Abstract
Purpose
Genetically engineered mouse models of sporadic cancers are critical for studying tumor biology and for preclinical testing of therapeutics. We present an MRI-based pipeline designed to produce high resolution, quantitative information about tumor progression and response to novel therapies in mouse models of medulloblastoma (MB).
Methods
Sporadic MB was modeled in mice by inducing expression of an activated form of the Smoothened gene (aSmo) in a small number of cerebellar granule cell precursors. aSmo mice were imaged and analyzed at defined time-points using a 3D manganese-enhanced MRI (MEMRI)-based pipeline optimized for high-throughput.
Results
A semi-automated segmentation protocol was established that estimates tumor volume in a time-frame compatible with a high-throughput pipeline. Both an empirical, volume-based classifier and a Linear Discriminant Analysis (LDA)-based classifier were tested to distinguish progressing from non-progressing lesions at early stages of tumorigenesis. Tumor centroids measured at early stages revealed that there is a very specific location of the probable origin of the aSmo MB tumors. The efficacy of the MEMRI pipeline was demonstrated with a small scale experimental drug trial designed to reduce the number of tumor associated macrophages and microglia (TAMs).
Conclusion
Our results revealed a high level of heterogeneity between tumors within and between aSmo MB models, indicating that meaningful studies of sporadic tumor progression and response to therapy could not be conducted without an imaging-based pipeline approach.
Keywords: Linear Discriminant Analysis, LDA; Magnetic Resonance Imaging, MRI; Segmentation; Sonic Hedgehog, SHH; Tumor Associated Macrophages and Microglia, TAMs
INTRODUCTION
Brain and other nervous system tumors are among the most prevalent and deadly cancer types in children1. Of these cancers, the cerebellar tumor Medulloblastoma (MB), is the most common malignant pediatric brain tumor1. Although survival rates in MB patients range between 60–80% following surgery, chemotherapy, and radiation therapy (for children over three years of age)2, the long-term prognosis for many MB patients includes moderate to severe cognitive and neurological deficits, often attributable to the therapies used to prolong survival3–7. Thus, there is a need for preclinical small animal studies to test drugs targeted to vulnerabilities in pediatric brain tumors.
Over the last decade, genetic profiling studies have classified four distinct molecular subgroups of MB (Wingless, WNT; Sonic Hedgehog, SHH; Group 3; and Group 4) based on their signaling pathways, activating mutations and cell types of origin8,9. As a result, novel targeted therapeutics are currently being designed and tested to exploit these pathways to mitigate off-target effects in MB survivors. Concurrently, in order to study MB and potential therapeutics, mouse models have been developed relevant to each of the four MB subtypes10–14. In efforts to genetically engineer mice that model sporadic MB, inducible genetic systems based on CreER technology are being used to initiate MB formation in small numbers of cells of origin at a desired developmental time-point15–17.
A key advantage of the genetically engineered mouse models of sporadic human MB compared to other models, where all the cells of origin are mutated or xenograft models in immunocompromised host mice, is the preservation of the clonal nature of tumor growth and the tumor microenvironment (i.e. stroma, host immune system including tumor associated macrophages/microglia (TAMs), and physical barriers to growth)18–21. Clonality in the cells of origin and preservation of the tumor microenvironment both drive tumor formation and progression in a stochastic manner, leading to heterogeneous disease presentation among tumors at a range of times after triggering the driver mutation16. While clonal models with heterogeneity in progression rate and high rates of regression can recapitulate human MB more accurately than xenograft models or use of constitutive Cre, they also introduce challenges for controlled studies of tumor development and the effects of anticancer therapeutics because of the heterogeneity 22,23.
Assessing heterogeneity in MB progression in sporadic mouse models is difficult, if not impossible without noninvasive imaging. Longitudinal changes in brain tumor size and morphology cannot be analyzed without in vivo imaging, and obvious clinical symptoms are not observed until animals have advanced MB. Fluorescence and bioluminescence imaging approaches have been used to gauge disease state for superficial or subcutaneous tumors24–27, but are subject to poor signal-to-noise ratio (SNR) for brain tumors and are unable to provide quantitative tumor volume data. X-ray and computed tomography (CT) images are useful for high-throughput screening, but exposure to ionizing radiation limits their application in longitudinal studies of cancer28,29. In contrast, magnetic resonance imaging (MRI) operates at non-ionizing radio-frequencies, provides unparalleled soft-tissue contrast for whole brain imaging, and produces images with sufficiently high spatial resolution to resolve small tissue features, including early-stage tumors. In addition, MRI has been used to study heterogeneity in clinical presentation across different subtypes of MB30,31. Although gadolinium-enhanced MRI is a prevalent method for detection of central nervous system tumors in the clinic, usually producing positive lesion contrast32, it does not effectively delineate MB tumor margins in genetically-engineered mouse models16,33. In contrast, T1-weighted manganese-enhanced MRI (MEMRI) has been shown to produce unparalleled contrast-to-noise ratio (CNR) images of the cerebellum34,35 and enables early detection and monitoring of MB from pre-neoplastic lesions to advanced tumors16 in mice. Short MEMRI acquisition times enable high-throughput screening and reduces anesthetic exposure time – a significant cause of attrition of animals with advanced tumors. We have previously demonstrated the utility of MEMRI for detailed analyses of longitudinal MB progression and normal cerebellar development in mice from early postnatal to adult stages16,34,35.
The goal of the current study was to design a practical, time-efficient MEMRI-based pipeline for studies of tumor progression in mouse models of sporadic MB (Fig. 1), including application of the pipeline to preclinical drug trials. To this end, we implemented and validated both short acquisition-time MEMRI sequences and semi-automated tumor segmentation methods. These methods were used to obtain longitudinal tumor volume data in two mouse MB models with the same underlying activating mutation in the Smoothened gene (aSmo) in different numbers of cells soon after birth. Longitudinal tumor volume data were analyzed to provide quantitative insights into the likely location where aSmo MBs originate, a fundamental question in MB tumor biology. To address the heterogeneity of sporadic MB models, we developed and tested both an empirical, volume-based classifier and a Linear Discriminant Analysis (LDA) based classifier to distinguish progressing from non-progressing lesions at early stages of tumorigenesis, and applied the resulting pipeline to study the effects of an experimental therapeutic agent on mouse MB progression.
Figure 1. Schematic overview of the imaging pipeline.

Animal models are bred in-house and recombination events are triggered at predetermined developmental time points to initiate tumor formation. Animals are imaged longitudinally after tumorigenesis to noninvasively probe disease state. Tumor margins found in imaging data are segmented to produce tumor volume measurements, determine tumor origin, and monitor progression quantitatively. Quantitative data, in addition to relevant categorical features, are used to classify tumor progression retrospectively. In addition, prediction of disease outcome at early tumor stage is possible after sufficient baseline classification efforts. Classification and prediction methods are validated and iteratively improved upon by feedback image acquisition and segmentation.
METHODS
Animals
All mice used in this study were maintained under protocols approved by the Institutional Animal Care and Use Committee at New York University School of Medicine. aSmo mice, which express an activated (oncogenic) form of the Smoothened gene (SmoM2) in rare cerebellar granule cell precursors after postnatal day 2 (P2), were generated by breeding Atoh1-CreER transgenic mice36 with homozygous R26loxPSTOPloxP-SmoM2 (R26LSL-SmoM2) mice37. Expression of the SmoM2 oncogene was induced by subcutaneous injection of low doses (1µg/g or 5µg/g) of Tamoxifen (TMX) at P2 to produce two models of sporadic MB: aSmo-1 or aSmo-5, respectively. The genotype of each mouse was confirmed by PCR of DNA from a tail or ear biopsy using primers for SmoM2 and Cre17.
Longitudinal MEMRI Data Acquisition
30 mM solution of manganese chloride (MnCl2) tetrahydrate (Sigma-Aldrich-221279) in isotonic saline was injected intraperitoneally (IP) 24 hours before each imaging session at a dose per weight of 0.5 mmol/kg (62.5 mg MnCl2 per kg body weight). This MnCl2 dose was similar to the dose documented in our previous work using MEMRI for brain and MB imaging16. At this dose, no chronic adverse effects were observed as a consequence of MnCl2 administration in this study.
aSmo mice were imaged between postnatal week (W)3 and W13, where each time point was the specified week ± 3 days. MRI was performed in a 7-Tesla, 200-mm diameter horizontal bore magnet (Magnex Scientific) interfaced to a Bruker Biospec Avance II console (Bruker BioSpin MRI) with actively shielded gradients (750 mT/m; BGA9S; Bruker) and using a 25-mm quadrature Litzcage coil (Doty Scientific). Animals were anesthetized using isofluorane at 1.5 L/minute compressed air flow rate −3% isoflurane for induction and 1–2% isoflurane for maintenance during each imaging session, delivered through a nose cone. Core body temperature and respiration rate were monitored using a MRI-compatible rectal thermocouple and respiratory pillow (SAII; SA Instruments). Molded plastic sleds customized to secure mice of varying size were used to minimize motion (Dazai Research Instruments). T1-weighted MEMRI sessions were conducted using the following protocol: 1 min low-resolution pilot, 21min 150µm isotropic resolution spoiled 3D gradient echo (GE) sequence (echo time/repetition time, TE/TR = 4/30 ms; flip angle, FA = 20°; field-of-view, FOV = 19.2 mm × 19.2 mm × 12 mm; Matrix = 128 × 128 × 64). Resultant images from online reconstruction followed by offline, image-space, bicubic interpolation to a final matrix size of 384 × 384 × 192 (50 µm isotropic resolution) were used for further analysis.
3D Segmentation of MB Tumors from MEMRI Images
3D MEMRI data were analyzed in Amira (v. 5.5.0, Visualization Sciences Group) after each imaging time point using a tablet and stylus (Wacom). Manual tumor segmentations required the paintbrush tool, using a 20-voxel spot size for first-pass tumor region-of-interest (ROI) drawing and 3-voxel brush size for cleanup. Initial segmentations were done in axial orientation from superior to inferior 2D sections. Semi-automated segmentations required use of the image histogram partitioning tool with slight modifications (Fig. 2). Freehand boundary ROIs were drawn around approximate tumor regions in each 2D axial section to generate a partitioned histogram. Seed voxels were selected within the apparent abnormal hypointensity within the drawn boundary, and the histogram range was truncated symmetrically about the seed voxel intensity value to produce 3D tumor segmentations using the Surface Rendering toolbox. Artifact or blood vessel-borne gaps in the segmentation were corrected by flood filling the ROI. Minor corrections (involving less than 1% of the segmented voxels) to both manual and semi-automated segmentations were done in sagittal orientation, including smoothing to remove jagged edges and section-to-section incongruities, and voxel counts from the 3D tumor ROIs were obtained using the Material Statistics function. In mice with bilateral tumors that grew together at the midline of the vermis, and were therefore indistinguishable, the midline was assigned to be a boundary between the two tumor ROIs. For visualization, these tumor ROIs were interpolated by a 3 × 3 × 3-voxel kernel smoothing pass. Whole brain segmentations were obtained by applying the 4-connected neighbor Magic Wand tool to known voxels within the skull to select all ‘skull’ voxels. Inverting this selection produces initial brain segmentation, which were corrected by whole-volume 2-voxel erosion and a 3 × 3 × 3-voxel kernel smoothing pass.
Figure 2. Semi-automated segmentation workflow.
For each 2D section in the 3D raw image, readers draw freehand ROIs (green contour) around the apparently hypointense tumor and select a seed voxel (yellow “x”) within this ROI to produce an initial segmentation (purple). Then, the ROI histogram is truncated symmetrically about the seed voxel value. The “holes” left due to extreme hypo- or hyperintensity excluded after histogram truncation are flood filled and the resulting segmentation is stored for analysis.
Tumor Origin Site Analysis
An average template was created from all images in the study using iterative linear and nonlinear image registration as previously described38,39 using Pydpiper40, MINC tools41, and ANTs42 for nonlinear registration. The centroid of each tumor was calculated and rendered using ITK-SNAP’s43 ‘Convert3D’ tool, which contains a list of functions for 3D image manipulation and format conversion. Specifically, the ‘-centroid’ and ‘-centroid-mark’ functions were used to calculate and render the tumor centroids. The corresponding centroid points in the average template were then computed using the subject-to-model transformations. It should be noted that registration between subjects lacking homology, such as tumor-bearing mouse brains, is ill-defined, so the resulting centroids are only approximate. The voxel-wise incidence of tumor was integrated over all images and normalized by the number of images to produce a tumor probability heatmap. The transformed centroids and heatmap were nonlinearly aligned to the template using ANTs, then overlaid on a control mouse image using Amira.
Classifier Analysis: Progressing vs Non-Progressing
Empirical classification of longitudinal tumor volume data employed an absolute volume threshold of 7-mm3 at W7. Tumors that exhibited positive volume change between W5 and W7, and met the volume threshold criterion were considered ‘Progressing’; all other tumors were considered ‘Non-progressing’. Classification criteria were validated on cohorts of aSmo-1 and aSmo-5 tumors by monitoring progression biweekly from W5 to W9–W13, retrospectively applying the volume threshold to these data, and comparing the predicted class (‘Progressing’ or ‘Non-progressing’) to the observed outcome.
In an attempt to improve the accuracy in early detection of ‘Progressing’ tumors, an LDA classifier was trained in R using these progression data44. W5 – W13 longitudinal 3D MEMRI data were preprocessed to obtain the following features:
‘W5 Tumor Volume’ - defined as volume measured on W5;
‘W7 Tumor Volume’ - defined as volume measured on W7;
‘Volume Change’ - defined as the volume change between W5 and W7;
‘Normalized Growth Rate’ - defined as , where Time is the (exact) elapsed time in weeks between the W5 and W7 time points (approximately two weeks).
The progression data were retrospectively classified with LDA, and predicted classes were compared to observed outcome.
Administration of PLX5622
Mice with progressing tumors, as determined by the empirical classifier, were fed ad libito with PLX5622 (Plexxikon) (PLX) or control chow (CTL) beginning as early as W7 and as late as W945. PLX was delivered at 1200 mg drug per kg chow. PLX or CTL feeding continued until W13 or time of sacrifice.
Statistical Analyses
Statistical analyses were performed in R (R, v3.5.0) and G*Power46,47. Linear mixed-effects models48 were used to assess the effects of tumor model (aSmo-1 or aSmo-5) and/or treatment arm (CTL or PLX in the drug trial data) on tumor volume while allowing for small variation between subjects – each subject having up to two tumors. The general form of the linear mixed effects-models used in our analysis can be written as follows:
More specifically: individual tumors were denoted by (i); the interaction between tumor model and treatment arm as well as pre-treatment tumor volume were fixed-effects regressors with corresponding fixed-effects coefficients ; we allowed for a fixed-effect offset ; accounted for subtle, subject-specific variation as a random-effect ; and assumed normally distributed error . Given these parameters, two models were built in order to:
check for potential pre-treatment tumor volume bias in tumor model and treatment arm assignment;
directly assess the effects of the interaction between tumor model and treatment arm , as well as pre-treatment tumor volume on post-treatment absolute tumor volume .
Raw data, and implementation of these models in R, are provided in the Supporting Information Data S1. The same analyses were applied to the reclassified data after LDA. T-tests of the fixed-effect coefficients after Kenward-Roger approximation49 were applied for pairwise comparison. Bland-Altman50 analysis was used to assess inter-reader agreement using semi-automated segmentation and comparative overestimation of tumor volume between semi-automated and manual segmentation.
Results
Development of a high-throughput tumor imaging pipeline
A pipeline approach was developed for studies of tumor development in mouse models of sporadic MB. The pipeline included four sequential stages, described below (Fig. 1): Model Generation; Imaging; Segmentation; and Analysis.
Model Generation
aSmo-1 (n=27) and aSmo-5 (n=21) mice were generated for longitudinal MEMRI and development of the pipeline. Disease progression, including symptoms at advanced stages of MB progression (ataxia, hunched posture and domed head), were similar to our previous studies of aSmo-1 mice17.
Imaging
As previously reported, MEMRI provides an effective method for detecting and monitoring cerebellar MBs, which appear hypointense (Fig. 3)16,17. Furthermore, these hypointense regions were shown previously to correlate well with pre-neoplastic lesions and MB tumors on matched histological sections15,17. At W3, most aSmo-1 animals imaged had widespread, manually-segmentable, pre-neoplastic lesions throughout the cerebellum, characteristic of persistent external granule cell layer thickening (n=22/27; Fig. 3). By W5, most of these pre-neoplastic lesions had regressed below detectable volume and focal early stage tumors were observed (n=20/22 mice), similar to our previous report17. We therefore chose W5 as the starting point for longitudinal MEMRI studies of the aSmo-1 and aSmo-5 mouse MB models.
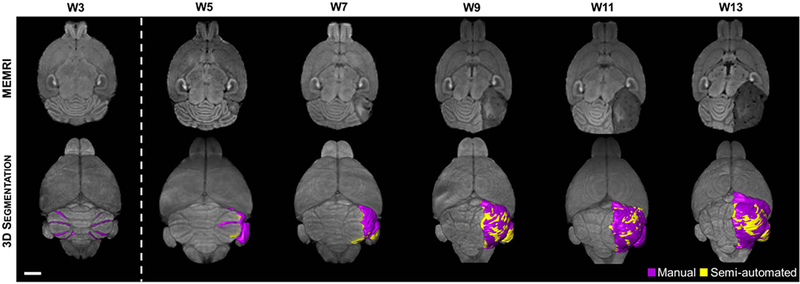
Figure 3. Semi-automated segmentation enables high throughput quantitative analysis, but overestimates tumor volume.
2D axial, T1-weighted MEMRI (top row) reveals preneoplastic lesion formation at W3 throughout the cerebellum. The majority of these lesions naturally regress. However, some lesions may accumulate secondary mutations which give rise to focal tumors apparent at W5. In this animal, the MB tumor continues to grow through W13 and displaces the remaining normal cerebellum. 3D manual segmentation (purple) of the hypointense lesions found in MEMRI (bottom row) shows the widespread preneoplastic lesion presentation at W3, followed by tumor growth, including anterior encroachment of this right-hemisphere tumor into the forebrain over time. Semi-automated segmentation (yellow) of these lesions illustrates the regions of overestimation in lesion volume. Scale bar – 2 mm.
aSmo-1 (n=14) and aSmo-5 (n=21) mice were followed with biweekly MEMRI sessions starting at W5 to assess tumor progression in the two MB models. In 12/14 of the aSmo-1 mice, MB tumors grew focally in the left cerebellar hemisphere (n=1), focally in the right hemisphere (n=3), and bilaterally (n=8) for a total of 20 aSmo-1 tumors. In contrast, MB tumors grew bilaterally in all 21 of the aSmo-5 mice for a total of 42 tumors. In both models, tumors were observed qualitatively to progress, to regress, and to both progress and regress at different time points, including some tumors that regressed below detectable levels. In mice that had progressing MB tumors, characteristic late-stage MB symptoms were often observable between W11 and W13, including displacement of normal cerebellar structures, expansion of the ventricles and skull deformation.
Segmentation - Semi-automated Approach
In order to quantify the tumor progression properties in the aSmo MB models, tumor volumes were segmented from MEMRI images. Fully manual segmentation, used in our previous studies of mouse MB16,17, require 4–5 hours per animal, which is not compatible with the needs of a high-throughput imaging pipeline. Therefore, a semi-automated approach was developed to significantly reduce the segmentation time to 10 ± 5 minutes per mouse (Figs. 2,3).
Visually, 3D renderings of the semi-automated segmentations (Fig. 3, yellow) tended to overestimate tumor volume compared to manual segmentation (Fig. 3, purple). Inter-reader agreement was generally strong and semi-automated segmentation quality was relatively reader skill independent (Fig. 4; n = 8 readers, n = 10 images; Bland-Altman coefficient of variation = 1.53, reproducibility coefficient = 9.49 mm3). Linear regression analysis of the inter-reader results showed a slightly higher slope compared to the unity line (1.13 vs 1.00), due to an overestimation of tumor volume by the semi-automated method. The cause of inter-reader discrepancy in small tumor volume estimation was inclusion by some readers of normal, hypointense white matter in the tumor segmentation. For segmentations of large tumors, untrained readers (<10 hours experience) applied the same histogram threshold for all axial sections, while skilled readers (black open circles) adjusted histogram threshold bounds to account for other sources of hypointensity at late tumor stage such as hydrocephalus, black blood artifact, and necrosis. Similarly, regions of apparent hyperintensity – generally produced by normal cerebellar lobules being compressed by large tumors – were included in the tumor segmentation by untrained readers while trained readers excluded these regions. The rapid time-to-result afforded by semi-automated segmentation enabled more efficient longitudinal quantitative analysis of aSmo tumors and justified use of semi-automated segmentation by readers with at least 10–20 hours of training to assess tumor volume.
Figure 4. Semi-automated segmentation generally overestimates tumor volume compared to manual segmentation, but is relatively reader skill independent.
Linear regression analysis shows relatively strong agreement between manual and semi-automated segmentations for small (<25 mm3) tumor volumes, and increasing overestimation for larger tumor volumes in a study of 10 aSmo-1 MEMRI datasets read by 8 readers. Dashed line – unity or 1:1 agreement line; Solid red line – modeled linear regression (y=1.133x-0.125, r2 = 0.960). Semi-automated segmentation by two skilled readers (R1, R4 - highlighted with black open circles) closely agreed with manual segmentation over all tumor volumes.
Using semi-automated segmentation, quantitative tumor volume data were extracted from the longitudinal MEMRI data acquired from aSmo-1 and aSmo-5 MBs (Figs. 5, 6).
Figure 5. Study of aSmo tumor heterogeneity in progression is made possible by MEMRI and semi-automated segmentation.
Mouse 1 (top row) presents distinct tumors in each cerebellar hemisphere at W5. The tumor in the left hemisphere decreased in volume whereas the tumor in the right hemisphere increased at W7. By W9, the tumor in the right hemisphere has encroached under the vermis and connected with the tumor in the left hemisphere. At W11, the large accumulated tumor mass has grown rapidly and infiltrated to the level of the brain stem. In cases with tumors of this size and location, hydrocephalus and deformation of the remaining normal cerebellum are common. Mouse 2 (bottom row) shows similar tumor presentation at W5 – distinct tumors are apparent in each cerebellar hemisphere. However, by W7 both tumors have reduced in volume while their morphology remains approximately the same. At W9, the tumor in the right hemisphere has reduced in volume while the tumor in the left hemisphere remains relatively unchanged. By W11, both tumors have regressed and are localized to the flocculae. Scale bar – 2 mm.
Figure 6. Raw progression data from aSmo-1 and aSmo-5 tumors.
Longitudinal volumetric data from semi-automated segmentation of aSmo-1 and aSmo-5 tumors quantitatively demonstrates the heterogeneity in progression apparent in these models.
Segmentation – Tumor Origin Sites
A fundamental question related to tumor biology, in MB and other cancers, is whether there are common locations for the origin of different tumor sub-types. Our previous studies revealed that all aSmo-1 MBs arise in the lateral cerebellum (hemispheres)17. To investigate this further, and to provide quantitative data related to aSmo-1 and aSmo-5 MB origin sites, tumor centroid and heatmap analyses were performed at W5, the earliest time point that overt tumors were reliably detected by MEMRI (Fig. 7). These analyses revealed that in both aSmo models the majority of tumor centroids (n=60/62), and the most probable tumor voxels in the heatmaps were localized specifically in the copula pyramidis underlying the lateral hemispheres.
Figure 7. Tumor centroid and heatmap analysis illustrates aSmo tumors originate in the posterior cerebellar hemispheres.
At W5, aSmo-1 (A) and aSmo-5 (B) tumors have centroids (green) closely clustered together in the lateral cerebellar hemispheres. (C,D) Heatmap analysis showed broad distribution of tumors in the posterior cerebellum. The peak voxel-wise tumor probability coincides with the localization of the centroids in the copula pyramidis. Scale bar – 2 mm.
Analysis – Empirical Classification of Tumor Progression
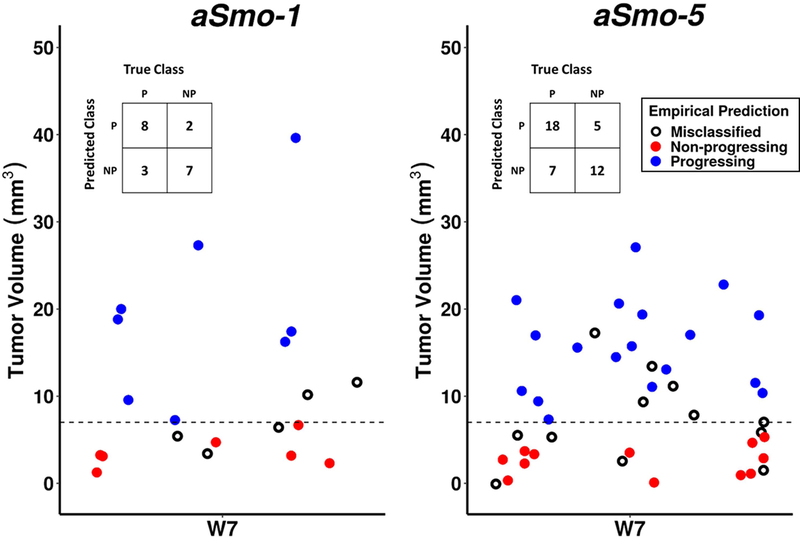
In order to utilize the imaging pipeline to assess tumor progression in the two MB models, an Analysis stage was required to classify individual tumors in terms of their early growth characteristics and to predict whether a given tumor was progressing or non-progressing. Given the heterogeneity in tumor growth characteristics in the aSmo MBs, it is important for studies testing experimental therapeutics to distinguish the ‘Progressing’ cohort early on and to eliminate or minimize the number of tumors from the ‘Non-progressing’ cohort. From our longitudinal MEMRI data, we determined empirically that MBs that grew in volume from W5 to W7, and which had tumor volume ≥ 7 mm3 at W7, were most likely to be ‘Progressing’. In aSmo-1 MBs, this method correctly identified ‘Progressing’ tumors in 72.7% (n=8/11) of cases and ‘Non-progressing’ tumors in 77.8% of cases (n=7/9). In aSmo-5 MBs, this method correctly identified ‘Progressing’ tumors in 72.0% of cases (n=18/25) and ‘Non-progressing’ tumors in 70.6% of cases (n=12/17) (Fig. 8).
Figure 8. Empirical classification of aSmo-1 and aSmo-5 longitudinal progression data.
Initial classification utilized positive volume change between W5 and W7, and tumor volume threshold of 7-mm3 at W7 (dashed line) as discrimination criteria. Tumors meeting these criteria were classified ‘Progressing’ (blue, P), and those not meeting these criteria were classified ‘Non-progressing’ (red, NP). These predictions were compared to observed outcome, and mismatches between prediction and outcome were considered ‘Misclassified’ (black, open circles). In the aSmo-1 MBs, the true positive rate was 72.7% and the true negative rate was 77.8%. In the aSmo-5 MBs, the true positive rate was 72.0% and the true negative rate was 70.6% (see confusion tables, insets).
Analysis – Linear Discriminant Analysis (LDA)
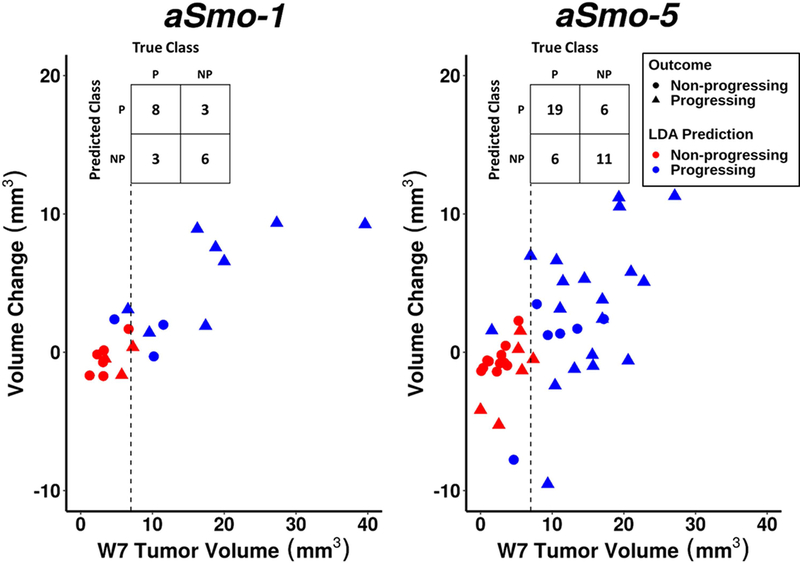
A linear discriminant classifier was trained in R over the aSmo progression data (Fig. 9). In the aSmo-1 model, ‘Progressing’ tumors were correctly identified with the same accuracy as the empirical classifier (n=8/11, 72.7%), and ‘Non-progressing’ tumors with slightly lower accuracy (n=6/9, 66.7%). In the aSmo-5 model, ‘Progressing’ tumors were identified with slightly higher accuracy than empirical classifier (n=19/25, 76.0%), and ‘Non-progressing’ tumors were identified with slightly lower accuracy (n=11/17, 64.7%). Overall, there were minimal changes in classification between the empirical classifier and LDA. Therefore, the empirical classification method was implemented as the ‘Analysis’ stage in the application of this pipeline.
Figure 9. LDA training over aSmo-1 and aSmo-5 longitudinal progression data.
A LDA classifier was trained over the longitudinal aSmo tumor progression data. Variables included ‘W5 Tumor Volume’, ‘W7 Tumor Volume’, ‘W5–W7 Volume Change’, and ‘Normalized W5–W7 Growth Rate’, and were used to predict ‘Progressing’ (blue, P) or ‘Non-progressing’ (red, NP) outcomes (triangles and circles, respectively). The dashed lines indicate the volume threshold used in the empirical classifier. In the aSmo-1 cohort, LDA had a 72.7% true positive rate and 66.7% true negative rate; in the aSmo-5 cohort, LDA had a 76.0% true positive rate and 64.7% true negative rate (see confusion tables, insets).
Application – Preclinical trial of PLX5622
A previous report found that CSF-1R inhibitors showed promise for treating glioma tumors in adult mice, suggesting that such TAM-targeted drugs can be effectively delivered through the blood-brain barrier and raising the possibility they could be effective in slowing the growth of other brain tumors51. We applied our MEMRI-based pipeline approach to test the efficacy of an experimental CSF-1R inhibitor, PLX5622, on aSmo MBs.
Model generation
Cohorts of 40 aSmo-1 and 25 aSmo-5 mice were generated for this study. Early time point imaging of these animals revealed 51 aSmo-1 tumors and 39 aSmo-5 tumors, for a total of 90 tumors. These tumors were monitored with MEMRI at W5 and W7 for outcome prediction.
Imaging, segmentation, and analysis of PLX5622 trial data
After pre-treatment imaging at W5 and W7, semi-automated tumor segmentation, and prediction using the empirical classification method, 26 aSmo-1 and 26 aSmo-5 tumors met the threshold criteria for ‘Progressing’ and were entered into the drug study. Mice with at least one tumor classified ‘Progressing’ were assigned to receive either control chow (CTL) or chow containing PLX5622 (PLX). Assignment of animals into PLX and CTL arms of the study was not biased in terms of pre-treatment tumor volume (Supporting Information Fig. S1). Animals entered into the study were monitored with MEMRI from treatment start to W13, or until time of sacrifice (Fig. 10A). Response to treatment was variable, and tumor model-dependent. It was also evident that some individual tumors did not stop progressing during PLX therapy, although it is impossible to know whether their growth was affected by the drug (Supporting Information Fig. S2). Interestingly, longitudinal data from the aSmo-5 MBs demonstrated a significant reduction in tumor volume during the time period between W11 and W13, comparing PLX tumors to CTL tumors, but that difference was no longer significant by W13 (Fig. 10A).
Figure 10. Results from the PLX5622 preclinical trial show response to therapy is heterogeneous and tumor model-dependent.
(A) From the aggregate longitudinal tumor volume data from the aSmo-1 model, it was apparent that growth of CTL-treated tumors (blue) did not differ significantly from the growth of PLX-treated tumors (red) after treatment start at W7. In contrast, in the aSmo-5 model, separation of the PLX and CTL-treated tumor volume trends was apparent between W11 and W13. The curves for the CTL and PLX arms show the mean volumes, fitted to a cubic spline and plotted as a function of postnatal week. For both models, there was substantial heterogeneity in tumor growth within treatment arms (95% confidence interval, gray). (B) Tumor centroids (green) from W5 aSmo-1 and aSmo-5 tumors were mostly localized to the posterior cerebellar hemispheres. The two exceptions were tumors in the more anterior hemispheres, found in the same animal, both of which regressed by W7. At the pre-treatment time point, tumor centroids (green) remained clustered together. After treatment start, tumor centroids from both PLX (red) and CTL (blue) groups spread medially into the vermis from W9–W11. At W13, the remaining tumors were again localized in the posterior hemispheres. Scale bar – 2 mm.
Longitudinal tumor centroid analysis showed that tumors were generally clustered in the same regions of the posterior cerebellar hemispheres at early pre-treatment time points (Fig. 10B), as observed in the previous measurements of aSmo-1 and aSmo-5 tumors. The two exceptions were from a single animal carrying tumors centered in more anterior lobules of the cerebellar hemispheres, however both tumors regressed below detectable levels by W7. The centroids spread medially from W9 through W11, as the MB tumors grew from hemispheres into the vermis. Many animals had been removed from the study by W13, and the remaining tumors were again centered in the hemispheres.
As expected, pre-treatment tumor volume had a statistically significant, positive effect on post-treatment tumor volume (effect size = 1.378, p = 0.003). The effect of CTL chow on final tumor volume was generally positive, but not statistically significant in either the aSmo-1 (effect size = 3.41, p = 0.765) or the aSmo-5 MBs (effect size = 9.27, p = 0.428). Interestingly, the effect of PLX chow was strongly negative in the aSmo-5 (effect size = −10.82, p = 0.384), but not in the aSmo-1 MBs (effect size = 1.17, p = 0.920). The overall effect of treatment was relatively small (Cohen’s d = 0.29). However, the apparent model-specific efficacy suggests an effect of PLX on the aSmo-5 MBs (Cohen’s d = 0.52), but not the aSmo-1 MBs (Cohen’s d= 0.04).
The pre-treatment time point data were also re-analyzed, retrospectively, using the trained LDA classifier to verify our preclinical trial entry criteria. The results of LDA re-analysis showed minimal differences in progression classification compared to the empirical classifier (Supporting Information Fig. S3).
DISCUSSION
Heterogeneous tumor progression is unavoidable in sporadic tumor models that mimic many important features of the human disease. We have developed a MEMRI-based pipeline and tested its utility to objectively characterize heterogeneous tumor progression in genetically-engineered mouse models of sporadic human MB. Our goal was to optimize each stage of the pipeline, including: i) production of mice with sporadic MB; ii) acquisition of time-efficient longitudinal 3D brain images through critical stages of disease progression; iii) time-efficient, semi-automated segmentation of tumor volumes from MEMRI images; and iv) unbiased classification of MB tumors based on their early stage progression features. We further applied this pipeline to test the effects of an experimental therapy on MB progression.
The aSmo MB model system was tunable (via TMX dose) in terms of the number of granule cell progenitors inheriting the activating SmoM2 mutation. This potentiated study of tumor biology and drug efficacy in two models of MB with the same initiating mutation: aSmo-1 and aSmo-5. Tumor centroid analysis reinforced that the aSmo-1 and aSmo-5 models share a common point of origin – the posterior cerebellar hemispheres. This feature could serve as specific metric for aberrant aSmo tumor formation in future tumor progression classification efforts. Although there were challenges in characterizing heterogeneity in tumor progression within each model, as well as between the two models, the MEMRI-pipeline was sensitive to differences in tumor presentation, both at the same time point, as well as analyzing trends over time. Indeed, the inherent heterogeneity observed in sporadic mouse models of MB makes it difficult or impossible to conduct studies of tumor progression and/or preclinical drug studies without an imaging-based pipeline approach. Moreover, it should be possible to apply our MEMRI-pipeline to other models of human MB.
A valid question is how generally applicable MEMRI will be for imaging other tumor types, both neural and non-neural. In the brain, T1-weighted Mn-enhancement is most obvious in the cerebellum, hippocampus and olfactory bulb, but there is also global enhancement of the brain and other organs, including heart, kidney and liver,52 after systemic administration of Mn. In MBs, the tumor cells take up significantly less Mn than the surrounding (enhanced) cerebellum cells, resulting in the negative contrast utilized in the current study. If future studies demonstrate that brain and other tumor cells are generally resistant to Mn uptake, then it is possible that MEMRI can be applied for imaging many types of tumors. Interestingly, we previously showed that by engineering glioma cells to overexpress the Divalent Metal Transporter, DMT1, we were able to convert negative to positive contrast using the same MEMRI protocol53. This implies that Mn uptake and storage is likely to be highly dependent on the set of proteins expressed by each tumor type, which is clearly beyond the scope of the current study.
Manual segmentation, although generally regarded to be accurate, is prohibitively time inefficient for pipelines and requires readers to be well trained. Efforts to employ atlas-based, fully-automated tumor segmentation techniques have shown promise54,55. However, deformation of normal tissue surrounding advanced tumors leads to significant error that requires reader correction. Our semi-automated segmentation protocol substantially reduced time-to-result and was generally agnostic to reader skill, but overestimated tumor volume compared to manual segmentation. Although the time-efficiency justified use of this method, ongoing efforts to develop a fully-automated segmentation technique – by means of trained convolutional neural networks56 or random forest classification57 – may have the potential to eliminate inter-reader error and further reduce time-to-result in the future.
Our empirical classification framework employed a simple volume threshold at a critical time point to predict whether a tumor would progress. We found that natural regression is a common feature in our mouse MB models, however the high propensity for tumors to naturally regress after early progression was surprising. Using the imaging pipeline, we discovered that a simple volume threshold was sometimes insufficient to parse the heterogeneity in progression present in this model of sporadic human cancer. The problem with objective characterization of longitudinal quantitative information is that large sample size is required to identify relevant, reproducible trends in the data58. Features used for classification must be carefully chosen as each additional degree of freedom geometrically increases requisite sample size to achieve sufficient statistical power. In addition, inclusion of unimportant features unnecessarily increases training time without providing meaningful axes for result interpretation59. Use of LDA with this relatively high-dimensional, low sample size dataset is prone to data piling, which usually results in failure to properly classify data60. However, classification methods that are more data-driven than our empirical classification method, including methods like LDA, will likely parse tumor progression heterogeneity more effectively after more data is acquired in future applications of this pipeline.
Statistical methods commonly employed in analysis of preclinical drug trial data61,62 are useful for understanding general trends in results. However quantitative comparisons between treatment arms are not feasible without more specific tests, nor can p-values alone be used to gauge effect size through direct comparison63. In addition, outcome measures are entangled with biological and experimental features or confounds e.g. sex, age, etc. It is not possible to correct for these effects using Student’s t-test or ANOVA. Because of this, incorrect result interpretation is common. For example, Kruskal-Wallis test (ANOVA using the ranks of the data) applied to the current PLX5622 trial data would suggest that statistically significant differences exist, comparing normalized tumor volume between the CTL and PLX groups in the aSmo-5 cohort (p = 0.043). However, this significance does not persist after Dunn’s Multiple Comparison correction (p = 0.072) nor does it account for multiple tumors from the same subject. Modeling outcome measures as a function of individual features, as performed in the current study, is a more appropriate method for hypothesis testing since pairwise comparisons can be made directly from individual fixed-effect coefficients.
The pipeline we developed is time-efficient, reproducible, and generates high-density quantitative information that has the potential to guide preclinical drug studies in other mouse models of sporadic human cancer in the future. It should be noted that we present a general framework for longitudinal brain tumor imaging and response-to-therapy monitoring, rather than a one-time implementation of a specific protocol. Each stage of the “modular” pipeline has potential to be further refined. There are several considerations that should be made for future studies. Thorough study of the span in heterogeneity of novel tumor models is essential before using this procedure de novo. Imaging modalities and parameters should be chosen carefully to effectively delineate tumor margins, if tumor volume is the only critical metric. As distributed computing and machine learning approaches continue to improve in efficiency, discriminator-driven segmentation and classification will likely become the norm for high-throughput analysis. With the emergence of such new imaging technologies, genetic engineering tools, and data analysis techniques, we propose that this pipeline provides a flexible scaffold for future longitudinal tumor progression characterization efforts.
Supplementary Material
Supporting Information Figure S1. After preliminary classification, for outcome prediction before entry into the PLX5622 preclinical trial, tumors were randomly assigned into the treatment (PLX) or control (CTL) arms of the study. Tukey’s HSD test of the W7 tumor volumes showed no significant differences between treatment arms nor across tumor models.
Supporting Information Figure S2. Raw progression aSmo tumor progression data from the PLX5622 preclinical trial. From these data, it was apparent there was substantial heterogeneity in response to therapy. In the aSmo-1 model, there are no apparent differences in progression between PLX (red) or CTL (blue) treated tumors. However, there appears to be a trend towards tumor volume reduction after treatment with PLX in the aSmo-5 model.
Supporting Information Figure S3. In an effort to reduce the number of false ‘Progressing’ tumors entered into the drug trial, the pre-treatment time point data were re-analyzed using the trained LDA classifier. The early time point data from tumors not meeting the empirical classification criteria (dashed line; which included sub-threshold tumors in animals with a second tumor that met the volume threshold criteria) were added to these retrospective analyses (10 aSmo-1 and 2 aSmo-5 tumors). It is apparent that LDA reclassification produced results very similar to those of the empirical classifier. After LDA reclassification, 5 aSmo-1 and 2 aSmo-5 MBs were removed from the CTL arm and 5 aSmo-1 tumors were removed from the PLX arm of the study. The statistically significant, positive effect of pre-treatment tumor volume on post-treatment tumor volume persisted after LDA reclassification (effect size = 1.43, p = 0.002). Also, the positive, statistically insignificant effects of CTL chow on post-treatment tumor volume remained in both the aSmo-1 (effect size = 2.5, p = 0.826) and aSmo-5 MBs (effect size = 8.33, p = 0.475). After LDA reclassification, a negative effect of PLX on post-treatment aSmo-1 tumor volume was observed (effect size = −1.16, p = .921), and a similar effect was observed in the aSmo-5 MBs (effect size = −11.88, p = 0.338). The effect of treatment nominally increased after LDA when compared to the results after empirical classification (Cohen’s d = 0.31), although the effect remained small. Also, the TMX dose-specific effects were relatively unchanged for both the aSmo-1 (Cohen’s d = 0.09) and aSmo-5 (Cohen’s d = 0.52) MBs. Retrospective LDA of early time point data from tumors excluded from the preclinical trial (U) suggests that 4 aSmo-1 tumors were in fact ‘Progressing’. This process did not change the classification for aSmo-5 tumors.
ACKNOWLEDGEMENTS
This work was supported by NIH grants R01CA192176 (to ALJ) and R01NS038461 (to DHT). We are grateful to Andrey Rymar and Parmveer Singh at Plexxikon for providing the PLX5622 used in these studies. We also thank Orlando Aristizábal, Joshua Frenster, Ivan Gando, Hannah Goldman, Steven Hoang-Phou, Helena Kim, and Nikola Otic for their help with the tumor segmentations, and the Preclinical Imaging Core at NYU School of Medicine for their assistance in implementing the MRI protocols and maintaining the MRI system used to acquire the MEMRI data. We thank Dr. Sungheon Kim for his critical review of this manuscript.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66(1):7–30. 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Pui C-H, Gajjar AJ, Kane JR, Qaddoumi IA, Pappo AS. Challenging issues in pediatric oncology. Nat Rev Clin Oncol 2011;8(9):540–549. 10.1038/nrclinonc.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rutkowski S, von Hoff K, Emser A, et al. Survival and prognostic factors of early childhood medulloblastoma: an international meta-analysis. J Clin Oncol 2010;28(33):4961–4968. 10.1200/JCO.2010.30.2299. [DOI] [PubMed] [Google Scholar]
- 4.Gajjar A, Chintagumpala M, Ashley D, et al. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): long-term results from a prospective, multicentre trial. Lancet Oncol 2006;7(10):813–820. 10.1016/S1470-2045(06)70867-1. [DOI] [PubMed] [Google Scholar]
- 5.Mabbott DJ, Penkman L, Witol A, Strother D, Bouffet E. Core neurocognitive functions in children treated for posterior fossa tumors. Neuropsychology 2008;22(2):159–168. 10.1037/0894-4105.22.2.159. [DOI] [PubMed] [Google Scholar]
- 6.Mabbott DJ, Spiegler BJ, Greenberg ML, Rutka JT, Hyder DJ, Bouffet E. Serial evaluation of academic and behavioral outcome after treatment with cranial radiation in childhood. J Clin Oncol 2005;23(10):2256–2263. 10.1200/JCO.2005.01.158. [DOI] [PubMed] [Google Scholar]
- 7.Spiegler BJ, Bouffet E, Greenberg ML, Rutka JT, Mabbott DJ. Change in neurocognitive functioning after treatment with cranial radiation in childhood. J Clin Oncol 2004;22(4):706–713. 10.1200/JCO.2004.05.186. [DOI] [PubMed] [Google Scholar]
- 8.Taylor MD, Northcott PA, Korshunov A, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol 2012;123(4):465–472. 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavalli FMG, Remke M, Rampasek L, et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell 2017;31(6):737–754. 10.1016/J.CCELL.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodrich LV, Milenković L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 1997;277(5329):1109–1113. 10.1126/SCIENCE.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 11.Gibson P, Tong Y, Robinson G, et al. Subtypes of medulloblastoma have distinct developmental origins. Nature 2010;468(7327):1095–1099. 10.1038/nature09587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swartling FJ, Grimmer MR, Hackett CS, et al. Pleiotropic role for MYCN in medulloblastoma. Genes Dev 2010;24(10):1059–1072. 10.1101/gad.1907510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau J, Schmidt C, Markant SL, Taylor MD, Wechsler-Reya RJ, Weiss WA. Matching mice to malignancy: molecular subgroups and models of medulloblastoma. Childs Nerv Syst 2012;28(4):521–532. 10.1007/s00381-012-1704-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawauchi D, Robinson G, Uziel T, et al. A Mouse Model of the Most Aggressive Subgroup of Human Medulloblastoma. Cancer Cell 2012;21(2):168–180. https://www.sciencedirect.com/science/article/pii/S1535610812000025?via%253Dihub. Accessed July 14, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci U S A 1988;85(14):5166–5170. http://www.ncbi.nlm.nih.gov/pubmed/2839833. Accessed April 26, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suero-Abreu GA, Praveen Raju G, Aristizábal O, et al. In vivo Mn-enhanced MRI for early tumor detection and growth rate analysis in a mouse medulloblastoma model. Neoplasia 2014;16(12):993–1006. 10.1016/j.neo.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan I-LI-L, Wojcinski A, Rallapalli H, et al. Lateral cerebellum is preferentially sensitive to high sonic hedgehog signaling and medulloblastoma formation. Proc Natl Acad Sci U S A 2018;115(13):3392–3397. 10.1073/pnas.1717815115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solinas G, Germano G, Mantovani A, Allavena P. Tumor-associated macrophages (TAM) as major players of the cancer-related inflammation. J Leukoc Biol 2009;86(5):1065–1073. 10.1189/jlb.0609385. [DOI] [PubMed] [Google Scholar]
- 19.Margol AS, Robison NJ, Gnanachandran J, et al. Tumor-associated macrophages in SHH subgroup of medulloblastomas. Clin Cancer Res 2015;21(6):1457–1465. 10.1158/1078-0432.CCR-14-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaelin WG. Common pitfalls in preclinical cancer target validation. Nat Publ Gr 2017;17 10.1038/nrc.2017.32. [DOI] [PubMed] [Google Scholar]
- 21.Blouw B, Song H, Tihan T, et al. The hypoxic response of tumors is dependent on their microenvironment. Cancer Cell 2003;4(2):133–146. 10.1016/S1535-6108(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 22.Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nat 2012 4837391. March 2012. [DOI] [PubMed]
- 23.Hutchinson L, Kirk R. High drug attrition rates—where are we going wrong? Nat Rev Clin Oncol 2011;8(4):189–190. 10.1038/nrclinonc.2011.34. [DOI] [PubMed] [Google Scholar]
- 24.Petrovsky A, Schellenberger E, Josephson L, Weissleder R, Bogdanov A. Near-infrared fluorescent imaging of tumor apoptosis. Cancer Res 2003;63(8):1936–1942. 10.1158/0008-5472.can-04-1956. [DOI] [PubMed] [Google Scholar]
- 25.Smith BR, Ghosn EEB, Rallapalli H, et al. Selective uptake of single-walled carbon nanotubes by circulating monocytes for enhanced tumour delivery. Nat Nanotechnol 2014;9(6):481–487. 10.1038/nnano.2014.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rehemtulla A, Stegman LD, Cardozo SJ, et al. Rapid and Quantitative Assessment of Cancer Treatment Response Using In Vivo Bioluminescence Imaging. Neoplasia 2000;2(6):491–495. 10.1038/SJ.NEO.7900121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jenkins DE, Oei Y, Hornig YS, et al. Bioluminescent imaging (BLI) to improve and refine traditional murine models of tumor growth and metastasis. Clin Exp Metastasis 2003;20:733–744. https://link.springer.com/content/pdf/10.1023%252FB%253ACLIN.0000006815.49932.98.pdf. Accessed April 26, 2018. [DOI] [PubMed] [Google Scholar]
- 28.Hasegawa M, Sone S, Takashima S, et al. Growth rate of small lung cancers detected on mass CT screening https://www.birpublications.org/doi/pdf/10.1259/bjr.73.876.11205667. Accessed April 26, 2018. [DOI] [PubMed]
- 29.Swensen SJ, Jett JR, Hartman TE, et al. Lung Cancer Screening with CT: Mayo Clinic Experience. Radiology 2003;226(3):756–761. 10.1148/radiol.2263020036. [DOI] [PubMed] [Google Scholar]
- 30.Perreault S, Ramaswamy V, Achrol AS, et al. MRI surrogates for molecular subgroups of medulloblastoma. AJNR Am J Neuroradiol 2014;35(7):1263–1269. 10.3174/ajnr.A3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fruehwald-Pallamar J, Puchner SB, Rossi A, et al. Magnetic resonance imaging spectrum of medulloblastoma 10.1007/s00234-010-0829-8. [DOI] [PubMed]
- 32.Valk J, M de Slegte RG, Crezee FC, Hazenberg GJ, Thjaha SI, P Nauta JJ. Contrast Enhanced Magnetic Resonance Imaging of the Brain Using Gadolinium-DTPA. Acta radiol 1987;28(6):659–665. 10.3109/02841858709177421. [DOI] [PubMed] [Google Scholar]
- 33.Nelson AL, Algon SA, Munasinghe J, et al. Magnetic Resonance Imaging of Patched Heterozygous and Xenografted Mouse Brain Tumors Vol 62; 2003. https://link.springer.com/content/pdf/10.1023%2FA%3A1023339902812.pdf. Accessed February 13, 2019. [DOI] [PubMed] [Google Scholar]
- 34.Wadghiri YZ, Blind JA, Duan X, et al. Manganese-enhanced magnetic resonance imaging (MEMRI) of mouse brain development. NMR Biomed 2004;17(8):613–619. 10.1002/nbm.932. [DOI] [PubMed] [Google Scholar]
- 35.Szulc KU, Lerch JP, Nieman BJ, et al. 4D MEMRI atlas of neonatal FVB/N mouse brain development. Neuroimage 2015;118:49–62. 10.1016/j.neuroimage.2015.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Machold R, Fishell G. Math1 Is Expressed in Temporally Discrete Pools of Cerebellar Rhombic-Lip Neural Progenitors Vol 48; 2005:17–24. 10.1016/j.neuron.2005.08.028. [DOI] [PubMed] [Google Scholar]
- 37.Mao J, Ligon KL, Rakhlin EY, et al. A Novel Somatic Mouse Model to Survey Tumorigenic Potential Applied to the Hedgehog Pathway. Cancer Res 2006;66(20):10171–10178. 10.1158/0008-5472.CAN-06-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lerch JP, Sled JG, Henkelman RM. MRI phenotyping of genetically altered mice. Methods Mol Biol 2010. 10.1007/978-1-61737-992-5_17. [DOI] [PubMed]
- 39.Nieman BJ, van Eede MC, Spring S, Dazai J, Henkelman RM, Lerch JP. MRI to Assess Neurological Function. Curr Protoc Mouse Biol 2018;8(2):e44 10.1002/cpmo.44. [DOI] [PubMed] [Google Scholar]
- 40.Friedel M, Van Eede MC, Pipitone J, et al. Pydpiper: a flexible toolkit for constructing novel registration pipelines 2014. 10.3389/fninf.2014.00067. [DOI] [PMC free article] [PubMed]
- 41.Vincent RD, Neelin P, Khalili-Mahani N, et al. MINC 2.0: A Flexible Format for Multi-Modal Images. Front Neuroinform 2016;10:35 10.3389/fninf.2016.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain 2007. 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed]
- 43.Yushkevich PA, Piven J, Hazlett HC, et al. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability 2006. 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed]
- 44.Fisher RA. THE USE OF MULTIPLE MEASUREMENTS IN TAXONOMIC PROBLEMS. Ann Eugen 1936;7(2):179–188. 10.1111/j.1469-1809.1936.tb02137.x. [DOI] [Google Scholar]
- 45.Huang Y, Xu Z, Xiong S, et al. Repopulated microglia are solely derived from the proliferation of residual microglia after acute depletion. Nat Neurosci 2018;21(4):530–540. 10.1038/s41593-018-0090-8. [DOI] [PubMed] [Google Scholar]
- 46.Faul F, Erdfelder E, Buchner A, Lang A-G. Statistical power analyses using G*Power 3.1: Tests for correlation and regression analyses. Behav Res Methods 2009;41(4):1149–1160. 10.3758/BRM.41.4.1149. [DOI] [PubMed] [Google Scholar]
- 47.Faul F, Erdfelder E, Lang A-G, Buchner A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods 2007;39(2):175–191. 10.3758/BF03193146. [DOI] [PubMed] [Google Scholar]
- 48.Bates D, Mächler M, Bolker B, Walker S. Fitting Linear Mixed-Effects Models Using lme4. J Stat Softw 2015;67(1):1–48. 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- 49.Kenward MG, Roger JH. An improved approximation to the precision of fixed effects from restricted maximum likelihood. Comput Stat Data Anal 2009;53(7):2583–2595. 10.1016/j.csda.2008.12.013. [DOI] [Google Scholar]
- 50.Altman DG, Bland JM. Measurement in Medicine: The Analysis of Method Comparison Studies. Stat 1983;32(3):307 10.2307/2987937. [DOI] [Google Scholar]
- 51.Pyonteck SM, Akkari L, Schuhmacher AJ, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med 2013;19(10):1264–1272. 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mendonça-Dias MH, Gaggelli E, Lauterbur PC. Paramagnetic contrast agents in nuclear magnetic resonance medical imaging. Semin Nucl Med 1983;13(4):364–376. 10.1016/S0001-2998(83)80048-8. [DOI] [PubMed] [Google Scholar]
- 53.Bartelle BB, Szulc KU, Suero-Abreu GA, Rodriguez JJ, Turnbull DH. Divalent metal transporter, DMT1: A novel MRI reporter protein. Magn Reson Med 2013;70(3):842–850. 10.1002/mrm.24509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young AV, Wortham A, Wernick I, Evans A, Ennis RD. Atlas-based segmentation improves consistency and decreases time required for contouring postoperative endometrial cancer nodal volumes. Int J Radiat Oncol Biol Phys 2011;79(3):943–947. 10.1016/j.ijrobp.2010.04.063. [DOI] [PubMed] [Google Scholar]
- 55.Liu Jin, Li Min, Wang Jianxin, Wu Fangxiang, Liu Tianming, Pan Yi. A survey of MRI-based brain tumor segmentation methods. Tsinghua Sci Technol 2014;19(6):578–595. 10.1109/TST.2014.6961028. [DOI] [Google Scholar]
- 56.Havaei M, Davy A, Warde-Farley D, et al. Brain tumor segmentation with Deep Neural Networks. Med Image Anal 2017;35:18–31. 10.1016/J.MEDIA.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 57.Bauer S, Fejes T, Slotboom J, Wiest R, Nolte L-P, Reyes M. Segmentation of Brain Tumor Images Based on Integrated Hierarchical Classification and Regularization; 2012. http://www2.imm.dtu.dk/projects/BRATS2012/. Accessed August 23, 2018.
- 58.Figueroa RL, Zeng-Treitler Q, Kandula S, Ngo LH. Predicting sample size required for classification performance. BMC Med Inform Decis Mak 2012;12(1):8 10.1186/1472-6947-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li J, Cheng K, Wang S, et al. Feature Selection: A Data Perspective. ACM Comput Surv 2017;50 10.1145/3136625. [DOI] [Google Scholar]
- 60.Marron JS, Todd MJ, Ahn J. Distance Weighted Discrimination. J Am Stat Assoc 2007;102(480):1267–1271. 10.1198/016214507000001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li D, Ambrogio L, Shimamura T, et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene 2008;27(34):4702–4711. 10.1038/onc.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Munoz R, Man S, Shaked Y, et al. Highly Efficacious Nontoxic Preclinical Treatment for Advanced Metastatic Breast Cancer Using Combination Oral UFT-Cyclophosphamide Metronomic Chemotherapy. Cancer Res 2006;66(7):3386–3391. 10.1158/0008-5472.CAN-05-4411. [DOI] [PubMed] [Google Scholar]
- 63.Johansson T Hail the impossible: P-values, evidence, and likelihood. Scand J Psychol 2011;52(2):113–125. 10.1111/j.1467-9450.2010.00852.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure S1. After preliminary classification, for outcome prediction before entry into the PLX5622 preclinical trial, tumors were randomly assigned into the treatment (PLX) or control (CTL) arms of the study. Tukey’s HSD test of the W7 tumor volumes showed no significant differences between treatment arms nor across tumor models.
Supporting Information Figure S2. Raw progression aSmo tumor progression data from the PLX5622 preclinical trial. From these data, it was apparent there was substantial heterogeneity in response to therapy. In the aSmo-1 model, there are no apparent differences in progression between PLX (red) or CTL (blue) treated tumors. However, there appears to be a trend towards tumor volume reduction after treatment with PLX in the aSmo-5 model.
Supporting Information Figure S3. In an effort to reduce the number of false ‘Progressing’ tumors entered into the drug trial, the pre-treatment time point data were re-analyzed using the trained LDA classifier. The early time point data from tumors not meeting the empirical classification criteria (dashed line; which included sub-threshold tumors in animals with a second tumor that met the volume threshold criteria) were added to these retrospective analyses (10 aSmo-1 and 2 aSmo-5 tumors). It is apparent that LDA reclassification produced results very similar to those of the empirical classifier. After LDA reclassification, 5 aSmo-1 and 2 aSmo-5 MBs were removed from the CTL arm and 5 aSmo-1 tumors were removed from the PLX arm of the study. The statistically significant, positive effect of pre-treatment tumor volume on post-treatment tumor volume persisted after LDA reclassification (effect size = 1.43, p = 0.002). Also, the positive, statistically insignificant effects of CTL chow on post-treatment tumor volume remained in both the aSmo-1 (effect size = 2.5, p = 0.826) and aSmo-5 MBs (effect size = 8.33, p = 0.475). After LDA reclassification, a negative effect of PLX on post-treatment aSmo-1 tumor volume was observed (effect size = −1.16, p = .921), and a similar effect was observed in the aSmo-5 MBs (effect size = −11.88, p = 0.338). The effect of treatment nominally increased after LDA when compared to the results after empirical classification (Cohen’s d = 0.31), although the effect remained small. Also, the TMX dose-specific effects were relatively unchanged for both the aSmo-1 (Cohen’s d = 0.09) and aSmo-5 (Cohen’s d = 0.52) MBs. Retrospective LDA of early time point data from tumors excluded from the preclinical trial (U) suggests that 4 aSmo-1 tumors were in fact ‘Progressing’. This process did not change the classification for aSmo-5 tumors.