Abstract
Solving high-resolution structures of membrane proteins has been an important challenge for decades, still lagging far behind that of soluble proteins even with the recent remarkable technological advances in X-ray crystallography and electron microscopy. Central to this challenge is the necessity to isolate and solubilize membrane proteins in a stable, natively folded and functional state, a process influenced by not only the proteins but also their surrounding chemical environment. This review highlights recent community efforts in the development and characterization of novel membrane agents and ligand tools to stabilize individual proteins and protein complexes, which together have accelerated progress in membrane protein structural biology.
Introduction
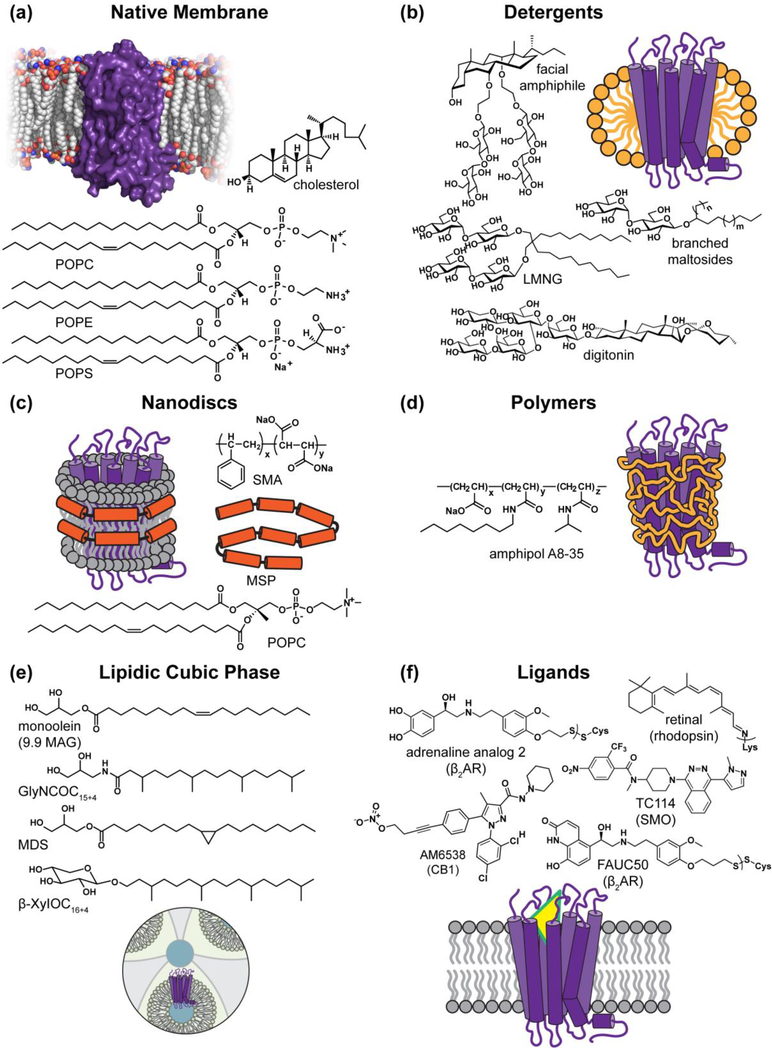
Much has been written about the critical biological and biomedical significance of membrane proteins (MPs). Structural knowledge is crucial for understanding the underlying biological function and mechanism, as well as for structure-based drug design. However, membrane protein structural biology lags far behind that of soluble proteins [1–3]. Currently, X-ray crystallography, electron cryomicroscopy (cryoEM), and nuclear magnetic resonance spectroscopy are the major biophysical techniques for solving high-resolution structures, with each method having its own advantages and limitations [4–6]. Regardless of the technique, sample preparation is the most significant challenge for MP structure determination. Important tasks throughout this process are engineering of protein constructs and selection of the best expression platform. Stabilization of individual MPs or their complexes in a solubilized state is essential, requiring optimization of the chemical environment, including lipids, detergents, membrane mimetics and ligands [7,8]. The development of novel chemical and protein-centric tools together and significant technological advancements in cryoEM and X-ray crystallography have accelerated progress in MP structural biology. This review describes chemical tools, including both membrane reagents and ligands (Figure 1), highlights recent achievements and discusses unmet challenges, with a perspective for more innovative tool development to impact both MP structural biology and drug discovery.
Figure 1.
Chemical tools for MP stabilization outside of their native membrane environment.
Detergents for MP Stabilization
MPs reside in cells in an anisotropic and heterogeneous membrane environment, where the lipid bilayer imposes geometric constraints on their structure and thermodynamic stability (Figure 1a). Specific lipid interactions are crucial for the function of many MPs [9,10]. Ideally, MP structures should be determined in their native biological membranes, where the proteins of interest are stable and fully functional, retaining their endogenous lipids, ligands and protein partners. However, very few MPs, such as bacteriorhodopsin [11], vertebrate rhodopsin [12], the acetylcholine receptor [13], and the sarcoplasmic reticulum Ca2+-ATPase [14], are sufficiently abundant in their native membranes for direct electron crystallography, typically at low to subnanometer resolution. Most MPs are present in low copy numbers within cell membranes, necessitating purification and enrichment from either natural sources or overexpression using recombinant methods. A common practice is to extract and solubilize MPs using small-molecule detergents that can partition into and effectively disperse the membrane. Detergent micelles provide an artificial and less than ideal membrane-mimetic environment, in which MPs are prone to denaturation, aggregation and loss of activity. For structural studies, use of detergents may cause additional problems, such as the difficulty in crystallization of MPs with small soluble ectodomains that mediate protein-protein interactions in 3D crystals. A common vexation is the growth of low-quality MP crystals that are difficult to improve [15]. A challenge of cryoEM is the exceedingly low signal-to-noise ratio between the protein and the surrounding vitreous ice. Empty detergent micelles contribute to the background and interfere with single-particle image analysis, especially for MPs of similar particle size to the micelles. Nevertheless, despite all the challenges associated with their use, detergents were the first and remain the most versatile and invaluable tools for MP sample preparation. For both crystallography and cryoEM, a large fraction of MP structures are still determined in detergent micelles. Most other types of membrane mimetics (e.g. amphipols, nanodiscs, lipidic cubic phase (LCP)), as will be discussed in this review, do not effectively disperse cell membranes, and their use requires detergent solubilization and purification of MPs prior to exchange into the mimetic. A brief comment on the advantages and limitations of various membrane-mimicking reagents is included in Table 1.
Table 1.
Advantages and limitations of different chemical tools for MP structural studies.
| Advantages | Limitations | |
|---|---|---|
| Detergents | - Highly versatile | - Often too harsh - Less than ideal membrane mimic - High background in cryoEM - Poor crystal quality for MPs with small soluble domains |
| Polymers | - Gentler alternative to detergents | - Most do not directly extract MPs |
| MSPs | - Used for encompassing nanodiscs - Most native-like membrane mimic - Great for cryoEM |
- Do not directly extract MPs - Not suitable for crystallography |
|
Lipid Mesophases |
- Membrane-like environment - Support crystallization |
- Not suitable for cryoEM - Limited range of host lipids |
| Ligands | - Provide synergistic stabilization - Can be covalently attached |
- Not available for many MPs |
Many new, less harsh detergents have been developed in recent years with improved properties, which enhance the stability of MPs for structural studies (Figure 1b). Of particular interest among these are molecules with improved properties as membrane mimetics, which include lipid-like branched or double alkyl chain detergents [15–19], as well as cholesterol-like, steroid-based detergents [20–24]. Although structurally distinct from each other, these detergents share common features, including enhanced hydrophobicity and compactness, allowing tighter association with MP surfaces than conventional single-chain detergents. New detergent designs that include an even larger hydrophobic surface or contain more than two alkyl chains further increase MP stability [25–27]. Related to the development of various steroid-based detergents, many MPs can be stabilized by a combination of cholesteryl hemisuccinate (CHS) with common detergents such as dodecyl-β-D-maltoside (DDM) or lauryl maltose neopentyl glycol (LMNG). CHS may impart this important benefit by mimicking direct interactions of cholesterol with some eukaryotic MPs [28]. Structurally, the incorporation of CHS into DDM micelles induces flattened bicelle-like structures that better mimic a membrane setting [29].
It is also worthwhile to note recent successful cryoEM MP structures using digitonin or similar molecules (glyco-diosgenin/GDN [23]). Digitonin has had a long history of use in membrane biochemistry, but to the best of our knowledge, it has yet to succeed in supporting crystallization of MPs. The reason for its lack of success in crystallography might be attributed to its mixed (and variable) chemical composition, its complex phase properties, or the large size of micelles that it forms. Digitonin is, nevertheless, among the best detergents for stabilization of challenging eukaryotic MPs or MP complexes, as it has been used in the structure determination of some notoriously difficult targets, such as γ-secretase [30], the cystic fibrosis transmembrane conductance regulator (CFTR) [31], and the multidrug resistance protein 1 (MRP1) [32]. Digitonin has also been demonstrated to retain phospholipids during the solubilization of the transporter associated with antigen processing (TAP) [33]. The unique steroid moiety of digitonin along with extreme structural rigidity may confer its particularly mild properties compared to other detergent types. Given the known limitations of digitonin for crystallographic use and the scarcity of digitonin analogs, it is worthwhile to expand this promising class of detergents by synthesizing new digitonin-like molecules with improved chemical and physical properties. To this end, DGN, a synthetic digitonin, has now been commercialized (Anatrace).
Detergent-free Solubilization of MPs
Dramatic advances made in cryoEM methodology over the last few years have greatly expanded the utility of non-detergent membrane-mimetic systems for high-resolution MP structural studies. Prominent examples for single-particle cryoEM are phospholipid bilayer nanodiscs [34] and amphipols [35] (Figures 1c and 1d), but they have thus far eluded applications in MP crystallization. In contrast to traditional detergent molecules, amphipols are amphiphilic polymers decorated with multiple alkyl chains that can wrap around the hydrophobic surfaces of MPs to form a less dynamic protein-polymer assembly (Figure 1d). Nanodiscs are usually prepared by mixing detergent-solubilized MPs, lipids, and membrane scaffold proteins (MSPs) at controlled ratios, followed by removal of detergent. Individual MPs or MP complexes become embedded in the resulting lipid bilayer discs, which are encircled by the MSP “belt” of defined size (Figure 1c). MSPs were originally designed based on the differently truncated sequences of high-density apolipoproteins [36–38]. More recent variations include covalently circularized MSPs, designed to achieve more precise control over the nanodisc size [39], and an alternative scaffold-protein, called saposin [40]. The various nanodisc systems offer a wide range of disc diameters (from 6 nm up to 80 nm), capable of accommodating small to large MPs or MP complexes at different stoichiometries, for structural and functional studies. We refer readers to more extensive reviews on the applications of protein-based nanodiscs [34,41].
Unlike MSPs and aforementioned amphipols, styrene-maleic acid (SMA) copolymers have been recently shown to have the ability to solubilize MPs directly from cell membranes [42,43]. The membrane solubilization efficacy of SMA polymers, which randomly display hydrophobic styrene and hydrophilic maleic acid moieties along their linear hydrocarbon chains, varies with the polymer length, styrene/maleic anhydride ratio, and pH [44]. An important feature of direct solubilization with SMAs is its co-solubilization of endogenous lipids together with MPs into nanosized lipid particles termed SMALPs (Figure 1c). Along with bypassing the use of conventional detergents, this is an ideal strategy to capture MPs in a nearly native environment for subsequent purification and structure determination. But at present, the solubilization efficacy and stabilizing benefits of SMAs remain less well established than many other widely adopted chemical tools for MP structural and functional studies. In some cases, SMAs can be difficult to utilize. For instance, UV absorbance of the styrene moieties in the polymer presents an inconvenience. Chelation of divalent cations (e.g. Ni2+, Ca2+ and Mg2+), owing to the density of carboxylate anions, constrains the use of SMAs for some chromatographic purifications (e.g. immobilized metal affinity chromatography) and for certain functional assays (e.g. ATPases). As such, various structural modifications in both polar and apolar segments of the polymer have been made in order to improve the properties of these polymers [45–47].
SMALP nanoparticles appear to have distinct properties compared to MSP nanodiscs. In MSP nanodiscs the lipid movement is largely confined within individual discs, providing a relatively static, stable lipid environment. In contrast, fast lipid transfer (occurring within seconds) has been observed between SMALP particles [48]. In this way SMALP nanoparticles are a dynamic system similar to detergent micelles. Direct crystallization of SMA-solubilized MPs will likely be challenging, although bacteriorhodopsin crystals have been grown in LCP after detergent-free SMA solubilization and purification [49]. In a recent marked success, SMA-solubilized nanoparticles yielded a 3.4-Å resolution cryoEM structure of the Flavobacterium johnsoniae alternative complex III (ACIII), together with a structure of its supercomplex with an aa3-type cytochrome c oxidase [50]. The cryoEM map revealed a surprisingly thin layer of density contributed by SMA and lipids, following the contours of the protein. The structural flexibility of SMALPs was also manifested in another recent cryoEM structure of the bacterial multidrug exporter AcrB, where a putative thin layer of lipid was described that followed the contours of the protein [51].
Membrane-Mimetic Mesophases for Crystallization
MPs can readily crystallize directly from lipid bilayers, provided that the bilayers are interconnected to form a 3D network, such as the arrangement that exists in LCP or in mixed lipid-detergent, perforated lamellar phases, often referred to as bicelles. Since the high-resolution structure determination of bacteriorhodopsin in 1996 [52], LCP has become one of the most successful membrane-mimetic matrices for stabilization and crystallization of MPs. MPs have also been crystallized in bicelle systems [53–55], but their popularity appears to be waning.
LCP spontaneously self-assemble upon mixing of specific lipids with an aqueous buffer to create a periodic structure with cubic symmetry. Topologically, LCP consists of a single lipid bilayer dividing the space into two networks of interwoven water channels (Figure 1e). The most commonly used and least expensive host lipid for LCP crystallization is monoolein, a monoacylglycerol with a double bond in the middle of its 18-hydrocarbon chain (9.9 MAG) [56]. An efficient synthesis of variable chain MAGs has been described [57,58]. In particular, the shorter chain MAGs that support larger diameter solvent channels were essential for crystallization of several challenging MPs, including GPCRs bound to their heterotrimeric G protein signaling partners [59,60], a proton-translocating transhydrogenase enzyme [61], and other MPs [62,63]. As an alternative to MAGs, a series of isoprenoid-chain lipids have been developed, and one of them (β-XylOC16+4) supports MP crystallization in LCP [64].
Native lipids of biological membranes do not spontaneously produce LCP. However, certain types of phospholipids, cholesterol, and other natural lipids can be doped into an LCP mixture to tune such properties as the membrane thickness and curvature, or to provide specific lipid-protein interactions. Despite the wide adoption of the LCP crystallization method, the number of available host lipids and their properties remain limited. Among recent developments to expand the LCP host lipid repertoire, the double bond of monoolein was replaced with a cyclopropyl group (monodihydrosterculin, MDS), extending the temperature range of LCP to enable low-temperature crystallization [65]. The relatively low chemical stability of MAGs was recently addressed by the development of non-hydrolysable lipids (e.g. GlyNCOC15+4), which also support MP crystallization at 20 °C and 4 °C [66].
Custom Ligands for Stabilization of MPs
In addition to engineering a membrane-like environment, selecting or designing tightly bound ligands can provide synergistic MP stabilization (Figure 1f). The early structural studies of bacteriorhodopsin and rhodopsin took advantage of the stability conferred by the covalently-bound retinal ligand. Similarly, many recent successful structural studies of GPCRs have benefited from wide-ranging medicinal chemistry efforts that produced a wealth of high-affinity antagonists and agonists. These ligands substantially enhance MP thermal stability and conformational homogeneity [67]. To overcome generally low affinity or a brief residence time of natural or synthetic agonists, covalent agonists have also been designed to trap receptors in active conformations [68,69]. The design and synthesis of stabilizing antagonists also contributed to the successful crystallization of full-length smoothened [70] and cannabinoid receptor 1 [71]. Nevertheless, there is a pressing need to generate ligand tools for many other GPCRs that have thus far defied high-resolution structural determination, such as the largest subfamily of ~400 olfactory receptors.
Unlike the rich pharmacology of GPCRs, there is a general lack of high-affinity ligands for transporters and channels. Conformational heterogeneity of transporters poses a significant challenge for their high-resolution structure determination, and obtaining ligand-bound structures will be especially important to define ligand binding sites in the context of a dynamic conformational pathway for substrate transport. For instance, several crystal and cryoEM structures of the multidrug resistance P-glycoprotein (ABCB1) were determined at 3.4–3.8 Å resolution in complexes with several ligands bound within a V-shaped transmembrane cavity [72–74]. These ligands have only moderate affinities (> 200 nM) and do not appear to stabilize P-glycoprotein in a single conformation. As such, development of higher affinity ligands may lead to higher-resolution structures of this multidrug transporter. To this end, a 2.9 Å-resolution crystal structure of the bacterial P-glycoprotein homolog MsbA was recently solved in a complex with both the lipopolysaccharide substrate and a potent inhibitor, which was discovered in a screen of ~ 3 million compounds [75]. In this case, the use of a novel facial amphiphile [21] also contributed to stabilization of MsbA.
Assaying a Large Chemical Space
It can be a daunting task to screen a vast number of chemical variables including membrane mimetics, ligands, as well as other buffer conditions and additives to identify a stabilizing matrix for structural studies of a given MP. Various stability assays, such as those based on protein activity, thermal stability, aggregation, or chemical denaturation, have been frequently employed. Among these, several fluorescence-based thermal stability assays, including, for example, fluorescent protein-based size-exclusion chromatography (FSEC) [76,77] and a protein unfolding assay using a cysteine-reactive fluorescent dye (CPM) [78] are appealing because of their high sensitivity and easy adaptability to different MPs. The use of only nanogram to microgram protein samples in these assays allows screening of many conditions typically in a medium-to-high throughput format. Despite these advances, it is still imperative for the community to develop MP-specific sparse matrix type high-throughput screens, such as those recently reported for studies of soluble proteins [79]. Complications involving a membrane matrix in MP assays limit the application of many fluorescent dye-based stability assays because of the high fluorescence background that is amplified in a hydrophobic environment. The advantage of using a CPM dye that becomes highly fluorescent upon conjugation with free cysteines partly addresses this issue. On the other hand, the application of CPM assays may require careful engineering of free cysteines embedded in the protein core, which could be especially challenging for multidomain proteins and complexes. Binding of CPM to cysteines in MP ectodomains may also complicate the interpretation. Lastly, non-specific CPM reactivity limits its application for broad screening of chemical libraries, as well as some buffer and high pH conditions. In this regard, miniaturized label-free differential scanning fluorimetry that measures the changes of intrinsic protein fluorescence represents a promising and relatively new development for thermal stability assays of MPs [80].
Conclusions and Future Perspectives
Preparing high-quality samples amenable to high-resolution structural studies continues to be a major challenge in MP structural biology. The fundamental issue is stabilization of MPs while faithfully maintaining their native activity through solubilization, purification and structural studies. Over the last two decades, many innovative reagents have been developed that solubilize and stabilize MPs in a more membrane-like environment. However, despite the exciting progress, the selection of the most efficient combinations of such chemical tools for a new MP target remains largely a process of trial-and-error.
Direct solubilization of MPs into a nearly native environment, as embodied in the concept of amphipols such as SMAs, can make the handling of MPs more convenient and economical, which would significantly impact a broad range of MP research beyond structural applications. In addition, access to large libraries of chemical or biological ligands, with high affinities and high propensity for thermostabilization would have an enormous impact on structural studies of MPs. New developments in synthetic chemistry could be applied to the discovery of such ligands. For example, the rapidly evolving DNA-encoded library (DEL) synthesis has enabled the generation of an unprecedented number of compounds (> 108), which can be screened efficiently [81]. In a recent example, the DEL strategy was used to identify the first allosteric antagonist of the β2-adrenergic receptor, which was subsequently co-crystallized with the receptor [82,83]. Covalent in situ attachment of a ligand, such as by biorthogonal SuFEx chemistry [84,85], may be attempted to stabilize the numerous MP targets that currently lack high-affinity binders. Looking forward, we envision exciting opportunities for synthetic chemists to play an important role in advancing MP structural biology and pharmacology.
Acknowledgements
This work was supported by NIH grants R01 GM118594 (QZ) and R35 GM127086 (VC).
Funding
No funding was received for this work.
Footnotes
Conflict of Interest Statement
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as
• of special interest
•• of outstanding interest
- 1.Imming P, Sinning C, Meyer A: Drugs, their targets and the nature and number of drug targets. Nat Rev Drug Discov 2006, 5:821–834. [DOI] [PubMed] [Google Scholar]
- 2.White SH: Membrane proteins of known 3D structure. http://blanco.biomol.uci.edu/mpstruc/.
- 3.Hendrickson WA: Atomic-level analysis of membrane-protein structure. Nat Struct Mol Biol 2016, 23:464–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng Y: Single-Particle Cryo-EM at Crystallographic Resolution. Cell 2015, 161:450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishchenko A, Gati C, Cherezov V: Structural biology of G protein-coupled receptors: new opportunities from XFELs and cryoEM. Curr Opin Struct Biol 2018, 51:44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ziarek JJ, Baptista D, Wagner G: Recent developments in solution nuclear magnetic resonance (NMR)-based molecular biology. J Mol Med (Berl) 2018, 96:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magnani F, Serrano-Vega MJ, Shibata Y, Abdul-Hussein S, Lebon G, Miller-Gallacher J, Singhal A, Strege A, Thomas JA, Tate CG: A mutagenesis and screening strategy to generate optimally thermostabilized membrane proteins for structural studies. Nat Protoc 2016, 11:1554–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bill RM, Henderson PJ, Iwata S, Kunji ER, Michel H, Neutze R, Newstead S, Poolman B, Tate CG, Vogel H: Overcoming barriers to membrane protein structure determination. Nat Biotechnol 2011, 29:335–340. [DOI] [PubMed] [Google Scholar]
- 9.Contreras FX, Ernst AM, Wieland F, Brugger B: Specificity of intramembrane protein-lipid interactions. Cold Spring Harb Perspect Biol 2011, 3:a004705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrera NP, Zhou M, Robinson CV: The role of lipids in defining membrane protein interactions: insights from mass spectrometry. Trends Cell Biol 2013, 23:1–8. [DOI] [PubMed] [Google Scholar]
- 11.Kimura Y, Vassylyev DG, Miyazawa A, Kidera A, Matsushima M, Mitsuoka K, Murata K, Hirai T, Fujiyoshi Y: Surface of bacteriorhodopsin revealed by high-resolution electron crystallography. Nature 1997, 389:206–211. [DOI] [PubMed] [Google Scholar]
- 12.Unger VM, Hargrave PA, Baldwin JM, Schertler GF: Arrangement of rhodopsin transmembrane alpha-helices. Nature 1997, 389:203–206. [DOI] [PubMed] [Google Scholar]
- 13.Miyazawa A, Fujiyoshi Y, Unwin N: Structure and gating mechanism of the acetylcholine receptor pore. Nature 2003, 423:949–955. [DOI] [PubMed] [Google Scholar]
- 14.Xu C, Rice WJ, He W, Stokes DL: A structural model for the catalytic cycle of Ca(2+)-ATPase. J Mol Biol 2002, 316:201–211. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Q, Tao H, Hong WX: New amphiphiles for membrane protein structural biology. Methods 2011, 55:318–323.• A review of amphiphile designs with a focus on crystallographic applications.
- 16.Hong WX, Baker KA, Ma X, Stevens RC, Yeager M, Zhang Q: Design, synthesis, and properties of branch-chained maltoside detergents for stabilization and crystallization of integral membrane proteins: human connexin 26. Langmuir 2010, 26:8690–8696.• A novel design of branch-chained maltosides for stabilization of MPs.
- 17.Chae PS, Rasmussen SG, Rana RR, Gotfryd K, Chandra R, Goren MA, Kruse AC, Nurva S,Loland CJ, Pierre Y, et al. : Maltose-neopentyl glycol (MNG) amphiphiles for solubilization, stabilization and crystallization of membrane proteins. Nat Methods 2010, 7:1003–1008.•• Introduction of maltose-neopentyl glycol detergents for stabilization of a broad range of MPs.
- 18.Nguyen KA, Peuchmaur M, Magnard S, Haudecoeur R, Boyere C, Mounien S, Benammar I,Zampieri V, Igonet S, Chaptal V, et al. : Glycosyl-Substituted Dicarboxylates as Detergents for the Extraction, Overstabilization, and Crystallization of Membrane Proteins. Angew Chem Int Ed Engl 2018, 57:2948–2952.• Apart from the most widely used sugar detergents, a series of glycosyl-substituted dicarboxylate detergents were designed to stabilize MPs.
- 19.Ehsan M, Du Y, Scull NJ, Tikhonova E, Tarrasch J, Mortensen JS, Loland CJ, Skiniotis G,Guan L, Byrne B, et al. : Highly Branched Pentasaccharide-Bearing Amphiphiles for Membrane Protein Studies. J Am Chem Soc 2016, 138:3789–3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q, Ma X, Ward A, Hong WX, Jaakola VP, Stevens RC, Finn MG, Chang G: Designing facial amphiphiles for the stabilization of integral membrane proteins. Angew Chem Int Ed Engl 2007, 46:7023–7025.
- 21.Lee SC, Bennett BC, Hong WX, Fu Y, Baker KA, Marcoux J, Robinson CV, Ward AB, Halpert JR, Stevens RC, et al. : Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins. Proceedings of the National Academy of Sciences of the United States of America 2013, 110:E1203–1211.•• The utility of facial amphiphiles is demonstrated by stabilization and crystallization of diverse MPs.
- 22.Chae PS, Gotfryd K, Pacyna J, Miercke LJ, Rasmussen SG, Robbins RA, Rana RR, Loland CJ, Kobilka B, Stroud R, et al. : Tandem facial amphiphiles for membrane protein stabilization. J Am Chem Soc 2010, 132:16750–16752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chae PS, Rasmussen SG, Rana RR, Gotfryd K, Kruse AC, Manglik A, Cho KH, Nurva S, Gether U, Guan L, et al. : A new class of amphiphiles bearing rigid hydrophobic groups for solubilization and stabilization of membrane proteins. Chemistry 2012, 18:9485–9490.• A simple modification of natural steroids yielded useful detergents, including glycol-diosgenin, for stabilization of MPs.
- 24.Chae PS, Das M, Du Y, Mortensen J, Bae HE, Byrne B, Loland C, Kobilka B: An engineered lithocholate-based facial amphiphile stabilizes membrane proteins: assessing the impact of detergent customizability on protein stability. Chemistry 2018, 24:9860–9868. [DOI] [PubMed] [Google Scholar]
- 25.Bae HE, Mortensen JS, Ribeiro O, Du Y, Ehsan M, Kobilka BK, Loland CJ, Byrne B, Chae PS: Tandem neopentyl glycol maltosides (TNMs) for membrane protein stabilisation. Chem Commun (Camb) 2016, 52:12104–12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadaf A, Du Y, Santillan C, Mortensen JS, Molist I, Seven AB, Hariharan P, Skiniotis G, Loland CJ, Kobilka BK, et al. : Dendronic trimaltoside amphiphiles (DTMs) for membrane protein study. Chem Sci 2017, 8:8315–8324.• New detergents synthesized containing an increased number (four) of alkyl chains compared to the most commonly used one or two-chained detergents.
- 27.Hussain H, Du Y, Tikhonova E, Mortensen JS, Ribeiro O, Santillan C, Das M, Ehsan M, Loland CJ, Guan L, et al. : Resorcinarene-Based Facial Glycosides: Implication of Detergent Flexibility on Membrane-Protein Stability. Chemistry 2017, 23:6724–6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EY, Velasquez J, Kuhn P, Stevens RC: A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor. Structure 2008, 16:897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson AA, Liu JJ, Chun E, Wacker D, Wu H, Cherezov V, Stevens RC: GPCR stabilization using the bicelle-like architecture of mixed sterol-detergent micelles. Methods 2011, 55:310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu P, Bai XC, Ma D, Xie T, Yan C, Sun L, Yang G, Zhao Y, Zhou R, Scheres SHW, et al. : Three-dimensional structure of human gamma-secretase. Nature 2014, 512:166–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu F, Zhang Z, Csanady L, Gadsby DC, Chen J: Molecular Structure of the Human CFTR Ion Channel. Cell 2017, 169:85–95 e88. [DOI] [PubMed] [Google Scholar]
- 32.Johnson ZL, Chen J: Structural Basis of Substrate Recognition by the Multidrug Resistance Protein MRP1. Cell 2017, 168:1075–1085 e1079. [DOI] [PubMed] [Google Scholar]
- 33.Scholz C, Parcej D, Ejsing CS, Robenek H, Urbatsch IL, Tampe R: Specific lipids modulate the transporter associated with antigen processing (TAP). J Biol Chem 2011, 286:13346–13356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denisov IG, Sligar SG: Nanodiscs in Membrane Biochemistry and Biophysics. Chem Rev 2017, 117:4669–4713.•• A comprehensive review of nanodiscs for their broad use in biochemical, biophysical and structural studies of MPs.
- 35.Popot JL, Berry EA, Charvolin D, Creuzenet C, Ebel C, Engelman DM, Flotenmeyer M, Giusti F, Gohon Y, Hong Q, et al. : Amphipols: polymeric surfactants for membrane biology research. Cell Mol Life Sci 2003, 60:1559–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG: Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J Am Chem Soc 2004, 126:3477–3487. [DOI] [PubMed] [Google Scholar]
- 37.Grinkova YV, Denisov IG, Sligar SG: Engineering extended membrane scaffold proteins for self-assembly of soluble nanoscale lipid bilayers. Protein Eng Des Sel 2010, 23:843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hagn F, Etzkorn M, Raschle T, Wagner G: Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J Am Chem Soc 2013, 135:1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nasr ML, Baptista D, Strauss M, Sun ZJ, Grigoriu S, Huser S, Pluckthun A, Hagn F, Walz T, Hogle JM, et al. : Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat Methods 2017, 14:49–52.•• Covalently circularized nanodiscs (cNDs) were engineered with enhanced stability and homogeneity, and the ability to generate larger nanodiscs (up to 80-nm diameters). cNDs are excellent developments to broaden the applications of nanodisc technology.
- 40.Frauenfeld J, Loving R, Armache JP, Sonnen AF, Guettou F, Moberg P, Zhu L, Jegerschold C, Flayhan A, Briggs JA, et al. : A saposin-lipoprotein nanoparticle system for membrane proteins. Nat Methods 2016, 13:345–351.•• Saponin-lipoprotein encircled nanodiscs (Salipro) were used for cryoEM of MPs. The self-assembly of Slipro nanodiscs was adaptable to the size of the incorporated MPs.
- 41.Rouck JE, Krapf JE, Roy J, Huff HC, Das A: Recent advances in nanodisc technology for membrane protein studies (2012–2017). FEBS Lett 2017, 591:2057–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee SC, Knowles TJ, Postis VL, Jamshad M, Parslow RA, Lin YP, Goldman A, Sridhar P, Overduin M, Muench SP, et al. : A method for detergent-free isolation of membrane proteins in their local lipid environment. Nat Protoc 2016, 11:1149–1162. [DOI] [PubMed] [Google Scholar]
- 43.Esmaili M, Overduin M: Membrane biology visualized in nanometer-sized discs formed by styrene maleic acid polymers. Biochim Biophys Acta 2018, 1860:257–263.• An updated overview of the development and appliation of SMA polymers for MP research.
- 44.Scheidelaar S, Koorengevel MC, van Walree CA, Dominguez JJ, Dorr JM, Killian JA: Effect of Polymer Composition and pH on Membrane Solubilization by Styrene-Maleic Acid Copolymers. Biophys J 2016, 111:1974–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oluwole AO, Danielczak B, Meister A, Babalola JO, Vargas C, Keller S: Solubilization of Membrane Proteins into Functional Lipid-Bilayer Nanodiscs Using a Diisobutylene/Maleic Acid Copolymer. Angew Chem Int Ed Engl 2017, 56:1919–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ravula T, Hardin NZ, Ramadugu SK, Ramamoorthy A: pH Tunable and Divalent Metal Ion Tolerant Polymer Lipid Nanodiscs. Langmuir 2017, 33:10655–10662. [DOI] [PubMed] [Google Scholar]
- 47.Fiori MC, Jiang Y, Altenberg GA, Liang H: Polymer-encased nanodiscs with improved buffer compatibility. Sci Rep 2017, 7:7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cuevas Arenas R, Danielczak B, Martel A, Porcar L, Breyton C, Ebel C, Keller S: Fast Collisional Lipid Transfer Among Polymer-Bounded Nanodiscs. Sci Rep 2017, 7:45875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Broecker J, Eger BT, Ernst OP: Crystallogenesis of Membrane Proteins Mediated by Polymer-Bounded Lipid Nanodiscs. Structure 2017, 25:384–392. [DOI] [PubMed] [Google Scholar]
- 50.Sun C, Benlekbir S, Venkatakrishnan P, Wang Y, Hong S, Hosler J, Tajkhorshid E, Rubinstein JL, Gennis RB: Structure of the alternative complex III in a supercomplex with cytochrome oxidase. Nature 2018, 557:123–126.•• The first near atomic resolution MP structure solved in SMA nanodiscs using cryoEM and single-particle analysis.
- 51.Qiu W, Fu Z, Xu GG, Grassucci RA, Zhang Y, Frank J, Hendrickson WA, Guo Y: Structure and activity of lipid bilayer within a membrane-protein transporter. Proc Natl Acad Sci U S A 2018, 115:12985–12990.• A cryoEM structure of AcrB purified in SMA revealed a distinct lipid belt around the transmembrane region.
- 52.Landau EM, Rosenbusch JP: Lipidic cubic phases: a novel concept for the crystallization of membrane proteins. Proc Natl Acad Sci U S A 1996, 93:14532–14535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, et al. : Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature 2007, 450:383–387. [DOI] [PubMed] [Google Scholar]
- 54.Lee JY, Kinch LN, Borek DM, Wang J, Wang J, Urbatsch IL, Xie XS, Grishin NV, Cohen JC, Otwinowski Z, et al. : Crystal structure of the human sterol transporter ABCG5/ABCG8. Nature 2016, 533:561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen S, Oldham ML, Davidson AL, Chen J: Carbon catabolite repression of the maltose transporter revealed by X-ray crystallography. Nature 2013, 499:364–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kulkarni CV, Wachter W, Iglesias-Salto G, Engelskirchen S, Ahualli S: Monoolein: a magic lipid? Phys Chem Chem Phys 2011, 13:3004–3021. [DOI] [PubMed] [Google Scholar]
- 57.Misquitta Y, Cherezov V, Havas F, Patterson S, Mohan JM, Wells AJ, Hart DJ, Caffrey M: Rational design of lipid for membrane protein crystallization. J Struct Biol 2004, 148:169–175. [DOI] [PubMed] [Google Scholar]
- 58.Fu Y, Weng Y, Hong WX, Zhang QH: Efficient Synthesis of Unsaturated 1-Monoacyl Glycerols for in meso Crystallization of Membrane Proteins. Synlett 2011:809–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, et al. : Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature 2011, 477:549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kang Y, Zhou XE, Gao X, He Y, Liu W, Ishchenko A, Barty A, White TA, Yefanov O, Han GW, et al. : Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 2015, 523:561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Padayatti PS, Leung JH, Mahinthichaichan P, Tajkhorshid E, Ishchenko A, Cherezov V, Soltis SM, Jackson JB, Stout CD, Gennis RB, et al. : Critical Role of Water Molecules in Proton Translocation by the Membrane-Bound Transhydrogenase. Structure 2017, 25:1111–1119 e1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Z, Tang Y, Wu Y, Zhao S, Bao J, Luo Y, Li D: Structural insights into the committed step of bacterial phospholipid biosynthesis. Nat Commun 2017, 8:1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li D, Lyons JA, Pye VE, Vogeley L, Aragao D, Kenyon CP, Shah ST, Doherty C, Aherne M, Caffrey M: Crystal structure of the integral membrane diacylglycerol kinase. Nature 2013, 497:521–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Borshchevskiy V, Moiseeva E, Kuklin A, Buldt G, Hato M, Gordeliy V: Isoprenoid-chained lipid beta-XylOC(16+4)-A novel molecule for in meso membrane protein crystallization. J Cryst Growth 2010, 312:3326–3330.• A new isoprenoid lipid was used for LCP crystallization of bacteriorhodopsin.
- 65.Salvati Manni L, Zabara A, Osornio YM, Schoppe J, Batyuk A, Pluckthun A, Siegel JS, Mezzenga R, Landau EM: Phase behavior of a designed cyclopropyl analogue of monoolein: implications for low-temperature membrane protein crystallization. Angew Chem Int Ed Engl 2015, 54:1027–1031.• Characterization of a new LCP lipid derived from monoolein, useful for crystallogenesis of MPs at low temperature.
- 66.Ishchenko A, Peng L, Zinovev E, Vlasov A, Lee SC, Kuklin A, Mishin A, Borshchevskiy V, Zhang Q, Cherezov V: Chemically Stable Lipids for Membrane Protein Crystallization. Cryst Growth Des 2017, 17:3502–3511.• Characterization of an amide-linked isoprenoid LCP lipid, which is resistant to hydrolysis at acidic and basic pH. The lipid was used for crystallization of MPs at both 20 °C and 4 °C.
- 67.Zhang X, Stevens RC, Xu F: The importance of ligands for G protein-coupled receptor stability. Trends Biochem Sci 2015, 40:79–87.•• This review summarizes the properties of stabilizing effects of ligands, leading to successful crystallization and structure determination of GPCRs.
- 68.Rosenbaum DM, Zhang C, Lyons JA, Holl R, Aragao D, Arlow DH, Rasmussen SG, Choi HJ, Devree BT, Sunahara RK, et al. : Structure and function of an irreversible agonist- beta(2) adrenoceptor complex. Nature 2011, 469:236–240.•• A covalent agonist was irreversibly attached to β2AR, and the complex activated G proteins. However, the structure of the covalent β2AR-agonist complex was in a relaxed, inactive conformation.
- 69.Weichert D, Kruse AC, Manglik A, Hiller C, Zhang C, Hubner H, Kobilka BK, Gmeiner P: Covalent agonists for studying G protein-coupled receptor activation. Proc Natl Acad Sci U S A 2014, 111:10744–10748.•• Another design of a covalent agonist for cysteine conjugation to β2AR.The structure was solved in an active conformation that was stabilized by a conformationally selective nanobody.
- 70.Zhang X, Zhao F, Wu Y, Yang J, Han GW, Zhao S, Ishchenko A, Ye L, Lin X, Ding K, et al. : Crystal structure of a multi-domain human smoothened receptor in complex with a super stabilizing ligand. Nat Commun 2017, 8:15383.•• An X-ray structure of human, multi-domain smoothened was determined in a complex with TC114, a super stabilizing ligand resulting from structure-based design.
- 71.Hua T, Vemuri K, Pu M, Qu L, Han GW, Wu Y, Zhao S, Shui W, Li S, Korde A, et al. : Crystal Structure of the Human Cannabinoid Receptor CB1. Cell 2016, 167:750–762 e714.•• An X-ray structure of CB1 determined in a complex with AM6538, a stabilizing antagonist designed from a known CB1 drug molecule.
- 72.Szewczyk P, Tao H, McGrath AP, Villaluz M, Rees SD, Lee SC, Doshi R, Urbatsch IL, Zhang Q, Chang G: Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein. Acta Crystallogr D Biol Crystallogr 2015, 71:732–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, et al. : Structure of P-glycoprotein reveals a molecular basis for poly- specific drug binding. Science 2009, 323:1718–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alam A, Kung R, Kowal J, McLeod RA, Tremp N, Broude EV, Roninson IB, Stahlberg H, Locher KP: Structure of a zosuquidar and UIC2-bound human-mouse chimeric ABCB1. Proc Natl Acad Sci U S A 2018, 115:E1973–E1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ho H, Miu A, Alexander MK, Garcia NK, Oh A, Zilberleyb I, Reichelt M, Austin CD, Tam C, Shriver S, et al. : Structural basis for dual-mode inhibition of the ABC transporter MsbA. Nature 2018, 557:196–201.•• The 2.9-Å resolution, inward-facing crystal structure of MsbA was solved in a complex with both a small molecule antagonist (G907) and a co-purified lipopolysaccharide. Binding of G907 uncoupled the distant nucleotide binding domains, suggesting a novel mechanism of MsbA inhibition.
- 76.Kawate T, Gouaux E: Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 2006, 14:673–681. [DOI] [PubMed] [Google Scholar]
- 77.Hattori M, Hibbs RE, Gouaux E: A fluorescence-detection size-exclusion chromatography-based thermostability assay for membrane protein precrystallization screening. Structure 2012, 20:1293–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alexandrov AI, Mileni M, Chien EY, Hanson MA, Stevens RC: Microscale fluorescent thermal stability assay for membrane proteins. Structure 2008, 16:351–359. [DOI] [PubMed] [Google Scholar]
- 79.Chari A, Haselbach D, Kirves JM, Ohmer J, Paknia E, Fischer N, Ganichkin O, Moller V, Frye JJ, Petzold G, et al. : ProteoPlex: stability optimization of macromolecular complexes by sparse-matrix screening of chemical space. Nat Methods 2015, 12:859–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boland C, Olatunji S, Bailey J, Howe N, Weichert D, Fetics SK, Yu X, Merino-Gracia J, Delsaut C, Caffrey M: Membrane (and Soluble) Protein Stability and Binding Measurements in the Lipid Cubic Phase Using Label-Free Differential Scanning Fluorimetry. Anal Chem 2018, 90:12152–12160. [DOI] [PubMed] [Google Scholar]
- 81.Neri D, Lerner RA: DNA-Encoded Chemical Libraries: A Selection System Based On Endowing Organic Compounds with Amplifiable Information. Annu Rev Biochem 2018, 87:479–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ahn S, Kahsai AW, Pani B, Wang QT, Zhao S, Wall AL, Strachan RT, Staus DP, Wingler LM, Sun LD, et al. : Allosteric “beta-blocker” isolated from a DNA-encoded small molecule library. Proc Natl Acad Sci U S A 2017, 114:1708–1713.•• An allosteric modulator ofβ2AR was identified by screening a ~190 million-compound, DNA-encoded library. The method should be broadly applicable to other MPs.
- 83.Liu X, Ahn S, Kahsai AW, Meng KC, Latorraca NR, Pani B, Venkatakrishnan AJ, Masoudi A, Weis WI, Dror RO, et al. : Mechanism of intracellular allosteric beta2AR antagonist revealed by X-ray crystal structure. Nature 2017, 548:480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen W, Dong J, Plate L, Mortenson DE, Brighty GJ, Li S, Liu Y, Galmozzi A, Lee PS, Hulce JJ, et al. : Arylfluorosulfates Inactivate Intracellular Lipid Binding Protein(s) through Chemoselective SuFEx Reaction with a Binding Site Tyr Residue. J Am Chem Soc 2016, 138:7353–7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Narayanan A, Jones LH: Sulfonyl fluorides as privileged warheads in chemical biology. Chem Sci 2015, 6:2650–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]