Hood, R.D. , Carl, M.A. , et al (2012) A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host & Microbe, 11, 538–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, A.B. , Peterson, S.B. and Mougous, J.D. (2014a) Type VI secretion system effectors: poisons with a purpose. Nature Reviews Microbiology, 12, 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, A.B. , Wexler, A.G. , Harding, B.N. , Whitney, J.C. , Bohn, A.J. , Goo, Y.A. , et al (2014b) A type VI secretion‐related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host & Microbe, 16, 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sana, T.G. , Hachani, A. , Bucior, I. , Soscia, C. , Garvis, S. , Termine, E. , et al (2012) The second type VI secretion system of Pseudomonas aeruginosa strain PAO1 is regulated by quorum sensing and Fur and modulates internalization in epithelial cells. The Journal of Biological Chemistry, 287, 27095–27105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sana, T.G. , Flaugnatti, N. , Lugo, K.A. , Lam, L.H. , Jacobson, A. , Baylot, V. , et al (2016) Salmonella Typhimurium utilizes a T6SS‐mediated antibacterial weapon to establish in the host gut. Proceedings of the National Academy of Sciences of the United States of America, 113, E5044–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng, L. , Lv, Y. , Liu, Q. , Wang, Q. and Zhang, Y. (2013) Connecting type VI secretion, quorum sensing, and c‐di‐GMP production in fish pathogen Vibrio alginolyticus through phosphatase PppA. Veterinary Microbiology, 162, 652–662. [DOI] [PubMed] [Google Scholar]
- Skotnicka, D. , Smaldone, G.T. , Petters, T. , Trampari, E. , Liang, J. , Kaever, V. , et al (2016) A minimal threshold of c‐di‐gmp is essential for fruiting body formation and sporulation in Myxococcus xanthus . PLoS Genetics, 12, e1006080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trampari, E. , Stevenson, C.E. , Little, R.H. , Wilhelm, T. , Lawson, D.M. and Malone, J.G. (2015) Bacterial rotary export ATPases are allosterically regulated by the nucleotide second messenger cyclic‐di‐GMP. The Journal of Biological Chemistry, 290, 24470–24483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trunk, K. , Peltier, J. , Liu, Y.C. , Dill, B.D. , Walker, L. , Gow, N.A.R. , et al (2018) The type VI secretion system deploys antifungal effectors against microbial competitors. Nature Microbiology, 3, 920–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, A. and Wood, T.K. (2010) Tyrosine Phosphatase TpbA of Pseudomonas aeruginosa controls extracellular DNA via cyclic diguanylic acid concentrations. Environmental Microbiology, 2, 449–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini, M. and Filloux, A. (2016) Biofilms and cyclic di‐GMP (c‐di‐GMP) signaling: lessons from Pseudomonas aeruginosa and other bacteria. The Journal of Biological Chemistry, 291, 12547–12555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini, M. , Laventie, B.J. , Moscoso, J. , Jenal, U. and Filloux, A. (2016) The diguanylate cyclase HsbD intersects with the HptB regulatory cascade to control Pseudomonas aeruginosa biofilm and motility. PLoS Genetics, 12, e1006354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney, J.C. , Beck, C.M. , Goo, Y.A. , Russell, A.B. , Harding, B.N. , De Leon, J.A. , et al (2014) Genetically distinct pathways guide effector export through the type VI secretion system. Molecular Microbiology, 92, 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney, J.C. , Quentin, D. , Sawai, S. , LeRoux, M. , Harding, B.N. , Ledvina, H.E. , et al (2015) An interbacterial NAD(P)(+) glycohydrolase toxin requires elongation factor Tu for delivery to target cells. Cell, 163, 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H.Y. , Chung, P.C. , Shih, H.W. , Wen, S.R. and Lai, E.M. (2008) Secretome analysis uncovers an Hcp‐family protein secreted via a type VI secretion system in Agrobacterium tumefaciens . Journal of Bacteriology, 190, 2841–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C.F. , Lin, J.S. , Shaw, G.C. and Lai, E.M. (2012) Acid‐induced type VI secretion system is regulated by ExoR‐ChvG/ChvI signaling cascade in Agrobacterium tumefaciens . PLoS Path, 8, e1002938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H.Y. , Liu, K.H. , Wang, Y.C. , Wu, J.F. , Chiu, W.L. , Chen, C.Y. , et al (2014) AGROBEST: an efficient Agrobacterium‐mediated transient expression method for versatile gene function analyses in Arabidopsis seedlings. Plant Methods, 10, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. , Kim, J. , Koestler, B.J. , Choi, J.H. , Waters, C.M. and Fuqua, C. (2013) Genetic analysis of Agrobacterium tumefaciens unipolar polysaccharide production reveals complex integrated control of the motile‐to‐sessile switch. Molecular Microbiology, 89, 929–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


