Abstract
Epithelial cells in the field of lung injury can give rise to distinct premalignant lesions that may bear unique genetic aberrations. A subset of these lesions may escape immune surveillance and progress to invasive cancer, however the mutational landscape that may predict progression has not been determined. Knowledge of premalignant lesion composition and the associated microenvironment are critical for understanding tumorigenesis and the development of effective preventive and interception strategies. To identify somatic mutations and the extent of immune cell infiltration in adenomatous premalignancy and associated lung adenocarcinomas, we sequenced exomes from 41 lung cancer resection specimens including 89 premalignant atypical adenomatous hyperplasia lesions, 15 adenocarcinomas in situ, and 55 invasive adenocarcinomas and their adjacent normal lung tissues. We defined non-synonymous somatic mutations occurring in both premalignancy and the associated tumor as progression-associated mutations whose predicted neoantigens were highly correlated with infiltration of CD8+ and CD4+ T cells as well as upregulation of PD-L1 in premalignant lesions, suggesting the presence of an adaptive immune response to these neoantigens. Each patient had a unique repertoire of somatic mutations and associated neoantigens. Collectively, these results provide evidence for mutational heterogeneity, pathway dysregulation, and immune recognition in pulmonary premalignancy.
Introduction
One of the major driving forces of carcinogenesis is somatic mutagenesis [1]. Atypical adenomatous hyperplasias (AAH), small focal proliferative lesions often found in the distal airways of patients with lung adenocarcinoma (ADC), as well as those at risk, are considered to be the earliest premalignant lesions in the progression from normal airway epithelium to ADC [2]. Targeted sequencing of AAH lesions identified mutations in several cancer-related genes and clonality between AAH and associated ADC [3]. As suggested by the clinical efficacy of checkpoint blockade immunotherapies for lung cancer [4, 5], non-synonymous mutations can yield neoepitopes resulting in immune recognition.
However, the earliest molecular events associated with lung carcinogenesis and the clinical evidence for neoepitope recognition in pulmonary premalignancy have not yet been defined. Here we report evidence for mutational heterogeneity, pathway dysregulation and immune recognition in pulmonary adenomatous premalignancy. We performed whole exome sequencing of AAH, the associated non-invasive adenocarcinoma in situ (AIS) and invasive adenocarcinoma in 41 surgical resection specimens and characterized the genomic relationship in the lung cancer continuum. We identified progression-associated somatic mutations and oncogenic pathways as well as the association between putative neoantigens and adaptive immune responses in AAH. High heterogeneity between premalignant lesions in different patients suggests that future therapies that target progression-associated neoantigens in cancer interception and immunoprevention may need to be tailored to individual patients. We anticipate that based on these findings, future studies will develop approaches for targeting clinically actionable neoepitopes across the spectrum of premalignancy to invasive disease, before the development of invasive cancer.
Materials and Methods
Specimen identification and processing.
FFPE tissue blocks from 41 patients with premalignant lesions and lung adenocarcinoma were obtained from the UCLA Lung Cancer Tissue Repository, and were subjected to pathology review by two independent pathologists to identify specific histologic areas for LCM. All patients provided written informed consent. The studies were approved by the UCLA institutional review board. Tissues were first sectioned at 7 μm thickness onto membrane PEN slides (Leica Microsystems), and serial sections were stained with haematoxylin and eosin. LCM was performed utilizing a Leica LMD7000 in the California NanoSystems Institute Advanced Light Microscopy/Spectroscopy (ALMS) Core at UCLA. The following regions were dissected from distal airways: a) at least one region of normal airway epithelial cells (type I and II pneumocytes) adjacent to but not contiguous with the tumor, b) a minimum of two premalignant AAH lesions, c) all AIS regions (if present), and d) at least one ADC region. The location of the resection specimens from which the regions of interest were excised is indicated in Supplementary Table S1.
Genomic DNA isolation and library preparation for DNA sequencing.
DNA was extracted from microdissected cells utilizing the HiPure FFPE DNA isolation kit (Roche). Sequencing libraries were constructed using NuGen Ovation Ultralow V2 system, followed by exome capture using the Roche SeqCap EZ kit as recommended by the manufacturers. The quality of each library preparation and exome capture reaction was evaluated by utilizing a Bioanalyzer instrument (Agilent), Quant-iT assay and qPCR. Sequencing was then performed on an Illumina HiSeq2000 instrument as 100 bp paired-end runs with the aim of ~50× per base (based on the Illumina Sequencing Coverage Calculation with an assumption of 35% PCR duplication and a minimum of 85% target coverage). Samples with an estimated library size < 2×107 based on Picard MarkDuplicates function were re-sequenced to achieve a higher depth of coverage.
Whole exome sequencing analysis and variant calling.
Sequencing Alignment.
Sequence reads were aligned to the human genome based on the NCBI human genome reference build 37 (GRCh37) by following the pipeline suggested by Genome Analysis Toolkit (GATK) [6]. In brief, raw reads were first pre-processed to remove adapter contamination by scythe adapter trimmer (https://github.com/vsbuffalo/scythe) and low quality base calls (Phred score Q <15) and short reads (length < 20) by sickle (https://github.com/najoshi/ sickle). Reads were mapped to the reference human genome by Burrows-Wheeler Aligner (v 0.7.7) [7], and then marked for PCR and optical duplicates with the Picard (v 1.77) MarkDuplicates tool. The GATK 2.7 was used for local indel realignment and base recalibration. For cases with multiple normal samples, their bam files from the bases recalibration step were combined and re-aligned to local indels before being subjected to variant calling analysis. In case samples were re-sequenced by multiple runs, raw reads in each run were first aligned and base recalibrated independently. Their bam files were then combined and re-aligned for indel realignment. Default values were set for the parameters unless noted otherwise.
Variant Calling and Annotation.
Somatic variants between pairs of abnormal regions (i.e. AAH, AIS, and ADC) and matched normal tissue were determined by VarScan2 [8]. Tumor and normal cells having exomes sequenced were obtained from LCM, and VarScan2 was performed with a) tumor purity set to 1, and b) minimum coverage for normal and abnormal exomes set to 4. Because multiple exomes from different areas were sequenced per patient, the p-value threshold was set to 0.1 in somatic variant calling of individual exomes, and adjusted further in the next step of mutation calling in which somatic variants from all regions were analyzed together to identify mutations for each patient. The remaining VarScan2 parameters were set at default values. The output single nucleotide variant (SNV) calls were filtered further to remove false positive calls due to sequencing- or alignment-related artefacts by utilizing VarScan2’s associated fpfilter.pl script. The resulting somatic SNV and indel calls were then annotated by ANNOVAR [9] to identify non-synonymous (n.s.) variants from silent variants and common SNPs.
Mutation calling.
For each patient, a n.s. somatic mutation was defined if a n.s. variant was: 1) supported by at least three reads, and 2) observed in either: a) more than one lesion with p-value ≤ 0.1, or b) a single lesion with p-value ≤ 0.01.
Genetic homogeneity analysis.
The similarity in n.s. somatic mutations between any pair of regions was assessed by Jaccard index, which was defined as the ratio between the number of shared mutations between the regions and the total number of mutation identified in the regions.
Phylogenetic analysis.
Non-synonymous somatic mutations were first converted into the format with 1 being mutated and 0 otherwise. For each patient, the analysis only considered n.s. somatic mutations that were present in more than one region to determine resemblance among AAH, AIS and ADC regions based on their mutation profiles. The analysis was performed in R by using ape and phangorn packages [10, 11]. In brief, the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) approach was utilized to cluster regions based on their mutation-defined binary format matrix. Unrooted phylogenetic trees were then drawn with relative branch lengths disproportionate to the number of shared mutations among corresponding regions.
Mutational architecture analysis.
For each individual patient, n.s. mutations in all regions were pooled together and categorized into three groups: premalignant mutations, progression-associated mutations (PAMs) and malignant-specific mutations (MSMs) based on their presence in different regions. A premalignant mutation was defined as n.s. mutation observed only in AAH lesion(s), while a MSM was only identified in AIS/ADC lesion(s), and finally a PAM was present in both AAH and AIS/ADC lesions. For each patient, the number of mutations in each category was then normalized to the total number of n.s. mutations observed in the corresponding patient. For each individual region, its PAM was normalized to the total number of mutations identified in the respective region.
Identification of patient HLA typing.
The OptiType algorithm [12] was applied to deduce a four-digit HLA genotype from whole exome sequencing data. Before applying the algorithm, raw reads were first pre-processed to a) remove adapter contamination by scythe, and b) remove low quality base calls (Phred score Q <20) by sickle, and c) keep reads that mapped on HLA reference regions by bwa and had a length of at least 50 bp by fastqutils [13]. For pair-end data, sequences from each end were pre-processed independently before subjecting them to the OptiType algorithm.
Identification of putative neoantigens.
For every patient, each n.s. single nucleotide mutation was able to generate a maximum of ten 10-mer peptides having the mutated amino acid at different locations. Similarly, for each indel which did not cause early termination, ten 10-mer peptides were also created that had from 1–9 amino acids altered from the reference sequence. MHC-I binding prediction tools downloaded from Immune Epitope Database (IEDB) [14] were utilized to predict the binding affinity of 10-mer peptides to the patient’s HLA germline alleles. IEDB protocol recommended using multiple algorithms including: a) Artificial Neural Network [15, 16], b) Stabilized Matrix Method [17], and c) NetMHCpan [18] for predicting binding strength to a given HLA allele due to the allele’s available database and preferred algorithms previously proven to have outstanding performance for such allele. The smallest IC50 value derived from multiple algorithms was used as the predicted binding affinity of each peptide to each HLA allele. Approximately 60 peptide-MHC combinations (i.e. 10 peptides × 6 MHC-I) were derived from a single n.s. mutation. The peptide-MHC pair with the lowest predicted IC50 was selected to represent the candidate mutant peptide and its binding MHC-I partner. Finally, candidate neoantigens were defined as those with the predicted binding strength IC50 < 500nM. Neoantigens were categorized as premalignant, progression-associated (PAN) and malignant-specific neoantigen in accord with their corresponding tissue mutation group.
Pathway analysis.
In pathway analysis, every affected gene should be counted once for each individual patient even though multiple n.s. mutation sites were identified on the same gene. Therefore, n.s. mutated sites were first consolidated to their corresponding gene identity. In our study, n.s. somatic mutations were categorized into three different groups based on their presence in various tissues. Thus, their affected genes should be assigned to the corresponding groups to evaluate their effects on molecular pathways, especially related to tumor initiation and development. To achieve this, for each patient, eligible genes were first labelled based on PAMs, which were then removed from the available gene list before labelling MSMs and premalignant mutations. The labelling procedure was then repeated for MSMs, followed by premalignant mutations. This meant that each patient had three mutually exclusive gene groups representing their PAMs, MSMs, and premalignant mutations.
For each individual patient, the enrichment of mutated genes in the group i involved in a specific the pathway j is measured by an enrichment score, ESij, defined as:
| Equation S1 |
where Hij is the number of mutated genes in the group i (e.g. PAM-, MSM-, and premalignant mutations-bearing genes) involved in the pathway j. Mi, Sj and P are the numbers genes in group i, pathway j, and the genome. In other words, the ES is the number of mutated genes involved in a pathway normalized by the estimated number based on the numbers of genes in the interested groups i, pathways j and the genome. Note that a non-zero ES requires a minimum of two mutated genes associated with the pathway of interest. Furthermore, for a given pathway j (i.e. denominator is constant in the right most side of Equation S1), the discrepancy in ES between two groups of interest is proportional to the difference of the percentage of genes that are associated with the pathway in those groups.
The false discovery rate of ES was estimated by the permutation approach in which mutated genes in each patient were first randomly sampled from the genome, and then assigned to PAM- and MSM-bearing groups. ES were then calculated according to the above equation for a total of 123 mutated gene groups (41 patients × 3 groups: PAM-, MSM-, and union of PAM- and MSM-bearing genes) based on 1341 canonical and hallmark pathways downloaded from the Molecular Signature Database [19]. A total of 100 permutations were executed.
Finally, a pathway was defined to be deregulated by a certain mutated gene group if the corresponding ES was greater or equal to 2 (FDR = 0.03). In each patient, the deregulation states of all pathways based on PAM- and MSM-bearing genes were represented in binary format with 1 being deregulated and 0 for otherwise. The pathway-based binary data from all patients were then combined into the matrix form and subjected to unsupervised clustering analysis to stratify patients into subgroups. The cluster analysis was performed in R by utilizing Ward’s clustering method (i.e. ward.D2).
Analyses using TCGA data sets (DNAseq, RNAseq and survival analysis).
Processed data sets from whole exome DNA and mRNA sequencing, as well as clinical information for lung adenocarcinoma (LUAD) samples were downloaded from the Cancer Genome Atlas (TCGA) data portal. The information of mutated genes in samples was extracted from somatic mutation calls (level 2 maf file), and organized into a table in which one was employed to indicate if the gene of interest had at least one non-silent mutation call located on its coding regions, and zero for otherwise in the specific sample. The frequency of how often a gene was mutated in the cohort was then calculated from the table.
In gene expression analysis, RSEM normalized gene expression (level 3 text files) files were utilized to build a data matrix of all samples. The expression data was processed by removing a) genes with low abundance (i.e. < 1 copies per million reads in >30% samples), and b) tumor samples without whole exome sequencing data. Pathway activities per individual sample were derived from its gene expression by using Gene Set Variation Analysis (GSVA) [20]. The information of gene sets involved in the immune regulated pathways was obtained from the Molecular Signature Database. To eliminate the effect of genes commonly shared among pathways, the original gene sets were modified such that the overlapping genes were kept in the “child” and removed from the “parent” set. A “child” set was defined as the one having more than 90% of members overlapping with the parent set. The GSVA scores of the interested pathways were then subjected to the unsupervised hierarchical cluster analysis to stratify samples into subgroups. The cluster analysis, which was performed in R, used Ward’s clustering method (i.e. ward.D2) and Spearman correlation coefficient as the metric measuring similarity between sample pairs. Finally, patient survival among the subgroups was compared by log-rank test.
Evaluation of lymphocytic infiltration.
For each lesion, a section stained with hematoxylin and eosin underwent an initial qualitative evaluation by a board-certified pathologist to assess the overall degree of lymphocytic infiltration. This assessment utilized a simple graded scale: 0 (absent), 1 (focal with <3 clusters of 3 lymphocytes), 2 (multifocal with 3 or more clusters) and 3 (diffuse). X2 test was used to compared distributions of scores in different histological lesions (normal, AAH, AIS and ADC).
Immunohistochemistry analyses.
For nine cases, additional serial sections of 5 μm thickness were obtained from FFPE tissue blocks. Single-color immunostaining was performed on the Leica Bond III autostainer using Bond Low (H1) and High (H2) heat retrieval solutions, wash buffer, and Refine Polymer Detection system. Heat-induced epitope retrieval was performed in the autostainer, except for PD1 and PD-L1, which were treated in a pressure cooker. Antibodies used for detection of a single marker per slide included: CD8 (Dako #M7103), CD4 (Cell Marque #104R-16), Granzyme B (Dako #M7236), PD1 (Cell Marque #315M), PD-L1 (Spring Bio M4420), and FOXP3 (Bio SB #BSB676).
All slides were scanned at an absolute magnification of 3200 (resolution of 0.5 mm per pixel). Bright field image analysis was performed using the Indica Labs Halo platform. With the assistance of a board-certified pathologist, each region of interest (AAH, AIS and ADC) was identified and outlined on the hematoxylin and eosin guide slide, excluding necrotic areas and stroma. The guide slide was aligned and synchronized with the corresponding serial sections immunostained for each marker. Existing Halo algorithms developed for detection of positive staining were accepted or modified based on the positive control slide for each marker. The final algorithm was then used to analyze the density (cells/mm2) and percentage cellularity (% positive cells/all nucleated cells) for each marker on each region of interest. This raw data was then exported for statistical analysis.
Statistical analyses.
All analyses were performed utilizing R 3.2. Appropriate rank-based statistical tests were applied according to the nature of variables. For instance, Kendal’s τ coefficient was used to assess association between the pairs of variables, such as percentage of PAMs, percentage of positively stained cells and log transformed neoantigen numbers, while the Kruskal-Wallis rank sum was applied to compare variables of interest between groups. R lmerTest package was utilized in linear mixed effects model, which incorporates individual patient variation.
Results
Pulmonary premalignant lesions reveal a spectrum of intra- and inter-patient genetic heterogeneity.
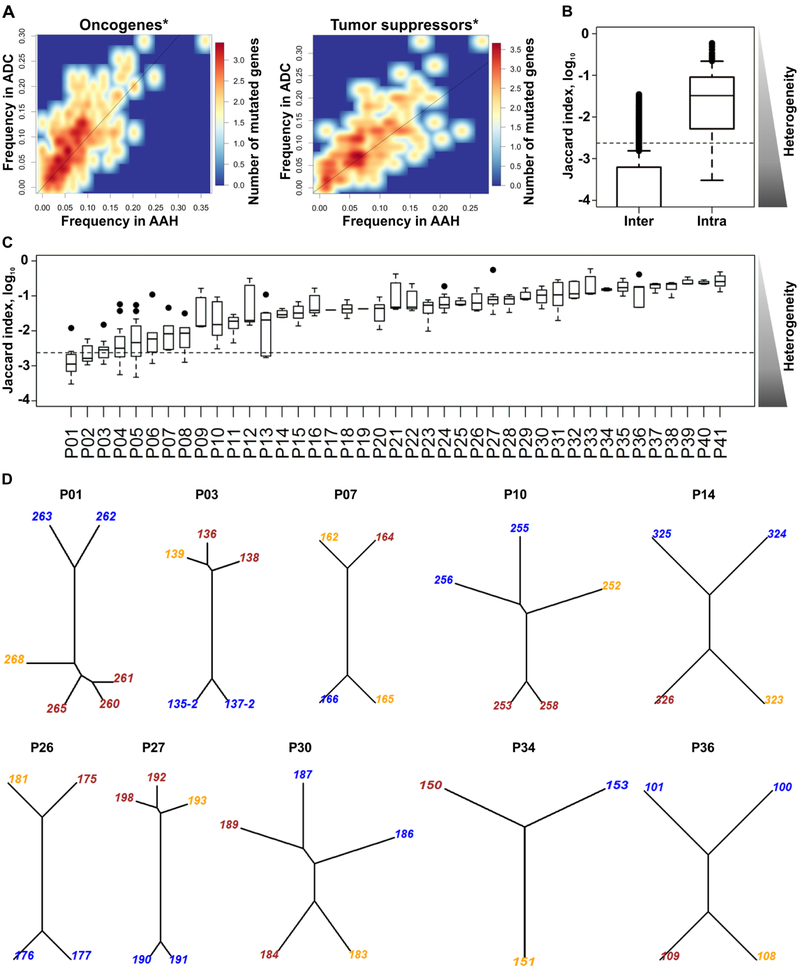
To identify the somatic mutations relevant for progression from premalignancy to cancer, we performed whole exome sequencing of 89 AAH, 15 AIS, and 55 ADC lesions (Supplementary Table S1) from lobectomy specimens from 41 patients who had undergone surgery for early stage ADC (Supplementary Tables S1 and S2). All patients provided written informed consent. The cells of interest were dissected from the following regions of distal airways utilizing Laser Capture Microdissection (LCM): a) normal airway epithelial cells (1–3 regions per patient), b) AAH lesions (1–4 anatomically independent lesions per patient), c) AIS (all independent lesions per patient where present), and d) ADC (all independent primary lung tumors per patient). Whole exome sequencing was conducted with at least 2×1010 bases sequenced per exome. The median number of unique mutations per patient was 1323, whereas in individual premalignant and malignant lesions it was 351 per lesion (Supplementary Table S1). The mutational load per patient did not increase significantly by the addition of more sequenced regions (Kruskal-Wallis rank sum test p = 0.20), and within individual patients it was independent of lesion type (linear mixed effects model F-test p = 0.46). Analysis of the mutations in oncogenes and tumor suppressor genes (from the UniProt database) demonstrated that somatic mutations in these genes are found more frequently in ADC than in AAH lesions (Figure 1A). Somatic variants between abnormal lesions and matched normal lung tissue were determined as described in the Materials and Methods section below. Recent studies demonstrated that normal lung epithelium can harbor oncogenic driver mutations [21, 22]. The mutation calling algorithm would not call the mutations if they were present in both normal and abnormal tissues. To avoid the oncogenic mutations in normal lung tissues mutations being undetected, we inspected whole exome sequencing data of the normal lung tissues aligned against the human genome reference for mutations in driver genes. This analysis did not reveal any additional known driver mutations. To characterize the genomic heterogeneity among sequenced lesions, we utilized the Jaccard index, which measures the similarity of non-synonymous (n.s.) somatic mutations between a pair of lesions, and is inversely proportional to the level of heterogeneity. We found that lesions obtained from within individual patients most often had significantly higher Jaccard indices and, thus, lower heterogeneity compared to lesions between different patients (Kruskal-Wallis rank sum test p < 10−16) (Figure 1B). With the exception of the first four patients (P01 — P04, Figure 1C), individual patients had higher indices (lower heterogeneity) among lesions compared to those from different patients. In some patients, certain lesion pairs had very low heterogeneity indicated by high (>95 percentile) Jaccard indices (black circles in Figure 1C) compared to the rest of the lesion pairs. Thus, the individual patients most often demonstrated unique repertoires of n.s. somatic mutations rarely shared with other patients.
Figure 1. Genetic heterogeneity of pulmonary lesions.
A) A density heat map of mutated oncogene and tumor suppressor gene frequencies in ADC (y-axis) or AAH (x-axis). Oncogenes (upper panel) and tumor suppressor genes (lower panel) are mutated at higher frequencies in ADC than in AAH (above diagonal line; Wilcoxon test *p < 7.2 × 10−9 and **p < 1.7 × 10−8). B) Distribution of Jaccard indices comparing n.s. somatic mutation heterogeneity between pairs of lesions from the same (intra-) or different (inter-) patients. C) Distribution of intra-patient Jaccard indices in 41 individual patients. The subjects are displayed in the low-to-high order based on their median values. Black circles indicate lesion pairs with >95 percentile Jaccard indices. In B and C, the side triangles represent the heterogeneity levels inversely proportional to Jaccard indices, and the dashed line marks the 90 percentile level of inter-subject Jaccard index. D) Phylogenetic trees for 10 patients with AAH (blue), AIS (orange) and ADC (brown). The numbers are lesion IDs. Phylogenetic trees for the entire cohort are shown in Supplementary Figure S1.
To explore the relationship between sequenced lesions for each individual patient, phylogenetic trees were constructed. AAH, AIS and ADC were all present in 10 out of 41 patients and their mutational profile-based phylogenetic trees are illustrated in Figure 1D. In the majority of cases, the mutational profiles of ADC (brown labels) were closely related to the profiles of AIS (orange labels), but not AAH (blue labels), except: a) case P30 where one of two primary ADCs was related to AAH, while another primary ADC — to AIS, b) case P34 where ADC clustered with AAH but not AIS, c) case P10 where AAH and AIS lesions were closely related to each other, but not to the ADC (Figure 1D). Phylogenetic trees for the remainder of the patients that had only AAH and ADC, but not AIS, are shown in Supplementary Figure S1.
Premalignant lesions bear somatic mutations associated with progression.
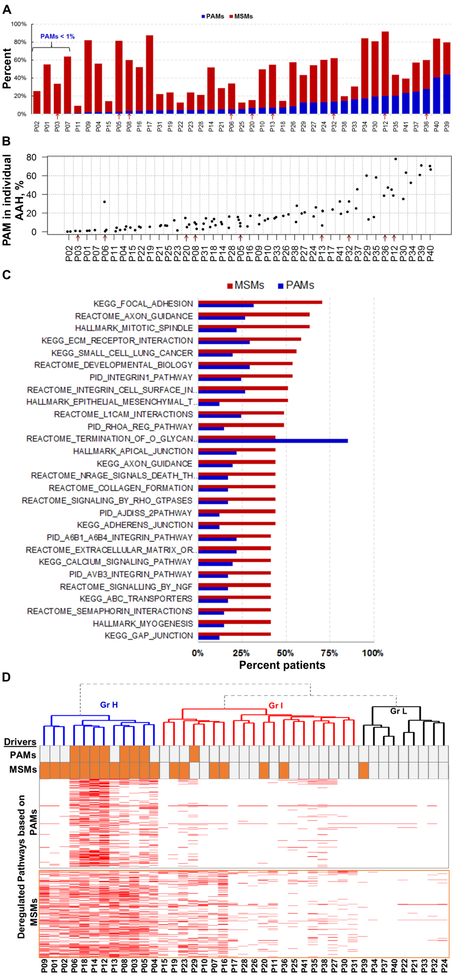
To determine how n.s. somatic mutations affect tumor development at various stages, we classified them into three different categories: a) premalignant mutations which were observed only in AAH lesions, b) progression-associated mutations (PAMs) which were located in both AAH and AIS/ADC lesions, and c) malignant-specific mutations (MSMs) which were only identified in AIS/ADC lesions (Supplementary Figure S2). Recent studies that have focused on tumor heterogeneity and cancer evolution, have classified mutations as trunk (or clonal), branch, and private (subclonal) mutations [23, 24]. Our classification takes into account the histology of the lesion in which the mutations are located. Thus, MSMs are composed of branch and private mutations, while PAMs are comprised of trunk and branch mutations and are indicative of the homogeneity among AAH and ADC within each patient. The distribution of mutation groups in 41 cases is summarized in Figure 2A (the cases are ordered based on the percentage of PAMs in the total number of somatic mutations identified in the corresponding patient). The percentage of PAMs per patient varied over a wide range (0.2% to 44%) (Figure 2A). In addition to the aggregated patient level analysis, PAMs were also characterized in each individual lesion. We found that the percentages of PAMs in the individual AAH lesions were similar to those in the associated ADC (Supplementary Table S1; linear mixed effects model F-test p = 0.25). The percentage of PAMs per individual AAH lesion varied over a wide range (0.2% to 77.8%) (Figure 2B). AAH lesions with high PAM percentages (Figure 2B rightmost patients) have higher homogeneity with the associated ADC, whereas those with low PAM percentages (Figure 2B leftmost patients) are distantly related to the associated ADC and may have independently accumulated additional mutations over time. For instance, patient P06 has three AAH lesions, of which one has significantly higher PAM percentage compared to two others, suggesting that the two AAH lesions with low PAM percentages might have originated from the same precursor, which was distinct from the third AAH (Supplementary Figure S1).
Figure 2. Progression-associated mutation (PAM) and malignant-specific mutation (MSM) distribution and the role in pathway deregulation.
A) Distribution of PAMs and MSMs in 41 study patients. The patients are displayed in the low-to-high order based on their percentages of PAMs. Red arrows in A and in B indicate nine patients whose cellular immune response was evaluated. B) Percentage of PAMs in individual AAH lesions from 41 patients. The cases are displayed in the low-to-high order based on their median levels, and not in the same order as those in A. C) The top 27 pathways frequently affected by MSM- (red) and PAM- (blue) bearing genes. D) Heatmap of the pathways affected (red) by PAM- (top) and MSM- (bottom) bearing genes. The mutations in the 29 driver genes observed in PAM and MSM are indicated by orange bars above the heatmap.
PAMs and MSMs lead to deregulation of distinct cancer-related pathways.
We next evaluated the role of somatic mutations in tumor development. We found that 49% of patients had somatic mutations in at least one of 29 driver genes known to be frequently mutated in lung ADC [1, 25]. Here, these driver mutations were predominantly found in ADC but rarely in AAH (Supplementary Table S3). Of note, oncogenic KRAS mutations were also found in ADC from 4 patients that were not included in Supplementary Table S3 because these mutations were present in low numbers of reads and our mutation calling algorithm could not classify them as true positives; nonetheless, these mutations produced a positive signal on allele-specific PCR. Oncogenic BRAF and KRAS mutations were found only in ADC, but not in AAH lesions from the same patients. Consistent with findings of Sivakumar et al. [26], BRAF and KRAS mutations were mutually exclusive within the lesions. Previous studies have shown that a significant percentage of lung ADCs lack mutations in known driver genes [1, 27]. Therefore, to assess the possible driver gene mutation-independent mechanisms of progression, we next investigated the mutations in the context of molecular pathways.
For the pathway analysis, enrichment scores (ES) of the mutated genes involved in each specific pathway were defined (see Methods). Deregulation of the 1341 well-defined hallmark gene sets and canonical pathways from the Molecular Signature Database [19] was evaluated in both the current cohort and TCGA LUAD. We found that these pathways were deregulated at similar frequencies in both data sets (Supplementary Figure S3A and Supplementary Table S4). We identified 59 and 42 frequently deregulated pathways for the current cohort and TCGA data sets, respectively (Supplementary Figure S3A). Twenty-four of these pathways involved in tumor proliferation and invasion were shared between the data sets (Fisher’s exact test p=3.2 × 10−24). Thus, patients in both cohorts demonstrated common affected pathways involved in carcinogenesis.
Because some genes in ADC were affected by PAMs and other genes by MSMs, it was essential to dissect the input of each of the gene groups in the pathway regulation context. The ES of each gene group was calculated for all 1341 pathways. The majority of the pathways were deregulated by MSMs at significantly higher frequencies than by PAMs. The recurrence rate of the top 27 pathways that are frequently deregulated by the MSM-bearing genes is shown in Figure 2C. The O-glycan biosynthesis pathway was the only pathway more frequently deregulated by PAMs then by MSMs.
To dissect the role of PAM- and MSM-deregulated pathways in tumor initiation and development, we performed unsupervised hierarchical cluster analysis and identified three patient groups designated as high (H, n=12), intermediate (I, n=20), and low (L, n=9) according to the number of pathways deregulated by PAM- and MSM-bearing genes (Figure 2D). Group H had the highest number of deregulated pathways among the three groups (Supplementary Figure S3B) and pathways and driver genes in this group were frequently affected by both PAMs and MSMs (Figures 2D and S3B). In Group H, mutations in KRAS, BRAF and EGFR genes were MSMs, whereas PI3KCA and PI3K/AKT pathway components were PAMs. However, these PAMs were present only in a subset of AAH lesions in each patient, appearing to be branch mutations and suggesting that deregulation of additional driver gene(s) was required for progression. Similarly, higher overall number of deregulated pathways in group H suggests higher genetic complexity of the tumors in this group. The intermediate group included the majority of study patients, in which MSMs (but not PAMs) were the predominant source of pathway deregulation and were frequently found in the driver genes (Figures 2D and S3A). Thus, in the H group, the somatic mutations in driver genes were likely essential for malignant progression. Group L, the smallest group, had infrequent pathway deregulation by either PAM- or MSM-bearing genes, suggesting that the transformation in this group could be caused by events other than the somatic driver mutations that were not readily detectable by whole exome sequencing, such as gene rearrangements, copy number variation, epigenetic changes, deregulation of gene expression or alternative splicing.
Cell-mediated immunity and adaptive responses in pulmonary premalignancy.
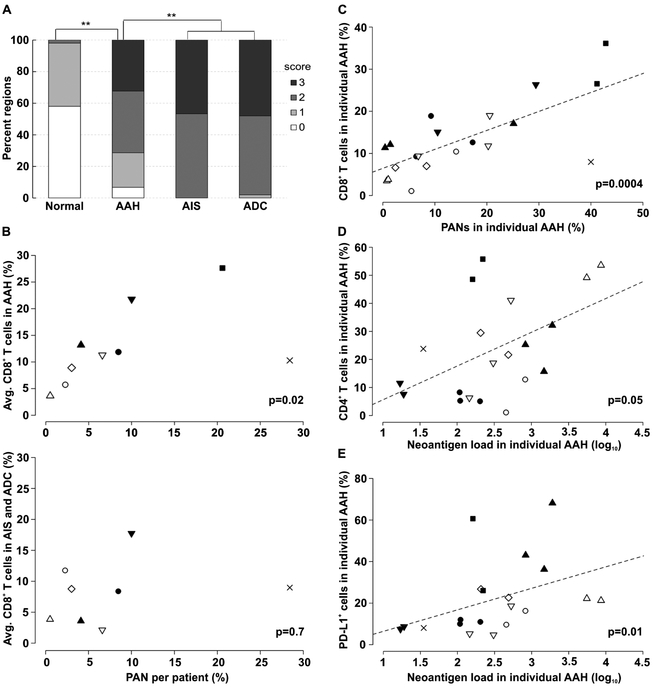
To evaluate the presence of early adaptive immune responses against pulmonary premalignancy, we first assessed the degree of lymphocyte infiltration in premalignant (n = 328) and malignant lesions (n = 15 AIS and 50 ADC), along with adjacent histologically normal areas (n = 50) in the entire cohort of study patients. The median number of lesions evaluated per patient was six for AAH and two for malignant lesions. Lymphocyte infiltration was graded 0-to-3 based on H&E staining (see Methods) and was significantly increased in AAH vs. normal areas (X2 test p<10−16), and became highest in AIS and ADC vs. AAH (X2 test p<10−14) (Figure 3A). We then assessed the expression of regulators of cell-mediated immunity, including CD4, CD8, FOXP3, PD-1 and PD-L1 in AAH and ADC by immunohistochemistry (Supplementary Figure S4). We found both infiltration of T effector and cytotoxic cells and expression of the PD-L1 checkpoint in premalignancy, suggesting that cell-mediated immunity and possible recognition of neoepitopes occur in pulmonary premalignancy.
Figure 3. Immune cell infiltration, neoantigens and the immune response in adenomatous premalignancy.
A) Local lymphocyte infiltration index (0 — lowest, 3 — highest) in adjacent normal tissue, AAH, AIS and ADC (**X2 test p < 10−10). B) Average percentages of infiltrating CD8+ T cells observed in AAH (upper panel) and ADC (lower panel) plotted against percentage of patient-wise PANs. Each patient is represented by a data point indicated by a unique symbol. ADC in one patient was not evaluated. C) Correlation between the percentage of infiltrating CD8+ T cells and the percentage of PANs in corresponding AAH lesions. D-E) Correlation between the percentage of infiltrating CD4+ T cells (D) and PD-L1+ cells (E) plotted against the corresponding log-transformed neoantigen number identified in AAHs. In C-E each region is represented by a point, while each patient is marked by the symbol identical to those in B. P-values are based on Kendall rank correlation coefficient. The trend line (dashed line) in C-E indicates the linear association between variables. Other pair-wise comparisons between immune marker levels and neoantigen-related variables were insignificant.
Somatic mutations produce putative neoantigens in pulmonary premalignancy.
We next sought to determine if somatic mutations and corresponding putative neoantigens were associated with immune responses observed in AAH lesions. Putative neoantigens were derived from n.s. somatic mutations as outlined in Supplementary Figure S2. Multiple algorithms were applied to predict binding affinity (IC50) between mutant proteins and patient HLAs based on the Immune Epitope Database recommendations [14]. Mutant peptides with predicted IC50 < 500 nM were considered neoantigens. In accordance with our mutation classification, the neoantigens were also categorized into three groups as premalignant, progression-associated (PANs) and malignant-specific neoantigens. The total number of aggregated putative neoantigens per lesion was highly correlated with the corresponding mutational load (Kendall’s τ = 0.9) (Supplementary Table S1).
We next evaluated the association of putative neoantigen load and the number and phenotypes of infiltrating immune cells by lesion- and patient-wise comparisons. The lesion-wise comparison evaluated neoantigens and infiltrating immune cell characteristics from the individual AAH lesions, while in the patient-wise analysis these endpoints were aggregated for the corresponding patient. At the patient level, the percentage of PANs significantly correlated with the average percentage of CD8+ T cells infiltrating AAH lesions (Kendall’s τ = 0.61, p = 0.02, Figure 3B upper panel) but not to those infiltrating AIS/ADC (Kendall’s τ = 0.14, p = 0.7, Figure 3B lower panel). At the lesion level, we found that the percentage of CD8+ T cells infiltrating AAH correlated strongly with the percentage of PANs in the respective lesions (Kendall’s τ = 0.56, p = 0.0003) (Figure 3C). Furthermore, AAH lesions with greater neoantigen loads had significantly more infiltrating CD4+ T cells (Kendall’s τ = 0.32, p = 0.05) (Figure 3D) and PD-L1-positive cells (Kendall’s τ = 0.44, p = 0.01) (Figure 3E). These results indicate that the high percentage of PANs promotes CD8+ T cell infiltration in AAH lesions, whereas the overall neoantigen load in AAH is associated with CD4+ T cell infiltration and PD-L1 expression.
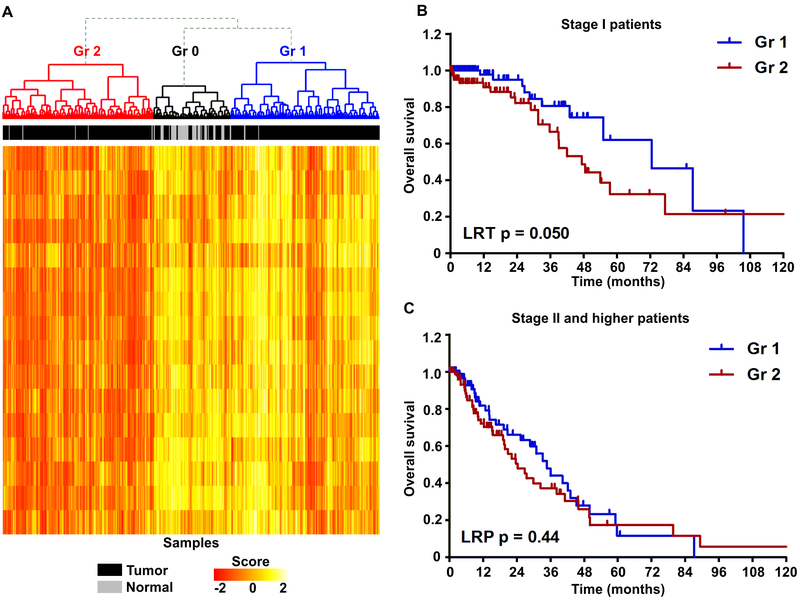
The evidence of apparent immune responses in lung cancer premalignancy and the notion that somatic mutations can contribute to modulation of the pathways regulating tumor immunity, prompted us to determine if the activity of such pathways was associated with outcomes in early stage ADC. The expression of genes involved in 16 pathways from the Molecular Signature Database [19] was analyzed in the TCGA LUAD cohort (444 tumors and 58 normal samples). Gene Set Variation Analysis [20] was utilized to estimate the activities of immune-modulating pathways in individual patients, and these were then subjected to unsupervised hierarchical cluster analysis to stratify samples. Based on the pathway activity, we identified three major groups (Figure 4A). Among them, group 0 (Gr0, annotated by black) had the highest levels of immune-related gene expression and included 51 tumors and the majority of normal samples (n = 52), whereas the other two groups included the remainder of the tumor samples (X2 test p<10−16): Gr1 (n = 198, blue) with intermediate and Gr2 (n = 201, red) with lowest expression of immune-related genes. These groups were not significantly associated with tumor stage (X2 test p = 0.14 for stage I vs. stage II and higher), however, the overall survival was marginally higher in Gr1 compared to Gr2 (log-rank test (LRT) p = 0.063). Remarkably, the difference in survival between Gr1 and Gr2 was most prominent for stage I patients (LRT p=0.05, Figure 4B), but not for stage II and higher patients (LRT p=0.44, Figure 4C). Together, these results suggest that modulation of the immune-related pathways, especially at the earliest stages of lung ADC, may have a significant impact on outcomes of lung cancer patients.
Figure 4. Analysis of immune pathway deregulation and patient outcomes in TCGA LUAD.
A) Heatmap of gene expression scores of 16 immune-related pathways in TCGA LUAD and normal lung samples. B-C) Kaplan-Meier survival curves of stage I (B) and stage II and higher (C) patients from the groups identified in (A).
Discussion
Recent studies suggest the immune response exerts selective pressure on tumor cells, as well as premalignant cells, throughout the course of carcinogenesis [28–30]. This process of immune editing may result in resolution of a premalignant lesion or, alternatively, progression with persistent or newly developed neoantigens in the context of a microenvironment hostile to effective cell-mediated immune responses [31]. Here we report that neoantigens are expressed in the earliest pulmonary premalignant lesions. Neoantigen load in these lesions correlates with the extent of CD8 T-cell infiltration and levels of PD-L1 expression. These findings suggest that specific immune recognition of neoepitopes can occur at the earliest points of pulmonary premalignancy and lung cancer development, indicating the potential for future strategies utilizing immunoprevention in lung cancer interception.
We sought to identify somatic mutations in adenomatous premalignancy and associated lung adenocarcinoma and also, to determine the extent of immune cell infiltration of premalignant lesions and the associated tumors. Our findings indicate that premalignant AAH lesions from within an individual patient may have distinct mutational profiles (Figures 1D and S1) and bear a range of PAMs (Figures 2A and B). Analysis of 29 driver genes, frequently mutated in ADC, demonstrated that driver mutations were predominantly found in ADC but rarely in AAH (Supplementary Table S3), suggesting that malignant progression was induced by the driver mutations occurring in some, but not all, premalignant lesions.
Furthermore, we demonstrate that heterogeneity between different lesions from an individual patient is significantly lower than that among lesions from different patients (Figure 1B and C). In the majority of cases the mutational profiles of AIS are distinct from those of AAH and highly overlap with those of ADC (Figure 1D). Previous studies suggest that passenger mutations can promote malignant progression by modulating the activity of oncogenic or tumor suppressor pathways [32, 33]. Therefore, beyond the individual mutations, we assessed the effect of premalignant somatic mutations in the context of pathways. One of the most frequently deregulated pathways in both the UCLA and TCGA cohorts is the O-glycan biosynthesis pathway that includes mucin proteins which protect epithelial cells from physical and chemical damage. Deregulated expression of mucins promotes tumor cell invasion and migration, and increases drug resistance in a variety of malignancies [34, 35]. Genetic variation of MUC4 has been associated with increased lung cancer risk [36], and here we find that PAMs of MUC4 were present in over 90% of patients. These mutations produced a total of 132 PANs in 31 of 41 patients. Two of the recurring PAN-producing mutations in MUC4 were found in 4 patients, 6 of these were in 3 patients and 18 in 2 patients. The functional significance of these and other recurring PANs will be assessed in our future studies. Also, focal adhesion, extracellular matrix-receptor interaction and calcium signaling pathways were frequently deregulated (Supplementary Table S4). These pathways have established roles in carcinogenesis, including proliferation, invasion and resistance to therapy [37, 38].
The analysis of an immune contexture of the lung cancer continuum revealed histologic evidence of immune recognition of AAH lesions, characterized by lymphocyte infiltration and checkpoint molecule upregulation consistent with adaptive immune responses. From the whole exome sequencing data, putative neoantigens were identified. We demonstrated that the neoantigen load in AAH lesions correlates significantly with CD4+ T cell infiltration and PD-L1 expression. Progression-associated neoantigens (PANs) were detected in all patients, with 37 out of 41 patients expressing them at greater than 1% frequency. CD8+ T cell infiltration was strongly correlated with the percentage of PANs in individual AAH lesions. These findings provide evidence of adaptive immunity in pulmonary premalignancy and are consistent with recent studies demonstrating that gene sets associated with suppressed antitumor and elevated pro-tumor immune signaling are enriched in AAH development and progression [26]. Furthermore, we identified frequent premalignancy-specific putative neoantigens. Consistent with the immunoediting concept of Schreiber [39], this suggests active immune- editing in the progression of adenomatous premalignancy to invasive adenocarcinoma.
Neoantigens, produced by PAMs, are potential immunotherapy targets, but these neoantigens do not necessarily correspond to known driver genes. Consistent with findings in melanoma [40] and colorectal cancer [41], our analysis of mutations in lung adenocarcinoma indicates that while there are many common driver mutations among tumors from different patients, mutations producing PANs are most often unique to individual patients. Due to high genomic plasticity, established cancers have highly heterogeneous mutational landscapes in different areas of the tumor due to potential parallel evolution and subclonal expansion [42–44]. This has been postulated to be one of the reasons for tumor resistance to therapies targeting actionable somatic events. Our data suggests that future therapies targeting PANs in cancer interception [45], as well as prevention strategies, may need to be tailored to individual patients.
The notion that genes bearing somatic mutations often encode tumor specific neoantigens capable of eliciting immunity and tumor rejection was first described in murine models sixty years ago [46]. Furthermore, the concept of immune surveillance first proposed by Burnet [47], suggests that the host immune response is able to recognize and destroy the incipient tumors at the earliest point of development before clinical recognition. While extensive data exists in laboratory models, the clinical evidence for the relevance of immune surveillance in human lung cancer has not yet been defined, nor is it yet known when an individual’s immune system begins to engage in the defense against the disease. Our findings warrant further investigations to evaluate the efficacy of persisting neoantigens, such as PANs, as interception targets for immunoprevention strategies. This approach may include “vaccination” of post-surgery lung cancer patients with the autologous T cell clones recognizing strong persisting neoantigens. Alternatively, autologous dendritic cells presenting persistent neoantigens could be administered in order to block the progression of remaining premalignant lesions.
In accord with the cancer immune surveillance theory, our current findings support the concept that the immune system is capable of recognizing cancer precursors [48, 49]. Because evasion of immune surveillance has been implicated as an emerging hallmark of cancer development, future investigations will focus on stimulating specific immune responses [50]. Thus, it has been suggested that unleashing the immune response against pulmonary premalignancy may facilitate a blockade of the progression of premalignancy to invasive cancer at the earliest stages of disease [51]. This will require a more complete understanding of the immune microenvironment of pulmonary premalignancy as well as the identification of premalignant markers that could be targeted in immunoprevention strategies.
Supplementary Material
Statement of Significance.
Findings identify progression-associated somatic mutations, oncogenic pathways, and association between the mutational landscape and adaptive immune responses in adenomatous premalignancy.
Acknowdgements
The authors thank Drs. Antoni Ribas and Siwen Hu-Lieskovan for the assistance with the HALO system, Lauren Winter for the administrative support, Francis Rosen for clinical coordination, the UCLA Jonsson Comprehensive Cancer Center for shared resources, the UCLA CNSI Advanced Light Microscopy/Spectroscopy Facility and, the UCLA Institute for Digital Research and Education for providing the Hoffman2 Cluster. Grant support: Research supported by a Stand Up To Cancer-LUNGevity-American Lung Association Lung Cancer Interception Dream Team Translational Cancer Research Grant (Grant Number: SU2C-AACR-DT23–17). Stand Up To Cancer is a division of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C. DOD W81XWH-16-1-0194 (Kostyantyn Krysan), UC Tobacco-Related Disease Research Program (TRDRP) 27IR-0036 (Kostyantyn Krysan), DOD W81XWH-17-1-0399 (Steven M. Dubinett), Stand Up To Cancer SU2C-AACR-DT-23–17 (Steven M. Dubinett, Avrum E. Spira), NCI HTAN (PCA) 1U2CCA233238–01 (Avrum E. Spira, Steven M. Dubinett), NIH/NCI Molecular Characterization Laboratory 5U01CA196408–04 (Steven M. Dubinett, Denise Aberle, Avrum E. Spira), NIH/NCI EDRN (Steven M. Dubinett, Avrum E. Spira, David A. Elashoff, Denise Aberle) 1U01CA214182, NIH/NCATS—UCLA Clinical and Translational Science Institute UL1TR001881 (Steven M. Dubinett), VA Merit Review 1I01CX000345–01 (Steven M. Dubinett), NIH/NHLBI T32HL072752 (Brandon S. Grimes, Ramin Salehi-rad).
Footnotes
Disclosure of Potential Conflicts of Interest
Steven M. Dubinett is an advisory board member for EarlyDx Inc., T-Cure Bioscience Inc., Cynvenio Biosystems Inc. and the Johnson and Johnson Lung Cancer Initiative; Avrum E. Spira is an employee of Johnson & Johnson.
References
- 1.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niho S, Yokose T, Suzuki K, Kodama T, Nishiwaki Y, Mukai K. Monoclonality of atypical adenomatous hyperplasia of the lung. Am J Pathol 1999;154:249–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Izumchenko E, Chang X, Brait M, Fertig E, Kagohara LT, Bedi A, et al. Targeted sequencing reveals clonal genetic changes in the progression of early lung neoplasms and paired circulating DNA. Nat Commun 2015;6:8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016;48:607–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 2015;372:2018–28. [DOI] [PubMed] [Google Scholar]
- 6.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarSca 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 2004;20:289–90. [DOI] [PubMed] [Google Scholar]
- 11.Schliep KP. phangorn: phylogenetic analysis in R. Bioinformatics 2011;27:592–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szolek A, Schubert B, Mohr C, Sturm M, Feldhahn M, Kohlbacher O. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics 2014;30:3310–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breese MR, Liu Y. NGSUtils: a software suite for analyzing and manipulating next-generation sequencing datasets. Bioinformatics 2013;29:494–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vita R, Overton JA, Greenbaum JA, Ponomarenko J, Clark JD, Cantrell JR, et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res 2015;43:D405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, Nielsen M. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res 2008;36:W509–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lundegaard C, Lund O, Nielsen M. Accurate approximation method for prediction of class I MHC affinities for peptides of length 8, 10 and 11 using prediction tools trained on 9mers. Bioinformatics 2008;24:1397–8. [DOI] [PubMed] [Google Scholar]
- 17.Peters B, Sette A. Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinformatics 2005;6:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O, et al. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 2009;61:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kadara H, Sivakumar S, Jakubek Y, San Lucas FA, Lang W, McDowell T, et al. Driver Mutations in Normal Airway Epithelium Elucidate Spatiotemporal Resolution of Lung Cancer. Am J Respir Crit Care Med 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadara H, Wistuba, II. Field cancerization in non-small cell lung cancer: implications in disease pathogenesis. Proc Am Thorac Soc 2012;9:38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016;351:1463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 2014;346:256–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berger AH, Brooks AN, Wu X, Shrestha Y, Chouinard C, Piccioni F, et al. High-throughput Phenotyping of Lung Cancer Somatic Mutations. Cancer Cell 2016;30:214–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sivakumar S, Lucas FAS, McDowell TL, Lang W, Xu L, Fujimoto J, et al. Genomic Landscape of Atypical Adenomatous Hyperplasia Reveals Divergent Modes to Lung Adenocarcinoma. Cancer Res 2017;77:6119–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sholl LM, Aisner DL, Varella-Garcia M, Berry LD, Dias-Santagata D, Wistuba II, et al. Multi-institutional Oncogenic Driver Mutation Analysis in Lung Adenocarcinoma: The Lung Cancer Mutation Consortium Experience. J Thorac Oncol 2015;10:768–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bremnes RM, Al-Shibli K, Donnem T, Sirera R, Al-Saad S, Andersen S, et al. The role of tumor-infiltrating immune cells and chronic inflammation at the tumor site on cancer development, progression, and prognosis: emphasis on non-small cell lung cancer. J Thorac Oncol 2011;6:824–33. [DOI] [PubMed] [Google Scholar]
- 29.McGranahan N, Swanton C. Cancer Evolution Constrained by the Immune Microenvironment. Cell 2017;170:825–7. [DOI] [PubMed] [Google Scholar]
- 30.McGranahan N, Rosenthal R, Hiley CT, Rowan AJ, Watkins TBK, Wilson GA, et al. Allele-Specific HLA Loss and Immune Escape in Lung Cancer Evolution. Cell 2017;171:1259–71 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jerby-Arnon L, Shah P, Cuoco MS, Rodman C, Su MJ, Melms JC, et al. A Cancer Cell Program Promotes T Cell Exclusion and Resistance to Checkpoint Blockade. Cell 2018;175:984–97 e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leedham S, Tomlinson I. The continuum model of selection in human tumors: general paradigm or niche product? Cancer Res 2012;72:3131–4. [DOI] [PubMed] [Google Scholar]
- 33.Muller FL, Colla S, Aquilanti E, Manzo VE, Genovese G, Lee J, et al. Passenger deletions generate therapeutic vulnerabilities in cancer. Nature 2012;488:337–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brockhausen I Pathways of O-glycan biosynthesis in cancer cells. Biochim Biophys Acta 1999;1473:67–95. [DOI] [PubMed] [Google Scholar]
- 35.Rao CV, Janakiram NB, Mohammed A. Molecular Pathways: Mucins and Drug Delivery in Cancer. Clin Cancer Res 2017;23:1373–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z, Wang J, He J, Zheng Z, Zeng X, Zhang C, et al. Genetic variants in MUC4 gene are associated with lung cancer risk in a Chinese population. PLoS One 2013;8:e77723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roderick HL, Cook SJ. Ca2+ signalling checkpoints in cancer: remodelling Ca2+ for cancer cell proliferation and survival. Nat Rev Cancer 2008;8:361–75. [DOI] [PubMed] [Google Scholar]
- 38.Zhao J, Guan JL. Signal transduction by focal adhesion kinase in cancer. Cancer Metastasis Rev 2009;28:35–49. [DOI] [PubMed] [Google Scholar]
- 39.Mittal D, Gubin MM, Schreiber RD, Smyth MJ. New insights into cancer immunoediting and its three component phases--elimination, equilibrium and escape. Curr Opin Immunol 2014;27:16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015;350:207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Angelova M, Charoentong P, Hackl H, Fischer ML, Snajder R, Krogsdam AM, et al. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol 2015;16:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Sousa EMF, Vermeulen L, Fessler E, Medema JP. Cancer heterogeneity--a multifaceted view. EMBO Rep 2013;14:686–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 2015;27:15–26. [DOI] [PubMed] [Google Scholar]
- 44.Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013;501:338–45. [DOI] [PubMed] [Google Scholar]
- 45.Blackburn EH. Cancer interception. Cancer Prev Res (Phila) 2011;4:787–92. [DOI] [PubMed] [Google Scholar]
- 46.Prehn RT, Main JM. Immunity to methylcholanthrene-induced sarcomas. J Natl Cancer Inst 1957;18:769–78. [PubMed] [Google Scholar]
- 47.Burnett FM. Immunological surveillance. Oxford: Pergamon Press; 1970. [Google Scholar]
- 48.Zitvogel L, Tesniere A, Kroemer G. Cancer despite immunosurveillance: immunoselection and immunosubversion. Nat Rev Immunol 2006;6:715–27. [DOI] [PubMed] [Google Scholar]
- 49.Galon J, Angell HK, Bedognetti D, Marincola FM. The continuum of cancer immunosurveillance: prognostic, predictive, and mechanistic signatures. Immunity 2013;39:11–26. [DOI] [PubMed] [Google Scholar]
- 50.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 51.Spira A, Yurgelun MB, Alexandrov L, Rao A, Bejar R, Polyak K, et al. Precancer Atlas to Drive Precision Prevention Trials. Cancer Res 2017;77:1510–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.