To improve simultaneous S-palmitoylation and S-nitrosylation site profiling, we developed a novel mass spectrometry-based strategy Palmitoylation And Nitrosylation Interplay Monitoring (PANIMoni). Using this approach we evaluated dynamic, site-specific changes in two important cysteine modifications, S-PALM and S-NO, in a mouse model of chronic stress. High-throughput analysis provided valuable insight into chronic, stress-induced remodeling of the synaptic proteome. Based on the obtained data, we postulate that the mechanism underlying chronic stress involves mutual competition between S-PALM and S-NO for modification sites.
Keywords: Post-translational modifications, Label-free quantification, Animal models, Imaging Visualization Tools, Neurobiology, Psychiatric disease, PLA, postsynaptic density, S-nitrosylation, S-palmitoylation, synapse
Graphical Abstract
Highlights
Using mass spectrometry-based proteomic approach, PANIMoni, we analyzed endogenous S-nitrosylation and S-palmitoylation of postsynaptic density proteins in mouse models of chronic stress.
We provided differential analysis of S-palmitoylation and S-nitrosylation at the level of exact sites of modifications.
We show that affected mechanism of S-palmitoylation and S-nitrosylation interplay of proteins involved in synaptic transmission, protein localization and regulation of synaptic plasticity is associated with chronic stress disorder.
Abstract
The precise regulation of synaptic integrity is critical for neuronal network connectivity and proper brain function. Essential aspects of the activity and localization of synaptic proteins are regulated by posttranslational modifications. S-palmitoylation is a reversible covalent modification of the cysteine with palmitate. It modulates affinity of the protein for cell membranes and membranous compartments. Intracellular palmitoylation dynamics are regulated by crosstalk with other posttranslational modifications, such as S-nitrosylation. S-nitrosylation is a covalent modification of cysteine thiol by nitric oxide and can modulate protein functions. Therefore, simultaneous identification of endogenous site-specific proteomes of both cysteine modifications under certain biological conditions offers new insights into the regulation of functional pathways. Still unclear, however, are the ways in which this crosstalk is affected in brain pathology, such as stress-related disorders. Using a newly developed mass spectrometry-based approach Palmitoylation And Nitrosylation Interplay Monitoring (PANIMoni), we analyzed the endogenous S-palmitoylation and S-nitrosylation of postsynaptic density proteins at the level of specific single cysteine in a mouse model of chronic stress. Among a total of 813 S-PALM and 620 S-NO cysteine sites that were characterized on 465 and 360 proteins, respectively, we sought to identify those that were differentially affected by stress. Our data show involvement of S-palmitoylation and S-nitrosylation crosstalk in the regulation of 122 proteins including receptors, scaffolding proteins, regulatory proteins and cytoskeletal components. Our results suggest that atypical crosstalk between the S-palmitoylation and S-nitrosylation interplay of proteins involved in synaptic transmission, protein localization and regulation of synaptic plasticity might be one of the main events associated with chronic stress disorder, leading to destabilization in synaptic networks.
Synaptic plasticity is a dynamic process that is driven by molecular changes at individual synapses that lead to the rapid and recurrent rewiring of neuronal circuitry (1, 2). Each synapse contains thousands of different proteins, including receptors, signaling molecules, scaffolding proteins, and cytoskeleton components that are directly involved in synaptic transmission (3, 4). Understanding the mechanisms by which functions of synaptic proteins are regulated is crucial for elucidating the molecular basis of synaptic plasticity.
Synaptic proteins are regulated by a wide range of posttranslational modifications (PTMs)1, including S-palmitoylation (S-PALM) and S-nitrosylation (S-NO). These chemical modifications affect the properties of target molecules, resulting in the modulation of synaptic function and plasticity (5–7). Recent evidence suggests that S-PALM may play a key role in regulating various synaptic proteins, including receptors, and ion channels (8–10). S-palmitoylation refers to the addition of palmitoyl moieties to selected cysteine via thioester bonds, which increases protein hydrophobicity and the affinity for plasma membranes (11, 12). One of the key features of S-PALM is its full reversibility, which makes it an exceptional mechanism for the rapid spatiotemporal control of intracellular and extracellular signaling. S-palmitoylation modulates multiple aspects of synaptic protein activity, including trafficking, conformational changes, stability, protein-protein interactions, and other PTMs (10, 13, 14). The regulation of S-PALM is mediated by two types of enzymes: (I) protein acyltransferases (PATs) that attach the palmitate group to protein side chains and (II) acyl-protein thioesterases (APTs) that depalmitoylate proteins that hydrolyze thioester bonds (10, 15). In addition to enzymatic control, the dynamics of S-PALM are regulated by other protein modifications that occur in the target protein, such as phosphorylation and S-NO (16, 17). Recently, the S-NO-dependent regulation of S-PALM dynamics was reported. The covalent modification of a protein's cysteine thiol by a nitric oxide group appears to directly compete for cysteine residues with the palmitate or even displace it from the palmitoylation site (17, 18). S-nitrosylation is involved in many intracellular signaling mechanisms. It is stimulus-evoked, precisely targeted, reversible, spatiotemporally restricted, and necessary for specific cellular responses (19, 20). Independent proteomic studies have shown that both S-NO and S-PALM occur at numerous neuronal proteins with equal frequency and site specificity on the target molecule (21–23). Ho et al. recently provided evidence of reciprocal regulation of the major postsynaptic protein - postsynaptic density protein 95 (PSD-95) by S-NO and S-PALM (24). They noted that the decrease in PSD-95 S-PALM increased its S-NO, suggesting that the specific sites of PSD-95 protein may undergo S-PALM or S-NO in a competitive manner. Interestingly, however, when S-palmitoylated PSD-95 localized in the synapse, it coordinated N-methyl-d-aspartate (NMDA) receptor stimulation and the subsequent production of nitric oxide by neuronal nitric oxide synthase. Nitric oxide, in turn, inhibited PSD-95 S-PALM and reduced the amount of this protein at the synapse.
A critical role for the crosstalk between S-PALM and S-NO in maintaining proper synaptic organization and function is further underscored by the widely reported imbalance in the dynamics of protein S-PALM and S-NO that are associated with the pathophysiology of neurodegenerative disorders, such as Alzheimer's disease, Huntington's disease, schizophrenia, mental disorders, and Parkinson's disease (25). Still unknown, however, is whether disruption of the crosstalk between S-PALM and S-NO leads to the development of these diseases.
Emerging evidence indicates that the chronic stress-related emotional and cognitive impairment is associated with divergent changes in brain plasticity. It has been shown, that chronic stress leads to structural changes in synapses complexity, volume and number (26–29). Moreover, it has been also demonstrated that chronic stress causes atrophy of dendrites and synapses and suppresses neurogenesis (30–32). The molecular mechanism underling these changes remains elusive. Among the best-described mechanisms governing synaptic proteins function is their regulation by post-translational modifications such as S-palmiotylation and S-nitrosylation.
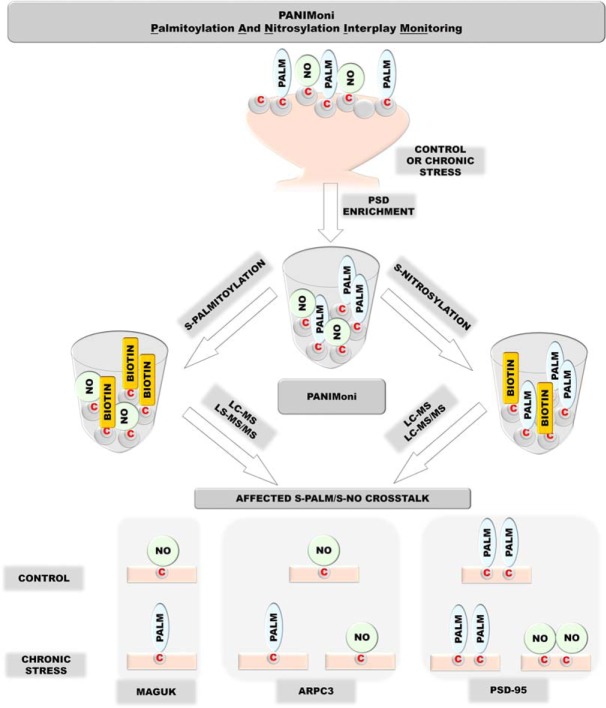
Considering recent insights into the role of rapid PTMs in the synaptic response to physiological stimuli and under pathological conditions, the present study investigated PTMs of endogenous synaptic proteins at the level of single cysteines. We employed a unique proteomic-based approach that integrates specific PSD proteins enrichment, the chemical derivatization of modified sites and specific resin-based enrichment, label-free mass spectrometry (MS), and the differential analysis of two-dimensional (2D) heat-maps that represented the mass-to-charge ratio (m/z) versus liquid chromatography (LC) retention times of each peptide ion (Fig. 1).
Fig. 1.
Schematic representation of PANIMoni proteomic approach for large-scale, site-specific S-palmitoylation and S-nitrosylation monitoring. Protein modifications were selectively replaced with biotin groups. After trypsin digestion and enrichment on avidin resin, the S-PALM and S-NO peptides were analyzed using MS in two independent runs (LC-MS and LC-MS/MS). MSparky software was used for site-specific data analysis (21).
To improve simultaneous S-palmitoylation and S-nitrosylation site profiling, we developed a novel mass spectrometry-based strategy - Palmitoylation And Nitrosylation Interplay Monitoring (PANIMoni). We used this approach to assess dynamic, site-specific changes in two important cysteine modifications, S-PALM and S-NO, in a mouse model of chronic stress. Our differential, high-throughput analysis of 813 palmitoylation and 620 nitrosylation sites that were characterized on 465 and 360 proteins, respectively, provides valuable insight into the chronic stress-induced remodeling of the synaptic proteome.
We postulate that the mechanism underlying chronic stress involves mutual competition between S-PALM and S-NO for the sites of modification. Our extensive analysis of datasets reveals unusual mechanism of crosstalk between the S-palmitoylation and S-nitrosylation of 122 known synaptic proteins, including major plasticity players, such as Gsk3b,GluR1 or CamKII. Moreover, we identified previously unknown palmitoylation and nitrosylation targets, underscoring the broad impact of the PTMs on brain functions. The mechanisms of crosstalk between S-PALM and S-NO of proteins that are involved in synaptic transmission, synaptic localization, and the regulation of synaptic plasticity appear to be a major event associated with chronic stress-related disorders leading to the rewiring of neuronal circuitry.
MATERIALS AND METHODS
Chemicals
Bradford reagent, sucrose, Ficoll, neocuproine, N-ethylmaleimide, corticosterone, S-Nitroso-N-acetyl-dl-penicillamine (SNAP), Nω-Nitro-l-arginine methyl ester hydrochloride (l-NAME), 2-bromohexadecanoic acid (2-Br), copper sulfate (CuSO4), Tris(2-carboxyethyl)phosphine hydrochloride (TCEP, Sigma Aldrich, Germany), hydroxylamine and sodium ascorbate were purchased from Sigma Aldrich. Neutravidin-agarose, N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide (biotin-HPDP), and Oregon Green 488 azide were purchased from Thermo Fisher Scientific. Biotin PEG Thiol 5kDa were purchased from Nanocs. Sequencing-grade modified trypsin was obtained from Promega. Complete protease inhibitor mixture was obtained from Roche Diagnostics, Germany. Enhanced chemiluminescence reagents were purchased from Amersham Biosciences, UK. Alkyne-palmitic acid was obtained from Iris Biotech, Germany. The corticosterone enzyme-linked immunosorbent assay (ELISA) kit was purchased from Enzo Life Sciences. All basic reagents and solvents (e.g. salts, buffers) were purchased from Sigma-Aldrich.
Chronic Restraint Stress
Twelve- to 14-week-old wild type male C57/BL6 mice were subjected to restraint stress as previously described (33). The procedures were performed during the light period of the circadian cycle. All of the animals were individually housed with free access to food and water. The mice were weighed every other day. After the initial 7 days, during which all of the mice were subjected to handling only, control mice were left undisturbed, and stressed animals were subjected to restraint stress for 6 h/day for 21 days in a separate room. The experiments were approved by the Local Bioethical Committee.
Tail Suspension Test
One day after the chronic restraint stress procedure, stressed and control mice were subjected to the tail suspension test by hanging them by their tails according to a previously described protocol (34). The test was recorded, and immobility time was measured for 6 min using a stopwatch. Mice were considered immobile only when they hung passively. The results were expressed as mean ± S.E. The statistical significance was determined by heteroscedastic two-tailed t test.
Measurement of Serum Corticosterone
At the end of the experiment (2 days after the last restraint session), stressed and control mice were euthanized by cervical dislocation and decapitated. Blood was collected and centrifuged. Corticosterone levels were measured using an ELISA kit (Enzo Life Sciences) according to the manufacturer's instructions. The results were expressed as mean ± S.E. The statistical significance was determined by heteroscedastic two-tailed t test.
Synaptoneurosomes Isolation
Synaptoneurosomes were prepared from the brains of control and stressed mice as previously described (21). After euthanasia by cervical dislocation, the mice were decapitated. The brains were immediately removed and homogenized using a Dounce homogenizer in 6 ml of buffer A that contained 5 mm HEPES (pH 7.4) (Sigma Aldrich), 0.32 m sucrose (Sigma Aldrich), 0.2 mm ethylenediaminetetraacetic acid (EDTA) (Sigma Aldrich), 50 mm N-ethylmaleimide (NEM) (Sigma Aldrich), and protease inhibitor mixture (Complete Roche). The homogenate was centrifuged at 2500 × g for 5 min, yielding a pellet and supernatant fractions. The supernatant was then centrifuged at 12,000 × g for 5 min. The pellet was resuspended in buffer A, placed on a discontinuous Ficoll (Sigma Aldrich) gradient (4%, 6%, and 13%), and centrifuged at 70,000 × g for 45 min. The synaptoneurosomal fraction was collected in buffer A and centrifuged at 20,000 × g for 20 min. The pellet corresponded to the synaptoneurosomes fraction. Workflow of synaptoneurosomes isolation and its visualization using electron microscopy are presented in the supplemental Fig. S1.
Postsynaptic Density Fraction Isolation
Isolated synaptoneurosomes were further diluted with 5 ml of 1% TritonX-100 (Sigma Aldrich) in 32 mm sucrose (Sigma Aldrich)and 12 mm Tris-HCl (pH 8.1) (Sigma Aldrich). The sample was stirred for 15 min in the same open-top tube in a 4 °C cold room and then centrifuged at 33,000 × g for 20 min. The pellet was resuspended with 500 μl of buffer solution and layered onto a sucrose gradient that contained 4 ml of 1.5 m sucrose and 4 ml of 1.0 m sucrose (Sigma Aldrich). The sample was spun in a swing rotor at 200,000 × g for 2 h. The streak-like cloudy band between 1.0 m sucrose and 1.5 m sucrose that containing PSDs was carefully removed and resuspended in 600 μl of buffer solution. An equal amount of 1% Triton X-100 and 150 mm KCl was added to the sample for resuspension. The sample was then centrifuged at 200,000 × g for 30 min. The resulting pellet contained the PSD fraction. Workflow of PSD isolation, the purity and reproducibility analysis are presented in the supplemental Fig. S1.
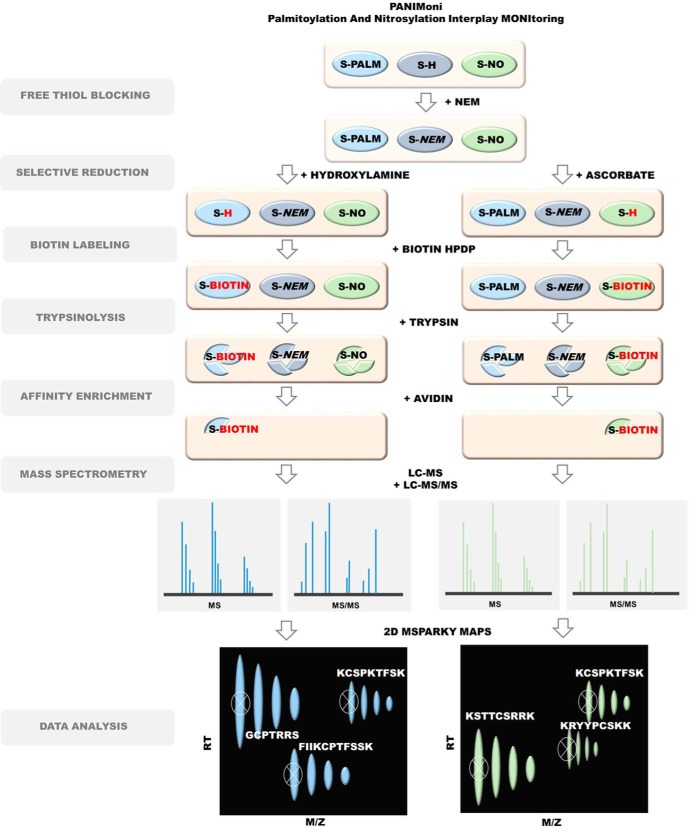
Acyl Biotin Exchange (ABE)/Biotin Switch Method (BSM)
The substitution of S-palmitoylated Cys (S-PALM-Cys sites or S-nitrosylated Cys (S-NO-Cys) sites with S-biotinylated Cys in PSD protein lysates was based on a previously described Acyl Biotin Exchange and Biotin Switch Method procedures (35, 36). Postsynaptic density protein fractions that were obtained in the previous steps were dissolved in HEN buffer that contained 250 mm HEPES (pH 7.7), 1 mm EDTA, and 0.1 mm neocuproine (Sigma Aldrich). To avoid rearrangements of thiol-modifying groups, the protein mixture was treated with blocking buffer solution that contained 250 mm HEPES (pH 7.7), 1 mm EDTA, 0.1 mm neocuproine, 5% sodium dodecyl sulfate (SDS), and 50 mm N-ethylmaleimide at 4 °C for 16 h with agitation. To remove excess reagent, the protein extracts were precipitated with 96% of ethanol and resuspended in the same volume of HEN buffer with 2.5% SDS. The obtained protein solutions were then divided into two equal parts. One part was treated with a mixture of 400 μm Biotin-HPDP and 1 m hydroxylamine (Sigma Aldrich) for S-PALM or 5 mm sodium ascorbate (Sigma Aldrich) for S-NO. The other part was used as a negative control for the all experiments and treated with 400 μm biotin-HPDP without hydroxylamine or sodium ascorbate. All the samples were incubated in the dark for 1.5 h at room temperature.
Palmitoyl Protein Identification and Site Characterization (PalmPISC)/SNO Site Identification (SNOSID)
The biotin labeling of S-palmitoylated and S-nitrosylated proteins in lysates was based on previously described procedures (namely ABE and BSM, respectively). Postsynaptic density protein fractions that contained biotinylated proteins were digested using sequencing-grade modified trypsin (Promega V 5111) for 16 h at 37 °C. Digestion was terminated using protease inhibitor mixture. The tryptic peptide mixture was incubated with 100 μl of neutravidin beads at room temperature for 1 h. The neutravidin beads were washed five times in 1 ml of wash buffer. Neutravidin-bound peptides were eluted with 150 μl of elution buffer (25 mm NH4CO3 [pH 8.2] and 5 mm TCEP (Sigma Aldrich)) and concentrated in a SpeedVac. Trifluoroacetic acid (Sigma Aldrich) was added to the peptide solution to achieve a final concentration of 0.1%. The samples were analyzed by nanoLC-MS and nanoLC-MS/MS.
Acyl-biotin-PEG Exchange (AbPE)
Obtained postsynaptic density protein fractions were dissolved in HEN buffer that contained 250 mm HEPES (pH 7.7), 1 mm EDTA, and 0.1 mm neocuproine. To avoid rearrangements of thiol-modifying groups, the protein mixture was treated with blocking buffer solution that contained 250 mm HEPES (pH 7.7), 1 mm EDTA, 0.1 mm neocuproine, 5% sodium dodecyl sulfate (SDS), and 50 mm N-ethylmaleimide at 4 °C for 16 h with agitation. To remove excess reagent, the protein extracts were precipitated with 96% of ethanol and resuspended in the same volume of HEN buffer with 2.5% SDS. The obtained protein solutions were then divided into two equal parts. One part was treated with a mixture of 400 μm Biotin PEG Thiol 5kDa (Nanocs) and 1 m hydroxylamine for S-PALM. The other part was used as a negative control for all experiments. All samples were incubated in the dark for 1.5 h at room temperature. To remove excess reagent, the protein extracts were once again precipitated with 96% of ethanol and resuspended in HEN buffer. Next, modified protein mixture was incubated with 100 μl of neutravidin beads at room temperature for 1 h. The neutravidin beads were washed five times in 1 ml of wash buffer. Neutravidin-bound peptides were eluted with 150 μl of elution buffer (25 mm NH4CO3 [pH 8.2] and 5 mm TCEP) The samples were analyzed using Western blotting method.
LC-MS and LC-MS/MS Analysis
For each enriched S-PALM peptide- or S-NO peptide-containing sample, and PSD proteome analysis, separate LC-MS (profile type, peak amplitude data) and LC-MS/MS (peptide identification data) runs were performed. Thermo Orbitrap Elite coupled with Thermo EASY-nLC 1000 was used. S-PALM or S-NO peptides in 0.1% formic acid/water were loaded from a cooled (10 °C) autosampler tray to a pre-column (Symmetry C18, 180 μm × 20 mm, 5 μm; Waters) and resolved on a BEH130 column (C18, 75 mm × 250 mm, 1.7 mm; Waters) in a gradient of 5–30% acetonitrile/water for 70 min at a flow rate of 0.3 μl/min. The ultra-performance LC system was directly connected to the ion source of the mass spectrometer. All MS runs were separated by blank runs to reduce the carry-over of peptides from previous samples. All MS runs were performed in triplicate (for both control and stressed mice). The spectrometer resolution was set to 50.000 for MS acquisitions, with an m/z measurement range of 300–2000 Th. In the LC-MS/MS runs, up to 10 fragmentation events were allowed for each parent ion. Datasets of parent and daughter ions were processed using MascotDistiller 2.5.1 software (MatrixScience).The Mascot search engine (version 2.4.1) was used to survey data against the UniProtKB/Swiss-Prot database (Swissprot 2017_02; 16,905 sequences). The Mascot search parameters were set to the following: taxonomy (Mus musculus), variable modifications (cysteine carbamidomethylation or N-malemideidation, methionine oxidation, peptide tolerance (5 ppm), fragment mass tolerance (5 ppm). Enzyme specificity was set to trypsin with one missed or nonspecific cleavages permitted. The lists of the peptide sequences that were identified in all of the LC-MS/MS runs from control (nsamples = 3) and stressed (nsamples = 3) PSD fractions were merged into one selected peptide list (SPL) using MascotScan software (http://proteom.ibb.waw.pl/mscan/). The SPL consists of sequences of peptides with Mascot scores exceeding the threshold value corresponding to <5% expectation value and FDR<1% calculated by Mascot procedure and unique m/z and LC retention time values. Proteins identified by a subset of peptides from another protein were excluded from the analysis. Proteins that exactly matched the same set of peptides were combined into a single group. Subsequently, contaminants such as keratins were removed from the list. The mass calibration and data filtering described above were also carried out with MScan software.
Raw LC-MS data were converted to the data format of NMRPipe software (http://spin.niddk.nih.gov/NMRPipe) using an finnigan2Pipe data conversion tool. MSparky (http://proteom.ibb.waw.pl/mscan/index.html), an modified version of Sparky NMR software (http://www.cgl.ucsf.edu/home/sparky), was used to convert the LC-MS data into 2D heat maps, in which the peptide's m/z is one dimension and its LC retention time is the other dimension. MSparky allows interactive validation and inspection of the data. The list of assigned MS sequences was simplified so that a single sequence was assigned for peptides with parent entries that differed only by the charge state (z). The SPL list of sequences was used to label peaks in all 2D heat maps (profile data) with the same LC, m/z, and z coordinates using TagProfile software. The software allows for the correction of differences in peptide retention times that are caused by changes in the quality of LC columns and slight differences in LC mobile phase content. Acceptance criteria included deviations in m/z values (<20 ppm), retention times (<2 min), envelope root mean squared error (i.e. a deviation between the expected isotopic envelope of the peak heights and their experimental values, 0.9), and charge state values in accordance with the SPL value. As a result, selected monoisotopic peaks on the 2D heat map were tagged with a peptide sequence. Peptides from the SPL list that were automatically assigned by TagProfile to appropriate signals on 2D heat maps or those that did not meet all of the acceptance criteria were manually verified using MSparky software.
For proteome differential analysis, PSD peptides intensities were determined as the surface of the isotopic envelope of the tagged isotopic envelopes. Lists of identified peptides with corresponding quantitative values were analyzed with Diffprot software (http://proteom.ibb.waw.pl/). Before the analysis, quantitative values were normalized with LOWESS. Proteins with >90% common peptides were clustered and only peptides unique for the cluster were used for statistical analysis. To compare the number of identified S-PALM or S-NO proteins and corresponding sites in experiments we utilized BioVenn software (37). The annotated spectra displaying sequence information of S-PALM and S-NO peptides proteins identified based on single peptide are presented in supplemental Fig. S2. All identified S-PALM and S-NO proteins are presented in supplemental Table S1. All data from PSD proteins analysis are presented in supplemental Table S2.
Cortical Neuronal Culture and Treatment
Dissociated cortical cultures were prepared from newborn (postnatal day 0) C57/6J mice as previously described (38). Cells were plated on six-well plates (for Western blotting) or 13-mm-diameter coverslips (for S-PALM visualization) coated with poly-d-lysine (50 μg/ml) and laminin (2.5 μg/ml) at a concentration of 5 × 104 cells/cm2. The cultures were used for the experiments on day 12 in vitro. Corticosterone, 2-Br, and SNAP were dissolved in ethanol. l-NAME was dissolved in water. Cells were pretreated with either corticosterone (1 μm) or 2-Br (30 μm) for 24 h, followed by 30 min exposure to l-NAME (500 μm) or SNAP (100 μm).
Single-cell In Situ Imaging of Total Protein Palmitoylation
To visualize the palmitoylated total proteome, neuronal cell cultures grown on coverslips were incubated overnight with Alkyne-palmitic acid (50 μm) dissolved in dimethyl sulfoxide (Sigma Aldrich). After washing with phosphate-buffered saline (PBS), the cells were fixed in pre-chilled methanol for 10 min, permeabilized in 0.1% Triton X-100 in PBS, and subjected to click chemistry reaction. Following 1 h incubation at room temperature with click reaction mixture that contained Oregon Green 488 azide (0.1 mm) (Thermo Fisher Scientific), TCEP (0.1 mm), and CuSO4 (0.1 mm), the cells were washed six times with PBS, and the coverslips were mounted in Fluoromount G anti-quenching medium. Images of neurons were acquired using a Zeiss LSM780 laser scanning confocal microscope with a Plan Apochromat 40×/1.4 oil immersion objective using a 488 nm diode-pumped solid-state laser at a pixel count 1024 × 1024. A series of z-stacks were collected for each stimulation with a 0.4 μm step size. The average green fluorescence intensity was determined using ImageJ software. After background subtraction, the results were presented as the mean ± S.E.
Western Blot Analysis of S-palmitoylation and S-nitrosylation Pattern in PSD from Control and Mouse Model of Chronic Stress
Total PSD protein fractions after ABE, or BSM were resolved using reducing 10% SDS-polyacrylamide gel electrophoresis. For analysis of S-PALM or S-NO pattern selectively biotinylated proteins were captured using streptavidin-horseradish peroxidase-conjugated antibodies and visualized by enhanced chemiluminescence (Amersham Biosciences). The level of GAPDH expression was used as a loading control. The membranes were incubated with specific antibody anti GAPDH (Millipore) in dilution 1:10,000, 2 h at room temperature. The membranes were then washed three times in TBST and incubated with anti-rabbit HRP-conjugated secondary antibodies (1:10,000).
Western Blotting Detection of Differentially S-palmitoylated PSDs Proteins
Total PSDs fractions after AbPE were resolved using reducing 10% SDS-polyacrylamide gel electrophoresis. The membranes were first blocked with caseine-based buffer (Sigma) and incubated with primary antibodies Strn4 (1:5000 Strn4 (Santa Cruz)), Shank3 (anti Shank3 1:1000 (Abcam), and PSD-95 (anti PSD-95 1:5000 (Millipore)) followed by secondary HRP-conjugated antibodies. Protein bands were detected using the ECL chemiluminescence system (Amersham Biosciences).
Functional Bioinformatics Analysis
Bioinformatics analyses were performed using Panther software, the BINGO plugin in Cytoscape software (release 3.2.0; http://www.cytoscape.org/), and the STRING database (39–41). Gene Ontology and REACTOME were performed with 122proteins that exhibited stress-induced changes in S-PALM/S-NO crosstalk (identified by the PANIMoni method) using the Panther Classification System (version 9.0; www.pantherdb.org). Proteins were classified based on biological process categories in GO annotations. Functional grouping was based on a Fisher Exact test p ≤ 0.05 and at least two counts. We used here the “synaptic reference set,” containing over 5772 NCBI unique gene identifiers of MS-measured mouse PSD proteins, enriched with non-redundant proteins from our experiments and two most comprehensive, expert - curated synaptic databases SynsysNet (42) and Synaptome DB (43).
Enriched GO terms (relative to our synaptic reference set) for selected protein subsets were retrieved using the BINGO Cytoscape plugin and default hypergeometric statistics with Benjamini-Hochberg FDR correction for multiple testing (39). For the identification of functional annotations, associations, interactions, and networks within our dataset, STRING 9.1 (Search Tool for the Retrieval of Interacting Genes/Proteins) was used (40). A threshold confidence level of 0.7 was used to ensure that only highly confident protein interactions were considered for inclusion in the network. Seven types of protein interactions were used for network generation, including neighborhood, gene fusion, co-occurrence, co-expression, experimental, database knowledge, and text mining.
Experimental Design and Statistical Rationale
The postsynaptic density proteins were isolated from 12- to 14-week-old wild type male C57/BL6 mice were subjected to restraint stress as previously described (33). Mice were sacrificed, brains were dissected, and synaptoneurosomes and postsynaptic density fractions were briefly isolated. For the proteome, three independent biological replicates of control (nsample = 3, Nmice = 3) and chronic stress mice (nsample = 3, Nmice = 3), were analyzed. To analyze S-PALM and S-NO we used a newly developed approach called PANIMoni based on selective cleavage of S-PALM and S-NO by hydroxylamine and ascorbate, respectively. Newly formed thiol was then blocked by biotin, proteins were trypsynized and specific peptides were enriched. With palmiotylproteome and nitrosoproteome, three independent biological replicates (nsample = 3, Nmice = 6, PSDs from two mouse brains were pooled into one) were performed.
For the proteome and each enriched S-PALM or S-NO peptide-containing sample, separate LC-MS (profile type, peak amplitude data) and LC-MS/MS (peptide identification data) runs were performed. The results from the replicates were combined when analyzing the data with MScan and MSparky software and Diffprot software. This number of samples were enough to perform the required tests for determining the statistical significance of the results analysis.
RESULTS
S-palmitoylation and S-nitrosylation Crosstalk in the Primary Neurons
Cysteines of a single protein may be either S-palmitoylated or S-nitrosylated, depending on the physiological context. This may represent a general mechanism of the modulation and fine-tuning of molecular signaling, but it has not yet been characterized in a global type of analysis.
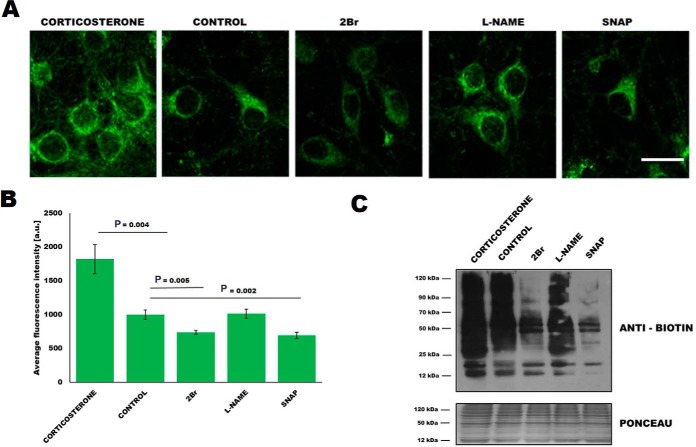
To explore this possibility, we analyzed changes in S-PALM in primary mouse cortical neurons that were treated with the stress hormone corticosterone. Western blotting and in situ cell imaging revealed that corticosterone increased the level of S-palmitoylated proteins in cultured neurons (Fig. 2A, 2B panel 1, Fig. 2C lane 1), which corresponded to our in vivo results. Interestingly, treatment of the cultures with the nitric oxide donor S-nitroso-N-acetyl-dl-penicillamine (SNAP) which caused rapid S-nitrosylation of proteins or S-PALM inhibitor 2-bromopalmitate (2Br) decreased the level of global S-PALM. On the other hand, an inhibitor of nitric oxide synthase, Nω-nitro-l-arginine methyl ester hydrochloride (l-NAME) does not affect protein palmitoylation (Fig. 2A, 2B panel 2–5, Fig. 2C lane 2–5), indicating that not the level of nitric oxide, but only protein S-nitrosylation mediates S-palmitoylation of synaptic proteins. Elevated S-PALM is thus an element of the neuronal response to high levels of corticosterone and can be regulated by S-nitrosylation.
Fig. 2.
Crosstalk between protein S-palmitoylation and S-nitrosylation. A, Imaging of total proteome palmitoylation in neuronal cultures subjected to click chemistry reaction with Oregon Green 488 dye. Scale bar represent 50 μm. B, Quantitation of average fluorescence intensity of Oregon Green under the same treatment conditions as in (A). C, Western blot analysis of ABE used to detect total S-palmitoylome in primary cortical neurons treated with corticosterone, 2Br, SNAP, and l-NAME. Results are presented as the mean ± S.E.
The Synaptic Palmitoyl-Proteome
To analyze the S-PALM of the synaptic proteome under physiological conditions, we used enriched fractions of PSD proteins that were obtained from brain synaptoneurosomes of control mice (supplemental Fig. S1A). Electron micrographs of synaptoneurosomes that were isolated by ultracentrifugation with a discontinuous Ficoll gradient revealed the well-preserved morphology of synaptic structures with a presynaptic mitochondrion, synaptic vesicles, and densely stained membranes that represented PSDs (supplemental Fig. S1B) (21). From the synaptoneurosomal fraction, we isolated enriched PSDs (350–450 μg/single brain) by sucrose density gradient centrifugation, which includes limited solubilization by the non-ionic surfactant Triton X-100. Using Western blotting and MS, we confirmed that all of the fractions were enriched in PSD-95 protein, a postsynaptic marker, whereas the exclusively presynaptic protein synaptophysin was present only in synaptoneurosomal and homogenate samples (supplemental Fig. S1C). We identified a total of 3258 peptides that were assigned to 1639 PSD proteins. The intensities of the peptides that were identified in the PSD fractions from two independent biological replicates were compared on a scatter plot (supplemental Fig. S1D). The correlation coefficient among biological replicates was 0.99, and the mean coefficient of variation (CV) was < 1%, indicating high reproducibility of the procedure. This optimized workflow for PSD isolation was used for all the subsequent PTMs analyses.
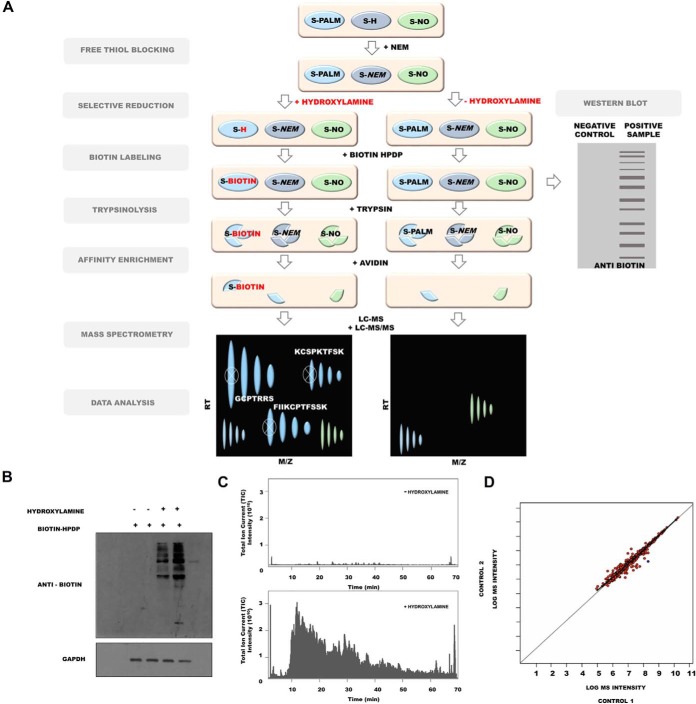
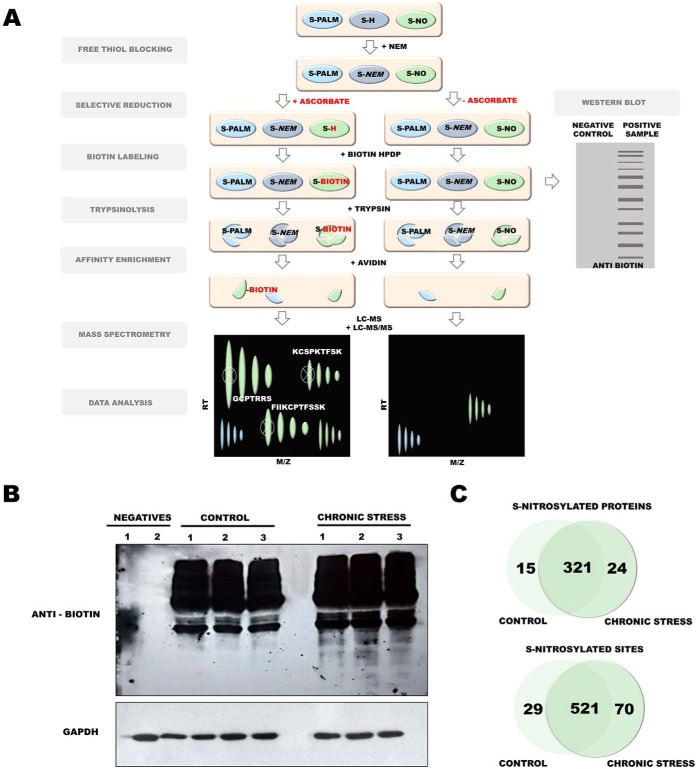
Because of an insufficient amount of protein for PTM analysis that could be obtained from one mouse brain, each sample contained PSDs from two freshly isolated mouse brains (nsamples = 3, Nmice = 6). To assess whether the PSD enrichment protocol enabled the identification of endogenously S-palmitoylated proteins, we used the Acyl-Biotin Exchange (ABE) assay, based on the labeling of S-PALM proteins with a biotin moiety at the site of a modified cysteine (Fig. 3A). Biotinylated proteins were visualized by immunoblotting with an anti-biotin antibody (Fig. 3B). This analysis revealed numerous protein bands across a broad mass range in the PSD fractions that were isolated from the brains of control mice, indicating the presence of endogenously S-PALM proteins. The selectivity of ABE was confirmed by the lack of a signal from biotinylated proteins in samples that were not treated with hydroxylamine, which reduced S-acyl bonds (Fig. 3B).
Fig. 3.
S-palmitoylome of mouse PSD proteins. A, Schematic representation of S-palmitoylation analysis. B, Western blot analysis of S-palmitoylation in the PSD protein fraction. C, Mass spectrometry chromatograms LC-MS/MS showing base peak chromatograms of eluted peptides ions (MS1 mode). Results depict PANIMoni analysis of PSDs proteins in negative control (- hydroxylamine) and positive sample (+ hydroxylamine). Note the detection of peptide ions at low levels in negative control and abundant levels in positive sample. D, Mass spectrometry-based analysis of S-palmitoylation that shows a high correlation between two biological replicates (CV < 1%).
We then sought to identify the exact sites of S-palmitoylated cysteines. To analyze the S-palmitoylome of the PSD fractions, we used a high-throughput proteomic PANIMoni approach. This approach utilizes the PalmPISC method (65) to simultaneously identify S-PALM sites and their cognate proteins in complex biological mixtures, combined with the analysis of 2D heat-maps that represent the m/z versus LC retention times of each peptide ion. Importantly, S-PALM proteins were distinguished from nonspecifically enriched proteins by comparing samples that were treated in parallel with or without hydroxylamine (Fig. 3A). This type of control was used in all experiments based on Western blotting and mass spectrometry. Representative chromatograms from negative (−hydroxylamine) and positive (+hydroxylamine) control samples are shown in Fig 3C. Using MSparky software, 2D heat-maps were generated, and the peptide signals on the 2D heat-maps that were obtained for different biological replicates and their negative controls were correlated and labeled with appropriate peptide sequences. The merged list of all S-PALM PSD peptide sequences that were acquired in the entire LC-MS and LC-MS/MS experiments was used for signal assignments in a 2D heat-map for a single biological replicate. Nonspecifically enriched proteins were excluded based on a simultaneous analysis of the hydroxylamine-free negative control.
Using the PANIMoni approach, 705 S-PALM cysteine sites were identified on 404 distinct proteins in PSD fractions that were isolated from the mouse brains. In all of the samples, we identified mainly cysteine-containing peptides. The MSparky 2D heat-map analysis showed a high correlation among biological replicates (CV < 1%, r = 0.98), indicating the high reproducibility of S-PALM peptide enrichment and measurement (Fig. 3D). The same sites of S-PALM were found in three independent biological replicates. Cysteine-containing peptides and assigned proteins that were identified with high stringency in the MS/MS experiments from three biological replicates are listed in supplemental Table S3. Among them, 344 S-PALM proteins have been previously shown to undergo S-PALM or to have their activity altered by this modification, whereas 60 proteins (from 404 listed) represented newly identified targets of S-PALM.
Dynamic Changes in S-palmitoylation In An Animal Model of Stress-related Diseases
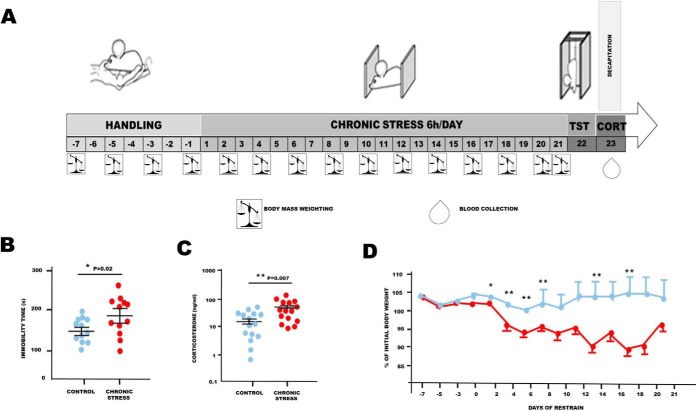
To determine whether the changes in protein S-PALM are associated with the chronic stress response, we used a mouse model of chronic restraint stress in which aberrant plasticity has been well documented (Fig. 4A) (32, 44–48). Mice that were subjected to restraint stress for 21 days (6 h/day) exhibited depressive-like behavior (i.e. an increase in immobility time in the tail suspension test (p < 0.05), elevated serum corticosterone levels (p < 0.01), and a decrease in body mass (p < 0.01), compared with control animals (Fig. 4B–4D).
Fig. 4.
Schematic representation of the mouse model of chronic restraint stress and behavioral tests. A, Schematic illustration of mouse treatment. B, Tail suspension test. Results are represented as mean ± S.E. with Nmice = 12 in each group. C, Corticosterone levels. Results are represented as mean ± S.E. with Nmice = 15 in each group. D, Changes in body mass recorded every other day throughout the experiment. Results are represented as mean ± S.E. with Nmice = 15 in each group. Values are significant at P * <0.05, ***<0.01 as compared with control group. Statistical significance was determined by the heteroscedastic two-tailed t test.
Because changes in protein expression may interfere with the palmitoylome profile, we first analyzed the effects of chronic restraint stress on the level of PSD proteins. By employing a quantitative proteomics approach (label-free method coupled with 2D heat-map analysis) we identified 1247 protein clusters containing 1639 proteins (single proteins or clusters of gene identifiers that matched the same set of peptides) that were represented by at least two peptides and met other inclusion criteria, such as the correct m/z, retention time, and peak envelope (supplemental Table S4).
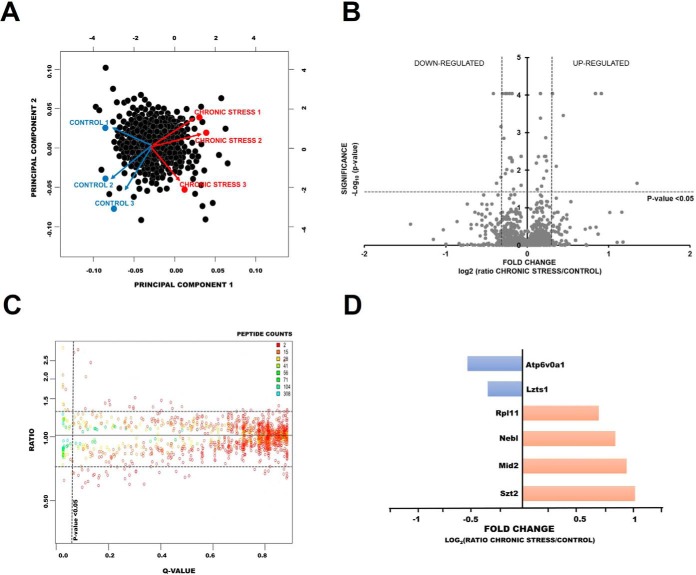
Principal component analysis (PCA) was used to assess the level of similarity of protein content. Within each group of control mice (Nmice = 3) and chronically stressed mice (Nmice = 3), we confirmed the reproducibility of the PSD proteome preparation and found a clear distinction between control and stressed mice Control samples separated from chronic stress samples along both PCA components PC1 and PC2 represent 32 and 11% of variance respectively. Therefore, the impact of individual variance is not strong enough to overcome chronic stress-specific differences (Fig. 5A). The results of the relative quantification experiment are summarized in Fig. 5B–5D. Volcano plot showing proteins differentially expressed between chronic stress and control mice is presented in Fig. 5B. Proteins with statistically significant differential expression (≥0,5-fold change, p < 0.05) are located in the right upper and left quadrants. Fold-change indicates the magnitude of the increase or decrease in abundance. Additionally, we performed the Diffprot analysis shown in Fig. 5C. The ratio indicates the direction and magnitude of change, with more than 1.0 indicating an increase, whereas less than 1 indicting a decrease in the protein abundance. Color coded peptides counts indicates the number of unique peptides for each protein used for identification and averaged abundance measurement. Using very stringent criteria (ratio > 1.25 and q < 0.05) we identified six significantly changed proteins: four up-regulated proteins, such SZT2 protein, LIM zinc-binding domain-containing Nebulette, probable E3 ubiquitin-protein ligase MID2, 60S ribosomal protein L11 and two downregulated proteins, V-type proton adenosine triphosphatase [ATPase] 116 kDa proteolipid subunit and Leucine zipper putative tumor suppressor. Detailed analysis of changes for these proteins (bar graphs) is presented in Fig. 5D. The raw protein ratios, fold changes and q values are listed in supplemental Table S4.
Fig. 5.
Analysis of changes in protein expression in mouse brains after chronic restraint stress. A, Principal Component Analysis of identified proteins in control and chronically stressed mice. Black points represent proteins; the color-coded points represent samples, either from control (blue) or stressed (red). The 2-dimensional graph represents the first two main components of the samples. As expected, the control/stressed status is the principal component responsible for the differences among samples. (nsamples = 3, Nmice = 3) B, Volcano plot showing proteins differentially expressed between control and chronic stress PSDs (nsamples = 3, Nmice = 3). Proteins with statistically significant differential expression (≥0.5-fold change <0.5, p < 0.05) are located in the right upper and left quadrants. C, Distributions of the log-ratios (vertical axis) and q values (horizontal axis) of proteins from control and chronically stressed mice (data analyzed using Diffprot). The numbers of peptides per protein are color-coded as shown in the inserts. In the Diffprot analysis, 6 protein clusters out of 1293 were assigned q-values below 0.05 (i.e. they are significantly regulated). D, Bar-graphs show detailed differential analysis of down and up-regulated proteins after chronic stress.
Next, to determine whether chronic stress affects protein S-PALM, we analyzed the changes in the patterns of this modification. Using the ABE method combined with Western blotting, we observed an increase in the number and intensity of bands that represented S-palmitoylated proteins in PSDs from stressed mice (Fig. 6A, lanes 6–8) compared with control mice (Fig. 6A, lanes 3- 5). In negative control experiments, no bands were detected confirming selectivity of hydroxylamine reduction of S-PALM-bonds. (Fig 6A, lanes 1–2).
Fig. 6.
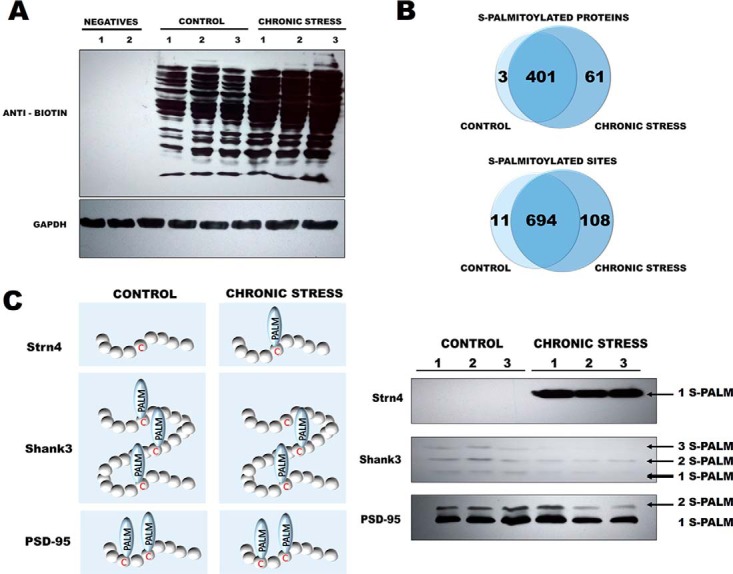
Analysis of changes in protein S-palmitoylation in mouse brains after chronic restraint stress. A, Western blot analysis of S-palmitoylation pattern in control mice and chronically stressed mice. S-palmitoylation sites were selectively labeled with S-S-biotin (ABE). The visualization of biotinylated proteins was achieved using anti-biotin antibodies. (nsamples = 3, Nmice = 6; pulled PSDs fractions from two mice per one sample). Negatives - negative controls without specific reduction of S-acyl bonds with hydroxylamine (pulled 3 negative controls into 1 control (negatives 1) and 1 chronic stress (negatives 2). B, Venn diagram comparisons of the numbers of S-PALM proteins and sites that were identified in synaptic protein fractions from control mice and chronically stressed mice. (nsamples = 3, Nmice = 6; pulled PSDs fractions from two mice per one sample). C, Schematically depicted results from mass spectrometry for the newly discovered targets of S-PALM, Striatin4 Shank3 and a well-known PSD-95. Differential analysis of S-PALM in control and chronic stress PSDs using the Acyl biotin-PEG exchange method (AbPE) reveals site-specific S-PALM of protein targets, Striatin-4 and Shank3. S-palmitoylation of PSD protein was used as an internal control (nsamples = 3, Nmice = 6).
To further confirm that stress increases the S-PALM of proteins, we performed a detailed, differential analysis of S-PALM at the level of single cysteines using mass-spectrometry-based the PANIMoni approach. The inclusion criteria used for selection of differential protein sets proposed in our work were very strict. The protein was defined as differentially S-palmitoylated only when it was repeatedly observed in PSDs after chronic stress and was not detected in age-matched controls. This led to highly selective identification of differentially S-PALM synaptic proteins after chronic stress. We identified 813 S-PALM cysteine sites that were assigned to 465 proteins in the PSD fractions (supplemental Table S5A–S5C). Among the 465 S-PALM PSD proteins that were identified, three were found only in control mice: Sn1-specific diacylglycerol lipase beta Peptidyl-prolyl cis-trans isomerase FKBP8 or d-beta-hydroxybutyrate dehydrogenase and 61 were exclusively found in stressed mice, e.g. striatin-4, MAGUK p55 or Catenin delta-2 (Fig. 6B). The number of differential S-PALM-peptides (i.e. S-PALM-sites) in control and stressed mice differs from that of differential proteins, suggesting that some proteins are possibly sequentially S-palmitoylated. A total of 813 sites were identified, including 11 that were uniquely detected in control PSD fractions and 108 sites that were found exclusively in PSDs in stressed mice (Fig. 6B). In addition, negative controls were used to distinguish putative S-PALM peptides from contaminating peptides; thus, the identified S-PALM sites were strictly evaluated. Subsequently, probable contaminants (keratin) were removed from the list. All identified peptides identified in the experiment contain cysteine residues (supplemental Table S1). Total ion chromatograms from negative controls and positive samples from all analyzed samples are shown in supplemental Fig. S3. Fig. 6C schematically depicts the possibilities for S-PALM-based regulation of PSDs proteins as a result of chronic stress in mice. Seventy-seven proteins were identified for the first time as the S-palmitoylation targets, where about 16% of the total, of which 15 were solely present in chronic stress PSDs (supplemental Table S5A–S5C). Next, Western blot analysis was used to validate the results of MS-based identifications of the newly identified targets of S-palmitoylation (Fig. 6D). Immunoreactivity was traced in enriched PSDs during Acyl biotin-PEG Exchange (AbPE) enrichment from control and chronic stress mice. Using the AbPE method, we demonstrated that Strn4, Shank3 and PSD-95 are endogenously S-PALM in the mice brain. Moreover, we confirmed that chronic stress affects S-PALM of Strn4 and Shank3. We showed differential S-PALM of Shank3 protein at the level of single specific sites of modification after chronic stress treatment. To confirm the utility of AbPE method, we analyzed PSD-95 protein S-palmitoylation, a well-characterized S-palmitoylated protein.
Mechanism of Dynamic S-palmitoylation Regulation
To characterize crosstalk between S-PALM and S-NO, we analyzed the global protein S-NO profile in chronic restraint stress model of aberrant synaptic plasticity in mice using S-NO part of the PANIMoni approach (Fig. 7A).
Fig. 7.
Differential protein S-nitrosylation after chronic stress. A, Schematic representation of S-nitrosylation analysis. B, Western blot analysis of S-nitrosylation pattern in control mice and chronically stressed mice. S-nitrosylation sites were selectively labeled with S-S-biotin (BSM). The visualization of biotinylated proteins was achieved using anti-biotin antibodies. (nsamples = 3, Nmice = 6; pulled PSDs fractions from two mice per one sample) Negatives - negative controls without specific reduction of S-NO bonds with ascorbate (pulled 3 negative controls into one (negatives 1) and chronic stress (negatives 2). C, Venn diagram comparisons of the numbers of S-NO proteins and sites that were identified in synaptic protein fractions from control mice and chronically stressed mice.
To assess whether the chronic stress causes changes in endogenous protein S-NO, we used a BSM, in which S-NO proteins are selectively labeled with biotin, followed by Western blotting detection with anti-biotin antibodies. We identified numerous protein bands across a broad mass range, both in the control and chronic stress derived PSDs fractions indicating the presence of endogenously S-NO proteins (Fig. 7B). The number and intensity of specific bands were changed after chronic stress. In negative control experiments, no bands were detected confirming selectivity of ascorbate reduction of SNO-bonds.
To obtain more detailed changes in S-nitrosylome, we performed an analysis based on mass spectrometry. The analysis generated lists of peptides that were obtained separately for stressed mice and age-matched controls. Similarly to S-PALM, negative controls (omitting of ascorbate) were used to distinguish S-NO peptides from contaminating peptides. Total ion chromatograms for all analyzed samples are shown in supplemental Fig. S4. Protein IDs were assigned to the appropriate peptide sequences and are listed in supplemental Table S6A–S6C.
A total of 620 S-NO cysteine sites that were assigned to 360 proteins were identified among PSD proteins (Fig. 7C). A total of 39 differential protein species were identified. Fifteen proteins appeared only in controls, e.g. Rho GTPase-activating protein 23, phosphatase and actin regulator 1, whereas 24 proteins were modified only in PSD fractions that were obtained from stressed mice, such as glutamate receptor ionotropic, NMDA 1 or neuronal proto-oncogene tyrosine-protein kinase Src. Of the 620 S-NO sites that were identified, 29 were found only in control PSDs, and 70 were detected only in stressed mice. These nitrosopeptides represented 63 uniquely regulated proteins after chronic stress (supplemental Table S6B and S6C). Of all identified S-NO proteins, 107 were identified for the first time as new S-nitrosylation targets.
Complete lists of S-palmitoylated and S-nitrosylated PSD proteins from the brains of chronically stressed and control mice enabled us to perform detailed comparisons of these PTM profiles at the level of single cysteines. The results of the high-throughput MS experiments revealed two sets of S-PALM proteins and two sets of S-NO proteins that corresponded to the chronic stress model and age-matched controls.
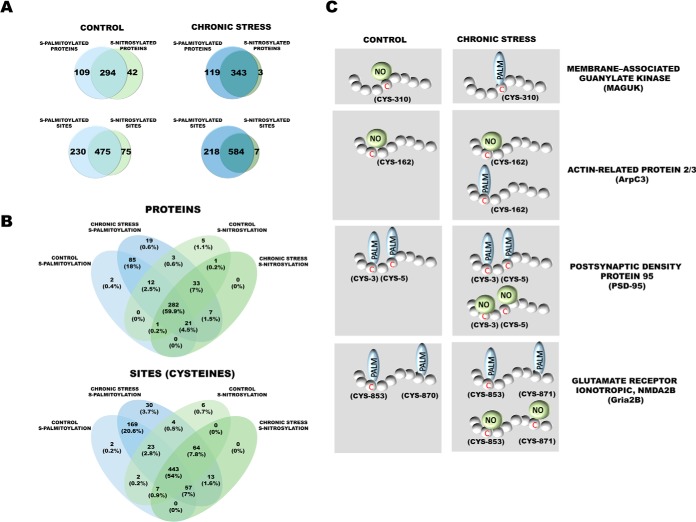
Of the 446 modified PSD proteins that were identified in control mice, 42 were exclusively S-nitrosylated, and 109 were exclusively S-palmitoylated (Fig. 8A). At the level of specific sites of modification (cysteines level), we observed that many cysteines exist only in one PTM form. Among 778 cysteines, 295 cysteines occur only in S-PALM form (assigned to 198 proteins) and 75 in S-NO form (assigned to 61 proteins). Summarizing proteins exclusively modified by one PTM at the level of proteins and sites we can conclude most of the protein cysteines (58% of the proteins that were identified) occurred only in one of the PTM types in control mice.
Fig. 8.
Aberrant S-palmitoylation/S-nitrosylation crosstalk caused by chronic stress. A, Venn diagram comparisons of the numbers of S-PALM and S-NO proteins and sites that were identified in synaptic protein fractions from control mice and chronically stressed mice. B, 4-Tailed Venn diagram comparison of all identified S-PALM and S-NO proteins. C, S-PALM pattern in control mice and chronically stressed mice that shows differential S-PALM for important synaptic proteins.
Chronic stress generally results in a greater number of modified cysteine sites that undergo both S-PALM and S-NO. Interestingly, in chronically stressed mice, among the 464 modified proteins that were identified, only two were exclusively associated with nitric oxide, and 119 were exclusively associated with palmitate (Fig. 8A). Notably, the differences between the sets of S-NO and S-PALM proteins at the cysteine (site) level were also much less pronounced, and only seven peptides were exclusively modified by nitric oxide. The remainder set of the cysteine sites and proteins that were identified existed in two modified forms after chronic stress. The crosstalk between the S-NO and S-PALM of brain PSD proteins appears to vary, depending on the physiological condition.
Finally, we compared all sets of proteins, among a total of 813 S-PALM and 620 S-NO cysteine sites that were characterized on 465 and 360 proteins, respectively, we sought to identify those that were differentially affected by stress.
We obtained evidence that 443 cysteine sites (54% of all the sites that were identified) that belonged to 297 proteins in all of the experimental conditions occurred in both S-PALM and S-NO forms. Interestingly, the remainder of the cysteines that were identified (46%) underwent regulation that strictly depended on the biological conditions (Fig. 8B).
One hundred twenty-two among all the modified proteins underwent changes in the patterns of S-NO and S-PALM that were induced by chronic stress. All these proteins grouped according to their molecular function are presented in Table I. Interestingly, for two synaptic proteins, Iqsec2 and Maguk, we observed an exchange of S-PALM to S-NO after chronic stress. Although, much more frequent were observed changes where cysteine sites modified exclusively by one PTM in control conditions, undergo both S-PALM and S-NO after chronic stress. All types of molecular switches that were found between these two PTMs that were triggered by chronic stress are shown in Fig. 8C.
Table I. Identified PSDs proteins with affected S-PALM/S-NO crosstalk.
| Protein class | Gene name | S-PALM studies | SNO studies |
|---|---|---|---|
| Bolded | Underlined | ||
| Cell adhesion proteins | |||
| Ctnna190,91,92, Cntnap193,94,111, Ncam192,93,94,99,100,111, Ctnnd190,91,92,94,95,97,98,111, Ctnnb190,91,97,111, Jup90,96,97,111, Jam398, Ctnnd290,91,93,112, Src96,112 | 90. Shen LF et al., 2017, 91. Collins MO et al., 2017, 92. Chesarino NM et al., 2014, 93. Wan J et al., 2013, 94. Ren W et al., 2013, 95. Sobocińska J et al., 2018, 96. Zhang X et al., 2018, 97. Gould NS et al., 2015, 98. Li Y et al., 2012, 99. Lievens PM et al., 2016, 100. Ponimaskin E et al., 2008 | 111. Chen, Y.J., et al., 2015, 112. Seneviratne, U., et al., 2016, 113. Ryu, I.H., K.Y. Lee, and S.I. Do, 2016 | |
| Cytoskeletal proteins | |||
| Tubb4a101,94,111, Map1a91,93,111, Ctnna190,91,92, Ablim196,111, Arpc392,96,114, Map1b93,96,111, Dync1h191,92,93,111, Ctnnd190,91,92,94,95,97,98,111, Tuba4a90,111, Plec90,92,94,111, Tuba1a93,108, Fscn192,93,94,111, Myo5a93, Coro1c90,95,96,102,111, Sept496, Opa1102,111, Ctnnd290,91,93,111, Epb41l393,114, Inf2103,115, Map6d1105, Iqsec2112, Ank291,93, Iqsec1112, Actn190,97,101,,104,111, Ank393,116, Sptbn192,93,94,96,97,111, Syne1114, Cyfip192,93,104,117, Phactr1 | 90. Shen LF et al., 2017, 91. Collins MO et al., 2017, 92. Chesarino NM et al., 2014, 93. Wan J et al., 2013, 94. Ren W et al., 2013, 95. Sobocińska J et al., 2018, 96. Zhang X et al., 2018, 97. Gould NS et al., 2015, 98. Li Y et al., 2012, 101. Thinon E et al., 2018, 102. Kang R et al., 2008, 103. Wei X et al., 2014, 104. Thinon (15) E et al., 2016, 105. Gory-Fauré S et al., 2006, 108. Martin BR et al., 2011 | 111. Chen, Y.J., et al., 2015, 114. Tooker, R.E. and J. Vigh, 2015, 112. Seneviratne, U., et al., 2016, 115. Ben-Lulu, S., et al., 2017, 116. Ulrich, C., et al., 2012, 117. Nicolas, F., et al., 2015 | |
| Structural proteins | |||
| Crocc106,107,114, Ina93,96,111, Bsn91,93,112, Pclo112 | 91. Collins MO et al., 2017, 93. Wan J et al., 2013, 96. Zhang X et al., 2018, 106. Pinner AL et al., 2016, 107. Hernandez JL et al., 2016 | 111. Chen, Y.J., et al., 2015, 112. Seneviratne, U., et al., 2016, 114. Tooker, R.E. and J. Vigh, 2015 | |
| Chaperonins | |||
| Ywhah92,95,101,111, Dnajc1190,91,94,108,111 | 90. Shen LF et al., 2017, 91. Collins MO et al., 2017, 92. Chesarino NM et al., 2014, 94. Ren W et al., 2013, 95. Sobocińska J et al., 2018,101. Thinon E et al., 2018,108. Martin BR et al., 2011 | 111. Chen, Y.J., et al., 2015 | |
| Cytokine | |||
| Cmtm4 | |||
| Receptors/channels | |||
| Adgrb2, Leprotl190,91, Gabbr2, Gria196,106, Atp1a391,93,111, Adam2296,111 | 90. Shen LF et al., 2017, 91. Collins MO et al., 2017, 93. Wan J et al., 2013, 96. Zhang X et al., 2018 106. Pinner AL et al., 2016 | 111. Chen, Y.J., et al., 2015 | |
| Ion/electron transporting proteins | |||
| Ndufs290,94,96,111, Ndufs190,91,93,94,97,101,111, Dld90,93,94,97,104,111, Slc25a194,97,114, Slc25a390,91,92,93,94,95,104,111, Slc12a593,114 | 90. Shen LF et al., 2017, 91. Collins MO et al., 2017, 92. Chesarino NM et al., 2014, 93. Wan J et al., 2013, 94. Ren W et al., 2013, 95. Sobocińska J et al., 2018, 96. Zhang X et al., 2018, 97. Gould NS et al., 2015, 101. Thinon E et al., 2018, 104. Thinon E et al., 2016 | 111. Chen, Y.J., et al., 2015, 114. Tooker, R.E. and J. Vigh, 2015 | |
| Receptor binding proteins/receptor modulators | |||
| Lgi1102,112, Ctnnb190,91,97,96,111, Prkcb91,93,111, Ly6h, Ank291,93 | 90. Shen LF et al., 2017, 91. Collins MO et al., 2017, 93. Wan J et al., 2013, 96. Zhang X et al., 2018, 97. Gould NS et al., 2015, 102. Kang R et al., 2008 | 111. Chen, Y.J., et al., 2015, 112. Seneviratne, U., et al., 2016 | |
| Transferases | |||
| Mecp296, Acly90,92,93,95,97,101,104,111, Crat96,111, Tst90,97,96,111, Acadsb111,90, Gsk3b96,111, Cdc42bpb, Comtd196,111, Prkcb91,93,111, Mpp291,93,96,111, Cs93,97,104,95,101,111, Gk90,96,97,111, Got290,92,93,95,97,104,109,111, Src96,113, Hk191,93,94,95,98,101,104,111, Pkm90,93,94,101,111, Ogt113, Kalrn96, Begain, Ngef | 90. Shen LF et al., 2017, 91. Collins MO et al., 2017, 92. Chesarino NM et al., 2014, 93. Wan J et al., 2013, 94. Ren W et al., 2013, 95. Sobocińska J et al., 2018, 96. Zhang X et al., 2018, 97. Gould NS et al., 2015, 101. Thinon E et al., 2018, 104. Thinon E et al., 2016, 109. Yount JS et al., 2010 | 111. Chen, Y.J., et al., 2015, 113. Ryu, I.H., K.Y. Lee, and S.I. Do, 2016 | |
| Protein transporters | |||
| Dync1h192,93,97,111, Ap1b193,97,96,111, Ap3d196,104, Exoc1, Exoc296, Exoc5, Mtx190, Ank291,93, Snph91,96,112 | 90. Shen LF et al., 2017, 91. Collins MO et al., 2017, 92. Chesarino NM et al., 2014, 93. Wan J et al., 2013, 96. Zhang X et al., 2018, 97. Gould NS et al., 2015, 104. Thinon E et al., 2016 | 111. Chen, Y.J., et al., 2015, 112. Seneviratne, U., et al., 2016 | |
| GTPases/signaling proteins | |||
| Homer196,117, Kalrn96, Plcb191,93,96,117, Sirpa114, Rasal191,93, Tubb4A94,101,111, Gnai290,91,92,93,94,95,98,104,108,109,110,111, Eef1a291,93,94,111, Tbc1d2496, Kras93,92,108,111, Sept496, Opa196,102,111, Rap1b92,101,104,108,111 | 90. Shen LF et al., 2017, 91. Collins MO et al., 2017, 92. Chesarino NM et al., 2014, 93. Wan J et al., 2013, 94. Ren W et al., 2013, 95. Sobocińska J et al., 2018, 96. Zhang X et al., 2018,98. Li Y et al., 2012,101. Thinon E et al., 2018,102. Kang R et al., 2008, 104. Thinon E et al., 2016,108. Martin BR et al., 2011, 109. Yount JS et al., 2010, 110. Merrick BA et al., 2011 | 111. Chen, Y.J., et al., 2015, 114. Tooker, R.E. and J. Vigh, 2015, 117. Nicolas, F., et al., 2015 | |
| GTPase modulating proteins | |||
| Rasa3, Sbf1, Ngef, Dock5 | |||
| Nucleic acid binding proteins | |||
| Rps3a90,92,94,97,109,111, Sirt291, Eef1a291,93,94,111, Mecp296, Tst90,96,97,111 | 90. Shen LF et al., 2017, 91. Collins MO et al., 2017, 92. Chesarino NM et al., 2014, 93. Wan J et al., 2013, 94. Ren W et al., 2013, 96. Zhang X et al., 2018, 97. Gould NS et al., 2015, 109. Yount JS et al., 2010 | 111. Chen, Y.J., et al., 2015 | |
| Enzymatic activity proteins | |||
| Plcb191,93,117, Prkcb91,93,111, Src96,113, Hk191,93,94,95,98,104,111, Adam2296,111, Bphl90,92,97,111, Ass190,95,97,111, Acly90,92,93,95,97,104,111, Aldoa90,92,93,94,95,101,111, Idh3a92,93,94,95,97,104,111, Pdha190,91,93,94,97,111, Mdh190,92,93,94,95,97,104,111, Acadsb90,111, Ogdh90,91,92,93,94,95,111, Aldh3b192,94,95,101, Marc290,97,112, Pdhb90,92,93,94,111, Got290,92,93,95,97,109,111, Bdh190,91,97,111 | 90. Shen LF et al., 2017, 91. Collins MO et al., 2017, 92. Chesarino NM et al., 2014, 93. Wan J et al., 2013, 94. Ren W et al., 2013, 95. Sobocińska J et al., 2018, 96. Zhang X et al., 2018, 97. Gould NS et al., 2015, 98. Li Y et al., 2012, 101. Thinon E et al., 2018, 104. Thinon E et al., 2016, 109. Yount JS et al., 2010 | 111. Chen, Y.J., et al., 2015, 112. Seneviratne (23), U., et al., 2016, 113. Ryu (24), I.H., K.Y. Lee, and S.I. Do, 2016, 117. Nicolas, F., et al., 2015 | |
| Scaffolding Synaptic proteins/ Synapse organization | |||
| Pclo112, Homer196,117, Mpp291,93,96,111, Shank2, Dlgap291,96, Ank393,116, Lrrtm291, Bsn91,93,112 | 91. Collins MO et al., 2017, 93. Wan J et al., 2013, 96. Zhang X et al., 2018 | 111. Chen, Y.J., et al., 2015, 112. Seneviratne, U., et al., 2016, 117. Nicolas, F., et al., 2015, 116. Ulrich, C., et al., 2012 | |
| Developmental proteins | |||
| Ctnnd290,91,93,112, Snph91,96,112, Cyfip192,93,104,117, Hapln1112, Dip2a, Tspan291,118 | 90. Shen LF et al., 2017, 91. Collins MO et al., 2017, 92. Chesarino NM et al., 2014, 93. Wan J et al., 2013, 96. Zhang X et al., 2018, 104. Thinon E et al., 2016, | 112. Seneviratne, U., et al., 2016, 117. Nicolas, F., et al., 2015 118. Gu, L. and R.A. Robinson, 2016 | |
| Other | |||
| Tmem50b91, Wdr793,117, Strn4111, Gm99696, Snph91,96,112, Dip2b, Amer101,104,112, Gbas91,96,111, Ncdn91,93,111 | 91. Collins MO et al., 2017, 93. Wan J et al., 2013, 96. Zhang X et al., 2018, 101. Thinon E et al., 2018, 104. Thinon E et al., 2016 | 111. Chen, Y.J., et al., 2015, 112. Seneviratne, U., et al., 2016, 117. Nicolas, F., et al., 2015 |
S-palmitoylation and S-nitrosylation Affect the Function of Various PSD Proteins
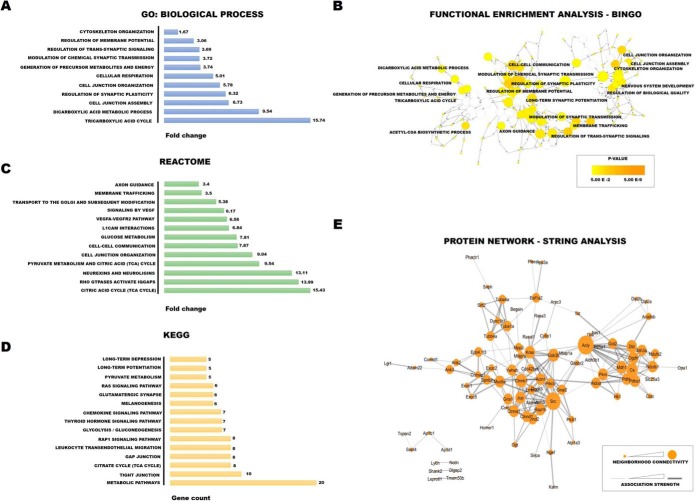
To investigate the biological importance and functional significance of PSD proteins that exhibited changes in S-PALM/S-NO crosstalk, we performed functional enrichment analysis using Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) (41, 49). Overrepresentation statistics were used to calculate the probability of highly populated protein classes, pathways, and GO terms among S-PALM and S-NO proteins that occurred in a non-random manner. Indeed, many of the protein categories were overrepresented in the data. As a reference, we used the set that comprised 5772 proteins that were associated with PSDs. The reference set was generated from our experimental data and comprehensive, expert - curated synaptic databases SynsysNet (42); and Synaptome DB (43).
Gene Ontology biological process (GO_BP) analysis linked a majority of the proteins with stress-induced changes in S-PALM/S-NO crosstalk to processes that are associated with synaptic plasticity, such as regulation of synaptic plasticity (fold change = 6.32, p = 0.032), regulation of trans-synaptic signaling (fold change = 3.69, p = 0.0002), modulation of chemical synaptic transmission (fold change = 3.72, p = 0.0002) (Fig. 9A). Further, energy metabolism (fold change = 15.74, p = 0.038, tricarboxylic acid cycle; fold change = 3.74, p = 0.003, generation of precursor metabolites and energy, fold change = 5.01, p = 0.047, cellular respiration) and protein localization (fold change = 1.87, p = 0.0001, actin cytoskeleton organization, fold change = 5.78, p = 0.013, cell junction organization) were also significantly overrepresented.
Fig. 9.
Bioinformatics tools for the analysis of proteins with aberrant S-PALM/S-NO crosstalk. A–C, Panther software analysis in terms of (A) biological processes (GO_BP), (B) REACTOME pathway analysis, (C) KEGG pathway analysis (D) STRING analysis of protein interactome. E, Bingo analysis of enriched biological classes. The analysis was done using the 'hyper geometric test', and all GO terms that were significant with p < 0.05 (after correcting multiple term testing by Benjamini and Hochberg false discovery rate corrections) were selected as overrepresented.
We visualized biological process overrepresentation using the BINGO plugin (Fig. 9B). To visualize GO terms and biological pathways that are associated with synaptic proteins that exhibited stress-induced changes, we used BINGO bioinformatics software in the integrative environment of Cytoscape 3.1. Significantly overrepresented biological processes, based on GO terms, were visualized in Cytoscape. The size of a node is proportional to the number of targets in the GO category. The color represents enrichment significance. A deeper color on the color scale indicates higher enrichment significance. p values were adjusted using Benjamini and Hochberg False Discovery Rate (FDR) correction.
In addition, we analyzed proteins that exhibited changes in PTM crosstalk using REACTOME pathways. Most of the significantly enriched (q < 0.05) proteins are related to energy metabolism (e.g. glucose metabolism, fold change = 7.81, p = 0.018, TCA cycle, fold change = 15.43, p = 0.033, and pyruvate metabolism, fold change = 9.54, p = 0.019), protein localization (e.g. fold change = 3.5, p = 0.000012, membrane trafficking and fold change = 5.38, p = 0.0002, transport to the GOLGI and subsequent modifications), and synapse signaling (e.g. fold change = 6.17, p = 0.017, signaling by vascular endothelial growth factor (VEGF), fold change = 13.11, p = 0.016, neurexins and neuroligins, and fold change = 13.99, p = 0.04, RHO GTPases activate IQGAP; Fig. 9C).
The KEGG pathway enrichment results associated the target proteins with both energy metabolism (citrate cycle [TCA cycle], glycolysis/gluconeogenesis, and pyruvate metabolism) and synaptic plasticity (long-term depression, long-term potentiation, and glutamatergic synapse-connected terms as enriched), which was consistent with the GO and KEGG analyses (Fig. 9D).
Most proteins exert their biological functions by interacting with each other. To uncover functional aspects that are associated with proteins that are affected by chronic stress, we used the STRING database. Overall, at a medium STRING confidence level > 0.7, 84.0% of the identified proteins were grouped in a single cluster, with an enrichment ratio of 3.15 over the randomly expected ratio. Further, two smaller connection clusters were identified (Fig. 9E). Likewise, proteins that had changes in S-PALM/S-NO balance were associated with synaptic plasticity regulation, protein localization, and energy metabolism.
Our dissected analysis uncovered a remarkable degree of the stress-induced disruption of S-PALM/S-NO crosstalk in proteins that play key roles in proper synaptic function. This phenomenon may be a major event that is involved in the development of stress-related aberrant plasticity.
DISCUSSION
Crosstalk among different PTMs of proteins is an emerging theme in neurobiology. Proteins can be modified by multiple PTMs at the same residue which implies that they are mutually exclusive at a given time. This direct crosstalk of PTMs acting as specific PTM code that modulates protein function. Proteins are frequently modified not only by a single type of PTM but also by several different PTMs. Therefore, to elucidate the role of individual PTMs in the context of specific protein activities, we must first understand their co-occurrence in the same protein or at the same protein site and their functional profile. However, studies of PTMs crosstalk on specific site have been hindered because of a lack of suitable methods. Most of proteomic studies concerns on one modification at a time what do not permit critical investigations into the interface, crosstalk and of these modifications at the proteome level.
The reciprocal dynamics between S-PALM and S-NO have been described in the context of synaptic proteins (24). These studies, however, were limited to single proteins and did not consider the global effects of alterations of S-PALM/S-NO crosstalk in complex physiological states.
For the purpose of the present study, we designed and developed a new proteomic approach called PANIMoni. It is an MS-based method that allows the differential, site-specific identification of PTMs of endogenous synaptic proteins. PANIMoni, in contrast to commonly used spectral counting methods, relies on peptide intensity quantification combined with protein identification. Its high sensitivity makes it suitable for comprehensively assessing changes in protein PTMs under different physiological and pathological conditions. Further, our approach can be extended to quantitatively analyze different types of PTMs, such as sulfenylation and glutathionylation, in which the selective liberation of modifications and switch to biotin are possible.
Using the PANIMoni approach, we were able to perform a large-scale analysis of cysteine PTMs, namely S-PALM and S-NO, within PSD proteins of the mouse brain. Moreover, the inclusion criteria used for selection of differential protein sets proposed in our work were very stringent. The protein was defined as differentially modified only when it was repeatedly observed in the postsynaptic density fraction of chronic stress, and not observed in age-matched controls. In our opinion, this led to the highly selective identification of differentially modified postsynaptic density proteins after chronic stress exposure.
Our goal was to test the hypothesis that stress-related aberrant plasticity is associated with global changes in the reciprocal interplay between these two PTMs. By reducing the complexity of the system that was analyzed and narrowing our research to analyzing PSD proteins in a well-defined mouse model of chronic stress, we obtained highly reproducible and informative results.
To better understand the stress-dependent alternations in synaptic signaling pathways, we reduced the complexity of the analyzed system and narrowed our research to the analysis enriched PSD proteins. The multistep enrichment procedure and LC-MS/MS analysis allow the identification of core proteins previously known to be biochemically enriched in PSD such as PSD-95, Maguk, or Shank. On the other hand, we have identified numerous proteins that have not previously been described at the PSD fraction, such as Gnao 3 or Rapgef4. The strong interaction beetween pre- and postsynaptic density proteins results in the presence of presynaptic proteins such as basson or picollo in our enriched PSD fraction. In the enriched PSD fraction we have also identified other proteins that seem to be contaminants, such as metabolic protein, mitochondrial myelin or astrocytic proteins. All these proteins are very abundant in the brain. Similar contaminations were noted in the previously described mass spectrometry analysis of PSDs (3, 50–52). On the other hand, some proteins considered for many years as contamination, such as glyceraldehyde-3-phosphate dehydrogenase, have been identified in postsynaptic density by immunocytochemistry and it has been shown to provide ATP to PSD (53).
The high-throughput analysis showed that postsynaptic proteins are strictly regulated by both cysteine modifications, S-PALM and S-NO, under physiological conditions. Our detailed analysis strictly defined 813 S-palmitoylated sites that were assigned to 465 proteins and 620 S-nitrosylated sites that were assigned to 365 proteins.
Importantly, the number of proteins were exclusively modified with one of the two PTMs. A competitive relationship between S-PALM and S-NO for neuronal proteins, including SNAP-25 and PSD-95, has been previously reported (24). Similar to previous studies, the proteins that we identified were associated with various signaling pathways (e.g. axon guidance, synaptic plasticity, and protein localization) and cell energy metabolism.
We also provide insights into the ways in which chronic stress affects S-PALM and S-NO. The analysis of PSD fractions from the brains of chronically stressed mice showed a substantial increase in the overall number of palmitoylated proteins compared with control animals. Interestingly, this elevation of protein S-PALM correlated with changes in the number of both nitrosylated cysteines and proteins.
Using PANIMoni approach, we defined a very stringent set of 122 proteins that exhibited alterations of S-PALM/S-NO crosstalk after chronic stress at the level of specific sites of modification. Using functional bioinformatics analysis with various tools (i.e. REACTOME, KEGG, and GO), we assigned these 122 proteins to molecular pathways that are involved in synaptic transmission, protein localization, and energy metabolism.
Notably, among this group of proteins, PSD-95, Striatin-4 (Strn4), CaMKII, Iqseq7, Shank, GluR1, glycogen synthase kinase-3β (GSK3β), and catenin delta play key roles in regulating synaptic plasticity. Synaptic plasticity is involved in various stress-related disorders, and changes in protein PTM profiles may be a particularly important mechanism in the development of neuropsychiatric disorders.
One of the proteins strongly involved into synaptic plasticity is GSK3β. GSK3β activity is altered by chronic stress and in schizophrenia and depression (54, 55). Previous studies have reported the S-NO-mediated activation of GSK3β (56). The present data suggest that GSK3β activity may also be modulated by S-PALM, and S-PALM/S-NO crosstalk is crucial for GSK3β activity.
We also found that chronic stress influenced the S-PALM/S-NO crosstalk of several neuronal kinases, including CaMKII and protein kinase C (PKC). CaMKII and PKC are activated by calcium influx in the synapse, resulting in an increase in synaptic strength (57, 58). The activity of these kinases is involved in actin polymerization, membrane trafficking, and glutamate receptor trafficking. CaMKII is up-regulated in numerous diseases, including stress-related disorders, and stress hormones have been shown to significantly alter their functions (59, 60). S-nitrosylation-dependent CaMKII regulation has been recently reported. The present study suggests a possible link between alterations of the S-PALM/S-NO crosstalk of CaMKII and the response to chronic stress (61). Like CaMKII, changes in PKC activity have been associated with behavioral effects of chronic stress, and these effects were mediated by the formation of nitric oxide, which is a substrate for S-NO (62). Alterations of PKC activity following chronic stress may thus be regulated by crosstalk between two cysteine modifications (S-PALM and S-NO).
The third kinase identified in the group of proteins with altered S-PLAM/S-NO interplay is Src. Src is strongly expressed in mature neurons and is implicated in neuronal proliferation and differentiation during CNS development (63). A large body of evidence indicates that the main function of Src is up-regulation of NMDA receptors and other ion channels activities (64–66). Thus, Src can be crucial to the processes underlying physiological plasticity, including learning and memory, and pathological plasticity, such as in chronic stress (67). It has been shown that Src activity is mainly regulated by the phosphorylation (68). However, recent evidence has demonstrated the importance of cysteine modifications in the regulation of Src kinase function. S-PALM of Src is responsible for proper subcellular trafficking and precise coupling of extracellular stimuli and intracellular signaling (69). On the other hand, S-NO leads to Src activation (70). We postulate that not only one modification, but the interaction among different PTMs is essential for the modulation of the Src kinase function.
Another component associated with synaptic plasticity and characterized by altered S-PALM/S-NO interplay after chronic stress is Striatin-4 (Strn4). Strn4 belongs to the striatin family of scaffold proteins located in dendritic spines (71, 72). One of the proteins identified here for the first time as S-palmitoylated is Striatin-4. It has been shown that the local expression of Strn4 protein depends on NMDA receptor activation (73). Moreover, it has been demonstrated that the expression of Strn4 is one of the mechanisms controlling dendritic spine morphology. Its changes in the S-PALM/S-NO interplay may be an important mechanism of dendritic spines alterations induced by chronic stress.
In the present study, cytoskeletal proteins were abnormally modified by chronic stress. The dynamics of the synaptic cytoskeleton are particularly important in synaptic plasticity. Rapid activity-dependent changes in synapse volume or shape are crucial for increasing synaptic strength (74). Disruptions of the synaptic cytoskeleton affect the stability and maturation of synapses and subsequently disturb neuronal communication (75).
One of the cytoskeleton proteins that were identified in our study is ArpC3. Alterations of ArpC3 activity in excitatory neurons lead to the asymmetric structural plasticity of dendritic spines, followed by the progressive loss of spine synapses (76). ArpC3 has been shown to be present in S-PALM and S-NO forms, but the molecular function of these modifications remains unclear (77).
Previous studies showed that many enzymes that are involved in cellular energy metabolism, including glycolysis and fatty acid oxidation, are susceptible to modifications by either palmitate or nitric oxide, and some are susceptible to both. Although these proteins seem to be contaminations in PSD fraction, many of them play an important role in chronic stress. In the present study, we observed chronic stress-induced alterations of PTM crosstalk of the following proteins that are involved in energy metabolism: malate dehydrogenase (Mdh1), citrate synthase (Cs), pyruvate dehydrogenase (Pdh1), and short/branched chain specific acyl-CoA dehydrogenase (Acadsb). Palmitate was shown to regulate the activity of such enzymes as malate dehydrogenase and ATP-citrate synthase by affecting the formation of their active ternary complexes (78). Stress-induced changes in the interplay among PTMs of proteins that are involved in energy metabolism may decrease the activity of metabolic pathways and thus decrease ATP production.
TCA cycle activation is directly involved in neurotransmission in the brain (79). Glutamate dehydrogenase (Glud1). S-NO inhibits glutamate oxidation and promotes its conversion to glutamine (80). Moreover, the absence of S-NO of this enzyme results in a higher ratio of glutamine to glutamate, consistent with the increase in glutamate oxidation (81). The association between chronic stress and dysfunction of the mechanisms that regulate glutamate metabolism have been very well described (30, 82). Lower levels of glutamate/glutamine uptake and cycling were found in the brains of chronically stressed mice (82). Further, higher glutamate levels and lower glutamine/glutamate ratios have been consistently detected in the plasma of depressed patients (83). The PTM interplay of enzymes that govern the glutamine-glutamate cycle may be involved in these changes.
In each of the functional groups, we can distinguish proteins with kinase and protein phosphatase activity. Among the various PTMs, protein phosphorylation is the most prevalent modification that regulates synaptic protein structures and functions in a wide range of cellular processes (84–86). The dysregulation of protein kinases and phosphatases has been widely reported to be associated with the development of different diseases (87). The reciprocal interplay between S-PALM and phosphorylation was recognized many years ago for the β-adrenergic G protein-coupled receptor (88, 89). Recent studies confirmed this molecular mechanism for other neuronal proteins, such as α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) and NMDA receptors (16). Our data indicate that S-PALM/S-NO crosstalk may also influence phosphorylation by regulating phosphorylating enzyme machinery.
Identification of the interplay among different types of PTMs of the same protein has increased our understanding of signal processing, but it has not yet been characterized on a larger scale. In the present study, we employed an MS approach, called PANIMoni that allows the site-specific analysis of two important cysteine modifications: S-PALM and S-NO. We applied this method to identify chronic stress-induced site-specific dynamic changes in PTMs of mouse brain PSD proteins. Our results demonstrated that the S-PALM/S-NO crosstalk of synaptic proteins under physiological conditions modulates almost all aspects of synaptic function. Our data indicate that this interplay is altered by chronic stress.
We hypothesize that alterations of S-PALM/S-NO crosstalk of proteins that are involved in synaptic transmission, protein localization, and the regulation of synaptic plasticity might be a major event that leads to the destabilization of synaptic networks in chronic stress-related disorders. Future work should focus on functional validating of the proteins identified here to better understand the molecular mechanisms of chronic stress.
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD010822. https://www.ebi.ac.uk/pride/archive/projects/PXD010822.
Supplementary Material
Acknowledgments
We would like to thank Mass Spectrometry Laboratory IBB PAN (http://mslab-ibb.pl/en/) where all MS measurements were performed. All authors read and approved the final manuscript.
Footnotes
* This research was supported by the National Science Centre grant 2015/17/B/NZ3/00557. I.F. was supported by the National Science Centre grant 2015/19/B/NZ3/01376. M.B. was supported by the Polish Ministry of Science (1342/1/MOB/IV/15/2016/0). The authors declare that they have no conflict of interest.
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- PTMs
- Post-translational modifications
- S-PALM
- S-palmitoylation
- S-NO
- S-nitrosylation
- PANIMoni
- Palmitoylation And Nitrosylation Interplay Monitoring
- PATs
- protein acyltransferases
- APTs
- acyl-protein thioesterases
- PSD-95
- postsynaptic density protein 95
- NMDA
- N-methyl-D-aspartate
- ABE
- acyl-biotin exchange
- GO
- Gene Ontology
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- MS
- mass spectrometry
- 2D
- two-dimensional
- m/z
- mass-to-charge ratio
- LC
- liquid chromatography
- PCA
- principal component analysis
- SNAP
- S-nitroso-N-acetyl-DL-penicillamine
- L-NAME
- Nω-nitro-L-arginine methyl ester hydrochloride
- 2Br
- 2-bromopalmitate
- STRING
- search tool for the retrieval of interacting genes
- FDR
- false discovery rate
- AbPE
- acyl–biotin-PEG exchange.
REFERENCES
- 1. Amtul Z., and Atta-Ur-Rahman (2015) Neural plasticity and memory: molecular mechanism. Rev. Neurosci. 26, 253–268 [DOI] [PubMed] [Google Scholar]
- 2. Holtmaat A., and Svoboda K. (2009) Experience-dependent structural synaptic plasticity in the mammalian brain. Nat. Rev. Neurosci. 10, 647–658 [DOI] [PubMed] [Google Scholar]
- 3. Bayés A., van de Lagemaat L. N., Collins M. O., Croning M. D., Whittle I. R., Choudhary J. S., and Grant S. G. (2011) Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat. Neurosci. 14, 19–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xu W. (2011) PSD-95-like membrane associated guanylate kinases (PSD-MAGUKs) and synaptic plasticity. Curr. Opin. Neurobiol. 21, 306–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Karve T. M., and Cheema A. K. (2011) Small changes huge impact: the role of protein posttranslational modifications in cellular homeostasis and disease. J. Amino Acids 2011, 207691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Okamoto S., Nakamura T., Cieplak P., Chan S. F., Kalashnikova E., Liao L., Saleem S., Han X., Clemente A., Nutter A., Sances S., Brechtel C., Haus D., Haun F., Sanz-Blasco S., Huang X., Li H., Zaremba J. D., Cui J., Gu Z., Nikzad R., Harrop A., McKercher S. R., Godzik A., Yates J. R., and Lipton S. A. (2014) S-nitrosylation-mediated redox transcriptional switch modulates neurogenesis and neuronal cell death. Cell Rep. 8, 217–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yokoi N., Fukata M., and Fukata Y. (2012) Synaptic plasticity regulated by protein-protein interactions and posttranslational modifications. Int. Rev. Cell Mol. Biol. 297, 1–43 [DOI] [PubMed] [Google Scholar]
- 8. el-Husseini A. l.-D., and Bredt D. S. (2002) Protein palmitoylation: a regulator of neuronal development and function. Nat. Rev. Neurosci. 3, 791–802 [DOI] [PubMed] [Google Scholar]
- 9. Fukata Y., and Fukata M. (2010) Protein palmitoylation in neuronal development and synaptic plasticity. Nat. Rev. Neurosci. 11, 161–175 [DOI] [PubMed] [Google Scholar]
- 10. Fukata Y., Dimitrov A., Boncompain G., Vielemeyer O., Perez F., and Fukata M. (2013) Local palmitoylation cycles define activity-regulated postsynaptic subdomains. J. Cell Biol. 202, 145–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Linder M. E., and Deschenes R. J. (2007) Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8, 74–84 [DOI] [PubMed] [Google Scholar]
- 12. Tortosa E., and Hoogenraad C. C. (2018) Polarized trafficking: the palmitoylation cycle distributes cytoplasmic proteins to distinct neuronal compartments. Curr. Opin. Cell Biol. 50, 64–71 [DOI] [PubMed] [Google Scholar]
- 13. Lussier M. P., Sanz-Clemente A., and Roche K. W. (2015) Dynamic Regulation of N-Methyl-d-aspartate (NMDA) and α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptors by Posttranslational Modifications. J. Biol. Chem. 290, 28596–28603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Conibear E., and Davis N. G. (2010) Palmitoylation and depalmitoylation dynamics at a glance. J. Cell Sci. 123, 4007–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Won S. J., Cheung See Kit M., and Martin B. R. (2018) Protein depalmitoylases. Crit Rev. Biochem. Mol. Biol. 53, 83–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gauthier-Kemper A., Igaev M., Sündermann F., Janning D., Brühmann J., Moschner K., Reyher H. J., Junge W., Glebov K., Walter J., Bakota L., and Brandt R. (2014) Interplay between phosphorylation and palmitoylation mediates plasma membrane targeting and sorting of GAP43. Mol. Biol. Cell 25, 3284–3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Salaun C., Greaves J., and Chamberlain L. H. (2010) The intracellular dynamic of protein palmitoylation. J. Cell Biol. 191, 1229–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stamler J. S., Simon D. I., Osborne J. A., Mullins M. E., Jaraki O., Michel T., Singel D. J., and Loscalzo J. (1992) S-nitrosylation of proteins with nitric oxide: synthesis and characterization of biologically active compounds. Proc. Natl. Acad. Sci. U.S.A. 89, 444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Seth D., Hess D. T., Hausladen A., Wang L., Wang Y. J., and Stamler J. S. (2018) A Multiplex Enzymatic Machinery for Cellular Protein S-nitrosylation. Mol. Cell 69, 451–464.e456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stamler J. S., and Hess D. T. (2010) Nascent nitrosylases. Nature Cell Biol. 12, 1024–1026 [DOI] [PubMed] [Google Scholar]
- 21. Zareba-Koziol M., Szwajda A., Dadlez M., Wyslouch-Cieszynska A., and Lalowski M. (2014) Global analysis of S-nitrosylation sites in the wild type (APP) transgenic mouse brain-clues for synaptic pathology. Mol. Cell. Proteomics 13, 2288–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Percher A., Thinon E., and Hang H. (2017) Mass-tag labeling using Acyl-PEG exchange for the determination of endogenous protein S-fatty acylation. Curr. Protoc. Protein Sci. 89, 14.17.11–14.17.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Forrester M. T., Thompson J. W., Foster M. W., Nogueira L., Moseley M. A., and Stamler J. S. (2009) Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat. Biotechnol. 27, 557–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ho G. P. H., Selvakumar B., Mukai J., Hester L. D., Wang Y., Gogos J. A., and Snyder S. H. (2011) S-nitrosylation and S-palmitoylation reciprocally regulate synaptic targeting of PSD-95. Neuron 71, 131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zaręba-Kozioł M., Figiel I., Bartkowiak-Kaczmarek A., and Włodarczyk J. (2018) Insights into protein. Front. Mol. Neurosci. 11, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Penzes P., Cahill M. E., Jones K. A., VanLeeuwen J. E., and Woolfrey K. M. (2011) Dendritic spine pathology in neuropsychiatric disorders. Nat. Neurosci. 14, 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Christoffel D. J., Golden S. A., and Russo S. J. (2011) Structural and synaptic plasticity in stress-related disorders. Rev. Neurosci. 22, 535–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sierakowiak A., Mattsson A., Gómez-Galán M., Feminía T., Graae L., Aski S. N., Damberg P., Lindskog M., Brené S., and Åberg E. (2014) Hippocampal morphology in a rat model of depression: the effects of physical activity. Open Neuroimag. J. 9, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sapolsky R. M. (2001) Depression, antidepressants, and the shrinking hippocampus. Proc. Natl. Acad. Sci. U.S.A. 98, 12320–12322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Popoli M., Yan Z., McEwen B. S., and Sanacora G. (2011) The stressed synapse: the impact of stress and glucocorticoids on glutamate transmission. Nat. Rev. Neurosci. 13, 22–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McEwen B. S. (2007) Physiology and neurobiology of stress and adaptation: central role of the brain. Physiol. Rev. 87, 873–904 [DOI] [PubMed] [Google Scholar]
- 32. Duman R. S., and Aghajanian G. K. (2012) Synaptic dysfunction in depression: potential therapeutic targets. Science 338, 68–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pawlak R., Magarinos A. M., Melchor J., McEwen B., and Strickland S. (2003) Tissue plasminogen activator in the amygdala is critical for stress-induced anxiety-like behavior. Nat. Neurosci. 6, 168–174 [DOI] [PubMed] [Google Scholar]
- 34. Can A., Blackwell R. A., Piantadosi S. C., Dao D. T., O'Donnell K. C., and Gould T. D. (2011) Antidepressant-like responses to lithium in genetically diverse mouse strains. Genes Brain Behav. 10, 434–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jaffrey S. R., and Snyder S. H. (2001) The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE 2001, pl1. [DOI] [PubMed] [Google Scholar]
- 36. Wan J., Roth A. F., Bailey A. O., and Davis N. G. (2007) Palmitoylated proteins: purification and identification. Nat. Protoc. 2, 1573–1584 [DOI] [PubMed] [Google Scholar]
- 37. Hulsen T., de Vlieg J., and Alkema W. (2008) BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9, 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Michaluk P., Kolodziej L., Mioduszewska B., Wilczynski G. M., Dzwonek J., Jaworski J., Gorecki D. C., Ottersen O. P., and Kaczmarek L. (2007) Beta-dystroglycan as a target for MMP-9, in response to enhanced neuronal activity. J. Biol. Chem. 282, 16036–16041 [DOI] [PubMed] [Google Scholar]
- 39. Maere S., Heymans K., and Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 40. Jensen L. J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., Simonovic M., Bork P., and von Mering C. (2009) STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37, D412–D416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mi H., Huang X., Muruganujan A., Tang H., Mills C., Kang D., and Thomas P. D. (2017) PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45, D183–D189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. von Eichborn J., Dunkel M., Gohlke B. O., Preissner S. C., Hoffmann M. F., Bauer J. M., Armstrong J. D., Schaefer M. H., Andrade-Navarro M. A., Le Novere N., Croning M. D., Grant S. G., van Nierop P., Smit A. B., and Preissner R. (2013) SynSysNet: integration of experimental data on synaptic protein-protein interactions with drug-target relations. Nucleic Acids Res. 41, D834–D840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pirooznia M., Wang T., Avramopoulos D., Valle D., Thomas G., Huganir R. L., Goes F. S., Potash J. B., and Zandi P. P. (2012) SynaptomeDB: an ontology-based knowledgebase for synaptic genes. Bioinformatics 28, 897–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Conrad C. D., Jackson J. L., and Wise L. S. (2004) Chronic stress enhances ibotenic acid-induced damage selectively within the hippocampal CA3 region of male, but not female rats. Neuroscience 125, 759–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Conrad C. D. (2008) Chronic stress-induced hippocampal vulnerability: the glucocorticoid vulnerability hypothesis. Rev. Neurosci. 19, 395–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McLaughlin K. J., Baran S. E., Wright R. L., and Conrad C. D. (2005) Chronic stress enhances spatial memory in ovariectomized female rats despite CA3 dendritic retraction: possible involvement of CA1 neurons. Neuroscience 135, 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McLaughlin K. J., Gomez J. L., Baran S. E., and Conrad C. D. (2007) The effects of chronic stress on hippocampal morphology and function: an evaluation of chronic restraint paradigms. Brain Res. 1161, 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wright R. L., and Conrad C. D. (2005) Chronic stress leaves novelty-seeking behavior intact while impairing spatial recognition memory in the Y-maze. Stress 8, 151–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kanehisa M., and Goto S. (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Distler U., Schmeisser M. J., Pelosi A., Reim D., Kuharev J., Weiczner R., Baumgart J., Boeckers T. M., Nitsch R., Vogt J., and Tenzer S. (2014) In-depth protein profiling of the postsynaptic density from mouse hippocampus using data-independent acquisition proteomics. Proteomics 14, 2607–2613 [DOI] [PubMed] [Google Scholar]
- 51. Shinohara Y. (2012) Quantification of postsynaptic density proteins: glutamate receptor subunits and scaffolding proteins. Hippocampus 22, 942–953 [DOI] [PubMed] [Google Scholar]
- 52. Trinidad J. C., Specht C. G., Thalhammer A., Schoepfer R., and Burlingame A. L. (2006) Comprehensive identification of phosphorylation sites in postsynaptic density preparations. Mol. Cell. Proteomics 5, 914–922 [DOI] [PubMed] [Google Scholar]
- 53. Walikonis R. S., Jensen O. N., Mann M., Provance D. W., Mercer J. A., and Kennedy M. B. (2000) Identification of proteins in the postsynaptic density fraction by mass spectrometry. J. Neurosci. 20, 4069–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jope R. S., and Roh M. S. (2006) Glycogen synthase kinase-3 (GSK3) in psychiatric diseases and therapeutic interventions. Curr. Drug Targets 7, 1421–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li X., and Jope R. S. (2010) Is glycogen synthase kinase-3 a central modulator in mood regulation? Neuropsychopharmacology 35, 2143–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ryu I. H., Lee K. Y., and Do S. I. (2016) Aβ-affected pathogenic induction of S-nitrosylation of OGT and identification of Cys-NO linkage triplet. Biochim. Biophys. Acta 1864, 609–621 [DOI] [PubMed] [Google Scholar]
- 57. Lisman J., Yasuda R., and Raghavachari S. (2012) Mechanisms of CaMKII action in long-term potentiation. Nat. Rev. Neurosci. 13, 169–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chu Y., Fioravante D., Leitges M., and Regehr W. G. (2014) Calcium-dependent PKC isoforms have specialized roles in short-term synaptic plasticity. Neuron 82, 859–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sanhueza M., Fernandez-Villalobos G., Stein I. S., Kasumova G., Zhang P., Bayer K. U., Otmakhov N., Hell J. W., and Lisman J. (2011) Role of the CaMKII/NMDA receptor complex in the maintenance of synaptic strength. J. Neurosci. 31, 9170–9178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ghosh A., and Giese K. P. (2015) Calcium/calmodulin-dependent kinase II and Alzheimer's disease. Mol. Brain 8, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Erickson J. R., Nichols C. B., Uchinoumi H., Stein M. L., Bossuyt J., and Bers D. M. (2015) S-Nitrosylation Induces Both Autonomous Activation and Inhibition of Calcium/Calmodulin-dependent Protein Kinase II δ. J. Biol. Chem. 290, 25646–25656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen G., Henter I. D., and Manji H. K. (2009) A role for PKC in mediating stress-induced prefrontal cortical structural plasticity and cognitive function. Proc. Natl. Acad. Sci. U.S.A. 106, 17613–17614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kalia L. V., Gingrich J. R., and Salter M. W. (2004) Src in synaptic transmission and plasticity. Oncogene 23, 8007–8016 [DOI] [PubMed] [Google Scholar]
- 64. Trepanier C. H., Jackson M. F., and MacDonald J. F. (2012) Regulation of NMDA receptors by the tyrosine kinase Fyn. FEBS J 279, 12–19 [DOI] [PubMed] [Google Scholar]
- 65. Gingrich J. R., Pelkey K. A., Fam S. R., Huang Y., Petralia R. S., Wenthold R. J., and Salter M. W. (2004) Unique domain anchoring of Src to synaptic NMDA receptors via the mitochondrial protein NADH dehydrogenase subunit 2. Proc. Natl. Acad. Sci. U.S.A. 101, 6237–6242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Salter M. W., and Kalia L. V. (2004) Src kinases: a hub for NMDA receptor regulation. Nat. Rev. Neurosci. 5, 317–328 [DOI] [PubMed] [Google Scholar]
- 67. Roskoski R. (2015) Src protein-tyrosine kinase structure, mechanism, and small molecule inhibitors. Pharmacol. Res. 94, 9–25 [DOI] [PubMed] [Google Scholar]
- 68. Roskoski R. (2005) Src kinase regulation by phosphorylation and dephosphorylation. Biochem. Biophys. Res. Commun. 331, 1–14 [DOI] [PubMed] [Google Scholar]
- 69. Sato I., Obata Y., Kasahara K., Nakayama Y., Fukumoto Y., Yamasaki T., Yokoyama K. K., Saito T., and Yamaguchi N. (2009) Differential trafficking of Src, Lyn, Yes and Fyn is specified by the state of palmitoylation in the SH4 domain. J. Cell Sci. 122, 965–975 [DOI] [PubMed] [Google Scholar]
- 70. Rahman M. A., Senga T., Ito S., Hyodo T., Hasegawa H., and Hamaguchi M. (2010) S-nitrosylation at cysteine 498 of c-Src tyrosine kinase regulates nitric oxide-mediated cell invasion. J. Biol. Chem. 285, 3806–3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Benoist M., Gaillard S., and Castets F. (2006) The striatin family: a new signaling platform in dendritic spines. J. Physiol. Paris 99, 146–153 [DOI] [PubMed] [Google Scholar]
- 72. Gaillard S., Bailly Y., Benoist M., Rakitina T., Kessler J. P., Fronzaroli-Molinières L., Dargent B., and Castets F. (2006) Targeting of proteins of the striatin family to dendritic spines: role of the coiled-coil domain. Traffic 7, 74–84 [DOI] [PubMed] [Google Scholar]
- 73. Lin L., Lo L. H., Lyu Q., and Lai K. O. (2017) Determination of dendritic spine morphology by the striatin scaffold protein STRN4 through interaction with the phosphatase PP2A. J. Biol. Chem. 292, 9451–9464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Spence E. F., and Soderling S. H. (2015) Actin Out: Regulation of the Synaptic Cytoskeleton. J. Biol. Chem. 290, 28613–28622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Goellner B., and Aberle H. (2012) The synaptic cytoskeleton in development and disease. Dev. Neurobiol. 72, 111–125 [DOI] [PubMed] [Google Scholar]
- 76. Kim I. H., Racz B., Wang H., Burianek L., Weinberg R., Yasuda R., Wetsel W. C., and Soderling S. H. (2013) Disruption of Arp2/3 results in asymmetric structural plasticity of dendritic spines and progressive synaptic and behavioral abnormalities. J. Neurosci. 33, 6081–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kang R., Wan J., Arstikaitis P., Takahashi H., Huang K., Bailey A. O., Thompson J. X., Roth A. F., Drisdel R. C., Mastro R., Green W. N., Yates J. R., Davis N. G., and El-Husseini A. (2008) Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature 456, 904–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kostiuk M. A., Corvi M. M., Keller B. O., Plummer G., Prescher J. A., Hangauer M. J., Bertozzi C. R., Rajaiah G., Falck J. R., and Berthiaume L. G. (2008) Identification of palmitoylated mitochondrial proteins using a bio-orthogonal azido-palmitate analogue. FASEB J. 22, 721–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hall C. N., Klein-Flügge M. C., Howarth C., and Attwell D. (2012) Oxidative phosphorylation, not glycolysis, powers presynaptic and postsynaptic mechanisms underlying brain information processing. J. Neurosci. 32, 8940–8951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cooper A. J. (2012) The role of glutamine synthetase and glutamate dehydrogenase in cerebral ammonia homeostasis. Neurochem. Res. 37, 2439–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Raju K., Doulias P. T., Evans P., Krizman E. N., Jackson J. G., Horyn O., Daikhin Y., Nissim I., Yudkoff M., Sharp K. A., Robinson M. B., and Ischiropoulos H. (2015) Regulation of brain glutamate metabolism by nitric oxide and S-nitrosylation. Sci. Signal. 8, ra68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sanacora G., Treccani G., and Popoli M. (2012) Towards a glutamate hypothesis of depression: an emerging frontier of neuropsychopharmacology for mood disorders. Neuropharmacology 62, 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Abdallah C. G., Jiang L., De Feyter H. M., Fasula M., Krystal J. H., Rothman D. L., Mason G. F., and Sanacora G. (2014) Glutamate metabolism in major depressive disorder. Am. J. Psychiatry 171, 1320–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zahid S., Oellerich M., Asif A. R., and Ahmed N. (2012) Phosphoproteome profiling of substantia nigra and cortex regions of Alzheimer's disease patients. J. Neurochem. 121, 954–963 [DOI] [PubMed] [Google Scholar]
- 85. Xia Q., Cheng D., Duong D. M., Gearing M., Lah J. J., Levey A. I., and Peng J. (2008) Phosphoproteomic analysis of human brain by calcium phosphate precipitation and mass spectrometry. J. Proteome Res. 7, 2845–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ubersax J. A., and Ferrell J. E. (2007) Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 8, 530–541 [DOI] [PubMed] [Google Scholar]
- 87. Ardito F., Giuliani M., Perrone D., Troiano G., and Lo Muzio L. (2017) The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review). Int. J. Mol. Med. 40, 271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Adachi N., Hess D. T., McLaughlin P., and Stamler J. S. (2016) S-Palmitoylation of a novel site in the β2-adrenergic receptor associated with a novel intracellular itinerary. J. Biol. Chem. 291, 20232–20246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Moritz A. E., Rastedt D. E., Stanislowski D. J., Shetty M., Smith M. A., Vaughan R. A., and Foster J. D. (2015) Reciprocal phosphorylation and palmitoylation control dopamine transporter kinetics. J. Biol. Chem. 290, 29095–29105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Shen L. F., Chen Y. J., Liu K .M., Haddad A. N. S., Song I. W., Roan H. Y., Chen L. Y., Yen J. J. Y., Chen Y. J., Wu J. Y., and Chen Y. T. (2017) Role of S-palmitoylation by ZDHHC13 in mitochondrial function and metabolism in liver. Sci. Rep. 7, 2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Collins M. O., Woodley K. T., and Choudhary J. S. (2017) Global, site-specific analysis of neuronal protein S-acylation. Sci. Rep. 7, 4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Chesarino N. M., Hach J. C., Chen J. L., Zaro B. W., Rajaram M. V., Turner J., Schlesinger L. S., Pratt M. R., Hang H. C., and Yount J. S. (2014) Chemoproteomics reveals Toll-like receptor fatty acylation. BMC Biol. 12, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wan J., Savas J. N., Roth A. F., Sanders S. S., Singaraja R. R., Hayden M. R., Yates J. R. 3rd, and Davis N. G. (2013) Tracking brain palmitoylation change: predominance of glial change in a mouse model of Huntington's disease. Chem. Biol. 20, 1421–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ren W., Jhala U. S. and Du K. (2013) Proteomic analysis of protein palmitoylation in adipocytes. Adipocyte 2, 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Sobocińska J., Roszczenko-Jasiñska P., Zarêba-Kozioł M., Hromada-Judycka A., Matveichuk O.V., Traczyk G., £ukasiuk K., and Kwiatkowska K. (2018) Lipopolysaccharide upregulates palmitoylated enzymes of the phosphatidylinositol cycle: an insight from proteomic studies. Mol. Cell. Proteomics. 17, 233–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zhang X., Zhang Y., Fang C., Zhang L., Yang P., Wang C., and Lu H. (2018) Ultradeep palmitoylomics enabled by dithiodipyridine-functionalized magnetic nanoparticles. Anal. Chem. 90, 6161–6168 [DOI] [PubMed] [Google Scholar]
- 97. Gould N. S., Evans P., Martínez-Acedo P., Marino S. M., Gladyshev V. N., Carroll K. S., and Ischiropoulos H. (2015) Site-specific proteomic mapping identifies selectively modified regulatory cysteine residues in functionally distinct protein networks. Chem. Biol. 22, 965–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Li Y., Martin B. R., Cravatt B. F., and Hofmann S. L. (2012) DHHC5 protein palmitoylates flotillin-2 and is rapidly degraded on induction of neuronal differentiation in cultured cells. J. Biol. Chem. 287, 523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lievens P. M., Kuznetsova T., Kochlamazashvili G., Cesca F., Gorinski N., Galil D.A., Cherkas V., Ronkina N., Lafera J., Gaestel M., Ponimaskin E., and Dityatev A. (2016) ZDHHC3 tyrosine phosphorylation regulates neural cell adhesion molecule palmitoylation. Mol. Cell Biol. 36, 2208–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ponimaskin E., Dityateva G., Ruonala M. O., Fukata M., Fukata Y., Kobe F., Wouters F. S., Delling M., Bredt D. S., Schachner M., and Dityatev A. (2008) Fibroblast growth factor-regulated palmitoylation of the neural cell adhesion molecule determines neuronal morphogenesis. J. Neurosci. 28, 8897–8907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Thinon E., Fernandez J. P., Molina H., and Hang H. C. (2018) Selective enrichment and direct analysis of protein s-palmitoylation sites. J. Proteome Res. 17, 1907–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kang R., Wan J., Arstikaitis P., Takahashi H., Huang K., Bailey A. O., Thompson J. X., Roth A. F., Drisdel R. C., Mastro R., Green W. N., Yates J. R. 3rd, Davis N. G., and El-Husseini A. (2008) Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature 456, 904–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Wei X., Song H., and Semenkovich C. F. (2014) Insulin-regulated protein palmitoylation impacts endothelial cell function. Arterioscler. Thromb. Vasc. Biol. 34, 346–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Thinon E., Percher A., and Hang H.C., (2016) Bioorthogonal chemical reporters for monitoring unsaturated fatty-acylated proteins. Chembiochem 17, 1800–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Gory-Fauré S., Windscheid V., Bosc C., Peris L., Proietto D., Franck R., Denarier E., Job D., and Andrieux A. (2006) STOP-like protein 21 is a novel member of the STOP family, revealing a Golgi localization of STOP proteins. J. Biol. Chem. 281, 28387–28396 [DOI] [PubMed] [Google Scholar]
- 106. Pinner A. L., Tucholski J., Haroutunian V., McCullumsmith R. E., and Meador-Woodruff J. H. (2016) Decreased protein S-palmitoylation in dorsolateral prefrontal cortex in schizophrenia. Schizophr. Res. 177, 78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Hernandez J. L., Davda D., Majmudar J. D., Won S. J., Prakash A., Choi A. I., and Martin B. R. (2016) Correlated S-palmitoylation profiling of Snail-induced epithelial to mesenchymal transition. Mol. Biosyst. 12, 1799–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Martin B. R., Wang C., Adibekian A., Tully S. E., and Cravatt B. F. (2011) Global profiling of dynamic protein palmitoylation. Nat. Methods 9, 84–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Yount J. S., Moltedo B., Yang Y. Y., Charron G., Moran T. M., López C. B., and Hang H. C. (2010) Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat. Chem. Biol. 6, 610–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Merrick B. A., Dhungana S., Williams J. G., Aloor J. J., Peddada S., Tomer K. B., and Fessler M. B. (2011) Proteomic profiling of S-acylated macrophage proteins identifies a role for palmitoylation in mitochondrial targeting of phospholipid scramblase 3. Mol. Cell. Proteomics 10, M110.006007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Chen Y. J., et al. , (2015) dbSNO 2.0: a resource for exploring structural environment, functional and disease association and regulatory network of protein S-nitrosylation. Nucleic Acids Res. 43, D503–D5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Seneviratne U., Nott A., Bhat V. B., Ravindra K. C., Wishnok J. S., Tsai L. H., and Tannenbaum S. R. (2016) S-nitrosation of proteins relevant to Alzheimer's disease during early stages of neurodegeneration. Proc. Natl. Acad. Sci. U. S. A. 113, 4152–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Ryu I. H., Lee K. Y., and Do S. I. (2016) Aβ-affected pathogenic induction of S-nitrosylation of OGT and identification of Cys-NO linkage triplet. Biochim. Biophys. Acta 1864, 609–621 [DOI] [PubMed] [Google Scholar]
- 114. Tooker R. E., and Vigh J. (2015) Light-evoked S-nitrosylation in the retina. J. Comp. Neurol. 523, 2082–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Ben-Lulu S., Ziv T., Weisman-Shomer P., and Benhar M. (2017) Correction: Nitrosothiol-trapping-based proteomic analysis of S-nitrosylation in human lung carcinoma cells. PLoS One 12, e0179803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Ulrich C., Quillici D. R., Schegg K., Woolsey R., Nordmeier A., and Buxton I. L. (2012) Uterine smooth muscle S-nitrosylproteome in pregnancy. Mol. Pharmacol. 81, 143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Nicolas F., Wu C., Bukhari S., de Toledo S. M., Li H., Shibata M., and Azzam E. I. (2015) - Nitrosylation in organs of mice exposed to low or high doses of γ-rays: the modulating effect of iodine contrast agent at a low radiation dose. Proteomes 3, 56–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Gu L., and Robinson R. A. (2016) High-throughput endogenous measurement of S-nitrosylation in Alzheimer's disease using oxidized cysteine-selective cPILOT. Analyst 141, 3904–3915 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD010822. https://www.ebi.ac.uk/pride/archive/projects/PXD010822.