Abstract
Chronic obstructive pulmonary disease (COPD) is a common and progressive disease that is influenced by both genetic and environmental factors. For many years, knowledge of the genetic basis of COPD was limited to Mendelian syndromes, such as alpha-1 antitrypsin deficiency and cutis laxa, caused by rare genetic variants. Over the past decade, the proliferation of genome-wide association studies, the accessibility of whole-genome sequencing, and the development of novel methods for analyzing genetic variation data have led to a substantial increase in the understanding of genetic variants that play a role in COPD susceptibility and COPD-related phenotypes. COPDGene (Genetic Epidemiology of COPD), a multicenter, longitudinal study of over 10,000 current and former cigarette smokers, has been pivotal to these breakthroughs in understanding the genetic basis of COPD. To date, over 20 genetic loci have been convincingly associated with COPD affection status, with additional loci demonstrating association with COPD-related phenotypes such as emphysema, chronic bronchitis, and hypoxemia. In this review, we discuss the contributions of the COPDGene study to the discovery of these genetic associations as well as the ongoing genetic investigations of COPD subtypes, protein biomarkers, and post–genome-wide association study analysis.
Keywords: chronic obstructive pulmonary disease, epidemiology, genetics
Contents
COPDGene Study Overview
COPD GWASs
GWASs of COPD-related Phenotypes
Genetic Association Studies of Lung Function Phenotypes
Genetic Association Studies of Imaging Phenotypes
Genetic Association Studies of Other COPD-related Phenotypes
Genetic Studies of COPD Subtypes
Applications of Genome Sequencing in COPDGene
Candidate Gene Studies: Alpha-1 Antitrypsin (SERPINA1)
Genetic Determinants of Protein Biomarkers
Additional Investigations after Genetic Association Analysis
Advances in Statistical Genetics
Conclusions and Future Directions
Chronic obstructive pulmonary disease (COPD) is a complex disease influenced by both environmental and genetic factors. Through research performed largely in the past decade, the genetic basis of COPD susceptibility is now beginning to be understood. Aside from rare genetic variants causing Mendelian syndromes, such as alpha-1 antitrypsin deficiency (caused by variants in SERPINA1 [serpin family A, member 1]) and cutis laxa (caused by mutations in ELN [elastin], FBLN5 [fibulin 5], and other genes), there were frustratingly few consistently replicated genetic variants associated with COPD reported during the era of candidate gene association studies from the 1970s to early 2000s (1). Soon after the COPDGene study (Genetic Epidemiology of COPD; www.COPDGene.org) was launched, two genome-wide association studies (GWASs) published in 2009 demonstrated significant associations with COPD. One study found associations with SNPs at the chromosome 15q25 CHRNA3 (cholinergic receptor nicotinic α3 subunit)/CHRNA5/IREB2 (iron-responsive element–binding protein 2) region in the GenKOLS study (Genetics of COPD, Norway) sample (a case–control study in Norway) that were also replicated using COPD cases in the NETT study (National Emphysema Treatment Trial) and control subjects from the NAS study (Normative Aging Study) (2). At the same time, another genome-wide significant locus near HHIP (hedgehog interacting protein) was shown to be associated with spirometric measures by Wilk and colleagues (3). In this review, we highlight genomic regions influencing COPD susceptibility and COPD-related phenotypes identified through collaborative studies involving COPDGene. We also discuss other study populations that have contributed to progress in COPD genetics (see Table E1 in the online supplement). Furthermore, we review contributions from the COPDGene study to the evolving understanding of COPD genetics based on studies of COPD subtypes, DNA sequencing, and proteomic data.
COPDGene Study Overview
The NHLBI-funded COPDGene study is one of the largest studies ever undertaken to investigate the genetic factors involved in COPD development. COPDGene participants were enrolled from 2008 to 2011; 5-year follow-up data collection was completed, and 10-year follow-up is now underway. The study includes 10,198 individuals with inclusion criteria of age 45 to 80 years old and at least a 10–pack-year smoking history. Individuals who self-identified as non-Hispanic white (NHW) or African American (AA) were enrolled. COPDGene has recruited one of the largest cohorts of AA current and former smoking subjects, enriched for COPD cases, comprising approximately one-third of the COPDGene study population. Significant findings within the AA population are highlighted throughout this review and summarized in Table E2. Additional details regarding the COPDGene study population are included in the online supplement.
COPD GWASs
GWASs detect associations between genetic variants and traits in study populations. To detect associations between variants and diseases such as COPD, a case–control design is often used. Genetic associations with greatest confidence are those that have met genome-wide statistical significance (typically with P < 5 × 10−8) in one study and then have been replicated in another study. Findings that meet genome-wide significance in one study but have not yet been replicated are intriguing, whereas findings below genome-wide statistical significance are of uncertain importance.
The first genetic association study using COPDGene subjects was based on GWAS of subjects from GenKOLS, NETT/NAS, and ECLIPSE (Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints), with the initial 1,006 subjects in COPDGene used to replicate the most statistically significant findings (4). This GWAS confirmed prior COPD-associated loci at 4q31 (HHIP) and 15q25 (CHRNA3/CHRNA5/IREB2) and found a new COPD locus at 4q22 near FAM13A (family with sequence similarity 13 member A), a locus contemporaneously identified as associated with quantitative measures of lung function (5, 6).
A follow-up case–control study used genome-wide SNP genotyping in the initial 1,006 COPDGene subjects for discovery, combining them with the previous cohorts from ECLIPSE, NAS, NETT, and GenKOLS. This study found and replicated a novel genome-wide significant association with COPD at 19q13 near RAB4B, EGLN2 (egl-9 family hypoxia-inducible factor 2), MIA, and CYP2A6 (cytochrome P450, family 2, subfamily A, member 6) (7) in a region previously identified as associated with smoking behavior (8, 9). Although this study and previous COPD GWASs had adjusted for pack-years of smoking, these results suggested that at least some of the genetic susceptibility to risk of COPD at this locus is mediated through genetic control of smoking behavior.
The contribution of genetic factors to COPD susceptibility had been demonstrated previously in family-based studies (10, 11). Genome-wide SNP genotyping data in COPDGene allowed the use of newer methods to estimate heritability. Zhou and colleagues applied these methods to demonstrate substantial heritability for risk to COPD (37.7% in NHW and 37.9% in AA), lung function (for FEV1, 38.4% in NHW and 50.9% in AA), and emphysema (28.2% in NHW and 31.3% in AA) using SNP genotyping array data from COPDGene (12).
When genome-wide SNP genotyping with the Illumina OmniExpress chip was available in the entire COPDGene study population, Cho and colleagues combined the populations in ECLIPSE, NETT/NAS, GenKOLS, and COPDGene, achieving a larger sample (total of 6,633) of moderate-to-severe COPD cases and 5,704 current and former smoking control subjects (13). In addition to confirming the three previously identified COPD susceptibility loci (FAM13A on 4q22, CHRNA3/CHRNA5/IREB2 on 15q25, and HHIP on 4q31), this study also discovered one additional novel locus at 14q32 near RIN3 (Ras and Rab interactor 3). The effect estimates for the loci near HHIP and CHRNA3 became significantly stronger when the same set of control subjects was compared with cases with only severe COPD. Two new genome-wide significant loci in this analysis of patients with severe COPD compared with control subjects were found at 11q22 (near MMP3 [matrix metalloproteinase 3], MMP12, and MMP1) and at 1q41 (near TGFB2 [transforming growth factor-β2]).
Although genome-wide SNP genotyping arrays provided reasonable coverage across the genome, more thorough investigations of the coding exons throughout the genome became possible with the commercial development of exome genotyping arrays covering both common and rare nonsynonymous coding variants. Using a combined analysis of two family-based cohorts and three case–control studies (including COPDGene) totaling 12,165 subjects, investigators performed an exome-wide genetic association analysis (14) and found one novel exome-wide significant association with a SNP in IL27 in the chromosome 16p11.2 locus. Noncoding variants in linkage disequilibrium with the IL27 variant were previously associated with other diseases, including early-onset inflammatory bowel disease and type 1 diabetes mellitus (15, 16). Subsequent COPD association studies found noncoding variants with stronger association at the 16p11.2 locus, suggesting that a noncoding variant, and not the IL27 nonsynonymous variant, explains the 16p11.2 locus COPD association.
Recognizing that further advances in COPD genetics would require larger sample sizes and well-defined phenotypes, a group of international experts met to develop plans for future collaborative work (17). Their recommendations led to the development of the International COPD Genetics Consortium (ICGC; www.copdconsortium.org) to encourage use of collaborative study populations to identify COPD subtypes and search for their possible genetic basis.
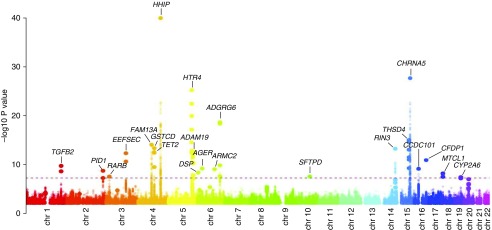
COPDGene was a key member in the first genetic projects of the ICGC, including the ICGC GWAS (18) and a concurrently published UK Biobank/UK BiLEVE (UK Biobank Lung Exome Evaluation) study (19). The ICGC GWAS examined COPD cases (defined as prebronchodilator moderate-to-severe airflow limitation) and control subjects with normal spirometry (18). The ICGC investigators combined data across 26 cohorts totaling over 83,000 subjects in a two-stage analysis followed by a meta-analysis. Twenty-two genome-wide significant loci were identified (Figure 1), including nine loci reaching genome-wide significance in previous GWASs of COPD, an additional nine loci not previously described as associated with risk of COPD but identified as influencing spirometric measures of lung function (FEV1 and/or FEV1/FVC), and four novel loci not previously achieving genome-wide significance for risk of COPD or quantitative measures related to COPD (EEFSEC [eukaryotic elongation factor, selenocysteine-tRNA specific], DSP [desmoplakin], MTCL1 [microtubule crosslinking factor 1], and SFTPD [surfactant protein D]). This study also implicated variants overlapping with genetic determinants of pulmonary fibrosis (but with opposite directions of effect) and an overall risk of COPD shared with lung function and asthma. The UK Biobank/UK BiLEVE study used a staged analytical approach for FEV1, FVC, and FEV1/FVC in general population studies to ultimately identify 97 independent risk signals for lung function, including many that were also associated with COPD in COPDGene and other COPD cohorts (19). These loci included potential targets for therapeutics.
Figure 1.
Manhattan plot of chronic obstructive pulmonary disease genome-wide association study by the International COPD Genetics Consortium. The dashed line corresponds to the threshold for genome-wide statistical significance (P < 5 × 10−8). Reprinted by permission from Reference 18.
Thus, through multiple collaborative studies, more than 20 genetic loci have been convincingly associated with risk of COPD. These regions of the genome likely contain functional genetic variants related to COPD pathogenesis. Table 1 summarizes the genome-wide significant loci for COPD susceptibility identified through collaborations with COPDGene.
Table 1.
Genomic Regions Significantly Associated with Chronic Obstructive Pulmonary Disease Affection Status in Meta-analyses That Have Included COPDGene
| Top SNPs | Odds Ratio (95% CI) or β-Coefficient (SE)* | P Value* | Reference |
|---|---|---|---|
| Chromosome 1q41: TGFB2 | |||
| rs10429950 | 1.11 (1.07–1.14) | 1.66 × 10−10 | 18 |
| Chromosome 2q26: PID1 | |||
| rs16825267 | 1.19 (1.12–1.25) | 1.68 × 10−9 | 18 |
| Chromosome 3p24: RARB | |||
| rs1529672 | 1.11 (1.07–1.15) | 2.47 × 10−8 | 18 |
| Chromosome 3q21: EEFSEC | |||
| rs2955083 | 1.18 (1.13–1.24) | 4.16 × 10−13 | 18 |
| Chromosome 4q22: FAM13A | |||
| rs1903003 | 0.78 (0.64–0.95)† | 7.19 × 10−3† | 4 |
| rs7671167 | 0.77 (0.63–0.93)‡ | 3.39 × 10−3‡ | 4 |
| rs7671167 | 0.73 (0.66–0.81) | 2.22 × 10−9 | 7 |
| rs1964516 | 0.73 (0.66–0.81) | 1.88 × 10−9 | 7 |
| rs4416442 | 1.28 (1.2–1.36) | 1.12 × 10−14 | 13 |
| rs6837671 | 1.12 (1.09–1.15) | 7.48 × 10−15 | 18 |
| Chromosome 4q24: GTSCD | |||
| rs11727735 | 1.26 (1.18–1.33) | 3.84 × 10−14 | 18 |
| Chromosome 4q24: TET2 | |||
| rs2047409 | 1.12 (1.08–1.15) | 2.46 × 10−13 | 18 |
| Chromosome 4q31: HHIP | |||
| rs13118928 | 0.77 (0.70–0.85) | 9.3 × 10−8 | 4 |
| rs13118928 | 0.76 (0.65–0.89) | 5.97 × 10−7 | 7 |
| rs13141641 | 0.76 (0.65–0.88) | 2.56 × 10−7 | 7 |
| rs13141641 | 1.27 (1.19–1.36) | 1.57 × 10−12 | 13 |
| rs145506456 | 1.22 (1.16–1.27) | 9.1 × 10−41 | 18 |
| Chromosome 5q32: HTR4 | |||
| rs7733088 | 1.18 (1.14–1.21) | 5.33 × 10−26 | 18 |
| Chromosome 5q33: ADAM19 | |||
| rs113897301 | 1.16 (1.12–1.21) | 1.58 × 10−13 | 18 |
| Chromosome 6p24: DSP | |||
| rs2076295 | 1.09 (1.06–1.12) | 3.97 × 10−9 | 18 |
| Chromosome 6p21: AGER | |||
| rs2070600 | 1.24 (1.16–1.32) | 5.94 × 10−10 | 18 |
| Chromosome 6q21: ARMC2 | |||
| rs2806356 | 1.12 (1.08–1.16) | 8.34 × 10−10 | 18 |
| Chromosome 6q24: ADGRG6 | |||
| rs9399401 | 1.15 (1.12–1.19) | 1.81 × 10−19 | 18 |
| Chromosome 10q22: SFTPD | |||
| rs721917 | 1.08 (1.05–1.11) | 2.49 × 10−8 | 18 |
| Chromosome 14q32: RIN3 | |||
| rs754388 | 1.28 (1.18–1.39) | 5.25 × 10−9 | 13 |
| rs754388 | 1.15 (1.11–1.20) | 4.96 × 10−14 | 18 |
| Chromosome 15q23: THSD4 | |||
| rs1441358 | 1.13 (1.10–1.16) | 8.22 × 10−16 | 18 |
| Chromosome 15q25: CHRNA3/5 and IREB2 | |||
| rs8034191 | 1.29 (1.16–1.41) | 8.32 × 10−8 | 4 |
| rs13180 | 0.77 (0.70–0.85) | 2.01 × 10−8 | 4 |
| rs12914385 | 1.28 (1.2–1.36) | 6.38 × 10−14 | 13 |
| rs16969968 | 0.27 (0.04) | 1.7 × 10−14 | 14 |
| rs754388 | 1.18 (1.15–1.22) | 1.77 × 10−28 | 18 |
| Chromosome 16p11: IL27, CCDC101 | |||
| rs181206§ | −0.16 (0.04) | 4.7 × 10−6 | 14 |
| rs17707300 | 1.10 (1.06–1.13) | 6.75 × 10−10 | 18 |
| Chromosome 16q23: CFDP1 | |||
| rs7186831 | 1.12 (1.08–1.16) | 1.12 × 10−11 | 18 |
| Chromosome 18p11: MTCL1 | |||
| rs647097 | 1.10 (1.06–1.13) | 6.14 × 10−9 | 18 |
| Chromosome 19q13: RAB4B/EGLN2/CYP2A6 | |||
| rs7937 | 0.73 (0.63–0.83) | 2.88 × 10−9 | 7 |
| rs2604894 | 0.74 (0.65–0.84) | 3.41 × 10−8 | 7 |
| rs12459249 | 1.10 (1.06–1.14) | 3.42 × 10−8 | 18 |
Definition of abbreviations: CI = confidence interval; COPDGene = Genetic Epidemiology of Chronic Obstructive Pulmonary Disease.
Genes located near top SNPs in each region are shown, but they may not be the genes influenced by functional variants in that region; odds ratios and confidence intervals were taken from the cited references.
For meta-analysis.
Odds ratio and confidence interval for COPDGene non-Hispanic whites only; genome-wide meta-analysis P value = 9.47 × 10−11.
Odds ratio and confidence interval for COPDGene non-Hispanic whites only; genome-wide meta-analysis P value = 1.22 × 10−11.
This study was an exome-wide study, so this SNP was considered significant, given this study design.
GWASs of COPD-related Phenotypes
GWASs using COPDGene subjects and other cohorts have identified multiple genetic loci associated with COPD susceptibility and severity. However, owing in part to the complex and heterogeneous nature of COPD, effect sizes of individual genetic loci remain modest. Thus, COPDGene investigators have sought to determine the heritable factors leading to clinical manifestations of COPD by examining the genetic associations with lung function phenotypes, imaging phenotypes, and other COPD-related phenotypes (summarized in Table E3).
Genetic Association Studies of Lung Function Phenotypes
Because spirometric lung function tests measuring FEV1 and FEV1/FVC are used to diagnose and determine the severity of COPD, investigators have reasoned that genetic loci associated with these phenotypes could also be associated with COPD susceptibility. Population-based studies that have been conducted before and concurrently with COPDGene have to date discovered nearly 100 different loci associated with FEV1 and FEV1/FVC ratio, beginning with the discovery of HHIP by Wilk and colleagues (3) and followed by the CHARGE (Cohorts for Heart and Aging Research in Genomic Epidemiology) and SpiroMeta consortiums and the UK BiLEVE and UK Biobank studies (5, 6, 19–21). Among the first studies to demonstrate the relevance of these measures in COPD (and COPDGene) was by Castaldi and colleagues, who hypothesized that other loci associated with FEV1 and FEV1/FVC were likely to be associated with COPD susceptibility (22). They tested 32 genome-wide significant SNPs in 17 genes for association with COPD status in four COPD case–control study samples (NETT/NAS, the GenKOLS case–control study from Norway, ECLIPSE, and the first 1,000 subjects in COPDGene), totaling 3,456 cases and 1,906 control subjects. Three of these loci were confirmed to contain SNPs associated with risk for COPD: 4q24 (FLJ20184/INTS12 [integrator complex subunit 12]/GSTCD [glutathione S-transferase C-terminal domain containing]/NPNT [nephronectin]), 6p21 (AGER [advanced glycosylation endproduct specific receptor]/PPT2 [palmitoyl-protein thioesterase 2]), and 5q33 (ADAM19 [ADAM metallopeptidase domain 19]). Subsequent larger studies by Wain and colleagues (19) and Hobbs and colleagues (18) reinforced the overlap between these lung function loci and COPD.
Examining genetic determinants of lung function specifically within COPD cohorts, Lutz and colleagues performed a GWAS of post-bronchodilator pulmonary function (FEV1 and FEV1/FVC) (23). NHW and AA COPDGene subjects were analyzed separately. In the NHW individuals, these investigators found that FEV1 and FEV1/FVC ratio were significantly associated with SNPs in the 15q25 region, near CHRNA3, CHRNA5, AGPHD1 (aminoglycoside phosphotransferase domain–containing protein 1), IREB2, and CHRNB4 (cholinergic receptor nicotinic β4 subunit), and that FEV1/FVC was associated with HHIP, replicating previously seen associations when analyzing COPD affection status as a categorical phenotype. Meta-analysis of COPDGene NHW subjects combined with ECLIPSE and GenKOLS subjects revealed that most of the genome-wide significant results for FEV1 and FEV1/FVC were at the 15q25 locus, chromosome 4q22 near FAM13A, and chromosome 4q31 near HHIP. In addition, genome-wide significant associations were identified for both FEV1 and FEV1/FVC with regions on chromosomes 1 (near TGFB2) and 19 (near CYP2A6). Genome-wide significant associations were found for FEV1/FVC on chromosomes 11 (near MMP3, MMP12, and MMP1) and 14 (near RIN3), as well as for FEV1 on chromosome 9 (near DBH). Analysis of COPDGene AA subjects for FEV1 and FEV1/FVC did not find any genome-wide significant associations using GWAS, but admixture mapping was subsequently applied in COPDGene AA subjects (24). African ancestry was associated with lower FEV1/FVC values, and a region of suggestive association to FEV1/FVC was identified in FAM19A2.
Genetic Association Studies of Imaging Phenotypes
Variable combinations of emphysema and small airway disease leading to chronic airflow obstruction contribute to the phenotypic heterogeneity of COPD. Several methods of computed tomography (CT) imaging quantification have been developed to assess both pathologic emphysema patterns and functional small airway disease, and they have been applied in genetic studies outside of COPDGene. For example, using these imaging parameters in a population-based GWAS, Manichaikul and colleagues identified several genetic determinants associated with percentage emphysema (SNRPF [small nuclear ribonucleoprotein polypeptide F], PPT2, MAN1C1 [mannosidase α class 1C member 1]) and (upper lobe emphysema)/(lower lobe emphysema) ratio (MAN2B1 [mannosidase α class 2B member 1], DHX15 [DEAH-box helicase 15], as well as MGAT5B [α-1,6-mannosylglycoprotein 6-β-N-acetylglucosaminyltransferase B]) (25). In individuals with COPD (from the ECLIPSE, NETT, and GenKOLS cohorts), Kong and colleagues discovered a SNP in BICD1 (BICD cargo adaptor 1) that was associated with the presence or absence of emphysema determined by radiologist visual assessments (26).
Densitometric measures of emphysema provide objective and quantitative assessments of emphysema severity. In a total sample size of 12,031 subjects from COPDGene, ECLIPSE, NETT, and GenKOLS, Cho and colleagues performed a GWAS on two quantitative emphysema phenotypes (percentage of lung density histogram below −950 Hounsfield units [HU] and 15th percentile point of the lung density histogram), two quantitative airway imaging phenotypes (wall area percentage and square root of airway wall area of 10 mm internal perimeter airway), and the combined emphysema/small airway phenotype of expiratory gas trapping in smokers with and without COPD (27). Five different loci were associated with emphysema phenotypes, including the previously reported HHIP, CHRNA3, and AGER loci, as well as novel associations near DLC1 (deleted in liver cancer 1) and SERPINA10 (serpin family A member 10). The SERPINA10 association is located near the SERPINA1 gene and appears to be driven largely by the low-frequency SERPINA1 Z allele, demonstrating that even relatively rare genetic variants such as alpha-1 antitrypsin deficiency can be confirmed using large samples of subjects with disease-specific phenotypes. Two loci were associated with gas trapping on expiratory CT scans, near AGER on chromosome 6p21 and LINC00310 (long intergenic non–protein-coding RNA 310)/KCNE2 (potassium voltage-gated channel subfamily E regulatory subunit 2) on chromosome 21q22.
Local histogram–based emphysema quantification is a texture-based method that captures emphysema patterns and is more strongly associated with COPD-related physiological and functional measures than densitometric measures (28). Castaldi and colleagues sought to identify genetic determinants associated with distinct patterns of emphysema on chest CT scans within the COPDGene study (29). They performed a GWAS using 9,614 total COPDGene subjects (6,456 NHW and 3,158 AA) for five emphysema patterns. Two novel genome-wide significant associations were identified for SNPs in MYO1D (myosin ID) and VMA8 with severe centrilobular and panlobular local histogram–based emphysema patterns, respectively. Five other genome-wide significant associations were found for loci previously associated with COPD susceptibility and quantitative measures of lung function (HHIP, IREB2/CHRNA3, CYP2A6/ADCK [aarF domain–containing kinase], TGFB2, and MMP12).
Boueiz and colleagues examined a total of 11,532 subjects from COPDGene, ECLIPSE, and GenKOLS in a GWAS to determine the genetic determinants of the anatomic distribution of emphysema (30). Two loci previously reported to be associated with COPD susceptibility (specifically, 4q31 near HHIP and 15q25 near CHRNA5) were associated with both measures of emphysema distribution used in this study: the difference and ratio of upper-third to lower-third percentage emphysema at −950 HU. The COPD-associated variants in these two regions were associated with greater amounts of upper lobe emphysema. For the emphysema ratio phenotype, three novel associations were reported, near SOWAHB (sosondowah ankyrin repeat domain family member B), TRAPPC9 (trafficking protein particle complex 9), and KIAA1462.
Halper-Stromberg and colleagues hypothesized that visual assessment of CT images may provide independent information for genetic association studies compared with the semiautomated CT imaging measures of emphysema and airway disease described above (31). They performed a GWAS for visual assessment of CT images in 1,540 NHW COPDGene subjects to determine the genetic determinants contributing to the presence and severity of radiographic features representing airway wall thickness and emphysema. This study identified a genome-wide significant association with visual emphysema at the 15q25 region (within IREB2) but found no significant associations for visual airway wall thickness. Furthermore, these authors identified several previously identified COPD susceptibility loci that were also nominally associated with either an emphysema or airway phenotype. To demonstrate how visual measures can provide independent information, these associations were adjusted for quantitative densitometric assessment of emphysema.
Genetic Association Studies of Other COPD-related Phenotypes
In addition to spirometry and CT measures of emphysema and airway disease, COPDGene investigators have sought to identify the genetic determinants of multiple other disease-related phenotypes contributing to the extensive heterogeneity of COPD. Before COPDGene, Pillai and colleagues demonstrated associations of known COPD GWAS loci with several COPD-related phenotypes using ECLIPSE study subjects, including spirometry (CHRNA3/CHRNA5, HHIP, FAM13A), smoking (CHRNA3/CHRNA5), body mass index (BMI) (FAM13A), fat-free mass index (HHIP), emphysema (CHRNA3/CHRNA5, HHIP), COPD exacerbations (HHIP), and BODE score (BMI, airflow obstruction, dyspnea, and exercise capacity; a metric of risk of death in patients with COPD) (CHRNA3/CHRNA5) (32). A meta-analysis of 2,950 individuals with COPD from ECLIPSE, GenKOLS, and NETT conducted by Wan and colleagues demonstrated an association of the well-known FTO (FTO α-ketoglutarate dependent dioxygenase) locus with measures of cachexia, BMI, and fat-free mass index (33). Replication of the most significantly associated SNP with BMI was achieved using 502 genotyped subjects from COPDGene with Global Initiative for Chronic Obstructive Lung Disease (GOLD) spirometry grade 2 or higher (moderate, severe, and very severe) COPD.
McDonald and colleagues performed a GWAS for hypoxemia, as measured by pulse oximetry, in subjects from COPDGene (2,810 NHW and 820 AA) and ECLIPSE (1,758 total) (34). Although no SNPs reached genome-wide significance in the GWAS of NHW subjects, several suggestive loci were identified. In the GWAS of AA COPDGene subjects, two regions were found to be significantly associated with resting oxygen saturation: an intergenic region on chromosome 14 bounded by FOXG1 (forkhead box G1) and LINC00645 and a region on chromosome 15 encompassing TICRR (DNA topoisomerase II binding protein 1 interacting checkpoint and replication regulator) and KIF7 (kinesin family member 7).
In previous studies, chronic bronchitis, defined as symptoms of chronic cough and chronic phlegm production, has been associated with accelerated lung function decline and an increase in risk of respiratory infections (see additional citations in Kim and colleagues [35, 36]). Using subjects from COPDGene, GenKOLS, and ECLIPSE, Lee and colleagues investigated the genetic variants associated with chronic bronchitis in COPD subjects relative to current and former smokers with normal spirometry or subjects with COPD without chronic bronchitis (37). Comparing COPD subjects with chronic bronchitis with current or former cigarette smokers without COPD (“smoking control subjects”), they identified a novel genome-wide significant locus on chromosome 11p15.5 near the genes EFCAB4A (EF-hand calcium-binding domain–containing protein 4A), CHID1 (chitinase domain–containing protein 1), and AP2A2 (adaptor-related protein complex 2 subunit α2). The four most significant SNPs were located in the FAM13A region. Comparing subjects with COPD who had chronic bronchitis with subjects with COPD who did not have chronic bronchitis, they identified a novel locus yielding suggestive evidence of association on chromosome 1q23 containing genes RPL31P11 (ribosomal protein L31 pseudogene 11) and ATF6 (activating transcription factor 6). Because chronic bronchitis was present in some of the smoking control subjects, further association analysis of chronic bronchitis in COPD was completed using smoking control subjects without chronic bronchitis, resulting in slightly reduced statistical significance of the top SNPs. Together, these analyses assessed the combined outcome of COPD with chronic bronchitis; genome-wide significant genetic determinants of chronic bronchitis independent of COPD status have not been reported.
Enlargement of the pulmonary arteries was found to be associated with increased COPD exacerbation frequency in COPDGene (38). To determine the genetic determinants of pulmonary arterial hypertension associated with advanced COPD, Lee and colleagues performed a GWAS of pulmonary artery enlargement (PAE) in subjects with COPD (39). This study included 1,266 subjects with COPD with PAE, 3,755 subjects with COPD without PAE, and 4,461 current and former smokers with normal spirometry from both the COPDGene and ECLIPSE cohorts. Relative to subjects with COPD without PAE, the GWAS of subjects with PAE showed significantly associated SNPs on chromosomes 15q25 (within IREB2) and 14q31 (near GALC [galactosylceramidase]). Relative to smokers with normal spirometry, the GWAS of subjects with PAE demonstrated genome-wide significant association for markers in the 15q25 region containing CHRNA5, PSMA4 (proteasome subunit α4), IREB2, AGPHD1, and CHRNA3, in which all SNPs were either identical to or in strong linkage disequilibrium with SNPs previously reported as associated with risk for COPD. These results emphasize the complexity of the chromosome 15q25 region in controlling risk for COPD, where multiple regional genes could influence COPD-related phenotypes.
History of childhood pneumonia was shown to be associated with increased risk of COPD in COPDGene subjects (40). In COPDGene, Hayden and colleagues hypothesized that genetic variants would be associated with development of pneumonia during childhood and over the course of the lifetime, which may contribute to reduced lung function and risk for COPD (41). Although there were no genome-wide significant findings, several loci with suggestive associations were identified for childhood pneumonia (in NGR1, PAK6 [p21 {RAC1} activated kinase 6], and near MATN1 [matrilin 1]) and for lifetime pneumonia (in LOC339862, RAPGEF2 [Rap guanine nucleotide exchange factor 2], PHACTR1 [phosphatase and actin regulator 1], and PRR27 [proline-rich 27] and near MCPH1 [microcephalin 1]).
Because cigarette smoking is the principal environmental risk factor for developing COPD, genetic factors associated with smoking behavior and nicotine dependence may contribute to COPD susceptibility and severity. Previous GWASs of smoking behavior have identified significant associations, including the 15q25 (CHRNA5–CHRNA3–CHRNB4) and 8p11 (CHRNB3–CHRNA6) loci, with nicotine dependence (8, 9, 42, 43). As noted above, the 15q25 locus was one of the first COPD GWAS loci identified (2). Kim and colleagues confirmed the association of 15q25 SNPs with nicotine dependence in COPDGene subjects and demonstrated association of CHRNA3/CHRNA5 genetic variants with severity of emphysema in former smokers (44). Hancock and colleagues conducted a GWAS meta-analysis including 17,074 subjects from COPDGene, deCODE, EAGLE (Environment and Genetics in Lung Cancer Etiology study), COGEND (Collaborative Genetic Study of Nicotine Dependence), and SAGE (Study of Addiction: Genetics and Environment) to identify novel loci associated with nicotine dependence in ever smokers (45). In addition to confirming the associations with 15q25 and 8p11, the authors identified a novel genome-wide significant association with a locus on chromosome 20q13, within the CHRNA4 gene.
In addition to COPD susceptibility, genetic determinants could also influence response to COPD treatment. To determine whether genetic variants contributed to the acute response to inhaled β2-agonists, Hardin and colleagues performed a GWAS of quantitative bronchodilator responsiveness using 5,789 current or former smokers with COPD from the COPDGene (both NHW and AA), ECLIPSE, GenKOLS, and NETT studies (46). Among NHW COPDGene subjects, a variant located upstream of the gene EPHA7 (EPH receptor A7) was significantly associated with the absolute difference in pre- versus post-bronchodilator FEV1 levels. Among AA COPDGene subjects, two low-frequency variants in the gene CDH13 were significantly associated with the absolute volume change with bronchodilator or the difference in pre- versus post-bronchodilator change expressed as a percentage of the predicted FEV1. A rare SNP in GOLGA8B (golgin A8 family member B) was associated with bronchodilator response expressed as change from baseline FEV1. The reliability of these associations to low-frequency variants in a relatively small study population is uncertain. In addition, a more common SNP in SGCD (sarcoglycan delta; minor allele frequency, 0.07) was significantly associated with the difference in pre- versus post-bronchodilator FEV1 as a percentage of baseline FEV1. In the meta-analysis of all five study populations, several SNPs upstream from the gene KCNJ2 (potassium voltage-gated channel subfamily J member 2) were found to be associated with several different bronchodilator responsiveness metrics, but none achieved genome-wide significance. Although most genetic association analyses of COPD-related phenotypes in COPDGene have focused on SNPs, Begum and colleagues assessed association of estimated larger structural variants (also known as copy number variants) with COPD-related phenotypes and found suggestive associations of a structural variant on chromosome 5q35 with TLC (from chest CT scans) among AA (47) and on chromosome 3p26 with smoking behavior among AA (48).
Thus, studies conducted by COPDGene investigators have identified numerous genetic associations with lung function, lung imaging phenotypes, and COPD-related clinical phenotypes. It is notable that a considerable number of genetic loci associated with COPD susceptibility are also associated with one or more of these additional COPD-related phenotypes. As reported by Hobbs and colleagues (18), nearly half of COPD susceptibility loci are also associated with measures of lung function in general population samples. Two COPD susceptibility loci (CYP2A6 and MMP1/3/12) exhibited overlap with local histogram imaging phenotypes and lung function. Furthermore, four COPD susceptibility loci (TGFB2, FAM13A, HHIP, and CHRNA3/CHRNA5/IREB2) overlapped with multiple imaging, lung function, and COPD-related phenotypes, demonstrating their likely importance to COPD etiology. These loci also represent the most frequently reported and replicated signals associated with COPD. However, other genetic associations, such as the DLC1 locus with quantitative emphysema and TRAPPC9 with emphysema distribution, have not been associated with other COPD-related phenotypes; these unique associations could also point to specific biological mechanisms relevant for COPD heterogeneity. A summary of the associations of COPD-related phenotypes with genome-wide significant COPD susceptibility loci is provided in Figure 2.
Figure 2.
Summary phenogram plot of genome-wide association studies for chronic obstructive pulmonary disease (COPD) susceptibility and COPD-related phenotypes reviewed in this article. Individual SNP associations have been collapsed into the nearest gene or locus where genes are densely clustered. Although multiple SNP associations may be reported in an individual locus for a given phenotype, only one association is depicted. COPD susceptibility associations (blue) include all 22 loci reported by Hobbs and colleagues (18). Lung function (FEV1, FVC, and FEV1/FVC) associations depicted here (red) include associations from the comprehensive study conducted by Wain and colleagues (19). BDR = bronchodilator response; BMI = body mass index; CB = chronic bronchitis; EMPH.DIST.RATIO = emphysema distribution ratio of upper divided by lower lung fields; FFMI = fat-free mass index; LAA.950 = percentage of lung density histogram below −950 Hounsfield units; LHE = local histogram–based emphysema; MOD = moderate centrilobular emphysema on local histogram–based emphysema analysis; NORM = normal/nonemphysematous on local histogram–based emphysema analysis; OS = resting oxygen saturation; PAE = pulmonary artery enlargement; PAN = panlobular emphysema on local histogram–based emphysema analysis; PB = post-bronchodilator; PCT.GAS = percent gas trapping at −856 Hounsfield units on expiratory computed tomographic scan; PERC15 = 15th percentile point of the lung density histogram; SEV = severe centrilobular emphysema on local histogram–based emphysema analysis; VIS.EMPH = visual emphysema (presence/absence).
Genetic Studies of COPD Subtypes
Because COPD is a heterogeneous clinical syndrome, COPDGene has aimed to define clinical subtypes of COPD to enable better elucidation of the pathophysiologic mechanisms of the disease. The clinical subtypes examined to date for genetic associations are preserved ratio impaired spirometry (PRISm), mild COPD (GOLD spirometry grade 1), and asthma–COPD overlap. In addition, genetic determinants of COPD subtypes determined by unsupervised clustering approaches have been assessed.
PRISm is defined as a reduced FEV1 with a preserved FEV1/FVC ratio. Subjects with PRISm have previously been shown to have increased respiratory symptoms, decreased exercise capacity, more difficulty with activities of daily living, and increased mortality (49, 50). The prevalence of PRISm among all COPDGene subjects was estimated to be 12.3%, and these patients showed increased dyspnea, reduced 6-minute walk distance, increased percentage emphysema, decreased TLC, and increased segmental bronchial wall area percentage. Cluster analysis suggested that a subset of PRISm subjects may have COPD (50). However, in a GWAS of PRISm, there were no common genetic variants significantly associated with this clinical subtype at a genome-wide level. Of interest, a nominally significant association of PRISm with Klinefelter’s syndrome (47XXY) was observed.
Because FEV1/FVC normally declines with advancing age, the GOLD 1 spirometry group (FEV1 ≥80% predicted with FEV1/FVC <0.70) includes subjects with mild COPD as well as healthy subjects with reduced FEV1/FVC due to advanced age. Lee and colleagues performed a GWAS of GOLD 1 subjects in COPDGene (51). Although no genome-wide significant associations were identified, the COPD GWAS region near FAM13A was nominally associated with GOLD 1 status. K-means cluster analysis was used to dissect GOLD 1 subjects into four groups, which showed nominally significant evidence for differential associations with previously identified COPD GWAS SNPs.
In the COPDGene cohort, 13% of the patients with COPD also reported a history of asthma. Compared with the population of patients with COPD alone, the subjects with COPD and asthma were younger, had lower lifetime smoking intensity, had worse disease-related quality of life, were more likely to have had a COPD exacerbation in the past year, and were more likely to report frequent exacerbations (52). A GWAS in the NHW group of patients with COPD–asthma overlap identified significant SNPs in CSMD1 (CUB and Sushi multiple domains 1) on chromosome 8p23 and SOX5 (SRY-box 5) on chromosome 12p12. A meta-analysis of the NHW and AA subjects identified a region containing GPR65 (G protein–coupled receptor 65) on chromosome 14q31 that was associated with the COPD–asthma overlap phenotype.
COPDGene has also assessed sex-specific genetic influences on COPD. Hardin and colleagues identified SNP-by-sex interactions for SNPs in the CELSR1 (cadherin EGF LAG seven-pass G-type receptor 1) gene (53). In a sex-stratified association analysis, a SNP in CELSR1 approached genome-wide significance in females but was not even nominally associated with COPD in males. Further research to identify sex-specific determinants of COPD and COPD-related phenotypes may provide insight into the variable susceptibility to COPD among men and women.
In addition to clinically defined subtypes, machine learning approaches have also been used to classify COPD into distinct subtypes. Castaldi and colleagues used k-means cluster analysis to group COPDGene subjects into four clusters that showed distinct characteristics: relatively resistant smokers, mild upper zone–predominant emphysema, airway-predominant disease, and severe emphysema (54). Among NHW from COPDGene, they found evidence of association (although not genome-wide significance) with a SNP near the HHIP gene for the mild upper zone–predominant emphysema group, and within the severe emphysema group, there was also evidence for association near HHIP and in the chromosome 15q25 locus containing CHRNA3, CHRNA5, and IREB2.
Applications of Genome Sequencing in COPDGene
Although genome-wide SNP panels can provide reasonable coverage of common genetic variants, whole-exome sequencing (WES) provides comprehensive coverage of both rare and common variants in the protein-coding portion of the genome (encompassing ∼1% of the total genome), whereas whole-genome sequencing provides similar coverage across essentially the entire genome. To focus on the extremes of COPD susceptibility and resistance, the WES studies in COPDGene have focused on highly resistant smokers and severe COPD cases. Qiao and colleagues used WES in COPDGene early-onset COPD cases and resistant smoking control subjects to attempt to replicate genes segregating with COPD in families from the Boston Early-Onset COPD study (55). Although nominally significant replication was observed for variants in DNAH8 (dynein axonemal heavy chain 8), ALCAM (activated leukocyte cell adhesion molecule), RARS (arginyl-tRNA synthetase), and GBF1 (Golgi brefeldin A–resistant guanine nucleotide exchange factor 1), rare coding genetic determinants of COPD other than SERPINA1 appear to be quite heterogeneous. Using WES from COPDGene, Stanley and colleagues identified rare functional variants in the TERT (telomerase reverse transcriptase) gene in severe COPD cases, demonstrating the importance of this gene previously reported to be associated with risk for pulmonary fibrosis (56). The entire COPDGene study population is undergoing whole-genome sequencing through the TOPMed (Trans-Omics for Precision Medicine) project, which will enhance opportunities for future identification of rare variants influencing COPD susceptibility.
Candidate Gene Studies: Alpha-1 Antitrypsin (SERPINA1)
Although earlier candidate gene association studies had focused on biologically plausible candidates, selection of candidate genes from Mendelian syndromes or large-scale GWAS gives a greater likelihood of finding valid associations. Alpha-1 antitrypsin deficiency, caused primarily by homozygosity for the Z allele of the SERPINA1 gene, is the most well-established genetic cause of COPD. In addition to candidate studies of variants identified through lung function GWAS noted above (22), COPDGene has also been important for studying Z allele heterozygotes (PI MZ). In COPDGene, NHW heterozygous Pi MZ patients had significantly lower FEV1 and FEV1/FVC, as well as significantly more radiographic emphysema, than subjects not carrying SERPINA1 Z alleles (57). Findings were similar in the substantially smaller AA PI MZ group of COPDGene. A larger collaborative study in severe COPD cases versus control subjects found a genome-wide significant association in SERPINA1, adding further support to the elevated risk for heterozygote carriers of this variant (58). These findings, combined with efforts identifying this locus in emphysema GWAS (27), highlight the contribution of COPDGene to the increasing evidence that PI MZ smokers are at significantly increased risk for COPD (59).
Genetic Determinants of Protein Biomarkers
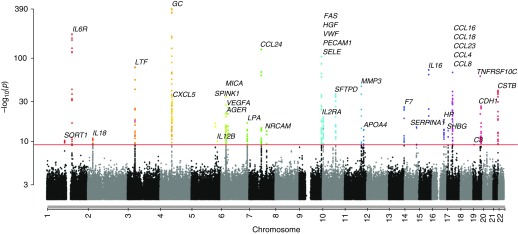
One of the major strengths of COPDGene is its abundance of proteomic data. These data have allowed COPDGene investigators to explore the relationships between genetic variation and blood biomarker concentrations using quantitative trait locus (QTL) analysis. In COPDGene and SPIROMICS (Subpopulations and Intermediate Outcome Measures in COPD Study), genome-wide SNP data were combined with proteomic data to identify hundreds of novel SNPs that were associated with blood protein concentrations of quantitative trait loci (pQTLs) (60) (Figure 3). These pQTLs are distinct from expression quantitative trait loci (eQTLs) identified from the transcriptome of RNA in an appropriate tissue sample. SNPs shown to be associated at genome-wide levels with disease status become more biologically interesting if they are also associated with RNA and/or protein expression within cells of relevant tissues (because it suggests the SNP may be directly functional and not just a “tagging” marker correlated with some unobserved causal SNP). pQTLs are enriched for missense mutations, which alter the amino acid sequence of the resulting protein. Although many of the strongest pQTL SNPs were within or near the gene encoding their respective proteins, several genetic “hot spots” were found where pQTLs for multiple proteins were detected. For instance, the ABO locus on chromosome 9, which encodes a glycosyltransferase, was a significant pQTL for six different proteins, all of which were coded by genes on different chromosomes.
Figure 3.
Many novel genome-wide associations between SNPs and blood protein biomarkers were found. Combined Manhattan plots show SNPs associated with blood protein levels quantitative trait loci (pQTL) by chromosomal location for 38 biomarkers with at least one SNP significant at genome-wide significance after adjustment for multiple testing (red line). The −log10 P values are shown using results from a meta-analysis of both SPIROMICS and COPDGene SNPs. More than 300 novel pQTLs were identified. COPD = chronic obstructive pulmonary disease; COPDGene = Genetic Epidemiology of COPD; SPIROMICS = Subpopulations and Intermediate Outcome Measures in COPD Study. Reprinted by permission from Reference 60.
Additional Investigations after Genetic Association Analysis
The discovery of genomic regions associated with COPD and COPD-related phenotypes needs to be followed by efforts to identify the functional variant(s) and gene(s) in these regions and mechanistic studies to determine how those variants and the genes that they influence impact COPD susceptibility and heterogeneity. A substantial amount of work has leveraged the genetic findings in COPDGene to help identify enriched cell and tissue types as well as potentially causal variants and genes (18). Genes identified with support from both COPD GWAS association evidence in COPDGene and murine emphysema models include FAM13A (61), HHIP (62), IREB2 (63), MMP12 (64), MMP1 (65), SFTPD (66), and AGER (67).
Although the impact of each common variant consistently and significantly associated with risk for COPD is modest, when combined into a genetic risk score reflecting the combined impact of multiple genetic risk factors, there is the potential for more clinically important findings. For example, Busch and colleagues showed that a genetic risk score based on seven COPD susceptibility SNPs conferred a 1.9% reduction in FEV1 percentage predicted for each risk allele in an independent study population (International COPD Genetics Network) (58). An expanded risk score using 97 variants associated with quantitative measures of lung function from COPD case–control studies (including COPDGene) was highly significant, with a 1-SD difference in risk score resulting in an odds ratio of 1.36 for COPD (19). Although these cumulative risk scores could be useful in identifying high-risk groups for clinical trials, their relevance for individual subjects remains uncertain.
Because complex diseases such as COPD are likely influenced by a network of interacting genes and proteins, efforts to identify the biologically relevant networks for COPD susceptibility have been pursued. McDonald and colleagues built a series of disease network modules within the protein–protein interaction network based on genetic association evidence in COPDGene and GenKOLS using the dmGWAS software (https://bioinfo.uth.edu/dmGWAS/); they identified genes that were involved in several novel biological pathways, including the IL-7 pathway, as influencing risk for COPD (68).
Advances in Statistical Genetics
The statistical analysis of genetic data for COPD studies is challenging because of the complex genetic architecture of COPD (i.e., multiple genes contributing to multiple correlated phenotypes) and the inclusion of skewed, nonnormally distributed traits such as lung function and/or cigarette smoking intensity. To deal with these challenges, multiple statistical genetic analysis approaches were developed and applied to COPDGene datasets.
To incorporate information from genetic and phenotypic sources, several statistical approaches were developed to jointly use this information. The multimarker association test by Qiao and colleagues exploited the polygenic architecture of COPD by testing multiple SNPs jointly for association with the outcome of interest (69). Zhou and colleagues developed an approach combining multiple phenotypes into a single-variant association test (70). Lutz and colleagues developed an approach and corresponding R package to test for pleiotropy (i.e., a SNP that is jointly associated with multiple phenotypes) (71). In addition, new methods to identify genetic markers related to imaging features have been developed by Batmanghelich and colleagues (72, 73).
For COPDGene genetic analysis, methods have also been developed for the adjustment of genetic heterogeneity and population substructure in GWAS and whole-genome sequencing studies. Prokopenko and colleagues developed new approaches based on the Jaccard index that are particularly well suited to detecting local/recent substructure captured in rare variant data from whole genome sequencing (74). Fier and colleagues proposed clustering approaches for the region-based analysis of rare variant data (75). Llinares-López and colleagues developed a new method to discover genetic heterogeneity and applied this approach to the COPDGene study (76).
Statistical methods for genetic association studies were also developed to account for issues relating to challenging phenotypic distributions and ascertainment bias. Lutz and colleagues derived the nonparametric population-based association test approach for the association of a SNP with the outcome of interest that is robust against phenotypic misspecifications (77). This method is implemented as an interactive software package for genetic association analyses in population-based studies using generalized estimating equations (sites.google.com/site/genenpbat/home/npbat). Lutz and colleagues also developed an approach to adjust for ascertainment bias when testing for genetic associations of secondary phenotypes in case–control studies (78).
Mediation analysis is an important tool for discriminating between direct and indirect effects of genetic markers on risk for COPD. To examine how a SNP affects an outcome of interest through a mediator, Siedlinski and colleagues used mediation analysis to examine how SNPs on chromosome 15q25 act on FEV1 through cigarette smoking (79). Because mediation analysis assumes no unmeasured confounding of the exposure–mediator–outcome relationship and most sensitivity analyses focus on measured confounding of the mediator–outcome relationship (80), Lutz and colleagues developed the R package Umediation to analyze how results of mediation analysis would change in the presence of unmeasured confounding of the exposure–mediator–outcome relationship (81).
The COPDGene study, as a large, well-phenotyped and genotyped resource, has been important for other studies requiring adult control subjects and for method development by external investigators. For example, the recent upgrade of the popular software package plink2 uses a highly efficient algorithm for logistic regression developed using COPDGene data (82).
Conclusions and Future Directions
Substantial progress in understanding of the underlying genes influencing risk for COPD has been made in the past decade, and COPDGene has played a major role in these advances. Although understanding of COPD genetics remains incomplete, collaborative studies involving COPDGene and other large, well-phenotyped study populations (e.g., ECLIPSE and SPIROMICS) will be essential to provide greater insight into the genetic architecture of COPD. For example, after this review article was drafted, a larger collaborative GWAS using the ICGC and UK Biobank identified 82 genome-wide significant loci for COPD (83). In addition, studies that include younger subjects than those in COPDGene will be necessary to differentiate the impact of genetic determinants on reduced maximally attained lung function versus accelerated lung function decline on the development of COPD. Furthermore, ongoing research using novel machine learning and deep learning methods to define clinically relevant subtypes of COPD may ultimately allow for targeted therapy and prognostication of clinical course. With the addition of longitudinal phenotypes and whole-genome sequencing, especially when combined with proteomic, transcriptomic, epigenetic, and metabolomic data, COPDGene will likely provide important new information regarding the biological pathways controlling COPD risk, phenotypic heterogeneity, and disease progression that may allow more accurate diagnosis and prognosis, as well as new targets for treatment intervention.
Supplementary Material
Footnotes
The COPDGene project described was supported by awards U01 HL089897 and U01 HL089856 from the NHLBI. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NHLBI or the NIH. The COPDGene project is also supported by the COPD Foundation through contributions made to an industry advisory board comprised of AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Novartis, Pfizer, Siemens, and Sunovion.
Author Contributions: Conceptualization of work: all authors. Acquisition, analysis, and interpretation: all authors. Manuscript drafting: M.F.R., C.J.B., S.M.L., R.P.B., and E.K.S. Revision and approval of the manuscript: all authors.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201808-1455SO on March 25, 2019
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Castaldi PJ, Cho MH, Cohn M, Langerman F, Moran S, Tarragona N, et al. The COPD genetic association compendium: a comprehensive online database of COPD genetic associations. Hum Mol Genet. 2010;19:526–534. doi: 10.1093/hmg/ddp519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pillai SG, Ge D, Zhu G, Kong X, Shianna KV, Need AC, et al. ICGN Investigators. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilk JB, Chen TH, Gottlieb DJ, Walter RE, Nagle MW, Brandler BJ, et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 2009;5:e1000429. doi: 10.1371/journal.pgen.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho MH, Boutaoui N, Klanderman BJ, Sylvia JS, Ziniti JP, Hersh CP, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet. 2010;42:200–202. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hancock DB, Eijgelsheim M, Wilk JB, Gharib SA, Loehr LR, Marciante KD, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet. 2010;42:45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Repapi E, Sayers I, Wain LV, Burton PR, Johnson T, Obeidat M, et al. Wellcome Trust Case Control Consortium; NSHD Respiratory Study Team. Genome-wide association study identifies five loci associated with lung function. Nat Genet. 2010;42:36–44. doi: 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho MH, Castaldi PJ, Wan ES, Siedlinski M, Hersh CP, Demeo DL, et al. ICGN Investigators; ECLIPSE Investigators; COPDGene Investigators. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum Mol Genet. 2012;21:947–957. doi: 10.1093/hmg/ddr524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, et al. ENGAGE Consortium. Sequence variants at CHRNB3–CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silverman EK, Chapman HA, Drazen JM, Weiss ST, Rosner B, Campbell EJ, et al. Genetic epidemiology of severe, early-onset chronic obstructive pulmonary disease: risk to relatives for airflow obstruction and chronic bronchitis. Am J Respir Crit Care Med. 1998;157:1770–1778. doi: 10.1164/ajrccm.157.6.9706014. [DOI] [PubMed] [Google Scholar]
- 11.McCloskey SC, Patel BD, Hinchliffe SJ, Reid ED, Wareham NJ, Lomas DA. Siblings of patients with severe chronic obstructive pulmonary disease have a significant risk of airflow obstruction. Am J Respir Crit Care Med. 2001;164:1419–1424. doi: 10.1164/ajrccm.164.8.2105002. [DOI] [PubMed] [Google Scholar]
- 12.Zhou JJ, Cho MH, Castaldi PJ, Hersh CP, Silverman EK, Laird NM. Heritability of chronic obstructive pulmonary disease and related phenotypes in smokers. Am J Respir Crit Care Med. 2013;188:941–947. doi: 10.1164/rccm.201302-0263OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho MH, McDonald ML, Zhou X, Mattheisen M, Castaldi PJ, Hersh CP, et al. NETT Genetics, ICGN, ECLIPSE and COPDGene Investigators. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir Med. 2014;2:214–225. doi: 10.1016/S2213-2600(14)70002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hobbs BD, Parker MM, Chen H, Lao T, Hardin M, Qiao D, et al. NETT Genetics Investigators; ECLIPSE Investigators; COPDGene Investigators; International COPD Genetics Network Investigators. Exome array analysis identifies a common variant in IL27 associated with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2016;194:48–57. doi: 10.1164/rccm.201510-2053OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imielinski M, Baldassano RN, Griffiths A, Russell RK, Annese V, Dubinsky M, et al. Western Regional Alliance for Pediatric IBD; International IBD Genetics Consortium; NIDDK IBD Genetics Consortium; Belgian-French IBD Consortium; Wellcome Trust Case Control Consortium. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat Genet. 2009;41:1335–1340. doi: 10.1038/ng.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, et al. Type 1 Diabetes Genetics Consortium. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silverman EK, Vestbo J, Agusti A, Anderson W, Bakke PS, Barnes KC, et al. Opportunities and challenges in the genetics of COPD 2010: an International COPD Genetics Conference report. COPD. 2011;8:121–135. doi: 10.3109/15412555.2011.558864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hobbs BD, de Jong K, Lamontagne M, Bosse Y, Shrine N, Artigas MS, et al. COPDGene Investigators; ECLIPSE Investigators; LifeLines Investigators; SPIROMICS Research Group; International COPD Genetics Network Investigators; UK BiLEVE Investigators; International COPD Genetics Consortium. Genetic loci associated with chronic obstructive pulmonary disease overlap with loci for lung function and pulmonary fibrosis. Nat Genet. 2017;49:426–432. doi: 10.1038/ng.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wain LV, Shrine N, Artigas MS, Erzurumluoglu AM, Noyvert B, Bossini-Castillo L, et al. Understanding Society Scientific Group; Geisinger-Regeneron DiscovEHR Collaboration. Genome-wide association analyses for lung function and chronic obstructive pulmonary disease identify new loci and potential druggable targets. Nat Genet. 2017;49:416–425. doi: 10.1038/ng.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soler Artigas M, Wain LV, Miller S, Kheirallah AK, Huffman JE, Ntalla I, et al. UK BiLEVE. Sixteen new lung function signals identified through 1000 Genomes Project reference panel imputation. Nat Commun. 2015;6:8658. doi: 10.1038/ncomms9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wain LV, Shrine N, Miller S, Jackson VE, Ntalla I, Soler Artigas M, et al. Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK Biobank. Lancet Respir Med. 2015;3:769–781. doi: 10.1016/S2213-2600(15)00283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castaldi PJ, Cho MH, Litonjua AA, Bakke P, Gulsvik A, Lomas DA, et al. The association of genome-wide significant spirometric loci with chronic obstructive pulmonary disease susceptibility. Am J Respir Cell Mol Biol. 2011;45:1147–1153. doi: 10.1165/rcmb.2011-0055OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lutz SM, Cho MH, Young K, Hersh CP, Castaldi PJ, McDonald ML, et al. A genome-wide association study identifies risk loci for spirometric measures among smokers of European and African ancestry. BMC Genet. 2015;16:138. doi: 10.1186/s12863-015-0299-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker MM, Foreman MG, Abel HJ, Mathias RA, Hetmanski JB, Crapo JD, et al. Admixture mapping identifies a quantitative trait locus associated with FEV1/FVC in the COPDGene Study. Genet Epidemiol. 2014;38:652–659. doi: 10.1002/gepi.21847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manichaikul A, Hoffman EA, Smolonska J, Gao W, Cho MH, Baumhauer H, et al. Genome-wide study of percent emphysema on computed tomography in the general population: the Multi-Ethnic Study of Atherosclerosis Lung/SNP Health Association Resource Study. Am J Respir Crit Care Med. 2014;189:408–418. doi: 10.1164/rccm.201306-1061OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong X, Cho MH, Anderson W, Coxson HO, Muller N, Washko G, et al. Genome-wide association study identifies BICD1 as a susceptibility gene for emphysema. Am J Respir Crit Care Med. 2011;183:43–49. doi: 10.1164/rccm.201004-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho MH, Castaldi PJ, Hersh CP, Hobbs BD, Barr RG, Tal-Singer R, et al. A genome-wide association study of emphysema and airway quantitative imaging phenotypes. Am J Respir Crit Care Med. 2015;192:559–569. doi: 10.1164/rccm.201501-0148OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castaldi PJ, San Jose Estepar R, Mendoza CS, Hersh CP, Laird N, Crapo JD, et al. Distinct quantitative computed tomography emphysema patterns are associated with physiology and function in smokers. Am J Respir Crit Care Med. 2013;188:1083–1090. doi: 10.1164/rccm.201305-0873OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castaldi PJ, Cho MH, San Jose Estepar R, McDonald ML, Laird N, Beaty TH, et al. Genome-wide association identifies regulatory loci associated with distinct local histogram emphysema patterns. Am J Respir Crit Care Med. 2014;190:399–409. doi: 10.1164/rccm.201403-0569OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boueiz A, Lutz SM, Cho MH, Hersh CP, Bowler RP, Washko GR, et al. COPDGene and ECLIPSE Investigators. Genome-wide association study of the genetic determinants of emphysema distribution. Am J Respir Crit Care Med. 2017;195:757–771. doi: 10.1164/rccm.201605-0997OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halper-Stromberg E, Cho MH, Wilson C, Nevrekar D, Crapo JD, Washko G, et al. Visual assessment of chest computed tomographic images is independently useful for genetic association analysis in studies of chronic obstructive pulmonary disease. Ann Am Thorac Soc. 2017;14:33–40. doi: 10.1513/AnnalsATS.201606-427OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pillai SG, Kong X, Edwards LD, Cho MH, Anderson WH, Coxson HO, et al. ECLIPSE and ICGN Investigators. Loci identified by genome-wide association studies influence different disease-related phenotypes in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;182:1498–1505. doi: 10.1164/rccm.201002-0151OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wan ES, Cho MH, Boutaoui N, Klanderman BJ, Sylvia JS, Ziniti JP, et al. Evaluation of Chronic Obstructive Pulmonary Disease Longitudinally to Identify Predictive Surrogate End-Points (ECLIPSE); Norway-Bergen Cohort; National Emphysema Treatment Trial; COPD Gene Investigators. Genome-wide association analysis of body mass in chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2011;45:304–310. doi: 10.1165/rcmb.2010-0294OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDonald ML, Cho MH, Sørheim IC, Lutz SM, Castaldi PJ, Lomas DA, et al. Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints and COPDGene Investigators. Common genetic variants associated with resting oxygenation in chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2014;51:678–687. doi: 10.1165/rcmb.2014-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim V, Han MK, Vance GB, Make BJ, Newell JD, Hokanson JE, et al. COPDGene Investigators. The chronic bronchitic phenotype of COPD: an analysis of the COPDGene Study. Chest. 2011;140:626–633. doi: 10.1378/chest.10-2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim V, Zhao H, Boriek AM, Anzueto A, Soler X, Bhatt SP, et al. COPDGene Investigators. Persistent and newly developed chronic bronchitis are associated with worse outcomes in chronic obstructive pulmonary disease. Ann Am Thorac Soc. 2016;13:1016–1025. doi: 10.1513/AnnalsATS.201512-800OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee JH, Cho MH, Hersh CP, McDonald ML, Crapo JD, Bakke PS, et al. COPDGene and ECLIPSE Investigators. Genetic susceptibility for chronic bronchitis in chronic obstructive pulmonary disease. Respir Res. 2014;15:113. doi: 10.1186/s12931-014-0113-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wells JM, Washko GR, Han MK, Abbas N, Nath H, Mamary AJ, et al. COPDGene Investigators; ECLIPSE Study Investigators. Pulmonary arterial enlargement and acute exacerbations of COPD. N Engl J Med. 2012;367:913–921. doi: 10.1056/NEJMoa1203830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee JH, Cho MH, Hersh CP, McDonald ML, Wells JM, Dransfield MT, et al. COPDGene and ECLIPSE Investigators. IREB2 and GALC are associated with pulmonary artery enlargement in chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2015;52:365–376. doi: 10.1165/rcmb.2014-0210OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hayden LP, Hobbs BD, Cohen RT, Wise RA, Checkley W, Crapo JD, et al. COPDGene Investigators. Childhood pneumonia increases risk for chronic obstructive pulmonary disease: the COPDGene study. Respir Res. 2015;16:115. doi: 10.1186/s12931-015-0273-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hayden LP, Cho MH, McDonald MN, Crapo JD, Beaty TH, Silverman EK, et al. COPDGene Investigators. Susceptibility to childhood pneumonia: a genome-wide analysis. Am J Respir Cell Mol Biol. 2017;56:20–28. doi: 10.1165/rcmb.2016-0101OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rice JP, Hartz SM, Agrawal A, Almasy L, Bennett S, Breslau N, et al. GENEVA Consortium. CHRNB3 is more strongly associated with Fagerström test for cigarette dependence-based nicotine dependence than cigarettes per day: phenotype definition changes genome-wide association studies results. Addiction. 2012;107:2019–2028. doi: 10.1111/j.1360-0443.2012.03922.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim DK, Hersh CP, Washko GR, Hokanson JE, Lynch DA, Newell JD, et al. COPD Gene Investigators. Epidemiology, radiology, and genetics of nicotine dependence in COPD. Respir Res. 2011;12:9. doi: 10.1186/1465-9921-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hancock DB, Reginsson GW, Gaddis NC, Chen X, Saccone NL, Lutz SM, et al. Genome-wide meta-analysis reveals common splice site acceptor variant in CHRNA4 associated with nicotine dependence. Transl Psychiatry. 2015;5:e651. doi: 10.1038/tp.2015.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hardin M, Cho MH, McDonald ML, Wan E, Lomas DA, Coxson HO, et al. ECLIPSE and COPDGene Investigators; COPDGene Investigators—Clinical Centers. A genome-wide analysis of the response to inhaled β2-agonists in chronic obstructive pulmonary disease. Pharmacogenomics J. 2016;16:326–335. doi: 10.1038/tpj.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Begum F, Ruczinski I, Li S, Silverman EK, Cho MH, Lynch DA, et al. Identifying a deletion affecting total lung capacity among subjects in the COPDGene study cohort. Genet Epidemiol. 2016;40:81–88. doi: 10.1002/gepi.21943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Begum F, Ruczinski I, Hokanson JE, Lutz SM, Parker MM, Cho MH, et al. Hemizygous deletion on chromosome 3p26.1 is associated with heavy smoking among African American subjects in the COPDGene study. PLoS One. 2016;11:e0164134. doi: 10.1371/journal.pone.0164134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wan ES, Hokanson JE, Murphy JR, Regan EA, Make BJ, Lynch DA, et al. COPDGene Investigators. Clinical and radiographic predictors of GOLD-unclassified smokers in the COPDGene study. Am J Respir Crit Care Med. 2011;184:57–63. doi: 10.1164/rccm.201101-0021OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wan ES, Castaldi PJ, Cho MH, Hokanson JE, Regan EA, Make BJ, et al. COPDGene Investigators. Epidemiology, genetics, and subtyping of preserved ratio impaired spirometry (PRISm) in COPDGene. Respir Res. 2014;15:89. doi: 10.1186/s12931-014-0089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee JH, Cho MH, McDonald ML, Hersh CP, Castaldi PJ, Crapo JD, et al. COPDGene Investigators. Phenotypic and genetic heterogeneity among subjects with mild airflow obstruction in COPDGene. Respir Med. 2014;108:1469–1480. doi: 10.1016/j.rmed.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hardin M, Cho M, McDonald ML, Beaty T, Ramsdell J, Bhatt S, et al. The clinical and genetic features of COPD-asthma overlap syndrome. Eur Respir J. 2014;44:341–350. doi: 10.1183/09031936.00216013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hardin M, Cho MH, Sharma S, Glass K, Castaldi PJ, McDonald ML, et al. COPDGene and Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-Points Investigators. Sex-based genetic association study identifies CELSR1 as a possible chronic obstructive pulmonary disease risk locus among women. Am J Respir Cell Mol Biol. 2017;56:332–341. doi: 10.1165/rcmb.2016-0172OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Castaldi PJ, Dy J, Ross J, Chang Y, Washko GR, Curran-Everett D, et al. Cluster analysis in the COPDGene study identifies subtypes of smokers with distinct patterns of airway disease and emphysema. Thorax. 2014;69:415–422. doi: 10.1136/thoraxjnl-2013-203601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qiao D, Lange C, Beaty TH, Crapo JD, Barnes KC, Bamshad M, et al. Lung GO; NHLBI Exome Sequencing Project; COPDGene Investigators. Exome sequencing analysis in severe, early-onset chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2016;193:1353–1363. doi: 10.1164/rccm.201506-1223OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stanley SE, Chen JJ, Podlevsky JD, Alder JK, Hansel NN, Mathias RA, et al. Telomerase mutations in smokers with severe emphysema. J Clin Invest. 2015;125:563–570. doi: 10.1172/JCI78554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Foreman MG, Wilson C, DeMeo DL, Hersh CP, Beaty TH, Cho MH, et al. Genetic Epidemiology of COPD (COPDGene) Investigators. Alpha-1 antitrypsin PiMZ genotype is associated with chronic obstructive pulmonary disease in two racial groups. Ann Am Thorac Soc. 2017;14:1280–1287. doi: 10.1513/AnnalsATS.201611-838OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Busch R, Hobbs BD, Zhou J, Castaldi PJ, McGeachie MJ, Hardin ME, et al. National Emphysema Treatment Trial Genetics; Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-Points; International COPD Genetics Network; COPDGene Investigators. Genetic association and risk scores in a chronic obstructive pulmonary disease meta-analysis of 16,707 subjects. Am J Respir Cell Mol Biol. 2017;57:35–46. doi: 10.1165/rcmb.2016-0331OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Silverman EK. Risk of lung disease in PI MZ heterozygotes: current status and future research directions. Ann Am Thorac Soc. 2016;13(Suppl 4):S341–S345. doi: 10.1513/AnnalsATS.201507-437KV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun W, Kechris K, Jacobson S, Drummond MB, Hawkins GA, Yang J, et al. SPIROMICS Research Group; COPDGene Investigators. Common genetic polymorphisms influence blood biomarker measurements in COPD. PLoS Genet. 2016;12:e1006011. doi: 10.1371/journal.pgen.1006011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang Z, Lao T, Qiu W, Polverino F, Gupta K, Guo F, et al. A chronic obstructive pulmonary disease susceptibility gene, FAM13A, regulates protein stability of β-catenin. Am J Respir Crit Care Med. 2016;194:185–197. doi: 10.1164/rccm.201505-0999OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lao T, Glass K, Qiu W, Polverino F, Gupta K, Morrow J, et al. Haploinsufficiency of Hedgehog interacting protein causes increased emphysema induced by cigarette smoke through network rewiring. Genome Med. 2015;7:12. doi: 10.1186/s13073-015-0137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cloonan SM, Glass K, Laucho-Contreras M, Bhashyam AR, Cervo M, Pabon MA, et al. Mitochondrial iron chelation ameliorates cigarette smoke–induced bronchitis and emphysema in mice. Nat Med. 2016;22:163–174. doi: 10.1038/nm.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hautamaki RD, Kobayashi DK, Senior RM, Shapiro SD. Requirement for macrophage elastase for cigarette smoke-induced emphysema in mice. Science. 1997;277:2002–2004. doi: 10.1126/science.277.5334.2002. [DOI] [PubMed] [Google Scholar]
- 65.D’Armiento J, Dalal SS, Okada Y, Berg RA, Chada K. Collagenase expression in the lungs of transgenic mice causes pulmonary emphysema. Cell. 1992;71:955–961. doi: 10.1016/0092-8674(92)90391-o. [DOI] [PubMed] [Google Scholar]
- 66.Wert SE, Yoshida M, LeVine AM, Ikegami M, Jones T, Ross GF, et al. Increased metalloproteinase activity, oxidant production, and emphysema in surfactant protein D gene-inactivated mice. Proc Natl Acad Sci USA. 2000;97:5972–5977. doi: 10.1073/pnas.100448997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sambamurthy N, Leme AS, Oury TD, Shapiro SD. The receptor for advanced glycation end products (RAGE) contributes to the progression of emphysema in mice. PLoS One. 2015;10:e0118979. doi: 10.1371/journal.pone.0118979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McDonald ML, Mattheisen M, Cho MH, Liu YY, Harshfield B, Hersh CP, et al. GenKOLS, COPDGene and ECLIPSE Study Investigators. Beyond GWAS in COPD: probing the landscape between gene-set associations, genome-wide associations and protein-protein interaction networks. Hum Hered. 2014;78:131–139. doi: 10.1159/000365589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qiao D, Cho MH, Fier H, Bakke PS, Gulsvik A, Silverman EK, et al. On the simultaneous association analysis of large genomic regions: a massive multi-locus association test. Bioinformatics. 2014;30:157–164. doi: 10.1093/bioinformatics/btt654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou JJ, Cho MH, Lange C, Lutz S, Silverman EK, Laird NM. Integrating multiple correlated phenotypes for genetic association analysis by maximizing heritability. Hum Hered. 2015;79:93–104. doi: 10.1159/000381641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lutz SM, Fingerlin TE, Hokanson JE, Lange C. A general approach to testing for pleiotropy with rare and common variants. Genet Epidemiol. 2017;41:163–170. doi: 10.1002/gepi.22011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Batmanghelich KN, Cho M, Jose RS, Golland P. Spherical topic models for imaging phenotype discovery in genetic studies. Bayesian Graph Models Biomed Imaging (2014) 2014;8677:107–117. doi: 10.1007/978-3-319-12289-2_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Batmanghelich NK, Saeedi A, Cho M, Estepar RS, Golland P. Generative method to discover genetically driven image biomarkers. Inf Process Med Imaging. 2015;24:30–42. doi: 10.1007/978-3-319-19992-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prokopenko D, Hecker J, Silverman EK, Pagano M, Nöthen MM, Dina C, et al. Utilizing the Jaccard index to reveal population stratification in sequencing data: a simulation study and an application to the 1000 Genomes Project. Bioinformatics. 2016;32:1366–1372. doi: 10.1093/bioinformatics/btv752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Loehlein Fier H, Prokopenko D, Hecker J, Cho MH, Silverman EK, Weiss ST, et al. On the association analysis of genome-sequencing data: a spatial clustering approach for partitioning the entire genome into nonoverlapping windows. Genet Epidemiol. 2017;41:332–340. doi: 10.1002/gepi.22040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Llinares-López F, Papaxanthos L, Bodenham D, Roqueiro D, Borgwardt K COPDGene Investigators. Genome-wide genetic heterogeneity discovery with categorical covariates. Bioinformatics. 2017;33:1820–1828. doi: 10.1093/bioinformatics/btx071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lutz S, Yip WK, Hokanson J, Laird N, Lange C. A general semi-parametric approach to the analysis of genetic association studies in population-based designs. BMC Genet. 2013;14:13. doi: 10.1186/1471-2156-14-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lutz SM, Hokanson JE, Lange C. An alternative hypothesis testing strategy for secondary phenotype data in case-control genetic association studies. Front Genet. 2014;5:188. doi: 10.3389/fgene.2014.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Siedlinski M, Tingley D, Lipman PJ, Cho MH, Litonjua AA, Sparrow D, et al. COPDGene and ECLIPSE Investigators. Dissecting direct and indirect genetic effects on chronic obstructive pulmonary disease (COPD) susceptibility. Hum Genet. 2013;132:431–441. doi: 10.1007/s00439-012-1262-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lutz SM, Hokanson JE. Genetic influences on smoking and clinical disease: understanding behavioral and biological pathways with mediation analysis. Ann Am Thorac Soc. 2014;11:1082–1083. doi: 10.1513/AnnalsATS.201407-315ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lutz SM, Thwing A, Schmiege S, Kroehl M, Baker CD, Starling AP, et al. Examining the role of unmeasured confounding in mediation analysis with genetic and genomic applications. BMC Bioinformatics. 2017;18:344. doi: 10.1186/s12859-017-1749-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hill A, Loh PR, Bharadwaj RB, Pons P, Shang J, Guinan E, et al. Stepwise distributed open innovation contests for software development: acceleration of genome-wide association analysis. Gigascience. 2017;6:1–10. doi: 10.1093/gigascience/gix009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sakornsakolpat P, Prokopenko D, Lamontagne M, Reeve N, Guyatt A, Jackson V, et al. SpiroMeta Consortium; International COPD Genetics Consortium. Genetic landscape of chronic obstructive pulmonary disease identifies heterogeneous cell-type and phenotype associations. Nat Genet. 2019;51:494–505. doi: 10.1038/s41588-018-0342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.