Abstract
The histone demethylase KDM6B (JMJD3) is upregulated in blood disorders, suggesting it may have important pathogenic functions. Here we examined the function of Kdm6b in hematopoietic stem cells (HSC) to evaluate its potential as a therapeutic target. Loss of Kdm6b lead to depletion of phenotypic and functional HSCs in adult mice, and Kdm6b is necessary for HSC self-renewal in response to inflammatory and proliferative stress. Loss of Kdm6b leads to a pro-differentiation poised state in HSCs due to the increased expression of the AP-1 transcription factor complex (Fos and Jun) and immediate early response (IER) genes. These gene expression changes occurred independently of chromatin modifications. Targeting AP-1 restored function of Kdm6b-deficient HSCs, suggesting Kdm6b regulates this complex during HSC stress response. We also show Kdm6b supports developmental context-dependent leukemogenesis for T-cell acute lymphoblastic leukemia (T-ALL) and M5 acute myeloid leukemia (AML). Kdm6b is required for effective fetal-derived T-ALL and adult-derived AML, but not vice versa. These studies identify a crucial role for Kdm6b in regulating HSC self-renewal in different contexts, and highlight the potential of KDM6B as a therapeutic target in different hematopoietic malignancies.
Keywords: Hematopoietic stem cell, epigenetics, cancer stem cell, self-renewal, AP-1
INTRODUCTION
KDM6B (JMJD3) is one of two epigenetic modifiers responsible for enzymatic removal of the repressive chromatin mark histone H3 lysine 27 trimethylation (H3K27me3)1. H3K27me3 is associated with transcriptional repression and gene silencing2. Demethylation of H3K27me3 by either KDM6B or UTX1 (KDM6A) is required for resolution of bivalent chromatin domains, lineage-specific gene expression and differentiation in embryonic stem cells3–6. In somatic cells, KDM6B plays a role in stress response in macrophages7–9, hippocampal neurons10, and fibroblasts11, 12, and may have demethylase-independent functions in response to different physiological perturbations7, 13. However, most of these studies have been performed in cell lines. As these molecular functions of epigenetic regulators are highly cell type-dependent, analysis of purified populations is required to deconvolute specific activities in primary cells.
Genome sequencing of hematopoietic malignancies has identified somatic mutations in many epigenetic modifiers14–16. While UTX1 mutations are recurrent in blood cancers16, 17, KDM6B mutations have not been identified apart from chromosomal deletions in Sezary syndrome18. In contrast, KDM6B is over-expressed in a myriad of blood disorders including myelodysplastic syndromes (MDS)19, Hodgkin’s lymphoma (HL)20, multiple myeloma (MM)13, and T-cell acute lymphoblastic leukemia (T-ALL)21. KDM6B seems to have opposing roles in leukemia development dependent on morphological subtype and genetic background. While UTX1 acts as a tumor suppressor in NOTCH1-driven T-ALL, KDM6B supports this leukemia by activating oncogenic gene expression21. In contrast, KDM6B has oncorepressor activity M2/M3 AML (granulocytic subtypes). Downregulation of KDM6B in these AMLs results in differentiation arrest and correlates with poor clinical outcomes22. The role of KDM6B in hematopoietic development must be defined to understand the mechanisms through which it acts in different leukemia subtypes.
A recent study showed constitutive over-expression of Kdm6b in the hematopoietic system leads to upregulation of genes involved in innate immune signaling, resulting in compromised hematopoiesis and pathologies reminiscent of human MDS23. But the function of Kdm6b specifically in hematopoietic stem cells (HSCs) has not been studied. Here, we developed a conditional genetic mouse model to study Kdm6b in hematopoietic development, HSC fate decisions and leukemogenesis. We show that loss of Kdm6b leads to the inability for HSCs to self-renew following proliferative stress, driven in part by H3K27me3-independent dysregulation of the AP-1 transcription factor complex. We also show that Kdm6b supports context-dependent leukemia initiation and maintenance, conditional upon disease subtype and developmental age.
METHODS
Mice and Transplantation
All animal procedures were approved by the Institutional Animal Care and Use Committee at Washington University. All mice were C57Bl/6 background. Kdm6bfl/fl24 and Utx1fl/fl 25 mice were crossed to Vav-CRE26, Mx1-CRE27 or ERT2-CRE28 strains. Six doses (300 μg/mouse) of polyinosinic:polycytidylic acid (pIpC; Sigma #p1530) were administered every other day via intraperitoneal injection to induce Mx1-CRE. Nine doses (4 mg) of Tamoxifen (Sigma #T5648) in corn oil (Sigma #C8267) were administered via oral gavage to induce ERT2-CRE. For transplantation, recipient mice (CD45.1) were given a split dose (~4-hours apart) of lethal irradiation (10.5 Gy). For secondary leukemia transplants, recipient mice were given a sublethal dose (6.0 Gy) of irradiation.
Flow Cytometry
Samples were incubated at 4°C for >20 minutes with appropriate antibodies. Bone marrow was enriched with anti-mouse CD117-conjugated microbeads (Miltenyi Biotec #130-091-224) prior to HSC purification. Donor cell chimerism (CD45.1 vs. CD45.2) and lineage contribution was determined by assessing myeloid (Gr-1+ Mac-1+), B-cell (B220+) and T-cell (CD3e+) populations in peripheral blood. For intracellular flow cytometry, samples were stained for surface markers overnight at 4°C, processed using the Cytofix/Cytoperm Kit (BD Biosciences #554714), then stained with intracellular antibodies for 20 minutes at room temperature.
Plasmids and Viral Transduction
293T (ATCC #CRL-3216) cells were co-transfected with packaging vector (pCL-Eco) and either empty vector control (MIG-GFP), MIG-NICD or MIG-MLL-AF9 using Lipofectamine 3000 (ThermoFisher Scientific #L3000008). 1.0×106 cells/500uL were plated in Stempro-34 medium (Gibco #10639011) supplemented with Pen-Strep (100 Units/mL), L-glutamine (2 mM), mSCF (100 ng/mL), mTPO (100ng/mL), mFlt3L (50ng/mL), mIL-3 (5 ng/mL), and polybrene (4 μg/mL; Sigma), and spinfected with retrovirus supernatant at 250g for two-hours.
RNA-SEQ
RNA was isolated using the NucleoSpin RNA XS kit (Macherey-Nagel #740902.250). The SMARTer Ultra Low RNA kit (Clontech) was used to prepare libraries. Sequencing was performed with an Illumina HiSeq-3000. RNA-seq reads were aligned to the Ensembl release 76 top-level assembly with STAR version 2.0.4b29. Gene counts were derived from the number of uniquely aligned reads by Subread:featureCount version 1.4.530, 31. Transcript counts were generated by Sailfish version 0.6.332. Gene set enrichment analysis (GSEA)33 was performed to identify dysregulated genesets. Primary data is available under GEO accession number GSE110378.
ChIP-mentation
ChIP-mentation was performed as previously described34. Chromatin was sheared using a Covaris E220 Focused-ultrasonicator and incubated overnight at 4°C with H3K27me3 antibody (Diagenode #pAb-069–050) or H3K4me3 antibody (Diagenode #pAb-003–050). Libraries were sequenced on an Illumina Hiseq 3000. Sequences were aligned to mm10 using Bowtie235. Peak calling was performed with hiddenDomains36, the R package ChIPQC37 was used for quality control including PCA plots. HOMER plots for H3K27me3 and H3K4me3 were made using deepTools38. BEDOPS39 was used to determine overlapping and unique peaks. Primary data is available under GEO accession number GSE110378.
ATAC-seq
Using a modified Omni-ATAC method40, 10,000 HSCs were sorted and the transposition reaction was carried out in 2x TD/Transposase buffer with transposase (Nextera) for 30 minutes at 37°C. Libraries were generated using NEBnext (NewEngland Biolabs) and run on an Illumina Hiseq 3000. Primary data is available under GEO accession number GSE110378.
Statistics
Student t-test, one-way, and two-way ANOVA’s were used for statistical comparisons where appropriate. Survival curves were analyzed using a Mantel-Cox logrank test. Significance is indicated using the following convention: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. All graphs represent mean ± S.E.M.
RESULTS
Loss of Kdm6b results in depletion of primitive hematopoietic progenitors
A conditional knockout mouse was generated by crossing the Vav-CRE driver26 to delete floxed exons 14–2024 of Kdm6b in hematopoietic cells. Complete floxed allele recombination was observed in HSCs (Lineage- Sca-1+ c-Kit+ CD48- CD150+ EPCR+; Supplementary Figure 1a). Complete protein ablation was observed in thymocytes (Supplementary Figure 1b), but global H3K27me3 was unchanged (Supplementary Figure 1c). Whole bone marrow (WBM) cellularity (Supplementary Figure 1d) and spleen weight (Supplementary Figure 1e) were unchanged in young adult (eight-week old) Vav-CRE:Kdm6bfl/+ (Kdm6b-HetVAV) and Vav-CRE:Kdm6bfl/fl (Kdm6b-KOVAV) mice. Both heterozygous- and homozygous mice showed increased B-cells and reduced myeloid cells in the blood (Supplementary Figure 1f) compared to control mice (Vav-CRE:Kdm6b+/+ = ControlVav). No hematological disease was observed in mutant mice by 80-weeks of age and blood counts remained relatively normal (Supplementary Figure 1g–k).
Flow cytometry analysis (Figure 1a) revealed slightly reduced frequency and absolute number of HSCs and multipotent progenitor cell (MPPs; Lineage- EPCR+ c-Kit+ Sca-1+ CD48- CD150-) in young adult Kdm6b-KOVAV mice (Figure 1b). This depletion of HSCs and MPPs was magnified in aged mice (Figure 1c). Colony-forming units were reduced in Kdm6b-KOVAV WBM (Figure 1d) and serial replating of purified heterozygous- and homozygous HSCs was defective (Figure 1e). Detailed analysis revealed minimal differences in myeloid (Supplementary Figure 2a–c), lymphoid (Supplementary Figure 2d–f), B-cell (Supplementary Figure 2g–i) and erythroid (Supplementary Figure 2j–l) progenitors with age. In agreement with previous studies41, there was increased CD4+ and CD8a+ cells in the thymus of Kdm6b-KOVAV mice (Supplementary Figure 2m–o).
Figure 1: Loss of Kdm6b results in depletion of primitive hematopoietic progenitors.
(a) Flow cytometry gating scheme to identify hematopoietic stem cells (HSCs) and multipotent progenitors (MPPs) in bone marrow (BM) of eight-week old ControlVAV (top) or Kdm6b-KOVAV mice (bottom). (b) Frequency and absolute number of HSCs and MPPs in BM of eight-week old ControlVAV (CNT, n=14), Kdm6b-HetVAV (HET, n=18), and Kdm6b-KOVAV (KO, n=16) mice. (c) Frequency and absolute number of HSCs and MPPs in BM of 80-week old ControlVAV (CNT, n=12), Kdm6b-HetVAV (HET, n=9), and Kdm6b-KOVAV (KO, n=14) mice. (d) Colony-forming units per 1×104 BM cells from young ControlVAV (n=6), Kdm6b-HetVAV (n=6) and Kdm6b-KOVAV (n=6) mice. (e) Colonies generated in Methocult serial replating. For plate 1, 100 HSCs were sorted from individual young mice (n=5–9), then 1×104 cells were plated for serial rounds. *p<0.05, **p<0.01, ***p<0.001, ****p<0.001. Mean ± S.E.M. values are shown.
Kdm6b is required for HSC self-renewal
To determine if loss of phenotypic HSCs in Kdm6b-KOVAV mice correlated with functional decline, limiting dilutions of WBM from ControlVAV, Kdm6b-HetVAV and Kdm6b-KOVAV mice were competitively transplanted. Recipients were classified as long-term multi-lineage reconstituted (LTMR) if B-cell, T-cell and myeloid blood lineages had >1% donor-derived engraftment at 16-weeks post-transplant (Figure 2a). The proportion of LTMR mice at each dose was used to calculate long-term repopulating cell frequency42. While ControlVAV and Kdm6b-HetVAV mice had a comparable frequency of long-term repopulating cells (1:49,984 and 1:55,422 respectively), Kdm6b-KOVAV WBM was estimated to have a two-fold reduction (1:108,101; p=0.054, Figure 2b). Kdm6b-KOVAV HSCs were reduced in recipient mice 18-weeks post-transplant (Figure 2c). To assess self-renewal, 3.0×106 WBM from LTMR mice was transferred to secondary recipients. Kdm6b-KOVAV secondary recipients had no long-term engraftment (Figure 2d). While overall engraftment was equivalent between ControlVAV and Kdm6b-HetVAV recipients, only 37.5% of Kdm6b-HetVAV recipient mice were LTMR compared to 83.3% of ControlVAV recipients (Figure 2e). Post-secondary transplant, Kdm6b-HetVAV HSCs were reduced in recipient bone marrow (Figure 2f).
Figure 2: Kdm6b is required for HSC self-renewal.
(a) Proportion of long-term multi-lineage reconstituted (LTMR) recipient mice at stated cell doses 16-weeks post-transplant (n=9–10 per genotype at each cell dose). (b) Frequency of long-term repopulating cells using a maximum likelihood estimate with extreme limiting dilution analysis (ELDA) software. (c) Frequency of donor-derived (CD45.2+) HSCs in BM of recipient mice 18-weeks post-transplant. (d) Peripheral blood engraftment in secondary transplants of 3.0×106 BM from LTMR primary recipient mice (n=6–8). (e) Proportion of LTMR secondary recipient mice transplanted with ControlVAV (n=6), Kdm6b-HetVAV (n=8), and Kdm6b-KOVAV (n=6) primary BM. (f) Frequency of donor-derived HSCs in BM of secondary recipient mice 18-weeks post-transplant. (g) Donor-derived peripheral blood engraftment from primary transplant of 200 HSCs from ControlVAV (n=10), Kdm6b-HetVAV (n=9), and Kdm6b-KOVAV (n=9) mice. (h) Donor-derived chimerism in blood lineages 16-weeks post-transplant of 200 HSCs. (i) Frequency of donor-derived HSCs in BM of recipient mice 18-weeks post-transplant of 200 HSCs. (j) Donor-derived peripheral blood engraftment from competitive transplant of BM from aged ControlVAV (n=3), Kdm6b-HetVAV (n=4), and Kdm6b-KOVAV (n=3) mice. (k) Donor-derived chimerism in blood lineages 16-weeks post-transplant of aged BM. (l) Frequency of donor-derived HSCs in recipient mice 18-weeks post-transplant of aged BM. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Mean ± S.E.M. values are shown.
To examine Kdm6b function specifically in HSCs, 200 HSCs were purified from ControlVAV, Kdm6b-HetVAV and Kdm6b-KOVAV mice and competitively transplanted. While initial engraftment was comparable, subsequent timepoints showed significantly decreased contribution from Kdm6b-KOVAV HSCs (Figure 2g). Kdm6b-HetVAV and Kdm6b-KOVAV HSCs contributed equally to all three blood lineages (Figure 2h). 18-weeks post-transplant, 200 HSCs from primary recipients were re-purified and transplanted into secondary recipients. The severe reduction in donor-derived HSCs (Figure 2i) in Kdm6b-KOVAV recipients precluded secondary transfer. Secondary transplant saw a mild reduction in donor-derived chimerism in Kdm6b-HetVAV recipients (Supplementary Figure 3a) across all blood lineages (Supplementary Figure 3b), but post-secondary transplant there was a significant reduction in Kdm6b-HetVAV donor-derived HSCs (Supplementary Figure 3c). The engraftment deficiency from Kdm6b-mutant mice could not be attributed to homing defects (Supplementary Figure 3d). To examine effect of age, 5.0×105 WBM from old (80-week) ControlVAV, Kdm6b-HetVAV, or Kdm6b-KOVAV mice was transplanted with 2.5×105 wild-type WBM competitor cells. Aged WBM from Kdm6b-HetVAV and Kdm6b-KOVAV mice showed a significant reduction in blood chimerism (Figure 2j), lymphoid engraftment (Figure 2k), and HSC regeneration (Figure 2l).
Kdm6b loss leads to a stress response gene expression signature in HSCs independent of chromatin changes
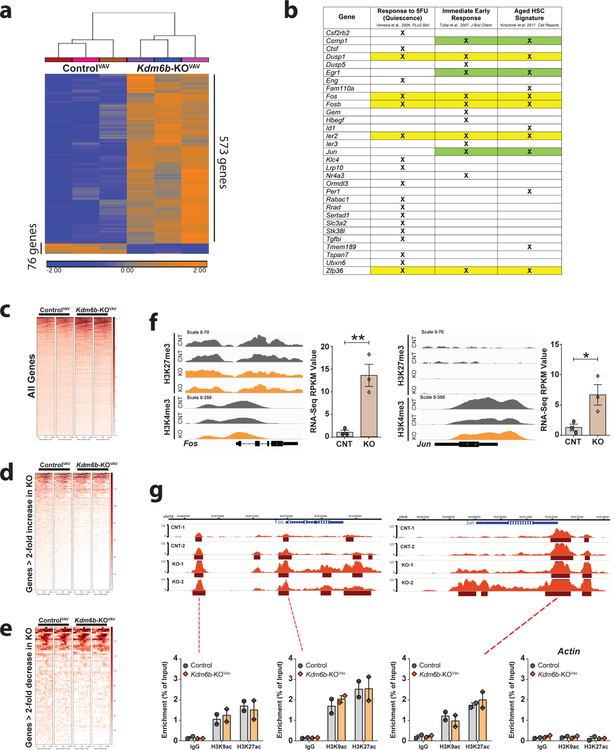
To elucidate mechanisms underlying self-renewal defect of Kdm6b-KOVAV HSCs, global transcriptomic analysis was performed. Comparison of RNA-seq profiles of ControlVAV and Kdm6b-KOVAV HSCs to define genes with >2-fold increased or decreased expression (adjusted p-value <0.05) identified 649 genes (Supplementary Table 1). The majority (88.3%) of these genes showed increased expression in Kdm6b-KOVAV HSCs (Figure 3a). Gene Set Enrichment Analysis (GSEA) identified several inflammatory response pathways upregulated in Kdm6b-KOVAV HSCs (Supplementary Figure 4a). The expression profile of Kdm6b-KOVAV HSCs overlapped with genesets enriched in aged HSCs (p=3.8655×10−8)43 and immediate early response (IER) genes (p=1.6495×10−9)44. Genes downregulated in HSCs in response to 5-Flurouracil (5-FU)45 were significantly upregulated in Kdm6b-KOVAV HSCs (p=0.000295; Figure 3b), suggesting loss of Kdm6b results in a state whereby HSCs are primed for differentiation. Of the overlapping genes, the IER master transcription factor AP-1 subunits Jun and Fos were consistently identified (Figure 3b).
Figure 3: Kdm6b loss leads to a stress response gene expression signature in HSCs independent of chromatin changes.
(a) Hierarchical clustering of genes with >2-fold expression change (adjusted p-value <0.05) in Kdm6b-KOVAV HSCs compared to ControlVAV HSCs. (b) Overlap of genes upregulated in Kdm6b-KOVAV HSCs which are downregulated in wild-type HSCs in response to 5-FU, immediate early response genes, and genes upregulated in aging HSCs. Yellow shading signifies genes identified in all comparisons. H3K27me3 distribution ± 5Kb from transcriptional start sites (TSS) in all genes (c), for 573 genes with >2-fold increased expression in Kdm6b-KOVAV HSCs (d), and for 76 genes with >2-fold decreased expression in Kdm6b-KOVAV HSCs (e). (f) H3K27me3 and H3K4me3 profiles (left) and RNA-seq expression values (right) of Fos and Jun in ControlVAV HSCs and Kdm6b-KOVAV HSCs. *p<0.05, **p<0.01. Mean ± S.E.M. values are shown. (g) ATAC-seq profiles of Fos and Jun in control (CNT) and Kdm6b-KOVav (KO) HSCs. Shown underneath is enrichment of other activating histone marks at relevant sites of open chromatin. Actin is shown as a negative control locus for ChIP-qPCR.
ChIP-mentation34 for H3K27me3 and H3K4me3 was performed on ControlVAV and Kdm6b-KOVAV HSCs to determine if gene expression differences were associated with chromatin changes. H3K4me3 peaks were highly overlapping (Supplementary Figure 4b) with 99.98% of the 16,163 peaks within 5kB of a transcriptional start site conserved between ControlVAV and Kdm6b-KOVAV HSCs (Supplementary Figure 4c–e). Similarly, of the 6,232 H3K27me3 domains within 5kB of a transcriptional start site, 99.94% were conserved between genotypes (Figure 3c). No differentially expressed genes in Kdm6b-KOVAV HSCs were associated with changes in chromatin methylation (Figure 3d,e), exemplified by Fos and Jun (Figure 3f). ATAC-seq was performed on ControlVAV and Kdm6b-KOVAV HSCs, but there were also minimal differences in chromatin accessibility with only 1,305 peaks enriched in ControlVav HSCs compared to 91,276 total peaks (1.43%; Supplementary Figure 4f). Similarly, only 1,688/81,203 ATAC-seq peaks (2.08%) were enriched in Kdm6b-KOVAV HSCs (Supplementary Figure 4g; Supplementary Table 2). Interestingly, one of the few loci that showed a change in chromatin accessibility in Kdm6b-KOVAV HSCs encompassed Fos (Figure 3g). The Jun locus also appeared more open in Kdm6b-KOVav HSCs although not statistically significant (Figure 3g). But like other results, the open chromatin at these loci in the mutant HSCs was not associated with increased marks of active chromatin like H3K9ac and H3K27ac (Figure 3g).
Inflammatory stress rapidly depletes Kdm6b-deficient HSCs
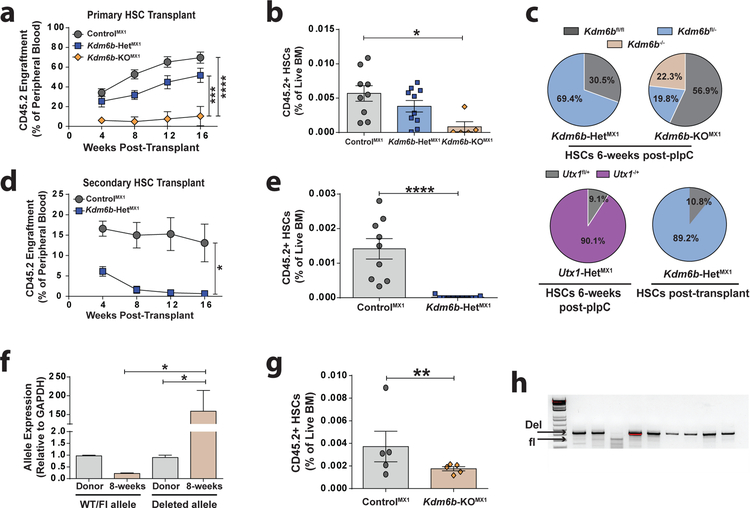
Gene expression analysis of Kdm6b-KOVAV HSCs enlightened a previous result. To initially study Kdm6b in HSCs, Kdm6bfl/fl mice were crossed to the Mx1-CRE driver27. Mx1-CRE is induced in hematopoietic cells by polyinosinic:polycytidylic (pIpC) acid, an immunostiumulant that elicits an IFNα response46. Mx1-CRE:Kdm6b+/+ (ControlMX1), Mx1-CRE:Kdm6bfl/+ (Kdm6b-HetMX1), and Mx1-CRE:Kdm6bfl/fl (Kdm6b-KOMX1) mice were treated with six-doses of pIpC, allowed to recover for six-weeks, then 200 HSCs were competitively transplanted. Kdm6b-KOMX1 HSCs showed severely compromised engraftment (Figure 4a) and HSC regeneration (Figure 4b). At the time of transplant, individual HSCs were concurrently sorted to determine floxing efficiency. 56.9% of Kdm6b-KOMX1 HSCs retained both floxed alleles, and only 22.3% of HSCs had deletion of both Kdm6b alleles (Figure 4c). While efficiency of recombination in Kdm6b-HetMX1 HSCs was higher, 30.5% were still intact (Figure 4c). To determine if this CRE-deficiency was specific to the Kdm6b allele, floxing efficiency of Mx1-CRE:Utx1fl/+ (Utx1-HetMX1) HSCs was assessed25. Only 9.1% of Utx1-HetMX1 HSCs were not recombined six-weeks post-pIpC (Figure 4c). Post-transplant, only 10.8% of Kdm6b-HetMX1 HSCs retained unrecombined alleles (Figure 4c). We were unable to recover Kdm6b-KOMX1 HSCs post-transplant to assess floxing efficiency. While primary Kdm6b-HetMX1 recipients showed no engraftment defects, secondary transplantation showed a significant decrease (Figure 4d). 18-weeks post-secondary transplant, donor-derived HSCs were virtually undetectable in Kdm6b-HetMX1 recipients (Figure 4e).
Figure 4: Inflammatory stress rapidly depletes Kdm6b-deficient HSCs.
(a) Peripheral blood engraftment of mice transplanted with 200 HSCs from ControlMX1 (n=9), Kdm6b-HETMX1 (n=10), and Kdm6b-KOMX1 (n=5) mice. (b) Frequency of donor-derived HSCs in BM of primary recipient mice at 18-weeks post-transplant. (c) Efficiency of Mx1-CRE driven floxed allele recombination in HSCs from mice six-weeks post-pIpC treatment, as well as post-primary transplant in Kdm6b-HETMX1 HSCs. (d) Peripheral blood engraftment of secondary recipients transplanted with 200 ControlMX1 (n=9) and Kdm6b-HetMX1 (n=11) HSCs from primary recipients. (e) Frequency of donor-derived HSCs in BM of secondary recipient mice 18-weeks post-transplant. (f) qPCR quantification of floxed:deleted Kdm6b allele ratio in Kdm6b-KOMX1 granulocytes pre- and post-pIpC treatment. (g) Donor-derived HSC frequency in the bone marrow of recipient mice two-weeks post-pIpC treatment. (h) Representative PCR analysis of genomic DNA from colonies derived from single HSCs showing floxed allele recombination efficiency in individual Kdm6b-KOMX1 HSCs two-weeks post-pIpC. *p<0.05, **p<0.01, ***p<0.001, ****p<0.001. Mean ± S.E.M. values are shown.
To determine why Kdm6b-deficient HSCs were rapidly depleted in response to pIpC, 200 HSCs from untreated Mx1-CRE:Kdm6b+/+ and Mx1-CRE:Kdm6bfl/fl mice were transplanted, then treated with pIpC four-weeks post-transplant. Granulocytes were purified from donor mice pre-transplant, and compared to donor-derived granulocytes from recipient mice to examine floxing efficiency. As granulocytes have a very short half-life, their generation reflects the output of active HSCs. Analysis of genomic DNA indicated granulocytes two-weeks post-pIpC were derived from efficiently recombined Kdm6b-KOMX1 HSCs (Figure 4f). Two-weeks post-pIpC, there was already significant depletion of Kdm6b-KOMX1 HSCs (Figure 4g). But of the residual Kdm6b-KOMX1 HSCs, ~90% did show recombination of both Kdm6b alleles (Figure 4h). This indicates that pIpC-driven recombination of Kdm6b flox alleles using Mx1-CRE is efficient, but those HSCs differentiate rapidly under inflammatory stress and become depleted from the HSC pool.
Kdm6b-deficient HSCs are primed for differentiation
To examine the role of Kdm6b in HSC function in response to other proliferative stress, mice were injected with 5-FU. 5-FU is a myeloablative agent that kills cycling hematopoietic cells, forcing the quiescent HSCs into cycle to repopulate the depleted bone marrow45. While Kdm6b-deficient mice were capable of initial hematopoietic recovery (Figure 5a), upon serial 5-FU injection both Kdm6b-HetVAV and Kdm6b-KOVAV mice showed significantly decreased survival (Figure 5b), indicative of impaired HSC regenerative capacity.
Figure 5: Kdm6b-deficient HSCs are primed for differentiation.
(a) Blood counts showing hematopoietic recovery of white blood cells (WBC), red blood cells (RBC) and platelets (PLT) in ControlVAV (CNT), Kdm6b-HETVAV (HET) and Kdm6b-KOVAV (KO) mice 10-days after injection with 5-FU (n=6–9). (b) Kaplan-Meier survival curve comparing time to morbidity of ControlVAV (n=8), Kdm6b-HetVAV (n=8), and Kdm6b-KOVAV (n=8) mice after serial 5-FU treatment (weekly injections). (c) Relative donor-derived peripheral blood chimerism following treatment (red arrows) of hematopoietic chimeras with pIpC (dashed line) or PBS (control). (d) HSC and progenitor cell frequency 24-hours after two doses of PBS (control) or pIpC in ControlVAV (CNT), Kdm6b-HETVAV (HET) and Kdm6b-KOVAV (KO) mice (n=6–16). (e) Apoptosis analysis showing proportion of PARP+ HSCs and progenitors 24-hours after two doses of either PBS or pIpC. (f) Cell cycle analysis showing proportion of quiescent (G0) HSCs and progenitors 24-hours after two doses of either PBS or pIpC. *p<0.05, **p<0.01, ***p<0.001, ****p<0.001. Mean ± S.E.M. values are shown.
To examine the inflammatory response of Kdm6b-deficient HSCs without the confounding effects of Mx1-CRE, hematopoietic chimeras were generated by transplanting mice with 2.5×105 WBM from ControlVAV, Kdm6b-HetVAV and Kdm6b-KOVAV donors along with 2.5×105 wild-type WBM competitor cells and treated with pIpC or PBS (control) five-weeks post-transplant. Kdm6b-KOVAV engraftment increased significantly in pIpC-treated recipients (Figure 5c), suggesting these HSCs differentiate more rapidly in response to inflammation. To investigate the acute kinetics of HSC response to inflammatory stress, mice were treated with two doses of pIpC and analyzed 24-hours after the second injection. While ControlVAV HSCs (Lineage- c-Kit+ EPCR+ CD48- CD150+; Sca-1 is excluded because it is IFN-responsive) increased post-pIpC, Kdm6b-HetVAV and Kdm6b-KOVAV HSCs showed no change (Figure 5d). But the total Kdm6b-KOVAV progenitor cell pool (Lineage- c-Kit+ EPCR+; Figure 5d) expanded after pIpC. The changes in HSC and progenitor cell frequency between the genotypes could not be attributed to altered apoptotic response to pIpC (Figure 5e). Cell cycle analysis showed reduced quiescence amongst all HSC and progenitor genotypes following pIpC (Figure 5f). This indicates that upon inflammatory stress, Kdm6b-KOVAV HSCs differentiate to downstream progenitors, but fail to self-renew. Cumulatively, these data suggest that Kdm6b acts to limit the differentiation response of HSCs after inflammatory stress to preserve the HSC pool.
Kdm6b regulates the HSC stress response through the AP-1 transcription factor complex
Downregulation of the AP-1 transcription factor complex downregulation is associated with differentiation arrest47–49. The AP-1 complex subunits Fos and Jun were over-expressed in Kdm6b-KOVAV HSCs at basal conditions and upon inflammatory or myeloablative stress, Fos and Jun became even more over-expressed in Kdm6b-KOVAV HSCs (Figure 6a,b), as did AP-1 target IER genes (Dusp1, Zfp36, and Ier2; Supplementary Figure 5). If downregulation of AP-1 inhibits differentiation, we hypothesized that AP-1 over-expression may enforce stem cell differentiation and drive depletion of Kdm6b-KOVAV HSCs following proliferative stress. As such, ectopic expression of Fos and Jun in wild-type HSCs may produce a phenotype similar to Kdm6b-KOVAV HSCs. ControlVAV c-Kit enriched WBM was co-transduced (Supplementary Figure 6a) with lentiviruses expressing either Fos-GFP or Jun-mCherry in parallel with empty vector control viruses and transplanted. Total donor-derived (CD45.2+) engraftment in both the control and over-expression recipients was comparable (Figure 6c). However, in the AP-1 over-expression recipients, there was a significant depletion of Fos-GFP and Jun-mCherry expressing cells in the blood (Figure 6d). By eight-weeks post-transplant, 90% of donor-derived cells were double negative (not expressing either Fos or Jun), despite representing only <25% of the initial transplanted population. BM analysis showed a significant reduction in AP-1 over-expressing progenitors (Figure 6e), which phenocopies loss of Kdm6b and implies upregulation of AP-1 forces HSC differentiation.
Figure 6: Kdm6b regulates the HSC stress response through the AP-1 transcription factor complex.
(a) Expression of AP-1 components Jun and Fos in ControlVAV and Kdm6b-KOVAV HSCs after PBS (control) or two-doses of pIpC (n=3). (b) Expression of Jun and Fos in ControlVAV and Kdm6b-KOVAV HSCs during recovery from a single dose of 5-FU (n=3–5). (c) Donor-derived peripheral blood engraftment of wild-type BM progenitors transduced with pMND-Fos-GFP and pMND-Jun-mCherry, or respective empty vector control lentiviruses (n=5). (d) Proportion of donor-derived cells expressing each lentivirus combination in peripheral blood at eight-weeks post-transplant compared to initial transduction efficiency. (e) Proportion of donor-derived progenitor cells (Lineage- c-Kit+ Sca-1+) expressing each lentivirus combination in BM eight-weeks post-transplant. (f) Donor-derived peripheral blood engraftment of ControlVAV (WT) and Kdm6b-KOVAV (KO) BM progenitor cells transduced with lentiviruses expressing shRNAs against Fos or Jun (shRNA), or respective LacZ-targeting lentiviruses (n=10). (g) Proportion of donor-derived cells expressing each lentivirus combination in peripheral blood at 16-weeks post-transplant. LacZ = LacZ-shRNA-mCherry / LacZ-shRNA-GFP; shRNA = Jun-shRNA-mCherry / Fos-shRNA-GFP. (h) Frequency of donor-derived progenitor cells in the BM of recipient mice 18-weeks post-transplant. (i) Proportion of donor-derived BM progenitor cells expressing each lentivirus combination. *p<0.05, **p<0.01. Mean ± S.E.M. values are shown.
To determine if inhibiting Fos and Jun could rescue Kdm6b-KOVAV HSC function, shRNAs were generated (Supplementary Figure 6b) and c-Kit enriched WBM from ControlVAV and Kdm6b-KOVAV mice was transduced with LacZ (control) or Fos- (marked by GFP) / Jun- (marked by mCherry) targeting shRNA viruses (Supplementary Figure 6c) and transplanted into lethally irradiated mice. Knockdown of Fos and Jun significantly increased engraftment of Kdm6b-KOVAV cells over all groups (Figure 6f), implying Kdm6b-deficient progenitor cells are extremely sensitive to modulation of AP-1 activity. There was no difference in peripheral blood engraftment between LacZ and Fos- / Jun-shRNA transduced ControlVAV cells, and there was equal transduction representation across all groups in donor-derived blood cells at 16-weeks post-transplant (Figure 6g). Fos- / Jun-knockdown also restored bone marrow progenitors in Kdm6b-KOVAV recipients (Figure 6h) without biases in shRNA representation (Figure 6i). Thus, inhibiting AP-1 in Kdm6b-KOVAV HSCs rescues their engraftment defects.
Kdm6b is required for self-renewal of MLL-AF9 leukemia-initiating cells
Over-expression of KDM6B can inhibit growth of M2/M3 AML cells, but has not M5 (granulocytic subtype) AML22. But as expression level does not always equate with activity, to clarify a function of Kdm6b in M5 AML, c-Kit enriched WBM from ControlVAV, Kdm6b-HetVAV and Kdm6b-KOVAV mice was transduced with the MLL-AF9 oncogene50 and transplanted. Transduction efficiency and frequency of MLL-AF9+ GMPs (the leukemia-initiating cells in this model) were comparable between genotypes (Supplementary Figure 7a). Four-weeks post-transplant there was a significant reduction in MLL-AF9 GFP+ cells in the blood in both Kdm6b-HetVAV and Kdm6b-KOVAV recipients (Supplementary Figure 7b). This correlated with increased survival (Figure 7a). There was no difference in spleen weights of moribund mice (Supplementary Figure 7c), but decreased leukemic GMPs in the bone marrow of Kdm6b-HetVAV and Kdm6b-KOVAV recipients (Figure 7b,c; Supplementary Figure 7d). Secondary transplant was performed with limiting doses of WBM from primary tumors to determine frequency of functional leukemia-initiating cells. Recipient mice were scored as positive if they developed AML (Figure 7d). Kdm6b-HetVAV and Kdm6b-KOVAV tumors had a three-fold reduction in leukemia-initiating cell frequency (Figure 7e).
Figure 7: Kdm6b is required for self-renewal of MLL-AF9 leukemia-initiating cells.
(a) Kaplan-Meier plot comparing time to morbidity between mice transplanted with ControlVAV (n=8), Kdm6b-HetVAV (n=7), and Kdm6b-KOVAV (n=6) c-Kit+ BM cells transduced with MLL-AF9. (b) Representative flow cytometry plots showing leukemia-initiating cells (GFP+ GMPs) in MLL-AF9 ControlVAV (top) and Kdm6b-KOVAV (bottom) recipient mouse BM. (c) Absolute number of leukemic GMPs in BM of moribund recipient mice. (d) Secondary limiting dilution transplantation response from transfer of stated doses of cells from primary moribund animals. (e) Leukemic-initiating cell frequency estimates calculated by ELDA software. (f) Frequency of GFP+ cells in blood of mice transplanted with ControlERT2, Kdm6b-HetERT2, and Kdm6b-KOERT2 c-Kit+ BM cells transduced with MLL-AF9 two-weeks post-transplant. (g) Frequency of GFP+ cells in blood of surviving mice transplanted with MLL-AF9-expressing cells two-days post-tamoxifen treatment. (h) Kaplan-Meier plot comparing time to morbidity between mice transplanted with ControlERT2 (n=6), Kdm6b-HetERT2 (n=6), and Kdm6b-KOERT2 (n=6) c-Kit+ BM cells transduced with MLL-AF9. Red arrows indicate timing of tamoxifen injections. (i) Dose response of ControlVAV, Kdm6b-HetVAV and Kdm6b-KOVAV MLL-AF9 AML cells to increasing concentrations of GSK-J4 (n=3). (j) Kaplan-Meier plot comparing time to morbidity between mice transplanted with Kdm6b-KOVAV MLL-AF9 AML cells transduced with lentiviruses expressing Kdm6b, Kdm6bH1388A, or empty vector (EV) control (n=7 recipient mice, n=3 independent AMLs). *p<0.05, **p<0.01. Mean ± S.E.M. values are shown.
To determine if Kdm6b is necessary for maintenance of MLL-AF9 AML, Kdm6bfl/fl mice were crossed to ERT2-CRE mice28. Post-tamoxifen analysis of Kdm6b-HetERT2 mice showed complete floxing in HSCs (Supplementary Figure 7e), and there was no leaky CRE activity (Supplementary Figure 7f). c-Kit+ WBM was transduced with MLL-AF9 and transplanted. Recipients were treated with tamoxifen when the blood comprised ~10% GFP+ cells (Figure 7f). Of the mice surviving two-days after the last dose, GFP+ cells were reduced in Kdm6b-KOERT2 recipients (Figure 7g). Either heterozygous or homozygous deletion of Kdm6b in pre-existing AML significantly extended survival (Figure 7h). GFP+ blasts two-days post-tamoxifen showed intact floxed alleles in both Kdm6b-HETERT2 and Kdm6b-KOERT2 cells (Supplementary Figure 7g). Analysis of moribund animals showed many mice succumbed to AML with intact floxed alleles (Supplementary Figure 7h), indicating a strong pressure to maintain Kdm6b. Fos and Jun were also over-expressed in Kdm6b-deficient MLL-AF9 cells (Supplemental Figure 7i).
As we observed gene expression changes in Kdm6b-KOVAV HSCs occurred without significant chromatin alterations, to determine if the role of Kdm6b in MLL-AF9 AML was also demethylase-independent, primary AML cells were cultured in GSK-J4, a small molecule that inhibits the demethylase domain of Kdm6b (and also Utx1)9. The dose response to GSK-J4 was identical between genotype (Figure 7i), indicating that inhibition Kdm6b demethylase activity has no effect MLL-AF9 AML. To examine this in vivo, primary Kdm6b-KOVAV MLL-AF9 AMLs were transduced with lentiviruses expressing either full-length Kdm6b or a catalytically-dead Kdm6b point mutant (Kdm6bH1388A)6, 51. While expression of full-length Kdm6b restored leukemic potential, so did Kdm6bH1388A (Figure 7j). Cumulatively, these data suggest the functions of Kdm6b in normal and malignant stem cells are largely H3K27me3-independent.
Kdm6b supports context-dependent leukemogenesis
To investigate Kdm6b in other leukemias, c-Kit+ WBM was transduced with a retrovirus expressing Notch1 Intra-Cellular Domain (NICD), a model that recapitulates human T-ALL52. 1.0×105 cells were transplanted (Figure 8a) with similar transduction efficiencies (Figure 8b). Four-weeks post-transplant, NICD-GFP+ cells were increased in the blood of Kdm6b-KOVAV recipients (Figure 8c). But despite this, and unlike MLL-AF9 AML, loss of Kdm6b did not produce a survival benefit in adult T-ALL (Figure 8d). Because these results were disparate with a previous NICD study using fetal liver-derived Kdm6b-deficient cells21, c-Kit+ cells from E14.5 fetal liver were transduced with NICD and transplanted (Figure 8e). Four-weeks post-transplant showed equivalent NICD-GFP+ in recipient blood (Figure 8f). However, Kdm6b-HetVAV and Kdm6b-KOVAV recipients showed a significant survival benefit (Figure 8g), and mice alive after 100-days showed a complete dearth of T-ALL cells (data not shown) despite being initially engrafted. This implies a unique requirement for Kdm6b in sustaining T-ALL cells of fetal origin. This result was reversed in fetal-derived MLL-AF9 AML (Figure 8h–j) where no survival benefit was observed in a Kdm6b-deficient background. The differences between fetal- versus adult-derived leukemias could not be explained by variation in fetal HSC content (Figure 8k,l).
Figure 8: Kdm6b supports context-dependent leukemogenesis.
(a) Schematic for transduction of adult c-Kit+ BM progenitors with NICD virus and transplantation. (b) Representative flow cytometry plots showing NICD transduction efficiencies in Lineage- c-Kit+ Sca-1+ cells 48-hours post-transduction. (c) Frequency of NICD-GFP+ cells in peripheral blood of recipient mice four-weeks post-transplant of adult BM progenitor cells. (d) Kaplan-Meier plot comparing time to morbidity between mice transplanted with ControlVAV (n=7), Kdm6b-HETVAV (n=5) and Kdm6b-KOVAV (n=7) adult c-Kit+ cells transduced with NICD. (e) Schematic for transduction of fetal liver c-Kit+ progenitors with NICD virus and transplantation. (f) Frequency of NICD-GFP+ cells in peripheral blood of recipient mice four-weeks post-transplant of fetal liver progenitor cells. (g) Kaplan-Meier plot comparing time to morbidity between mice transplanted with ControlVAV (n=10), Kdm6b-HETVAV (n=5) and Kdm6b-KOVAV (n=10) fetal liver c-Kit+ cells transduced with NICD. (h) Schematic for transduction of fetal liver c-Kit+ progenitors with MLL-AF9 virus and transplantation. (i) Frequency of MLL-AF9+ cells in peripheral blood of recipient mice four-weeks post-transplant of fetal liver progenitor cells. (j) Kaplan-Meier plot comparing time to morbidity between mice transplanted with ControlVAV (n=5) and Kdm6b-KOVAV (n=5) fetal liver c-Kit+ cells transduced with MLL-AF9. (k) Flow cytometry gating scheme to identify HSCs in E14.5 fetal liver. (l) Quantification of HSC frequency in ControlVAV, Kdm6b-HETVAV and Kdm6b-KOVAV E14.5 fetal liver. **p<0.01, ****p<0.001. Mean ± S.E.M. values are shown.
DISCUSSION
We show loss of Kdm6b compromises self-renewal of both normal and leukemic stem cells (dependent on lineage subtype and developmental stage). Our results suggest KDM6B mutations are not observed in hematopoietic malignancies (or clonal hematopoiesis) as this would result in loss of that clone due to competitive disadvantage. This was observed experimentally when Kdm6b was deleted after the initiation of MLL-AF9 AML and the “wild-type” blasts outcompeted those that lost Kdm6b.
We show the major functions of Kdm6b in HSCs appear unrelated to chromatin regulation, but rather demethylase-independent regulation of stress response gene expression programs. As chromatin architecture in Kdm6b-KOVav HSCs is largely unaltered, the gene expression differences may be secondary effects downstream of master regulators like AP-1. Upon proliferative stress, Kdm6b-deficient HSCs differentiate rapidly and self-renew inefficiently, due at least in part to upregulation of Fos and Jun. Over-expression of AP-1 in wild-type progenitors phenocopied loss of Kdm6b, and reducing their expression in Kdm6b-deficient HSCs restored engraftment potential. Enforced expression of Fos in M1 leukemia cells mimics what was observed here, with increased differentiation and reduced disease aggressiveness53. Moreover, prolonged expression of Fos in transgenic mouse HSCs decreases functional potential54. This implicates Fos over-expression as a major determinant of the Kdm6b-deficient HSC phenotype. Indeed, the Fos locus was one of the only genomic regions that showed a difference in chromatin accessibility in Kdm6b-KOVav HSCs. But the mechanisms through which AP-1 and Kdm6b interact in HSCs and regulate each other and their target genes remains unclear. Given improving genomic technologies in regards to low cell inputs, it is possible this can be directly tested in the future.
Our data suggest the potential of KDM6B as a therapeutic target for leukemia depends on developmental and cellular context. GSK-J4, which binds to the catalytic domain and inhibits demethylase activity of both KDM6B and UTX19, has been cited as a potential clinical agent in blood cancers21, 55. It is possible the activity observed in other studies occurs primarily through UTX1 inhibition, which is a potent hematopoietic tumor suppressor56. Our data suggest inhibiting demethylase activity of KDM6B would likely be ineffective, and that more specific effects could be achieved by targeting the domains that regulate its unique functions in HSCs. Nevertheless, KDM6B may still represent an attractive therapeutic target in certain contexts as haploinsufficiency had modest effects for normal HSCs, but both heterozygous and homozygous loss of Kdm6b impeded adult-derived MLL-AF9 AML with similar kinetics. Thus, a 50% inhibition could be deleterious for leukemia-initiating cells, without being toxic to normal HSCs. Cumulatively, our data show that Kdm6b is necessary for HSC maintenance in response to proliferative stress, and present KDM6B as a potential therapeutic target in leukemias depending on the subtype and developmental context.
Supplementary Material
ACKNOWLEDGEMENTS
We thank and Dr. Martin Matzuk (Baylor College of Medicine) for providing Kdm6bfl/fl mice, Dr. Lukas Wartman (Washington University) for providing Utx1fl/fl mice, and Dr. Jeff Magee (Washington University) for providing MLL-AF9 retroviral plasmid. We thank the Alvin J. Siteman Cancer Center at Washington University for use of the Siteman Flow Cytometry Core, supported in part by NCI Grant CA91842. We thank the Genome Technology Access Center at Washington University for genomic analysis, partially supported by NCI Grant CA91842 and by ICTS/CTSA Grant UL1TR000448 NIH.
This work was supported by the National Institutes of Health (R01DK102428), the Edward Mallinckrodt Jr. Foundation, the American Society of Hematology, the V Foundation, Gabrielle’s Angel Foundation, and the Sidney Kimmel Foundation (all to G.A.C.). Bioinformatics analysis was supported by the Washington University Center for Regenerative Medicine. C.M. was supported by NIH T32HL007088, and NIH DK111058-01. E.L.O. was supported by NIH 5T32CA113275-10 and NIH F31DK114951. H.C. was supported by a post-doctoral scholar award from the American Society of Hematology and an Edward P. Evans Foundation Young Investigator Award. G.A.C. is a Leukemia and Lymphoma Society scholar.
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci U S A 2007;104(47):18439–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res 2011;21(3):381–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 2007;449(7163):689–94. [DOI] [PubMed] [Google Scholar]
- 4.Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev 2008;22(14):1865–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burgold T, Voituron N, Caganova M, Tripathi PP, Menuet C, Tusi BK, et al. The H3K27 demethylase JMJD3 is required for maintenance of the embryonic respiratory neuronal network, neonatal breathing, and survival. Cell Rep 2012;2(5):1244–58. [DOI] [PubMed] [Google Scholar]
- 6.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 2007;449(7163):731–4. [DOI] [PubMed] [Google Scholar]
- 7.De Santa F, Narang V, Yap ZH, Tusi BK, Burgold T, Austenaa L, et al. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. Embo J 2009;28(21):3341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 2007;130(6):1083–94. [DOI] [PubMed] [Google Scholar]
- 9.Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 2012;488(7411):404–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wijayatunge R, Chen LF, Cha YM, Zannas AS, Frank CL, West AE. The histone lysine demethylase Kdm6b is required for activity-dependent preconditioning of hippocampal neuronal survival. Mol Cell Neurosci 2014;61:187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, et al. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev 2009;23(10):1171–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barradas M, Anderton E, Acosta JC, Li S, Banito A, Rodriguez-Niedenfuhr M, et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev 2009;23(10):1177–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohguchi H, Harada T, Sagawa M, Kikuchi S, Tai YT, Richardson PG, et al. KDM6B modulates MAPK pathway mediating multiple myeloma cell growth and survival. Leukemia 2017;31(12):2661–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenblatt SM, Nimer SD. Chromatin modifiers and the promise of epigenetic therapy in acute leukemia. Leukemia 2014;28(7):1396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdel-Wahab O, Levine RL. Mutations in epigenetic modifiers in the pathogenesis and therapy of acute myeloid leukemia. Blood 2013;121(18):3563–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas Research N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013;368(22):2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mar BG, Bullinger L, Basu E, Schlis K, Silverman LB, Dohner K, et al. Sequencing histone-modifying enzymes identifies UTX mutations in acute lymphoblastic leukemia. Leukemia 2012;26(8):1881–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiel MJ, Sahasrabuddhe AA, Rolland DC, Velusamy T, Chung F, Schaller M, et al. Genomic analyses reveal recurrent mutations in epigenetic modifiers and the JAK-STAT pathway in Sezary syndrome. Nat Commun 2015;6:8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei Y, Chen R, Dimicoli S, Bueso-Ramos C, Neuberg D, Pierce S, et al. Global H3K4me3 genome mapping reveals alterations of innate immunity signaling and overexpression of JMJD3 in human myelodysplastic syndrome CD34+ cells. Leukemia 2013;27(11):2177–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderton JA, Bose S, Vockerodt M, Vrzalikova K, Wei W, Kuo M, et al. The H3K27me3 demethylase, KDM6B, is induced by Epstein-Barr virus and over-expressed in Hodgkin’s Lymphoma. Oncogene 2011;30(17):2037–43. [DOI] [PubMed] [Google Scholar]
- 21.Ntziachristos P, Tsirigos A, Welstead GG, Trimarchi T, Bakogianni S, Xu L, et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu SH, Zhu KY, Chen J, Liu XZ, Xu PF, Zhang W, et al. JMJD3 facilitates C/EBPbeta-centered transcriptional program to exert oncorepressor activity in AML. Nat Commun 2018;9(1):3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei Y, Zheng H, Bao N, Jiang S, Bueso-Ramos CE, Khoury J, et al. KDM6B overexpression activates innate immune signaling and impairs hematopoiesis in mice. Blood Adv 2018;2(19):2491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwamori N, Iwamori T, Matzuk MM. H3K27 demethylase, JMJD3, regulates fragmentation of spermatogonial cysts. PLoS ONE 2013;8(8):e72689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang C, Lee JE, Cho YW, Xiao Y, Jin Q, Liu C, et al. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc Natl Acad Sci U S A 2012;109(38):15324–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Georgiades P, Ogilvy S, Duval H, Licence DR, Charnock-Jones DS, Smith SK, et al. VavCre transgenic mice: a tool for mutagenesis in hematopoietic and endothelial lineages. Genesis 2002;34(4):251–6. [DOI] [PubMed] [Google Scholar]
- 27.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible Gene Targeting in Mice. Science 1995;269(5229):1427–9. [DOI] [PubMed] [Google Scholar]
- 28.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol 2002;244(2):305–18. [DOI] [PubMed] [Google Scholar]
- 29.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30(7):923–30. [DOI] [PubMed] [Google Scholar]
- 31.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 2013;41(10):e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patro R, Mount SM, Kingsford C. Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat Biotechnol 2014;32(5):462–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidl C, Rendeiro AF, Sheffield NC, Bock C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat Methods 2015;12(10):963–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9(4):357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Starmer J, Magnuson T. Detecting broad domains and narrow peaks in ChIP-seq data with hiddenDomains. BMC Bioinformatics 2016;17:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carroll TS, Liang Z, Salama R, Stark R, de Santiago I. Impact of artifact removal on ChIP quality metrics in ChIP-seq and ChIP-exo data. Front Genet 2014;5:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 2016;44(W1):W160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neph S, Kuehn MS, Reynolds AP, Haugen E, Thurman RE, Johnson AK, et al. BEDOPS: high-performance genomic feature operations. Bioinformatics 2012;28(14):1919–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 2017;14(10):959–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Q, Zou J, Wang M, Ding X, Chepelev I, Zhou X, et al. Critical role of histone demethylase Jmjd3 in the regulation of CD4+ T-cell differentiation. Nat Commun 2014;5:5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 2009;347(1–2):70–8. [DOI] [PubMed] [Google Scholar]
- 43.Kirschner K, Chandra T, Kiselev V, Flores-Santa Cruz D, Macaulay IC, Park HJ, et al. Proliferation Drives Aging-Related Functional Decline in a Subpopulation of the Hematopoietic Stem Cell Compartment. Cell Rep 2017;19(8):1503–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tullai JW, Schaffer ME, Mullenbrock S, Sholder G, Kasif S, Cooper GM. Immediate-early and delayed primary response genes are distinct in function and genomic architecture. J Biol Chem 2007;282(33):23981–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Venezia TA, Merchant AA, Ramos CA, Whitehouse NL, Young AS, Shaw CA, et al. Molecular signatures of proliferation and quiescence in hematopoietic stem cells. PLoS Biol 2004;2(10):e301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fortier ME, Kent S, Ashdown H, Poole S, Boksa P, Luheshi GN. The viral mimic, polyinosinic:polycytidylic acid, induces fever in rats via an interleukin-1-dependent mechanism. Am J Physiol Regul Integr Comp Physiol 2004;287(4):R759–66. [DOI] [PubMed] [Google Scholar]
- 47.Yamazaki S, Tanaka Y, Araki H, Kohda A, Sanematsu F, Arasaki T, et al. The AP-1 transcription factor JunB is required for Th17 cell differentiation. Sci Rep 2017;7(1):17402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leppa S, Eriksson M, Saffrich R, Ansorge W, Bohmann D. Complex functions of AP-1 transcription factors in differentiation and survival of PC12 cells. Mol Cell Biol 2001;21(13):4369–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han B, Rorke EA, Adhikary G, Chew YC, Xu W, Eckert RL. Suppression of AP1 transcription factor function in keratinocyte suppresses differentiation. PLoS One 2012;7(5):e36941. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 2006;442(7104):818–22. [DOI] [PubMed] [Google Scholar]
- 51.Xiang Y, Zhu Z, Han G, Lin H, Xu L, Chen CD. JMJD3 is a histone H3K27 demethylase. Cell Res 2007;17(10):850–7. [DOI] [PubMed] [Google Scholar]
- 52.Pear WS, Aster JC, Scott ML, Hasserjian RP, Soffer B, Sklar J, et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med 1996;183(5):2283–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lord KA, Abdollahi A, Hoffman-Liebermann B, Liebermann DA. Proto-oncogenes of the fos/jun family of transcription factors are positive regulators of myeloid differentiation. Mol Cell Biol 1993;13(2):841–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okada S, Fukuda T, Inada K, Tokuhisa T. Prolonged expression of c-fos suppresses cell cycle entry of dormant hematopoietic stem cells. Blood 1999;93(3):816–25. [PubMed] [Google Scholar]
- 55.Li Y, Zhang M, Sheng M, Zhang P, Chen Z, Xing W, et al. Therapeutic potential of GSK-J4, a histone demethylase KDM6B/JMJD3 inhibitor, for acute myeloid leukemia. J Cancer Res Clin Oncol 2018;144(6):1065–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van der Meulen J, Sanghvi V, Mavrakis K, Durinck K, Fang F, Matthijssens F, et al. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood 2015;125(1):13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.