Abstract
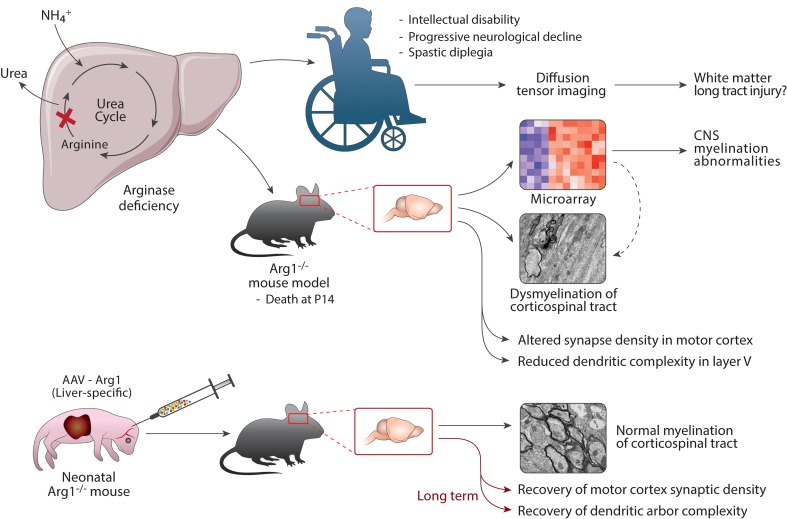
Deficiency of arginase is associated with hyperargininemia, and prominent features include spastic diplegia/tetraplegia, clonus, and hyperreflexia; loss of ambulation, intellectual disability and progressive neurological decline are other signs. To gain greater insight into the unique neuromotor features, we performed gene expression profiling of the motor cortex of a murine model of the disorder. Coexpression network analysis suggested an abnormality with myelination, which was supported by limited existing human data. Utilizing electron microscopy, marked dysmyelination was detected in 2-week-old homozygous Arg1-KO mice. The corticospinal tract was found to be adversely affected, supporting dysmyelination as the cause of the unique neuromotor features and implicating oligodendrocyte impairment in a deficiency of hepatic Arg1. Following neonatal hepatic gene therapy to express Arg1, the subcortical white matter, pyramidal tract, and corticospinal tract all showed a remarkable recovery in terms of myelinated axon density and ultrastructural integrity with active wrapping of axons by nearby oligodendrocyte processes. These findings support the following conclusions: arginase deficiency is a leukodystrophy affecting the brain and spinal cord while sparing the peripheral nervous system, and neonatal AAV hepatic gene therapy can rescue the defects associated with myelinated axons, strongly implicating the functional recovery of oligodendrocytes after restoration of hepatic arginase activity.
Keywords: Neuroscience
Keywords: Gene therapy, Genetic diseases, Neurodevelopment
Neuromotor abnormalities that develop in arginase deficiency are the result of dysmyelination during neurodevelopment, indicating this disorder is a leukodystrophy.
Introduction
The urea cycle is responsible for the detoxification of ammonia by conversion to urea, a nontoxic nitrogen-containing compound that is excreted in the urine. Disorders can occur in any of the 6 enzymes of the cycle or of the 2 mitochondrial transporters and are typically hallmarked by hyperammonemia. The prognosis for patients can be mild encephalopathy to substantial developmental disability, often depending upon the severity, duration, and the frequency of episodes of hyperammonemia (1), and it can be fatal. Ammonia has been shown to compromise potassium buffering of astrocytic membranes and to cause the clinical neurological abnormalities of hyperammonemic encephalopathy by impairing cortical inhibition (2).
Arginase is the final enzyme of the urea cycle, catalyzing the hydrolysis of arginine to ornithine, which is necessary for the cycle to continue. Biallelic mutations in arginase 1 (ARG1) result in its deficiency, an autosomal recessive disorder with an incidence of 1 in 350,000 to 1 in 1,100,000 births (3); the exact incidence is unknown and is thought to be underestimated. Unlike the other urea cycle disorders, hyperammonemia is uncommon. Thus, the clinical presentation is unique: the typical presentation usually occurs later in life, beginning in late infancy to the second year after birth, and includes spastic diplegia along with spasticity, clonus, and/or hyperreflexia (4). These signs can be indistinguishable from those of cerebral palsy in some patients (5). Later findings include microcephaly, seizures, loss of ambulation, growth retardation, intellectual disabilities, and progressive neurological decline (4, 6, 7).
Our laboratory has previously developed a mouse model of arginase deficiency (8), characterizing the biochemical phenotype and its similarities with humans afflicted with the disorder (9–12). Hyperargininemia and the presence of guanidino compounds are found in the model (13–16), as they are in afflicted patients (6, 12, 17). With these biochemical similarities, we sought to examine the neurodevelopment of the CNS of arginase-deficient mice to gain an improved understanding of the underlying neuropathology.
In these studies, we have found striking abnormalities in myelination of the brain and spinal cord, sparing the peripheral nervous system, of homozygous arginase-deficient mice, leading us to suggest that this disorder be classified as a leukodystrophy. In addition, we have found that gene replacement with an adeno-associated viral (AAV) vector containing a liver-specific transgene encoding arginase 1 in the neonatal mouse leads to normalization of myelination. These studies have not only elucidated the likely cause of spastic diplegia/tetraplegia and corticospinal tract (CST) dysfunction in arginase deficiency, but also support the continued neonatal screening for arginase deficiency, initiation of early intervention to normalize plasma arginine and its metabolites (as they likely play a role in oligodendrocyte dysfunction), and initiation of an early postnatal gene therapy approach to prevent the underlying neurodevelopmental abnormalities including that of dysmyelination.
Results
Two hypotheses have been tested in this quantitative microscopic study.
Hypothesis I: Deficiency of arginase 1 results in CNS dysmyelination.
In our previous study (18), we demonstrated that, in motor cortex layer V of Arg1-KO mice at P15, a significant decrease in both the density of asymmetrical synapses and AMPA receptor–enriched (AMPAR-enriched) perforated synapses (19–21) corresponded with a malfunction in glutamatergic excitatory synaptic transmission. We also found in that study that intrinsic excitability is altered in proportion to the copy number loss of arginase and that the circuit connectivity is altered in KO neurons, as shown in the electrophysiological studies by decreased frequencies of miniature excitatory postsynaptic currents (mEPSCs) and the decrease in amplitude of miniature inhibitory postsynaptic currents (mIPSCs). In addition, we found a marked decrease in complexity of the basal dendritic arbor of layer V motor cortical neurons.
Clinically, the unique neuromotor findings in patients have suggested a different origin of disease pathogenesis compared with the more proximal urea cycle disorders where hyperammonemia is prominent. MRI of 1 patient demonstrated, on T2 weighted and inversion recovery images, evidence of patchy CNS demyelination (22). In a second patient, a limited investigation by diffusion tensor MRI (23) revealed a reduction in diffusion tensor imaging fiber count at the level of the decussation of the CST (Figure 1); such findings were not detected in either control subjects or in those with ornithine transcarbamylase deficiency. Alteration in white matter integrity in upper motor neuron function could impact the motor cortex and the CST being the cause, at least in part, of this unique clinical phenotype. The nature of this abnormality suggested by imaging is not clear and, to our knowledge, has not been further investigated until now.
Figure 1. White matter tractography demonstrates alteration in neuronal fibers with arginase deficiency but not ornithine transcarbamylase deficiency, a proximal urea cycle disorder.
Diffusion tensor imaging was used to evaluate the white matter tracts in a patient with arginase deficiency (A) and ornithine transcarbamylase deficiency (B). (C) Age-matched control. Arrows indicate the corticospinal tract. Colors denote the dominant direction of the fibers. Reduction in the volume of fibers is visualized in the arginase deficient patient.
Taken together, along with our gene expression data, this leads to a hypothesis that the output pathway containing axon fibers originating from layer V pyramidal neurons in KO mice and in an ARG1 deficiency patient may be compromised in postnatal development due to defects of cortical excitatory synaptic function and/or malfunction of myelination of CNS axons. Therefore, the subcortical white matter below the motor cortex, which contains these axonal fibers, as well as the pyramidal tract in the medulla and in the cervical spinal cord, which contains CST axons, may display significant alterations such as axonal degeneration and demyelination and may ultimately show a decrease in density and myelin compactness of axons in these regions. We theorized that P2 gene therapy may rescue these ultrastructural defects in terms of recovery of myelinated axon density and in regaining myelination of axons in the subcortical white matter, pyramidal tract, and spinal cord of P14 mice.
Hypothesis II: Hepatic Arg1 gene therapy results in long-term restoration of synaptic density and dendritic arbor complexity.
Following AAV-based Arg1 hepatic gene therapy, long-term recovery in the density of asymmetrical synapses, as well as the number of perforated synapses in motor cortex layer V, may be detected in treated 4-month-old Arg1-KO mice. In addition, we also expected to find a similar level of complexity of the basal dendritic arbor of layer V cortical neurons.
To test Hypothesis I, we carried out light microscopy and extensive electron microscopic (EM) imaging on the subcortical white matter of the motor cortex, pyramidal tract, and dorsal funiculus of the cervical spinal cord in P14–P15 WT, homozygous Arg1-KO, heterozygotes (Het), and hepatic AAV-gene therapy P2 gene therapy–treated KO mice (treated KO mice). These studies focused on visualizing the ultrastructural features of CST axonal fibers in these regions. Based on large-scale EM imaging, we counted myelinated axons in each image field (defined as a unit area), and then calculated and compared the density of myelinated axons in the different genotypes. Finally, we measured and calculated myelin sheath layer thickness of the axons (which are reflected by myelin sheath thickness analysis [G-ratio] values) in the pyramidal tract and CST in the spinal cord to detect any change in compactness of the myelin sheath layers in KO mice, along with possible recovery after hepatic gene therapy of KO mice initiated at P2.
Analysis of Arg1-deficient cortex suggests dysregulation of myelinating oligodendrocytes.
Overexpression of Arg1 in the cortex was found in homozygous KO, Het, and treated KO mice when compared with the WT (P = 5 × 10–13 by one-way ANOVA) (Figure 2A). Further analysis revealed that there is upregulation of a faulty Arg1 transcript, demonstrating that regulatory mechanisms are activated to produce Arg1 in the CNS when either reduced (Het) or absent (KO, treated KO). In humans, ARG1 is expressed in the cortex and subcortical regions as early as the 8th postconception week, peaking near birth and continuing throughout childhood, while declining in middle age (Figure 2B).
Figure 2. Microarray expression studies of the brain in arginase deficiency reveal evidence of dysregulation of myelinating oligodendrocytes.
(A) Arg1 graph: P values for Arg1 (adjusted P value from Tukey HSD test from ANOVA [P = 5 × 10−13] of Arg1 normalized expression by genotype/treatment) are plotted for WT, homozygous Arg1-KO, Arg1 heterozygote (Het), and AAV-treated hepatic Arg1-expressing mice (treated KO) at P14 (n = 6 per group). Individual samples are also plotted. Overexpression of Arg1 indicates the upregulation of a faulty transcript, demonstrating that regulatory mechanisms are activated to produce Arg1 in the CNS in the KO, Het, and treated KO groups. (B) ARG1 in Allen Developing Human Brain Atlas: BrainSpan data was acquired (http://www.brainspan.org/static/download.html) and RNA-Seq (reads per kilobase of transcript, per million mapped reads; RPKM) Gencode v.10 summarized to genes. Gray shading indicates 95% CI. Evident expression of ARG1 in the cortical and subcortical regions of the human brain is present with upregulation around birth. (C) Myelinating oligodendrocyte (MO) dysregulated module graph. This module contains 374 coexpressed genes that share a cell type expression signature with MOs. The first principal component of expression is plotted on the y axis. P values for this PC1 (adjusted P value from Tukey HSD test from ANOVA of MO dysregulated genes PC1 by genotype/treatment [P = 0.024]) plotted above graph. Individual samples are also plotted. Cell type enrichment (D) and gene ontology (E) for MO module. Cell type markers for MOs that were specific for MOs with P < 0.01 (pSI method), utilizing neural cell type specific gene expression (61) were used for cell type enrichment analysis. Cell type enrichment P values were calculated with overrepresentation analysis. Gene ontology overlap was established with the gProfileR method, which implements overrepresentation analysis to obtain enrichment P values. Bonferroni adjusted P values plotted for both.
We used weighted gene coexpression network analysis (WGCNA) (24) to analyze our gene expression data after it was normalized and adjusted to remove technical variability. Briefly, WGCNA uses gene-to-gene connectivity measures to identify groups of genes (modules) that are coexpressed across all samples. This is an advantageous approach for analyzing differential gene expression in a disease model, since the data can be reduced from thousands of discrete genes to a handful of coexpressed gene modules in which genes share some common biological function. The first principal component of expression for each gene module (considered the consensus expression of all module genes, referred to as the module eigengene [ME] in WGCNA) can be associated with covariates of interest to look for interesting associations (e.g., to determine if disease is associated with any of the modules).
In our WGCNA analysis, 19 modules were identified, 6 of which were dysregulated in at least 1 of our groups (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.130260DS1). One of these modules, module 2 (M2), was upregulated in all genetically modified mice and contained genes involved with reactivity to lowered oxygen levels in endothelial cells and astrocytes. Of note, this module also contained Arg1. Another module, M9, was upregulated in the KO mice and contained genes involved with neuron and oligodendrocyte development. Four other modules were downregulated in KO mice: M5, M6, M13, and M7. M5 genes were involved with endothelial and astrocyte development; M6 genes were involved with neural cell adhesion; and M13 genes were active in astrocyte actin projection assembly. M7 (which will subsequently be referred to as the myelinating oligodendrocyte [MO] dysregulated genes) included genes highly expressed in MO and active in ion transport activity (Figure 2, C and D). Several studies support an important role for ion transporters (Figure 2E) and channels in maintaining healthy MOs and in promoting mature differentiation processes (via calcium signaling) that are required for myelinating function (25–27). This data implies that mature oligodendrocyte function was possibly disrupted in the Arg1-KO mouse, leading us to conduct an investigation into myelination in these mice.
Subcortical white matter shows marked myelinated axon loss and recovery with treatment.
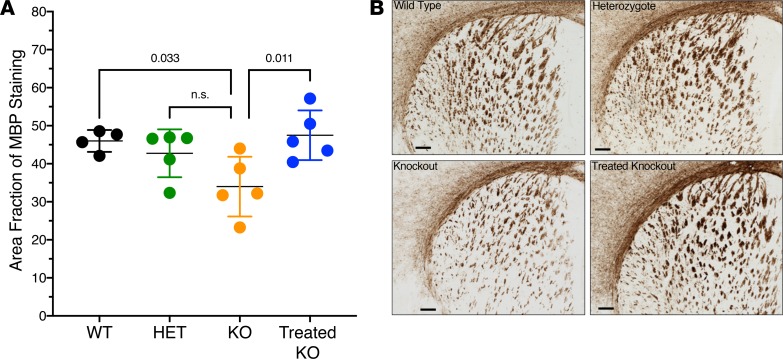
Initial studies were conducted to examine myelination in the corpus callosum, the collection of white matter fibers connecting both cerebral hemispheres, below the motor and primary sensory cortices, and the anterio-dorsal caudatus putamen — a key structure for motor function, which is traversed by myelinated fiber bundles including the ones connecting the motor cortex to the lower motor neurons (28). Utilizing a sampling region between bregma 1.10 mm and bregma 0.26 mm, quantification of myelin basic protein (MBP) staining (Figure 3A) and examination of immunostained sections (Figure 3B) revealed a reduction of myelination in these regions in the homozygous Arg1-KO compared with WT mice (P = 0.033). AAV hepatic Arg1 gene therapy initiated on P2 restored the level of myelination as measured by MBP staining in the treated KO mice.
Figure 3. Myelin basic protein (MBP) IHC in corpus callosum and dorsal striatum demonstrates reduced expression in arginase 1 deficiency.
(A) Area fraction of MBP staining for each group (n = 5 per genotype). P values were determined by 1-way ANOVA with Dunnett’s Multiple Comparisons test. All groups were compared with the KO group. Significant difference is seen between KO and treated KO groups, and KO and WT groups. (B) High-magnification images of approximate bregma level 0.86 mm for each group. Scale bars: 200 μm. Error bars in graph represent ± SD. Het, Arg1 heterozygote; KO, homozygous Arg1-KO.
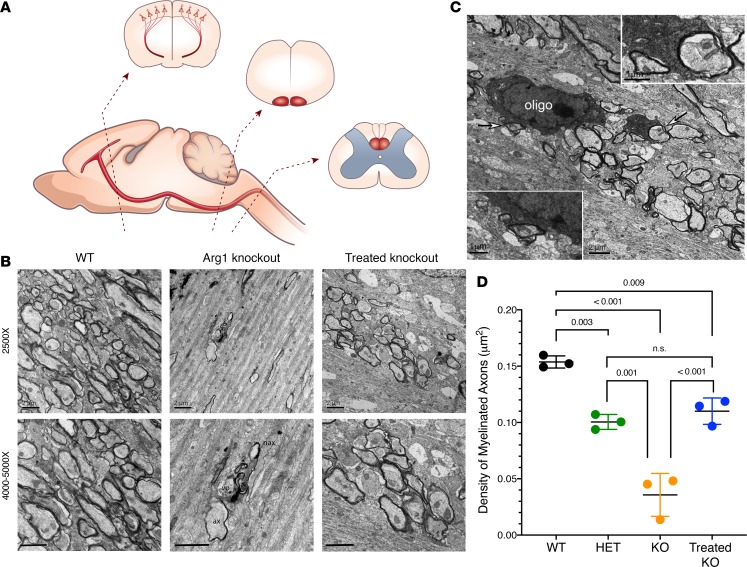
As shown in a schematic (Figure 4A), under EM, the P15 murine brain was examined in the subcortical white matter (motor cortex level), pyramidal tract (medulla level), and CST of the cervical spinal cord (cervical segment of spinal cord level). Compared with WT mice (Figure 4B), the KO subcortical white matter had few myelinated axons and, when present, had thinner myelin sheath layers. In fact, many unmyelinated axons were present. Some degenerated axons filled with fragmented electron-dense material were found; in addition, vacuolar structures were often associated with the degenerating debris. In Het samples (Supplemental Figure 2), myelinated axons were found, but few degenerated axons were detected. Following P2 hepatic gene therapy of homozygous Arg1-KO mice, the subcortical white matter showed a remarkable recovery in terms of myelinated axon density and ultrastructural integrity, with active wrapping of axons by nearby oligodendrocyte processes being frequently identified (Figure 4C); no degenerated axons were found.
Figure 4. Analysis demonstrates distribution of myelinated axon fibers in the subcortical white matter of the motor cortex of P15 Arg1-deficient mice to be greatly reduced with Arg1 deficiency and restored with hepatic gene therapy.
(A) Diagram with corticospinal tract illustrated. Representative regions that were examined are displayed: motor cortex with layer V pyramidal neurons and their descending axons; medulla, pyramidal tract; cervical segment of spinal cord, corticospinal tract in dorsal funiculus. (B) Assessment of myelination in subcortical white matter of the motor cortex. WT (top) image shows many differently sized myelinated axons; myelin sheath thickness is variable among fibers. WT (bottom) shows higher-magnification image (4,000×). Arg1-KO (top) Image shows only a few axon profiles. Note in the middle is a degenerated axon; another axon with a thin myelin sheath layer is adjacent. Arg1-KO (bottom) shows higher-power view with some fragments of myelin sheath debris; a vacuolar structure (vo) is associated with a degenerated axon indicating debris engulfment (5,000×). One thin myelinated axon (ax) and unmyelinated axon (nax) are closely associated with the degenerated axon. Treated Arg1-KO (top) image shows a significant number of myelinated axons. Treated Arg1-KO (bottom) shows high-power (5,000×) normal axonal features and myelin sheath layers. Scale bars: 2 μm. (C) Micrograph showing electron-dense oligodendrocyte soma (oligo) and a major process wrapping axon fibers (arrows); insets demonstrate the processes tightly enclosing axons. Scale bar: 2 μm and 1 μm [insets]) (D) Density comparison of myelinated axons in subcortical white matter of the Arg1 genotypes. WT has the highest density; untreated KO has the lowest. Treated KO shows remarkable recovery and has the second highest density. Het group shows lower density compared with the WT and treated KO group. P values determined by one-way ANOVA with Tukey’s multiple comparisons. Error bars represent SD. (KO = knockout, Het = heterozygote) (n = 3 per genotype group)
Quantitatively, a comparison of myelinated axon density was performed (Figure 4D) (n = 3 mice per genotype). Although some variability is present between animals in each genotype, we consistently found that the WT group showed the highest mean value in density (0.154 ± 0.005 per μm2), while the untreated KO group showed the lowest value (0.036 ± 0.019 per μm2; P < 0.0001); the Het (0.101 ± 0.007 per μm2) and AAV-treated KO groups (0.110 ± 0.012 per μm2) exhibited similar mean values (P = 0.765), which were slightly lower than WT (WT vs. Het, P = 0.003; WT vs. treated KO, P = 0.009), as a dramatic recovery in the treated KO is demonstrated (KO vs. treated KO, P = 0.0003).
Pyramidal tract and CST show a markedly reduced number of myelinated axons and recovery with therapy.
Consistent with findings in previous studies (29–33), a significant number of myelinated axons in the P15 WT pyramidal tract (0.187 ± 0.027 per μm2) and CST (0.289 ± 0.023 per μm2) in the spinal cord were identified, with some having much thicker myelin sheath layers (Figure 5). Many unmyelinated axon fibers were also present at this stage in WT mice. A drastic reduction in the number of myelinated axons was demonstrated in the untreated P15 KO pyramidal tract (0.080 ± 0.007 per μm2; P < 0.001 vs. WT) (Figure 5, A and B) and spinal cord (0.075 ± 0.001 per μm2; P < 0.001 vs. WT) (Figure 5, D and E). Some advanced degenerated axonal fragments were found in the KO in a small number of myelinated axons (Figure 5A). Interestingly, a few electron-dense oligodendrocytes (Figure 5A) were localized near the axons, but very few processes were derived from the soma, indicating less active axonal wrapping in the untreated KO. Degenerated axonal debris was often seen associated with vacuolar processes, which may be derived from nearby astrocytes, indicating engagement of active glial engulfment in the region (Figure 5A). As in the subcortical white matter, Het had reduced myelin density in the pyramidal tract (0.140 ± 0.0157 per μm2; P = 0.080 vs. WT; Figure 5B) and in the CST of cervical spinal cord (0.221 ± 0.018 per μm2; P = 0.002 vs. WT; Figure 5E).
Figure 5. Analysis of axons in pyramidal tract and spinal cord of P15 mice demonstrate markedly reduced myelination with Arg1 deficiency and recovery with hepatic Arg1 expression.
(A) Assessment of myelination. WT top: myelinated axons with differently sizes distributed in pyramidal tract. WT, bottom: high-power image (4,000×). Arg1-KO, top: Image showing a few axons (ax) with 1 advanced degenerated axon (arrow) engulfed by a vacuolar structure (vo). Electron-dense oligodendrocyte (oligo) is seen; no processes derive from the soma, indicating inactivity. Arg1-KO, bottom: Higher-power (6,500×, serial thin section) showing degenerated axon with electron-dense material in axoplasm (arrow); myelin is fragmented/engulfed by a vacuolar process (vo). AAV-treated KO, top: Image showing complete recovery of myelination, similar morphological features as WT. AAV-treated KO, bottom: High-power (4,000×) of myelinated axons; one is wrapped by an oligodendrocyte process. (B) Quantitative comparison of myelinated axon densities. (C) Image showing portions of 2 oligodendrocyte somata in treated KO; several processes wrap the axons (arrows). Scale bar: 2 μm. (D) Myelination assessment in spinal cord. WT, top: Image showing a portion of corticospinal tract (CST) in the dorsal funiculus. WT, bottom: Higher-power image (6,000×) showing myelinated axons. Arg1-KO, top: Image showing a majority of axons are unmyelinated; only a few myelinated axons present in CST region. Myelin sheaths are thinner, and 2 axons show signs of degeneration (arrow). Arg1-KO, bottom: Higher-magnification (8,000×). AAV-treated KO, top: Image showing recovery of myelinated axons. Distribution pattern is similar to WT. AAV-treated KO, bottom: High-magnification (6,500×). (E) Quantitative comparison of myelinated axon density in CST. (F) Image shows an electron-dense oligodendrocyte. Processes are seen to wrap multiple axon profiles with multiple myelinated axons distributed around the soma (arrows). P values determined by 1-way ANOVA with Tukey’s multiple comparisons. Error bars represent ± SD. Scale bars: 2 μm (top images); 1μm (bottom images) (n = 3 per genotype).
Following P2 hepatic gene therapy, recovery of myelination is demonstrated in P15-treated KO pyramidal tract (0.200 ± 0.024 per μm2; P = 0.850 vs. WT) and spinal cord samples (0.2611 ± 0.006 per μm2; P = 0.180 vs. WT) (Figure 5, B and E). Many oligodendrocyte processes are seen to wrap the axons (Figure 5, C and F).
G-ratio analysis demonstrates increased ratio in the KO with recovery in AAV-treated mice.
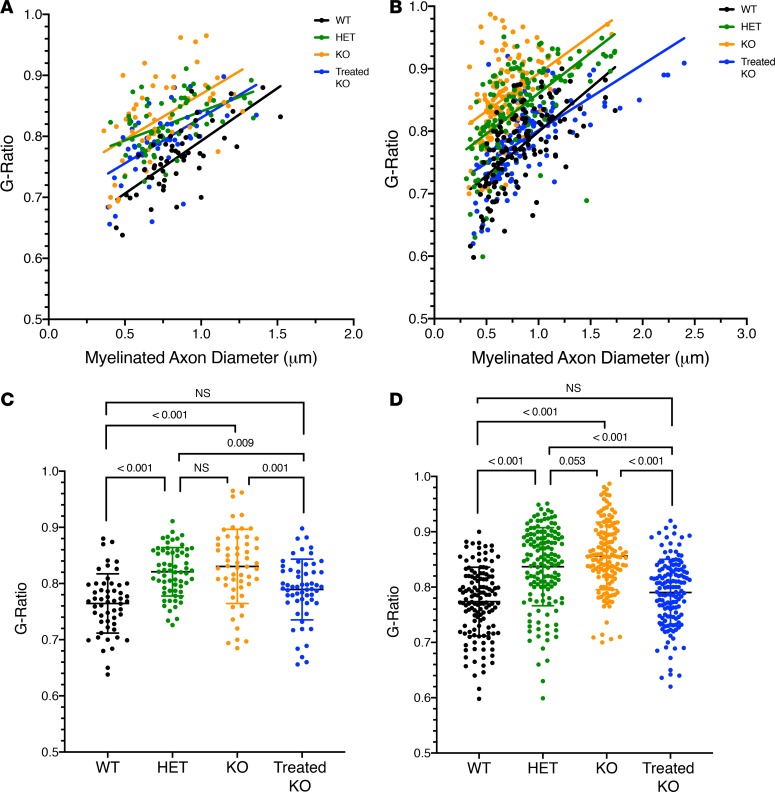
To quantitatively investigate myelination of CST axons in the pyramidal tract and spinal cord, and to further confirm the recovery of myelination after gene therapy at P2, at least 50 myelinated axons were randomly selected from each genotype for measuring the diameter of axon fibers and myelinated fibers; thus, the G-ratio was calculated for each axon (34–37). For the first set of data (Figure 6, A and B), we sought to reveal the relationship between G-ratio and myelinated axon diameter and to determine if there was a correlation in both pyramidal tract (Figure 6A) and spinal cord (Figure 6B). WT mice showed positive linear correlations in the pyramidal (R2 = 0.517) and CSTs (R2 = 0.398) (compared with mature myelinated fibers in the CNS, R2 = 0.54) (38). Pyramidal and CSTs from untreated homozygous KOs were markedly reduced (R2 = 0.308 and R2 =0.230, respectively), indicating that the majority of the axons, when myelinated, have thinner myelin sheath layers as their axon diameters increase. However, with AAV-based hepatic gene therapy, the correlation in both the pyramidal tract (R2 = 0.360) and CST (R2 = 0.405) increased compared with the KO group. Interestingly, both heterozygote groups were reduced compared with WT (pyramidal tract, R2 = 0.232; CST, R2 = 0.385).
Figure 6. Quantification of myelin sheath thickness of P15 genotypes demonstrated increased G-ratio in pyramidal tract and spinal cord in Arg1 deficiency and recovery with gene therapy.
For quantification, each genotype includes at least 50 myelinated axons. The upper scatterplots (A for pyramidal tract, B for corticospinal tract [CST] in cervical spinal cord) demonstrate the G-ratio of individual axons as a function of axonal diameter (μm). Regression analysis (represented by the solid line for each genotype) demonstrates that a positive linear relationship is present between the G-ratio value and the myelinated axon diameter. Consistently, in the lower scatter plots (C for pyramidal tract, D for CST in cervical spinal cord) the WT and treated KO groups show the lowest G-ratio values, indicative of thicker myelin sheath layers; KO and Het groups show higher G-ratio values, indicative of thinner myelin sheath layers. Complete recovery in myelination is evidenced in the treated KO group. P values determined by 1-way ANOVA with Tukey’s multiple comparisons (n = 3 per genotype).
For the second set of data (Figure 6, C and D), the distribution of G-ratio values was compared across the genotypes. Both the WT (Figure 6C, [pyramidal tract, 0.765 ± 0.053, n = 54], and Figure 6D [CST, 0.774 ± 0.062, n = 132]) and treated KO (pyramidal tract, 0.789 ± 0.054, n = 57; CST, 0.790 ± 0.060, n = 144) groups have the smallest G-ratio values or the thickest myelin sheath layers (Figure 6C [pyramidal tract, WT vs. KO, P < 0.001; WT vs. treated KO, P = 0.076] and Figure 6D [CST, WT vs. KO, P < 0.001; WT vs. treated KO, P = 0.142]). KO (pyramidal tract, 0.830 ± 0.066, n = 55; CST, 0.856 ± 0.061, n = 138) and Het (pyramidal tract, 0.821 ± 0.043, n = 63; CTS, 0.837 ± 0.071, n = 145) groups had the largest G-ratio values with thinner myelin layers (pyramidal tract, WT vs. Het, P < 0.001; CST, WT vs. Het, P < 0.001). Again, these data strongly indicate that P2 gene therapy can significantly enhance the myelination of CST axons both in the pyramidal tract and spinal cord. To test that the defect in myelination is caused by malfunction of oligodendrocytes confined to the CNS, we examined the branches of sciatic nerves from P15 WT and KO mice at EM level, as shown in Supplemental Data and Supplemental Figure 3. The myelination in axons was normal in both genotypes, indicating no dysmyelination detected in the peripheral nerves where myelination occurs by Schwann cells (39).
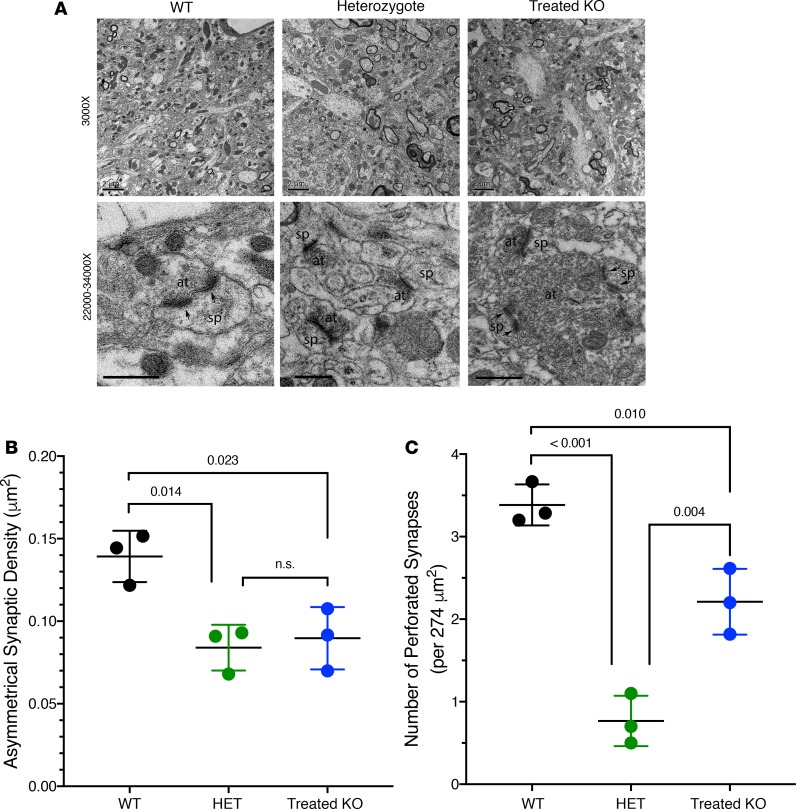
To test Hypothesis II regarding recovery in density of excitatory synapses and number of perforated synapses in layer V motor cortex following gene therapy as treated 4-month-old KO mice, we carried out EM imaging of the layer V motor cortex in 4-month-old WT, heterozygote, and P2 AAV hepatic gene therapy–treated KO mice (n = 3 mice per genotype); untreated KO mice were not included, as they typically perish at P14. We counted the total number of asymmetrical synapses (excitatory synapse) and perforated synapses in each image (defined as the unit area) and compared the density of the synapses in the 3 different genotypes, generating quantitative data showing a recovery of excitatory synapse density following gene therapy treatment.
As demonstrated in Figure 7A, low-magnification EM images show a portion of layer V in WT, heterozygote, and treated KO. Overall, the synaptic distribution pattern was similar in the 3 genotypes: asymmetrical synapses were clearly found in these regions, and no degeneration was detected in Het and treated KO samples. A quantitative comparison regarding the density of asymmetrical synapses in layer V (Figure 7B) clearly demonstrates that the WT group had the highest density (0.139 ± 0.016 per μm2), with some variability among animals with treated KO mice showing a remarkable recovery in the density values (0.090 ± 0.019 per μm2; P = 0.023 WT vs. treated KO) being comparable with the Het group (green) (0.084 ± 0.014 per μm2; P = 0.904 Het vs. treated KO).
Figure 7. Electron microscopic analysis and quantification of asymmetrical and perforated synapse density in the motor cortex of 4-month-old mice demonstrates long-term recovery in AAV-treated KO mice.
(A) WT, top: Low-power magnification image showing fine structure of neuropils in motor cortex layer V; many asymmetrical synapses are distributed in the region, and some myelinated axons are also visualized. WT, bottom: High-power image showing a perforated asymmetrical synapse at center, which has 2 separated postsynaptic density segments (indicated by 2 arrows); the presynaptic terminal (at) is making an asymmetrical synapse with a dendritic spine head (sp). Heterozygote, top: Low-power image showing layer V of the motor cortex with some asymmetrical synapses distributed among the neuropil structures; there are some myelinated axons present as well. Heterozygote, bottom: High-power image showing several asymmetrical synapses (at) contacting dendritic spines (sp). Note that few perforated synaptic contacts are found. Treated KO, top: Low-power image showing many asymmetrical synapses in the region; there are also myelinated axons present. Treated KO, bottom: High-power image showing an axon terminal (at) forming 2 perforated asymmetrical synapses with 2 dendritic spines (sp). Note the segments of postsynaptic densities (arrows). Toward the top of the image, another terminal (at) is forming an asymmetrical synaptic contact with a spine (sp). (B) The density of asymmetrical synapses in layer V of the different genotypes. WT is highest in density; Het is of lower density, while the treated KO shows partial recovery. (C) The comparison of the number of perforated synapses per unit area in layer V of different genotypes. The WT has the highest number; Het shows a much lower number, while the treated KO shows a high number and evidence of recovery. P values were determined by 1-way ANOVA with Tukey’s multiple comparisons. Error bars represent ± SD. Scale bars: 2 μm.
When carefully examined, the perforated synapses in layer V (Figure 7A) were found to be significantly lower in the heterozygote group (0.767 ± 0.306 per 274 μm2; P = 0.0002 vs. WT) (Figure 7C). The perforated synapses in WT (3.384 ± 0.248 per 274 μm2) and treated KO (2.211 ± 0.399 per 274 μm2) groups showed similar ultrastructural features: each presynaptic axon terminal contacted 2 split postsynaptic density segments associated with a spine head. Interestingly, in this treated KO image, 2 adjacent axon terminals form 2 perforated synapses with spine heads. Thus, recovery of perforated synapses in gene therapy–treated KO mice in 4-month-old samples is clearly present.
Taken together, we have two major findings from the EM study. Firstly, At P15, in Arg1 KO mice, the density of myelinated axons was significantly reduced in subcortical white matter of the motor cortex, pyramidal tract in medulla, and CST in the cervical spinal cord; degenerated axons were detected in these regions. G-ratio analysis further confirmed the defects of myelination in CST axons in the pyramidal tract and spinal cord of KO mice. P2 AAV hepatic gene therapy rescued the defects associated with myelinated axons in the regions examined and strongly implicates the functional recovery of oligodendrocytes after gene therapy. Secondly, by extending our previous studies (18), we further demonstrated that P2 AAV hepatic gene therapy can rescue the defects of excitatory synaptic density and perforated synapses in layer V of the Arg1-KO motor cortex up to 4 months of age, strongly indicating that early postnatal treatment can drastically change the synaptic organization in layer V and ultimately lead to recovery of function in the corticospinal pathway.
Dendritic arborization and soma size are altered in Arg1 deficiency.
Previously, we were able to demonstrate that the basal dendritic arbor at P14 of layer V cortical neurons had markedly less complexity than WT or Het (18); this was markedly improved with P2 AAV hepatic-based gene therapy. To examine dendritic complexity and the dendritic arbor long-term, we performed Sholl analysis to examine the dendritic complexity of the basal dendritic arbors of Golgi-stained layer V cortical neurons in WT, heterozygote, and treated KO mice at 4 months of age (Figure 8, A–C). Sholl analysis (Figure 8D) showed that the WT neurons had the greatest dendritic branching complexity; both heterozygote neurons and AAV hepatic-treated Arg1-deficient neurons were reduced in comparison (repeated measures ANOVA with Bonferroni’s multiple comparison test for Sholl analysis, P < 0.0001). The greatest differences in the Sholl analysis were detected as the distance increased in relationship to the cell body; most distally, there were relatively less differences between the 3 groups. Thus, the basal dendritic complexity, which was shown to be dramatically decreased by loss of Arg1 in a graded manner (18), can be substantially rescued and maintained long-term with neonatal hepatic-based Arg1 gene therapy.
Figure 8. Analysis of the layer V pyramidal tract neurons demonstrate differences in dendritic complexity and soma size.
Micrographs show the basal dendritic arbor in the 3 groups at P4 months: (A) WT, (B) heterozygote, and (C) treated KO. Assessment of the dendritic basilar tree of the Golgi-stained layer V pyramids of the parietal cortex was done from camera lucida drawings taken from randomly selected neurons from coded slides (n = 5 mice per group, with 7-layer V pyramids randomly selected for analysis; a total of 35 neurons from each group were studied). A comparison of the amount and distribution of the basilar dendritic arbor and branching of the layer V pyramids showed that the neurons from the WT mice had significantly more dendritic complexity (D) than neurons from the age-matched heterozygotes or the age-matched AAV-treated KOs. (There were no 4-month-old untreated KO mice, as these did not survive). The soma size of the WTs also was significantly larger than that of the heterozygotes and the AAV-treated KO mice (n = 25 neurons per group) (E). ***P < 0.0001, Sholl analysis, repeated measures ANOVA; Bonferroni’s multiple comparisons test where error bars represent ± SEM. For soma size, 1-way ANOVA with Tukey’s multiple comparisons test was performed where error bars represent ± SD.
Differences were also detected in soma size (n = 5 mice, 25 neurons per genotype) (Figure 8E). Compared with WT neurons (238.00 ± 63.47 μm2 [mean ± SD]), heterozygote soma size (172.2 ± 42.71 μm2) was decreased by 27.6%. Treated homozygous Arg1-KO layer V pyramidal soma (145.10 ± 37.15 μm2) were smaller, at 61% of the size of WT soma (WT vs. Het, P < 0.0001; WT vs. treated KO, P < 0.0001; 1-way ANOVA with Tukey’s multiple comparisons test).
Discussion
While it appears that the proximal urea cycle disorders may be caused by the same toxin and through a similar mechanism of hyperammonemic encephalopathy (2), the clinical phenotype of the patient with arginase deficiency suggests that the toxins and mechanism are different. It is not known what the proximate toxin is, but it is believed to be arginine or a metabolite produced directly from it that leads to the neurological phenotype, including spastic diplegia/tetraplegia and progressive neurological decline. Guanidino compounds have been found in the plasma and cerebrospinal fluid of patients (17) and mice (15) with arginase deficiency and have been suggested to be responsible for the neurological dysfunction.
Until recently, little was known of the neurological compromise that occurs in arginase deficiency. Our group had previously found decreased dendritic complexity, synapse number, and intrinsic excitability, as well as the presence of functional synaptic deficits in homozygous Arg1-KO mice; remarkably, hepatic-based gene therapy on P2 led to substantial improvement in these abnormalities when examined at P14 (18). However, those studies did not explore the etiology of the unique neuromotor findings, nor did they examine the long-term effectiveness of a therapy initiated during the neonatal period, when neurodevelopment is incomplete.
Two hypotheses had emerged from these prior studies and are addressed in this series of investigations. In our first hypothesis, we speculated that the output pathway containing axonal fibers originating from layer V pyramidal neurons in arginase deficiency may be compromised in postnatal development due to defects of (a) cortical excitatory synaptic function and/or (b) malfunction of oligodendrocytes and myelination. In fact, these present studies demonstrate that dysmyelination is a prominent feature of arginase deficiency. As was suggested by the limited human investigations (22, 23), the myelination of the CST was found to be adversely affected and appears to be the cause of the unique neuromotor features. While still primarily a metabolic disorder, arginase deficiency should now also be classified as a leukodystrophy affecting the brain and spinal cord but sparing the peripheral nervous system; Schwann cells and their ability to myelinate peripheral neurons do not appear to be affected. Although our analyses only found strong biological evidence for a demyelination phenotype, future studies into the biological correlates of the other dysregulated WGCNA gene modules identified in this study will likely discover additional relevant pathology in this disease.
A study by Wang and colleagues (37) on myelination in a hypoxia mouse model has some relevant findings to our results. They quantified the myelinated axons in the corpus callosum in mice at P15. While the corpus callosum contains the axons from cortico-cortical projection neurons (cortical layers II–III and V–VI) and the CST contains the axons from layer V cortico-spinal projection neurons, the WT axon count is relevant to the ones reported in the study conducted herein; this region is very close to the subcortical white matter, which we targeted for EM analysis. Wang and colleagues’ results showed the myelinated axon density was 12 × 104 axons/mm2 (12 × 104 axons/106 μm2 or 0.12 axons/μm2). Our calculations from the P15 WT mouse subcortical white matter is approximately 0.15–0.16 axons/μm2. Thus, the myelinated axon density at P15 is consistent with the reported data in this previous study, indicating that the myelinated axon density may be tightly regulated and similar across different brain regions at certain postnatal time periods.
Our myelinated axon density analysis of the pyramidal tract and CST in the cervical spinal cord is the most detailed quantitative EM analysis of this kind in P15 mice, to our knowledge. The results demonstrate that the density was drastically reduced in the homozygous Arg1-KO mice compared with the WT and Het mice in both regions, indicating that the myelination of CST axons is severely affected in homozygous Arg1 deficiency. Furthermore, these findings implicate the function of oligodendrocytes, which appear to be significantly impaired in a deficiency of arginase 1. Interestingly, the density of myelinated axons in the spinal cord region is significantly higher than in the pyramidal tract, suggesting that CST axons are much more compact at the cervical spinal cord level; this localized area has been vigorously confirmed by many previous studies using different labeling techniques (29, 30, 33, 37).
In addition, we calculated the G-ratio in P15 mice in the pyramidal tract and CST in the spinal cord. In WT mice, the average value was approximately 0.765 in the pyramidal tract and 0.774 in the CST — very close to other studies in similar regions of the rodent CNS (40–42). This finding confirms a previous theoretical model, which predicted that the optimal value of the G-ratio in rat CNS is about 0.77 (34). In comparing the G-ratio of the KO with the WT mice, a difference is found in both, with a greater difference observed in the spinal cord, confirming that the CST myelinated axons in the spinal cord are much more localized and specifically revealed by EM imaging. The Het group showed an increased G-ratio in both pyramidal tract (0.821) and spinal cord (0.837) neurons, indicating that much thinner myelin sheath layers are present in these mice with only 1 functional copy of Arg1; however, the myelinated axon numbers in the Het group did not change drastically when compared with WT and treated KO groups. Together, these findings implicate the wrapping of axons by oligodendrocytes as being also compromised in the Arg1 heterozygote but not as severely as in the homozygous KO mice. These changes in myelination (i.e., thin sheaths) in the Het is of further support for a primary myelination phenotype.
In our second hypothesis, we theorized that AAV-based hepatic gene therapy initiated during early postnatal development could lead to long-term recovery in the density of asymmetrical synapses, the number of perforated synapses in layer V, and the basal dendritic arbor in the motor cortical neurons of treated KO mice. There are several comments to be made about our present data. First, previous studies found that there is postnatal refinement in the cerebral cortex regarding excitatory synaptic density (43–45). It is generally agreed upon that a gradual decline in excitatory synapse density is found in different cortical regions (including layer V during postnatal development), suggesting that pruning of synaptic contacts is significant for establishing mature functional brain circuits. Glynn et al. (46) compared the excitatory synapse density in layer V of the visual cortex at P15 and P60 (adult). They found a density of 40 synapses/100 μm2 (or 0.4 synapse/μm2) at P15 that decreased to 20 synapses/100 μm2 (or 0.2 synapse/μm2) at P60. Interestingly, our data in layer V of the motor cortex in the 4-month-old adult WT mouse indicate that the density is approximately 0.15 synapse/μm2, decreased from 0.21 synapse/μm2 at P15 (18), indicating a similar trend when compared with this previous study (18). Interestingly, decreased excitatory synapses in layer V of the motor cortex were detected in homozygous Arg1-KO mice at P15 (18); there is a drastic 3-fold reduction when compared with WT samples (density was reduced to about 0.07 synapse/μm2) whether from reduced synaptogenesis or excessive removal.
While not excluding other cellular processes, this change in excitatory synapse number suggests an enhanced active removal during this period, which may be mediated by engulfment by microglia the macrophage population of the CNS (47), or in synapse maturation or elimination (48). Interestingly, arginase 1 is one of a group of uniquely expressed microglial genes at P4/P5 (49). Could the absence of CNS arginase 1 in microglia lead to overpruning in the normal refinement of synaptic circuits? While speculative, some preexisting data suggest that this hypothesis is plausible: arginase 1–deficient macrophages are linked with enhanced activity in the control of certain infections (50), and arginase 1 is an essential suppressive mediator of alternatively activated macrophages (51). Further investigation to link the arginase 1 signaling to immunoregulatory pathways could provide the answer to this speculation.
Perforated synapses are frequently found in brain regions associated with synaptic plasticity, such as the CA1 region of the hippocampus and the cerebral cortex engaged in learning and memory (19, 20, 52). These synapses exhibit a complicated postsynaptic density organization, which contain GluR2 subunits that are impermeable to calcium influx and enhance AMPAR-mediated synaptic transmission. Our finding of long-term recovery in perforated synapse number in layer V of the motor cortex in gene therapy–treated 4-month-old Arg1-KO mice — as we found a reduction of perforated synapses in untreated KO mice (18) — strongly suggests that the regaining of AMPAR at excitatory synapses is critical for restoring synaptic functions and that excitatory synaptic transmission may be compromised in the untreated Arg1-KO mice.
Anatomically, we had previously demonstrated by quantitative analysis that the dendritic complexity of the basal dendritic arbor in homozygous Arg1-deficient mice was lowest in the KO and that, with AAV-based neonatal gene therapy, it was completely recovered to that of the WT when examined at P14 (18). Herein, we demonstrate that, in adult mice, WT neurons have the greatest dendritic complexity and that, in AAV hepatic-treated Arg1-deficient neurons, this is slightly reduced. Similarly, we now find that overall dendritic branching and soma size is also reduced with a loss of functional arginase copy number.
These studies have now shown by multiple modalities — and again unexpectedly (18) — that, in arginase deficiency, a gradation of abnormalities exist in neurons based on the functional copy number of arginase, with heterozygote animals showing an intermediate phenotype. While biochemically these mice are not different than WT mice (8, 14, 15, 18), Het had increased faulty transcript arginase 1 CNS expression by microarray, fewer perforated and asymmetrical synapses, a reduction in myelinated axon density with thinner myelin sheaths, less complex basal dendritic arbor, and smaller soma in layer V pyramidal neurons. Furthermore, hepatic gene transfer with Arg1 expression administered on P2 to homozygous Arg1-KO mice typically resulted in recovery but incomplete correction (i.e., to that of the heterozygote) in most of these measures. The significance of this incomplete equivalence with WT mice is unknown in that, cognitively and behaviorally, Arg1 heterozygote mice and treated homozygous KOs are normal (15). A possible explanation relates to the fact that arginase expression in WT mice (and in humans) is not limited to hepatocytes; it is also expressed in neurons (53) and glia (49) of both the cortex and subcortical structures throughout life. While its role and function in the CNS have not been elucidated, this assemblage of differences may be related to the absence of Arg1 and will be the source of future investigation. Whether human single copy carriers without clinically definable phenotypic abnormalities demonstrate such differences at the neural cellular level is not known.
These studies support intervention in ARG1 deficiency in the early postnatal period, when neurodevelopment is incomplete. Given the ability to attain near-normal neurological development, we believe that ARG1 AAV hepatic-based gene therapy presents an important consideration to prevent the neurodevelopmental compromise that occurs in the deficiency of arginase.
Methods
Mouse procedures
Exon 4 of the Arg1 allele was replaced by the neomycin resistance gene to generate the arginase 1–KO mice, as was previously described (8). Arginase Het were backcrossed more than 10 times to achieve a homogeneous NIH-Swiss strain (14) and were housed in our UCLA colony under specific pathogen-free conditions with food and water provided ad libitum. Gene therapy of homozygous Arg1-deficient pups on P2 (treated KO) was performed by injecting 1.0 × 1014 genome copies (gc)/kg of AAVrh10 with the thyroxine-binding globulin (TBG) promoter driving murine Arg1 expression (previously described in ref. 14). The virus was diluted in pharmaceutical-grade saline and administered by the superficial facial vein as previously described (16). Mice underwent genotyping by PCR at birth and by a second freshly collected independent sample at the time of study to ensure that the initial genotype was correct. All attempts were made to include equal numbers of male and female mice and littermates. Mice were fed standard mouse chow (Labdiet/PMI Nutrition International, Picolab Rodent diet 20, #5053).
PCR genotyping
Genomic DNA was prepared from tail tip or ear clip by standard methods, and genotyping was performed by PCR amplification as previously described (14, 54).
Microarray procedure and analysis
A total of 20 mg of the frontal lobe of P14 mice of WT, Het, KO, and treated KO (n = 6 per genotype) was immediately removed after euthanizing with isoflurane. RNA extraction was performed following the manufacturer’s instructions (RNeasy mini kit, Qiagen), and DNase treatment (Roche Diagnostics) was applied. RNA was transcribed to double-stranded complementary RNA (cRNA) using MessageAmp Premier RNA Amplification Kit (Invitrogen) with an oligo(dT) primer following the manufacturer’s instructions. Following fragmentation, 11 μg of biotin-labeled cRNA were hybridized for 16 hours at 45°C on Affymetrix Mouse Genome 430 Plus 2.0 Arrays. GeneChips were washed and stained in the Affymetrix Fluidics Station 450 and then scanned with the Affymetrix GeneChip Scanner 3000 G7.
All WGCNA calculations were done in R using WGCNA R package. All preprocessing of raw data was done in R (55) (www.r-project.org/). Raw fluorescence data was stored as .CEL.gz files and were loaded into R as an AffyBatch object (using the affy package in R) (56). This object was then processed using robust multiarray average (RMA) normalization, which removed background noise and converted data to a log2 scale. ComBat sva (57) R package was then applied to the normalized data to adjust for batch effects. Samples were split across 3 batches – 3 KO and 3 WT were in the first batch, 3 treated KO and 3 Het were in the second batch, and the remaining samples (3 of each genotype) were in the final batch.
No outlier samples were found in the data (outlier investigation was done using a WGCNA sample network connectivity approach; ref. 24). Affymetrix probes were summarized and reannotated with Ensembl gene names from the Ensembl July 2015 archive (Ensembl version 81) (58). This annotation dataset was chosen because it most closely matched the time of the Affymetrix microarray manufacture date and following hybridization run. Finally, the dataset was filtered so that genes with a standard deviation in the 5th quantile or lower were removed. This step was performed to maximize the connectivity that could be found in our subsequent WGCNA analysis. This normalized, batch-adjusted, reannotated, and filtered gene expression data matrix was our input for all later WGCNA and differential expression analyses.
The limma package (59) in R was used for differential gene expression analysis. Our linear model only included group (Gene Expression stratified by Group). Differentially expressed genes were those with an FDR-adjusted P value of less than 0.05.
All WGCNA (24) calculations were done in R, using the WGCNA R package. In our WGCNA analysis, we chose a soft-threshold value of 14, since this value best accentuated the scale-free topology in our dataset. We required that all modules contain at least 200 genes. Further user-specified module cutting parameters included deepSplit=2 for module identification and, for close module merging, cutHeight=0.1. The pSI (60) R library was used with data from Zhang et al. (61) for cell-type enrichment analysis. The gProfileR (62) R library was used for gene ontology analysis. All module dysregulation was established with ANOVA followed by a Tukey post hoc test. The first principal component of module gene expression (referred to as the ME in WGCNA analysis) was associated with group (WT, KO, HET, and treated KO) for all modules. P < 0.05 was required for a significant result. For M7 (MO dysregulated genes), P = 0.024, by ANOVA; WT vs. KO, P = 0.014, by Tukey’s post hoc test.
All processed and unprocessed expression microarray data utilized in this publication is freely available online through the NCBI Gene Expression Omnibus (GEO) repository (accession number GSE132058).
Immunostaining for MBP
Mice P14–P15 were transcardially perfused with 5–10 ml of cold 4% paraformaldehyde (PFA) (Electron Microscopy Services). Whole brains were removed and then post-fixed in 4% PFA overnight at 4°C. Immunostaining for MBP was performed by free floating staining with a 1:200 dilution of rat anti-MBP antibody (MilliporeSigma, MAB386]) and with a 1:200 dilution of goat anti-rat biotinylated antibody (Vector Laboratories, BA-9401). For quantification, 50× magnification images were acquired on a Zeiss Axio Imager M2 microscope equipped with a color digital camera (Axiocam 506 color), using Stereo Investigator software v.2018 (MBF Bioscience) within the defined sampling region by setting the cingulum bundle as a reference point in the top corner of the frame (Supplemental Figure 4). MBP-immunostaining quantification was performed using ImageJ (NIH). Complete details are described in the Supplemental Methods.
Electron microscopy
Animals.
For P15 mice, 4 groups of genotypes (n = 3 per group) were included: WT, KO, Het, and P2 gene therapy–treated KO (treated KO). Each group included at least 3 mice (in-house). For 4-month-old mice, 3 groups of genotypes (n = 3 mice per group) were included: WT, Het, and treated KO. Each group had at least 3 mice.
Mice were anesthetized with isoflurane and cardiac perfused with cold 1× PBS solution for 1–2 minutes until clear. Mice were then perfused with 4% PFA plus 2.5% glutaraldehyde (MilliporeSigma) in 1× PBS for at least 5 minutes. Brains were removed and stored in the fixative solution at 4°C.
Brain sample preparation.
For each brain, somatosensory and motor cortex, pyramidal tract in the medulla, and cervical segment (C1–C7) in the spinal cord were identified based on a standard reference (63). Under a dissecting microscope, the sections approximately 1 mm in thickness were cut coronally with a sharp surgical blade and saved in cold 1× PBS. The motor cortex containing layer V, subcortical white matter under layer VI, the ventral portion of medulla containing pyramidal tract, and the dorsal funiculus of cervical spinal cord were further identified under the dissecting microscope and carefully dissected out for embedding (Figure 4A).
To compare the myelination of peripheral nerves with CNS, branches of sciatic nerves from 2 perfused P15 WTs and 2 perfused P15 KO mice were identified and collected under dissecting microscope and immediately saved in the cold 1× PBS solution.
Electron microscopy procedure.
For each brain, sections containing motor cortex, the pyramidal tract, and the spinal cord were processed for EM analysis, as previously described (18). Briefly, sections were fixed in 2% OsO4 PBS for 30 minutes; after washing in PBS, sections were dehydrated in 70%, 90%, 95%, and 100% ethanol and then in 100% acetone. Sections were processed in 1:1 pure acetone and Araldite (Ted Pella) for 1 hour and were then placed in freshly made pure Araldite overnight at 4°C. The next day, sections were transferred to Araldite for 3–4 hours and then mounted on siliconized slides with glass coverslips (Sigmacote, MilliporeSigma). For sciatic nerves, samples were carefully oriented and placed in silicone rubber molds (PELCO Flat Embedding Mold, Ted Pella) filled with pure Araldite and polymerized in a 60°C–70°C oven for 48 hours. Polymerized sections were examined under a dissecting microscope to confirm that the motor cortex, pyramidal tract, and spinal cord regions were included; subregions were further dissected to layer V of motor cortex, subcortical white matter of motor cortex, pyramidal tract region, and dorsal funiculus of spinal cord containing CST using a sharp surgical blade. Sections containing these regions were glued to the blank Araldite blocks using super glue and dried in air. Sections were trimmed on an UCT Ultramicrotome (Leica); a typical cutting surface of the section was about 2 × 2 mm2, including cortical layer V–VI, subcortical white matter, and partial corpus callosum; the pyramidal tract and some surrounding medulla structures; and dorsal funiculus in spinal cord. To further confirm the selected regions, the semithin sections (about 0.4–0.6 μm in thickness) were cut on the ultramicrotome using a diamond knife (DiATOME); sections were collected on clean glass slides in drops of water and then dried on a hot plate. The slides were then stained with Toluidine blue; semithin sections were examined with a light microscope to confirm regions, including cortical layer V and subcortical white matter, pyramidal tract, and CST in the spinal cord.
The blocks were trimmed and 60–70 nm ultrathin sections cut on the ultramicrotome using the diamond knife. Thin sections were collected on Formvar-coated single slot copper grids (Ted Pella) and were stained with uranyl acetate and lead citrate to enhance contrast.
EM imaging.
Stained sections were examined in a JEM1200-EX (JEOL Ltd.) electron microscope at 80 kV. Images were acquired using a wide angle (top mount) BioScan 600W 1K × 1K digital camera (Gatan). For axon analysis and synaptic counting, EM images were taken at 8,000× — each image covering approximately 274 μm2, which is the unit area for myelinated axons and asymmetrical synaptic contact number counting and density calculations. For visualizing the fine structure of axon terminals, postsynaptic elements and myelin sheaths, some images were taken at 20,000× or 50,000×. All images were saved as Gatan 3 and TIFF formats, with the files in TIFF format imported to ImageJ (NIH) for counting and measuring, or to Adobe Photoshop CC (Adobe System) for image editing, adjusting size, contrast and brightness, and figure composition and saved at a final resolution of 300 dpi. All the figures were composed in Adobe Photoshop CC.
EM data quantitative analysis.
From EM imaging, we have generated 4 sets of data, including myelinated axon density analysis and comparison in subcortical white matter of the motor cortex, pyramidal tract, and cervical spinal cord in 4 genotype groups. The subcortical white matter, pyramidal tract, and spinal cord regions were first examined at low magnification (3,000×) comparing with semithin sections and confirming targeted regions to be imaged. The regions containing myelinated and nonmyelinated axons were randomly selected for EM photographing at 8,000×. Myelinated axons are defined as the axon caliber covered by at least 1 layer of electron-dense myelin sheath, regardless of the sagittal or coronal sectioned plane. In each genotype group, for each animal, about 15–25 images were taken from each region; each image covers approximately 274 μm2. Total myelinated axons were identified and counted in ImageJ; thus, the density of myelinated axons in each unit area is calculated as total number of myelinated axons counted/274 μm2 or number of myelinated axons/μm2. The myelinated axon density of each region was scatter-plotted and compared in genotype groups.
Regarding myelin sheath thickness (G-ratio) analysis and comparison in pyramidal tract and CST in spinal cord of 4 genotype groups, the myelinated axon G-ratio analysis was carried out using EM images taken at 8,000×. In each animal, about 5–10 EM images were imported to ImageJ; for each image, approximately 5–15 coronally cut myelinated axons were chosen for measurement based on the method detailed in previous publications (36, 64). Briefly, for each myelinated axon, its axon diameter (a) and entire myelinated fiber diameter (A) were measured and recorded (G-ratio = a/A). Thus, at least 50 myelinated axons from each genotype were measured, and G-ratios were calculated. To analyze the relationship between G-ratio and myelinated axon diameters, the G-ratio and axon diameters of myelinated axons from each genotype were plotted; a regression analysis was applied to determine their linear correlation.
Quantitative analysis of excitatory synapse density in layer V of motor cortex of 4-month -old mice was performed. As described in the previous study (18), this set of data was generated from the quantification of density of asymmetrical synapses in layer V of the motor cortex of the 3 genotype groups of 4-month-old mice (WT, heterozygote, and treated KO). An asymmetrical synapse is defined as its presynaptic terminal and contains oval or round-shaped synaptic vesicles with a clear synaptic cleft and prominent postsynaptic density associated with postsynaptic membranes (39). Synapses fitting these features were identified and counted (each image covers the unit area of approximately 274 μm2) using ImageJ based on a method described in detail previously (46). For each genotype, a total of 10–22 images were analyzed. The density of asymmetrical synapses was calculated as the number of asymmetrical synapses per μm2; the numbers were scatter-plotted, and a comparison was made between genotypes.
The number of perforated synapses in layer V of the motor cortex was counted in the 3 genotype groups of 4-month-old mice. Perforated asymmetrical synapses are distinguished visually by having 2 juxtaposed prominent postsynaptic densities from 1 dendritic spine drawing the same pool of synaptic vesicles in the presynaptic axon terminals (21). In each image (unit area of approximately 274 μm2), total perforated synapses were counted (at least 10 images assessed per genotype); thus, the average number of perforated synapses in each unit area was generated in each genotype and compared.
Morphology studies (Golgi staining)
Tissue blocks from 4-month-old WT, Het, and treated KO mice (n = 5 per genotype) were Golgi stained (65, 66) and Sholl analysis was used to quantify the amount and distribution of basilar dendritic branches at increasing distances from the soma (67) as previously described (18).
Statistics
Data analysis was performed with Prism 8 statistical package (GraphPad Software). Results are expressed as mean ± SD, and P values were determined using 1-way ANOVA with Tukey’s or Dunnett’s multiple comparisons test. For the Sholl analyses, we used repeated measures ANOVA followed by Bonferroni’s multiple comparisons test, where error bars represent ± SEM. P ≤ 0.05 was considered significant.
Study approval
All experimental procedures were approved by the UCLA Chancellor’s Animal Research Committee and were conducted in accordance with guidelines for the care and use of research animals. Mice were housed according to the NIH guidelines.
Author contributions
XBL conducted studies and wrote the paper. JRH analyzed and interpreted the microarray studies and wrote the paper. GC and JRL conducted studies and analyzed data. MOG conducted studies and critically reviewed the manuscript. BT performed the statistical analysis, constructed the graphical data, and critically reviewed the paper. AG provided data and critically reviewed the paper. SDC and IC provided expertise and critically reviewed the paper. GSL conceived of the studies, conducted studies, analyzed data, and wrote the paper.
Supplementary Material
Acknowledgments
The authors thank the Semel Institute for Neuroscience and the Intellectual and Developmental Disabilities Research Center at UCLA for their support, Neurostructural Research Labs for collecting data for the Sholl and soma size study, and Masha Stoianova BA for assistance in preparation of Figure 1. GSL has served as a consultant for Audentes Therapeutics in an area unrelated to these studies. This work was supported by grants from the NIH to GSL (R01NS110596 and R03NS101682). The authors acknowledge the use of instruments at the Electron Imaging Center for NanoMachines supported by NIH (1S10RR23057 to Z. Hong Zhou) and CNSI at UCLA.
Version 1. 09/05/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(17):e130260. https://doi.org/10.1172/jci.insight.130260.
Contributor Information
Xiao-Bo Liu, Email: xiaoboliu2015@gmail.com.
Gloria Cantero, Email: gcanteronieto@gmail.com.
Jenna R. Lambert, Email: jennarosel@g.ucla.edu.
Marcos Otero-Garcia, Email: marcos.otero@stanford.edu.
Brian Truong, Email: btruong92@ucla.edu.
Andrea Gropman, Email: AGropman@childrensnational.org.
Inma Cobos, Email: icobos@stanford.edu.
References
- 1.Helman G, Pacheco-Colón I, Gropman AL. The urea cycle disorders. Semin Neurol. 2014;34(3):341–349. doi: 10.1055/s-0034-1386771. [DOI] [PubMed] [Google Scholar]
- 2.Rangroo Thrane V, et al. Ammonia triggers neuronal disinhibition and seizures by impairing astrocyte potassium buffering. Nat Med. 2013;19(12):1643–1648. doi: 10.1038/nm.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Summar ML, et al. The incidence of urea cycle disorders. Mol Genet Metab. 2013;110(1-2):179–180. doi: 10.1016/j.ymgme.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prasad AN, Breen JC, Ampola MG, Rosman NP. Argininemia: a treatable genetic cause of progressive spastic diplegia simulating cerebral palsy: case reports and literature review. J Child Neurol. 1997;12(5):301–309. doi: 10.1177/088307389701200502. [DOI] [PubMed] [Google Scholar]
- 5.Jichlinski A, Clarke L, Whitehead MT, Gropman A. “Cerebral Palsy” in a Patient With Arginase Deficiency. Semin Pediatr Neurol. 2018;26:110–114. doi: 10.1016/j.spen.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 6.Cederbaum SD, Shaw KN, Spector EB, Verity MA, Snodgrass PJ, Sugarman GI. Hyperargininemia with arginase deficiency. Pediatr Res. 1979;13(7):827–833. doi: 10.1203/00006450-197907000-00007. [DOI] [PubMed] [Google Scholar]
- 7.Sin YY, Baron G, Schulze A, Funk CD. Arginase-1 deficiency. J Mol Med. 2015;93(12):1287–1296. doi: 10.1007/s00109-015-1354-3. [DOI] [PubMed] [Google Scholar]
- 8.Iyer RK, et al. Mouse model for human arginase deficiency. Mol Cell Biol. 2002;22(13):4491–4498. doi: 10.1128/MCB.22.13.4491-4498.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crombez EA, Cederbaum SD. Hyperargininemia due to liver arginase deficiency. Mol Genet Metab. 2005;84(3):243–251. doi: 10.1016/j.ymgme.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 10.Iyer R, Jenkinson CP, Vockley JG, Kern RM, Grody WW, Cederbaum S. The human arginases and arginase deficiency. J Inherit Metab Dis. 1998;21 Suppl 1:86–100. doi: 10.1023/a:1005313809037. [DOI] [PubMed] [Google Scholar]
- 11.Scaglia F, Lee B. Clinical, biochemical, and molecular spectrum of hyperargininemia due to arginase I deficiency. Am J Med Genet C Semin Med Genet. 2006;142C(2):113–120. doi: 10.1002/ajmg.c.30091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deignan JL, et al. Guanidino compound levels in blood, cerebrospinal fluid, and post-mortem brain material of patients with argininemia. Mol Genet Metab. 2010;100 Suppl 1:S31–S36. doi: 10.1016/j.ymgme.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 13.Deignan JL, et al. Increased plasma and tissue guanidino compounds in a mouse model of hyperargininemia. Mol Genet Metab. 2008;93(2):172–178. doi: 10.1016/j.ymgme.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 14.Lee EK, et al. Long-term survival of the juvenile lethal arginase-deficient mouse with AAV gene therapy. Mol Ther. 2012;20(10):1844–1851. doi: 10.1038/mt.2012.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee EK, et al. AAV-based gene therapy prevents neuropathology and results in normal cognitive development in the hyperargininemic mouse. Gene Ther. 2013;20(8):785–796. doi: 10.1038/gt.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu C, et al. Myocyte-mediated arginase expression controls hyperargininemia but not hyperammonemia in arginase-deficient mice. Mol Ther. 2014;22(10):1792–1802. doi: 10.1038/mt.2014.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marescau B, Qureshi IA, De Deyn P, Letarte J, Ryba R, Lowenthal A. Guanidino compounds in plasma, urine and cerebrospinal fluid of hyperargininemic patients during therapy. Clin Chim Acta. 1985;146(1):21–27. doi: 10.1016/0009-8981(85)90120-2. [DOI] [PubMed] [Google Scholar]
- 18.Cantero G, et al. Rescue of the Functional Alterations of Motor Cortical Circuits in Arginase Deficiency by Neonatal Gene Therapy. J Neurosci. 2016;36(25):6680–6690. doi: 10.1523/JNEUROSCI.0897-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goddard CA, Butts DA, Shatz CJ. Regulation of CNS synapses by neuronal MHC class I. Proc Natl Acad Sci USA. 2007;104(16):6828–6833. doi: 10.1073/pnas.0702023104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medalla M, Luebke JI. Diversity of glutamatergic synaptic strength in lateral prefrontal versus primary visual cortices in the rhesus monkey. J Neurosci. 2015;35(1):112–127. doi: 10.1523/JNEUROSCI.3426-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chenaux G, et al. Loss of SynDIG1 Reduces Excitatory Synapse Maturation But Not Formation In Vivo. eNeuro. 2016;3(5):ENEURO.0130-16.2016. doi: 10.1523/ENEURO.0130-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brockstedt M, Smit LM, de Grauw AJ, van der Klei-van Moorsel JM, Jakobs C. A new case of hyperargininaemia: neurological and biochemical findings prior to and during dietary treatment. Eur J Pediatr. 1990;149(5):341–343. doi: 10.1007/BF02171562. [DOI] [PubMed] [Google Scholar]
- 23.Oldham MS, VanMeter JW, Shattuck KF, Cederbaum SD, Gropman AL. Diffusion tensor imaging in arginase deficiency reveals damage to corticospinal tracts. Pediatr Neurol. 2010;42(1):49–52. doi: 10.1016/j.pediatrneurol.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Annunziato L, Boscia F, Pignataro G. Ionic transporter activity in astrocytes, microglia, and oligodendrocytes during brain ischemia. J Cereb Blood Flow Metab. 2013;33(7):969–982. doi: 10.1038/jcbfm.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheli VT, Santiago González DA, Spreuer V, Paez PM. Voltage-gated Ca2+ entry promotes oligodendrocyte progenitor cell maturation and myelination in vitro. Exp Neurol. 2015;265:69–83. doi: 10.1016/j.expneurol.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fink D, Knapp PE, Mata M. Differential expression of Na,K-ATPase isoforms in oligodendrocytes and astrocytes. Dev Neurosci. 1996;18(4):319–326. doi: 10.1159/000111422. [DOI] [PubMed] [Google Scholar]
- 28.Yager LM, Garcia AF, Wunsch AM, Ferguson SM. The ins and outs of the striatum: role in drug addiction. Neuroscience. 2015;301:529–541. doi: 10.1016/j.neuroscience.2015.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faulkner RL, Low LK, Liu XB, Coble J, Jones EG, Cheng HJ. Dorsal turning of motor corticospinal axons at the pyramidal decussation requires plexin signaling. Neural Dev. 2008;3:21. doi: 10.1186/1749-8104-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Low LK, Liu XB, Faulkner RL, Coble J, Cheng HJ. Plexin signaling selectively regulates the stereotyped pruning of corticospinal axons from visual cortex. Proc Natl Acad Sci USA. 2008;105(23):8136–8141. doi: 10.1073/pnas.0803849105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rünker AE, Little GE, Suto F, Fujisawa H, Mitchell KJ. Semaphorin-6A controls guidance of corticospinal tract axons at multiple choice points. Neural Dev. 2008;3:34. doi: 10.1186/1749-8104-3-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welniarz Q, Dusart I, Roze E. The corticospinal tract: Evolution, development, and human disorders. Dev Neurobiol. 2017;77(7):810–829. doi: 10.1002/dneu.22455. [DOI] [PubMed] [Google Scholar]
- 33.Nakanishi T, Fujita Y, Yamashita T. Neuropilin-1-mediated pruning of corticospinal tract fibers is required for motor recovery after spinal cord injury. Cell Death Dis. 2019;10(2):67. doi: 10.1038/s41419-019-1338-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chomiak T, Hu B. What is the optimal value of the g-ratio for myelinated fibers in the rat CNS? A theoretical approach. PLoS ONE. 2009;4(11):e7754. doi: 10.1371/journal.pone.0007754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang C, Tao Y, Shen C, Tan Z, Xiong WC, Mei L. Erbin is required for myelination in regenerated axons after injury. J Neurosci. 2012;32(43):15169–15180. doi: 10.1523/JNEUROSCI.2466-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu XB, Schumann CM. Optimization of electron microscopy for human brains with long-term fixation and fixed-frozen sections. Acta Neuropathol Commun. 2014;2:42. doi: 10.1186/2051-5960-2-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang F, et al. Enhancing Oligodendrocyte Myelination Rescues Synaptic Loss and Improves Functional Recovery after Chronic Hypoxia. Neuron. 2018;99(4):689–701.e5. doi: 10.1016/j.neuron.2018.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fraher J, Dockery P. Injury-induced Changes in Spinal Root Transitional Zones: Morphometric Ultrastructural Studies. In: Aldskogius H, Fraher J, eds. Glial Interfaces in the Nervous System: Role in Repair and Plasticity. Amsterdam, Netherlands: IOS Press; 2001:41–59. [Google Scholar]
- 39. Peters A, Palay SL, Webster HD. The Fine Structure of the Nervous System: Neurons and Their Supporting Cells. New York, New York, USA: Oxford University Press; 1991. [Google Scholar]
- 40.Arnett HA, Mason J, Marino M, Suzuki K, Matsushima GK, Ting JP. TNF alpha promotes proliferation of oligodendrocyte progenitors and remyelination. Nat Neurosci. 2001;4(11):1116–1122. doi: 10.1038/nn738. [DOI] [PubMed] [Google Scholar]
- 41.Benninger Y, et al. Beta1-integrin signaling mediates premyelinating oligodendrocyte survival but is not required for CNS myelination and remyelination. J Neurosci. 2006;26(29):7665–7673. doi: 10.1523/JNEUROSCI.0444-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mason JL, Langaman C, Morell P, Suzuki K, Matsushima GK. Episodic demyelination and subsequent remyelination within the murine central nervous system: changes in axonal calibre. Neuropathol Appl Neurobiol. 2001;27(1):50–58. doi: 10.1046/j.0305-1846.2001.00301.x. [DOI] [PubMed] [Google Scholar]
- 43.Rakic P, Bourgeois JP, Eckenhoff MF, Zecevic N, Goldman-Rakic PS. Concurrent overproduction of synapses in diverse regions of the primate cerebral cortex. Science. 1986;232(4747):232–235. doi: 10.1126/science.3952506. [DOI] [PubMed] [Google Scholar]
- 44.Huttenlocher PR, Dabholkar AS. Regional differences in synaptogenesis in human cerebral cortex. J Comp Neurol. 1997;387(2):167–178. doi: 10.1002/(SICI)1096-9861(19971020)387:2<167::AID-CNE1>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 45.Gonzalez-Burgos G, et al. Functional maturation of excitatory synapses in layer 3 pyramidal neurons during postnatal development of the primate prefrontal cortex. Cereb Cortex. 2008;18(3):626–637. doi: 10.1093/cercor/bhm095. [DOI] [PubMed] [Google Scholar]
- 46.Glynn MW, et al. MHCI negatively regulates synapse density during the establishment of cortical connections. Nat Neurosci. 2011;14(4):442–451. doi: 10.1038/nn.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perry VH, Teeling J. Microglia and macrophages of the central nervous system: the contribution of microglia priming and systemic inflammation to chronic neurodegeneration. Semin Immunopathol. 2013;35(5):601–612. doi: 10.1007/s00281-013-0382-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Graeber MB. Changing face of microglia. Science. 2010;330(6005):783–788. doi: 10.1126/science.1190929. [DOI] [PubMed] [Google Scholar]
- 49.Hammond TR, et al. Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity. 2019;50(1):253–271.e6. doi: 10.1016/j.immuni.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El Kasmi KC, et al. Toll-like receptor-induced arginase 1 in macrophages thwarts effective immunity against intracellular pathogens. Nat Immunol. 2008;9(12):1399–1406. doi: 10.1038/ni.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pesce JT, et al. Arginase-1-expressing macrophages suppress Th2 cytokine-driven inflammation and fibrosis. PLoS Pathog. 2009;5(4):e1000371. doi: 10.1371/journal.ppat.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toni N, Buchs PA, Nikonenko I, Povilaitite P, Parisi L, Muller D. Remodeling of synaptic membranes after induction of long-term potentiation. J Neurosci. 2001;21(16):6245–6251. doi: 10.1523/JNEUROSCI.21-16-06245.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu H, Iyer RK, Kern RM, Rodriguez WI, Grody WW, Cederbaum SD. Expression of arginase isozymes in mouse brain. J Neurosci Res. 2001;66(3):406–422. doi: 10.1002/jnr.1233. [DOI] [PubMed] [Google Scholar]
- 54.Gau CL, et al. Short-term correction of arginase deficiency in a neonatal murine model with a helper-dependent adenoviral vector. Mol Ther. 2009;17(7):1155–1163. doi: 10.1038/mt.2009.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/ Accessed August 30, 2019.
- 56.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 57. Leek JT, et al. sva: Surrogate Variable Analysis. R package version 3.26.0. Bioconductor. https://bioconductor.org/packages/release/bioc/html/sva.html Accessed August 30, 2019.
- 58.Zerbino DR, et al. Ensembl 2018. Nucleic Acids Res. 2018;46(D1):D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xu X, Wells AB, OBrien D, Nehorai A, Dougherty JD. pSI: Specificity Index Statistic. R package version 1.1. 2014. CRAN. https://cran.r-project.org/web/packages/pSI/pSI.pdf Published January 30, 2014. Accessed August 30, 2019.
- 61.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34(36):11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Reimand J, Kolde R, Tambet A. Interface to ‘gProfiler’ Toolkit. R package version 0.6.6. 2018. CRAN. https://cran.r-project.org/web/packages/gProfileR/gProfileR.pdf Published October 24, 2018. Accessed August 30, 2019.
- 63. Dong HW. Allen Reference Atlas: A Color Brain Atlas of the C57Bl/6J Male Mouse. Hoboken, New Jersey, USA: John Wiley & Sons; 2008. [Google Scholar]
- 64.Cheli VT, et al. Conditional Deletion of the L-Type Calcium Channel Cav1.2 in Oligodendrocyte Progenitor Cells Affects Postnatal Myelination in Mice. J Neurosci. 2016;36(42):10853–10869. doi: 10.1523/JNEUROSCI.1770-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Morest DK. The Golgi Methods. In: Heym C, Fresmann WG, eds. Techniques in Neuroanatomical Research. Heidelberg, Germany: Springer-Verlag; 1981:124–38. [Google Scholar]
- 66. Valverde F. The rapid Golgi technique for staining CNS neurons: light microscopy. In: Wouterlood FG, ed. Neuroscience Protocols. Amsterdam, Netherlands and New York, New York, USA: Elsevier; 1993:1–9. [Google Scholar]
- 67.Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat. 1953;87(4):387–406. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.