Abstract
Pancreatic ductal adenocarcinoma (PDA) is a major cause of cancer-related death, with limited therapeutic options available. This highlights the need for improved understanding of the biology of PDA progression, a highly complex and dynamic process, featuring changes in cancer cells and stromal cells. A comprehensive characterization of PDA cancer cell and stromal cell heterogeneity during disease progression is lacking. In this study, we aimed to profile cell populations and understand their phenotypic changes during PDA progression. To that end, we used single-cell RNA–sequencing technology to agnostically profile cell heterogeneity during different stages of PDA progression in genetically engineered mouse models. Our data indicate that an epithelial-mesenchymal transition of cancer cells accompanies tumor progression in addition to distinct populations of macrophages with increasing inflammatory features. We also noted the existence of 3 distinct molecular subtypes of fibroblasts in the normal mouse pancreas, which ultimately gave rise to 2 distinct populations of fibroblasts in advanced PDA, supporting recent reports on intratumor fibroblast heterogeneity. Our data also suggest that cancer cells and fibroblasts may be dynamically regulated by epigenetic mechanisms. This study systematically describes the landscape of cellular heterogeneity during the progression of PDA and has the potential to act as a resource in the development of therapeutic strategies against specific cell populations of the disease.
Keywords: Gastroenterology, Oncology
Keywords: Cancer
Single-cell RNA sequencing of mouse models of pancreatic ductal adenocarcinoma reveals the molecular subtypes of cancer cells, fibroblasts, and macrophages.
Introduction
Pancreatic ductal adenocarcinoma (PDA) carries the highest mortality rate of all major malignancies in industrialized countries, with a 5-year survival of 8.5%. Patients are faced with limited treatment options that achieve poor durable response rates, highlighting the need for an improved understanding of PDA disease biology (1). PDA progression is a complex and dynamic process that requires interaction between cancer cells and stromal cells (2). It is characterized by the formation of a unique microenvironment, consisting of heterogeneous stromal cell populations that include fibroblasts, macrophages, lymphocytes, and endothelial cells. These stromal compartments are critical in driving PDA biology (3).
The dynamic phenotypic changes in different cell populations during PDA progression are not fully understood. Gene expression profiling of bulk tissues provides a limited picture of the cellular complexity of the heterogeneous cell populations in PDA. In contrast, single-cell RNA–sequencing (scRNA-Seq) has the potential to enable gene expression profiling at the level of the individual cell (4) and provides a powerful tool to understand the cellular heterogeneity of PDA. We applied scRNA-Seq to investigate gene expression changes of cancer cells and stromal cells during PDA progression in genetically engineered mouse models (GEMMs). This unbiased approach provided evidence of considerable intratumor cellular heterogeneity, including molecular insights into epithelial and mesenchymal populations of cancer cells and distinct molecular subtypes of macrophages and cancer-associated fibroblasts (CAFs). These data will provide a resource for future studies aimed at further characterizing and targeting specific cell populations in PDA.
Results
Cellular heterogeneity during PDA progression.
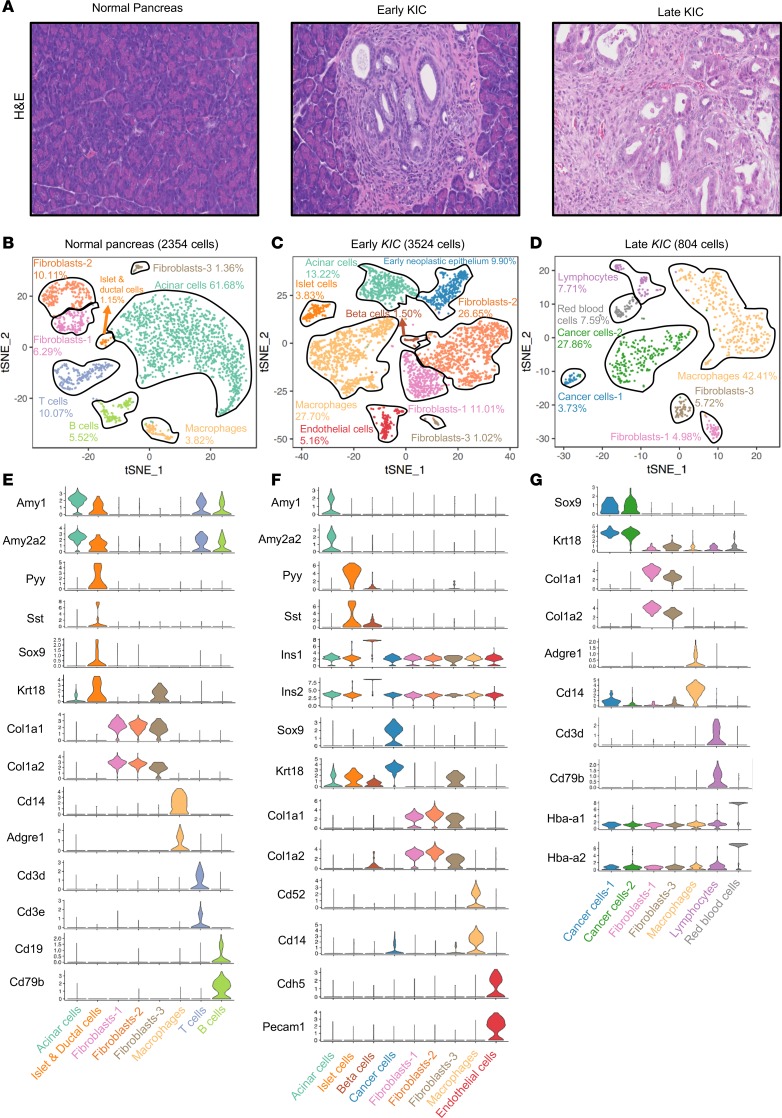
We sought to determine the composition of single cells during the progression of PDA GEMMs. Normal mouse pancreas; 40-day-old KrasLSL−G12D/+Ink4afl/flPtf1aCre/+ (KIC) (5) mouse pancreas, termed “early KIC” (with the early lesion initially confirmed by ultrasound; Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.129212DS1); and 60-day-old KIC pancreas, termed “late KIC” (Figure 1A) were freshly isolated and enzymatically digested followed by single-cell cDNA library generation using the 10× Genomics platform (6). Libraries were subsequently sequenced at a depth of more than 105 reads per cell. We performed stringent filtering, normalization, and graph-based clustering, which identified distinct cell populations in the normal pancreas and both stages of PDA.
Figure 1. Cellular heterogeneity during PDA progression.
(A) Representative H&E sections of the normal pancreas, early KIC lesion (which shows pancreatic intraepithelial neoplasia), and late KIC lesion (original magnification, ×20). (B) t-distributed stochastic neighbor embedding (tSNE) plot of the normal pancreas displaying 2354 cells comprising 8 distinct cell populations (pancreas pooled from 2 mice). (C) tSNE plot of the early KIC lesion displaying 3524 cells containing 9 cell types with the emergence of the cancer cell population (lesions pooled from 2 mice). (D) tSNE plot of the late KIC tumor showing 804 cells and 7 distinct populations (tumors pooled from 3 mice). Stacked violin plots of representative marker gene expression for each of the cell populations seen in the (E) normal pancreas, (F) early KIC lesions, and (G) late KIC lesion.
In the normal mouse pancreas, 2354 cells were sequenced and classified into appropriate cell types based on the gene expression of known markers: acinar cells, islet and ductal cells (Supplemental Figure 2), macrophages, T cells, and B cells, as well as 3 distinct populations of fibroblasts (Figure 1, B and E) were noted. In the early KIC lesion (3524 cells sequenced), the emergence of an expanded ductal population was observed (9.9% of cells), expressing known ductal markers, such as Krt18 and Sox9 (7), and displaying early neoplastic changes (Figure 1, A, C, and F, and Supplemental Figure 3). The acinar cell population was substantially reduced, while there was a marked increase in total macrophages and fibroblasts. Of note, the same 3 populations of fibroblasts seen in the normal pancreas were identified in the early KIC lesion. Additionally, endothelial cells were observed at this stage. This indicates that the expansion of fibroblasts and macrophages is an early event during PDA development. We next characterized the late KIC pancreas (804 cells sequenced) and noted the absence of normal exocrine (acinar) and endocrine (islet) cells (Figure 1, D and G). Instead, 2 distinct populations of cancer cells were present, suggesting phenotypic cancer cell heterogeneity as a late event in the course of the disease. We also observed the presence of only 2 distinct fibroblast populations, which had a similar percentage in relation to total cells. Noticeably, macrophages became a predominant cell population in the late KIC tumor. Moreover, we observed lymphocytes at this stage. The cellular heterogeneity in cancer cells and stromal cells in the early and late KIC lesions highlighted the dynamic cellular changes that occur during PDA progression.
Cancer cells enriched with mesenchymal markers emerge in advanced PDA.
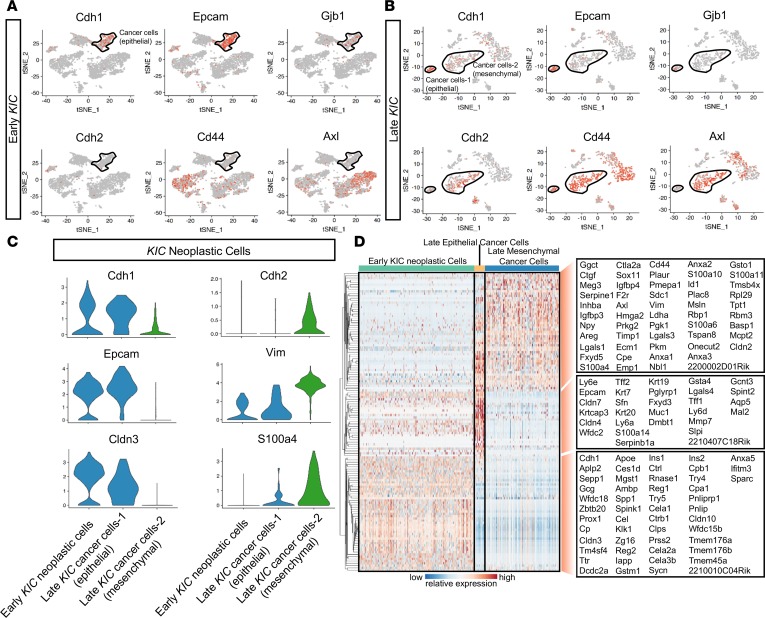
Gene expression analysis of epithelial markers (Cdh1, Epcam, Gjb1, and Cldn3) and mesenchymal markers (Cdh2, Cd44, Axl, Vim, and S100a4) revealed that the early KIC neoplastic cell population assumed an epithelial expression profile (Figure 2, A and C). This is in contrast with tumor cell populations in the late KIC tumors, where we identified 2 distinct cancer cell populations: 1 enriched for epithelial markers and the other, more abundant population enriched for mesenchymal markers (Figure 2, B and C). These scRNA-Seq data were confirmed by costaining immunohistochemistry of the late KIC tumor, which showed Sox9+Vim+ cancer cells invading the stroma whereas Sox9+Vim– cancer appeared in more ordered cell clusters (Supplemental Figure 4). These data support the notion that tumor cell epithelial plasticity may contribute to cancer cell heterogeneity during the progression of KIC tumors.
Figure 2. Analysis of early and late KIC neoplastic cell populations demonstrate the emergence of the mesenchymal cancer cell population as a late event.
(A) tSNE plots of the early KIC lesion demonstrated the expression of known epithelial markers in the early neoplastic cell population (black outline). Mesenchymal markers were absent in this population. (B) tSNE plots demonstrating the emergence of 2 cancer cell populations in the late KIC tumor. One cancer cell population expressed the epithelial markers (smaller population outlined in black), and a second expressed the mesenchymal markers (larger population outlined in black). (C) Violin plots showing the high expression of epithelial markers (Cdh1, Epcam, and Cldn3) in the early neoplastic KIC cell population and late KIC epithelial cancer cell population but not in the mesenchymal population. Mesenchymal markers (Cdh2, Vim, and S100a4) were overexpressed in the mesenchymal cancer cell population but not in the early KIC neoplastic or late KIC epithelial cancer cell population. (D) Single-cell profiling heatmap of all early and late KIC neoplastic cells displaying differentially expressed genes among the 3 cell populations. Gene names are listed in the boxes on the far right of the heatmap. Each column represents an individual cell, and each row is the gene expression value for a single gene.
The hierarchical clustering of the top significant genes in each of the 3 putative cancer/neoplastic cell populations (early neoplastic cells in early KIC, epithelial and mesenchymal cancer cell populations in late KIC) was performed (Figure 2D). In addition, gene clusters from the cancer cell populations were subjected to pathway and Gene Ontology (GO) analysis. First, we compared neoplastic cells of the early KIC population with the total cancer cells of the late KIC and found that the most downregulated genes in late KIC cancer cells were associated with normal pancreatic function, such as pancreatic secretion, digestion and absorption, and insulin secretion (Supplemental Figure 5, A and B). Moreover, normal pancreatic acinar genes, such as Try4, Try5, Cela2a, Cela3b, Reg2, and Rnase1, were expressed at higher levels in early neoplastic KIC cells (Figure 2D). This is suggestive of ongoing acinar-to-ductal metaplasia (ADM) during tumor progression in this GEMM, which was supported by histological analysis (Supplemental Figure 3). In contrast, the most upregulated genes in late KIC cancer cells were associated with ribosome, glycolysis/gluconeogenesis, and amino acid biosynthesis, which is highly suggestive of increased translation and metabolically active cancer cells in established KIC tumors. Interestingly, pathways previously reported to be closely associated with the stroma and progression of PDA were also highlighted, such as ECM-receptor interaction (8), TGF-β (9), and hippo signaling pathways (10). We then compared early neoplastic KIC cells with the late KIC epithelial cancer cell population to understand processes that promoted the progression of PDA in the epithelial cancer cell compartment. Interestingly, similar cell functions and signaling pathways were identified by comparing the 2 epithelial cancer cell populations (Supplemental Figure 5, C and D). Taken together, these analyses demonstrate an ongoing ADM state during the progression of KIC tumors.
Cancer cells enriched for mesenchymal markers exist in advanced PDA GEMMs with different driver mutations.
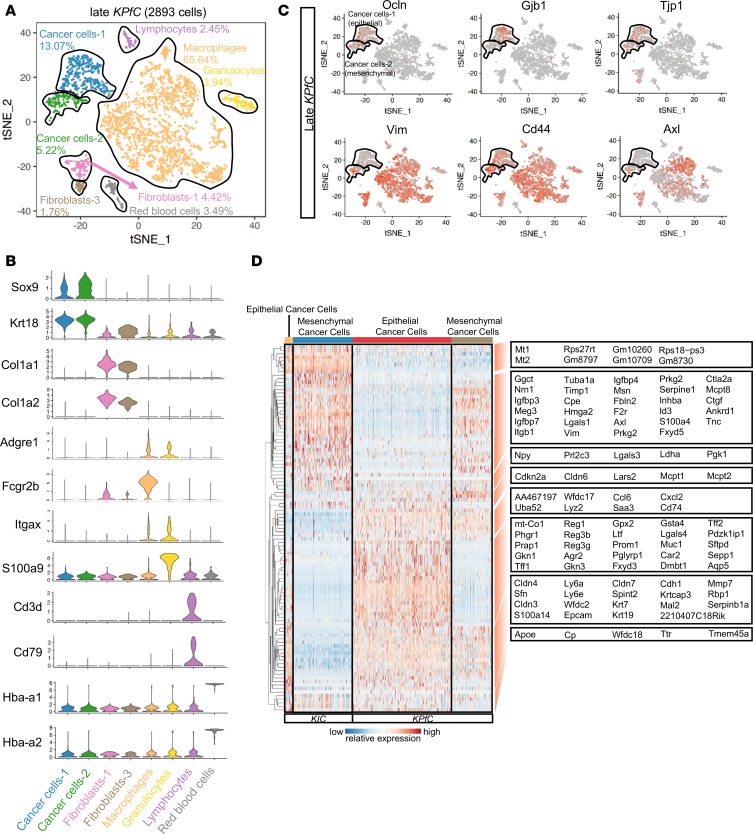
In addition to KRAS mutations, additional driver events are required for PDA progression (8), with TP53 and INK4A being the second and third most commonly mutated genes in human PDA, respectively. As such, we sought to understand the effect of different secondary driver mutations on the phenotypes and heterogeneity of cancer cells. We performed scRNA-Seq in another PDA GEMM, KrasLSL−G12D/+Trp53fl/flPdx1Cre/+ (KPfC) (Figure 3, A and B). Consistent with late KIC tumors, 2 distinct cancer cell populations expressing Krt18 and Sox9 were noted in late KPfC (60-day-old) tumors: 1 was marked by epithelial markers, such as Gjb1, Tjb1, Ocln, and Cldn3, while the other was marked by mesenchymal markers, such as Vim, Cd44, Axl, S100a4, and Fbln2 (Figure 3C and Supplemental Figure 6A). Epithelial and mesenchymal cancer cell populations in KPfC mice shared many genes in common with the corresponding populations in KIC; however, they also expressed unique gene signatures (Figure 3D).
Figure 3. Comparison between cancer cells of KIC and KPfC tumors.
(A) tSNE plot of the late KPfC lesion displaying 2893 cells and 8 distinct cell populations (tumor analyzed from 1 mouse). (B) Stacked violin plots showing representative marker gene expression for each of the cell populations seen in the late KPfC lesion. (C) Single-gene tSNE plots of the KPfC tumor displaying the presence of epithelial markers (Ocln, Gjb1, and Tjp1) in the epithelial cancer cell population (top black-outlined population) and mesenchymal markers (Vim, Cd44, and Axl) in the mesenchymal cancer cell population (bottom black-outlined population). (D) Single-cell profiling heatmap comparing all cancer cells in late KIC versus all cancer cells in late KPfC. Each column represents an individual cell, and each row is the gene expression value for a single gene.
We then compared the total cancer cell gene signatures between late KIC and late KPfC mice by KEGG and Biocarta pathway analysis methods, to identify potential differences in cancer cell signaling pathways caused by the different secondary driver mutations. As expected, the p53 signaling pathway was upregulated in the KIC model in comparison with the KPfC model (Supplemental Figure 6B). The analyses of late KIC and late KPfC mice suggest that cancer cell heterogeneity is a late-stage tumor event that occurs in the setting of multiple secondary driver mutations. However, under the same oncogenic Kras mutation, different secondary driver mutations can potentially lead to different signaling pathways that drive PDA progression.
Macrophage heterogeneity during PDA progression.
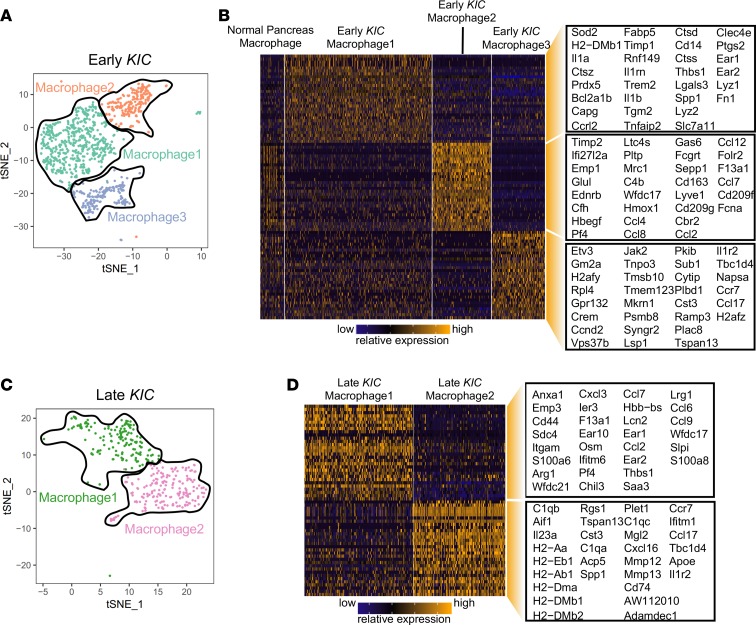
We found a marked increase in the size of the macrophage population as PDA progressed from normal pancreas to early KIC and eventually late KIC tumors (Figure 1, B–D). We further characterized the macrophage compartment during PDA progression by subclustering macrophages in early and late KIC lesions, which revealed 3 transcriptionally distinct macrophage clusters in early KIC and 2 in late KIC (Figure 4, A and C).
Figure 4. scRNA-Seq analysis of KIC tumor progression reveals multiple subpopulations of macrophages.
(A) tSNE plot of 3 macrophage subpopulations in the early KIC lesion. (B) Heatmap depicting the 30 top significantly overexpressed genes in each of the 3 early KIC macrophage subpopulations. Macrophages from the normal pancreas are displayed (far left group). Each column represents an individual cell, and each row is the gene expression value for a single gene. (C) tSNE plot representation of 2 macrophage subpopulations in late KIC. (D) Heatmap depicting the top 30 significantly overexpressed genes in each of the 2 late KIC macrophage subpopulations. Each column represents an individual cell, and each row is the gene expression value for a single gene.
Macrophage population 1 in early KIC tumors was characterized by the expression of Fn1, Lyz1, Lyz2, Ear1, and Ear2 as well as Cd14 (Figure 4B). Moreover, these macrophages specifically expressed high levels of the IL-1 receptor ligands: Il1a, Il1b, and Il1rn. GO analysis suggested that this macrophage population was involved in healing during inflammation, the regulation of type I and III hypersensitivities, and antigen processing and presentation (Supplemental Figure 7A). In contrast, macrophage population 2 was noted to express an abundance of chemokines, including Ccl2, Ccl4, Ccl7, Ccl8, and Ccl12, as well as many complement-associated genes (Figure 4B). Indeed, leukocyte activation, complement activation, and humoral response genes were the most significantly enriched GO categories in this macrophage population (Supplemental Figure 7A). The third macrophage population expressed Ccl17 and Ccr7 and was enriched in ribosomal small-unit biogenesis, translation, and antigen-processing functions (Figure 4B and Supplemental Figure 7A). Importantly, macrophages in normal mouse pancreas weakly expressed genes found in macrophage populations 2 and 3 from early KIC mice, suggesting that the normal pancreas macrophages could be noncommitted macrophages residing in tissue in the normal organ that are induced to adopt a distinct phenotype upon tumor initiation.
The late KIC tumor featured 2 macrophage subpopulations (Figure 4C). Macrophage population 1 highly expressed genes such as S100a8 and Saa3, which have been shown to be expressed in lipopolysaccharide-treated monocytes (11). Moreover, numerous chemokines were elevated in this population, such as Ccl2, Ccl7, Ccl9, Ccl6, Cxcl3, and Pf4 (Figure 4D). GO analysis revealed this population is likely associated with Stat3 activation, leukocyte chemotaxis, and response to lipopolysaccharide and inflammatory stimuli (Supplemental Figure 7B). These data suggest that macrophage population 1 was inflammatory in nature. Macrophage population 2 of late KIC tumors was rich in MHC-II antigen presentation molecules: Cd74, H2-Aa, H1-Ab1, H2-Dma, H2-Dmb1, H2-Dmb2, and H2-Eb1 (Figure 4D), and GO analysis highlighted antigen presentation and adaptive immune response pathways as being elevated (Supplemental Figure 7B). Consistently, in late KPfC tumors, we also observed 2 distinct populations of macrophages with similar features (Supplemental Figure 8). Interestingly, we did not observe a macrophage population in late tumors that correlated with macrophage population 1 from the early tumors, suggesting that this population might undergo negative selection or differentiation into inflammatory or MHC-II–rich macrophages during tumor progression.
We also compared the features of the total macrophage clusters between early and late KIC tumors and observed a substantially enhanced macrophage inflammatory signature as the tumor progressed (Supplemental Figure 7, C and D). A wide variety of inflammatory genes increased, including Il1a, Il1b, Il1r2, and Il6. GO analysis of this gene list highlighted leukocyte chemotaxis and inflammatory response functions as increased in advanced KIC tumors. These data suggest that PDA progression is characterized by an increase in inflammatory features in macrophages.
Fibroblast heterogeneity during PDA progression.
In normal pancreas and the early KIC lesion, we identified 3 distinct populations of fibroblasts, while in late KIC only 2 fibroblast populations were noted (Figure 1, B–D). To ascertain the relationship between these fibroblast populations and the dynamics of their phenotypic changes during PDA progression, we projected fibroblasts from the 3 analyses onto a single tSNE plot and applied a graph-based clustering algorithm (Figure 5A), which revealed 3 distinct molecular subtypes of fibroblasts among the normal pancreas, early KIC, and late KIC. The overlay demonstrates that the normal pancreas and early KIC contained all 3 fibroblast subtypes while late KIC contained only 2, confirming our initial analysis (Figure 1, B–D). Specifically, this analysis demonstrated that fibroblast population 1 (FB1) and fibroblast population 3 (FB3) found in normal and early KIC pancreas were present in the late KIC tumor whereas fibroblast population 2 (FB2) was absent in late KIC.
Figure 5. Analysis of fibroblasts during PDA progression reveals multiple molecular subtypes.
(A) All fibroblasts from the normal pancreas and early and late KIC lesions were projected onto a single tSNE plot with the FB1, FB2, and FB3 populations distinguished by pink, orange, and brown, respectively (top left). Normal pancreas fibroblasts were highlighted in red (top right), early KIC fibroblasts in green (bottom left) and late KIC fibroblasts in blue (bottom right). Normal pancreas and early KIC contained fibroblasts in all 3 groups whereas the late KIC only contained FB1 and FB3. (B) Heatmap displaying the top significant genes (cutoff: P < 10–40) for each of the 3 fibroblast populations. Thirty random cells from each fibroblast population are displayed. All 3 late-cancer GEMMs (late KIC, KPfC, and KPC) display only FB1 and FB3 populations. (C) Violin plots demonstrating representative marker genes for each fibroblast subtype: FB1 overexpressed cytokines and Pdgfra. FB3 overexpressed mesothelial markers, myofibroblast markers, MHC-II molecules, and Cdh11.
In the normal pancreas, FB1, FB2, and FB3 made up 35.4%, 56.9%, and 7.7% of total fibroblasts, respectively (Supplemental Figure 9). In early KIC, although the total fibroblasts expanded (Figure 1C), the ratios of each fibroblast population remained similar. Furthermore, in the late KIC tumor, FB1 and FB3 were present in nearly equal proportions of 46.5% and 53.5%, respectively (Supplemental Figure 9). Each fibroblast population was characterized by distinct marker genes. For example, FB1 markedly expressed Cxcl14, Ptn, and several genes mediating insulin-like growth factor signaling, such as Igf1, Igfbp7, and Igfbp4 (Figure 5B). FB2 specifically expressed Nov, a member of the CCN family of secreted matricellular proteins (12) as well as Pi16, which has been shown to be expressed in fibroblast populations in various tissue types (13), in addition to Ly6a and Ly6c1. FB3 showed distinct expression of mesothelial markers, such as Lrrn4, Gpm6a, Nkain4, Lgals7, and Msln (14), in addition to other genes previously shown to be expressed in fibroblasts, such as Cav1, Cdh11, and Gas6 (15–17).
We also performed scRNA-Seq of the KrasLSL−G12D/+Trp53LSL−R172H/+Ptf1aCre/+ (KPC) mouse model (18), which also displayed 2 subtypes of CAFs (Supplemental Figure 10) as did the KPfC GEMM (Figure 3A). Hierarchical clustering of the most significant genes for each fibroblast subtype showed the persistence of FB1 and FB3 during the progression of PDA (Figure 5B) and that they exist across different advanced-stage PDA GEMMs (KIC, KPC, and KPfC), suggesting a consistent cell of origin. Interestingly, the gene expression heatmap also indicated that the FB2 population started to move toward an FB1-like expression profile in the early KIC lesion, suggesting FB1 and FB2 might converge into a single CAF population with FB1-predominant features by late invasive disease. Of note, Il6, Ccl2, Ccl7, Cxcl12, and Pdgfra were expressed in FB1 and FB2 in the normal pancreas and early KIC lesion and showed greater expression in FB1 of late KIC (Figure 5C). In contrast, the myofibroblast markers Acta2 and Tagln were expressed by a portion of FB3. These data support the presence of previously described, mutually exclusive, inflammatory (FB1) and myofibroblastic (FB3) CAF subtypes (19–21). Interestingly, FB3 also expressed numerous MHC-II–associated genes (Figure 5C). GO analysis suggested that FB1 was involved in an acute phase response and inflammatory response, FB2 was more associated with physiological functions of fibroblasts, and FB3 had antigen processing and presentation through the MHC-II pathway and had complement activation functions (Supplemental Figure 11A). Furthermore, we analyzed genes that increased in FB1 and FB3 during PDA progression and found that FB1 showed a progressive increase in the expression of genes associated with inflammatory response and chemotaxis while FB3 genes displayed increased function on translation during disease progression, possibly due to enhanced antigen-processing activity (Supplemental Figure 11, B and C). These data suggest that FB1 is an inflammatory population and the inflammatory feature increases during PDA progression, while FB3 consists of the well-studied myofibroblast population and displays an enrichment for MHC-II genes.
We also found that some genes essentially exclusive to FB3 in the normal and early KIC pancreas became expressed in FB1 and FB3 populations in late KIC, marking these genes as potential global fibroblast markers in advanced PDA. One such gene was Cdh11 (Figure 5C). We supported these data by immunohistochemistry. We found in late KIC and KPC tumors, stromal staining for α–smooth muscle actin (α-SMA) and PDGFR-α were nearly mutually exclusive, whereas CDH11 showed uniform staining across all morphologically discernable fibroblasts (Supplemental Figure 11D). Taken together, these data provide the first in vivo description to our knowledge of all CAF populations during PDA progression.
Mesenchymal cancer cells and CAFs show evidence of increased epigenetic regulation and transcriptional activity in advanced PDA.
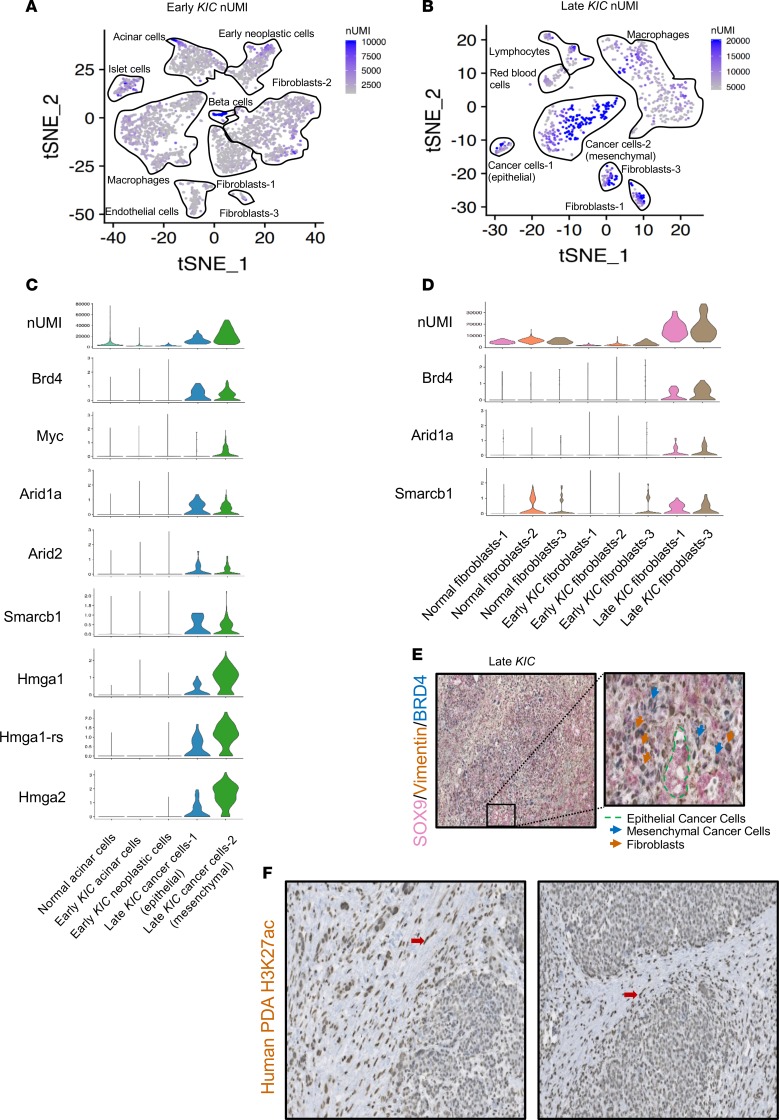
Unique molecular identifiers (UMIs) serve to place a bar code on each input mRNA molecule during cDNA library generation, enabling the determination of initial transcript number even after cDNA library amplification (22). We compared UMI counts across all cell types between early and late KIC lesions (Figure 6, A and B). In early lesions, there was a marked increase in UMI in the beta islet cells (median: 2849, range: 1322–12,857), which might indicate that increased transcriptional activity is a means by which the endocrine requirements of these cells are met. No other cell population in early KIC displayed this level of UMIs. The early neoplastic KIC cells displayed a relatively low UMI count (median: 1979, range: 1163–7735). In contrast, the mesenchymal cancer cell population in the late KIC tumor displayed a marked increase in total UMI count with a median count of 18,334 and range of 4433–50,061 (Figure 6C). The epithelial cancer cells in the late KIC also displayed an increased UMI, albeit to a far lesser degree than the mesenchymal cancer cell population (median: 10,368, range: 4940–30,440).
Figure 6. Analysis of transcriptional activity in different stages of PDA reveals differential epigenetic and transcriptional activity in distinct tissue compartments.
tSNE plot of number of unique molecular identifiers (nUMIs) in the (A) early and (B) late KIC. (C) Violin plots of epigenetic regulatory genes in the epithelial cell populations of the normal pancreas, early KIC, and late KIC. (D) Violin plots of epigenetic regulator genes in the normal, early fibroblast, and late fibroblast populations showing their upregulation in CAFs. (E) Sequential triple immunohistochemical staining on the same late KIC tumor section for cancer cells (SOX9, shown in pink), mesenchymal cells (vimentin, shown in brown), and increased enhancer activity (BRD4, shown in blue). Well-differentiated ductal epithelium stained solely for SOX9 (green outline). Mesenchymal cancer cells (blue arrows) and CAFs (brown arrows). Original magnification ×10. Inset magnification ×30. (F) Immunohistochemical analysis of human PDA whole tissue sections using the H3K27ac antibody. These representative figures from 2 human PDAs demonstrate the 3+/3+ staining in the stromal fibroblasts (red arrows) with 1+–2+ staining in the cancer epithelium (original magnification, ×20).
We reasoned that the increased transcriptional activity may be at least in part regulated by increased activity of epigenetic-associated genes as well as increased enhancer activity (23). BRD4 belongs to the bromodomain family of transcriptional regulators and is a key regulator of super-enhancer activity (24). Moreover, prior studies have shown that MYC activity is promoted by super-enhancer activity in PDA (25). We found that in late KIC and KPfC tumors, Brd4 was expressed highly in epithelial and mesenchymal cancer cells while Myc was expressed mainly in the mesenchymal cancer cell population (Figure 6C and Supplemental Figure 12). In addition, several genes encoding high-mobility group A proteins (Hmga1, Hmga1-rs, and Hmga2) were markedly expressed in late KIC and KPfC mesenchymal cancer cells. HMGA proteins are chromatin-associated proteins that regulate transcriptional activity, including enhancesome formation (26). Last, critical components of the SWI/SNF complex (Smarcb1, Arid1a, Arid2), which are essential in nucleosome remodeling and transcriptional regulation (27), were also expressed highly in epithelial and mesenchymal cancer cells of the late KIC but not early neoplastic cells in the early KIC lesion. Taken together, these data provide multiple lines of evidence to suggest that the transcript load of a more aggressive mesenchymal cancer cell population is increased relative to cancer cells in early lesions or epithelial cancer cells in advanced PDA. We also noted that fibroblasts in late KIC tumors showed increased UMIs (median: 14,538, range: 4461–37,497). They also displayed an increased expression of epigenetic transcriptional regulator genes in contrast with fibroblasts from normal mouse pancreas or early KIC pancreas (Figure 6D).
We supported these scRNA expression data using 3-color immunohistochemical analysis of late KIC tumors: SOX9 was used as a pan-cancer cell marker, vimentin as a mesenchymal marker, and BRD4 as a surrogate marker for increased enhancer activity. We identified positive costaining for vimentin and BRD4 in CAFs, positive triple staining (Vimentin+/SOX9+/BRD4+) in mesenchymal cancer cells, and single staining of SOX9 in epithelial cancer cells that localized to more differentiated, duct-like structures in the advanced tumors (Figure 6E). Next, we performed immunohistochemical analysis on 16 whole-tumor human pancreatic cancer sections using an antibody against H3K27ac, a commonly accepted marker of increased gene regulatory element activity (23, 28). The malignant epithelium and stromal fibroblasts were scored separately. These analyses showed markedly positive 3+/3+ staining in the stromal fibroblasts of all whole-tumor sections (Figure 6F). In 6 of 16 cancer epithelia the score was 1+, and 10 of 16 scored 2+, with no samples showing a cancer epithelial scoring of 3+. These data warrant further study of epigenetic regulatory mechanisms in fibroblasts as normal pancreas fibroblasts progress to CAFs in PDA.
Discussion
We have carried out scRNA-Seq of different stages of the KIC GEMM, in addition to late KPfC and KPC tumors, to agnostically profile the phenotypic changes of cancer and stromal cells during PDA progression. We have established the emergence of a mesenchymal cancer cell population as a late-stage tumor event and have identified potentially novel features of different macrophage and fibroblast populations. Although intertumor heterogeneity can exist between mice with the same GEMM genotype (29, 30), analyses of late KIC and late KPfC revealed the same 2 populations of cancer cells (epithelial and mesenchymal), macrophages (inflammatory and MHC-II rich), and fibroblasts (FB1 and FB3), illustrating consistent intratumor cellular heterogeneity between PDA GEMMs with distinct secondary driver mutations. These populations were consistent between the pooled (late KIC) and nonpooled (late KPfC) experiments. Nonetheless, 3 animals were assayed for the late KIC GEMM and 1 for each of the late KPfC and KPC GEMMs, which may have caused intertumor population variants to be missed. Future studies should aim to compare multiple mice of the same genotype to refine the definition of intertumor heterogeneity in these GEMMs. Endothelial cells in particular warrant further investigation in PDA GEMMs. We found that the early KIC lesion featured an expanded endothelial cell population that dissipated by the late KIC lesion, thus limiting the utility of the current data set regarding endothelial heterogeneity in PDA. It is likely that pre–scRNA-Seq cellular enrichment techniques will be required to identify an endothelial signature in late-stage PDA GEMMs. These findings are an important resource and improve our understanding of PDA progression while laying the foundation for mechanistic studies aimed at dissecting the function of specific cell populations in PDA progression.
PDA pathogenesis involves metaplasia of normal acinar cells to ductal epithelium, which in turn undergo neoplastic transformation in a KRAS-driven manner (31). Malignant ductal epithelium may then assume more aggressive, mesenchymal features as the disease progresses. In this study, mesenchymal cancer cell populations were noted in late-stage tumors. Moreover, although human scRNA-Seq data are limited in PDA, analyses of a previously sequenced human PDA tumor (32) also suggest the possibility of distinct epithelial-like and mesenchymal-like cancer cell populations (Supplemental Figure 13), although to make firm conclusions requires a greatly expanded cohort. Our data support a model in which mesenchymal features of cancer cells are acquired later in the disease process, although others have argued that this can be one of the earliest events in PDA (33). Mesenchymal cancer cell populations have been studied extensively in pancreatic cancer mouse models and have been shown to be critical to chemotherapeutic resistance while their contribution to metastasis has been more controversial (34, 35). Mesenchymal cancer cells have previously demonstrated an increased protein anabolism and activation of the endoplasmic reticulum stress–induced survival pathways in a PDA GEMM (36).
Indeed, in the late KIC model, ribosomal pathways were the most significantly upregulated pathways in cancer cells (Supplemental Figure 5, A and C). It is likely that the demand for increased ribosomal activity stems from high transcriptional activity governed by epigenetic mechanisms in the mesenchymal cancer cells because we also saw markedly increased UMI counts in this population (Figure 6B). The bromodomain and extraterminal (BET) family of proteins, such as BRD4, which is markedly upregulated in cancer cells and fibroblasts of late-stage PDA (Figure 6, C and D), serve to recruit regulatory complexes to acetylated histones at enhancer sites, resulting in increased transcription (37). Previously, a combination approach using a BET protein inhibitor and a histone deacetylase inhibitor led to near-complete tumor regression and improved animal survival in a PDA GEMM (25). Super-enhancer activation has recently been shown to be fundamental in the pathophysiology of a variety of neoplasms (38) and is intimately associated with Hmg2a in PDA because super-enhancer attenuation has been demonstrated to downregulate Hmg2a expression and the growth of PDA cells in a 3-dimensional in vitro model (39). It has been shown that enhancer activity is critical to PDA metastasis (40) and may even be involved in class switching between classical and basal molecular subtypes in human PDA samples (41). Nonetheless, future efforts to target increased transcriptional activity in PDA should consider distinct tissue compartments governing the sensitivity and resistance to novel therapeutics.
Our data revealed 2 molecular subtypes of macrophages in advanced PDA (Figure 4). One expressed numerous chemokine- and inflammation-associated genes while the other was rich in MHC-II–associated genes. In a previous study, MHC-II+ macrophages were isolated from orthotopic breast tumors and highly expressed CCL17, consistent with our data (42). In parallel with our study, MHC-IIlo macrophages were found to be highly enriched for numerous chemokines. Moreover, in an orthotopic hepatoma mouse model, an early MHC-II+ macrophage population appeared to suppress tumor growth, but an MHC-IIlo macrophage population became the predominant macrophage population as the tumor progressed, resulting in a protumor phenotype (43). Nonetheless, to confirm their pathophysiological significance, functional studies are required in which inducible selective ablation (44) is performed on the 2 late-stage PDA macrophage subpopulations using specific markers we have identified in this study. Zhu and colleagues (45) have shown that bone marrow–derived monocytes make up approximately 80% of MHC-II+ macrophages in a PDA GEMM whereas MHC-II– macrophages in normal pancreas and PDA were shown to be maintained independently of monocyte contributions. Monocyte-independent MHC-IIlo tissue resident macrophages expanded during tumor progression and contributed to PDA growth and survival. Conversely, Sanford and colleagues (46) have shown that monocytes can give rise to a proinflammatory macrophage population in a PDA mouse model, which, when antagonized with neutralizing antibodies against CCR2, resulted in decreased tumor growth and reduced metastases in vivo (46). These data highlight the need for an scRNA-Seq study on macrophage populations in PDA GEMMs with labeled bone marrow replacement to reconcile these discrepancies.
More importantly, in the studies of tumor-associated macrophages, inflammatory chemokines are commonly used to indicate an M1 type of macrophage, which is normally associated with immune-stimulatory functions. Nevertheless, our study indicates that a distinct M1/M2 macrophage phenotype is not readily discernable at the single-cell level. Instead, as PDA progresses, an inflammatory feature is substantially increased, and this accompanies an increase of an important M2 macrophage marker, ARG1 (Supplemental Figure 7, A–D). This raises questions on the M1/M2 classification system because the inflammatory feature is associated with the progression of PDA. Future studies should focus on the function of these inflammatory macrophages in PDA in addition to validating markers for macrophage classification.
Although numerous studies have generally shown that CAFs are tumor promoting in the biology of PDA and other carcinomas (47, 48), recent studies have found that the functions of CAFs in PDA biology are more varied. Özdemir and colleagues (44) demonstrated that the depletion of α-SMA+ cells from the microenvironment in a PDA GEMM resulted in shortened survival and poorly differentiated tumors (44), and low myofibroblast tumor content was shown to be associated with worse survival in human PDA sections. These data prompted a paradigm shift whereby certain CAFs may function to constrain, rather than promote, PDA. Moreover, until recently, the molecular heterogeneity of CAFs in PDA has not been well appreciated. The primary attempt to characterize fibroblast heterogeneity in PDA demonstrated that mouse pancreatic stellate cells (PSCs) could be induced to express α-SMA in vitro when directly cocultured with primary mouse PDA cells in an organoid coculture system (19). These myofibroblastic CAFs were designated as “myCAFs.” This was distinct from IL-6+ fibroblasts that were produced in vitro when PSCs were indirectly cocultured with mouse PDA organoids through a semipermeable membrane. The IL-6+ fibroblasts were also positive for PDGFR-α and numerous other cytokines and therefore termed inflammatory CAFs or “iCAFs.” Immunohistochemistry of human and mouse PDA tissue showed distal IL-6+ stroma as a distinct population from the peritumoral α-SMA+ stroma (32). Subsequent studies in PDA GEMMs demonstrated that the iCAF population can mediate protumorigenic properties and is a potential therapeutic target to sensitize PDA to immunotherapeutic strategies (20, 21).
Our study is the first to our knowledge to demonstrate the existence of 3 distinct molecular subtypes of fibroblasts in the normal mouse pancreas, which in turn gave rise to 2 distinct subtypes of CAFs that were largely conserved across 3 PDA GEMMs. We noted that FB1 expressed insulin-like growth factor signaling genes (Igfbp7, Igfbp4, and Igf1) in addition to Pdgfra, Cxcl12, Il6, and several other cytokines (Ccl11, Ccl7, Ccl2, and Csf1). We propose that our FB1 population is the previously described iCAF population and hence likely protumorigenic. Conversely, the FB3 population was positive for the myofibroblast markers Acta2 and Tagln and therefore most closely represents the previously described myCAF population. Importantly, our agnostic approach did not identify any further putative CAF populations, and so we support the 2-CAF model Öhlund and colleagues proposed (19).
In summary, this report systematically outlines the cellular landscape during the progression of PDA and highlights the cellular heterogeneity in PDA pathogenesis. As such, future targeted therapeutic strategies should be developed with their intended target subpopulation in mind.
Methods
Animal studies.
KIC (KrasLSL−G12D/+Ink4afl/flPtf1aCre/+), KPC (KrasLSL−G12D/+Trp53LSL−R172H/+Ptf1aCre/+), and KPfC (KrasLSL−G12D/+Trp53fl/flPdx1Cre/+) mice were generated as previously described (18, 49, 50). Mice were sacrificed when they were moribund: 60 days old for the KIC (n = 3, late KIC) and KPfC (n = 1) or 6 months old for the KPC (n = 1). Early KIC mice were sacrificed at 40 days old (n = 2), and normal pancreas mice (n = 2) were sacrificed at 60 days old. Normal pancreas was obtained from Cre-negative littermates of the KIC mice. In experiments using more than 1 mouse, tissues were pooled before enzymatic digestion. The KPfC mouse had a pure C57BL/6 genetic background, and all others had a mixed background (C57BL/6 with FVB). Ultrasound imaging was carried out under general anesthesia with isoflurane. Mice were euthanized by cervical dislocation under anesthesia. Tissues were either fixed in 10% formalin for immunohistochemistry or enzymatically digested for single-cell analysis.
Tissue digestion.
A 10× digestion buffer was prepared in PBS: collagenase type I (450 units/mL, Worthington Biochemical), collagenase type II (150 units/mL, Worthington Biochemical), collagenase type III (450 units/mL, Worthington Biochemical), collagenase type IV (450 units/mL, Gibco, Thermo Fisher Scientific), elastase (0.8 units/mL, Worthington Biochemical), hyaluronidase (300 units/mL, MilliporeSigma), and DNAse type I (250 units/mL, MilliporeSigma). Tumors and pancreas were enzymatically digested into a single-cell suspension. Briefly, freshly dissected tissue was placed into a 10-cm tissue culture dish, and a sterile razor blade was used to cut the tissue into fine pieces. Samples were resuspended in PBS and washed twice by centrifuge at 480 g for 3 minutes and added to a 50-mL tube containing 1× digestion buffer (Worthington Biochemical) containing 1% FBS. The tube was incubated on a shaker at 37°C for 60 minutes. Then 35 mL of PBS was added, and cells were washed 3 times before filtering out debris using a 70-μm mesh filter (MilliporeSigma). Single cells were resuspended in 100 μL of PBS in preparation for single-cell library creation. Cell viability was measured by trypan blue. Viability was 80% for the normal pancreas and late KIC samples, 75% for the early KIC and KPfC, and 90% for the KPC.
Single-cell cDNA library preparation and sequencing.
Library generation was performed using the 10× Chromium System (10× Genomics Inc.). Single-cell suspensions were washed in 1× PBS (calcium and magnesium free) containing 0.04% w/v bovine serum albumin (400 μg/mL) and brought to a concentration of 200–700 cells/μL. The appropriate volume of cells was loaded with Single Cell 3′ gel beads into a Single Cell A Chip and run on the Chromium Controller (10x Genomics, Inc.). Gel bead in emulsion (GEM) was incubated and then broken. Dynabeads MyOne Silane magnetic beads (Thermo Fisher Scientific) were used to clean up the GEM reaction mixture. Read 1 primer sequence (10X Genomics’ Chromium Single Cell 3ʹ Reagent Kits User Guide; https://support.10xgenomics.com/single-cell-gene-expression/library-prep) was added during incubation, and full-length, barcoded cDNA was amplified by PCR after cleanup. Sample size was checked on an Agilent Tapestation 4200 using DNA HS 5000 tape and concentration determined by a Qubit 4 Fluorometer (Thermo Fisher Scientific) using the DNA HS assay. Samples were enzymatically fragmented and underwent size selection before proceeding to library construction. During library preparation, Read 2 primer sequence, sample index, and both Illumina adapter sequences were added. Samples were cleaned up using AMPure XP beads (Beckman Coulter), and after library preparation quality control was performed using DNA 1000 tape on the Agilent Tapestation 4200. The final concentration was ascertained using the Qubit 4 Fluorometer DNA HS assay. Samples were loaded at 1.5 pM and run on the Illumina NextSeq500 High Output Flowcell using V2.5 chemistry. The run configuration was 26 base pairs (bp) × 98 bp × 8.
Bioinformatics analyses.
We used Cell Ranger version 1.3.1 (10x Genomics) to process raw sequencing data. Briefly, raw base call (BCL) files were converted to FASTQ files and aligned to mouse mm10/GRCm38 reference transcriptome. Transcript counts in each cell were quantified using barcoded UMIs and 10× cell barcode sequences. The gene-by-cell-expression matrices were loaded to the R package Seurat version 2.3.1 for downstream analyses (51). Low-quality cells were further filtered out based on number of genes detected, nUMIs, and mitochondrial gene content. Data were scaled by regressing out the nUMIs and percentage of mitochondrial gene content and were subjected to dimensional reduction by principal component analysis and visualization using tSNE. Cell clusters were identified via the FindClusters function using a resolution of 0.6 for all samples, based on a graph-based clustering algorithm implemented in Seurat. A likelihood ratio–based test or an AUC-based scoring algorithm (implemented in Seurat) was used to compute marker genes for each cluster, and expression levels of several known marker genes were examined. Different clusters expressing known marker genes for a given cell type were selected and combined as 1 for each cell type. GO and pathway analyses were performed using the Database for Annotation, Visualization and Integrated Discovery bioinformatics suite, version 6.8 (52).
Human scRNA-Seq data were obtained from a previously published report (32). UMAPs were generated following Seurat’s recommended practices as previously described (51). Analysis and plots were done through Seurat 3.0.
Histological analysis.
Formalin-fixed tissues were embedded in paraffin and cut in 5-μm sections. Sections were evaluated by H&E and immunohistochemical analysis using antibodies against vimentin (catalog 5741, Cell Signaling Technology), BRD4 (catalog AB128874, clone EPR5150, Abcam; ref. 2), SOX9 (catalog AB5535, MilliporeSigma), CDH11 (catalog NBP2-15661, Novus Biologicals), and H3K27ac (catalog AB4729, Abcam). Following an initial antigen retrieval with Tris-EDTA-glycerol (10%) buffer (Thermo Fisher Scientific) and inhibition of endogenous peroxidase activity, the slides were incubated with primary antibody overnight at 4°C. Slides were then incubated with horseradish peroxidase– or alkaline phosphatase–conjugated secondary antibody (ImmPRESS-AP anti-rabbit IgG alkaline phosphatase [catalog MP-5401] and ImmPRESS HRP anti-rabbit IgG peroxidase [catalog MP-7451] Vector Laboratories) for 1 hour at 25°C. This was followed by development using the appropriate chromogenic substrate: DAB, Warp Red, or Ferangi Blue (Biocare Medical). In the case of multichannel immunohistochemistry, slides were subsequently stripped by using a sodium citrate buffer and by boiling at 110°C for 3 minutes. The procedure was then repeated as above using a different-colored chromogen for development. All human PDA samples were provided by the University of Texas (UT) Southwestern Tissue Management Shared Resource, and their use was approved by the UT Southwestern institutional review board for research. All patient samples were deidentified and interpreted by a board-certified pathologist.
Data sharing statement.
There are no additional unpublished data from this study.
Data availability.
All raw and processed scRNA-Seq data described in this manuscript have been uploaded to the National Center for Biotechnology Information’s Gene Expression Omnibus database repository (https://www.ncbi.nlm.nih.gov/geo) under accession number GSE125588.
Statistics.
For KEGG pathway analysis, an analysis of variance was performed across different pathways and P < 0.05 was considered significant. In the analysis of previously published human PDA scRNA-Seq data (32), a Z score cutoff of 1 was used to denote cells in which the gene of interest was overexpressed.
Study approval.
Mice were used in this study according to UT Southwestern institutional guidelines, and protocols were approved by the institutional animal care and use committee at UT Southwestern Medical Center. American Veterinary Medical Association Guidelines for the Euthanasia of Animals were strictly followed. All human samples were procured through the UT Southwestern Tissue Management Shared Resource and approved through the UT Southwestern institutional review board.
Author contributions
ANH and HH were responsible for study concept and design, acquisition of data, analysis and interpretation of data, and drafting of the manuscript. ZW performed bioinformatics analyses and interpreted data. KP performed pathological interpretation and scoring of human tissue samples. WD acquired data. JH performed bioinformatics analyses and critical review of the manuscript. AM and EO performed bioinformatics analyses, interpretation of data, and critical review of the manuscript. UV was responsible for study concept and design and critical revision of the manuscript. RAB was responsible for study concept and design, interpretation of data, and drafting of the manuscript.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01 CA192381 and U54 CA210181 Project 2 to RAB; the Effie Marie Cain Fellowship to RAB; the H. Ray and Paula Calvert Pancreatic Cancer Research Fund to UV; NIH grants U01 CA200468, U01 CA196403, R01 CA218004, R01 CA204969, and P01 CA117696 to AM; and grants from the NIH and Welch Foundation to EO. ZW was supported by a predoctoral fellowship from the American Heart Association and the Harry S. Moss Heart Trust (19PRE34380436).
We thank the McDermott Center Next-Generation Sequencing Core at UT Southwestern for preparing and sequencing the scRNA-Seq libraries. We thank Jeon Lee of the UT Southwestern Bioinformatics Core for assistance in preprocessing of scRNA-Seq data. The UT Southwestern Bioinformatics core is funded by Cancer Prevention and Research Institute of Texas (RP150596). We also thank Dave Primm of the UT Southwestern Department of Surgery for help in editing this article.
Version 1. 07/23/2019
In-Press Preview
Version 2. 08/22/2019
Electronic publication
Funding Statement
This work was supported by NIH grants R01 CA192381 and U54 CA210181 Project 2 to RAB, the Effie Marie Cain Fellowship to RAB, the H. Ray and Paula Calvert Pancreatic Cancer Research Fund to UV, NIH grants U01 CA200468, U01 CA196403, R01 CA218004, R01 CA204969, P01 CA117696 to AM and grants from the NIH and Welch Foundation to EO.
Footnotes
Conflict of interest: AM has received royalties from Cosmos Wisdom Biotechnology for a biomarker assay related to early detection of pancreatic cancer.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(16):e129212.https://doi.org/10.1172/jci.insight.129212.
Contributor Information
Abdel Nasser Hosein, Email: abdel.hosein@utsouthwestern.edu.
Huocong Huang, Email: huocong.huang@utsouthwestern.edu.
Zhaoning Wang, Email: Zhaoning.wang@utsouthwestern.edu.
Kamalpreet Parmar, Email: kamalpreet.parmar@gmail.com.
Wenting Du, Email: wenting.du@utsouthwestern.edu.
Jonathan Huang, Email: Huang.j@wustl.edu.
Anirban Maitra, Email: amaitra@mdanderson.org.
Eric Olson, Email: Eric.Olson@utsouthwestern.edu.
Udit Verma, Email: udit.verma@utsouthwestern.edu.
Rolf A. Brekken, Email: rolf.brekken@utsouthwestern.edu.
References
- 1.Kleeff J, et al. Pancreatic cancer. Nat Rev Dis Primers. 2016;2:16022. doi: 10.1038/nrdp.2016.22. [DOI] [PubMed] [Google Scholar]
- 2.Mei L, Du W, Ma WW. Targeting stromal microenvironment in pancreatic ductal adenocarcinoma: controversies and promises. J Gastrointest Oncol. 2016;7(3):487–494. doi: 10.21037/jgo.2016.03.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valkenburg KC, de Groot AE, Pienta KJ. Targeting the tumour stroma to improve cancer therapy. Nat Rev Clin Oncol. 2018;15(6):366–381. doi: 10.1038/s41571-018-0007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Potter SS. Single-cell RNA sequencing for the study of development, physiology and disease. Nat Rev Nephrol. 2018;14(8):479–492. doi: 10.1038/s41581-018-0021-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aguirre AJ, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17(24):3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng GX, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kopp JL, et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22(6):737–750. doi: 10.1016/j.ccr.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weniger M, Honselmann KC, Liss AS. The extracellular matrix and pancreatic cancer: a complex relationship. Cancers (Basel) 2018;10(9):E316. doi: 10.3390/cancers10090316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen W, Tao GQ, Zhang Y, Cai B, Sun J, Tian ZQ. TGF-β in pancreatic cancer initiation and progression: two sides of the same coin. Cell Biosci. 2017;7:39. doi: 10.1186/s13578-017-0168-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruber R, Panayiotou R, Nye E, Spencer-Dene B, Stamp G, Behrens A. YAP1 and TAZ control pancreatic cancer initiation in mice by direct up-regulation of JAK-STAT3 signaling. Gastroenterology. 2016;151(3):526–539. doi: 10.1053/j.gastro.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao JJ, et al. Regulation of gene expression in mouse macrophages stimulated with bacterial CpG-DNA and lipopolysaccharide. J Leukoc Biol. 2002;72(6):1234–1245. [PubMed] [Google Scholar]
- 12.Leask A, Abraham DJ. All in the CCN family: essential matricellular signaling modulators emerge from the bunker. J Cell Sci. 2006;119(Pt 23):4803–4810. doi: 10.1242/jcs.03270. [DOI] [PubMed] [Google Scholar]
- 13.Regn M, et al. Peptidase inhibitor 16 is a membrane-tethered regulator of chemerin processing in the myocardium. J Mol Cell Cardiol. 2016;99:57–64. doi: 10.1016/j.yjmcc.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Wang J, Asahina K. Mesothelial cells give rise to hepatic stellate cells and myofibroblasts via mesothelial-mesenchymal transition in liver injury. Proc Natl Acad Sci USA. 2013;110(6):2324–2329. doi: 10.1073/pnas.1214136110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simpkins SA, Hanby AM, Holliday DL, Speirs V. Clinical and functional significance of loss of caveolin-1 expression in breast cancer-associated fibroblasts. J Pathol. 2012;227(4):490–498. doi: 10.1002/path.4034. [DOI] [PubMed] [Google Scholar]
- 16.Chang SK, et al. Cadherin-11 regulates fibroblast inflammation. Proc Natl Acad Sci USA. 2011;108(20):8402–8407. doi: 10.1073/pnas.1019437108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanzaki R, et al. Gas6 derived from cancer-associated fibroblasts promotes migration of Axl-expressing lung cancer cells during chemotherapy. Sci Rep. 2017;7(1):10613. doi: 10.1038/s41598-017-10873-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hingorani SR, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7(5):469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 19.Öhlund D, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. 2017;214(3):579–596. doi: 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feig C, et al. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci USA. 2013;110(50):20212–20217. doi: 10.1073/pnas.1320318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Djurec M, et al. Saa3 is a key mediator of the protumorigenic properties of cancer-associated fibroblasts in pancreatic tumors. Proc Natl Acad Sci USA. 2018;115(6):E1147–E1156. doi: 10.1073/pnas.1717802115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kivioja T, et al. Counting absolute numbers of molecules using unique molecular identifiers. Nat Methods. 2011;9(1):72–74. doi: 10.1038/nmeth.1778. [DOI] [PubMed] [Google Scholar]
- 23.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lovén J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153(2):320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazur PK, et al. Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nat Med. 2015;21(10):1163–1171. doi: 10.1038/nm.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozturk N, Singh I, Mehta A, Braun T, Barreto G. HMGA proteins as modulators of chromatin structure during transcriptional activation. Front Cell Dev Biol. 2014;2:5. doi: 10.3389/fcell.2014.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masliah-Planchon J, Bièche I, Guinebretière JM, Bourdeaut F, Delattre O. SWI/SNF chromatin remodeling and human malignancies. Annu Rev Pathol. 2015;10:145–171. doi: 10.1146/annurev-pathol-012414-040445. [DOI] [PubMed] [Google Scholar]
- 28.Mack SC, et al. Therapeutic targeting of ependymoma as informed by oncogenic enhancer profiling. Nature. 2018;553(7686):101–105. doi: 10.1038/nature25169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark CE, Hingorani SR, Mick R, Combs C, Tuveson DA, Vonderheide RH. Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer Res. 2007;67(19):9518–9527. doi: 10.1158/0008-5472.CAN-07-0175. [DOI] [PubMed] [Google Scholar]
- 30.Chung WJ, et al. Kras mutant genetically engineered mouse models of human cancers are genomically heterogeneous. Proc Natl Acad Sci USA. 2017;114(51):E10947–E10955. doi: 10.1073/pnas.1708391114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reichert M, Rustgi AK. Pancreatic ductal cells in development, regeneration, and neoplasia. J Clin Invest. 2011;121(12):4572–4578. doi: 10.1172/JCI57131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernard V, et al. Single-Cell transcriptomics of pancreatic cancer precursors demonstrates epithelial and microenvironmental heterogeneity as an early event in neoplastic progression. Clin Cancer Res. 2019;25(7):2194–2205. doi: 10.1158/1078-0432.CCR-18-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rhim AD, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148(1-2):349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Izumiya M, et al. Chemoresistance is associated with cancer stem cell-like properties and epithelial-to-mesenchymal transition in pancreatic cancer cells. Anticancer Res. 2012;32(9):3847–3853. [PubMed] [Google Scholar]
- 35.Zheng X, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527(7579):525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Genovese G, et al. Synthetic vulnerabilities of mesenchymal subpopulations in pancreatic cancer. Nature. 2017;542(7641):362–366. doi: 10.1038/nature21064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell. 2014;54(5):728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sengupta S, George RE. Super-Enhancer-Driven transcriptional dependencies in cancer. Trends Cancer. 2017;3(4):269–281. doi: 10.1016/j.trecan.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sahai V, et al. BET bromodomain inhibitors block growth of pancreatic cancer cells in three-dimensional collagen. Mol Cancer Ther. 2014;13(7):1907–1917. doi: 10.1158/1535-7163.MCT-13-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roe JS, et al. Enhancer reprogramming promotes pancreatic cancer metastasis. Cell. 2017;170(5):875–888.e20. doi: 10.1016/j.cell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lomberk G, et al. Distinct epigenetic landscapes underlie the pathobiology of pancreatic cancer subtypes. Nat Commun. 2018;9(1):1978. doi: 10.1038/s41467-018-04383-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Movahedi K, et al. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res. 2010;70(14):5728–5739. doi: 10.1158/0008-5472.CAN-09-4672. [DOI] [PubMed] [Google Scholar]
- 43.Wang B, Li Q, Qin L, Zhao S, Wang J, Chen X. Transition of tumor-associated macrophages from MHC class II(hi) to MHC class II(low) mediates tumor progression in mice. BMC Immunol. 2011;12:43. doi: 10.1186/1471-2172-12-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Özdemir BC, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2015;28(6):831–833. doi: 10.1016/j.ccell.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 45.Zhu Y, et al. Tissue-Resident macrophages in pancreatic ductal adenocarcinoma originate from embryonic hematopoiesis and promote tumor progression. Immunity. 2017;47(3):597. doi: 10.1016/j.immuni.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanford DE, et al. Inflammatory monocyte mobilization decreases patient survival in pancreatic cancer: a role for targeting the CCL2/CCR2 axis. Clin Cancer Res. 2013;19(13):3404–3415. doi: 10.1158/1078-0432.CCR-13-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olumi AF, Grossfeld GD, Hayward SW, Carroll PR, Tlsty TD, Cunha GR. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999;59(19):5002–5011. doi: 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hwang RF, et al. Cancer-associated stromal fibroblasts promote pancreatic tumor progression. Cancer Res. 2008;68(3):918–926. doi: 10.1158/0008-5472.CAN-07-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ostapoff KT, et al. Neutralizing murine TGFβR2 promotes a differentiated tumor cell phenotype and inhibits pancreatic cancer metastasis. Cancer Res. 2014;74(18):4996–5007. doi: 10.1158/0008-5472.CAN-13-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hingorani SR, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4(6):437–450. doi: 10.1016/S1535-6108(03)00309-X. [DOI] [PubMed] [Google Scholar]
- 51.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw and processed scRNA-Seq data described in this manuscript have been uploaded to the National Center for Biotechnology Information’s Gene Expression Omnibus database repository (https://www.ncbi.nlm.nih.gov/geo) under accession number GSE125588.