SUMMARY
Eukaryotic chromatin is highly condensed yet dynamically accessible to regulation, and organized into subdomains. We demonstrate that reconstituted chromatin undergoes histone tail-driven liquid-liquid phase separation (LLPS) in physiologic salt and when microinjected into cell nuclei, producing dense and dynamic droplets. Linker histone H1 and internucleosome linker lengths shared across eukaryotes promote phase separation of chromatin, tune droplet properties, and coordinate to form condensates of consistent density, in manners that parallel chromatin behaviors in cells. Histone acetylation by p300 antagonizes chromatin phase separation, dissolving droplets in vitro and decreasing droplet formation in nuclei. In the presence of multi-bromodomain proteins, such as BRD4, highly acetylated chromatin forms a new phase separated state, with droplets of distinct physical properties, which can be immiscible with unmodified chromatin droplets, mimicking nuclear chromatin subdomains. Our data suggest a framework, based on intrinsic phase separation of the chromatin polymer, for understanding organization and regulation of eukaryotic genomes.
Graphical Abstract

INTRODUCTION
Genomic DNA is highly compacted in the nucleus of eukaryotic cells into a nucleoprotein assembly called chromatin (Olins and Olins, 2003). The basic unit of chromatin is the nucleosome, where ~146 base pair increments of the genome are wrapped ~1.65 times around an octameric assembly of histone proteins (Kornberg, 1974; Luger et al., 1997). Emanating from the nucleosome core particle are positively charged histone tails that serve as hubs for a variety of post-translational modifications, such as acetylation, methylation, and phosphorylation. Histone post-translational modifications can in turn be recognized by specific histone tail “reader” proteins, which play important roles in the control of myriad nuclear processes such as transcription, replication, and DNA repair (Jenuwein and Allis, 2001).
The exact organization of chromatin in cells remains unclear. However, it is known that chromatin is structured across a range of length scales in cells. Within the nucleus, each chromosome is retained within a sub-region of the nucleoplasm known as a chromosome territory (Meaburn and Misteli, 2007). Chromosomes are then further organized into self-associating regions (e.g. topologically associated domains), constrained in a cohesin-dependent manner (Bintu et al., 2018; Lieberman-Aiden et al., 2009; Rao et al., 2017), enriched in particular post-translational modifications and chromatin-binding proteins. Beyond these spatial definitions for higher order chromatin organization, regions of the genome can be characterized by their function (e.g. enhancer, insulator, promoter, etc.), accessibility to exogeneous factors (e.g. “open” or “closed”), or extent of compaction (e.g. euchromatin or heterochromatin). These frameworks of understanding are useful, but ultimately serve as placeholders for a more physical description of the underlying molecular features that give rise to these different properties.
In vitro, various cations promote self-association of chromatin, resulting in its precipitation from solution (Hansen, 2002). Initial analyses of precipitates of purified native chromatin using small angle x-ray scattering (Widom, 1986), in addition to contemporary structural analyses (Bednar et al., 2017; Schalch et al., 2005), suggested that nucleosomes are organized into a two-start polynucleosome fiber with ~30 nm diameter. However, subsequent experimentation has revealed contaminants in native chromatin preparations spaced at 30 nm increments (Nishino et al., 2012) and a lack of higher-order structure in electron tomograms of nuclei (Chen et al., 2016; Ou et al., 2017), calling into question 30 nm fiber-based models of chromatin organization. These confounding data, plus recent observations of cation-induced spherical aggregates of nucleosome arrays (Maeshima et al., 2016b), views of nuclear chromatin as liquid-like (Dubochet et al., 1986; Maeshima et al., 2016a), and theoretical treatments of chromatin compartmentalization by phase separation (Di Pierro et al., 2016; Falk et al., 2019; Lieberman-Aiden et al., 2009) suggest that alternative models for chromatin organization should be explored.
Liquid-liquid phase separation (LLPS) driven by weak, multivalent interactions between macromolecules is believed to play an important role in organizing eukaryotic cells on nanometer to micrometer length scales (Banani et al., 2017; Brangwynne et al., 2009; Li et al., 2012; Shin and Brangwynne, 2017). Chromatin resembles many macromolecules known to undergo LLPS; it is a highly-valent array of nucleosomes that can interact in diverse ways with multiple partners including itself (Banani et al., 2017). In this light we asked here what the physical nature of cation-driven chromatin precipitates might be. We found that physiologic concentrations of cations induce reversible LLPS of reconstituted nucleosomal arrays (chromatin, hereafter), producing dynamic histone tail-dependent droplets with nucleosome concentrations similar to those estimated in cells (Hihara et al., 2012). The linker histone H1 promotes phase separation, increasing the concentration of nucleosomes within droplets and decreasing droplet dynamics, consistent with the role of this protein as a repressive chromatin architectural factor that promotes chromatin condensation in cells. The spacing of nucleosomes every 10n+5 base pairs, which is predominant to 10n spacing in cells, strongly favors phase separation of chromatin, further suggesting that cells use this nucleosome-driven phase separation in regulation and organization of the genome. Histone H1 is more effective at concentrating chromatin with longer linkers, whereas short linkers produce high-concentration condensates in the absence of linker histone, suggesting a potential physical mechanism for the known interplay between nucleosome repeat length and linker histone content across eukaryotes. Histone acetylation causes dissolution of chromatin droplets. Microinjected chromatin forms droplets in the nucleus of cells, which are largely abrogated by chromatin acetylation prior to injection. Highly acetylated chromatin can be induced to re-phase separate by multi-bromodomain proteins, including the transcriptional regulator, BRD4. Droplets formed by acetylated chromatin plus a penta-bromodomain protein associate, but do not coalesce, with droplets of non-acetylated chromatin. These observations illustrate how LLPS could produce functionally distinct but physically adjacent chromatin regions in vivo, and suggest that the intrinsic capacity of the chromatin polymer to undergo LLPS may play important roles in organization and regulation of eukaryotic genomes.
RESULTS
Liquid-liquid Phase Separation of Chromatin in Physiologic Salt
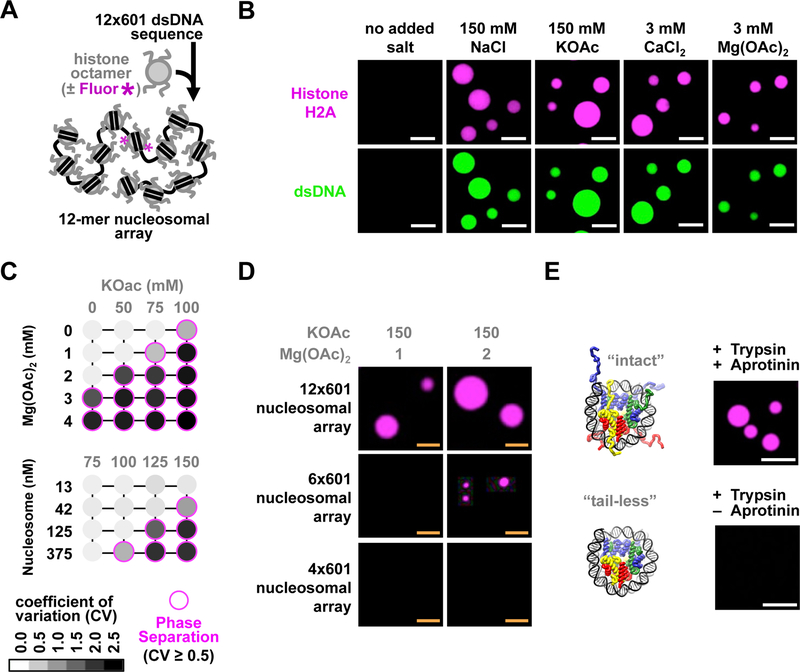
We began by reconstituting chromatin composed of recombinant purified and fluorophore-labeled histone octamers and a defined DNA template containing 12 repeats of Widom’s 601 nucleosome positioning sequence (Figs. 1A, S1A, and S1B). Using confocal fluorescence microscopy, we found that addition of mono- or divalent cations at physiologically relevant concentrations to otherwise homogeneous solutions of chromatin resulted in the formation of round droplets of phase-separated chromatin (Fig. 1B, C). These droplets were dependent on the assembly of DNA and histone octamers into chromatin, and formed without crowding agents, independent of the presence or type of fluorophore label, histone octamer species of origin, and treatment of the microscopy glass (Figs. S1C–E). A characteristic trait of molecules that undergo phase separation is their sharp and reversible transition from a homogeneous solution to immiscible phases at defined threshold concentrations that depend on buffer conditions and are favored by higher molecular valency (Banani et al., 2017). In this regard, we titrated monovalent (KOAc) and divalent (Mg[OAc]2) salts, and varied the number of nucleosomes in each array as well as array concentration. This revealed behavior consistent with phase separation: droplets appeared sharply as salt or chromatin concentrations were increased, and were favored by increasing nucleosome number in the arrays (Figs. 1C, D, and S2AF). To ensure that phase separation of chromatin was not a peculiarity of the assembly process, we designed an alternative approach wherein nucleosomal arrays were generated through ligation of pre-assembled mononucleosomes (Fig. S2D and S2E). Ligation of mononucleosomes by T4 DNA ligase produced cation-dependent phase separated droplets similar to those seen using dodecameric nucleosomal arrays (Fig. S2F).
Figure 1. Phase separation of reconstituted chromatin in physiologic salt.
(A) Assembly of a dodecameric nucleosomal array (chromatin, unless otherwise stated) labeled with a fluorophore (magenta). (B) Fluorescence microscopy images of chromatin labeled on histone H2A with Atto565 (magenta) and dsDNA stained with YOYO-1 (green), following addition of cationic salts. (C) Phase diagram of chromatin (46 base pair internucleosome linker length) under varying conditions. Magenta circles indicate LLPS. Gray scale in each circle indicates coefficient of variation (CV) value calculated from representative images following titration of potassium acetate (KOAc) and either Mg(OAc)2 (top) or chromatin (bottom). (D) Fluorescence microscopy images of chromatin labeled with Alexa Fluor 594 (AF594) with different numbers of nucleosomes at identical total nucleosome concentration (100 nM). (E) (left) Structure of “intact” and “tail-less” nucleosome core particles (PDBID: 1AOI), with and without N-terminal histone tails, respectively. (right) Fluorescence microscopy images of AF594-labeled chromatin following 30 minutes of trypsin digestion. Scale bars, in orange and white, are 4 and 10 μm, respectively.
Nucleosomes associate with one another through a variety of mechanisms, including histone tail-DNA interactions and contacts between the “acidic” and “basic” patches of the nucleosome (Davey et al., 2002; Kan et al., 2009). We asked whether these mechanisms also contribute to LLPS. Similar to previous observations of chromatin precipitation (Fletcher and Hansen, 1995), we found that chromatin without histone tails, generated by partial proteolysis with trypsin (Fig. S2G), do not undergo LLPS in the presence of physiologic salts (Fig. 1E). Additionally, neutralizing mutations of the histone H4 basic patch (H4K16, R17, R19, and K20), but not the H2A/H2B acidic patch (H2A E61, E64, D90, and E92) resulted in chromatin defective in droplet formation (Fig. S3A), suggesting that interactions between the histone H4 basic patch and DNA is an important determinant for LLPS of chromatin.
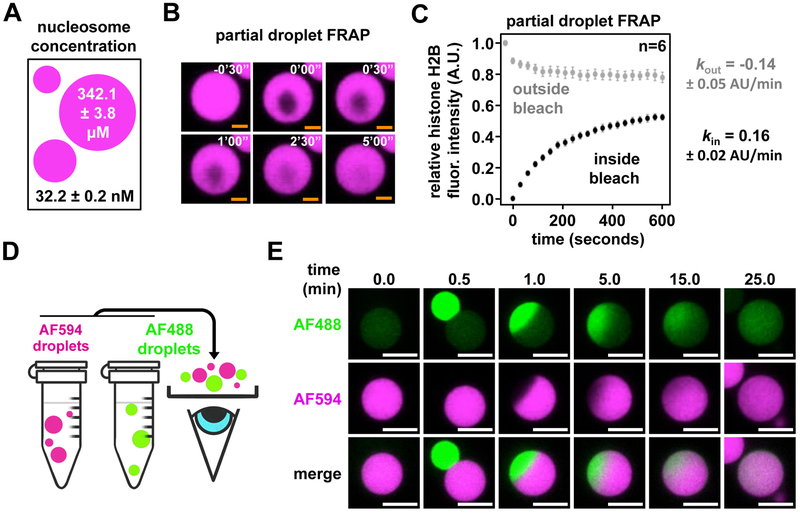
Phase separated polymers can exhibit a variety of material properties, from rigid solids to dynamic liquid-like structures (Shin and Brangwynne, 2017). Chelation of free magnesium by super-stoichiometric addition of EDTA resulted in rapid dispersion of magnesium-dependent chromatin droplets (Fig. S3B), suggesting the droplets are dynamic and exchange cations rapidly with solution. In contrast, however, photobleaching of the labeled histone in entire chromatin droplets resulted in very slow recovery of fluorescence (Figs. S3C, D), an observation more often found in biological molecules that produce solid-like phases. We wondered if the slow fluorescence recovery was due, not to solid-like properties, but rather an absence of material in bulk solution from which to recover. Quantitation of nucleosome concentration in chromatin droplets (~340 μM) and in bulk solution (~30 nM) revealed a >10,000-fold increase in concentration following LLPS, indicating that the inability to recover fluorescence of entire droplets following their photobleaching likely results from a dearth of free material in solution (Fig. 2A and S3E–G). Short time-scale recovery of fluorescence following photobleaching of a portion of the chromatin droplets confirmed the dynamic and liquid-like properties of phase-separated chromatin (Figs. 2B, C). Internal droplet dynamics drive this fluorescence recovery, as fluorescence was lost outside of, and gained within, the irradiated volume following partial photobleaching of fluorescence of droplets at an equal and opposite initial rate (Fig. 2C). Given the very slow exchange of fluorescence with bulk solution, we could directly image droplet fusion and subsequent internal mixing of materials by co-incubating differentially labeled chromatin droplets (Fig. 2D, E, and Movie S1). This revealed that fusion occurred rapidly, with droplets changing from an initial hourglass shape to spherical within ~30 seconds. However, internal mixing was much slower, occurring on timescales of 10–20 minutes. Together, these behaviors indicate that chromatin droplets have high surface tension and high viscosity.
Figure 2. Chromatin droplets are highly concentrated and liquid-like.
(A) Graphical depiction of nucleosome concentrations within chromatin droplets formed by chromatin with 46 base pair internucleosomal linker lengths. See Figs. S3E–G for details. (B) Microscopy images of fluorescence recovery following partial photobleaching of AF594-labeled chromatin droplets. (C) Quantification of average relative fluorescence intensity and its initial rate of change both inside (black, kin) and outside (grey, kout) the area of photobleach across 6 individual chromatin droplets. Error bars and ± error are standard deviation. (D) Experimental workflow for two-color droplet mixing assay. (E) Fluorescence microscopy images of chromatin droplets labeled with either Alexa Fluor 488 (AF488) or AF594 fusing. Scale bars, in orange and white, are 4 and 10 μm, respectively.
Linker Histone H1 Promotes Phase Separation, and Slows Dynamics and Increases Concentration of Droplets
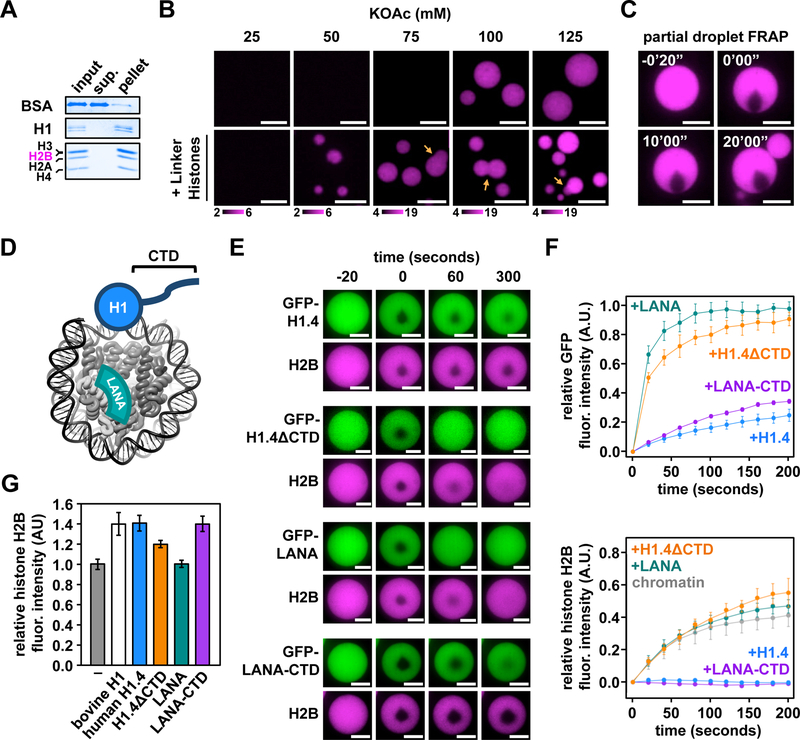
The most abundant chromatin-binding protein in the majority of eukaryotes is the general architectural protein linker histone H1. Histone H1 binds at the dyad axis of the nucleosome and regulates genomic access, gene regulation, and condensation of the genome in cells (Bednar et al., 2017; Carruthers et al., 1998; Hergeth and Schneider, 2015; Shen et al., 1995). Given recent reports that the lysine-rich C-terminal tail of histone H1 can phase separate when mixed with DNA (Turner et al., 2018), we wondered how binding of histone H1 (Fig. 3A) might affect the phase separation of chromatin and material properties of the resulting droplets. Addition of purified calf thymus histone H1 to chromatin promoted phase separation at half the concentration of monovalent salt compared to chromatin alone (Fig. 3B). Imaging of histone H1-bound chromatin droplets revealed the presence of unresolved fusion intermediates (Fig. 3B) and no observable recovery from photobleaching (Fig. 3C), indicating that linker histone binding results in decreased dynamics within chromatin droplets.
Figure 3. The C-terminal domain of histone H1 promotes phase separation of chromatin with altered material properties.
(A) Coomassie brilliant blue-stained SDS-PAGE gel of proteins in supernatant (sup.) or pellet following sedimentation of chromatin droplets containing bovine linker histones. (B) Fluorescence microscopy of AF594-labeled chromatin following titration of potassium acetate with (bottom) or without (top) bovine linker histones. Enumeration of pixel intensities is indicated below each buffering condition and orange arrows indicate stalled droplet fusion intermediates. (C) Microscopy images of fluorescence recovery of AF594-labeled chromatin in the presence of bovine linker histones following partial droplet photobleaching. (D) Schematic depicting approximate nucleosome binding sites of LANA peptide (teal) and histone H1 (blue) relative to the nucleosome core particle (PDBID: 1AOI). (E) Microscopy images of GFP fusion proteins of human histone H1.4, LANA peptide, and H1.4 fragments bound to AF594-labeled chromatin droplets before and after partial droplet photobleaching. Images were processed separately for each experimental condition. So unlike data for panel G, relative brightness is not comparable between conditions. (F) Quantitation of relative fluorescence recovery of GFP fusion proteins (above) and AF594-labeled H2B (below) following partial droplet photobleaching. (G) Quantification of relative fluorescence intensity of droplets of chromatin alone and in the presence of bovine linker histones, or unlabeled recombinant human histone H1.4, LANA peptide, or H1.4-fragments. Error bars indicate standard deviation (N = 6 droplets in each case). Scale bars are 10 m.
To better understand the mechanisms underlying these effects we examined the largely globular N-terminal region and disordered lysine-rich C-terminal domain (CTD) of histone H1 independently (Fig. 3D). We purified recombinant proteins with and without monomeric green fluorescent protein (GFP) fused to: human histone H1.4 (GFPH1.4), H1.4 lacking the CTD (H1.4ΔCTD), a nucleosome targeting peptide from the N-terminus of Kaposi’s sarcoma-associated herpesvirus protein LANA (GFP-LANA), and a fusion protein between the LANA peptide and the CTD of H1.4 (GFP-LANA-CTD) (Figs. S3H,I). Structural analyses of histone H1-nucleosome complexes show that the globular portion of the N-terminal domain is well ordered at the nucleosome dyad axis, where linker DNA emanates from the nucleosome core particle. The CTD of histone H1 binds linker DNA more distal to the dyad axis, but may not adopt a discrete bound conformation (Bednar et al., 2017).
Both calf thymus histone H1 and unlabeled recombinant human H1.4 cause increased concentration (~1.4-fold) and decreased dynamics of chromatin droplets independent of fluorophore labeling strategy (Figs. 3C, 3E–G, S3J). H1.4ΔCTD causes a modest increase in concentration of chromatin droplets (~1.2-fold), but does not affect their photobleaching recovery rate. In contrast, a LANA-CTD fusion protein (but not LANA peptide alone) increases the density of the resultant droplets and greatly decreases the rate of droplet recovery after photobleaching (Figs. 3E–G). Thus, it appears that the effects of histone H1 derive largely from the disordered CTD, with the folded domain serving in this context primarily to recruit this lysine-rich region of the molecule to the nucleosome surface. Note that we do not yet understand how the structural and dynamic changes in individual nucleosomes induced by histone H1 binding (Bednar et al., 2017), assuming they occur in the phase separation context, translate to the changes in macroscopic properties of chromatin droplets observed here.
Together, these data indicate that histone H1 promotes phase separation of chromatin, with increased concentration of material and decreased dynamics in the droplets. Our biochemical data parallel cellular observations that histone H1 depletion in eukaryotic cells results in a loss of chromatin cohesion and increased nuclear volume (Shen et al., 1995).
Nucleosome Spacing Controls Phase Separation and Chromatin Condensation
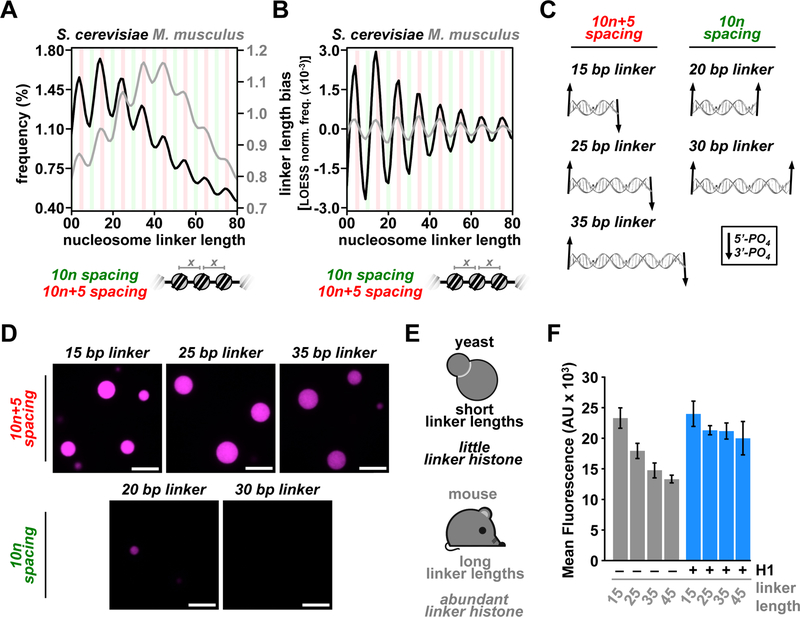
The exact location of nucleosomes across the genome are the combined result of sequence-dependent deposition, movement by ATP-dependent remodeling enzymes, and spatial constraint exerted by adjacent DNA binding factors (Struhl and Segal, 2013). While it remains unclear how these distinct mechanisms contribute, it was demonstrated 40 years ago that across a wide variety of eukaryotes, internucleosomal distances are biased toward 10n+5 (e.g. 5, 15, 25) base pair spacing, and depleted for 10n (e.g. 10, 20, 30) spacing (Lohr and Van Holde, 1979). Figure 4A shows the frequency of internucleosome linker lengths in yeast and mouse (Brogaard et al., 2012; Voong et al., 2016). Figure 4B shows the same data following LOcally Estimated Scatterplot Smoothing (LOESS) normalization (Fig. S4A), highlighting the extent of 10n+5 bias irrespective of differences in average linker length between these organisms. Structural analyses have shown that 10n linker lengths orient every other nucleosome in arrays such that they engage in face-to-face nucleosome “stacking” interactions (Schalch et al., 2005; Song et al., 2014). This orientation results in the formation of a two-start helix that is consistent with the proposed and canonical 30 nm fiber (Hansen, 2002). Because double-stranded DNA rotates 360° every ~10.4 base pairs, the 10n+5 spacing results in a ~180° difference in the orientation of DNA ends, and consequently in the orientation of nucleosomes (Fig. 4C). Thus, 10n+5 linker length spacing disfavors the two-start helical arrangements found in 30 nm-like structures (Bass et al., 2019; Nikitina et al., 2017; Schalch et al., 2005; Song et al., 2014), suggesting that a different molecular association might arise from the linker lengths preferred in cells.
Figure 4. Physiologic spacing of nucleosomes drives LLPS of chromatin and modulates chromatin droplet density.
(A) Genome-wide analyses of internucleosome linker lengths in yeast (black) and mouse ES cells (gray) at base pair resolution. (B) Data in (A) following LOESS normalization to quantify the extent of linker length bias. (C) End orientation trajectories (5’ to 3’ of terminal phosphates) of idealized B-form DNA with 10n+5 or 10n base pair distances. (D) Fluorescence microscopy images of AF594-labelled chromatin (0.5 μM nucleosome) with the indicated internucleosome linker lengths (10n+5 series above, 10n series, below), following addition of 150 mM KOAc. (E) Graphical depiction of internucleosome linker length and linker histone expression differences between mouse and yeast. (F) Fluorescence intensity within droplets (n=6) composed of 10n+5-spaced chromatin with different internucleosome linker lengths both with and without binding of bovine linker histone H1. Nucleosome length for analysis is assumed to be 147 bp and scale bars are 10 μm.
To understand how internucleosome linker lengths affect LLPS of chromatin, we compared the propensity of chromatin with either 10n+5 or 10n nucleosome spacing to phase separate. We found that 10n+5 chromatin forms droplets at appreciably lower salt than 10n chromatin (Figs. 4D and S4B). Thus, the 10n+5 spacing favors phase separation, while the 10n spacing disfavors it. Droplets formed from 10n+5-spaced chromatin also had ~1.8-fold higher fluorescence intensity than 10n chromatin (15 vs 20 bp linker lengths; Figs. 4D, S4B, C), indicating that 10n+5 internucleosome spacing results in a different intermolecular arrangement and an appreciably higher intrinsic condensation activity. These observations indicate that 10n and 10n+5 spacing of nucleosomes, the latter of which is preferred in cells, encode distinct archetypes of higher order nucleosome organization, inherent to the chromatin polymer.
Across a number of organisms, cell types, and biochemical reconstitutions, the expression level of linker histone and internucleosome linker length are linearly related (Woodcock et al., 2006). For example, yeast express little linker histone and have short internucleosome spacing, while mammals such as mice express linker histone at levels nearly stoichiometric with nucleosomes, and have long internucleosome linkers (Figs. 4A, E), (Hergeth and Schneider, 2015). We asked whether there are biochemical relationships between linker length and histone H1 in the context of phase separation, which might be associated with these biological relationships. In the 10n+5 linker series, we found that shorter linkers produce higher concentrations of chromatin within phase-separated droplets (Fig. 4D). Above, we showed that linker histone H1 also increases the droplet concentration of chromatin formed with 46 base pair linker lengths (Fig. 3G). We examined whether linker histone H1 has the same effect on chromatin formed with shorter linker lengths. We found that addition of histone H1 to chromatin with different internucleosomal linker lengths causes the density of all droplets to increase to a common value (Figs. 4F and S4D), with no significant increase in condensation for chromatin with 15 base pair linkers and ~1.5-fold increase for 45 base pair linkers. These results suggest that cell types or organisms with short linkers might require less linker histone, since high levels of condensation are achieved in an intrinsic manner through nucleosome spacing. However, higher eukaryotes with longer linkers might rely more on histone H1 to achieve high chromatin condensation and regulate compaction properly. Such a model could also explain observations that histone H1 depletion results in a global shortening of linker lengths and expanded nuclear volume (Fan et al., 2005; Shen et al., 1995).
Disruption of Chromatin Droplets by Histone Acetylation
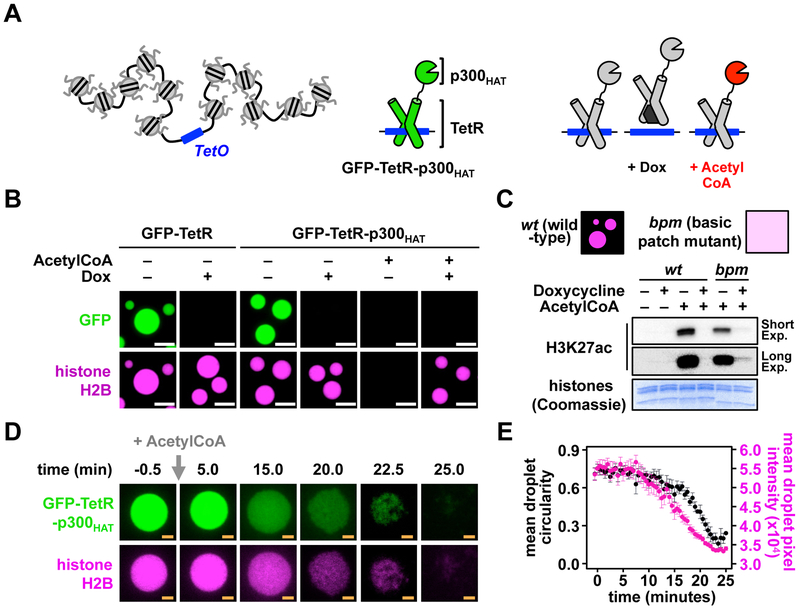
Nucleosome histone tails are acetylated in vivo, often by histone acetyltransferase enzymes recruited by transcription factors to specific loci, in order to regulate gene expression (Brownell et al., 1996; Grunstein, 1997). These modifications impair self-interaction and precipitation of chromatin in vitro (Allahverdi et al., 2011; Shogren-Knaak et al., 2006), similar to basic patch mutations. To examine how acetylation might alter the formation and material properties of chromatin droplets, we devised a model system to mimic transcription factor-driven histone acetylation. We genetically linked the model E. coli transcription factor, Tet Repressor (TetR), to the catalytic domain of the relatively promiscuous histone acetyltransferase, p300 (p300HAT) and GFP, and combined this fusion protein (GFP-TetR-p300HAT) with chromatin containing a central Tet Operator (TetO) (Fig. 5A). In this system, the tetracycline analog, doxycycline (Dox), inhibits transcription factor binding to the chromatin, and Acetyl-CoA is necessary for histone acetylation by p300HAT. GFP-TetR and GFP-TetR-p300HAT were both strongly recruited to TetO-containing chromatin droplets, an effect that was blocked by Dox (Fig. 5B). Addition of Acetyl-CoA caused dissolution of chromatin droplets containing GFP-TetR-p300HAT, concomitant with acetylation of H3K27 and likely other sites as well (Fig. 5C). This effect required recruitment of GFP-TetR-p300HAT into the droplets, as acetylation and droplet dissolution were both blocked by Dox (Figs. 5B, C). By comparing GFPTetR-p300HAT-mediated histone acetylation of phase separated wild type chromatin and non-phase separating basic patch mutant chromatin we found that wild type has both increased transcription factor-dependent acetylation (-Dox), and decreased transcription factor-independent acetylation (+Dox). While we cannot exclude phase separation-independent alterations in histone acetylation as a result basic patch mutation, or that other histone lysines might exhibit different patterns of acetylation, these results suggest that condensation by LLPS enhances the fidelity of this signaling pathway (Fig. 5C). Time-resolved imaging of acetylation-mediated dissolution of chromatin droplets (Fig. 5D and Movie S2) shows that following a short delay after reaction initiation, the density of droplets (assessed by fluorescence intensity) progressively decreases until the structures disappear. Droplets maintain their size and approximate shape through the early stages of this process until density (i.e. fluorescence intensity) decreases to roughly half its initial value, at which point they begin to crumple and lose circularity (Fig. 5E). These behaviors show that acetylation of histone tails can tune the density and material properties of chromatin droplets in vitro, illustrating a potential physical mechanism by which “open” chromatin might be formed in cells (see also below for the effects of bromodomain proteins). It remains unknown which tail sites are most important for these effects, and whether more specific histone acetyl transferases might have greater or lesser effects on chromatin LLPS.
Figure 5. Histone acetylation dissolves chromatin droplets.
(A) A TetO-containing chromatin and small-molecule modulated model transactivating protein GFP-TetR-p300HAT. (B) Fluorescence microscopy images of AF594-labeled chromatin (magenta) and GFP fused to either the model transcription factor TetR (GFP-TetR) or TetR fused to the catalytic domain of p300 (GFP-TetR-p300HAT) (both green) including doxycycline (Dox) and/or AcetylCoA. (C) (top) Western blot of histone H3K27 acetylation following addition of doxycycline and/or AcetylCoA to TetO-containing chromatin composed of wild-type or basic-patch mutant histones in the presence of GFP-TetR-p300HAT and 150 mM KOAC. (bottom) Coomassie brilliant blue-stained SDS-PAGE gel of core histone proteins. (D) Fluorescence microscopy of AF594-labeled chromatin (magenta) and GFPTetR-p300HAT (green) following addition of AcetylCoA. (E) Mean droplet circularity and pixel intensity of AF594-labeled chromatin droplets in the presence of GFP-TetR-p300HAT following addition of AcetylCoA. Error bars indicate standard error. Scale bars, in orange and white, are 4 and 10 μm, respectively.
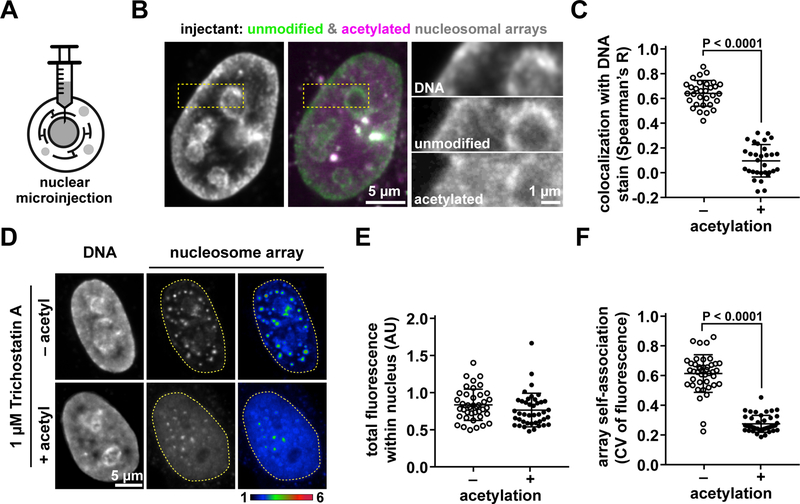
We next investigated how liquid phase separation of nucleosome arrays might relate to chromatin organization in cells. Co-injection of unmodified and pre-acetylated nucleosome arrays carrying different fluorescent labels, respectively, into nuclei of live HeLa cells (Fig. 6A) resulted in accumulation of unmodified nucleosome arrays in DNA-dense areas near the nucleolus and nuclear envelope (Figs. 6B, C, S5A, B). Acetylated nucleosome arrays distributed much more homogenously throughout the nucleus, enriching only slightly in DNA-dense regions (Figs. 6B, C). Thus, non-acetylated nucleosome arrays adhere more strongly than acetylated arrays to chromatin in cells, consistent with their greater propensity to self-associate and phase separate in vitro.
Figure 6. Nucleosomal arrays form condensates in the nucleus of cells.
(A) Nuclear microinjection of fluorophore-labeled nucleosomal arrays into cultured cells. (B) (left) Confocal live-cell fluorescence microscopy of Hoechst 33342 DNA stained HeLa cell nuclei injected with (middle) unmodified nucleosomal arrays (green) and acetylated nucleosomal arrays (magenta). (right) Close up view of DNA, unmodified arrays, and acetylated nucleosomal arrays from orange dotted box of confocal fluorescence microscopy image. (C) Spatial correlation of mean fluorescence intensity from 31 cells across two biological replicates between Hoechst 33342 DNA stain and either unmodified AF488-labeled arrays or acetylated AF594-labeled arrays. (D) Confocal live-cell fluorescence microscopy of unmodified nucleosomal arrays and acetylated nucleosomal arrays injected into Hoechst 33342-stained HeLa cell nuclei following 3 hours of treatment with Trichostatin A. Quantitation from 42 cells (mean fluor. > 0.5 AU) across two biological replicates (E) mean nuclear fluorescence and coefficient of variation (CV) (F) for injected unmodified and acetylated nucleosomal arrays in the nuclei.
Nucleosome arrays injected into the nuclei of untreated cells did not self-associate into detectable droplets. We suspected that this might be due to their rapid adsorption into existing chromatin architecture. If that were the case, then suppression of the interactions and architecture of endogenous chromatin compaction might enable self-association of injected nucleosome arrays into separate droplets. To test this, we incubated cells in Trichostatin A to induce histone hyperacetylation and chromatin decondensation (Toth et al., 2004) before injecting nucleosome arrays (Figs. S5C, D). Under these conditions, unmodified nucleosome arrays formed condensed foci with low levels of soluble background (Figs. 6D–F). Nucleosome arrays that were acetylated before injection distributed more uniformly throughout the nucleus, forming few and dim condensates. Following Trichostatin A treatment, microinjected unmodified nucleosome arrays remained more significantly correlated with nuclear DNA than acetylated arrays, though to a lesser extent than in untreated cells (Fig. S5E, F). This is consistent with our interpretation that acetylation of lysines on histone tails modifies the adherent properties of chromatin both in vitro and in cells. Thus, nucleosome arrays retain their intrinsic ability to phase separate, in an acetylation-dependent manner, within the complex environment of the nucleus.
Multi-Bromodomain Proteins Can Induce a New Liquid Phase of Acetylated Chromatin
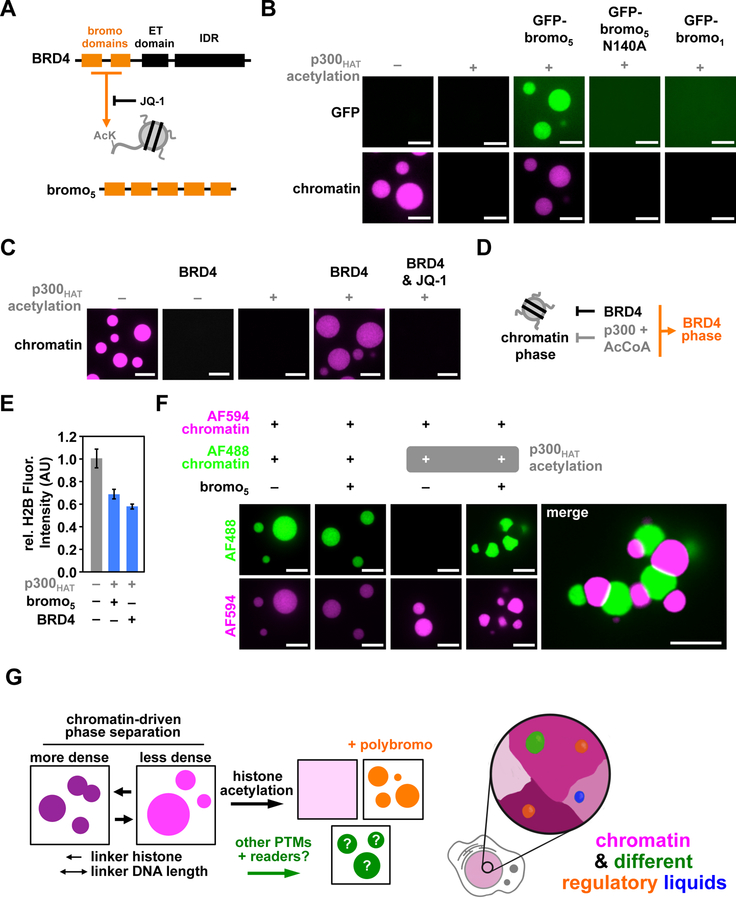
Acetyllysine-modified histone tails can be recognized specifically by the bromodomain, a modular ~12 kDa bundle of four α-helices that is found in numerous chromatin associated proteins of diverse functions (Fujisawa and Filippakopoulos, 2017). Many such proteins contain multiple bromo domains and/or are oligomeric, suggesting that they could interact with acetylated chromatin in multivalent fashion. Since interactions between multivalent macromolecules canonically drive LLPS, we asked whether multi-bromodomain proteins could induce LLPS of acetylated chromatin. We examined both an engineered protein consisting of five copies of the first bromodomain of BRD4 separated by flexible linkers, bromo5, and BRD4, a protein with two bromodomains that plays important roles in organizing chromatin and regulating transcription (Figs. 7A, S6A, B) (Fujisawa and Filippakopoulos, 2017). Addition of either bromo5 or BRD4 to acetylated chromatin (that cannot phase separate on its own) in physiologic buffer indeed induces LLPS (Figs. 7B, C). This effect depends on binding of bromodomains to acetyl lysine, since a mutant bromo5 that cannot bind this moiety does not cause phase separation (Fig. 7B), and BRD4-induced phase separation is blocked by the bromodomain inhibitor drug, JQ1 (Filippakopoulos et al., 2010) (Fig. 7C). LLPS also depends on multivalency, as a mono-bromodomain protein does not induce LLPS (Fig. 7B). The resulting droplets are liquid-like, showing fusion (Figs. S6C, D) and FRAP recovery (Figs. S6E, F), but have lower chromatin density and altered FRAP recovery compared to droplets of unacetylated chromatin (Fig. 7E, S6F). Thus, multivalent bromodomain proteins can induce LLPS of acetylated chromatin, producing liquid phases with different compositions and dynamics, which could enable different functions.
Figure 7. BRD4 promotes a new liquid phase of acetylated chromatin.
(A) Schematic illustration of the domain organization of BRD4 and bromo5 and their JQ-1-sensitive interactions with acetyllysine. (B) and (C) Fluorescence microscopy images of AF594-labeled chromatin (magenta) without and with acetylation by the catalytic domain of p300HAT and (B) with synthetic GFP-labeled bromodomain-containing proteins and (C) with BRD4 without and with JQ-1. (D) Schematic depicting the effects of BRD4 on nonacetylated and acetylated chromatin. (E) Relative H2B fluorescence intensity of chromatin droplets composed of chromatin alone and BRD4 or bromo5 and acetylated chromatin. (F) Fluorescence microscopy of unmodified AF594-labeled chromatin mixed in stoichiometric quantities of either unmodified or acetylated AF488-labeled chromatin with and without unlabeled bromo5. Scale bars are 10 μm. (G) Model for phase separation-based organization of chromatin in nuclei.
We next asked how these bromodomain-induced droplets of acetylated chromatin relate to those produced by unmodified chromatin. When bromo5 is added to a solution of acetylated and non-acetylated chromatin labeled with different fluorophores, two distinct phases are formed (Fig. 7F). One phase is enriched in the acetylated chromatin and the other is enriched in the non-acetylated chromatin. These distinct droplets adhere to each other, but do not coalesce. An analogous experiment with BRD4 yielded only a single droplet phase containing both acetylated and non-acetylated chromatin (Fig S6G). This difference is likely due to the ability of BRD4 to dissolve droplets of non-acetylated chromatin (Fig. 7C). Although we do not yet fully understand the molecular basis for these differences, the data on bromo5 demonstrate how, in principle, spatially and functionally distinct chromatin regions could be created by LLPS in vivo based on the inherent properties of chromatin, covalent modifications of histone tails and the actions of histone tail reader proteins.
DISCUSSION
We have demonstrated that in the presence of physiologic salt, reconstituted chromatin has an intrinsic ability to form a highly compact, yet dynamic, liquid phase. The density of the chromatin droplets (~340 μM nucleosome concentration for 46 base pair linkers) is similar to estimates of nucleosome density in cells (~80–520 μM) (Hihara et al., 2012), indicating that LLPS is sufficient produce the degree of compaction necessary to organize the genome in the nucleus. Factors known to affect chromatin properties in cells have parallel effects on properties of the droplets. Histone H1 increases the density of droplets and decreases their dynamics, mirroring its chromatin compaction activity in cells (Hergeth and Schneider, 2015). 10n+5 base pair internucleosome spacing, which predominates in cells (Brogaard et al., 2012; Lohr and Van Holde, 1979; Voong et al., 2016), favors chromatin LLPS and produces droplets whose densities decrease with linker length. Histone H1 addition increases all droplets to a common density, with a greater effect on droplets with longer linkers. These data suggest that correlations between nucleosome linker length and histone H1 expression levels across many eukaryotes (Fan et al., 2005; Woodcock et al., 2006), may reflect cellular efforts to maintain homeostasis of genome compaction. Finally, acetylation of histone tails, which can cause decompaction of chromatin in cells and is associated with “open” chromatin, decreases droplet density and abrogates LLPS at high levels. Multi-bromodomain proteins can induce a distinct droplet phase of acetylated chromatin, consistent with reports that transcriptional regulators recruited to acetylated enhancers form a phase separated structure important for gene expression (Boija et al., 2018; Cho et al., 2018; Sabari et al., 2018). Together, these observations suggest eukaryotic cells may utilize LLPS to dynamically control the compaction, spatial organization and function of the genome. This idea is consistent with various models of liquid-like organization of the genome (Dubochet et al., 1986; Maeshima et al., 2016a; Mirny et al., 2019; Nozaki et al., 2017), and suggest that previous reports of in vitro chromatin aggregates (Maeshima et al., 2016b; Shogren-Knaak et al., 2006) can now be described as dynamic liquid-liquid phase separated droplets.
Compartmentalization on various length scales is a hallmark of eukaryotic genomes. While long-range interactions within the chromatin fiber and short-range interactions by ATP-dependent loop extrusion have emerged as key organizing principles of the genome (Di Pierro et al., 2016; Mirny et al., 2019), it has remained unclear how the intrinsic physical properties of chromatin govern compartmentalization. We have shown that liquid chromatin droplets fuse rapidly, but the rate of content mixing is very slow. As phase separated polymers increase in length, the intrinsic viscosity of their solutions increases as well (Rubinstein and Colby, 2003). This suggests that chromatin with length scales longer than the 12-nucleosome arrays used here would occupy distinct regions from one another for longer periods of time following fusion. The slow merging of chromatin phases could enable epigenetic processes (e.g. covalent modifications and chromatin binding proteins) to reinforce differences in organization/dynamics/etc. between them, allowing establishment and maintenance of chromatin states. At shorter length scales, the liquid chromatin state might facilitate the lateral mobility of the chromatin fiber during cohesin-mediated loop formation, thus contributing to the formation of topologically associated chromatin sub-domains (Bintu et al., 2018; Mirny et al., 2019; Schwarzer et al., 2017; Uhlmann, 2016). At the length scale of a chromosome, a highly viscous and liquid-like organization could contribute to the maintenance of individual chromosome territories within the nucleus (Meaburn and Misteli, 2007). In this way regions of the genome would remain dynamic on shorter length scales while maintaining their spatial integrity on longer length scales within the nucleus.
The relationship between nucleosome spacing and density in chromatin droplets, and the model that nucleosome spacing tunes chromatin condensation, has implications for genome organization and sequence content in eukaryotes. DNA sequences phased in TA:AT:TT:AA dinucleotide base pairs position nucleosomes in vitro and in vivo (Kaplan et al., 2009; Lowary and Widom, 1998; Struhl and Segal, 2013). In simpler eukaryotes such as yeast, and to a lesser extent in higher eukaryotes, this pattern underlies the position of nucleosomes in cells (Brogaard et al., 2012; Kaplan et al., 2009; Voong et al., 2016). Our data on the biochemical behaviors afforded by 10n+5 vs 10n spacing, coupled with the genome-wide preference for 10n+5 spacing, suggests that chromatin condensation might also be encoded into genomic sequence, especially in simpler organisms. It is thus likely that nucleosome positioning sequences across the genome, which produce and tune length-dependent condensation, are under evolutionary selection and constrain genomic sequence. Moreover, our data highlight the need to understand how genomic sequence and ATP-dependent processes coordinate to give rise to 10n+5-spaced nucleosome spacing, and how disruption of this organization effects chromatin regulation and structure in cells.
In the presence of the multivalent bromodomain-containing reader protein, BRD4, or the engineered protein, bromo5, histone acetylation can give rise to a phase separated liquid distinct from that of unmodified chromatin. These data have specific implications for acetylation-dependent processes such as transcription and general implications for the functional organization of the genome (see below). In the former, high levels of lysine acetylation are a hallmark of genomic sites of active transcription (Brownell et al., 1996; Consortium, 2012; Whyte et al., 2013). These sites form 100–500 nm foci in the nucleus where the transcriptional machinery is concentrated, including RNA polymerase II, mediator, transcription factors and BRD4. Smaller foci form and dissolve rapidly, with average lifetimes of ~10 seconds. Larger foci are > 10-fold longer lived, and appear to be super enhancers. While interpretations differ (Chong et al., 2018), recent data have suggested models for transcriptional focus assembly through LLPS, driven by networks of weak associations among transcription factors and coactivator proteins (Cho et al., 2016; Cho et al., 2018; Sabari et al., 2018). These models have cast chromatin itself as a largely passive platform on which phase separated puncta assemble. Our findings would rather suggest that acetylated chromatin may also play a direct role in the sharp formation and dissolution of transcriptional condensates through phase separating with bromodomain-containing proteins such as BRD4. It remains unknown whether acetylation and deacetylation of chromatin at active loci occur on the same timescale as formation and dissolution of transcriptional foci, or whether the active loci remain persistently acetylated as the transcriptional apparatus assembles and dissolves. In both scenarios, our data suggest that the structural organization of the genome could be changing dynamically at sites of transcription through chromatin-based LLPS.
In a phase separation-based model for cellular control of chromatin organization and regulation (Fig. 7G), compaction of the genome and the drive to phase separate can be tuned through engagement of cellular factors such as linker histone binding, histone acetylation, interactions with histone tail readers, and spacing of nucleosomes. In cells, there are defined functional chromatin sub-types (e.g. promoters, enhancers, insulators, polycomb group regions, etc.) that are enriched in specific signaling molecules and chromatin binding proteins (Consortium, 2012; Filion et al., 2010). Analysis of genome-wide datasets reveals differing degrees of 10n+5 nucleosome positioning bias, histone H1 binding, and histone acetylation at these regions (Fig. S7). This suggests that these chromatin sub-types may adopt different phase separated states, with specifically tuned structural and dynamic properties important to their unique functions in cells. Our results demonstrate that chromatin can form adherent but non-coalescing chromatin phases based on differential tail modifications and binding partners, and illustrate how such distinct phases could form in the nucleus. Various classes of histone modifications— acetylation, methylation, phosphorylation, ubiquitination, SUMOylation, etc.—with or without cognate reader proteins, as well as the numerous histone variants, may give rise to different liquid phases with unique physical properties, compositions and consequent functions. This chromatin phase separation then could act together with the recently demonstrated phase separation of proteins that are unique to different chromatin sub-types (Larson et al., 2017; Plys et al., 2019; Sabari et al., 2018; Strom et al., 2017) to produce the full regulatory capacity and functionality of these regions. Consistent with this conceptual framework, recent efforts to recapitulate whole-genome chromatin interaction maps in silico have invoked phase separation, driven by weak but differential interactions of chromatin and associated factors, to account for the formation and segregation of chromatin sub-domains (Di Pierro et al., 2016; Falk et al., 2019). Thus, phase separation is a mechanism that could produce a compact yet dynamic “ground state” of chromatin, and enable generation of numerous “excited” structural states through covalent modifications and regulatory factors, providing the functional diversity of the genome.
STAR METHODS
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michael K. Rosen (michael.rosen@utsouthwestern.edu). Plasmids generated in this study are available by contacting the Lead Author with no restrictions. Requests for cell lines described in Cuylen et al. (2016) should be directed to Daniel Gerlich (daniel.gerlich@imba.oeaw.ac.at) and are available with no restrictions.
Experimental Model and Subject Details
Bacterial Strains
DH5α (Invitrogen) and MACH1 (Invitrogen) E. coli strains were used for passage during cloning of plasmid DNA. Large-scale preparations of plasmid DNA for isolation of nucleosome assembly sequences were passaged through and grown to scale in the ER2925 (dam-/dcm-) E. coli strain (NEB).
Insect Cell Line
Sf9 cells were passaged in SF-900 II serum free medium (GIBCO), which was supplemented with 10% FBS and penicillin/streptomycin during viral expansion and recombinant protein expression.
Mammalian Cell Line
The HeLa Kyoto cell line, which has been regularly tested negatively for mycoplasm contamination, was used for these experiments. HeLa cells were cultivated in Dulbecco’s Modified Eagle Medium (DMEM, produced in-house by the IMP/IMBA media kitchen) supplemented with 10 % (v/v) fetal bovine serum (FBS, Gibco), 1 % (v/v) penicillin-streptomycin (Sigma-Aldrich) and 1 % (v/v) GlutaMAX™ supplement (Thermo Fisher Scientific). For microinjection, cells were cultivated to 80 % confluency in a high-wall 35 mm μ-Dish (IBIDI). Prior to microinjection and imaging, cells were transferred into imaging medium (Gibco, custom-made: DMEM without phenol red and riboflavin, supplemented as described above). 2 hours before imaging, cells were treated with 1.6 μM Hoechst 33342 (Invitrogen) with or without 1 μM trichostatin A (Sigma).
Method Details
Molecular Biology and Cloning
Construction of Bacterial Protein Expression Vectors.
H. sapiens Core Histones:
Synthetic open reading frames (ORFs) encoding H. sapiens histone H3C111A and H2BT116C were amplified from a dsDNA synthesized by Integrated DNA Technologies (IDT) using polymerase chain reaction (PCR) and primers adding a 5’-proximal NcoI restriction endonuclease recognition site and a 3’-proximal stop codon and BamHI restriction endonuclease recognition site. NcoI and BamHI restriction endonucleases from New England Biolabs (NEB) were used to directionally clone the H3C111A and H2BT116C ORFs into pET19b (Novagen). The sequence content of the protein expression vectors pET19b_H3C111A and pET19b_H2BT116C were confirmed by Sanger sequencing.
X. laveis Core Histones:
pET-based protein expression constructs for the expression of wild-type and H3T33C and H2AK120C histone proteins from X. laevis were a generous gift from Dr. Geeta Narlikar. pET-based protein expression constructs for the expression of acidic patch mutant X. laevis histone H2A (H2A E61A, E64A, D90A, and E92A) and basic patch mutant X. laevis histone H4 (H4 K16A, R17A, R19A, K20A) were a generous gift from Dr. Song Tan.
ySIR2:
A synthetic ORF encoding the histone deacetylase domain of S. cerevisiae protein SIR2 was amplified from a dsDNA synthesized by IDT using PCR and primers adding a 5’-proximal translation start codon and a 3’-proximal translation stop codon and XhoI restriction endonuclease recognition site. XhoI restriction endonuclease (NEB) digested PCR product encoding amino acids 87–562 of wild-type SIR2 was cloned into the pETduet-1 expression vector (Novagen) using a XhoI and a blunted NdeI restriction endonuclease digestion site (NEB). The sequence content of the protein expression vector pETduet_ySIR2 was confirmed by sanger sequencing.
p300HAT, GFP-TetR, and GFP-TetR-p300HAT:
p300HAT, GFP-TetR, and GFP TetR-p300HAT were amplified using a one or two-step PCR method from (1) pEB1-sfGFP (Addgene #103983), (2) a synthetic sequence encoding E. coli TetR synthesized by IDT, and (3) a natural sequence encoding for amino acids 649–1026 of H. sapiens p300 (p300HAT) synthesized by IDT (Balleza et al., 2018). Primers used for PCR amplification of ORFs encoding the GFP-TetR and GFP-TetR-p300HAT fusion proteins added (1) a 5’-proximal BamHI restriction endonuclease recognition site, (2) a 3’-proximal translation stop codon followed by a NotI restriction endonuclease recognition site, and (3) a sequence encoding for flexible (GGS)3 linkers in between the domains of fusion proteins. BamHI-HF and NotI-HF restriction endonucleases (NEB) were used to directionally clone restriction endonuclease digested PCR products into the pETduet_ySIR2 expression vector (New England Biolabs) to encode for p300HAT, GFP-TetR, and GFP-TetR-p300HAT with an N-terminal HHHHHHHHHHENLYFQGS sequence. The sequence content of the protein expression vectors pETduet_p300HAT, pETduet_GFPTetR, and pETduet_GFP-TetR-p300HAT were confirmed by Sanger sequencing.
mEGFP-tagged and Untagged Histone H1.4-based Expression Constructs:
dsDNA of a natural sequence encoding of H. sapiens histone H1.4 and a synthetic sequence encoding the first 22 amino acids of Karposi’s Sarcoma Herpesvirus protein LANA were ordered as gene blocks from IDT. One or two-step PCR methods were used to amplify dsDNA that encoding for histone H1.4, histone H1.4 without amino acids 112–219 (H1Δ4 CTD), the LANA peptide (1–22), and a LANA-H1.4CTD chimera with an 11 amino acid glycine and serine-rich linker. Monomeric eGFP was subcloned using PCR into a pMAL-based vector (pMTTH) that produces recombinant produces recombinant proteins containing an N-terminal MBP tag and a C-terminal His6 tag, each separated by a TEV protease recognition sequence (ENLYFQG). H1.4-derived sequences (H1.4, H1.4ΔCTD, LANA, and LANA-CTD) were sub-cloned into both pMTTH and pMTTH+mEGFP and sanger sequenced to confirm correct sequence content.
EGFP-tagged and Untagged Synthetic Bromodomain-containing Expression Constructs:
dsDNA of a synthetic sequence encoding 1 or 5 copies of the wild type or acetyllysine-binding deficient sequence (N140A) of the first bromodomain (AAs 40–168) of H. sapiens BRD4, each separated by (GGS)5 linkers, was ordered as gene blocks from IDT. bromo1, bromo5, and bromo5(N140A) encoding sequences were amplified by PCR from gene block sequences and cloned into the pMTTH vector (described above) with and without an N-terminal EGFP protein tag and sanger sequenced to confirm correct sequence content.
Construction of Insect Cell Protein Expression Vectors.
BRD4:
An ORF encoding the natural sequence of “long” H. sapiens BRD4 was amplified by PCR and cloned into a pFastbac1-derived vector (pAV5B) that encodes for an N-terminal His6 tag and TEV protease recognition sequence.
Construction of 12×601 dsDNA Array-Producing Bacterial Vectors.
193bp Repeat Length TetO-containing 12×601:
The p601 plasmid (Larson et al., 2017), containing a 12×601 array with Tet Operator (TetO) inserted between 601 sequences 6 and 7 cloned in a pCR-Blunt II-TOPO vector background was modified by (1) adding an EcoRV site 3’-proximal to the 12×601 sequence and (2) adding a 3,697 bp SpeI-released fragment from pMD2.G 5’-proximal to the 12×601 sequence using an SpeI restriction endonuclease recognition site. This plasmid was called p12×601.
10n and 10n+5 NRL 12×601 Arrays:
Widom’s 601 sequence DNA was PCR amplified using primers outlined in Table S1. Initial 3×601 DNA arrays were cloned into the BamHI and HindIII sites of pUC19, containing an NheI site between 601–1 and 601–2 and an XbaI site between 601–2 and 601–3. Larger repeat arrays (4×601, 6×601, 10×601, and 12×601) were generated using iterative rounds of digestion, ligation, transformation, and plasmid isolation using the isoschizomer NheI and XbaI restriction sites. 12×601 arrays with 15, 20, 25, 30, 35, or 45 bp linker DNA lengths were then subcloned from pUC19 into the WM530 plasmid (a generous gift from Dr. Tom Muir). Final plasmids for production of 12×601 DNA were named pWM_12×601_15bpLinker, pWM_12×601_20bpLinker, pWM_12×601_25bpLinker, pWM_12×601_30bpLinker, pWM_12×601_35bpLinker, and pWM_12×601_45bpLinker.
Expression and Purification of Recombinant Proteins
Purification of X. laeviss Core Histone Proteins Expressed in E. coli.
Expression:
Recombinant histones from X. Laevis were expressed in E. coli as previously described (Luger et al., 1999).
Purification:
Recombinant histones from X. Laevis were purified from E. coli as previously described (Luger et al., 1999), with some modification. Briefly, washed inclusion bodies containing E. coli-expressed histone proteins were solubilized with XL Unfolding Buffer (20 mM Tris•HCl, pH 7.5, 7 M Guanidinium•HCl, 5 mM ß-Mercaptoethanol) and dialyzed into XL Dialysis Buffer (20 mM Tris•HCl, pH 7.5, 6 M Urea, 5 mM ß-Mercaptoethanol). Histone proteins were purified from soluble dialysate in XL Dialysis buffer by denaturing cation exchange chromatography using a TSKgelSP-5PW (TOROH) column, eluting histone proteins with a linear gradient of 0–1 M NaCl. Fractions containing histones were dialyzed into >18 MΩ H2O and lyophilized prior to histone octamer reconstitution.
Purification of H. sapiens Histones H3C111A, H4, and H2A Expressed in E. coli.
Expression:
An overnight culture of Rosetta 2 (pLysS) E. coli (Novagen) transformed with pET19b_H3C111A, pET28a_H4 (Addgene #42633), or pET28A_H2A.1 (Addgene #42634) plasmids encoding wild-type H. sapiens histones H4 or H2A or mutant H. sapiens histones H3C111A, were grown on an agar plate by re-plating a single transformant on LB supplemented with 100 ng/μL of ampicillin (pET19b_H3C111A) or 35 μg/mL Kanamycin (pET28a-based expression) and 25 ng/μL of chloramphenicol at 37°C (Wilson et al., 2016). The bacterial lawn was suspended in LB supplemented with the appropriate antibiotics, as above, and grown to a density (OD600 nm) of 0.4. Recombinant protein expression was then induced by addition of IPTG to 1 mM for 3 hours at 37°C. The cells were collected by centrifugation, resuspended in Histone Lysis Buffer (50 mM Tris•HCl, pH 8, 150 mM NaCl, 5 mM ß-mercaptoethanol, 1 mM Benzamidine, 100 μM Leupeptin, 100 μM Antipain, 1 μM Pepstatin), and the cellular suspension flash frozen in liquid N2, and stored at −80°C.
Purification:
Histones were purified essentially as previously described (Luger et al., 1999), with some modifications. E. coli expressing histone H4, H2A, or H3C111A resuspended in Histone Lysis Buffer were thawed on wet ice and lysed by multiple passages through an Avestin Emulsiflex-C5 high pressure homogenizer at ~10,000 PSI. Histone-containing inclusion bodies were separated from soluble bacterial lysate by centrifugation in a Beckman Avanti J-26 XPI centrifuge in a JA25.5 rotor at 19,500 RPM. Soluble bacterial lysate was discarded, and inclusion bodies were washed by resuspension and in 25 mL of Inclusion Body Wash Buffer (50 mM Tris•HCl, pH 7.5, 100 mM NaCl, 1% Triton X-100, 1 mM EDTA, 1 mM Benzamidine, 5 mM ß-mercaptoethanol) per liter of bacterial expression followed by pelleting by centrifugation in a Beckman Avanti J-26 XPI centrifuge in a JA25.5 rotor at 19,500 RPM. Inclusion bodies were washed once more with Inclusion Body Wash Buffer and twice more with Inclusion Body Wash Buffer omitting Triton X-100. Inclusion Bodies were soaked for 30 minutes with 167μL DMSO per liter bacterial expression, minced with a spatula, and proteins were extracted for 1 hour by stirring in 5 mL of Histone Unfolding Buffer (20 mM Tris•HCl, pH 7.5, 7M Guanidinium-HCl, 10 mM DTT) per liter of bacterial expression. Extracted unfolded proteins were separated from precipitate by centrifugation in a Beckman Avanti 6 XPI centrifuge in a JA25.5 rotor at 19,500 RPM. Inclusion body proteins were extracted once more for 40 minutes with stirring in 1.7 mL of Histone Unfolding Buffer per liter bacterial expression. Extracted unfolded proteins were separated once more from precipitate by centrifugation in a Beckman Avanti J-26 XPI centrifuge in a JA25.5 rotor at 19,500 RPM.
Pooled unfolded soluble inclusion body proteins were filtered through a 0.45 μm membrane (GE Healthcare) and run in Histone Unfolding Buffer over a HiLoad 26/60 Superdex 200 pg size exclusion column. Fractions containing histone proteins were dialyzed twice against 5 mM ß-mercaptoethanol in >18 MΩ H2O and once in 50% [w/v] glycerol. Precipitated proteins were pelleted by centrifugation in a Beckman Avanti J-26 centrifuge in a JA25.5 rotor at 19,500 RPM. Histone proteins in Histone Storage Buffer (50% [w/v] glycerol, 5 mM ß-mercaptoethanol) were concentrated in centrifugal concentrators (Amicon) with a 3,000 dalton molecular weight cutoff (MWCO) and stored at −20°C.
8 M Urea was deionized by 3 passages through Amberlite MB-20 resin (SIGMA) and used to make SAU200 (20 mM NaOAc, pH 5.2, 7 M Urea, 200 mM NaCl, 1 mM EDTA, 5 mM ß-mercaptoethanol) and SAU600 (20 mM NaOAc, pH 5.2, 7 M Urea, 600 mM NaCl, 1 mM EDTA, 5 mM ß-mercaptoethanol) Buffer. Histone proteins in Histone Storage Buffer stored at −20°C were diluted in >20 volumes of SAU200, filtered through a 0.45 μm membrane (GE Healthcare), and applied to Source 15S Chromatography Resin (GE Healthcare) equilibrated in SAU200. After washing with SAU200 and a 5 column volume gradient of 0–30% SAU600, histone proteins were eluted over a 35 column volume gradient of 30–100% SAU600. Fractions containing histone proteins were pooled and dialyzed three times against 5 mM ß-mercaptoethanol in >18 MΩ H2O. Histone proteins were concentrated in centrifugal concentrators with a 3,000 dalton MWCO to less than 500 μL per liter bacterial expression and concentration quantified by measuring histone protein absorbance at 280 nm and the calculated molar extinction coefficients (https://web.expasy.org/protparam/) for histones H3C111A, H4, and H2A of 4470/M•cm, 5960/M•cm, and 4470/M•cm, respectively. Purified histone proteins were aliquoted in 200 (histones H3C111A or H4) or 240 nmol (histone H2A) quantities, flash frozen with liquid N2, and stored at −80 °C.
Purification of Fluorophore-labeled H. sapiens Histone H2B Expressed in E. coli.
Expression:
An overnight culture of Rosetta 2 (pLysS) E. coli (Novagen) transformed with pET19b_H2BT116C plasmids encoding H. sapiens histone H2B with an unnatural cysteine introduced at threonine position 116, were grown on an agar plate by re-plating a single transformant on LB supplemented with 100 ng/μL of ampicillin and 25 ng/μL of chloramphenicol at 37°C. The bacterial lawn was suspended in LB supplemented antibiotics, as above, and grown to a density (OD600 nm) of 0.4. Recombinant protein expression was then induced by addition of IPTG to 1 mM for 3 hours at 37°C. The cells were collected by centrifugation, resuspended in Histone Lysis Buffer (50 mM Tris•HCl, pH 8, 150 mM NaCl, 5 mM ß-mercaptoethanol, 1 mM Benzamidine, 100 μM Leupeptin, 100 μM Antipain, 1 μM Pepstatin), and the cellular suspension was flash frozen in liquid N2, and stored at −80°C.
Purification:
Histone H2B was purified similar to a previously described protocol (Klinker et al., 2014), with modification. E. coli expressing histone H2BT116C resuspended in Histone Lysis Buffer were thawed on wet ice and lysed by multiple passages through an Avestin Emulsiflex-C5 high pressure homogenizer at ~10,000 PSI. Soluble bacterial lysate was isolated by centrifugation of cellular debris in a Beckman Avanti J-26 XPI centrifuge in a JA25.5 rotor at 19,500 RPM. Urea was added to soluble bacterial lysate until achieving a concentration of 7 M, and 3 M NaOAc at pH 5.2 was added to lysate until the lysate was acidified to ~ pH 5.2. Lysate was filtered by passage through a 0.45 μm membrane (GE Healthcare), and applied to Source 15S Chromatography Resin (GE Healthcare) equilibrated in SAU200. After washing with SAU200 and a 5 column volume gradient of 0–30% SAU600, histone H2BT116C was eluted over a 35 column volume gradient of 30–100% SAU600. Fractions containing histone proteins were pooled and dialyzed twice against 5 mM ß-mercaptoethanol in >18 MΩ H2O and once against Histone Storage Buffer. Histone H2BT116C was concentrated in a centrifugal concentrator with a 3,000 dalton MWCO quantified by measuring histone protein absorbance at 280 nm and the calculated molar extinction coefficient (https://web.expasy.org/protparam/) for histones H2BT116C of 7450/M•cm. Purified histone H2BT116C was stored prior to labeling at −20°C in Histone Storage Buffer.
Labeling:
Tris-neutralized TCEP (500 mM Tris•HCl, pH 8, 100 mM TCEP) was added to 1 mM final concentration to histone H2BT116C in Histone Storage Buffer and incubated at room temperature for 1 hour. Histone H2BT116C with fully reduced cysteines was moved into Phosphate Buffered Saline (8 mM Na2HPO4, 2 mM KH2PO4, pH 7.4, 137 mM NaCl, 2.7 mM KCl) using 2×5 mL HiTrap Desalting Columns (GE Healthcare) and labeled by addition of 1.5 molar excess Alexa Fluor 488 (AF488)-C5- maleimide or Alexa Fluor 594 (AF594)-C5-maleimide followed by incubation in the dark for 4 hours at room temperature. Addition of DMSO alone was used to generate unlabeled H2BT116C protein. Conjugation reactions were quenched by addition of 10 mM DTT. Removal of free fluorophore and any free dsDNA was achieved by flowing unlabeled, AF488-labeled, or AF594-labeled histone H2B through 2×5mL desalting columns and Source 15Q resin (GE Healthcare) equilibrated in Histone CleanUp Buffer (20 mM Tris•HCl, pH 7.5, 150 mM NaCl, 1 mM DTT). Fractions containing unlabeled, AF488-labeled, or AF594-labeled histone H2B proteins were pooled and dialyzed three times against 5 mM ß-mercaptoethanol in >18 MΩ H2O. The different Fluor-labeled or unlabeled proteins were concentrated in centrifugal concentrators with a 3,000 dalton MWCO to less than 500 μL per liter bacterial expression and both concentration and percent labeling were quantified by measuring absorbance at 280, 495, and 590 as well as absorbance at and the calculated molar extinction coefficients (https://web.expasy.org/protparam/ or Thermo Scientific) for histone H2BT116C, AF488, and AF594 of 4470/M•cm, 73000/M•cm, and 92000/M•cm, respectively. 100% labeling was confirmed, and the purified and differentially labeled histone H2B proteins were aliquoted in 240 nmol quantities, flash frozen with liquid N2, and stored at −80 °C.
Fluorophore Labeling of X. laevis Histones H2A and H3 Expressed in E. coli.
Lyophilized purified X. laevis histone proteins H2A and H3 with engineered cysteines at positions 120 and 33, respectively, were resuspended in XL Labeling Buffer (20 mM Tris•HCl, pH 7.5, 6 M Guanidinium•HCl, 5 mM EDTA, 0.7 mM TCEP) and Atto565-C5-maleimide (SIGMA) or Cy5-C5-maleimide (GE) were added at a 1:5 molar ratio of histone protein: maleimide dye. Conjugation reactions were carried out for 12 hours at room temperature in the dark. Free unconjugated dye was removed from labeled histone proteins with a 5 mL HiTrap Desalting Column (GE) followed by dialysis into water. After percent conjugation and protein concentration of dialyzed histone proteins were quantified, histones were aliquoted and lyophilized for histone octamer reconstitution.
Purification of H1.4 and H1.4-based Proteins Expressed in E. coli.
Expression:
An overnight culture of Rosetta 2 (pLysS) E. coli (Novagen) transformed with pMTTH-based plasmids encoding mEGFP-tagged or untagged H. sapiens H1.4 and H1.4-based (H1.4ΔCTD and LANA-CTD) recombinant proteins were grown on an agar plate by re-plating a single transformant on LB supplemented with 100 ng/μL of ampicillin and 25 ng/μL of chloramphenicol at 37°C. The bacterial lawn was suspended in LB supplemented antibiotics, as above, and grown to a density (OD600 nm) of 0.4, cooled over 1 hour to 18°C and recombinant protein expression was induced by addition of IPTG to 0.5 mM for 18 hours at 18°C. The cells were collected by centrifugation, resuspended in NiNTA Lysis Buffer (50 mM HEPES•NaOH, pH 7, 150 mM NaCl, 10% [w/v] glycerol, 5 mM Imidazole, 5 mM ß-mercaptoethanol, 1 mM Benzamidine, 100 μM Leupeptin, 100 μM Antipain, 1 μM Pepstatin), the cellular suspension was flash frozen in liquid N2, and stored at −80°C.
Purification:
mEGFP-tagged or untagged H. sapiens H1.4 and H1.4-based (H1.4ΔCTD and LANA-CTD) recombinant proteins in NiNTA Lysis Buffer were thawed in a water bath and lysed by multiple passages through an Avestin Emulsiflex-C5 high pressure homogenizer at ~10,000 PSI. An equal volume of NiNTA Dilution Buffer (50 mM HEPES•NaOH, pH 7, 1.85 M NaCl, 10% [w/v] glycerol, 5 mM Imidazole, 5 mM ß-mercaptoethanol, 1 mM Benzamidine, 100 μM Leupeptin, 100 μM Antipain, 1 μM Pepstatin) was added to the lysate to increase NaCl concentration to 1 M. Soluble bacterial lysate was isolated by centrifugation of cellular debris in a Beckman Avanti J-26 XPI centrifuge in a JA25.5 rotor at 19,500 RPM. Soluble lysate was incubated with NiNTA resin (Qiagen) equilibrated in NiNTA Wash Buffer NiNTA Dilution Buffer (50 mM HEPES•NaOH, pH 7, 1 M NaCl, 10% [w/v] glycerol, 10 mM Imidazole, 5 mM ß-mercaptoethanol, 1 mM Benzamidine, 100 μM Leupeptin, 100 μM Antipain, 1 μM Pepstatin) for 2 hours in batch with end-over-end mixing. NiNTA resin was poured into a BioRad EconoColumn and resin was washed with at least 20 column volumes of NiNTA Wash Buffer before elution in NiNTA Elution Buffer (50 mM HEPES•NaOH, pH 7, 1.85 M NaCl, 10% [w/v] glycerol, 350 mM Imidazole, 5 mM ß-mercaptoethanol, 1 mM Benzamidine, 100 μM Leupeptin, 100 μM Antipain, 1 μM Pepstatin). Fractions with recombinant protein of interest were applied to Amylose resin equilibrated in NiNTA Elution Buffer for 1 hour in batch with end-over-end mixing. Amylose resin was poured into a BioRad EconoColumn and resin was washed with at least 20 column volumes of Amylose Wash Buffer (50 mM HEPES•NaOH, pH 7, 150 mM NaCl, 10% [w/v] glycerol, 1 mM dithiothreitol, 1 mM Benzamidine, 100 μM Leupeptin, 100 μM Antipain, 1 μM Pepstatin) before elution in Amylose Wash Buffer containing 1% maltose. Fractions with recombinant proteins of interest were cleaved overnight with TEV protease at 4°C.
TEV-cleaved recombinant H1.4-based proteins were diluted with 9 volumes of Buffer SA (20 mM HEPES•NaOH, pH 7, 10% [w/v] glycerol, 1 mM DTT) and applied to Source15S resin (GE Healthcare) equilibrated in 98.5% Buffer SA and 1.5 % Buffer SB (20 mM HEPES•NaOH, pH 7, 1 M NaCl, 10% [w/v] glycerol, 1 mM DTT) and eluted with a linear gradient to 100% Buffer SB. Fractions containing TEV-cleaved recombinant H1.4-based proteins were concentrated in a centrifugal concentrator with a 3,000 dalton MWCO and purified further by size exclusion chromatography using a Superdex 75 10/300 GL gel filtration column equilibrated with Gel Filtration Buffer (20 mM Tris•HCl, pH 8, 150 mM NaCl, 10% [w/v] glycerol, 1 mM DTT). Peak fractions of the protein of interest were concentrated and quantified by measuring protein absorbance at 280 nm and their calculated molar extinction coefficient (https://web.expasy.org/protparam/). Purified proteins were flash frozen in liquid N2 and stored at −80°C in single use aliquots.
Purification of bromo5, eGFP-bromo1, eGFP-bromo5, and mEGFP-tagged LANA Peptide Fusion Proteins Expressed in E. coli.
Expression:
An overnight culture of Rosetta 2 (pLysS) E. coli (Novagen) transformed with pMTTH-based plasmids encoding bromo5, EGFP-tagged bromo1, bromo5, bromo5(N140A), or mEGFP-tagged LANA peptide recombinant proteins were grown on an agar plate by re-plating a single transformant on LB supplemented with 100 ng/μL of ampicillin and 25 ng/μL of chloramphenicol at 37°C. The bacterial lawn was suspended in LB supplemented antibiotics, as above, and grown to a density (OD600 nm) of 0.4, cooled over 1 hour to 18°C and recombinant protein expression was induced by addition of IPTG to 0.5 mM for 18 hours at 18°C. The cells were collected by centrifugation, resuspended in NiNTA Lysis Buffer (50 mM HEPES•NaOH, pH 7, 150 mM NaCl, 10% [w/v] glycerol, 5 mM Imidazole, 5 mM ß-mercaptoethanol, 1 mM Benzamidine, 100 μM Leupeptin, 100 μM Antipain, 1 μM Pepstatin), the cellular suspension flash frozen in liquid N2, and stored at −80°C.
Purification:
Recombinant proteins in NiNTA Lysis Buffer were thawed in a water bath, lysed, and affinity purified with NiNTA and Amylose resins exactly as described above for H1.4-based recombinant proteins. Amylose elution fractions with recombinant protein of interest were cleaved overnight with TEV protease at 4°C. TEV-cleaved recombinant proteins were diluted with 9 volumes of Buffer QA (20 mM Tris•HCl, pH 8, 10% [w/v] glycerol, 1 mM DTT) and applied to Source15Q resin (GE Healthcare) equilibrated in 98.5% Buffer QA and 1.5 % Buffer QB (20 mM Tris•HCl, pH 8, 1 M NaCl, 10% [w/v] glycerol, 1 mM DTT) and eluted with a linear gradient to 100% Buffer SB. Fractions containing TEV-cleaved proteins of interest were concentrated in a centrifugal concentrator and purified further by size exclusion chromatography using a Superdex 75 10/300 GL (EGFP-bromo1), Superdex 200 10/300 GL (EGFP-bromo5 and EGFP-bromo5[N140A]), or HiLoad 26/60 superdex 75 pg (mEGFP-LANA) gel filtration column equilibrated with Gel Filtration Buffer (20 mM Tris•HCl, pH 8, 150 mM NaCl, 10% [w/v] glycerol, 1 mM DTT). Peak fractions of protein of interest were concentrated and quantified by measuring protein absorbance at 280 nm and their calculated molar extinction coefficient (https://web.expasy.org/protparam/). Purified proteins were flash frozen in liquid N2 and stored at −80°C in single use aliquots.
Purification of Calf Thymus Histone H1.
Calf thymus histone H1 (14–155; EMD Millipore) was diluted in 20 volumes of Histone H1 Buffer A (25 mM K+PO−4, pH 6.8, 100 mM NaCl, 1 mM DTT) and flowed through a Source 15Q column to remove any free DNA and applied to a Source 15S column equilibrated in Histone H1 Buffer A. The Source 15S column was washed with 2 column volumes of Histone H1 Buffer A, and 0–20% Histone H1 Buffer B (25 mM K+PO−4, pH 6.8, 2 M NaCl, 1 mM DTT) over 5 column volumes. Fractions containing histone H1 from a linear elution gradient of 20–100% Histone H1 Buffer B were concentrated with a 3,000 Dalton MWCO centrifugal concentrator and applied to a Superdex 75 Increase 10/300 GL column (GE Healthcare) equilibrated in Histone H1 Buffer A. Fractions containing purified histone H1 were concentrated with a 3,000 Dalton MWCO centrifugal concentrator and quantified by measuring histone H1 protein absorbance at 280 nm and the calculated molar extinction coefficient (https://web.expasy.org/protparam/) for histone H1.5 of 1490/M•cm. Purified histone H1 was aliquoted in single use quantities, flash frozen with liquid N2, and stored at −80 °C.
Purification of p300HAT, GFP-TetR and GFP-TetR-p300HAT Expressed in E. coli.
Expression:
Rosetta 2 (pLysS) E. coli (Novagen) were transformed with either pETduet+p300HAT, pETduet+sfGFP-TetR, or pETduet+sfGFP-TetR-p300HAT plasmids encoding the histone acetyltransferase domain of p300 (p300HAT), and fusion proteins composed of superfolder GFP, the E. coli Tetracycline Receptor (TetR), and for GFPTetR-p300HAT with an N-terminal Tobacco Etch Virus protease cleavable 10xHIS tag and grown overnight on an agar plate with LB medium supplemented with 100 ng/μL Ampicillin and 25 ng/μL Chloramphenicol. A single colony was used to inoculate a liquid preculture of LB with Ampicillin and Chloramphenicol, as above, with growth for ~8 hours at 37°C. The liquid preculture was centrifuged at 4000 RPM for 10 minutes in a benchtop centrifuge and resuspended in a small volume of LB medium to inoculate 3 L of Terrific Broth (Difco) containing 100 ng/μL Ampicillin and 25 ng/μL of Chloramphenicol with growth position 116, were grown on an agar plate by re-plating a single transformant on LB supplemented with Antifoam 204, 100 ng/μL of ampicillin, and 25 ng/μL of chloramphenicol with growth at 37°C to a density (OD600 nm) of 1.0. Bacterial Cultures were cooled over 1 hour to 18 degrees Celsius and recombinant protein expression was induced by addition of 1 mM IPTG and incubation at 18°C for 18 hours. The cells were collected by centrifugation, resuspended in IMAC Lysis Buffer (50 mM HEPES, pH 7, 150 mM NaCl, 5 mM ß-mercaptoethanol, 10% [w/v] glycerol, 1 mM Benzamidine, 100 μM Leupeptin, 100 μM Antipain, 1 μM Pepstatin), the cellular suspension flash frozen in liquid N2, and stored at −80°C.
Purification:
E. coli expressing recombinant proteins resuspended in IMAC Lysis Buffer were thawed on wet ice, diluted with an equal volume of IMAC Dilution Buffer (50 mM HEPES, pH 7, 1.85 M NaCl, 10 mM Imidazole, 5 mM ß-mercaptoethanol, 10% [w/v] glycerol, 1 mM Benzamidine, 100 μM Leupeptin, 100 μM Antipain, 1 μM Pepstatin), and lysed with multiple passages through an Avestin Emulsiflex-C5 high pressure homogenizer at ~10,000 PSI. Soluble bacterial lysate was isolated by centrifugation of cellular debris in a Beckman Avanti J-26 XPI centrifuge in a JA25.5 rotor at 4°C for 40 minutes at 19,500 RPM. Clarified soluble bacterial lysate was applied to 15 mL of NiNTA resin (Qiagen) for 2 hours in batch with end-over-end mixing and washed in a glass Econo-Column (BioRad) with 500 mL of IMAC Wash Buffer A (50 mM HEPES, pH 7, 1 M NaCl, 25 mM Imidazole, 5 mM ß-mercaptoethanol, 10% [w/v] glycerol, 1 mM Benzamidine) and 100 mL of IMAC Wash Buffer B (50 mM HEPES, pH 7, 150 mM NaCl, 30 mM Imidazole, 5 mM ß-mercaptoethanol, 10% [w/v] glycerol, 1 mM Benzamidine). Recombinant proteins were eluted from NiNTA resin with 100 mL of IMAC Elution Buffer (50 mM HEPES, pH 7, 150 mM NaCl, 300 mM Imidazole, 5 mM ß-mercaptoethanol, 10% [w/v] glycerol, 1 mM Benzamidine). NiNTA eluate was cleaved with Tobacco Etch Virus protease overnight (≥ 16 hours) at 4°C, diluted with 9 volumes of Ion Exchange Buffer A (20 mM Tris•HCl, pH 8, 1 mM DTT), and applied to Source 15Q resin (GE Healthcare). Recombinant proteins were eluted from the Source 15Q resin with 5–60% Ion Exchange Buffer B over 40 column volumes (20 mM Tris•HCl, pH 8, 1 M NaCl, 1 mM DTT). For GFP-TetR-p300HAT, fractions containing the fusion protein as determined by SDS-PAGE with Coomassie Brilliant Blue Staining, were pooled, diluted with 9 volumes Ion Exchange Buffer A, applied to Source 15S resin (GE Healthcare), and eluted with 5–55% Ion Exchange Buffer B over 50 column volumes. p300HAT and GFP-TetR proteins from Source 15Q cation exchange chromatography and GFP-TetR-p300 protein from Source 15S anion exchange chromatography were further purified by size exclusion chromatography using either a HiLoad SD200 26/60 pg column (GFP-TetR and p300HAT) or a Superdex 200 Increase 10/300 GL column (GFP-TetR-p300) in Gel Filtration Buffer (20 mM Tris•HCl, pH 8, 150 mM NaCl, 10% [w/v] glycerol, 1 mM DTT). The purest peak fractions of either each protein, as determined by SDS-PAGE with Coomassie Brilliant Blue Staining, were pooled, concentrated, and quantified using the calculated extinction coeefficient and absorbance at 280 nm for p300HAT and the known molar extinction coefficient for sfGFP at 485 nm (83300/M•cm). Purified p300HAT, GFP-TetR and GFP-TetR-p300HAT were aliquoted, flash frozen with liquid N2, and stored at −80°C.
Purification of Cy5-labeled BRD4 Expressed in Insect Cells.
Expression:
Bacmids for BRD4 baculovirus generation were produced using the Bac-to-Bac method, where DH10bac cells were transformed with the pAV5B+BRD4 plasmid followed by blue/white colony screening. Bacmids were transformed into Sf9 cells to generate baculovirus. High titer baculovirus was generated by multiple rounds of infection and medium collection. Sf9 cells were infected with high titer virus and supplemented with 10% FBS and penicillin/streptomycin to induce large-scale expression of BRD4. After 2 days, cells were collected by centrifugation and resuspended in Sf9 Harvest Buffer (50 mM Tris, pH 8, 150 mM NaCl, 5 mM ß-mercaptoethanol, 10% [w/v] glycerol, 1 mM Benzamidine, 100 μM Leupeptin, 100 μM Antipain, 1 μM Pepstatin).
Purification:
2 liters of Sf9 cells expressing BRD4 resuspended in Sf9 Harvest Buffer were thawed in a water bath, diluted with an equal volume of Sf9 Dilution Buffer (50 mM HEPES, pH 7, 1.85 M NaCl, 10 mM Imidazole, 20 mM NaPPi, 0.01% NP-40, 10 mM ß-mercaptoethanol, 10% [w/v] glycerol, 1 mM Benzamidine, 100 μM Leupeptin, 100 μM Antipain, 1 μM Pepstatin, 100ng/mL aprotinin, Complete EDTA-free protease inhibitors [Roche]), and lysed by sonication for 3 minutes with a Branson digital sonifier. Soluble insect cell lysate was isolated by centrifugation of cellular debris in a Beckman Avanti J-26 XPI centrifuge in a JA25.5 rotor at 4°C for 1 hour at 19,500 RPM. Clarified lysate was filtered twice through a 0.22 μm syringe filter and applied to 5 mL of Talon resin (Clontech) for 2 hours in batch with end-over-end mixing and washed in a glass Econo-Column (BioRad) with 250 mL of Talon Wash Buffer (50 mM HEPES, pH 7, 1 M NaCl, 10 mM Imidazole, 15 mM NaPPi, 0.01% NP-40, 10 mM ß-mercaptoethanol, 10% [w/v] glycerol, 1 mM Benzamidine, 100 μM Leupeptin, 100 μM Antipain, 1 μM Pepstatin, 100ng/mL aprotinin, Complete EDTA-free protease inhibitors [Roche]). BRD4 was fractionated with 3 column volumes from Talon resin using Talon Elution Buffer (50 mM HEPES, pH 7, 150 mM NaCl, 500 mM Imidazole, 10 mM ß-mercaptoethanol, 10% [w/v] glycerol, 0.01% NP-40, 1 mM Benzamidine, 100 μM Leupeptin, 100 μM Antipain, 1 μM Pepstatin, 100ng/mL aprotinin, Complete EDTA-free protease inhibitors [Roche]). BRD4-containing fractions were diluted with 9 volumes of Heparin Buffer A (20 mM Bis-Tris, pH 6, 2 mM DTT, 10% [w/v] glycerol, 1 mM Benzamidine, 100 μM Leupeptin, 100 μM Antipain, 1 μM Pepstatin, 100ng/mL aprotinin) and applied to a HiTrap Heparin chromatography column equilibrated in 95% Heparin Buffer A and 5% Heparin Buffer B (20 mM Bis-Tris, pH 6, 1 M NaCl, 2 mM DTT, 10% [w/v] glycerol, 1 mM Benzamidine, 100 μM Leupeptin, 100 μM Antipain, 1 μM Pepstatin, 100ng/mL aprotinin). Following loading of BRD4, the Heparin column was washed with 5% Heparin Buffer B in Heparin Buffer A, and eluted with a linear gradient from 5–100% Heparin Buffer B. Fractions containing BRD4 were cleaved with Tobacco Etch Virus protease overnight (≥ 16 hours) at 4°C, concentrated with a centrifugal concentrator, and further purified by size exclusion chromatography using a Superdex 200 Increase 10/300 GL column in Gel Filtration Buffer (20 mM Tris•HCl, pH 8, 150 mM NaCl, 10% [w/v] glycerol, 1 mM DTT). The purest peak fractions as determined by SDS-PAGE with Coomassie Brilliant Blue Staining were pooled, concentrated, and quantified using absorbance at 280 nm using the molar extinction coefficient for BRD4. Purified BRD4 not used for labeling was aliquoted, flash frozen with liquid N2, and stored at −80°C in single use aliquots.
Labeling:
To 22 μM purified BRD4, 50 μM Cy5-labeled depsipeptide sortase recognition sequence (GenScript) (Williamson et al., 2014) and 3 μM eSrtA (Chen et al., 2011) was added. Sortase-mediated attachment of the Cy5-labeled peptide was initiated by addition of 10 mM CaCl2 to the reaction. Following overnight labeling of BRD4 with Cy5-conjugated peptide at 4°C, reaction products were diluted in 9 volumes of MonoQ Buffer A (10 mM TRIS, pH 8, 10% [w/v] glycerol, 1 mM DTT) and applied to a 0.5 mL MonoQ column (GE Healthcare) equilibrated in 98.5% MonoQ Buffer A and 1.5% MonoQ Buffer B (10 mM TRIS, pH 8, 1 M NaCl, 10% [w/v] glycerol, 1 mM DTT). Cy5-labeled BRD4 was separated from unreacted depsipeptide and eSrtA over a linear gradient from 1.5–100% MonoQ Buffer B. The purest fractions of BRD4, as determined by SDS-PAGE with Coomassie Brilliant Blue Staining, were pooled, concentrated, and quantified using absorbance at 280 nm using the molar extinction coefficient for BRD4. Labeling was confirmed by in-gel fluorescence of BRD4 following separation by SDS-PAGE. Purified Cy5-BRD4 was aliquoted, flash frozen with liquid N2, and stored at −80°C in single use aliquots.
Reconstitution of Histone Octamers
Aliquots of histone H4 and H3 or histone H2A and fluorophore labeled or unlabeled H2B at 200 or 240 nmol quantities, respectively, were thawed on wet ice and diluted to 3 mL volume each in Histone Unfolding Buffer for 12 mL of combined total volume. The histone mix was dialyzed in 1000 Dalton MWCO dialysis tubing (Spectra/Por) three times against 2 liters of Refolding Buffer (10 mM Tris•HCl, pH 7.5, 2 M NaCl, 1 mM EDTA, 5 mM ß-mercaptoethanol) with the second and/or third dialysis steps proceeding overnight. The dialysate was filtered by passage through a 0.45 μm Whatman GD/XP syringe filter. Refolded histone octamer was isolated by size exclusion chromatography of dialysate with a HiLoad SD200 26/60 pg column. Peak Fractions were analyzed by 15% PAGE-SDS analysis for stoichiometry of core histone proteins and those with clear histone octamers were pooled, concentrated by with a 10,000 Dalton MWCO centrifugal concentrator, and quantified by measuring absorbance at 280, 495, and 590 as well as absorbance at and the calculated molar extinction coefficients (https://web.expasy.org/protparam/ or Thermo Scientific) for histone octamer, AF488, and AF594 of 44700/M•cm, 73000/M•cm, and 92000/M•cm, respectively. 100% labeled histone octamers were confirmed by the presence of 2:1 stoichiometric excess of fluor to histone octamer. Purified and differentially labeled histone octamers were aliquoted, flash frozen with liquid N2, and stored at −80 °C.
Isolation and Purification of 12×601, 6×601, and 4×601 Array DNA.
p12×601 was transformed into dam−/dcm− E. coli strain ER2925 and plated onto LB agar plates supplemented with 20 ng/μL Zeocin for growth overnight. 6 liters of LB with 20 ng/μL of Zeocin were incubated with shaking overnight at 37 °C following their inoculation with an 8 hour liquid LB and 20 ng/μL Zeocin preculture started from a single colony on the LB agar plate. The 6 liters of turbid culture were harvested the next morning by centrifugation in a Sorvall RC3C Plus centrifuge, and plasmid DNA was purified using a Qiagen Plasmid Giga Kit according to the manufacturers instructions. 12×601 array DNA was cut from carrier plasmid DNA by incubation overnight (≥ 16 hours) with 5,000 units of EcoRV-HF restriction endonuclease (New England Biolabs) in 1x CutSmart Buffer (20 mM Tris•OAc, pH 7.9, 50 mM KOAc, 10 mM Mg[OAc]2, 0.1 mg/mL BSA). Reaction was stopped by addition of 20 mM EDTA and DNA was purified by Phenol:Chloroform:Isoamyl Alcohol (25:24:1) Extraction and Ethanol Precipitation. DNA was resuspended in TE500 (10 mM Tris•HCl, pH 7.5, 1 mM EDTA, 500 mM NaCl) and 12×601 was size-fractionated by PEG precipitation with drop-by-drop addition of 30% PEG-8000 in TE500 with DNA solution under vortex. Before use in nucleosome assembly, >99% purity of 12×601 DNA array was confirmed by 1% agarose gel electrophoresis in 1xTAE (40 mM Tris•OAc, pH 8.6, 20 mM OAc, 1 mM EDTA) and staining with ethidium bromide (Bio-Rad). 6×601 or 4×601 array DNA were prepared by digesting 12×601 array DNA with SacI or NcoI and KpnI restriction endonucleases in 1x CutSmart Buffer. DNA was purified by Phenol:Chloroform:Isoamyl Alcohol (25:24:1) Extraction and Ethanol Precipitation. >99% purity of 6×601 or 4×601 DNA array was confirmed by 1% agarose gel electrophoresis in 1xTAE (40 mM Tris•OAc, pH 8.6, 20 mM OAc, 1 mM EDTA) and staining with ethidium bromide (Bio-Rad). 6×601 or 4×601 array DNA were prepared by digesting 12×601 array DNA with SacI or NcoI and KpnI restriction endonucleases in 1x CutSmart Buffer. DNA was purified by Phenol:Chloroform:Isoamyl Alcohol (25:24:1) Extraction and Ethanol Precipitation. >99% purity of 6×601 or 4×601 DNA array was confirmed by 1% agarose gel electrophoresis in 1xTAE (40 mM Tris•OAc, pH 8.6, 20 mM OAc, 1 mM EDTA) and staining with ethidium bromide (Bio-Rad).
pWM_12×601 arrays with different repeat lengths were generated as described above for p12×601 preparation, with omission of PEG purification of 12×601 DNA. EcoRV-digested pWM backbone was included during assembly as a source of “carrier” DNA.
Preparation of Ligation-competent 601 DNA
98×102 μL polymerase chain reactions (20 mM Tris•HCl, pH 8.8, 10 mM [NH4]2SO4, 10 mM KCl, 2 mM MgSO4, 0.1% Triton®-X-100, 500 nM oligonucleotides, 25 pg/μL 601 template, 25 mU/μL TAQ DNA polymerase [NEB], 250 μM each dNTPs, 1% DMSO) were performed with (1) 2 minutes at 95°C, (2) 40 cycles of 30 seconds at 95°C, 30 seconds at 60°C, and 1 minute at 72°C, (3) 2 minutes at 72°C, and (4) cooling to 4°C. Separate reactions were pooled in a 3,000 Dalton MWCO centrifugal concentrator (Amicon) and the 10 mL PCR reaction was stopped by addition of 400 μL of 0.5 M EDTA. Unincorporated dNTPS and oligonucleotides were removed, and PCR amplified DNA was washed by repeated centrifugal concentration and subsequent dilution of PCR reaction with 1xTE (10 mM Tris•HCl, pH 7.5, 1 mM EDTA). Concentrated and 1xTE-washed PCR product was purified by Phenol:Chloroform:Isoamyl Alcohol (25:24:1) Extraction and Ethanol Precipitation. Amplified DNA was digested with 1,000 units of restriction endonuclease BstXI (NEB) overnight (≥ 16 hours). Cut PCR Product was purified by Phenol:Chloroform:Isoamyl Alcohol (25:24:1) Extraction and Ethanol Precipitation and resuspended in TE50 (10 mM Tris•HCl, pH 7.5, 1 mM EDTA, 50 mM NaCl). Ligation-competent dsDNA was isolated by size exclusion chromatography of BstXI-digested DNA in TE50 using a Superdex 200 Increase 10/300GL column and concentrated for nucleosome assembly using 3,000 Dalton MWCO centrifugal concentrator (Amicon).
Preparation of Polynucleosomal Arrays and Mononucleosomes
Setup of Nucleosomal Assemblies.
Quantified DNA template and histone octamers of choice were thawed on wet ice. 601 sequence-containing DNA dissolved in 1xTE and an equal volume of 4M Assembly Buffer (10 mM Tris•HCl, pH 7.5, 1 mM EDTA, 4 M NaCl, 2 mM DTT) were mixed thoroughly on ice, followed by addition equimolar quantities of histone octamer ratio relative to 601 nucleosome positioning sequences in the template. Final concentrations of octamer/601 varied between 1–5 μM, and the percent of fluorophore-labeled to unlabeled histome octamer varied between 1 and 100% labeled nucleosomes with no detectable difference in assembly efficiency. Assembly of mutant X. laevis histone octamers into nucleosomes was aided by addition of 0.2 mol ratio labeled histone H2A/H2B dimers. Histone octamers and 601-containing DNA templates were moved into <8,000 Dalton MWCO dialysis chambers equilibrated in High Salt Assembly Buffer (10 mM Tris•HCl, pH 7.5, 1 mM EDTA, 2 M KCl, 1 mM DTT).
Salt Dialysis-mediated Assembly of Nucleosomes.
First, dialysis chambers were placed in 2 L of High Salt Assembly Buffer and salt concentration was lowered by continuous dilution with 2 L of Low Salt Assembly Buffer (10 mM Tris•HCl, pH 7.5, 1 mM EDTA, 200 mM KCl, 1 mM DTT) using a peristaltic pump at 0.8 mL/min and vigorous stirring at 4°C. Second, after exhaustion of Low Salt Assembly Buffer, the dialyzing volume was reduced to 500 mL of liquid and the salt concentration was lowered further by continuous dilution with 1 L of No Salt Assembly Buffer (10 mM Tris•HCl, pH 7.5, 1 mM EDTA, 1 mM DTT) at 0.8 mL/min at 4°C with constant vigorous stirring. Last, the dialysis chambers were dialyzed against No Salt Assembly Buffer for at least for hours at 4°C.
Sucrose Gradient-mediated Purification of Nucleosomes.
Following salt-mediated dialysis, assembled polynucleosomal arrays or mononucleosomes in No Salt Assembly Buffer were applied to linear 15–40% or 5–20% sucrose gradients in No Salt Assembly Buffer. Sucrose gradient fractions containing assembled nucleosomes were concentrated in 10,000 Dalton MWCO centrifugal concentrators (Amicon). For NRL series 12×601 arrays, all “carrier” DNA was removed from nucleosomal assemblies after sucrose gradient-mediated purification of nucleosomes.
Quantitation of Nucleosome Concentration.
To quantitate final chromatin concentrations, 1 μL of assembled polynucleosomal arrays or mononucleosomes or their DNA templates of known DNA concentration were added to 99 μL of SDS/PK Buffer (45 mM Tris•HCl, pH 7.5, 9 mM EDTA, 1% SDS) and incubated for 30 minutes at room temperature. DNA was purified by either Phenol:Chloroform:Isoamyl alcohol extraction and ethanol precipitation or processing samples with a Qiagen PCR purification Kit. The quantity of DNA in assembled mononucleosomes was determined according to a standard curve generated by serial dilution of DNA purified from unassembled template.
Quality Assurance of Nucleosomal Assembly.
Mononucleosomal assemblies and linker DNA digested polynucleosomal assemblies were assessed for quality by electrophoretic mobility shift assay. Additionally, polynucleosomal assemblies were assessed for quality by differential digestion relative to purified unassembled 12×601 DNA with either EcoRI-HF (linker DNA sites) or BsiWI-HF (nucleosomal DNA sites) in 1x CutSmart Buffer. For this assay, DNA fragments were purified with a Qiagen PCR purification Kit and analyzed on a 1% Agarose gel in 1xTAE.
Preparation of 384-well Microscopy Plates
mPEGylation of Silica.
384-well microscopy plates (Brooks Life Science Systems Matriplate) were washed with 5% Hellmanex at 37°C for 4 hours and then rinsed copiously with ≥ 18 MΩ H2O. Silica was etched with 1 M NaOH for 1 hour at room temperature and then rinsed copiously with ≥ 18 MΩ H2O. Depolymerized Silica was covalently bonded overnight (≥ 18 hours) at room temperature to 25 mg/mL 5K mPEG-silane (PEGWorks) suspended in 95% Ethanol. Plate was washed once with 95% Ethanol, rinsed with copious amounts of ≥ 18 MΩ H2O, and completely dried in a chemical hood over 3–4 hours. PEGylated microscopy plate was sealed until individual wells’ use with an adhesive PCR plate foil (Thermo).
Passivation of Well with Bovine Serum Albumin.
Following PEGylation, foil was cut above individual wells prior to their use and both plastic and PEGylated glass were passivated by incubation with freshly prepared 100 mg/mL BSA for 30 minutes. Wells were rinsed once with ≥ 18 MΩ H2O to remove excess BSA, and microscopy samples (15–40 μL in volume) were immediately added to the empty well. Desiccation of microscopy samples was limited following their addition to the plate by sealing with transparent scotch tape.
Phase Separation of Polynucleosomal Arrays
Of note, we have observed that nucleosomal arrays with vanishing quantities of plasmid DNA or hexanucleosomal contamination can result in modest differences in absolute quantities of salt (eg ± 25 mM KOAC, ±1 mM MgOAc) and nucleosome arrays (eg ± 50 nM) that result in observable phase separation. Despite these modest quantitative differences, which are irrelevant in magnitude in regards to our interpretation of the data, qualitative differences (eg more phase separation with linker histone or 10n+5 linker DNA lengths) remain unaffected by minute differences in nucleosome array production.
Phase Separation of Polynucleosomal Arrays.
Nucleosomal arrays, with 1 in 100 histone H2B proteins labeled with a fluorophore, was first equilibrated in Chromatin Dilution Buffer (25 mM Tris•OAc, pH 7.5, 5 mM DTT, 0.1 mM EDTA, 0.1 mg/mL BSA, 5%[w/v] glycerol) and incubated for 5 minutes at room temperature. Phase separation was induced, unless otherwised indicated, by addition of 1 volume of Phase Separation Buffer (25 mM Tris•OAc, pH 7.5, 0.1 mM EDTA, 5 mM DTT, 0.1 mg/mL BSA, 5%[w/v] glycerol, 300 mM KOAc, 1 mM Mg[OAc]2, 2 μg/mL Glucose Oxidase [SIGMA cat. no. G2133], 350 ng/mL Catalase [SIGMA cat. no. C1345], 4 mM Glucose) to 750 nM chromatin equilibrated in Chromatin Dilution Buffer. 30 minutes after addition of Phase Separation Buffer reactions were gently mixed and added to the well of a PEGylated and BSA passivated microscopy plate.
In-phase Chromatin Binding and Microscopy.
Calf thymus Histone H1 and H1.4-based Recombinant Proteins:
Following a 30 minute incubation period after addition of Phase Separation Buffer, 10x concentrated histone H1 in Histone H1 Buffer A, 10x concentrated H1.4-based recombinant proteins, or their gel filtration buffer alone was added to phase separated chromatin at a 1:1 stoichiometric ratio relative to nucleosomes. Samples were immediately added to a PEGylated and BSA passivated microscopy plate. Spinning disk confocal microscopy images were acquired after 1 hour of incubation in the 384-well microscopy plate.
GFP-TetR and GFP-TetR-p300 Fusion Proteins:
Following a 30 minute incubation period after addition of Phase Separation Buffer either supplemented with or without 2 μg/mL Doxycycline, 4 μM GFP-TetR or GFP-TetR-p300HAT diluted in Gel Filtration Buffer was added in 0.333 reaction volumes to phase-separated chromatin and incubated for 30 minutes at room temperature. Samples were then moved to a PEGylated and BSA passivated microscopy plate. Spinning disk confocal microscopy images were acquired after 1 hour of incubation in the 384-well microscopy plate. For time-resolved microscopy of GFP-TetR-p300HAT-dependent histone acetylation and droplet dissolution, 1 μL of 10 mM AcetylCoA was added to the microscopy well and allowed to mix by diffusion.
Trypsinization of Core Histone Tails
Sequencing grade Trypsin (Promega) was solubilized at a concentration of 100 ng/μL in 50 mM Acetic Acid, flash frozen, and stored at −80°C prior to use in trypsinization reactions. Just prior to proteolysis, Trypsin was diluted in 9 volumes of 25 mM Tris•HCl, pH 8 to neutralize 50 mM Acetic Acid. Nucleosomes were trypsinized in Trypsin Digestion Buffer (25 mM Tris•OAc, pH 7.5, 0.1 mM EDTA, 5 mM DTT, 5% [w/v] glycerol, 0.25 ng/μL Trypsin) for 30 minutes at room temperature and reactions were stopped by addition of Aprotinin and BSA at a final concentration of 50 ng/μL and 0.25 mg/mL, respectively. Digestion was confirmed by 15% PAGE-SDS and Coomassie Brilliant Blue staining of core histone proteins and stopped reactions were added to phase separation assays exactly as described above.
Ligation-dependent Assembly of Nucleosomal Arrays from Mononucleosomes.
1 μM mononucleosomes with BstXI-digested DNA were ligated in Ligation Assembly Buffer (50 mM Tris•OAc, pH 7.5, 5 mM DTT, 200 μM ATP, 3 mM Mg[OAc]2, 100 mM KOAc, 0.1 mg/mL BSA, 5% [w/v] glycerol, 1 U/μL T4 DNA Ligase [Enzymatics]) for 30 minutes at room temperature, and reactions were stopped by addition of 0.2 reaction volumes of 6x Reaction Stop Buffer (60 mM EDTA, pH 8, 400 mM KOAc). After quenching ligation by chelation of free magnesium, ligation reactions were moved into the well of an mPEGylated and BSA passivated microscopy plate. DNA was extracted by addition of SDS/PK buffer to 100 μL of final volume and purified with a PCR Purification Kit (Qiagen) according to the manufacturer’s instructions. BsiWI-HF digestion of 601 DNA sequence within ligation products was used as a quantitative measure of the extent of ligation by T4 DNA Ligase.
Absolute Quantitation of Nucleosome Concentration in Condensates and In Solution
In-phase Quantitation.
Unlabeled and 8.3% (1:12) AF594-labeled 12×601 polynucleosomal arrays were assembled using the salt dialysis method and sucrose gradient purification as described above. Nucleosomal arrays were combined to achieve 0.1% AF594- or AF488-labeled octamers and phase separation of chromatin was triggered as described above, omitting Mg(OAc)2 in the Phase Separation Buffer. Droplets with or without addition of equimolar histone H1 relative to nucleosomes were moved into an mPEGylated and BSA passivated microscopy well. Mean fluorophore intensity within droplets was measured after 1 hour of incubation relative to a standard curve of free AF594 or AF488 dye.
In-solution Quantitation.
12×601 polynucleosomal arrays were assembled with 100% AF594-labeled histone octamers using the salt dialysis method and sucrose gradient purification as described above. Phase separated was triggered exactly as described above and droplets were pelleting by 10 × 1 minute at maximum speed in a microcentrifuge. After equilibration for 10 minutes at room temperature supernatant above the droplets was moved into an mPEGylated and BSA passivated microscopy well and fluorophore intensity was measured relative to a standard curve of free AF594 dye.
Elecrophoretic Mobility Shift Assay
Glycerol and NaCl were added to purified DNA, mononucleosomes, or BstXI-digested 12×601 arrays at final concentrations of 40% w/v and 5 mM, respectively, and loaded into the wells of 6% polyacrylamide gels buffered with 0.5x TAE. Nucleic acid-containing samples were separated by electrophoresis at 100 volts for 40 minutes before imaging fluorescence of labeled nucleosomes with a ChemiDoc Imaging Station (BioRad) and staining of DNA using ethidium bromide.
Histone Acetylation by GFP-TetR-p300HAT
12×601 nucleosomal arrays assembled with wild-type X. laevis histone octamers labeled with Atto565 fluorophores or basic-patch mutant X. laevis histone octamers labeled with Cy5 fluorophores were equilibrated in Chromatin Dilution Buffer (25 mM Tris•OAc, pH 7.5, 5 mM DTT, 0.1 mM EDTA, 0.1 mg/mL BSA, 5% [w/v] glycerol) and incubated for 5 minutes at room temperature. Phase separation was induced by addition of 1 volume of Phase Separation Buffer (25 mM Tris•OAc, pH 7.5, 0.1 mM EDTA, 5 mM DTT, 0.1 mg/mL BSA, 5% [w/v] glycerol, 300 mM KOAc, 1 mM Mg[OAc]2, 2 μg/mL Glucose Oxidase [SIGMA cat. no. G2133], 350 ng/mL Catalase [SIGMA cat. no. C1345], 4 mM Glucose, ± 2 μg/mL Doxycycline) to chromatin (1μM nucleosomes) equilibrated in Chromatin Dilution Buffer. 30 minutes after addition of Phase Separation Buffer, GFP-TetR-p300HAT was added at a final concentration of 250 nM and incubated for 30 minutes at room temperature. In vitro acetylation was triggered by addition of 400 μM AcetylCoA and reactions proceeded for 30 minutes at room temperature before stopping by addition of 1 reaction volume of 2x SDS-PAGE Sample Buffer (65.8 mM Tris•HCl, pH 6.8, 0.71 mM ß-Mercaptoethanol, 26.3% [w/v] glycerol, 2.1% [w/v] SDS, 0.01% bromophenol blue)
Reaction products from the in vitro acetylation reaction were run on a 15% PAGE-SDS gel and transferred to a PVDF membrane for analysis by western blotting. The PVDF membrane was blocked with 5% Milk in TBST (20 mM Tris•HCl, pH 7.4, 150 mM NaCl, 0.05% Tween) and blotted with a 1:5,000 dilution of Rabbit polyclonal antibody against histone H3K27 acetylation (Abcam ab4729) in 1% milk in TBST overnight at 4°C. Primary antibody was washed off of the membrane with multiple washes of TBST and 1:10,000 mouse anti-rabbit HRP was incubated with the membrane for 1 hour at room temperature in 1% milk in TBST. Excess secondary antibody was washed from the membrane and the western blot signal was developed with Millipore Immobilon HRP substrate using a ChemiDoc Imaging System (Bio-Rad).
Histone Acetylation and Microinjection.
For the microinjection of dodecameric nucleosome arrays, arrays were acetylated in vitro prior to injection. Nucleosome arrays with a concentration of 5.5 μM, labeled with Alexa Fluor 488 or Alexa Fluor 594 were incubated with 2.3 μM recombinant p300 histone-acetyl transferase domain in the presence of 0.8 mM acetyl coenzyme A. Control reactions were conducted using only buffer without enzyme and coenzyme A added to the same amount of nucleosome arrays. After 2 hours at room temperature the reaction was quenched by addition of A-485 (100 μM in 10 % DMSO, Tocris) to 10 μM final concentration. Nucleosome arrays were wither injected directly or acetylated and nonacetylated arrays with differential fluor labels were mixed 1:1 for microinjection. The microinjection was performed using an Eppendorf InjectMan® 4 controller with FemtoJet® 4i, mounted on a Zeiss LSM780. For nuclear injections of nucleosome arrays commercial Femtotips™ Microinjection capillaries were used. Injection parameters were 150 hPa, 0.7 to 1 s, 20 hPa for delivery of medium to high volumes into nuclei.
Histone Acetylation Reactions with Bromodomain-containing Proteins.
12×601 nucleosome arrays (2 μM nucleosome concentration) were incubated with 1 μM recombinant p300 histone-acetyl transferase domain in Acetylation Buffer (10 mM Tris, pH 7.5, 13.6 mM KOAc, 5% [w/v] glycerol, 5 mM dithiothreitol, 0.1 mM EDTA, 0.1 mg/mL BSA, 700 μM AcetylCoA). Control reactions were conducted using only buffer without p300HAT enzyme. After 1 hour at room temperature 5.5 μM bromo5, EGFP-bromo5, EGFP-bromo5N140A, 27.5 μM EGFP-bromo1, or 10 μM BRD4 were added to nucleosomal arrays for a final concentration of 1 μM nucleosomes and Bromophase Buffer (25 mM Tris, pH 7.5, 150 mM KOAc, 1 mM MgOAc, 5% [w/v] glycerol, 5 mM dithiothreitol, 0.1 mM EDTA, 0.1 mg/mL BSA) composition. Chemical inhibition of BRD4- and acetyllysine-driven phase separation was performed by addition of 40 μM JQ-1. For synthetic bromodomain-containing proteins, 10 μM C646 inhibitor (sigma) was added to quench acetylation by p300HAT.
Microscopy
Chromatin Droplets.
Confocal fluorescence microscopy images were captured on a Nikon Eclipse Ti microscope base equipped with a Yokogawa CSU-X1 spinning disk confocal scanner unit, 100 X 1.49 NA objective, and Andor EM-CCD camera. Fluorescence Recovery After Photobleaching (FRAP) was achieved with a TIRF/iLAS2 FRAP Module (Biovision) and Rapp UGA-40 Phototargeter.
Live-cell Microscopy.
Confocal microscopy was performed on a customized Zeiss LSM780 microscope using a × 40, 1.4 NA, Oil DIC Plan-Apochromat objective (Zeiss), controlled by ZEN 2011.The microscope was equipped with an incubation chamber (European Molecular Biology Laboratory (EMBL), Heidelberg, Germany), providing a humidified atmosphere at 37 °C with 5% CO2.
Immunofluorescence.
Anti-acetyl histone antibodies and fluorescent secondary anti-mouse or anti-rabbit antibodies were added to a 2% fatty acid free BSA solution at a 1:500 and 1:1000 dilution, respectively. Immunofluorescence was imaged with a customized Zeiss LSM780 microscope using a × 40, 1.4 NA, Oil DIC Plan-Apochromat objective (Zeiss), controlled by ZEN 2011.
Quantification and Statistical Analyses
Statistical tests performed on experimental data and their representations are noted in figure legends.
Chromatin Droplets Generated In Vitro
Image analysis was performed using ImageJ (Version 1.51) (Schneider et al., 2012). Unless otherwise described, equivalent brightness and contrast were used when depicting microscopy images in a given panel. Microscopy data processed by ImageJ was graphed using the R Statistical Package (Team, 2013).
Phase Diagrams.
Different concentrations of monovalent and/or divalent salt were titrated against a concentration gradient of a dodecameric nucleosome array with 194 bp nucleosome repeat length composed of Atto565-labeled X. laevis histones and imaged at the glass bottom of a 384-well plate using confocal fluorescence microscopy. Each well was acquired with identical microscopy settings. Resultant .tif files were baseline corrected with the minimum fluorescence intensity value across the experiment. Mean pixel intensity and standard deviation were calculated in ImageJ for representative images for each experimental condition. Greyscale intensities of coefficient of variation values for each condition were determined using Java TreeView (Saldanha, 2004).
Quantitation of Relative Fluorescence Intensity.
Mean fluorescence values of chromatin droplets were determined from nucleosome arrays with 1 in 100 fluor-labeled nucleosomes or less. For determination of factor-driven fluorescence intensity differences, non fluorophore-containing proteins were used to avoid fluor-quenching artifacts. All mean fluorescence intensities were measured in ImageJ from .tif files using the center of similar-sized chromatin droplets without observable fluorescence degradation arising from without the focal plane.
Microinjected Nucleosomal Arrays
Image analysis was performed with Fiji (version ImageJ 1.51w with Java 1.8.0_66, 64-bit), using custom scripts to avoid user-based bias.
For determination of colocalization between injected microarrays and endogeneous chromatin, images of Hoechst-stained DNA was used to generate a mask in which pixels were binned in a 3 by 3 neighborhood. For each pixel, coordinates and pixel values were extracted and subsequently correlated using Graphpad PRISM version 7.04. For the analysis of coefficient of variation, the DNA-channel was used to generate a mask in which the mean fluorescence and coefficient of variation of the nucleosome array channel was determined. Cells with a nuclear mean fluorescence above 5000 AU were considered. Normality tests and statistical tests were performed in Graphpad PRISM version 7.04.
For the line profile measurements of nucleosome arrays in Ki67-eGFP expressing cells (Cuylen et al., 2016), an 8 pixel wide section of the microscopy images were measured through the nucleolar periphery. DNA (Hoechst), Ki67-eGFP and nucleosome array intensities along the line profiles were measured, aligned at the highest value for DNA (Hoechst) and normalized relative to cytoplasmic background for each channel, respectively.
For quantification of acetylation levels, a nuclear mask was generated by automatic thresholding using the DNA (Hoechst) channel. Within this nuclear mask, the mean fluorescence intensity value per nucleus was determined for each antibody and normalized relative to non-TSA treated cells.
End Trajectory of B-form DNA
A PDB structure file was generated by the make-na server (http://structure.usc.edu/make-na/) using the DNA sequence 5’ – CTAGATAGCTCGCGCTATCGATGCTAGCTAGCTGC – 3’. This PDB file was loaded into Chimera (Pettersen et al., 2004) and a graphical arrow was generated using a custom bild script, through linear extension of the xyz coordinates from 5’ to 3’ terminal phosphates at 0, 15, 20, 25, 30 and 35 base pairs within the sequence.
Genomic Analyses
Genomic analyses were performed with a combination of the published software analysis packages, referenced below, and custom-generated scripts, which can be provided upon request.
Processing of High Density Microarray and High-throughput Sequencing Data.
All datasets not already mapped to the mm10 build of the mouse genome (https://www.encodeproject.org) were mapped to the mm9 genome using bowtie (Langmead et al., 2009). Mapped bowtie reads were updated to mm10 using CrossMap (Zhao et al., 2014). Peaks of enrichment and bedGraph files were made using MACS (Zhang et al., 2008).
Processing of Base Pair Resolution Nucleosome Mapping Datasets.
Nucleosome mapping data from S. cerevisiae (Brogaard et al., 2012) and M. musculus (Voong et al., 2016) were downloaded from the gene expression omnibus and NPS values of greater than 0.8 for yeast and 0.5 for mouse were retained as calls for a well-positioned nucleosome. Mouse nucleosome positions were converted to the mm10 build of genome using CrossMap (Zhao et al., 2014), and internucleosome distances (140–250 bp) were determined using Bedtools (Quinlan, 2014; Quinlan and Hall, 2010).
Non-parametric Normalization of NRL Quantization.
NRL quantization was calculated as the difference between the per base frequency of internucleosome distances and it’s LOESS-smoothed envelope using the R statistical package (Team, 2013). An individual nucleosome was assumed to occupy 147 base pairs of DNA.
Definitions for Sub-types of the Chromatin Landscape.
Promoters:
Promoters were defined as the H3K4me3 ChIP-seq peak overlap from ESE14 and E14tga2 mouse ES cells (Consortium, 2012) that coincided with (I) RNA pol II Ser5 phosphorylation ChIP-seq peaks (Kagey et al., 2010), and overlapped with (II) annotated transcription start sites (http://genome.ucsc.edu), (III) H3K27ac ChIP-seq peaks, and (IV) DNase hypersensitivity maps of ESE14 mouse ES cells (Consortium, 2012).
Typical Enhancers:
“Typical” enhancers were defined as the H3K27ac ChIP-seq peak overlap from ESE14 and Bruce mouse ES cells (Consortium, 2012) that coincided with (I) MED1 ChIP-seq peaks in mouse ES cells (Kagey et al., 2010), as well as (II) p300 ChIP-seq peaks (Consortium, 2012), and (III) DNase hypersensitivity maps of ESE14 mouse ES cells (Consortium, 2012) that did not overlap with (IV) annotated transcription start sites (http://genome.ucsc.edu) or (V) super-enhancers (Whyte et al., 2013).
Super Enhancers:
“Super” enhancers were defined as previously outlined for mouse ES cells (Whyte et al., 2013) and moved to mm10 mouse build coordinates using CrossMap (Zhao et al., 2014).
Heterochromatin:
Heterochromatin regions were defined as H3K9me3 ChIP-seq peaks from both E14tga2 (Consortium, 2012) and mouse ES cells from (Kundu et al., 2017) overlapping (I) H3K9me3 ChIP-seq peaks from Bruce mES cells (Consortium, 2012), (II) suv39H1 ChIP-seq peaks from mouse ES cells (Bulut-Karslioglu et al., 2014), and (III) HP1 ChIP-seq peaks from mouse ES cells (Bulut-Karslioglu et al., 2014).
Polycomb Group Regions:
Polycomb group regions were defined as H3K27me3 ChIP-seq peaks from E14tga2 mouse ES cells (Consortium, 2012) that overlap with H3K27me3, H2Aub, EZH2, and Ring1b ChIP-seq peaks from mouse ES cells (Kundu et al., 2017).
Lamin-associated Domains:
Lamin-associated domains (LADs) were defined using previously published genomic coordinates for LADs from mouse ES cells (Peric-Hupkes et al., 2010) with values of LOESS-normalized log2 signal greater than 0.5 for Lamin A (Amendola and van Steensel, 2015), LaminB1 (Peric-Hupkes et al., 2010), and Emerin (Amendola and van Steensel, 2015). These genomic positions were moved to the mm10 build of the mouse genome using CrossMap (Zhao et al., 2014).
Insulators:
Insulators were defined as 1 kilobase pairs of sequence flanking CTCF ChIP-seq peak overlap from ES14tga2 and Bruce mouse ES cells (Consortium, 2012) that coincided with Smc1 and Smc3 ChIP-seq peaks (Kagey et al., 2010).
ChIP-seq Enrichment at Chromatin Sub-types.
Boxplots of log2 read depth normalized enrichment of Histone H1d ChIP-seq (Cao et al., 2013) and H3K27ac ChIP-seq in ESE14 cells (Consortium, 2012) relative to input were calculated at each chromatin sub-type or the genome at large following its division into 1 megabase pair segments.
Data and Code Availability
All custom code is available from the authors upon request.
Supplementary Material
Figure S1. Qualitative controls for LLPS of chromatin following nucleosome array assembly, related to Figure 1. (A) (top left) Diagram depticitng EcoRI and BsiWI restriction endonuclease recognition sites relative to a well-positioned nucleosome within a 12×601 nucleosomal array. (bottom right) Differential digestion of naked 12×601 dsDNA and 12×601 DNA assembled into chromatin using EcoRI-HF and BsiWI-HF restriction endonucleases. Cut DNA was extracted using a Qiagen PCR Purification Kit and run on a 1% Agarose Gel in 1xTAE. (B) (top left) Diagram depticitng BstXI restriction endonuclease recognition sites relative to a well-positioned nucleosome within a 12×601 nucleosomal array. (bottom right) Ethidium Bromide stained Native PAGE electrophoretic mobility shift assay of 12×601 array DNA with and without its assembly into chromatin. (C) Confocal fluorescence microscopy images of 12×601 array DNA alone, histone octamer alone, free histones mixed with 12×601 array DNA, and chromatin prepared by salt dialysis-mediated assembly. DNA, in green, was labeled with YOYO-1 and histones, in magenta, are labeled on histone H2B with AF594. (D) Confocal fluorescence microscopy images of phase separated chromatin assembled with X. laevis or H. Sapiens histone octamers labelled on histone H2A with Atto565, or histone H2B with AF488 or AF594 dyes (top to middle bottom). Differential interference contrast microscopy images of chromatin assembled with H. Sapiens histone octamers without a conjugated dye (bottom). (E) Microscopy images of chromatin droplets labeled on histone H2B with AF594 and with dsDNA labeled using YOYO-1. Droplets were added to the well of a microscopy plate treated with and without mPEGylation and with and without passivation with BSA. Scale bars are 4 and 10 μm, in orange and white, respectively.
Figure S2. Modulation of chromatin droplet formation by titration of monovalent salt, divalent salt, nucleosome concentrations, chromatin length, and histone tails, related to Figure 1. Fluorescence microscopy images of chromatin assembled with X. laevis histone octamers labelled on histone H2A with Atto565 following titration of (A) KOAc and Mg(OAc)2 at or (B) KOAc and chromatin. (C) 1% agarose gel electrophoresis of 12×601 array DNA, SacI-digested 6×601 array DNA, and KpnI- and NcoI-digested 4×601 array DNA with staining by ethidium bromide. (D) (above) Schematic depicting the molecular features of a DNA template for assembling mononucleosomes containing a 601 nucleosome positioning sequence, directionally ligate-able ends, and a BsiWI restriction site. (below) In-gel imaging of ethidium bromide-stained DNA separated by electrophoresis on a 6% native PAGE gel, with (mn lanes) and without assembly into mononucleosomes. Left gel shows ethidium bromide fluorescence, right gel shows AF488-histone H2B fluorescence. (E) Following T4 DNA ligase-mediated joining of mononucleosomes, isolated ligation products with (bottom) and without (top) digestion by BsiWI were separated by agarose gel electrophoresis and visualized with ethidium bromide. (F) Fluorescence imaging of AF488-labeled chromatin droplets (upper left, false-colored in magenta). In the absence of magnesium (upper right), ATP (lower left) or T4 DNA ligase (lower right) ligation does not occur (panel E) and droplets do not form. Scale bars, in white, are 10 μm. (G) Coomassie Brilliant Blue-stained 15% PAGE-SDS gel following proteolysis of chromatin with Trypsin. * Denotes a trypsin-resistant band that appears following digestion of chromatin, increases in intensity over time, and is retained for the second 15 minutes of digestion, suggesting successful digestion of histone tails by trypsin and retention of the histone octamer core.
Figure S3. Droplet dynamics, dependence on the histone H4 tail, and density in the presence and absence of histone H1, related to Figures 1–3. (A) Fluorescence microscopy images of chromatin at 375 nM nucleosome concentration in a buffered solution with 150 mM KOAc, assembled with wild-type, basic patch mutant, or acidic patch mutant X. laevis histone octamers. (B) Microscopy images of AF594-labeled chromatin before and 30 seconds after addition of 10 mM EDTA. (C) Microscopy images of fluorescence recovery after photobleaching of a whole chromatin droplet composed of AF594-labeled nucleosome arrays. (D) Quantitation from 6 individual photobleaching experiments from panel C. (E) Mean droplet intensity of chromatin droplets with 1 in 1000 nucleosomes (0.1%) doubly-labeled with Alexa Fluor 594 (AF594) relative to a standard curve of free AF594 dye. Mean droplet intensity of 10 individual chromatin droplets are depicted with horizontal red lines. (F) Three independent experiments quantitating fluorophore content in supernatant above pelleted chromatin condensates with nucleosomes all (100%) doubly-labeled with AF594 relative to a standard curve of free AF594 fluorescent dye (filled black circle). (G) Diagram depicting labeling scheme of histone octamers highlighting 2 dyes per octamer. Scale bars, in white, are 4 and 10 μm. (H) Coomassie brilliant blue-stained 15% PAGE-SDS gel of recombinant purified monomeric eGFP and H1.4-derived fusion proteins. (I) Coomassie brilliant blue-stained 15% PAGE-SDS gel of recombinant purified histone H1.4-derived proteins. (J) Bar graph representation of relative mean fluorophore intensity of histone H1-bound and unbound chromatin droplets labeled with of 0.1% Alexa Fluor 488 (AF488) or 0.1% Alexa Fluor 594 (AF594).
Figure S4. LOESS normalization of per base internucleosome frequencies and chromatin condensation by 10n+5 arrays, related to Figure 4. (A) Internucleosome linker lengths of chromatin from the S. cerevisiae genome at base pair resolution (Brogaard et al., 2012) (left) and signal difference of nucleosome density and its LOESS-smoothed envelope reveals strong bias toward linker lengths of 10n+5 spacing (right). (B) Fluorescence microscopy images of chromatin droplets with either 10n or 10n+5 internucleosome linker lengths following addition of 150 mM KOAc with and without 1 mM MgOAc. (C) Quantification of mean fluorescence intensity of chromatin droplets (n=6) from lower panel in B. (D) Microscopy images of chromatin droplets with shorter and longer 10n+5 internucleosome linker lengths with and without addition of linker histone H1. Scale bars, in white, are 10 μm.
Figure S5. Quantitation of nucleosome array mincroinjections, related to Figure 6. (A) Microinjection of Ki67-eGFP cells with non-modified nucleosomal arrays (Panel A). Semi-transparent masks indicate the line profile measurement regions. Scalebars: 5 μm (Overview), 2 μm (Insets) (B) Quantification of line profiles as indicated in Insets in panel A. Position is relative to DNA (Hoechst) peak value. Cell number n=21, one line profile per nucleus, 2 biological replicates. (C) Immunofluorescence against indicated acetylated histones, without/with Trichostain A treatment. Scalebar 5 μm. (D) Quantifications of histone acetylation levels shown in panel C. Data from 2 independent biological replicates, cell numbers 25 cells for each condition from 2 biological replicates. (E) Quantification of Spearman r of unmodified nucleosome arrays into TSA-treated cells (goes with main figure panel C upper row) (F) Quantification of Spearman r of acetylated nucleosome arrays into TSA-treated cells (goes with main figure panel C lower row).
Figure S6. Droplet formation by acetylated chromatin and multibromodomain proteins, related to Figure 7. (A) Coomassie brilliant blue-stained 10% PAGE-SDS gel of recombinant purified synthetic bromodomain-containing proteins. (B) Coomassie brilliant blue-stained 10% PAGE-SDS gel of recombinant purified BRD4. Time-resolved microscopy of acetyllysine-modified chromatin and (C) bromo5 and (D) BRD4. Microscopy images were processed separately for each condition and BRD4 fluorescence was corrected for photobleaching. (E) Fluorescence microscopy of fluorescence recovery after photobleaching of bromo5 and BRD4-bound chromatin droplets. (F) Quantitation from 6 individual photobleaching experiments from panel E. (G) Fluorescence microscopy of bromo5 and BRD4 bound chromatin droplets following acetylation of AF488-labeled chromatin in the presence of unacetylated AF594-labelled chromatin.
Figure S7. Facets that modulate phase separation of chromatin and their distributions at chromatin sub-types, related to Figures 3–7. LOESS-normalized internucleosome linker length bias (left) and box plot representations of histone H1 occupancy (middle) and histone acetylation (right) both genome-wide and at chromatin sub-types, in grey and color, respectively, reveal differential regulation of cellular factors that tune LLPS of chromatin.
Movie S1. Imaging of two-color droplet-droplet fusion, related to Figure 2. Following separate phase separation, chromatin labeled with Alexa Fluor 488 (AF488) in green, or Alexa Fluor 594 (AF594) in magenta were mixed in a microscopy well and microscopy images were captured every 15 seconds using a spinning disk confocal microscope.
Movie S2. Histone acetylation-dependent dissolution of chromatin droplets, related to Figure 5. Following in-phase binding of GFP-TetR-p300HAT (in green) to AF594-labeled chromatin droplets (in magenta) composed of 12×601 TetO-containing nucleosomal arrays, AcetylCoA was added to the microscopy well, between frames 1 and 2, and mixing at 400 μM final concentration achieved by diffusion. Microscopy images were captured every 15 seconds at a using a spinning disk confocal microscope.
Highlights.
Chromatin undergoes liquid-liquid phase separation (LLPS) in physiologic conditions
Linker DNA length and patterning, histone H1, and acetylation modulate chromatin LLPS
Acetylated chromatin only phase separates upon binding multi-bromodomain proteins
LLPS could enable establishment and maintenance of distinct chromatin compartments
Properties inherent to chromatin, including nucleosomal spacing, allow it to phase separate within the nucleoplasm, and this ability can be further modulated by regulatory factors.
ACKNOWLEDGEMENTS
We thank Geeta Narlikar for advice and sharing unpublished results. Research was supported by the Howard Hughes Medical Institute, a Paul G. Allen Frontiers Distinguished Investigator Award (to M.K.R.) and grants from the NIH (F32GM129925 to B.A.G.), the Welch Foundation (I-1544 to M.K.R.), the European Research Council (281198 to D.W.G.), the Austrian Science Fund (SFB F34–06 and DK W1238 to D.W.G), the Wiener Wissenschafts-, Forschungs- und Technologiefonds (LS14–009 and LS17–003 to D.W.G), a Boehringer Ingelheim Fonds PhD fellowship to M.W.G.S., and funding from the UCSF Program for Breakthrough Biomedical Research (PBBR) provided by the Sandler Foundation to S.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
M.K.R. is a consultant with Third Rock Ventures.
REFERENCES
- Allahverdi A, Yang R, Korolev N, Fan Y, Davey CA, Liu CF, and Nordenskiold L (2011). The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association. Nucleic acids research 39, 1680–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amendola M, and van Steensel B (2015). Nuclear lamins are not required for lamina-associated domain organization in mouse embryonic stem cells. EMBO reports 16, 610–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balleza E, Kim JM, and Cluzel P (2018). Systematic characterization of maturation time of fluorescent proteins in living cells. Nature methods 15, 47–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, and Rosen MK (2017). Biomolecular condensates: organizers of cellular biochemistry. Nature reviews Molecular cell biology 18, 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass MV, Nikitina T, Norouzi D, Zhurkin VB, and Grigoryev SA (2019). Nucleosome spacing periodically modulates nucleosome chain folding and DNA topology in circular nucleosome arrays. The Journal of biological chemistry 294, 4233–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednar J, Garcia-Saez I, Boopathi R, Cutter AR, Papai G, Reymer A, Syed SH, Lone IN, Tonchev O, Crucifix C, et al. (2017). Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1. Molecular cell 66, 384–397 e388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu B, Mateo LJ, Su JH, Sinnott-Armstrong NA, Parker M, Kinrot S, Yamaya K, Boettiger AN, and Zhuang X (2018). Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, Zamudio AV, Li CH, Shrinivas K, Manteiga JC, Hannett NM, et al. (2018). Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 175, 1842–1855 e1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, and Hyman AA (2009). Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732. [DOI] [PubMed] [Google Scholar]
- Brogaard K, Xi L, Wang JP, and Widom J (2012). A map of nucleosome positions in yeast at base-pair resolution. Nature 486, 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, and Allis CD (1996). Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84, 843–851. [DOI] [PubMed] [Google Scholar]
- Bulut-Karslioglu A, De La Rosa-Velazquez IA, Ramirez F, Barenboim M, Onishi-Seebacher M, Arand J, Galan C, Winter GE, Engist B, Gerle B, et al. (2014). Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Molecular cell 55, 277–290. [DOI] [PubMed] [Google Scholar]
- Cao K, Lailler N, Zhang Y, Kumar A, Uppal K, Liu Z, Lee EK, Wu H, Medrzycki M, Pan C, et al. (2013). High-resolution mapping of h1 linker histone variants in embryonic stem cells. PLoS genetics 9, e1003417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carruthers LM, Bednar J, Woodcock CL, and Hansen JC (1998). Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: mechanistic ramifications for higher-order chromatin folding. Biochemistry 37, 14776–14787. [DOI] [PubMed] [Google Scholar]
- Chen C, Lim HH, Shi J, Tamura S, Maeshima K, Surana U, and Gan L (2016). Budding yeast chromatin is dispersed in a crowded nucleoplasm in vivo. Molecular biology of the cell 27, 3357–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I, Dorr BM, and Liu DR (2011). A general strategy for the evolution of bond-forming enzymes using yeast display. Proceedings of the National Academy of Sciences of the United States of America 108, 11399–11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho WK, Jayanth N, English BP, Inoue T, Andrews JO, Conway W, Grimm JB, Spille JH, Lavis LD, Lionnet T, et al. (2016). RNA Polymerase II cluster dynamics predict mRNA output in living cells. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho WK, Spille JH, Hecht M, Lee C, Li C, Grube V, and Cisse II (2018). Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S, Dugast-Darzacq C, Liu Z, Dong P, Dailey GM, Cattoglio C, Heckert A, Banala S, Lavis L, Darzacq X, et al. (2018). Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EP (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuylen S, Blaukopf C, Politi AZ, Muller-Reichert T, Neumann B, Poser I, Ellenberg J, Hyman AA, and Gerlich DW (2016). Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature 535, 308–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey CA, Sargent DF, Luger K, Maeder AW, and Richmond TJ (2002). Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. Journal of molecular biology 319, 1097–1113. [DOI] [PubMed] [Google Scholar]
- Di Pierro M, Zhang B, Aiden EL, Wolynes PG, and Onuchic JN (2016). Transferable model for chromosome architecture. Proceedings of the National Academy of Sciences of the United States of America 113, 12168–12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubochet J, Adrian M, Schultz P, and Oudet P (1986). Cryo-electron microscopy of vitrified SV40 minichromosomes: the liquid drop model. The EMBO journal 5, 519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk M, Feodorova Y, Naumova N, Imakaev M, Lajoie BR, Leonhardt H, Joffe B, Dekker J, Fudenberg G, Solovei I, et al. (2019). Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature 570, 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, and Skoultchi AI (2005). Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 123, 1199–1212. [DOI] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. (2010). Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143, 212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. (2010). Selective inhibition of BET bromodomains. Nature 468, 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher TM, and Hansen JC (1995). Core histone tail domains mediate oligonucleosome folding and nucleosomal DNA organization through distinct molecular mechanisms. The Journal of biological chemistry 270, 25359–25362. [DOI] [PubMed] [Google Scholar]
- Fujisawa T, and Filippakopoulos P (2017). Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nature reviews Molecular cell biology 18, 246–262. [DOI] [PubMed] [Google Scholar]
- Grunstein M (1997). Histone acetylation in chromatin structure and transcription. Nature 389, 349–352. [DOI] [PubMed] [Google Scholar]
- Hansen JC (2002). Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annual review of biophysics and biomolecular structure 31, 361–392. [DOI] [PubMed] [Google Scholar]
- Hergeth SP, and Schneider R (2015). The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO reports 16, 1439–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hihara S, Pack CG, Kaizu K, Tani T, Hanafusa T, Nozaki T, Takemoto S, Yoshimi T, Yokota H, Imamoto N, et al. (2012). Local nucleosome dynamics facilitate chromatin accessibility in living mammalian cells. Cell reports 2, 1645–1656. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, and Allis CD (2001). Translating the histone code. Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. (2010). Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan PY, Caterino TL, and Hayes JJ (2009). The H4 tail domain participates in intra- and internucleosome interactions with protein and DNA during folding and oligomerization of nucleosome arrays. Molecular and cellular biology 29, 538–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. (2009). The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458, 362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinker H, Haas C, Harrer N, Becker PB, and Mueller-Planitz F (2014). Rapid purification of recombinant histones. PloS one 9, e104029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD (1974). Chromatin structure: a repeating unit of histones and DNA. Science 184, 868–871. [DOI] [PubMed] [Google Scholar]
- Kundu S, Ji F, Sunwoo H, Jain G, Lee JT, Sadreyev RI, Dekker J, and Kingston RE (2017). Polycomb Repressive Complex 1 Generates Discrete Compacted Domains that Change during Differentiation. Molecular cell 65, 432–446 e435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, and Narlikar GJ (2017). Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature 547, 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. (2012). Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D, and Van Holde KE (1979). Organization of spacer DNA in chromatin. Proceedings of the National Academy of Sciences of the United States of America 76, 6326–6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowary PT, and Widom J (1998). New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. Journal of molecular biology 276, 19–42. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, and Richmond TJ (1997). Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260. [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, and Richmond TJ (1999). Expression and purification of recombinant histones and nucleosome reconstitution. Methods in molecular biology 119, 1–16. [DOI] [PubMed] [Google Scholar]
- Maeshima K, Ide S, Hibino K, and Sasai M (2016a). Liquid-like behavior of chromatin. Current opinion in genetics & development 37, 36–45. [DOI] [PubMed] [Google Scholar]
- Maeshima K, Rogge R, Tamura S, Joti Y, Hikima T, Szerlong H, Krause C, Herman J, Seidel E, DeLuca J, et al. (2016b). Nucleosomal arrays self-assemble into supramolecular globular structures lacking 30-nm fibers. The EMBO journal 35, 1115–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn KJ, and Misteli T (2007). Cell biology: chromosome territories. Nature 445, 379–781. [DOI] [PubMed] [Google Scholar]
- Mirny LA, Imakaev M, and Abdennur N (2019). Two major mechanisms of chromosome organization. Current opinion in cell biology 58, 142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitina T, Norouzi D, Grigoryev SA, and Zhurkin VB (2017). DNA topology in chromatin is defined by nucleosome spacing. Science advances 3, e1700957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino Y, Eltsov M, Joti Y, Ito K, Takata H, Takahashi Y, Hihara S, Frangakis AS, Imamoto N, Ishikawa T, et al. (2012). Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure. The EMBO journal 31, 1644–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki T, Imai R, Tanbo M, Nagashima R, Tamura S, Tani T, Joti Y, Tomita M, Hibino K, Kanemaki MT, et al. (2017). Dynamic Organization of Chromatin Domains Revealed by Super-Resolution Live-Cell Imaging. Molecular cell 67, 282–293 e287. [DOI] [PubMed] [Google Scholar]
- Olins DE, and Olins AL (2003). Chromatin history: our view from the bridge. Nature reviews Molecular cell biology 4, 809–814. [DOI] [PubMed] [Google Scholar]
- Ou HD, Phan S, Deerinck TJ, Thor A, Ellisman MH, and O’Shea CC (2017). ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. (2010). Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Molecular cell 38, 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Plys AJ, Davis CP, Kim J, Rizki G, Keenen MM, Marr SK, and Kingston RE (2019). Phase separation of Polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes & development 33, 799–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR (2014). BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Current protocols in bioinformatics 47, 11 12 11–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huang SC, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon KR, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, et al. (2017). Cohesin Loss Eliminates All Loop Domains. Cell 171, 305–320 e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein M, and Colby RH (2003). Polymer Physics (OUP Oxford).
- Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, et al. (2018). Coactivator condensation at super-enhancers links phase separation and gene control. Science 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ (2004). Java Treeview--extensible visualization of microarray data. Bioinformatics 20, 3246–3248. [DOI] [PubMed] [Google Scholar]
- Schalch T, Duda S, Sargent DF, and Richmond TJ (2005). X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature 436, 138–141. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nature methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, Fonseca NA, Huber W, C HH, Mirny L, et al. (2017). Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Yu L, Weir JW, and Gorovsky MA (1995). Linker histones are not essential and affect chromatin condensation in vivo. Cell 82, 47–56. [DOI] [PubMed] [Google Scholar]
- Shin Y, and Brangwynne CP (2017). Liquid phase condensation in cell physiology and disease. Science 357. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, and Peterson CL (2006). Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847. [DOI] [PubMed] [Google Scholar]
- Song F, Chen P, Sun D, Wang M, Dong L, Liang D, Xu RM, Zhu P, and Li G (2014). Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science 344, 376–380. [DOI] [PubMed] [Google Scholar]
- Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, and Karpen GH (2017). Phase separation drives heterochromatin domain formation. Nature 547, 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K, and Segal E (2013). Determinants of nucleosome positioning. Nature structural & molecular biology 20, 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team RC (2013). R: A language and environment for statistical computing.
- Toth KF, Knoch TA, Wachsmuth M, Frank-Stohr M, Stohr M, Bacher CP, Muller G, and Rippe K (2004). Trichostatin A-induced histone acetylation causes decondensation of interphase chromatin. Journal of cell science 117, 4277–4287. [DOI] [PubMed] [Google Scholar]
- Turner AL, Watson M, Wilkins OG, Cato L, Travers A, Thomas JO, and Stott K (2018). Highly disordered histone H1-DNA model complexes and their condensates. Proceedings of the National Academy of Sciences of the United States of America. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F (2016). SMC complexes: from DNA to chromosomes. Nature reviews Molecular cell biology 17, 399–412. [DOI] [PubMed] [Google Scholar]
- Voong LN, Xi L, Sebeson AC, Xiong B, Wang JP, and Wang X (2016). Insights into Nucleosome Organization in Mouse Embryonic Stem Cells through Chemical Mapping. Cell 167, 1555–1570 e1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, and Young RA (2013). Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widom J (1986). Physicochemical studies of the folding of the 100 A nucleosome filament into the 300 A filament. Cation dependence. Journal of molecular biology 190, 411–424. [DOI] [PubMed] [Google Scholar]
- Williamson DJ, Webb ME, and Turnbull WB (2014). Depsipeptide substrates for sortase-mediated N-terminal protein ligation. Nature protocols 9, 253–262. [DOI] [PubMed] [Google Scholar]
- Wilson MD, Benlekbir S, Fradet-Turcotte A, Sherker A, Julien JP, McEwan A, Noordermeer SM, Sicheri F, Rubinstein JL, and Durocher D (2016). The structural basis of modified nucleosome recognition by 53BP1. Nature 536, 100–103. [DOI] [PubMed] [Google Scholar]
- Woodcock CL, Skoultchi AI, and Fan Y (2006). Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology 14, 17–25. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome biology 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sun Z, Wang J, Huang H, Kocher JP, and Wang L (2014). CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics 30, 1006–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Qualitative controls for LLPS of chromatin following nucleosome array assembly, related to Figure 1. (A) (top left) Diagram depticitng EcoRI and BsiWI restriction endonuclease recognition sites relative to a well-positioned nucleosome within a 12×601 nucleosomal array. (bottom right) Differential digestion of naked 12×601 dsDNA and 12×601 DNA assembled into chromatin using EcoRI-HF and BsiWI-HF restriction endonucleases. Cut DNA was extracted using a Qiagen PCR Purification Kit and run on a 1% Agarose Gel in 1xTAE. (B) (top left) Diagram depticitng BstXI restriction endonuclease recognition sites relative to a well-positioned nucleosome within a 12×601 nucleosomal array. (bottom right) Ethidium Bromide stained Native PAGE electrophoretic mobility shift assay of 12×601 array DNA with and without its assembly into chromatin. (C) Confocal fluorescence microscopy images of 12×601 array DNA alone, histone octamer alone, free histones mixed with 12×601 array DNA, and chromatin prepared by salt dialysis-mediated assembly. DNA, in green, was labeled with YOYO-1 and histones, in magenta, are labeled on histone H2B with AF594. (D) Confocal fluorescence microscopy images of phase separated chromatin assembled with X. laevis or H. Sapiens histone octamers labelled on histone H2A with Atto565, or histone H2B with AF488 or AF594 dyes (top to middle bottom). Differential interference contrast microscopy images of chromatin assembled with H. Sapiens histone octamers without a conjugated dye (bottom). (E) Microscopy images of chromatin droplets labeled on histone H2B with AF594 and with dsDNA labeled using YOYO-1. Droplets were added to the well of a microscopy plate treated with and without mPEGylation and with and without passivation with BSA. Scale bars are 4 and 10 μm, in orange and white, respectively.
Figure S2. Modulation of chromatin droplet formation by titration of monovalent salt, divalent salt, nucleosome concentrations, chromatin length, and histone tails, related to Figure 1. Fluorescence microscopy images of chromatin assembled with X. laevis histone octamers labelled on histone H2A with Atto565 following titration of (A) KOAc and Mg(OAc)2 at or (B) KOAc and chromatin. (C) 1% agarose gel electrophoresis of 12×601 array DNA, SacI-digested 6×601 array DNA, and KpnI- and NcoI-digested 4×601 array DNA with staining by ethidium bromide. (D) (above) Schematic depicting the molecular features of a DNA template for assembling mononucleosomes containing a 601 nucleosome positioning sequence, directionally ligate-able ends, and a BsiWI restriction site. (below) In-gel imaging of ethidium bromide-stained DNA separated by electrophoresis on a 6% native PAGE gel, with (mn lanes) and without assembly into mononucleosomes. Left gel shows ethidium bromide fluorescence, right gel shows AF488-histone H2B fluorescence. (E) Following T4 DNA ligase-mediated joining of mononucleosomes, isolated ligation products with (bottom) and without (top) digestion by BsiWI were separated by agarose gel electrophoresis and visualized with ethidium bromide. (F) Fluorescence imaging of AF488-labeled chromatin droplets (upper left, false-colored in magenta). In the absence of magnesium (upper right), ATP (lower left) or T4 DNA ligase (lower right) ligation does not occur (panel E) and droplets do not form. Scale bars, in white, are 10 μm. (G) Coomassie Brilliant Blue-stained 15% PAGE-SDS gel following proteolysis of chromatin with Trypsin. * Denotes a trypsin-resistant band that appears following digestion of chromatin, increases in intensity over time, and is retained for the second 15 minutes of digestion, suggesting successful digestion of histone tails by trypsin and retention of the histone octamer core.
Figure S3. Droplet dynamics, dependence on the histone H4 tail, and density in the presence and absence of histone H1, related to Figures 1–3. (A) Fluorescence microscopy images of chromatin at 375 nM nucleosome concentration in a buffered solution with 150 mM KOAc, assembled with wild-type, basic patch mutant, or acidic patch mutant X. laevis histone octamers. (B) Microscopy images of AF594-labeled chromatin before and 30 seconds after addition of 10 mM EDTA. (C) Microscopy images of fluorescence recovery after photobleaching of a whole chromatin droplet composed of AF594-labeled nucleosome arrays. (D) Quantitation from 6 individual photobleaching experiments from panel C. (E) Mean droplet intensity of chromatin droplets with 1 in 1000 nucleosomes (0.1%) doubly-labeled with Alexa Fluor 594 (AF594) relative to a standard curve of free AF594 dye. Mean droplet intensity of 10 individual chromatin droplets are depicted with horizontal red lines. (F) Three independent experiments quantitating fluorophore content in supernatant above pelleted chromatin condensates with nucleosomes all (100%) doubly-labeled with AF594 relative to a standard curve of free AF594 fluorescent dye (filled black circle). (G) Diagram depicting labeling scheme of histone octamers highlighting 2 dyes per octamer. Scale bars, in white, are 4 and 10 μm. (H) Coomassie brilliant blue-stained 15% PAGE-SDS gel of recombinant purified monomeric eGFP and H1.4-derived fusion proteins. (I) Coomassie brilliant blue-stained 15% PAGE-SDS gel of recombinant purified histone H1.4-derived proteins. (J) Bar graph representation of relative mean fluorophore intensity of histone H1-bound and unbound chromatin droplets labeled with of 0.1% Alexa Fluor 488 (AF488) or 0.1% Alexa Fluor 594 (AF594).
Figure S4. LOESS normalization of per base internucleosome frequencies and chromatin condensation by 10n+5 arrays, related to Figure 4. (A) Internucleosome linker lengths of chromatin from the S. cerevisiae genome at base pair resolution (Brogaard et al., 2012) (left) and signal difference of nucleosome density and its LOESS-smoothed envelope reveals strong bias toward linker lengths of 10n+5 spacing (right). (B) Fluorescence microscopy images of chromatin droplets with either 10n or 10n+5 internucleosome linker lengths following addition of 150 mM KOAc with and without 1 mM MgOAc. (C) Quantification of mean fluorescence intensity of chromatin droplets (n=6) from lower panel in B. (D) Microscopy images of chromatin droplets with shorter and longer 10n+5 internucleosome linker lengths with and without addition of linker histone H1. Scale bars, in white, are 10 μm.
Figure S5. Quantitation of nucleosome array mincroinjections, related to Figure 6. (A) Microinjection of Ki67-eGFP cells with non-modified nucleosomal arrays (Panel A). Semi-transparent masks indicate the line profile measurement regions. Scalebars: 5 μm (Overview), 2 μm (Insets) (B) Quantification of line profiles as indicated in Insets in panel A. Position is relative to DNA (Hoechst) peak value. Cell number n=21, one line profile per nucleus, 2 biological replicates. (C) Immunofluorescence against indicated acetylated histones, without/with Trichostain A treatment. Scalebar 5 μm. (D) Quantifications of histone acetylation levels shown in panel C. Data from 2 independent biological replicates, cell numbers 25 cells for each condition from 2 biological replicates. (E) Quantification of Spearman r of unmodified nucleosome arrays into TSA-treated cells (goes with main figure panel C upper row) (F) Quantification of Spearman r of acetylated nucleosome arrays into TSA-treated cells (goes with main figure panel C lower row).
Figure S6. Droplet formation by acetylated chromatin and multibromodomain proteins, related to Figure 7. (A) Coomassie brilliant blue-stained 10% PAGE-SDS gel of recombinant purified synthetic bromodomain-containing proteins. (B) Coomassie brilliant blue-stained 10% PAGE-SDS gel of recombinant purified BRD4. Time-resolved microscopy of acetyllysine-modified chromatin and (C) bromo5 and (D) BRD4. Microscopy images were processed separately for each condition and BRD4 fluorescence was corrected for photobleaching. (E) Fluorescence microscopy of fluorescence recovery after photobleaching of bromo5 and BRD4-bound chromatin droplets. (F) Quantitation from 6 individual photobleaching experiments from panel E. (G) Fluorescence microscopy of bromo5 and BRD4 bound chromatin droplets following acetylation of AF488-labeled chromatin in the presence of unacetylated AF594-labelled chromatin.
Figure S7. Facets that modulate phase separation of chromatin and their distributions at chromatin sub-types, related to Figures 3–7. LOESS-normalized internucleosome linker length bias (left) and box plot representations of histone H1 occupancy (middle) and histone acetylation (right) both genome-wide and at chromatin sub-types, in grey and color, respectively, reveal differential regulation of cellular factors that tune LLPS of chromatin.
Movie S1. Imaging of two-color droplet-droplet fusion, related to Figure 2. Following separate phase separation, chromatin labeled with Alexa Fluor 488 (AF488) in green, or Alexa Fluor 594 (AF594) in magenta were mixed in a microscopy well and microscopy images were captured every 15 seconds using a spinning disk confocal microscope.
Movie S2. Histone acetylation-dependent dissolution of chromatin droplets, related to Figure 5. Following in-phase binding of GFP-TetR-p300HAT (in green) to AF594-labeled chromatin droplets (in magenta) composed of 12×601 TetO-containing nucleosomal arrays, AcetylCoA was added to the microscopy well, between frames 1 and 2, and mixing at 400 μM final concentration achieved by diffusion. Microscopy images were captured every 15 seconds at a using a spinning disk confocal microscope.
Data Availability Statement
All custom code is available from the authors upon request.