The reverse genetics method of influenza virus generation has enabled us to generate recombinant viruses bearing modified viral proteins. Recombinant influenza viruses expressing foreign genes have become useful tools in basic research, and such viruses can be utilized as efficient virus vectors or multivalent vaccines. However, the insertion of a foreign gene into the influenza virus genome often impairs virus replication, and the inserted genes are unstable. Elucidation of the mechanisms of foreign gene stabilization will help us to establish useful recombinant influenza viruses.
KEYWORDS: foreign gene insertion, genetic stability, influenza, recombinant virus, virus polymerase
ABSTRACT
We previously attempted to establish a reporter influenza virus by inserting the gene for the Venus fluorescent protein into the NS segment of influenza A/Puerto Rico/8/34 (PR8, H1N1) virus to yield WT-Venus-PR8. Although the inserted Venus gene was deleted during serial passages of WT-Venus-PR8, we discovered that the PB2-E712D mutation stabilizes the Venus gene. Here, we explored the mechanisms by which Venus gene deletion occurs and how the polymerase mutation stabilizes the Venus gene. Deep sequencing analysis revealed that PB2-E712D does not cause an appreciable change in the mutation rate, suggesting that the stability of the Venus gene is not affected by polymerase fidelity. We found by using quantitative real-time PCR that WT-Venus-PR8 induces high-level interferon beta (IFN-β) expression. The induction of IFN-β expression seemed to result from the reduced transcription/replication efficiency of the modified NS segment in WT-Venus-PR8. In contrast, the transcription/replication efficiency of the modified NS segment was enhanced by the PB2-E712D mutation. Loss of the Venus gene in WT-Venus-PR8 appeared to be caused by internal deletions in the NS segment. Moreover, to further our understanding of the Venus stabilization mechanisms, we identified additional amino acid mutations in the virus polymerase complex that stabilize the Venus gene. We found that some of these amino acids are located near the template exit or the product exit of the viral polymerase, suggesting that these amino acids contribute to the stability of the Venus gene by affecting the binding affinity between the polymerase complex and the RNA template and product.
INTRODUCTION
Influenza A viruses are segmented negative-strand RNA viruses that belong to the family Orthomyxoviridae. The genome of influenza A virus consists of eight RNA segments encoding different viral proteins. With the development of reverse genetics for influenza viruses (1), it became possible to generate recombinant viruses bearing modified viral proteins. Recombinant viruses expressing foreign proteins such as reporter proteins are now useful tools in basic research. Moreover, influenza viruses stably expressing foreign genes may be used as virus vectors or multivalent vaccines. For example, an influenza virus expressing a modified hemagglutinin (HA) segment, in which polypeptides of the Bacillus anthracis protective antigen were inserted, expresses chimeric HA proteins stably and induces antibody responses against both the HA and B. anthracis antigens (2). In another study, insertion of the human interleukin-2 gene into the influenza NS segment enhanced the CD8+ immune response to viral antigens (3). However, insertion of foreign genes into virus genomes often impairs virus replication (4, 5), and inserted sequences are not stable during the replication cycle (6).
Previously, we attempted to establish a reporter influenza virus that would allow us to visualize virus-infected cells as a tool to understand influenza virus-induced pathology (7). The gene of the Venus fluorescent protein was inserted into the NS segment of influenza A/Puerto Rico/8/34 (PR8, H1N1) virus to yield WT-Venus-PR8. However, WT-Venus-PR8 was significantly attenuated, and the inserted Venus gene was deleted during serial virus passages. We found that an E-to-D mutation at position 712 of the polymerase subunit PB2 (PB2-E712D) stabilized the inserted Venus gene (7, 8). Furthermore, we also established H5N1 virus carrying the Venus gene, which was inserted into the NS segment from PR8 (Venus-H5N1) (7). Although, like WT-Venus-PR8, WT-Venus-H5N1 showed moderate virulence and low Venus expression, we acquired a variant that became more lethal to mice and stably expressed Venus after mouse adaptation. We found that a V-to-A mutation at position 25 of the polymerase subunit PB2 and a R-to-K mutation at position 443 of the polymerase subunit PA contributed to the stable maintenance of the Venus gene (9). These results indicate that the composition of the viral polymerase plays an important role in the stabilization of the inserted foreign gene. However, the mechanisms by which the Venus gene can be deleted and how polymerase mutations stabilize the Venus gene have remained unknown.
In this study, we explored the mechanisms of Venus gene stabilization by comparing events upon infection with WT-Venus-PR8 and Venus-PR8 possessing the PB2-E712D mutation (Venus-PR8-PB2-E712D). We examined polymerase fidelity and RNA and protein expression in infected cells, and we performed sequencing analysis coupled with coinfection experiments to determine how the Venus gene is deleted. Moreover, we identified additional mutations that contribute to the stabilization of the Venus gene to further our understanding of the stabilization mechanisms.
RESULTS
Loss of Venus expression in WT-Venus-PR8 restores replication efficiency.
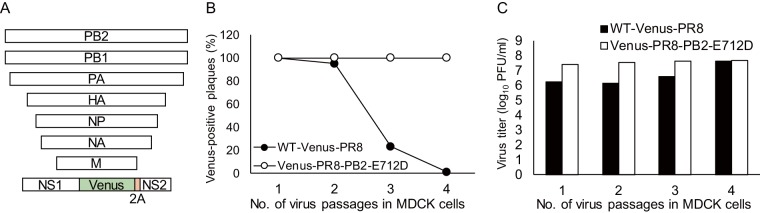
We prepared WT-Venus-PR8 and Venus-PR8-PB2-E712D by using reverse genetics as previously described (1). The gene of the Venus fluorescent protein was inserted into the NS segment as illustrated in Fig. 1A (7). First, we confirmed how quickly Venus expression was lost in WT-Venus-PR8 and the relationship between Venus deletion and virus titer. We passaged the viruses in MDCK cells at a multiplicity of infection (MOI) of 0.001 and measured the proportion of Venus-positive plaques (Fig. 1B). We confirmed that the expression of Venus was lost immediately in WT-Venus-PR8, whereas all plaques of Venus-PR8-PB2-E712D showed Venus expression after four passages. Although WT-Venus-PR8 showed a lower titer than Venus-PR8-PB2-E712D in MDCK cells, as described previously (8), the virus titer increased during virus passages as the proportion of Venus-positive plaques decreased (Fig. 1C). This result suggests that the loss of the Venus gene in the mutated WT-Venus-PR8 restored the replicative efficiency of the virus.
FIG 1.
(A) Schematic structure of the eight viral RNA segments contained in WT-Venus-PR8. 2A, protease 2A autoproteolytic site. (B) Each virus was passaged in MDCK cells. The proportion of Venus-expressing plaques in virus stocks from different passages was determined in MDCK cells by using fluorescence microscopy. (C) The virus stocks from different passages were titrated by use of plaque assays in MDCK cells.
The PB2-E712D mutation does not cause an appreciable change in polymerase fidelity.
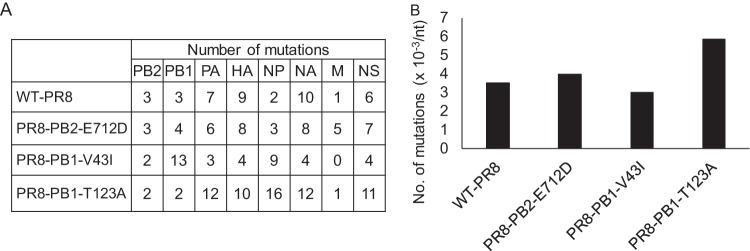
We hypothesized that the PB2-E712D mutation increases viral polymerase fidelity in order to retain the inserted Venus gene during passages. To test this hypothesis, we generated WT-PR8, as well as PR8-PB2-E712D, which possesses aspartic acid at position 712 of PB2 and therefore differs from WT-PR8 by only this amino acid, by reverse genetics and compared their mutation rates. Here, we used viruses that did not contain the Venus gene to make it easier to measure the mutation rates. PR8-PB1-V43I, which has been reported to be a high-fidelity mutant virus (10, 11), and PR8-PB1-T123A, which has been reported to be a low-fidelity mutant virus (12), were also generated by reverse genetics and used as controls. To estimate the mutation rates, we passaged these viruses in MDCK cells at an MOI of 0.001 and performed deep sequencing of the entire genome; the sequencing data for the five-times passaged viruses were compared to those of viruses before passaging. We counted the number of nucleotide changes in the five-times passaged viruses that were not present before passaging. The number of mutations introduced during the five passages are shown in Fig. 2A by segment. Also, the number of mutations per nucleotide was calculated for normalization, and the mean values for all eight segments were compared (Fig. 2B). We confirmed that PR8-PB1-V43I, the high-fidelity control, had fewer mutations, and that PR8-PB1-T123A, the low-fidelity control, had more mutations than WT-PR8. Although PR8-PB1-V43I had fewer mutations than WT-PR8, the difference between PR8-PB1-V43I and WT-PR8 was small. Since a previous report suggested that PB1-V43I does not alter the mutation rate (12), the influence of PB1-V43I on the mutation rate might be dependent on the virus strain or experimental conditions. Moreover, there was no clear difference in mutation number between WT-PR8 and PR8-PB2-E712D. Although the possibility that the PB2-E712D mutation affects virus polymerase fidelity cannot be excluded, the effect does not seem to be large enough to cause an appreciable difference in Venus stability. Therefore, this result suggests that the stability of the Venus gene is not influenced by the fidelity of the virus polymerase.
FIG 2.
Effect of PB2-E712D on the mutation rate. (A) Each virus was passaged five times in MDCK cells, and the mutations introduced into each segment during the passages were counted. (B) The mutation number per nucleotide in each segment was calculated, and the mean values for all eight segments are shown.
Transcription/replication of the modified RNA segment is impaired in WT-Venus-PR8.
Some reports suggest that recombinant viruses containing a foreign gene insertion in their NS segment can propagate more efficiently in interferon (IFN)-deficient Vero cells than in IFN-competent cells such as MDCK cells (4, 13, 14). Therefore, we quantified the expression levels of IFN-β in virus-infected cells by using quantitative real-time PCR. MDCK cells were infected with WT-Venus-PR8 or Venus-PR8-PB2-E712D at an MOI of 1 or mock infected with medium only, and the relative expression levels of IFN-β in infected cells were quantified at 9 h postinfection. WT-Venus-PR8 induced a higher level of IFN-β expression than did Venus-PR8-PB2-E712D (Fig. 3A). This result suggests that WT-Venus-PR8 does not efficiently inhibit IFN-β expression. Given that NS1 plays a key role in suppressing IFN expression and IFN-mediated antiviral responses in the host (15, 16), we next quantified the NS vRNA in infected cells by using influenza virus strand-specific real-time PCR (17, 18). The amount of NS vRNA in WT-Venus-PR8-infected cells was 90% lower than that in Venus-PR8-PB2-E712D-infected cells (Fig. 3B), whereas there was no significant difference in their NP vRNA expression levels (Fig. 3C). The NS vRNA/NP vRNA ratio in WT-Venus-PR8-infected cells was 80% lower than that in Venus-PR8-PB2-E712D-infected cells (Fig. 3D). This result suggests that the transcription/replication of the NS segment is specifically impaired in WT-Venus-PR8. The sequences of all of the plasmids used to generate viruses by reverse genetics were confirmed before use, and the NS segments of WT-Venus-PR8 and Venus-PR8-PB2-E712D were derived from the same NS-Venus plasmid. Therefore, it is unlikely that either WT-Venus-PR8 or Venus-PR8-PB2-E712D has a mutated promoter sequence in its NS segment. Accordingly, the difference in the transcription/replication efficiency of the NS segment was likely caused by the PB2-E712D. Moreover, we confirmed the expression level of the NS1 protein by Western blotting (Fig. 3E). Due to the reduced transcription/replication efficiency of the NS segment, the expression level of the NS1 protein in WT-Venus-PR8-infected cells was much lower than that in Venus-PR8-PB2-E712D-infected cells. In contrast, the expression level of NP was almost the same. The NS1/NP ratio, quantified based on the band intensity, was significantly reduced in WT-Venus-PR8-infected cells (Fig. 3F). It therefore appears that the low level of NS1 expression leads to the high expression of IFN-β in WT-Venus-PR8-infected cells. Furthermore, the high expression of IFN-β causes attenuation of WT-Venus-PR8, although it is possible that other factors are involved.
FIG 3.
RNA and protein expression in infected cells. (A to C) MDCK cells were infected with each virus at an MOI of 1. The relative expression levels of IFN-β mRNA (A), NS vRNA (B), and NP vRNA (C) were determined by quantitative real-time PCR at 9 h postinfection. (D) The NS vRNA/NP vRNA ratio was calculated. (E) MDCK cells were infected with WT-Venus-PR8 (WT) or Venus-PR8-PB2-E712D (712) at an MOI of 1. Cells were lysed at the indicated time points, and the expression of NS1, NP, and β-actin was detected by Western blotting. (F) NS1/NP ratios were determined based on the band intensity of the Western blotting. Means ± the standard deviations of triplicate experiments, taking each value in Venus-PR8-PB2-E712D-infected cells as 1, are shown in panels A, B, C, D, and F. **, P < 0.01; ns, not significant (Student t test); hpi, hours postinfection.
The inserted Venus gene is deleted via internal deletion.
To explore how deletion of the Venus gene occurs, we determined the sequence of the NS segment in WT-Venus-PR8 that lost Venus expression after serial passages. We performed plaque assays using three independently passaged WT-Venus-PR8 virus stocks and found that the majority of the plaques were Venus negative. We then sequenced more than five plaques from each stock and found that one or two deletion patterns in each virus stock. We found that large deletions occurred in the NS segment and most of the Venus sequence was lost (Fig. 4A). However, we could not identify any specific patterns with regard to the deletion, such as the number of nucleotide deletions, the site of the deletion(s), or specific sequences at which deletions occurred. We hypothesized that the large deletion resulted from internal deletion caused by polymerase jumping, which is a known mechanism of defective interfering viral RNA production (19, 20), or by gene recombination, which plays an important role in RNA virus adaptation through rearrangement of the virus genome (21–24). We introduced synonymous mutations into the 3′ or 5′ region of the NS segment of WT-Venus-PR8 (Fig. 4B) and then infected MDCK cells at an MOI of 0.001 or 5 with each of these mutant viruses; supernatants were collected at 2 days or 8 h postinfection, respectively. The supernatant was then incubated with MDCK cells. Venus-negative plaques were picked up and amplified in MDCK cells, and then we determined the NS segment sequence in the viruses that lost Venus expression. We found that the truncated NS segments had a synonymous mutation on only one side (Fig. 4C). We found no viruses that had NS segments with synonymous mutations on both sides or without a synonymous mutation in both low- and high-MOI coinfections. This result indicates that the large deletion in the NS segment resulted from internal deletion in each NS segment and not gene recombination between NS segments.
FIG 4.
Internal deletions occurred in the NS segment of WT-Venus-PR8. (A) Schematic sequence of the NS segment in WT-Venus-PR8 viruses that lost Venus expression after serial passages in MDCK cells. Selected examples are shown. (B) The procedure for the coinfection experiment is illustrated. The synonymous mutation was introduced into the 3′ or 5′ region of the NS segment of WT-Venus-PR8. The viruses were then used to coinfect MDCK cells. Viruses not expressing Venus were plaque purified, and the sequences of their NS segments were analyzed. (C) Examples of the sequences of the NS segment of Venus-negative viruses obtained after coinfection experiments. The red “X” indicates an introduced synonymous mutation.
Additional mutations stabilize the Venus gene.
To further understand the mechanisms of the Venus deletion and stabilization, we attempted to obtain additional mutations in the polymerase complex that stabilize the Venus gene. MDCK cells were infected with WT-Venus-PR8 at an MOI of 0.001, and then Venus-positive plaques were picked and amplified in MDCK cells. After consecutive passaging and plaque purification of Venus-positive viruses, we obtained mutants that stably expressed enhanced Venus fluorescence. Sequence analysis revealed that mutations were introduced into the polymerase genes PB2, PB1, and PA of each mutant (Fig. 5A). PA-180 and PA-200 are located on the surface of the polymerase complex, as is PB2-712, whereas PB2-540, PB1-149, and PB1-684 are located inside the complex. PA-180 and PA-200 are located in the endonuclease domain, PB1-149 and PB1-684 are located near the exit of the RNA template, and PB2-540 is located near the exit of newly synthesized RNA products (Fig. 5B), while the function of the region around PB2-712 has remained unclear (25–27). To determine whether these mutations contribute to the stabilization of the Venus gene, we generated mutant viruses containing each of the mutations by using reverse genetics and measured the Venus retention ratio after four passages in MDCK cells (Fig. 5C). The mutant viruses showed enhanced Venus stability compared to WT-Venus-PR8, indicating that these amino acids play important roles in the stabilization of the Venus gene. Although further analysis is needed to clarify how these amino acids contribute to the stability of the Venus gene, considering that PB2-540, PB1-149, and PB1-684 are located near the polymerase internal tunnels (25–27) that the template and product go through during the transcription/replication reaction, these amino acids may affect the binding stability of the RNA template, product, and polymerase complex. Moreover, when we examined whether these mutations were found in previously isolated influenza A viruses in the Influenza Research Database (Fig. 5D), we found that these amino acids are extremely rare, suggesting that they are not evolutionarily beneficial.
FIG 5.
Additional mutations that stabilizes the Venus gene inserted into the NS segment. (A) Identified amino acid mutations were mapped onto the influenza polymerase complex (PDB ID 4WSB). (B) Polymerase internal tunnels (shown as yellow tubes). The vRNA promoter binds to the polymerase, and the template vRNA enters the polymerase complex. The template vRNA passes through the active site, where RNA synthesis occurs, and then leaves via the template exit. The RNA products synthesized at the active site, leave via the product exit. (C) Each mutant Venus-PR8 virus was passaged four times in MDCK cells, and the proportion of Venus-expressing plaques after passaging was determined in MDCK cells by using fluorescence microscopy. (D) Percentages of influenza A virus strains containing mutations that stabilize the Venus gene in Venus-PR8 (i.e., the number of strains containing the indicated amino acid/total number of strains available in the Influenza Research Database).
DISCUSSION
Recombinant influenza viruses expressing foreign genes would be useful tools; however, long insertions in virus genomes are often unstable and cause attenuation of the recombinant viruses. We previously found that amino acids in the influenza virus polymerase complex play crucial roles in the stabilization of foreign gene insertions; the Venus gene inserted into the NS segment was stabilized by PB2-E712D in an H1N1 virus (7, 8) and by PB2-V25A and PA-R443K in an H5N1 virus (9). However, the mechanisms by which these amino acids contribute to the stabilization remained unclear. In the present study, we explored the mechanism of PB2-E712D-induced stabilization of the Venus gene inserted into the NS segment of an H1N1 virus. We found that the transcription/replication efficiency of the modified segment was significantly reduced in WT-Venus-PR8 compared to Venus-PR8-PB2-E712D. This finding suggests that the PB2-E712D mutation stabilizes the inserted foreign gene due to the enhanced transcription/replication efficiency of the modified RNA segment. In contrast, the transcription/replication efficiency of segments that do not contain additional sequences is not changed in the presence or absence of the PB2-E712D mutation. Moreover, we previously showed that polymerase activity is reduced, not enhanced, by the PB2-E712D mutation in a minireplicon assay (8). These results indicate that the alteration of the transcription/replication efficiency caused by PB2-E712D is specific to modified RNA segments. The insertion of foreign genes appears to impair the transcription/replication of the modified segments, and the polymerase overcomes this impairment in the presence of the PB2-E712D mutation.
In WT-Venus-PR8, in which the Venus gene is inserted into the NS segment, the transcription/replication efficiency of this segment is significantly reduced. As a result, the expression of the NS1 protein is also reduced. Since NS1 plays an important role in inhibiting IFN-mediated antiviral responses (15, 16), WT-Venus-PR8 cannot inhibit IFN-β expression efficiently, which may lead to virus attenuation. The viral titer of WT-Venus-PR8 increases during serial passages in MDCK cells as the virus loses Venus expression (Fig. 1B and C), suggesting that mutated WT-Venus-PR8 that does not contain the Venus gene propagates more efficiently than the original WT-Venus-PR8. Therefore, it is likely that the immediate loss of Venus expression in WT-Venus-PR8 results from the selection of variants without the Venus gene during serial passaging. Venus-PR8-PB2-E712D restores the transcription/replication efficiency of the NS segment, leading to efficient virus replication. Therefore, viruses expressing Venus are not purged by selective pressure in the presence of the PB2-E712D mutation, which enables Venus-PR8-PB2-E712D to stably maintain the inserted Venus gene.
How is the transcription/replication efficiency reduced on modified RNA segments specifically, and how is it enhanced by the PB2-E712D mutation? The RNA secondary structure and the binding affinity between the polymerase complex and the RNA templates likely hold the answer to these questions. Insertion of a foreign gene must change the RNA secondary structure, and transcription/replication by the viral polymerase complex may be negatively influenced by this unusual RNA secondary structure. Although we do not conclusively know how the PB2-E712D mutation overcomes the impairment of transcription/replication, one possible explanation is that the binding affinity between the polymerase complex and the RNA templates is increased.
Our sequence analysis of WT-Venus-PR8 that lost Venus expression, coupled with the coinfection experiments (Fig. 4B and C), suggested that the inserted sequence is deleted due to an internal deletion. Internal deletions often occur during influenza virus replication cycles regardless of the presence of a foreign gene insertion and have been reported to play roles in virus adaptation (28–30) and the generation of defective interfering viral RNA (19, 20). Internal deletion is believed to be caused by polymerase complex dissociation from RNA templates during transcription/replication (20, 31–33). Amino acid mutations in the polymerase complex affect the frequency of occurrence of internal deletions (34–37). PB2-E712D may also be involved in the occurrence of internal deletions. Therefore, the stabilization of the Venus gene in Venus-PR8-PB2-E712D may be caused not only by the enhancement of the transcription/replication on the modified segment but also by the reduced frequency of internal deletions.
We also identified additional mutations in the influenza virus polymerase complex that stabilize the inserted Venus gene, which may help us to further understand the stabilization mechanisms based on the positions of these mutations in the viral polymerase complex. Some of the identified amino acids are located near the polymerase internal tunnels, which are near the RNA template or newly synthesized RNA product during the transcription/replication reactions (25–27). These amino acids might directly affect the binding affinity between the polymerase complex, template, and product. A previous report, which showed that PB2 amino acids located at the template exit channel are involved in the formation of short aberrant RNAs (37), supports the possibility that amino acids near the polymerase internal tunnels affect the binding affinity between the polymerase complex, template, and product. However, PA-180 and PA-200, which are located at the endonuclease domain, are not near the polymerase internal tunnels, which is also true for PB2-712. Therefore, these amino acids may affect the binding affinity indirectly, or there may be other mechanisms involved in the stabilization of the Venus gene. These mutations could be used to establish recombinant influenza viruses expressing a foreign gene. However, these amino acids may not necessarily cause the stabilization of a foreign gene in all influenza virus strains, since PB2-V25A, which stabilizes the Venus gene in Venus-H5N1, had a negative effect on virus replication in Venus-PR8 and did not cause Venus stabilization (our unpublished data).
Although the identified amino acids seem to enhance the genetic stability of virus genomes, they have been rarely found in virus isolates (Fig. 5D). It seems likely that mutations that support the maintenance of inserted sequences are not evolutionarily beneficial to the virus. Insertions of additional sequences into virus genomes are often deleterious for virus replication. These mutations are probably rare in virus populations to avoid the accumulation of deleterious insertions. Viruses may purge deleterious insertions by reducing the transcription/replication efficiency of RNA segments that contain insertions that form abnormal secondary structures. In conclusion, although the amino acid mutations we identified in this study are useful for generating recombinant viruses, they do not seem to be beneficial to the virus in nature in the long run.
MATERIALS AND METHODS
Cells and viruses.
Madin-Darby canine kidney (MDCK) cells were cultured in minimal essential medium (Gibco) with 5% newborn calf serum at 37°C in 5% CO2. Human embryonic kidney 293T (HEK293T) cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum. WT-Venus-PR8 and Venus-PR8 mutants with NS segments encoding the Venus fluorescent protein (7) were generated by using reverse genetics (1) and propagated in MDCK cells at 37°C.
Venus stability.
MDCK cells were infected with WT-Venus-PR8 or each Venus-PR8 mutant at an MOI of 0.001. The supernatants were collected at 48 h postinfection and titrated by using plaque assays in MDCK cells. Obtained viruses were similarly passaged four times. The proportion of Venus-expressing plaques in virus stocks from different passages was determined in MDCK cells by observing more than 65 plaques in each virus stock using fluorescence microscopy. To exclude false-positive plaques, Venus-negative plaques were picked up, amplified in MDCK cells, and reassessed for Venus expression.
Deep sequencing analysis.
WT-PR8, PR8-PB2-E712D, PR8-PB1-V43I (10, 11), and PR8-PB1-T123A (12) were generated by reverse genetics (1), and MDCK cells were infected at an MOI of 0.001. The supernatants were collected at 48 h postinfection and titrated by using plaque assays in MDCK cells. The obtained viruses were passaged five times in the same way. Virus RNA was extracted from viruses before passaging and from viruses passaged five times by using a QIAamp viral RNA minikit (Qiagen). Reverse transcription-PCR (RT-PCR) was performed by using a Superscript III high-fidelity RT-PCR kit (Invitrogen). DNA amplicons were purified by 0.45× of Agencourt AMpure XP magnetic beads (Beckman Coulter), and 1 ng was used for barcoded library preparation with a Nextera XT DNA kit (Illumina). After bead-based normalization (Illumina), libraries were sequenced on the MiSeq platform in a paired-end run using the MiSeq v2, 300 cycle reagent kit (Illumina). The raw sequence reads were analyzed by using the ViVan pipeline (38). Here, a cutoff of 1% as the minimum frequency was used. Moreover, we defined an empirical cutoff for the minimum read coverage: for a variant with 1% frequency, at least 1,000 reads should cover that region. Likewise, for a variant with 0.1% frequency, 10,000 reads should cover that region. Namely, if the coverage was <1,000/(frequency), the variant was removed. The sequencing data of the five-times passaged viruses were compared to those of the viruses before passaging. The number of nucleotide mutations that were not observed before passaging but observed only after passaging was counted. The number of mutations per nucleotide was calculated for each segment and the mean values for all eight segments in each virus were compared.
Quantitative real-time PCR.
MDCK cells were infected with WT-Venus-PR8 or Venus-PR8-PB2-E712D at an MOI of 1 or mock infected with medium. The total RNA was extracted from cells at 9 h postinfection by using an RNeasy minikit (Qiagen). Quantification of RNA was performed as described previously (17). The primers for IFN-β, NS vRNA, NP vRNA, and β-actin were described previously (17, 18, 39). Data were analyzed with the 2–ΔΔCT method (40) and normalized to the expression of β-actin mRNA.
Western blotting.
MDCK cells were infected with each virus at an MOI of 1 or mock infected with medium. Cells were lysed at the indicated time points with Tris-glycine SDS sample buffer (Invitrogen). The cell lysates were sonicated, heated for 10 min at 95°C, and then subjected to SDS-PAGE. SDS-PAGE was performed on Any kD Mini-PROTEAN TGX precast protein gels (Bio-Rad). Proteins on SDS-PAGE gels were transferred to a polyvinylidene fluoride membrane (Millipore) and detected by using the indicated primary antibodies (rabbit anti-NS1 [GeneTex], mouse anti-Aichi NP [2S 347/4], mouse anti-β-actin [Sigma-Aldrich]), followed by secondary antibodies (sheep horseradish peroxidase [HRP]-conjugated anti-mouse IgG [GE Healthcare] or donkey HRP-conjugated anti-rabbit IgG [GE Healthcare]). Signals of specific proteins were detected by using ECL Prime Western blotting detection reagent (GE Healthcare). Images were captured with a ChemiDoc Touch imaging system (Bio-Rad) and quantified by using Image Lab software (Bio-Rad).
Coinfection analysis.
Three nucleotides in the 3′ or 5′ region, which does not overlap the packaging signal sequence (41) of the NS segment of WT-Venus-PR8, were substituted synonymously. These modified viruses were used to coinfect MDCK cells at an MOI of 0.001 each or an MOI of 5 each. The supernatant was collected at 2 days or 8 h postinfection, respectively, and infected to MDCK cells. Venus-negative plaques were picked up and amplified in MDCK cells, and then the sequences of the NS segments in the obtained viruses were analyzed.
Identification of additional mutations that stabilize the Venus gene.
WT-Venus-PR8 was infected to MDCK cells at an MOI of 0.001. The supernatant was collected at 2 days postinfection and infected to MDCK cells. Next, Venus-positive plaques were picked up and amplified in MDCK cells repeatedly until mutants stably expressing Venus fluorescence were obtained. The sequences of the mutants were analyzed to identify amino acid mutations in PB2, PB1, and PA. To determine whether these mutations contributed to the stability of the Venus gene, mutants containing each of the identified amino acid mutations were generated by reverse genetics (1), and the Venus stability of each mutant was examined as described above. Confirmed amino acid positions were plotted on the crystal structure of the influenza virus polymerase complex (PDB ID 4WSB) by using the PyMOL molecular graphics system. In Fig. 5B, the polymerase internal tunnels were visualized by using the MOLEonline web interface (42), and the information was deposited in ChannelsDB (43). The percentage of strains that contained the identified amino acid was determined by using the “Sequencing Feature Variant Type” tool in the Influenza Research Database (44, 45).
Statistical analysis.
Statistically significant differences between WT-Venus-PR8 and Venus-PR8-PB2-E712D were assessed by using a two-tailed unpaired Student t test. A P value of <0.05 was considered significantly different.
ACKNOWLEDGMENTS
We thank Susan Watson for editing the manuscript.
This research was supported by Strategic Basic Research Programs from the Japan Science and Technology Agency; by Leading Advanced Projects for Medical Innovation from the Japan Agency for Medical Research and Development (AMED; JP18am001007 and JP18am001008); by Grants-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Science, Sports, and Technology (MEXT) of Japan (16H06429, 16K21723, and 16H06434); by the Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) from AMED (JP19fm0108006); by e-ASIA Joint Research Program from AMED (JP17jm0210042); by a Research Program on Emerging and Re-emerging Infectious Diseases from AMED (JP19fk0108104; by a fund for the Promotion of Joint International Research (Fostering Joint International Research [B]) from the Japan Society for the Promotion of Science (JSPS; JP18KK0225); by the NIAID-funded Center for Research on Influenza Pathogenesis (HHSN272201400008C); and by JSPS KAKENHI grant 18J21238.
Y.F., S.Y., and Y.K. designed the study. Y.F., J.D., Z.K., D.K., and H.V.B. performed the experiments. Y.F., S.Y., T.J.D.S.L., and Y.K. analyzed the data. Y.F., S.Y., and Y.K. wrote the manuscript. All authors reviewed and approved the manuscript.
Y.K. has received speaker’s honoraria from Toyama Chemical and Astellas, Inc. Y.K. has received grant support from Chugai Pharmaceuticals; Daiichi Sankyo Pharmaceutical; Toyama Chemical; Tauns Laboratories, Inc.; Otsuka Pharmaceutical Co., Ltd.; and Denka Seiken Co., Ltd. Y.K. is a cofounder of FluGen.
Footnotes
Citation Furusawa Y, Yamada S, da Silva Lopes TJ, Dutta J, Khan Z, Kriti D, van Bakel H, Kawaoka Y. 2019. Influenza virus polymerase mutation stabilizes a foreign gene inserted into the virus genome by enhancing the transcription/replication efficiency of the modified segment. mBio 10:e01794-19. https://doi.org/10.1128/mBio.01794-19.
REFERENCES
- 1.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci U S A 96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Z-N, Mueller SN, Ye L, Bu Z, Yang C, Ahmed R, Steinhauer DA. 2005. Chimeric influenza virus hemagglutinin proteins containing large domains of the Bacillus anthracis protective antigen: protein characterization, incorporation into infectious influenza viruses, and antigenicity. J Virol 79:10003–10012. doi: 10.1128/JVI.79.15.10003-10012.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kittel C, Ferko B, Kurz M, Voglauer R, Sereinig S, Romanova J, Stiegler G, Katinger H, Egorov A. 2005. Generation of an influenza A virus vector expressing biologically active human interleukin-2 from the NS gene segment. J Virol 79:10672–10677. doi: 10.1128/JVI.79.16.10672-10677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kittel C, Sereinig S, Ferko B, Stasakova J, Romanova J, Wolkerstorfer A, Katinger H, Egorov A. 2004. Rescue of influenza virus expressing GFP from the NS1 reading frame. Virology 324:67–73. doi: 10.1016/j.virol.2004.03.035. [DOI] [PubMed] [Google Scholar]
- 5.Shinya K, Fujii Y, Ito H, Ito T, Kawaoka Y. 2004. Characterization of a neuraminidase-deficient influenza A virus as a potential gene delivery vector and a live vaccine. J Virol 78:3083–3088. doi: 10.1128/JVI.78.6.3083-3088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manicassamy B, Manicassamy S, Belicha-Villanueva A, Pisanelli G, Pulendran B, García-Sastre A. 2010. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc Natl Acad Sci U S A 107:11531–11536. doi: 10.1073/pnas.0914994107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukuyama S, Katsura H, Zhao D, Ozawa M, Ando T, Shoemaker JE, Ishikawa I, Yamada S, Neumann G, Watanabe S, Kitano H, Kawaoka Y. 2015. Multi-spectral fluorescent reporter influenza viruses (Color-flu) as powerful tools for in vivo studies. Nat Commun 6:6600. doi: 10.1038/ncomms7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katsura H, Fukuyama S, Watanabe S, Ozawa M, Neumann G, Kawaoka Y. 2016. Amino acid changes in PB2 and HA affect the growth of a recombinant influenza virus expressing a fluorescent reporter protein. Sci Rep 6:19933. doi: 10.1038/srep19933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao D, Fukuyama S, Yamada S, Lopes TJS, Maemura T, Katsura H, Ozawa M, Watanabe S, Neumann G, Kawaoka Y. 2015. Molecular determinants of virulence and stability of a reporter-expressing H5N1 influenza A virus. J Virol 89:11337–11346. doi: 10.1128/JVI.01886-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung PPH, Watson SJ, Choy K-T, Fun Sia S, Wong DDY, Poon LLM, Kellam P, Guan Y, Malik Peiris JS, Yen H-L. 2014. Generation and characterization of influenza A viruses with altered polymerase fidelity. Nat Commun 5:4794. doi: 10.1038/ncomms5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naito T, Mori K, Ushirogawa H, Takizawa N, Nobusawa E, Odagiri T, Tashiro M, Ohniwa RL, Nagata K, Saito M. 2017. Generation of a genetically stable high-fidelity influenza vaccine strain. J Virol 91:e01073-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pauly MD, Lyons DM, Fitzsimmons WJ, Lauring AS. 2017. Epistatic interactions within the influenza A virus polymerase complex mediate mutagen resistance and replication fidelity. mSphere 2:e00323-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferko B, Stasakova J, Sereinig S, Romanova J, Katinger D, Niebler B, Katinger H, Egorov A. 2001. Hyperattenuated recombinant influenza A virus nonstructural-protein-encoding vectors induce human immunodeficiency virus type 1 Nef-specific systemic and mucosal immune responses in mice. J Virol 75:8899–8908. doi: 10.1128/JVI.75.19.8899-8908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuznetsova I, Shurygina A-P, Wolf B, Wolschek M, Enzmann F, Sansyzbay A, Khairullin B, Sandybayev N, Stukova M, Kiselev O, Egorov A, Bergmann M. 2014. Adaptive mutation in nuclear export protein allows stable transgene expression in a chimeric influenza A virus vector. J Gen Virol 95:337–349. doi: 10.1099/vir.0.056036-0. [DOI] [PubMed] [Google Scholar]
- 15.García-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 16.Opitz B, Rejaibi A, Dauber B, Eckhard J, Vinzing M, Schmeck B, Hippenstiel S, Suttorp N, Wolff T. 2007. IFNβ induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell Microbiol 9:930–938. doi: 10.1111/j.1462-5822.2006.00841.x. [DOI] [PubMed] [Google Scholar]
- 17.Kawakami E, Watanabe T, Fujii K, Goto H, Watanabe S, Noda T, Kawaoka Y. 2011. Strand-specific real-time RT-PCR for distinguishing influenza vRNA, cRNA, and mRNA. J Virol Methods 173:1–6. doi: 10.1016/j.jviromet.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kupke SY, Riedel D, Frensing T, Zmora P, Reichl U. 2018. A novel type of influenza A virus-derived defective interfering particle with nucleotide substitutions in its genome. J Virol 93:e01786-18. doi: 10.1128/JVI.01786-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis AR, Hiti AL, Nayak DP. 1980. Influenza defective interfering viral RNA is formed by internal deletion of genomic RNA. Proc Natl Acad Sci U S A 77:215–219. doi: 10.1073/pnas.77.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jennings PA, Finch JT, Winter G, Robertson JS. 1983. Does the higher order structure of the influenza virus ribonucleoprotein guide sequence rearrangements in influenza viral RNA? Cell 34:619–627. doi: 10.1016/0092-8674(83)90394-x. [DOI] [PubMed] [Google Scholar]
- 21.Xiao Y, Rouzine IM, Bianco S, Acevedo A, Goldstein EF, Farkov M, Brodsky L, Andino R. 2016. RNA recombination enhances adaptability and is required for virus spread and virulence. Cell Host Microbe 19:493–503. doi: 10.1016/j.chom.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simon-Loriere E, Holmes EC. 2011. Why do RNA viruses recombine? Nat Rev Microbiol 9:617–626. doi: 10.1038/nrmicro2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitnaul LJ, Matrosovich MN, Castrucci MR, Tuzikov AB, Bovin NV, Kobasa D, Kawaoka Y. 2000. Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza A virus. J Virol 74:6015–6020. doi: 10.1128/jvi.74.13.6015-6020.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khatchikian D, Orlich M, Rott R. 1989. Increased viral pathogenicity after insertion of a 28S ribosomal RNA sequence into the haemagglutinin gene of an influenza virus. Nature 340:156. doi: 10.1038/340156a0. [DOI] [PubMed] [Google Scholar]
- 25.Reich S, Guilligay D, Pflug A, Malet H, Berger I, Crépin T, Hart D, Lunardi T, Nanao M, Ruigrok RWH, Cusack S. 2014. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature 516:361. doi: 10.1038/nature14009. [DOI] [PubMed] [Google Scholar]
- 26.Pflug A, Lukarska M, Resa-Infante P, Reich S, Cusack S. 2017. Structural insights into RNA synthesis by the influenza virus transcription-replication machine. Virus Res 234:103–117. doi: 10.1016/j.virusres.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 27.Gerlach P, Malet H, Cusack S, Reguera J. 2015. Structural insights into bunyavirus replication and its regulation by the vRNA promoter. Cell 161:1267–1279. doi: 10.1016/j.cell.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu C, Air GM. 1993. Selection and characterization of a neuraminidase-minus mutant of influenza virus and its rescue by cloned neuraminidase genes. Virology 194:403–407. doi: 10.1006/viro.1993.1276. [DOI] [PubMed] [Google Scholar]
- 29.Liu C, Eichelberger MC, Compans RW, Air GM. 1995. Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding. J Virol 69:1099–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang P, Bansal A, Liu C, Air GM. 1997. Hemagglutinin specificity and neuraminidase coding capacity of neuraminidase-deficient influenza viruses. Virology 229:155–165. doi: 10.1006/viro.1996.8421. [DOI] [PubMed] [Google Scholar]
- 31.Lazzarini RA, Keene JD, Schubert M. 1981. The origins of defective interfering particles of the negative-strand RNA viruses. Cell 26:145–154. doi: 10.1016/0092-8674(81)90298-1. [DOI] [PubMed] [Google Scholar]
- 32.Dimmock NJ, Easton AJ. 2014. Defective interfering influenza virus RNAs: time to reevaluate their clinical potential as broad-spectrum antivirals? J Virol 88:5217–5227. doi: 10.1128/JVI.03193-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.López CB. 2014. Defective viral genomes: critical danger signals of viral infections. J Virol 88:8720–8723. doi: 10.1128/JVI.00707-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fodor E, Mingay LJ, Crow M, Deng T, Brownlee GG. 2003. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase promotes the generation of defective interfering RNAs. J Virol 77:5017–5020. doi: 10.1128/jvi.77.8.5017-5020.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vasilijevic J, Zamarreño N, Oliveros JC, Rodriguez-Frandsen A, Gómez G, Rodriguez G, Pérez-Ruiz M, Rey S, Barba I, Pozo F, Casas I, Nieto A, Falcón A. 2017. Reduced accumulation of defective viral genomes contributes to severe outcome in influenza virus-infected patients. PLoS Pathog 13:e1006650. doi: 10.1371/journal.ppat.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slaine PD, MacRae C, Kleer M, Lamoureux E, McAlpine S, Warhuus M, Comeau AM, McCormick C, Hatchette T, Khaperskyy DA. 2018. Adaptive mutations in influenza A/California/07/2009 enhance polymerase activity and infectious virion production. Viruses 10:272. doi: 10.3390/v10050272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Te Velthuis AJW, Long JC, Bauer DLV, Fan RLY, Yen H-L, Sharps J, Siegers JY, Killip MJ, French H, Oliva-Martín MJ, Randall RE, de Wit E, van Riel D, Poon LLM, Fodor E. 2018. Mini viral RNAs act as innate immune agonists during influenza virus infection. Nat Microbiol 3:1234–1242. doi: 10.1038/s41564-018-0240-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Isakov O, Bordería AV, Golan D, Hamenahem A, Celniker G, Yoffe L, Blanc H, Vignuzzi M, Shomron N. 2015. Deep sequencing analysis of viral infection and evolution allows rapid and detailed characterization of viral mutant spectrum. Bioinformatics 31:2141–2150. doi: 10.1093/bioinformatics/btv101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park W-J, Park B-J, Song Y-J, Lee D-H, Yuk S-S, Lee J-B, Park S-Y, Song C-S, Lee S-W, Choi I-S. 2015. Analysis of cytokine production in a newly developed canine tracheal epithelial cell line infected with H3N2 canine influenza virus. Arch Virol 160:1397–1405. doi: 10.1007/s00705-015-2395-1. [DOI] [PubMed] [Google Scholar]
- 40.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 41.Fujii K, Fujii Y, Noda T, Muramoto Y, Watanabe T, Takada A, Goto H, Horimoto T, Kawaoka Y. 2005. Importance of both the coding and the segment-specific noncoding regions of the influenza A virus NS segment for its efficient incorporation into virions. J Virol 79:3766–3774. doi: 10.1128/JVI.79.6.3766-3774.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pravda L, Sehnal D, Toušek D, Navrátilová V, Bazgier V, Berka K, Svobodová Vareková R, Koca J, Otyepka M. 2018. MOLEonline: a web-based tool for analyzing channels, tunnels and pores (2018 update). Nucleic Acids Res 46:W368–W373. doi: 10.1093/nar/gky309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pravda L, Sehnal D, Svobodová Vareková R, Navrátilová V, Toušek D, Berka K, Otyepka M, Koca J. 2018. ChannelsDB: database of biomacromolecular tunnels and pores. Nucleic Acids Res 46:D399–D405. doi: 10.1093/nar/gkx868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Aevermann BD, Anderson TK, Burke DF, Dauphin G, Gu Z, He S, Kumar S, Larsen CN, Lee AJ, Li X, Macken C, Mahaffey C, Pickett BE, Reardon B, Smith T, Stewart L, Suloway C, Sun G, Tong L, Vincent AL, Walters B, Zaremba S, Zhao H, Zhou L, Zmasek C, Klem EB, Scheuermann RH. 2017. Influenza Research Database: an integrated bioinformatics resource for influenza virus research. Nucleic Acids Res 45:D466–D474. doi: 10.1093/nar/gkw857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noronha JM, Liu M, Squires RB, Pickett BE, Hale BG, Air GM, Galloway SE, Takimoto T, Schmolke M, Hunt V, Klem E, García-Sastre A, McGee M, Scheuermann RH. 2012. Influenza virus sequence feature variant type analysis: evidence of a role for NS1 in influenza virus host range restriction. J Virol 86:5857–5866. doi: 10.1128/JVI.06901-11. [DOI] [PMC free article] [PubMed] [Google Scholar]