Significance
Over the last few thousand years, the seemingly inconspicuous cabbage white butterfly, Pieris rapae, has become one of the most abundant and destructive butterflies in the world. Here, we assessed variation at thousands of genetic markers from butterflies collected across 32 countries by over 150 volunteer scientists and citizens to reconstruct the global spread of this agricultural pest. Our results suggest this butterfly spread out from eastern Europe to occupy every continent except South America and Antarctica, with the timing of many of these events coinciding with human activities—migration, trade, and the development of crop cultivars that serve as food plants for the butterfly larvae. Interestingly, many of these invasions were hugely successful despite repeated losses of genetic diversity.
Keywords: invasive, agricultural pest, genomics, citizen science, approximate Bayesian computation
Abstract
The small cabbage white butterfly, Pieris rapae, is a major agricultural pest of cruciferous crops and has been introduced to every continent except South America and Antarctica as a result of human activities. In an effort to reconstruct the near-global invasion history of P. rapae, we developed a citizen science project, the “Pieris Project,” and successfully amassed thousands of specimens from 32 countries worldwide. We then generated and analyzed nuclear (double-digest restriction site-associated DNA fragment procedure [ddRAD]) and mitochondrial DNA sequence data for these samples to reconstruct and compare different global invasion history scenarios. Our results bolster historical accounts of the global spread and timing of P. rapae introductions. We provide molecular evidence supporting the hypothesis that the ongoing divergence of the European and Asian subspecies of P. rapae (∼1,200 y B.P.) coincides with the diversification of brassicaceous crops and the development of human trade routes such as the Silk Route (Silk Road). The further spread of P. rapae over the last ∼160 y was facilitated by human movement and trade, resulting in an almost linear series of at least 4 founding events, with each introduced population going through a severe bottleneck and serving as the source for the next introduction. Management efforts of this agricultural pest may need to consider the current existence of multiple genetically distinct populations. Finally, the international success of the Pieris Project demonstrates the power of the public to aid scientists in collections-based research addressing important questions in invasion biology, and in ecology and evolutionary biology more broadly.
Invasive species—species spread to places beyond their natural range, where they generate a negative impact [e.g., extirpate or displace native fauna, spread disease, destroy agricultural crops (1)]—continue to increase in number, with no signs of saturation (2). The spread of invasive species often is driven by (human) migration, global trade, and transportation networks (3), and, in some cases, domestication of wild plants and animals (4). A critical and often first step to mitigating the spread and impacts of invasive species is to understand their invasion history, including assessing source populations, routes of spread, number of independent invasions, and the effects of genetic bottlenecks, among other factors. Such detailed knowledge is crucial from an applied perspective (e.g., developing an effective biological control program) as well as for addressing basic questions associated with the invasion process (e.g., genetic changes, adaptation to novel environments) (5).
Unraveling a species’ invasion history often requires sampling across large spatial and temporal scales, which can be challenging and costly, particularly for many invasive species found on multiple continents. Citizen science—research in which the public plays a role in project development, data collection, or discovery and which is subject to the same system of peer review as conventional science—is a potentially powerful means to overcome some of these challenges. A major strength of citizen science is that it can greatly enhance the scale and scope of science and its impact on society (6). Consequently, there are now thousands of citizen science projects worldwide (https://scistarter.org/), although they are not always described as such (7). Yet, still very few involve agricultural pests (8), and nearly all rely on observations (e.g., sightings, photographs), limiting their capacity to address some fundamental questions in ecology and evolution, for example, those requiring physical material for molecular analyses.
Pieris rapae, the small cabbage white butterfly, is arguably one of the world’s most widespread and abundant pest butterflies. Caterpillars of this species are a serious agricultural pest of crops in the Brassicaceae family (e.g., cabbage, canola, bok choy, turnips) (9). This butterfly is believed to have originated in Europe and to have subsequently undergone a range expansion into Asia several thousand years ago as a result of domestication and trade of its host plants (10, 11). The Europe and Asia populations recognized today are believed to represent separate subspecies—P. rapae rapae and P. rapae crucivora, respectively.
This butterfly has been introduced to many other parts of the world over the last ∼160 y. These invasions are unique in that there is a wealth of historical records (observations and collections) documenting the putative dates of first introduction [North America in the 1860s (12), New Zealand in 1930 (13), and Australia in 1937 (14, 15)]. Detailed accounts and observations from what was essentially a 19th century citizen science project led by the entomologist Samuel Scudder provide a chronology of the rapid spread of P. rapae across North America and suggest that there were multiple independent introductions (12, 16). While the small cabbage white butterfly ranks as one of the most successful and abundant invasive species, a detailed analysis of its invasion history has never been undertaken (10, 17). In addition, the consequences of this rapid invasion on the population genetic structure and diversity are unknown.
Here, we employ a collection-based citizen science approach to obtain range-wide, long-term, population-level sampling of this globally distributed invasive agricultural pest. Molecular genomics tools are then applied to this global collection of specimens to reconstruct the global invasion history of P. rapae and assess historical and contemporary patterns of genetic structure and diversity.
Results
Citizen Scientist-Assisted Sampling.
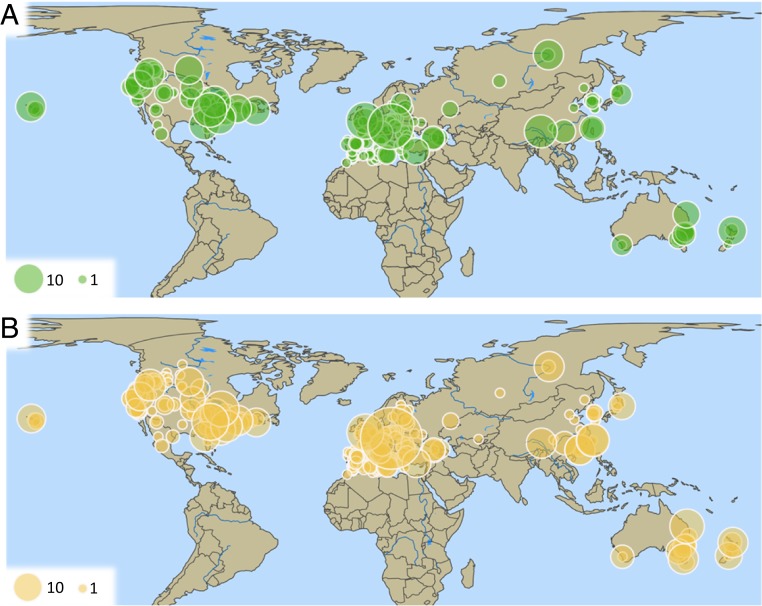
The international citizen science project—Pieris Project—recruited more than 150 participants, primarily through entomological and lepidopterist societies and other organizations related to nature and science. These citizen scientist collections were supplemented with collections we (authors of this paper) made, bringing the total to >3,000 P. rapae from the period from 2002 through 2017 (median collection year: 2014; SI Appendix, Fig. S1). Most people have a reasonable idea of what a citizen scientist embodies—someone who contributes to a scientific study but who is not a professional within the field of study (in this case, someone who contributes but does not have an advanced degree in biology). Using that definition, 44% of all specimens used to generate mitochondrial DNA (mtDNA) or double-digest restriction site-associated DNA fragment procedure sequencing (ddRADseq) data would have come from citizen scientists (SI Appendix, Table S1). If we apply a broader, more inclusive, definition (i.e., a citizen scientist is anyone contributing to this project who is not a formal collaborator), we find that 64% of all specimens were from citizen scientists. Collectively, these samples cover nearly the entire native and invaded ranges, comprising 293 localities spanning 32 countries (Fig. 1, up-to-date collections map; http://www.pierisproject.org/); it should be noted that we do not have collections from South America because there are no (known) populations of P. rapae on that continent.
Fig. 1.
Sample size by location and dataset. (A) ddRADseq (n = 559). (B) mtDNA (n = 1,002). The size of the points corresponds to the sample size. Explore these data further through interactive data visualizations (http://www.pierisproject.org/ResultsInvasionHistory.html).
A total of 22,059 autosomal (ddRADseq) single-nucleotide polymorphisms (SNPs) for 559 individuals (average depth: 74× ± 28 SD; average missingness: 2.9% ± 4.3 SD) passed quality filtering (Fig. 1A). We also sequenced a 502-base pair (bp) region of the mitochondrial gene cytochrome c oxidase subunit 1 (COI) from 751 individuals (559 of these individuals were also used to generate ddRADseq data) and supplemented these sequences with 251 additional sequences from various online databases (total individuals with COI sequence = 1,002; Fig. 1B).
Global Patterns of Autosomal Genetic Differentiation and Diversity.
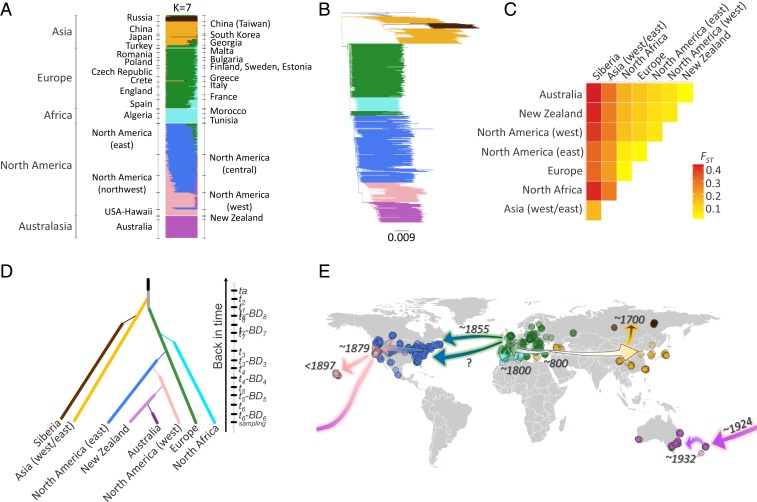
We filtered the ddRADseq data for autosomal markers and found evidence for at least 7 genetically distinct clusters (ADMIXTURE lowest cross-validation error: 0.25 for subpopulations [K] = 7) (Fig. 2A). These genetic clusters largely correspond to the continental regions sampled, and we refer to them henceforth as populations, named based on their sampling region: Europe, North Africa, Asia (west/east; including Crete, Georgia, China, Taiwan, Japan, and South Korea), Siberia, North America (east), North America (west), and Australia/New Zealand (Fig. 2E). The greatest genetic differentiation was between Asia (west/east, including Siberia) and all other populations (average fixation index [FST] = 0.26 ± 0.03 SD) (Fig. 2C). Visual inspection of ancestry assignments (at higher values of K) suggests additional hierarchical levels of structure, primarily in Asia (west/east), but also within North America, and between Australia and New Zealand (SI Appendix, Fig. S2 A and B). Surprisingly, we were unable to detect (geographically coherent) structure within Europe (except for Malta being distinct from the rest of Europe) or within Australia.
Fig. 2.
Global invasion history and patterns of genetic structure and diversity of P. rapae. (A) Genetic ancestry assignments based on the program ADMIXTURE. (B) Rooted neighbor-joining tree based on Nei’s genetic distance. (C) Among population genetic differentiation based on Weir and Cockerham’s FST (64), New Zealand and Australia are treated separately. (D) Graphical illustration of divergence scenario chosen in ABC-RF analysis (Table 1). (E) Geographic representation of divergence scenario with the highest likelihood based on ABC-RF analysis; points are colored based on their population assignment using ADMIXTURE as in A, and dates represent median estimates from ABC-RF analysis. All analyses are based on 558 individuals genotyped for 17,917 ddRADseq SNPs. Explore these data further through interactive data visualizations (http://www.pierisproject.org/ResultsInvasionHistory.html).
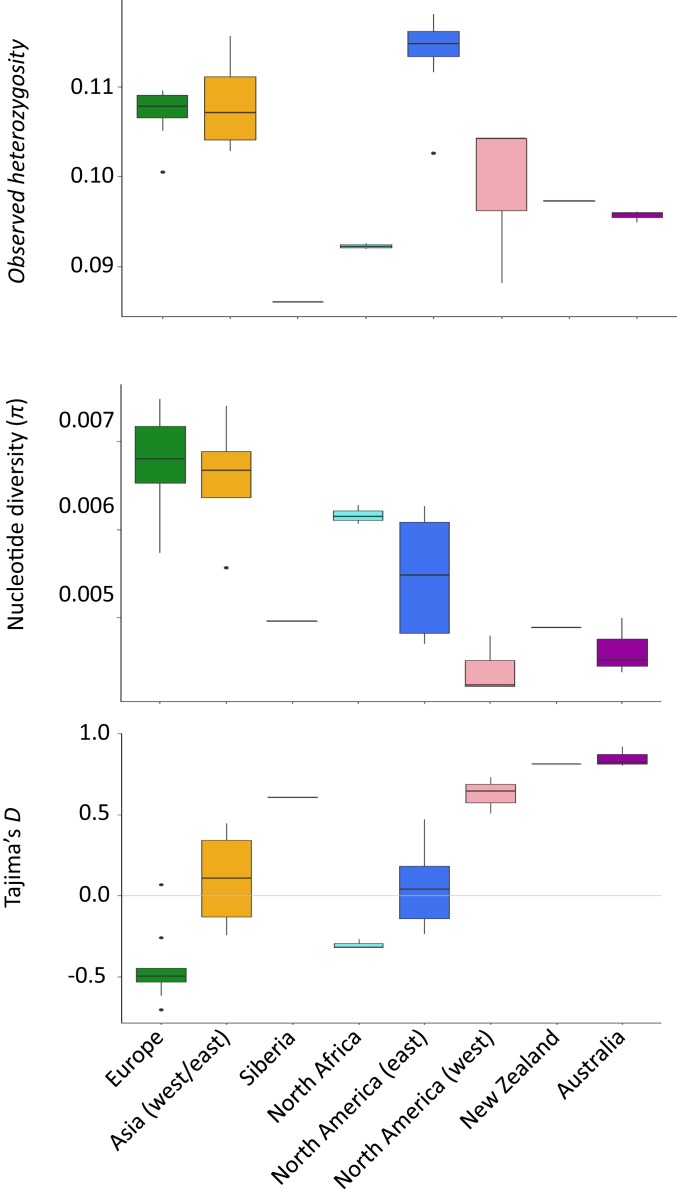
Almost all recently introduced populations (i.e., North America, Australia, New Zealand) exhibit lower observed heterozygosity and nucleotide diversity compared with populations in the native range (i.e., Europe), consistent with population bottlenecks associated with these introductions (Fig. 3). North America (east) was a notable exception among the introduced populations, with observed heterozygosity higher than populations found in the native range (Europe). All estimates of Tajima’s D fell within the range of −1 to 1, suggesting most populations are near equilibrium. However, there is a negative relationship between estimates of Tajima’s D and time since introduction (i.e., more recent introductions have higher [positive] estimates of Tajima’s D), suggesting that North America (west), New Zealand, and Australia are still recovering from repeated population bottlenecks.
Fig. 3.
Patterns of autosomal genetic diversity—observed heterozygosity, pairwise nucleotide diversity, and Tajima’s D—by population.
Global Invasion History.
We compared a number of alternative invasion history scenarios for both the native and introduced populations using ddRADseq autosomal data within an approximate Bayesian computation random forest (ABC-RF) framework. We used an iterative process for selecting each bifurcation event, beginning with the most ancestral populations and then adding each introduction event. The chronological order and timing of these bifurcation events were informed by historic records and previous population genetic analyses (Fig. 2A). Specifically, we first assessed the bifurcation event between Europe and Asia (west/east); then Siberia and North Africa; followed by the recently introduced populations in North America (east and west), New Zealand, and Australia (Table 1). We then simulated a full model that incorporated all of the best supported scenarios to get final parameter estimates (Fig. 2E and SI Appendix, Tables S2 and S3). This approach was done using 3 datasets that each contained a different combination of countries (Dataset S2) from which individuals were sampled to minimize the potential impact of within-population genetic structure and to provide a form of replication for the ABC-RF analyses (SI Appendix, Table S2). More details are provided in Methods.
Table 1.
Description of the competing scenarios and results of the 6 successive ABC analyses to infer the invasion history of P. rapae
| Step scenario | Prior error rate, % | RF votes | Posterior probability |
| Analysis 1: Europe and Asia (west/east), 18 summary statistics | 14 | ||
| S1: Asia is source of Europe | 57 | — | |
| S2: Europe is source of Asia | 132 | — | |
| S3: Asia and Europe derived from ancestral population | 811 | 0.85 | |
| Analysis 2: Siberia and North Africa, 115 summary statistics | 16 | ||
| S1: Asia is source of Siberia, Europe is source of Africa | 602 | 0.70 | |
| S2: Asia is source of Siberia; Africa is source of Europe | 162 | — | |
| S3: Africa is source of Europe; Siberia is source of Asia | 56 | — | |
| S4: Europe is source of Africa; Siberia is source of Asia | 180 | — | |
| Analysis 3: North America east (NAE), 51 summary statistics | 33 | ||
| S1: Asia is source of NAE, 1 introduction | 0 | — | |
| S2: Europe is source of NAE, 1 introduction | 576 | 0.50 | |
| S3: Asia is source of NAE, 2 introductions | 6 | — | |
| S4: Europe is source of NAE, 2 introductions | 418 | — | |
| Analysis 4: North America west (NAW), 116 summary statistics | 11 | ||
| S1: Asia is source of NAW | 19 | — | |
| S2: Europe is source of NAW | 144 | — | |
| S3: NAE is source of NAW | 721 | 0.85 | |
| S4: Europe is source of NAW ∼1600 CE | 85 | — | |
| S5: Europe is source of NAW ∼1600 CE; NAW is source of NAE | 31 | — | |
| Analysis 5: New Zealand, 223 summary statistics | 2 | ||
| S1: Asia is source of New Zealand | 2 | — | |
| S2: Europe is source of New Zealand | 14 | — | |
| S3: NAE is source of New Zealand | 16 | — | |
| S4: NAW is source of New Zealand | 968 | 0.97 | |
| Analysis 6: Australia, 388 summary statistics | 15 | ||
| S1: New Zealand is source of Australia | 631 | 0.78 | |
| S2: NAW is source of Australia | 63 | — | |
| S3: New Zealand and Europe are sources of Australia (admixture) | 15 | — | |
| S4: New Zealand and Asia are sources of Australia (admixture) | 15 | — | |
| S5: New Zealand and NAW are sources of Australia (admixture) | 276 | — |
Results are provided for only Dataset S2, dataset 1 (results for all datasets are provided in SI Appendix, Table S2). For each ABC analysis, a forest of 1,000 trees was grown. The boldfaced scenarios correspond to the selected (most likely) scenarios. Analyses 1, 2, 3, 4, 5, and 6 used 13,974, 15,533, 16,753, 17,049, 17,100, and 17,116 SNPs, respectively.
Based on the (full) final scenario (Fig. 2D), posterior model checking revealed that the observed values of only 6 summary statistics out of 928 (i.e., 0.6%) fall in the tail of the probability distribution of statistics calculated from the posterior simulation (i.e., P < 0.05 or P > 0.95), which indicates that the chosen model fitted the observed genetic data well. We found the greatest support for a scenario with an ancestral population undergoing a demographic expansion ca. 20,000 (32,000 to 4,900) y B.P. (SI Appendix, Table S3). In evaluating the source for the Europe and Asia (west/east) populations, we found the strongest support for the scenario of an ancestral population giving rise to both the Europe and Asia (west/east) populations (∼85% posterior probability), ca. 1,200 (2,900 to 300) y B.P., over scenarios with Europe as the source for Asia (west/east) or Asia (west/east) as the source for Europe (Table 1 and SI Appendix, Fig. S3A). We evaluated multiple scenarios to determine the source for the Siberia and North Africa populations and found the strongest support for a scenario with Asia (west/east) giving rise to the Siberia population ca. 300 (800 to 200) y B.P. and the Europe population giving rise to the North Africa population ca. 200 (600 to 200) y B.P. (Table 1 and SI Appendix, Fig. S3B).
We found strong support (total of 994 RF votes out of 1,000) for Europe being the source of introduction to North America (east) (Table 1 and SI Appendix, Fig. S3C). The scenario of a single introduction had only slightly better support than the scenario with multiple (2) introductions, and both have a similar number of RF votes (576 and 418, respectively, out of 1,000; Dataset S2, dataset 1). Thus, we cannot clearly distinguish between these 2 scenarios, and prior error rate was consequently relatively high (∼33%). However, subsequent analyses performed by considering multiple introductions for the formation of North America (east) do not qualitatively change any of the following results.
We found the strongest support for North America (east) serving as the source for the genetically distinct North America (west) population when compared with alternative scenarios with Asia (west/east) or Europe (for the scenarios with an ∼400 or 200 y B.P. prior estimate for date of introduction) as the source (Table 1 and SI Appendix, Fig. S3D). This introduction was estimated to have occurred ca. 137 y B.P. For the introduction into New Zealand, we found strong support for North America (west) being the source, when compared with Europe, Asia (west/east), or North America (east) as the source (Table 1 and SI Appendix, Fig. S3E). The New Zealand population was found to have the greatest support as being the source for the introduction to Australia (Table 1 and SI Appendix, Fig. S3F). All of these results were obtained with Dataset S2, dataset 1 but were qualitatively confirmed by the analyses of Dataset S2, datasets 2 and 3 (Table 1).
Demographic parameter estimates from ABC-RF analyses suggest each introduced population underwent a severe bottleneck, but the intensity (duration and number of founders with respect to the effective size of the source population) differed among populations (SI Appendix, Table S3). Specifically, New Zealand and North America (west) were estimated to have undergone the most intense bottlenecks, whereas North America (east) and, to a lesser extent, Australia suffered less intense bottlenecks.
Global Patterns of mtDNA Haplotype Diversity and Distribution.
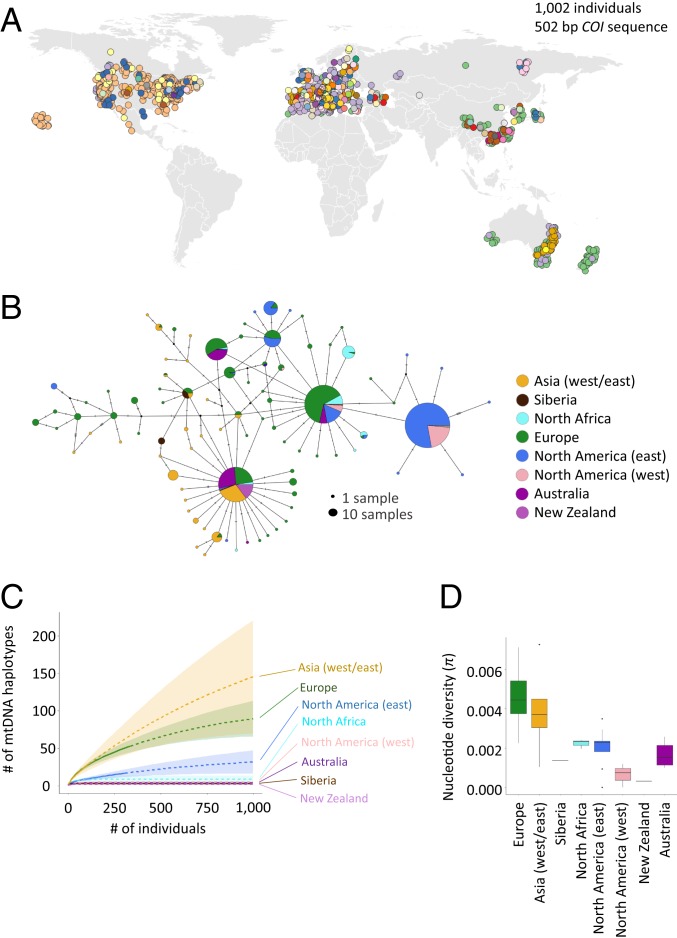
A total of 88 COI haplotypes were identified from 1,002 individuals, and 85% of these individuals harbored one of the 11 most common haplotypes (Fig. 4, SI Appendix, Fig. S4, and Dataset S1). The geographic distribution of mtDNA haplotypes is consistent with the invasion routes identified from ABC-RF analyses: Haplotypes in introduced populations are largely a subset of those from putative source populations or differ by only 1 to 2 mutations from haplotypes in high frequency in the putative source populations (Fig. 4A; an interactive figure to plot haplotypes individually is available at http://www.pierisproject.org/ResultsInvasionHistory.html).
Fig. 4.
Global patterns of mitochondrial haplotype diversity. (A) Geographic distribution of all 88 mtDNA haplotypes discovered (unique color for each haplotype; explore individual haplotypes further through interactive data visualizations [http://www.pierisproject.org/ResultsInvasionHistory.html]). Note points jittered to avoid overlapping (hidden) points; thus, coordinates are approximate, and the colors used for haplotypes are unrelated to those used in other panels. (B) Haplotype network inferred using median-joining algorithm and colored by population. Hash marks between haplotypes represent base changes (mutations). (C) Number of unique mtDNA haplotypes by population as well as subpopulation estimated using a rarefaction approach (Methods) and plotted by geographic location. (D) Pairwise nucleotide diversity by population.
Estimates of mtDNA haplotype diversity (richness) were highest in Asia (west/east) and Europe and had large confidence intervals (based on rarefaction curve analysis), indicating these populations were likely undersampled (Fig. 4C). All introduced populations had substantially lower estimates of mtDNA haplotype diversity. New Zealand, Australia, North America (west), North Africa, and Siberia were estimated to have less than a dozen mtDNA haplotypes, whereas North America (east) had substantially (∼3-fold) more mtDNA haplotypes and was significantly greater than all other recently introduced populations (based on nonoverlapping confidence intervals). Estimates of mtDNA haplotype diversity are similar to nucleotide diversity observed for autosomal markers using ddRADseq data, with the exception of Australia, which had higher mtDNA haplotype diversity than New Zealand and North America (west) (Fig. 4D). From a global perspective, there appears to be a general trend of decreasing mtDNA haplotype (and autosomal nucleotide) diversity with increasing distance from southern Europe and the eastern Mediterranean region (SI Appendix, Fig. S5).
Considering the mtDNA haplotypes found in North America and their frequencies in subpopulations of Europe, we estimate that the minimum number of individuals that would need to have been sampled from the subpopulation of England (2.3) is 23 ± 12 SD individuals or 123 ± 88 SD individuals for Spain/southern France (2.4) to account for all haplotypes in North America.
Discussion
Geographic Spread and Divergence of P. rapae Driven by Host Plant Diversification and Trade.
The Europe and Asia (west/east) populations of P. rapae are believed to represent distinct subspecies—P. rapae rapae and P. rapae crucivora, respectively—based on phenotypic differences (10) and evidence for reproductive isolation (18). Our study provides further support for this by revealing 2 main genetic lineages recovered by ddRADseq for P. rapae worldwide corresponding to the Europe (P. rapae rapae) and Asia (west/east) (P. rapae crucivora) populations. The Europe and Asia (west/east) populations appear to have diverged from an ancestral population within the last ∼3,000 y, supporting previous estimates of P. rapae being introduced to Asia within the last 2,000 to 3,000 y (10). The timing of these divergence events coincides with 2 major human activities—the diversification of Brassica crops and the development of the “Silk Road” trade routes.
Our median estimate of ∼1,200 y B.P. for the divergence of the Europe and Asia (west/east) populations coincides more with the diversification of Brassica crops than the early stages of domestication (e.g., domestication of Brassica nigra and Brassica rapa began >6,000 y B.P., domestication of Brassica oleracea began >4,000 y B.P.) (19). Selection for morphologically diverse varieties, specifically the selection for leafier varieties (20–23), may have inadvertently facilitated the growth (fitness) of P. rapae populations by providing more biomass for the P. rapae caterpillars that feed primarily on leaf tissue. Given that P. rapae larvae can feed successfully on many (if not all) species and varieties of Brassica crops, it is difficult to determine the relative role that any single diversification event contributed to the spread and/or divergence of Europe and Asia (west/east) populations. A comprehensive study that evaluates the adult (oviposition) preference and larval performance among representatives of each major species of Brassica could help tease this apart.
If Brassica crops have been cultivated across Eurasia for at least 7,000 y, why was P. rapae not introduced to Asia until a few thousand years ago? The simple answer may be that without the establishment of heavily trafficked trade routes, the natural and human-mediated dispersal of P. rapae would have been greatly restricted by geographic barriers (i.e., deserts, mountain ranges) found throughout much of western Asia. Our results suggest that development of the Silk Route facilitated the introduction of P. rapae to Asia (west/east), although the details remain unclear. The estimated timing of the divergence of the Asia (west/east) population (∼800 common era [CE]) coincides with peak trade along the Silk Route during the Tang Dynasty (∼600 to 900 CE). Further, the geographic distribution of the Silk Route largely overlaps with the range boundary of the Asia (west/east) population (P. rapae crucivora). As we would expect, Georgia and the island of Crete contain admixed populations connecting Europe and Asia (west/east), with the latter historically connected through maritime silk routes in the Mediterranean Sea (SI Appendix, Fig. S2). The general decrease in genetic diversity of P. rapae with distance from the eastern Mediterranean region further suggests that this region (or the Levant) is the source for the introduction to Asia (west/east). A pattern of eastern movement from the eastern Mediterranean or the Levant region to Georgia, followed by further spread eastward to China, reflects both human admixture and migration patterns beginning ca. 1,500 y B.P. (24). The spread of P. rapae across Asia likely consisted of multiple introductions and some level of ongoing gene flow during the initial stages. Unfortunately, our lack of samples from western/central/southern Asia limits our ability to assess fully these possibilities. Future sampling in countries located along the historical Silk Route may help uncover this part of the invasion history in more detail.
Trade appears to have facilitated the further spread of P. rapae to Siberia and North Africa. We estimate the divergence of the Siberia population occurred ca. 300 y B.P., which corresponds to the time period when many cities within the sampled regions were founded (e.g., Yakutsk in 1632 CE) and with the establishment of formal trade routes between China and Russia (i.e., the “Siberian Route,” specifically the Treaty of Kyakhta in 1727 CE) (25). The basis for introduction into North Africa is less clear, but our estimate for this event to have occurred ca. 200 y B.P. does coincide with a period of increased colonial imperialism in Africa (i.e., French colonization in Algeria began in 1830 CE) (26). It also appears there may be ongoing gene flow from North Africa populations to Europe, particularly in Spain and southern France.
Interestingly, the putative ancestral population that gave rise to the Europe and Asia (west/east) populations appears to have undergone a rapid increase in effective population size ca. 7,000 to 28,000 y B.P. This time period overlaps with early human development of agriculture. However, our median estimate for the date of this expansion is ca. 20,000 y B.P., placing it at the end of the last glacial maximum ca. 23,000 to 19,000 y B.P. (27). Changes in the distribution and demography of species in response to glacial–interglacial cycles is well documented (28, 29), and may be more likely to have facilitated a major demographic shift in P. rapae, as the earliest domestication of brassicaceous crops was relatively recent (with the earliest evidence being ca. 7,000 y B.P.) (21).
Recent Invasion History Largely Reflects Historical Records, but with a Few Unexpected Findings.
Although historical records of species invasions can be misleading (30), our molecular genomics-based reconstruction of the P. rapae global invasion history is largely consistent with historically documented observations. As expected, we found Europe to be the most likely source of this butterfly’s introduction into North America. However, we unexpectedly found that there was no discernable nuclear genetic structure within Europe (even when K = 30), making it impossible to narrow down with confidence the source population to a specific locality or country (e.g., England vs. Spain). However, mtDNA haplotype distributions and frequencies in European countries suggest England as the most likely source (i.e., fewer individuals would be required to produce the mtDNA haplotypes found in North America if England were the source rather than Spain and southern France). We do not know what specific factors account for the lack of genetic structure in Europe. One possibility is that long-distance dispersal of this species (31), coupled with historic and/or ongoing human-assisted dispersal, has led to high levels of gene flow. Another interesting possibility supported by some evidence (31–34) is that the Europe P. rapae population is migratory or undergoes migratory-like events, which would act to homogenize subpopulations across Europe. This butterfly has been shown to have no mitochondrial genetic spatial structure at the boundary between Africa and Europe (Italy-Sicily-Maghreb), as was also the case for other migratory species (35).
Historical records from Scudder (12) pointed to possible multiple introductions of P. rapae into North America, occurring shortly (<15 y) after the initial invasion. Confirming multiple introductions from the same source population early in an invasion, particularly one that quickly underwent a rapid expansion, is extremely difficult. The best-fit model to our data suggests a scenario with a single introduction, but it seems reasonable there were multiple introductions for a couple of reasons. First, both competing scenarios—one vs. multiple introductions from Europe—had a similar number of RF votes (576 vs. 418 out of 1,000) and the selected scenario (i.e., a single introduction from Europe) had a low posterior probability estimate (∼0.50; in contrast, all scenarios that were chosen in the other analyses had posterior probability estimates >0.70). Second, the estimated bottleneck intensity is rather low, with a founding population size of ∼50 to 100 individuals. This estimate is much higher than a previous estimate of 1 to 4 individuals (36). This rather large estimated founding population size and the reasonable assumption that no more than a few dozen (unrelated) butterflies would be transported on any one ship would suggest it is unlikely that North America was founded from a single introduction. Third, we observed higher heterozygosity in North America (east) than in the native range, a pattern potentially explained by multiple introduction events (from Europe) (i.e., multiple introductions aided in the rebound in genetic diversity as the introduced populations merged as they spread across eastern North America).
A second unexpected finding was the presence of a genetically distinct population within North America that is restricted to the western United States. We found evidence of admixture in areas where the 2 North America (east and west) populations come into contact, suggesting that these genetically distinct populations are neither geographically nor reproductively isolated from each other. The geographic extent of this admixture zone and the consequences of gene flow between these populations are not clear from our sampling. One hypothesis is that the western population represents an early introduction brought by Spaniards during the 1600s. However, our data indicate that it most likely results instead from a secondary founder event from the North America (east) population brought over during the period from ∼1860 to the 1880s as a result of the rapid development of railroad lines (37), specifically from the eastern United States to central California (Movie S1).
Our results confirm previous speculation that North America (west), likely San Francisco, California, was the source of introduction to the Hawaiian Islands, based on individuals from Hawaii being assigned to the North America (west) cluster but not being reported in the Hawaiian Islands until 1897 (38) (after the arrival of P. rapae in central California). Also, unexpectedly, our results suggest that the introduction of P. rapae into New Zealand came from North America (west), specifically San Francisco, and not from Europe, as was assumed, given the United Kingdom was the largest exporter into New Zealand at the time (39). Lastly, previous speculation that New Zealand is the immediate source of P. rapae in Australia (14, 15) is supported by our data.
Collections-Based Citizen Science Greatly Expands Range-Wide Sampling.
We show the public can contribute in a meaningful way to studies of species invasions through a collection-based citizen science project. Collections made by citizen scientists substantially, both qualitatively and quantitatively, expanded the geographic scope of our study. For example, the majority of countries in our study were almost entirely sampled by citizen scientists, including the entire region of Australasia. Moreover, these contributions allowed us to increase substantially the total number of individuals in each of the populations studied, with roughly half of all specimens sequenced in our study coming from citizen scientists. We estimate that the use of citizen science to aid in the collection of P. rapae from across its near-global range resulted in tens of thousands of US dollars in cost-savings that would have been required to cover salary and travel costs. We believe our collections-based citizen science approach can be applied to other systems, particularly to organisms that are easily identifiable (e.g., spotted lantern fly, Giant African snail) and easy to transport (e.g., dead invertebrates), to address questions in invasion biology as well as a broad range of questions in ecology and evolutionary biology. As examples, we currently are leveraging our large collection to address questions concerning the effects of climate and land use changes on wing pigmentation of this butterfly and to identify genomic regions underlying ecological selection.
The development, implementation, and maintenance of this project were not trivial, as is the case with many citizen science projects, and required considerable time and effort engaging the public (e.g., contacting organizations, using social media, responding to emails) and processing samples. Collections-based citizen science projects that focus on less charismatic species or incorporate nonlethal forms of sampling (e.g., environmental DNA) and are easy to collect (slow-moving or sessile) may have the greatest success. We suggest that those interested in applying a collections-based citizen science approach seek advice from, or build collaborations with, individuals with experience in the field of citizen science.
Implications for Invasion Biology.
Growing evidence shows many invasive populations are able to flourish and adapt to new environments despite substantial loss of genetic diversity—a phenomenon termed the genetic paradox of invasions (40). P. rapae is a remarkable example of this paradox. The spread of P. rapae to new continents was facilitated by long-distance jump dispersal events linked together through an almost linear (and branching) series of invasions (41, 42), with each new founding population the product of a previously bottlenecked population (i.e., multiple serial founding events). Whether introduced P. rapae populations maintained high genetic variation in ecologically relevant traits following each founding event remains unclear. Evidence of local adaption for thermal tolerances among populations in North America (43) suggests such variation exists. Further, the observation that nonmigratory populations in Australia are “preadapted” for migration (exhibit biased directionality in dispersal) suggests this trait has been retained despite each successive bottleneck and may have aided within-continent dispersal (e.g., rapid spread across North America and Australia). Resolving this paradox and the persistent puzzle of how this butterfly has been an extremely successful invader into new environments will require future studies to assess the relative contributions of factors such as adaptive evolution, phenotypic plasticity, natural enemy escape, and the spread and diversification of its Brassica host plants, as well as use of native hosts or feral populations of brassicaceous species.
The elucidation of routes of invasion and patterns of genetic structure also has implications for development of practical management strategies. In the case of P. rapae, the finding that all introductions originated in Europe provides further support that this region, or perhaps more specifically the eastern Mediterranean or even Levant region, may harbor the greatest diversity of natural enemies. On the other hand, the genetic structure detected at both the continent and subcontinent levels, particularly in Asia and North America, suggests that management efforts of P. rapae should consider how these genetic differences influence responses and outcomes to specific practices. Interestingly, no known P. rapae populations have evolved resistance to Bacillus thuringiensis, whereas the diamondback moth (Plutella xylostella) and cabbage looper (Trichoplusia ni), also pests of Brassica, have been able to evolve resistance in the field or laboratory (44). While the cause of such differences is unclear, one possibility is P. rapae populations may be at a disadvantage to evolving resistance because of loss of genetic diversity as a result of recent and repeated bottlenecks. Research focused on understanding the genetic underpinnings of ecologically and physiologically relevant phenotypic traits of P. rapae and assessing whether and how genetic variation in these traits have changed as a result of invasion history may shed light on future management efforts of P. rapae and our understanding of invasion biology more broadly.
Methods
Specimens Collection and DNA Extractions.
The P. rapae specimens were collected as part of an international citizen science project—Pieris Project—that was launched in June 2014 and through collections by researchers. A website—www.pierisproject.org—was created in 2014 for the Pieris Project that included a description of the research goals and collection protocol: Specimens were to be individually placed in handmade or glassine envelopes, labeled with the location and date collected, and placed in a freezer overnight; they were then air-dried for at least 2 d and shipped using standard mail. The project was advertised through social media (Twitter [@PierisProject] and Facebook [https://www.facebook.com/pierisproject/]) and through listservs, social media, and blogs of entomological and lepidopterists societies and nature/science/citizen science-related organizations (e.g., YourWildLife, eButterfly, National Geographic, SciStarter). Once received, specimens were stored in 95% ethanol and kept at −20 °C; depending on the collector, specimens were air-dried for a few days to years prior to being placed in ethanol. Genomic DNA was isolated from tissue from the prothorax or (2 to 3) legs using DNeasy Blood and Tissue Kit spin-columns (catalog no./ID: 6950; Qiagen).
To estimate the contributions by scientists, we binned the collector of each specimen into one of 2 categories: 1) researcher and 2) citizen scientist. There is a great deal of debate as to what does or does not constitute being a citizen scientist. Therefore, we used both an inclusive definition (a citizen scientist is anyone contributing to this project who is not a formal collaborator) and a restrictive definition (a citizen scientist is anyone contributing who is not a formal collaborator and not a professional scientist [i.e., has an advanced degree in biology]).
ddRADseq Sequencing and Filtering.
Nine reduced-complexity libraries were generated using ddRADseq (45) following the method of Ryan et al. (46). Briefly, genomic DNA (∼400 ng) was digested with the restriction enzymes EcoR1 and Mse1 and a universal Mse1 and barcoded EcoR1 adapter ligated to the digested DNA. Ligated products were diluted 10 times with 0.1× TE buffer prior to PCR enrichment. Amplified products with unique barcodes were pooled into a single mixture prior to purification. The library was purified 3 times with a 0.8× volume of Agencourt Ampure XP beads (A63881; Beckman Coulter). At the end of each round of purification, the elution volume was reduced to 0.25- to 0.5-fold of the beginning sample volume. After 3 rounds of purification, each library (1.0 μg) was size-selected for 400- to 600-bp fragment length using a 1.5% DF Cassette and BluePippin System (Sage Science). Libraries were evaluated using a Bioanalyzer 2100 system and sequenced across 1 lane using an Illumina MiSeq system (Genomics & Bioinformatics Core Facility, University of Notre Dame) and 14 lanes of an Illumina HiSeq 4000 system (12 at the University of Illinois and 2 at the Beijing Genomics Institute); most samples were sequenced on 2 (some on 3) independent lanes.
Raw reads were demultiplexed, and barcodes/cutsite were removed using a custom Python script. Reads were further trimmed and cleaned with the program Trimmomatic (v0.32) (47) using default settings. The first 5 bp and any after 80 bp were then trimmed from all reads, and only reads at least 76 bp in length were retained, resulting in all reads being exactly 76 bp.
Reads were then aligned to the P. rapae genome v1 (36) using BWA-aln (v0.7.15) (48). Variant calling was performed using the Genome Analysis Toolkit’s (GATK’s) Haplotypecaller (v3.8) (49, 50) with the default settings. Filters were applied in the following order: kept only biallelic SNPs, applied GATK’s “hard filtering” (quality by depth < 2.0 || mapping quality < 40.0 || mapping quality Rank Sum test < −12.5 || read position Rank Sum test < −8.0), SNPs with a genotype quality <20 were converted to missing data, removed SNPs with minor allele frequency less than 0.01, kept SNPs with minimum of 1-fold coverage for 50% of individuals, removed SNPs with coverage >95th percentile (112.8-fold coverage), removed individuals with >75% missing data, kept only SNPs with a minimum of 10-fold coverage in 90% of individuals, and removed individuals with >25% missing data. Finally, SNPs with heterozygosity >0.6 were considered potential paralogs and were discarded.
As there is no linkage map for P. rapae and the genome is not assembled into chromosomes, we applied a simple heterozygosity method to determine whether SNPs were autosomal or sex (Z)-linked. To do this, we used the expectation that females should be homozygous at all SNPs on the Z-chromosome—females are the heterogametic sex (ZW) in Lepidoptera. Using 231 females, we calculated the percentage that were heterozygous or homozygous at each site (SNP) using the is_het function from the R package vcfR (v1.6.0) (51) and custom scripts in R. SNPs with greater than 25% missing data were removed. A scaffold (and all SNPs within) was considered putatively Z-linked if >60% of the SNPs fell below the threshold of having less than 1% of the females being heterozygous (average number of SNPs for each scaffold was 84 ± 101, mode = 4).
To complement the heterozygosity method, we also inferred the chromosome assignment of each P. rapae scaffold using the approach by Ryan et al. (52). Briefly, we used BlastX (ncbi-blast-2.2.30+) (53, 54) to blast all peptide sequences within each scaffold of the P. rapae assembly against the Bombyx mori genome (silkdb v2.0) (55). The B. mori scaffold with the most significant BLAST hits (based on bit scores) was retained and used to determine the putative chromosome of each P. rapae scaffold. All, except 1 scaffold, of those we identified as Z-linked using the heterozygosity method mapped to chromosomes 1 (Z) and 2 (W) of B. mori (SI Appendix, Fig. S6). That we found some regions of P. rapae mapping to the B. mori chromosome 2 (W) suggests they are not completely syntenic: The P. rapae genome was assembled from males; thus, there should be no scaffold mapping to this chromosome. Some P. rapae scaffolds mapping to chromosome 2 (W) of B. mori were recovered as actually being on chromosome 1 (Z) based on the heterozygosity method.
Using a subset of the putative Z-linked markers—SNPs where <1% of females had a heterozygous call (i.e., SNPs with a high likelihood of being Z-linked), we validated the sex of each individual. Females and males with >20% or <20% of these SNPs being heterozygous were considered possibly mislabeled males and females, respectively. These individuals were flagged, and the specimens were double-checked visually; in all cases, visual identification confirmed that these individuals were mislabeled.
Inference of Population Structure and Diversity.
Population structure was investigated with the model-based clustering algorithm ADMIXTURE (56) using default settings and a cross-validation = 10 for K 1 to 30 using a modified SNP dataset (i.e., pruned for linkage disequilibrium [LD] [r2 > 0.2; calculated using VCFtools geno-r2], for a total of 17,917 SNPs). The optimal K was that with the lowest cross-validation error. The Bayesian program fastSTRUCTURE (57) and a non–model-based multivariate approach—discriminant analysis of principal components (SI Appendix, Fig. S2C) (58)—were also used to confirm the results from ADMIXTURE (more details are provided in SI Appendix) using the R package adegenet (v2.1.1) (59). Genetic assignments were plotted using custom scripts and the R package pophelper (v2.2.3) (60). A neighbor-joining tree based on genetic distance was constructed in the poppr (v2.8.0) (61) and ape (v5.1) (62) packages in R, which included the species Pieris napi, Pieris brassicae, and Pieris canidia as outgroups, using only sites with at least 15-fold coverage in 90% of individuals from this new dataset. Trees were visualized using FigTree (v1.4.4) (63). Population differentiation was estimated between all populations using the Weir and Cockerham’s estimator of FST (64) implemented in VCFtools (v0.1.15) using 10-kilobase (kb) windows and a window step size of 5 kb.
All measures of genetic diversity (observed heterozygosity, pairwise nucleotide diversity [π], and Tajima’s D) were calculated using SNPs restricted to scaffolds longer than 100 kb (22,059 SNPs). In an attempt to minimize the Wahlund effect (i.e., reduction of heterozygosity caused by subpopulation structure), individuals were split into spatially contiguous subpopulations from within the 7 identified by ADMIXTURE (n = 34; 1 subpopulation from Mexico was not included because it contained only 3 individuals); these were the same subpopulations used for the ABC-RF analyses. To control for differences in sample size, we computed each statistic 1,000 times using a random subset (without replacement) of 7 individuals (size of smallest population). Heterozygosity was estimated using the R package adegenet v2.1.1. Calculations for π and Tajima’s D were estimated using a dataset containing invariant sites (i.e., vcf [variant call format] files were created using gatk-4.0.4.0 with the flag -allSites true, and the same filters as described above were then applied) with VCFtools (v0.1.15) using 10-kb windows (and a window step size of 5 kb used for estimating π).
ABC-RF–Based Inferences of Global Invasion History.
An ABC analysis (65) was carried out to infer the global invasion history of P. rapae. The 8 populations considered in the ABC analysis corresponded to the 7 identified by ADMIXTURE, with an additional separation of New Zealand and Australia for geographic reasons. Each population was represented in the analysis by a single subpopulation (individuals sampled within the same subregion and within a 3-y period) (Dataset S2, dataset 1). ABC is a model-based Bayesian method allowing posterior probabilities of historical scenarios to be computed, based on historical data and massive simulations of genetic data. We used historical information (e.g., dates of first observation of invasive populations) to define 6 sets of competing introduction scenarios that were analyzed sequentially (Table 1 and SI Appendix, Fig. S3). Step by step, subsequent analyses used the results obtained from the previous analyses, until the most recent invasive populations were considered. The first set of competing scenarios (3 scenarios) considered the evolutionary relationship between the Asia (west/east) and Europe populations. In the second analysis (4 scenarios), we explored the links between Asia (west/east), Europe, North Africa, and Siberia. In the third analysis (4 scenarios), we set North America (east) as the target and determined whether it originated from Asia (west/east) or Europe, through either 1 or 2 introductions. In the fourth analysis (5 scenarios), North America (west) could be originating from Europe, Asia (west/east), or North America (east), and the introduction could be ancient (400 y B.P.) in the case of Europe. In the fifth analysis (4 scenarios), the New Zealand population could be originating from either Europe, Asia (west/east), North America (east), or North America (west). Finally, the sixth analysis (5 scenarios) aimed at deciphering the origin of the Australian population by testing as the source population New Zealand; North America (west); and admixtures between New Zealand and Europe, Asia (west/east), or North America (west). All scenarios of all analyses are detailed in Table 1 and SI Appendix, Fig. S3.
In our ABC analyses, historical and demographic parameter values for simulations were drawn from prior distributions defined from historical data and demographic parameter values available from empirical studies on P. rapae (12–14, 38), as described in SI Appendix, Table S4. Simulated and observed datasets were summarized using the whole set of summary statistics proposed by DIYABC (66) for SNP markers, describing genetic variation per population (e.g., mean gene diversity across loci), per pair (e.g., mean across loci of FST distances), or per triplet (e.g., mean across loci of admixture estimates) of populations (details about statistics are provided in DIYABC v2.1.0), plus the linear discriminant analysis axes (67) as additional summary statistics (SI Appendix, Table S5). The total number of summary statistics ranged from 18 to 388 depending on the analysis (Table 1).
To compare the scenarios, we used a RF process (68, 69). RF is a machine-learning algorithm that uses hundreds of bootstrapped decision trees to perform classification using a set of predictor variables, the summary statistics here. Some simulations are not used in tree building at each bootstrap (i.e., the out-of-bag simulations), and can thus be used to compute the “prior error rate,” which provides a direct method for cross-validation. We simulated a 10,000-SNP dataset for each competing scenario using the standard Hudson’s algorithm for minor allele frequency (i.e., only polymorphic SNPs over the entire dataset are considered), so the number of used SNP markers ranged between 13,974 and 17,116 depending on the analysis (Table 1). We then grew a classification forest of 1,000 trees based on the simulated datasets. The RF computation applied to the observed dataset provides a classification vote (i.e., the number of times a model is selected among the 1,000 decision trees). The scenario with the highest classification vote was selected as the most likely scenario, and we then estimated its posterior probability by way of a second RF procedure of 1,000 trees (69). To evaluate the global performance of our ABC-RF scenario choice, we computed the prior error rate based on the available out-of-bag simulations and conducted the complete scenario selection analysis with 2 additional datasets with different subpopulations (Dataset S2, datasets 2 and 3) representative of the same populations as Dataset S2, dataset 1 (70).
We then performed a posterior model checking analysis on a full final scenario, including all 8 populations (Dataset S2, dataset 1), to determine whether this scenario matches well with the observed genetic data. Briefly, if a model fits the observed data correctly, then data simulated under this model with parameters drawn from their posterior distribution should be close to the observed data. The lack of fit of the model to the data with respect to the posterior predictive distribution can be measured by determining the frequency at which the observed summary statistics are extreme with respect to the simulated summary statistics distribution (hence, defining a tail-area probability, or P value, for each summary statistic). We simulated 100,000 datasets under the full final scenario (17,609 SNP and 928 summary statistics), and then obtained a “posterior sample” of 5,000 values of the posterior distributions of parameters through a rejection step based on Euclidean distances and a linear regression posttreatment (65). We simulated 1,000 new datasets with parameter values drawn from this “posterior sample,” and each observed summary statistic was compared with the distribution of the 1,000 simulated test statistics, and its P value, corrected for multiple comparisons with the false discovery rate procedure (71), was computed.
Finally, 10,000 simulated datasets of the full final scenario were used to infer posterior distribution values of all parameters, and some relevant composite parameters under a regression by RF methodology (72), with classification forests of 1,000 trees. The simulation steps, the computation of summary statistics, and the model checking analysis were performed using DIYABC v2.1.0. All scenario comparisons and parameter estimations were carried out in R using the package abcrf (v1.7.1) (69).
mtDNA Sequencing and Analysis.
A 1,600-bp region of COI was amplified using primers optimized to work with multiple species within the genera Pieris (Pieridae_COI_F 5-AAATTTACAATYTATCGCTTA-3, Pieridae_COI_R 5-TGGGGTTTAAATCCATTACATATW-3). When these primers failed, we amplified a 658-bp region of COI using previously published primers (73). PCR amplicons were purified using magnetic beads and amplified using standard fluorescent cycle sequencing PCR reactions (ABI Prism Big Dye terminator chemistry; Applied Biosystems). Sequencing reactions were purified using Agencourt CleanSeq magnetic beads (Beckman Coulter) and run on an ABI-3730XL-96 capillary sequencer (Applied Biosystems) at the University of Florida biotechnology facility (ICBR) or Macrogen, Inc. Individuals with both forward and reverse reads were assembled in Geneious 11.0.4 using the De Novo Assemble tool with default settings. The find heterozygotes tool (peak similarity set to 50%) was used to find and discard any sequences found to be heterozygous. Reads were trimmed to 502 bp and aligned (error probability limit of 0.001) with sequences from the GenBank and Barcode of Life databases using MUSCLE Alignment in Geneious with default settings.
To evaluate whether we were adequately sampling mtDNA haplotype diversity, we plotted rarefaction curves (estimates of haplotype richness by sampling effort) for each population using iNEXT (74) and predicted the total haplotypes for each population assuming 1,000 sampled individuals. A median-joining haplotype network was created using PopART (75) for all populations and for each population separately.
In an effort to further pinpoint whether the introductions in North America came from western (i.e., United Kingdom) or southwestern (i.e., Spain, France) Europe, we estimated the minimum number of individuals that would need to be sampled from each of these native populations to generate the mtDNA diversity found in North America. Specifically, for each native subpopulation, we randomly sampled (with replacement) a haplotype from each subpopulation based on their haplotype frequencies until all haplotypes represented in North America were sampled and simulated this procedure 10,000 times for each subpopulation. This approach assumes that the true source population will be the most parsimonious (i.e., require sampling of fewer individuals to create the diversity found in North America).
Data Availability.
Demultiplexed ddRADseq reads generated in this study are available through the National Center for Biotechnology Information’s Sequence Read Archive associated with Bioproject PRJNA542919 (76). All COI sequences were deposited in the Barcode of Life Database under the project “Pieris rapae Global Invasion History [PRA]” (77) and in the GenBank database (BankIt2244911: accession nos. MN181608 to MN182331). All metadata and scripts associated with analyses in this study have been deposited on GitHub (https://github.com/citscisean/PierisrapaeInvasionHistory) (78).
Supplementary Material
Acknowledgments
We thank all the participants in the Pieris Project; without their help, this research would not have been possible. We also thank Arthur Shapiro for his extraordinary insights into this system and the many researchers who contributed specimens, including Brent Sinclair, Jantina Toxopeus, Dmitry Musolin, Tatsuro Konagaya, John Peters, Chuan-Kai Ho, Michael Braby, Siobhan Leachman, and many more. We thank Sang-guy Park for donating specimens from his private collection and Robert Cullenbine for his support of this research. We also thank Jacqueline Lopez and Melissa Stephens in the Notre Dame Genomics & Bioinformatics Core Facility for ddRADseq library preparations. This research was funded by US Department of Agriculture-National Institute of Food and Agriculture Postdoctoral Fellowship Grant 2017-67012-26999 (to S.F.R.). A.E. is supported by a National Science Foundation Graduate Research Fellowship under Grant NSF-1447692. R.V. was supported by project CGL2016-76322-P (Spanish Agencia Estatal de Investigación and European Union Regional Development Fund). G.T. is supported by Ministerio de Economia y Competitividad Programme IJCI-2016-29083 and by the National Geographic Society (Grant WW1-300R-18). E.A.H. was supported by Marie Curie Actions International Outgoing Fellowship 330136.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Demultiplexed double-digest restriction site-associated DNA fragment procedure sequencing reads generated in this study are available through the National Center for Biotechnology Information’s Sequence Read Archive associated with Bioproject PRJNA542919. All gene cytochrome c oxidase subunit 1 (COI) sequences were deposited in the Barcode of Life Database under the project “Pieris rapae Global Invasion History [PRA]” and in the GenBank database (BankIt2244911: accession nos. MN181608 to MN182331). All metadata and scripts associated with analyses in this study have been deposited on GitHub (https://github.com/citscisean/PierisrapaeInvasionHistory).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907492116/-/DCSupplemental.
References
- 1.Meyerson L. A., Mooney H. A., Invasive alien species in an era of globalization. Front. Ecol. Environ. 5, 199–208 (2007). [Google Scholar]
- 2.Seebens H., et al. , No saturation in the accumulation of alien species worldwide. Nat. Commun. 8, 14435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Westphal M. I., Browne M., MacKinnon K., Noble I., The link between international trade and the global distribution of invasive alien species. Biol. Invasions 10, 391–398 (2008). [Google Scholar]
- 4.Chen Y. H., Crop domestication, global human-mediated migration, and the unresolved role of geography in pest control. Elem. Sci. Anth. 4, 000106 (2016). [Google Scholar]
- 5.Estoup A., Guillemaud T., Reconstructing routes of invasion using genetic data: Why, how and so what? Mol. Ecol. 19, 4113–4130 (2010). [DOI] [PubMed] [Google Scholar]
- 6.McKinley D. C., et al. , Citizen science can improve conservation science, natural resource management, and environmental protection. Biol. Conserv. 208, 15–28 (2017). [Google Scholar]
- 7.Cooper C. B., Shirk J., Zuckerberg B., The invisible prevalence of citizen science in global research: Migratory birds and climate change. PLoS One 9, e106508 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryan S. F., et al. , The role of citizen science in addressing grand challenges in food and agriculture research. Proc. Biol. Sci. 285, 20181977 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hely P. C., Gellatley J. G., Pasfield G., Agriculture NSWD of Insect Pests of Fruit and Vegetables in NSW (Inkata Press, Clayton, VIC, 1982). https://trove.nla.gov.au/version/25802846?q&versionId=45531505. Accessed 30 August 2018.
- 10.Fukano Y., Satoh T., Hirota T., Nishide Y., Obara Y., Geographic expansion of the cabbage butterfly (Pieris rapae) and the evolution of highly UV-reflecting females. Insect Sci. 19, 239–246 (2012). [Google Scholar]
- 11.Hiura I., Monshirochou-zoku no Rekishi. Konchu Shizen 3, 9–15 (1968). [Google Scholar]
- 12.Scudder S. H., The Introduction and Spread of Pieris rapae in North America, 1860-1885 [i.e., 1886] (Boston Society of Natural History, Boston, 1887). https://www.biodiversitylibrary.org/bibliography/38374. Accessed 14 September 2016.
- 13.Ashby J. W., Pottinger R. P., Natural regulation of Pieris rapae Linnaeus (Lepidoptera : Pieridae) in Canterbury, New Zealand. N. Z. J. Agric. Res. 17, 229–239 (1974). [Google Scholar]
- 14.Braby M. F., The Butterflies of Australia: Their Identification, Biology and Distribution (CSIRO Publishing, Melbourne, 2000). [Google Scholar]
- 15.Peters J. V., The cabbage white butterfly. Aust. Nat. Hist. 16, 300–303 (1970). [Google Scholar]
- 16.Andow D. A., Kareiva P. M., Levin S. A., Okubo A., Spread of invading organisms. Landsc. Ecol. 4, 177–188 (1990). [Google Scholar]
- 17.Seiter S., Kingsolver J., Environmental determinants of population divergence in life-history traits for an invasive species: Climate, seasonality and natural enemies. J. Evol. Biol. 26, 1634–1645 (2013). [DOI] [PubMed] [Google Scholar]
- 18.McQueen E. W., Morehouse N. I., Rapid divergence of wing volatile profiles between subspecies of the butterfly Pieris rapae (Lepidoptera: Pieridae). J. Insect Sci. 18, 33 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maggioni L., von Bothmer R., Poulsen G., Branca F., Origin and domestication of cole crops (Brassica oleracea L.): Linguistic and literary considerations1. Econ. Bot. 64, 109–123 (2010). [Google Scholar]
- 20.Qi X., et al. , Genomic inferences of domestication events are corroborated by written records in Brassica rapa. Mol. Ecol. 26, 3373–3388 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Prakash S., Wu X.-M., Bhat S. R., “History, evolution, and domestication of brassica crops” in Plant Breeding Reviews, Janick J., Ed. (Wiley–Blackwell, 2011), pp. 19–84. [Google Scholar]
- 22.Maggioni L., von Bothmer R., Poulsen G., Lipman E., Domestication, diversity and use of Brassica oleracea L., based on ancient Greek and Latin texts. Genet. Resour. Crop Evol. 65, 137–159 (2018). [Google Scholar]
- 23.Maggioni L., “Domestication of Brassica oleracea L,” PhD thesis, Acta Universitatis Agriculturae Sueciae, Uppsala, Sweden (2015). https://pub.epsilon.slu.se/12424/. Accessed 15 November 2018.
- 24.Mezzavilla M., et al. , Genetic landscape of populations along the Silk Road: Admixture and migration patterns. BMC Genet. 15, 131 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noda J., The Kazakh Khanates between the Russian and Qing Empires: Central Eurasian International Relations during the Eighteenth and Nineteenth Centuries (Islamic Area Studies, Brill, 2016), vol. 3. https://brill.com/view/title/32948. Accessed 25 April 2019.
- 26.Sessions J. E., By Sword and Plow: France and the Conquest of Algeria (Cornell University Press, 2011), ed. 1. [Google Scholar]
- 27.Clark P. U., et al. , The last glacial maximum. Science 325, 710–714 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Hewitt G. M., Genetic consequences of climatic oscillations in the Quaternary. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359, 183–195, discussion 195 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hewitt G., The genetic legacy of the Quaternary ice ages. Nature 405, 907–913 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Fischer M. L., et al. , Historical invasion records can be misleading: Genetic evidence for multiple introductions of invasive Raccoons (Procyon lotor) in Germany. PLoS One 10, e0125441 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones R. E., Gilbert N., Guppy M., Nealis V., Long-distance movement of Pieris rapae. J. Anim. Ecol. 49, 629–642 (1980). [Google Scholar]
- 32.Williams C., The Migration of Butterflies (Oliver & Boyd, Edinburgh, 1930). [Google Scholar]
- 33.John E., Cottle N., McArthur A., Markis C., Eastern Mediterranean migrations of Pieris rapae (Linnaeus, 1758) (Lepidoptera: Pieridae): Observations in Cyprus, 2001 and 2007. Entomol. Gaz. 59, 71–78 (2008). [Google Scholar]
- 34.Gilbert N., Raworth D. A., Movement and migration patterns in Pieris rapae (Pieridae). J. Lepid. Soc. 59, 10–18 (2005). [Google Scholar]
- 35.Vodă R., et al. , Historical and contemporary factors generate unique butterfly communities on islands. Sci. Rep. 6, 28828 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen J., et al. , Complete genome of Pieris rapae, a resilient alien, a cabbage pest, and a source of anti-cancer proteins. F1000 Res. 5, 2631 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atack J., Historical geographic information systems (GIS) database of U.S. Railroads for 1830-1972 (2016). https://my.vanderbilt.edu/jeremyatack/data-downloads/. Accessed 1 November 2018.
- 38.Opler P. A., Krizek G. O., Butterflies East of the Great Plain: An Illustrated Natural History (Johns Hopkins University Press, Baltimore, 1984). [Google Scholar]
- 39.Census and Statistics Office , New Zealand Official Yearbook (1930). https://www3.stats.govt.nz/New_Zealand_Official_Yearbooks/1930/NZOYB_1930.html. Accessed 15 November 2018.
- 40.Allendorf F. W., Lundquist L. L., Introduction: Population biology, evolution, and control of invasive species. Conserv. Biol. 17, 24–30 (2003). [Google Scholar]
- 41.Suarez A. V., Holway D. A., Case T. J., Patterns of spread in biological invasions dominated by long-distance jump dispersal: Insights from Argentine ants. Proc. Natl. Acad. Sci. U.S.A. 98, 1095–1100 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Floerl O., Inglis G. J., Dey K., Smith A., The importance of transport hubs in stepping-stone invasions. J. Appl. Ecol. 46, 37–45 (2009). [Google Scholar]
- 43.Kingsolver J. G., Massie K. R., Ragland G. J., Smith M. H., Rapid population divergence in thermal reaction norms for an invading species: Breaking the temperature-size rule. J. Evol. Biol. 20, 892–900 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Tabashnik B. E., Evolution of resistance to Bacillus thuringiensis. Annu. Rev. Entomol. 39, 47–79 (1994). [DOI] [PubMed] [Google Scholar]
- 45.Peterson B. K., Weber J. N., Kay E. H., Fisher H. S., Hoekstra H. E., Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One 7, e37135 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ryan S. F., et al. , Climate-mediated hybrid zone movement revealed with genomics, museum collection, and simulation modeling. Proc. Natl. Acad. Sci. U.S.A. 115, E2284–E2291 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DePristo M. A., et al. , A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKenna A., et al. , The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knaus B. J., Grünwald N. J., vcfr: A package to manipulate and visualize variant call format data in R. Mol. Ecol. Resour. 17, 44–53 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Ryan S. F., et al. , Patterns of divergence across the geographic and genomic landscape of a butterfly hybrid zone associated with a climatic gradient. Mol. Ecol. 26, 4725–4742 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 54.Shiryev S. A., Papadopoulos J. S., Schäffer A. A., Agarwala R., Improved BLAST searches using longer words for protein seeding. Bioinformatics 23, 2949–2951 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Wang J., et al. , SilkDB: A knowledgebase for silkworm biology and genomics. Nucleic Acids Res. 33, D399–D402 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alexander D. H., Novembre J., Lange K., Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raj A., Stephens M., Pritchard J. K., fastSTRUCTURE: Variational inference of population structure in large SNP data sets. Genetics 197, 573–589 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jombart T., Devillard S., Balloux F., Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 11, 94 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jombart T., adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Francis R. M., pophelper: An R package and web app to analyse and visualize population structure. Mol. Ecol. Resour. 17, 27–32 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Kamvar Z. N., Tabima J. F., Grünwald N. J., Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2, e281 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paradis E., Claude J., Strimmer K., APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Rambaut A., FigTree v1.4.4: Tree Figure Drawing Tool (2009). http://tree.bio.ed.ac.uk/software/figtre?e/. Accessed 3 December 2018.
- 64.Weir B. S., Cockerham C. C., Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370 (1984). [DOI] [PubMed] [Google Scholar]
- 65.Beaumont M. A., Zhang W., Balding D. J., Approximate Bayesian computation in population genetics. Genetics 162, 2025–2035 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cornuet J.-M., et al. , DIYABC v2.0: A software to make approximate bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 30, 1187–1189 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Estoup A., et al. , Estimation of demo-genetic model probabilities with approximate Bayesian computation using linear discriminant analysis on summary statistics. Mol. Ecol. Resour. 12, 846–855 (2012). [DOI] [PubMed] [Google Scholar]
- 68.Breiman L., Random forests. Mach. Learn. 45, 5–32 (2001). [Google Scholar]
- 69.Pudlo P., et al. , Reliable ABC model choice via random forests. Bioinformatics 32, 859–866 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Lombaert E., et al. , Complementarity of statistical treatments to reconstruct worldwide routes of invasion: The case of the Asian ladybird Harmonia axyridis. Mol. Ecol. 23, 5979–5997 (2014). [DOI] [PubMed] [Google Scholar]
- 71.Cornuet J.-M., Ravigné V., Estoup A., Inference on population history and model checking using DNA sequence and microsatellite data with the software DIYABC (v1.0). BMC Bioinformatics 11, 401 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Raynal L., et al. , ABC random forests for Bayesian parameter inference. Bioinformatics 35, 1720–1728, (2019). [DOI] [PubMed] [Google Scholar]
- 73.Hebert P. D. N., Stoeckle M. Y., Zemlak T. S., Francis C. M., Identification of birds through DNA barcodes. PLoS Biol. 2, e312 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hsieh T. C., Ma K. H., Chao A., iNEXT: An R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol. Evol. 7, 1451–1456 (2016). [Google Scholar]
- 75.Leigh J. W., Bryant D., popart: Full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116 (2015). [Google Scholar]
- 76.Ryan S. F., et al. , Global invasion history of the agricultural pest butterfly Pieris rapae: A citizen science population genomics study. Sequence Read Archive. https://www.ncbi.nlm.nih.gov/bioproject?term=PRJNA542919. Deposited 6 May 2019. [DOI] [PMC free article] [PubMed]
- 77.Ryan S. F., et al. , Pieris rapae Global Invasion History [PRA]. Barcode of Life Database. http://v3.boldsystems.org/index.php/MAS_Management_OpenProject?code=PRA. Deposited 14 May 2019.
- 78.Ryan S. F., et al. , PierisrapaeInvasionHistory. GitHub. https://github.com/citscisean/PierisrapaeInvasionHistory. Deposited 18 June 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Demultiplexed ddRADseq reads generated in this study are available through the National Center for Biotechnology Information’s Sequence Read Archive associated with Bioproject PRJNA542919 (76). All COI sequences were deposited in the Barcode of Life Database under the project “Pieris rapae Global Invasion History [PRA]” (77) and in the GenBank database (BankIt2244911: accession nos. MN181608 to MN182331). All metadata and scripts associated with analyses in this study have been deposited on GitHub (https://github.com/citscisean/PierisrapaeInvasionHistory) (78).