Abstract
Antimicrobial drug concentrations in the gastrointestinal tract likely drive antimicrobial resistance in enteric bacteria. Our objective was to determine the concentration of ceftiofur and its metabolites in the gastrointestinal tract of steers treated with ceftiofur crystalline-free acid (CCFA) or ceftiofur hydrochloride (CHCL), determine the effect of these drugs on the minimum inhibitory concentration (MIC) of fecal Escherichia coli, and evaluate shifts in the microbiome. Steers were administered either a single dose (6.6 mg/kg) of CCFA or 2.2 mg/kg of CHCL every 24 hours for 3 days. Ceftiofur and its metabolites were measured in the plasma, interstitium, ileum and colon. The concentration and MIC of fecal E. coli and the fecal microbiota composition were assessed after treatment. The maximum concentration of ceftiofur was higher in all sampled locations of steers treated with CHCL. Measurable drug persisted longer in the intestine of CCFA-treated steers. There was a significant decrease in E. coli concentration (P = 0.002) within 24 hours that persisted for 2 weeks after CCFA treatment. In CHCL-treated steers, the mean MIC of ceftiofur in E. coli peaked at 48 hours (mean MIC = 20.45 ug/ml, 95% CI = 10.29–40.63 ug/ml), and in CCFA-treated steers, mean MIC peaked at 96 hours (mean MIC = 10.68 ug/ml, 95% CI = 5.47–20.85 ug/ml). Shifts in the microbiome of steers in both groups were due to reductions in Firmicutes and increases in Bacteroidetes. CCFA leads to prolonged, low intestinal drug concentrations, and is associated with decreased E. coli concentration, an increased MIC of ceftiofur in E. coli at specific time points, and shifts in the fecal microbiota. CHCL led to higher intestinal drug concentrations over a shorter duration. Effects on E. coli concentration and the microbiome were smaller in this group, but the increase in the MIC of ceftiofur in fecal E. coli was similar.
Introduction
Ceftiofur, a third generation cephalosporin, is one of the most common antimicrobials administered to feedlot cattle and lactating dairy cows in the United States for treatment of respiratory disease, [1,2] metritis, [3] and is also used in an extralabel manner to treat enteric disease [4]. This use has led to widespread concern over selection for bacteria with antimicrobial resistance (AMR) in the feces of treated cattle that could be transferred to humans through the food chain [5]. To determine this risk, studies have determined the presence of AMR genes in dairy cattle feces immediately after ceftiofur treatment [6], tested the susceptibility of E. coli isolates from preweaned calves and cows on farms that use ceftiofur [7,8], and quantified bla(CMY-2) and/or bla(CTX-M) in feedlot steers [9–12] and dairy cattle [13] treated with ceftiofur. Due to the variability in the populations studied and the outcome measures, the conclusions of these studies have varied widely, precluding any clear recommendations for prudent use of ceftiofur in cattle.
Further complicating the association between ceftiofur use and AMR outcomes is the availability of different drug formulations, which varied across previous studies. Ceftiofur hydrochloride (CHCL) is a 50 mg/ml oil-based suspension that is FDA-approved for daily administration for 3–5 days at 1.1–2.2 mg/kg by subcutaneous or intramuscular route. Cattle cannot be slaughtered for human consumption within 4 days of the last treatment with CHCL (Zoetis). Ceftiofur crystalline-free acid (CCFA) is a 200 mg/ml oil-based suspension that is administered as single-dose therapy at 6.6 mg/kg with a meat withdrawal time of 14 days. Due to the slow-release formulation, CCFA must be administered subcutaneously on the posterior aspect of the ear or at the base of the ear (Zoetis). Some producers may use CHCL because of the shorter withdrawal time and easier route of administration, while others may prefer CCFA due to the ease of single-dose therapy. Although both products are FDA-approved for similar conditions in cattle and used somewhat interchangeably, they produce different exposure profiles, which could affect selection of antibiotic-resistant bacteria. To our knowledge, a comparison of the effect of these formulations on AMR in fecal bacteria of treated cattle has not been performed.
We hypothesize that the concentration of antimicrobials within the gastrointestinal tract (GIT) is significantly associated with the risk of AMR in fecal bacteria as measured by an increase in MIC. Surprisingly, this association remains unknown because it has been difficult to directly obtain intestinal drug concentration data. We showed in our previous studies that continuous collection of luminal fluid from ileum and colon is possible, and allowed for measurement of drug concentrations in intestinal fluid and pharmacokinetic modelling of the active drug concentrations during the time after drug injection [14–16]. The objective of the current study was to compare the active antimicrobial concentrations in the GIT of steers treated with either CHCL or CCFA, and correlate those concentrations with changes in fecal bacteria, including changes in the minimum inhibitory concentration of ceftiofur in E. coli. In addition to serving as indicator organism for foodborne pathogens, E. coli has human health importance and can acquire relevant resistance to third-generation cephalosporins.
Materials and methods
Animals and treatments
This study was approved by the North Carolina State University Institutional Animal Care and Use Committee (protocol # 18-020A). This study took place May through July of 2016. Twelve six-month-old Holstein steers (186 to 288 kg) were obtained from the North Carolina State University Dairy Educational Unit as was done in previous studies [15,17–19]. Sample size was determined based on the number needed for appropriate pharmacokinetic modeling from our previous studies in cattle [14,15,19]. Investigators were not blinded to the treatment groups of the steers. Steers were fitted for placement of ultrafiltration probes in the ileum and spiral colon as described below. At 24–48 hours post-probe placement, animals received one of two treatments (n = 6 steers per treatment)—subcutaneous injection of ceftiofur crystalline-free acid (CCFA; Excede®; 6.6 mg/kg) as a single dose at the base of the ear, or ceftiofur hydrochloride (CHCL; Excenel®; 2.2 mg/kg) subcutaneously in the neck every 24 hours for 3 treatments. Using a parallel study design, all procedures and sample collection with CCFA steers were completed first, and then treatment and sampling of CHCL steers were completed, so steers were not randomly allocated to treatment groups. Steers were housed in pairs that received the same treatment in stalls bedded with shavings and were fed grass hay with free access to water for the duration of the study. At the conclusion of the study and observation of the appropriate meat withdrawal time, all ultrafiltration probes and catheters were removed, and the steers were sold.
Plasma collection
Prior to drug administration, a jugular catheter (Intracath®, Becton Dickinson, Franklin Lakes, NJ) was inserted into the jugular vein. Blood samples (6 ml) were collected in lithium heparin tubes at appropriate intervals for optimum pharmacokinetic modeling of each drug for at least 3 drug half-lives, accounting for 90% of drug elimination from the plasma. These were time 0, 15 min, 30 min, 1, 2, 4, 8, 12, 24, 32, 48, 72, 96, 120, 144, 168, 192 hours for CCFA and time 0, 15 min, 30 min, 1, 2, 4, 8, 12, 24, 25, 26, 28, 30, 32, 36, 48, 48.25, 48.5, 49, 50, 52, 54, 56, 60, 72, 74, 78, 96 hr after the initial dose for CHCL. The tubes were immediately centrifuged at 1,000 x g for 10 minutes to collect plasma and stored at -80°C until assayed.
Placement of intestinal ultrafiltration probes
Surgical procedures took place over 4 days with 3 surgeries per day as previously described [16] with the following variation in the anesthesia procedure. Briefly, food and water were withheld from all steers for 12 hours prior to surgery. The steers were restrained standing in a conventional chute. The right flank was anesthetized by infiltration of 2% lidocaine dorsally and ventrally to the lateral vertebral processes of L1-L4 (approximately 80 ml per steer). After entering the abdomen, the ileum and colon were identified by first retracting the cecum through the flank incision. The collecting loops of an ultrafiltration probe (UF-3-12, BAS; Bioanalytical Systems, West Lafayette, IN, USA) were inserted into the lumen of the ileum and spiral colon, and sutured into place. The free ends of the probes were exteriorized cranial to the skin incision. The calves received 2 mg/kg of flunixin meglumine intravenously prior to surgery and 24 hours after surgery according to the IACUC protocol. There is no evidence of any impact on ceftiofur pharmacokinetics due to flunixin administration [20].
Gastrointestinal fluid collection
After surgery, the probes were prepared to collect samples of fluid from the ileum and spiral colon of each animal. The tubing exiting the body cavity was connected to a needle within a vacuum vial needle holder using flexible tubing and secured. The vial holder was sutured to the skin over the transverse processes of the lumbar spine using 2–0 nylon (Ethicon; Somerville, NJ) and white tape butterfly tags. To collect the ultrafiltrate, a 3-ml evacuated tube with no additive (Becton-Dickinson) was inserted onto the needle of the vacuum vial needle holder. The ultrafiltrate collected is free of protein and other intestinal contents that could potentially bind to the antibiotic. Drug administration and sample collection began 24–48 hours after surgery. Steers were allowed free-choice grass hay and water after recovery from surgery until the completion of the study. Samples from probes placed in the ileum and spiral colon were collected 0, 2, 4, 8, 12, 24, 32, 48, 72, 96, 120, 144, 168, and 192 hours post administration of CCFA and 0, 2, 4, 6, 8, 12, 24, 26, 28, 30, 32, 36, 48, 50, 52, 54, 56, 60, 72, 74, 78, and 96 hours post administration of initial CHCL dose by changing the tubes at the predetermined time points.
Interstitial fluid collection
An in-vivo ultrafiltration probe was also inserted in the subcutaneous space above the shoulders in a manner described in previous studies (Davis et al., 2007; Messenger et al., 2012). The interstitial fluid (ISF) was collected at time 0 (pre-treatment) and at the same time points as for the GI fluid collection. The collected fluid was immediately frozen at -80°C for further analysis.
Determining active drug concentration
Plasma and tissue fluid samples were analyzed by reverse-phase high pressure liquid chromatography (HPLC) with ultraviolet detection to determine the active concentrations of ceftiofur and its metabolites as previously described [21,22]. Ceftiofur is rapidly metabolized to the active metabolite desfuroylceftiofur in cattle, which is the predominant metabolite responsible for antibacterial effects. The assay converts all ceftiofur and desfuroylceftiofur conjugates to a single stable derivative, desfuroylceftiofur acetamide, which is measured by HPLC ultraviolet detection. All drug concentrations were determined from calibration curves made from fortified (spiked) blank plasma, intestinal and interstitial fluid collected from the experimental calves prior to antibiotic administration. Calibration curves were prepared from fortifying the blank matrix with reference drug standards of ceftiofur (United States Pharmacopeia {USP}, Rockville, MD) to validate the HPLC analysis and perform Quality Control (QC) assessments during the assay.
Pharmacokinetic analysis
The drug concentrations were analyzed using standard pharmacokinetic methods to examine the drug disposition for each calf. A computer program (Phoenix® WinNonlin®, V. 8.0; Pharsight Corporation, Certara, St. Louis MO) was used to determine pharmacokinetic (PK) parameters.
Plasma, ISF, and intestinal drug concentrations were plotted on linear and semi-logarithmic graphs for analysis and for visual assessment of the best model for pharmacokinetic analysis. Specific models (e.g., one, two, etc. compartments) were determined on the basis of visual analysis for goodness of fit and by visual inspection of residual plots. The best model fit was based on the equation described in the following formula:
Where C is the plasma concentration, t is time, k01 is the non-IV absorption rate, assuming first-order absorption, k10 is the elimination rate constant, V is the apparent volume of distribution, F is the fraction of drug absorbed, and D is the non-IV dose. Secondary parameters calculated from the model included the peak concentration (CMAX), time to peak concentration (TMAX), area under the plasma-concentration vs time profile (AUC), and the respective absorption and terminal half-lives (t½).
A compartmental model could not be fit to all of the concentrations from the intestinal fluid samples because of sparse sampling (incomplete collection) in some calves. Therefore, data from some calves were analyzed using noncompartmental analysis (NCA) in the same pharmacokinetic program described above. For the NCA, the area under the plasma concentration vs time curve (AUC) from time 0 to the last measured concentration, (defined by the limit of quantification) was calculated using the log-linear trapezoidal method. The AUC from time 0 to infinity was calculated by adding the terminal portion of the curve, estimated from the relationship Cn/ λZ, to the AUC0 Cn, where λZ is the terminal slope of the curve, and Cn is the last measured concentration point.
The relative drug transfer from the plasma compartment to the ISF and intestinal fluids was measured by calculation of a penetration factor. The penetration factor was determined by the ratio of AUC for the intestinal fluid to the AUC for plasma:
Fecal sampling
Individual fecal samples (>50 g) from each steer were manually collected from the rectum using a clean rectal sleeve and sterile lubricant immediately prior to treatment, and at 24, 36, and 48 hours after treatment. Following this period, fecal samples were collected approximately every day through 7 days and again at 14 days after drug administration.
Quantification of E. coli from feces
One gram of feces was inoculated into 9 ml EC broth (Oxoid Ltd., Basingstoke, Hampshire, England) and vortexed. One ml was immediately removed and serially diluted ten-fold in sterile phosphate-buffered saline, and 100 μl was plated in triplicate onto HardyCHROMTM ECC Media (Hardy Diagnostics, Santa Maria, CA). Plates were incubated overnight at 37°C and dilutions that yielded between 30 and 300 pink-violet colonies on each of the 3 plates were counted and counts were averaged to determine the concentration (CFU/g) of E. coli at each time point. The remaining EC broth was incubated overnight at 37°C, and if no growth was observed on direct plates, the enrichment was streaked for isolation on ECC plates and incubated overnight at 37°C. From the quantified or enrichment plate, eight colonies were randomly picked, streaked onto Columbia agar with 5% sheep blood (Remel, Lenexa, KS) and incubated overnight at 37°C. Following incubation, each isolate was transferred to a 2-ml cryogenic vial containing LB Broth (Sigma-Aldrich, St. Louis, MO) supplemented with 25% glycerol, vortexed, and frozen at -80°C.
Determination of minimum inhibitory concentration
The minimum inhibitory concentration (MIC) of ceftiofur for each E. coli isolate was determined using a broth microdilution method and following Clinical and Laboratory Standards Institute standards [23]. Isolates were grown overnight on blood agar. A single colony was inoculated into 3 ml of sterile phosphate-buffered saline and brought to a 0.5 McFarland Standard, then 10 μl of this Standard were further diluted in 990 μl sterile phosphate-buffered saline. This final bacterial suspension (50 μl) was inoculated into 50 μl of two-fold serial dilutions of ceftiofur (USP, Rockville, MD) in Mueller Hinton broth (BD, Sparks, MD) ranging in concentration from 0.03 to 32 mg/L. Each bacterial suspension (50 μl) was also inoculated into a control well containing 50 μl Mueller Hinton broth (BD, Sparks, MD), with no antibiotic. The 96-well plates were incubated 18 hours at 37°C. The drug concentration in the first well with no visible growth was determined to be the MIC.
DNA extraction and 16S rRNA gene sequencing
Fresh feces collected at each time point were frozen in 1 gram aliquots for future analysis. From these samples, 50 mg of feces were extracted individually using a MoBio PowerMag Microbiome kit (Qiagen, Inc., Germanton, MD) optimized for the epMotion 5075 TMX (Eppendorf, Hauppauge, NY). The DNA libraries were prepared as described previously [24].
Illumina MiSeq sequencing of bacterial communities
The V4 region of the 16S rRNA gene was amplified from each sample using the Dual indexing sequencing strategy [25]. Sequencing was done on the Illumina MiSeq platform, using a MiSeq Reagent Kit V2 500 cycles (2 x 250bp) (Illumina cat# MS102-2003), according to the manufacturer's instructions with modifications [25]. Accuprime High Fidelity Taq (Life Technologies cat # 12346094) was used. PCR was performed using the conditions (Standard or Touch Down) shown by Seekatz [24]. If additional template was used, the water volume was changed accordingly. PCR products were visualized using an E-Gel 96 with SYBR Safe DNA Gel Stain, 2% (Life technologies cat# G7208-02). Libraries were normalized using SequalPrep Normalization Plate Kit (Life technologies cat #A10510-01) following the manufacturer’s protocol for sequential elution. The concentration of the pooled samples was determined using Kapa Biosystems Library Quantification kit for Illumina platforms (KapaBiosystems KK4824). The sizes of the amplicons in the library were determined using the Agilent Bioanalyzer High Sensitivity DNA analysis kit (cat# 5067–4626). The final library consisted of equal molar amounts from each of the plates, normalized to the pooled plate at the lowest concentration. Sequencing libraries were prepared according to Illumina’s protocol for Preparing Libraries for Sequencing on the MiSeq (part# 15039740 Rev. D) for 2 nM or 4 nM libraries. If the library concentration was below 1 nM, an alternative method was used for denaturation [26]. PhiX and genomes were added in 16S amplicon sequencing to create diversity. Sequencing reagents were prepared according to the Schloss SOP [25], and custom read 1, read 2 and index primers were added to the reagent cartridge. FASTQ files were generated for paired end reads.
Microbiota analysis
Analysis of the V4 region of the 16S rRNA gene was performed using the DADA2 method for the inference of exact amplicon sequence variants (ASVs) [27,28]. Our analysis protocol generally followed the DADA2 tutorial (https://benjjneb.github.io/dada2/tutorial.html). Samples were filtered based on a maximum expected error threshold of 2. ASVs were inferred using the dada function with default parameters except that samples were pooled in order to increase sensitivity to rare sequence variants. Chimeras were removed using the default consensus removal method [27]. Taxonomy was assigned using the implementation of the naive Bayesian classifier available in the dada2 R package, and the Silva v128 reference database [29,30].
Statistical analysis
Mean pharmacokinetic parameters were compared using a Student’s t test. Mean E. coli concentration was compared over time using one-way analysis of variance with the Holm-Sidak method for comparison of individual time points to time 0 [31,32] (SigmaPlot 14.0, Systat Software Inc., San Jose, CA). Descriptive statistics (mean, median, standard deviation, and range) for MIC were computed overall, by treatment and by time point. The dependent variable consisted of the MIC values of isolates (up to 8 isolates per sample and per time point) and independent variables included treatment (CHCL vs CCFA), time point (0, 24, 36, 48, 60, 72, 96, 120, 168, 240, and 336 hours) and a two-way interaction term between treatment and time point. Several family distributions and transformations of the outcome were modeled including normal, lognormal, tobit, poisson, and negative binomial. After checking assumptions, model fit was assessed using information criteria (AIC, BIC) and residual plots. The effect of treatment and time period on MICs (log10-based transformed) was estimated using generalized linear mixed models (GLMMs) fitted with a Gaussian distribution, identity link, residual pseudo-likelihood, Newton-Raphson with ridging optimization and Kenward Rogers adjustment for denominator degrees of freedom using Proc Glimmix in SAS (SAS 9.4, SAS Institute Inc., Cary, NC). A random intercept for animal and an unstructured covariance structure were included to account for the design structure of the study (isolates nested within samples (animals) and repeated measures at the animal level). Model assumptions were tested and residuals were investigated using graphical tools. Mean MIC values and their 95% confidence intervals were computed by drug and sample time. P-values < 0.05 were considered statistically significant. The Tukey-Kramer procedure was used to prevent inflation of Type I error due to multiple comparisons [33,34].
Results
Pharmacokinetic modeling
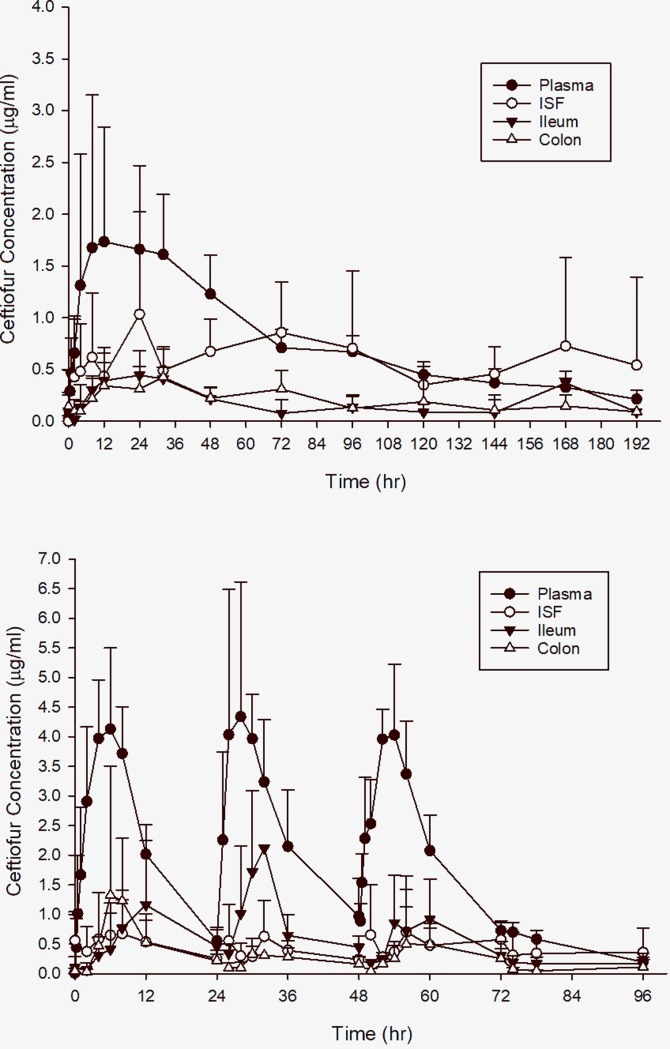
The values for pharmacokinetic parameters for both CHCL and CCFA are presented in Table 1. Because of the slow release of ceftiofur from CCFA, the maximum concentrations (CMAX) of ceftiofur and metabolites in the plasma were significantly lower than from injection of CHCL (Fig 1; P = 0.01), while the TMAX (P<0.001), half-life (P<0.001), and AUC (P<0.001) were greater for steers receiving CCFA. Low drug concentrations in the GIT were noted over a longer time as the half-life of ceftiofur and metabolites were 2–3 times greater in the ileum and colon for CCFA than for CHCL, but due to the variability between animals this difference was not statistically significant. Similarly, the TMAX was later in both the ileum and colon of CCFA treated calves, although this difference was only significant in the colon (P = 0.03). The penetration of ceftiofur and metabolites into the ileum and colon were similar for both drugs at approximately 20% of plasma AUC. The penetration of CCFA into the ISF was significantly higher for CCFA (86 ± 62%) compared to CHCL (24 ± 16%; P = 0.009).
Table 1. Pharmacokinetic parameters for ceftiofur crystalline free acid (CCFA) and ceftiofur hydrochlroide (CHCL) in the plasma, interstitial fluid (ISF), ileum and colon of steers.
| CCFA Plasma | CHCL Plasma | ||||||||
| Parameter | Units | Mean | Std Dev | CV% | Parameter | Units | Mean | Std Dev | CV% |
| AUC | hr*ug/ml | 182.26 | 22.97 | 12.60 | AUC | hr*ug/ml | 61.63* | 11.63 | 18.88 |
| CMAX | ug/ml | 1.80 | 0.95 | 52.70 | CMAX | ug/mL | 3.29* | 0.74 | 22.43 |
| k01 | 1/hr | 0.26 | 0.10 | 37.50 | k01 | 1/hr | 0.53 | 0.45 | 84.82 |
| Absorption T½ | hr | 3.07 | 1.16 | 37.81 | Absorption T½ | hr | 1.80* | 0.74 | 40.92 |
| k10 | 1/hr | 0.01 | 0.01 | 49.66 | k10 | 1/hr | 0.08* | 0.01 | 12.30 |
| Elimination T½ | hr | 73.25 | 33.54 | 45.79 | Elimination T½ | hr | 8.79* | 1.12 | 12.79 |
| TMAX | hr | 14.13 | 4.56 | 32.25 | TMAX | hr | 4.98* | 1.51 | 30.23 |
| CCFA ISF | CHCL ISF | ||||||||
| Parameter | Units | Mean | Std Dev | CV% | Parameter | Units | Mean | Std Dev | CV% |
| AUC infinity | hr*ug/ml | 91.54 | 35.98 | 39.31 | AUC_TAU | hr*ug/ml | 12.26 | 11.61 | 94.66 |
| AUC 0 to Cn | hr*ug/ml | 90.81 | 28.50 | 31.39 | AUC infinity | hr*ug/ml | 73.32 | 84.09 | 114.68 |
| CMAX | ug/ml | 1.62 | 0.61 | 37.68 | AUC 0 to Cn | hr*ug/ml | 43.36* | 35.24 | 81.28 |
| Half-life | hr | NA | NA | NA | CAVE | ug/ml | 0.51 | 0.48 | 94.66 |
| Lambda z | 1/hr | NA | NA | NA | CMAX | ug/ml | 0.72* | 0.61 | 83.88 |
| MRT | hr | NA | NA | NA | CMIN | ug/ml | 0.40 | 0.48 | 119.17 |
| TMAX | hr | 76.67 | 65.99 | 86.07 | Half-life | hr | NA | NA | NA |
| Penetration | 0.86 | 0.62 | 71.44 | Lambda z | 1/hr | NA | NA | NA | |
| MRT | hr | NA | NA | NA | |||||
| TMAX | hr | 5.00 | 4.69 | 93.81 | |||||
| Penetration | 0.24* | 0.16 | 65.59 | ||||||
| CCFA Ileum | CHCL Ileum | ||||||||
| Parameter | Units | Mean | Std Dev | CV% | Parameter | Units | Mean | Std Dev | CV% |
| AUC infinity | hr*ug/ml | 55.19 | 6.86 | 12.43 | AUC_TAU | hr*ug/ml | 13.82 | 9.27 | 67.08 |
| AUC 0 to Cn | hr*ug/ml | 27.67 | 7.36 | 26.59 | AUC infinity | hr*ug/ml | 62.98 | 21.22 | 33.70 |
| CMAX | ug/ml | 0.54 | 0.16 | 30.37 | AUC 0 to Cn | hr*ug/ml | 39.05 | 19.70 | 50.43 |
| Half-life | hr | 127.74 | 16.46 | 12.89 | CAVE | ug/mL | 0.58 | 0.39 | 67.08 |
| Lambda z | 1/hr | 0.01 | 0.00 | 12.16 | CMAX | ug/mL | 1.20 | 1.00 | 82.75 |
| MRT | hr | 192.91 | 36.54 | 18.94 | CMIN | ug/mL | 0.06 | 0.10 | 154.92 |
| TMAX | hr | 45.33 | 61.12 | 134.82 | Half-life | hr | 66.27 | 79.51 | 119.97 |
| Penetration | 0.20 | 0.07 | 37.79 | Lambda z | 1/hr | 0.06 | 0.06 | 97.53 | |
| MRT | hr | 204.42 | 175.78 | 85.99 | |||||
| TMAX | hr | 8.33 | 4.80 | 57.63 | |||||
| Penetration | 0.23 | 0.10 | 44.87 | ||||||
| CCFA Colon | CHCL Colon | ||||||||
| Parameter | Units | Mean | Std Dev | CV% | Parameter | Units | Mean | Std Dev | CV% |
| AUC infinity | hr*ug/ml | 53.34 | 43.33 | 81.22 | AUC TAU | hr*ug/ml | 12.07 | 8.09 | 67.00 |
| AUC 0 to Cn | hr*ug/ml | 32.38 | 28.26 | 87.27 | AUC infinity | hr*ug/ml | 41.09 | 25.25 | 61.46 |
| CMAX | ug/ml | 0.44 | 0.24 | 54.82 | AUC 0 to Cn | hr*ug/ml | 27.21 | 11.25 | 41.36 |
| Half-life | hr | 94.85 | 39.34 | 41.47 | CAVE | ug/ml | 0.50 | 0.34 | 67.00 |
| Lambda z | 1/hr | 0.01 | 0.01 | 64.74 | CMAX | ug/ml | 1.55 | 2.07 | 133.04 |
| MRT | hr | 144.43 | 55.28 | 38.27 | CMIN | ug/ml | 0.02 | 0.03 | 137.15 |
| TMAX | hr | 25.60 | 28.37 | 110.82 | Half-life | hr | 39.45 | 42.39 | 107.45 |
| Penetration | 0.24 | 0.27 | 111.16 | Lambda z | 1/hr | 0.04 | 0.04 | 80.86 | |
| MRT | hr | 98.83 | 78.43 | 79.37 | |||||
| TMAX | hr | 8.00* | 2.45 | 30.62 | |||||
| Penetration | 0.15 | 0.07 | 47.90 | ||||||
k01, and k10, rates for absorption and elimination processes, respectively, and accompanying half-lives (T½); AUC, area under the curve; AUC infinity, area under the curve from time zero to infinity; AUC 0 to Cn, area under the curve from time zero to the last measured time point (Cn); AUCTAU , AUC (τ) for the dose interval (tau = 24 hours) for ceftiofur administered 3 times; CMAX, maximum drug concentration; TMAX, time to maximum drug concentration; CMIN , minimum drug concentration; CAVE , average drug concentration; Lambda-z (λ Z), terminal slope; MRT, mean residence time; penetration factor, calculated from the AUC ratios of tissue fluid/plasma; NA indicates that there was insufficient sample collection to calculate these values;
* indicates that values are significantly different between the two formulations, P<0.05.
Fig 1.
Total concentration of ceftiofur equivalents in plasma, interstitial fluid (ISF), ileum, and colon for steers treated with (A) ceftiofur crystalline free acid (CCFA) and (B) ceftiofur hydrochloride (CHCL).
Concentration of E. coli
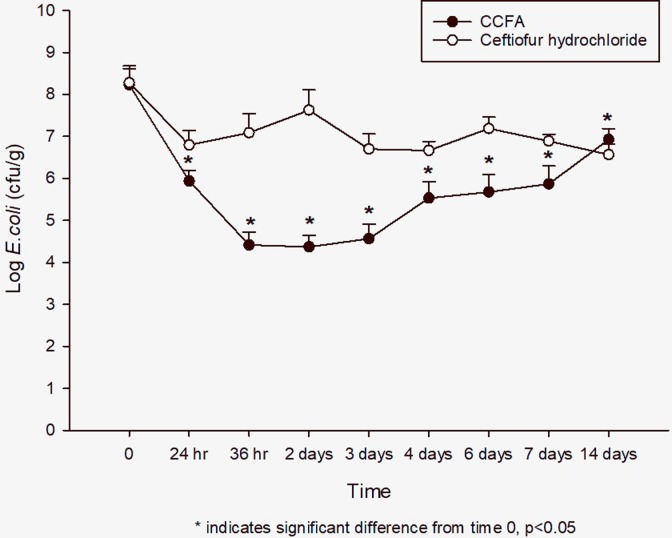
The fecal concentration of E. coli was not different between CHCL (7.8 ± 0.25 log10 CFU/g) and CCFA (7.6 ± 0.35 log10 CFU/g) treatment groups at time 0 (P = 0.9). In the CHCL group, the mean concentration decreased by 1.7 log10 by 24 hours, but the change at this or any other time point was not significantly different compared to time 0 (P = 0.52). In the CCFA group, the mean concentration significantly decreased within 24 hours (5.4 + 0.38 log10 CFU/g, P = 0.002), and ultimately decreased by 3.4 log10 by 48 hours (P = 0.007). The concentration slowly increased after this point, but never returned to baseline (Fig 2).
Fig 2. Fecal E. coli concentration over time after treatment with either ceftiofur crystalline free acid (CCFA) or ceftiofur hydrochloride (CHCL).
Mean ± SD. * indicates a significant difference from time 0, P ≤0.05.
E. coli minimum inhibitory concentration
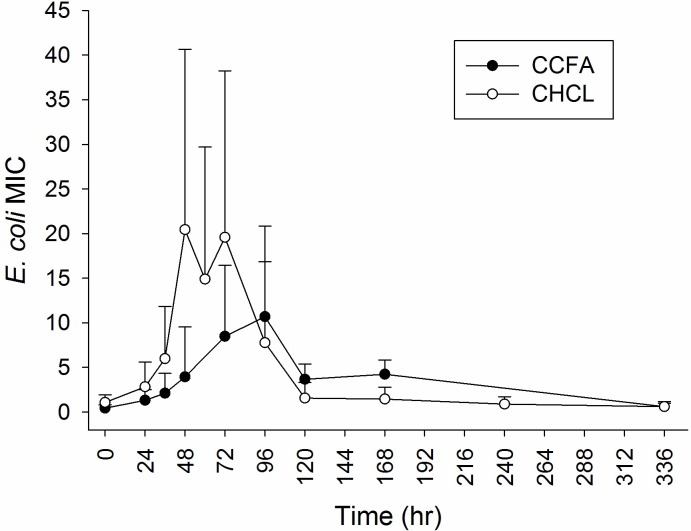
Descriptive statistics (mean, median, SD and range) for MIC values by drug and sample time are presented in Table 2. Table 3 depicts the mean MIC estimates from multivariable models including fixed effects for drug, sample time and drug by sample time. The interaction between drug formulation and sample time was significantly (P < 0.001) associated with MIC values, indicating that the effect of drug formulation on MIC values depended on the time point. When CCFA was administered, MIC values significantly increased at 24 hours (mean MIC = 1.32 ug/ml, 95% CI = 0.60–1.93 ug/ml) and continued to increase up to 96 hours, when MIC peaked (mean MIC = 10.68 ug/ml, 95% CI = 5.47–20.85 ug/ml), followed by a decrease in MIC values to baseline levels, and the MIC50 was within the wild-type distribution at 14 days after treatment. The mean MIC was significantly greater than the MIC at 0 hours from 24 hours through 168 hours after treatment (Table 3). When CHCL was administered, MIC values peaked at 48 hours (mean MIC = 20.45 ug/ml, 95% CI = 10.29–40.63 ug/ml) and then again at 72 hours (mean MIC = 19.58 ug/ml, 95% CI = 10.03–38.24 ug/ml), followed by a steady decrease to baseline levels, and the MIC50 was within the wild-type distribution by 120 hours after the initial dose (Table 3 and Fig 3). In this group, the mean MIC was significantly greater than the MIC at 0 hours from 24 hours to 96 hours after initial treatment (Table 3). At no time was there a significant difference between the two treatment groups at the same time point.
Table 2. Descriptive statistics for minimum inhibitory concentration (MIC) by drug and time point.
| MIC | ||||||
|---|---|---|---|---|---|---|
| Drug | Time Point | n | Mean | Median | SD | Range |
| CCFA | ||||||
| 0 h | 48 | 0.46 | 0.50 | 0.13 | 0.25–1.00 | |
| 24 h | 48 | 14.5 | 0.50 | 51.33 | 0.25–256.00 | |
| 36 h | 48 | 13.16 | 0.50 | 38.41 | 0.25–256.00 | |
| 48 h | 48 | 23.63 | 8.00 | 61.14 | 0.25–256.00 | |
| 60 h | - | - | - | - | - | |
| 72 h | 48 | 35.58 | 12.00 | 76.26 | 0.50–256.00 | |
| 96 h | 48 | 37.16 | 16.00 | 75.75 | 0.25–256.00 | |
| 120 h | 48 | 23.57 | 8.00 | 61.12 | 0.25–256.00 | |
| 168 h | 48 | 32.33 | 8.00 | 77.30 | 0.25–256.00 | |
| 240 h | - | - | - | - | - | |
| 336 h | 48 | 1.05 | 0.50 | 1.84 | 0.25–8.00 | |
| CHCL | ||||||
| 0 h | 48 | 8.75 | 0.50 | 37.03 | 0.25–256.00 | |
| 24 h | 48 | 8.71 | 8.00 | 9.20 | 0.25–32.00 | |
| 36 h | 48 | 12.31 | 16.00 | 8.53 | 0.25–32.00 | |
| 48 h | 48 | 41.85 | 16.00 | 74.09 | 1.00–256.00 | |
| 60 h | 48 | 32.52 | 16.00 | 59.70 | 0.25–256.00 | |
| 72 h | 48 | 50.85 | 16.00 | 85.99 | 0.25–256.00 | |
| 96 h | 48 | 51.05 | 8.00 | 93.11 | 0.25–256.00 | |
| 120 h | 48 | 5.31 | 0.50 | 6.70 | 0.25–16.00 | |
| 168 h | 48 | 10.47 | 0.50 | 37.10 | 0.25–256.00 | |
| 240 h | 48 | 7.89 | 0.50 | 36.91 | 0.25–256.00 | |
| 336 h | 48 | 2.17 | 0.50 | 4.61 | 0.25–16.00 | |
CCFA = ceftiofur crystalline free acid; CHCL = ceftiofur hydrochloride; n = number of observations; SD = standard deviation.
Table 3. Model-adjusted mean*minimum inhibitory concentration (MIC) estimates, 95% confidence intervals and P-values by drug, sample time and drug by sample time.
| Variable | Mean MIC | 95% CI mean MIC | P-value† |
|---|---|---|---|
| Drug | 0.425 | ||
| CCFA | NA | NA | |
| CHCL | 3.53 | 1.91–6.52 | |
| Sample Time | <0.001 | ||
| 0 | 0.69 | 0.46–1.05 | |
| 24 | 1.93 | 1.19–3.14 | |
| 36 | 3.56 | 2.20–5.77 | |
| 48 | 8.98 | 5.53–14.58 | |
| 60 | - | - | |
| 72 | 12.88 | 7.97–20.84 | |
| 96 | 9.11 | 5.44–15.26 | |
| 120 | 2.40 | 1.39–4.12 | |
| 168 | 2.48 | 1.51–4.08 | |
| 240 | - | - | |
| 336 | 0.61 | 0.39–0.97 | |
| Drug x Sample Time | < 0.001 | ||
| CCFA 0 | 0.44 | 0.25–0.80 | |
| CCFA 24 | 1.32 | 0.60–1.93 | |
| CCFA 36 | 2.12 | 1.07–4.19 | |
| CCFA 48 | 3.94 | 1.98–7.83 | |
| CCFA 60 | - | - | |
| CCFA 72 | 8.48 | 4.25–16.91 | |
| CCFA 96 | 10.68 | 5.47–20.85 | |
| CCFA 120 | 3.67 | 1.67–8.04 | |
| CCFA 168 | 4.24 | 2.01–8.96 | |
| CCFA 240 | - | - | |
| CCFA 336 | 0.61 | 0.32–1.70 | |
| CHCL 0 | 1.08 | 0.60–1.93 | |
| CHCL 24 | 2.83 | 1.42–5.62 | |
| CHCL 36 | 5.99 | 3.03–11.84 | |
| CHCL 48 | 20.45 | 10.29–40.63 | |
| CHCL 60 | 14.89 | 7.46–29.70 | |
| CHCL 72 | 19.58 | 10.03–38.24 | |
| CHCL 96 | 7.77 | 3.55–20.85 | |
| CHCL 120 | 1.57 | 0.74–3.31 | |
| CHCL 168 | 1.46 | 0.76–2.79 | |
| CHCL 240 | 0.90 | 0.48–1.70 | |
| CHCL 336 | 0.61 | 0.32–1.18 | |
| Significant contrasts for Drug x Sample Time interaction | |||
| Contrast | P-value‡ | ||
| CCFA 0 vs CCFA 24 | 0.009 | ||
| CCFA 0 vs CCFA 36 | <0.001 | ||
| CCFA 0 vs CCFA 48 | <0.001 | ||
| CCFA 0 vs CCFA 72 | <0.001 | ||
| CCFA 0 vs CCFA 96 | <0.001 | ||
| CCFA 0 vs CCFA 120 | <0.001 | ||
| CCFA 0 vs CCFA 168 | <0.001 | ||
| CHCL 0 vs CHCL 24 | 0.041 | ||
| CHCL 0 vs CHCL 36 | <0.001 | ||
| CHCL 0 vs CHCL 48 | <0.001 | ||
| CHCL 0 vs CHCL 60 | <0.001 | ||
| CHCL 0 vs CHCL 72 | <0.001 | ||
| CHCL 0 vs CHCL 96 | <0.001 | ||
CCFA = ceftiofur crystalline free acid; CHCL = ceftiofur hydrochloride; NA = Not available.
† Overall significance test (F-test) .
‡ P-values represent Tukey-Kramer’s adjustment for multiple comparisons.
*These estimates are from GLMM models including drug, sample time and a two-way. interaction between drug and sample time, after accounting for isolates nested within samples and repeated measures at the animal level.
Fig 3. Mean minimum inhibitory concentration (MIC, ± 95% CI) of ceftiofur in E. coli isolates from steers treated with ceftiofur crystalline free acid (CCFA) and ceftiofur hydrochloride (CHCL).
Alterations in the fecal microbiota
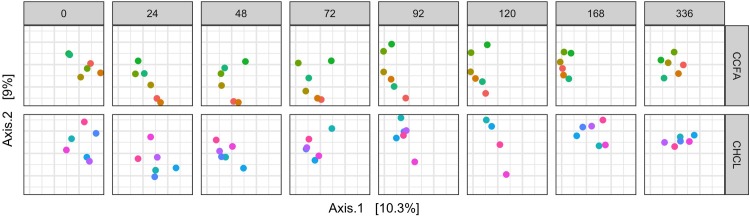
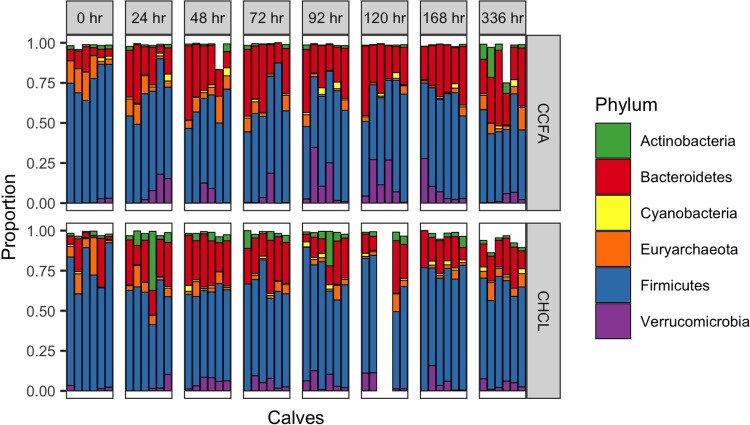
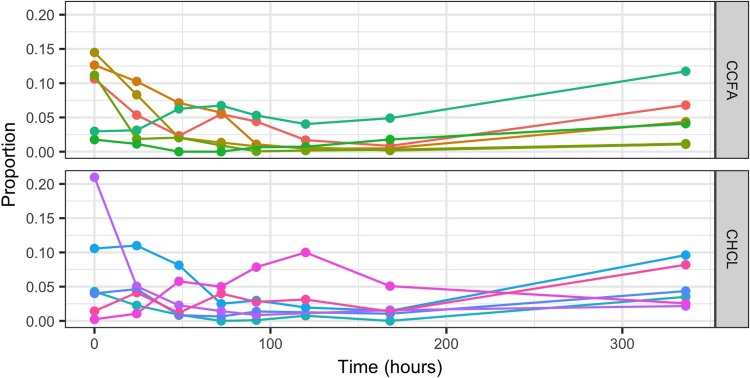
As seen in Fig 4, there was a shift in the microbial communities after treatment with either CHCL or CCFA. This shift appears to be slower, but more pronounced and persistent in the steers treated with CCFA. Yet, in neither group does the community return to its initial structure at 2 weeks after treatment. These shifts are largely due to a reduction in Firmicutes and an increase in Bacteroidetes (Fig 5). The Archea, primarily composed of Methanobrevibacter, follow a similar trajectory of initial decline with a slow, incomplete recovery over time (Fig 6).
Fig 4. Principal coordinate analysis of the microbial communities in each steer over time.
Fig 5. The relative abundance of each phylum present in steers after treatment with either ceftiofur hydrochloride (CHCL) or ceftiofur crystalline free acid (CCFA).
Each bar represents an individual calf.
Fig 6. Changes in the proportion of Methanobrevibacter over time in both the ceftiofur hydrochloride (CHCL) and ceftiofur crystalline free acid (CCFA) groups.
Discussion
Because of its broad spectrum of activity, short slaughter withdrawal time, and zero milk withdrawal time, ceftiofur is one of the most commonly used antimicrobials in cattle in the United States. As it is available in multiple formulations for use in cattle, we investigated the gastrointestinal PK of two different formulations and their impact on enteric bacteria to determine if selection of one formulation over another could be a viable means to mitigate selection of AMR enteric bacteria.
Because of the slow release of ceftiofur from CCFA, the maximum concentrations (CMAX) of ceftiofur and metabolites in the plasma were significantly lower than from injection of CHCL (Fig 1). The slow release formulation also impacted the plasma TMAX as concentrations from CCFA peaked later than that of CHCL, but the prolonged half-life significantly increased the plasma AUC. These findings are similar to a comparative PK study in neonatal calves [35]. ISF fluid concentrations for CHCL were similar to those of ceftiofur sodium [15], and reflects the high protein binding (93%) of the metabolite measured in our previous study [22]. However, the ISF concentrations after CCFA injection were much higher. This likely occurred because of a longer time for equilibration between plasma and interstitial tissue fluid for CCFA. This also produced longer persistence of ceftiofur and its metabolites in intestinal fluids. These observations are consistent with previous evidence from tissue cages that showed that with a prolonged half-life, penetration into interstitial fluids can increase due to the additional time for diffusion [36,37].
The concentrations measured in the GIT were lower than previously reported [15], which may be due to differences in formulation, but we cannot confirm this without additional study. In the previous study, steers were administered ceftiofur sodium, compared to CHCL and CCFA that were administered in this study. Interestingly, there minimal significant differences in the GI PK parameters between the two formulations in this study. This may be explained by the large variability between calves and the relatively small numbers in each group. The penetration into the ileum and colon was similar for both formulations, yet the numerical differences in CMAX and half-life suggest prolonged, low drug concentrations in the ileum and colon of steers receiving CCFA. Because there were differences in the effect on E. coli and the microbiome, it suggests to us that this property produced clinically relevant differences in the GI PK parameters. Further, these concentrations were dramatically lower than predicted through mathematical models [38], demonstrating the need for empirical data. Here, the intestinal concentrations were above MIC 90 for E. coli after CHCL administration, but only briefly. For CCFA, concentrations never reached the MIC 90, but the concentrations were above the MIC for the most susceptible wild-type strains.
According to EUCAST (www.EUCAST.org) wild-type E. coli have ceftiofur MIC values that range from 0.12–1.0 μg/ml; thus, the more susceptible strains were exposed to ceftiofur and metabolites longer after injection of CCFA compared to CHCL. This may explain why CCFA had a more significant impact on the concentration of E. coli in the feces compared to CHCL. As ceftiofur is a time-dependent drug, one can speculate that longer drug exposure of E. coli in the GIT produced a greater reduction in the E. coli populations in spite of the low drug concentrations. This reduction is similar to what has been described previously in feedlot cattle [10,39]. CHCL peak concentrations in GIT were higher than CCFA, but with a much shorter half-life. As demonstrated in our previous study with ceftiofur sodium and by others, at this high concentration ceftiofur appears to be rapidly degraded by enteric bacteria, which shortens the exposure time [15,40]. This may mitigate the impact of the drug on the concentration of E. coli in the feces in our study. In a previous study of dairy cattle treated daily for 5 days with ceftiofur, there was a significant decrease in fecal shedding of E. coli by these cows [41]. It is unclear in that study if the cows received CHCL or ceftiofur sodium. We found higher concentrations of ceftiofur and metabolites in the intestine after injections of ceftiofur sodium [15] compared to CHCL in this study, which could explain this difference.
The relative impact of the two formulations on microbiota of the steers was similar to their impact on E. coli. CCFA appears to have a more significant and prolonged effect on the bacterial communities overall. Specifically, there is a greater reduction in Firmicutes and increase in Bacteroidetes in steers treated with CCFA compared to CHCL. The clinical impact of these changes is undetermined, because the normal microbiota is undefined in this population. Both phyla are commonly found in the feces of adult cattle, with Firmicutes commonly being the predominate phylum [6,42]. Most studies of Methanobrevibacter have shown that it is the most common methanogen in the rumen of cattle [43,44], but its role in the fecal microbiota is unclear. Interestingly, reducing this organism could reduce methane production in treated animals and improve feed efficiency [43]. It is not known if this reduction in Methanobrevibacter is found in the rumen as well. Antimicrobial concentrations in the rumen after injection of these formulations have not been reported.
Though CCFA had a greater impact on the concentration of E. coli and the microbiome, the changes in E. coli ceftiofur MIC depended on the time of sampling, with no evidence of statistical differences in MIC between treatments at the same time point. Over time, the increase in mean MIC was greater in the CHCL group than in CCFA. The increase in mean MIC persisted longer for CCFA than CHCL (168 hours vs 96 hours), which is not surprising as the drug in intestinal fluids persisted longer from CCFA than CHCL (Fig 1). Nonetheless, in both groups, the MIC values returned to baseline prior to the end of meat residue withdrawal time for each drug, suggesting that persistence of E. coli with an MIC above the wild-type cutoff (www.EUCAST.org) in an animal at slaughter would be relatively unlikely. These findings are similar to previous results in dairy cattle [13,41] and beef cattle [39] demonstrating short-lived resistance to third-generation cephalosporins. Resistance to third-generation cephalosporin among E. coli isolates found in cattle at slaughter [45] is likely caused by other factors, rather than single uses of ceftiofur in cattle. Two studies in feedlot cattle have demonstrated a significant increase in resistant fecal E. coli [10] and carriage of cephalosporin resistance genes [11] in association with combined treatment with CCFA and oral chlortetracycline. This suggests that co-selection of resistance mechanisms may play a greater role in maintaining these resistance elements within the fecal microbiota once the initial selection pressure associated with ceftiofur administration has waned.
While this is the first study to associate intestinal pharmacokinetics with changes in AMR in enteric E. coli and with changes in the microbiome, our conclusions are limited by the size of the study. The sample size was determined based on the numbers needed to assess the pharmacokinetics of the drugs and this may not have been adequate for the microbial analyses. When analyzing MIC values, challenges in terms of the nature of the distribution of these data, which are not truly continuous and may be truncated, arise. Although several family distributions and transformations of the outcome were attempted, the chosen logarithmic transformation and statistical model provided a better fit by improving the skewness of the underlying distribution while accounting for the design structure (lack of independence due to multiple isolates per sample and repeated measures) of the study. Statistical comparisons of the changes in the microbiome are limited due to the high variability and small sample size. Our observations on antimicrobial resistance are limited to the changes in E. coli. Although this organism is commonly used as an indicator organism, it is unclear how generalizable these findings are to changes in the susceptibility profile of other enteric bacteria.
In conclusion, the relatively long persistence of active drug in the intestine of cattle treated with CCFA has a significant and prolonged effect on the concentration of E. coli in the feces and the microbiome. Repeated injections of CHCL did not have the same effect on fecal E. coli concentrations or the microbiome. CCFA increased the mean MIC of ceftiofur in fecal E. coli for a longer period of time, but this returned to baseline with two weeks after treatment.
Acknowledgments
This research was supported by work performed by The University of Michigan Microbial Systems Molecular Biology Laboratory. The authors thank Delta R. Dise of the NC State University Clinical Pharmacology Laboratory for her expertise performing the drug assays.
Data Availability
All sequence files are available from the Bioproject database (ID PRJNA560079).
Funding Statement
DMF: This work is/was supported by the USDA National Institute of Food and Agriculture, project 1010130. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.National Animal Health Monitoring System. Beef Feedlot. Unites States Department of Agriculture Animal and Plant Health Inspection Service; 2011. [Google Scholar]
- 2.USDA:APHIS:VS:CEAH. Dairy 2007:Part II: Changes in the U.S. Dairy Cattle Industry, 1991–2007.
- 3.USDA–APHIS–VS–CEAH–NAHMS. Dairy 2014:Milk Quality, Milking Procedures, and Mastitis on U.S. Dairies, 2014.
- 4.Pereira RV, Siler JD, Ng JC, Davis MA, Grohn YT, Warnick LD. Effect of on-farm use of antimicrobial drugs on resistance in fecal Escherichia coli of preweaned dairy calves. J Dairy Sci. 2014;97: 7644–7654. 10.3168/jds.2014-8521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoelzer K, Wong N, Thomas J, Talkington K, Jungman E, Coukell A. Antimicrobial drug use in food-producing animals and associated human health risks: what, and how strong, is the evidence? BMC Vet Res. 2017;13: 211 10.1186/s12917-017-1131-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers L, Yang Y, Littier H, Ray P, Zhang T, Pruden A, et al. Metagenomic Analysis of Antibiotic Resistance Genes in Dairy Cow Feces following Therapeutic Administration of Third Generation Cephalosporin. PLoS ONE. 2015;10: e0133764 10.1371/journal.pone.0133764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pereira RV, Siler JD, Ng JC, Davis MA, Grohn YT, Warnick LD. Effect of on-farm use of antimicrobial drugs on resistance in fecal Escherichia coli of preweaned dairy calves. J Dairy Sci. 2014;97: 7644–7654. 10.3168/jds.2014-8521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mann S, Siler JD, Jordan D, Warnick LD. Antimicrobial susceptibility of fecal Escherichia coli isolates in dairy cows following systemic treatment with ceftiofur or penicillin. Foodborne Pathog Dis. 2011;8: 861–867. 10.1089/fpd.2010.0751 [DOI] [PubMed] [Google Scholar]
- 9.Alali WQ, Scott HM, Norby B, Gebreyes W, Loneragan GH. Quantification of the bla(CMY-2) in feces from beef feedlot cattle administered three different doses of ceftiofur in a longitudinal controlled field trial. Foodborne Pathog Dis. 2009;6: 917–924. 10.1089/fpd.2009.0271 [DOI] [PubMed] [Google Scholar]
- 10.Kanwar N, Scott HM, Norby B, Loneragan GH, Vinasco J, McGowan M, et al. Effects of ceftiofur and chlortetracycline treatment strategies on antimicrobial susceptibility and on tet(A), tet(B), and bla CMY-2 resistance genes among E. coli isolated from the feces of feedlot cattle. PLoS ONE. 2013;8: e80575 10.1371/journal.pone.0080575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanwar N, Scott HM, Norby B, Loneragan GH, Vinasco J, Cottell JL, et al. Impact of treatment strategies on cephalosporin and tetracycline resistance gene quantities in the bovine fecal metagenome. Sci Rep. 2014;4: 5100 10.1038/srep05100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt JW, Griffin D, Kuehn LA, Brichta-Harhay DM. Influence of therapeutic ceftiofur treatments of feedlot cattle on fecal and hide prevalences of commensal Escherichia coli resistant to expanded-spectrum cephalosporins, and molecular characterization of resistant isolates. Appl Environ Microbiol. 2013;79: 2273–2283. 10.1128/AEM.03592-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyer TC, Singer RS. Quantitative measurement of blaCMY-2 in a longitudinal observational study of dairy cattle treated with ceftiofur. Foodborne Pathog Dis. 2012;9: 1022–1027. 10.1089/fpd.2012.1198 [DOI] [PubMed] [Google Scholar]
- 14.Ferguson KM, Jacob ME, Theriot CM, Callahan BJ, Prange T, Papich MG, et al. Dosing Regimen of Enrofloxacin Impacts Intestinal Pharmacokinetics and the Fecal Microbiota in Steers. Front Microbiol. 2018;9: 2190 10.3389/fmicb.2018.02190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster DM, Jacob ME, Warren CD, Papich MG. Pharmacokinetics of enrofloxacin and ceftiofur in plasma, interstitial fluid, and gastrointestinal tract of calves after subcutaneous injection, and bactericidal impacts on representative enteric bacteria. Journal of Veterinary Pharmacology and Therapeutics. 2016;39: 62–71. 10.1111/jvp.12236 [DOI] [PubMed] [Google Scholar]
- 16.Warren CD, Prange T, Campbell NB, Gerard MP, Martin LG, Jacob ME, et al. Implantation of an ultrafiltration device in the ileum and spiral colon of steers to continuously collect intestinal fluid. Research in veterinary science. 2014;97: 611–615. 10.1016/j.rvsc.2014.10.012 [DOI] [PubMed] [Google Scholar]
- 17.McKellar Q, Gibson I, Monteiro A, Bregante M. Pharmacokinetics of enrofloxacin and danofloxacin in plasma, inflammatory exudate, and bronchial secretions of calves following subcutaneous administration. Antimicrob Agents Chemother. 1999;43: 1988–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.TerHune TN, Skogerboe TL, Shostrom VK, Weigel DJ. Comparison of pharmacokinetics of danofloxacin and enrofloxacin in calves challenged with Mannheimia haemolytica. Am J Vet Res. 2005;66: 342–349. 10.2460/ajvr.2005.66.342 [DOI] [PubMed] [Google Scholar]
- 19.Davis JL, Foster DM, Papich MG. Pharmacokinetics and tissue distribution of enrofloxacin and its active metabolite ciprofloxacin in calves. Journal of Veterinary Pharmacology and Therapeutics. 2007;30: 564–571. 10.1111/j.1365-2885.2007.00914.x [DOI] [PubMed] [Google Scholar]
- 20.Gorden PJ, Kleinhenz MD, Wulf LW, Rajewski SJ, Wang C, Gehring R, et al. Comparative plasma and interstitial fluid pharmacokinetics of flunixin meglumine and ceftiofur hydrochloride following individual and co-administration in dairy cows. J Vet Pharmacol Ther. 2018;41: 76–82. 10.1111/jvp.12437 [DOI] [PubMed] [Google Scholar]
- 21.Jaglan PS, Cox BL, Arnold TS, Kubicek MF, Stuart DJ, Gilbertson TJ. Liquid chromatographic determination of desfuroylceftiofur metabolite of ceftiofur as residue in cattle plasma. J Assoc Off Anal Chem. 1990;73: 26–30. [PubMed] [Google Scholar]
- 22.Foster DM, Martin LG, Papich MG. Comparison of Active Drug Concentrations in the Pulmonary Epithelial Lining Fluid and Interstitial Fluid of Calves Injected with Enrofloxacin, Florfenicol, Ceftiofur, or Tulathromycin. PLoS ONE. 2016;11: e0149100 10.1371/journal.pone.0149100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clinical Laboratory Standards Institute, CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals; Approved Standard—Fourth Edition. Wayne, PA: Clinical Laboratory Standards Institute, CLSI; 2018. [Google Scholar]
- 24.Seekatz AM, Theriot CM, Molloy CT, Wozniak KL, Bergin IL, Young VB. Fecal Microbiota Transplantation Eliminates Clostridium difficile in a Murine Model of Relapsing Disease. Infect Immun. 2015;83: 3838–3846. 10.1128/IAI.00459-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79: 5112–5120. 10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, et al. A large genome center's improvements to the Illumina sequencing system. Nat Methods. 2008;5: 1005–1010. 10.1038/nmeth.1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13: 581–583. 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Callahan BJ, McMurdie PJ, Holmes SP. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017;11: 2639–2643. 10.1038/ismej.2017.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41: 590 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73: 5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holm Sture. A Simple Sequentially Rejective Multiple Test Procedure. Scandinavian Journal of Statistics. 1979;6: 65–70. 10.2307/4615733 [DOI] [Google Scholar]
- 32.Šidák Z. Rectangular Confidence Regions for the Means of Multivariate Normal Distributions. Journal of the American Statistical Association. 1967;62: 626–633. 10.1080/01621459.1967.10482935 [DOI] [Google Scholar]
- 33.Tukey JW. The Collected Works of John W. Tukey VIII. Multiple Comparisons: 1948–1933. New York: Chapman and Hall; 1953. [Google Scholar]
- 34.Kramer Clyde Young. Extension of Multiple Range Tests to Group Means with Unequal Numbers of Replications. Biometrics. 1956;12: 307–310. 10.2307/3001469 [DOI] [Google Scholar]
- 35.Woodrow JS, Caldwell M, Cox S, Hines M, Credille BC. Comparative plasma pharmacokinetics of ceftiofur sodium and ceftiofur crystalline-free acid in neonatal calves. J Vet Pharmacol Ther. 2016;39: 271–276. 10.1111/jvp.12275 [DOI] [PubMed] [Google Scholar]
- 36.Washburn K, Johnson R, Clarke CR, Anderson K, Lucas M, Bryson W, et al. Penetration of ceftiofur into sterile vs. Mannheimia haemolytica-infected tissue chambers in beef calves after subcutaneous administration of ceftiofur crystalline free acid sterile suspension in the ear pinna. J Vet Pharmacol Ther. 2005;28: 247–251. 10.1111/j.1365-2885.2005.00642.x [DOI] [PubMed] [Google Scholar]
- 37.Clarke CR. Tissue-chamber modeling systems—applications in veterinary medicine. J Vet Pharmacol Ther. 1989;12: 349–368. 10.1111/j.1365-2885.1989.tb00686.x [DOI] [PubMed] [Google Scholar]
- 38.Volkova VV, Cazer CL, Gröhn YT. Models of antimicrobial pressure on intestinal bacteria of the treated host populations. Epidemiol Infect. 2017;145: 2081–2094. 10.1017/S095026881700084X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lowrance TC, Loneragan GH, Kunze DJ, Platt TM, Ives SE, Scott HM, et al. Changes in antimicrobial susceptibility in a population of Escherichia coli isolated from feedlot cattle administered ceftiofur crystalline-free acid. Am J Vet Res. 2007;68: 501–507. 10.2460/ajvr.68.5.501 [DOI] [PubMed] [Google Scholar]
- 40.Wagner RD, Johnson SJ, Cerniglia CE, Erickson BD. Bovine intestinal bacteria inactivate and degrade ceftiofur and ceftriaxone with multiple beta-lactamases. Antimicrob Agents Chemother. 2011;55: 4990–4998. 10.1128/AAC.00008-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singer RS, Patterson SK, Wallace RL. Effects of therapeutic ceftiofur administration to dairy cattle on Escherichia coli dynamics in the intestinal tract. Appl Environ Microbiol. 2008;74: 6956–6962. 10.1128/AEM.01241-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muñoz-Vargas L, Opiyo SO, Digianantonio R, Williams ML, Wijeratne A, Habing G. Fecal microbiome of periparturient dairy cattle and associations with the onset of Salmonella shedding. PLoS ONE. 2018;13: e0196171 10.1371/journal.pone.0196171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cunha CS, Marcondes MI, Veloso CM, Mantovani HC, Pereira LGR, Tomich TR, et al. Compositional and structural dynamics of the ruminal microbiota in dairy heifers and its relationship to methane production. Journal of the Science of Food and Agriculture. 2019;99: 210–218. 10.1002/jsfa.9162 [DOI] [PubMed] [Google Scholar]
- 44.Li J, Hao H, Cheng G, Liu C, Ahmed S, Shabbir MAB, et al. Microbial Shifts in the Intestinal Microbiota of Salmonella Infected Chickens in Response to Enrofloxacin. Front Microbiol. 2017;8: 1711 10.3389/fmicb.2017.01711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Food and Drug Administration, (FDA), Rockville, MD: U S Department of Health and Human Services. NARMS Now.