Significance
The realized threats of global warming to biodiversity have catalyzed the search for a solution to protect and conserve extant plant genetic resources. Part of the solution, however, is dependent on the knowledge of how plant populations respond genetically to these threats, which is largely lacking. We conducted a unique genomic characterization of genetic responses in 10 wild emmer wheat populations in Israel that were sampled twice in 1980 and 2008. After the 28 y of global warming, these populations displayed elevated selection, reduced diversity and temporal divergence, and carried increased mutational burdens forward. However, some populations still showed the ability to acquire beneficial alleles for future adaptation. The patterns of genetic response to rainfall and temperature were complex.
Keywords: wild emmer wheat, mutation, selection, exome capture, global warming
Abstract
Global warming has been documented to threaten wild plants with strong selection pressures, but how plant populations respond genetically to the threats remains poorly understood. We characterized the genetic responses of 10 wild emmer wheat (Triticum dicoccoides Koern.; WEW) populations in Israel, sampling them in 1980 and again in 2008, through an exome capture analysis. It was found that these WEW populations were under elevated selection, displayed reduced diversity and temporal divergence, and carried increased mutational burdens forward. However, some populations still showed the ability to acquire beneficial alleles via selection or de novo mutation for future adaptation. Grouping populations with mean annual rainfall and temperature revealed significant differences in most of the 14 genetic estimates in either sampling year or over the 28 y. The patterns of genetic response to rainfall and temperature varied and were complex. In general, temperature groups displayed more temporal differences in genetic response than rainfall groups. The highest temperature group had more deleterious single nucleotide polymorphisms (dSNPs), higher nucleotide diversity, fewer selective sweeps, lower differentiation, and lower mutational burden. The least rainfall group had more dSNPs, higher nucleotide diversity, lower differentiation and higher mutational burden. These characterized genetic responses are significant, allowing not only for better understanding of evolutionary changes in the threatened populations, but also for realistic modeling of plant population adaptability and vulnerability to global warming.
Global warming is one of the major environmental stresses threatening plant populations in the wild (e.g., ref. 1). However, how these threatened populations respond ecologically and evolutionarily to these stresses for adaptation to avoid extinction remains elusive (e.g., ref. 2). Population genetic theory predicts that a plant population will respond genetically to directional selection such as global warming via selection on standing genetic variation before deleterious mutations are accumulated sufficiently large to drive the population toward extinction (e.g., ref. 3). However, empirical genetic data to support the theoretical prediction on population vulnerability under threats are largely lacking, as characterizing deleterious and beneficial mutations and analyzing genome-wide selections were technically limited (4) until recent advances in genome sequencing (5–8). Little is known about the interplay of selection and mutation in plant natural populations under stresses (9, 10), particularly from global warming.
The wild relative species of domesticated crops harbor abundant and useful genetic diversity (11) and are the best genetic hope for improving genetically impoverished cultivars for human food production (12–17). However, these valuable genetic resources are found to be highly underconserved (18), and concerns for losing these genetic resources are mounting (19). Also, many studies have revealed increasing threats for crop wild relatives in natural populations, particularly from global warming (1, 20, 21), but few studies have characterized genetic responses of wild relative populations to the threats of global warming (22–24).
Wild emmer wheat (Triticum dicoccoides Koern.; WEW) (25) is an important wild progenitor of cultivated wheat. It represents useful genetic resources with adaptation to abiotic (e.g., solar radiation, temperature, drought, and mineral poverty) and biotic (e.g., pathogens and parasites) stresses. However, these wild cereals become eroded by urbanization and agriculture (14) and affected by climate changes such as rising temperature and less rainfall (26). For example, our previous study showed the shortening of flowering time 8.5 and 10.9 d in 10 WEW and 10 wild barley populations, respectively, after 28 y of global warming (23). Thus, it is important to assess the evolutionary responses of the wild relative populations to the severe ongoing threats (22). Also, the advances in emmer wheat genome sequencing (27–29) open new opportunities to characterize genome-wide genetic variations and to make genetic inferences in wild emmer populations.
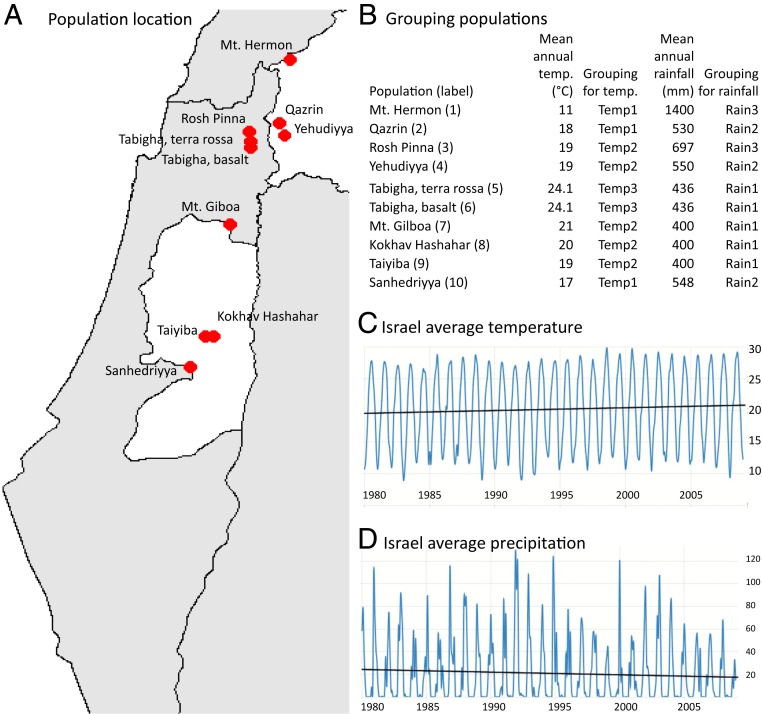
To understand genetic responses of wild crop relatives under global warming, we conducted a comprehensive characterization of genome-wide genetic variations using advanced sequencing technologies and assessed the evolutionary responses in WEW natural populations. Specifically, we selected 10 WEW populations in Israel, sampling them in 1980 and 2008 (Fig. 1A and SI Appendix, Table S1), genotyped them using exome capture (27), and analyzed the changes in mutation, selection, diversity, and population differentiation with respect to climate-specific groups (Fig. 1B). We found elevated mutation and selection in these wild emmer populations over the 28 y of global warming (Fig. 1 C and D). Remarkably, some populations were still capable of generating adaptive mutations for adaptation potential. Temperature groups displayed more temporal differences in genetic response than rainfall groups. The highest temperature group had more deleterious single nucleotide polymorphisms (dSNPs), higher nucleotide diversity, fewer selective sweeps, lower differentiation, and lower mutational burden.
Fig. 1.
Sampling location, population grouping, temperature, and precipitation in Israel. A shows the locations of the 10 WEW populations studied; B displays the population grouping for climate-specific groups based on mean annual rainfall and temperature in each location; C illustrates the changes in Israel from 1980 to 2008 of average temperature from below 20 °C to above 20 °C; and D shows the changes in Israel from 1980 to 2008 of average precipitation from above 20 mm to below 20 mm. Note that the weather data were acquired from the World Bank website Tradingeconomics.com.
Results
Sequencing, SNP Identification, and Annotation.
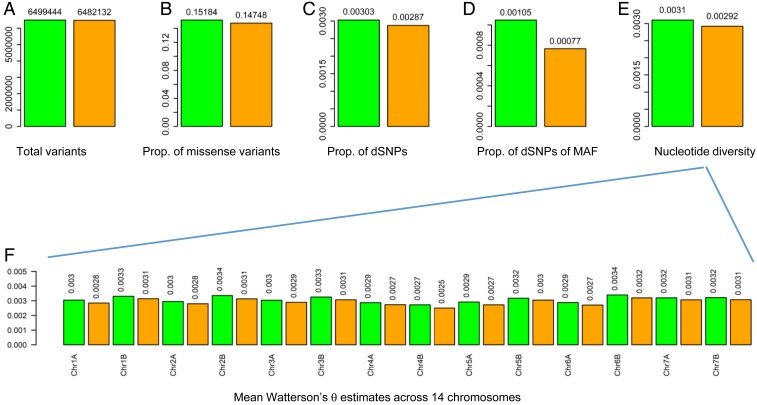
We conducted exome capture sequencing based on materials derived from ref. 23 with a total of 184 WEW samples with 6–10 individuals per population in each sampling year (Fig. 1A and SI Appendix, Table S1). The sequencing generated 4.3 million mapped sequence reads per WEW sample (SI Appendix, Table S2). SNP calling using ANGSD (30) identified 6,499,444 and 6,482,132 SNPs across the WEW genome for the 1980 and 2008 samples, respectively (Fig. 2A and SI Appendix, Table S4). SNP annotation using Ensembl-Variant Effect Predictor (31) allowed for classification of detected SNPs into 17 different classes (SI Appendix, Table S4). Most SNPs were located on intergenic, intron, upstream, and downstream genic regions. A total of 899,855 and 874,833 SNPs were identified as synonymous variants and 986,903 and 956,002 SNPs as missense variants for 1980 and 2008 samples, respectively. Weighting by the total SNPs detected, the proportional missense SNPs were higher in the 1980 (0.152), than 2008 (0.147), samples (Fig. 2B and SI Appendix, Table S4).
Fig. 2.
SNP detection, SNP characterization, and nucleotide diversity in the samples of WEW collected in 1980 and in 2008. A shows the total variants detected for each sample group; B the proportion of the missense variants detected over the total SNPs; C the proportion of the dSNPs; D the proportion of dSNPs with minor allelic frequencies (MAF) smaller than 0.05; E mean Watterson’s θ estimates; and F mean Watterson’s θ estimates across 14 chromosomes. In each graph, the 1980 and 2008 samples are labeled in green and orange, respectively, and the sample mean values are shown above the bars.
Genetic Diversity.
We inferred nucleotide diversity based on the estimates of Watterson’s θ and Tajima’s π (SI Appendix, Fig. S2). Overall, significantly lower estimate of Watterson’s θ was found in the 2008, than 1980, samples (Fig. 2E and SI Appendix, Table S5). Such diversity reduction was more obvious across the 14 chromosomes (Fig. 2F). Also, there were 6 populations displaying significant diversity reduction (SI Appendix, Table S5). In contrast, the estimates of Tajima’s π were not significantly different between samples of the 2 sampling years, but they were significantly reduced in 5 populations (SI Appendix, Table S6). Moreover, the estimates of individual inbreeding coefficient in a population were generally reduced over the 28 y (SI Appendix, Table S7). Quantifying the population differentiation (Fst) over the 28 y revealed an overall Fst of 0.376 at the population level (ranging from 0.143 to 0.684) and of 0.021 for all combined populations (SI Appendix, Table S7). These results suggested that the populations had reduced diversity and were diverged genetically.
Selective Sweep.
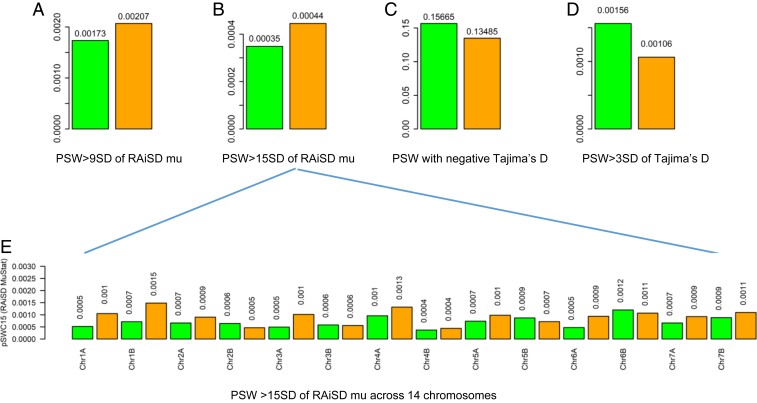
We applied 2 methods to screen selective sweeps across the WEW genome. Applying RAiSD (32) revealed more sliding windows with selective sweeps across the 14 chromosomes in the 2008, than 1980, samples, based on the outliers of MuStat estimates being 9 or 15 SDs (Figs. 2B and 3A and SI Appendix, Tables S8 and S9). Such patterns hold with respect to sampling year, population and chromosome. The detected selective sweeps on each chromosome were illustrated in SI Appendix, Fig. S3 for the 1980 and 2008 samples, and summarized in Fig. 3E, where the 2008 samples displayed more selective sweeps in 11 chromosomes and fewer sweeps in 3 chromosomes (Chr2B, Chr5B, and Chr6B) than the 1980 samples. These 3 chromosomes might carry fewer genes sensitive to the rising temperature than the other 11 chromosomes. Evidently, these wild emmer populations had elevated selection over the 28 y.
Fig. 3.
Selection signals detected by RAiSD and Tajima’s D in the samples of WEW collected in 1980 and in 2008. A shows the proportion of sliding windows (PSW) with RAiSD MuStat estimates greater than 9 SDs; B the proportion of sliding windows with RAiSD MuStat estimates greater than 15 SDs; C the proportion of sliding windows with negative Tajima’s D estimates; D the proportion of sliding windows with positive Tajima’s D estimates greater than 3 SDs; and E the proportion of sliding windows with RAiSD MuStat estimates greater than 15 SDs across 14 chromosomes. In each graph, the 1980 and 2008 samples are labeled in green and orange, respectively, and the sample mean values are shown above the bars.
Less accurate Tajima’s D analysis was also made to acquire indirect selection signals across the genome (33). The analysis revealed the dominance of balancing selection for the WEW samples with average Tajima’s D greater than zero across the WEW genome (SI Appendix, Fig. S4). However, the 2008 samples displayed lower counts of nonoverlapping sliding windows with Tajima’s D greater than 3 SDs per chromosome than the 1980 samples (Fig. 3D), while showing higher estimates of mean Tajima’s D per chromosome (SI Appendix, Fig. S4). Similarly, the 2008 samples also displayed reduced nonoverlapping sliding windows with Tajima’s D smaller than zero per chromosome (Fig. 3C) and over the chromosomal regions representing different functional classes (SI Appendix, Fig. S5). The reductions in the counts of balanced selection (Tajima’s D > 0) and purging selection (Tajima’s D < 0) in the 2008, relative to 1980, samples were also observed with respect to population and sampling year, as summarized in SI Appendix, Tables S10 and S11. It is highly possible that such patterns of reduction, particularly in purging selection, were partly confounded with demographic factors.
Deleterious Mutation.
We identified dSNPs based on the scores of both Sorting Intolerant From Tolerant (SIFT; ref. 34) and Genomic Evolutionary Rate Profiling (GERP; ref. 35). The SIFT score presents a prediction on the impact of an amino acid substitution and can distinguish between functionally neutral and deleterious amino acid changes. An amino acid substitution with a SIFT score of 0.05 or less is considered to be deleterious. GERP produces a “rejected substitution” (RS) score to quantify the conservation of each nucleotide in multispecies alignment. A positive score (RS > 0) at a substitution site means fewer substitutions than expected. Thus, a substitution occurring in a site with RS > 0 is predicted to be deleterious; the larger the RS score, the more deleterious the substitution. The identification generated 19,672 and 18,627 dSNPs for the 1980 and 2008 samples (SI Appendix, Table S4), respectively. For ease of comparison, SI Appendix, Table S12 summarized all of the dSNP detections with respect to population, climate group, and sampling year. Weighting by the total detected SNPs, we found that the 2008 samples displayed lower proportional dSNPs (Fig. 2C), but such reduction was not statistically significant, at least at the chromosomal level (SI Appendix, Table S13). Examining the chromosomal distributions of the detected dSNPs revealed that more dSNPs were located toward both ends of a chromosome, and such pattern of dSNP distribution was similar for both 1980 and 2008 samples (SI Appendix, Fig. S6).
To understand these dSNPs better, we assessed the deleterious allele frequency distributions (SI Appendix, Fig. S7) and found 105 and 104 dSNPs were fixed in the 1980 and 2008 samples, respectively (SI Appendix, Table S4). Comparing the extreme frequencies of these dSNPs revealed that the 1980 samples had more dSNPs of allelic frequency <0.1 or >0.90 than the 2008 samples (Fig. 2D and SI Appendix, Fig. S8). This finding helped to explain why the 2008 samples had fewer dSNPs than the 1980 samples. Further distribution analysis of allelic frequencies for the dSNPs revealed marked differences between sampling years at the population level, particularly for populations 4, 6, and 10 (SI Appendix, Fig. S9). These patterns were compatible with allelic frequency differences between sampling years at the population level for all of the detected SNPs (SI Appendix, Fig. S10), showing marked population divergences.
Beneficial Mutation.
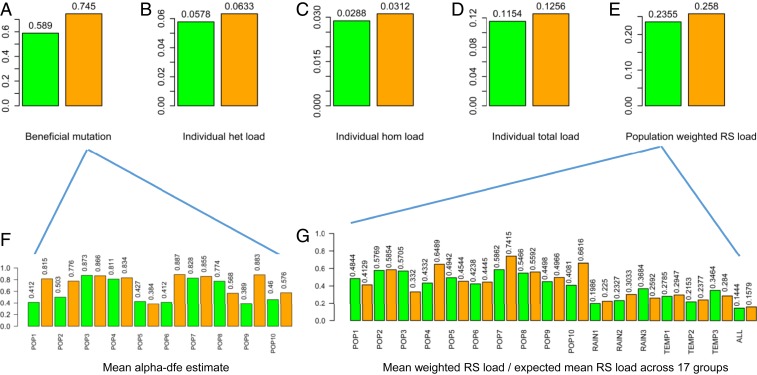
We also inferred beneficial mutations using PolyDFE (8). PolyDFE generates alpha-dfe statistics as the proportion of adaptive substitutions (with selection coefficient greater than zero) from site frequency spectrum data. Higher alpha-dfe estimates mean relatively more advantageous mutations. The estimation of alpha-dfe following model A showed an overall increase in adaptive mutations in the 2008 samples at the population level (Fig. 4A and SI Appendix, Table S14). Seven populations showed a significant increase in alpha-dfe estimates over the 28 y, while 3 populations (Rosh Pinna; Tabigha, terra rossa; Kokhav Hashahar) displayed a significant decrease (Fig. 4F and SI Appendix, Table S14).
Fig. 4.
Estimates of adaptive mutations and mutational burdens in the samples of WEW collected in 1980 and in 2008. A–E show the estimates of adaptive mutations, mean individual heterozygous (het), homozygous (hom) and total mutational burden, and the population weighted GERP++ RS mutational burdens. F and G display the mean alpha-dfe estimate for each population and the ratios of mean weighted RS load vs. expected mean RS load across 17 sample groups. In each graph, the 1980 and 2008 samples are labeled in green and orange, respectively, and the sample mean values are shown above the bars.
Mutational Burden.
We estimated mutational burdens for individual samples by counting deleterious heterozygotes and homozygotes for each deleterious SNP. The estimates and their tests of significance were summarized in SI Appendix, Tables S15–S17 for individual heterozygous load, homozygous load, and total load, respectively. On average, the 2008 samples displayed higher individual mutational burdens than the 1980 samples (Fig. 4 B–D). Interestingly, individual total loads were not associated with the WEW population latitudes (SI Appendix, Fig. S11).
We also inferred mutational burden at the population level following the method of Wang et al. (9) by weighting GERP++ RS scores with the deleterious allelic frequencies for all of the dSNPs. Seven populations displayed higher RS-based mutational burdens over the sampling years, while 3 populations showed a reduction in RS-based mutational burden (SI Appendix, Table S18). Overall, the 2008 samples displayed more population-weighted RS load inferred than the 1980 samples (Fig. 4 E and G). Further distribution analyses of RS and weighted RS scores for all of the dSNPs in the 1980 and 2008 samples (SI Appendix, Figs. S12 and S13) support the finding that the 2008 samples carried an increased RS-based mutational burden.
Gene Ontology Analysis.
We characterized further the detected deleterious genes by performing Gene Ontology (GO) analysis via REVIGO (36). The analysis revealed that inferred genes were mainly associated with the biological processes of protein phosphorylation, organic substance metabolism, lipid metabolism, and organic substance catabolism. However, considering the GO terms extracted from deleterious genes unique to 1980 or 2008 samples, we found more unique clusters of biological processes in the 1980, than 2008, samples (SI Appendix, Fig. S14A). Specifically, there were 28 and 18 unique REVIGO GO biological processes for the 1980 and 2008 samples (SI Appendix, Fig. S14B), respectively. Similarly, considering only 93 fixed deleterious genes unique to 1980 or 2008 samples, we found 3 and 14 GO terms for the 1980 and 2008 samples, respectively, and more biological processes present in the 2008 samples (SI Appendix, Fig. S15). Interestingly, REVIGO also generated tag clouds with the keywords that correlated with the values based on 1,044 and 1,022 GO terms identified from all of the deleterious genes. The tag clouds consistently displayed the word “temperature” in both 1980 and 2008 samples (SI Appendix, Fig. S16), indicating many of these deleterious genes had functions associated with temperature.
We also performed GO analysis of the selective genes detected in the selective chromosomal regions identified by RAiSD MuStat estimates of 20 or larger SDs. A total of 497 and 789 chromosomal segments across the WEW genome having 66 and 80 nonredundant genes were identified, and a total of 159 and 336 GO terms were extracted, for the 1980 and 2008 samples, respectively. More genes were underrepresented with smaller log10pvalue in the 2008, than 1980, samples (SI Appendix, Fig. S17A), but there were 16 and 77 unique REVIGO GO biological processes identified for the 1980 and 2008 samples, respectively (SI Appendix, Fig. S17B). These results further confirmed the elevated selection in the 2008 samples.
Variation Analysis for Climate-Specific Groups.
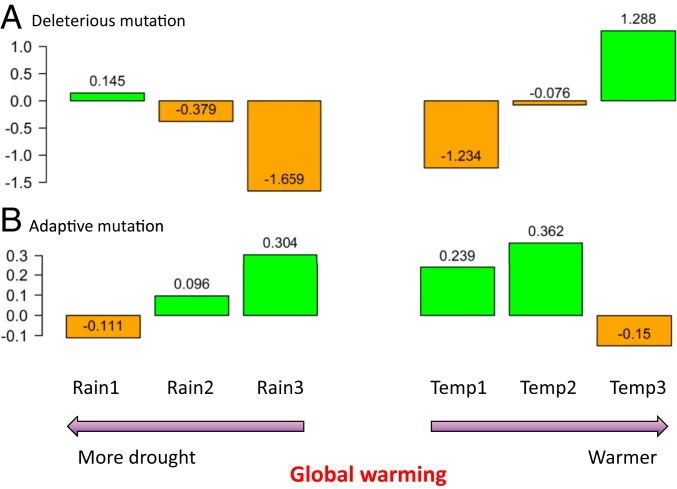
We evaluated the impacts of rainfall and temperature on genetic responses (or estimates of genetic parameters) in WEW by grouping the 10 populations with climate factor profiles to 3 rainfall and 3 temperature population groups (Fig. 1B), estimating 14 genetic parameters in each group, and testing the differences in genetic estimates among climate-specific groups by Kruskal–Wallis one-way ANOVA. The results are tabulated in SI Appendix, Tables S5–S18 and summarized in Table 1 with respect to global warming (less rainfall and higher temperature). It was found that these climate-specific groups displayed significant differences in most of the 14 genetic estimates in either sampling year or over the 28 y (Table 1). However, their impacts on genetic responses seemed to be varied and more complex than previously anticipated, conditional on the nature of genetic parameter and its estimate. In general, the temperature groups showed more temporal differences in genetic response than the rainfall groups. The highest temperature group had more dSNPs, higher nucleotide diversity, fewer selective sweeps, lower differentiation, and lower mutational burden. The least rainfall group had more dSNPs, higher nucleotide diversity, lower differentiation, and higher mutational burden. More specifically, more deleterious base-substitution mutations per sample (×10−8) and fewer adaptive mutations were found in the most drought and highest temperature groups of WEW after 28 y (Fig. 5).
Table 1.
Genetic impacts of rainfall and temperature in the samples of WEW collected in 1980 and in 2008, as illustrated with increase or decrease and with their statistical significance
| R1 vs. R3 | R1 vs. R3 | R1 over | T3 vs. T1 | T3 vs. T1 | T3 over | From | |
| Genetic parameter | in 1980 | in 2008 | 28 y | in 1980 | in 2008 | 28 y | table |
| Selection | |||||||
| RAiSD muStat 9SD (selective sweep) | INC*** | DEC | DEC* | INC* | INC | DEC** | S8 |
| RAiSD muStat 15SD (selective sweep) | INC*** | INC** | INC | INC | DEC | DEC | S9 |
| PSW (negative Tajima's D) (purging) | DEC*** | DEC*** | DEC*** | INC*** | DEC*** | DEC*** | S11 |
| PSW (3SD of Tajima's D) (balancing) | DEC*** | DEC*** | DEC | INC*** | INC*** | DEC*** | S10 |
| Mutation | |||||||
| Proportion of dSNP count | INC*** | INC*** | INC | DEC*** | DEC*** | INC*** | S13 |
| Individual heterozygous load | DEC*** | DEC*** | INC | INC*** | DEC*** | DEC* | S15 |
| Individual homozygous load | DEC*** | DEC*** | INC*** | INC*** | INC | DEC** | S16 |
| Individual total load | DEC*** | DEC*** | INC*** | INC*** | DEC* | DEC** | S17 |
| Population weighted RS load | DEC*** | DEC*** | INC*** | INC*** | DEC*** | DEC*** | S18 |
| Adaptive mutation | INC*** | DEC*** | DEC*** | INC*** | DEC*** | DEC*** | S14 |
| Diversity | |||||||
| Watterson's θ | DEC*** | INC*** | INC** | DEC*** | DEC*** | INC*** | S5 |
| Tajima's π | INC*** | INC*** | INC*** | DEC*** | DEC*** | INC*** | S6 |
| Individual Fis | INC*** | DEC | DEC* | DEC*** | INC** | INC*** | S7 |
| Differentiation | |||||||
| Fst | — | — | DEC*** | — | — | DEC*** | S7 |
The impact of an increase (INC) or decrease (DEC) is defined if an estimated genetic response was higher or lower for the least, than the most, rainfall group (R1 vs. R3) in a given sampling year, or for R1 over the 2 sampling years, respectively. Also similarly defined was for temperature with the highest temperature group (T3) vs. the lowest temperature group (T1) in a sampling year, or with T3 over the 28 y. All of the estimated genetic responses are listed in SI Appendix, Tables S5–S18. *P < 0.05, **P < 0.01, ***P < 0.001.
Fig. 5.
Changes in the estimates of deleterious base-substitution mutations per sample (×10−8; A) and adaptive mutations (B) in 6 climate-specific groups in response to 28 y of global warming. Positive and negative changes are highlighted in green and orange, respectively, and the differences are shown above or within the bars.
To understand the complexity of genetic responses to rainfall and temperature, we compared the extent of selective sweeps identified across the 14 WEW chromosomes in each climate-specific group and confirmed the complexity in the selection response to rainfall and temperature (SI Appendix, Figs. S18 and S19). Similar conclusion can also be drawn when allele frequency distributions were compared between 2 sampling years for all detected SNPs and for all dSNPs in 6 climate-specific groups (SI Appendix, Figs. S20 and S21).
Discussion
This study represents a comprehensive characterization of genetic responses in crop wild relative populations through a comparative genomic analysis of diversity, selection, and mutation. After the 28 y of global warming, the assayed 10 WEW populations were under elevated selection, displayed reduced diversity and temporal divergence, and carried increased mutational burdens forward. However, some populations were still capable of selecting beneficial alleles from existing genetic variations for adaptation. Grouping the populations with mean annual rainfall and temperature revealed significant differences in most of the 14 genetic estimates in either sampling year or over the 28 y. The patterns of genetic response to rainfall and temperature varied and were complex. Temperature groups generally displayed more temporal differences in genetic response than rainfall groups. These findings not only allow for better understanding of evolutionary changes in threatened populations, but also provide valuable empirical data for realistic modeling of plant population adaptability and vulnerability to global warming.
Our study has some limit in the association analyses to establish the immediate links of the global warming to all of these genetic responses, as other climate changes such as CO2 increase and their interactions may have also played a role in these genetic changes. However, our GO analysis (SI Appendix, Fig. S16) and climate group analysis (Table 1) seemed to favor the temperature as the major driver of the detected temporal genetic changes. Also, our genomic and genetic analyses may have suffered from the small sample size, sampling unbalance, and WEW polyploidy. Despite these limitations, the characterized genetic responses have helped to paint a picture in a resolution unachievable before on the evolutionary changes occurring at the levels of gene, chromosome, individual, and population in response to global warming, and allowed us to understand better the evolution of the threatened populations. First, our characterization confirmed the expectation generated from our early study that the WEW populations under global warming were under strong selection and had reduced diversity (23). Second, these populations overall will carry increased mutational burdens forward, but some populations also showed their ability to generate more adaptive mutations via selection of existing variations or de novo mutation (Fig. 4), which is also consistent with the early observation of adaptive SSR alleles present in WEW samples (23). These findings together are encouraging, as some WEW populations such as populations 1 and 9 will have the genetic potential of adaptation to the ongoing global warming. Third, we could also reason empirically that some populations may be more vulnerable genetically than the others. For example, the populations 4, 7, and 10 had some feature of becoming genetically vulnerable, as they had the highest mutational burdens (SI Appendix, Table S18 and Fig. S11) with accumulations of fewer adaptive mutations (SI Appendix, Table S14) and displayed the strongest temporal differentiations with reduced individual inbreeding coefficients (SI Appendix, Table S7) and marked allelic changes (SI Appendix, Figs. S9 and S10).
With these empirical genetic responses, we move closer toward the reliable prediction of population adaptability and vulnerability to climate changes (37, 38), as realistic modeling of a threatened population with deep learning tools is possible (39, 40). The main advantage of such population modeling is its ability to incorporate genetic responses (including adaptive mutations), demographics, climate factors, and environmental conditions for an integrated projection of threatened population dynamics (41, 42). With the incorporation of adaptive mutations into modeling, the projection may be more realistic and accurate than before. We believe the population modeling will be a fruitful area of research, enhancing our understanding of the adaptability and vulnerability projection in threatened populations and assisting in the development of effective conservation strategies and guidelines, particularly for those threatened populations of crop wild relatives. Conserving valuable crop wild relatives has now become more critical than before to secure valuable genetic resources for improving food production (13), as many crop wild relatives are not properly protected and under conservation (18).
Our research also demonstrates a feasible approach to monitor evolutionary responses of some plant populations under environmental stress in the wild (4) through a comparative analysis of selection, mutation, and diversity (SI Appendix, Fig. S1). The approach can be applied to characterize genome-wide variations of other plant species or organisms and to assess selection and mutation in the wild (43). The threatened populations of crop wild relatives, however, naturally have become an attractive model of research, as characterizing genetic responses in crop wild relative populations is more feasible than before with the availability of sequenced genomes and gene annotations in the related crops (27–29, 44). Also, many crop wild relatives have been collected and conserved in seed genebanks worldwide over the last 60 y with precise GIS information, allowing for population resampling (45–47). Thus, it is technically possible to acquire more temporal data on genetic responses for more reliable population adaptability and vulnerability modeling, allowing better understanding of the evolutionary processes and potential of crop wild relative populations under the threats of global warming.
Materials and Methods
Materials used for this study and methods used for collecting samples, DNA extractions, sequencing, SNP calling, population genetic analysis, GO analysis, variation analyses for climate groups, and data and code availability are available in the SI Appendix. The SI Appendix has several components: A, Supplemental materials and methods; B, References for materials and methods; C, Grouping of supplementary tables and figures; D, Tables S1 to S18; and E, Figs. S1 to S21.
Supplementary Material
Acknowledgments
We acknowledge helpful assistance in sequencing from Janet Condie, Christine Sidebottom, and Brian Boyle, and in bioinformatics analysis from Punna Ramu, Nikolaos Alachiotis, Paula Tataru, Thomas Bataillon, Thorfinn Sand Korneliussen, Logan Kistler, Jennifer Hillman-Jackson, Pankaj Jaiswal, Thomas Kono, Peter Morrell, Tal Pupko, Chris Benner, Martin Mascher, Jeffrey Ross-Ibarra, and Frank You. Thanks also go to Fengqun Yu and Qilin Chen for providing access to Blast2GO Pro software. This research was financially supported by AAFC Genomics Research and Development Initiative funding (to Y.-B.F.), the National Research Council Canada’s Canadian Wheat Improvement program (D.J.K.), and the Ancell-Teichert Research foundation for Genetics and Molecular Evolution (to E.N.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Sequence data were deposited to NCBI. Meta output data were deposited to FigShare.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909564116/-/DCSupplemental.
References
- 1.Root T. L., et al. , Fingerprints of global warming on wild animals and plants. Nature 421, 57–60 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Ravenscroft C. H., Whitlock R., Fridley J. D., Rapid genetic divergence in response to 15 years of simulated climate change. Glob. Change Biol. 21, 4165–4176 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynch M., Lande R., “Evolution and extinction in response to environmental change” in Biotic Interactions and Global Change, Kareiva P., Kingsolver J., Huey R., Eds. (Sinauer Assocs., Inc., Sunderland, MA, 1993), pp. 234–250. [Google Scholar]
- 4.Hoffmann A. A., Willi Y., Detecting genetic responses to environmental change. Nat. Rev. Genet. 9, 421–432 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Renaut S., Rieseberg L. H., The accumulation of deleterious mutations as a consequence of domestication and improvement in sunflowers and other Compositae crops. Mol. Biol. Evol. 32, 2273–2283 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Kono T. J., et al. , The role of deleterious substitutions in crop genomes. Mol. Biol. Evol. 33, 2307–2317 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramu P., et al. , Cassava haplotype map highlights fixation of deleterious mutations during clonal propagation. Nat. Genet. 49, 959–963 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Tataru P., Mollion M., Glémin S., Bataillon T., Inference of distribution of fitness effects and proportion of adaptive substitutions from polymorphism data. Genetics 207, 1103–1119 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L., et al. , The interplay of demography and selection during maize domestication and expansion. Genome Biol. 18, 215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaut B. S., Seymour D. K., Liu Q., Zhou Y., Demography and its effects on genomic variation in crop domestication. Nat. Plants 4, 512–520 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Krugman T., Nevo E., Beharav A., Sela H., Fahima T., The Institute of Evolution wild cereal gene bank at the University of Haifa. Isr. J. Plant Sci. 65, 129–146 (2018). [Google Scholar]
- 12.Nevo E., “Origin, evolution, population genetics and resources for breeding of wild barley, Hordeum spontaneum, in the Fertile Crescent” in Barley, Shewry P. R., Ed. (C.A.B. International, 1992), pp. 19–43. [Google Scholar]
- 13.Nevo E., Global warming and future food production. Geoinfor Geostat An Overview 4, 4 (2016). [Google Scholar]
- 14.Saranga Y., A century of wheat research–from wild emmer discovery to genome analysis. Isr. J. Plant Sci. 55, 3–4 (2007). [Google Scholar]
- 15.Hajjar R., Hodgkin T., The use of wild relatives in crop improvement: A survey of developments over the last 20 years. Euphytica 156, 1–13 (2007). [Google Scholar]
- 16.Fu Y. B., Somers D. J., Genome-wide reduction of genetic diversity in wheat breeding. Crop Sci. 49, 161–168 (2009). [Google Scholar]
- 17.Lobell D. B., Schlenker W., Costa-Roberts J., Climate trends and global crop production since 1980. Science 333, 616–620 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Khoury C. K., et al. , Comprehensiveness of conservation of useful wild plants: An operational indicator for biodiversity and sustainable development targets. Ecol. Indic. 98, 420–429 (2019). [Google Scholar]
- 19.Castañeda-Álvarez N. P., et al. , Global conservation priorities for crop wild relatives. Nat. Plants 2, 16022 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Thuiller W., Lavorel S., Araújo M. B., Sykes M. T., Prentice I. C., Climate change threats to plant diversity in Europe. Proc. Natl. Acad. Sci. U.S.A. 102, 8245–8250 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen I.-C., Hill J. K., Ohlemüller R., Roy D. B., Thomas C. D., Rapid range shifts of species associated with high levels of climate warming. Science 333, 1024–1026 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Jarvis A., Lane A., Hijmans R. J., The effect of climate change on crop wild relatives. Agric. Ecosyst. Environ. 126, 13–23 (2008). [Google Scholar]
- 23.Nevo E., et al. , Evolution of wild cereals during 28 years of global warming in Israel. Proc. Natl. Acad. Sci. U.S.A. 109, 3412–3415 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitney K. D., et al. , Experimental drought reduces genetic diversity in the grassland foundation species Bouteloua eriopoda. Oecologia 189, 1107–1120 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nevo E., Korol A. B., Beiles A., Fahima T., Evolution of Wild Emmer and Wheat Improvement. Population Genetics, Genetic Resources, and Genome Organization of Wheat’s Progenitor, Triticum Dicoccoides (Springer-Verlag, 2002), p. 364. [Google Scholar]
- 26.Parmesan C., Ecological and evolutionary responses to recent climate change. Annu. Rev. Ecol. Evol. Syst. 37, 637–669 (2006). [Google Scholar]
- 27.Henry I. M., et al. , Efficient genome-wide detection and cataloging of EMS-induced mutations using exome capture and next-generation sequencing. Plant Cell 26, 1382–1397 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avni R., et al. , Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357, 93–97 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Ling H.-Q., et al. , Genome sequence of the progenitor of wheat A subgenome Triticum urartu. Nature 557, 424–428 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korneliussen T. S., Albrechtsen A., Nielsen R., ANGSD: Analysis of next generation sequencing data. BMC Bioinformatics 15, 356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naithani S., Geniza M., Jaiswal P., Variant effect prediction analysis using resources available at Gramene Database. Methods Mol. Biol. 1533, 279–297 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Alachiotis N., Pavlidis P., RAiSD detects positive selection based on multiple signatures of a selective sweep and SNP vectors. Commun. Biol. 1, 79 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korneliussen T. S., Moltke I., Albrechtsen A., Nielsen R., Calculation of Tajima’s D and other neutrality test statistics from low depth next-generation sequencing data. BMC Bioinformatics 14, 289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaser R., Adusumalli S., Leng S. N., Sikic M., Ng P. C., SIFT missense predictions for genomes. Nat. Protoc. 11, 1–9 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Cooper G. M., et al. ; NISC Comparative Sequencing Program , Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 15, 901–913 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Supek F., Bošnjak M., Škunca N., Šmuc T., REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.IPCC , “Assessing key vulnerabilities and the risk from climate change” in Climate Change 2007: Impacts, Adaptation and Vulnerability. Contribution of Working Group II to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change, Parry M. L., Canziani O. F., Palutikof J. P., van der Linden P. J., Hanson C. E., Eds. (Cambridge University Press, Cambridge, 2007), vol. 4, pp. 779–810. [Google Scholar]
- 38.Parmesan C., Hanley M. E., Plants and climate change: Complexities and surprises. Ann. Bot. 116, 849–864 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grimm V., Wyszomirski T., Aikman D., Uchmański J., Individual-based modelling and ecological theory: Synthesis of a workshop. Ecol. Modell. 115, 275–282 (1999). [Google Scholar]
- 40.Erickson R. A., et al. , Assessing local population vulnerability with branching process models: An application to wind energy development. Ecosphere 6, 254 (2015). [Google Scholar]
- 41.Hoffmann A. A., Sgrò C. M., Climate change and evolutionary adaptation. Nature 470, 479–485 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Landguth E. L., et al. , Combining demographic and genetic factors to assess population vulnerability in stream species. Ecol. Appl. 24, 1505–1524 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Mousseau T. A., Sinervo B., Endler J., Eds., Adaptive Genetic Variation in the Wild (Oxford University Press, New York, 2000), p. 265. [Google Scholar]
- 44.Mascher M., et al. , Barley whole exome capture: A tool for genomic research in the genus Hordeum and beyond. Plant J. 76, 494–505 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Franks S. J., Weber J. J., Aitken S. N., Evolutionary and plastic responses to climate change in terrestrial plant populations. Evol. Appl. 7, 123–139 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Etterson J. R., et al. , Project Baseline: An unprecedented resource to study plant evolution across space and time. Am. J. Bot. 103, 164–173 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Thormann I., et al. , Genotypic and phenotypic changes in wild barley (Hordeum vulgare subsp. spontaneum) during a period of climate change in Jordan. Genet. Resour. Crop Evol. 64, 1295–1312 (2017). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.