Abstract
A clonal line of Camellia taliensis, ‘Taliensis-akeme’ has a recessive caffeine-less gene. To accelerate breeding of caffeine-less tea cultivars using this gene, DNA markers are indispensable for selecting heterozygotes that do not show a caffeine-less phenotype as parental lines. Therefore, we tried to determine the sequence of the six tea caffeine synthase (TCS) genes to search for polymorphisms and to prepare one of the TCS genes as a selection marker. Six TCS genes and the caffeine-less trait were mapped on the reference linkage map of tea. Strong linkage between the caffeine-less phenotype and TCS1 indicate that it is a promising candidate as a causative gene of the caffeine-less trait. We decided to use a three-nucleotide insertion in TCS1 that can be distinguished by sequencing as a selection marker named ‘CafLess-TCS1’. Caffeine-less individuals appeared in the progeny population of caffeine-less heterozygous individuals selected using ‘CafLess-TCS1’. These results confirmed that the developed ‘CafLess-TCS1’ will be an effective selection marker for breeding of caffeine-less tea cultivars.
Keywords: caffeine, caffeine-less, Camellia sinensis, DNA marker, tea
Introduction
Caffeine is a methylxanthine found in tea plants (Camellia sinensis (L.) O. Kuntze), coffee trees (Coffea arabica) and in other plants (Kuribara 2016). It is an important ingredient of tea as a beverage. The stimulatory effects of caffeine on the brain neocortex and the limbic system (central nervous system stimulant action) increases ability to think and brain activity, and its stimulatory effect on the heart (cardiac action) mildly increases the cardiac rate and so increases heart contraction (Loke 1988, Smits et al. 1985). Furthermore, caffeine has a relaxation effect on vascular smooth muscle, increases circulating blood flow rate in many organs, increases skeletal muscle contraction force and fatigue resistance and, as a result, leads to maintenance of exercise ability (Hibino et al. 1997, Tazzeo et al. 2012). However, excessive caffeine intake has various negative effects including palpitations and insomnia. Side-effects of caffeine are marked in children and the elderly, and there are also adults who are highly susceptible, due to genotype of the genes for enzymes related to caffeine metabolism (Kuribara 2016). To expand the consumption of green tea for people susceptible to caffeine, it is necessary to make low-caffeine green tea. Industrial methods for removing caffeine (P2014-140351A, P2014-140348A and P2013-121335A) are costly and of low quality. To produce green tea by existing methods that are beneficial to producers and customers, it is necessary to breed naturally caffeine-less cultivars.
Our research group has already started breeding caffeine-less tea cultivars. As breeding material for caffeine-less tea cultivars, we detected a caffeine-less line of C. taliensis, a wild relative of tea, which contains a large amount of the caffeine precursor theobromine. Genetic analysis showed that the caffeine-less trait might be controlled by one recessive locus (Ogino et al. 2009). Since no other breeding materials are available for caffeine-less tea breeding in Japan, we backcrossed ‘Taliensis-akame’ with several elite tea cultivars in order to improve agronomic traits, e.g. tea quality, tree shape and cold tolerance. We need to select heterozygotes for the caffeine-less locus during backcross breeding, but these are not phenotypically distinguishable. Therefore, we require a DNA marker to detect the caffeine-less gene and so accelerate breeding.
We have developed simple sequence repeat (SSR) markers and constructed a high-density reference linkage map for tea (Taniguchi et al. 2012a, 2012b).
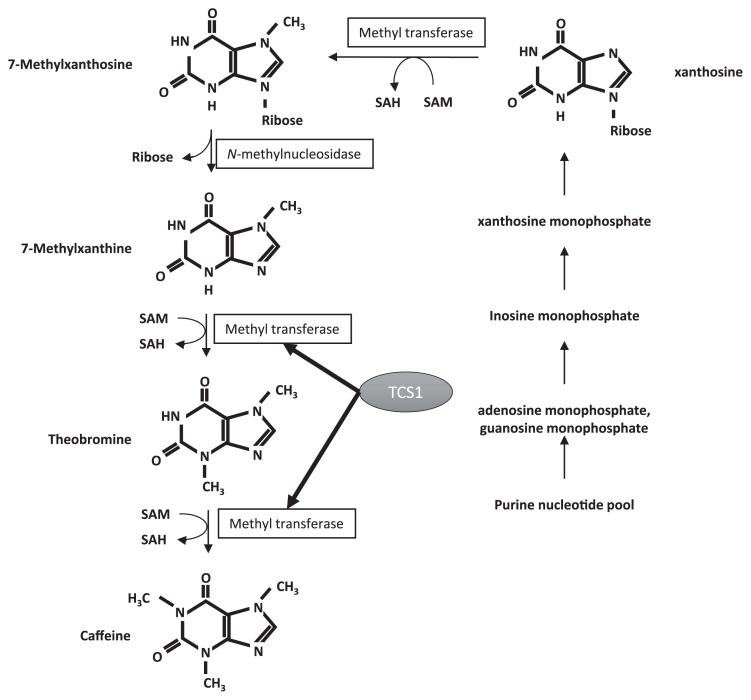
Furthermore, research on caffeine biosynthesis of tea has advanced since the 1990s, providing much information. In tea plants, caffeine is synthesized in chloroplasts of new shoots (Kato et al. 1998). The caffeine biosynthesis pathway is described in Fig. 1. Theobromine is the immediate precursor of caffeine (Kato et al. 1996). This latter two-step methylation is catalyzed by the enzyme tea caffeine synthase (TCS) which has been isolated (Kato et al. 1999) (Fig. 1).
Fig. 1.
Caffeine biosynthesis pathway. Caffeine is synthesized in young leaf from purine nucleotide. Latter two-step methylation is catalyzed by the enzyme tea caffeine synthase (TCS). SAM: S-adenosyl-methionine SAH: S-adenosyl-homocysteine.
The cDNA sequences of TCS1 and TCS2, the genes encoding the enzyme and its homolog were cloned (Kato et al. 2000). The functions of TCS1 and TCS2 have been reported—in vitro experiments using recombinant protein demonstrated that TCS1 encoded the methyltransferase which catalyzes the reactions of theobromine and caffeine synthesis but TCS2 did not encode methyltransferase. Equivalents of both TCS1 and TCS2 are also present in other Camellia spp.: the equivalent to TCS1 encodes theobromine synthase, but the equivalent to TCS2 does not (Yoneyama et al. 2006). In coffee tree, there are multiple synthetic pathways for caffeine, with multiple enzymes involved (Mizuno et al. 2003). There are no reports on multiple caffeine biosynthesis pathways in tea plants, and possible presence of genes other than TCS1 and TCS2 is unknown. However, in 2012 the genomic sequences of TCS1–TCS6 were registered in GenBank (Accession Nos. JX647690–JX647695) by a Chinese research group, and low expression levels of TCS3–TCS6 were indicated in new shoots (Jin et al. 2016). These six TCS genes, TCS1–TCS6, are all similar, with homology ranging within 88.08–93.91%. Their evolutionary relationships are shown in Supplemental Fig. 1. These findings indicate that the six TCS genes are candidates for the caffeine-less causative gene, but this has not been clarified. In this study, we conducted a detailed analysis to narrow down the leading candidates for the caffeine-less causative gene and caffeine-less traits and also developed a caffeine-less selection marker.
Materials and Methods
Plant materials and DNA extraction
Details of the plants used in this study are summarized in Table 1. ‘Taliensis-akame’ (C. taliensis), ‘Cha Chukanbohon Nou 6’, ‘Makura F1-95180’ and ‘Yabukita’ were used in the search for polymorphism of TCS genes. ‘Taliensis-akame’ is the caffeine-less wild relative of tea, and the others contain caffeine. Although the seed parent of ‘Cha Chukanbohon Nou 6’ and ‘Makura F1-95180’ is ‘Taliensis-akame’, their pollen parents are unknown. It is clear they are heterozygous for the caffeine-less locus (Ogino et al. 2009). ‘Yabukita’ is a leading cultivar for green tea production in Japan and was used as the standard of tea lines. A mapping population derived from ‘Cha Chukanbohon Nou 6’ and ‘Makura F1-95180’ consisting of 101 individuals was used for linkage analysis. ‘Makura Ko 03-1342’, ‘Yacha Ko10-1269’ and ‘Yacha Ko 11-4027’ are heterozygous lines derived from ‘Cha Chukanbohon Nou 6’, and their progenies were used for confirming the segregation ratio of the generated marker. We used 78 Japanese tea cultivars (Supplemental Table 1) to confirm the usefulness of the generated marker, CafLess-TCS1. The DNA of these plant materials was extracted from leaves using a Plant Genomic DNA Extraction Mini Kit (Favorgen, Tokyo, Japan).
Table 1.
List of plants used in this study
| Cultivar or line name | Pedigree | Purpose of use | Caffeine content | CafLess-TCS1 genotype |
|---|---|---|---|---|
| Taliensis-akame | Unknown | Search for polymorphism of TCS genes | Caffeine-less | Taliensis-type homozygote |
| Cha Chukanbohon Nou 6 | Taliensis-akame’ natural cross | Search for polymorphism of TCS genes; create a mapping population | Caffeine-containing | Heterozygote |
| Makura F1-95180 | Taliensis-akame’ natural cross | Search for polymorphism of TCS genes; create a mapping population | Caffeine-containing | Heterozygote |
| Yabukita | Unknown | Search for polymorphism of TCS genes | Caffeine-containing | Tea-type homozygote |
|
| ||||
| Makura Ko 03-1342 | Cha Chukanbohon Nou 6’ natural cross | Confirm usefulness of generated marker | Caffeine-containing | Heterozygote |
| Yacha Ko 10-1267 | Cha Chukanbohon Nou 6 × Makura Kei 54-13 (Marishi × Yumekaori) | Confirm usefulness of generated marker | Caffeine-containing | Heterozygote |
| Yacha Ko 11-4027 | Yachaken 03 {Yumekaori × Makura F1-102882 (Yutakamidori × Yamanoibuki)} × Makura Ko 03-1342 | Confirm usefulness of generated marker | Caffeine-containing | Heterozygote |
Yutakamidori, Marishi, Yumekaori and Yamanoibuki are green tea cultivars containing caffeine.
Analysis of caffeine content and determining the caffeine-less trait
Caffeine and theobromine contents of individuals were analyzed according to Ogino et al. (2009). We classified individuals with caffeine content of 0.2% dry matter weight or less as caffeine-less. Other individuals were classified as normal caffeine-containing tea.
Detecting polymorphism of TCS genes
We designed primers for amplification of TCS genes by Primer3web (http://primer3.ut.ee/) using the mutual different sequence regions among six genes. The DNA fragments of each gene were amplified by PCR using the primers listed in Table 2. The PCR condition for TCS1 was the same as for the detection of the tea SSR markers except that the extension time was 2 min (Taniguchi et al. 2012b). Platinum Taq DNA Polymerase High Fidelity (Thermo Fisher Scientific Inc., Tokyo, Japan) was used for amplification of TCS2–TCS6; and the reaction solution composition and PCR temperature conditions were set according to the corresponding manual. The reaction solution was 25 μL total volume containing 1 unit/μL Platinum Taq DNA Polymerase High Fidelity, the attached reaction buffer, 2.0 mM MgCl2, 0.2 mM of each dNTP, 2 ng/μL template DNA and 2 μM of each primer DNA. The PCR program was as follows: 30 s at 94°C to completely denature the DNA; followed by 35 cycles of 15 s at 94°C, 30 s at 55°C and 90 s or 4 min at 68°C; and held at 4°C. Extension time was 90 s for TCS5 and 4 min for the four other TCS genes. Amplified fragments of TCS genes were sequenced directly or purified using EASYTRAP Ver. 2 (Takara Bio Inc., Shiga, Japan), then ligated into pGEM T-Easy Vector (Promega, Tokyo, Japan) and cloned into E. coli DH5α (Toyobo Co., Ltd., Osaka, Japan). Each clone was sequenced with T7 (5′-TAATACGACTCACTATAGGG-3′) or SP6 (5′-CAT ACGATTTAGGTGACACTATAG-3′) primer. Primers for direct sequencing were the same as used for amplifying DNA fragments. Cycle sequencing reactions were performed using a BigDye Terminator Cycle Sequencing Kit (Thermo Fisher Scientific Inc.) and capillary electrophoresis performed using an ABI 3130xl sequencer (Thermo Fisher Scientific Inc.). The determined nucleotide sequences were analyzed using DNA Sequence Analysis Software to distinguish those alleles derived from ‘Taliensis-akame’ from among the other alleles. Polymorphisms were detected for each TCS gene and were used as markers.
Table 2.
Primer information for TCS genes
| Gene | Forward primer (5′→3′) | Reverse primer (5′→3′) | Amplified region of the registered sequence | |
|---|---|---|---|---|
| TCS1 | ACAGCCAGCGCTAGAAAATG | CATGGCATGGTCAATGAAAA | 4509–5036 bp: part of 2nd exon and 2nd intron | |
|
| ||||
| TCS2 | 1st PCR | TTTGTTCAAATGAATCGAAATGA | CAAGTCCGCTGCGTTAAGAG | |
| 2nd PCR | AAATAATGAAATGTGTGAACCAAA | CACTGGCATTGTCATTGAGG | 3029–3884 bp: part of 1st intron and 2nd exon | |
|
| ||||
| TCS3 | CATGAACAGAGGAGAAGGAGAAA | ATAGGGTGCCAACGTGACAT | 425–1251 bp: part of putative 1st exon and 1st intron | |
|
| ||||
| TCS4 | AATGCTAGTGGATGGGTTGG | CATGGCATGGTCAATGAAAA | 3293–4230 bp: part of 1st intron, 2nd exon and part of 2nd intron | |
|
| ||||
| TCS5 | 1st PCR | CATGAACAGAGGAGAAGGAGAAA | CAACTGCATTTTCTAGCGCTGGC | |
| 2nd PCR | CATGAACAGAGGAGAAGGAGAAA | CCCAACCATCCACTAGCATT | 569–1419 bp: part of 1st exon and 1st intron | |
|
| ||||
| TCS6 | CATGAACAGAGGAGAAGGAGAAA | AGTTCCAGTGTTTGGCAATTC | 608–3248 bp: part of 1st exon, 1st intron and part of 2nd exon | |
GenBank Accession Nos of TCS1–TCS6 are JX647690–JX647695.
Genotyping of TCS genes in the F1 mapping population
We used the mapping population derived from ‘Cha Chukanbohon Nou 6’ and ‘Makura F1-95180’ to detect polymorphisms of TCS genes. Polymorphisms of TCS1, TCS2 and TCS4 [single nucleotide polymorphisms (SNPs) or indel polymorphisms] were detected by direct sequencing under the same conditions as for polymorphism detection. The segregating patterns for TCS3 and TCS5 were detected by Cleaved Amplified Polymorphic Sequence (CAPS). The CAPS markers were derived from two SNPs in TCS3 and TCS5. Restriction enzymes Cla I (TaKaRa Bio Inc.), Sty I (Nippon Gene Co., Ltd., Tokyo, Japan), and Sfc I and BsrD I (New England Biolabs Japan Inc., Tokyo, Japan) were used for polymorphism detection. The PCR products were treated overnight under the reaction conditions specified for each enzyme and electrophoresed on a 2% agarose gel (Nippon Gene Co., Ltd.) to detect the presence or absence of cleavage as polymorphisms. Polymorphism of TCS6 was examined using Sequence Characterized Amplified Region (SCAR), and electrophoresis on a 2% agarose gel because only one allele of ‘Taliensis-akame’ was amplified by the PCR conditions used.
Determining haplotypes of TCS genes
In order to map TCS genes using JoinMap 4.1 (Kyazma) in CP mode, the sequence polymorphisms which were heterozygous in one or both parent cultivars were selected. Most of them could discriminate only one or two alleles. Although single locus SNPs or indel polymorphisms can be used in linkage analysis, it was expected that haplotype information combining several SNPs or indel polymorphisms could discriminate three or four alleles of each marker locus for improved mapping accuracy. Therefore, we used the haplotype information deduced from the segregation data of the F1 mapping population of SNPs or indel polymorphisms as markers for linkage analysis of TCS genes. However, we could not find polymorphisms that provided haplotype information for TCS2 and TCS6, and so single polymorphism was used for mapping.
Linkage analysis of TCS genes, SSR markers and caffeine-less traits
The linkage of the caffeine-less trait, the six TCS gene loci and polymorphism of the SSR markers, investigated in the mapping population derived from ‘Cha Chukanbohon Nou 6’ and ‘Makura F1-95180’, were analyzed using JoinMap 4.1 software for construction of linkage maps. Genotyping of SSR markers was performed according to Taniguchi et al. (2012b). Information on the SSR markers used is listed in Supplemental Table 2. The SSR markers were used in two steps. We first selected two or three markers on each linkage group from the tea reference linkage map (Taniguchi et al. 2012b) and confirmed the linkage group on which the caffeine-less trait and TCS genes sat. Then we analyzed linkage in detail by increasing the number of markers in the linkage group in which the caffeine-less trait and each TCS gene were present.
Ascertaining marker usefulness
The usefulness of the marker was ascertained as follows. (1) Crossing using heterozygous individuals for caffeine-less trait was performed, and some populations were created in which caffeine-less heterozygous individuals might appear. (2) The genotype of the populations was determined with the developed marker, and this confirmed the separation ratio; simultaneously, caffeine-less heterozygotes were selected. (3) Crossing between individuals selected by the developed marker was performed, and some populations were created in which caffeine-less individuals might appear. (4) We investigated the caffeine and theobromine contents and genotypes of the populations and confirmed that caffeine-less individuals appeared. Furthermore, 78 Japanese tea cultivars were investigated to confirm whether the developed marker could be used for genotypes other than ‘Taliensis-akame’ and its progeny.
Results
Detected polymorphism of TCS genes
Indel polymorphisms, SNPs and SCAR were detected on the sequence of TCS1–TCS6 (Table 3). In analysis of the F1 mapping population between ‘Cha Chukanbohon Nou 6’ and ‘Makura F1-95180’, these polymorphisms were classified into three segregation types: hk × hk type, heterozygous locus in the both parents with two alleles; lm × ll type, heterozygous locus in the seed parent; and nn × np type, heterozygous locus in the pollen parent. In TCS1 and TCS3–TCS5, segregation data of two or three polymorphisms in the mapping population revealed that the four alleles of their loci all differed (Supplemental Table 3). This haplotype information from two or three polymorphisms could be used as one marker, and was heterozygous in both parents and had four alleles. However, in TCS2, due to the simultaneous amplification of TCS6, it was not possible to discriminate the four alleles. Therefore, we decided to use SNP 3774 for mapping of TCS2. In TCS6, due to finding only one segregation type, we decided to use SCAR.
Table 3.
Detected polymorphisms and segregation type on TCS1–TCS6
| Gene | Indel | SNP | SCAR | |||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| hk × hk | nn × np | hk × hk | lm × ll | nn × np | lm × ll | |
| TCS1 | 1 | 0 | 11 | 2 | 1 | 0 |
| TCS2 | 0 | 0 | 2 | 0 | 21 | 0 |
| TCS3 | 0 | 0 | 6 | 6 | 2 | 0 |
| TCS4 | 0 | 0 | 19 | 3 | 5 | 0 |
| TCS5 | 1 | 3 | 1 | 20 | 15 | 0 |
| TCS6 | 0 | 0 | 0 | 10 | 0 | 1 |
hk × hk: heterozygous in both parents with two alleles,
lm × ll: heterozygous in the seed parent,
nn × np: heterozygous in the pollen parent.
Mapping of caffeine-less trait and six TCS genes
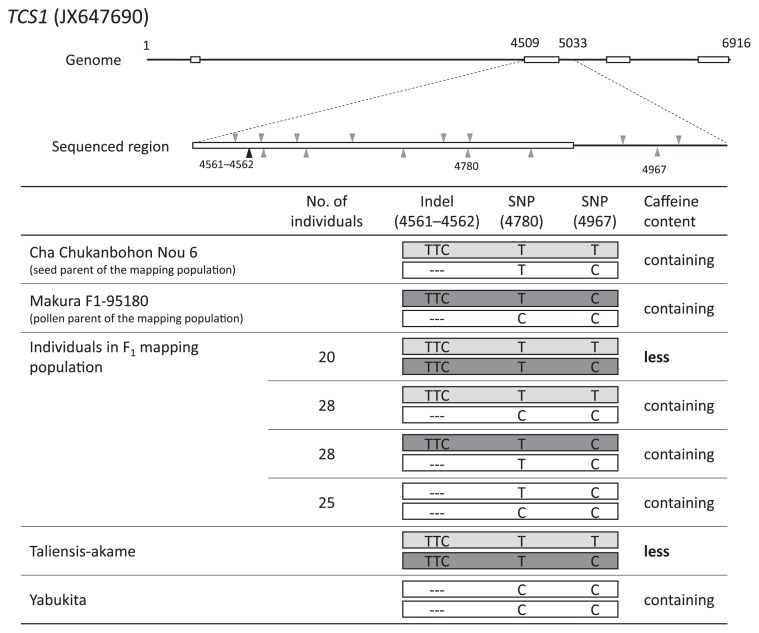
Two or three SSR markers were selected from each linkage group of the reference linkage map (core map) of tea, and the genotypes of the SSR markers were investigated in the mapping population derived from ‘Cha Chukanbohon Nou 6’ and ‘Makura F1-95180’. Then we analyzed caffeine and theobromine contents and TCS gene genotypes in the mapping population (Supplemental Table 3). The result of the first linkage analysis showed that the caffeine-less trait and six TCS genes were located on linkage group 3 (LG03). Next, we reanalyzed using more SSR markers located on LG03 with the same mapping population (Fig. 2). Genes TCS3 and TCS4 were mapped to positions distant from the caffeine-less trait and the other TCS genes. Genes TCS1, TCS2, TCS5 and TCS6 were mapped near the caffeine-less trait—TCS1 was closest to the caffeine-less trait, at 0.3 cM distance. Linkage analysis of the genotype of TCS1 and the caffeine-less phenotype in the mapping population showed that all caffeine-less individuals were homozygous with the alleles having a three nucleotide insertion of TTC (TTC-insertion) between bases 4561 and 4562, and the caffeine-containing individuals were of other genotypes (Supplemental Table 3). In addition, it was revealed that alleles with TTC-insertion were derived from ‘Taliensis-akame’ (Fig. 3). We considered that the TTC-insertion could be used as a selection marker for the caffeine-less trait.
Fig. 2.
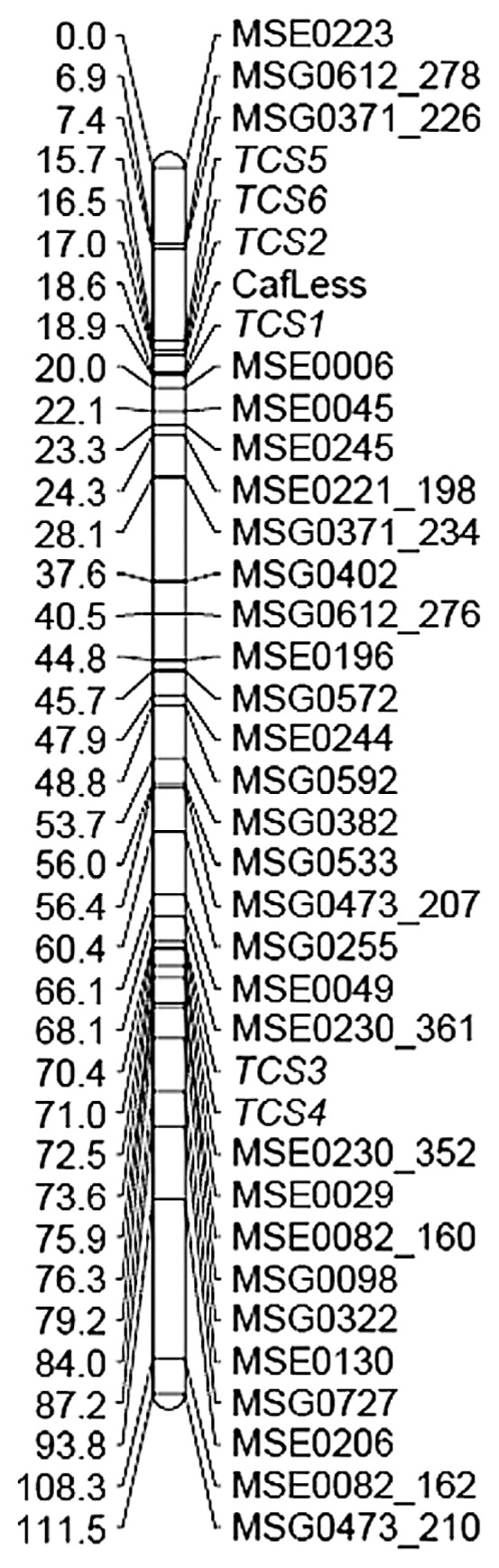
Mapping of caffeine-less trait and six TCS genes to LG03. ‘CafLess’ indicates caffeine-less trait.
Fig. 3.
Sequence region, position of polymorphisms and haplotypes of TCS1. White box indicates exon. Black triangles are indel polymorphisms, gray triangles are SNPs. Light and dark gray colored alleles in haplotypes indicate that it is derived from ‘Taliensis-akame’.
Caffeine-less individuals obtained from crossing heterozygous individuals selected by TCS1 polymorphism
We determined whether caffeine-less individuals occurred among progeny of individuals selected using the developed TCS1 polymorphism, to confirm its usefulness. Some caffeine-less heterozygotes were used to cross with tea cultivars and the genotype of progeny populations was investigated using TTC-insertion of TCS1 to select the parental lines. Of the 1031 individual progeny derived from 30 crosses, 497 were heterozygous for caffeine-less, 467 were homozygous for tea-type and 67 individuals could not be determined due to poor growth and other reasons (data not shown). We selected several well-grown individuals from the heterozygous groups and carried out two crosses, using them to confirm that caffeine-less individuals appeared from the cross (Table 4). These crossings produced 127 seedlings—of these, 77 individuals, excluding those that died or grew poorly, were analyzed for component composition and genotype, and resulted in four individuals identified as caffeine-less.
Table 4.
Genotyping and component composition of progenies derived from two crossings using TCS1 heterozygous individuals as parents
| Crossings | Caffeine-less phenotype/TCS1 genotype | Total | |||
|---|---|---|---|---|---|
|
| |||||
| Caffeine-less/taliensis-type homozygote | Caffeine-containing/heterozygote | Caffeine-containing/tea-type homozygote | Unknown | ||
| Makura Ko 03-1342 × Yacha Ko 10-1267 | 2 | 22 | 13 | 21 | 58 |
| Makura Ko 03-1342 × Yacha Ko 11-4027 | 2 | 26 | 17 | 24 | 69 |
|
| |||||
| Total | 4 | 48 | 30 | 45 | 127 |
Bold: TCS1 heterozygote. ‘Unknown’ means individuals for which genotype and caffeine contents could not be determined, due to dying or growing poorly.
TCS1 genotyping of Japanese tea cultivars
We examined the genotype of TCS1 for 78 Japanese tea varieties and found that 75 cultivars did not have the TTC-insertion in alleles of TCS1 (Supplemental Table 1). ‘Fushun’, ‘Miyamakaori’ and ‘Harunonagori’ were excluded because we could not determine their sequences.
Discussion
It is most efficient to use the causative gene, or a sequence located nearby, of a target trait as a DNA marker for selection. Mapping of the caffeine-less trait and six TCS genes to the reference linkage map of SSR markers placed them all in the same linkage group (Fig. 2). Because of the significant distance from the caffeine-less trait, TCS3 and TCS4 were thought to be unrelated to the caffeine-less trait; however, TCS1, TCS2, TCS5 and TCS6 were located close to the caffeine-less trait. However, some caffeine-less individuals in the mapping population derived from ‘Cha Chukanbohon Nou 6’ and ‘Makura F1-95180’ were not of Taliensis-type homozygote for TCS2, TCS5 and TCS6. Thus, they were likely unrelated to the caffeine-less trait. For example, caffeine-less plant individual No. 84 showed Taliensis-type homozygote TCS1, heterozygote or tea-type homozygote TCS2, and tea-type homozygote TCS5 and TCS6. The gene TCS1 was located closest to the caffeine-less trait. Although caffeine-less individuals with tea-type TCS1 were not found in the mapping population (Supplemental Table 3), JoinMap calculated a distance of 0.3 cM between TCS1 and the caffeine-less trait. According to the software developer, the distances between co-dominant and dominant markers with the same segregation pattern are estimated larger than 0.0 cM in the JoinMap software, because the pairwise recombination frequency will slightly differ for the two markers. According to past reports, TCS1 of tea encodes caffeine synthase (Kato et al. 2000). Our results and those of past reports indicate that TCS1 derived from ‘Taliensis-akame’ is a candidate causative gene of the caffeine-less trait. Furthermore, most Japanese tea cultivars were TCS1 homozygotes without the TTC-insertion (Supplemental Table 1). Therefore, the TTC-insertion in the TCS1 fragment is considered to be an efficient selection marker for detecting caffeine-less heterozygous individuals within the caffeine-less tea breeding process. We named this polymorphism ‘CafLess-TCS1’ for use as a selection marker.
In coffee, multiple methyltransferase genes involved in caffeine synthesis have been found (Mizuno et al. 2003, Ogawa et al. 2001) and the presence of a pathway to synthesize caffeine via paraxanthine was also demonstrated (Uefuji et al. 2003). Our results indicated that TCS2–TCS6 of tea did not participate in the synthesis step from theobromine to caffeine and that caffeine synthesis in tea is a single pathway without a biosynthetic pathway via paraxanthine, unlike the multiple caffeine synthetic pathways of coffee. This result will be useful for future research on caffeine biosynthesis in plants.
Selection using the developed marker CafLess-TCS1 showed that heterozygous individuals and tea-type homozygous individuals segregated almost 1:1 in the ‘Taliensis-akame’ progeny population (data not shown). Therefore, the TCS1 gene was inherited according to a theoretical value, and it was confirmed that progeny could be selected using the marker. Use of CafLess-TCS1 showed some caffeine-less individuals among progeny of crossed heterozygous individuals. Thus, it was also shown that caffeine-less individuals were produced from crossing of caffeine-less heterozygous individuals selected using CafLess-TCS1 (Table 4). The CafLess-TCS1 developed in this study proved to be a highly effective selection marker for caffeine-less tea breeding.
In our mapping population derived from the crossing of ‘Cha Chukanbohon Nou 6’ and ‘Makura F1-95180’, the appearance ratio of caffeine-less and caffeine-containing individuals was near to the theoretical value (Supplemental Table 3). However, in crossing between caffeine-less heterozygous individuals carried out so far, the appearance ratio of caffeine-less individuals was extremely low. The appearance ratio of tea-type homozygous and heterozygous individuals was also far from the theoretical value. This may be a bias due to the small numbers of crossings and of analyzed individuals. During the crossing to create parental lines, when the number of seeds was very low (i.s. tens of seeds), the segregation ratio of heterozygous individuals and tea-type homozygous individuals often deviated from the theoretical ratio; however, when the number of seeds was several hundreds, it become almost 1:1 (data not shown). Furthermore, nearly half of the seedlings obtained by this crossing died during the 10 months in the nursery and ‘Taliensis-akame’ also grew poorly. The parental lines used to create the population in Table 4 were the F3 or F4 generation of ‘Taliensis-akame’, but these were of a sibling or parent–child relationship. Therefore, it is highly likely that these parental lines inherited many poor traits from ‘Taliensis-akame’ via ‘Cha Chukanbohon Nou 6’ and that they died as a result, suggesting that linkage drag or some other process occurred. If we can accelerate the generation advance of parental lines using CafLess-TCS1, we can create multiple parental lines with few of the poor traits of ‘Taliensis-akame’. Crossings between such parental lines will produce caffeine-less progeny with greatly improved agronomic traits.
Supplementary Information
Acknowledgments
The authors thank Mrs. K. Ogawa, Mrs. M. Iwata and Mrs. Y. Kuramae, assistants at the Makurazaki Tea Research Station of NARO. This research was supported by grants from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomic-based Technology for Agriculture Improvement, DHR) and the Project of the Bio-oriented Technology Research Advancement Institution, NARO (the special scheme project on advanced research and development for next-generation technology).
Literature Cited
- Hibino, G., Moritani, T., Kawada, T. and Fushiki, T. (1997) Caffeine enhances modulation of parasympathetic nerve activity in humans: quantification using power spectral analysis. J. Nutr. 127: 1422–1427. [DOI] [PubMed] [Google Scholar]
- Jin, J.-Q., Yao, M.-Z., Ma, C.-L., Ma, J.-Q. and Chen, L. (2016) Natural allelic variations of TCS1 play a crucial role in caffeine biosynthesis of tea plant and its related species. Plant Physiol. Biochem. 100: 18–26. [DOI] [PubMed] [Google Scholar]
- Kato, A., Crozier, A. and Ashihara, H. (1998) Subcellular localization of the N-methyltransferase involved in caffeine biosynthesis in tea. Phytochemistry 48: 777–779. [Google Scholar]
- Kato, M., Kanehara, T., Shimizu, H., Suzuki, T., Gillies, F.M., Crozier, A. and Ashihara, H. (1996) Caffeine biosynthesis in young leaves of Camellia sinensis: in vitro studies on N-methyltransferase activity involved in the conversion of xanthosine to caffeine. Physiol. Plant. 98: 629–636. [Google Scholar]
- Kato, M., Mizuno, K., Fujimura, T., Iwama, M., Irie, M., Crozier, A. and Ashihara, H. (1999) Purification and characterization of caffeine synthase from tea leaves. Plant Physiol. 120: 579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, M., Mizuno, K., Crozier, A., Fujimura, T. and Ashihara, H. (2000) Caffeine synthase gene from tea leaves. Nature 406: 956–957. [DOI] [PubMed] [Google Scholar]
- Kuribara, H. (2016) Caffeine intake in the daily life: mechanism of action and safety assessment. Bulletin of Tokyo Univ. and Grad. School of Social Welfare 6: 109–125. [Google Scholar]
- Loke, W.H. (1988) Effects of caffeine on mood and memory. Physiol. Behav. 44: 367–372. [DOI] [PubMed] [Google Scholar]
- Mizuno, K., Okuda, A., Kato, M., Yoneyama, N., Tanaka, H., Ashihara, H. and Fujimura, T. (2003) Isolation of a new dual-functional caffeine synthase gene encoding an enzyme for the conversion of 7-methylxanthine to caffeine from coffee (Coffea arabica L.). FEBS Lett. 534: 75–81. [DOI] [PubMed] [Google Scholar]
- Ogawa, M., Herai, Y., Koizumi, N., Kusano, T. and Sano, H. (2001) 7-Methylxanthine methyltransferase of coffee plants. J. Biol. Chem. 276: 8213–8218. [DOI] [PubMed] [Google Scholar]
- Ogino, A., Tanaka, J., Taniguchi, F., Yamamoto, M.P. and Yamada, K. (2009) Detection and characterization of caffeine-less tea plants originated from interspecific hybridization. Breed. Sci. 59: 277–283. [Google Scholar]
- Smits, P., Thien, T. and Van’t Laar, A. (1985) The cardiovascular effects of regular and decaffeinated coffee. Br. J. Clin. Pharmacol. 19: 852–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi, F., Fukuoka, H. and Tanaka, J. (2012a) Expressed sequence tags from organ-specific cDNA libraries of tea (Camellia sinensis) and polymorphisms and transferability of EST-SSRs across Camellia species. Breed. Sci. 62: 186–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi, F., Furukawa, K., Ota-Metoku, S., Yamaguchi, N., Ujihara, T., Kono, I., Fukuoka, H. and Tanaka, J. (2012b) Construction of a high-density reference linkage map of tea (Camellia sinensis). Breed. Sci. 62: 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazzeo, T., Bates, G., Roman, H.N., Lauzon, A.-M., Khasnis, M.D., Eto, M. and Janssen, L.J. (2012) Caffeine relaxes smooth muscle through actin depolymerization. Am. J. Physiol. Lung Cell. Mol. Physiol. 303: L334–L342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uefuji, H., Ogita, S., Yamaguchi, Y., Koizumi, N. and Sano, H. (2003) Molecular cloning and functional characterization of three distinct N-methyltransferases involved in the caffeine biosynthetic pathway in coffee plants. Plant Physiol. 132: 372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama, N., Morimoto, H., Ye, C.X., Ashihara, H., Mizuno, K. and Kato, M. (2006) Substrate specificity of N-methyltransferase involved in purine alkaloids synthesis is dependent upon one amino acid residue of the enzyme. Mol. Genet. Genomics 275: 125–135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.