Abstract
The hyperpolarization-activated cation current (termedIh,Iq, orIf) was recently shown to be encoded by a new family of genes, named HCN forhyperpolarization-activated cyclicnucleotide-sensitive cationnonselective. When expressed in heterologous cells, each HCN isoform generates channels with distinct activation kinetics, mirroring the range of biophysical properties of nativeIh currents recorded in different classes of neurons. To determine whether the functional diversity ofIh currents is attributable to different patterns of HCN gene expression, we determined the mRNA distribution across different regions of the mouse CNS of the three mouse HCN genes that are prominently expressed there (mHCN1, 2 and 4). We observe distinct patterns of distribution for each of the three genes. Whereas mHCN2 shows a widespread expression throughout the CNS, the expression of mHCN1 and mHCN4 is more limited, and generally complementary. mHCN1 is primarily expressed within neurons of the neocortex, hippocampus, and cerebellar cortex, but also in selected nuclei of the brainstem. mHCN4 is most highly expressed within neurons of the medial habenula, thalamus, and olfactory bulb, but also in distinct neuronal populations of the basal ganglia. Based on a comparison of mRNA expression with an electrophysiological characterization of nativeIh currents in hippocampal and thalamic neurons, our data support the idea that the functional heterogeneity ofIh channels is attributable, in part, to differential isoform expression. Moreover, in some neurons, specific functional roles can be proposed for Ihchannels with defined subunit composition.
Keywords: Ih, pacemaker channel, hippocampus, thalamus, in situ hybridization, HCN
Hyperpolarization-activated cation channels, first identified in cardiac sinoatrial node cells, are found in a variety of peripheral and central neurons (for review, seeDiFrancesco, 1993; Pape, 1996). These channels slowly activate in response to hyperpolarization to generate inward currents, termedIf in cardiac cells andIh in neurons. In spontaneously firing nerve and muscle cells, Ih contributes to the pacemaker depolarization that generates rhythmic activity (McCormick and Bal, 1997). In nonpacing cells,Ih helps determine resting membrane properties and limits the extent of hyperpolarizing or depolarizing responses (Pape, 1996). Ih channels are also present in dendrites (Magee, 1998, 1999) and presynaptic terminals (Fletcher and Chiappinelli, 1992; Beaumont and Zucker, 2000), where they regulate synaptic transmission. The rate ofIh activation is enhanced by the direct binding of cAMP to the channel, providing a powerful means to regulate excitability (DiFrancesco and Tortora, 1991; Ingram and Williams, 1996).
The diverse roles of Ih are mirrored in the heterogeneous biophysical properties observed in the various cells in which this current is expressed (Santoro and Tibbs, 1999). Thus, two important questions arise: What is the molecular basis for this diversity? And how are the dynamic and modulatory properties ofIh tuned to specific neuronal functions? The recent cloning of four mammalianIh genes, termed HCN1–4 forhyperpolarization-activated cyclicnucleotide-sensitive cationnonselective channels (Santoro et al., 1997, 1998; Ludwig et al., 1998, 1999; Ishii et al., 1999; Seifert et al., 1999; Vaccari et al., 1999; Moroni et al., 2000) (for review, see Clapham, 1998; Santoro and Tibbs, 1999), now allows such questions to be studied.
Three of the four HCN genes have been heterologously expressed, where they generate hyperpolarization-activated currents with distinct biophysical characteristics. HCN1 channels activate relatively rapidly on hyperpolarization (in tens of milliseconds) and show a minimal response to cAMP (Santoro et al., 1998). HCN2 channels activate more slowly (hundreds of milliseconds) and are modulated strongly by cAMP (Ludwig et al., 1998, 1999; Santoro and Tibbs, 1999). Finally HCN4 channels activate very slowly (seconds) and respond strongly to cAMP (Ishii et al., 1999; Ludwig et al., 1999; Seifert et al., 1999). This functional diversity of recombinant HCN channels suggests that differential HCN gene expression may generate the heterogeneity in native Ih currents.
Initial studies of mRNA distribution showed that all four mouse HCN isoforms are expressed in the brain, although with different regional distributions and at different levels (Santoro et al., 1997, 1998;Ludwig et al., 1998; Moosmang et al., 1999). mHCN1 is strongly expressed in cortical areas; mHCN2 is widely expressed at a high level throughout the brain; mHCN3 is also widely expressed, but at very low levels; and mHCN4 shows strong subcortical expression. mHCN2 and mHCN4 are also strongly expressed in the heart. However, many of the details of the cellular distribution of mHCN in regions that exhibit prominentIh are lacking. In this study we present a detailed mapping of HCN isoform expression in the brain and spinal cord for the three mouse HCN genes that are strongly expressed in the CNS (mHCN1, mHCN2, and mHCN4). By comparing recombinant HCN channel properties with native Ih from thalamic and hippocampal neurons, we suggest specific functional roles for the different mHCN isoforms.
MATERIALS AND METHODS
In situ hybridization
In situ hybridization was performed essentially as described (Schaeren-Wiemers and Gerfin-Moser, 1993). Adult male C57Bl/6 mice were killed by cervical dislocation, and the brains removed and quickly frozen in OCT (Tissue-Tek) compound. Coronal cryostat sections (20 μm) were prepared on Superfrost Plus (VWR Scientific) slides, fixed in 4% paraformaldehyde–PBS, and acetylated (triethanolamine–acetic anhydride). Coordinates for coronal sections shown in Figures 1-7 are according to Franklin and Paxinos (1997).
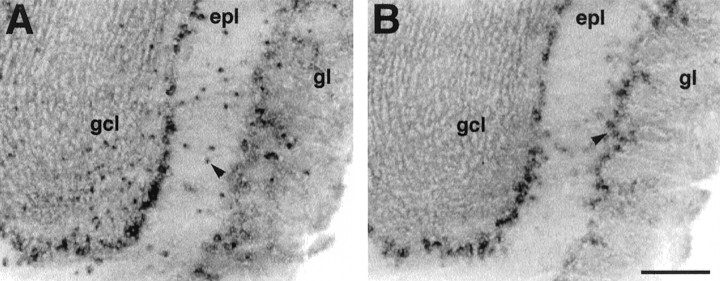
Fig. 1.
mHCN2 and mHCN4 are expressed in olfactory bulb.A, Coronal section (bregma, +4.28 mm) showing labeling by mHCN2 probe of mitral cells, tufted cells, and short-axon cells (arrowhead). B, mHCN4 transcripts are present in mitral cells and tufted cells (arrowheadshows externally placed tufted cells), but not in short-axon cells.gcl, Granule cell layer; epl, external plexiform layer; gl, glomerular layer. Scale bar, 200 μm.
Fig. 2.
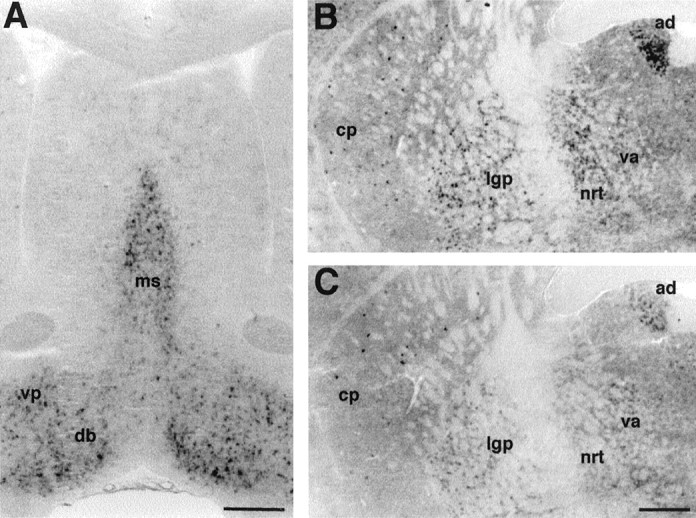
Expression of mHCN2 and mHCN4 mRNA in the mouse basal forebrain. A, Coronal section (bregma, +0.74) showing mHCN2 labeling in the medial septum (ms), vertical and horizontal limb of the diagonal band (db), and ventral pallidum (vp). B, Coronal section (bregma, −0.82) showing staining by the mHCN2 probe of cells in the caudate putamen (cp), lateral globus pallidus (lgp), thalamus nucleus reticularis (nrt), ventral anterior thalamic nucleus (va), and anterodorsal thalamic nucleus (ad).C, Coronal section (bregma, −0.82) showing staining by the mHCN4 probe of cells in the caudate putamen, lateral globus pallidus, and anterodorsal thalamic nucleus. mHCN4 is not present in thalamus nucleus reticularis. Scale bars, 500 μm.
Fig. 3.
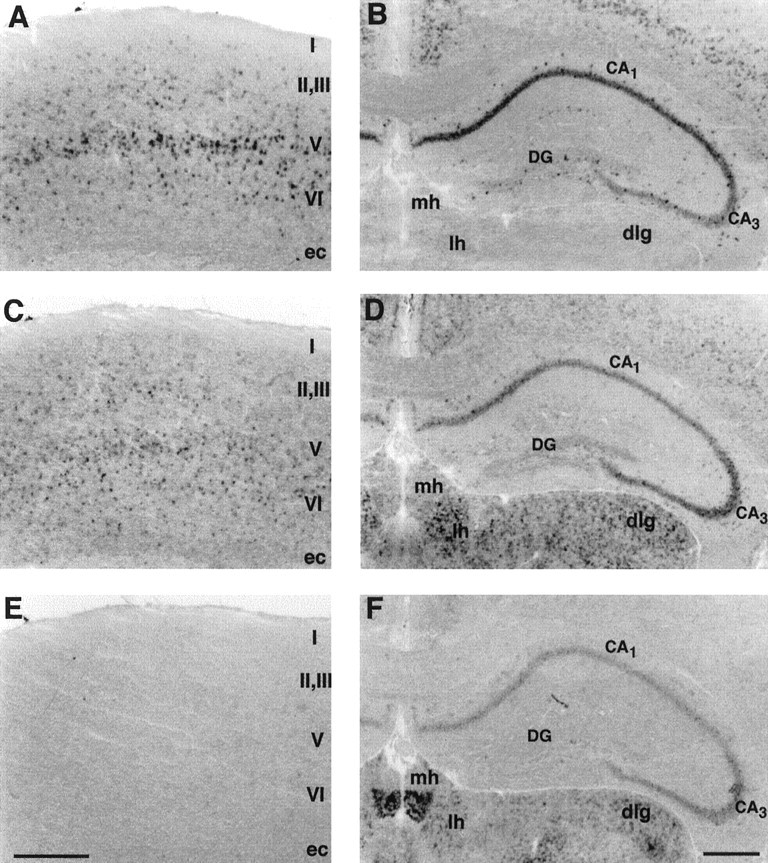
Differential distribution of mHCN1, mHCN2, and mHCN4 in the mouse cerebral and hippocampal cortex. A,Coronal section (bregma, +0.74) showing mHCN1 labeling in the motor cortex. B, Coronal section (bregma, −2.06) showing mHCN1 labeling in the hippocampus. C, mHCN2 labeling in the motor cortex. D, mHCN2 labeling in the hippocampus, lateral habenula, and dorsal lateral geniculate nucleus.E, mHCN4 labeling is absent in the motor cortex.F, mHCN4 labeling in the hippocampus, medial and lateral habenula, and dorsal lateral geniculate nucleus. ec, External capusle; mh, medial habenula;lh, lateral habenula; dlg, dorsal lateral geniculate nucleus; DG, dentate gyrus;CA1, CA3,cornus ammonis fields CA1 and CA3. Scale bars, 500 μm.
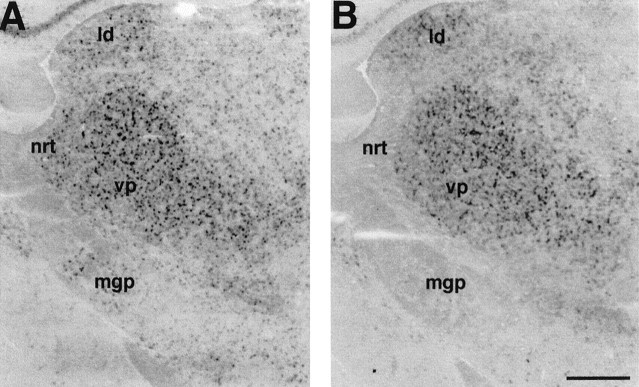
Fig. 4.
Expression of mHCN2 and mHCN4 in the mouse thalamus. A, Coronal section (bregma, −1.58) showing labeling of laterodorsal (ld), ventroposterior (vp), and reticular (nrt) thalamic nuclei by mHCN2 probe. Also visible is the labeling of cells in the medial globus pallidus (mgp). B, Labeling by mHCN4 probe of laterodorsal and ventroposterior thalamic nuclei. Weak labeling is also visible in the medial globus pallidus. Scale bar, 500 μm.
Fig. 5.
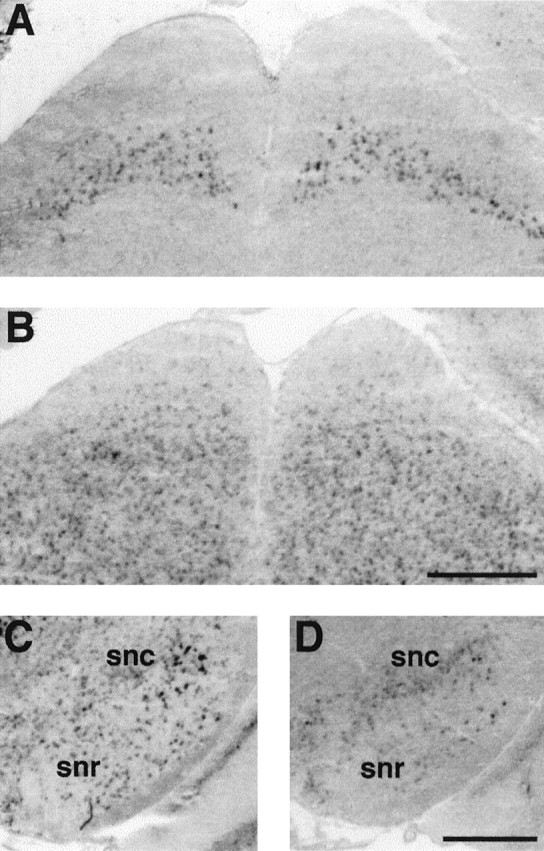
Expression of mHCN1, mHCN2, and mHCN4 transcripts in midbrain. A, Coronal section (bregma, −4.04) showing mHCN1 labeling in the superior colliculus. B, mHCN2 labeling in the superior colliculus. C, Coronal section (bregma, −3.28) showing staining for mHCN2 in substantia nigra, pars reticulata (snr). D, Staining for mHCN4 in substantia nigra, mostly pars compacta (snc). Scale bars, 500 μm.
Fig. 6.
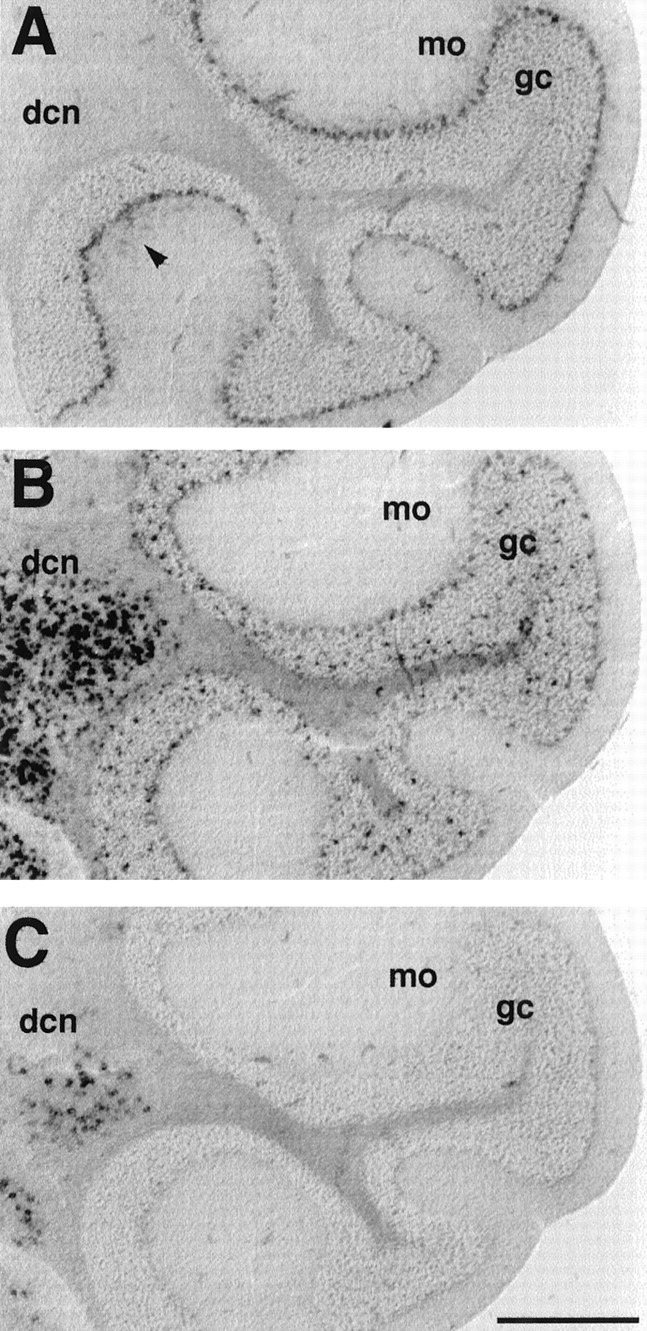
Differential expression of mHCN1, mHCN2, and mHCN4 transcripts in cerebellum. A, Coronal section (bregma, −6.24) showing labeling of Purkinje cells by mHCN1 probe. Weak labeling of basket cells (arrowheads) is also visible in the molecular layer (mo). B, Labeling of Purkinje cell layer, granule cell layer (gc), and deep cerebellar nuclei (dcn) by mHCN2 probe.C, mHCN4 labeling in deep cerebellar nuclei. Scale bar, 500 μm.
Fig. 7.
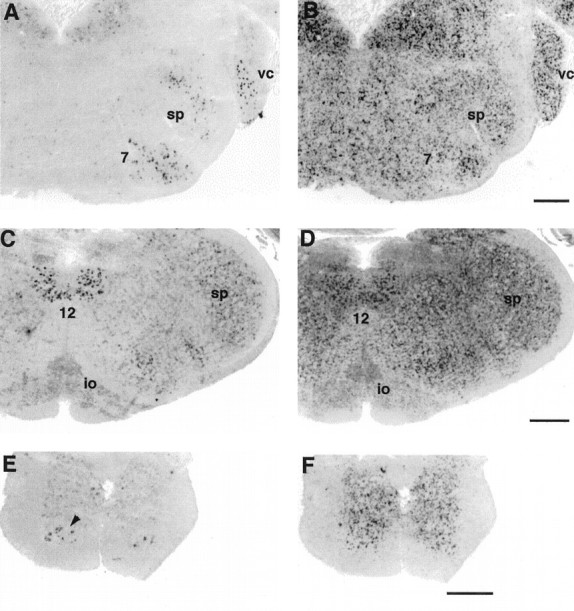
mHCN1 is expressed in selected nuclei in the brainstem and spinal cord. A, Coronal section (bregma, −6.24) showing labeling of ventral cochlear nucleus (vc), spinal trigeminal nucleus (sp), and facial nucleus (7) by mHCN1 probe. Weak labeling of vestibular nuclei is also visible. B, Serial section showing labeling by mHCN2 probe. C, Coronal section (bregma, −7.08) showing labeling of hypoglossal nucleus (12) by mHCN1 probe. Weak labeling of spinal trigeminal and lateral reticular nuclei, as well as of inferior olive (io) is also visible. D, Serial section showing labeling by mHCN2 probe. E, Coronal section through thoracic spinal cord showing labeling by mHCN1 probe; notice strong labeling of the motorneurons in the ventral horn (arrowhead). F, Serial section showing labeling by mHCN2 probe. Scale bars, 500 μm.
Prehybridization was performed for 1 hr at room temperature (RT) in 50% formamide, 5× SSC, 5× Denhardt's solution, 0.25 mg/ml tRNA, and 0.5 mg/ml salmon sperm DNA, followed by hybridization overnight at 68°C in the same buffer including 200–400 ng/ml riboprobe (see below). Slices were washed at 68°C in 0.2× SSC, blocked for 1 hr at RT in 0.1 m Tris, pH 7.5, 0.15 m NaCl + 10% inactivated goat serum (Sigma, St. Louis, MO), and incubated overnight at 4°C in the same buffer + 1% goat serum and 1:5000 dilution of anti-DIG antibody coupled to alkaline phosphatase (Boehringer Mannheim, Mannheim, Germany). The staining reaction was performed for 24–48 hr at RT in 0.1 m Tris, pH 9.5, 0.1 m NaCl, 50 mm MgCl2, 0.24 mg/ml levamisole, 0.225 mg/ml nitroblue tetrazolium, and 0.175 mg/ml 5-bromo-4-chloro-3-indolyl-phosphate (Promega, Madison, WI). Slides were washed in 10 mm Tris, pH 7.5, 1 mmEDTA and mounted in Fluoromount-G (Southern Biotechnology Associates, Birmingham, AL).
Antisense and sense riboprobes corresponding to amino acids 777–910 of mHCN1, 322–612 of mHCN2, and 400–690 of mHCN4 were in vitro transcribed in the presence of 3.5:6.5 digoxigenin-UTP:UTP (Boehringer Mannheim) according to the manufacturer's instructions. Control hybridizations performed in the presence of sense RNA probes did not yield any detectable signals with staining reactions of up to 72 hr (data not shown).
Electrophysiology
Native currents. Hippocampal or thalamic slices (400 μm) were prepared from 2.5- to 4-week-old C57Bl/6 mice using a gravity-driven chopper (hippocampal slices) or a Vibratome (thalamic slices). After at least 1 hr of recovery, slices were transferred to a submerged recording chamber perfused (1–2 ml/min) with artificial CSF (ACSF) containing (mm): NaCl 119; KCl 5; MgSO4 1; CaCl2 2; NaHCO3 26.2; NaH2PO4 1; and dextrose 11; bubbled with 95% O2 and 5% CO2. The temperature of the chamber was maintained at 32–34°C.
Whole-cell recordings were obtained from visually identified pyramidal cells, interneurons, thalamocortical cells, and reticular cells using infrared-differential interference contrast microscopy (Dodt and Zieglgansberger, 1990). For hippocampal recordings, electrodes were pulled from borosilicate glass to a resistance of 3–5 MΩ and filled with (in mm): 120 KMeSO4, 20 KCl, 10 HEPES, 4.0 Mg2-ATP, 0.3 Na2-GTP, 14 phosphocreatine, 4 NaCl, and 0.5 mm EGTA, pH 7.25 with KOH. All current records were made within 10 min of attaining whole-cell access. Interneurons were identified by the position and morphology of the cell body as well as by electrophysiological characteristics, including a relative lack of action potential accommodation and short, brief action potentials as compared to pyramidal cells. Thalamic recordings were done with pipettes filled with (in mm): 110 K gluconate, 10 KCl, 10 HEPES, 2 MgCl2, 2 Na2ATP, 0.2 NaGTP, 0.02 calmodulin, and 290 mOsm, pH 7.25, as previously described (Lüthi and McCormick, 1999). Series resistance of 5–12 MΩ was electronically compensated 32–80%. A liquid junction potential of 7–8 mV measured as described (Neher, 1992) was taken into account for all the data. Thalamocortical cells were identified electrophysiologically by their robust rebound burst firing after transient membrane hyperpolarization, whereas nRT neurons displayed a burst-tonic action potential discharge.
Oocyte recordings. mHCN1 and mHCN2 (Ludwig et al., 1998;Santoro et al., 1998) were subcloned into the pGHE expression vector. RNA was transcribed from NheI-linearized DNA (mHCN1) orSphI-linearized DNA (mHCN2) using T7 RNA polymerase (Message Machine; Ambion, Houston, TX) and injected into Xenopusoocytes prepared as previously described (Santoro et al., 1998). Two microelectrode voltage-clamp recordings were obtained 1–2 d after cRNA injection using a Warner Instruments (Hamden, CT) OC-725B amplifier. Data were digitized and acquired using an ITC-18 interface (Instrutech) and acquired and analyzed with Pulse and PulseFit software (Heka Electronics). Data were filtered at 250 Hz and sampled at 500 Hz for most experiments. For some mHCN1 data, currents were filtered at 1 kHz and sampled at 2 kHz. Custom analysis routines were written with IgorPro. We used a high-KCl extracellular solution to maximize the amplitude of the Ih currents. The solution contained (in mm): 96 KCl, 2 NaCl, 10 HEPES, and 2 MgCl2 , pH 7.5, with KOH. The microelectrodes were filled with 3 m KCl and had resistances of 0.5–1.5 MΩ. All recordings were obtained at room temperature (23–25°C).
Analysis
“Steady-state” activation curves were determined from the amplitude of tail currents observed after hyperpolarizing voltage steps on return to −40 mV for recombinant currents and the given holding potential for native currents. Tail current amplitudes were measured after the decay of the capacitive transient by averaging the current during the plateau of the tail (between 35 and 50 msec after the return to −40 mV for mHCN1 and between 80 and 100 msec for mHCN2). Changing the time window had no effect on activation curves. Current values were plotted versus the hyperpolarization step voltage and fitted with the Boltzmann equation:
where A1 is an offset caused by a nonzero holding current, A2 is the maximal tail current amplitude, V is voltage during the hyperpolarizing test pulse in millivolts, andV1/2 is the activation midpoint voltage. For each experiment, the tail current data were fitted with the above equation. To average the activation data from different experiments, the tail current amplitudes,I(V), from each individual experiment were normalized by first subtracting the derivedA1 parameter and then dividing byA2. The normalized data at each voltage were then averaged, and the averaged data were fitted by the Boltzmann equation (with A1 set at 0 and A2 set at 1). These normalized curves have been plotted in the indicated figures.
The rate of channel activation of Ihin the various cell types was determined by fitting the current evoked during hyperpolarizing voltage steps to single or double exponential functions in IgorPro. For all cell types where there was measurableIh, simultaneous fitting with two exponential components yielded fits that were significantly better than single exponential terms for all currents activated in response to steps to potentials that were negative to the steady-state midpoint voltage of activation (V1/2); the fit was not improved after addition of a third component.Ih currents in certain types of native neurons and for certain isoforms exhibit a distinct lag in their activation time course that is not described by the one or two exponential function fits. We have therefore excluded the initial lag from the fitting procedure. The purpose of these fits was solely to provide an empirical description of current kinetics that allows us to compare directly our results with previous studies on native and recombinant channels and do not imply a particular model for current activation (Moroni et al., 2000).
To exclude the uncompensated capacitive transients and activation delay of Ih currents in different cell types, it was necessary to use slightly different windows for the fits. For the rapidly activating HCN1 currents, we excluded the first 10–20 msec of the current traces. For the slowly activating HCN2 currents, we excluded the first 180–250 msec of the current traces. For nativeIh currents, we excluded the first 20 msec of the current trace for CA1 pyramidal and stratum oriens neurons; the first 40–100 msec of the records were excluded for stratum radiatum neurons, and the first 44 msec were excluded for thalamic neurons. All traces were then fit in a window extending from this initial point to within a few percent of the end of the current record. For CA3 pyramidal neurons, the Ihcurrents were generally too small to fit. Data are presented as mean ± SE.
RESULTS
Three members of the mHCN gene family show a differential and complementary pattern of expression in neurons across the mouse CNS
Digoxigenin-labeled riboprobes were generated for each of the mouse mHCN1, mHCN2, and mHCN4 cDNAs, and in situhybridization was performed on coronal sections taken from mouse brain and spinal cord. As previously reported, all three isoforms are expressed throughout the CNS, but each one exhibits a distinct and often complementary pattern of distribution (Santoro et al., 1997;Ludwig et al., 1998; Moosmang et al., 1999). Below we present detailed localization of mHCN transcripts in cell types within regions of the brain that show distinctive patterns of HCN mRNA expression and/or in which prominent Ih currents have been reported (see Figs. 1-7). In general, considerable overlap in expression is found between mHCN1 and mHCN2, as well as between mHCN2 and mHCN4; however, very little overlap exists between mHCN1 and mHCN4 (Table 1).
Table 1.
HCN mRNA expression in mouse CNS
| Brain region | mHCN1 | mHCN2 | mHCN4 |
|---|---|---|---|
| Olfactory bulb | |||
| Glomerular layer/EPL | +/− | ++ | ++ |
| Mitral cell layer | +/− | ++++ | ++++ |
| Granule cell layer/IPL | +/− | ++ | +/− |
| Basal forebrain | |||
| Striatum | − | ++ | + |
| Globus pallidus | − | ++ | + |
| Septum/diagonal band | + | +++ | + |
| Amygdala | |||
| Basomedial ant. nucl. | − | +/− | ++ |
| Basolateral nucleus | + | + | + |
| Central nucleus | + | +/− | +/− |
| Lateral nucleus | +/− | +/− | + |
| Cerebral cortex | |||
| Neocortex | +++ | ++ | +/− |
| Piriform cortex | + | + | + |
| Hippocampus | |||
| Pyramidal cells (CA1) | +++ | ++ | +/− |
| Pyramidal cells (CA3) | ++ | +++ | + |
| Stratum oriens | ++ | ++ | +/− |
| Stratum radiatum/L-M | ++ | ++ | +/− |
| Granule cells (DG) | +/− | + | − |
| Hilus of dentate gyrus | ++ | ++ | − |
| Thalamus | |||
| Anterodorsal nucleus | + | +++ | ++ |
| Principal relay nuclei | − | ++++ | +++ |
| Geniculate nuclei | − | ++++ | ++ |
| Nucleus reticularis | − | +++ | +/− |
| Lateral habenula | − | ++ | ++ |
| Medial habenula | − | +/− | ++++ |
| Subthalamic nucleus | +/− | +++ | +/− |
| Hypothalamus | |||
| Preoptic nuclei | − | +++ | ++ |
| Ventromedial nuclei | ++ | +/− | +/− |
| Mammillary nuclei | + | + | + |
| Midbrain | |||
| Superior colliculus | ++ | ++ | +/− |
| Inferior colliculus | ++ | ++ | +/− |
| Substantia nigra | +/− | ++ | + |
| Cerebellum | |||
| Molecular cell layer | ++ | − | − |
| Purkinje cell layer | +++ | + | − |
| Granule cell layer | − | ++ | − |
| Deep cerebellar nuclei | +/− | ++++ | ++ |
| Brainstem | |||
| Vestibular nucleus | +/− | +++ | + |
| Cochlear nucleus | ++ | +++ | + |
| Spinal trigem. nucl. | ++ | +++ | + |
| Facial nucleus | +++ | +++ | + |
| Hypoglossal nucleus | +++ | +++ | + |
| Reticular fields | + | +++ | + |
| Inferior olive | + | + | − |
| Spinal cord | |||
| Dorsal horn | + | ++ | +/− |
| Ventral horn | +++ | ++ | +/− |
++++, Very high expression.
+++, High expression.
++, Moderate expression.
+, Low expression.
+/−, Barely above background/background.
−, No detectable signal.
Olfactory bulb
Prominent labeling was found in this region for the mHCN2 and mHCN4 transcripts (Fig. 1). Both isoforms are highly expressed in type I and type II (displaced) mitral cells, and labeling is also found in tufted cells. However, mHCN4 probes appear to label a larger number of externally placed tufted cells (Fig.1B) compared to mHCN2 transcripts (Fig.1A). Finally, mHCN2 probes also distinctively label a scattered population of small cells present throughout the granule, internal and external plexiform, and glomerulal layers, which are likely to correspond to the short-axon cells, a type of inhibitory intrinsic neuron (Fig. 1A).
Basal forebrain
We were particularly interested in examining the patterns of expression of mHCN isoforms in regions of the brain that control oscillatory electrical activity. It is thus intriguing that high levels of mHCN2 expression are present in the medial septum, as well as in the vertical and horizontal limb of the diagonal band (Fig.2A). These structures are important for generating the theta rhythm in the hippocampus, a prominent 5–10 Hz synchronous electrical oscillation associated with exploratory activity (for review, see Bland and Oddie, 1998;Vinogradova et al., 1998).
Notable Ih currents have been described in the giant cholinergic interneurons of the striatum. These cells exhibit pacemaker properties in vitro and are tonically active in vivo (Jiang and North, 1991; Kawaguchi, 1992, 1993). mHCN2 and mHCN4 transcripts are, in fact, found throughout the striatum, although in a relatively small number of cells (Fig.2B,C). This sparse pattern of labeling is consistent with expression of mHCN transcripts within striatal interneurons, which represent ∼3–4% of the total cell population. In particular, mHCN4 appears to label only a restricted number of very large neurons, suggesting its presence in the giant cholinergic pacemaking interneurons (Fig. 2C).
Staining for mHCN2 and mHCN4 transcripts was found in the globus pallidus, in both the lateral (Fig. 2B,C) and medial (see Fig. 4A,B) segment. Globus pallidus neurons comprise a class of neurons (type II) characterized by the presence of time- and voltage-dependent inward rectification of the membrane potential evoked by hyperpolarizing current steps, and anodal break rebound depolarization, a hallmark ofIh currents (Stanford and Cooper, 1999). Labeling for mHCN2 and mHCN4 transcripts is also present in the other main nuclei of the basal ganglia system, namely the subthalamic nucleus (mHCN2; Table 1) and the substantia nigra (mHCN2 and mHCN4; see Fig. 5C,D).
Staining of the anterodorsal thalamic nucleus by both transcripts is also noticeable in Figure 2, B and C. Furthermore, the reticular thalamic nucleus appears to be strongly labeled by mHCN2, but not by mHCN4 probes (see also below, Thalamus; see Fig. 4A,B).
Cerebral cortex and hippocampus
mHCN1 and mHCN2 transcripts are expressed at moderate to high levels in the neocortex, whereas mHCN4 appears to be essentially absent (Fig. 3A,C,E). The staining of mHCN1 shows a distinctively layered distribution, with most prominent labeling of the pyramidal cells in layer V (Fig. 3A; shown is sensorimotor cortex). This is consistent with the previous finding that the apical dendrites of layer V pyramidal neurons are strongly labeled by anti-mHCN1 antibodies (Santoro et al., 1997). In contrast, mHCN2 transcripts appear to have a more scattered distribution throughout the cerebral cortex, with a labeling of large and small neurons (Fig. 3C).
Similar to neocortex, both mHCN1 and mHCN2 transcripts are prominently expressed in the hippocampus, whereas mHCN4 is expressed at very low levels (Fig. 3B,D,F). Again, the staining patterns appear to be somewhat different for the two isoforms. Labeling for mHCN1 is strongest in the CA1 pyramidal cell layer, moderate in CA3, and almost absent in dentate granule cells (Fig. 3B). Labeling for mHCN2 is strongest in the CA3 pyramidal cell layer, moderate in CA1, and very low in dentate granule cells (Fig. 3D). mHCN4 shows a very modest labeling, principally in area CA3 (Fig.3F). These findings are in excellent agreement with the results reported by Moosmang et al. (1999), who used radioactivein situ labeling to characterize the expression of HCN mRNA isoforms in the hippocampus. Both mHCN1 and mHCN2 probes also label cells outside of the pyramidal cell layers, consistent with the reported presence of Ih in various types of hippocampal interneurons (see Fig. 10B;Williams et al., 1994; Maccaferri and McBain, 1996). mHCN1 shows a distinct labeling of a group of larger interneurons in the stratum oriens, which are most likely basket cells (Fig. 3B), consistent with the observed labeling of basket cell axon terminals by anti-mHCN1 antibodies (Santoro et al., 1997). Neurons at the stratum lacunosum-moleculare/radiatum border of area CA1 and larger cells in the hilus of the dentate gyrus also appear to be labeled by the mHCN1 probe (Fig. 3B). A scattered labeling of smaller interneurons located in stratum oriens, radiatum, lacunosum-moleculare (L-M), and the hilus of the dentate gyrus (DG) is observed for mHCN2 transcripts (Fig. 3D).
Fig. 10.
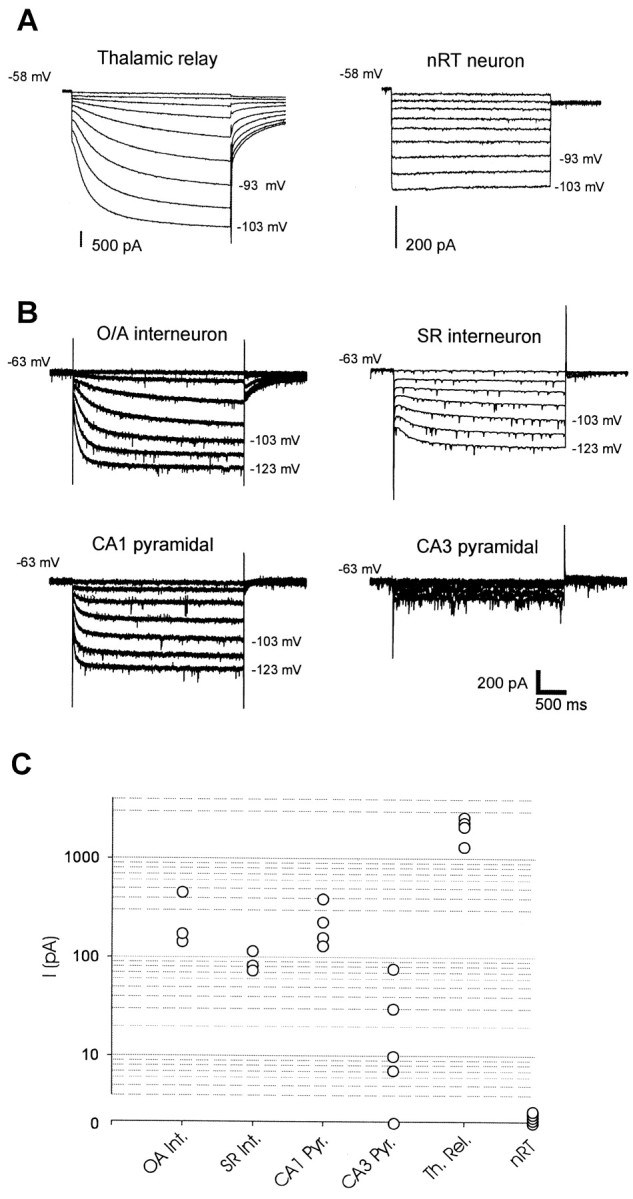
Differential functional properties ofIh in thalamic and hippocampal neurons as assessed by whole-cell patch-clamp recordings. A,Current recording from a thalamocortical relay cell from the ventral posterior nucleus (left) and from a nucleus reticularis neuron (right). Holding potential was −58 mV, and the voltage was stepped to negative potentials for 2.8 sec in 5 mV increments to −103 mV. Scale bars as indicated or as inB for time. B, Recordings from hippocampal neurons. Top left, Stratum oriens/alveus (O/A) interneuron; top right, stratum radiatum (SR) interneuron; bottom left, CA1 pyramidal neuron; bottom right, CA3 pyramidal neuron. In each case, the holding potential was −63, and the voltage was stepped to negative potentials for 3 sec in 10 mV increments down to −123 mV. Calibration: 200 pA, 0.5 sec. C, Comparison of peak current in the six cell types shown in A andB. Each point is an individual experiment. TheIh current at the end of 2.8- to 3-sec-long voltage steps to −103 mV was measured as the difference between the net current record and the leakage current, measured from the initial current level after the capacitative transient at the beginning of each voltage step.
Strong labeling of the medial habenula is obtained with the mHCN4 probe (Fig. 3F), whereas mHCN1 and mHCN2 probes show little staining in this region. This observation is particularly interesting in view of the fact that medial habenula neurons are spontaneously active, firing in a regular 2–6 Hz repetitive manner, while completely lacking the low-threshold Ca conductance that often interacts withIh to generate spontaneous firing (McCormick and Prince, 1987). Both mHCN2 and mHCN4 are expressed in the lateral habenula (Fig. 3D,F).
Thalamus
A physiologically important role forIh has been particularly well characterized in the thalamus. Thalamocortical relay neurons undergo transitions from burst firing modes to tonic firing modes during the sleep–wake cycle (for review, see McCormick and Bal, 1997; Lüthi and McCormick, 1998). This change in excitability results in part from a cAMP-dependent speeding of the kinetics ofIh activation. Consistent with the presence of high levels of Ihcurrents, a strong pattern of mHCN2 and mHCN4 staining was found in the principal relay nuclei of the thalamus, including the dorsal lateral geniculate nucleus (Figs. 3D,F,4A,B; Moosmang et al., 1999). In contrast, mHCN1 expression was not detected to any significant extent. In rodents, the ventral posterior nucleus of the thalamus does not contain local interneurons, thus, it is likely that the observed staining represents entirely thalamocortical relay neurons. As noted above (see Basal Forebrain; Fig.2B,C), GABAergic neurons in the reticular nucleus thalami are positive for mHCN2 transcripts, whereas mHCN4 appears to be essentially absent from this region (Figs. 2B,C,4A,B).
Labeling for mHCN transcripts was also found in several nuclei of the hypothalamus (Table 1).
Amygdala
mHCN1, 2, and 4 are all expressed in the amygdala, but they show a very distinct distribution across different nuclei (Table 1). Thus, mHCN4 is present at a moderate level in the rostral portion of the basomedial nucleus and at a low to moderate level in the lateral and basolateral amygdaloid nuclei. A low level of expression of mHCN2 is found in the basolateral nucleus, whereas mHCN1 is expressed in both the basolateral and central nuclei (Table 1).
Midbrain
Labeling of the superior colliculus by the mHCN1 and mHCN2 probes is shown in Figure 5, A andB, respectively. Whereas mHCN2 is expressed throughout this region, mHCN1 transcripts appear to be restricted to one layer in the superior colliculus, most likely the intermediate gray layer. Cells in the intermediate layer send descending motor commands to the brainstem and are involved in the control of saccadic eye movement. Electrophysiological data indicate the presence of a time-dependent inward rectifier current with properties consistent withIh in a particular class of cells within this layer, namely the wide-field vertical cells (Saito and Isa, 1999). These cells exhibit extensive dendritic arborization into the superficial and optic nerve layer, where they receive the incoming visual information.
Labeling of the substantia nigra pars reticulata, a nucleus of the basal ganglia with a very similar cellular organization to the globus pallidus, was found for mHCN2 and at lower levels for mHCN4 transcripts (Fig. 5C,D). The mHCN4 probe also showed significant staining of the substantia nigra pars compacta (Fig.5D).
Cerebellum
In the cerebellar cortex, a strong labeling for mHCN1 was found in the Purkinje cell layer (Fig.6A). Labeling for mHCN1 is also present in the molecular layer (weak labeling is visible in the experiment shown in Fig. 6A; Moosmang et al., 1999), most likely within the basket cells, whose axon terminals were previously found to be heavily stained by mHCN1 antibodies (Santoro et al., 1997). A low amount of mHCN2 is also present in Purkinje cells; the mHCN2 probe mainly labels a population of scattered cells within the granule cell layer. These cells may correspond to the Golgi cells (interneurons), but could also represent glial cells, because the labeling is also observed in the underlying white matter (Fig.6B). The latter conclusion is supported by the presence of high levels of mHCN2 mRNA within the corpus callosum, as was previously determined by Northern blot analysis (Santoro et al., 1998). mHCN4 labeling is completely absent from the cerebellar cortex. In contrast, cells within the deep cerebellar nuclei exhibit strong labeling with both mHCN2 and mHCN4 probes (Fig.6B,C).
Brainstem and spinal cord
The highest levels of mHCN2 expression in the CNS are found in the brainstem and this strong staining extends into the spinal cord (Fig.7B,D,F). All areas within these structures contain cells that are strongly labeled for mHCN2 transcripts, with the notable exception of the inferior olive, which exhibits only a low amount of staining (Fig. 7D). Labeling for mHCN2 is often accompanied by a low level of mHCN4 staining (data not shown). In contrast, mHCN1 probes display a strikingly restricted pattern of labeling, with strong staining seen in selected nuclei (Fig. 7A,C,E). High levels of mHCN1 expression are found in the ventral cochlear nucleus and spinal trigeminal nucleus (Fig. 7A), as well as in the motor nuclei of the facial (Fig. 7A) and hypoglossal (Fig. 7C) cranial nerves, and in the α-motorneurons of the ventral horn of the spinal cord (Fig. 7E).
Different neurons display Ih currents with diverse biophysical properties
Given the differential and complementary staining pattern of mHCN transcripts in different neurons within certain brain regions, we next investigated whether there was a similar heterogeneity in the functional properties of Ih currents in these cells as determined using whole-cell recordings. In addition, given the marked difference in the properties of recombinant mHCN1, 2, and 4 channels expressed in heterologous cells as previously reported by different investigators and further delineated below, we also were interested in whether there was any correlation between channel properties in native cells and the properties expected based on mRNA expression.
Biophysical properties of recombinant mHCN1 and mHCN2 channels
We first determined the kinetics and steady-state activation properties for recombinant mouse mHCN1 and mHCN2 channels expressed inXenopus oocytes. Although we had previously reported on activation parameters of mHCN1 channels expressed in Xenopusoocytes using cell-free patches (Santoro et al., 1998) and others had reported on mHCN2 channel properties using whole-cell recordings from mammalian cell lines (Ludwig et al., 1998), changes in the local environment can exert a marked effect on the kinetics and steady-state voltage dependence of Ih. For example, submicromolar concentrations of cAMP, patch excision, or whole-cell dialysis affect the kinetics and voltage dependence ofIh (DiFrancesco and Mangoni, 1994) (G. R. Tibbs, B. Santoro, and S. A. Siegelbaum, unpublished data). Thus, experimental differences between different laboratories could conceivably contribute to differences in reported channel properties. We have therefore reexamined the kinetics and steady-state activation properties of mHCN1 and mHCN2 channels recorded under identical conditions using a two microelectrode voltage-clamp ofXenopus oocytes so as not to perturb the intracellular environment.
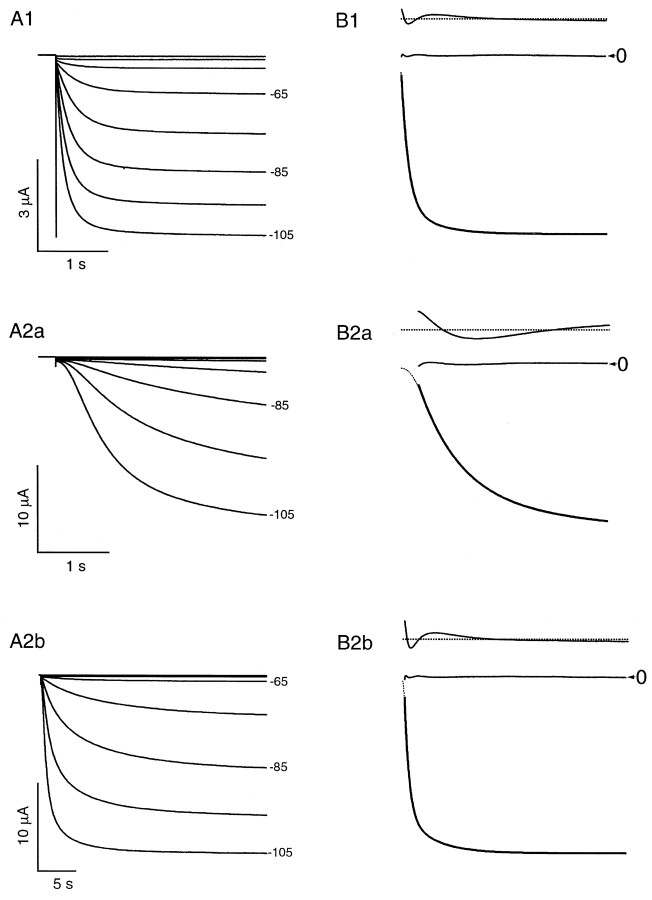
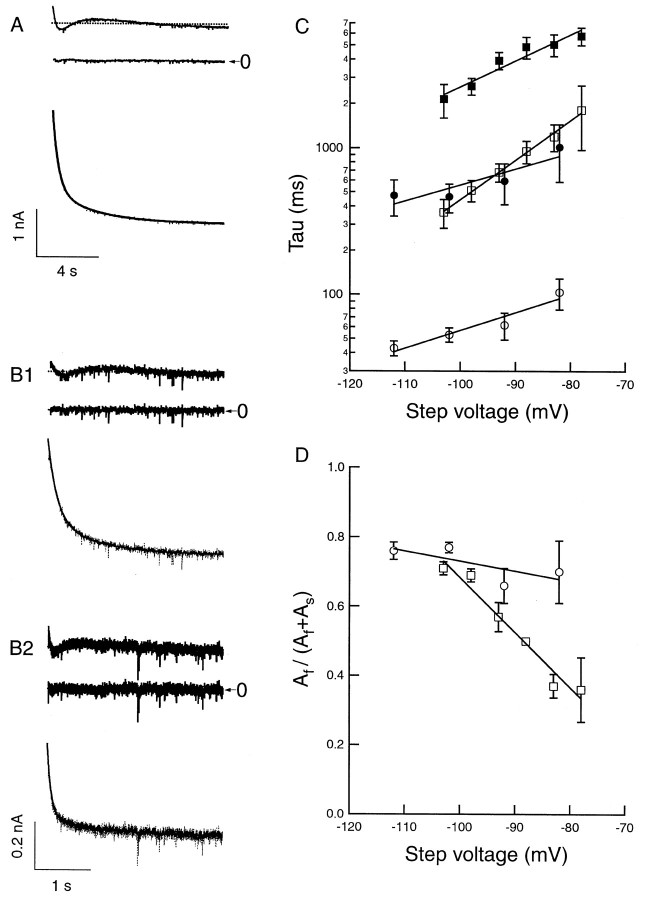
Recombinant mHCN1 and mHCN2 channels clearly display distinct kinetics of activation in response to hyperpolarizing voltage-clamp steps (Fig.8). The mHCN2 currents show a distinct, sigmoidal onset of activation after the hyperpolarizing step, contributing an apparent delay. In contrast, mHCN1 currents show a much less pronounced delay in their onset of activation. Even after the initial delay, mHCN2 currents activate with a much slower time course compared to mHCN1 currents (Fig. 8A). The kinetics of mHCN1 and mHNC2 current activation after the initial delay have been previously described by single exponential functions (Ludwig et al., 1998; Santoro et al., 1998). However, fits with two exponential components (after the initial lag) provide a more accurate description of the kinetics of both mHCN1 (Fig. 8B1) and mHCN2 (Fig. 8B2) currents. For each mHCN isoform, the fast time constant is ∼5- to 10-fold more rapid than the slow time constant of activation, with both fast and slow components becoming more rapid with steps to more hyperpolarized potentials (Fig.9A). Over the entire voltage range of activation, the fast and slow time constants of activation for mHCN1 are approximately 10-fold faster than the respective time constants for mHCN2. Moreover, for mHCN1, the fast component of activation accounts for the majority of the current amplitude over the entire voltage range, representing 70–90% of the total current amplitude relative to the slow component (Fig. 9B). In contrast, for mHCN2, the slow component is predominant for voltage steps to potentials near the midpoint for current activation. With steps to more hyperpolarized voltages, the contribution of the slow component rapidly diminishes, and the fast component becomes predominant (Fig. 9B).
Fig. 8.
Distinct functional properties ofIh currents generated by mHCN1 and mHCN2.A, Currents generated in response to hyperpolarizing voltage steps in Xenopus oocytes expressing mHCN1 (A1) or mHCN2 (A2), respectively.A1, mHCN1 currents shown during 3-sec-long hyperpolarizing voltage steps. A2a,b, mHCN2 currents shown during initial 3 sec (A2a) and entire 30 sec time course (A2b) in response to hyperpolarizing voltage steps. For both A1 and A2, membrane held at −30 mV and stepped from −35 to −105 mV in 10 mV increments (selected voltages indicated to right of current traces). B, Two exponential components are required to adequately fit activation time course of mHCN currents. Time and current scales as in corresponding panels in A. ForB1 and B2, bottom tracesshow current during hyperpolarizing step to −105 mV with superimposed fit using two exponential components. The middle andtop traces show the residuals of difference between the recorded current and the fitted single (top trace) or double (middle trace) exponential functions. Zero current is indicated by the arrowhead (labeled 0). The residuals from the single exponential fits are displaced from zero for clarity; zero current for these traces is indicated by thedashed line. To facilitate comparison between mHCN1 and mHCN2, the first 3 sec of the mHCN2 activation time course and fits to this are shown on an expanded time scale in B2a.
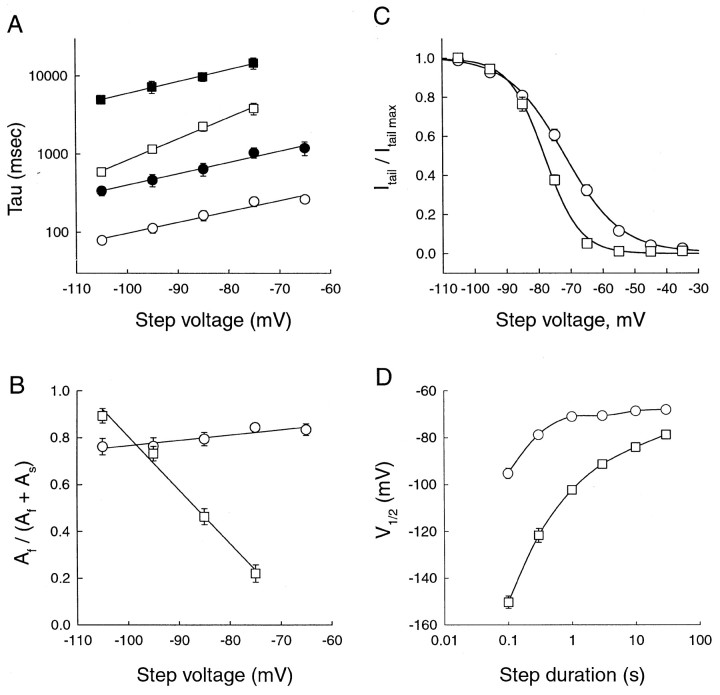
Fig. 9.
Comparison of kinetic and steady-state activation properties between mHCN1 and mHCN2. A, Fast (open symbols) and slow (filled symbols) exponential time constants as function of voltage during hyperpolarizing step. For all panels: circles, mHCN1;squares, mHCN2. B, Relative amplitude of fast exponential component as function of hyperpolarizing voltage step. Af and As are the amplitudes of the fast and slow exponential components, respectively.C, Steady-state tail current activation curves obtained using 30 sec hyperpolarizing steps for mHCN2 channels and 3 sec steps for mHCN1 channels. Curves fit with Boltzmann relation (see Materials and Methods for details). D, Relation between values of V1/2 determined from activation curves using hyperpolarizing steps of different durations for mHCN1 and mHCN2 channels.
To determine how the extent of Ihactivation depends on voltage, we measured tail current amplitudes at a fixed membrane potential (−40 mV) after hyperpolarizing voltage steps to different test potentials. The duration of the hyperpolarization was varied between 0.1 and 30 sec to determine the length of time required for activation to approach its steady-state value. Activation curves were then fit by a Boltzmann distribution to determine the midpoint voltage of activation (V1/2) as well as the slopes of the relation (see Materials and Methods). Both mHCN1 and mHCN2 channels display typical S-shaped activation curves after hyperpolarization (Fig. 9C). However, the midpoint activation of mHCN1 (−71.6 ± 1.0 mV; n = 8) is ∼7 mV more positive than the value for mHCN2 (−78.3 ± 0.8 mV;n = 3). These values for both mHCN1 and mHCN2 are 30–40 mV more positive than the initial reported values (Ludwig et al., 1998; Santoro et al., 1998). This discrepancy is attributable to negative shifts in steady-state activation curves associated with patch excision and whole-cell dialysis (DiFrancesco and Mangoni, 1994) (Tibbs, Santoro, and Siegelbaum, unpublished data). Estimates of theV1/2 in previous studies may have also been shifted to negative voltages by the use of relatively brief hyperpolarizing test pulses that do not permit activation to approach steady-state values. A plot of V1/2 as a function of test pulse duration clearly shows the time dependence of this parameter which, because of the differences in kinetics between mHCN1 and mHCN2, is distinct for mHCN1 and mHCN2 (Fig. 9D). Test pulses of 3 sec in duration are evidently sufficient for mHCN1 to reach equilibrium, whereas even 30 sec test pulses may not be sufficient for mHCN2 to reach its equilibrium condition.
Because of the voltage dependence of activation kinetics, we have compared the time constants of activation of recombinant mHCN channel isoforms at a fixed voltage of −105 mV, at the peak of the activation curve. The mean fast and slow time constants for mHCN1, determined from the biexponential fits, were 79 ± 9 and 339 ± 47 msec, respectively, with the fast component accounting for 76 ± 4% of the amplitude. For mHCN2 channels, the kinetics of the fast component of activation were approximately eightfold slower than those of mHCN1, with a time constant of 591 ± 8 msec. The slow component of mHCN2 was ∼12-fold slower than that of mHCN1, with a time constant of 4970 ± 540 msec, with the fast component accounting for 89 ± 3% of the amplitude. We have also approximated the time course of activation of the recombinant channels using single exponential fits, to allow comparison with previous studies. In these fits, mHCN1 channels activate with a time constant of 125 ± 12.6 msec (n = 8), whereas mHCN2 channels activate with a time constant of 1194 ± 276 msec (n = 3).
Native Ih currents in thalamus and hippocampus
Given the marked differences in kinetics of the mHCN isoforms and distinct patterns of gene expression, we next examined the biophysical properties of native Ih currents in some detail. Our analysis first focused on the thalamus, a region that presents strong expression of mHCN transcripts. Although previous studies have characterized Ih currents from thalamic neurons in other species (McCormick and Pape, 1990;Williams et al., 1997), no detailed studies have been reported for mice, the species in which we characterized mHCN mRNA expression patterns. We measured currents during hyperpolarizing voltage-clamp steps from both thalamocortical relay neurons, which express mHCN2 and mHCN4 transcripts at high levels, and neurons from the nucleus reticularis, an area of the thalamus that displays labeling for mHCN2, but no detectable labeling for mHCN4 (Fig. 4A,B). Consistent with recordings from thalamic neurons in guinea pig and cat (McCormick and Pape, 1990; Williams et al., 1997), principal relay neurons of the mouse thalamus display very large hyperpolarization-activated currents (1000–2000 pA at −103 mV; Fig.10A, left traces), which activate with distinctly slow kinetics after an initial pronounced sigmoidal lag. Indeed, for voltage steps near the threshold for Ih activation, steady-state activation is reached only after tens of seconds.
In contrast to the thalamocortical relay neurons, we detected noIh currents in recordings from five separate neurons from the reticular nucleus (Fig. 10A, right traces). Hyperpolarizing steps to −103 mV failed to activate any time-dependent inward current. This lack of current is surprising, given the prominent expression of mHCN2 in this cell type (Fig. 4A).
The second brain region that we examined was the hippocampus, wherein situ hybridization revealed distinct patterns of mHCN1, 2, and 4 labeling in CA1 and CA3 pyramidal neurons and stratum oriens and stratum radiatum/lacunosum-moleculare interneurons (Fig. 3). Whole-cell recordings were performed on each of these neuronal cell types, and current traces in response to hyperpolarizing voltage steps are shown for representative cells in Figure 10B. It is apparent that the different types of neurons display markedly different Ih current magnitudes, albeit never as high as found in the relay cells of the thalamus (Fig.10C). Thus, CA1 pyramidal cells and stratum oriens interneurons display moderately largeIh currents (100–400 pA at −103 mV), whereas CA1 and CA3 stratum radiatum interneurons revealed smaller, but reproducible levels of Ih (∼100 pA). The time course of Ih activation in the hippocampal neurons was generally more rapid than that seen in the thalamus. In particular, Ih in CA1 pyramidal neurons activates very rapidly and with little initial lag. Surprisingly, despite the presence of high levels of mHCN1 and mHCN2 transcripts, CA3 pyramidal neurons showed either only very smallIh currents (<100 pA; two of five cells) or no detectable Ih (<10 pA; three of five cells; Fig. 10B,C).
These data from the thalamus and the hippocampus suggest that the simple presence of an mHCN channel transcript in a cell is not a reliable predictor of significant functional expression of the channel, at least within compartments that are electrically accessible to somatic whole-cell recording. It should be noted that the lack of current in nucleus reticularis and CA3 pyramidal neurons was not attributable to the poor health of those particular slices. In several experiments, robust Ih currents could be measured in either thalamic relay cells or CA1 pyramidal neurons from the same slices in which either nucleus reticularis or CA3 pyramidal neurons were found to lack detectableIh.
We next asked, for those cells where mHCN transcripts were present andIh was detected, whether there is any correlation between the pattern of isoform expression and the biophysical parameters of Ihactivation predicted from the properties of the recombinant channels. Although the V1/2 for activation of thalamocortical relay neurons (−82 ± 1.5 mV) is similar to that observed in hippocampal neurons (approximately −85 mV), the kinetics of Ih activation are markedly different. The activation time course ofIh currents for both thalamic (Fig.11A) and hippocampal (Fig. 11B) neurons requires two exponential components (after an initial lag), whose time constants decrease as the membrane test voltage becomes more negative (Fig. 11C). Over the entire voltage range, the fast and slow exponential components for hippocampal pyramidal neurons are ∼10-fold more rapid than the respective fast and slow exponential components for the thalamic relay neurons. In the thalamic relay cells, the slow component ofIh activation is predominant with voltage steps to the middle of the activation range. However, with steps to more hyperpolarized potentials, the relative contribution from the fast component increases (Fig. 11D). In contrast, in hippocampal CA1 neurons the fast component is predominant over the entire activation voltage range (Fig. 11D).
Fig. 11.
Comparison of biexponential activation kinetics of Ih current in native neurons. A, B, Double exponential fit of current traces for thalamic relay neuron (A), hippocampal OA interneuron (B1), and hippocampal CA1 pyramidal neuron (B2) during hyperpolarizing voltage step to −103 mV. Step length was 11.2 sec for thalamocortical relay cell and 3 sec for hippocampal cells. From bottom, traces show superimposed currents and biexponential fits, residuals from biexponential fits, and residuals from single exponential fits. C, “Fast” (open symbols) and “slow” (filled symbols) time constants of activation as function of voltage for thalamic relay neurons (squares) and hippocampal CA1 pyramidal neurons (circles); n = 4.D, Relative amplitude of the fast exponential component as function of voltage. Circles, Hippocampal CA1 neurons. Squares, Thalamic relay neurons.
Comparison of Ih kinetics for native and recombinant channels
In Figure 12, we compare the activation kinetics of recombinant and nativeIh currents. Because of the voltage dependence of channel activation, we determined time constants ofIh at a fixed membrane potential of −103 mV, similar to that used to study recombinantIh kinetics (−105 mV; the slight difference, arising from junction potentials, does not significantly affect the comparison; see Figs. 9, 11). For thalamic relay cells, the mean time constants of activation were 364 ± 81 msec and 2.14 ± 0.55 sec with the fast component accounting for 71 ± 1.9% of the amplitude (n = 4). In CA1 pyramidal neurons, the mean time constants for activation were 53 ± 6 msec and 464 ± 103 msec with the fast component representing 77 ± 1.5% of the amplitude (n = 4). For stratum oriens interneurons, the mean time constants of activation (216 ± 101 msec and 1084 ± 487 msec with 59 ± 5.5% fast component;n = 3) fall between those of the CA1 pyramidal and thalamic relay cells.
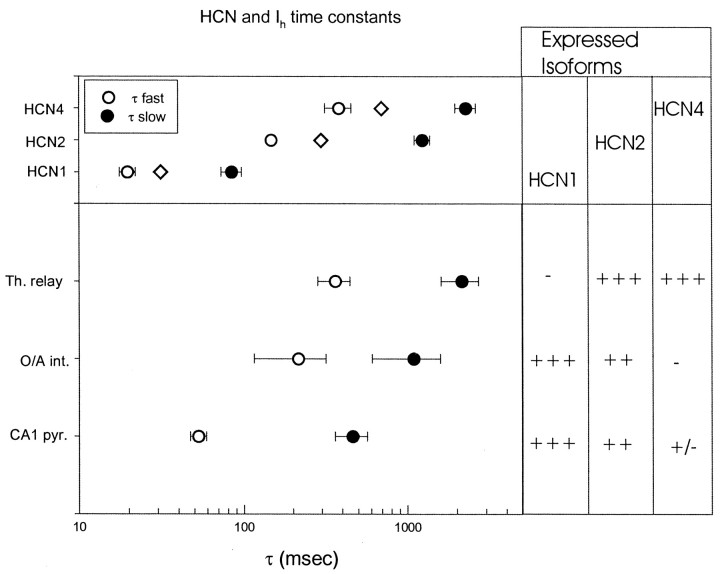
Fig. 12.
Summary comparing HCN isoform mRNA expression with time constants of activation for recombinant HCN isoforms and native Ih currents. Top, Fast (open circles) and slow (filled circles) exponential time constants of activation for different HCN isoforms from biexponential fits during voltage step to −105 mV.Diamonds show time constants from single exponential fits, reflecting predominant fast component. Bottom, Fast and slow exponential time constants for Ihin thalamocortical relay neurons, hippocampal O/A interneurons, and hippocampal CA1 pyramidal neurons. Data show mean ± SE for fast and slow components. Panel on right summarizes relative expression levels for mHCN1, 2, and 4 mRNA. Data for HCN4:circles, at −110 mV and 35°C (Ishii et al., 1999);diamonds, extrapolated to −105 mV (Seifert et al., 1999) and scaled to 34°C.
To compare the native Ih kinetics, determined at 34°C, with the mHCN1 and mHCN2Ih kinetics, studied at 23–25°C, we have scaled the recombinant kinetics using a Q10of 4 (Santoro and Tibbs, 1999). Although we have not succeeded in expressing full-length mouse HCN4, rabbit (Ishii et al., 1999) and human (Seifert et al., 1999) HCN4 have been expressed and studied in mammalian cell lines. HCN4 activates even more slowly than mHCN2.Seifert et al. (1999) report a single exponential time constant of activation for human HCN4, whereas Ishii et al. (1999) report two exponential components of activation for rabbit HCN4. The kinetic data for HCN4 (at 34–35°C) are plotted with our data for mHCN1 and mHCN2 in Figure 12 (top panel), and compared to nativeIh kinetics (bottom panel). The implications of these comparisons are discussed below.
DISCUSSION
The results presented above, together with data from a number of previous studies (Santoro et al., 1997; Ludwig et al., 1998; Moosmang et al., 1999), indicate that each mHCN channel isoform has a unique distribution within the CNS. The distribution of mHCN isoforms is associated with differences in Ihkinetics recorded from different brain regions. Comparison of isoform expression patterns with the kinetic properties of recombinant mHCN channel currents suggests a potential molecular basis for the heterogeneity in native Ih channel properties.
We find that mHCN1 currents show the fastest kinetics, mHCN2 currents show intermediate kinetics, and HCN4 channels show the slowest kinetics. In native cells, hippocampal CA1 neurons, which express high levels of mHCN1 and somewhat lower levels of mHCN2, show very rapid kinetics. Thalamic relay cells, which express mHCN2 and mHCN4, show very slow kinetics. Moreover, the kinetics of activation ofIh in CA1 neurons span the time scale of activation of mHCN1 and mHCN2, whereas the kinetics ofIh activation in thalamic relay neurons span the time scale of activation of mHCN2 and HCN4. Therefore, it appears likely that at least some of the differences in kinetics among neuronal types reflect the differential expression of mHCN isoforms in the different neuronal types. Such comparisons, however, may be complicated by differences in posttranslational modification between native neurons and heterologous expression systems, potential formation of heteromultimeric mHCN channels, and subtle differences in recording conditions in the two types of experiments. Finally, because homomeric recombinant HCN channels display biexponential activation kinetics, the biexponential kinetics ofIh in native neurons does not necessarily imply the expression of two different HCN isoforms. Thus, the biexponential kinetics of Ihcurrents measured in CA1 pyramidal neurons or in thalamocortical relay cells could reflect the biexponential kinetics of a uniform population of channels composed of a single HCN isoform (i.e., mHCN1 for pyramidal and mHCN4 for thalamic neurons). Alternatively, the native kinetics could reflect coexpression of mHCN1 channels and mHCN2 channels for CA1 neurons or mHCN2 channels and mHCN4 channels for thalamic relay neurons. Definitive proof as to the role of specific mHCN isoforms in determining native Ih properties will have to await studies in mutant mice with deletions in specific mHCN genes.
Because we have not performed co-staining for expression of different isoforms in a single section, we also cannot definitively conclude whether multiple isoforms are indeed expressed in a single cell. However, the high levels of uniform staining for mHCN1 and mHCN2 in the hippocampal pyramidal CA1 and CA3 layers and for mHCN2 and mHCN4 in thalamocortical relay nuclei strongly suggest that multiple isoforms are likely to be expressed in the same cells. In contrast, the low density of the stratum oriens and stratum radiatum interneurons and resultant sparse punctate mRNA staining patterns for mHCN1 and mHCN2 isoforms make it difficult to determine whether any single interneuron expresses both isoforms.
It is puzzling why certain cells express mHCN mRNA at high levels but do not exhibit detectable Ih-type currents. For example, hippocampal CA3 pyramidal cells display significant levels of all three mHCN transcripts (Fig. 3), yet little or no Ih current is recorded (Fig.10). Similarly, thalamic reticular cells, which express mHCN2 transcripts (Fig. 4), do not appear to exhibit any nativeIh currents (Fig. 10). It is possible that mHCN channels are targeted to distal regions of the cell that are electrically remote from the cell body, where our recordings were performed. Indeed, we know that the mHCN1 protein can be localized to the distal ends of dendritic and axonal processes (Nicoll et al., 1993;Santoro et al., 1997; Magee, 1998, 1999; Stuart and Spruston, 1998). It is also possible that certain mHCN mRNA is not stably translated or that the mHCN channels are present but functionally inactivated. This latter view is consistent with findings in adult cardiac ventricular muscle (Yu et al., 1993; Robinson et al., 1997), in which the threshold of activation of Ih is shifted to very negative potentials that are not reached during typical whole-cell recordings.
Physiological function of mHCN isoforms
Can we associate any particular mHCN isoform with a specific physiological function, based on the observed patterns of distributions? On a general note, whereas mHCN2 has a very widespread distribution in the brain, mHCN1 and mHCN4 show predominantly cortical and subcortical distributions, respectively. Thus, mHCN1 is most abundant in the cerebral, hippocampal, and cerebellar cortices, whereas mHCN4 is primarily expressed in the thalamus. mHCN4 is also expressed, together with mHCN2, throughout the basal ganglia and in the deep cerebellar nuclei, structures in which mHCN1 is absent. As a consequence, there is very little overlap in the localization of the fastest (mHCN1) and slowest (mHCN4) of the mHCN channel isoforms. Nevertheless, all combinations occasionally do occur in different parts of the brain, including each isoform being expressed alone (e.g., mHCN1 in cerebellar basket cells, mHCN2 in thalamic reticular nucleus cells, and mHCN4 in medial habenula). The latter observation is consistent with the idea that the mHCN subunits can be expressed as homomeric channels in vivo. A determination of the specific subcellular patterns of mHCN protein localization within neurons containing multiple mHCN transcripts will help establish whether theIh channels may assemble as heteromultimers under physiological conditions.
One interesting recurrent finding is the presence of high levels of mHCN1 transcripts in large, principal, output neurons that display extensive dendritic arborizations and often have far-reaching axons. Such cells include the layer V pyramidal neurons in the cerebral cortex, the motorneurons of the cranial and spinal nerves, as well as the Purkinje neurons in the cerebellar cortex. Superior colliculus neurons of the intermediate gray layer may also fall into this category. A potential functional role for this pattern of expression is suggested by the finding that Ihchannels are distributed along a gradient in the apical dendrites of pyramidal cells, being present at higher density at increasing distances from the soma, and are important for the regulation of signal integration and propagation along the dendrosomatic axis (Nicoll et al., 1993; Magee, 1998, 1999; Stuart and Spruston, 1998). This might be an important feature of Ih channels in principal neurons, which have to integrate numerous incoming signals on a spatially extended field. The mHCN1 channels, by virtue of their rapid kinetics, would be better suited to rapidly activate during fast IPSPs and EPSPs and shape the postsynaptic response.
Another cell population in which mHCN1 appears consistently in different brain regions is basket cells, in which anti-mHCN1 antibody labeling suggests that the channel is confined to axon terminals (Santoro et al., 1997). It has been proposed that theIh conductance acts to regulate the frequency of tonic inhibitory input from basket cells to principal neurons and provides the substrate for neuromodulatory transmitter control of the inhibition (Saitow and Konishi, 1999).
mHCN2 transcripts have a widespread distribution and are likely to subserve diverse roles. Because of the lack of mHCN2-specific antibodies, the subcellular localization of the respective protein is unknown. However, mHCN2 transcripts appear consistently in inhibitory GABAergic neurons, such as the short-axon cells in the olfactory bulb, hippocampal interneurons, and neurons of the thalamic reticular nucleus, globus pallidus, and substantia nigra pars reticulata. mHCN2 transcripts are found coupled with mHCN4 transcripts in neurons that display prominent oscillations, such as thalamocortical relay neurons. Moreover, the mHCN2 and mHCN4 isoforms are also found in the heart, suggesting their association with a pacemaking function. However, HCN4 is the predominant isoform expressed in the adult rabbit sinoatrial node, whereas mHCN2 is virtually absent from these cells but is predominant in quiescent ventricular muscle (Shi et al., 1999).
On a final note, interesting patterns of association can be observed between Ih channels and low voltage-activated T-type Ca channels (Talley et al., 1999), which participate in pacemaking by providing a rapid inward current that activates near threshold and contributes to the rising phase of the action potential burst (Pape, 1996). Specific associations between expression of T-type and HCN channel isoforms may help to further define the diverse physiological functions ofIh channels. For example, in inhibitory glomerular short-axon cells and reticular thalamic neurons, mHCN2 is expressed with the α1I isoform of the T-type Ca channel. In contrast, excitatory thalamocortical relay neurons express mHCN2 and mHCN4 in association with α1G T-type channels (Talley et al., 1999). In turn, mHCN1 occurs in combination with α1H in layer V pyramidal cells and in ventromedial hypothalamic cells, as well as in spinal motorneurons (Talley et al., 1999). The particular combination of conductances that operate in the subthreshold range of potentials will define the unique firing properties of a neuron and the function of that neuron in the context of its higher order circuitry. Understanding the relationship between the different HCN channel isoforms, their association with other components of the neuronal machinery, and finally their association with specific neuronal types and functions, should open the way to the rational targeting of subtypes of these channels to correct specific cardiovascular and neurological diseases.
Footnotes
This work was partially supported by a grant from the Whitehall Foundation (G.R.T.) and National Institutes of Health Grant NS-36658 (S.A.S.).We thank Huan Yao, John Riley, Matthew DeGennaro, and Eric Odell for technical assistance and Cliff Kentros, Eric Kandel, and David McCormick for helpful comments on this manuscript.
Correspondence should be addressed to Bina Santoro, Center for Neurobiology and Behavior, 722 West 168th Street, New York, NY 10032. E-mail: bs73@columbia.edu.
REFERENCES
- 1.Bland BH, Oddie SD. Anatomical, electrophysiological and pharmacological studies of ascending brainstem hippocampal synchronizing pathways. Neurosci Biobehav Rev. 1998;22:259–273. doi: 10.1016/s0149-7634(97)00013-4. [DOI] [PubMed] [Google Scholar]
- 2.Beaumont V, Zucker RS. Enhancement of synaptic transmission by cyclic AMP modulation of presynaptic Ih channels. Nat Neurosci. 2000;3:133–141. doi: 10.1038/72072. [DOI] [PubMed] [Google Scholar]
- 3.Clapham DE. Not so funny anymore: pacing channels are cloned. Neuron. 1998;21:5–7. doi: 10.1016/s0896-6273(00)80508-5. [DOI] [PubMed] [Google Scholar]
- 4.DiFrancesco D. Pacemaker mechanisms in cardiac tissue. Annu Rev Physiol. 1993;55:455–472. doi: 10.1146/annurev.ph.55.030193.002323. [DOI] [PubMed] [Google Scholar]
- 5.DiFrancesco D, Mangoni M. Modulation of single hyperpolarization-activated channels (i(f)) by cAMP in the rabbit sino-atrial node. J Physiol (Lond) 1994;474:473–482. doi: 10.1113/jphysiol.1994.sp020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiFrancesco D, Tortora P. Direct activation of cardiac pacemaker channels by intracellular cyclic AMP. Nature. 1991;351:145–147. doi: 10.1038/351145a0. [DOI] [PubMed] [Google Scholar]
- 7.Dodt HU, Zieglgansberger W. Visualizing unstained neurons in living brain slices by infrared DIC-videomicroscopy. Brain Res. 1990;537:333–336. doi: 10.1016/0006-8993(90)90380-t. [DOI] [PubMed] [Google Scholar]
- 8.Fletcher GH, Chiappinelli VA. An inward rectifier is present in presynaptic nerve terminals in the chick ciliary ganglion. Brain Res. 1992;575:103–112. doi: 10.1016/0006-8993(92)90429-d. [DOI] [PubMed] [Google Scholar]
- 9.Franklin KBJ, Paxinos G. The mouse brain in stereotaxic coordinates. Academic; San Diego: 1997. [Google Scholar]
- 10.Ingram SL, Williams JT. Modulation of the hyperpolarization-activated current (Ih) by cyclic nucleotides in guinea-pig primary afferent neurons. J Physiol (Lond) 1996;492:97–106. doi: 10.1113/jphysiol.1996.sp021292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishii TM, Takano M, Xie LH, Noma A, Ohmori H. Molecular characterization of the hyperpolarization-activated cation channel in rabbit heart sinoatrial node. J Biol Chem. 1999;274:12835–12839. doi: 10.1074/jbc.274.18.12835. [DOI] [PubMed] [Google Scholar]
- 12.Kawaguchi Y. Large aspiny cells in the matrix of the rat neostriatum in vitro: physiological identification, relation to the compartments and excitatory postsynaptic currents. J Neurophysiol. 1992;67:1669–1682. doi: 10.1152/jn.1992.67.6.1669. [DOI] [PubMed] [Google Scholar]
- 13.Kawaguchi Y. Physiological, morphological, and histochemical characterization of three classes of interneurons in rat neostriatum. J Neurosci. 1993;13:4908–4923. doi: 10.1523/JNEUROSCI.13-11-04908.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang ZG, North RA. Membrane properties and synaptic responses of rat striatal neurones in vitro. J Physiol (Lond) 1991;443:533–553. doi: 10.1113/jphysiol.1991.sp018850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ludwig A, Zong X, Jeglitsch M, Hofmann F, Biel M. A family of hyperpolarization-activated mammalian cation channels. Nature. 1998;393:587–591. doi: 10.1038/31255. [DOI] [PubMed] [Google Scholar]
- 16.Ludwig A, Zong X, Stieber J, Hullin R, Hofmann F, Biel M. Two pacemaker channels from human heart with profoundly different activation kinetics. EMBO J. 1999;18:2323–2329. doi: 10.1093/emboj/18.9.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lüthi A, McCormick DA. H-current: properties of a neuronal and network pacemaker. Neuron. 1998;21:9–12. doi: 10.1016/s0896-6273(00)80509-7. [DOI] [PubMed] [Google Scholar]
- 18.Lüthi A, McCormick DA. Modulation of a pacemaker current through Ca(2+)-induced stimulation of cAMP production. Nat Neurosci. 1999;2:634–641. doi: 10.1038/10189. [DOI] [PubMed] [Google Scholar]
- 19.Maccaferri G, McBain CJ. The hyperpolarization-activated current (Ih) and its contribution to pacemaker activity in rat CA1 hippocampal stratum oriens-alveus interneurones. J Physiol (Lond) 1996;497:119–130. doi: 10.1113/jphysiol.1996.sp021754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magee JC. Dendritic lh normalizes temporal summation in hippocampal CA1 neurons. Nat Neurosci. 1999;2:508–514. doi: 10.1038/9158. [DOI] [PubMed] [Google Scholar]
- 21.Magee JC. Dendritic hyperpolarization-activated currents modify the integrative properties of hippocampal CA1 pyramidal neurons. J Neurosci. 1998;18:7613–7624. doi: 10.1523/JNEUROSCI.18-19-07613.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCormick DA, Bal T. Sleep and arousal: thalamocortical mechanisms. Annu Rev Neurosci. 1997;20:185–215. doi: 10.1146/annurev.neuro.20.1.185. [DOI] [PubMed] [Google Scholar]
- 23.McCormick DA, Pape HC. Properties of a hyperpolarization-activated cation current and its role in rhythmic oscillation in thalamic relay neurones. J Physiol (Lond) 1990;431:291–318. doi: 10.1113/jphysiol.1990.sp018331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCormick DA, Prince DA. Acetylcholine causes rapid nicotinic excitation in the medial habenular nucleus of guinea pig, in vitro. J Neurosci. 1987;7:742–752. doi: 10.1523/JNEUROSCI.07-03-00742.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moosmang S, Biel M, Hofmann F, Ludwig A. Differential distribution of four hyperpolarization-activated cation channels in mouse brain. J Biol Chem. 1999;380:975–980. doi: 10.1515/BC.1999.121. [DOI] [PubMed] [Google Scholar]
- 26.Moroni A, Barbuti A, Altomare C, Viscomi C, Morgan J, Baruscotti M, DiFrancesco D. Kinetic and ionic properties of the human HCN2 pacemaker channel. Pflügers Arch. 2000;439:618–626. doi: 10.1007/s004249900225. [DOI] [PubMed] [Google Scholar]
- 27.Neher E. Correction for liquid junction potentials in patch clamp experiments. Methods Enzymol. 1992;207:123–131. doi: 10.1016/0076-6879(92)07008-c. [DOI] [PubMed] [Google Scholar]
- 28.Nicoll A, Larkman A, Blakemore C. Modulation of EPSP shape and efficacy by intrinsic membrane conductances in rat neocortical pyramidal neurons in vitro. J Physiol (Lond) 1993;468:693–710. doi: 10.1113/jphysiol.1993.sp019795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pape HC. Queer current and pacemaker: the hyperpolarization-activated cation current in neurons. Annu Rev Physiol. 1996;58:299–327. doi: 10.1146/annurev.ph.58.030196.001503. [DOI] [PubMed] [Google Scholar]
- 30.Robinson RB, Yu H, Chang F, Cohen IS. Developmental change in the voltage-dependence of the pacemaker current, if, in rat ventricle cells. Pflügers Arch. 1997;433:533–535. doi: 10.1007/s004240050309. [DOI] [PubMed] [Google Scholar]
- 31.Saito Y, Isa T. Electrophysiological and morphological properties of neurons in the rat superior colliculus. I Neurons in the intermediate layer. J Neurophysiol. 1999;82:754–767. doi: 10.1152/jn.1999.82.2.754. [DOI] [PubMed] [Google Scholar]
- 32.Saitow F, Konishi S. β-adrenergic receptor mediated excitability increase via activation of a hyperpolarization activated current (Ih) in rat cerebellar basket cells. Soc Neurosci Abstr. 1999;25:1255. doi: 10.1152/jn.2000.84.4.2026. [DOI] [PubMed] [Google Scholar]
- 33.Santoro B, Tibbs GR. The HCN gene family: molecular basis of the hyperpolarization-activated pacemaker channels. Ann NY Acad Sci. 1999;868:741–764. doi: 10.1111/j.1749-6632.1999.tb11353.x. [DOI] [PubMed] [Google Scholar]
- 34.Santoro B, Grant SG, Bartsch D, Kandel ER. Interactive cloning with the SH3 domain of N-src identifies a new brain specific ion channel protein, with homology to eag and cyclic nucleotide-gated channels. Proc Natl Acad Sci USA. 1997;94:14815–14820. doi: 10.1073/pnas.94.26.14815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santoro B, Liu DT, Yao H, Bartsch D, Kandel ER, Siegelbaum SA, Tibbs GR. Identification of a gene encoding a hyperpolarization-activated pacemaker channel of brain. Cell. 1998;93:717–729. doi: 10.1016/s0092-8674(00)81434-8. [DOI] [PubMed] [Google Scholar]
- 36.Schaeren-Wiemers N, Gerfin-Moser A. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 37.Seifert R, Scholten A, Gauss R, Mincheva A, Lichter P, Kaupp UB. Molecular characterization of a slowly gating human hyperpolarization-activated channel predominantly expressed in thalamus, heart, and testis. Proc Natl Acad Sci USA. 1999;96:9391–9396. doi: 10.1073/pnas.96.16.9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi W, Wymore R, Yu H, Wu J, Wymore RT, Pan Z, Robinson RB, Dixon JE, McKinnon D, Cohen IS. Distribution and prevalence of hyperpolarization-activated cation channel (HCN) mRNA expression in cardiac tissues. Circ Res. 1999;85:E1–E6. doi: 10.1161/01.res.85.1.e1. [DOI] [PubMed] [Google Scholar]
- 39.Stanford IM, Cooper AJ. Presynaptic mu and delta opioid receptor modulation of GABAA IPSCs in the rat globus pallidus in vitro. J Neurosci. 1999;19:4796–4803. doi: 10.1523/JNEUROSCI.19-12-04796.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stuart G, Spruston N. Determinants of voltage attenuation in neocortical pyramidal neuron dendrites. J Neurosci. 1998;18:3501–3510. doi: 10.1523/JNEUROSCI.18-10-03501.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Talley EM, Cribbs LL, Lee JH, Daud A, Perez-Reyes E, Bayliss DA. Differential distribution of three members of a gene family encoding low voltage-activated (T-type) calcium channels. J Neurosci. 1999;19:1895–1911. doi: 10.1523/JNEUROSCI.19-06-01895.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaccari T, Moroni A, Rocchi M, Gorza L, Bianchi ME, Beltrame M, DiFrancesco D. The human gene coding for HCN2, a pacemaker channel of the heart. Biochim Biophys Acta. 1999;1446:419–425. doi: 10.1016/s0167-4781(99)00092-5. [DOI] [PubMed] [Google Scholar]
- 43.Vinogradova OS, Kitchigina VF, Zenchenko CI. Pacemaker neurons of the forebrain medical septal area and theta rhythm of the hippocampus. Membr Cell Biol. 1998;11:715–725. [PubMed] [Google Scholar]
- 44.Williams S, Samulack DD, Beaulieu C, LaCaille JC. Membrane properties and synaptic responses of interneurons located near the stratum lacunosum-moleculare/radiatum border of area CA1 in whole-cell recordings from rat hippocampal slices. J Neurophysiol. 1994;71:2217–2235. doi: 10.1152/jn.1994.71.6.2217. [DOI] [PubMed] [Google Scholar]
- 45.Williams SR, Turner JP, Hughes SW, Crunelli V. On the nature of anomalous rectification in thalamocortical neurones of the cat ventrobasal thalamus in vitro. J Physiol (Lond) 1997;505:727–747. doi: 10.1111/j.1469-7793.1997.727ba.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu H, Chang F, Cohen IS. Pacemaker current exists in ventricular myocytes. Circ Res. 1993;72:232–236. doi: 10.1161/01.res.72.1.232. [DOI] [PubMed] [Google Scholar]