Abstract
Objective
Recent studies have demonstrated that gut microbiota was closely related to metabolic disorders such as type 2 diabetes. Oral antidiabetic medications including metformin, acarbose and sitagliptin lowered blood glucose levels via acting on the gastrointestinal tract. The aim of the study was to observe the comparisons among those medications on gut microbiota composition.
Research design and methods
Zucker diabetic fatty rats (n=32) were randomly divided into four groups, and had respectively gastric administration of normal saline (control), metformin (215.15 mg/kg/day), acarbose (32.27 mg/kg/day), or sitagliptin (10.76 mg/kg/day) for 4 weeks. Blood glucose levels were measured during an intragastric starch tolerance test after the treatments. 16S rRNA gene sequencing was used to access the microbiota in the fecal samples.
Results
Metformin, acarbose, and sitagliptin monotherapy effectively decreased fasting and postprandial blood glucose levels (p<0.001). Acarbose group displayed specific cluster and enterotype mainly composed by Ruminococcus 2 while Lactobacillus was the dominant bacterium in the enterotype of the other three groups. The relative abundance of genera Ruminococcus 2 and Bifidobacterium was dramatically higher in acarbose group. Metformin and sitagliptin increased the relative abundance of genus Lactobacillus. Metagenomic prediction showed that the functional profiles of carbohydrate metabolism were enriched in acarbose group.
Conclusions
Metformin, acarbose and sitagliptin exerted different effects on the composition of gut microbiota and selectively increased the beneficial bacteria. Supplementation with specific probiotics may further improve the hypoglycemic effects of the antidiabetic drugs.
Keywords: Metformin, acarbose, sitagliptin, type 2 diabetes, gut microbiota
Significance of this study.
What is already known about this subject?
Antidiabetic drugs, metformin, acarbose, and sitagliptin monotherapy may respectively alter the gut microbiota to improve inflammation levels and glucose homeostasis compared with placebo.
What are the new findings?
Metformin, acarbose and sitagliptin at commonly used dosages exerted different effects on the composition of gut microbiota and selectively increased the beneficial bacteria.
Gut microbiota after acarbose treatment clustered into a specific enterotype mainly composed by Ruminococcus 2 at genus level, and Lactobacillus was the dominant bacterium in the enterotype of the other three groups.
The relative abundance of genera Ruminococcus 2 and Bifidobacterium was increased after acarbose treatment, and metformin and sitagliptin increased the relative abundance of genus Lactobacillus.
How might these results change the focus of research or clinical practice?
It is important to observe the interaction between gut microbiota and oral antidiabetic drugs.
Combination of antidiabetic drugs and specific probiotics may further improve the hypoglycemic effects.
Introduction
Microbiota colonized in gastrointestinal tract is closely associated with the occurrence of metabolic disorders such as obesity and type 2 diabetes.1 There were notable differences in composition and functions of microbiota among healthy women, those with impaired glucose tolerance and type 2 diabetes.2 A metagenome-wide association study in Chinese population showed that patients with type 2 diabetes had lower amount of beneficial butyrate-producing bacteria in the gut, and higher number of opportunistic pathogens compared with healthy subjects.3
Various interventions, including dietary modification, hypoglycemic medications and bariatric surgeries, change the bacterial community structures to improve glycemic control in type 2 diabetes mellitus.4 5 The effects of oral antidiabetic drugs via acting on the gastrointestinal tract include metformin, α-glycosidase inhibitors, and dipeptidyl peptidase 4 (DPP-4) inhibitors. Metformin, a first-line hypoglycemic drug, is widely used to reduce hyperglycemia by inhibiting intestinal glucose absorption and hepatic glycogen output via AMPK-dependent and non-AMPK-dependent pathways.6 Emerging evidence suggests that alterations in gut microbiota are involved in the antidiabetic effects of metformin.7 Metformin increased short-chain fatty acid (SCFA)-producing and mucin-degrading bacteria in patients with type 2 diabetes.8 Transplantation of intestinal microbiota from metformin-treated patients into germ-free mice improved glucose intolerance.9
Acarbose, a complex oligosaccharide produced by Actinoplanes, plays an important role in reducing postprandial hyperglycemia, by competitively inhibiting the activity of α-glycosidase in the brush border of the small intestine, and further reducing the absorption of monosaccharides by intestinal epithelial cells.10 The effects of acarbose on weight loss and HbA1c were similar to those of metformin in Chinese patients with type 2 diabetes.11 12 The reduction of carbohydrate hydrolysis induced by acarbose probably altered the microbial fermentation and further reduced the levels of lipopolysaccharides and inflammation cytokines in type 2 diabetes.13
Sitagliptin, a DPP-4 inhibitor, decreases postprandial blood glucose levels by preventing the degradation of incretin hormones. However, the effects of DPP-4 inhibitors on gut microbiota remain unclear. There were two animal studies implied that sitagliptin and vildagliptin might correct the microbial diversity in diabetic rats.14 15
Studies about the effects of hypoglycemic drugs on gut microbiota were mostly restricted to comparisons between monotherapy and placebo, while there were few studies focused on the differences among drugs. Therefore, we aimed to compare the effects of acarbose, metformin and sitagliptin on intestinal microbiota in Zucker diabetic fatty fa/fa (ZDF) rats.
Methods
Animals
Thirty-two 7-week-old male ZDF rats were acquired from Vital River Laboratory Animal Technology (Beijing, China). They were maintained on a 12-hour dark-light cycle with controlled temperature (20°C–23°C) and humidity (40%–60%) in Shanghai SLAC Laboratory Animal (Shanghai, China, SYXK2017-0008). Purina 5008 diet (containing 16.7% fat, 26.8% protein and 56.5% carbohydrate) and water were given ad libitum except the specific fasting time. After 4 weeks of induced diet, ZDF rats were considered as the model of type 2 diabetes mellitus. The Animal Research: Reporting of In Vivo Experiments guidelines and the National Institutes of Health guide for the care and use of laboratory animals were compliant in the animal experiment.
Drugs treatment
The ZDF rats were randomly divided into four groups according to different treatments: normal saline (control group), acarbose, metformin and sitagliptin. After induced diet, rats were administrated with certain solution via oral gavage: normal saline, acarbose (Bayer Health Care, Germany, H19990205, 32.27 mg/kg/day), metformin (Sino-American Shanghai Squibb Pharmaceuticals, H20023370, 215.15 mg/kg/day) and sitagliptin (Merck, Sharp & Dohme, UK, J20140095, 10.76 mg/kg/day) daily for 4 weeks. We employed commonly used dosages in the clinic, the dosage of acarbose is 100 mg, three times, daily; the dosage of metformin is 2000 mg, daily, and sitagliptin is 100 mg, daily. The dose of antidiabetic drugs for ZDF rats was calculated according to the formula: drat=dhuman×0.71/0.11.16
Intragastric starch tolerance test
The intragastric starch tolerance tests (IGSTT) were carried out after 4 weeks of intervention. After fasting for 12 hours, animals were given a gavage starch load (soluble starch, Sangon Biotech, Shanghai, China, 2 g starch/kg body weight dissolved in deionized water, total volume: 3 mL). Blood glucose levels were monitored by a glucometer (Accu-Chek, Roche, Swiss) at 0, 30, 60, 120 and 180 min.
Fecal sample collection, DNA extraction and PCR amplification
One week after IGSTT, with a 12-hour fasting, fresh fecal pellets were obtained directly by holding individual rat and collected separately into a sterile tube. The fecal samples were immediately stored at −80℃. Total microbial DNA was extracted from fecal sample by the EZNA soil DNA Kit (Omega Bio-tek, Norcross, GA, USA). The purification and concentration of DNA were detected by NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Wilmington, USA), and DNA quality was assessed by 1% (w/v) agarose gel electrophoresis. Total DNA was used as a template for PCR amplification (95°C for 3 min, followed by 27 cycles of 30 s at 95°C, 30 s at 55°C, and 45 s at 72°C, and a final extension at 72°C for 10 min). The V3–V4 hypervariable regions of 16S rRNA gene were amplified with specific bacterial primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) by thermocycler PCR system (GeneAmp 9700, ABI, USA). PCR products were extracted from a 2% (w/v) agarose gel and further purified by using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) and quantified using QuantiFluor-ST (Promega, USA) in accordance with protocols of manufacturers.
Illumina MiSeq sequencing and processing of sequencing data
Purified amplicons from PCR were pooled in equimolar and paired-end sequence (2×300) and based on an Illumina MiSeq platform (Illumina, San Diego, USA). The procedure followed the standard protocols of Majorbio Bio-Pharm Technology (Shanghai, China).
Raw fastq files were demultiplexed, and quality filtered by Trimmomatic. They were merged by Fast Length Adjustment of Short reads (FLASH) according to the following criteria: (1) the reads were truncated at any sites, when receiving an average quality score <20 over a 50 bp sliding window; (2) primers were completely matched (allowing two nucleotides mismatching), and reads containing ambiguous bases were removed; (3) sequences, overlapped longer than 10 bp, were merged by the overlap sequence.
Operational taxonomic units (OTU) were clustered with 97% sequence similarity cut-off by using UPARSE (V.7.1; http://drive5.com/uparse/). Chimeric sequences were identified and removed via UCHIME. The taxonomy of each sequence was analyzed according to RDP Classifier algorithm (http://rdp.cme.msu.edu/) against the Silva (SSU123) 16S rRNA database with confidence threshold of 70%.
Clustering of microbial community types
To investigate OTU-level microbial beta diversity, the principal coordinates analysis (PCoA) and non-metrical multidimensional scaling (NMDS) were performed on the basis of Bray-Curtis distances. For enterotype analysis, the Jensen-Shannon divergence distance was used for clustering on both genus and OTU levels according to the methods described previously.17
Metabolic function prediction
Metagenomic prediction of functional composition was developed by using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt). The clusters of orthologous groups (COG) and Kyoto Encyclopedia of Genes and Genomes (KEGG) ortholog abundances were predicted for each sample and they were annotated in the certain database including evolutionary genealogy of genes: Non-supervised Orthologous Groups (eggNOG) (http://eggnog.embl.de/) and KEGG database (http://www.genome.jp/kegg/). The statistical analysis of taxonomic and functional profiles was performed by using the online interface Galaxy (http://huttenhower.sph.harvard.edu/galaxy/root). The linear discriminant analysis (LDA) effect size (LEfSe) algorithm was used to identify the functional pathways of microbiome within different treatments.
Statistical analysis
Statistical differences were analyzed by using SPSS Statistics V.23 software (IBM) and the free online platform of Majorbio I-Sanger Cloud Platform (https://www.i-sanger.com/). Blood glucose levels during IGSTT were analyzed by repeated measures analysis of variance (ANOVA) with Bonferroni post hoc test. Data were presented as mean±SEM.
Alpha diversity indices including the observed richness (sobs), the ACE estimator (ace), the Chao1 estimator (chao1) and Shannon diversity index (shannon) were measured. The significance of beta diversity was determined by using permutational multivariate ANOVA (pMANOVA) (adonis function). The relative abundance of phylum, genus and species was compared by using the Kruskal-Wallis rank-sum test followed by Scheffe post hoc analysis and adjusted by the false discovery rate (FDR) correction. P<0.05 was considered as significant.
LEfSe algorithm was used to investigate taxonomic differences in gut microbiota. It was based on the Kruskal-Wallis sum-rank test followed by the pairwise Wilcoxon test. The alpha values were both set less than 0.05. The strategy of multiple comparisons among four groups was defined as all-against-all (more strict). Only functional categories reaching a log LDA significant threshold value of >3.0 were shown.
Results
Blood glucose levels
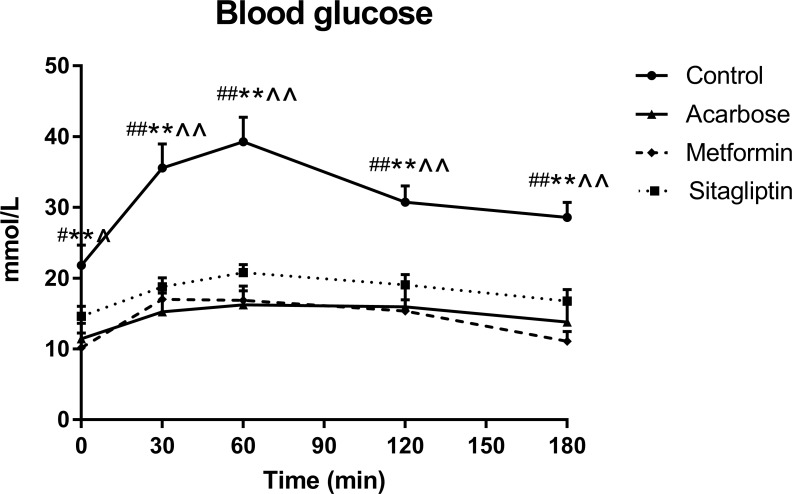
Fasting blood glucose levels were significantly lower after metformin, acarbose, and sitagliptin treatments compared with control group (figure 1, time * treatment interaction p≤0.001, respectively), with no significant differences among three treatment groups. Blood glucose levels rose after starch load in all groups. Postload blood glucose levels were lower at t=30, 60, 120 and 180 min following acarbose, metformin and sitagliptin treatments than control (p<0.05, respectively), with no differences among three treatment groups.
Figure 1.
Blood glucose levels during the intragastric starch tolerance tests (IGSTT) in control, acarbose, metformin and sitagliptin groups. Data were all expressed as mean±SEM. Values were assessed by one-way analysis of variance (ANOVA). The statistical significance was presented by special characters as #control versus acarbose; *control versus metformin; ∧control versus sitagliptin. One special character meant p<0.05 and two meant p<0.01.
Characteristics of sequencing data and alpha diversity of microbiota
Totally, 1 467 509 high-quality sequences were obtained from 32 samples, with an average length of 441.99 bp per sequence. The rarefaction curves indicated that the sequencing depth was adequate, and the coverage indices were >0.996. Alpha diversity indices showed that the richness (sobs, ace and chao1) and diversity (shannon) were decreased after acarbose monotherapy compared with control group (p≤0.001). The shannon of acarbose group was slightly lower than metformin (p<0.05) and sitagliptin group (p<0.01) (online supplementary figure S1).
bmjdrc-2019-000717supp002.pdf (764.5KB, pdf)
Bacterial community structure based on beta diversity and enterotypes
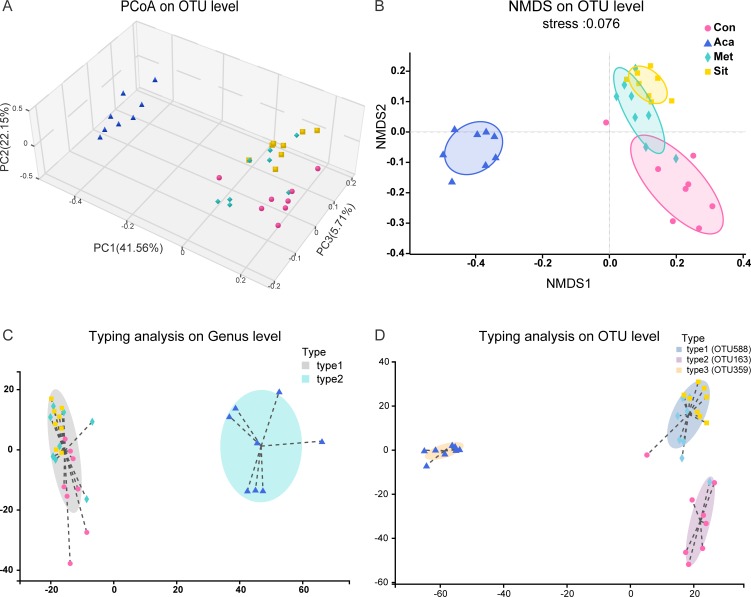
Beta diversity was used to assess the differences and similarities in fecal microbiota composition and further species complexity among the samples. The PCoA (figure 2A) and NMDS (figure 2B) exhibited that cluster of acarbose group was significantly specific compared with placebo, metformin or sitagliptin (PCoA, PC1 41.56%, PC2 22.15% and PC3 5.71%). The sample distances between acarbose and the other three groups were much greater (online supplementary figure S2). The pMANOVA based on Bray-Curtis distances revealed that there were significant differences among four groups (p=0.001 and p adjust=0.001).
Figure 2.
Beta diversity and enterotypes analysis in four groups. (A) Principal coordinates analysis (PCoA). The three principal coordinates (PC1–PC3) explain 41.56%, 22.15% and 5.71%, respectively, and the analysis of permutational multivariate analysis of variance (pMANOVA) was p=0.001 and p adjust=0.001. (B) Non-metrical multidimensional scaling (NMDS). The stress was 0.076. (C) Clustering of all 32 samples into two enterotypes at genus level. Type 1 was represented by Lactobacillus and type 2 was represented by Ruminococcus 2. (D) Typing analysis at OTU level. Types 1–3 were driven by OTU588 (Lactobacillus johnsonii sp) OTU163 (s_unclassified_g_Lactobacillus) and OTU359 (s_uncultured_bacterium_g_Ruminococcus_2), respectively. One dot represents one individual (n=8). Circles=control. Squares=acarbose. Diamond=metformin. Regular triangle=sitagliptin. OTU, operational taxonomic units.
bmjdrc-2019-000717supp003.pdf (1.1MB, pdf)
The composition of enterotypes was analyzed at genus (figure 2C) and OTU (figure 2D) levels. At genus level, the composition of intestinal microbiota was classified into two distinct enterotypes. Acarbose group displayed specific enterotype mainly composed by Ruminococcus 2, while Lactobacillus was the dominant bacterium in the enterotype of the other three groups. Furthermore, three enterotype clusters were found on the analysis at OTU level. The primary component in enterotype of acarbose group was OTU359 which was uncultured bacteria in Ruminococcus 2 genus. Samples from metformin or sitagliptin monotherapy were clustered together. The OTU588 representing Lactobacillus johnsonii sp was the most abundant taxon. OTU163, which formed unclassified species in Lactobacillus, was dominant in the enterotype of control group.
Characterization of core microbial communities
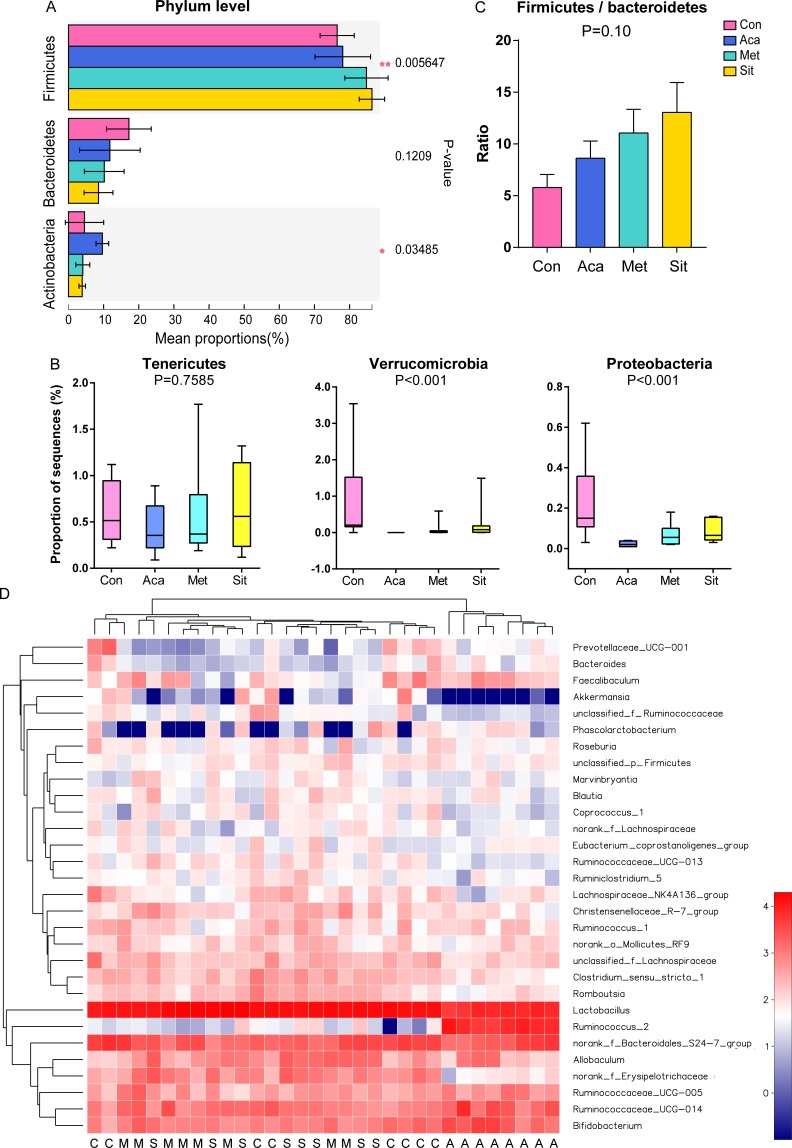
Out of eight bacterial phyla, phylum Firmicutes was the most abundant proportion in feces as predicted (figure 3A). It was increased after acarbose, metformin and sitagliptin treatment (p<0.05). Acarbose treatment increased the relative abundance of phylum Actinobacteria (p<0.05), especially comparing with the sitagliptin group. After the monotherapy of antidiabetic drugs, the relative abundance of phyla Verrucomicrobia and Proteobacteria was significantly decreased (figure 3B, p<0.001 and FDR <0.01, respectively). The phylum Cyanobacteria had the least proportion and it was slightly decreased after treatment of antidiabetic drugs (p<0.05, FDR=0.036, data did not show). There was no difference in phyla Bacteroidetes, Tenericutes and Saccharibacteria among four groups. Of note, the ratios of Firmicutes to Bacteroidetes in ZDF rats were increased by sitagliptin. The differences were not statistically significant (figure 3C).
Figure 3.
Characterization of core microbial communities. (A) Relative abundance of the domain phyla Firmicutes, Bacteroidetes and Actinobacteria. (B) Relative abundance of phyla Tenericutes, Verrucomicrobia and Proteobacteria. (C) Ratio of Firmicutes to Bacteroidetes. Kruskal-Wallis rank-sum test with adjusting by the false discovery rate (FDR) correction was used to assess the Firmicutes/Bacteroidetes ratio. (D) Heatmap showing top 30 abundant microbes at genus level (log10 transformation). One column represents one sample. A, acarbose group; C, control group; M, metformin group; S, sitagliptin group.
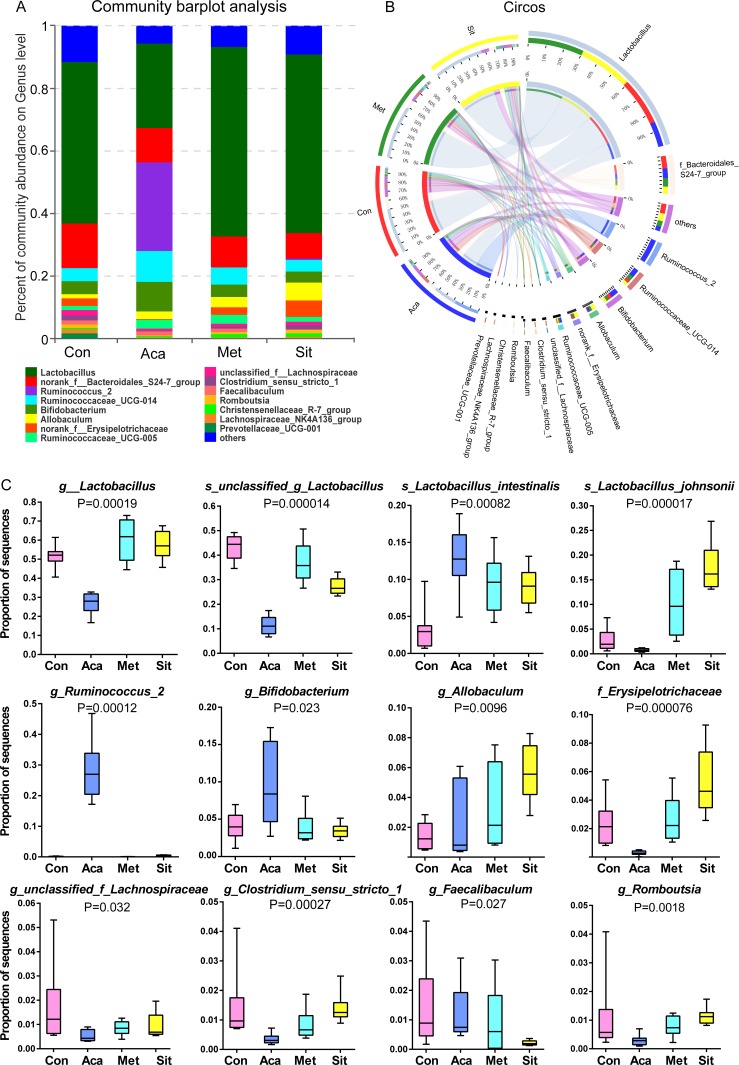
On the analysis of community abundance at genus level, Lactobacillus was dominated in intestinal microbiota of ZDF rats (figures 3D and 4A,B). It was significantly reduced in acarbose group while it was higher in metformin and sitagliptin groups (figure 4C). However, among the main species of genus Lactobacillus, acarbose has significantly increased the relative abundance of Lactobacillus intestinalis with no effects on L. johnsonii. Conversely, both L. intestinalis and L. johnsonii were significantly increased in response to metformin and sitagliptin therapy.
Figure 4.
Bacterial community abundance on specific taxa of each group. (A) Community barplot analysis showing the relative abundance on top 15 abundance genera. (B) Circos plot displaying the relationship between samples and bacterial genera. (C) Relative abundance of genus and specific species significantly altered by hypoglycemic agents.
As the decrease of unclassified species of Lactobacillus, the relative abundance of genera Ruminococcus 2 and Bifidobacterium was dramatically higher in acarbose group than the others (figure 4C). Ruminococcus 2 was hardly detected in the microbial communities of control, metformin and sitagliptin group.
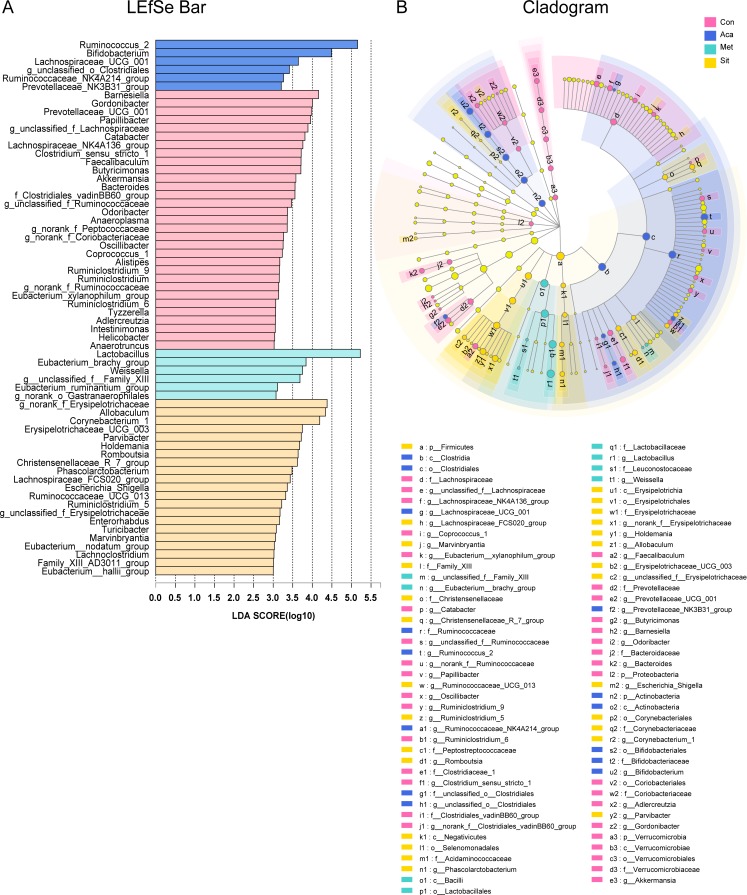
A LEfSe analysis with LDA measurement was performed to identify the taxonomic levels in the community composition among four groups (figure 5). At genus level, a differential enrichment of Ruminococcus 2, Bifidobacterium, Lachnospiraceae_UCG_001, g_unclassified_o_Clostridiales, Ruminococcaceae_NK4A214_group and Prevotellaceae_NK3B31_group was identified in acarbose group. Genus Lactobacillus was more abundant in metformin group. It was also observed that g_norank_f_Erysipelotrichaceae, Allobaculum, Corynebacterium 1 and Turicibacter were enriched in sitagliptin group.
Figure 5.
LDA effect size (LEfSe) analysis at genus level among control (Con), acarbose (Aca), metformin (Met) and sitagliptin (Sit) groups. (A) LEfSe barplot showing different abundance at bacterial genus level. (B) Cladogram representing the taxonomic levels by rings with phyla in the outermost and genera in the innermost ring. Only categories meeting a log linear discriminant analysis (LDA) significant threshold >3 are shown. The strategy of multiple comparison among four groups was defined as all-against-all. The prefixes ‘p’, ‘c’, ‘o’, ‘f’ and ‘g’ represent the level of phylum, class, order, family, and genus.
Metagenomic functional predictions
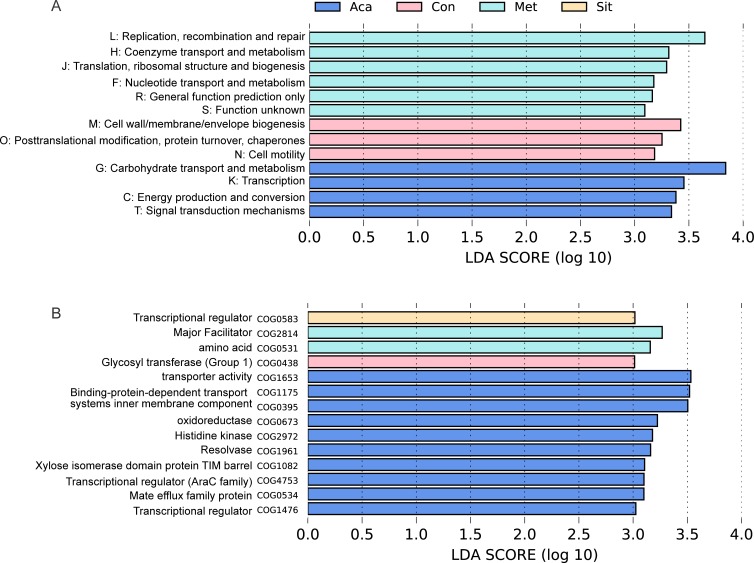
The metagenomic analysis revealed functional profiles of the gut microbiome in four groups. Functional COG categories assigned to the carbohydrate transport and metabolism [G] were the most abundant in these fecal samples (online supplementary figure S3). Notably, the reads [G] were enriched in acarbose group compared with those in metformin and sitagliptin groups (figure 6). Acarbose group also had more reads which were involved in energy production and conversion [C]. The COG hits, transporter activity (COG1653) and binding-protein-dependent transport systems inner membrane component (COG1175 and COG0395), were enriched in acarbose group. Moreover, acarbose treatment also elevated carbohydrate transport and metabolism [G] through xylose isomerase domain protein TIM barrel (COG1082).
Figure 6.
Functional profilings in metagenomic predictions by using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) and LDA effect size (LEfSe) analysis. LEfSe barplot showing different abundance of (A) clusters of orthologous groups (COG) functional classifications and (B) COG secondary classifications among control (Con), acarbose (Aca), metformin (Met) and sitagliptin (Sit) groups. They were annotated in evolutionary genealogy of genes: Non-supervised Orthologous Groups (eggNOG) database. Only log linear discriminant analysis (LDA) >3 was shown.
bmjdrc-2019-000717supp004.pdf (946.6KB, pdf)
With using PICRUSt and KEGG database on KEGG level 3 (online supplementary figure S4), nine functional categories were enriched in acarbose group including sporulation, amino acid metabolism (phenylalanine, tyrosine and tryptophan biosynthesis; valine, leucine and isoleucine biosynthesis; arginine and proline metabolism), transcription factors, glucose metabolism (pentose and glucuronate interconversions; pentose phosphate pathway; starch and sucrose metabolism) and DNA repair and recombination proteins. KEGG associated with purine metabolism was enriched in metformin group, while sitagliptin group upregulated the KEGG pathways of transporters, phosphotransferase system, and ATP-binding cassette transporters.
bmjdrc-2019-000717supp005.pdf (906.5KB, pdf)
Discussion
We have demonstrated that metformin, acarbose, and sitagliptin monotherapy similarly lowered blood glucose levels, but differently affected the diversities, composition and functions of gut microbiota in ZDF rats.
In our study, alpha diversity analysis was used to detect the diversity of microbiota after the treatments. We found that acarbose reduced the microbial richness and diversity, but there were no changes after metformin and sitagliptin treatments. It has been previously shown acarbose strikingly increased the relative abundance of beneficial bacteria in patients with type 2 diabetes, along with the decreased diversity.18 19 Other antidiabetic medications, such as glucagon-like peptide-1 agonists, liraglutide decreased the alpha diversity in obese and diabetic rats.20 Interestingly, it has been reported recently that alpha diversity was reduced after 8 weeks of treatment with dapagliflozin, a sodium-glucose co-transporter-2 inhibitor, which mainly improved blood glucose levels by inhibiting reabsorption of glucose filtered from the renal glomerulus.21 22 The indices of alpha diversity normally serve as a summary measure tool and might not be accurate to detect the components of gut microbiota. Therefore, we did further analysis on core microbial composition to clarify the effects of interventions.
We observed that the relative abundance of phylum Firmicutes and the ratio of Firmicutes to Bacteroidetes were increased after using three hypoglycemic agents. The ratio of Firmicutes to Bacteroidetes was positively correlated with fecal SCFAs.23 Although Larsen et al24 found the opposite results, it has been shown that the ratio of Firmicutes to Bacteroidetes was decreased in diabetic rats.25 After 4-week treatments of metformin, acarbose and sitagliptin, the ratio was reversed in our study. The effect of sitagliptin was the most notable among the three medications. Another, an abnormal expansion of phylum Proteobacteria plays an important role in the inflammation. In our current study, metformin, acarbose and sitagliptin dramatically decreased the abundance of Proteobacteria. It was striking to find that Proteobacteria tended to decline after thiazolidinediones (TZD), pioglitazone treatment in the high-fat diet-fed rats.5 26 The interaction between TZD and gut microbiota needs to be clarified in the future study.
As a predominant genus in phylum Firmicutes, Lactobacillus exhibits prominent antidiabetic effects via stimulating incretin hormones secretion and reducing endotoxemia.27 28 This study first proposed that different hypoglycemic agents could selectively regulate the abundance of Lactobacillus sp. Metformin and sitagliptin rose the overall abundance of Lactobacillus genus including L. johnsonii and L. intestinalis spp. Acarbose dramatically increased L. intestinalis, but reduced the species of unclassified Lactobacillus and L. johnsonii. It was validated by previous studies that acarbose18 and metformin7 promoted the level of Lactobacillus genus. Wang et al showed that the genera Lactobacillus, Allobaculum and Turicibacter were also enriched following liraglutide and saxagliptin dosage.29 30 There was little research on the taxonomic composition of sitagliptin. Only one study in a diabetic rat model showed that sitagliptin did not change Lactobacillus genus.14 In fact, the genus level of Lactobacillus is unable to encompass the entire species due to the various fermentations of Lactobacillus, which include homofermentation or heterofermentation.31
In addition to Lactobacillus, Bifidobacterium has anti-inflammatory effects and the relative abundance is reduced in type 2 diabetes.32 We demonstrated that acarbose increased the relative abundance of Bifidobacterium which was not changed in the other three groups. High carbohydrates intake could result in higher abundance of Bifidobacterium.33 Thus, the elevated abundance of Bifidobacterium might be due to the more exposure of carbohydrates to the distal gut by acarbose.
In 2011, the MetaHIT team proposed the concept of ‘Enterotypes’, which divided the intestinal microbiota into three types: Bacteroides, Prevotella and Ruminococcus.34 Enterotypes are mainly determined by the dominant bacteria and relatively stable, independent of age, gender, race, body mass index, and nutritional status.35 Although three medications similarly lowered blood glucose levels, the bacterial cluster of acarbose group was notably different from the other three groups in terms of enterotypes and beta diversity. At genus level, Ruminococcus 2 was the main taxonomic driver after acarbose treatment whereas Lactobacillus was the dominant genus in the other three groups. Further analysis of enterotypes showed that there were differences between antidiabetic treatment and control at OTU level. L. johnsonii (OTU588) was the dominant taxon followed by metformin and sitagliptin treatment while the control group was dominated by unclassified Lactobacillus sp (OTU163).
Ruminococcus 2 is a genus of bacteria in the family Ruminococcaceae and class Clostridia. Ruminococcus was originally isolated from bovine rumen and then found in various mammalian hosts, including humans, rodents, and so on. Fermentable carbohydrates are required in growth of all Ruminococcus.36 Therefore, it is not surprising that acarbose increases the carbohydrates in the distal gut and may specifically promote the growth of Ruminococcus 2. Blaubia (Ruminococcus) obeum shares the same class Clostridia with Ruminococcus 2 and expresses α-glucosidases (Ro-αG1). Recently, a theory proposed that α-glycosidase inhibitors (acarbose, voglibose, miglitol) could affect the bacterial Ro-αG1 in human gut and exerted antidiabetic effects or created adverse gastrointestinal symptoms.37 Furthermore, Rumenococcus, SCFA-producing bacteria, mainly produced acetate and propionate38 to improve metabolic abnormalities and intestinal inflammation.39 Further functional predictions indicated that acarbose increased abundance of profiles in carbohydrate transport and metabolism [G], which mainly included transporter activity (COG1653) and binding-protein-dependent transport systems inner membrane component (COG1175 and COG0395). KEGG analysis confirmed that the enzyme function associated with carbohydrate metabolism was active in the intestinal flora after acarbose therapy, compared with metformin and sitagliptin.
There are some limitations in this study. First, the distribution of the flora significantly varies with the changes of pH and intestinal metabolites in different intestinal segments. There is comparatively less abundance of microbiota in the small intestine, and the dominant bacteria such as Ruminococcus and Clostridium mainly localize in the large intestine.40 We collected fecal samples to assess the composition of gut microbiota. Hence, it is hard to clarify the distribution characteristics of microbiota in the small intestine and proximal colon. Second, the animal model used in this study was a diabetic model, ZDF rat, which was mainly caused by genetic defects. It might be different in the distribution of the gut microbiota from that of the human body. Lastly, we used soluble starch which was extracted from corn during the IGSTT test. The type of starch, in particular, resistant starch, interacts differently with the microbiota, and the interaction between starch and drugs may also have a significant impact on it.41 In fact, we did collect the fresh fecal samples 1 week after the IGSTT. We believe that interaction between soluble starch and the different drugs would have little influence on the results.
In conclusion, our study found that metformin, acarbose and sitagliptin differently affected intestinal bacteria. Acarbose selectively increased the bacteria including genera Ruminococcus 2 and Bifidobacterium. Metformin and sitagliptin increased the relative abundance of Lactobacillus. Supplementation with specific probiotic may further improve the hypoglycemic effects of the antidiabetic drugs.
bmjdrc-2019-000717supp001.pdf (95.3KB, pdf)
Footnotes
MZ and RF contributed equally.
Contributors: MCZ analyzed the data, prepared the figures and wrote the manuscript. RLF conducted the animal experiments and evaluated the data. MY performed the animal experiments. CQ performed the fecal DNA extraction. WL and ZW contributed to the study design and reviewed the manuscript. JM is the guarantor of this study who has full access to all the data to guarantee the accuracy and integrity of the data.
Funding: This work was supported by the National Natural Science Foundation of China (grant number NSFC81670728), the Shanghai Municipal Education Commission–Gaofeng Clinical Medicine Grant Support (grant number 20181807) and the Clinical Research Funding in Renji Hospital affiliated to Shanghai Jiao Tong University (grant number PYZY16-020).
Competing interests: None declared.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Musso G, Gambino R, Cassader M. Obesity, diabetes, and gut microbiota: the hygiene hypothesis expanded? Diabetes Care 2010;33:2277–84. 10.2337/dc10-0556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karlsson FH, Tremaroli V, Nookaew I, et al. . Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013;498:99–103. 10.1038/nature12198 [DOI] [PubMed] [Google Scholar]
- 3.Qin J, Li Y, Cai Z, et al. . A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012;490:55–60. 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- 4.Aron-Wisnewsky J, Doré J, Clement K. The importance of the gut microbiota after bariatric surgery. Nat Rev Gastroenterol Hepatol 2012;9:590–8. 10.1038/nrgastro.2012.161 [DOI] [PubMed] [Google Scholar]
- 5.Montandon SA, Jornayvaz FR. Effects of antidiabetic drugs on gut microbiota composition. Genes 2017;8. doi: 10.3390/genes8100250. [Epub ahead of print: 30 Sep 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Viollet B, Guigas B, Sanz Garcia N, et al. . Cellular and molecular mechanisms of metformin: an overview. Clin Sci 2012;122:253–70. 10.1042/CS20110386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauer PV, Duca FA, Waise TMZ, et al. . Metformin alters upper small intestinal microbiota that impact a Glucose-SGLT1-Sensing glucoregulatory pathway. Cell Metab 2018;27:101–17. 10.1016/j.cmet.2017.09.019 [DOI] [PubMed] [Google Scholar]
- 8.de la Cuesta-Zuluaga J, Mueller NT, Corrales-Agudelo V, et al. . Metformin is associated with higher relative abundance of mucin-degrading Akkermansia muciniphila and several short-chain fatty acid-producing microbiota in the gut. Diabetes Care 2017;40:54–62. 10.2337/dc16-1324 [DOI] [PubMed] [Google Scholar]
- 9.Wu H, Esteve E, Tremaroli V, et al. . Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med 2017;23:850–8. 10.1038/nm.4345 [DOI] [PubMed] [Google Scholar]
- 10.Lebovitz HE, inhibitors alpha-G. Alpha-Glucosidase inhibitors.. Endocrinol Metab Clin North Am 1997;26:539–51. 10.1016/S0889-8529(05)70266-8 [DOI] [PubMed] [Google Scholar]
- 11.Weng J, Ji L, Jia W, et al. . Standards of care for type 2 diabetes in China. Diabetes Metab Res Rev 2016;32:442–58. 10.1002/dmrr.2827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J-P, Wang N, Xing X-Y, et al. . Efficacy of acarbose and metformin in newly diagnosed type 2 diabetes patients stratified by HbA1c levels. J Diabetes 2016;8:559–67. 10.1111/1753-0407.12337 [DOI] [PubMed] [Google Scholar]
- 13.Su B, Liu H, Li J, et al. . Acarbose treatment affects the serum levels of inflammatory cytokines and the gut content of bifidobacteria in Chinese patients with type 2 diabetes mellitus. J Diabetes 2015;7:729–39. 10.1111/1753-0407.12232 [DOI] [PubMed] [Google Scholar]
- 14.Yan X, Feng B, Li P, et al. . Microflora disturbance during progression of glucose intolerance and effect of sitagliptin: an animal study. J Diabetes Res 2016;2016:1–10. 10.1155/2016/2093171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Q, Xiao X, Li M, et al. . Vildagliptin increases butyrate-producing bacteria in the gut of diabetic rats. PLoS One 2017;12:e0184735 10.1371/journal.pone.0184735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Q, Xiao X, Li M, et al. . Acarbose reduces blood glucose by activating miR-10a-5p and miR-664 in diabetic rats. PLoS One 2013;8:e79697 10.1371/journal.pone.0079697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De-Souza DA, Greene LJ. Intestinal permeability and systemic infections in critically ill patients: effect of glutamine. Crit Care Med 2005;33:1125–35. 10.1097/01.CCM.0000162680.52397.97 [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, Fang Z, Zhang C, et al. . Effects of acarbose on the gut microbiota of prediabetic patients: a randomized, double-blind, controlled crossover trial. Diabetes Ther 2017;8:293–307. 10.1007/s13300-017-0226-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu Y, Wang X, Li J, et al. . Analyses of gut microbiota and plasma bile acids enable stratification of patients for antidiabetic treatment. Nat Commun 2017;8:1785 10.1038/s41467-017-01682-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao L, Chen Y, Xia F, et al. . A glucagon-like peptide-1 receptor agonist lowers weight by modulating the structure of gut microbiota. Front Endocrinol 2018;9:233 10.3389/fendo.2018.00233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee DM, Battson ML, Jarrell DK, et al. . Sglt2 inhibition via dapagliflozin improves generalized vascular dysfunction and alters the gut microbiota in type 2 diabetic mice. Cardiovasc Diabetol 2018;17:62 10.1186/s12933-018-0708-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whang A, Nagpal R, Yadav H. Bi-Directional drug-microbiome interactions of anti-diabetics. EBioMedicine 2019;39:591–602. 10.1016/j.ebiom.2018.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandes J, Su W, Rahat-Rozenbloom S, et al. . Adiposity, gut microbiota and faecal short chain fatty acids are linked in adult humans. Nutr Diabetes 2014;4:e121 10.1038/nutd.2014.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsen N, Vogensen FK, van den Berg FWJ, et al. . Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 2010;5:e9085 10.1371/journal.pone.0009085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou W, Xu H, Zhan L, et al. . Dynamic development of fecal microbiome during the progression of diabetes mellitus in Zucker diabetic fatty rats. Front Microbiol 2019;10:232 10.3389/fmicb.2019.00232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bai J, Zhu Y, Dong Y. Response of gut microbiota and inflammatory status to bitter melon (Momordica charantia L.) in high fat diet induced obese rats. J Ethnopharmacol 2016;194:717–26. 10.1016/j.jep.2016.10.043 [DOI] [PubMed] [Google Scholar]
- 27.Yadav H, Jain S, Sinha PR. Antidiabetic effect of probiotic Dahi containing Lactobacillus acidophilus and Lactobacillus casei in high fructose fed rats. Nutrition 2007;23:62–8. 10.1016/j.nut.2006.09.002 [DOI] [PubMed] [Google Scholar]
- 28.Simon M-C, Strassburger K, Nowotny B, et al. . Intake of Lactobacillus reuteri improves incretin and insulin secretion in glucose-tolerant humans: a proof of concept. Diabetes Care 2015;38:1827–34. 10.2337/dc14-2690 [DOI] [PubMed] [Google Scholar]
- 29.Wang L, Li P, Tang Z, et al. . Structural modulation of the gut microbiota and the relationship with body weight: compared evaluation of liraglutide and saxagliptin treatment. Sci Rep 2016;6:33251 10.1038/srep33251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kyriachenko Y, Falalyeyeva T, Korotkyi O, et al. . Crosstalk between gut microbiota and antidiabetic drug action. World J Diabetes 2019;10:154–68. 10.4239/wjd.v10.i3.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaunmüller T, Eichert M, Richter H, et al. . Variations in the energy metabolism of biotechnologically relevant heterofermentative lactic acid bacteria during growth on sugars and organic acids. Appl Microbiol Biotechnol 2006;72:421–9. 10.1007/s00253-006-0514-3 [DOI] [PubMed] [Google Scholar]
- 32.Lê K-A, Li Y, Xu X, et al. . Alterations in fecal Lactobacillus and Bifidobacterium species in type 2 diabetic patients in southern China population. Front Physiol 2012;3:496 10.3389/fphys.2012.00496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duncan SH, Belenguer A, Holtrop G, et al. . Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol 2007;73:1073–8. 10.1128/AEM.02340-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arumugam M, Raes J, Pelletier E, et al. . Enterotypes of the human gut microbiome. Nature 2011;473:174–80. 10.1038/nature09944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhute SS, Suryavanshi MV, Joshi SM, et al. . Gut microbial diversity assessment of Indian Type-2-Diabetics reveals alterations in eubacteria, archaea, and eukaryotes. Front Microbiol 2017;8:214 10.3389/fmicb.2017.00214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.La Reau AJ, Meier-Kolthoff JP, Suen G. Sequence-based analysis of the genus Ruminococcus resolves its phylogeny and reveals strong host association. Microb Genom 2016;2:e000099 10.1099/mgen.0.000099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan K, Tesar C, Wilton R, et al. . Interaction of antidiabetic α-glucosidase inhibitors and gut bacteria α-glucosidase. Protein Sci 2018;27:1498–508. 10.1002/pro.3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koh A, De Vadder F, Kovatcheva-Datchary P, et al. . From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 2016;165:1332–45. 10.1016/j.cell.2016.05.041 [DOI] [PubMed] [Google Scholar]
- 39.Sivaprakasam S, Prasad PD, Singh N. Benefits of short-chain fatty acids and their receptors in inflammation and carcinogenesis. Pharmacol Ther 2016;164:144–51. 10.1016/j.pharmthera.2016.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartstra AV, Bouter KEC, Bäckhed F, et al. . Insights into the role of the microbiome in obesity and type 2 diabetes. Diabetes Care 2015;38:159–65. 10.2337/dc14-0769 [DOI] [PubMed] [Google Scholar]
- 41.Warren FJ, Fukuma NM, Mikkelsen D, et al. . Food starch structure impacts gut microbiome composition. mSphere 2018;3. doi: 10.1128/mSphere.00086-18. [Epub ahead of print: 16 05 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjdrc-2019-000717supp002.pdf (764.5KB, pdf)
bmjdrc-2019-000717supp003.pdf (1.1MB, pdf)
bmjdrc-2019-000717supp004.pdf (946.6KB, pdf)
bmjdrc-2019-000717supp005.pdf (906.5KB, pdf)
bmjdrc-2019-000717supp001.pdf (95.3KB, pdf)