Abstract
By examination of the cancer genomics database we identified a new set of mutations in core histones that frequently recur in cancer patient samples and are predicted to disrupt nucleosome stability. In support of this idea, we characterized a glutamate to lysine mutation of histone H2B at amino acid 76 (H2B-E76K) found particularly in bladder and head and neck cancer that disrupts the interaction between H2B and H4. Although H2B-E76K forms dimers with H2A, it does not form stable histone octamers with H3 and H4 in vitro and when reconstituted with DNA forms unstable nucleosomes with increased sensitivity to nuclease. Expression of the equivalent H2B mutant in yeast restricted growth at high temperature and led to defective nucleosome-mediated gene repression. Significantly, H2B-E76K expression in the normal mammary epithelial cell line MCF10A increased cellular proliferation, cooperated with PIK3CA to promote colony formation, caused a significant drift in gene expression and fundamental changes in chromatin accessibility particularly at gene regulatory elements. Taken together, these data demonstrate that mutations in the globular domains of core histones may give rise to an oncogenic program due to nucleosome dysfunction and deregulation of gene expression.
INTRODUCTION
With the introduction of next-generation sequencing and the elucidation of now tens of thousands of cancer genomes, it has become evident that among the commonest class of genes recurrently mutated in human cancers are those encoding components of the gene expression machinery including: transcription factors, DNA methyl transferases, histone methyl transferases and demethylases, histone acetyl transferases, components of the SWI/SNF chromatin remodeling system required for nucleosome movement, and components of the cohesin complex necessary for promoter/enhancer looping interactions (1). Collectively these mutations may affect chromatin modification states and accessibility of DNA to transcription factors, DNA replication and DNA repair machinery.
Epigenetic regulation of gene expression occurs by modifying the accessibility of DNA wrapped around the histone octamer consisting of two molecules each of histone H2A, H2B, H3, and H4 to sequence specific transcription factors and RNA polymerase (2,3). Formation of a nucleosomal array inhibits gene activity, and effective transcriptional activation requires the movement of nucleosomes at key sites in response to physiological stimuli (4,5). Histones are among the most highly conserved proteins across species, and the genes encoding the canonical histones are located in four clusters encompassing a total of 72 histone genes that includes 16 genes for histone H2A, 22 genes for histone H2B, 14 genes for histone H3, 14 genes for histone H4 and 6 genes for histone H1 (6). The canonical histones are synthesized during S phase to package newly synthesized DNA. In addition, each core histone has specialized replication-independent variant genes dispersed throughout the genome that can differ significantly in sequence from their canonical counterparts and substitute for canonical histones in context-dependent instances to form alternative nucleosome conformations (7). These alternative nucleosome configurations provide functional heterogeneity to chromatin dynamic processes such as DNA repair or regulation of gene expression.
Chromatin packaging and gene expression is controlled in part by the nature of chemical modifications on histone, particularly on “tail” residues which are unstructured and do not directly interact with DNA, but rather serve as recognition sites for epigenetic writers of chromatin marks, erasers of such marks and readers that bind to such marks to further modify chromatin. For example, the Histone H3 lysine 27 trimethyl (H3K27me3) modification created by the PRC2/EZH2 complex is recognized by the PRC1 complex which contains chromatin remodeling ATPases that actively condense and close chromatin (8). Mutation of the H3K27 to methionine, inhibits the activity of PRC2, leads to global decrease of H3K27me3, aberrantly open chromatin and dysregulation of gene expression that contributes to the pathogenesis of diffuse intrinsic pontine glioma in children (9–11). Missense mutations of H3 at amino acid position 34 and 36 are associated with gliomas, sarcomas and head and neck cancers, and lead to inhibition of methyl transferases that create H3K36 methyl marks, which are associated with open and transcribed chromatin (12–15). Hence, loss of the H3K36me2 or H3K36me3 may lead to aberrantly closed chromatin.
To gain a more complete understanding of histone mutations in cancer we used the cBioPortal of cancer patient genomics data to identify mutations in the four core histones that recur with high frequency in cancer (16). In agreement with a recent report (17), we observed that the most common histone mutations in cancer were often found in the globular histone fold domains of canonical histones rather than the tail region, and many of these mutations were located in discrete structural regions important for histone octamer and nucleosome integrity. To analyze the functional impact of histone mutations we characterized the most frequent canonical histone mutation observed in cancer, a glutamate to lysine missense mutation of histone H2B at amino acid position 76 (H2B-E76K). The H2B-E76K mutation destabilized the histone octamer and disrupted nucleosome-mediated transcriptional repression. These findings suggest that missense mutations in the body of core histones are a new and important mode of global epigenetic dysfunction that occurs frequently in human cancers.
RESULTS
Missense mutations at specific residues of core histones frequently occur in cancer patient samples
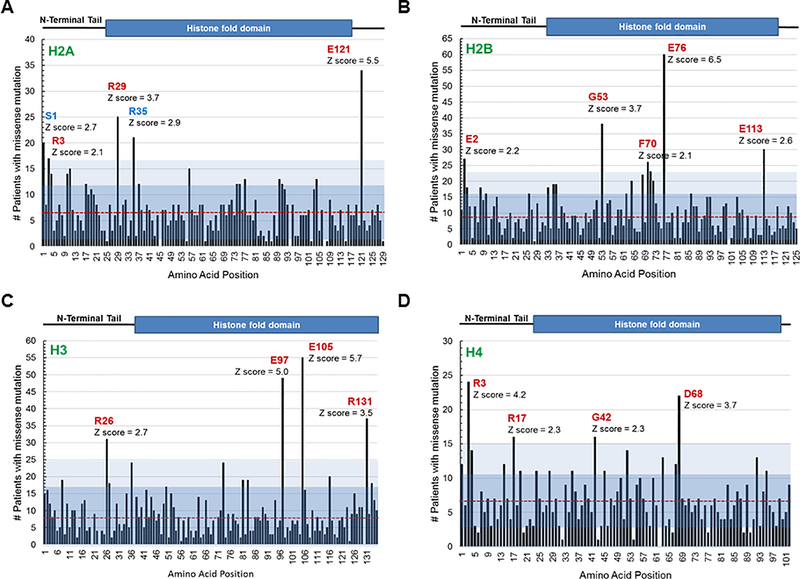
We analyzed a curated set of 159 non-redundant cancer studies with a total of 41,738 sequenced patient samples using cBioPortal to identify mutations that frequently occur in genes encoding H2A, H2B, H3 and H4, in patient samples of all cancer types (16). Because there are multiple gene paralogs for each histone, we determined the cumulative number of patients with a mutation at each conserved amino acid residue across all canonical gene paralogs for each histone (Figure 1). Across the four core histones, ~90% of the mutations reported were missense due to a one base substitution with frameshift or indels much less common. Generally, most residues of histones were reported mutated at least once and on average each residue was reported mutated in 6–10 patients. Across all 4 histones there were clear hot spots where a recurrent mutation was noted at a much higher frequency. A Z score was calculated for the number of patients with mutations at each amino acid, and a score of >2 was considered significant.
Figure 1. Survey of the most frequently found canonical histone mutations in cancer patient samples.
A cross cancer mutation summary was performed using the cBioPortal to search a total of 41,738 non-redundant patient samples across all cancer types. The number of patients reported to have a missense mutation at each amino acid residue position across histone paralogs was graphed. A red line denotes the average number of mutations for each histone across all paralogs, and blue shaded regions indicate the first two standard deviations from the average. Location of the cumulative missense mutations found in: (A) 15 canonical H2A genes (HIST1H2AA/B/C/D/E/G/H/I/J/K/L/M, HIST2H2AB/C, HIST3H2A), (B) 18 canonical H2B genes (HIST1H2BA/B/C/D/E/F/G/H/I/J/K/L/M/N/O, HIST2H2BE/F, HIST3H2BB), (C) 12 canonical H3 genes (HIST1H3A/B/C/D/E/F/G/H/I/J, HIST2H3D, HIST3H3), and (D) 14 canonical H4 genes (HIST1H4A/B/C/D/E/F/G/H/I/J/K/L, HIST2H4A, HIST4H4) across all cancer patient samples. The Z scores indicate the most frequently found mutations (Z > 2) in each histone. Amino acids displayed in red were significantly more frequent in cBioPortal than the general SNP database.
Mutations in 15 canonical histone paralog genes encoding H2A were observed in 864 of 40,317 sequenced patients (2.1%) of all cancer types (Figure 1A). The overall somatic mutation frequency in each individual H2A paralog ranged from 0.1 to 0.2%. When the H2A mutations were sorted by cancer type using cancer studies with at least 50 cases, H2A mutations were commonly found in bladder cancer (10.7%), endometrial carcinomas (10.1%), head and neck carcinomas (8.2%), esophagogastric adenocarcinomas (7.8%), and cervical squamous cell carcinomas (7.2%). The most frequently found missense mutation in H2A was a glutamate to lysine mutation at amino acid position 121 (Figure 1A and Supplemental Table I). Mutation at E121 was the only mutation observed across all H2A histone cluster 1 paralog genes.
Mutations in 18 canonical H2B paralogs were observed in 1224 of 40,317 sequenced patient samples (Figure 1B, 2.8%). The mutation frequency in individual H2B paralog genes ranged from 0.2–0.3%. When sorted by cancer type using studies with at least 50 cases, the 5 cancers with the most frequently observed H2B mutations were: Endometrial carcinomas (13.8%), bladder urothelial carcinomas (13.4%), cervical squamous cell carcinomas (10.8%), head and neck squamous cell carcinomas (10.3%) and esophageal squamous cell carcinomas (9.5%). The most common missense mutation in H2B was a glutamate to lysine mutation at amino acid position 76, H2B-E76K, and this was also the most frequently observed canonical histone gene mutation across all cancer types (Figure 1B and Supplemental Table 2).
Histone H3 mutations in 12 canonical paralog genes were observed in 1533 of 40,317 cancer patient samples (3.8%) and the frequency of mutation in individual paralogues H3 genes ranged from 0.1 to 0.5% (Figure 1C and Supplemental Table 3). When sorted by cancer type using studies with at least 50 cases, the five cancer types with the most frequently observed H3 mutations were: high grade glioma (19.3%), bladder cancer, NOS (13.7%), esophageal squamous cell carcinoma (13.7%), endometrial carcinoma (11.4%) and non-melanoma skin cancers (9.2%). The most frequent amino acid found mutated in the canonical H3 histone genes was the glutamate at amino acid position 105 (Figure 1C and Supplementary Table 3). Importantly, our cross-cancer survey of histone mutations identified missense mutations at amino acid K27 and G34 in histone variant H3.3 (Supplemental Figure 1). Mutations at these latter residues were well characterized to be significant in cancer, suggesting that other frequently occurring histone mutations identified by this study may be of similar importance. Along these lines we also uncovered a recurrent change A114G in H3.3. The H3F3A gene that encodes H3.3 uses the sequence AAG to encode lysine at amino acid position 27 and is found frequently mutated to ATG (K→M) in cancers. Although more than half of the genes encoding canonical histone H3.1 and H3.2 use AAG to encode K27, the K27M missense mutation is not observed frequently among these proteins, suggesting that the significance of the H3K27M mutation may be specific to histone H3.3 function. Also of note although few mutations have been catalogued for the centrosome specific H3 histone CENPA, two mutations R69C/L and R130Q/W/L were among the most frequent (Supplemental Figure 1 and Table 3).
Mutations in 14 canonical histone H4 paralogs were observed in 687 of 40,317 queried cancer patients (1.7%) and the frequency of mutation in individual paralogs H4 genes ranged from 0.1 to 0.2% (Figure 1D and Supplemental Table 4). When sorted by cancer type using studies with at least 50 cases, the 5 cancer types with the most frequently observed H4 mutations were: endometrial carcinomas (9%), bladder urothelial carcinomas (7.3%), head and neck squamous cell carcinomas (6.3%), esophagogastric adenocarcinomas (5.6%) and colorectal adenocarcinomas (5.3%). The most frequently observed mutation across H4 paralogs was the arginine at amino acid position 3 (Figure 1D and Supplemental Table 4).
Missense mutations were found to occur beyond two standard deviations of the average frequency at only eighteen amino acid residues across all four core canonical histones, and patients usually only displayed one histone gene mutation (Figure 1 and Supplemental Tables 1–4). We examined dbSNP the database of short genomic variation (18) to determine if some mutations could actually represent rare polymorphisms. This revealed that in general specific histone protein changes were much more common in the cancer genomics database than in the general population (Supplemental Figure 2 and Supplemental Tables 1–4). Only H2A amino acids S1 and R35 had polymorphism frequencies between the general population and cancer patient databases, indicating they may not be pathogenic. By contrast, polymorphisms at H2B-E76 are found to be approximately 100-fold more common in cancer patients than the general population (Supplemental Figure 2). Furthermore, the average variant allele frequency of these eighteen mutations, including H2A S1 and R35 was in the 20–30% range in diploid tumor samples, suggesting that histone mutations are subclonal and acquired rather than germ line mutation or polymorphisms (Supplemental Tables 1–4).
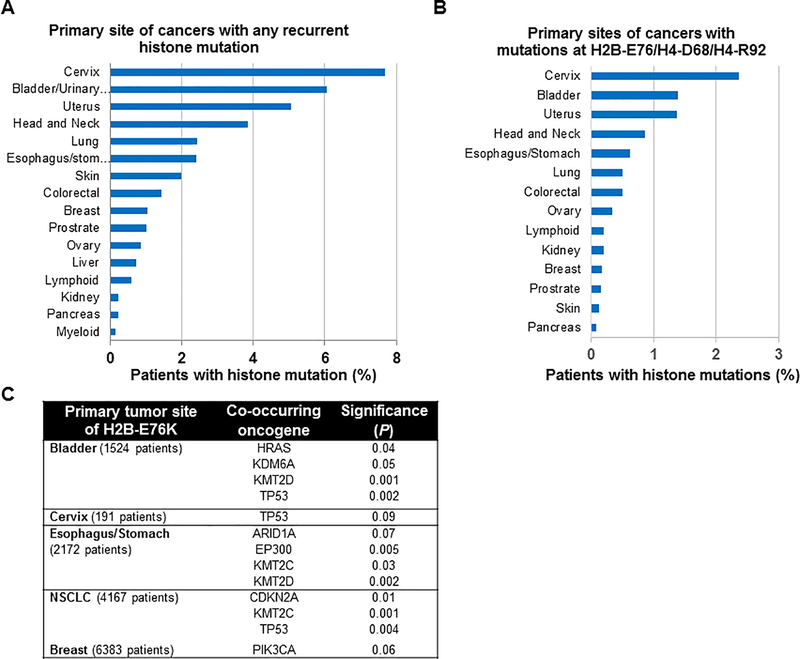
A mutation in one of the 18 recurrently mutated histone residues was were found in up to 8% of some solid tumors (Figure 2A). Most of these mutations lie within the histone fold of each protein rather than the tail regions and several map to locations important for nucleosome integrity, suggesting that nucleosome destabilization may play a role in cancer. For instance, E76 of H2B and R92 of H4 (the seventh most frequent mutation in H4) are predicted to form salt bridges bolstered by hydrogen bonds with each other at the H2B-H4 interface and H4-D68 is also in this vicinity in the nucleosome crystal structure (Supplemental Figure 3A). In addition, E105 of H3 interacts with G42 of H4 in the nucleosome (Supplemental Figure 3B). Molecular dynamics simulations and a recently solved structure indicate that mutation of H2B at E76 disrupts the association of H2B with H4 (Supplemental Figure 3C, D and (19)). Missense mutations at H2B-E76, H4-D68 or H4-R92, are found in about 1–2% of cervical bladder and uterine cancer patient samples (Figure 2B). Furthermore, these mutations are often found to occur together with common oncogenes in cancer patient samples. For instance, the most frequent histone mutation was at E76 of H2B, and mutations at this residue tends to co-occur with common oncogenes such as RAS, KDM6A, KMT2C and TP53 at a rate greater than chance (Figure 2C). Taken together these findings suggest that mutations at any of these eighteen residues may contribute to oncogenesis potentially by destabilizing the histone octamer and nucleosome structure in such a way to favor the cancer phenotype without being lethal to the cell.
Figure 2. Frequency of recurrent histone missense mutations in cancer.
The cBioPortal was queried to determine the frequency of histone mutations in cancer patient samples at each primary site. (A) The frequency for any of the eighteen most significantly recurring histone missense mutations (Z score > 2) in each primary cancer site. (B) The frequency of missense mutations at either H2B glutamate 76 (H2B-E76), H4 aspartate (H4-D68) or H4 arginine (H4-R92), which destabilizes the histone octamer at a specific structural region, in cancers at each primary site. (C) The co-occurrence of the H2B-E76K mutation with mutations in the most common oncogenes found in each cancer type (for cancers with 3 or more patients with H2B-E76K) was assessed by a randomization test through 10,000 rounds of randomization.
Mutation of H2B at E76 disrupts histone octamer formation and nucleosome structure
To test the effect of histone mutations on nucleosome structure, gene expression and cellular phenotype we chose to fully characterize the effect of H2B mutation at glutamate 76 (E76) since this mutation occurred at the highest frequency among cancer patient samples and previous reports indicated that it destabilized nucleosome structure (19,20). Alteration at H2B at residue E76 was rarely reported as a polymorphism in dbSNP (18), and the average variant allele frequency of H2B-E76K in tumors was 0.23 ± 0.13, suggesting it is not a founding “driver” lesion but rather an acquired subclonal mutation that may increase cancer development (Supplemental Figure 2).
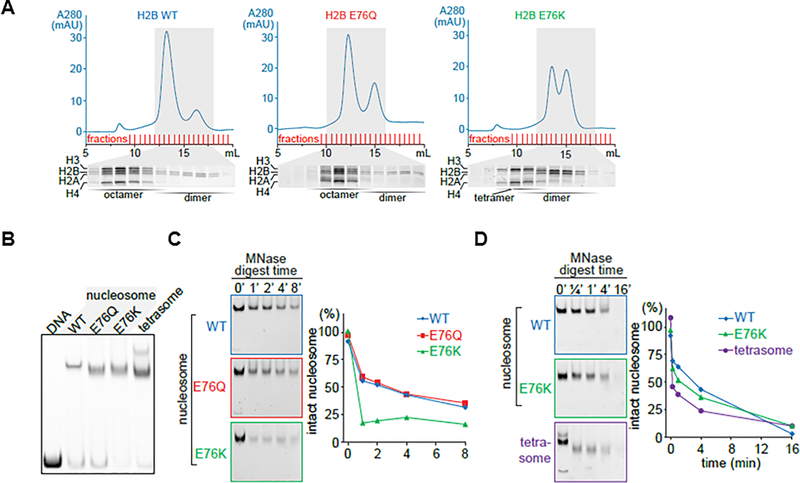
To measure the extent to which the H2B-E76K mutation could affect nucleosome structure or stability we reconstituted nucleosomes using recombinant histones. Gel filtration chromatography showed that wild-type (WT) H2B yielded mostly a peak of histone octamer and a small peak of H2A-H2B dimer when the dimer is added in 10% excess (Figure 3A, left). By contrast, the H2B-E76K mutant failed to form histone octamers and two peaks were resolved, one consisting of H3-H4 tetramers and a second of H2A-H2B dimers (Figure 3A, right). In addition, the H2B-E76Q mutant which was less frequent that H2B-E76K but was recurrent in cancer, formed histone octamers less efficiently than WT as a greater fraction of H2A-H2B dimers was observed than when WT-H2B was used (Figure 3A, middle). Despite octamer instability, H2B-E76K and H2B-E76Q histones can be reconstituted into nucleosomes from H2A-H2B dimers and (H3-H4)2 tetramers with a strong positioning DNA sequence. However, nucleosomes containing H2B-E76K or H2B-E76Q displayed faster electrophoretic mobility in a native gel, suggesting a qualitative change in the complex. The DNA-protein complex formed by nucleosomes containing H2B-E76K was significantly more sensitive to micrococcal nuclease (MNase) digestion than nucleosomes reconstituted with WT-H2B or H2B-E76Q (Figure 3B and C). However, DNA- protein complexes containing H2B-E76K nucleosomes were more resistant to MNase than tetrasomes of H3/H4 (Figure 3D). Collectively these data suggest that H2B-E76K forms unstable nucleosomes with the full complement of histones, while H2B-E76Q more closely resembles wild-type histone H2B.
Figure 3: The E76K mutation in H2B destabilizes the histone octamer and fails to protect the nucleosome from nuclease treatment in vitro.
(A) The H2B-E76K mutant was unable to form stable octamers in vitro. Recombinant human histones (H2A, H2B, H3, and H4) were mixed and histone octamers resolved from (H3/H4)2 tetramers, H2A/H2B dimers and free histones by gel filtration chromatography. (B) Nucleosomes were reconstituted by mixing equimolar amounts of DNA (147bp) and octamers or in the case of E76K of tetramers and dimers (1:2 molar ratio) and resolved by Native PAGE. Nucleosomes containing H2B-E76K and E76Q have an altered migration pattern, intermediate between a tetrasome and a WT nucleosome. (C) Micrococcal nuclease (MNase) sensitivity assay performed on nucleosomes made with WT, E76Q and E76K H2B mutants shows more rapid digestion of E76K containing nucleosomes than those with WT H2B. A time course by gel (left) and densitometry quantification (right) of intact nucleosomes following MNase treatment. (D) The MNase susceptibility of E76K nucleosomes is distinct from nucleosomes formed only with tetrasomes.
H2B mutation deregulates growth and gene expression in yeast
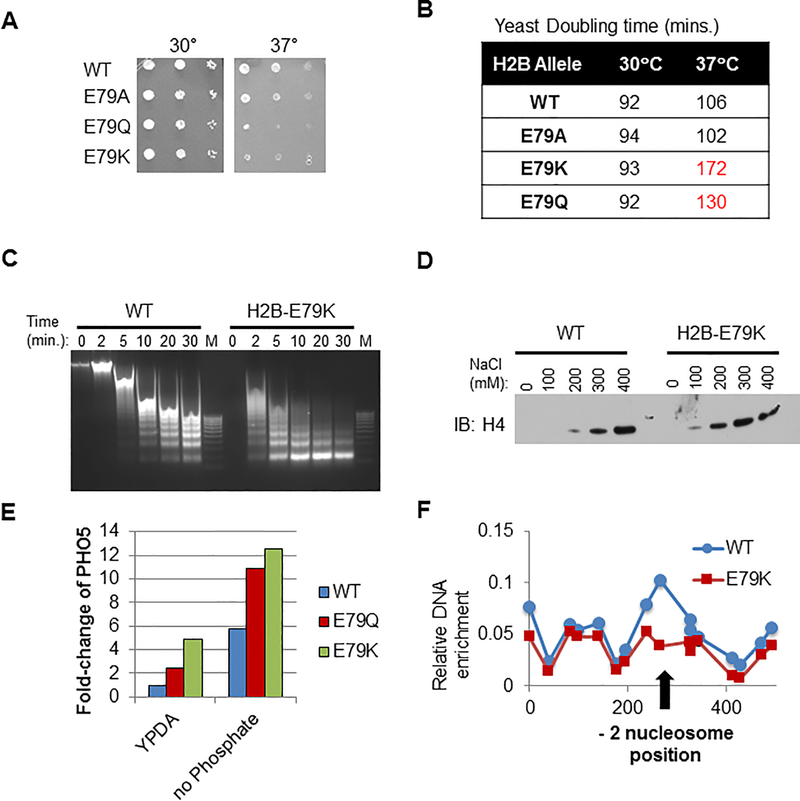
To begin to characterize the biological effects of the mutant histones we expressed them in S. cerevisiae, which have only one gene for each of the core histones. In yeast, one of the two endogenous wild-type H2B alleles was replaced with mutant H2B. Conversion to H2B-E79K (equivalent to human E76K, (21)) or H2B-E79Q caused temperature sensitivity while conversion to alanine did not have an effect (Figures 4A and B), suggesting that E76K or E76Q changes protein folding/octamer stability at higher temperatures. Chromatin from yeast heterozygous for E79K had a global change in accessibility as exhibited by increased sensitivity to MNase (Figure 4C). In addition, H4 was more readily extracted from chromatin by salt from yeast expressing H2B-E79K (Figure 4D), indicating that the nucleosomes in yeast with H2B-E79K have decreased stability.
Figure 4. Expression of mutant H2B in yeast destabilizes nucleosomes, deregulates gene expression and reduces nucleosome occupancy at the PHO5 promoter.
WT or E79K H2B (analogous to human H2B-E76K) was expressed in S. Cerevisiae. (A) Yeast cells expressing H2B-E79K are temperature sensitive. Limiting dilutions of yeast expressing WT, E79A, E79Q or E79K were plated and incubated at 30°C or 37°C. Cell growth was evaluated after 1 day. (B) Yeast doubling time is significantly increased in cells expressing E79K-H2B at 37°C. (C) Time course of MNase sensitivity from spheroplasted yeast grown in rich media. M, marker. (D) Chromatin pellets were extracted with increasing concentrations of salt as indicated. Immuno-blotting of the soluble fraction was performed with antibody to H4. (E) Cells expressing WT, E79Q or E79K H2B were maintained in either rich media (YPDA) or phosphate-free media and expression of the phosphate-inducible PHO5 gene was measured by RT-PCR. (F) Nucleosome scanning assay of the PHO5 promoter from cells expressing either WT or E79K grown in rich media. Chromatin was digested with MNase, mononucleosomal DNA was purified and MNase protection was determined by qPCR. H2B occupancy at −2 nucleosome position of PHO5 is reduced in E79K cells as indicated by the arrow.
To test if nucleosome instability may disrupt nucleosome-mediated gene repression, we examined the PHO5 gene that is inactive when yeast are grown in complete media and induced when grown in phosphate-free media. Compared to wild-type yeast, cells expressing the E79K allele displayed a 4-fold increase in PHO5 expression under non-permissive conditions and yeast with the E79Q allele demonstrated a two-fold increase (Figure 4E). The expression of PHO5 in yeast harboring E79K in nutrient-replete media was comparable to the level of expression of this gene in wild-type yeast in phosphate-free media and when E79K or E79Q yeast was grown without phosphate there was a further increase in gene expression (Figure 4E). Using a nucleosome scanning assay, we mapped the presence of a well-positioned nucleosome present at the “−2” position relative to the transcription start site (TSS), which occludes critical transcription factor binding sites (22). The qPCR readout of this assay showed the presence of positioned nucleosomes at the −1, −2 and −3 position in yeast with wild-type H2B but the loss of the −2 nucleosome in yeast expressing H2B E79K (Figure 4F). Thus, nucleosome instability and loss of nucleosome-mediated gene repression may cause aberrant gene expression and growth phenotype of cells harboring mutant H2B.
Expression of H2B-E76K in mammary epithelial cells alters cellular growth and gene expression
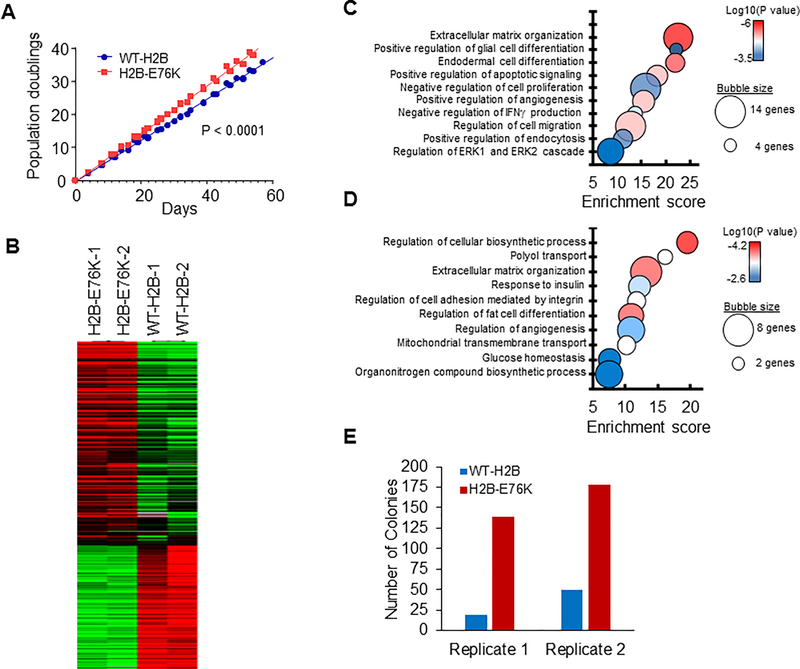
Since H2B-E76K was observed in a number of cancers we sought to characterize its ability to confer oncogenic properties to a human cell line model. To measure the effects of H2B mutation in the absence of other oncogenic mutations, wild-type H2B (WT-H2B) or H2B-E76K was expressed as either a FLAG-epitope tagged version or as an inducible GFP fusion in the immortalized but non-transformed mammary epithelial cell line MCF10A. Exogenously expressed H2B accounted for 5–10% of total H2B and was incorporated into chromatin as evidenced by co-precipitation with H4, presence in salt extracted chromatin, or GFP signal in the chromosomes of dividing cells (Supplemental Figure 4A–D and data not shown). To determine how H2B mutation might contribute to malignancy, we measured the growth of MCF10A cells stably transduced with WT-H2B or H2B-E76K. MCF10A cells were grown continuously and cumulative cell counts determined. After several weeks, cells expressing the mutant H2B displayed significant increased growth (Figure 5A).
Figure 5. Expression of H2B-E76K alters growth properties and gene expression in human mammary epithelial cells.
(A) Proliferation of MCF10A transduced with lentivirus expressing either WT or H2B-E76K was measured every 3 to 4 days. The results from three different infections are shown. Linear regression analysis was used to calculate statistical significance. (B) Heat maps depicting differential gene expression between MCF10A cells expressing either WT or H2B-E76K. (C and D) Bubble plots of gene ontology analysis identify biological processes significantly different in cells expressing H2B-E76K compared to cells expressing WT H2B. Bubble area is relative to the number of genes identified in each classification. Ontology of genes with increased (C) or decreased (D) expression in cells expressing H2B-E76K. (E) MCF10A cells expressing WT H2B or H2B-E76K and PIK3CA(H1047R) were grown in soft agar for 14 days and colonies counted.
In order to characterize the pathways that may be altered by mutant histone, we performed expression profiling of MCF10A cells expressing either WT-H2B or H2B-E76K by RNA-Seq. Relative to MCF10A cells expressing WT-H2B, expression of 247 genes was increased while expression of 149 genes was decreased in cells expressing H2B-E76K. (Figure 5B). Gene ontology analysis (Enrichr (23), Revigo (24)) revealed that genes upregulated by H2B-E76K tended to regulate differentiation, apoptosis, proliferation, migration and cellular signaling (Figure 5C). Genes decreased by expression of H2B-E76K were primarily involved in regulation of cellular biosynthetic processes, response to growth factors and adhesion, mitochondrial membrane transport and glucose homeostasis (Figure 5D).
To determine the relevance of the H2B mutation to cancer, we tested its ability to cooperate with mutant PIK3CA which tends to co-occur with H2B E76K in breast cancer (Figure 2C). Expression of oncogenic PI3KCA-H1047R along with mutant but not WT H2B led to increased colony formation in soft agar (Figure 5E and Supplemental Figure 4E). Taken together these results demonstrate that even a small fraction (<10%) of mutant histone causes altered cellular phenotypes that may collaborate with other oncogenes during tumorigenesis.
Expression of H2B-E76K relaxes chromatin structure and alters H2A/H2B dimer dynamics
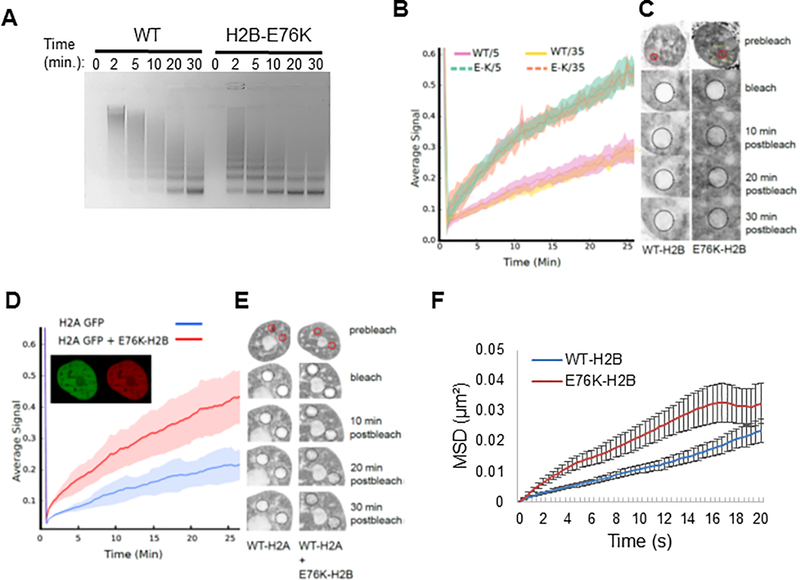
We next tested the effect of H2B-E76K on chromatin structure. Mass spectrometry analysis demonstrated that expression of mutant histone did not alter global histone methylation patterns, suggesting that the effect of H2B-E76K on gene expression was not due to altered histone post-translation modification states (Supplemental Figure 5). However, expression of H2B-E76K in MCF10A cells significantly increased sensitivity of genomic DNA to MNase, indicative of relaxed chromatin structure (Figure 6A). Fluorescence activated photobleaching (FRAP) experiments in cells expressing GFP-tagged WT or H2B-E76K showed that mutant H2B was incorporated into chromatin but was highly mobile, as the recovery time after photobleaching for H2B-E76K was considerably faster than that of WT-H2B (Figure 6B and C). In addition, when GFP-H2A was expressed in cells containing WT or H2B-E76K, FRAP was significantly faster in cells expressing H2B-E76K than WT-H2B, suggesting that E76K promotes increased mobility of the H2A/H2B dimer (Figure 6D and E). By contrast, the expression of H2B-E76K had no effect on the mobility of GFP-tagged histone H1 (Supplemental Figure 6A and B). To further test the effect H2B-E76K had on nuclear structure, we performed nuclear particle tracking experiments. In MCF10A cells expressing E76K, nuclear nanoparticle travel was significantly increased compared to WT cells indicating either an increase in active forces on the particles, or a decrease in resistance to particle motion in the mutant nucleus (Figure 6F and Supplemental Figure 6C).
Figure 6. H2B-E76K fundamentally alters chromatin structure and dynamics.
(A) Micrococcal nuclease (MNase) assay with nuclei of MCF10A cells stably expressing either WT or H2B-E76K demonstrate that E76K expression significantly increases sensitivity to MNase. Digest efficiency was visualized by agarose gel. (B) Fluorescence recovery after photobleaching (FRAP) analysis demonstrates that H2B-E76K has significantly faster chromatin dynamics than WT. FRAP analysis was carried out after induction of either WT or E76K mutant H2B-GFP fusions for 5 or 35 days in MCF10A cells. Dashed lines represent cells expressing H2B-E76K, solid lines represent data from cells expressing inducible GFP-tagged WT H2B (n = 10 cells). (C) Representative FRAP assay pre-bleach, bleach and post-bleach images of nuclei expressing GFP-tagged WT or H2B-E76K indicate faster fluorescent recovery in cells expressing E76K. (D) FRAP analysis of histone H2A-GFP dynamics in MCF10A cells expressing only H2A-GFP (n = 20 cells) or both H2A-GFP and a mutant H2B E76K mCherry fusion (n = 25 cells) with standard deviation envelopes. Inset demonstrates co-expression of H2A GFP and E76K mCherry. (E) Representative pre-bleach, bleach and post-bleach images of H2A GFP in WT MCF10A cells or in cells co-expressing mCherry tagged H2B-E76K. (F) 100nm particles injected into the nucleus had significantly increased mean square displacement (MSD) over time in MCF10A cells stably expressing exogenous H2B-E76K (red) compared to cells expressing WT H2B (blue). N=68 (WT) and N=67 (E76K) cells were analyzed. Error bars represent SEM.
The H2B-E76K mutant fundamentally alters chromatin accessibility
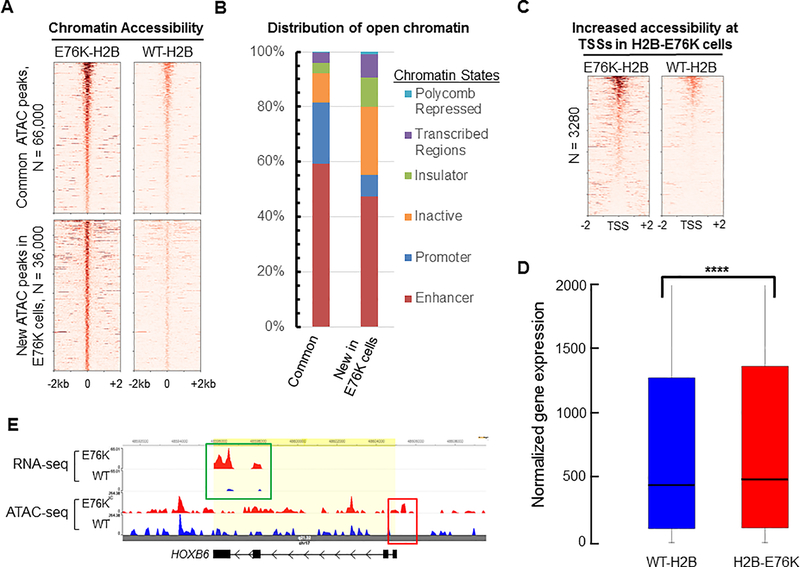
We performed ATAC-Seq on MCF10A cells that had expressed WT or mutant H2B to directly measure the extent to which mutant H2B altered chromatin accessibility in MCF10A cells. ATAC-Seq reads were categorized into common peaks present in both H2B-E76K and WT-H2B expressing cells or new peaks that displayed significantly increased chromatin accessibility in H2B-E76K compared to WT-H2B expressing cells (Figure 7A). In addition, a small subset of peaks were observed to have increased signal in cells expressing WT-H2B compared to H2B-E76K (Supplemental Figure 7A). In new open chromatin regions of H2B-E76K cells, the frequency of GC-rich (>60% GC content) peaks was decreased and the frequency of AT-rich (> 60% AT content) peaks increased compared to the frequency of GC- or AT-rich peaks observed in the common peaks shared by WT-H2B and H2B-E76K expressing cells, suggesting nucleosome formation is preferentially decreased by H2B-E76K in AT-rich genomic regions (Supplemental Figure 7B). When the distribution of ATAC peaks in various chromatin states was analyzed (25), open chromatin regions in common to WT-H2B and H2B-E76K expressing MCF10A cells were predominantly found in promoter and enhancer regions while more than 20% of new peaks caused by expression of mutant histone appeared in normally transcriptional-inactive chromatin regions such as heterochromatin (Figure 7B). In addition, for those ATAC-Seq peaks in common between WT and E76K cells, the peaks in E76K cells tended to have an average increased width of about 50bp compared to the same peak in WT cells (Supplemental Figure 7C). Expression of mutant H2B caused significantly increased chromatin accessibility in the promoter regions of more than 3200 genes (Figure 7C), and these genes also displayed increased gene expression while genes without an increased ATAC-Seq signal around the TSS had no significant difference in expression (Figure 7D). For example, expression of H2B-E76K promoted both a significant increase in chromatin accessibility at the normally repressed promoter region of the HOXB6 gene in MCF10A cells and a corresponding significant increase in gene expression (Figure 7E). Thus, histone mutations that affect nucleosome stability may significantly alter chromatin structure and cause aberrant gene expression.
Figure 7. Expression of H2B E76K alters chromatin accessibility in MCF10A cells.
(A) Heat maps of ATAC-Seq peaks from MCF10A cells expressing either WT H2B or H2B-E76K found within a ± 2kb window, ordered by highest ATAC-Seq signal to lowest. Peaks found in both cell lines are listed as common peaks. Peaks significantly enriched in cells expressing H2B-E76K are denoted as new peaks in E76K. (B) The distribution of ATAC-Seq peaks among six broad classes of chromatin states was performed using the encode HMEC ChromHMM data set. (C) Heat maps of ATAC-Seq reads within +/− 2 kb of transcription start sites (TSS) for genes with significantly increased chromatin accessibility at promoter regions of MCF10A cells expressing H2B-E76K compared to cells expressing WT-H2B. (D) Box and whiskers plot for gene expression of the 3280 genes that have increased ATAC-Seq signal at the TSS in cells expressing H2B-E76K compared to expression of those genes in cells expressing WT-H2B. *** indicates that P < 0.001. (E) Genome browser tracks of a representative gene (HOXB6) with increased chromatin accessibility at the TSS in cells expressing H2B-E76K (red box) that corresponds with increased expression (green box).
Discussion
Whole genome sequencing of human patient samples has revealed that dysfunction of almost every mechanism governing chromatin structure and regulation of gene expression occurs in cancer (1). Mutations found in tumors include those that affect the function of transcription factors, chromatin modifying enzymes, chromatin remodeling factors and mutations that disrupt the covalent modification of histone “tail” residues. Now, by examination of the cBioPortal patient genomics database we have identified a new class of mutations in the core histones (H2A, H2B, H3 and H4) that frequently occur in cancer and may function to destabilize the histone octamer and nucleosome. While some amino acids changes in histones we identified in our survey of cancer genomes can be found as polymorphisms in the general population, for most sites the frequency of specific amino acid changes was 10–100x more common in the cancer cases that in normal populations and many of the recurrent histone amino acid changes we identified were not observed in databases of >200,000 genomes. This plus the fact that histone mutations were found to have a variant allele frequency of 20–30% suggested that these variations are acquired somatic mutations that may contribute to carcinogenesis and not passenger polymorphisms. Such oncohistone mutations may by themselves or in combination with other oncogenic drivers change the dynamics of chromatin access that alters gene expression to a state that favors tumorigenesis. Thus, our data suggests that histones are directly affected in malignancy, and chromatin may assume a global pathogenic state.
Support for histone alterations as cancer drivers is growing and the prevalence of histone mutations in cancer was recently noted by others (17). Mutations have also been reported in the H1 “linker” histone, which couples nucleosomes to facilitate chromatin compaction (26). H1 has diverse roles in gene expression, and loss of H1 has pronounced effects on heterochromatin structure that alters gene expression, sensitivity to DNA damage and embryonic development (27). Downregulation of Histone H1.0 was proposed to alter gene expression in glioblastoma and cause more heterogeneity of gene expression in cell populations that could contribute to oncogenesis (28). In addition, aberrant histone gene expression has been reported to have an oncogenic role in cancer (29,30).
Mutations in the core histones were previously reported among a compilation of other cancer mutations in tumors but not in the systematic manner presented here using such a large set of patient data (17,19,20,30–32). Histone H2A and H2B were reported to be mutated in carcinosarcoma of the ovary and predicted to affect histone structure (32). Expression of these mutant histones in cells yielded biological effects but no molecular correlates in chromatin configuration or gene expression were assayed. Notably these mutations were not among the 18 recurrent mutations we found to be significantly elevated in this survey of cancer genomes.
Our analysis and those of Nacev et al. reveal that the most common histone mutations in human tumor samples tend to occur in the globular, histone fold domains of the core histone proteins rather than in N-terminal tail regions (17). Furthermore, among the most frequent mutations in the four core histones, two specific structural regions of the nucleosome were significantly impacted. Missense mutations altering H2B-E76, H4-R92 or H4-D68 affect a region of the nucleosome important for H2B/H4 interaction while missense mutations affecting H3-E105 and H4-G42 affect a region important for H3/H4 interaction (Supplemental Figure 3A and B). Thus, we propose that core histone mutations that disrupt nucleosome integrity may represent a new class of driver mutations in cancer. To test this concept, we evaluated the H2B-E76K mutation both in vitro and in cell models. This mutation was the most common cancer-associated missense mutation in core histones and often coincides with gain of function mutations of oncogenes including RAS and PIK3CA (Figure 2C and (31)). The subclonal nature of the H2B E76K and other histone mutations suggest that the mutations may not be initial drivers of the cancer phenotype but instead contribute to cancer progression. In accordance with this idea, expression of mutant H2B in MCF10A cells caused a modest increase in cell growth and a shift of gene expression. However, when co-expressed with well characterized driver mutations of PiK3CA, enhanced colony formation was noted, consistent with the idea that histone mutations may contribute to the progression of cancer.
Our results and those of others who crystalized the histone octamer in the presence of the H2B-E76K mutation indicate that H2B-E76K inhibits histone octamer formation by destabilizing the interaction of H2B with H4 (19). Furthermore, our results demonstrate that expression of even a limited amount of mutant histone led to widespread change in chromatin accessibility, nucleosome function, gene expression and cell proliferation. However, the impact of histone core mutations such as H2B-E76K may be more far reaching, for instance future studies must now include determining the impact of these mutations on histone deposition and chromatin structure during DNA replication, DNA damage response and DNA repair. Importantly, our results indicate that H2B-E76K can cooperate with oncogenes such as PIK3CA that tend to co-occur in cancer to alter cellular growth, suggesting that loss of nucleosome integrity synergizes with activated oncogenic signaling to propel the cancer program. What chromatin structures and configurations and specific genes and pathways are affected by such collaboration in the pathogenesis or progression of cancer remains to be uncovered.
MATERIALS AND METHODS
Cells lines and reagents
MCF10A cells were purchased from ATCC and maintained in MEBM (Lonza) supplemented with bovine pituitary extract, hydrocortisone, hEGF, insulin and 100ng/ml cholera toxin. The HIST1H2BD open reading frame was cloned into pCDH-EF1-MCS- Puro (SBI, Palo Alto, CA) and the E76K mutant made using a QuikChange Site directed mutagenesis kit (Agilent, Santa Clara, CA). Lentivirus was packaged in 293T cells and supernatant used to generate MCF10A cells expressing either WT-H2B or H2B-E76K. In addition, WT-H2B or H2B-E76K were cloned into a custom pLenti cumate switch vector to create cumate-inducible GFP-tagged constructs. To induce expression, cells were grown in media containing 1X cumate (SBI). For generating dual expression H2A-GFP and E76K H2B mCherry cell lines, we custom generated TRE3G-H2A-mGFP-rTTA3-T2A-Hygromycin and TRE3G-H2B-E76K-mCherry-rTTA3-T2A-Blasticidin vectors by modifying the commercially available cumate switch vector. To induce dual expression, the cells were induced with Doxycycline. Western blotting was performed to evaluate protein expression using antibodies to H2B (Cat# 12364, Cell signaling technologies), H4 (Cat# D2X4V, Cell signaling technologies), FLAG (Cat# F3165, Sigma), alpha-tubulin (cat#7291, Abcam). All cell lines generated were screened for mycoplasma monthly and authenticated to match MCF10A cells once a year by short tandem repeat analysis (Genetica DNA laboratories, Burlington, NC). Cells were used in experiments within 10–20 passages after thawing.
Structural modeling and molecular dynamics simulation
The atomistic structure of the simulated system was taken from the crystal structure of a nucleosome complex (Protein Data Bank [PDB] entry: 1AOI), and only the chains B and D, corresponding to the H4 and H2B units were used in the following MD simulations. The E76K mutation was generated using a method implemented in VMD (33). The missing H-atoms were added using H++ package. The H4 and H2B chains were first solvated in a water box filled with TIP3P water molecules. Periodic boundary conditions were applied in the MD simulations, and the dimensions of the water box were set at 110, 98, and 78 Å in x, y, and z directions, separately. In addition, Na+ and Cl− ions were added to neutralize the system and make the concentration of Na+ be 0.15 M. The force field parameters for the protein were obtained from the ff14 SB force field (34). Molecular dynamics simulations were carried out using NAMD (35). A 10 ns molecular dynamics simulation at 310 K with an NPT ensemble was performed to equilibrate the system. In the production period, the system was simulated for 20 ns using the NPT ensemble. Pressure was maintained at 1 atm using the Langevin piston method with a piston period of 100 fs, a damping time constant of 50 fs, and a piston temperature of 310 K (36). No atomic coordinates were restrained during the production period. Full electrostatics was employed using the particle-mesh Ewald method with a 1 Å grid width (37). Nonbonded interactions were calculated using a group-based cutoff with a switching function and updated every 10 time steps. Covalent bonds involving hydrogen were held rigid using the SHAKE algorithm, allowing a 2 fs time step (38).
Fluorescence recovery after photobleaching (FRAP) assay
Five single imaging scans were acquired followed by an iterative bleaching (5 pulses of 200ms each) using a spot of 1 μm in diameter without scanning. Z stack images were then collected at 20-second intervals. For imaging, the laser power was attenuated to 0.6% of the bleach intensity (25mW 488 nM laser excitation). FRAP recovery curves were generated from background-subtracted images. Spontaneous photobleaching was corrected by tracking changes in background fluorescence at every time point. The level of autofluorescence in cells not expressing the fusion proteins was 1% or less than in expressing cells. All quantitative values represent averages from at least 10 cells from 3 independent experiments.
Micrococcal nuclease digestion assay
Micrococcal nuclease (MNase) was purchased from Worthington. To assay for MNase accessibility, 2×105 cells in one well of a 6 well plate were washed with PBS and incubated in 1 ml of NP40 buffer (10mM TrisCl pH 7.4, 10mM NaCl, 3mM MgCl, 0.5% NP-40, 0.15 mM spermine, 0.5 mM spermidine) on ice for 5 minutes. Cells were washed with MNase buffer (10mM TrisCl pH 7.4, 15mM NaCl, 60mM KCl, 0.15 mM spermine, 0.5 mM spermidine, 1mM CaCl2) twice and 0.75ml of MNase buffer containing 0.015U MNase was added for the indicated times. Reactions were stopped by adding equal volume 10% SDS, 0.5M EDTA. Samples were harvested by scraping cells and transferring to a 15 ml tube. Cells were incubated with Proteinase K at 65°C overnight. Next, NaCl was added and a phenol chloroform extraction was performed. The nucleic acid fraction was incubated with RNase A at 37°C for 1 hour and DNA isolated by chloroform extraction and ethanol precipitation. The final DNA product was resuspended in 0.1X TE buffer and MNase sensitivity measured by resolving 100ng of each sample on a 2.5% agarose gel in TAE buffer.
Recombinant nucleosome assembly and stability assay
Recombinant human core histones (H2A, H2B, H3, and H4) were prepared and mixed at a 1.2:1.2:1:1 ratio in denaturing buffer and subsequently dialyzed into high salt native buffer to promote protein refolding and histone octamer formation (39,40). Histone octamers were resolved from (H3/H4)2 tetramers, H2A/H2B dimers and free histones by gel filtration chromatography (Superdex 200 10/30GL, GE Healthcare), and selected fractions visualized by SDS-PAGE. Equimolar amounts of 601 DNA sequence (147bp) and octamers or in the case of E76K of tetramers and dimers (1:2 molar ratio) were mixed in high salt buffer, then dialyzed against a salt gradient to reconstitute nucleosomes (41). Nucleosomes were than resolved by Native PAGE. MNase susceptibility assays were performed on nucleosomes made with WT, E76Q and E76K H2B mutants by mixing each nucleosome (0.4 pmol) with 9.6U of MNase (Worthington) and incubating for indicated amount of time. Reactions were stopped with a 20mM EGTA + 20mM EDTA solution and resolved by native PAGE. Relative amounts of nucleosomes at each time point were measured by densitometry and plotted.
Nuclear nanoparticle tracking
MCF10A nuclei were microinjected with polyethylene glycol (PEG) modified 100 nm red fluorescent Carboxylate-Modified nanoparticles (Invitrogen). We followed established protocols to coat carboxylate-modified nanoparticles (Invitrogen) with amine-terminated PEG (42,43). 10 μl PEGylated particles (3.6×1010 particles/ml) were sonicated for 30mins before loaded to Femtotip II (0.5 μm, Eppendorf). The particles were then microinjected into the nuclei using an Eppendorf InjetMan microjection system. Cells were incubated at least 6 hrs. before imaging. Time lapse images of fluctuating particles embedded in the nucleus were acquired on a Nikon TE2000 inverted epifluorescent microscope with 60x/1.49 NA oil immersion objective (Nikon) and CCD camera (CoolSNAP, HQ; Photometrics). Cells were maintained at 37ºC in 5% CO2. Tracking of particles was achieved by using TrackMate plugin of ImageJ (44). The trajectories of the particles were then used to calculate time-averaged mean squared displacements using @msdanalyzer Matlab tool (45).
Next Generation Sequencing and Analysis
For gene expression analysis cells were collected at approximately 50–70% confluency and stored in RNAlater (Thermo Fisher Scientific, Waltham MA) until RNA could be prepared using the Aurum total RNA isolation kit (Bio-Rad, Hercules, CA). RNA sequencing libraries were prepared from 0.5 μg of total RNA and 1:100 ERCC spike in control using a KAPA RNA Seq kit with RiboErase (Roche, Pleasanton CA). Sequencing was performed on a Nextseq500 with a 2 × 150 bp read length kit to achieve approximately 30–40 million reads per sample (Illumina, San Diego, CA). Sequences were trimmed using trimmomatic with quality control performed before and after trimming using FastQC. Input sequences were aligned to the transcriptome using STAR_2.5.2a, and differential gene expression was analyzed using RSEM v1.2.31. Differentially expressed genes in each contrast were defined as abs(log2(FC)) >= 1.0 and FDR-corrected P-value <= 0.01. Gene ontology analysis of biological processes was performed using the Enrichr and Revigo web tools (24,46).
The Assay for transposase accessible chromatin using sequencing (ATAC-Seq) was performed as described (47). Libraries were size selected by SPRIselect and library QC was performed by measuring the ratio of GAPDH promoter to a genomic desert region by qPCR prior to sequencing using a NextSeq500 (2 × 150 bp) to achieve approximately 60 million reads per sample. Short reads were trimmed using trimmomatic (v 0.36), and QC on the original and trimmed reads was performed using FastQC (v 0.11.4) and MultiQC (v 1.1). The reads were aligned to the human genome (version GRCh38) using Bowtie2 (v. 2.3.3). ATAC peak calling and differential peak analysis were performed using the MACS2 (v. 2.0.10) `callpeak’ command. Insert size distribution with analyzed with the CollectInsertSizeMetrics tool (Picard v 2.9.2). Custom scripts were used to produce heatmaps and enrichment plots. The MAnorm script with custom modifications, was used to compare differences in ATAC seq peak size. The ChromHMM database was used to assess enrichment of open chromatin at broad classes of chromatin states (25). Sequencing data can be accessed through the sequence read archive (SRA), Bioproject ID: PRJNA544800.
Proliferation and cell growth assays
A cumulative cell count method was used to measure cell proliferation differences between cultures expressing either WT or mutant histone. Briefly, 1.5×105 cells were plated into a 6 well dish. After 3 days growth cells were harvested, counted and 1.5×105 cells replated. Experiments were done in triplicate and population doubling was calculated using the formula n = 3.32*(log CC - log l) + X, where n = the final population doubling number at end of a given subculture, CC = the cell count at that point, l = the number of cells used as inoculum to begin that subculture, and X = the doubling level of the inoculum used to initiate the subculture being quantitated (48). A linear regression analysis with GraphPad Prism 7.03 was used to measure the significance of proliferation differences. Colony formation assay was performed using a soft agar assay. Briefly, 10,000 cells were embedded in growth medium containing 0.3% Agar into each well of a 6 well dish. After 3 weeks, colonies were fixed and stained with crystal violet.
Measurement of growth and nucleosome occupancy in yeast
The yeast H2B gene was PCR amplified from BY4741 and cloned into pRS316. Mutations were introduced using the Quikchange mutagenesis kit (Stratagene). The WT or mutant H2B was incorporated into BY4741 by integrative transformation of the SacII XhoI digested plasmid. For rich media growth, cells were grown in YPD. For phosphate starvation growth, the cells were washed with PBS and transferred to complete synthetic dropout media without potassium phosphate for three hours. A nucleosome scanning assay was used to examine the nucleosome architecture of the yeast PHO5 promoter (22). Briefly, yeast grown in rich media were converted to spheroplasts, chromatin was digested with MNase, mononucleosomal DNA was purified, and MNase protection of the PHO5 promoter was determined by quantitative PCR as previously described (22).
Supplementary Material
Significance Statement.
Mutations in the core histones frequently occur in cancer and represent a new mechanism of epigenetic dysfunction that destabilizes the nucleosome, deregulates chromatin accessibility and alters gene expression to drive cellular transformation.
Acknowledgments
Supported by Physical Science of Oncology grant U54CA143869 (JDL), R01GM115945 (AJR), American Cancer Society 130230-RSG-16-248-01-DMC (AJR), R01EB014869 (TPL), American Cancer Society Postdoctoral Fellowship PF-17-010-01-CDD (BN). Harper Dissertation Prize and Dean’s International Student Fellowship from the University of Chicago (ATG).
Footnotes
Disclosures: The authors have no conflicts of interest to declare
References
- 1.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014;505(7484):495–501 doi 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kornberg RD, Thomas JO. Chromatin structure; oligomers of the histones. Science 1974;184(4139):865–8. [DOI] [PubMed] [Google Scholar]
- 3.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science 1974;184(4139):868–71. [DOI] [PubMed] [Google Scholar]
- 4.Lorch Y, LaPointe JW, Kornberg RD. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell 1987;49(2):203–10. [DOI] [PubMed] [Google Scholar]
- 5.Struhl K, Segal E. Determinants of nucleosome positioning. Nature structural & molecular biology 2013;20(3):267–73 doi 10.1038/nsmb.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh R, Bassett E, Chakravarti A, Parthun MR. Replication-dependent histone isoforms: a new source of complexity in chromatin structure and function. Nucleic acids research 2018;46(17):8665–78 doi 10.1093/nar/gky768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Talbert PB, Henikoff S. Histone variants on the move: substrates for chromatin dynamics. Nature reviews Molecular cell biology 2017;18(2):115–26 doi 10.1038/nrm.2016.148. [DOI] [PubMed] [Google Scholar]
- 8.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature 2011;469(7330):343–9 doi 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khuong-Quang DA, Buczkowicz P, Rakopoulos P, Liu XY, Fontebasso AM, Bouffet E, et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta neuropathologica 2012;124(3):439–47 doi 10.1007/s00401-012-0998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Justin N, Zhang Y, Tarricone C, Martin SR, Chen S, Underwood E, et al. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nature communications 2016;7:11316 doi 10.1038/ncomms11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 2013;340(6134):857–61 doi 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu C, Jain SU, Hoelper D, Bechet D, Molden RC, Ran L, et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 2016;352(6287):844–9 doi 10.1126/science.aac7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papillon-Cavanagh S, Lu C, Gayden T, Mikael LG, Bechet D, Karamboulas C, et al. Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nature genetics 2017;49(2):180–5 doi 10.1038/ng.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Behjati S, Tarpey PS, Presneau N, Scheipl S, Pillay N, Van Loo P, et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nature genetics 2013;45(12):1479–82 doi 10.1038/ng.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012;482(7384):226–31 doi 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 16.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery 2012;2(5):401–4 doi 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nacev BA, Feng L, Bagert JD, Lemiesz AE, Gao J, Soshnev AA, et al. The expanding landscape of ‘oncohistone’ mutations in human cancers. Nature 2019;567(7749):473–8 doi 10.1038/s41586-019-1038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitts A, Phan L, Ward M, Holmes J. The Database of Short Genetic Variation (dbSNP). The NCBI Handbook; [Internet], 2nd ed2014. [Google Scholar]
- 19.Arimura Y, Ikura M, Fujita R, Noda M, Kobayashi W, Horikoshi N, et al. Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome. Nucleic acids research 2018;46(19):10007–18 doi 10.1093/nar/gky661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martins MD, Castilho RM. Histones: Controlling Tumor Signaling Circuitry. Journal of carcinogenesis & mutagenesis 2013;1(Suppl 5):1–12 doi 10.4172/2157-2518.S5-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wallis JW, Hereford L, Grunstein M. Histone H2B genes of yeast encode two different proteins. Cell 1980;22(3):799–805. [DOI] [PubMed] [Google Scholar]
- 22.Small EC, Xi L, Wang JP, Widom J, Licht JD. Single-cell nucleosome mapping reveals the molecular basis of gene expression heterogeneity. Proceedings of the National Academy of Sciences of the United States of America 2014;111(24):E2462–71 doi 10.1073/pnas.1400517111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic acids research 2016;44(W1):W90–7 doi 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PloS one 2011;6(7):e21800 doi 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 2011;473(7345):43–9 doi 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pruss D, Bartholomew B, Persinger J, Hayes J, Arents G, Moudrianakis EN, et al. An asymmetric model for the nucleosome: a binding site for linker histones inside the DNA gyres. Science 1996;274(5287):614–7. [DOI] [PubMed] [Google Scholar]
- 27.Hergeth SP, Schneider R. The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO reports 2015;16(11):1439–53 doi 10.15252/embr.201540749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torres CM, Biran A, Burney MJ, Patel H, Henser-Brownhill T, Cohen AS, et al. The linker histone H1.0 generates epigenetic and functional intratumor heterogeneity. Science 2016;353(6307) doi 10.1126/science.aaf1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monteiro FL, Vitorino R, Wang J, Cardoso H, Laranjeira H, Simoes J, et al. The histone H2A isoform Hist2h2ac is a novel regulator of proliferation and epithelial-mesenchymal transition in mammary epithelial and in breast cancer cells. Cancer letters 2017;396:42–52 doi 10.1016/j.canlet.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 30.Bhattacharya S, Reddy D, Jani V, Gadewal N, Shah S, Reddy R, et al. Histone isoform H2A1H promotes attainment of distinct physiological states by altering chromatin dynamics. Epigenetics & chromatin 2017;10(1):48 doi 10.1186/s13072-017-0155-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011;333(6046):1157–60 doi 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao S, Bellone S, Lopez S, Thakral D, Schwab C, English DP, et al. Mutational landscape of uterine and ovarian carcinosarcomas implicates histone genes in epithelial-mesenchymal transition. Proceedings of the National Academy of Sciences of the United States of America 2016;113(43):12238–43 doi 10.1073/pnas.1614120113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. Journal of molecular graphics 1996;14(1):33–8, 27–8. [DOI] [PubMed] [Google Scholar]
- 34.Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. Journal of chemical theory and computation 2015;11(8):3696–713 doi 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang W, Phillips JC, Huang L, Fajer M, Meng Y, Gumbart JC, et al. Generalized Scalable Multiple Copy Algorithms for Molecular Dynamics Simulations in NAMD. Computer physics communications 2014;185(3):908–16 doi 10.1016/j.cpc.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feller SE, Zhang YH, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: The langevin piston method. J Chem Phys 1995;103(11). [Google Scholar]
- 37.Darden T, York DM, Pedersen L. Particle mesh Ewald: An N log(N) method for Ewald sums in large systems. J Chem Phys 1993;98:10089. [Google Scholar]
- 38.Andersen HC. Rattle: A “velocity” version of the shake algorithm for molecular dynamics calculations. J Comput Phys 1983;52(1):24–34. [Google Scholar]
- 39.Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods in enzymology 2004;375:23–44. [DOI] [PubMed] [Google Scholar]
- 40.Grzybowski AT, Chen Z, Ruthenburg AJ. Calibrating ChIP-Seq with Nucleosomal Internal Standards to Measure Histone Modification Density Genome Wide. Molecular cell 2015;58(5):886–99 doi 10.1016/j.molcel.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. Journal of molecular biology 1998;276(1):19–42 doi 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 42.Valentine MT, Perlman ZE, Gardel ML, Shin JH, Matsudaira P, Mitchison TJ, et al. Colloid surface chemistry critically affects multiple particle tracking measurements of biomaterials. Biophysical journal 2004;86(6):4004–14 doi 10.1529/biophysj.103.037812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kole TP, Tseng Y, Wirtz D. Intracellular microrheology as a tool for the measurement of the local mechanical properties of live cells. Methods in cell biology 2004;78:45–64. [DOI] [PubMed] [Google Scholar]
- 44.Tinevez JY, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, et al. TrackMate: An open and extensible platform for single-particle tracking. Methods 2017;115:80–90 doi 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 45.Tarantino N, Tinevez JY, Crowell EF, Boisson B, Henriques R, Mhlanga M, et al. TNF and IL-1 exhibit distinct ubiquitin requirements for inducing NEMO-IKK supramolecular structures. The Journal of cell biology 2014;204(2):231–45 doi 10.1083/jcb.201307172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC bioinformatics 2013;14:128 doi 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods 2013;10(12):1213–8 doi 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dell’Orco RT, Mertens JG, Kruse PF, Jr. Doubling potential, calendar time, and senescence of human diploid cells in culture. Experimental cell research 1973;77(1):356–60. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.