Significance
MITA/STING is an essential adaptor protein for innate immune response to DNA virus or damaged cellular DNA. In this study, we identified 2 protein phosphatases, PTPN1 and PTPN2, as negative regulators of MITA/STING. After the onset of innate immune response to DNA virus, PTPN1/2 dephosphorylate MITA/STING, which subsequently causes its degradation by the ubiquitin-independent 20S proteasomes. This study reveals a striking mechanism on how innate immune response to DNA virus is attenuated and therefore would help for drug or vaccine development against DNA virus infection.
Keywords: DNA virus, PTPN1, PTPN2, 20S proteasome, innate immune response
Abstract
Upon cytosolic viral DNA stimulation, cGMP-AMP synthase (cGAS) catalyzes synthesis of 2′3′cGMP-AMP (cGAMP), which binds to the adaptor protein MITA (mediator of IRF3 activation, also called STING, stimulator of IFN genes) and induces innate antiviral response. How the activity of MITA/STING is regulated to avoid excessive innate immune response is not fully understood. Here we identified the tyrosine-protein phosphatase nonreceptor type (PTPN) 1 and 2 as MITA/STING-associated proteins. PTPN1 and PTPN2 are associated with MITA/STING following viral infection and dephosphorylate MITA/STING at Y245. Dephosphorylation of MITA/STING leads to its degradation via the ubiquitin-independent 20S proteasomal pathway, which is dependent on the intrinsically disordered region (IDR) of MITA/STING. Deficiencies of PTPN1 and PTPN2 enhance viral DNA-induced transcription of downstream antiviral genes and innate antiviral response. Our findings reveal that PTPN1/2-mediated dephosphorylation of MITA/STING and its degradation by the 20S proteasomal pathway is an important regulatory mechanism of innate immune response to DNA virus.
The innate immune system utilizes a limited number of pattern-recognition receptors (PRRs) to recognize pathogen-associated molecular patterns (PAMPs) of microbes. Virus-derived nucleic acids are major PAMPs for initiation of innate antiviral immunity as well as subsequent adaptive immune response (1). It is well established that viral DNA is mostly sensed by the cGMP-AMP synthase (cGAS). Upon binding of viral DNA, cGAS catalyzes synthesis of the second messenger molecule 2′3′cGMP-AMP (cGAMP), which subsequently binds to the endoplasmic reticulum (ER)-associated membrane protein mediator of IRF3 Activation (MITA, also known as STING, stimulator of IFN genes) (2, 3). MITA consists of 4 N-terminal transmembrane domains, a cyclic dinucleotide binding domain (CBD), and a C-terminal tail (CTT). In unstimulated cells, MITA exists in an autoinhibited status with an intramolecular interaction between its CBD and CTT. Following DNA virus infection, binding of cGAMP to MITA displaces its CTT and induces its oligomerization and activation (1). Activated MITA is translocated from the ER via ER-Golgi intermediate compartment (ERGIC) to perinuclear punctate structures by iRhom2-TRAPβ translocon complexes (4). In these processes, the kinase TBK1 is recruited to MITA and phosphorylates MITA at Ser366. This causes further recruitment of the transcription factor IRF3 to MITA and its phosphorylation by TBK1 (2, 3). The phosphorylated IRF3 dimerizes and then enters the nucleus, where it collaborates with NF-κB and other transcription factors to induce transcription of type I interferons (IFNs), inflammatory cytokines, and other downstream antiviral genes.
Although transient activation of the cGAS-MITA axis is essential for host defense to DNA pathogens, sustained or chronic inflammatory response to pathogenic DNA causes autoimmune diseases. Therefore, the cGAS-MITA axis is heavily regulated by various mechanisms (5). For example, MITA is regulated by distinct posttranslational modifications, including serine phosphorylation, polyubiquitination, and sumoylation (1, 6). Recently, it has been shown that MITA is phosphorylated by the tyrosine kinase SRC, which is important for its activation (7). However, how the tyrosine phosphorylation of MITA is regulated remains unknown.
The tyrosine-protein phosphatase nonreceptor type (PTPN) 1 and 2 (also known as PTP1B and TC-PTP, respectively) are 2 closely related members of the class I nonreceptor protein tyrosine phosphatase family (8). Previously, it has been shown that both PTPN1 and PTPN2 are ubiquitously expressed with relatively high levels in immune cells (9). PTPN1 and PTPN2 are involved in regulation of signaling triggered by certain growth factor and cytokine receptors, such as epidermal growth factor receptor (EGFR), platelet-derived growth factor receptor (PDGFR), and insulin receptor (IR) (10). Despite their similarity, the studies with PTPN1- and PTPN2-deficient mice suggest that their functions are not redundant. Ptpn1−/− mice are more sensitive to insulin and leptin and resistant to diet-induced obesity (11). However, Ptpn2−/− mice die within 3 to 5 wk after birth as a result of hematopoietic defects and the development of progressive systemic inflammatory diseases (12). Furthermore, PTPN1/2 double-deficiency is lethal during embryonic development (13).
In this study, we identified PTPN1 and PTPN2 as MITA-associated proteins. We found that after DNA virus infection, PTPN1 and PTPN2 mediated dephosphorylation of MITA at Y245, leading to its degradation via the ubiquitin-independent 20S proteasomal pathway. Our findings suggest that PTPN1/2-mediated dephosphorylation of MITA and its subsequent ubiquitin-independent 20S proteasomal degradation is an important regulatory mechanism of innate immune response to DNA virus.
Results
Identification of PTPN1 and PTPN2 as MITA-Associated Proteins.
To investigate the mechanisms on DNA virus-triggered innate immune response, we attempted to identify MITA-associated proteins using an affinity enrichment coupled with label-free quantitative MS (AE-LFQ-MS) strategy. THP1 cells mock-infected or infected with herpes simplex virus-1 (HSV-1) were used for affinity enrichments of proteins associated with endogenous MITA. For identification of high-confidence interactors, the fold-changes and P values of identified proteins between MITA and control pull-downs were calculated and plotted on a volcano plot (SI Appendix, Fig. S1A). In these experiments, 8 MITA-associated proteins were identified in mock-infected cells, and 2 were identified in HSV-1–infected cells (SI Appendix, Fig. S1B). Among the candidate proteins, SURF4 has been reported to be involved in MITA trafficking (14). Previously, it has been shown that tyrosine phosphorylation of MITA is critically involved in innate antiviral response (7). Therefore, we first attempted to address the roles of PTPN1 in innate antiviral response.
PTPN1 contains an N-terminal catalytic domain and a C-terminal ER-targeting hydrophobic stretch. It has been reported that PTPN1 is highly homologous with PTPN2, and they act together in regulation of signaling triggered by certain growth factors and cytokines (10). We therefore examined whether both PTPN1 and PTPN2 are involved in innate immune response to DNA virus. Confocal microscopy confirmed that PTPN1 and PTPN2 were localized to the ER, while PTPN2 also colocalized with ERGIC and Golgi markers (SI Appendix, Fig. S1C). In addition, MITA was colocalized with PTPN1 and PTPN2 at the ER in uninfected cells (Fig. 1A). After HSV-1 infection, MITA was translocated from the ER to perinuclear punctate structures, where it also colocalized with PTPN1 and PTPN2 (Fig. 1 B and C). Endogenous coimmunoprecipitation experiments indicated that MITA was barely associated with PTPN1 and PTPN2 in uninfected cells. However, HSV-1 infection induced the association of MITA with PTPN1 or PTPN2 which peaked at 6 h postinfection and then decreased (Fig. 1D). Interestingly, although serine phosphorylation and total levels of MITA were decreased at 6 h postinfection in comparison with the earlier phase of infection, tyrosine phosphorylation of MITA was increased at 9 or 12 h postinfection, which was correlated with the decrease of MITA-associated PTPN1 and PTPN2 levels at these late phases of infection (Fig. 1D). These results suggest that PTPN1 and PTPN2 are associated with MITA at the early phase of HSV-1 infection, and their associations are correlated with the decreased tyrosine phosphorylation of MITA.
Fig. 1.
Identification of PTPN1 and PTPN2 as MITA-associated proteins. (A) MITA is colocalized with PTPN1 and PTPN2 in the ER. Confocal microscopy of MLF cells transfected with MITA-Cherry and GFP-PTPN1 or GFP-PTPN2 for 24 h. The ER was stained with ER-Tracker Blue-White DPX for 30 min. (B) MITA is colocalized with PTPN1 upon HSV-1 infection. Mita−/− MLFs stably transduced with MITA and PTPN1 were infected with HSV-1 [multiplicity of infection (MOI) = 1] for 4 h before immunostaining was performed with anti-MITA and anti-PTPN1. (C) MITA is colocalized with PTPN2 upon HSV-1 infection. Mita−/− MLFs stably transduced with MITA and Flag-PTPN2 were infected with HSV-1 (MOI = 1) for 4 h before immunostaining was performed with anti-MITA and anti-Flag. (D) Endogenous PTPN1 and PTPN2 are associated with MITA. THP1 cells were left uninfected or infected with HSV-1 for the indicated times before endogenous coimmunoprecipitation and immunoblot analysis with the indicated antibodies.
PTPN1 and PTPN2 Negatively Regulate dsDNA-Triggered Signaling.
To determine the roles of endogenous PTPN1 and PTPN2, we generated PTPN1-deficient (PTPN1-KO), PTPN2-deficient (PTPN2-KO), and PTPN1/2 double-deficient (PTPN1/2-DKO) human monocytic THP1 cell pools by the CRISPR/Cas9 method (Fig. 2A). qPCR analysis indicated that HSV-1–induced transcription of downstream genes such as IFNB1, CXCL10, ISG56, and IL6 was dramatically increased in PTPN1/2-DKO but only slightly increased in PTPN1-KO and PTPN2-KO cells in comparison to control cells (Fig. 2B). In similar experiments, transcription of IFNB1 and ISG56 genes induced by Sendai virus (SeV, an RNA virus), as well as transcription of TNFA and IKBA genes induced by TNFα or IL-1β, was comparable between PTPN1/2-DKO and control THP1 cells (SI Appendix, Fig. S2 A and B). We next determined the effects of PTPN1/2 deficiency on transcription of downstream genes induced by synthetic dsDNA, including the IFN stimulatory DNA of 45 bp (ISD45), dsDNA of 90 bp (DNA90), and 120-mer dsDNA representing the genome of HSV-1 (HSV120) (15). PTPN1/2 deficiency increased transcription of IFNB1, CXCL10, and IL6 genes induced by transfection of these dsDNAs in THP1 cells (SI Appendix, Fig. S2C). These results suggest that PTPN1 and PTPN2 play redundant roles in negative regulation of viral DNA-induced transcription of downstream effector genes.
Fig. 2.
PTPN1 and PTPN2 negatively regulate dsDNA-triggered signaling. (A) Knockout efficiencies of PTPN1 and PTPN2. The infected THP1 cells were selected with puromycin (0.5 μg/mL) for a week before immunoblotting analysis with the indicated antibodies. (B) Effects of PTPN1-, PTPN2- and double-deficiency on transcription of downstream genes induced by HSV-1 in THP1 cells. PTPN1-KO, PTPN2-KO, and DKO THP1 cells were generated by the CRISPR-Cas9 method. The KO and control THP1 cells were left uninfected or infected with HSV-1 for 8 h before qPCR analysis. (C) Effects of PTPN1/2 double-deficiency on cGAMP production. The DKO and control THP1 cells were left untreated or treated with HT-DNA (0.5 mg/mL) for 4 h, and then cell extracts containing cGAMP were delivered to digitonin-permeabilized MLF for 4 h before qPCR. (D) Effects of PTPN1/2 double-deficiency on transcription of downstream genes induced by 2′3′-cGAMP. The DKO and control THP1 cells were treated with 2′3′-cGAMP (0.2 mg/mL) for 4 h before qPCR analysis. (E) Effects of PTPN1/2 double-deficiency on production of IFN-β and IL-6 induced by 2′3′-cGAMP. The DKO and control THP1 cells were treated with 2′3′-cGAMP (0.2 mg/mL) for 10 h. The culture media were collected for ELISA. (F) Effects of PTPN1-, PTPN2-, and double-deficiency on HSV-1 replication. The KO and control THP1 cells were infected with HSV-1 (MOI = 0.01) for 48 h before plaque assay. Graphs show mean ± SEM, n = 3. **P < 0.01, *P < 0.05.
We next examined the effects of PTPN1/2 deficiency on DNA-triggered synthesis of cGAMP. The results indicated that PTPN1/2 deficiency had no marked effects on DNA-induced cGAMP production in THP1 cells (Fig. 2C). However, PTPN1/2 deficiency increased transcription of downstream genes induced by cGAMP in THP1 cells (Fig. 2D). In addition, PTPN1/2 deficiency increased production of IFN-β and IL-6 cytokines induced by cGAMP in THP1 cells (Fig. 2E). These results suggest that PTPN1/2 regulates components downstream of cGAMP.
Because PTPN1 and PTPN2 regulate DNA virus-triggered transcription of downstream effector genes, we next determined whether they play a role in cellular antiviral response. In plaque assays, PTPN1/2 deficiency inhibited HSV-1 replication in THP1 cells (Fig. 2F). These data suggest that PTPN1 and PTPN2 negatively regulate innate immune response to DNA virus.
PTPN1 and PTPN2 Dephosphorylate MITA at Y245.
Recently, it has been demonstrated that several key components of the cGAS-MITA signaling pathway, including cGAS, TBK1, MITA, and IRF3, are regulated by tyrosine phosphorylation (7, 16, 17). To determine whether MITA is a specific substrate of PTPN1 and PTPN2, we constructed their “substrate-trapping” mutants, PTPN1(D181A) and PTPN2(D182A), which maintain a high affinity for their substrates but are unable to effectively dephosphorylate their substrates (18). Coimmunoprecipitation experiments indicated that wild-type PTPN1 and PTPN2 barely interacted with MITA, whereas PTPN1(D181A) and PTPN2(D182A) strongly interacted with MITA. In these experiments, both wild-type and mutant PTPN1 and PTPN2 failed to interact with cGAS, TBK1, and IRF3 (SI Appendix, Fig. S3). These results suggest that MITA is a candidate substrate of PTPN1 and PTPN2.
The interactions between PTPs and their substrate are mostly mediated by their catalytic domains. The binding of phosphosubstrates to the active sites of PTPs leads to the formation of the enzyme-substrate complexes (19). Domain-mapping experiments indicated that the N-terminal catalytic domain of PTPN1 and the cyclic dinucleotide binding domain (CBD) of MITA mediate their interaction (SI Appendix, Fig. S4). Interestingly, the N-terminal catalytic domain and the C-terminal ER-targeting domain of PTPN2 could independently interact with the CBD and/or transmembrane domains (TMs) of MITA (SI Appendix, Fig. S5).
There are 7 conserved tyrosine residues in the CDB of MITA (SI Appendix, Fig. S6A). Reporter assays indicated that mutation of Y167, Y240, or Y245 of MITA abolished its ability to activate ISRE (SI Appendix, Fig. S6B). Previously, it has been shown that Y167 and Y240 were important for MITA binding to cGAMP (20, 21). In addition, we have found that the tyrosine kinase SRC phosphorylates MITA at Y245 (7). To determine whether PTPN1 and PTPN2 dephosphorylate MITA at Y245, we generated a rabbit polyclonal antibody specific for Y245-phosphorylated human MITA (p-Y245). Coimmunoprecipitation experiments indicated that mutation of Y245 of MITA to phenylalanine (F) reduced its interaction with PTPN1(D181A) or PTPN2(D182A) (SI Appendix, Fig. S6C). Consistently, overexpression of wild-type PTPN1 or PTPN2 dephosphorylated MITA at Y245, but their enzyme-inactive mutants, PTPN1(C215S) and PTPN2(C216S), and their “substrate-trapping” mutants had marked reduced ability to dephosphorylate MITA (Fig. 3A). Furthermore, endogenous MITA was phosphorylated at Y245 after HSV-1 infection, and PTPN1/2 deficiency increased HSV-1–induced phosphorylation of MITA at Y245, as well as phosphorylation of MITA at S366 and TBK1 at S172 (Fig. 3B), which are hallmarks of activation of MITA and TBK1, respectively (22). However, PTPN1/2 deficiency did not affect SeV-induced phosphorylation of TBK1 and IRF3 (SI Appendix, Fig. S7). Previously, it has been shown that viral infection causes down-regulation of MITA levels (23, 24). It is noticeable that HSV-1–induced down-regulation of MITA levels was markedly inhibited in PTPN1/2-DKO cells (Fig. 3B). Taken together, these results suggest that PTPN1 and PTPN2 dephosphorylate MITA at Y245, which is correlated with its down-regulation and inactivation after DNA virus infection.
Fig. 3.
PTPN1 and PTPN2 dephosphorylate MITA at Y245. (A) PTPN1 and PTPN2 dephosphorylate MITA at Y245. HEK293 cells were transfected with HA-MITA and the indicated expression plasmids for 20 h followed by coimmunoprecipitation and immunoblot analysis with the indicated antibodies. (B) PTPN1/2 double-deficiency enhances MITA phosphorylation at Y245 after HSV-1 infection. THP1 cells were infected with HSV-1 (MOI = 4) for the indicated times. Cell lysates were subjected to immunoblotting analysis with the indicated antibodies.
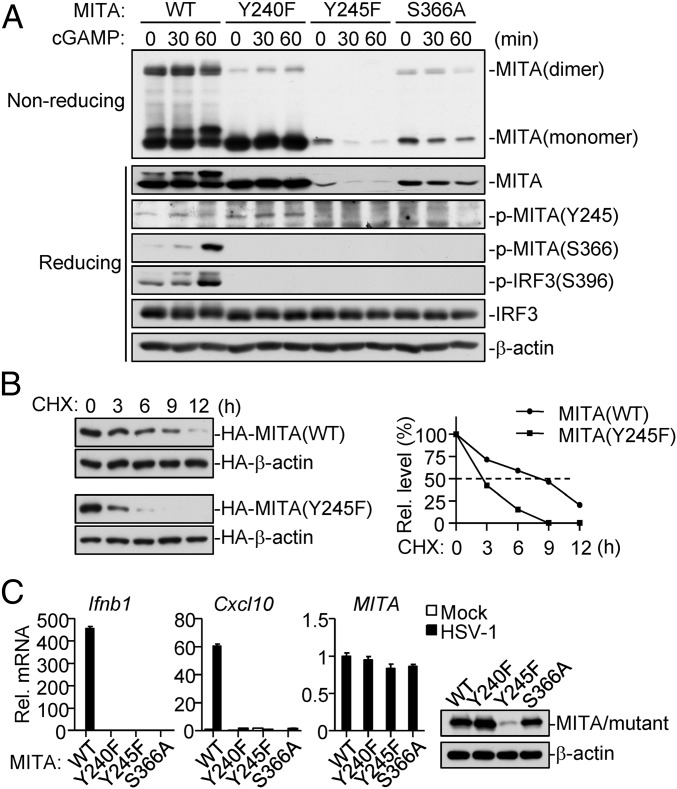
Dephosphorylation of MITA at Y245 Facilitates Its Degradation.
It has been demonstrated that binding of cGAMP to MITA causes MITA dimerization and subsequent phosphorylation at S366 and activation, leading to transcription of downstream antiviral genes (21). We next investigated whether phosphorylation of MITA at Y245 affects cGAMP-induced MITA dimerization. We reconstituted human MITA and its mutants into Mita−/− mouse lung fibroblast (MLF) cells via a pseudotyped retroviral-mediated gene transfer approach. Reconstitution experiments indicated that MITA(Y245F) was expressed at a dramatically lower level in comparison to wild-type MITA (Fig. 4A). Interestingly, MITA(Y240F) was expressed to similar levels as wild-type MITA, but had markedly reduced ability to dimerize upon cGAMP stimulation. On the other hand, although MITA(S366A) was expressed at lower levels in comparison to wild-type MITA, its dimerization after cGAMP stimulation was not affected (Fig. 4A). To exclude the possibility that the decreased expression of MITA(Y245F) is due to reduced protein synthesis, we measured its half-life time with cycloheximide inhibition experiments. The results indicated that MITA(Y245F) had a short intracellular half-life of ∼3 h, whereas wild-type MITA had a longer half-life of ∼9 h (Fig. 4B). Consistently, PTPN1/2 deficiency markedly inhibited HSV-1–induced down-regulation of MITA levels, as shown in Fig. 3B. As expected, MITA(Y245F), MITA(Y240F), and MITA(S366A) all lost their abilities to mediate HSV-1–triggered induction of downstream Ifnb1 and Cxcl10 genes (Fig. 4C). These data suggest that dephosphorylation of MITA at Y245 facilitates its degradation and termination of innate antiviral response.
Fig. 4.
Dephosphorylation of MITA at Y245 promotes its degradation. (A) Mutation of MITA at Y245 causes its down-regulation. Mita−/− MLFs reconstituted with MITA and its mutants were treated with 2′3′-cGAMP (0.2 mg/mL) for the indicated times. The lysates were fractionated by nonreducing SDS/PAGE and then analyzed by immunoblots with the indicated antibodies. (B) Half-lives of wild-type MITA and MITA(Y245F). HEK293 cells transfected with MITA or MITA(Y245F) were untreated or treated with cyclohexamide (CHX) (0.1 mM) for the indicated times before immunoblotting analysis (Left blots). The MITA or MITA(Y245F) band intensities relative to their respective HA–β-actin bands were shown in the Left histograph. (C) Mutation of Y245 of MITA abolishes its activity. Mita−/− MLFs reconstituted with human MITA or its mutants were left uninfected or infected with HSV-1 for 6 h before qPCR analysis.
Dephosphorylation of MITA Promotes Its 20S Proteasomal Degradation.
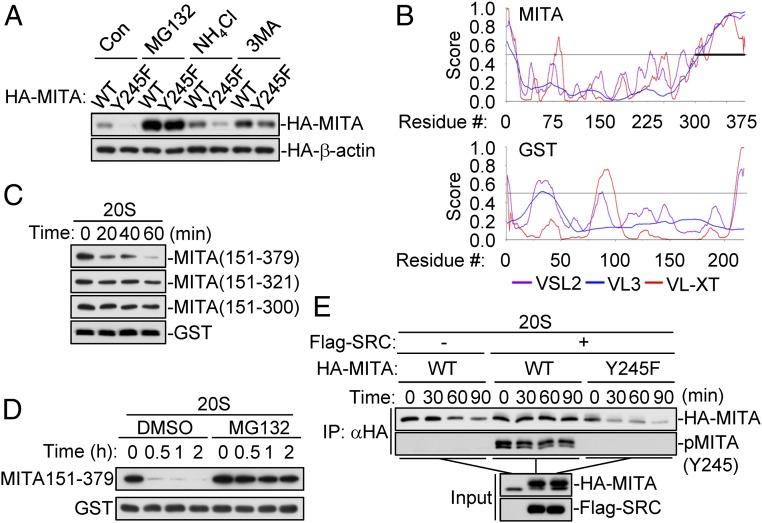
To investigate the mechanisms responsible for the degradation of MITA caused by dephosphorylation of Y245, we treated wild-type and Y245F MITA-transfected 293 cells with various inhibitors for protein degradation pathways. MG132, a proteasome inhibitor, but not the lysosome inhibitor ammonium chloride (NH4Cl) or autophagosome inhibitor 3-methyladenine (3-MA), markedly restored MITA(Y245F) to a similar level as wild-type MITA (Fig. 5A). Previously, it has been shown that HSV-1–induced degradation of endogenous MITA is blocked by MG132 treatment (4). These results suggest that Y245-unphosphorylated MITA is more susceptible to proteasomal-dependent degradation.
Fig. 5.
Dephosphorylation of MITA at Y245 leads to its degradation via the 20S proteasomal pathway. (A) MG132 inhibits the degradation of MITA(Y245F). HEK293 cells were transfected with HA-tagged MITA or MITA(Y245F) for 20 h. The cells were then treated with MG132 (100 μM), NH4Cl (25 mM), or 3-MA (500 ng/mL) for 6 h before immunoblotting analysis. (B) The disorder propensities of MITA and GST analyzed by PONDR (http://www.pondr.com/). High PONDR scores (above 0.5) for all 3 predictors [VSL2 (purple), VL3 (blue), VL-XT (red)] are characteristic of regions with high propensity to be disordered. The IDR of MITA (aa300-379) is indicated by a bold line. (C) The IDR of MITA is required for its degradation by 20S proteasomes. The indicated recombinant proteins were incubated with the 20S proteasomes for the indicated times in vitro before immunoblotting analysis. (D) The degradation of MITA(151-379) by 20S proteasomes is inhibited by MG132. The indicated recombinant proteins were incubated with 20S proteasomes with or without MG132 (50 μM) for the indicated times. (E) Phosphorylation of MITA at Y245 by SRC renders it resistant to 20S proteasomal degradation. HEK239 cells were transfected with Flag-SRC and MITA or MITA(Y245F) as indicated. HA-tagged MITA and MITA(Y245F) were pulled down by anti-HA beads and then incubated with 20S proteasomes at 37 °C for the indicated times before immunoblotting analysis.
It has been reported that the E3 ubiquitin ligase RNF5 and TRIM30α facilitate K48-linked polyubiquitination and subsequent proteasomal degradation of MITA, respectively (25). We determined whether dephosphorylation of Y245 affects K48-linked polyubiquitination of MITA. Unexpectedly, although MITA(Y245F) was restored to similar levels as wild-type MITA after MG132 treatment, the levels of their total polyubiquitination or K48- or K63-linked polyubiquitination were comparable (SI Appendix, Fig. S8A). Reconstitution experiments indicated that HSV-1–induced total or K48-linked polyubiquitination of MITA(Y245F) was similar to that of wild-type MITA (SI Appendix, Fig. S8B). These results suggest that the increased degradation of MITA(Y245F) is independent of its polyubiquitination.
Recently, it has been shown that some proteins can be degraded by proteasomes in an ubiquitin-independent manner (26). Unlike the ubiquitin-dependent 26S proteasomal pathway, the ubiquitin-independent proteasomal pathway is mediated by 20S proteasomes in an ATP-independent process (26). The primary requirement for degradation by 20S proteasomes is the presence of a large unstructured region (>30 amino acids in length) referred to as intrinsically disordered regions (IDRs), or proteins with entirely disordered sequences (27). We analyzed the disorder propensity of MITA by PONDR program, which predicts that aa300-379 is an IDR of MITA. However, no IDRs with appropriate length are found in GST protein (Fig. 5B). To investigate whether MITA can be degraded by the ubiquitin-independent 20S proteasomes, we prepared recombinant MITA mutant proteins for in vitro degradation experiments with 20S proteasomes. We were not able to make soluble recombinant full-length MITA because of the existence of 4 N-terminal transmembrane domains. The in vitro experiments indicated that MITA(151-379), but not MITA(151-300), MITA(151-321), and GST, was degraded by the 20S proteasomes (Fig. 5C). Moreover, the degradation of MITA(151-379) by 20S proteasomes was inhibited by MG132 treatment (Fig. 5D). These data suggest that MITA can be degraded by 20S proteasomes in an ubiquitin-independent manner.
To determine whether phosphorylation of Y245 has any effects on the degradation of MITA by the 20S proteasomes, wild-type MITA and MITA(Y245F) were cotransfected with SRC into HEK293 cells, and then the expressed proteins were purified for 20S proteasome assays in vitro. The results indicated that SRC-mediated phosphorylation of MITA inhibited its degradation by the 20S proteasomes, whereas mutation of MITA Y245 to phenylalanine increased its degradation by the 20S proteasomes (Fig. 5E). Collectively, these results suggest that dephosphorylation of MITA at Y245 promotes its degradation by the ubiquitin-independent 20S proteasomal pathway.
Effects of PTPN1 Deficiency in Mice.
We attempted to investigate the roles of PTPN1/2 in host defense against viral infection in vivo. The serum cytokine levels induced by HSV-1 infection, including IFN-β and CXCL10, showed no significant differences between Ptpn1−/− and wild-type mice (SI Appendix, Fig. S9A). In addition, HSV-1–induced deaths were comparable between Ptpn1−/− and their wild-type littermates (SI Appendix, Fig. S9B). These results suggest that PTPN1 deficiency has no marked effects on antiviral response in vivo. As described previously (12), the Ptpn2−/− mice developed systemic inflammatory diseases and died 2 wk after birth, which limited our test of the roles of PTPN2 in antiviral response in vivo.
Discussion
MITA is a rucial component in the innate immune response to cytosolic DNA. In this study, we found that PTPN1 and PTPN2 dephosphorylated MITA following DNA virus infection, leading to its ubiquitin-independent 20S proteasomal degradation and attenuation of innate immune response to DNA virus.
PTPN1 was identified as a MITA-associated protein in THP1 cells by MS. Endogenous coimmunoprecipitation experiments indicated that both PTPN1 and PTPN2 were barely associated with MITA in uninfected cells, and their associations were markedly increased following HSV-1 infection. Confocal microscopy indicated that PTPN1 and PTPN2 were colocalized with MITA both in uninfected and HSV-1–infected cells. These results suggest that PTPN1 and PTPN2 are involved in regulation of MITA-mediated signaling.
PTPN1/2 double-deficiency dramatically potentiated HSV-1–triggered induction of downstream genes, whereas deficiency of either PTPN1 or PTPN2 had only moderate effects, suggesting that PTPN1 and PTPN2 play redundant roles in regulation of innate immune response to DNA virus. Deficiency of PTPN2 had a relatively stronger effect on HSV-1–triggered transcription of downstream antiviral genes than that of PTPN1, which could be explained by the broader distribution of PTPN2 in organelles. Consistently, PTPN1 deficiency had no marked effects on antiviral response in vivo. The Ptpn2−/− mice had systemic inflammatory diseases and died 2 wk after birth. These results indicated that PTPN1 and PTPN2 are functionally complementary in vivo. Therefore, the development of specific inhibitors of both PTPN1 and PTPN2 will contribute to the further study of their functions in vivo.
It has been previously demonstrated that MITA is phosphorylated by SRC at Y245 (7). Our experiments suggest that PTPN1 and PTPN2 target MITA for dephosphorylation at Y245. The “substrate-trapping” mutants of PTPN1 and PTPN2 had increased association with MITA. Phosphorylation of wild-type MITA but not MITA(Y245F) by SRC increased its association with the PTPN1 and PTPN2 substrate-trapping mutants. PTPN1/2 double-deficiency increased HSV-1–induced MITA phosphorylation at Y245 and S366, as well as TBK1 phosphorylation at S172 and IRF3 phosphorylation at S396, leading to enhanced innate antiviral response. These results suggest that dephosphorylation of MITA at Y245 by PTPN1/2 after viral infection attenuates innate antiviral response.
In uninfected cells, PTPN1 weakly bound to MITA, and MITA Y245 is not phosphorylated. The low-level association of PTPN1 with MITA is probably a mechanism for the cells to keep the innate immune response inactive in uninfected cells. Upon infection, MITA Y245 is phosphorylated and activated, and then PTPN1 is able to dephosphorylate MITA. It is also possible that association of PTPN1 with MITA is not sufficient for PTPN1 to dephosphorylate MITA. The function of PTPN1 requires virus-triggered signals such as additional posttranslational modifications of either PTPN1 or MITA.
Mutation of Y245 of MITA to phenylalanine caused dramatic down-regulation of its protein level, which was reversed by the proteasomal inhibitor MG132. These results suggest that dephosphorylation of MITA at Y245 by PTPN1/2 causes its proteasomal degradation. K48-linked polyubiquitination of MITA and MITA(Y245F) had no marked differences either before or after HSV-1 infection, suggesting that the proteosomal degradation of MITA following its dephosphorylation by PTPN1/2 is not dependent on its polyubiquitination. The ubiquitin-26S proteasomal degradation pathway has been considered the primary route for proteasomal degradation. However, it has been demonstrated in recent years that proteins with IDRs can be targeted for degradation by the core 20S proteasomes, such as p21, p53, and IκBα (28). Interestingly, MITA contains an IDR at its C terminus by sequence analysis. Protein IDRs exist as highly dynamic structural ensembles, either at the secondary or at the tertiary level, and fail to form specific 3D structures. The C-terminal tail (CTT) is invisible in all of the available crystal structures of MITA (29), which is consistent with our prediction that the CTT of MITA contains an IDR. Our experiments indicated that MITA, but not its mutant lack of the C-terminal IDR, could be degraded by the 20S proteasomes in vitro. Further studies showed that phosphorylation of MITA at Y245 was necessary for its resistance to proteasomal degradation by the 20S proteasomes. These results suggest that dephosphorylation of MITA at Y245 by PTPN1/2 leads to its degradation by the ubiquitin-independent 20S proteasomal pathway. Since Y245 is not included in the IDR, how the phosphorylation of Y245 of MITA affects the disorder propensity of its IDR and its degradation by 20S proteasomes is unclear. Previous study has shown that MITA has an intramolecular interaction between its CTT and CBD (30). One possible explanation is that the dephosphorylation of Y245 may change the intramolecular interaction of MITA, which in turn changes the disorder propensity of its C terminus.
The majority of proteasomes in mammalian cells are 20S proteasomes, whereas only about 20 to 30% are 26S proteasomes (31). Protein degradation by 20S proteasomes is not only more efficient, but also spares energy costs (26). Previously, it has been demonstrated that activated MITA is translocated from the ER via Golgi apparatus to perinuclear punctate structures (2). In addition, it has also been shown that the proteasomes form perinuclear aggregates under certain conditions (32). In light of these observations, it is possible that MITA is degraded by the 20S proteasomes after trafficking to perinuclear punctate structures. Various studies have shown that activated MITA can also be degraded by the ubiquitin-proteasomal and lysosomal pathways after viral infection (6, 24). How these MITA degradation pathways are spatial and temporally regulated remains an open question. Additionally, it would be interesting to further investigate whether PTPN1 and PTPN2 regulate MITA-mediated innate immune responses in a cell- and tissue-specific manner.
In conclusion, our results indicate that after the onset of innate immune responses to DNA virus, the 2 tyrosine phosphatases PTPN1/2 dephosphorylate the central adaptor protein MITA, which subsequently causes its degradation by the ubiquitin-independent 20S proteasomes (SI Appendix, Fig. S10). Our findings reveal a striking mechanism on how MITA-mediated innate immune responses are attenuated by its tyrosine dephosphorylation. In addition, this study also provides an example on how tyrosine dephosphorylation of a substrate promotes its degradation by the 20S proteasomes, leading to attenuation of innate immune responses. Further investigations of these delicate regulatory mechanisms may help for drug or vaccine development against DNA virus infection.
Materials and Methods
All animal experiments were performed in accordance with the Wuhan University Animal Care and Use Committee guidelines. The information on reagents, antibodies, cells, constructs, PCR primers, RNAi target sequences, knockout mice, and various methods are described in SI Appendix. The proteomics sample preparation and nano-LC-MS analysis were performed as previously described (33).
Supplementary Material
Acknowledgments
This work was supported by grants from the State Key R&D Program of China (2017YFA0505800, 2016YFA0502102) and the National Natural Science Foundation of China (31830024, 31630045).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1906431116/-/DCSupplemental.
References
- 1.Hu M. M., Shu H. B., Cytoplasmic mechanisms of recognition and defense of microbial nucleic acids. Annu. Rev. Cell Dev. Biol. 34, 357–379 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Ishikawa H., Ma Z., Barber G. N., STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhong B., et al. , The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Luo W. W., et al. , iRhom2 is essential for innate immunity to DNA viruses by mediating trafficking and stability of the adaptor STING. Nat. Immunol. 17, 1057–1066 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Luo W. W., Shu H. B., Delicate regulation of the cGAS-MITA-mediated innate immune response. Cell. Mol. Immunol. 15, 666–675 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu M. M., et al. , Sumoylation promotes the stability of the DNA sensor cGAS and the adaptor STING to regulate the kinetics of response to DNA virus. Immunity 45, 555–569 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Hu M. M., et al. , Virus-induced accumulation of intracellular bile acids activates the TGR5-beta-arrestin-SRC axis to enable innate antiviral immunity. Cell Res. 29, 193–205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alonso A., et al. , Protein tyrosine phosphatases in the human genome. Cell 117, 699–711 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Arimura Y., Yagi J., Comprehensive expression profiles of genes for protein tyrosine phosphatases in immune cells. Sci. Signal. 3, rs1 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Dubé N., Tremblay M. L., Involvement of the small protein tyrosine phosphatases TC-PTP and PTP1B in signal transduction and diseases: From diabetes, obesity to cell cycle, and cancer. Biochim. Biophys. Acta 1754, 108–117 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Elchebly M., et al. , Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science 283, 1544–1548 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Heinonen K. M., et al. , T-cell protein tyrosine phosphatase deletion results in progressive systemic inflammatory disease. Blood 103, 3457–3464 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Heinonen K. M., Bourdeau A., Doody K. M., Tremblay M. L., Protein tyrosine phosphatases PTP-1B and TC-PTP play nonredundant roles in macrophage development and IFN-gamma signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 9368–9372 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S., Wang L., Berman M., Kong Y. Y., Dorf M. E., Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity 35, 426–440 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abe T., et al. , STING recognition of cytoplasmic DNA instigates cellular defense. Mol. Cell 50, 5–15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu H., et al. , Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature 563, 131–136 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Liu S., et al. , Lck/Hck/Fgr-mediated tyrosine phosphorylation negatively regulates TBK1 to restrain innate antiviral responses. Cell Host. Microbe. 21, 754–768.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Flint A. J., Tiganis T., Barford D., Tonks N. K., Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc. Natl. Acad. Sci. U.S.A. 94, 1680–1685 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao P., et al. , The second-sphere residue T263 is important for the function and catalytic activity of PTP1B via interaction with the WPD-loop. Int. J. Biochem. Cell Biol. 57, 84–95 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Ouyang S., et al. , Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity 36, 1073–1086 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin Q., et al. , Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol. Cell 46, 735–745 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S., et al. , Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Konno H., Konno K., Barber G. N., Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 155, 688–698 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong B., et al. , The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity 30, 397–407 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Chen Q., Sun L., Chen Z. J., Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 17, 1142–1149 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Ben-Nissan G., Sharon M., Regulating the 20S proteasome ubiquitin-independent degradation pathway. Biomolecules 4, 862–884 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Habchi J., Tompa P., Longhi S., Uversky V. N., Introducing protein intrinsic disorder. Chem. Rev. 114, 6561–6588 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Jariel-Encontre I., Bossis G., Piechaczyk M., Ubiquitin-independent degradation of proteins by the proteasome. Biochim. Biophys. Acta 1786, 153–177 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Shang G., Zhang C., Chen Z. J., Bai X. C., Zhang X., Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP. Nature 567, 389–393 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato K., Omura H., Ishitani R., Nureki O., Cyclic GMP-AMP as an endogenous second messenger in innate immune signaling by cytosolic DNA. Annu. Rev. Biochem. 86, 541–566 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Fabre B., et al. , Subcellular distribution and dynamics of active proteasome complexes unraveled by a workflow combining in vivo complex cross-linking and quantitative proteomics. Mol. Cell. Proteomics 12, 687–699 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wójcik C., DeMartino G. N., Intracellular localization of proteasomes. Int. J. Biochem. Cell Biol. 35, 579–589 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Shang J., et al. , Quantitative proteomics identified TTC4 as a TBK1 interactor and a positive regulator of SeV-induced innate immunity. Proteomics 18, 1700403 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.