Abstract
Kinesin superfamily proteins (KIFs) are the molecular motors conveying cargos along microtubules. KIF5s, the heavy chains of conventional kinesin (KHC), are originally identified members of KIFs, and neuronal KIF5A and ubiquitous KIF5B have been identified so far. In the present work, we cloned a novel member of KIF5, KIF5C, and generated specific antibodies against three KIF5s to investigate their distribution and functions. KIF5A showed pan-neuronal distribution in the nervous system. KIF5B showed a glial cell distribution pattern in general; however, interestingly, its expression was strongly upregulated in axon-elongating neurons, such as olfactory primary neurons and mossy fibers. KIF5C was also a neuronal KIF5 like KIF5A but was highly expressed in lower motor neurons in 2-week-old or older mice, suggesting its important roles in the maintenance of motor neurons rather than in their formation, such as axonal elongation. Because a large part of KIF5s in adult motor neurons were expected to be KIF5C, we generated mice lacking the kif5Cgene to investigate the functions of KIF5C in neurons in living animals. The mutant mice showed smaller brain size but were viable and did not show gross changes in the nervous system. Closer examinations revealed the relative loss of motor neurons to sensory neurons. Because three KIF5s showed high similarity in the amino acid sequence, could rescue the KIF5B mutant cells, and could form heterodimers, we think that there are functional redundancy among the three KIF5s and that KIF5A and KIF5B prevented the KIF5C null mice from the severe phenotype.
Keywords: kinesin, KIFs, KIF5A, KIF5B, KIF5C, motor neuron, axonal transport, brain, cloning, gene targeting
Kinesin superfamily proteins are the molecular motors acting in the microtubule-based motilities of organelles in eukaryotic cells. More than 10 members of the murine kinesin superfamily proteins (KIFs) have been identified in the nervous system (Aizawa et al., 1992; Goldstein, 1993; Hirokawa, 1993, 1998;Kondo et al., 1994; Nangaku et al., 1994; Sekine et al., 1994; Noda et al., 1995; Okada et al., 1995; Yamazaki et al., 1995; Saito et al., 1997; Nakagawa et al., 1997; Yang et al., 1997). Among them, kinesin heavy chain (KHC) was identified originally (Brady, 1985; Vale et al., 1985), with a microtubule plus-end directed motor domain in its N terminus, so that it is believed to contribute to the motility of organelles toward the cell periphery in general. A conventional kinesin is composed of two KHCs and two kinesin light chains (KLCs) and binds directly to microtubules, as revealed by electron microscopic analyses (Hirokawa et al., 1989; Scholey et al., 1989). AlthoughCaenorhabditis elegans (Patel et al., 1993), squid (Kosik et al., 1990), sea urchin (Wright et al., 1993), and fruit fly (Yang et al., 1988) have single conventional KHC genes, KHC has diverged into a neuron-specific form and a ubiquitous form in vertebrates (Kato, 1991; Navone et al., 1992; Gudkov et al., 1994;Niclas et al., 1994; Vignali et al., 1996; Bost-Usinger et al., 1997;Meng et al., 1997). In mice, we have identified a distinct murine brain-specific KHC, KIF5A (5A) (Aizawa et al., 1992), in addition to the ubiquitous KHC (uKHC) or KIF5B (5B) (Gudkov et al., 1994;Meng et al., 1997).
Here, we report the cloning of KIF5C (5C), the third member of KHC in mouse brain. Specific antibodies revealed their specialized localization in the nervous system. Ubiquitous KIF5B showed a glial cell distribution pattern in general; however, interestingly, its expression was strongly upregulated in axon-elongating neurons, such as olfactory primary neurons and mossy fibers. Although both KIF5A and KIF5C were expressed in neurons, they showed different patterns of distribution. Expression of KIF5A was observed in various kinds of neurons at similar levels, whereas KIF5C was enriched in motor neurons. Upregulation of KIF5C in motor neurons was observed in the 2-week-old or older mice, which was developmentally increased, indicating the important roles of KIF5C in maintenance of motor neurons rather than in axonal formation.
Because most of the KIF5 in the motor neurons of adult mice was suggested to be KIF5C, we disrupted the kif5C gene in mice to investigate the functions of KIF5C and the possible redundancy among the three KIF5s in neurons in living animals. Surprisingly, mutant mice were viable and did not show gross changes in the nervous system. The mutant mice only showed smaller brain size and relative loss of motor neurons to sensory neurons. Three KIF5s showed high similarity, could rescue KIF5B mutant cells, and could form heterodimers. These data strongly indicate that there is functional redundancy among the three KIF5s.
MATERIALS AND METHODS
cDNA cloning of mouse kif5C gene and construction of the expression vectors. We obtained a cDNA clone of KIF5C from the mouse brain library in accordance with a standard cloning method (Kanai et al., 1989; Sambrook et al., 1989) using the motor domain sequence as the probe (Nakagawa et al., 1997). A full-length and several partial-length clones were isolated, transferred to pBluescript SK(+), and carefully sequenced. We constructed deleted fragments of the clones by the conventional 3′ deletion method using Exonuclease III/mung bean nuclease (TaKaRa, Tokyo, Japan) to read the entire sequence (Kanai et al., 1992;Kanai and Hirokawa, 1995). All sequences were read bidirectionally at least four times with the ABI autosequencer 377 (Perkin-Elmer, Emeryville, CA). In expectation of the efficient expression of the three KIF5s in the transfection study, we changed their sequences upstream of the ATG to the Kozak sequence (Kozak, 1987) using PCR as described previously (Tanaka et al., 1998). The fragments were inserted into the cytomegalovirus (CMV) promoter-driven expression vector [pcDNA3.1/Zeo(−); Invitrogen, San Diego, CA], for the rescue study using kif5B−/− cells.
Production of recombinant KIF5 proteins. The full-length cDNA of each KIF5 was subcloned into the pET32a expression vector (Novagen, Madison, WI) to be expressed as thioredoxin/His-tag fusion proteins. Bacterial strain BL21(DE3) was transformed with these plasmids, and fusion protein expression was induced with 0.1 mmisopropyl-β-d-thiogalactopyranoside at 18°C for 12 hr. The bacteria were disrupted with a French Press (Otake Corp.), and the fusion proteins were recovered on Ni-Chelating-Sepharose beads (Amersham Pharmacia Biotech, Uppsala, Sweden). The proteins were conjugated to a cyanogen bromide (CNBr) Sepharose (Amersham Pharmacia Biotech) column to purify the antibodies or were used as standards in the immunoblot analysis.
To determine the concentration of the recombinant KIF5 proteins, serial dilutions of standard BSA and recombinant proteins were loaded on the same SDS-PAGE gel. We scanned the Coomassie blue-stained gel with an image scanner (GT-9500; Epson) and measured the staining intensity of the bands using NIH Image. According to this estimate, we diluted the recombinant proteins to match the standard and repeated the quantitation until the differences in the concentration among the standard and the three KIF5s were within 10%. After compensation for the thioredoxin/His-tag sequence, we determined their concentrations and used them as standard KIF5 proteins.
Antibody production. We have raised six independent antisera against KIF5A, KIF5B, and KIF5C (Fig. 1). Anti-KIF5A antibodies were raised against two synthetic peptides, CTERLAGEDSALGAELCEETPVNDNSSIVVRIAPE (5A-n) and CEAEDQAKLFPLHQETAAS (5A-c). Anti-KIF5B antibodies were raised against CQFDKEKANLEAFTADKDIAITSDKGAAAVGMAGSFTD (5B-n) and CGSFVQNNQPVGLRGG- GGKQS (5B-c), and anti-KIF5C antibodies were raised against CQISAKDQKSLEPCDNTPIIDNITPVVDGIS (5C-n) and CAVHAVRGGGGGSSNSTHYQK (5C-c). These peptides were conjugated to keyhole lympet hemocyanin and were injected into rabbits. Antibodies were then affinity-purified using the recombinant KIF5 protein-conjugated CNBr Sepharose columns.
Fig. 1.
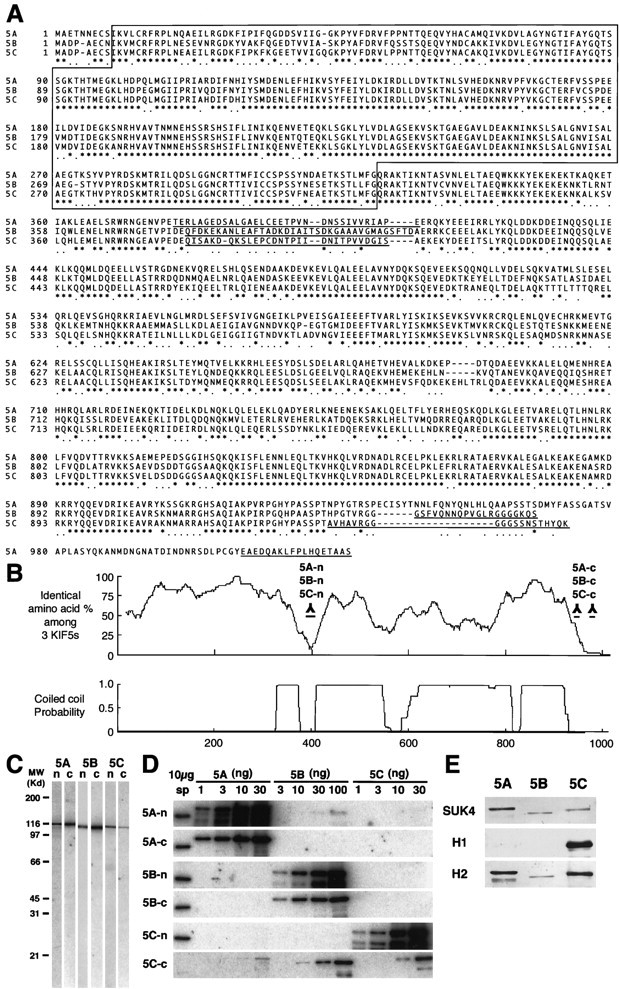
Alignment of the three KIF5s and antibody characterization. A, Alignment of the sequences of the three KIF5s. Amino acids are numbered in theleft margin. Asterisks represent the identical amino acids among the three KIF5s, and dotsshow the same residues between two KIF5s. The motor domain, located in the N-terminal region, is boxed. The amino acid sequences used for the antibody generation areunderlined. B, Homology among the three KIF5s and coiled-coil probability of KIF5C. There were two regions that showed very low similarity among the three KIF5s, with a small coiled-coil probability. We used these sequences for generating antibodies specific for each of the KIF5s (5A-n, 5B-n, 5C-n, 5A-c, 5B-c, and 5C-c). C, Immunoblot of anti-KIF5 antibodies (5A-n, 5B-n, 5C-n, 5A-c, 5B-c, and 5C-c). Ten micrograms of crude extract from an adult mouse brain were loaded on eachlane. D, Examination of the specificity of the six antibodies using recombinant KIF5 proteins. Filters transferred with 10 μg of crude extract from adult mouse spinal cord (sp) and various amounts of recombinant KIF5 proteins (KIF5A and KIF5C, 1, 3, 10, and 30 ng; KIF5B, 3, 10, 30, and 100 ng) were used. 5A-n showed weak cross-reactivity for KIF5B. 5A-c, 5B-n, 5B-c, and 5C-n did not cross-react with the other members of KIF5. 5C-c was not a specific antibody. E, Immunoblot analysis to characterize the widely used monoclonal anti-kinesin antibodies (SUK4, H1, and H2), using equal amount of recombinant KIF5 proteins (10 ng for SUK4 and H2; 30 ng for H1). SUK4 predominantly stained KIF5A; H1 detected only KIF5C; H2 preferred KIF5A and KIF5C to KIF5B. H1 did not recognize KIF5A or KIF5B, even when 100 ng of proteins were loaded (data not shown).
For immunostaining, the samples were incubated with these antibodies at 10 μg/ml and then with Cy2-labeled anti-rabbit IgG secondary antibody (Amersham Pharmacia Biotech). We also used monoclonal antibodies against GFAP (clone G-A-5; Sigma, St. Louis, MO) and S-100 (clone SH-B1; Sigma) to identify the glial cell types with Cy3-labeled anti-mouse IgG second antibody (Amersham Pharmacia Biotech). The samples were observed under a confocal laser scanning microscope (MRC1000; Bio-Rad, Hercules, CA).
During immunoblotting, the polyvinylidene difluoride membrane loaded with proteins were stained with 10 μg/ml anti-KIF5 antibody or the 1:100-diluted monoclonal anti-kinesin antibodies [SUK4 (Developmental Studies Hybridoma Bank), H1, and H2 (Chemicon, Temecula, CA)] and then with 125I-labeled protein A or125I-labeled goat anti-mouse IgG (ICN Biochemicals, Costa Mesa, CA).
As for KIF5A and KIF5B, the two anti-peptide antibodies gave the same results in both immunostaining and immunoblotting experiments. Because 5C-c cross-reacted with KIF5A and KIF5B, we used only 5C-n to investigate KIF5C in this study. We confirmed the specificity of this antibody using KIF5C-deficient mice as the control.
Quantitative analysis of the three KIF5s. The dilution chain of the standard KIF5 proteins and the crude extracts loaded on the SDS-PAGE gel were transferred to filters. We then blotted them separately using anti-KIF5 antibodies and analyzed the bands using the MacBas analyzer (Fuji). We then diluted the crude extracts to match the standard. After repeated quantitation, we determined the amount of each of the KIF5s in the crude extracts.
Gene targeting of KIF5C. Fragments of the mousekif5C gene were screened from a lambda EMBL3 genomic library of embryonic stem (ES) cell line J1 in accordance with a standard method (Sambrook et al., 1989), using a cDNA probe encoding the P-loop. The resulting two independent clones were transferred into pBluescriptII SK(+) and carefully mapped with restriction enzymes. These fragments were subjected to cytogenetic mapping to determine their loci on 2C as described previously (Nakagawa et al., 1997). The exon–intron structure was also determined by partial sequencing. Targeting vectors were constructed using the genomic clones pMCI-DTA (Yagi et al., 1993) and pPGKneopA (kindly provided by T. Noda), as shown in Figure 6A. pPGKneopA was applied in the reversed direction, flanked by 9 kb of 5′ and 1 kb of 3′ homologous region. Reversed pMCI-DTA without poly(A+) signal was ligated at the 5′ extremity. A splicing acceptor sequence (Watanabe et al., 1993) and a transcription termination signal sequence of the human gastrin gene (Sato et al., 1986) was inserted for reducing the downstream gene transcription. The linearized vectors were electroporated into ES cells J1 and screened by genomic Southern blotting as described previously (Harada et al., 1990; Takei et al., 1995, Tanaka et al., 1998). Three homologous recombinant clones with reversed neo were recovered from 407 clones, amounting to a targeting efficiency of 0.74%. Mice with the kif5C+/−genotype were successfully produced from D8 and L8 clones and maintained by backcrossing to C57BL/6J females under a specific pathogen-free environment.
Fig. 6.
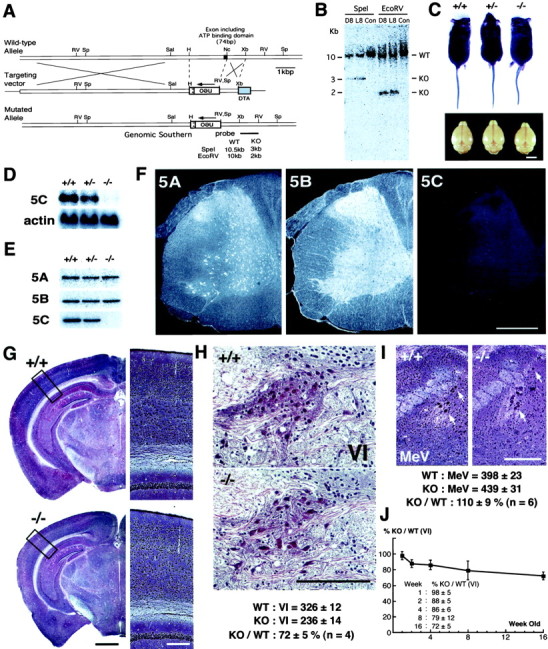
Targeted disruption of the mousekif5C gene. A, Targeting strategy with conventional positive-negative selection. The strategy of genomic Southern blotting for the screening of homologous recombinant ES clones is included. SA, Splicing acceptor; filled box, exon including ATP-binding motif (P-loop);Sp, SpeI; RV,EcoRV; Sal, SalI;H, HindIII; Nc,NcoI; Xb, XbaI.B, Southern blotting analysis of targeted ES clones digested, respectively, by SpeI andEcoRV. Clones D8 and L8 were homologous recombinants.Con was a nonhomologous control. C, Comparison of body and brain size among adult kif5C+/+,+/−, and −/− mice. Although the body size of null mutants was not significantly altered, they showed smaller brains than wild-type and heterozygote mice (∼5% decrease in weight). Scale bar, 5 mm. D, E, Northern blotting and immunoblotting analysis of KIF5C knock-out mice. Total brain RNAs and brain crude extracts were prepared from adult mice. The total RNAs were hybridized, respectively, with probes for KIF5C and β-actin (D), and the crude extracts (10 μg) were blotted using anti-KIF5 antibodies (E). In both cases, no KIF5C was detected in the KIF5C null mice. As for the effects of the absence of KIF5C on the other KIF5s, KIF5A and KIF5B showed little change in their expression levels in the brains of knock-out mice. F, Immunofluorescence analysis of KIF5C knock-out mice. Spinal cords of the wild-type and KIF5C null mutants were cross-sectioned at the C7 level and stained with anti-KIF5 antibodies. The staining pattern of the wild-type mouse spinal cord was shown previously in Figure 4,c, f, and i. KIF5A and KIF5B showed similar expression pattern as in the wild-type mice. Staining intensity of the anti-KIF5C antibody in the null mutants was at a background level. Scale bar, 0.4 mm. G, Bodian staining of frontal sections of the wild-type (+/+) and mutant (−/−) mice. The panels on the right represent high magnifications of the boxed areas. The brains of mutant mice were smaller but did not show any gross abnormality in their structures. Scale bars: left, 1 mm;right, 0.2 mm. H, Comparison of motor neurons (VI, abducens nucleus) between the wild-type (+/+) and mutant (−/−) mice 16 weeks of age using serial sections. The number of neurons in the abducens nucleus in the mutant mice (236 ± 14) was decreased to 72 ± 5% of that in the wild-type mice (326 ± 12) (n = 4). Scale bar, 0.1 mm. I, Comparison of sensory neurons in the sensory trigeminal neurons between wild-type (+/+) and mutant (−/−) 8-week-old mice (arrows). The numbers of sensory neurons in the wild-type and mutant mice were 398 ± 23 and 439 ± 31, respectively. Sensory trigeminal neurons did not decrease but rather showed the tendency of slight increase (110 ± 9%,n = 6). Scale bar, 0.2 mm. J, Developmental comparison of motor neurons (VI) between the wild-type and mutant mice (1-, 2-, 4-, 8-, and 16-week-old). The number of neurons in the abducens nucleus in the mutant mice showed little difference from that in the wild-type 1-week-old mice (98 ± 5%) but was decreased developmentally in those 2 weeks or later after birth (2 weeks, 88 ± 5% to 16 weeks, 72 ± 5%).
PCR genotyping. An animal tissue sample was processed in a lysis buffer [10 mm Tris-HCl, pH 8.0, 25 mm EDTA, 1% SDS, 75 mmNaCl (Wako Neuss, Germany), and 100 ng/ml of proteinase K (Merck, Darmstadt, Germany)] at 55°C, purified using phenol-chloroform extraction and ethanol precipitation, and amplified for 25 cycles (15 sec at 98°C, 2 sec at 65°C, 30 sec at 74°C) with the following primer sets on the GeneAmp thermal cycler (Perkin-Elmer). The neo transgene was amplified using 5′-TGGGCACAACAGACAATCGG-3′ and 5′-ACTTCGCCCAATAGCAGCCAG-3′. Intronic sequences from the deleted region of the kif5C gene were detected with 5′-CGAAAGCTTCACGTAGCCGAAAAAGAC-3′ and 5′-GACACCGACAGTGCAAACTTACCTCCA-3′ as primers.
Quantitation of motor and sensory neurons. Serial sections of the wild-type and KIF5C knock-out adult mouse brains were silver-stained according to the method of Bodian (1936). In counting the number of neurons in the abducens nucleus and mesencephalic trigeminal nucleus, all sections including the nuclei were counted (∼20 and 60 sections for the abducens nucleus and the mesencephalic trigeminal nucleus, respectively). The counting was performed without notification of their genotypes.
Rescue of kif5B−/− cells by plasmid injection.We described the methods of primary culture of extraembryonic membrane and plasmid injection previously (Tanaka et al., 1998). Twelve hours after injection, we vitally stained the cells with 330 nm Mitotracker (Molecular Probes, Eugene, OR) for 5 min and then fixed them with 2% paraformaldehyde (Merck) in 0.1m phosphate buffer at 37°C for 15 min. The samples were then treated with 0.1% Triton X-100 in PBS for 10 min, stained using anti-KIF5 antibodies, and observed as described above.
Immunoprecipitation. Mouse brain was dissected in ice-cold PBS and homogenized in buffer A (PBS with 1% Triton X-100 supplemented with protenase inhibitors, DTT 10 mm). The homogenate was clarified twice with centrifugation at 1000 ×g for 10 min. This postnuclear supernatant (PNS) fraction was used for the immunoprecipitation assays. KIF5 immunobeads and control beads were prepared as described previously (Okada et al., 1995). PNS was incubated with immunobeads for 12 hr at 4°C. The beads were then washed with buffer A, which were analyzed by immunoblotting.
RESULTS
Cloning and sequencing of the third conventional KHC, KIF5C, in mouse brain
We previously cloned a mouse neuronal KHC, KIF5A (Aizawa et al., 1992). Several years later, Meng et al. (1997) isolated a ubiquitous KHC from mouse pancreatic β-cells, which we termed KIF5B. We have now cloned the third member of KIF5, KIF5C, from a mouse brain cDNA library using the motor domain sequence as the probe (Nakagawa et al., 1997).
KIF5C consists of 956 amino acids and shows a high degree of similarity to KIF5A or KIF5B (60% identical), especially in its motor domain (KIF5C: 9–321, 80%) and in the C-terminal coiled-coil region (KIF5C: 829–908, 90%) (Fig. 1A,B). On the other hand, there were two short sequences showing very few similarities and low coiled-coil probabilities at the region joining the motor and coiled-coil rod domains and at the C-terminal; these regions were used for antibody production.
Generation of antibodies specific for each KIF5
To investigate the distribution and function of KIF5A, KIF5B and KIF5C, the use of specific antibodies that can clearly distinguish each of the KIF5s from the others was indispensable. However, because the KIF5s show a very high degree of homology in their entire length, we expected difficulties in using recombinant fragments as antigens. We thus generated antibodies using synthetic peptides corresponding to the two low-homology regions described above (Fig.1A,B). The antibodies against these sequences at the N-terminal junction and the C terminal were named 5A-n, 5B-n, 5C-n, and 5A-c, 5B-c, 5C-c, respectively. The antisera were then affinity-purified using recombinant KIF5 proteins.
We then characterized these anti-KIF5 antibodies. First, the antibodies were subjected to immunoblotting on filters loaded with 10 μg of brain crude extract (Fig. 1C). Each antibody revealed a single band nearly corresponding to the predicted molecular weight (5A, 117 kDa; 5B, 109 kDa; 5C, 107 kDa). We then confirmed their specificity using recombinant KIF5 proteins to exclude possible cross-reactivity among them (Fig. 1D). 5A-n showed slight cross-reactivity for KIF5B (the lane loaded with 100 ng of KIF5B protein). 5A-c, 5B-n, 5B-c, and 5C-n did not recognize the other members of KIF5. However, 5C-c cross-reacted with KIF5A and KIF5B and was therefore excluded from the following experiments. As for the antibodies against KIF5A and KIF5B, we used both antibodies for the following experiments and obtained similar results in both immunoblotting and immunofluorescence experiments. In the case of KIF5C, we confirmed the specificity of 5C-n using KIF5C knock-out mice as the negative controls (see Fig.6E,F).
Because a significant number of studies on kinesin have been performed using the monoclonal anti-kinesin antibodies SUK4, H1, and H2 (Ingold et al., 1988; Pfister et al., 1989), it is very important to elucidate their specificity for each KIF5. For this purpose, we prepared filters loaded with equal amounts of recombinant KIF5 proteins (10 ng for SUK4 and H2; 30 ng for H1) and subjected them to immunoblotting using the monoclonal anti-kinesin antibodies (Fig. 1E). The antibodies showed different staining patterns. SUK4 primary stained KIF5A. H1 recognized KIF5C but did not detect KIF5A or KIF5B, even when 100 ng of proteins were loaded (data not shown). H2 preferred KIF5A and KIF5C to KIF5B. These indicate that the characteristics of these antibodies should be considered before using them.
Different distributions of the three KIF5s in the nervous system
To elucidate the distribution of the three KIF5s, we analyzed their expression by immunoblotting. As shown in Figure2A, KIF5B was detected in every tissue investigated, whereas KIF5A and KIF5C were expressed exclusively in the brain and spinal cord. Further immunoblotting studies using cultured hippocampal neurons and glial cells revealed KIF5A and KIF5C to be neuronal.
Fig. 2.
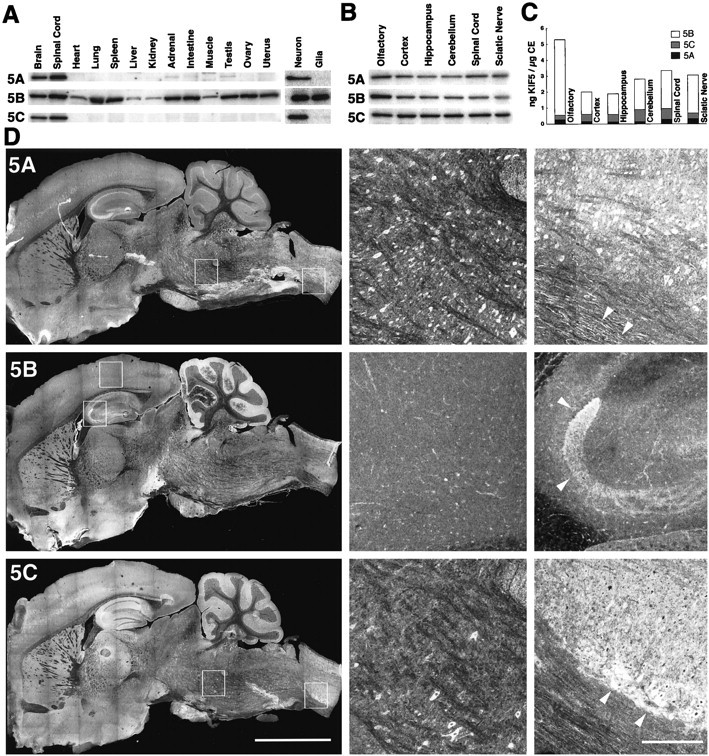
KIF5A and KIF5C are neuronal kinesins but showed differential distribution in the nervous system.A, Left, Tissue distribution of the three KIF5s by immunoblotting. KIF5B was detected in every tissue investigated, whereas KIF5A and KIF5C were expressed exclusively in the tissues of nervous system (Brain, Spinal Cord). A, Right, Immunoblot of cultured hippocampal neurons (Neuron) and glial cells (Glia). KIF5B was expressed in both neurons and glial cells, whereas the expression of KIF5A and KIF5C was limited to neurons and was not found in glial cells. B, C, Expression of the three KIF5s in nervous system as determined by quantitative immunoblotting. The olfactory bulb, cortex, hippocampus, cerebellum, spinal cord, and sciatic nerve were investigated. KIF5B was the most abundantly expressed KIF5 throughout the nervous system (70–90% of the total KIF5). As for the neuronal KIF5s, the expression ratio of KIF5C to KIF5A varied from one to five among different regions of the nervous system. Note the prominent expression of KIF5B in the olfactory bulb. D, Distribution of each of the KIF5s in the brain as shown by immunofluorescence analysis using anti-KIF5 antibodies (left, low magnification;middle and right, high magnification of the boxed areas in the left panels).5A, KIF5A stained various kinds of neurons. It was found in cell bodies, dendrites, and axons. Arrowheads in theright panel indicate the axons in the anterior funiculus of the spinal cord. 5B, Although KIF5B was highly expressed in the mossy fibers of the hippocampus (arrowheads in the right panel), anti-KIF5B antibody primary stained glial cells (e.g., astroglias in cortex; middle panel). 5C, KIF5C was expressed only in the subpopulation of neurons, especially in medulla (middle panel) and spinal cord (right panel). In spinal cord, KIF5C was highly expressed in the neurons located at the anterior horn (arrowheads). Note the tendency of a similar staining pattern in the cortex, hippocampus, cerebellum, and spinal cord to the immunoblotting data. Scale bars: left, 5 mm;middle, right, 0.5 mm.
We then performed a quantitative immunoblot analysis to measure the amount of each KIF5 in the nervous system (Fig.2B,C). We investigated the expression in the olfactory bulb, cortex, hippocampus, cerebellum, spinal cord, and sciatic nerve. The total amount of the three KIF5s constituted 0.2–0.5% of the total crude extract proteins, which matched the data of a previous study (brain, 0.29%; Hollenbeck, 1989). KIF5B was the most abundantly expressed KIF5, especially in the olfactory bulb (90% of total KIF5s and 0.5% of total crude extract proteins). Although both KIF5A and KIF5C were neuronal, they showed different distribution in the nervous system. In the olfactory bulb and sciatic nerve, they were expressed at a similar level, whereas the expression of KIF5C was three to five times higher than that of KIF5A in the cortex, hippocampus, cerebellum, and spinal cord.
For further analysis, we performed immunofluorescence using mouse brain sagittal sections (Fig. 2D). Detailed examination revealed that each KIF5 showed a unique distribution. Ubiquitous KIF5B showed a glial cell pattern of expression in general (e.g., astrocytes in cortex) (Fig. 2D, middle panel of5B). It was also highly expressed in the mossy fibers of the hippocampus (arrowheads in the right panel of5B). KIF5A and KIF5C showed neuronal but different distribution to each other. It was apparent particularly in the medulla and spinal cord (middle and right panels in5A and 5C). Expression of KIF5A was observed in various kinds of neurons, whereas KIF5C was found only in a subpopulation of neurons. As for the subcellular distribution of KIF5A and KIF5C in neurons, they were observed in the cell bodies, dendrites, and axons.
KIF5B is highly expressed in olfactory sensory neurons
Because the quantitative immunoblot analysis revealed high expression of KIF5B in the olfactory bulb (Fig. 2C), we performed an immunofluorescence study on its frontal sections (Fig.3). KIF5B was highly expressed in olfactory primary axons (arrows in 5B). However, in the rest of the olfactory bulb, anti-KIF5B antibodies predominantly stained glial cells (e.g., astrocytes; arrowheads in5B). As for KIF5A and KIF5C, they were localized in neurons (e.g., mitral cells; arrowheads in 5A), but higher expression of KIF5C was observed in the granule cells (arrows in 5C).
Fig. 3.
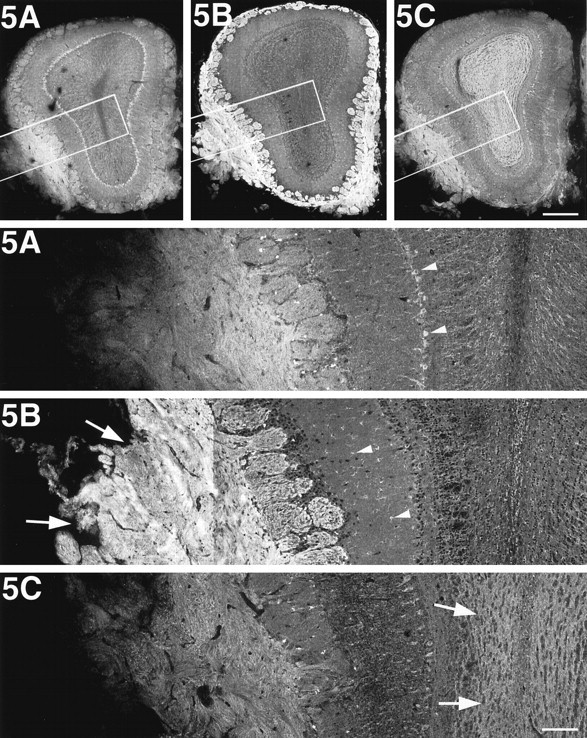
KIF5B is highly expressed in olfactory primary axons. Immunofluorescence of the olfactory bulb after staining with anti-KIF5 antibodies. The bottom panels represent high magnifications of the boxed areas of the top panels, respectively. KIF5B was highly expressed in the olfactory primary axons (arrows in 5B); however, glial cells, mainly astrocytes (arrowheads in5B), were stained predominantly in the rest of the olfactory bulb. On the other hand, KIF5A and KIF5C were localized only to the neuronal cells (e.g., mitral cell; arrowheads in5A). Note the strong staining of KIF5C in the granule cells (arrows in 5C). Scale bars:top, 0.4 mm (low magnifications); bottom, 0.1 mm (high magnifications).
KIF5C is highly expressed in motor neurons
Because each KIF5 showed unique patterns of distribution in the brain sagittal sections, we performed intensive immunofluorescence studies on a series of brain frontal and spinal cord cross-sections (Fig. 4). KIF5A showed pan-neuronal distribution with similar expression level among various types of neurons. KIF5B showed a glial cell pattern in general. [We confirmed the expression of KIF5B in both astrocytes and oligodendrocytes using the antibodies against GFAP (astrocyte marker) and S100 protein (oligodendrocyte marker) (data not shown).] However, interestingly, KIF5C was expressed prominently in motor neurons in both cranial nerves and spinal cord. Among the cranial nerves investigated, the oculomotor (III), trochlear (IV; data not shown), motor trigeminal (MoV), abducens (VI), and facial (VII) nerves and neurons showed high expression of KIF5C (Fig. 4g,h,o–s), but lower levels of expression of KIF5C was observed in the optic (II; data not shown), mesencephalic trigeminal (MeV), and vestibulocochlear (VIII) nerves and neurons (Fig. 4r,s). In the spinal cord, anti-KIF5C antibody strongly stained the motor neurons in the anterior horn (Fig. 4i).
Fig. 4.
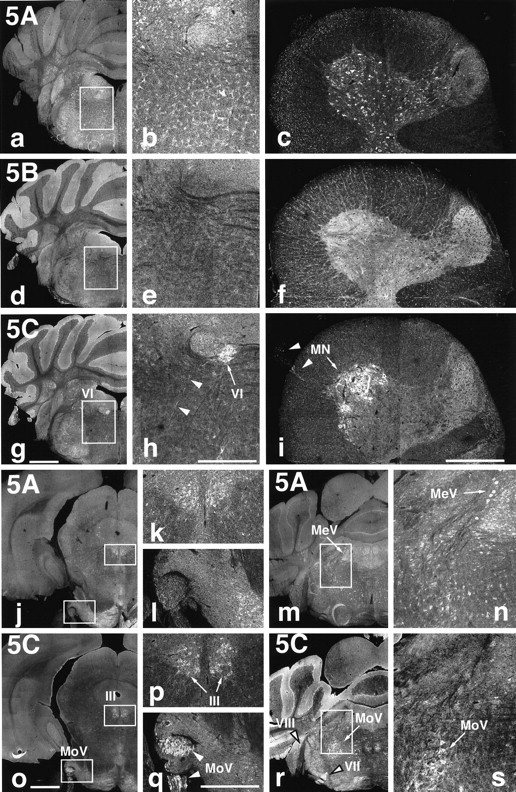
KIF5C is highly expressed in motor neurons. Localization of the three KIF5s in the brain and spinal cord.a–i, Immunofluorescence of brain frontal sections including the abducens nucleus (VI) [low magnification (a, d, g) and high magnification (b, e, h) of the boxed areas] and cross-sections of the spinal cord at the C7 level (c, f, i) after staining with anti-KIF5 antibodies (KIF5A, a–c; KIF5B, d–f; KIF5C, g–i). KIF5B was highly expressed in glial cells [mainly astrocytes (brain) and oligodendrocytes (spinal cord)] but was less abundant in neurons. KIF5A and KIF5C were localized to neurons but showed different expression patterns. KIF5A was found at similar levels of expression in various kinds of neurons, whereas KIF5C was highly expressed in motor neurons both in the brain (VI) and in the spinal cord (MN). j–s, Distribution of KIF5A (j–n) and KIF5C (o–s) in the sections containing several cranial nerves. KIF5A was expressed equally among neurons as described above. However, prominent expression of KIF5C was observed in motor [oculomotor (III), motor trigeminal (MoV), and facial (VII) nerves], but the expression was weaker in the sensory [mesencephalic trigeminal (MeV) and vestibulocochlear (VIII)] neurons. k,l, n, p, q, and s represent higher magnifications of theboxed areas in their left panels, respectively. Note the higher expression of KIF5A in sensory (MeV) than in motor (MoV) neurons, in contrast to KIF5C. Arrows andarrowheads indicate cell bodies and axons, respectively. Scale bars: a, d, g,j, m, o, r, 1 mm (low magnifications); b, c,e, f, h, i,k, l, n, p,q, s, 0.4 mm (high magnifications).
Because motor neurons have large cell bodies, it is possible that the strong staining of KIF5C in motor neurons was attributable to the large sizes of the cells. However, the findings that (1) axons of the motor trigeminal and facial nerves showed strong KIF5C staining and (2) KIF5A showed sensory neuron dominant staining in the trigeminal nerve (Fig. 4n,s), indicated that KIF5C was highly expressed in motor neurons regardless of the size of their cell bodies.
KIF5C is increased developmentally in motor neurons
KIF5C showed prominent expression in motor neurons. The next question is how KIF5C is highly expressed in motor neurons. To answer this question, we performed developmental immunofluorescence using mouse spinal cords 1, 2, 4, and 8 weeks after birth (Fig.5). KIF5A showed pan-neuronal distribution (arrows in 5A), and KIF5B was expressed dominantly in glial cells (e.g., oligodendrocytes;arrowheads in 5B) throughout the period. However, the expression pattern of KIF5C changed developmentally; the prominent expression of KIF5C in motor neurons was not found in 1-week-old mice but was observed in 2-week-old or older mice in which KIF5C was increased developmentally (arrows in 5C).
Fig. 5.
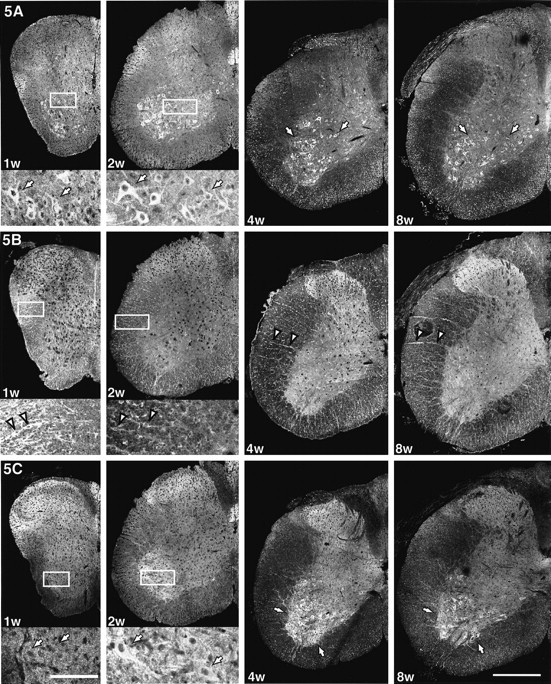
KIF5C is increased developmentally in motor neurons. Distribution of the three KIF5s in the mouse spinal cords 1, 2, 4, and 8 weeks after birth. Bottom panels of the spinal cords at 1 or 2 weeks old are the high magnifications of theboxed areas of the top panels. KIF5A showed pan-neuronal distribution (arrows in5A), and KIF5B was expressed dominantly in glial cells (arrowheads in 5B; radial glias are oligodendroglias) throughout this period. However, the expression pattern of KIF5C changed developmentally; the prominent expression of KIF5C in motor neurons was not found in 1-week-old mice but was observed in 2-week-old or older mice, which was increased developmentally (arrows in 5C). Scale bars: bottom left, 0.4 mm (low magnifications);bottom right, 0.1 mm (high magnifications).
To know when KIF5A and KIF5C are first expressed in the spinal cord, we prepared the crude extracts of spinal cords from embryonic day 13 (E13), E16, postnatal day 0 (P0), P7, P14, and P28 mice, and 20 μg of crude extract proteins were subjected to the immunoblot. However, each KIF5 was already expressed, even in the spinal cord at E13 and could not determine the start point of expression. We tried to prepare the sample at E10, but the embryo was too small to isolate spinal cords at certain amount. The expression levels of the three KIF5s did not change apparently during this period (data not shown).
Targeted disruption of the kif5C gene
KIF5C was highly expressed in motor neurons, whereas KIF5A was a pan-neuronal KIF5 and its expression level was lower than that of KIF5C. Indeed, the amount of KIF5A was approximately one-fifth to one-third of that of KIF5C in the cortex, hippocampus, cerebellum, and spinal cord (Fig. 2C). On the other hand, the most abundant KIF5, KIF5B, showed a glial cell pattern in general (Figs. 2-5). These findings collectively indicate the high contribution of KIF5C in motor neurons and the disruption of KIF5C will cause a dramatic decrease of kinesin in these cells. We then generated KIF5C null mice to elucidate its function in the neurons of living animals.
Mouse kif5C gene was disrupted by homologous recombination in ES cells. As shown in Figure6A, a genomic region encoding the ATP-binding motif P-loop was disrupted, and mouse strains derived from two independent ES clones (Fig. 6B) were used for the subsequent analyses. The homozygous offsprings were viable, fertile, and normal in body size, but their brains were slightly smaller than those of wild-type or heterozygous mice (−5% in weight) (Fig. 6C). The complete loss of KIF5C expression in the mutant mice was confirmed by Northern blotting and immunoblotting analysis (Fig. 6D,E).
To study the effect of KIF5C depletion on the expression of the other members of KIF5, we investigated the expression levels of the other KIF5s between the wild-type and mutant mice. First, we examined the gross changes by immunoblotting studies on crude brain extract, but neither KIF5A nor KIF5B expression changed significantly in the KIF5C knock-out mice (Fig. 6E). We then performed immunofluorescence analysis to detect the regional changes in expression. Spinal cords from the wild-type and mutant mice were sectioned at the C7 level. These sections were rich in motor neurons and were stained separately with the anti-KIF5 antibodies (Figs.4c,f,i, 6F). We compared the staining pattern between the wild-type and mutant mice, especially the staining intensity between the motor neurons in the anterior horn and the rest neurons of the spinal cord. Again, the expression of neither KIF5A nor KIF5B showed any apparent increase or decrease in motor neurons. These data indicated that the expression of neither KIF5A nor KIF5B was upregulated in the KIF5C null mutants.
Motor neurons decreased relatively to sensory neurons in KIF5C-deficient mice
For further investigation, we performed histological studies on KIF5C-deficient mice. We prepared frontal brain serial sections of both the wild-type and mutant mice. Samples were silver-stained by the Bodian method (Bodian, 1936) with slight modifications (Fig.6G–I). Mutant mice did not show any gross abnormality in brain structure except for the reduction in brain size (Fig. 6G).
Next, to investigate the effect of the loss of KIF5C on neurons, we compared the number of neurons expressing KIF5C in high or low levels between the wild and mutant adult mice (Fig.6H,I). First, we chose the abducens nucleus (motor neuron) for the example of KIF5C highly expressing neurons, because (1) the nucleus was easily identified and the neurons were clearly distinguished from the others (the nucleus was isolated from other large neurons and the density of the motor neurons was high) and (2) the number of neurons was enough for comparison but not too large to count (∼20 serial sections covered the whole nucleus and the total number of neurons was ∼300) (Figs. 4h,6H). The numbers of the motor neurons in the wild-type and mutant mice were 326 ± 12 and 236 ± 14, respectively, indicating that the neurons were decreased by 28 ± 5% in the mutant mice in this nucleus (n = 4). Next, we counted the number of mesencephalic trigeminal neurons (sensory neuron) for the example of low KIF5C-expressing neurons, because they had large and round cell bodies stained deeply in Bodian staining, and ∼60 sections covered the nucleus (Figs. 4n,s,6I). The numbers of the sensory neurons were 398 ± 23 and 439 ± 31 in the wild-type and mutant mice, respectively (n = 6). The sensory neurons did not decrease in the mutant mice, but rather showed the tendency to increase slightly (110 ± 9%). These data indicate that neurons with high KIF5C expression relatively decreased to the low KIF5C-expressing neurons. Of course, the overall reduction in brain size suggests that loss of KIF5C might affect other populations of neurons as well.
KIF5C in motor neurons was developmentally increased and its prominent expression was found in 2-week-old or older mice (Fig. 5). We then compared the number of neurons in the abducens nucleus between the 1-, 2-, 4-, 8-, and 16-week-old wild-type and mutant mice (Fig.6J). The number of neurons in the mutant mice showed little difference from that in the 1-week-old wild-type mice (98 ± 5%) but was decreased developmentally in those 2 weeks or later after birth (2 weeks, 88 ± 5% to 16 weeks, 72 ± 5%).
KIF5A and KIF5C, in addition to KIF5B, can rescue abnormal distribution of mitochondria in KIF5B-deficient cells
Although KIF5C knock-out mice showed a decrease in the number of motor neurons, they were viable and their brain structure was normal. This means that the fundamental role of KIF5C in most neurons might be taken over by other proteins. The most probable candidates for these proteins are KIF5A and KIF5B. Indeed, (1) KIF5A and KIF5B showed very high degree of homology to KIF5C, especially at the C-terminal coiled-coil region, which is thought to bind to a cargo via KLC and (2) KIF5A was expressed in neurons and KIF5B ubiquitously. To test this hypothesis, we performed a rescue study using culturedkif5B−/− cells (Fig. 7). Our previous study revealed that KIF5B-deficient mice were embryonic lethal, and their cultured cells showed abnormal perinuclear condensation of mitochondria (Tanaka et al., 1998). We thus injected the CMV promoter-driven cDNA of KIF5s or green fluorescent protein (GFP) into cultured mutant cells and investigated the distribution of mitochondria. The mitochondria in kif5B−/−cells and kif5B−/− cells transfected with GFP were aggregated at the cell center. However, cells transfected with KIF5A, KIF5B, and KIF5C showed dispersed distribution of mitochondria, similar to the wild-type control cells. These data strongly suggest that the three KIF5s had functional redundancy among them, at least in the terms of the subcellular mitochondrial localization.
Fig. 7.
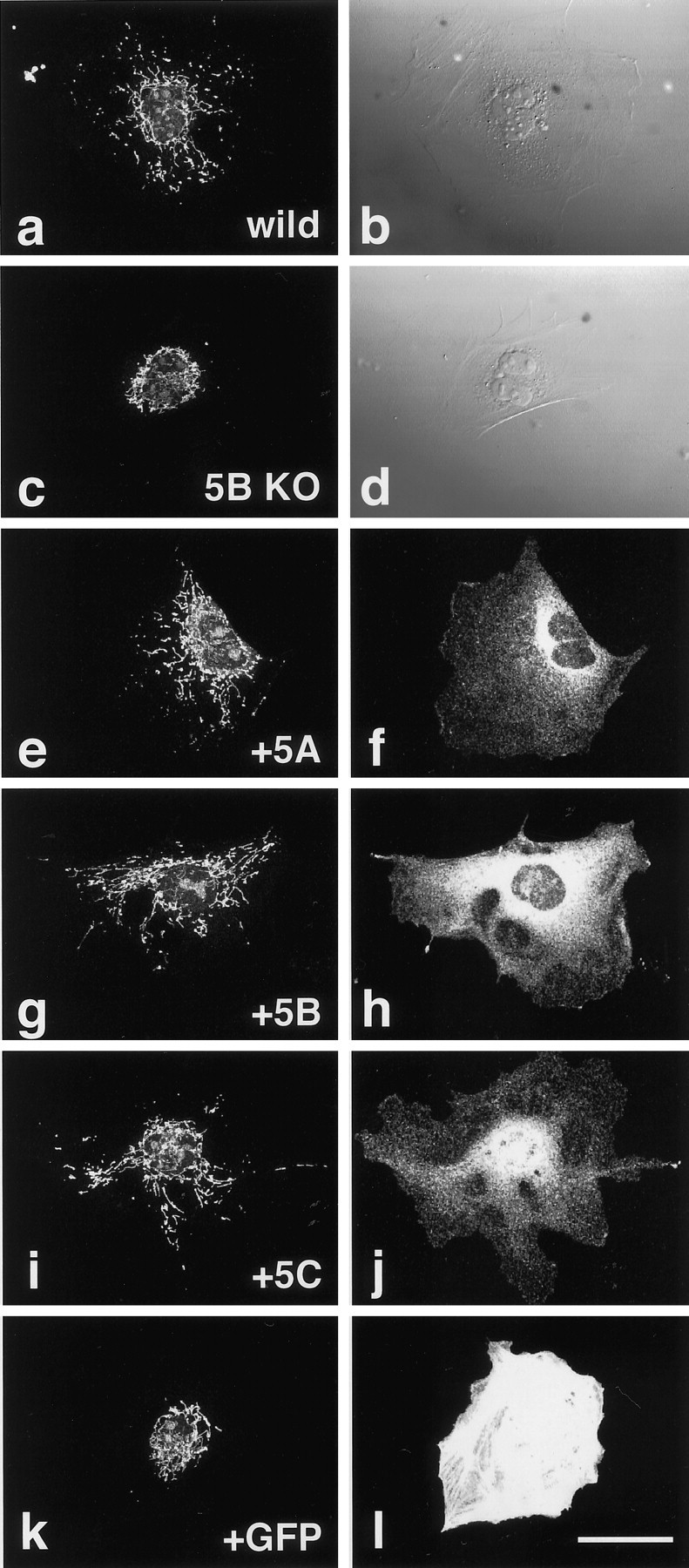
Rescue of abnormal mitochondrial localization in the KIF5B null mutant cells by each of the KIF5s.a, b, Wild-type control.c–l, KIF5B null mutant cells. Cells were injected with the expression vectors of KIF5A (e,f), KIF5B (g,h), KIF5C (i, j), or GFP (k, l). Mitochondrial staining using Mitotracker (a, c,e, g, i,k), Nomarski images (b,d), immunofluorescence after staining with each of the KIF5s (f, h, j), and fluorescence by exogenous GFP (l). Note the recovery of mitochondrial dispersion radiating from the cell center as in wild-type cells after microinjection of KIF5 but not after that of GFP plasmids. Scale bar, 50 μm.
KIF5s can form heterodimers
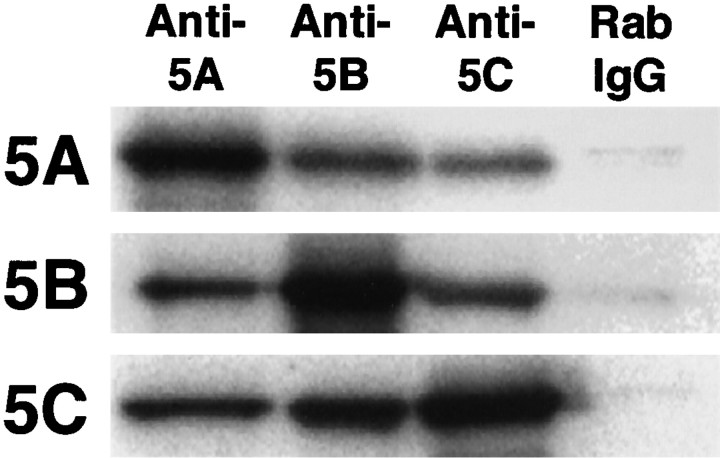
Kinesin consists of two KHCs (KIF5) and two KLCs. In view of the functions of kinesin, it is very important to know whether KIF5s function as homodimers or heterodimers. To answer this question, we performed an immunoprecipitation study on mouse brain crude extract using anti-KIF5 antibodies (Fig. 8). The result showed that the immunoprecipitated sample using one anti-KIF5 antibody also included the other KIF5 proteins to some extent. The control normal rabbit IgG did not precipitate any KIF5. We also performed an immunoprecipitation study using a cultured cell line, Neuro 2A, which endogenously expressed three KIF5s, and obtained the same result (data not shown). These data suggested that KIF5s can also form heterodimers in addition to forming homodimers, and we believe that the functions of the three KIF5s overlap each other.
Fig. 8.
Coimmunoprecipitation of three KIF5s with each of the anti-KIF5 antibodies. Mouse brain crude extracts were immunoprecipitated separately by anti-KIF5 antibodies or normal rabbit IgG (Anti-5A, Anti-5B,Anti-5C, and Rab IgG, respectively) and then blotted with each of the anti-KIF5 antibodies (5A,5B, and 5C, respectively). The sample precipitated by antibodies directed against one KIF5 also included the other KIF5s. The control normal rabbit IgG did not precipitate any KIF5.
DISCUSSION
Different preference of monoclonal anti-kinesin antibodies for each KIF5
Our member-specific anti-KIF5 antibodies revealed an interesting distribution of each KIF5 in the nervous system. In previous kinesin studies, SUK4, H1, and H2 (Ingold et al., 1988; Pfister et al., 1989) have been used widely and were established as the standard anti-kinesin antibodies. However, for the use of these antibodies, the nature of kinesin could not have been elucidated in such great detail, and the significance of these antibodies is truly admirable. However, our study revealed that these monoclonal antibodies had different preferences for each KIF5 (SUK4 predominantly recognized KIF5A; H1 detected only KIF5C; H2 preferred KIF5A and KIF5C to KIF5B) and that the three KIF5s showed different distribution patterns in the nervous system. Although the antibodies were tested only on the KIF5s of mouse origin and their preferences may differ on the samples from other species, our data indicate that one should consider their characteristics in using them. It is suggested that H1 antibody should be used carefully on non-neuronal cells, because H1 did not recognize ubiquitous kinesin in some species [mouse KIF5B, our current data; Xenopusoocyte, Robertson and Allan (2000)], H1 and H2 showed different staining pattern in fibroblasts (Lin et al., 1996), and H1 recognized other microtubule-based motor protein(s) in addition to uKHC in rat liver (Robertson and Allan, 2000).
Roles of the three KIF5s in neurons
KIF5B was expressed both in neuronal and non-neuronal cells (Fig.2A) but showed a glial cell distribution pattern in the nervous system in general in postnatal mice (Figs.2D, 3-5). However, very interestingly, we found high expression of KIF5B in the primary olfactory axons (Figs.2C, 3). It was also highly expressed in the mossy fibers of the hippocampus (Fig. 2D). These neurons are “regenerating,” even in the adult brain (Paxinos, 1995). We observed high expression of KIF5B in cultured hippocampal and spinal cord neurons (data not shown), and previous studies have revealed that the expression of kinesin was upregulated in the developing rat CNS (Vignali et al., 1997) and disruption of KIF5B using antisense oligonucleotides reduced the neurite length and inhibited vesicle transport in cultured hippocampal neurons (Ferreira et al., 1992). These findings collectively suggest that the expression of KIF5B is highly upregulated in neurons at the stage of axonal elongation. Why is KIF5B required for axonal elongation? Although there may be some functional specialization of different kinesin isoforms, one of the possible roles of KIF5B is to convey myosin V to growth cones. Unconventional myosin, myosin-V (Cheney et al., 1993), binds specifically to KIF5B but not to neuronal KIF5 (Huang et al., 1999) and is concentrated in the growth cone and plays important roles in filopodial extension in cultured neurons (Wang et al., 1996). We then consider that a high level of expression of KIF5B, but not KIF5A or KIF5C, may be required to convey myosin-V to the growth cones in neurons during axonal elongation.
KIF5A was expressed evenly among neurons, with similar pattern of distribution to its human homolog, nKHC (Navone et al., 1992; Niclas et al., 1994; Vignali et al., 1996, 1997). On the other hand, expression of KIF5C was highly upregulated, especially in mature lower motor neurons, indicating its important roles in the maintenance of motor neurons rather than in their formation. The length and caliber of the axons vary among neurons. We think that neurons with A-fiber axons (thick axons), e.g., motor neurons, are required to transport a larger amount of vesicles and that the strong expression of KIF5C satisfies this demand. However, considering the facts that the expression level of KIF5C in the primary motor neurons in the cortex was not so high as in the lower motor neurons and that some sensory neurons have A-fiber axons but did not show high expression of KIF5C, it is possible that KIF5C has some functions specialized in the lower motor neurons.
Overlapping functions of the three KIF5s
Our knock-out and rescue studies revealed the functional redundancy among the three KIF5s, and the immunoprecipitation study showed their ability to form heterodimers. Kinesin transports various kinds of cargos (Nakata and Hirokawa, 1995; Khodjakov et al., 1998;Tanaka et al., 1998), and kinesin cargo binding is speculated to be via the KLC (Hirokawa et al., 1989; Gindhart et al., 1998; Khodjakov et al., 1998; Stenoien and Brady, 1998). Indeed, several isoforms of KLC have been identified so far, indicating their important role in cargo selection (Cyr et al., 1991; Beushausen et al., 1993; Cabeza-Arvelaiz et al., 1993; Gauger and Goldstein 1993; Wedaman et al., 1993; Fan and Amos, 1994). Because the KLC-binding regions among the three KIF5s are highly conserved (∼90%) (Fig. 1), we assume that the fundamental functions of each KIF5 overlap, although the KIF5-KLC binding affinities may vary among the combinations.
On the other hand, the three KIF5s showed different patterns of distribution in the CNS. Indeed, only KIF5B binds to myosin-V, even if sequences are highly conserved among them (Huang et al., 1999). Therefore, the possibility of specialized functions for each KIF5 should not be excluded.
KIF5C works for the maintenance of motor neurons rather than for their axon formation
Our previous KIF5B knock-out study revealed the function of kinesins from the point of cargo determination. However, because KIF5B is the only KIF5 in the non-neuronal cells, the null mutants were embryonic lethal, and we could not analyze the effects of the lack of kinesin in neurons. Our current study showed that the prominent expression of KIF5C in motor neurons was found in 2-week-old or older mice and that the number of motor neurons in the KIF5C null mice was decreased after 2 week of age. These indicate that KIF5C works for the maintenance of mature motor neurons rather than for their formation such as axonal elongation. For further investigation, we performed the primary cultures of spinal cord neurons. However, the expression level of KIF5B was high in the cultured neurons, and there was few, if any, difference in the mitochondrial distribution in axons of motor neurons among the wild-type and mutant mice (data not shown). We also performed the electron microscopic studies on spinal cord but could not find apparent changes in the remaining motor neurons (data not shown).
Because the KIF5C null mice were viable but were highly reduced in the amount of kinesin specifically in motor neurons, these mutant mice will be a good tool for investigating the functions of kinesin in neurons in living animals.
Footnotes
This work was supported by a Center of Excellence grant from the Ministry of Education, Science, and Culture of Japan to N.H. We thank Tetsuo Noda (Cancer Institute, Tokyo, Japan) for the valuable suggestions on gene targeting, Hiroshi Hamada (Osaka University, Osaka, Japan) and his colleagues for technical assistance in genomic library construction, Sen Takeda, Junlin Teng, Harukata Miki, Chunjie Zhao, Nobuhisa Onouchi, Hiromi Sato, and Haruyo Fukuda in our laboratory for technical assistance, and the other staffs for their valuable suggestions. The SUK4 developed by Ingold et al. (1988) was obtained from the Developmental Studies Hybridoma Bank maintained by the Department of Pharmacology and Molecular Science, Johns Hopkins University School of Medicine, Baltimore, MD 21205, and the Department of Biological Science, University of Iowa, Iowa City, IA 52242, under Contract NO1-HD-2–3144 from the National Institute of Child Health and Human Development.
Correspondence should be addressed to Dr. Nobutaka Hirokawa, Department of Cell Biology, Graduate School of Medicine, University of Tokyo, 7-3-1, Hongo, Bunkyo-ku, Tokyo 113–0033, Japan. E-mail:hirokawa@m.u-tokyo.ac.jp.
REFERENCES
- 1.Aizawa H, Sekine Y, Takemura R, Zhang Z, Nangaku M, Hirokawa N. Kinesin family in murine central nervous system. J Cell Biol. 1992;119:1287–1296. doi: 10.1083/jcb.119.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beushausen S, Kladakis A, Jaffe H. Kinesin light chains: identification and characterization of a family of proteins from the optic lobe of the squid Loligo pealii. DNA Cell Biol. 1993;12:901–909. doi: 10.1089/dna.1993.12.901. [DOI] [PubMed] [Google Scholar]
- 3.Bodian P. A new method for staining nerve fibers and nerve ending in mounted paraffin sections. Anat Rec. 1936;65:89–96. [Google Scholar]
- 4.Bost-Usinger L, Chen RJ, Hillman D, Park H, Burnside B. Multiple kinesin family members expressed in teleost retina and RPE include a novel C-terminal kinesin. Exp Eye Res. 1997;64:781–794. doi: 10.1006/exer.1996.0271. [DOI] [PubMed] [Google Scholar]
- 5.Brady ST. A novel brain ATPase with properties expected for the fast axonal transport motor. Nature. 1985;317:73–75. doi: 10.1038/317073a0. [DOI] [PubMed] [Google Scholar]
- 6.Cabeza-Arvelaiz Y, Shih L, Hardman N, Asselbergs F, Bilbe G, Schmitz A, White B, Siciliano M, Lachman L. Cloning and genetic characterization of the human kinesin light-chain (KLC) gene. DNA Cell Biol. 1993;12:881–892. doi: 10.1089/dna.1993.12.881. [DOI] [PubMed] [Google Scholar]
- 7.Cheney RE, O'Shea MK, Heuser JE, Coelho MV, Wolenski JS, Espreafico EM, Forscher P, Larson RE, Mooseker MS. Brain Myosin-V is a two headed unconventional Myosin with motor activity. Cell. 1993;75:13–23. doi: 10.1016/S0092-8674(05)80080-7. [DOI] [PubMed] [Google Scholar]
- 8.Cyr J, Pfister K, Bloom G, Slaughter C, Brady S. Molecular genetics of kinesin light chains: generation of isoforms by alternative splicing. Proc Natl Acad Sci USA. 1991;88:10114–10118. doi: 10.1073/pnas.88.22.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan J, Amos L. Kinesin light chain isoforms in Caenorhabditis elegans. J Mol Biol. 1994;240:507–512. doi: 10.1006/jmbi.1994.1465. [DOI] [PubMed] [Google Scholar]
- 10.Ferreira A, Niclas J, Vale R, Banker G, Kosik K. Suppression of kinesin in cultured hippocampal neurons using antisense oligonucleotides. J Cell Biol. 1992;117:595–606. doi: 10.1083/jcb.117.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gauger A, Goldstein L. The Drosophila kinesin light chain. Primary structure and interaction with kinesin heavy chain. J Biol Chem. 1993;268:13657–13666. [PubMed] [Google Scholar]
- 12.Gindhart JG, Desai CJ, Beushausen S, Zinn S, Goldstein LS. Kinesin light chains are essential for axonal transport in Drosophila. J Cell Biol. 1998;141:443–454. doi: 10.1083/jcb.141.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldstein LS. With apologies to scheherazade: tails of 1001 kinesin motors. Annu Rev Genet. 1993;27:319–351. doi: 10.1146/annurev.ge.27.120193.001535. [DOI] [PubMed] [Google Scholar]
- 14.Gudkov AV, Kazarov AR, Thimmapaya R, Axenovich S, Mazo IA, Roninson IB. Cloning mammalian genes by expression selection of genetic suppressor elements: association of kinesin with drug resistance and cell immortalization. Proc Natl Acad Sci USA. 1994;91:3744–3748. doi: 10.1073/pnas.91.9.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harada A, Oguchi K, Okabe S, Kuno J, Terada S, Ohshima T, Sato-Yoshitake R, Takei Y, Noda N, Hirokawa N. Altered microtubule organization in small-calibre axons of mice lacking tau protein. Nature. 1990;369:488–491. doi: 10.1038/369488a0. [DOI] [PubMed] [Google Scholar]
- 16.Hirokawa N. Axonal transport and the cytoskeleton. Curr Opin Neurobiol. 1993;3:724–731. doi: 10.1016/0959-4388(93)90144-n. [DOI] [PubMed] [Google Scholar]
- 17.Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- 18.Hirokawa N, Pfister KK, Yorifuji H, Wagner MC, Brady ST, Bloom GS. Submolecular domains of bovine brain kinesin identified by electron microscopy and monoclonal antibody decoration. Cell. 1989;56:867–878. doi: 10.1016/0092-8674(89)90691-0. [DOI] [PubMed] [Google Scholar]
- 19.Hollenbeck P. The distribution, abundance and subcellular localization of kinesin. J Cell Biol. 1989;108:2335–2342. doi: 10.1083/jcb.108.6.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang JD, Brady ST, Richards BW, Stenoien D, Resau JH, Copeland NG, Jenkins NA. Direct interaction of microtubule- and actin-based transport motors. Nature. 1999;397:267–270. doi: 10.1038/16722. [DOI] [PubMed] [Google Scholar]
- 21.Ingold A, Cohn S, Scholey J. Inhibition of kinesin-driven microtubule motility by monoclonal antibodies to kinesin heavy chains. J Cell Biol. 1988;107:2657–2667. doi: 10.1083/jcb.107.6.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanai Y, Hirokawa N. Sorting mechanisms of tau and MAP2 in neurons: suppressed axonal transit of MAP2 and locally regulated microtubule binding. Neuron. 1995;14:421–432. doi: 10.1016/0896-6273(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 23.Kanai Y, Takemura R, Oshima T, Mori H, Ihara Y, Yanagisawa M, Masaki T, Hirokawa N. Expression of multiple tau isoforms and microtubule bundle formation in fibroblasts transfected with a single tau cDNA. J Cell Biol. 1989;109:1173–1184. doi: 10.1083/jcb.109.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanai Y, Chen J, Hirokawa N. Microtubule bundling by tau proteins in vivo: analysis of the functional domains. EMBO J. 1992;11:3953–3961. doi: 10.1002/j.1460-2075.1992.tb05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato K. Sequential analysis of twenty mouse brain cDNA clones selected by specific expression patterns. Eur J Neurosci. 1991;2:704–711. doi: 10.1111/j.1460-9568.1990.tb00460.x. [DOI] [PubMed] [Google Scholar]
- 26.Khodjakov A, Lizunova EM, Minin AA, Koonce MP, Gyoeva FK. A specific light chain of kinesin associates with mitochondria in cultured cells. Mol Biol Cell. 1998;9:333–343. doi: 10.1091/mbc.9.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kondo S, Sato-Yoshitake R, Noda Y, Aizawa H, Nakata T, Matsuura Y, Hirokawa N. KIF3A is a new microtubule-based anterograde motor in the nerve axon. J Cell Biol. 1994;125:1095–1107. doi: 10.1083/jcb.125.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kosik KS, Orecchio LD, Schnapp B, Inouye H, Neve RL. The primary structure and analysis of the squid kinesin heavy chain. J Biol Chem. 1990;265:3278–3283. [PubMed] [Google Scholar]
- 29.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin SX, Pfister KK, Collins CA. Comparison of the intracellular distribution of cytoplasmic dynein and kinesin in cultured cells: motor protein localization does not reliably predict function. Cell Motil Cytoskel. 1996;34:299–312. doi: 10.1002/(SICI)1097-0169(1996)34:4<299::AID-CM5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 31.Meng Y, Wilson G, Avery M, Varden C, Balczon R. Suppression of the expression of a pancreatic beta-cell form of the kinesin heavy chain by antisense oligonucleotides inhibits insulin secretion from primary cultures of mouse beta-cells. Endocrinology. 1997;138:1979–1987. doi: 10.1210/endo.138.5.5139. [DOI] [PubMed] [Google Scholar]
- 32.Nakagawa T, Tanaka Y, Matsuoka E, Kondo S, Okada Y, Noda Y, Kanai Y, Hirokawa N. Identification and classification of 16 new kinesin superfamily (KIF) proteins in mouse genome. Proc Natl Acad Sci USA. 1997;94:9654–9659. doi: 10.1073/pnas.94.18.9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakata T, Hirokawa N. Point mutation of adenosine triphosphate-binding motif generated rigor kinesin that selectively blocks anterograde lysosome membrane transport. J Cell Biol. 1995;131:1039–1053. doi: 10.1083/jcb.131.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nangaku M, Sato-Yoshitake R, Okada Y, Noda Y, Takemura R, Yamazaki H, Hirokawa N. KIF1B: a new microtubule plus-end directed monomeric motor for mitochondria transport. Cell. 1994;39:1209–1220. doi: 10.1016/0092-8674(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 35.Navone F, Niclas J, Hom-Booher N, Sparks L, Bernstein H, McCaffrey G, Vale R. Cloning and expression of a human kinesin heavy chain gene: interaction of the COOH-terminal domain with cytoplasmic microtubules in transfected CV-1 cells. J Cell Biol. 1992;117:1263–1275. doi: 10.1083/jcb.117.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niclas J, Navone F, Hom-Booher N, Vale R. Cloning and localization of a conventional kinesin motor expressed exclusively in neurons. Neuron. 1994;12:1059–1072. doi: 10.1016/0896-6273(94)90314-x. [DOI] [PubMed] [Google Scholar]
- 37.Noda Y, Sato-Yoshitake R, Kondo S, Nangaku M, Hirokawa N. KIF2 is a new anterograde microtubule based motor which transports membranous organelles distinct form those carried by KHC or KIF3A/B. J Cell Biol. 1995;129:157–167. doi: 10.1083/jcb.129.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okada Y, Yamazaki H, Sekine A, Hirokawa N. The neuron-specific kinesin superfamily KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell. 1995;81:769–780. doi: 10.1016/0092-8674(95)90538-3. [DOI] [PubMed] [Google Scholar]
- 39.Patel N, Thierry-Mieg D, Mancillas JR. Cloning by insertional mutagenesis of a cDNA encoding Caenorhabditis elegans kinesin heavy chain. Proc Natl Acad Sci USA. 1993;90:9181–9185. doi: 10.1073/pnas.90.19.9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paxinos G. The rat nervous system, Edition 2. Academic; San Diego: 1995. [Google Scholar]
- 41.Pfister K, Wagner M, Stenoien D, Brady S, Bloom G. Monoclonal antibodies to kinesin heavy and light chains stain vesicle-like structures, but not microtubules, in cultured cells. J Cell Biol. 1989;108:1453–1463. doi: 10.1083/jcb.108.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robertson A, Allan V. Brefeldin A-dependent membrane tubule formation reconstituted in vitro is driven by a cell-cycle-regulated microtubule motor. Mol Biol Cell. 2000;11:941–955. doi: 10.1091/mbc.11.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saito N, Okada Y, Noda Y, Kinoshita Y, Kondo S, Hirokawa N. KIFC2 is a novel neuron-specific C-terminal type kinesin superfamily motor for dendritic transport of multivesicular body-like organelles. Neuron. 1997;18:425–438. doi: 10.1016/s0896-6273(00)81243-x. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory; New York: 1989. [Google Scholar]
- 45.Sato K, Ito R, Baek KH, Agarwal K. A specific DNA sequence controls termination of transcription in the gastrin gene. Mol Cell Biol. 1986;6:1032–1043. doi: 10.1128/mcb.6.4.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scholey JM, Heuser J, Yang JT, Goldstein LS. Identification of globular mechanochemical heads of kinesin. Nature. 1989;338:355–357. doi: 10.1038/338355a0. [DOI] [PubMed] [Google Scholar]
- 47.Sekine Y, Okada Y, Noda Y, Kondo S, Aizawa H, Takemura R, Hirokawa N. A novel microtubule-based motor protein (KIF4) for organelle transports whose expression is regulated developmentally. J Cell Biol. 1994;127:187–201. doi: 10.1083/jcb.127.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stenoien DL, Brady ST. Immunochemical analysis of kinesin light chain function. Mol Biol Cell. 1998;8:675–689. doi: 10.1091/mbc.8.4.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takei Y, Harada A, Takeda S, Kobayashi K, Terada S, Noda T, Takahashi T, Hirokawa N. Synapsin I deficiency results in the structural change in the presynaptic terminals in the murine nervous system. J Cell Biol. 1995;131:1789–1800. doi: 10.1083/jcb.131.6.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanaka Y, Kanai Y, Okada Y, Nonaka S, Takeda S, Harada A, Hirokawa N. Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell. 1998;93:1147–1158. doi: 10.1016/s0092-8674(00)81459-2. [DOI] [PubMed] [Google Scholar]
- 51.Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vignali G, Niclas J, Sprocati MT, Vale RD, Sirtori C, Navone F. Differential expression of ubiquitous and neuronal kinesin heavy chains during differentiation of human neuroblastoma and PC12 cells. Eur J Neurosci. 1996;8:536–544. doi: 10.1111/j.1460-9568.1996.tb01238.x. [DOI] [PubMed] [Google Scholar]
- 53.Vignali G, Lizier C, Sprocati M, Sirtori C, Battaglia G, Navone F. Expression of neuronal kinesin heavy chain is developmentally regulated in the central nervous system of the rat. J Neurochem. 1997;69:1840–1849. doi: 10.1046/j.1471-4159.1997.69051840.x. [DOI] [PubMed] [Google Scholar]
- 54.Wang FS, Wolenski JS, Cheney RE, Mooseker MS, Jay DG. Function of Myosin-V in filopodial extension of neuronal growth cones. Science. 1996;273:660–663. doi: 10.1126/science.273.5275.660. [DOI] [PubMed] [Google Scholar]
- 55.Watanabe A, Tanaka K, Shimura Y. The role of exon sequences in splice site selection. Genes Dev. 1993;7:403–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- 56.Wedaman K, Knight A, Kendrick-Jones J, Scholey J. Sequences of sea urchin kinesin light chain isoforms. J Mol Biol. 1993;231:155–158. doi: 10.1006/jmbi.1993.1267. [DOI] [PubMed] [Google Scholar]
- 57.Wright BD, Terasaki M, Scholey JM. Roles of kinesin and kinesin-like proteins in sea urchin embryonic cell division: evaluation using antibody microinjection. J Cell Biol. 1993;123:681–689. doi: 10.1083/jcb.123.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yagi T, Nada S, Watanabe N, Tamemoto H, Kohmura N, Ikawa Y, Aizawa S. A novel negative selection for homologous recombinants using diphtheria toxin A fragment gene. Anal Biochem. 1993;214:77–86. doi: 10.1006/abio.1993.1459. [DOI] [PubMed] [Google Scholar]
- 59.Yamazaki H, Nakata T, Okada Y, Hirokawa N. KIF3A/B: a new heterodimer that works as microtubule-plus-end directed motor for fast axonal transport. J Cell Biol. 1995;130:1387–1400. doi: 10.1083/jcb.130.6.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang JT, Saxton WM, Goldstein LS. Isolation and characterization of the gene encoding the heavy chain of Drosophila kinesin. Proc Natl Acad Sci USA. 1988;85:1864–1868. doi: 10.1073/pnas.85.6.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang Z, Hanlon DW, Marszalek JR, Goldstein LSB. Identification, partial characterization, and genetic mapping of kinesin-like protein genes in mouse. Genomics. 1997;45:123–131. doi: 10.1006/geno.1997.4901. [DOI] [PubMed] [Google Scholar]