Abstract
Patient-derived xenografts (PDXs) are widely recognised as a more physiologically relevant preclinical model than standard cell lines, but are expensive and low throughput, have low engraftment rate and take a long time to develop. Our newly developed conditional reprogramming (CR) technology addresses many PDX drawbacks, but lacks many in vivo factors. Here we determined whether PDXs and CRCs of the same cancer origin maintain the biological fidelity and complement each for translational research and drug development. Four CRC lines were generated from bladder cancer PDXs. Short tandem repeat (STR) analyses revealed that CRCs and their corresponding parental PDXs shared the same STRs, suggesting common cancer origins. CRCs and their corresponding parental PDXs contained the same genetic alterations. Importantly, CRCs retained the same drug sensitivity with the corresponding downstream signalling activity as their corresponding parental PDXs. This suggests that CRCs and PDXs can complement each other, and that CRCs can be used for in vitro fast, high throughput and low cost screening while PDXs can be used for in vivo validation and study of the in vivo factors during translational research and drug development.
Keywords: Conditional reprogramming, Patient derived xenograft, Cell line models, Drug discovery, Bladder cancer
Introduction
Cell lines, their derived animal models and genetically engineered mouse models (GEMMs) have been commonly used in drug development, basic and translational cancer research. Cell lines and their derived models have been selected and cultured in vitro for a long time, acquired additional genetic and epigenetic alterations, and can behave dramatically differently from cancers in patients. After a few generations, there was a great irreversible genetic divergence between primary tumor and the cell line derived from that tumor [1]. GEMMs usually harbour one or a few genetic alterations, and cancers usually develop within a few weeks to months with uniform genetic alterations in contrary to cancers in human patients which usually develop after years of exposure of carcinogens, and harbour dozens to hundreds of genetic alterations [2]. Hence, it is not surprising that findings from these models are often not translated into clinical applications. For example, models to predict chemoresistance based on studies in cell lines are rarely translated into clinical practice [3–5] while only approximately 5% of oncology drugs that have gone through all preclinical studies and entered clinical trials are finally approved by the Food and Drug Administration [6].
To overcome the shortcomings of these models, patient-derived models of cancer (PDMCs) have recently been proposed to better replicate human cancers because they are directly derived from clinical patient cancer specimens with little and short out-of-patient intervention [7]. Not surprisingly, as we previously reported, patient-derived xenografts (PDXs) retained 92–97% of genetic alterations of their parental patient cancers [8]. Both positive and negative clinical outcomes of patient cancers to treatment can be reproduced in PDXs and retained over time [9].
There are several types of PDMCs currently being actively pursued for translational research and drug development. Patient-derived organoids (PDOs) and patient-derived xenografts (PDXs) are two robust preclinical models [10, 11]. Organoid cultures fairly maintain genetic stability, can be grown for a long period of time and are not clonal in selection, but rather capture partial heterogeneity of the original tumor [12]. Additionally, organoid cultures are more suitable for low-throughput rather than high throughput drug screening. On the other hand, PDXs are biologically stable when passaged in mice in terms of global gene-expression patterns, mutational status, metastatic potential, drug responsiveness and tumor architecture [13, 14]. Although preclinical PDX models closely recapitulate the human tumor heterogeneity that is needed for more-efficient oncology drug development, the major challenges include the immense resources needed to establish and maintain such living biobanks; high cost; low engraftment rate; interference of mouse stroma in the model-associated data generated from PDX tumor; low stringency in ‘response’ criteria that may not translate to benefit cancer patients and the lack of high throughput drug screening [14]. In addition, considering that it takes a long time (months) to establish the first (Passage 0) PDXs and more time to expand in order to generate sufficient numbers of PDXs for drug testing, individual patients with a rapidly progressing disease may not benefit from PDX studies. Clearly, PDX models need to be viewed as complementary to other preclinical models.
Recently, our group established CR technology that enables normal and tumor primary epithelial cells to be propagated indefinitely in vitro while maintaining their original karyotype, genetic integrity and differentiation ability [15, 16]. Unlike other cell models using viral [17–21] or cellular oncogenes [22], the CR protocol uses irradiated mouse fibroblasts and a ROCK inhibitor (Y-27632) to propagate epithelial cells indefinitely without any genetic manipulation. Interestingly, the induction of CR is rapid (within 2 days) and results from reprogramming of the whole cell population rather than clonal selection, as is the case with conventional cell lines, and thus tumor CRCs retain morphological features and intra-tumor heterogeneity [23]. Unlike embryonic stem cells and induced pluripotent stem (iPS) cells, CRCs from normal tissue do not express high levels of Sox2, Oct4, Nanog or Klf4 [24] and do not form teratomas in mice [16]. In tumor CRCs, the phenotypic and genotypic features of primary tumors are maintained and have recently been used to identify appropriate therapy for various cancers [25].
Even with multiple PDMCs being actively pursued, there is little direct comparison between these PDMCs on their genetic alterations and response to therapeutic intervention. In this study, we conducted biological comparisons of PDXs and CRCs, and determined whether CR technology can overcome the drawbacks associated with PDXs. We used our PDXs in bladder cancer for this comparison study as we have conducted extensive research in bladder cancer PDXs [8, 26–30], and are the only NCI-funded center (U54) to develop bladder cancer PDXs for health disparity, translational research and drug development.
Methods
Development of patient-derived bladder cancer xenografts and CR cell culture
The protocol for collecting cancer specimens and clinical information was approved by the Institutional Review Board of the University of California (UC) Davis (Protocol No. 218204). Written informed consents from all the participants were taken before any clinical information or specimens were collected. The protocol for animal work was approved by the UC Davis Institutional Animal Care and Use Committee (Protocol No. 17794). The PDX tumors were developed in 4–5 weeks old NOD. Cg-Prkdcscid Il2rgtm1Wjl/SzJ (aka, NSG) mice by subcutaneous implantation of fresh bladder cancer specimens (3–5mm3) as described previously [8]. Four CRC lines were generated from the above bladder PDX tumors according to an established protocol as described previously [15, 16]. CRCs were passaged at 1:8 when reached 80–90% confluent. The viability of the cells was measured using trypan blue staining before every passaging.
Differential trypsinization to separate epithelial cells from feeders
The epithelial cells were harvested from co-culture with irradiated feeder 3T3-J2 fibroblasts using two-step trypsinization [15, 16].
Genomic DNA isolation and PCR amplification
Total genomic DNA was isolated from the harvested cell pellets of CRCs using the DNeasy Blood and Tissue Kit (Qiagen). PCR amplifications of the genomic DNA samples from CRCs were performed using PCR master mix from Promega (Cat. no. M7502) using the primers for specific targets as shown in supplemental Table.
STR Profiling
Genomic authentication of the bladder PDX CRCs was conducted to ensure donor identity. This analysis was performed by short tandem repeat (STR) profiling using Genetica PowerPlex 16HS Cell line PCR kit (Genetica DNA Laboratories, a LabCorp brand).
DNA-sequencing
PCR products that showed single-band amplification were purified using the QIAquick PCR Purification Kit (Qiagen, Cat. No. 28104). Sanger sequencing of the purified PCR products were performed in GENEWIZ according to manufacturer’s instructions. Sequencing for each PCR samples were performed in both the directions using forward and reverse primers (Table 1). Sanger sequencing results were compared with whole-exome sequencing (WES) of the parental tumor PDXs [8].
Table 1.
Primers used for PCR amplification and sequencing.
| PDX CRC ID | Gene Symbol | Primer | Sequence 5’−−3’ |
|---|---|---|---|
| CRC269 and CRC269 R |
FAT4 | Forward | GTGTCTGTGGTTGAGAATGC |
| Reverse | TGGACCGGGTAGAAGACAGG | ||
| KMT2C | Forward | TGAAGAGTGGCTCCAGGAGA | |
| Reverse | CCTGTTCTAGCTGTTTCTGAACC | ||
| PIK3CA (for c.2176) | Forward | ACAGTTAGCCAGAGGTTTGG | |
| Reverse | AAACAACTCTGCCCCACTGC | ||
| PIK3CA (for c.3140) | Forward | AGCTATTCGACAGCATGCCA | |
| Reverse | GGTCTTTGCCTGCTGAGAGT | ||
| KMT2D (for c.2438) | Forward | CTATGCGCTGTGCCTGAGG | |
| Reverse | CTTCTCAAGCTCAGGGGAC | ||
| KMT2D (for c.7411) | Forward | GTTACCCCTCGCTTCCAGTC | |
| Reverse | CTTGGGACCTTGGCATGGAG | ||
| FOXA1 | Forward | GGCTTCTTCACTCGCTGTCT | |
| Reverse | CTACTGCGCCGGGACTCAG | ||
| SYNE1 | Forward | TGTTGTGGGGTTTCATTTCGT | |
| Reverse | TGCATTTTCCCTGGCTCACA | ||
| ZFR2 | Forward | GGGGCTTTGGTGTGTGTTTG | |
| Reverse | GTAGGTAGCCATGGTGGTGG | ||
| ADCY2 | Forward | GGAGGCTCTTAGAAACCAGAA | |
| Reverse | CCTGGTGGGATGTGGAAAGT | ||
| CRC293 | ARID1A | Forward | GCCCTGAACAATAACCTCACG |
| Reverse | GGTTGCCCGAAGCCGTAG | ||
| TP53 (for c.626) | Forward | GCTGGGGCTGGAGAGACG | |
| Reverse | GCACCACCACACTATGTCGA | ||
| TP53 (for c.743) | Forward | TGGCTCTGACTGTACCACCA | |
| Reverse | CTG GAG TCT TCC AGT GTG ATG | ||
| SYNE2 | Forward | TTGGCGTCTCTCAGAACAGC | |
| Reverse | CGATGTTTCACAGCTGGAACA | ||
| EP300 | Forward | TGGTGATTCCAGTCTGAATGAGT | |
| Reverse | ACAAATCCGGAGCTAGCCAC | ||
| CDKN2A | Forward | GCTTCCTTTCCGTCATGCC | |
| Reverse | TGGAAGCTCTCAGGGTACAA | ||
| MTOR | Forward | TGATGAACTTCGAAGCTGTGC | |
| Reverse | CTCAGTGACCTTCTTCTGCA | ||
| CRC382 | ATM | Forward | CTCAAACTATTGGGTGGATTTGT |
| Reverse | TCGTTTGCGAGAAGTGTCGA | ||
| TP53 (for c.610) | Forward | GCTGGGGCTGGAGAGACG | |
| Reverse | GCACCACCACACTATGTCGA | ||
| TP53 (for c.25) | Forward | TGCTGGATCCCCACTTTTCC | |
| Reverse | AGACAAGAGCAGAAAGTCAGTC | ||
| KLF5 | Forward | CGTTGTCACAGGTGAAAAGCC | |
| Reverse | GTGGTCAGAGCGCGAGAAG | ||
| MERTK | Forward | TGTGTGTGTATGTGTGTGTGT | |
| Reverse | ATGCTGCAATTCCTGAACGG | ||
| MYH10 | Forward | CTGTGTACGTATGTAATAGGGGCA | |
| Reverse | CCACTTGAAAATCCAAAATATGCTTCT | ||
| NUP98 | Forward | GCCTAGTCCCTCGTGAAAAGTC | |
| Reverse | GCATCAAGAAATGTGACTCACGG | ||
| SETD2 | Forward | GCCTATGTGGATCCCAGCAA | |
| Reverse | GCACTGGCAAGACAGCAAC |
Drug sensitivity assay
CRCs were cultured at 25% density taking same number of cells that examined. Irradiated 3T3-J2 feeders were taken at 10% in the CRC cultures. Low density of feeders compared to CRCs (approx. 25%) were used to reduce drug interference by the feeders. Next day, culture media were replaced with fresh media containing PI3K inhibitor GDC-0941 at 10 nM, 100 nM and 1000 nM. DMSO only was used as a control without adding drug. To quantify cell proliferation, cultures were monitored using the IncuCyte live-cell analysis system with IncuCyte ZOOM software (Essen BioScience).
Immunoblot analysis
Epithelial cells were harvested as discussed above and performed immunoblotting as described previously [31, 32]. The primary antibodies Phospho-AKT (S473, Cat. no. 9271), total-AKT (Cat. no. 2920), Phospho-ERK (Thr202/Tyr204, Cat. no. 9101), total-ERK (Cat. no. 9107) and GAPDH (Cat. no. 5174) were purchased from the Cell Signaling Technology (Danvers, MA). The secondary antibodies used were goat anti-rabbit-HRP (Bio-Rad Cat. no. 1706515) and goat anti-mouse-HRP (Bio-Rad Cat. no. 1706516).
Results
Establishment of CRCs from bladder PDX tumors
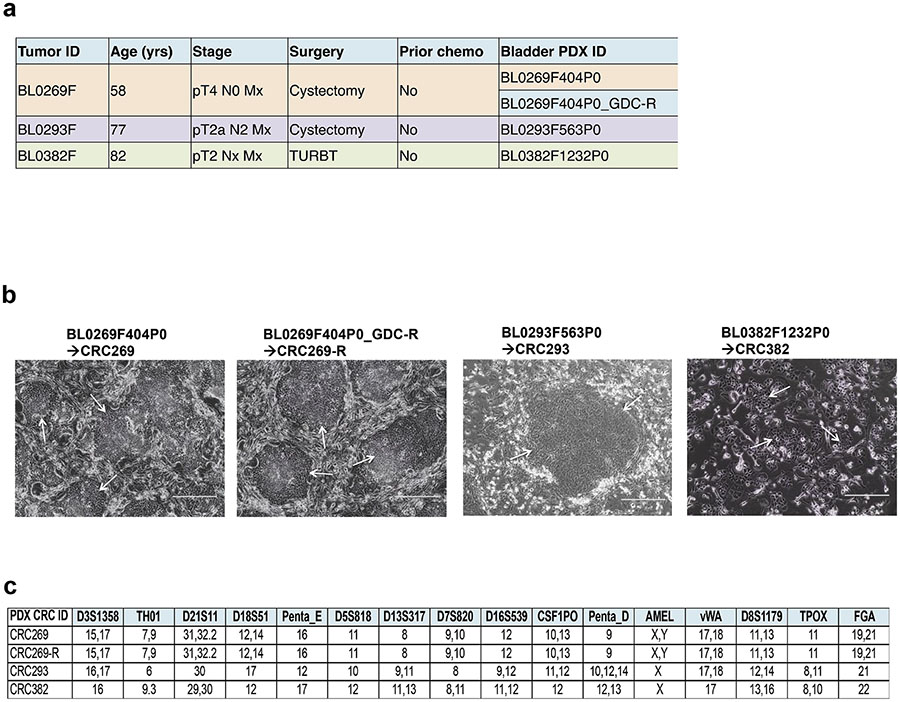
The PDX tumors used in this study were originally developed through subcutaneous implantation of patient tumors into immunocompromised NSG mice and consecutively passaged in vivo. None of the patients received any prior neoadjuvant chemotherapy (Fig. 1a). Among the four PDX tumors, two (BL0269 and BL0269_GDC-R) were from the same patient and two others were from two different patient donors (Fig. 1a). PDX tumor BL0269_GDC-R was developed from the parental BL0269 PDX after mice were treated with a PI3K inhibitor GDC-0941, and developed resistance to this drug. We have generated 4 CRC lines using the CR technology from the parental PDXs. Although the CRCs showed different cellular morphologies and varying doubling times, they all were growing efficiently and maintained steady rates of proliferation in the CRC conditions (Fig. 1b). CRC lines were tested and authenticated by STR profiling from Genetica DNA laboratories (Fig. 1c). As the CRC269 and CRC269-R were originated from the same donor’s PDXs, they showed the same allelic distribution of the STR markers.
Fig. 1.
Establishment of CRCs from 4 bladder PDX tumors. a Clinical characteristics of the 3 donor patients. Four PDXs were developed from the 3 advanced bladder cancer patients. BL0269_GDC-R was developed from a BL0269 PDX after with the host mouse was treated with a PI3K inhibitor GDC-0941 and developed resistant PDXs. b Representative images of CRCs grown (arrow) in co-culture with irradiated 3T3-J2 mouse fibroblasts. c STR analysis of the PDX CRCs after 3 passages in CR culture. Electropherograms are shown in Additional files 1–4: Figures S1–S4.
CRC lines maintain genetic mutations of the parental PDX tumors
The cancer cell lines do not accurately represent the genetics and heterogeneity of primary tumors and are therefore limited to their applicability in translational medicine [24]. Whole exome sequencing of the PDX tumors revealed mutations of known functionally active and significantly mutated genes previously identified in bladder cancer [7, 33, 34] and many of them are involved in transcriptional regulation, signal transduction and tumor suppression [35]. To determine if CRCs maintain the parental mutations identified in PDX tumors [8], targeted sequence analysis were performed for 20 genes (FAT4, KMT2C, KMT2D, PIK3CA, FOXA1, SYNE1, SYNE2, ZFR2, ADCY2, ARID1A, TP53, EP300, CDKN2A, MTOR, ATM, KLF5, MERTK, MYH10, NUP98 AND SETD2) in the cell lines of CRCs after 3 passages in CR culture conditions (Table 2). The genes considered in this study exhibit at least one single nucleotide variation leading to missense or nonsense mutations. All the CRCs consistently maintained genetic mutations, as demonstrated in the parental PDX tumors [8], in all the genes analysed in this study (Table 2).
Table 2.
Mutation statuses of 20 genes in the CRC lines in comparison with PDX tumors.
| PDX CRC ID | Bladder PDX ID | PDX CRC ID | Gene Symbol | Transcript ID | CDS mutation (in PDX) | AA Mutation (in PDX) | CDS mutation (in PDX CRCs) |
|---|---|---|---|---|---|---|---|
| CRC958-1 | BL0269F404P0 | CRC269 | FAT4 | NM_001291303.1 | c.1855C>T | p.R619C | c.1855C>T |
| KMT2C | NM_170606.2 | c.9694G>T | p.E3232★ | c.9694G>T | |||
| PIK3CA | NM_006218.2 | c.2176G>A | p.E726K | c.2176G>A | |||
| c.3140A>G | p.H1047R | c.3140A>G | |||||
| KMT2D | NM_003482.3 | c.2438C>T | p.P813L | c.2438C>T | |||
| c.7411C>T | p.R2471★ | c.7411C>T | |||||
| FOXA1 | NM_004496.3 | c.150G>A | p.M50I | c.150G>A | |||
| SYNE1 | NM_182961.3 | c.2653T>G | p.L885V | c.2653T>G | |||
| ZFR2 | NM_015174.1 | c.172G>A | p.G58S | c.172G>A | |||
| ADCY2 | NM_020546.2 | c.439G>T | p.V147L | c.439G>T | |||
| CRC958-2 | BL0269F404P0_GDC-R | CRC269-R | FAT4 | NM_001291303.1 | c.1855C>T | p.R619C | c.1855C>T |
| KMT2C | NM_170606.2 | c.9694G>T | p.E3232★ | c.9694G>T | |||
| PIK3CA | NM_006218.2 | c.2176G>A | p.E726K | c.2176G>A | |||
| c.3140A>G | p.H1047R | c.3140A>G | |||||
| KMT2D | NM_003482.3 | c.2438C>T | p.P813L | c.2438C>T | |||
| c.7411C>T | p.R2471 ★ | c.7411C>T | |||||
| FOXA1 | NM_004496.3 | c.150G>A | p.M50I | c.150G>A | |||
| SYNE1 | NM_182961.3 | c.2653T>G | p.L885V | c.2653T>G | |||
| ZFR2 | NM_015174.1 | c.172G>A | p.G58S | c.172G>A | |||
| ADCY2 | NM_020546.2 | c.439G>T | p.V147L | c.439G>T | |||
| CRC958-3 | BL0293F563P0 | CRC293 | ARID1A | NM_006015.4 | c.413C>G | p.S138★ | c.413C>G |
| TP53 | NM_001126112.2 | c.626G>A | p.R209Q | c.626G # | |||
| c.743G>A | p.R248Q | c.743G>A | |||||
| SYNE2 | NM_015180.4 | c.4177C>T | p.R1393W | c.4177C>T | |||
| EP300 | NM_001429.3 | c.4040G>T | p.G1347V | c.4040G>T | |||
| CDKN2A | NM_000077.4 | c.442G>A | p.A148T | c.442G>A | |||
| MTOR | NM_004958.3 | c.5533G>A | p.E1845K | c.5533G>A | |||
| CRC958-4 | BL0382F1232P0 | CRC382 | ATM | NM 000051.3 | c.5557G>A | p.D1853N | c.5557G>A |
| TP53 | NM_001126112.2 | c.610G>T | p.E204★ | c.610G>T | |||
| c.25G>T | Sense mutation | c.25G>T | |||||
| KLF5 | NM_001730.4 | c.1255G>C | p.E419Q | c.1255G>C | |||
| MERTK | NM_006343.2 | c.878G>A | p.R293H | c.878G>A | |||
| MYH10 | NM_001256012.1 | c.2879C>G | p.S960C | c.2879C>G | |||
| NUP98 | NM_016320.4 | c.3424C>G | p.Q1142E | c.3424C>G | |||
| SETD2 | NM_014159.6 | c.6694C>T | p.P2232S | c.6694C>T |
STOP codon
no mutataion observed
CRCs retain drug resistance as the parental PDX tumors
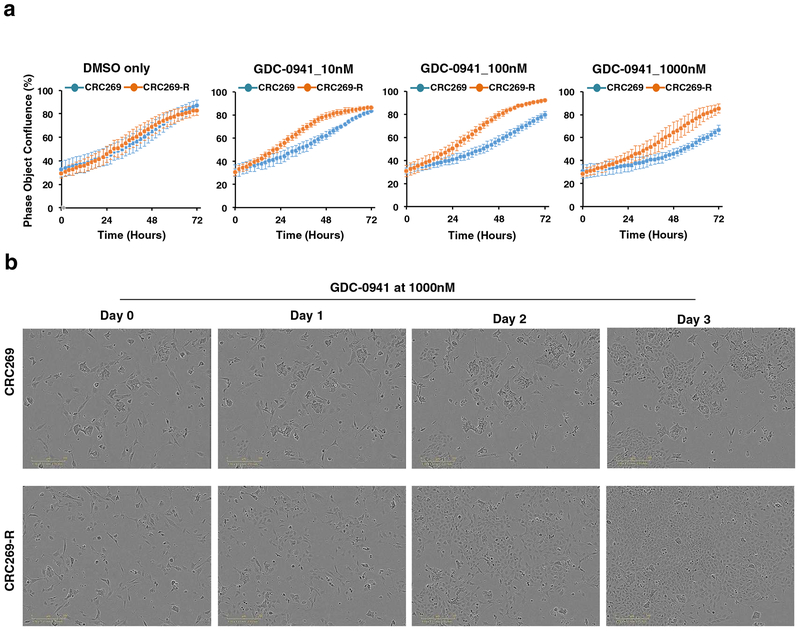
Next, we sought to determine if the CRCs could be used for in vitro drug sensitivity screening and if so, whether the cells maintain sensitivity to the targeted inhibitor in vitro as observed by the parental PDX tumors in vivo. PIK3CA, which encodes the p110a catalytic isoform of class I PI3K, is one of the most commonly mutated or amplified kinases in a variety of tumors [36]. The frequency of mutation and amplification of PIK3CA in bladder cancer ranged from 15.2% to 26.0% (http://www.cbioportal.org/). We previously reported that GDC-0941, a PI3K inhibitor, showed in vivo antitumor activity in bladder cancer PDXs [29]. We examined CRC269 and CRC269-R cells that were established from the PDX tumor BL0269 and BL0269_GDC-R, respectively. Although, these two PDXs were originated from the same patient donor, but BL0269_GDC-R showed in vivo resistance to the PI3K inhibitor GDC-0941 [29]. In this study, when the CRC lines were treated at 10–1000 nM of GDC-0941, we observed a dose dependent sensitivity of the CRC269 cells, whereas CRC269-R cells were mostly resistant to GDC-0941 (Fig. 2a and 2b). These results suggest that the drug sensitivity of the tumor PDX in vivo is retained in the CRCs in vitro.
Fig. 2.
In vitro drug sensitivity of the CRC lines in response to a PI3K inhibitor GDC-0941. a CRC269 and CRC269-R cells were treated with GDC-0941 at 10 nM, 100 nM and 1000 nM final concentration (n = 3, mean ± SD shown). DMSO only was the control cells. b Representative cell images of the CRC269 and CRC269-R cells treated with 1000 nM of GDC-0941 at Day 0, Day 1, Day 2 and Day 3.
Increased phosphorylation of AKT and ERK in the GDC-0941 resistant CRCs
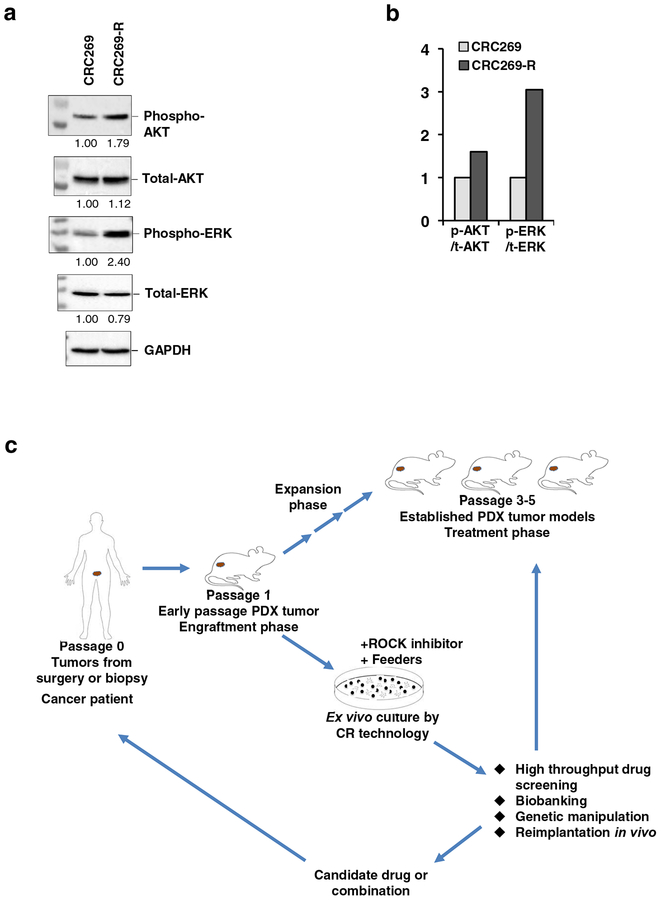
It is well evident that patients with solid tumors show marked increase of AKT and ERK activation [37–40]. In line with this mechanism, we previously [29] reported that the expression of phospho-AKT and phospho-ERK were upregulated when tumors become resistant to GDC-0941 treatment in the PDX BL0269 model. To explore the mechanism potentially underlying the development of drug resistance to the PI3K inhibitor in bladder cancer, and whether that is maintained in the CRC lines, we investigated the CRC269 and CRC269-R cells for AKT and ERK protein expression. CRC269-R cells showed increased expression of both the phosphorylated form of AKT and ERK as compared with the CRC269 cells, when the total levels were unchanged (Fig. 3a and 3b). These data suggest that alternative oncogenic pathways became activated to compensate for the PI3K/AKT pathway in the GDC-0941 resistant CRCs, similar to the previous report for the treatment resistant PDX tumors in vivo [29]. These findings implicate the importance of rational combinations of therapeutic agents to overcome drug resistance that was induced by compensatory activation of other pathways.
Fig. 3.
Increased phosphorylation of AKT and ERK in the GDC-0941 resistant CRCs and workflow. a Immunoblot analysis of phospho-AKT, total-AKT, phospho-ERK and total-ERK in the CRC269 and CRC-269-R cells. GAPDH was used as a loading control for quantification. Normalized densitometric values for expression levels are indicated below each lane relative to CRC269 (defined as 1.0). Uncropped blot images of the blots are shown in Additional file 5: Figure S5. b Bar graph of phospho-AKT and phospho-ERK levels compared with their total-AKT and total-ERK, respectively, in the cells shown in a. ‘p’ represents phospho, ‘t’ represents total. c Workflow for the establishment of CRCs from bladder PDXs and comparative molecular analysis of the CRCs.
Discussion
This is the first study showing that two patient-derived models of cancer retained the genetic alterations, response to targeted therapy matched to the underlying genetic alterations and the underlying signalling activities. Therefore, CRCs can potentially complement the in vivo PDX models, and be used to translational research and development of targeted therapies, but can be achieved at a much faster pace, and much lower cost.
Several in vitro patient-derived models of cancer (PDMC), such as the CR technology we developed [15], organoid [41–46], and induced pluripotent cells (iPS) [47, 48], have been widely employed in cancer research. Because of the in vitro culture system and lack of tumor microenvironment, these models are considered physiologically distinct from patient cancers in vivo. Hence, the in vivo PDXs are considered more physiologically relevant to cancers in clinical patients. We showed that the in vivo PDXs not only retained the pathohistological features, but also 92–97% of genetic alterations of parental patient cancers [8]. However, some intrinsic factors associated with PDXs, such as long engraftment time, low engraftment rate, and high cost, preclude its widespread use in translational research.
In this study we established four cell lines from the PDX tumors using the CR technology and assessed the genomic compositions and compared with their parental tumors. It has been always a challenge to establish a single model system that is rapid, simple to perform, and has a high rate of success. Conventional cell culture allows the least differential cells to thrive, resulting in distinct and irreversible losses of important biological properties, such as tumor heterogeneity and gene expression [1]. The CR technology meets all these needs and the CRCs retain cell lineage commitment with sustained expansion and maintain the heterogeneity of the cells present in a biopsy. This method offers the ability to expand PDX cells in vitro for subsequent high throughput drug screening assays, ex vivo genetic manipulation as well as for use in vivo to reduce animal usage, variability and study costs.
Overall, CRCs, like PDO and PDX, are able to capture the heterogeneity of the tumor of origin, but it still remains to be seen the level and similarity of heterogeneity captured by all these methods. Each of these above models has both merits and flaws with regard to their utility and in faithful representation of tumor architecture, microenvironment, cellular composition and heterogeneity, stem-differentiation states, growth patterns and responses to perturbagens, with respect to the patient specimen from which the model was initially derived. Combination of these models, for example, PDX-derived CRCs and CRC-derived xenografts (CDX,) will enable rapid, low cost expansion of the tumor cells, genetic manipulation, high-throughput analysis of drug sensitivity and comparisons with the genetic/cellular heterogeneity of these model systems (Fig. 3c).
Even though we studied only four PDX-derived CRCs, we previously showed that the success rate of CRC from primary patient cancer specimens is very high. For example, CRCs were established in 37/37 (100%) of primary prostate cancer specimens [15], and 18/18 (100%) with high-grade bladder cancer (unpublished data), compared to less than 5% with prostate cancer PDXs. Our results of retention of genetic alteration and drug response fidelity of PDXs by CRCs suggest CRCs can serve as a fast and low-cost alternative of the in vivo PDX model for translational research and precision medicine. One drawback of CR technology is lack of the in vivo factors. Hence drug absorption, distribution, metabolism and elimination or excretion (ADME), vascularization and local drug delivery can be assessed with the PDX platform, but CR. Combination of these two platforms can accelerate translational research and drug development, and readily translate patient-derived models of cancer for clinical applications.
In summary, we demonstrated that CRCs retained the fidelity of genetic alteration, drug response and signalling activity of the parental in vivo PDXs, and can potentially function as an alternative of the more expensive PDXs in the research of precision medicine, drug development and translational research.
Supplementary Material
Highlights:
Conditional Reprogramming Cells (CRCs) were established from PDX bladder tumors.
CRCs maintained same genetic alterations as those in the original PDX tumors.
CRCs retained the same drug sensitivity with the corresponding downstream signalling activity as their corresponding parental PDXs.
Funding
This study was supported by grants from NIH to X.L. (R21 CA180524–01) and to R.S. (R33 CA177466), and internal support from CCR at GUMC. C-X.P. was supported by Merit Review Award (I01 BX001784) from the U.S. Department of Veterans Affairs Biomedical Laboratory Research and Development Program, a U54 grant (1U54CA233306–01) and R01 grant (1R01CA176803–01A1) The contents do not represent the views of the U.S. Department of Veterans Affairs or the United States Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
Six patents for conditional reprogramming technology has been award to Georgetown University by the United States Patent Office. The license for this technology has been given to Propagenix for commercialization. The inventors (R.S. and X.L.) and Georgetown University receive potential royalties and payments from Propagenix.
References
- [1].Daniel VC, et al. , A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro, Cancer research, 69 (2009) 3364–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Vogelstein B, et al. , Cancer genome landscapes, Science (New York, N.Y.), 339 (2013) 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].McDermott U, et al. , Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling, Proc Natl Acad Sci U S A, 104 (2007) 19936–19941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Johnson JI, et al. , Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials, Br J Cancer, 84 (2001) 1424–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Barretina J, et al. , The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity, Nature, 483 (2012) 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kola I, et al. , Can the pharmaceutical industry reduce attrition rates?, Nature reviews. Drug discovery, 3 (2004) 711–715. [DOI] [PubMed] [Google Scholar]
- [7].Comprehensive molecular characterization of urothelial bladder carcinoma, Nature, 507 (2014) 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Pan CX, et al. , Development and Characterization of Bladder Cancer Patient-Derived Xenografts for Molecularly Guided Targeted Therapy, PloS one, 10 (2015) e0134346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Izumchenko E, et al. , Patient-derived xenografts effectively capture responses to oncology therapy in a heterogeneous cohort of patients with solid tumors, Ann Oncol, 28 (2017) 2595–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Okuyama H, et al. , Involvement of heregulin/HER3 in the primary culture of human urothelial cancer, The Journal of urology, 190 (2013) 302–310. [DOI] [PubMed] [Google Scholar]
- [11].Yoshida T, et al. , Organoid culture of bladder cancer cells, Investigative and clinical urology, 59 (2018) 149–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fujii M, et al. , A Colorectal Tumor Organoid Library Demonstrates Progressive Loss of Niche Factor Requirements during Tumorigenesis, Cell Stem Cell, 18 (2016) 827–838. [DOI] [PubMed] [Google Scholar]
- [13].Pompili L, et al. , Patient-derived xenografts: a relevant preclinical model for drug development, Journal of experimental & clinical cancer research: CR, 35 (2016) 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tentler JJ, et al. , Patient-derived tumour xenografts as models for oncology drug development, Nature reviews. Clinical oncology, 9 (2012) 338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Liu X, et al. , Conditional reprogramming and long-term expansion of normal and tumor cells from human biospecimens, Nature protocols, 12 (2017) 439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Liu X, et al. , ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells, Am J Pathol, 180 (2012) 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Van der Haegen BA, et al. , Immortalization of human mammary epithelial cells by SV40 large T-antigen involves a two step mechanism, In vitro cellular & developmental biology: journal of the Tissue Culture Association, 29a (1993) 180–182. [DOI] [PubMed] [Google Scholar]
- [18].Schlegel R, et al. , Quantitative keratinocyte assay detects two biological activities of human papillomavirus DNA and identifies viral types associated with cervical carcinoma, The EMBO journal, 7 (1988) 3181–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hawley-Nelson P, et al. , HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes, The EMBO journal, 8 (1989) 3905–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hudson JB, et al. , Immortalization and altered differentiation of human keratinocytes in vitro by the E6 and E7 open reading frames of human papillomavirus type 18, Journal of virology, 64 (1990) 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Munger K, et al. , The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes, Journal of virology, 63 (1989) 4417–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Roig AI, et al. , Immortalized epithelial cells derived from human colon biopsies express stem cell markers and differentiate in vitro, Gastroenterology, 138 (2010) 1012–1021.e1011–1015. [DOI] [PubMed] [Google Scholar]
- [23].Correa BRS, et al. , Patient-derived conditionally reprogrammed cells maintain intra-tumor genetic heterogeneity, Scientific reports, 8 (2018) 4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Suprynowicz FA, et al. , Conditionally reprogrammed cells represent a stem-like state of adult epithelial cells, Proc Natl Acad Sci U S A, 109 (2012) 20035–20040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yuan H, et al. , Use of reprogrammed cells to identify therapy for respiratory papillomatosis, The New England journal of medicine, 367 (2012) 1220–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gheibi P, et al. , Microchamber Cultures of Bladder Cancer: A Platform for Characterizing Drug Responsiveness and Resistance in PDX and Primary Cancer Cells, Scientific reports, 7 (2017) 12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Long Q, et al. , Imaging-guided photo-therapeutic nanoporphyrin synergized HSP90 inhibitor in patient-derived xenograft bladder cancer model, Nanomedicine: Nanotechnology, Biology, and Medicine, In press (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wang M, et al. , Humanized mice in studying efficacy and mechanisms of PD-1-targeted cancer immunotherapy, FASEB J, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zeng SX, et al. , The Phosphatidylinositol 3-Kinase Pathway as a Potential Therapeutic Target in Bladder Cancer, Clinical cancer research: an official journal of the American Association for Cancer Research, 23 (2017) 6580–6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zimmermann M, et al. , Microdose-Induced Drug-DNA Adducts as Biomarkers of Chemotherapy Resistance in Humans and Mice, Molecular cancer therapeutics, 16 (2017) 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mondal AM, et al. , p53 isoforms regulate aging- and tumor-associated replicative senescence in T lymphocytes, The Journal of clinical investigation, 123 (2013) 5247–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Fujita K, et al. , p53 isoforms Delta133p53 and p53beta are endogenous regulators of replicative cellular senescence, Nature cell biology, 11 (2009) 1135–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gui Y, et al. , Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder, Nature genetics, 43 (2011) 875–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Guo G, et al. , Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation, Nature genetics, 45 (2013) 1459–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Tamborero D, et al. , Comprehensive identification of mutational cancer driver genes across 12 tumor types, Scientific reports, 3 (2013) 2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Workman P, et al. , Drugging the PI3 kinome: from chemical tools to drugs in the clinic, Cancer Res, 70 (2010) 2146–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Carracedo A, et al. , Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer, J Clin Invest, 118 (2008) 3065–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Rozengurt E, et al. , Suppression of feedback loops mediated by PI3K/mTOR induces multiple overactivation of compensatory pathways: an unintended consequence leading to drug resistance, Molecular cancer therapeutics, 13 (2014) 2477–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yang X, et al. , Akt-mediated cisplatin resistance in ovarian cancer: modulation of p53 action on caspase-dependent mitochondrial death pathway, Cancer Res, 66 (2006) 3126–3136. [DOI] [PubMed] [Google Scholar]
- [40].Arjumand W, et al. , Phosphatidyl inositol-3 kinase (PIK3CA) E545K mutation confers cisplatin resistance and a migratory phenotype in cervical cancer cells, Oncotarget, 7 (2016) 82424–82439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Boj SF, et al. , Organoid models of human and mouse ductal pancreatic cancer, Cell, 160 (2015) 324–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Drost J, et al. , Sequential cancer mutations in cultured human intestinal stem cells, Nature, 521 (2015) 43–47. [DOI] [PubMed] [Google Scholar]
- [43].Li X, et al. , Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture, Nature medicine, 20 (2014) 769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nadauld LD, et al. , Metastatic tumor evolution and organoid modeling implicate TGFBR2 as a cancer driver in diffuse gastric cancer, Genome biology, 15 (2014) 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sachs N, et al. , Organoid cultures for the analysis of cancer phenotypes, Current opinion in genetics & development, 24 (2014) 68–73. [DOI] [PubMed] [Google Scholar]
- [46].van de Wetering M, et al. , Prospective derivation of a living organoid biobank of colorectal cancer patients, Cell, 161 (2015) 933–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Curry EL, et al. , Using induced pluripotent stem cells as a tool for modelling carcinogenesis, World journal of stem cells, 7 (2015) 461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Seki T, et al. , Methods of induced pluripotent stem cells for clinical application, World journal of stem cells, 7 (2015) 116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.