Abstract
As glutamate dehydrogenases (GDHs) of microorganisms usually have higher affinity for NH4+ than do those of higher plants, it is expected that ectopic expression of these GDHs can improve nitrogen assimilation in higher plants. Here, a novel NADP(H)-GDH gene (TrGDH) was isolated from the fungus Trichurus and introduced into rice (Oryza sativa L.). Investigation of kinetic properties in vitro showed that, compared with the rice GDH (OsGDH4), TrGDH exhibited higher affinity for NH4+ (Km = 1.48 ± 0.11 mM). Measurements of the NH4+ assimilation rate demonstrated that the NADP(H)-GDH activities of TrGDH transgenic lines were significantly higher than those of the controls. Hydroponic experiments revealed that the fresh weight, dry weight and nitrogen content significantly increased in the TrGDH transgenic lines. Field trials further demonstrated that the number of effective panicles, 1,000-grain weight and grain weight per plant of the transgenic lines were significantly higher than those of the controls, especially under low-nitrogen levels. Moreover, glutelin and prolamine were found to be markedly increased in seeds from the transgenic rice plants. These results sufficiently confirm that overexpression of TrGDH in rice can improve the growth status and grain weight per plant by enhancing nitrogen assimilation. Thus, TrGDH is a promising candidate gene for maintaining yields in crop plants via genetic engineering.
Keywords: glutamate dehydrogenase, Trichurus, ammonium metabolism, rice (Oryza sativa L.)
Introduction
Because of the deficiency and low utilization efficiency of nitrogen in most cultivated soils, nitrogen has become one of the limiting dominant factors in agricultural production (Frink et al. 1999, Stulen et al. 1998). However, a strong dependence on fertilizers is not economically sustainable, and even worse, more than half of the nitrogen in fertilizers applied to crops is not actually taken up by crop plants, giving rise to serious environmental problems (Socolow 1999). Thus, there is considerable interest in cultivating low-nitrogen-dependent crop varieties that present stable yields (Giles 2005). Nitrogen for plants mainly originates from in-organic nitrides such as ammonium (NH4+) and nitrate (NO3−). For crop species such as rice (Oryza sativa L.), the main form of nitrogen available in irrigated paddy fields is NH4+. Two pathways involved in the assimilation of NH4+ in higher plants have been identified: one is the glutamine synthetase (GS)/glutamate synthase (GOGAT) pathway (Lea and Miflin 1974), and the other is the glutamate dehydrogenase (GDH) pathway (Skopelitis et al. 2007). In the GS/GOGAT pathway, GS is the first enzyme responsible for ammonium assimilation to produce glutamine, and its activity requires energy in the form of adenosine triphosphate (ATP). GOGAT then transfers the amide nitrogen of glutamine to 2-oxoglutarate (2-OG) to form two glutamate molecules (Hodges 2002). In the GDH pathway, glutamate dehydrogenase (GDH, EC 1.4.1.2 and EC 1.4.1.4) catalyzes the reversible amination of 2-OG with NH4+ to form glutamate, which requires NAD(P)H as a cofactor but not ATP (Wootton 1983). Therefore, ammonia assimilation via the GDH pathway is purported to be more energy efficient than the GS/GOGAT pathway is (Helling 1998, Windass et al. 1980).
Although the GS/GOGAT pathways in plants were discovered in the 1970s (Lea and Miflin 1974) and are considered to constitute the main ammonium assimilation pathway, the role of GDH in ammonium assimilation remains controversial (Abiko et al. 2010, Helling 1998). There are at least two distinct GDH enzymes: NAD(H)-dependent glutamate dehydrogenase [NAD(H)-GDH; EC 1.4.1.2] and NADP(H)-dependent glutamate dehydrogenase [NADP(H)-GDH; EC 1.4.1.4]. In comparison with NAD(H)-GDH, NADP(H)-GDH has received very little attention in plants, and its role in plant metabolism remains obscure. Recently, NADP(H)-GDHs from microorganisms have been attracting widespread attention because they have been identified to play important roles in carbon and nitrogen metabolism in microorganisms, especially in fungi. For example, a mutant of Aspergillus nidulans deficient in NADP(H)-GDH (gdhA) showed poor growth when provided with ammonium as the sole nitrogen source (Kinghorn and Pateman 1973, Macheda et al. 1999). In particular, NADP(H)-GDHs from microorganisms such as bacteria and fungi exhibit a higher affinity for NH4+ (Km = 0.2–4.5 mM; Du et al. 2014, Zhou et al. 2015b) than do those from higher plants (Km = 10–80 mM; Oaks 1994), suggesting that NADP(H)-GDHs from microorganisms assimilate ammonium more efficiently. To date, several NADP(H)-GDHs from lower organisms have been introduced into crop plants to improve ammonium assimilation (Abiko et al. 2010, Ameziane et al. 2000, Du et al. 2014, Noor and Punekar. 2005, Tang et al. 2018, Zhou et al. 2014, 2015a, 2015b). For example, when an NADP(H)-GDH gene (gdhA) from Aspergillus niger was introduced into rice, the transgenic lines exhibited increased nitrogen utilization efficiency, and their dry weight, nitrogen content and grain weight per plant increased significantly compared with those of nontransformed plants (Abiko et al. 2010). Similarly, ectopic expression of another NADP(H)-GDH gene (EcGDH) from Eurotium chevalieri markedly improved the nitrogen assimilation and grain weight per plant in rice, especially under low-nitrogen conditions (Tang et al. 2018). To date, only CeGDH from Cylindrocarpon ehrenbergii and EcGDH from Eurotium chevalieri have been reported to be capable of improving grain yields in rice under low-nitrogen conditions (Tang et al. 2018, Zhou et al. 2015a).
GDHs are ubiquitous and present in both microorganisms and higher plants. It was reported that four GDH genes (OsGDH1, OsGDH2, OsGDH3, and OsGDH4) are present in the rice genome (Qiu et al. 2009). The NADP(H)-GDHs from fungi seem to be promising candidate genes for crop improvement and breeding due to their enhancement of nitrogen utilization efficiency in crops. However, except for the improvement of nitrogen assimilation, not all GDHs can always improve growth quality and grain yield, and even some GDHs severely inhibit seedling growth. For example, overexpression of PcGDH from Pleurotus cystidiosus showed improved nitrogen assimilation in rice, but no statistical enhancement in grain yields was detected between transgenic rice and wild-type plants (Zhou et al. 2014). Similarly, MgGDH from Magnaporthe grisea did not improve growth quality but did enhance tolerance to dehydration stress in transgenic rice (Zhou et al. 2015b). Moreover, overexpression of SsGDH from Sclerotinia sclerotiorum did not improve the growth quality or grain yield of rice, and even worse, it severely inhibited seedling growth (Du et al. 2014). Thus, to develop environmentally friendly crops with stable yields even under low-nitrogen fertility, we sought to isolate and identify new fungal NADP(H)-GDHs that can improve ammonium assimilation and production in crops.
The fungus Trichurus is a common soil saprophyte and usually causes some harm to plants and animals. When Trichurus infestation develops because of favorable environmental conditions, losses can be great, and control measures should be considered. However, the role of glutamate dehydrogenase from Trichurus in nitrogen metabolism remains unclear. In this study, an NADP(H)-GDH gene (TrGDH) was isolated from Trichurus, and the kinetic properties of TrGDH were characterized. To further evaluate whether TrGDH has a positive effect on ammonium assimilation efficiency as do other microorganism GDHs in rice, we tried to modify the ammonium assimilation pathway by overexpressing TrGDH in rice as an additional enzyme for NH4+ assimilation and biochemical characterization. Changes in the nitrogen metabolism and growth status of rice following the introduction of fungal TrGDH were reported. Our results strongly demonstrated that ectopic expression of fungal TrGDH in rice alters the pathway for ammonium assimilation and, consequently, leads to a number of growth alterations in nitrogen metabolism, growth status and grain yield. These results will enrich the development of strategies to engineer enhanced nitrogen utilization efficiency via exogenous GDHs from fungal microorganisms in higher plant species such as rice.
Materials and Methods
Fungus and plant materials
The fungus Trichurus was kindly provided by Dr. Hong Liu (Fujian Agriculture and Forestry University, Fuzhou, China). Notably, the species name of this fungus has not been identified, so its generic name Trichurus is used to denote this fungus. The model rice (Oryza sativa sub. japonica) cultivar Kitaake was used to generate transgenic rice.
Phylogenetic analysis
To evaluate the evolutionary relationships between TrGDH and previously characterized GDHs from various plants, fungi, and bacteria, a phylogenetic tree was constructed using MEGA 7.0 software and Clustal X based on their amino acid sequences (detailed information of all the GDHs is shown in Supplemental Table 2). Sequence information on the GDHs was obtained from the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/), TAIR (http://www.arabidopsis.org/) and GenBank (http://www.ncbi.nlm.nih.gov/).
Cloning of TrGDH from Trichurus and transformation of TrGDH in rice
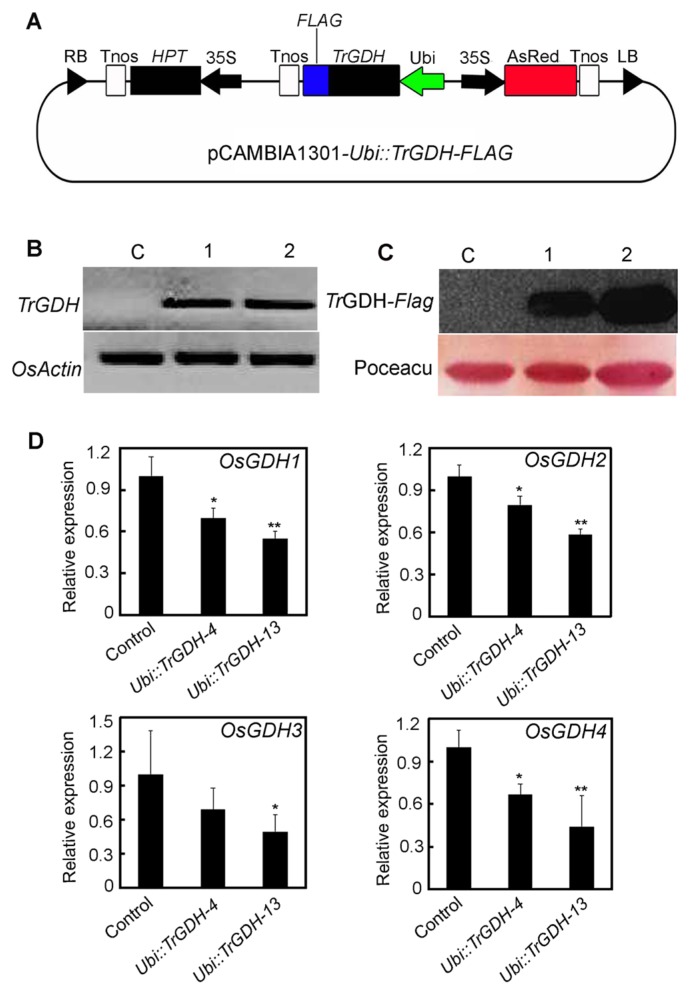
Total RNA was extracted from Trichurus mycelia using Trizol reagent (Invitrogen, USA). The full-length cDNA of the glutamate dehydrogenase gene was amplified by PCR with degenerate primers (Supplemental Table 1) as described previously (Zhou et al. 2014). The sequence of the PCR product was verified and denoted as TrGDH. To assess the effects of TrGDH in rice, TrGDH fused to a FLAG tag was introduced into a pCAMBIA1301 binary vector (slightly modified) with the specific primers (Supplemental Table 1) as described previously (Zhou et al. 2014), and the resulting vector was designated as pCAMBIA1301-Ubi::TrGDH-FLAG (Fig. 3A). To investigate the effects of TrGDH on the nitrogen utilization efficiency in rice, the recombinant plasmid was introduced into O. sativa cv. Kitaake by the Agrobacterium-mediated (EHA105) transformation method (Zhou et al. 2018); hygromycin-resistant plants from calli were designated as composing the T0 generation. The T2 progeny that were from the transgenic rice plants and that expressed high levels of TrGDH were used in subsequent experiments. The TrGDH transcription level in transgenic plants was investigated by semi-RT-PCR using TrGDH-specific primers (Supplemental Table 1).
Fig. 3.
Molecular confirmation of transgenic rice lines expressing TrGDH-FLAG. (A) Schematics of plant expression vector pCAMBIA1301-Ubi::TrGDH-FLAG. HPT hygromycin resistance gene, Tnos terminator of nopaline synthase gene (nos), FLAG tag sequence, AsRed red fluorescence protein gene, 35S CaMV 35S promoter, Ubi ubiquitin promoter derived from Zea mays, LB left border, RB right border, and TrGDH NADP(H)-GDH gene derived from Trichurus. (B) Relative expression level analysis of TrGDH in the control and TrGDH transgenic rice lines by semi-quantitative RT-PCR. Two independent transgenic rice lines, Ubi::TrGDH-4 and Ubi::TrGDH-13, were used for analysis. OsActin1 was amplified as a control for mRNA levels. (C) Western blotting analysis of TrGDH-FLAG in the control and TrGDH transgenic rice lines using anti-FLAG antibody (1:5000). Poceacu staining was used as a loading control. (D) Real-time quantitative PCR analysis of OsGDH1, OsGDH2, OsGDH3, OsGDH4 in the control and TrGDH transgenic rice lines. OsActin1 was amplified as a control for mRNA levels. Triplicate quantitative assays were performed on each cDNA sample. * indicates significant differences at p < 0.05. ** indicates significant differences at p < 0.01.
Expression and purification of His6-TF-TrGDH
The full sequence of TrGDH was amplified by PCR with specific primers (Supplemental Table 1), and the PCR products were digested with BamHI and HindIII and then inserted into a pCold-TF vector with the same sites to yield the recombinant prokaryotic expression vector pCold-TF-TrGDH with a trigger factor (TF) and a His-tag sequence at the 5′-end (Takara, Japan). In addition, the rice homogenous NADP(H)-GDH gene OsGDH4 was also cloned and inserted into the same prokaryotic expression vector, which served as a control. The pCold-TF-TrGDH vector was then transformed into E. coli BL21 (DE3), and positive clones were isolated for His6-TF-TrGDH expression. The recombinant protein His6-TF-TrGDH was affinity purified with nickel-nitrilotriacetic acid (Ni-NTA) resin (Invitrogen, USA) according to previously described methods (Zhou et al. 2015a). To verify the His6-TF-TrGDH, a western blot was used to examine the purified fusion proteins using anti-His monoclonal antibodies (1:5,000; Abmart, China) as described previously (Zhou et al. 2015b).
NADP(H)-GDH enzyme activity assays
The NADP(H)-GDH enzyme activity was measured in vitro following the method described by Noor and Punekar (2005). The aminating activity of His6-TF-TrGDH was determined in a reaction mixture containing various NH4Cl concentrations (1, 2.5, 5, and 7.5 mM) or various 2-oxoglutarate (2-OG) concentrations (1, 2.5, 5, and 10 mM) and 100 μM β-NADP(H) in Tris buffer (pH 8.0). The deaminating activity of His6-TF-TrGDH was determined in a reaction mixture containing various L-glutamate concentrations (25, 50, 75, and 100 mM) and 200 μM β-NADP+ in Tris buffer (pH 9.3). The reactions were preincubated at 25°C before adding His6-TF-TrGDH protein and routinely started by the addition of a coenzyme [β-NADP(H) or β-NADP+]. The enzyme activity assays were measured by monitoring the absorption change at 340 nm with a UV-1800 spectrophotometer (Shimadzu, Japan). One unit of enzyme activity is defined as the reduction or oxidation of 1 μM NADP(H)/second at 25°C. The kinetic constant (Km) for different substrates was calculated by using the Lineweaver–Burk equation. The purified His6-TF-OsGDH4 (as the control) was also used for the NADP(H)-GDH enzyme activity assay.
For the NADP(H)-GDH enzyme activity assay in vivo, the soluble proteins were extracted from the shoots and roots of TrGDH transgenic rice plants and nontransgenic plants (as controls) at the two-leaf stage according to the method described by Abiko et al. (2010), and their respective NADP(H)-GDH activities were tested in a reaction mixture containing NH4Cl (5 mM), 2-oxoglutarate (2-OG; 5 mM), and β-NADP(H) (100 μM) in a Tris buffer (pH 8.0). The reactions were preincubated at 25°C before adding soluble proteins and were started by the addition of coenzyme β-NADP(H). The absorption change was monitored as described above. The data points represent the averages of three replicates and are consistent.
Reverse transcription-PCR (RT-PCR) analysis
For RNA expression level analysis, semiquantitative RT-PCR was used to investigate the expression of TrGDH in the T2 progeny of the transgenic homozygous rice plants. RT-PCR was performed using a SYBR PremixEx Taq kit (Takara, Japan) according to the manufacturer’s instructions with an Mx3000P instrument. Triplicate quantitative assays were performed on each cDNA sample. OsActin1 was used as an internal control. The relative expression levels were measured as described by Livak and Schmittgen (2001). Each data point represents the average of three replicates. Three experiments were performed with consistent results. The primers used are shown in Supplemental Table 1.
Characterization of TrGDH transgenic rice plants at the seedling stage
Two T2 progeny seeds of transgenic lines (Ubi::TrGDH-4 and Ubi::TrGDH-13) and the controls were sterilized with 3% NaClO and then germinated, after which they were grown in an incubator at 28°C/26°C (day/night) and normally cultured in 1/2-strength MS liquid medium for 7 d. The seedlings were then cultured in IRRI nutrient solution (0.3 mM KH2PO4, 0.35 mM K2SO4, 1 mM MgSO4·7H2O, 0.5 mM Na2SiO3·9H2O, 1 mM CaCl2·2H2O, 9 μM MnCl2·4H2O, 20 μM H3BO3, 0.77 μM ZnSO4·7H2O, 0.32 μM CuSO4·5H2O, 20 μM NaFeEDTA, and 0.39 μM Na2MoO4·2H2O, pH 5.5) with different concentrations of NH4+ (50 μM, 0.500 μM) as the only nitrogen source as described previously (Zhou et al. 2014) for 15 d to analyze the phenotype and physiology. The nutrient solution was maintained at pH 5.6 and refreshed every 3 d. After treatment for 15 d, phenotypic images were taken, and the fresh weights, dry weights and total nitrogen contents of the transgenic and control plants were measured by Kjeldahl analysis (Zhou et al. 2015a). For the above parameters, each data point represents the average of three replicates, with consistent results.
Field trial
A field trial was performed as described previously (Zhou et al. 2014). TrGDH transgenic and control plants were planted in paddy fields with different nitrogen gradient levels (0, 37.5, 112.5, and 187.5 kg N/ha; in the form of urea) in Changsha, Hunan Province, China. The nitrogen level of the paddy field, which was planted with early rice and did not receive any nitrogen fertilizer, was defined as 0 kg/ha (the actual fertility was 0.65 g N/kg soil). Fertilizer was applied in three split doses: one dose was half that of the total nitrogen fertilizer of each gradient and was applied as the basal dose [50 kg phosphorus (P), 50 kg potash (K), 25 kg zinc sulfate (ZnSO4) per hectare], whereas the other two doses were applied at the active tillering and panicle initiation stages. All rice plants in this study were grown as late rice in July. Two rows of control plants were planted as guard rows around the planted field. At harvest, agriculture traits including plant height, effective panicle number, filled grain rate and 1,000-seed weight were analyzed. The grain weight per plant represents the total weight of grain per plant, and after harvest, the date of grain weight per plant was measured from 20–30 rice plants in the middle two rows. The average value obtained through variance and statistical analyses of these data was evaluated using Student’s t-test.
Glutelin and prolamine content analysis
Glutelin and prolamine contents were extracted according to the method described by Krishnan and Okita (1986) from the seeds of the transgenic and control plants. The protein concentrations of glutelin and prolamine were measured by the Cai et al. (2009). Protein assay using Coomassie Plus Protein Assay Reagent (Tiangen, China). Bovine serum albumin (BSA) was used as the standard protein.
Statistical analysis
All the data were subjected to analysis of variance (ANOVA) or Student’s t-test using SPSS 13.0 (SPSS Co., USA).
Results
Phylogenetic analysis of TrGDH
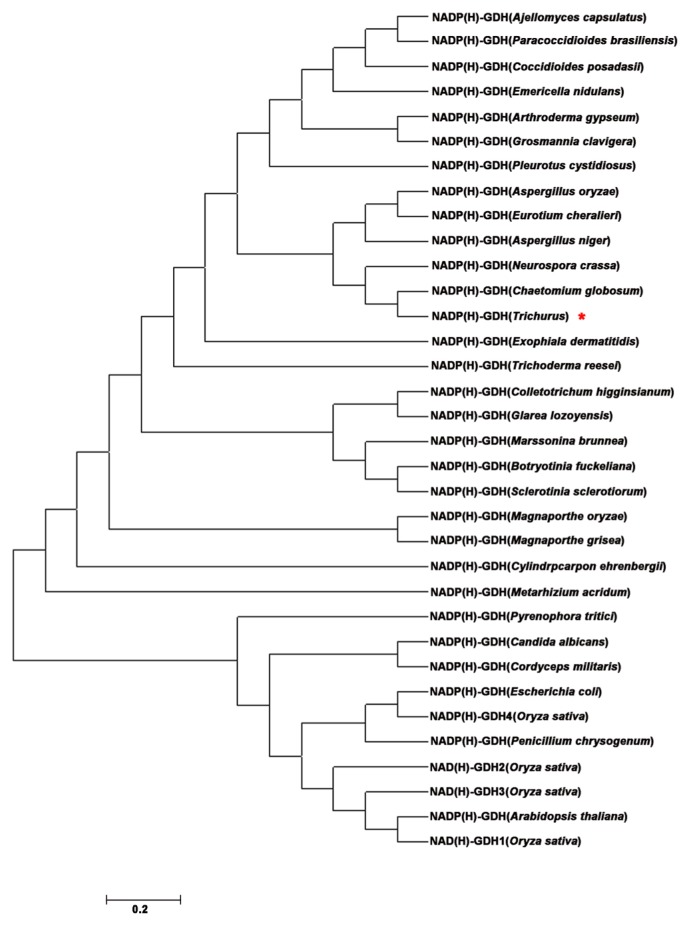
Based on the conserved regions in GDHs among fungi and bacteria, a full-length cDNA encoding glutamate dehydrogenase was isolated from the fungus Trichurus and designated as TrGDH. The full-length TrGDH sequence consists of 1,359 nucleotides, which was predicted to encode 452 amino acids with a molecular mass of 49.0 kDa and a calculated pI of 5.7 (Supplemental Fig. 1). Notably, neither the nucleotide nor the amino acid sequence of TrGDH has been deposited in the NCBI database or GenBank, so TrGDH was considered a novel GDH gene. The phylogenetic tree shows that NADP(H)-GDH proteins form a separate cluster from the group containing plant NAD(H)-GDH proteins; moreover, TrGDH belongs to the NADP(H)-GDH subgroup and exhibits high homology with GDHs in bacteria and fungi but low homology with GDHs in higher plants such as rice and Arabidopsis thaliana (Fig. 1). Thus, we speculate that the fungal TrGDH also may play an important role in enhancing ammonium assimilation efficiency, similar to other microorganism GDHs, when introduced into rice (Abiko et al. 2010, Tang et al. 2018, Zhou et al. 2015a). It is notable that three NAD(H)-GDHs (OsGDH1, OsGDH2 and OsGDH3) and one NADP(H)-GDH (OsGDH4) are present in the rice (Qiu et al. 2009, Supplemental Table 2), whereas only OsGDH4 exhibits the highest homology with TrGDH (Fig. 1). Moreover, we analyzed the protein sequence similarity between TrGDH and other representative reported GDHs from fungi (Magnaporthe oryzae, Chaetomium globosum and Neurospora crassa), which shows a higher homology with TrGDH (Fig. 1), bacteria (Escherichia coli), and rice (Oryza sativa) according to MEGA 7.0 and GENEDOC (Supplemental Fig. 2). As expected, TrGDH contains a Glu/2-OG binding site and an NADP(H) binding site, which are the characteristic conserved domains in NADP(H)-GDH proteins. Taken together, these results demonstrated that the NADP(H)-GDH gene TrGDH was successfully isolated from the fungus Trichurus, which allowed for its functional characterization and application in rice.
Fig. 1.
Phylogenetic relationship of GDHs among bacterial, fungal, and higher plants. Phylogenetic tree was constructed by neighbor-joining method using Clustal X and MEGA 7.0. The information on the deduced amino acid sequences of the NAD(P)(H)-GDHs was obtained from NCBI or GeneBank. The length of the branch is proportional to the degree of divergence. TrGDH was red asterisk marked.
TrGDH exhibits higher affinity for ammonium than does OsGDH4 in vitro
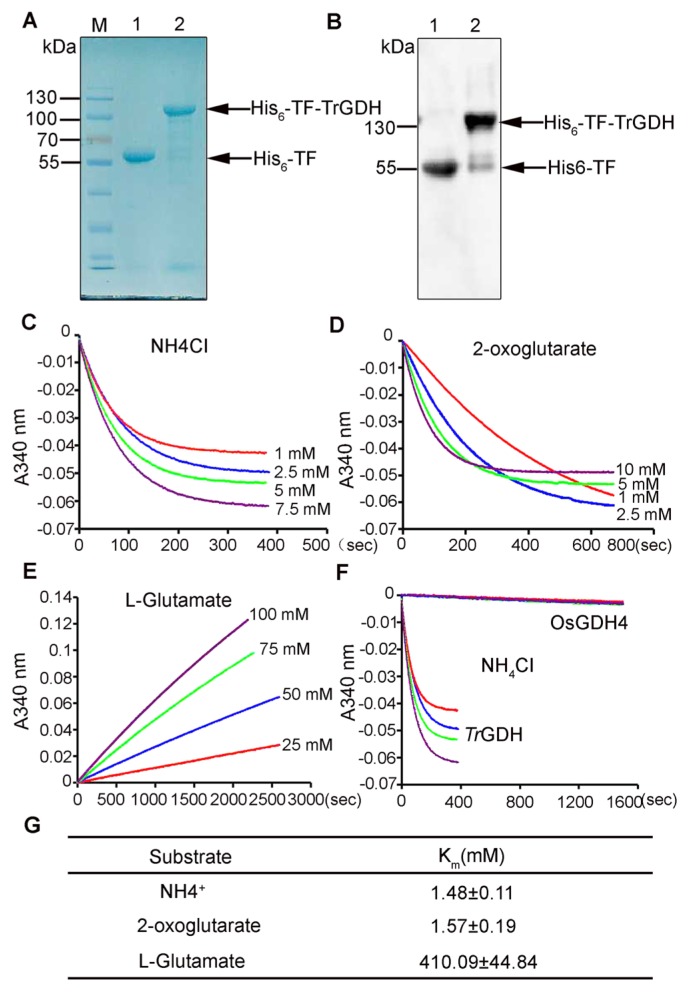
To characterize the properties of TrGDH in vitro, a pCold-TF-TrGDH recombinant plasmid was constructed and introduced into E. coli BL21 for His-tagged His6-TF-TrGDH recombinant protein expression. The recombinant protein was then affinity purified and subjected to western blotting and Coomassie Brilliant Blue staining. A single polypeptide band that corresponded to the deduced molecular size (104.0 kD) of His6-TF-TrGDH was detected, while the control showed only a 55.0 kDa band corresponding to that of the His6-TF chaperone protein in the original vector (Fig. 2A). Western blotting analysis also confirmed the presence of His6-TF-TrGDH protein via anti-His monoclonal antibodies (Fig. 2B). The purified His6-TF-TrGDH recombinant protein was then used for glutamate dehydrogenase activity analysis.
Fig. 2.
Purification and enzymatic kinetic assay of recombinant protein His6-TF-TrGDH in vitro. (A) SDS-PAGE detection of purified His6-TF-TrGDH protein. M: protein marker; 1: His6-TF-tag control; 2: His6-TF-TrGDH protein. The proteins were stained with Coomassie Blue dye after SDS-PAGE. (B) Western blot assay of purified His6-T-FTrGDH protein. 1: His6-TF-tag control; 2: His6-TF-TrGDH protein. The proteins were detected using anti-His antibody (1:5,000). (C) Time course of changes in absorbance at 340 nm in amination reaction using different NH4Cl concentrations (1, 2.5, 5, 7.5 mM). (D) Time course of changes in absorbance at 340 nm in amination reaction using different 2-OG concentrations (1, 2.5, 5, 10 mM). (E) Time course of changes in absorbance at 340 nm in deamination reaction using different glutamate concentrations (25, 50, 75, 100 mM). (F) Time course of changes comparison of His6-TF-TrGDH and His6-TF-OsGDH4 in absorbance at 340 nm in amination reaction using different NH4Cl concentrations (1, 2.5, 5, 7.5 mM). Three experiments were conducted with consistent results. The result from one set of experiments is presented here. (G) Kinetic property (Km) of purified His6-TF-TrGDH protein. One unit of enzyme activity is defined as the reduction or oxidation of 1 μM of NADP(H) per second at 25°C. The calculation was conducted by using the Lineweaver–Burk double-reciprocal protocol. Values represents the means of three independent experiments. Data represents the mean values ± SD (n = 3).
To study the enzymatic properties of TrGDH, the kinetic properties of the affinity of His6-TF-TrGDH for its substrates, including ammonium (NH4+), 2-oxoglutarate (2-OG), and L-glutamate, were determined in vitro. The absorption changes in the amination/deamination enzymatic reactions with different concentrations of NH4Cl, 2-OG and L-glutamate were monitored at 340 nm (Fig. 2C–2E). Via the Lineweaver–Burk equation, the Km values for NH4+, 2-OG and L-glutamate were then calculated. As shown in Fig. 2G, the affinity of the His6-TF-TrGDH protein for NH4+ (Km = 1.48 ± 0.11 mM) and 2-OG (Km = 1.57 ± 0.19 mM) was significantly higher than that for L-glutamate (Km = 410.09 ± 44.84 mM), indicating that TrGDH prefers to utilize ammonium to produce glutamate (Supplemental Fig. 3). As described above, among four OsGDHs, OsGDH4 has the highest homology with TrGDH (Fig. 1) and thus was selected for the comparison of enzyme activity with TrGDH in vitro. As expected, the time course of the comparison between His6-TF-TrGDH and His6-TF-OsGDH4 in absorbance at 340 nm in the amination reaction in conjunction with different NH4Cl concentrations showed that the affinity of His6-TF-TrGDH for NH4+ was prominently higher than that of His6-TF-OsGDH4 (Fig. 2F). Thus, these results indicate that, compared with the OsGDHs, TrGDH has a markedly stronger affinity for ammonium, which makes it possible that TrGDH can efficiently assimilate NH4+, even at very low ammonium concentrations.
Identification of TrGDH transgenic rice plants
The results showed that a higher TrGDH transcription level was identified in two TrGDH transgenic lines, Ubi::TrGDH-4 and Ubi::TrGDH-13, whereas no TrGDH was detected in the nontrangenic plants (negative controls) (Fig. 3B), indicating that TrGDH was successfully overexpressed in the transgenic rice lines. Furthermore, the TrGDH-FLAG fusion proteins in the two transgenic lines were also confirmed by western blot analysis using anti-FLAG monoclonal antibodies. As shown in Fig. 3C, target bands of TrGDH-FLAG were detected in the two transgenic lines but not in the control plants. This result further confirmed that transgenic lines constitutively expressing TrGDH were successfully obtained. Thus, these two transgenic lines, Ubi::TrGDH-4 and Ubi::TrGDH-13, were used for subsequent physiological and phenotypic analysis.
TrGDH improved the ammonium assimilation efficiency of rice seedlings
To evaluate the effects of TrGDH on rice homogenous GDHs, quantitative real-time PCR (qPCR) was performed to analyze the transcription levels of OsGDH1, OsGDH2, OsGDH3 and OsGDH4. The results showed that the expression levels of OsGDH1, OsGDH2, OsGDH3 and OsGDH4 were downregulated (Fig. 3D), indicating that the heterologous TrGDH competitively disrupted the transcription of their endogenous GDHs in the transgenic rice plants.
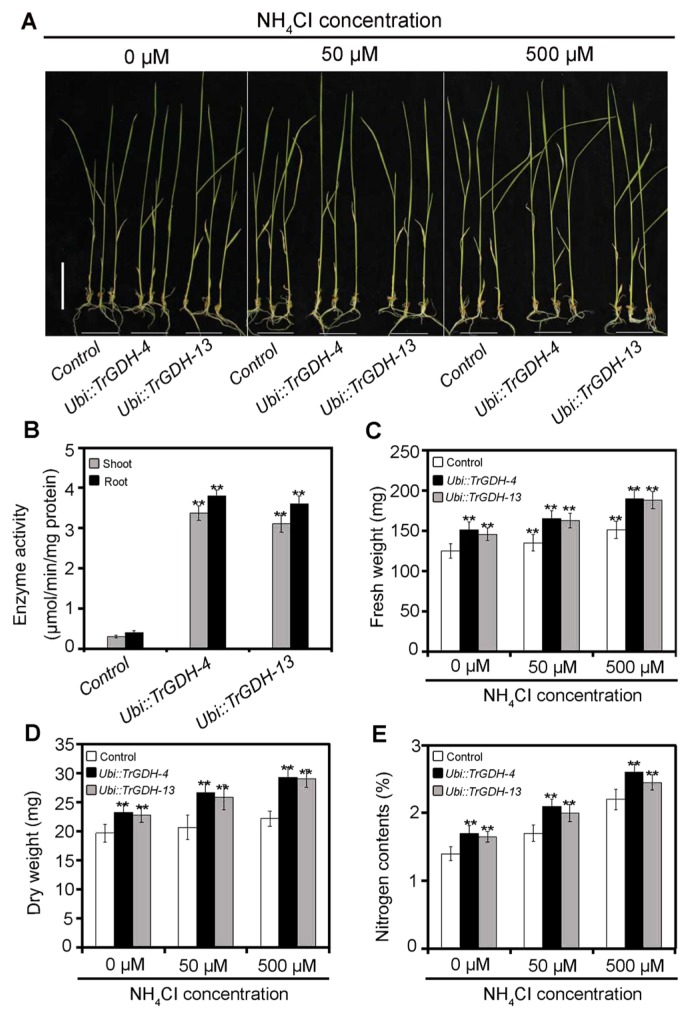
To detect whether the heterologously expressed TrGDH in the transgenic plants retained its function, the enzymatic activity (i.e., amination activity) of NADP(H)-GDH in the shoots and roots of the control and transgenic plants was measured, with NH4+ used as a substrate. The results showed that the NADP(H)-GDH activities of both the shoots and roots were significantly higher in the two transgenic lines than in the control plants (Fig. 4B), indicating that the heterologous TrGDH was still functional and that it indeed enhanced the ammonium assimilation efficiency in rice. To further investigate the effects of TrGDH on the ammonium assimilation efficiency of rice plants at the seedling stage, the phenotypic changes of seedlings of the controls and TrGDH transgenic lines were examined after treatment with different concentrations of ammonium (0, 50, and 500 μM NH4Cl) (Fig. 4A), and their fresh weights, dry weights and total nitrogen contents were analyzed. The results showed that, under the three ammonium concentrations, both the fresh weight and dry weight of TrGDH transgenic seedlings were significantly greater than those of the control seedlings (Fig. 4C, 4D), indicating that the heterologous TrGDH enhanced biomass accumulation in rice. Moreover, the transgenic seedlings also accumulated more nitrogen than did the control seedlings (Fig. 4E), which was consistent with our hypothesis that TrGDH can improve the ammonium assimilation efficiency in rice. Thus, we speculate that the marked increase in the absorption of nitrogen may give rise to improvement in rice growth status.
Fig. 4.
Characteristics of transgenic rice lines expressing TrGDH grow under different NH4Cl concentrations (0, 50, and 500 μM). (A) Images of control and TrGDH transgenic seedlings grown under different NH4Cl concentrations (0, 50, and 500 μM) for 15 d. Two independent transgenic rice lines, Ubi::TrGDH-4 and Ubi::TrGDH-13, were used for analysis. Bar = 5 cm. (B) Amination activity assay of NADP(H)-GDH in the shoots and roots of control and TrGDH transgenic seedlings. Data are presented as average values ± SD (n = 3). (C, D) Fresh weight and dry weight of control and TrGDH transgenic seedlings grow under different NH4Cl concentrations (0, 50, and 500 μM). Average values ± SD are shown (n = 20–30). From B–E, statistical differences were calculated by t-test. ** indicates significant differences at p < 0.01. (E) Nitrogen contents of control and TrGDH transgenic seedlings grow under different NH4Cl concentrations (0, 50, and 500 μM). Data represent the mean values ± SD (n = 3).
TrGDH improved the grain weight per plant and protein accumulation in seeds
To evaluate the effects of the TrGDH introduction on rice productivity, transgenic lines (Ubi::TrGDH-4 and Ubi::TrGDH-13) and control plants were planted and grown to maturity in paddy fields with different nitrogen concentrations (0, 37.5, 112.5 and 187.5 kg N/ha) in accordance with local standard practices for rice cultivation. Agronomic traits including plant height, effective panicle number, percentage of filled grains, 1,000-grain weight, and grain weight per plant were scored at harvest. Under different nitrogen concentrations, there was no difference in plant height between the control and transgenic plants (Table 1). However, the number of effective panicles, 1,000-grain weight and grain weight per plant of the transgenic lines were significantly higher than those of the control plants (Table 1). Surprisingly, an obvious decrease in the percentage of filled grains was observed in the transgenic rice lines compared with the control plants. In addition, the degree of increase in effective panicle number, 1,000-seed weight and grain weight per plant were reduced with the increase in the nitrogen gradient concentration, indicating that TrGDH can effectively promote biomass accumulation in grain weight per plant, especially under low-nitrogen conditions (0 and 37.5 kg N/ha).
Table 1.
Agronomic traits of the control and transgenic rice plants grown in paddy filed
| Nitrogen fertilizer concentration (kg N/ha) | Lines | Plant height (cm) | Number of effective panicles (per) | Filled grain rate (%) | Thousand grain weight (g) | Grain weight per plant (g) |
|---|---|---|---|---|---|---|
| 0 | Control | 73.3 ± 2.0 | 16.2 ± 2.3 | 58.2 ± 2.1 | 21.4 ± 0.8 | 14.05 ± 1.5 |
| Ubi::TrGDH-4 | 70.7 ± 2.8 | 20.6 ± 2.1** | 48.1 ± 1.3** | 25.3 ± 0.7** | 17.04 ± 0.9** | |
| Ubi::TrGDH-13 | 71.4 ± 3.2 | 21.5 ± 1.5** | 50.2 ± 2.0** | 25.7 ± 1.0** | 16.64 ± 0.9** | |
| 37.5 | Control | 73.6 ± 3.2 | 20.6 ± 1.5 | 62.3 ± 1.1 | 24.8 ± 0.7 | 16.70 ± 0.7 |
| Ubi::TrGDH-4 | 73.0 ± 1.9 | 25.1 ± 1.2** | 53.0 ± 1.4** | 28.3 ± 0.4** | 19.76 ± 1.2** | |
| Ubi::TrGDH-13 | 72.0 ± 3.4 | 26.6 ± 2.1** | 54.2 ± 1.2** | 28.9 ± 0.6** | 21.95 ± 0.7** | |
| 112.5 | Control | 75.2 ± 3.1 | 25.7 ± 2.2 | 66.3 ± 1.3 | 26.1 ± 0.6 | 22.34 ± 0.9 |
| Ubi::TrGDH-4 | 73.8 ± 2.5 | 28.6 ± 1.3** | 58.0 ± 1.5** | 29.6 ± 0.5** | 25.77 ± 1.0** | |
| Ubi::TrGDH-13 | 74.4 ± 3.2 | 29.7 ± 1.6** | 59.1 ± 1.1** | 30.0 ± 0.4** | 26.64 ± 0.7** | |
| 187.5 | Control | 76.3 ± 2.1 | 29.8 ± 3.2 | 68.6 ± 1.6 | 28.3 ± 0.7 | 29.36 ± 1.2 |
| Ubi::TrGDH-4 | 74.1 ± 3.3 | 31.5 ± 2.2** | 62.0 ± 1.7** | 30.6 ± 0.4** | 33.37 ± 1.2** | |
| Ubi::TrGDH-13 | 75.5 ± 2.3 | 32.7 ± 1.6** | 61.6 ± 1.3** | 30.8 ± 0.5** | 32.57 ± 0.9** |
A collection of quantitative data represent the mean values ± SE (n = 20–30). Statistical analysis of the data was performed by t-test. Asterisks indicate significantly differences at p < 0.01 (**).
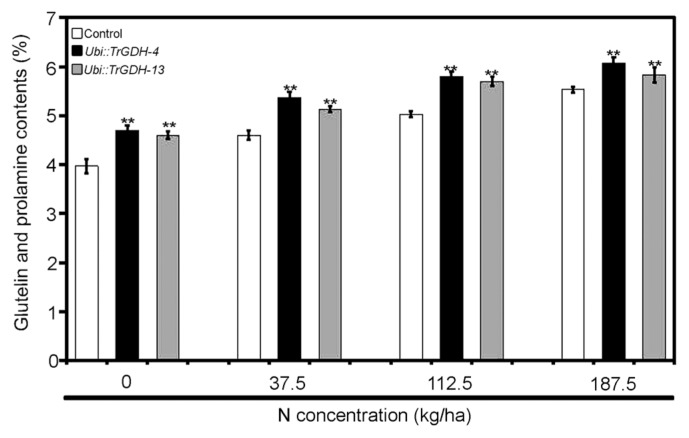
To investigate whether TrGDH introduction also affects protein accumulation in rice seeds, the glutelin and prolamine contents were further analyzed in the seeds of the control and transgenic plants. The results showed that the glutelin and prolamine contents of the seeds of transgenic plants were markedly higher than those of the control plants grown under the same field conditions (Fig. 5), indicating that TrGDH can promote protein accumulation in rice seeds and improve their nutritional value. These results confirmed that ectopic expression of TrGDH can improve nitrogen assimilation efficiency and grain weight per plant in rice.
Fig. 5.
Glutelin and prolamine contents of control and transgenic lines (Ubi::TrGDH-4 and Ubi::TrGDH-13) seeds. Average values ± SD are shown (n = 3). Statistical differences were calculated by t-test. ** indicates significant differences at p < 0.01.
Discussion
Using NAD+ and NADP+ as cofactors, glutamate dehydrogenase (GDH) catalyzes the reversible amination of 2-OG with NH4+ to form glutamate (Wootton 1983). In organisms, GDH contributes to important processes such as amino acid and carbohydrate metabolism, energy production and ammonia management through the Krebs cycle, interconnecting in parallel amino acid and carbohydrate metabolism (Hutson et al. 2011). Therefore, GDH is considered a key enzyme for metabolism and is essential for cell survival (Hudson and Daniel 1993). In plants, since the discovery of the glutamine synthetase (GS)/glutamate synthase (GOGAT) pathway in the 1970s, the role of GDH in ammonium assimilation has been a matter of controversy (Abiko et al. 2010). For example, a study involving feeding with both 15NH4+ and MSX in GDH1-null mutants of maize suggested that endogenous GDH does not catalyze the ammonium-assimilating reaction (Magalhães et al. 1990). By catalyzing the ATP-dependent condensation of ammonium with glutamate to produce glutamine, GS is therefore considered the key enzyme involved in the primary assimilation of ammonium in plants (Ireland and Lea 1999). Conversely, in microorganisms such as bacteria and fungi, both NADP(H)-dependent glutamate dehydrogenase (GDH) and GS play important roles in ammonium assimilation. In particular, in comparison with GDHs in plants, NADP(H)-GDHs in microorganisms assimilate ammonium more efficiently, as the affinity of the latter for ammonium is relatively stronger. Furthermore, Zhou et al. (2015b) reported that the Km value of OsGDH4 for NH4+ is 12.43 ± 1.84 mM, which is markedly higher than that of TrGDH (1.48 ± 0.11 mM). Therefore, the genes encoding fungal NADP(H)-GDHs have been proposed to be promising candidates for ectopic expression in crop plants for the production of an additional enzyme for ammonium assimilation (Abiko et al. 2010, Noor and Punekar 2005, Tang et al. 2018, Zhou et al. 2015a).
In this study, a novel NADP(H)-dependent glutamate dehydrogenase gene (TrGDH) was first isolated from the fungus Trichurus. Evolutionary relationship analysis demonstrated that TrGDH shows high homology with the NADP(H)-GDHs of other microorganisms (Fig. 1). Glutamate dehydrogenase activity assay in vitro showed that the amination activity of His6-TF-TrGDH was significantly higher than the deamination activity (Fig. 2C–2F), and the Km value for NH4+ (1.48 ± 0.11 mM) was markedly lower than that for L-glutamate (410.09 ± 44.84 mM) (Fig. 2G), which further demonstrated that TrGDH tends to catalyze the amination of 2-OG with NH4+ to form glutamate. Additionally, Zhou et al. (2015b) reported that the Km value of OsGDH4 from rice for NH4+ is 12.43 ± 1.84 mM, which is markedly higher than that of TrGDH (1.48 ± 0.11 mM). Similarly, His6-TF-OsGDH4 also exhibited a significantly lower affinity for NH4+ than did His6-TF-TrGDH (Fig. 2F). These results indicate that ectopic expression of TrGDH for the production of as an additional enzyme for ammonium assimilation might improve nitrogen assimilation efficiency in rice. Hydroponic experiments subsequently revealed that overexpression of TrGDH actually improved the nitrogen assimilation efficiency and growth status and increased biomass production at the seedling stage in rice (Fig. 4). Surprisingly, overexpression of exogenous TrGDH resulted in the downregulation of the transcription of endogenous GDHs such as OsGDH1, OsGDH2 and OsGDH3 in rice (Fig. 3D). Although the endogenous GDHs of higher plants were reported not to catalyze the ammonium-assimilating reaction (Magalhães et al. 1990), their transcription was still competitively disrupted by the exogenous TrGDH in transgenic rice plants. Furthermore, the endogenous GDH transcript levels were downregulated, but the TrGDH transgenic rice plants still exhibited higher amination activity (Fig. 4B) and accumulated more nitrogen than did the control plants (Fig. 4E). These results further verified, at least in part, that the endogenous GDHs of higher plants are not involved in ammonium assimilation but that fungal GDHs such as TrGDH play important roles in this physiological process.
As described above, GDHs from fungi are currently receiving increased amounts of attention for the improvement of ammonium assimilation in crops by genetic engineering. Except for their ability to enhance nitrogen assimilation, not all GDHs from fungi can always improve growth quality and grain yield (Abiko et al. 2010, Du et al. 2014, Zhou et al. 2014, 2015a, 2015b). Even worse, SsGDH from Sclerotinia sclerotiorum severely inhibited seedling growth in rice, but surprisingly, SsGDH transgenic rice plants exhibited a marked tolerance to Basta, which is an herbicide commonly used in agricultural production (Du et al. 2014). In this study, the field trial demonstrated that the number of effective panicles, 1,000-grain weight and grain weight per plant of the TrGDH transgenic lines were higher than those of the control plants at both low- and high-nitrogen levels, but the degrees of increase were more distinct at low-nitrogen levels (Table 1). Compared to that of the control plants, the percentage of filled grains of the transgenic rice plants obviously declined, but their increase in the number of effective panicles and 1,000-grain weight compensated or even surpassed the yield loss by the decrease in the percentage of filled grains, ultimately leading to an obvious increase in grain yield. These results indicated that the ectopic expression of TrGDH for the production of an additional enzyme for ammonium assimilation could indeed enhance the nitrogen utilization efficiency and grain weight per plant in rice. It is notable that gdhA from Aspergillus niger improved the grain yield of forage rice under low-nitrogen conditions (Zhang et al. 2016); however, it could increase the grain yield of a food rice cultivar only under high-nitrogen conditions and not low-nitrogen conditions (Abiko et al. 2010). Because TrGDH can enhance the grain weight per plant in rice under both low and high-nitrogen conditions, it is speculated that TrGDH might be a better fungal GDH gene than CeGDH, EcGDH and gdhA and has the potential for maintaining yields of crop species under various nitrogen fertility regimens. Interestingly, compared with the seeds of the control plants, the seeds of the TrGDH transgenic rice plants presented a greater accumulation of proteins such as glutelin and prolamine (Fig. 5), which means that TrGDH is a promising candidate gene for improving nutritional value as well as nitrogen utilization efficiency.
In conclusion, overexpression of TrGDH from Trichurus can improve nitrogen assimilation, growth status and grain weight per plant in rice, especially under low-nitrogen conditions. Thus, TrGDH is a promising candidate gene for maintaining yields of crop plants via genetic engineering.
Supplementary Information
Acknowledgments
The authors thank Dr. Hong Liu for providing the fungus Trichurus and for the assistance with cloning TrGDH (Fujian Agriculture and Forestry University, Fuzhou, China). This research is financially supported by the National Science Foundation of China (Nos. 31571635, 31871595 and 31170172), Hunan Provincial Important Science and Technology Specific Projects (No. 2018NK1010), Hunan Provincial Natural Science Foundation of China (No. 2017JJ2042), Planned Science and Technology Project of Hunan Province (No. 2017WK2012), and Planned Science and Technology Project of Changsha City (Nos. kq1801001 and kq1701028).
Footnotes
Author Contribution Statement
JZL and XML conceived the project and designed the research. CQD, LAD, DYT and JZL conducted the experiments and analyzed the data. CL, LY, MDC and SL partly participated in the experiments and analyzed the data. JZL, CQD and CL wrote the manuscript. All authors read and approved the manuscript.
Literature Cited
- Abiko, T., Wakayama, M., Kawakami, A., Obara, M., Kisaka, H., Miwa, T., Aoki, N. and Ohsugi, R. (2010) Changes in nitrogen assimilation, metabolism, and growth in transgenic rice plants expressing a fungal NADP(H)-dependent glutamate dehydrogenase (gdhA). Planta 232: 299–311. [DOI] [PubMed] [Google Scholar]
- Ameziane, R., Bernhard, K. and Lightfoot, D. (2000) Expression of the bacterial gdhA gene encoding a NADPH glutamate dehydrogenase in tobacco affects plant growth and development. Plant Soil 221: 47–57. [Google Scholar]
- Cai, H.M., Zhou, Y., Xiao, X.H., Li, X.H., Zhang, Q.F. and Lian, X.M. (2009) Overexpressed glutamine synthetase gene modifies nitrogen metabolism and abiotic stress responses in rice. Plant Cell Rep. 28: 527–537. [DOI] [PubMed] [Google Scholar]
- Du, C., Lin, J., Yang, Y., Liu, H., Li, C., Zhou, Y., Li, Y., Tang, D., Zhao, X., Zhu, Y.et al. (2014) Molecular cloning, characterization and function analysis of a GDH gene from Sclerotinia sclerotiorum in rice. Mol. Biol. Rep. 41: 3683–3693. [DOI] [PubMed] [Google Scholar]
- Frink, C.R., Waggoner, P.E. and Ausubel, J.H. (1999) Nitrogen fertilizer: Retrospect and prospect. Proc. Natl. Acad. Sci. USA 96: 1175–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles, J. (2005) Nitrogen study fertilizes fears of pollution. Nature 433: 791. [DOI] [PubMed] [Google Scholar]
- Helling, R.B. (1998) Pathway choice in glutamate synthesis in Escherichia coli. J. Bacteriol. 180: 4571–4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges, M. (2002) Enzyme redundancy and the importance of 2-oxoglutarate in plant ammonium assimilation. J. Exp. Bot. 53: 905–916. [DOI] [PubMed] [Google Scholar]
- Hudson, R. and Daniel, R. (1993) L-glutamate dehydrogenases: distribution, properties and mechanism. Comp. Biochem. Physiol. B. 106: 767–792. [DOI] [PubMed] [Google Scholar]
- Hutson, S.M., Islam, M.M. and Zaganas, I. (2011) Interaction between glutamate dehydrogenase (GDH) and L-leucine catabolic enzymes: intersecting metabolic pathways. Neurochem. Int. 59: 518–524. [DOI] [PubMed] [Google Scholar]
- Ireland, R.J. and Lea, P.J. (1999) The enzymes of glutamine, glutamate, asparagine, and aspartate metabolism. In: Singh, B.K. (ed.) Plant amino acids: biochemistry and biotechnology. Marcel Dekker, New York, pp. 49–109. [Google Scholar]
- Krishnan, H.B. and Okita, T.W. (1986) Structural relationship among the rice glutelin polypeptides. Plant Physiol. 81: 748–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea, P.J. and Miflin, B.J. (1974) Alternative route for nitrogen assimilation in higher plants. Nature 18: 614–616. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2[−Δ ΔC(T)] method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Magalhães, J.R., Ju, G.C., Rich, P.J. and Rhodes, D. (1990) Kinetics of 15NH4+ assimilation in Zea mays. Plant Physiol. 94: 647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor, S. and Punekar, N.S. (2005) Allosteric NADP-glutamate dehydrogenase from Aspergilli: purification, characterization and implications for metabolic regulation at the carbon–nitrogen interface. Microbiology 151: 1409–1419. [DOI] [PubMed] [Google Scholar]
- Oaks, A. (1994) Primary nitrogen assimilation in higher plants and its regulation. Can. J. Bot. 72: 739–750. [Google Scholar]
- Qiu, X., Xie, W., Lian, X. and Zhang, Q. (2009) Molecular analyses of the rice glutamate dehydrogenase gene family and their response to nitrogen and phosphorous deprivation. Plant Cell Rep. 28: 1115–1126. [DOI] [PubMed] [Google Scholar]
- Skopelitis, D.S., Paranychianakis, N.V., Kouvarakis, A., Spyros, A., Stephanou, E.G. and Roubelakis-Angelakis, K.A. (2007) The isoenzyme 7 of tobacco NAD (H)-dependent glutamate dehydrogenase exhibits high deaminating and low aminating activities in vivo. Plant Physiol. 145: 1726–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socolow, R.H. (1999) Nitrogen management and the future of food: lessons from the management of energy and carbon. Proc. Natl. Acad. Sci. USA 96: 6001–6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stulen, I., Perez-soba, M., Dekok, L.J. and Van Der Eerden, L. (1998) Impact of gaseous nitrogen deposition on plant functioning. New Phytol. 139: 61–70. [Google Scholar]
- Tang, D., Peng, Y., Lin, J., Du, C., Yang, Y., Wang, D., Liu, C., Yan, L., Zhao, X., Li, X.et al. (2018) Ectopic expression of fungal EcGDH improves nitrogen assimilation and grain yield in rice. J. Integr. Plant Biol. 60: 85–88. [DOI] [PubMed] [Google Scholar]
- Windass, J.D., Worsey, M.J., Pioli, E.M., Pioli, D., Barth, P.T., Atherton, K.T., Dart, E.C., Byrom, D., Powell, K. and Senior, P.J. (1980) Improved conversion of methanol to single-cell protein by Methylophilus methylotrophus. Nature 287: 396–401. [DOI] [PubMed] [Google Scholar]
- Wootton, J.C. (1983) Re-assessment of ammonium-ion affinities of NADP-specific glutamate dehydrogenases. Biochem. J. 209: 527–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Liang, C., Aoki, N., Kawai, K., Takane, K.I. and Ohsugi, R. (2016) Introduction of a fungal NADP (H)-dependent glutamate dehydrogenase (gdhA) improves growth, grain weight and salt resistance by enhancing the nitrogen uptake efficiency in forage rice. Plant Prod. Sci. 19: 267–278. [Google Scholar]
- Zhou, X., Lin, J., Zhou, Y., Yang, Y., Liu, H., Zhang, C., Tang, D., Zhao, X., Zhu, Y. and Liu, X. (2015a) Overexpressing a fungal gene improves nitrogen utilization and growth in rice. Crop Sci. 55: 811–820. [Google Scholar]
- Zhou, Y., Liu, H., Zhou, X., Yan, Y., Du, C., Li, Y., Liu, D., Zhang, C., Deng, X., Tang, D.et al. (2014) Over-expression of a fungal NADP(H)-dependent glutamate dehydrogenase PcGDH improves nitrogen assimilation and growth quality in rice. Mol. Breed. 34: 335–349. [Google Scholar]
- Zhou, Y., Zhang, C., Lin, J., Yang, Y., Peng, Y., Tang, D., Zhao, X., Zhu, Y. and Liu, X. (2015b) Over-expression of a glutamate dehydrogenase gene, MgGDH, from Magnaporthe grisea confers tolerance to dehydration stress in transgenic rice. Planta 241: 727–740. [DOI] [PubMed] [Google Scholar]
- Zhou, Y., Liu, C., Tang, D., Yan, L., Wang, D., Yang, Y., Gui, J., Zhao, X.Y., Li, L., Tang, X.D.et al. (2018) The receptor-like cytoplasmic kinase STRK1 phosphorylates and activates CatC, thereby regulating H2O2 homeostasis and improving salt tolerance in rice. Plant Cell 30: 1100–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.