Abstract
Immuno-oncology has rapidly grown in the last thirty years, and immunotherapeutic agents are now approved to treat many disparate cancers. Immune checkpoint inhibitors (ICIs) are employed to augment cytotoxic anti-cancer activity by inhibiting negative regulatory elements of the immune system. Modulating the immune system to target neoplasms has improved survivability of numerous cancers in many individuals, but forecasting outcomes post therapy is difficult due to insufficient predictive biomarkers. Recently, the tumor and gastrointestinal microbiome and immune milieu have been investigated as predictors and influencers of cancer immune therapy. In this review, we discuss: (1) ways to measure the microbiome including relevant bioinformatic analyses, (2) recent developments in animal studies and human clinical trials utilizing gut microbial composition and function as biomarkers of cancer immune therapy response and toxicity, and (3) using prebiotics, probiotics, postbiotics, antibiotics, and fecal microbiota transplant (FMT) to modulate immune therapy. We discuss the respective benefits of 16S ribosomal RNA (rRNA) gene and shotgun metagenomic sequencing including important considerations in obtaining samples and in designing and interpreting human and animal microbiome studies. We then focus on studies discussing the differences in response to ICIs in relation to the microbiome and inflammatory mediators. ICIs cause colitis in up to 25% of individuals, and colitis is often refractory to common immunosuppressive medications. Researchers have measured microbiota composition prior to ICI therapy and correlated baseline microbiota composition with efficacy and colitis. Certain bacterial taxa that appear to enhance therapeutic benefit are also implicated in increased susceptibility to colitis, alluding to a delicate balance between pro-inflammatory tumor killing and anti-inflammatory protection from colitis. Pre-clinical and clinical models have trialed probiotic administration, e.g. Bifidobacterium spp. or FMT, to treat colitis when immune suppressive agents fail. We are excited about the future of modulating the microbiome to predict and influence cancer outcomes. Furthermore, novel therapies employed for other illnesses including bacteriophage and genetically-engineered microbes can be adapted in the future to promote increased advancements in cancer treatment and side effect management.
Introduction
Patient survival for most cancers has increased dramatically with earlier diagnosis, improved treatments, and faster and more effective management of side effects [1]. Novel cancer therapies include medications that augment elements of the immune response to directly target and kill cancer cells or to replace and boost immune system functions; these collectively are referred to as cancer immunotherapy [2]. Commensurate with increased survival from new standards of care are increased efforts to understand and predict each individual’s response to and toxicity from these therapies [2]. For reasons not fully understood, variability exists in both efficacy and toxicity-related complications among patients receiving immune therapies [3]. Recently, gut microbiome and immune profiles have been identified as contributing factors to this inter-individual variability (reviewed in [4, 5, 6, 7]).
Microorganisms inhabit our skin and respiratory and gastrointestinal tracts and comprise a large portion of our overall cellular, metabolic, and genetic mass [8]. The organisms that comprise this community are collectively referred to as the microbiota while their collective genetic material is termed the microbiome [9]. The intimate relationship among microbiota composition, metabolic function, and the development and regulation of the immune system has been correlated with entities as disparate as inflammatory bowel disease, autism, asthma, and obesity [10, 11, 12, 13, 14]. Several microbes directly inflame tissues, and certain viruses and bacteria have been well described as direct causes of cancer, for example, Epstein-Barr virus (EBV) with Burkitt’s and Hodgkin’s lymphoma, human papilloma viruses (HPV) with cervical, head, neck, and anal cancers, and Helicobacter pylori with gastric cancers, among others [15]. Given the connection between gut microbiota and inflammatory diseases, researchers are exploring whether differences in the human microbiome have the potential to impact individual responses to cancer immune therapies (additionally reviewed in [4, 5, 6, 7]). Further, recent advances in sequencing technology have provided fine granular detail of a potential tumor microbiota in certain cancers [16, 17, 18]. With decreasing metagenomic sequencing costs, large scale observational studies and placebo-controlled trials can now be analyzed for longitudinal changes in microbial structure and gene content [19]. These techniques have revealed correlations between the microbiome and cancer immunotherapy outcomes in pre-clinical animal models and human clinical trials [20].
In this review, we first discuss methodologies for characterizing the microbiome, including technical limitations and challenges. Next, we describe ways in which the commensal gastrointestinal microbiota has been shown to interact directly with the immune system and how these interactions influence oncogenesis. We then focus on interactions among cancer immunotherapies and the microbiome and ways in which the microbiome can be modulated using prebiotics, probiotics, postbiotics, antibiotics and fecal microbiota transplantation (FMT) to improve survival and limit toxicity.
Measuring the microbiome
Traditionally, clinical assessment of bacterial populations is achieved by sampling the site of interest and culturing organisms in broth and agar plates, followed by phenotypic and molecular identification [21]. Although we have made significant advances in our ability to culture various organisms, a large proportion of human-associated microbes are time- and labor-intensive to grow [22]. Additionally, bacterial enrichment through culture can obscure abundance differences in the original sample because of differential and exclusionary growth [23]. Cell-free methods to measure bacterial presence and activity have become more popular and have allowed measurement of microbial populations at previously impossible scales, including in diagnostic microbiology [24]. Nucleic-acid based methods for detecting viruses and bacteria initially focused exclusively on their presence or absence [25], but recent advances have incorporated relative abundance measures to determine bacterial presence in clinical samples quantitatively to better understand the burden of disease [26].
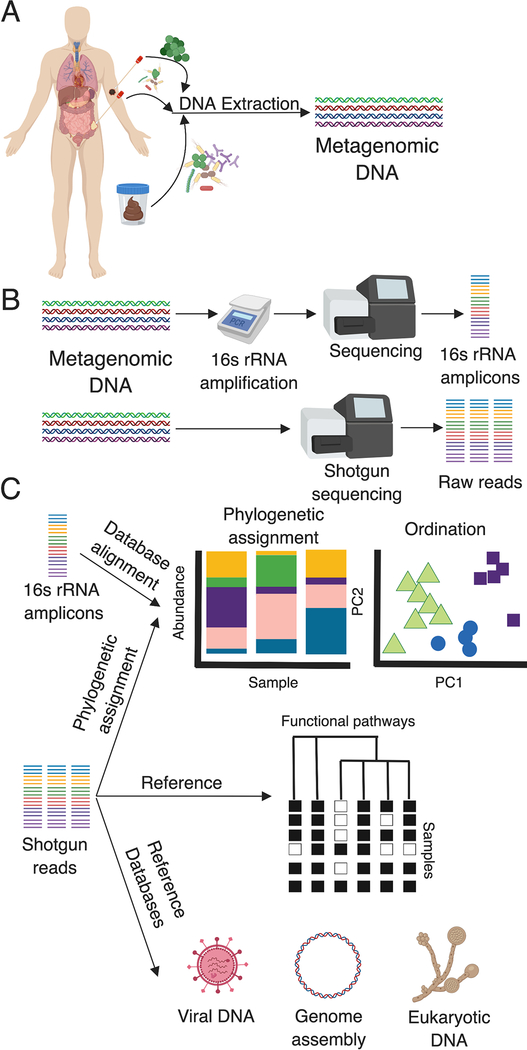
The first step in determining microbial colonization of a body site by sequencing is proper sampling with sterile technique and preservation from nucleases [27]. Once a sample is obtained, genomic DNA is extracted using phenol-chloroform or commercially-available kits with yield and purity dependent on sample type [28, 29] (Figure 1A). The 16S ribosomal RNA (rRNA) gene is highly conserved and is nearly universally present in bacteria, and its sequence can be used to distinguish bacterial genera or species [30]. 16S rRNA gene amplicon sequencing is a cheap, quick, and reliable method for determining bacterial taxonomy after computational processing with publicly-available tools such as QIIME 2 or DADA2 [31, 32] (Figure 1B). Although 16S rRNA gene sequencing is a useful tool, it has several limitations. Precision is often limited to the genus level, especially when assigning operational taxonomic units (OTUs) in lieu of amplicon sequence variants (ASVs), which are preferred due to enhanced specificity [33, 34]. OTU-picking methods cluster reads that differ by less than 3% from a standard, but often group several species into a common genus. For example, OTU methods would assign the genus Neisseria to both pathogenic (e.g. Neisseria gonorrhea and N. meningitidis) and commensal Neisseria spp., which is clearly an important clinical distinction depending on the sample type [34]. Therefore, methods that assign ASVs to each sequence are preferred, thereby allowing resolution of single nucleotide polymorphisms (SNPs) and strain-level differences [34]. Further, 16S rRNA gene sequencing is limited to genomic DNA, does not account for variable copy number within bacterial genera, which could artificially increase relative abundance[35], and ignores plasmids and genomic DNA that encode antibiotic resistance, toxin production, and other virulence factors of clinical importance.
Figure 1. Schematic for metagenomic analysis.
A) The first step in microbiome analysis is selecting the sample, obtaining it with sterile technique, and preserving it with consideration of DNAses and RNAses. Samples can be obtained directly from tumors, skin, or fecal samples. DNA extraction via phenol chloroform or commercially available kits yields metagenomic DNA. B) The 16S rRNA gene can be amplified and sequenced. Alternatively, metagenomic DNA can be shotgun sequenced to yield DNA fragments of varying sizes. C) 16S rRNA reads can be computationally processed with publicly available pipelines yielding relative abundance of taxa per sample. Ordination methods such as PCA and PCoA are utilized to identify similarities and differences in diversity metrics (alpha, beta, gamma) between samples or between groups. Reads from metagenomic shotgun sequencing can be processed to yield the same information. In addition to taxonomy, shotgun reads can be processed and mapped to databases to determine functional pathways, assemble bacterial genomes, and understand the presence of viral or eukaryotic DNA depending on appropriate sequencing depth. Created with BioRender.
Alternatively, DNA from all organisms in the sample, collectively referred to as the metagenome, can be shotgun sequenced using commercial technology, and the taxonomy can be obtained using clade-specific markers with bioinformatic tools such as Kraken or MetaPhlAn2 [36, 37, 38] (Figure 1C). Additionally, shotgun metagenomic sequencing can be used to infer whole community microbial functional information and annotate genes that confer antibiotic resistance, produce toxins, or provide metabolic pathway information via comparison to known databases, assuming appropriate sequencing depth [27, 39, 40, 41] (Figure 1C). Metagenomic sequencing can also identify non-bacterial components of the microbiota, such as fungi, viruses, and bacteriophages, which are gaining attention for their roles in the microbiome and human health [42, 43]. Therefore, desired analyses necessarily determine whether 16S rRNA or metagenomic shotgun sequencing is appropriate to profile the microbiota or microbiome, respectively.
After the sequencing data is obtained, the first question asked is ‘who is there?’ Both 16S rRNA and shotgun metagenomic sequencing can answer this question (Figure 1C). Data from sequencing analysis can be visualized in terms of relative abundance of taxa within a sample or as a similarity or dissimilarity ordination plot based on measures of alpha and beta diversity (Figure 1C). Alpha diversity measurements describe intra-sample diversity: measurements include Shannon diversity and species richness. Beta diversity measures are pairwise comparisons of dissimilarity between all sample pairs and are typically viewed as an ordination plot with principal coordinate analysis (PCoA). Measures of beta diversity include Bray-Curtis, binary-Jaccard, and weighted UniFrac (Figure 1C) (reviewed extensively in [44]). In addition to overall assessments of similarity and dissimilarity between samples, these same measures can be used to examine differences in microbial gene content (e.g. functional pathways and resistance genes) from metagenomic shotgun sequencing (Figure 1C) (Reviewed in [27]). Although shotgun metagenomic sequencing is more expensive than 16S rRNA gene sequencing per sample, when performed at a depth of 2.5 – 5 million reads per sample, the validation of shallow shotgun sequencing with as few as 500,000 reads per sample increases the affordability of this method for large datasets [33]. Thus, in designing any study interrogating the microbiome, it is vital to determine the desired downstream analyses so that sampling schema and sequencing depth can be planned appropriately.
After sequencing and characterizing a microbial community, correlations can be discerned by connecting individual taxa, groups of taxa, or overall trends in community composition (e.g. diversity) to host metadata. However, this observation-driven strategy is frequently insufficient to draw concrete conclusions or to elucidate underlying mechanisms in the absence of manipulation or recapitulation in animal models. To achieve mechanistic understanding, microbiota-humanized mice are produced by gavaging germ-free or antibiotic-treated, conventional mice with stool samples from healthy humans, patient cohorts, or pre-defined consortia [45, 46]. Different microbes colonize mice with varying success, necessitating quality checks to ensure the input is comparable to the output community with at least 65% uptake at the phylum level, though the definition of “successful” microbiota-colonization depends on the richness and taxonomic abundance of the input community [45, 46, 47].
Designing in vivo microbiome-related experiments is accompanied by its own set of challenges and considerations. If humanized mice are used, will they be derived from germ-free mice or antibiotic-treated mice? Germ-free mice are free of all currently detectable microorganisms and allow complete recolonization, but they are expensive to maintain, have severely underdeveloped immune systems that are not always restorable, and must be individually derived for unique genotypes [48, 49]. If antibiotic-treated mice are used, which antibiotic(s) will be used to deplete the conventional mouse microbiota, and what confounding effects may they produce? Individual antibiotics and antibiotic cocktails affect different microbial populations and have varying efficacy and tolerability in mice [48]. Even seemingly arbitrary decisions require due diligence. Considering the appropriate vendor and housing conditions for isogenic conventional mice is vital because microbiota compositions differ among mice that are obtained from different suppliers and that are housed in different facilities [50]. Once mice are obtained and appropriately humanized, it is important to ensure they are consistently colonized and that their microbiota are stably established over time with minimal strain dropout [45, 46, 47]. For short experiments, this can be done by cohousing animals, wherein they will engage in coprophagy and exchange microbiota content [51]. For longer studies, littermate crossing can aid in microbiome stability between cages [51, 52]. All these factors must be taken into consideration not only when designing experiments, but also when interpreting results, reading the literature, and extrapolating conclusions about the role of the microbiota in human health from animal studies.
An additional concern, especially when considering environments previously presumed to be sterile or when tissue is isolated, is to include proper negative controls. Sequencing contamination can strongly influence results; therefore, researchers should include the extraction kit alone with no sample added or a skin swab control obtained at the same time and processed in the same way as the tumor sample. With proper controls, several bioinformatic tools exist to minimize the impact of contamination [53, 54]. Because of the advances in sequencing and computational techniques, it is vital to consider contamination when designing experiments to examine relationships between cancer and the local or gut microbiota. Indeed, Robinson et al. analyzed a large public repository, Cancer Genome Atlas, for microbial DNA and identified numerous skin contaminant taxa such as Staphylococcus epidermidis and Cutibacterium acnes. They also identified sequencing center-specific contaminants, reinforcing the importance of including appropriate negative controls [16]. To uncover associations between microbes and cancer, consideration of experimental design, including negative controls and sterile sample collection, is therefore vital.
Local tumor microbiota
Up to 15% of cancer malignancies worldwide can be attributed to infectious diseases [55]. Oncogenic viruses, such as EBV and HPV carry genes that directly interfere with cell cycle checkpoints and inhibit the activity of tumor suppressors, and thus can be considered initiators of cancer [56]. In contrast, bacteria tend to be considered cancer promoters; they contribute to oncogenesis by inducing inflammation and recruiting immune cells, thus increasing the production of reactive oxygen species [56]. However, this paradigm is shifting, and recent studies show that bacteria can directly interact with host genes that regulate the cell cycle, apoptosis, and other mechanisms of tumor suppression [57].
Helicobacter pylori-associated gastric cancer is a classic example of inflammation-induced carcinogenesis. Prolonged or dysregulated inflammation can lead to uncontrolled cellular proliferation and subsequent tumorigenesis [56]. Several groups have found that H. pylori-associated gastritis and peptic ulcer disease, a common precursor to stomach cancer, are often accompanied by upregulated Th17 activity [58, 59]. Further, H. pylori-infected tissues and associated gastric tumor microenvironments exhibit similarly upregulated Th17 activity and an increase in interleukin (IL)-17 production [60] (Figure 2A). Other bacterial taxa have been associated with increased tumorigenesis accompanied by upregulation of Th17 cells. Enterotoxigenic Bacteroides fragilis (ETBF) enhances colonic tumor formation in a mouse model in an enterotoxin and Stat3/Th17-dependent manner [61] (Figure 2A). This increase of pro-inflammatory Th17 cells in both H. pylori-associated gastric cancer and ETBF-associated colon cancer suggests that certain bacterial taxa have oncogenic potential.
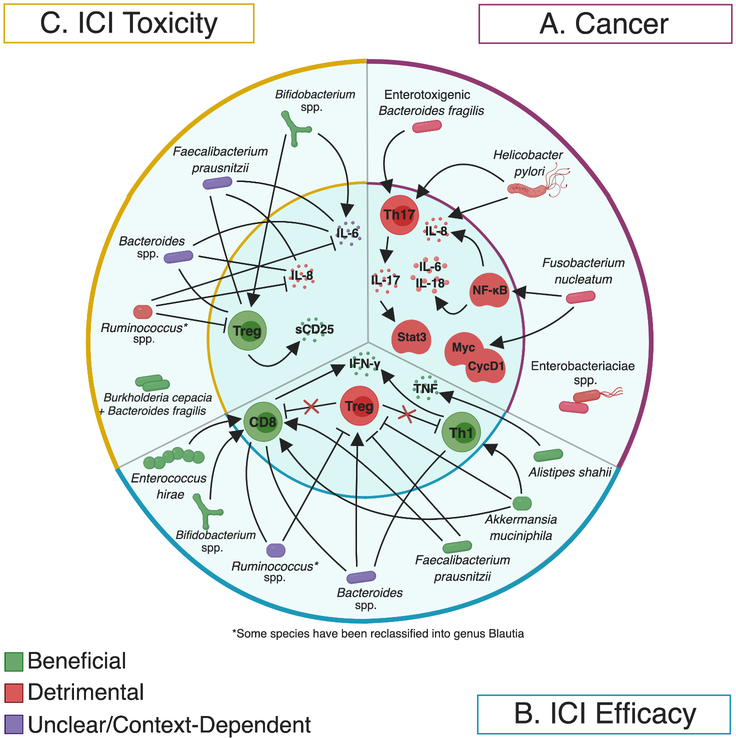
Figure 2. Interconnected relationships of the microbiota and immune system to affect cancer (A), immune checkpoint inhibitor efficacy (B), and colitis (C).
A line with an arrowhead indicates a positive association. A line with a perpendicular end indicates a negative association. A straight line indicates context- or species-specific association. An organism with no line indicates mechanism not yet elucidated.
A) Enterotoxigenic Bacteroides fragilis (ETBF), Helicobacter pylori, and Fusobacterium nucleatum have been shown to increase expression of inflammatory cytokines and/or host oncogenes resulting in cancer promotion [58, 62, 139]. Bacteria from the family Enterobacteriaceae have been detected in the pancreatic adenocarcinoma microenvironment [18].
B) Alistipes shahii, Akkermansia muciniphila, Enterococcus hirae, Faecalibacterium prausnitzii, Bifidobacterium spp., are associated with beneficial immunophenotypes in the context of immune checkpoint inhibitor therapy. Bacteroides spp. have mixed effects; they have been cited as detrimental, different strains enriched in both responders or nonresponders depending on the species, or beneficial. Ruminococcus spp. are associated with beneficial immunophenotypes in the context of therapy. Blautia obeum is associated with a detrimental immunophenotype [20, 83, 84, 85, 88, 89, 90].
C) Bacteroides fragilis, Burkholderia cepacia, and Bifidobacterium spp. may be protective against colitis [20, 104, 133]. Blautia obeum is associated with an increased risk of developing colitis. Different OTUs identified as Faecalibacterium prausnitzii and Bacteroides spp. were enriched in patients who did and did not develop ICI-associated colitis [85, 94]. Created with BioRender.
Inflammation is not the only mechanism by which bacteria can contribute to tumorigenesis. Fusobacterium nucleatum has been shown to induce inflammation and upregulate host oncogenes through interactions between its virulence factor FadA and E-cadherin expressed on epithelial cells, including colorectal cancer cells [62]. This binding triggers clathrin-mediated endocytosis of the bacterium, leading to phosphorylation of β-catenin and subsequent activation of NF-κB, which upregulates inflammatory cytokine production. Downstream activation of WNT, Myc, and CyclinD1 promote cellular proliferation and override cell cycle checkpoints (Figure 2A). Inhibition of clathrin prevented internalization and NF-κB activation, but the oncogenes were still upregulated [62]. This illustrates that bacteria can promote carcinogenesis simultaneously in multiple ways; inhibition of a single mechanism may be insufficient to treat or prevent disease.
Enrichment of certain bacteria has been identified in tumors without elucidated oncogenic mechanisms. For example, bacteria from the Enterobacteriaceae family were found by 16S rRNA gene sequencing and fluorescence in situ hybridization in 76% (86/113) of pancreatic adenocarcinoma tumor samples relative to 15% (3/20) of normal pancreas control samples [18]. In a mouse model of subcutaneous colon carcinoma, bacterial presence within tumors led to increased tumor size, and antibiotic treatment enhanced the efficacy of the chemotherapy drug, gemcitabine [18] (Figure 2A). It is unclear whether bacteria are present prior to cancer development or whether they colonize opportunistically, but local bacterial presence is an important consideration in cancer diagnosis and treatment. These data suggest the existence of a local tumor microbiota in certain cancers that can modulate therapy and also that the bacteria-immune interaction may be adjusted to fight cancer.
In addition to the presence of specific bacteria leading to immunological consequences connected to cancer, large scale case-control studies have demonstrated an association with frequent antibiotic courses in the ten-year interval prior to certain cancer diagnoses [63, 64]. Because cancer can affect the immune system for an interval prior to diagnosis, these authors excluded all antibiotics administered in the year prior to diagnosis, again suggesting that long-term bacterial dysbiosis could affect cancer predisposition [63]. More mechanistic studies are required to determine whether this association is driven by gastrointestinal dysbiosis or local, tumor-promoting communities and/or whether it is a direct oncogenic effect of antibiotics in humans. Animal studies have demonstrated that certain antibiotics enrich for pathobionts capable of stimulating the immune system, leading to bacterial translocation into the mesenteric lymph nodes and disruption of the local inflammatory balance [65, 66, 67]. Certain antibiotics have also been shown to directly inhibit immune cells [67], but the mechanistic interplay among specific antibiotics, the immune system, and the microbiota is not fully elucidated.
Cancer immune therapy
Immune checkpoint inhibitors (ICIs) are monoclonal antibodies that block inhibition of T cell function. ICIs have been developed to block programmed cell death protein 1 (PD-1; nivolumab, pembrolizumab), programmed death ligand 1 (PD-L1; atezolimumab, avelumab, durvalumab), or cytotoxic T lymphocyte antigen 4 (CTLA-4; ipilimumab). The inhibition of these proteins, which negatively regulate immune function, enhances the cytotoxic activity of T cells to increase anti-tumor immunity [61]. The proteins natively suppress T cells at different steps; therefore, inhibitors each have a unique mechanism of action. CTLA-4 is an inhibitory receptor found on T cells that competes with cluster of differentiation (CD)28, a constitutively expressed receptor critical for T cell activation [61]. CTLA-4 is not constitutively expressed on resting T cells but is upregulated following T cell activation [68, 69]. Therefore, CTLA-4 plays an important role as an “off” switch to suppress immune activity, but it can detrimentally affect control of cancer growth by inhibiting cytotoxic CD8+ T cells. Antibody blockade of CTLA-4 prevents this inhibition, enabling anti-tumor cytotoxic lymphocyte activity to resume. Similarly, PD-1 also plays an important role in immune suppression. It is expressed on activated T cells, and upon binding to PD-L1 or PD-L2 on the surface of various immune cells, it inhibits T cell activity [70]. Like CTLA-4, it is upregulated upon T cell activation, particularly in regulatory T cells (Tregs) [61]. The PD-1 signaling pathway is essential for preventing autoimmune disease by limiting peripheral T cell activity [70]. Although these two pathways vary somewhat in the mechanisms by which they inhibit T cell activation, both separately and combined, they have been demonstrated to be valuable targets for ICIs in the control of cancer progression [71, 72, 73, 74].
Fundamentally, all ICIs upregulate immune activity that may have been suppressed by Tregs and other immunomodulatory cells. Tregs inhibit immune activity, which can be particularly detrimental in the tumor microenvironment where they have been implicated in the suppression of the anti-tumor inflammatory response that leads to immune tolerance of cancer [61, 75]. However, Treg activity can be beneficial in mouse models of microbially-induced cancer, suggesting that context and the local microbiome-immune milieu is crucial [76, 77, 78, 79]. The gastrointestinal microbiota is essential for the induction of Tregs that tolerate commensal organisms. In germ-free mouse models, induction of Tregs is significantly impaired, but it can be restored by colonization with a defined, nonpathogenic microbial consortium [80]. Furthermore, short-chain fatty acids (SCFAs), abundant metabolites produced by intestinal microbiota, have been demonstrated to be important drivers in the induction and activity of Tregs [81]. Overall, this evidence establishes the intimate relationship shared between commensal microbiota and the immunomodulatory activity of Tregs. This implies that the microbiota has the potential to act as a complicating factor in the ongoing battle among cancer, the immune system, and ICIs by acting on the same immune cells synergistically or antagonistically.
Microbiota and immune checkpoint inhibitor response
Recently, several groups have begun to characterize the role of the microbiota in cancer immunotherapy treatment outcomes to better understand the dichotomy between responders and non-responders (Figure 2B). Anti-IL-10R/CpG therapy, which works synergistically with PD-1 inhibitors [82] to allow proliferation of tumor-specific CD8+ T cells, is rendered less effective in the presence of antibiotics, thereby providing evidence that the microbiota plays a crucial role in modulating immune therapy efficacy in a mouse model [83]. Conventional mice treated with vancomycin, imipenem, and neomycin in drinking water for three weeks before tumor injection showed decreased tumor necrosis factor alpha (TNF-α) secretion and increased tumor volume [83]. Further suggesting a role of the microbiota, administration of probiotic Alistipes shahii via oral gavage restored TNF-α production in conventional mice, a promising proof-of-concept supporting the therapeutic potential of probiotics (Figure 2B) [83]. TNF-α was similarly low in tumor-bearing germ-free mice, again implicating the commensal microbiota in the anti-cancer immune response. Germ-free mice were not treated with antibiotics, so the additional effect of antibiotics in the absence of the microbiota is not clear. It is also unknown whether the replenishment of TNF-α levels was sufficient to restore the efficacy of the immune therapy to reduce tumor volume and increase survival. TNF-α secretion was not restored until four weeks after antibiotic therapy, suggesting there is an important pre-chemotherapy window in which to consider microbial composition during which antibiotics might be particularly detrimental.
ICI efficacy can be manipulated by altering microbiota composition [20, 84]. Anti-PD-L1 efficacy differed in C57BL/6 mice from two different vendors; although these mice are considered to be genetically identical, they have unique microbiota compositions, which the authors hypothesized explained the differential efficacy that was observed [50, 84]. Further implicating the microbiota, co-housing mice or orally gavaging feces between groups to obtain similar microbiota compositions reduced the differential treatment response between mice [84]. Alternatively, a transferred, soluble metabolite could manipulate gastrointestinal microbiota composition indirectly through an altered immune milieu. Further evidence supporting the microbiota as a factor mediating tumor regression was the correlation with Bifidobacterium spp. abundance determined by 16s rRNA sequencing and increased tumor infiltration by CD8+ T cells, a hallmark of anticancer activity. Administration of a Bifidobacterium spp. cocktail was sufficient to significantly increase the anti-PD-L1 efficacy in the non-responder group of mice [84] (Figure 2B). In a similar study, mice were colonized via FMT from patient samples, followed by tumor inoculation, and then treated with anti-CTLA-4 therapy [20]. They found that donor stool could be clustered according to taxonomy using principal component analysis (PCA), and the cluster correlating with highest relative abundance, Bacteroides spp., was most predictive of tumor decrease after anti-CTLA-4 treatment. Although Bacteroides spp. abundance correlated with response, a causal relationship could have been solidified following probiotic administration of a bacterial consortium. The correlation between microbial abundance and ICI response provides exciting avenues of research interrogation and suggests a possible intervention to adjust the patient microbiome to influence cancer therapy outcomes.
These preclinical studies invoked closer examination of human cohorts and lead to a series of prospective studies in search of microbial and immune signatures that could accurately predict patient response to treatment. Patients with the longest overall survival (>18 months) after ipilimumab treatment for metastatic melanoma had gut microbiota strongly enriched for Faecalibacterium prausnitzii, Gemmiger formicilis, and butyrate-producing bacterium SS2–1 [85] (Figure 2B). They found that certain baseline immune signatures, particularly lower frequencies of Tregs and α4+β7+-expressing T cells (an integrin which directs T cells to the intestine during periods of inflammation), are correlated with long term benefit (Figure 2B) [85, 86]. It is plausible that the baseline decrease in Tregs systemically could imply a local tumor-microenvironment with fewer Tregs that could be more easily be inhibited by ipilimumab [87]. Finally, in a PCA identifying different microbial compositions between responders and non-responders, the authors found significantly increased inducible T cell activation in response to the CTLA-4 blockade [85]. This study was the first in-depth analysis of the relationship among the microbiota, immunotherapy efficacy and toxicity (discussed in the following section), but the cohort size was small, only one type of cancer and drug therapy combination was examined, and the study was strictly observational without any recapitulation in an animal model. It is thus promising to link microbiota composition to ICI response (Figure 2B), and several clinical trials are currently investigating this relationship (Table 1).
Table 1.
Clinical trials currently recruiting to study microbiota with cancer immune therapy
| National Clinical Trial | Number Enrolled | Malignancy | Intervention | Objective | Outcomes | Analysis |
|---|---|---|---|---|---|---|
| 03196609 | 55 (single group) | NSCLC, hepatocellular, colorectal, breast, prostate, glioblastoma | ICI | Catalog of microbial genes in patient treated with immune checkpoint inhibitors | Microbial changes after one month of ICI | Stool sample prior to therapy; NSCLC subset will have 1 month post-treatment sample |
| 03819296 | 800 | Cutaneous melanoma, genitourinary cancer | ICI, infliximab, vedolizumab | Role of gut microbiota on immune checkpoint toxicity; effectiveness of FMT to treat colitis; Incidence of adverse events and toxicity | Adverse events and toxicities | Microbiome and immune comparison between before and after colitis and between those with toxicity versus those without |
| 03445858 | 15 | Pediatric/adult solid tumor, lymphoma | pembrolizumab, decitabine, and radiotherapy | Feasibility and response of combination therapy | Toxicity, tolerability, and anti-tumor activity | Immune response kinetics and stool microbiota diversity |
| 03331562 | 24 | pancreas | PD1 inhibitor pembrolizumab with or without vitamin D | Microbiota differences between groups | Disease progression | 16S rRNA sequencing with alpha and beta diversity metrics |
| 03341143 | 20 | PD-1 resistant melanoma | Responder- FMT prior to start of pembrolizumab | Response rates to therapy | compare gut microbiota between responders and non-responders and before and after treatment | OTU and bacterial diversity metrics |
| 03353402 | 40 | PD-1 resistant melanoma | Responder-FMT after PD-1 blockade failure | Response rate to therapy | Adverse events and FMT engraftment; immune cell composition | Stool OTU abundance; immune cell proportions and cytokines |
| 03358511 | 20 | Breast adenocarcinoma | Probiotics | Increase CD8+ T cell abundance | Number of cytotoxic CD8+ T cells | Flow cytometry for CD8+ T cells |
| 03290651 | 40 | Breast cancer predisposition | Lactobacillus probiotics | Change local breast microbiota | Breast microbiota diversity | Inflammatory cytokine analysis |
Abbreviations: NSCLC: non-small cell lung cancer; FMT: fecal microbiota transplant; OTU: operational taxonomic unit
Less than a year later, three large scale studies characterized the relationship between the gut microbiome and immune efficacy [88, 89, 90]. These three landmark papers have been the subject of several reviews by the authors as well as by third parties [4, 6, 7, 91, 92]. Importantly, all three studies identified key bacterial species associated with responders and non-responders (Figure 2B) and immunophenotypes associated with responders, and recapitulated these findings in a mouse model.
Routy et al. evaluated the microbiome and outcomes of a cohort of 100 patients diagnosed with either non-small cell lung cancer or renal cell carcinoma being treated with PD-1 blockade [88]. They discovered that responders had increased microbiota diversity that was enriched for Akkermansia muciniphila and Enterococcus hirae, among other species within the Firmicutes phylum (Figure 2B). Upon FMT of responder and non-responder stool samples into mice, they recapitulated clinical outcomes following tumor inoculation and anti-PD-1 treatment. To illustrate the role of the microbiota, mice that received a non-responder microbiota were supplemented with A. muciniphila and E. hirae, resulting in a favorable anticancer immune phenotype characterized by an increased CD4+ T cell: Treg ratio (Figure 2B) [88]. This study illustrates the correlation among microbiota, cancer immune therapy efficacy, and local immune response, and demonstrates the possibility of microbial supplementation to augment ICI activity. Gopalakrishnan et al. conducted a similar study in a cohort of 43 patients being treated with PD-1 blockade for metastatic melanoma. They too found responders had increased microbiota diversity compared to non-responders [89]. However, rather than identify species-specific enrichments between the strata, they characterized responders to have higher proportions of taxa belonging to the order Clostridiales (phylum Firmicutes) compared to non-responders, which had gut microbiota dominated by Bacteroidales (phylum Bacteroides) (Figure 2B). Mice colonized with responder fecal microbiota had significantly lower tumor growth accompanied by higher CD8+ tumor infiltration, an immune phenotype consistent with responder patients (Figure 2B) [89]. Matson et al. identified distinct microbiota compositions driven by increased Bifidobacterium longum proportions in responders within their cohort of 42 patients receiving anti-PD-1 or anti-CTLA-4 treatments for metastatic melanoma (Figure 2B) [90]. In agreement with Routy et al., A. muciniphila was detected in four patients who were all classified as responders, but it was undetected in the other survivors and therefore it was not considered a significant influencer of response in this cohort [88]. Matson et al. also found unique microbial signatures associated with modulating the speed of tumor growth upon introduction into a mouse model in the absence of ICI treatment but did not describe which key taxa were associated with fast or slow tumor growth. Mice transplanted with responder microbiota had significantly smaller tumors than mice transplanted with non-responder microbiota even in the absence of PD-1 blockade. This finding implies an anti-cancer effect of the specific bacteria-immune interaction. Mice colonized with responder microbiota had increased relative abundance of Collinsella aerofaciens, B. longum, Lactobacillus spp., Klebsiella pneumoniae, and Parabacterioides merdae, and their tumors were characterized by increased CD8+ T cells and IFNγ production (Figure 2B) [90].
Each of these studies identified different microbial signatures, with some overlap between them, that were correlated with ICI efficacy (Figure 2B). This may be explained by differences in cohorts (e.g. cancer diagnosis, co-morbidities, prior or concurrent exposure to antibiotics) or experimental design (e.g. microbiota analysis pipeline, production of humanized mice, in vivo cancer model). Alternatively, these findings could imply that different bacterial species or genera are accomplishing the same metabolic role or that some unifying feature (e.g. membrane composition, lipopolysaccharide, or other molecular signal) is being recognized by the gut mucosal immune system. These data encourage the use of deeper sequencing methods, such as shotgun metagenomic sequencing, as was done by some of the studies described above, instead of 16S rRNA sequencing, and they highlight the utility of performing metatranscriptomics, metaproteomics, and metabolic pathway analysis (Figure 1) to identify unifying signatures indicative of protective and detrimental microbiota.
Microbiome and immune checkpoint inhibitor toxicity
By removing inhibitory checkpoints on immune cell activity, ICIs activate effector T cells to target tumors, but have adverse effects against numerous other tissue types [93]. The most common sites of adverse effects (gut and skin) are characterized by rapid cellular turnover and have close association with bacteria, suggesting a role of the microbiota in toxicity. The relationship between the microbiota and host biology is particularly complex in the context of inflammation. Microbes can modulate inflammation, which can in turn shape the microbiota, leading to a positive feedback loop characterized by dysbiosis and disease. Although there is strong evidence that both the microbiota and the immune system play critical roles, it is unclear which is the driving force. It is likely a combination of both, with dynamics that vary with host background, microbial composition and function, and cancer diagnosis and treatments.
Up to 25% of patients treated with ipilimumab (CTLA-4 inhibitor) experience colitis, so clinicians and researchers have undertaken studies to identify associations between the microbiota and toxicity [94]. In their 2015 mouse experiments, Vétizou et al. showed that toll-like receptor (TLR) agonists (i.e. the microbiota), intraepithelial lymphocytes, and ICI treatment were all required to induce apoptosis and ICI-associated colitis [20]. Further, they demonstrated that a prophylactic gavage of Burkholderia cepacia and Bacteroides fragilis provided protection against colitis during anti-CTLA-4 treatment (Figure 2C). To the best of our knowledge, this was the first demonstration, not only that ICI-associated colitis was microbiota-dependent, but that prophylactic probiotic treatment can protect against colitis in mice [20].
Human cohorts have also been studied to determine whether gut microbiota composition could predict or modulate colitis in addition to ICI efficacy. In a prospective study, Dubin et al. obtained fecal samples at the start of ipilimumab therapy for metastatic melanoma and stratified patients who did or did not develop colitis thereafter [94]. The authors used both 16S rRNA and shotgun sequencing to identify an increased relative abundance of Bacteroidetes in patients who did not develop colitis (Figure 2C). Similarly, they identified functional pathways using HUMAnN to show polyamine transport and riboflavin, pantothenate, thiamine, and biotin biosynthetic pathways were enriched in those protected from colitis [39, 94]. It is unclear whether these pathways are simply Bacteroides-associated functional pathways, directly inhibit cytotoxic activity of T cells, or otherwise provide local immunomodulatory effects, but it is intriguing to be able to utilize patient stool at therapy onset to predict toxicity.
In a similar cohort, increased abundance of Bacteroidetes relative to Firmicutes correlated with a longer colitis-free therapy period [85] (Figure 2C). However, the observed decreased incidence of colitis was offset by decreased survival. The authors initially show that ipilimumab, a CTLA-4 antibody, did not affect microbiota composition at the phylum level during treatment in the absence of colitis. They also conclude that ipilimumab did not decrease Shannon or Simpson diversity throughout treatment, but because of the small numbers of patients (only four patients sampled at the end of therapy), the null hypothesis of no difference between timepoints could be secondary to high variance in microbial composition. During colitis, however, they found decreased Shannon diversity as well as specific reductions in members of the Firmicutes. These authors proceeded to show that in addition to better clinical response, increased relative abundance of Firmicutes at baseline correlated with colitis. In a linear discriminant analysis to identify taxa associated with colitis, two OTUs designated as Faecalibacterium prausnitzii were strongly correlated with differential response (Figure 2C). This discrepancy, which may be artifactual based on OTU picking or may represent different metabolic contributions, highlights the importance of identifying strain level differences using ASV-based methods for 16S rRNA sequencing [31, 34] or with shotgun metagenomic sequencing [41, 95]. To explain the balance between ICI response and toxicity, they performed flow cytometry on whole blood at baseline and correlated immune populations with microbiota composition. They found that patients who ultimately developed colitis had decreased baseline levels of blood IL-6, IL-8, and sCD-25 relative to those that did not develop colitis [85] (Figure 2C). IL-6 and IL-8 are pro-inflammatory cytokines and sCD-25 is important for IL-2 function in promoting Treg development [96]. Paradoxically, patients with a pro-inflammatory state at baseline had lower incidence of developing colitis during treatment. Although preliminary and requiring further replication, it is interesting that microbial and immune biomarkers potentially exist that could stratify patients at the start of treatment such that probiotics or fecal microbiota transplant could alleviate or lessen toxicity while maintaining appropriate therapeutic efficacy.
Modulation of the microbiota to improve therapeutic outcomes
Antibiotics
Antibiotic treatment of patients before and during ICI therapy is associated with worse outcomes in the majority of studies [88, 97, 98, 99]. Antibiotic prophylaxis is common with many chemotherapeutic regimens, and antibiotics are given during periods of febrile neutropenia, which is accompanied by bacteremia in up to 25% of episodes [100, 101]. An important consideration in interpreting the findings discussed in the section above and incorporating changes into clinical practice is the spectrum of antibiotic activity. Gram-positive anaerobes are responsible for many effects discussed previously, but not all antibiotics have anti-anaerobic activity. Effect depends not only on antibiotic activity and duration, but also on whether they are delivered during a critical developmental period [102, 103]. Most literature on antibiotic administration in the context of cancer immunotherapy is binary, that is, it only indicates treatment or lack thereof. When the antibiotic spectrum was accounted for, only broad-spectrum antibiotics, defined as active against gram-negative, gram-positive, and anaerobic bacteria, were associated with the greatest relative risk of ICI non-response in a multivariate analysis of a small cohort [97]. Oral vancomycin, which has minimal systemic absorption and targets only gram-positive bacteria, enhanced anti-CTLA-4 anti-tumor activity potentially by increasing Bacteroidales and local gut inflammation in mice [20]. The observed ~25% decrease in tumor size was offset by increased colitis, which implied a delicate balance between pro-inflammatory and anti-cancer effects. The increase in colitis observed with anti-CTLA-4 antibodies and vancomycin was since replicated in a dextran sulfate sodium mouse model with added rescue by a Bifidobacterium consortium administration [104]. Antibiotic prescribing practices are at the discretion of the clinician; antibiotics with differential spectrum of activity and bioavailability could be theoretically administered for the same disease [98, 105, 106]. Antibiotic resistant infections are common in patients with cancer, with both antibiotic administration and chemotherapeutics being likely contributing factors [107]. It is therefore crucial to consider antimicrobial stewardship to obtain the best therapeutic response from ICIs while maintaining a low incidence of antibiotic-resistant infections that complicate cancer therapy [108].
Prebiotics
Prebiotics are various fibers that are digestible by individual commensal species, promoting expansion of bacteria that can metabolize them and produce secondary metabolites such as SCFAs. Preclinical studies employing the use of prebiotics to alter the microbiota to inhibit oncogenesis have shown promising results. In 2015, Hu et al. tested the effects of a diet enriched with breakdown-resistant starch in a colitis-associated colorectal cancer model in rats. They found that rats fed the prebiotic-supplemented food had increased relative abundances of Ruminococcus spp. and Bifidobacterium spp., which correlated with significantly decreased tumorigenesis, increased cecal and fecal SCFA concentrations, and decreased expression of the inflammatory markers, NF-κB and TNF-α [109].
While numerous studies have linked high fiber diets to decreased cancer incidence [110, 111, 112], most failed to characterize the corresponding change in microbiota or posit an underlying mechanism. A high-fiber diet in xenograft mice led to an increase in serum butyrate, a microbially-derived SCFA, and a decrease in tumor volume, an effect that is lost with antibiotic treatment [112]. They also observed increased expression of Fas, a host-derived apoptosis-inducing receptor, as well as increased mRNA expression of tumor suppressor genes CDKN1A (p21) and CDKN1B (p27), but did not propose any bacterial taxa that could be regulating these changes [112]. A retrospective study of healthcare professionals discovered that a prudent (whole grain and dietary fiber rich) diet was associated with a lower risk of F. nucleatum-positive cancers compared to a Western (red and processed meat, refined grains, and sugar-rich) diet [111]. Further mechanistic studies need to be performed to fully elucidate these relationships and the dynamics among diet, microbiota, and cancer. If certain taxa are identified and validated as biomarkers predictive of patient response to immunotherapy, patient diet could be adjusted to encourage the flourishing of responder-associated taxa and the reduction of non-responder taxa. Prebiotics are generally accepted as a safe, noninvasive method to modulate the microbiota.
Probiotics
Probiotics are an increasingly promising tool to manipulate the gut microbiota to fight disease. The ingestion of live bacterial strains can help correct microbial dysbiosis, exclude pathogenic bacteria, and modulate the host immune status by directly interacting with immune cells or through the production of secondary metabolites [113]. Unlike prebiotics, which require bacteria of interest to exist already in the microbiota, probiotics can introduce exogenous bacteria and establish either transient or stable colonization. In addition to the aforementioned experiments demonstrating the potential for administration of probiotic bacteria to differentially affect ICI efficacy and toxicity, research is ongoing to find specific taxa that predictably modulate the immune system and shape the microbiota so as to preclude cancer development.
Dissecting complex interactions from prospective studies in which patients have distinct microbiome compositions (during a disease state and at baseline), co-morbidities, and treatments is a challenging endeavor. In 2017, Geva-Zatorsky et al. performed an ambitious screen of the human microbiota for immunomodulatory bacteria. They monocolonized germfree mice with 53 different bacterial species that broadly represented the genetic diversity of the human gut microbiota and quantified their ability to influence immune cell populations in the colon [114]. In concordance with other literature, segmented filamentous bacteria induced an increase in Th17 cells [115]. Interestingly, they also demonstrated that Bifidobacterium longum increased Th1 cells, further contributing to an already incongruous body of evidence describing the immunomodulatory potential of B. longum, which has previously been described as an inducer of Th2 response [116]. This further emphasizes the importance of considering strain level differences, which could be an underlying factor in these discrepant findings [117].
The use of probiotics to improve cancer outcomes and ICI efficacy and toxicity is still in experimental stages and is being evaluated in ongoing clinical trials (Table 1). One recent study characterized the effects of probiotic supplementation of Bifidobacterium lactis Bl-04 and Lactobacillus acidophilus NCFM on the gut microbiota in a small cohort of patients with colon cancer [118]. They began by comparing the microbiota of mucosal biopsies from healthy controls and mucosal and tumor biopsies from patients with colon cancer. They observed several taxa, including Fusobacterium spp., with significantly different relative abundances in the mucosa and tumor biopsies of cancer patients compared to healthy controls, demonstrating that the microbiota within the tumor microenvironment could be contributing to oncogenesis. Intriguingly, they showed that the relative abundances of several taxa were substantially affected by the probiotic intervention. Bacteria belonging to the genus Faecalibacterium expanded, whereas bacteria belonging to the Clostridiales order and Fusobacterium genus decreased in relative abundance [118]. Although these results did not reach statistical significance following false discovery rate correction, the small sample size (N=15 colon cancer patients) and range of probiotic treatment length (8–78 days) could be contributing to excessive variation. These results demonstrate an important proof of concept for the ability of orally administered probiotics to alter the tumor microenvironment. It remains to be seen whether this change in the microbiota contributes to an altered immune phenotype or improved patient outcomes.
In addition to modulating the gut microbiota to increase therapy responsiveness, researchers have trialed direct instillation of bacteria to promote tumor killing. Uropathogenic E. coli strain CP1, a chronic prostatitis isolate, was instilled into the urethra of mice with prostate cancer [119]. Mice treated with anti-PD-1 inhibitor and CP1 had increased survival over either treatment alone. The immunomodulatory effect of CP1 related to its local increase in cytotoxic T cells and decrease of Tregs. Given the interconnectivity of the gut microbiota and urogenital tract and the systemic immune system in the context of Uropathogenic E. coli infection, future experiments should consider the potential change in the gut microbiota and immune response resultant from bacteria instillation into the urinary tract [120, 121, 122]. Instillation of microbes to elicit anti-cancer activity has been demonstrated by Bacillus Calmette-Guerin treatment for bladder cancer, but engineering tumor-homing microbes that persist therein has the potential added benefit of preventing recurrence by maintaining an anti-cancer environment [119]. Long-term effects of this increased inflammatory state and the safety of these treatments needs to be evaluated before widespread use given possible side effects of local or systemic infection or secondary cancer development [123].
Postbiotics
In addition to prebiotics and probiotics, postbiotics are yet another approach to modulating the microbiome and immune milieu using microbial metabolites. Rather than relying on bacteria supported by prebiotics or introduced through probiotics, postbiotics surpasses the bacteria entirely by providing the microbial product directly [124]. Although they have distinct advantages over prebiotics and probiotics by not being dependent on the cultivation of specific microbiota compositions with complex community dynamics, they require elucidation of a clear mechanism of action, which can be challenging. Once effector metabolites are identified, their increased shelf life, dosing reliability, safety profile, and decreased biological regulations compared to probiotics make postbiotics an appealing avenue for developing microbiota-based therapies [124].
One example of an effector molecule that can induce an immune phenotype in the absence of the viable microorganism is polysaccharide A (PSA) derived from B. fragilis. Round & Mazmanian showed that PSA administration alone was sufficient to induce expansion of Tregs and increase production of anti-inflammatory IL-10 in mice via TLR2 activation. Furthermore, PSA treatment in a 2,4,6-trinitrobenzene sulfonic acid model of colitis in mice was both protective and curative. PSA treatment one day post-induction of colitis resulted in decreased weight loss and improved colitis scores comparable to prophylactic treatment [125]. Although microbial antigens are often considered to act as adjuvants to stimulate an immune response, this study provides evidence that the immune cells affected by these antigens can promote immune suppression as well.
Propionate, acetate, butyrate, and succinate, collectively known as SCFAs, represent a class of metabolites produced by several bacteria in the gut [124, 126]. Because of their ubiquitous presence despite vast differences in gut microbiota composition from person to person, SCFAs are being investigated for their potential as universal metabolic regulators of the immune system. Smith et al. demonstrated their ability to induce colonic Treg proliferation in germfree mice in a manner dependent on free fatty acid receptor 2 [81]. Another study by Arpaia et al. showed butyrate similarly induced peripheral Treg proliferation [127]. These experiments are critical demonstrations that even in the absence of bacteria, exogenous bacterial metabolites can be utilized to influence immune activity.
Fecal microbiota transplantation
FMT is a process whereby healthy donor stools from another individual (allo-) or the same individual (auto-) are delivered via oral gavage or direct delivery into the gastrointestinal tract to balance or restore gut microbial composition. The largest use of FMT has been in the treatment of Clostridiodes difficile colitis and it has recently been employed for immunocompromised individuals, including in patients after hematopoietic stem cell transplantation (HSCT) [128, 129, 130]. Induction prior to HSCT is accompanied by broad-spectrum antibiotic treatment for prophylaxis and to treat infections. Mortality is increased in patients with decreased gut microbial diversity after HSCT relative to those with more diverse microbiota [130]. 16S rRNA sequencing of pre-allo-HSCT fecal samples and division of an 80-patient cohort into 3 groups based on inverse Simpson index showed that patients in the lowest diversity group had a 36% survival at 3 years whereas the highest diversity group had 67% survival [130]. The lower diversity group had received significantly more courses of vancomycin, metronidazole, and β-lactam antibiotics and had a 10-fold higher rate of C. difficile infection. Therefore, the negative correlation between gut microbial diversity at time of allo-HSCT and 3-year mortality may result from a loss of colonization resistance to pathobionts. Patients who died during the observation period were more likely to die from graft versus host disease or infection, both linked to decreased microbial diversity [130, 131]. Taur et al. additionally identified Enterobacteriaceae and Actinomycetaceae as biomarkers for death and survival, respectively [130]. Members of the Enterobacteriaceae such as Klebsiella pneumoniae, Escherichia coli, Citrobacter freundii, and Enterobacter cloacae are common causes of bacteremia in this population, and gram-negative bacteremia was associated with 45% mortality in a multi-center prospective cohort of patients after HSCT [132]. Thus, low diversity gut microbiota enriched with Enterobacteriaceae, which are often antibiotic resistant, may predispose patients to subsequent infection. It is therefore reasonable to hypothesize that increasing microbial diversity after HSCT or immunotherapy-induced dysbiosis via FMT could protect recipients from microbiota- and immune-related complications.
FMT was recently employed for the first time to treat two patients with steroid- and immune-suppressive refractory ICI-induced colitis [133]. After FMT, both patients had decreased systemic inflammatory cytotoxic T cells. Using 16S rRNA sequencing, the authors demonstrated differential microbial changes between the patients despite both becoming colitis-free. One patient had an expansion of Bifidobacterium in her gut after FMT, which is intriguing given a recent demonstration of its effect in abrogating colitis in mice [104, 133]. Although preliminary, these successes are promising and need to be confirmed with larger cohorts replete with frequent microbiome sampling and analysis to determine the protective species and functional pathways indicative of a therapeutic response.
Future directions
Baseline immunologic, gut microbial taxonomic, and functional pathway abundance could be used to develop personalized cancer immunotherapy treatments to maximize progression-free survival while minimizing toxicity. In the near future, fecal samples could be sequenced by clinical and molecular microbiology labs and directed microbiome adjustments could proceed simultaneously with or prior to initiating ICI treatment to augment response and limit toxicity. It is therefore vital to maximally replicate the findings described above and elsewhere in the literature in different cohorts with numerous analytical techniques (Figure 1). Studies are currently ongoing to identify microbial and immune differences correlating with toxicity and response to ICI (Table 1). Research studies with apparently conflicting taxonomic correlations discussed above could be related to insufficient resolution or different metabolic contributions of strain-level differences among the same species [134, 135]. We encourage researchers to consider incorporating functional metabolic profiling and strain tracking into longitudinal analysis to more precisely define the changes accompanying ICI therapy, toxicity, and response to microbial intervention.
In addition to potential microbiome adjustments with prebiotics, probiotics, postbiotics, antibiotics, and FMT, ample novel therapeutic strategies are emerging. An exciting technique would be to selectively kill pro-oncogenic or pathogenic bacteria with specific bacteriophages as has been recently achieved in other immunocompromised populations [136]. Genetically engineered microbes can also be used in vivo to diagnose and treat cancers [137]. It is compelling to consider other non-bacterial members of the microbiota including viruses and fungi and their role in modulating microbial composition and immune function as well as producing anti-cancer metabolites [138]. The era of personalized medicine is fast approaching due to the decreased cost of next-generation sequencing and the increased understanding researchers and clinicians share regarding the importance of the gut microbiome in regulating the immune system and disease.
Conclusions
Understanding microbiome compositions, manipulations, and interventions is now vital to understanding cancer progression and immunotherapy-based treatments. The details underlying the intricate relationship among the host immune system, cancer progression, the commensal and tumor microbiota, and therapeutic interventions continue to emerge. In addition, the tools and techniques used to characterize the microbiome are continually evolving to provide more data at lower costs. Although we have collectively learned much about commensal microorganisms and the modulatory role in local and systemic homeostasis, novel cancer immunotherapies have brought microbiome correlations and manipulations to the forefront. To progress beyond mere association in order to elucidate causation, we need to characterize precise molecular mechanisms by which the microbiota mediates cancer immunotherapy response and toxicity. Only after these molecular mechanisms are uncovered can interventions to modulate the microbiome to affect response and toxicity be widely employed. Continued partnerships among clinical scientists, laboratory physicians, and other translational researchers will help to usher in the era of personalized medicine with improved outcomes of many diseases while limiting often intolerable side effects.
Acknowledgements
This work was supported in part by awards to GD through the National Institute of Allergy and Infectious Diseases (NIAID: https://www.niaid.nih.gov/), the Eunice Kennedy Shriver National Institute of Child Health & Human Development (https://www.nichd.nih.gov/), and the National Center for Complementary and Integrative Health (https://nccih.nih.gov/) of the National Institutes of Health (NIH) under award numbers R01AI123394, R01HD092414, and R01AT009741, respectively. DJS is supported by a Pediatric Infectious Disease Society St. Jude Children’s Hospital Fellowship in Basic Research. ONR is supported by R25 GM 103757 IMSD Grant. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Abbreviations
- ASVs
amplicon sequence variants
- CD
cluster of differentiation
- CTLA-4
cytotoxic T lymphocyte antigen 4
- EBV
Epstein-Barr virus
- ETBF
enterotoxigenic Bacteroides fragilis
- FMT
fecal microbiota transplant
- HPV
human papilloma viruses
- HSCT
hematopoietic stem cell transplantation
- ICI
immune checkpoint inhibitors
- IL
interleukin
- OTUs
operational taxonomic units
- PCA
principal component analysis
- PCoA
principal coordinate analysis
- PD-1
programmed cell death protein 1
- PD-L1
programmed death ligand 1
- PSA
polysaccharide A
- rRNA
ribosomal RNA
- SCFAs
short-chain fatty acids
- SNPs
single nucleotide polymorphisms
- TLR
toll-like receptor
- TNF-α
tumor necrosis factor alpha
- Tregs
regulatory T cells
Footnotes
Conflict of interest
The authors report no conflict of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Nakamura K, Smyth MJ. Targeting cancer-related inflammation in the era of immunotherapy. Immunol Cell Biol. 2017. April;95(4):325–332. doi: 10.1038/icb.2016.126. [DOI] [PubMed] [Google Scholar]
- 3.Sambi M, Bagheri L, Szewczuk MR. Current challenges in cancer immunotherapy: Multimodal approaches to improve efficacy and patient response rates. J Oncol. 2019;2019:4508794. doi: 10.1155/2019/4508794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gopalakrishnan V, Helmink BA, Spencer CN, et al. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell. 2018. April 9;33(4):570–580. doi: 10.1016/j.ccell.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helmink BA, Khan MAW, Hermann A, et al. The microbiome, cancer, and cancer therapy. Nature Med. 2019. March;25(3):377–388. doi: 10.1038/s41591-019-0377-7. [DOI] [PubMed] [Google Scholar]
- 6.Elkrief A, Derosa L, Zitvogel L, et al. The intimate relationship between gut microbiota and cancer immunotherapy. Gut Microbes. 2018:1–5–5. doi: 10.1080/19490976.2018.1527167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fessler J, Matson V, Gajewski TF. Exploring the emerging role of the microbiome in cancer immunotherapy. J Immunother Cancer. 2019. April 17;7(1):108. doi: 10.1186/s40425-019-0574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012. June 13;486(7402):207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turnbaugh PJ, Ley RE, Hamady M, et al. The human microbiome project [10.1038/nature06244]. Nature. 2007. 10/18/print;449(7164):804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Young VB. The role of the microbiome in human health and disease: an introduction for clinicians. BMJ. 2017. March 15;356:j831. doi: 10.1136/bmj.j831. [DOI] [PubMed] [Google Scholar]
- 11.Sharon G, Sampson TR, Geschwind DH, et al. The central nervous system and the gut microbiome. Cell. 2016. November 3;167(4):915–932. doi: 10.1016/j.cell.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson NG, Hernandez-Leyva A, Kau AL. The ABCs of wheeze: Asthma and bacterial communities. PLoS Pathog. 2019. April;15(4):e1007645. doi: 10.1371/journal.ppat.1007645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ridaura VK, Faith JJ, Rey FE, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013. September 6;341(6150):1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly JR, Minuto C, Cryan JF, et al. Cross talk: The Microbiota and neurodevelopmental disorders. Front Neurosci. 2017;11:490. doi: 10.3389/fnins.2017.00490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garrett WS. Cancer and the microbiota. Science. 2015;348(6230):80–86–86. doi: 10.1126/science.aaa4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson KM, Crabtree J, Mattick JS, et al. Distinguishing potential bacteria-tumor associations from contamination in a secondary data analysis of public cancer genome sequence data. Microbiome. 2017. January 25;5(1):9. doi: 10.1186/s40168-016-0224-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flemer B, Lynch DB, Brown JM, et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017. April;66(4):633–643. doi: 10.1136/gutjnl-2015-309595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geller LT, Barzily-Rokni M, Danino T, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017. September 15;357(6356):1156–1160. doi: 10.1126/science.aah5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vijay P, McIntyre AB, Mason CE, et al. Clinical genomics: Challenges and opportunities. Crit Rev Eukaryot Gene Expr. 2016;26(2):97–113. doi: 10.1615/CritRevEukaryotGeneExpr.2016015724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vetizou M, Pitt JM, Daillere R, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015. November 27;350(6264):1079–84. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark AE, Kaleta EJ, Arora A, et al. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin Microbiol Rev. 2013. July;26(3):547–603. doi: 10.1128/CMR.00072-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martiny AC. High proportions of bacteria are culturable across major biomes. ISME J. 2019. April 5. doi: 10.1038/s41396-019-0410-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hugon P, Lagier JC, Robert C, et al. Molecular studies neglect apparently gram-negative populations in the human gut microbiota. JClin Microbiol. 2013. October;51(10):3286–93. doi: 10.1128/JCM.00473-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rossen JWA, Friedrich AW, Moran-Gilad J, et al. Practical issues in implementing whole-genome-sequencing in routine diagnostic microbiology. Clin Microbiol Infect:. 2018. April;24(4):355–360. doi: 10.1016/j.cmi.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Whelen AC, Persing DH. The role of nucleic acid amplification and detection in the clinical microbiology laboratory. Annu Rev Microbiol. 1996;50:349–73. doi: 10.1146/annurev.micro.50.1.349. [DOI] [PubMed] [Google Scholar]
- 26.Blauwkamp TA, Thair S, Rosen MJ, et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nature Microbiol. 2019. April;4(4):663–674. doi: 10.1038/s41564-018-0349-6. [DOI] [PubMed] [Google Scholar]
- 27.Boolchandani M, D’Souza AW, Dantas G. Sequencing-based methods and resources to study antimicrobial resistance. Nature Rev Genet. 2019:1. doi: 10.1038/s41576-019-0108-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.You HS, Lee SH, Ok YJ, et al. Influence of swabbing solution and swab type on DNA recovery from rigid environmental surfaces. J Microbiol Methods. 2019. June;161:12–17. doi: 10.1016/j.mimet.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 29.Hermans SM, Buckley HL, Lear G. Optimal extraction methods for the simultaneous analysis of DNA from diverse organisms and sample types. Mol Ecol Resour. 2018. May;18(3):557–569. doi: 10.1111/1755-0998.12762. [DOI] [PubMed] [Google Scholar]
- 30.Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol. 2007. September;45(9):2761–4. doi: 10.1128/JCM.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Callahan BJ, McMurdie PJ, Rosen MJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods. 2016. July;13(7):581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010. May;7(5):335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hillmann B, Al-Ghalith GA, Shields-Cutler RR, et al. Evaluating the information content of shallow shotgun metagenomics. mSystems. 2018;3(6):e00069–18–18. doi: 10.1128/msystems.00069-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Callahan BJ, McMurdie PJ, Holmes SP. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017. December;11(12):2639–2643. doi: 10.1038/ismej.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vetrovsky T, Baldrian P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PloS One. 2013;8(2):e57923. doi: 10.1371/journal.pone.0057923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segata N, Waldron L, Ballarini A, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nature Methods. 2012. August;9(8):811–4. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Truong DT, Franzosa EA, Tickle TL, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nature Methods. 2015. October;12(10):902–3. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 38.Davis C, Kota K, Baldhandapani V, et al. mBLAST: Keeping up with the sequencing explosion for (meta)genome analysis. J Data Mining Genomics Proteomics. 2015. August;4(3). doi: 10.4172/2153-0602.1000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abubucker S, Segata N, Goll J, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012;8(6):e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaminski J, Gibson MK, Franzosa EA, et al. High-Specificity Targeted Functional Profiling in Microbial Communities with ShortBRED. PLoS Comput Biol 2015. December;11(12):e1004557. doi: 10.1371/journal.pcbi.1004557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franzosa EA, McIver LJ, Rahnavard G, et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nature Methods. 2018;15(11):962–968–968. doi: 10.1038/s41592-018-0176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim ES, Zhou Y, Zhao G, et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nature Med. 2015. October;21(10):1228–34. doi: 10.1038/nm.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keen EC, Dantas G. Close Encounters of Three Kinds: Bacteriophages, Commensal Bacteria, and Host Immunity. Trends Microbiol. 2018. November;26(11):943–954. doi: 10.1016/j.tim.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knight R, Vrbanac A, Taylor BC, et al. Best practices for analysing microbiomes. Nature Rev Microbiol. 2018;16(7):410. doi: 10.1038/s41579-018-0029-9. [DOI] [PubMed] [Google Scholar]
- 45.Turnbaugh PJ, Ridaura VK, Faith JJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Science Transl Med. 2009. November 11;1(6):6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staley C, Kaiser T, Beura LK, et al. Stable engraftment of human microbiota into mice with a single oral gavage following antibiotic conditioning. Microbiome. 2017. August 1;5(1):87. doi: 10.1186/s40168-017-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seedorf H, Griffin NW, Ridaura VK, et al. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell. 2014. October 09;159(2):253–66. doi: 10.1016/j.cell.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kennedy EA, King KY, Baldridge MT. Mouse microbiota models: Comparing germ-free mice and antibiotics treatment as tools for modifying gut bacteria. Front Physiol. 2018;9:1534. doi: 10.3389/fphys.2018.01534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamamoto M, Yamaguchi R, Munakata K, et al. A microarray analysis of gnotobiotic mice indicating that microbial exposure during the neonatal period plays an essential role in immune system development. BMC Genomics. 2012. July 23;13:335. doi: 10.1186/1471-2164-13-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ericsson AC, Davis JW, Spollen W, et al. Effects of vendor and genetic background on the composition of the fecal microbiota of inbred mice. PloS One. 2015;10(2):e0116704. doi: 10.1371/journal.pone.0116704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robertson SJ, Lemire P, Maughan H, et al. Comparison of co-housing and littermate methods for microbiota standardization in mouse models. Cell Rep. 2019. May 7;27(6):1910–1919 e2. doi: 10.1016/j.celrep.2019.04.023. [DOI] [PubMed] [Google Scholar]
- 52.Stappenbeck TS, Virgin HW. Accounting for reciprocal host-microbiome interactions in experimental science. Nature. 2016. June 9;534(7606):191–9. doi: 10.1038/nature18285. [DOI] [PubMed] [Google Scholar]
- 53.Tanner MA, Goebel BM, Dojka MA, et al. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl Environ Microbiol. 1998. August;64(8):3110–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davis NM, Proctor DM, Holmes SP, et al. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6(1):226. doi: 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuper H, Adami HO, Trichopoulos D. Infections as a major preventable cause of human cancer. J Intern Med. 2000. September;248(3):171–83. [DOI] [PubMed] [Google Scholar]
- 56.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002. December 19-26;420(6917):860–7. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang AH, Parsonnet J. Role of bacteria in oncogenesis. Clinl Microbiol Rev. 2010. October;23(4):837–57. doi: 10.1128/CMR.00012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bagheri N, Razavi A, Pourgheysari B, et al. Up-regulated Th17 cell function is associated with increased peptic ulcer disease in Helicobacter pylori-infection. Infect Genet Evol :. 2018. June;60:117–125. doi: 10.1016/j.meegid.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 59.Serrano C, Wright SW, Bimczok D, et al. Downregulated Th17 responses are associated with reduced gastritis in Helicobacter pylori-infected children. Mucosal Immunol. 2013. September;6(5):950–959. doi: 10.1038/mi.2012.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pinchuk IV, Morris KT, Nofchissey RA, et al. Stromal cells induce Th17 during Helicobacter pylori infection and in the gastric tumor microenvironment. PloS One. 2013;8(1):e53798. doi: 10.1371/journal.pone.0053798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cancer Pardoll D. and the Immune system: Basic concepts and targets for intervention. Semin Oncol. 2015. Aug;42(4):523–38. doi: 10.1053/j.seminoncol.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rubinstein MR, Wang X, Liu W, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013. August 14;14(2):195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boursi B, Mamtani R, Haynes K, et al. Recurrent antibiotic exposure may promote cancer formation--Another step in understanding the role of the human microbiota? Eur J Cancer. 2015. November;51(17):2655–64. doi: 10.1016/j.ejca.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dik VK, van Oijen MG, Smeets HM, et al. Frequent use of antibiotics is associated with Colorectal cancer risk: Results of a nested case-control study. Dig Dis Sci. 2016. January;61(1):255–64. doi: 10.1007/s10620-015-3828-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kulkarni DH, McDonald KG, Knoop KA, et al. Goblet cell associated antigen passages are inhibited during Salmonella typhimurium infection to prevent pathogen dissemination and limit responses to dietary antigens. Mucosal Immunol. 2018. July;11(4):1103–1113. doi: 10.1038/s41385-018-0007-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Knoop KA, McDonald KG, Kulkarni DH, et al. Antibiotics promote inflammation through the translocation of native commensal colonic bacteria. Gut. 2016. July;65(7):1100–9. doi: 10.1136/gutjnl-2014-309059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scott NA, Andrusaite A, Andersen P, et al. Antibiotics induce sustained dysregulation of intestinal T cell immunity by perturbing macrophage homeostasis. Science Transl Med. 2018;10(464):eaao4755. doi: 10.1126/scitranslmed.aao4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lenschow DJ, Herold KC, Rhee L, et al. CD28/B7 regulation of Th1 and Th2 subsets in the development of autoimmune diabetes. Immunity. 1996. September;5(3):285–93. [DOI] [PubMed] [Google Scholar]
- 69.Freeman GJ, Lombard DB, Gimmi CD, et al. CTLA-4 and CD28 mRNA are coexpressed in most T cells after activation. Expression of CTLA-4 and CD28 mRNA does not correlate with the pattern of lymphokine production. J Immunol. 1992. December 15;149(12):3795–801. [PubMed] [Google Scholar]
- 70.Fife BT, Pauken KE, Eagar TN, et al. Interactions between PD-1 and PD-L1 promote tolerance by blocking the TCR-induced stop signal. Nature Immunol. 2009. November;10(11):1185–92. doi: 10.1038/ni.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O’Day SJ, Hamid O, Urba WJ. Targeting cytotoxic T-lymphocyte antigen-4 (CTLA-4): a novel strategy for the treatment of melanoma and other malignancies. Cancer. 2007. December 15;110(12):2614–27. doi: 10.1002/cncr.23086. [DOI] [PubMed] [Google Scholar]
- 72.Homet Moreno B, Ribas A. Anti-programmed cell death protein-1/ligand-1 therapy in different cancers. Br J Cancer. 2015. April 28;112(9):1421–7. doi: 10.1038/bjc.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Homet Moreno B, Mok S, Comin-Anduix B, et al. Combined treatment with dabrafenib and trametinib with immune-stimulating antibodies for BRAF mutant melanoma. Oncoimmunology. 2016. July;5(7):e1052212. doi: 10.1080/2162402X.2015.1052212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Curran MA, Montalvo W, Yagita H, et al. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc Natl Acad Sci USA. 2010. March 2;107(9):4275–80. doi: 10.1073/pnas.0915174107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Poutahidis T, Kleinewietfeld M, Erdman SE. Gut microbiota and the paradox of cancer immunotherapy. Front Immunol. 2014;5:157. doi: 10.3389/fimmu.2014.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Erdman SE, Poutahidis T, Tomczak M, et al. CD4+ CD25+ regulatory T lymphocytes inhibit microbially induced colon cancer in Rag2-deficient mice. Am J Pathol. 2003. February;162(2):691–702. doi: 10.1016/S0002-9440(10)63863-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Erdman SE, Rao VP, Poutahidis T, et al. CD4(+)CD25(+) regulatory lymphocytes require interleukin 10 to interrupt colon carcinogenesis in mice. Cancer Res. 2003. September 15;63(18):6042–50. [PubMed] [Google Scholar]
- 78.Shang B, Liu Y, Jiang SJ, et al. Prognostic value of tumor-infiltrating FoxP3+ regulatory T cells in cancers: a systematic review and meta-analysis. Sci Rep. 2015. October 14;5:15179. doi: 10.1038/srep15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Salama P, Phillips M, Grieu F, et al. Tumor-infiltrating FOXP3+ T regulatory cells show strong prognostic significance in colorectal cancer. J Clin Oncol. 2009. January 10;27(2):186–92. doi: 10.1200/JCO.2008.18.7229. [DOI] [PubMed] [Google Scholar]
- 80.Geuking MB, Cahenzli J, Lawson MA, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011. May 27;34(5):794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 81.Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013. August 2;341(6145):569–73. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun Z, Fourcade J, Pagliano O, et al. IL10 and PD-1 Cooperate to Limit the Activity of Tumor-Specific CD8+ T Cells. Cancer Res. 2015. April 15;75(8):1635–44. doi: 10.1158/0008-5472.CAN-14-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Iida N, Dzutsev A, Stewart CA, et al. Commensal Bacteria Control cancer response to therapy by modulating the tumor microenvironment. Science 2013;342(6161):967–970–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015. November 27;350(6264):1084–9. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chaput N, Lepage P, Coutzac C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. 2017. June 1;28(6):1368–1379. doi: 10.1093/annonc/mdx108. [DOI] [PubMed] [Google Scholar]
- 86.Erle DJ, Briskin MJ, Butcher EC, et al. Expression and function of the MAdCAM-1 receptor, integrin alpha 4 beta 7, on human leukocytes. J Immunol. 1994. July 15;153(2):517–28. [PubMed] [Google Scholar]
- 87.Tang F, Du X, Liu M, et al. Anti-CTLA-4 antibodies in cancer immunotherapy: selective depletion of intratumoral regulatory T cells or checkpoint blockade? Cell Biosci. 2018;8:30. doi: 10.1186/s13578-018-0229-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018. January 5;359(6371):91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 89.Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018. January 5;359(6371):97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients. Science. 2018;359(6371):104–108–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Keen EC, Crofts TS, Dantas G. Checkpoint Checkmate: Microbiota Modulation of Cancer Immunotherapy. Clin Chem. 2018. September;64(9):1280–1283. doi: 10.1373/clinchem.2017.286229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gharaibeh RZ, Jobin C. Microbiota and cancer immunotherapy: in search of microbial signals. Gut. 2018. December 8. doi: 10.1136/gutjnl-2018-317220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Johnson DB, Chandra S, Sosman JA. Immune checkpoint inhibitor toxicity in 2018. JAMA. 2018. October 23;320(16):1702–1703. doi: 10.1001/jama.2018.13995. [DOI] [PubMed] [Google Scholar]
- 94.Dubin K, Callahan MK, Ren B, et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nature Communic. 2016;7(1):10391. doi: 10.1038/ncomms10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Brito IL, Alm EJ. Tracking strains in the microbiome: Insights from metagenomics and models. Front Microbiol. 2016;7:712. doi: 10.3389/fmicb.2016.00712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bell CJ, Sun Y, Nowak UM, et al. Sustained in vivo signaling by long-lived IL-2 induces prolonged increases of regulatory T cells. J Autoimmun. 2015. January;56:66–80. doi: 10.1016/j.jaut.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ahmed J, Kumar A, Parikh K, et al. Use of broad-spectrum antibiotics impacts outcome in patients treated with immune checkpoint inhibitors. Oncoimmunology. 2018;7(11):1–6–6. doi: 10.1080/2162402x.2018.1507670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Elkrief A, Raichani LE, Richard C, et al. Antibiotics are associated with decreased progression-free survival of advanced melanoma patients treated with immune checkpoint inhibitors. Oncoimmunology. 2019;8(4):1–6–6. doi: 10.1080/2162402x.2019.1568812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Derosa L, Hellmann MD, Spaziano M, et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small cell lung cancer. Ann Oncol. 2018. doi: 10.1093/annonc/mdy103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bow EJ. There should be no ESKAPE for febrile neutropenic cancer patients: the dearth of effective antibacterial drugs threatens anticancer efficacy. J Antimicrob Chemother. 2013. March;68(3):492–5. doi: 10.1093/jac/dks512. [DOI] [PubMed] [Google Scholar]
- 101.Teillant A, Gandra S, Barter D, et al. Potential burden of antibiotic resistance on surgery and cancer chemotherapy antibiotic prophylaxis in the USA: a literature review and modelling study. Lancet Infect Dis. 2015. December;15(12):1429–37. doi: 10.1016/S1473-3099(15)00270-4. [DOI] [PubMed] [Google Scholar]
- 102.Perez-Cobas AE, Artacho A, Knecht H, et al. Differential effects of antibiotic therapy on the structure and function of human gut microbiota. PloS One. 2013;8(11):e80201. doi: 10.1371/journal.pone.0080201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cho I, Yamanishi S, Cox L, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012. August 30;488(7413):621–6. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang F, Yin Q, Chen L, et al. Bifidobacterium can mitigate intestinal immunopathology in the context of CTLA-4 blockade. Proc Nat Acad Sci. 2018;115(1):157–161–161. doi: 10.1073/pnas.1712901115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Trifilio S, Mehta J. Antimicrobial prophylaxis in hematopoietic stem cell transplantation recipients: 10 years after. Transpl Infect Dis. 2014. August;16(4):548–55. doi: 10.1111/tid.12237. [DOI] [PubMed] [Google Scholar]
- 106.Fisher BT, Gerber JS, Leckerman KH, et al. Variation in hospital antibiotic prescribing practices for children with acute lymphoblastic leukemia. Leuk Lymphoma. 2013. August;54(8):1633–9. doi: 10.3109/10428194.2012.750722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Papanicolas LE, Gordon DL, Wesselingh SL, et al. Not just antibiotics: Is cancer chemotherapy driving antimicrobial resistance? Trends Microbiol. 2018. May;26(5):393–400. doi: 10.1016/j.tim.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 108.Robilotti E, Holubar M, Seo SK, et al. Feasibility and applicability of antimicrobial stewardship in immunocompromised patients. Curr Opin Infect Dis. 2017. August;30(4):346–353. doi: 10.1097/QCO.0000000000000380. [DOI] [PubMed] [Google Scholar]
- 109.Hu Y, Le Leu RK, Christophersen CT, et al. Manipulation of the gut microbiota using resistant starch is associated with protection against colitis-associated colorectal cancer in rats. Carcinogenesis. 2016. April;37(4):366–375. doi: 10.1093/carcin/bgw019. [DOI] [PubMed] [Google Scholar]
- 110.De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Nat Acad Sci USA. 2010. August 17;107(33):14691–6. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mehta RS, Nishihara R, Cao Y, et al. Association of Dietary patterns with risk of colorectal cancer subtypes classified by Fusobacterium nucleatum in tumor tissue. JAMA Oncol. 2017. July 1;3(7):921–927. doi: 10.1001/jamaoncol.2016.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wei W, Sun W, Yu S, et al. Butyrate production from high-fiber diet protects against lymphoma tumor. Leuk Lymphoma. 2016. October;57(10):2401–8. doi: 10.3109/10428194.2016.1144879. [DOI] [PubMed] [Google Scholar]
- 113.Hill C, Guarner F, Reid G, et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nature Rev Gastroenterol Hepatol. 2014. August;11(8):506–14. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 114.Geva-Zatorsky N, Sefik E, Kua L, et al. Mining the human gut microbiota for immunomodulatory organisms. Cell. 2017. February 23;168(5):928–943 e11. doi: 10.1016/j.cell.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009. October 30;139(3):485–98. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lopez P, Gonzalez-Rodriguez I, Gueimonde M, et al. Immune response to Bifidobacterium bifidum strains support Treg/Th17 plasticity. PloS One. 2011;6(9):e24776. doi: 10.1371/journal.pone.0024776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ruiz L, Delgado S, Ruas-Madiedo P, et al. Bifidobacteria and Their molecular communication with the immune system. Front Microbiol. 2017;8:2345. doi: 10.3389/fmicb.2017.02345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hibberd AA, Lyra A, Ouwehand AC, et al. Intestinal microbiota is altered in patients with colon cancer and modified by probiotic intervention. BMJ Open Gastroenterol. 2017;4(1):e000145. doi: 10.1136/bmjgast-2017-000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Anker JF, Naseem AF, Mok H, et al. Multi-faceted immunomodulatory and tissue-tropic clinical bacterial isolate potentiates prostate cancer immunotherapy. Nature Communic. 2018;9(1):1591. doi: 10.1038/s41467-018-03900-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Spaulding CN, Klein RD, Ruer S, et al. Selective depletion of uropathogenic E. coli from the gut by a FimH antagonist. Nature. 2017;546(7659):528. doi: 10.1038/nature22972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schilling JD, Lorenz RG, Hultgren SJ. Effect of trimethoprim-sulfamethoxazole on recurrent bacteriuria and bacterial persistence in mice infected with uropathogenic Escherichia coli. Infect Immun. 2002;70(12):7042–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schwartz DJ, Conover MS, Hannan TJ, et al. Uropathogenic Escherichia coli superinfection enhances the severity of mouse bladder infection. PLoS Pathog. 2015. January;11(1):e1004599. doi: 10.1371/journal.ppat.1004599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Larsen ES, Nordholm AC, Lillebaek T, et al. The epidemiology of Bacillus Calmette-Guerin infections after bladder instillation from 2002 through 2017: A Nationwide Retrospective Cohort Study. BJU Int. 2019. May 4. doi: 10.1111/bju.14793. [DOI] [PubMed] [Google Scholar]
- 124.Aguilar-Toalá JE, Garcia-Varela R, Garcia HS, et al. Postbiotics: An evolving term within the functional foods field. Trends Food Science Technol. 2018;75:105–114. doi: 10.1016/j.tifs.2018.03.009. [DOI] [Google Scholar]
- 125.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Nat Acad Sciences USA. 2010. July 6;107(27):12204–9. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jacobson A, Lam L, Rajendram M, et al. A Gut Commensal-Produced Metabolite Mediates Colonization Resistance to Salmonella Infection. Cell Host Microbe. 2018. August 8;24(2):296–307 e7. doi: 10.1016/j.chom.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013. December 19;504(7480):451–5. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Battipaglia G, Malard F, Rubio MT, et al. Fecal microbiota transplantation before or after allogeneic hematopoietic transplantation in patients with hematological malignancies carrying multidrug-resistance bacteria. Haematologica. 2019. February 7. doi: 10.3324/haematol.2018.198549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.DeFilipp Z, Peled JU, Li S, et al. Third-party fecal microbiota transplantation following allo-HCT reconstitutes microbiome diversity. Blood Adv. 2018. April 10;2(7):745–753. doi: 10.1182/bloodadvances.2018017731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Taur Y, Coyte K, Schluter J, et al. Reconstitution of the gut microbiota of antibiotic-treated patients by autologous fecal microbiota transplant. Sci Transl Med. 2018;10(460):eaap9489. doi: 10.1126/scitranslmed.aap9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jenq RR, Ubeda C, Taur Y, et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J Exp Med. 2012. May 7;209(5):903–11. doi: 10.1084/jem.20112408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schuster MG, Cleveland AA, Dubberke ER, et al. Infections in hematopoietic cell transplant recipients: Results from the organ transplant infection project, a multicenter, prospective, cohort study. Open Forum Infect Dis. 2017. Spring;4(2):ofx050. doi: 10.1093/ofid/ofx050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang Y, Wiesnoski DH, Helmink BA, et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nature Med. 2018. December;24(12):1804–1808. doi: 10.1038/s41591-018-0238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yassour M, Jason E, Hogstrom LJ, et al. Strain-level analysis of mother-to-child bacterial transmission during the first few months of life. Cell Host Microbe. 2018. July 11;24(1):146–154 e4. doi: 10.1016/j.chom.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Vatanen T, Plichta DR, Somani J, et al. Genomic variation and strain-specific functional adaptation in the human gut microbiome during early life. Nature Microbiol. 2018. December 17. doi: 10.1038/s41564-018-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dedrick RM, Guerrero-Bustamante CA, Garlena RA, et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nature Med. 2019. May;25(5):730–733. doi: 10.1038/s41591-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Landry BP, Tabor JJ. Engineering diagnostic and therapeutic gut bacteria. Microbiol Spectrum. 2017. October;5(5). doi: 10.1128/microbiolspec.BAD-0020-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Saber A, Alipour B, Faghfoori Z, et al. Cellular and molecular effects of yeast probiotics on cancer. Crit Rev Microbiol. 2017. February;43(1):96–115. doi: 10.1080/1040841X.2016.1179622. [DOI] [PubMed] [Google Scholar]
- 139.Wu C, Wang S, Wang F, et al. Increased frequencies of T helper type 17 cells in the peripheral blood of patients with acute myeloid leukaemia. Clin Exp Immunol. 2009. November;158(2):199–204. doi: 10.1111/j.1365-2249.2009.04011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]