Abstract
Alcohol use disorder (AUD) and major depressive disorder (MDD) are prevalent, debilitating, and highly comorbid disorders. The molecular changes that underlie their comorbidity are beginning to emerge. For example, recent evidence showed that acute ethanol exposure produces rapid antidepressant-like biochemical and behavioral responses. Both ethanol and fast-acting antidepressants block N-methyl-D-aspartate receptor (NMDAR) activity, leading to synaptic changes and long-lasting antidepressant-like behavioral effects. We used RNA sequencing to analyze changes in the synaptic transcriptome after acute treatment with ethanol or the NMDAR antagonist, Ro 25–6981. Ethanol and Ro 25–6981 induced differential, independent changes in gene expression. In contrast with gene-level expression, ethanol and Ro 25–6981 produced overlapping changes in exons, as measured by analysis of differentially expressed exons (DEEs). A prominent overlap in genes with DEEs indicated that changes in exon usage were important for both ethanol and Ro 25–6981 action. Structural modeling provided evidence that ethanol-induced exon expression in the NMDAR1 amino-terminal domain could induce conformational changes and thus alter NMDAR function. These findings suggest that the rapid antidepressant effects of ethanol and NMDAR antagonists reported previously may depend on synaptic exon usage rather than gene expression.
Keywords: Alcohol Use Disorder, Major Depressive Disorder, Synaptic transcriptome, Alternative Splicing, Differential Exon Usage
1. Introduction
Alcohol use disorder (AUD) has a high comorbidity with other psychiatric disorders, particularly major depressive disorder (MDD). These two disorders are closely linked through epidemiological and clinical studies(Boden and Fergusson, 2011; Grant and Harford, 1995; Hasin and Grant, 2002). The self-medication hypothesis proposes that depressed individuals drink alcohol to initially alleviate symptoms of depression, and then chronic alcohol misuse leads to AUD (Boden and Fergusson, 2011; Koob and Le Moal, 2008).
Rapid-acting antidepressants, such as ketamine and Ro 25–6981, are N-methyl-D-aspartate receptor (NMDAR) antagonists; and a single injection of these compounds produced antidepressant-like behaviors lasting up to two weeks (Abdallah et al., 2015). Ethanol also blocks NMDARs, which play an important role in AUD(Abrahao et al., 2017; Zhao et al., 2015). Furthermore, a single injection of ethanol produced long-lasting antidepressant-like behaviors, similar to that of rapid-acting antidepressants (Wolfe et al., 2016). We previously showed that acute treatment with ethanol or a rapid antidepressant targets the same synaptic signaling pathway(Neasta et al., 2014; Wolfe et al., 2016). This pathway is also altered in AUD and is thought to involve activation of mammalian target of rapamycin and downstream protein synthesis. We focused on alterations in the hippocampus due to previous studies of ethanol and antidepressants affecting this brain region. Studies of human hippocampus demonstrate that this brain region shows changes in transcription patterns with chronic drinking(Enoch et al., 2012; Zhou et al., 2011). Animal studies of rapid antidepressants, including ketamine, identified altered gene expression after as little as one dose in the hippocampus and other brain regions (Bagot et al., 2017; Ficek et al., 2016). Additionally, our previous work identified shared molecular mechanisms induced by both ethanol and rapid antidepressant in the hippocampus (Wolfe et al., 2016).
To better understand the antidepressant-like effects of ethanol, we used RNA sequencing (RNA-Seq) to compare the synaptic transcriptome after treatment with ethanol and the rapid-acting antidepressant, Ro 25–6981. RNA-Seq allows for the unbiased detection of most mRNAs, including alternatively spliced variants(Trapnell et al., 2012). Pairing RNA-Seq with differential expression tools like DEXSeq provides information on differential exon usage of gene products(Anders et al., 2012). We identified the differentially expressed genes (DEGs) and differentially expressed exons (DEEs) in the synaptic transcriptome of mice treated acutely with ethanol or the rapid-acting antidepressant, Ro 25–6981. In contrast with changes in overall gene expression induced by these drugs, there was remarkable overlap in the number of genes with common DEEs. Collectively, these data propose the intriguing hypothesis that acute antidepressant action depends on exon selection rather than on gene expression.
2. Materials and Methods
2.1. Animals
Male C57BL/6NCrl mice at least 8 weeks old were given intraperitoneal (i.p.) injections of 200 μl of vehicle (saline, n=8), ethanol (2.5 g/kg in saline, n=8), or Ro 25–6981 (10 mg/kg in saline, n=8; 1594 Tocris Bioscience, Ellisville, MO). Mice were group-housed 4 per cage. Mice were sacrificed 45 minutes after injection, and hippocampi were dissected and immediately flash frozen in liquid nitrogen. All experiments were carried out in accordance with the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals and approved by the Wake Forest University of Health Sciences Institutional Animal Care and Use Committee.
2.2. Synaptoneurosome preparation and RNA isolation
Synaptoneurosomes were prepared by homogenizing hippocampal tissue in homogenization buffer (20 mM HEPES pH 7.4, 5 μM EDTA pH 8.0, and RNase inhibitor). Homogenates were filtered through 100-μm nylon filters followed by 5-μm filters and centrifuged at 14000 x g for 20 minutes at 4°C. Pellets were resuspended in buffer RLT and samples processed using an RNeasy Micro Kit (74004, Qiagen, Germantown, MD), following the manufacturer’s instructions. Samples were homogenized in buffer RLT. One volume of 70% ethanol was added to precipitate RNA, and samples were centrifuged for 15 seconds at 8000 x g through RNeasy MinElute spin columns. Buffer RW1 was added to the column and centrifuged for 15 seconds at 8000 x g. Samples were DNAse treated and centrifuged for 15 seconds at 8000 x g with buffer RW1. Columns were then washed in buffer RPE and centrifuged for 15 seconds at 8000 x g. Columns were dried by adding 80% ethanol and centrifuging for 2 minutes at 8000 x g and centrifuging again with the column lid open for 5 minutes at full speed. RNA was then eluted in nuclease-free water. RNA quality was determined using a 2200 Agilent TapeStation Instrument (G2965AA, Agilent Technologies, Santa Clara, CA). RNA integrity numbers ranged from 5.3 to 7.8, with the average being 7.0. Yield was determined using Qubit fluorometric quantitation (Q33216, Thermo Fisher Scientific, Pittsburgh, PA).
2.3. RNA-Sequencing
RNA samples were sequenced by the Genome Sequencing and Analysis Facility at the University of Texas at Austin. Poly-A mRNA was captured using the MicroPoly(A) Purist Kit (AM1919, Life Technologies, Carlsbad, CA). Samples were processed using a whole transcriptome library preparation kit (FC-404–2002, NextSeq 500/550 High Output Kit, (Illumina Inc., San Diego, CA) and sequenced on a NextSeq 500 system (Illumina Inc., San Diego, CA) at a depth of ~20 million reads per sample (paired-end 75 bp reads). Read quality was assessed with FASTQC (version 0.11.5). One ethanol-treated sample was discarded due to low sequencing reads. Reads were mapped to the mouse genome (Mus musculus version GRCm38 rel.80, Ensemble) using the alignment tool Splice Transcripts Alignment to a Reference (STAR, version 2.5.0a). HTSeq (version 8.0) was utilized to quantify raw counts of mapped sequence reads using Python (Anders et al., 2015). DESeq2 (version 1.14.1) identified differentially-expressed genes between treatments (vehicle and ethanol; vehicle and Ro 25–6981) from normalized quantified read counts(Love et al., 2014). DEXSeq (version 1.20.2) identified differential exon usage by quantifying exon ID counts between treatments (saline and ethanol; saline and Ro 25–6981) (Anders et al., 2012; Reyes et al., 2013). DESeq2 and DEXSeq are statistical tests based on a model that uses the negative binomial distribution, and provide the necessary functions for analysis and statistics as well as some packages for visualization and exploration of results. Enrichment analysis for gene ontologies (GOs) of the differentially expressed genes and exons was performed using the web-based enrichment analysis software Enrichr (Chen et al., 2013; Kuleshov et al., 2016). All tools used for analysis followed the recommended methods provided by the developers. Statistical significance was assessed using a nominal p-value less than or equal to 0.05 to minimize type I and type II error. DEGs and DEEs below a nominal p-value threshold of 0.05 were used to include enough genes for comparisons and enrichment analysis. Data processing and statistics including χ2 -test for statistical comparison with venn diagrams were performed using R programming unless otherwise noted (R Core Team, 2018).
2.4. Structural Analysis
DEXSeq exon ID sequences were aligned to genomic DNA sequences of the mouse ionotropic NMDAR, Grin1 (NCBI Gene ID: 14810). From this alignment, exon IDs mapping to the consensus coding DNA sequence (CCDS) for mRNA: NM_008169.3 (NCBI CCDS ID: CCDS15764.1) were identified and included DEXseq exon IDs E022, E026, E029, E030, E023, and E031. These aligned with exons 4, 5, 6, and 8 of the CCDS. Exon IDs E041 and E006 contained portions of CCDS exons 1 and 20, but also contained large non-coding portions and were not included in this analysis. Exons 4, 5, 6, and 8 were also found in the available structural data for the Rattus novergicus GluN1/GluN2B (NMDAR protein subunits) heterotetramer (PDB ID: 4PE5) and the Xenopus laevis NMDAR1/NMDAR2B amino-terminal domain heterodimer (PDB IDs: 3QEL and 5B3J). The rat and Xenopus Grin1 CCDS amino acid sequences have 99% and 92% sequence identity with mouse, respectively (UniProt entries: rat, P35439; mouse, P35438; Xenopus, A0A1L8F5J9). Alignments were performed in ApE, a plasmid editor (personal communication M. Wayne Davis, University of Utah). Structures were visualized in PyMOL (Schrodinger LLC, New York).
2.5. Data Availability
Sequencing data is publicly available at SRA Accession numbers TBD.
3. Results
3.1. Ethanol and Ro 25–6981 induce differential gene expression
Ethanol and Ro 25–6981 share similar functional and behavioral properties that are related to synaptic responses. We reasoned that the molecular changes that occur within an hour at hippocampal synapses to mediate long-lasting antidepressant effects since rodent models show antidepressant-like behaviors and humans report reduced depressive symptoms with a single injection of NMDAR antagonists within this time frame(Workman et al., 2015; Workman et al., 2013; Zarate et al., 2006). While ethanol treatment within this time frame produces intoxication (2.5mg/kg estimated to produce an alcohol concentration of 0.215g/dl)(Ginsburg et al., 2008), we predict that similar molecular changes that support the antidepressant and reduced anxiety-like behaviors observed at 24 hours in Wolfe et al., are initiated similarly. We used RNA-Seq to analyze gene expression in hippocampal synaptoneurosomes isolated from mice treated with ethanol or Ro 25–6981. We first assessed the purity of the synaptoneurosome preparation and found that the RNA was enriched for primarily neuronal and synaptic-related ontologies (Fig. S1), consistent with our previous findings (Most et al., 2015). No batch effects were identified by principle component analysis (Fig. S2), and no sample outliers were detected by hierarchical clustering (Fig. S3) or correlation plots (Fig. S4).
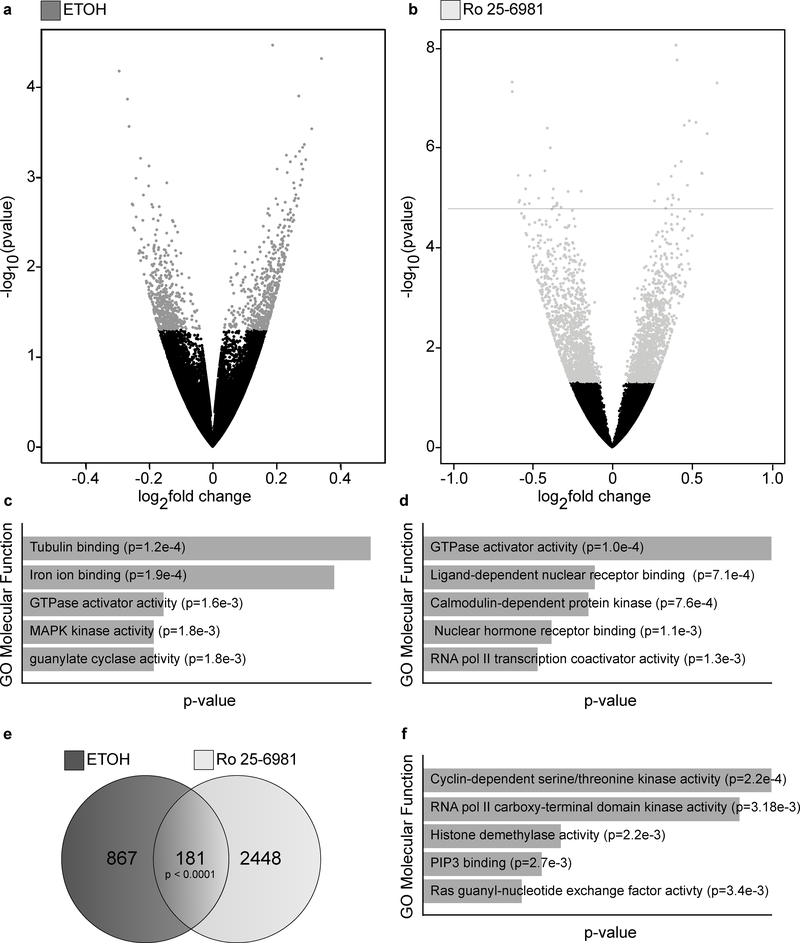
Next, we identified the differentially expressed genes following acute ethanol or Ro 25–6981 treatment. Unlike Ro 25–6981, which specifically inhibits NR2B-containing NMDA receptors(Fischer et al., 1997; Malherbe et al., 2003a; Malherbe et al., 2003b), ethanol has many targets, including inhibition of NMDARs(Lovinger and Roberto, 2013). Thus, we predicted that ethanol treatment would produce a greater number of differentially expressed genes compared with Ro 25–6981 treatment. Volcano plots indicating fold-changes and statistical significance (p-value) between saline control and treatment groups are shown in Figure 1. Significant genes are shown in grey (nominal p-value ≤ 0.05) for ethanol (Fig. 1A) and Ro 25–6981 (Fig. 1B) treatment. Following ethanol treatment, 1048 genes were differentially expressed. More than twice as many genes (2629) changed in response to Ro 25–6981 treatment.
Figure 1.
Acute ethanol and Ro 25–6981 treatment induced differentially expressed genes (DEGs). C57BL/6 male mice were treated (45 minutes) with vehicle (saline, n=8), ethanol (ETOH, 2.4 g/kg, n=7), or Ro 25–6981 (10 mg/kg, n=8). RNA-Seq and DEG analyses were performed using polyA-enriched RNA isolated from hippocampal synaptoneurosomes. Volcano plots for (A) ethanol and (B) Ro 25–6981, indicating up- and downregulation (log2 fold-change; x-axis) and statistical significance (-log nominal p-value; y-axis). Grey points indicate significant (p ≤ 0.05) DEGs after (A) ethanol and (B) Ro 25–6981 treatment. Molecular function gene ontology (GO) enrichment results are represented in the bar graphs, depicting the top five highest-ranking GOs for the DEGs after (C) ethanol and (D) Ro 25–6981 treatment. Of the 1048 DEGs induced by ethanol (p ≤ 0.05) and the 2629 DEGs induced by Ro 25–6981 (p ≤ 0.05), there were 181 overlapping DEGs (E). The top five molecular function GOs for the overlapping genes are shown (F).
The five highest-ranking molecular function gene ontologies (GOs) were identified for ethanol and Ro 25–6981 treatment (Fig. 1C and Fig. 1D, respectively) using the enrichment ontology tool Enrichr(Chen et al., 2013; Kuleshov et al., 2016). Only the category GTPase activator activity was a common function of the DEGs and was ranked highest for Ro 25–6981.
3.2. Minimal overlap in synaptic gene expression after ethanol and Ro 25–6981
There were 181 highly significant genes overlapping DEGs with treatment, comprising ~17% of the ethanol-induced DEGs and ~7% of the Ro 25–6981 DEGs (Fig. 1E). Molecular function GOs were then identified from the overlapping DEGs, and the top five categories are shown in Figure 1f.
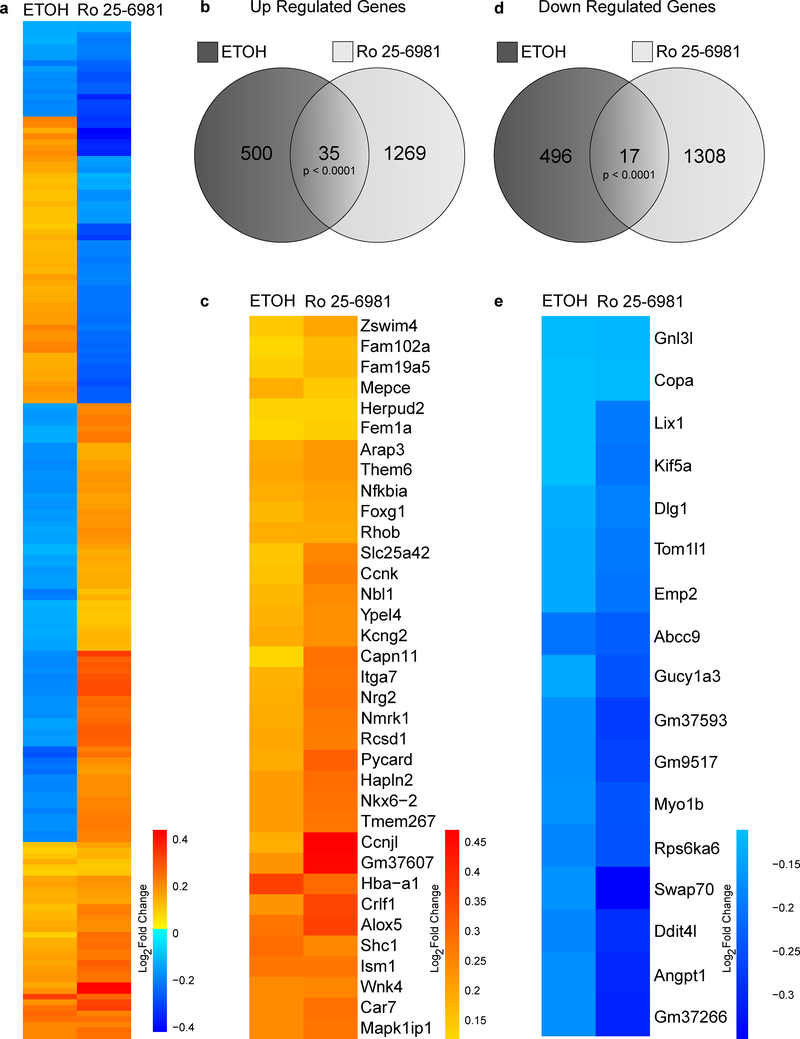
Of these overlapping genes, we identified those that were coordinately up- and downregulated by both treatments, which might suggest a similar underlying functional role for the proteins coded by the DEGs. A heat map of the fold-changes of the 181 overlapping DEGs was generated. Upregulated genes are shown in red and downregulated genes are in blue (Fig. 2A). Approximately 29% (52 genes) changed in the same direction in both treatment groups. For the upregulated DEGs, 535 and 1304 were present in the ethanol and Ro 25–6981 treatment groups, respectively, with 35 overlapping DEGs (Fig. 2B). These 35 upregulated genes are displayed in a heat map with increasing fold-change shown by the red color scale (Fig. 2C). For the downregulated DEGs, 513 and 1325 were identified in the ethanol and Ro 25–6981 treatment groups, respectively, with 17 overlapping DEGs (Fig. 2D). These 17 downregulated genes are displayed in a heat map with decreasing fold-change shown by the blue color scale (Fig. 2E). Our data suggest that few genes are co-regulated by acute treatment with ethanol or Ro 25–6981.
Figure 2.
Overlapping differentially expressed genes (DEGs) after ethanol and Ro 25–6981 treatment. The overlapping genes (181) were assessed for coordinated directional fold-changes (up- or downregulation). (A) The log2 fold-changes for these overlapping DEGs were plotted as a heat map with the ethanol column on the left and Ro 25–6981 column on the right and individual genes as rows. The scale bar indicates positive fold-changes or upregulated DEGs increasing from yellow to red, whereas negative fold-changes or downregulated DEGs decrease from light to dark blue; 52 genes showed directional fold-changes. (B) Venn diagram of upregulated DEGs after each treatment. Ethanol (ETOH) and Ro 25–6981 induced upregulation of 535 and 1304 genes, respectively, with 35 overlapping genes. (C) Heat map of the 35 overlapping upregulated DEGs, showing ethanol on the right and Ro 25–6981 on the left and the gene symbols for each row (colors indicate increasing log2 fold- change from yellow to red). (D) Venn diagram of downregulated DEGs after each treatment. ETOH and Ro 25–6981 induced downregulation of 513 and 1325 DEGs, respectively, with 17 overlapping genes. (E) Heat map of the 17 overlapping downregulated DEGs, showing ethanol on the right and Ro 25–6981 on the left and the gene symbols for each row (colors indicate decreasing log2 fold-change from light to dark blue).
3.3. Ethanol and Ro 25–6981 induce differential exon usage
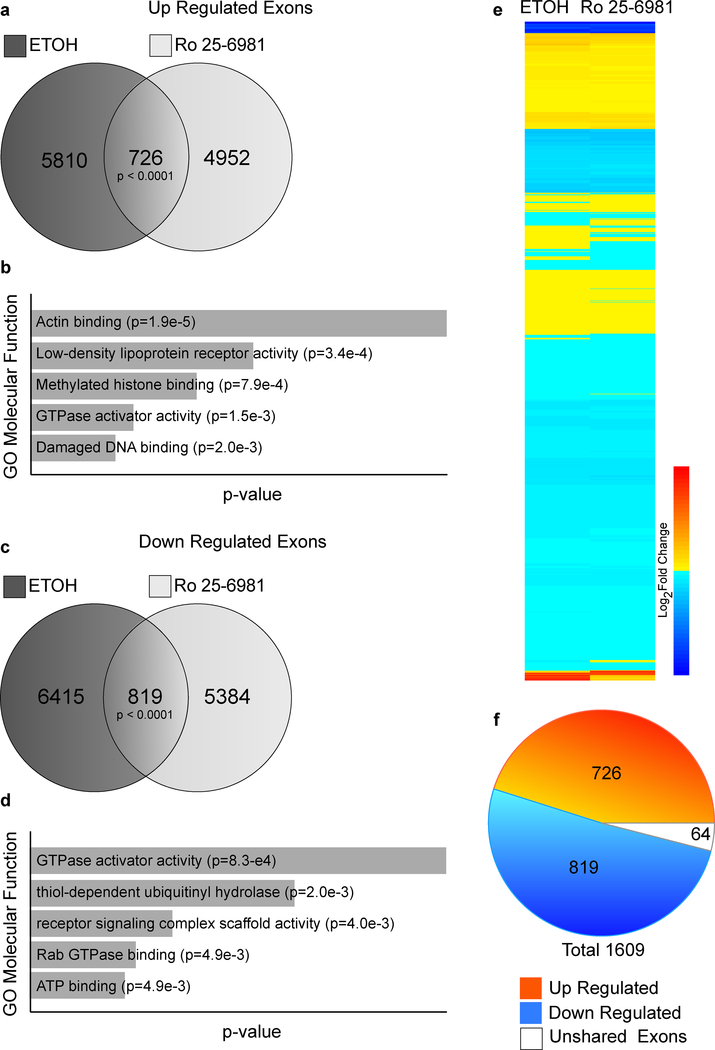
Fast-acting antidepressants are thought to mediate long-lasting effects through increased glutamatergic synapse formation and an increase in excitatory drive(Li et al., 2010). Furthermore, alternative splicing regulates the properties of glutamatergic synapses in the hippocampus(Traunmuller et al., 2016). To study exon usage after treatment with ethanol and Ro 25–6981, we utilized DEXSeq and found that large numbers of exons were differentially expressed (p < 0.05) following treatment with ethanol (13,770; Table S1) and Ro 25–6981 (11,881; Table S2). Volcano plots indicating fold-changes and statistical significance for these exons between control and treatment are shown in Figure S5. Significant DEEs (nominal p-value ≤ 0.05) are shown in grey for ethanol (Fig. S5A) and Ro 25–6981 (Fig. S5B) treatment. The DEEs identified by treatment responses were compared, yielding 1609 co-occurring DEEs (Fig. 3A). These overlapping exons were enriched for the molecular function GO associated GTPase activity, consistent with the GO analysis of the DEGs shown in Figure 1 (Fig.3B).
Figure 3.
Differentially expressed exons (DEEs) after ethanol and Ro 25–6981 treatment. C57BL/6 male mice were treated (45 minutes) with vehicle (saline, n=8), ethanol (ETOH, 2.4 g/kg, n=7), or Ro 25–6981 (10 mg/kg, n=8). RNA-Seq and DEE analysis (DEXSeq) were performed using RNA isolated from hippocampal synaptoneurosomes. (A) Venn diagram comparing DEEs in ethanol- and Ro 25–6981-treatment groups identified 1609 overlapping exons (p ≤ 0.0001). (B) The top five molecular function gene ontologies (GOs) for the overlapping DEEs are shown.
We predicted that if antidepressant mechanisms common to both drugs were related to exon usage, we would observe co-regulation of DEEs with both treatment groups. We thus identified coordinately up- and downregulated DEEs and found 6536 and 5678 upregulated exons following ethanol and Ro 25–6981 treatment, respectively, with 726 overlapping exons (Fig. 4A). There were 7234 and 6203 downregulated DEEs after ethanol and Ro 25–6981 treatment, respectively, with 819 overlapping DEEs (Fig. 4C). Enrichment analysis of the co-regulated exons revealed that for both the up- and downregulated exons, molecular function GOs were again enriched with genes associated with GTPase activity (Fig. 4B and D). A heat map of the fold-changes associated with the 1609 overlapping DEEs was generated (the yellow-red color scale indicates upregulation and the light-dark blue color scale indicates downregulation(Fig. 4E). Directional changes common to both treatments were remarkably similar for the majority of DEEs, with only 64 exons exhibiting differing expression patterns (Fig. 4F). This contrasts with the limited number of overlapping changes in DEGs between treatments, suggesting a specific role for exon usage in the acute antidepressant effects of ethanol and Ro 25–6981.
Figure 4.
Overlapping differentially expressed exons (DEEs) after ethanol and Ro 25–6981 treatment. The 1609 overlapping exons in both treatment groups were assessed for positive or negative fold-changes to identify coordinately up- or downregulated exons. (A) Venn diagram of upregulated DEEs in each treatment group. Ethanol (ETOH) and Ro 25–6981 treatment induced upregulation of 6536 and 5678 DEEs, respectively; of these, there were 726 overlapping genes. (B) The top five molecular function gene ontologies (GOs) for the overlapping upregulated exons. (C) Venn diagram of downregulated DEEs in each treatment group. ETOH and Ro 25–6981 treatment induced downregulation of 7234 and 13437 DEEs, respectively; of these, there were 819 overlapping genes. (D) The top five molecular function GOs for the overlapping downregulated exons. (E) Log2 fold-changes for the overlapping DEEs were plotted as a heat map with the ethanol column on the right and Ro 25–6981 column on the left. The scale bar indicates positive fold-changes or upregulated DEGs increasing from yellow to red, whereas negative fold-changes or downregulated DEGs decrease from light to dark blue; 1545 genes showed directional fold-changes. (F) Pie chart summarizing the up- and downregulated exons induced by both treatments compared with exons showing differential fold-changes (unshared; white). About 95% of the overlapping DEEs between the treatments showed coordinated up- or downregulated fold-changes.
A large number of DEEs common to both ethanol and Ro 25–6981 treatment were identified that were not detectable in the gene-level analysis. A large number of genes that contained single or multiple exon-specific differences was identified after ethanol (6572; Table S3) and Ro 25–6981 treatment (6082; Table S4). We found 3765 overlapping genes that had any DEEs located in the gene in both treatment groups, comprising ~57% of the ethanol-induced genes and ~62% of the Ro 25–6981-induced genes with DEEs (Fig. 5A). These 3765 overlapping genes were enriched for molecular function GOs associated with guanyl-nucleotide exchange factor and kinase activity (Fig. 5B). These data suggest that exon usage and splice variants may be important in mediating acute antidepressant effects, as the majority of genes containing DEEs were expressed with both treatments.
Figure 5.
Genes with differentially expressed exons (DEEs) after acute ethanol and rapid Ro 25–6981 treatment. (A) Venn diagram of genes with DEEs found in ethanol (dark grey) and Ro 25–6981(light grey) treatment groups, showing 3765 overlapping genes with DEEs (p ≤ 0.0001). (B) The top five molecular function gene ontologies (GOs) for the overlapping exons.
3.4. Ethanol-induced DEEs may target NMDAR1s
The prominent overlap in genes with DEEs induced by ethanol and Ro 25–6981 led us to consider the potential functional relevance of changes in exon usage among individual genes. Because both drugs can inhibit NMDAR function and NMDAR function is critical for induction of synaptic plasticity, we examined exons associated with mouse Grin1. Several exons in Grin1 were differentially expressed after treatment with ethanol or Ro 25–6981 (Fig. 6A). Although the Grin1 transcript contained alterations in exon usage for both ethanol and Ro 25–6981 they were not necessarily the same DEEs within Grin1, however, alterations in exon usage occurred in similar gene regions. We mapped the exon IDs to the CCDS (Fig. S6) of Grin1 and high-resolution x-ray crystal structures of the NMDAR1/NMDAR2B heterotetramer (Fig. 6B). The NMDAR is composed of three functionally distinct domains (transmembrane, ligand binding, and amino-terminal domains). Exon IDs located in exons 4, 5, 6, and 8 of the CCDS sequence of Grin1 displayed differential expression in the ethanol-treated group. These exons are localized to the amino-terminal domain and cover ~40% of the domain sequence, suggesting that they are critical to this domain’s function.
Figure 6.
Ethanol and Ro 25–6981 induce exon usage in Grin1 that may affect NMDAR activity. (A) DEXSeq graph of exon usage for the NMDAR1 gene, Grin1, showing each ensemble exon ID. Corresponding Grin1 transcript with gene locations is displayed below graph where lines indicate introns and grey bars indicate exons. Dotted lines indicate saline- (blue), ethanol- (ETOH; dark grey), or Ro 25–6981- (light grey) induced exon usage. Significant differences from the saline group are indicated by asterisks (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001). (B) NMDAR1 (white) /NMDAR2B (grey) heterotetramer (left side) and NMDAR1 monomer (right side). Peptides encoded by exons 4, 5, 6, and 8 are indicated in red. Domains are represented by the backdrop color, including the amino-terminal domain (ATD, green), ligand binding domain (LBD, yellow), and transmembrane domain (TMD, blue; PDB ID: 4PE5). (C) Alignment of ATD domains of the unbound structure model apo (white/red; PDB ID: 5B3J) and ifenprodil-bound (grey, PDB ID: 3QEL) structures. Exons 4, 5, 6, and 8 are indicated in red. The ATD has a bi-lobed structure and contains two subdomains, indicated as R1 (top) and R2 (bottom). Three “hinge” loops separating these domains are also shown. Exon numbers represent only the first 8 of 20 exons present along the Grin1 gene that encompass the ATD coding region (Note: panel c is a recreation of a figure previously published; see Figure 1c in Tajima et al., 2016)(Tajima et al., 2016). Subdomain R2 undergoes a 5° rotation from the open to closed conformation. Secondary structure was simplified to clarify the conformational change. (D) Representation of the ATD portion of the NMDAR subunit GluN1 (PDB ID: 4PE5) with peptides encoding DEEs shown in red and clinically observed point mutations shown as yellow spheres. Point mutations occurring in and outside of the DEEs are shown in red and black, respectively. Gold stars in the exon diagram indicate the approximate location of the point mutations.
The amino-terminal domain of NMDAR1 is conformationally dynamic and acts in trans with the NMDAR2 amino-terminal domain to modulate receptor activity(Zhu et al., 2013). Phenylethanolamines, like Ro 25–6981 and ifenprodil, inhibit NMDAR function by inducing conformational changes in the amino-terminal domain(Karakas et al., 2011; Zhu et al., 2016). Crystallographic studies have captured NMDAR1/NMDAR2B in an active “open” conformation relative to a “closed” inactive, ifenprodil-bound state(Tajima et al., 2016). The transition from open to closed conformation requires major rearrangement of the NMDAR2B amino-terminal domain accompanied by smaller rearrangements in the NMDAR1 amino-terminal domain(Tajima et al., 2016). Regions encoded by ethanol-induced DEEs within the R2 subdomain of the NMDAR1 amino-terminal domain are particularly mobile, suggesting alterations in exon usage at this region may regulate these dynamics (Fig. 6C).
We examined potential roles for the identified DEEs using publicly available human data retrieved from the Center for Functional Evaluation of Rare Variants, CFERV (http://functionalvariants.emory.edu). Four of the five Grin1 amino-terminal domain mutations indexed here localize to the DEEs; these mutations are associated with developmental delay, hypotonia, intellectual disability, movement disorder, schizophrenia, and epilepsy (Fig. 6D).
4. Discussion
MDD increases the risk of AUD by ~2 fold, and it has been suggested that some patients initially use alcohol as a self-prescribed antidepressant. NMDAR antagonists have gained acceptance as rapid-acting antidepressants. Their rapid but long-lasting effects are due, in part, to alterations in synaptic plasticity and synaptic protein composition (Autry et al., 2011; Li et al., 2010; Workman et al., 2015). Similar to rapid-acting antidepressants, ethanol induces antidepressant-like effects in behavioral tests, inhibits NMDAR function, and induces synaptic signaling pathways that trigger de novo protein synthesis (Wolfe et al., 2016). In this study, we hypothesized that similar changes in synaptic gene expression would occur following acute ethanol and rapid-acting antidepressant treatment. However, the two treatments induced high levels of overlap in exon expression and comparatively fewer changes in gene expression.
We observed distinct transcriptional changes with both treatments. The differences may be due to the potent and selective actions of Ro 25–6981(Fischer et al., 1997; Mutel et al., 1998) or the acute time point that we measured, which was based on previous work(Wolfe et al., 2016). Ethanol may induce greater and more pronounced gene expression changes with increased treatment time, as chronic ethanol consumption (Most et al., 2015) and repeated ethanol treatment to generate sensitization (O’Brien et al., 2018) is known to produce more changes in gene expression and exon usage in synaptoneurosomes than were identified here. Additional studies comparing ethanol and rapid-acting antidepressants will be necessary to identify the lasting effects of these drugs on the transcriptome.
One of our key findings was the abundance of genes with DEEs that were commonly expressed by both treatments. Alternative splicing and exon composition are essential mechanisms that expand the complexity of the proteome(Johnson et al., 2003; Vuong et al., 2016; Warden and Mayfield, 2017), generating isoforms that differ in function and localization and that regulate cellular development, differentiation, and in some instances progression to disease(Katz et al., 2010). Changes in exon expression and mRNA composition allow for translation of new protein isoforms, although alterations in basal mRNA or protein expression are not necessarily detected(Johnson et al., 2003; Lander et al., 2001; Modrek et al., 2001). Splicing induction can also occur rapidly and may be a means to alter neuron function (Carmo-Fonseca and Kirchhausen, 2014; Mauger et al., 2016); for example, splicing occurred within 30 seconds in mammalian cells46 and within 15 minutes after neuronal stimulation47. Splicing can also be much faster than transcription (Carmo-Fonseca and Kirchhausen, 2014; Mauger et al., 2016), which may explain the abundance of genes with DEEs compared with DEGs identified in this study. Alternatively, nonconical forms of transcriptional regulation such as cytoplasmic splicing or extranuclear splicing may explain the abundance of DEEs found in our data. New evidence for extranuclear splicing has recently begun to identify the biological relevance of endogenous extranuclear splicing systems that occur in mammals(Buckley et al., 2014). Notably, it was reported that splicing machinery was located in dendrites and that isolated dendrites were capable of splicing reporter RNA constructs (Glanzer et al., 2005). This provided evidence that splicing can occur not only in the cytoplasm, but also peripherally in dendrites. Further research is necessary to investigate the possibility of dendritic splicing in our model of acute ethanol and Ro 25–6981 treatment. The striking enrichment of DEEs in the amino-terminal domain suggests that ethanol’s effect on NMDAR function may be partly elicited through this domain. Ethanol and Ro 25–6981 may alter the conformation of the amino-terminal domain, albeit through different mechanisms, potentially contributing to their similar responses (Wolfe et al., 2016).
The GO molecular function linked with the activity of GTPase activator was identified in DEGs after ethanol or Ro 25–6981 treatment. However, this group was not in the top five GO terms when analyzing the molecular function of the overlapping DEGs from the two treatments. In contrast, the DEE results indicated that there were overlapping changes in GTPase activity following ethanol and Ro 25–6981 treatment. Most of the DEEs are within genes that code for proteins that are small GTPases or affect downstream G-protein signaling and have functional roles in synaptic transmission or synapse maturation. Interestingly, GTPase Rac1 was required for the antidepressant efficacy of the NMDAR agonist, rapastinel, a novel glutamatergic compound that enhances synaptic plasticity and shows promise in clinical settings (Burgdorf et al., 2013; Kato et al., 2017; Murrough et al., 2017; Preskorn et al., 2015). We identified the gene coding for Rac1 as one of the common DEEs induced by ethanol and Ro 25–6981 treatment. Rac1 is well characterized for its role in synapse formation, a requirement for rapid antidepressant efficacy(Kato et al., 2017). Rac1 is an mRNA target of fragile X mental retardation protein (FMRP), and we previously showed that FMRP was required for the rapid-acting antidepressant-like behaviors induced by ethanol(Wolfe et al., 2016). Additional research is needed to determine if Rac1-mediated antidepressant efficacy depends on synaptic isoform specificity.
We showed that alternative exon use in Grin1 in response to acute ethanol treatment may change NMDAR1 conformational dynamics through the amino-terminal domain, potentially altering synaptic function. In addition, clinical data from CFERV suggested that point mutations, which may also regulate conformation of the amino-terminal domain, alter NMDAR function and are associated with neurological disorders. Like ethanol, Ro 25–6981 increased utilization of an exon located in the amino-terminal domain in some transcripts, suggesting it may also regulate function via an exon-selection mechanism. However, this DEE was not contained in the CCDS and further research is necessary to identify its role at the transcriptional and protein levels. The DEEs in this region of NMDAR1 indicate that ethanol may elicit antidepressant properties mediated through conformational changes in the amino-terminal domain, similar to how Ro 25–6981 functions (Karakas et al., 2011; Zhu et al., 2016). Ethanol and Ro 25–6981 may thus utilize the same mechanism to produce their antidepressant-like phenotype. Exon usage and alternative splicing may control synaptic activity by targeting protein domains without necessarily altering gene expression. This mechanism could effectively localize specific mRNA isoforms to the synapse, differentiating their expression and function from somatic transcripts(Ouwenga et al., 2017; Taliaferro et al., 2016).
5. Conclusion
This study provides a foundation to explore the functional implications of exon usage by ethanol and rapid-acting antidepressants. Alternative splicing may be critical for molecular, synaptic, and behavioral changes common to acute ethanol and Ro 25–6981 treatments. The stability of these changes with continued alcohol or rapid-antidepressant use is unknown, and continued investigation of these changes over time and their implication in function and behavior is necessary. Although we observed limited treatment-dependent overlap in DEGs, there were remarkable similarities in exon usage and genes containing DEEs. More research is necessary to examine the role of alternative splicing on protein function in response to ethanol and rapid-acting antidepressants, and future studies may uncover co-regulated exons that are involved in functional domains common to disease pathologies in AUD and MDD. Understanding alternative splicing of key regions may drive drug discovery for these comorbid disorders.
Supplementary Material
Acknowledgements
We thank Jody Mayfield (Waggoner Center for Alcohol and Addiction Research, University of Texas at Austin) for thoughtful critiques and assistance with writing and editing the manuscript. This work was supported by an NIH-NIAAA pilot grant provided by the Integrated Neuroscience Initiative on Alcoholism (KRG), NSF grant IOS-1355158 (KRG), Department of Defense USAMRMC Award W81XWH-14-10061 (KRG), and NIH-NIAAA grants AA025117 and AA026551 (KRG), AA020683 (RDM), AA012404 and AA013517 (RAH), T32AA007565 (CFH), 5T32AA007471-29 (SAW).
Abbreviations
- AUD
Alcohol Use Disorder
- MDD
Major Depressive Disorder
- DEE
differentially expressed exons
- NMDAR
N-methyl-D-aspartate receptor
- RNA-Seq
RNA sequencing
- DEG
differentially expressed genes
- GO
gene ontologies
- FMRP
Fragile-X Mental Retardation Protein
Footnotes
Declarations of interest
The authors report no biomedical financial interests or potential conflicts of interests.
References
- Abdallah CG, Sanacora G, Duman RS, Krystal JH, 2015. Ketamine and rapid-acting antidepressants: a window into a new neurobiology for mood disorder therapeutics. Annu Rev Med 66, 509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahao KP, Salinas AG, Lovinger DM, 2017. Alcohol and the Brain: Neuronal Molecular Targets, Synapses, and Circuits. Neuron 96, 1223–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Reyes A, Huber W, 2012. Detecting differential usage of exons from RNA-seq data. Genome Res 22, 2008–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autry AE, Adachi M, Nosyreva E, Na ES, Los MF, Cheng PF, Kavalali ET, Monteggia LM, 2011. NMDA receptor blockade at rest triggers rapid behavioural antidepressant responses. Nature 475, 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagot RC, Cates HM, Purushothaman I, Vialou V, Heller EA, Yieh L, LaBonte B, Pena CJ, Shen L, Wittenberg GM, Nestler EJ, 2017. Ketamine and Imipramine Reverse Transcriptional Signatures of Susceptibility and Induce Resilience-Specific Gene Expression Profiles. Biol Psychiatry 81, 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden JM, Fergusson DM, 2011. Alcohol and depression. Addiction 106, 906–914. [DOI] [PubMed] [Google Scholar]
- Buckley PT, Khaladkar M, Kim J, Eberwine J, 2014. Cytoplasmic intron retention, function, splicing, and the sentinel RNA hypothesis. Wiley Interdiscip Rev RNA 5, 223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgdorf J, Zhang XL, Nicholson KL, Balster RL, Leander JD, Stanton PK, Gross AL, Kroes RA, Moskal JR, 2013. GLYX-13, a NMDA receptor glycine-site functional partial agonist, induces antidepressant-like effects without ketamine-like side effects. Neuropsychopharmacology 38, 729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Kirchhausen T, 2014. The timing of pre-mRNA splicing visualized in real-time. Nucleus 5, 11–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A, 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, Zhou Z, Kimura M, Mash DC, Yuan Q, Goldman D, 2012. GABAergic gene expression in postmortem hippocampus from alcoholics and cocaine addicts; corresponding findings in alcohol-naive P and NP rats. PLoS One 7, e29369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficek J, Zygmunt M, Piechota M, Hoinkis D, Rodriguez Parkitna J, Przewlocki R, Korostynski M, 2016. Molecular profile of dissociative drug ketamine in relation to its rapid antidepressant action. BMC Genomics 17, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer G, Mutel V, Trube G, Malherbe P, Kew JN, Mohacsi E, Heitz MP, Kemp JA, 1997. Ro 25–6981, a highly potent and selective blocker of N-methyl-D-aspartate receptors containing the NR2B subunit. Characterization in vitro. J Pharmacol Exp Ther 283, 1285–1292. [PubMed] [Google Scholar]
- Ginsburg BC, Javors MA, Friesenhahn G, Frontz M, Martinez G, Hite T, Lamb RJ, 2008. Mouse breathalyzer. Alcohol Clin Exp Res 32, 1181–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanzer J, Miyashiro KY, Sul JY, Barrett L, Belt B, Haydon P, Eberwine J, 2005. RNA splicing capability of live neuronal dendrites. Proc Natl Acad Sci U S A 102, 16859–16864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BF, Harford TC, 1995. Comorbidity between DSM-IV alcohol use disorders and major depression: results of a national survey. Drug Alcohol Depend 39, 197–206. [DOI] [PubMed] [Google Scholar]
- Hasin DS, Grant BF, 2002. Major depression in 6050 former drinkers: association with past alcohol dependence. Arch Gen Psychiatry 59, 794–800. [DOI] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD, 2003. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302, 2141–2144. [DOI] [PubMed] [Google Scholar]
- Karakas E, Simorowski N, Furukawa H, 2011. Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature 475, 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Fogaca MV, Deyama S, Li XY, Fukumoto K, Duman RS, 2017. BDNF release and signaling are required for the antidepressant actions of GLYX-13. Mol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB, 2010. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7, 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Le Moal M, 2008. Addiction and the brain antireward system. Annu Rev Psychol 59, 29–53. [DOI] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A, 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44, W90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowki J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ, Szustakowki J, International Human Genome Sequencing, C., 2001. Initial sequencing and analysis of the human genome. Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- Li N, Lee B, Liu RJ, Banasr M, Dwyer JM, Iwata M, Li XY, Aghajanian G, Duman RS, 2010. mTOR-dependent synapse formation underlies the rapid antidepressant effects of NMDA antagonists. Science 329, 959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S, 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovinger DM, Roberto M, 2013. Synaptic effects induced by alcohol. Curr Top Behav Neurosci 13, 31–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malherbe P, Kratochwil N, Knoflach F, Zenner MT, Kew JN, Kratzeisen C, Maerki HP, Adam G, Mutel V, 2003a. Mutational analysis and molecular modeling of the allosteric binding site of a novel, selective, noncompetitive antagonist of the metabotropic glutamate 1 receptor. J Biol Chem 278, 8340–8347. [DOI] [PubMed] [Google Scholar]
- Malherbe P, Mutel V, Broger C, Perin-Dureau F, Kemp JA, Neyton J, Paoletti P, Kew JN, 2003b. Identification of critical residues in the amino terminal domain of the human NR2B subunit involved in the RO 25–6981 binding pocket. J Pharmacol Exp Ther 307, 897–905. [DOI] [PubMed] [Google Scholar]
- Mauger O, Lemoine F, Scheiffele P, 2016. Targeted Intron Retention and Excision for Rapid Gene Regulation in Response to Neuronal Activity. Neuron 92, 1266–1278. [DOI] [PubMed] [Google Scholar]
- Modrek B, Resch A, Grasso C, Lee C, 2001. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res 29, 2850–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Most D, Ferguson L, Blednov Y, Mayfield RD, Harris RA, 2015. The synaptoneurosome transcriptome: a model for profiling the emolecular effects of alcohol. Pharmacogenomics J 15, 177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrough JW, Abdallah CG, Mathew SJ, 2017. Targeting glutamate signalling in depression: progress and prospects. Nat Rev Drug Discov 16, 472–486. [DOI] [PubMed] [Google Scholar]
- Mutel V, Buchy D, Klingelschmidt A, Messer J, Bleuel Z, Kemp JA, Richards JG, 1998. In vitro binding properties in rat brain of [3H]Ro 25–6981, a potent and selective antagonist of NMDA receptors containing NR2B subunits. J Neurochem 70, 2147–2155. [DOI] [PubMed] [Google Scholar]
- Neasta J, Barak S, Hamida SB, Ron D, 2014. mTOR complex 1: a key player in neuroadaptations induced by drugs of abuse. J Neurochem 130, 172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien Megan A, W. R. M., Sheth Nihar U, Steven Bradley, John Bigbee, Ashutosh Pandey, Williams Robert W, Wolstenholme Jennifer T, Miles Michael F, 2018. Ethanol-Induced Behavioral Sensitization Alters the Synaptic Transcriptome and Exon Utilization in DBA/2J Mice Frontiers in Genetics 9, 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouwenga R, Lake AM, O’Brien D, Mogha A, Dani A, Dougherty JD, 2017. Transcriptomic Analysis of Ribosome-Bound mRNA in Cortical Neurites In Vivo. J Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preskorn S, Macaluso M, Mehra DO, Zammit G, Moskal JR, Burch RM, Group G-CS, 2015. Randomized proof of concept trial of GLYX-13, an N-methyl-D-aspartate receptor glycine site partial agonist, in major depressive disorder nonresponsive to a previous antidepressant agent. J Psychiatr Pract 21, 140–149. [DOI] [PubMed] [Google Scholar]
- Reyes A, Anders S, Weatheritt RJ, Gibson TJ, Steinmetz LM, Huber W, 2013. Drift and conservation of differential exon usage across tissues in primate species. Proc Natl Acad Sci U S A 110, 15377–15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima N, Karakas E, Grant T, Simorowski N, Diaz-Avalos R, Grigorieff N, Furukawa H, 2016. Activation of NMDA receptors and the mechanism of inhibition by ifenprodil. Nature 534, 63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliaferro JM, Vidaki M, Oliveira R, Olson S, Zhan L, Saxena T, Wang ET, Graveley BR, Gertler FB, Swanson MS, Burge CB, 2016. Distal Alternative Last Exons Localize mRNAs to Neural Projections. Mol Cell 61, 821–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team RC, 2018. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L, 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traunmuller L, Gomez AM, Nguyen TM, Scheiffele P, 2016. Control of neuronal synapse specification by a highly dedicated alternative splicing program. Science 352, 982–986. [DOI] [PubMed] [Google Scholar]
- Vuong CK, Black DL, Zheng S, 2016. The neurogenetics of alternative splicing. Nat Rev Neurosci 17, 265–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warden AS, Mayfield RD, 2017. Gene expression profiling in the human alcoholic brain. Neuropharmacology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe SA, Workman ER, Heaney CF, Niere F, Namjoshi S, Cacheaux LP, Farris SP, Drew MR, Zemelman BV, Harris RA, Raab-Graham KF, 2016. FMRP regulates an ethanol-dependent shift in GABABR function and expression with rapid antidepressant properties. Nat Commun 7, 12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman ER, Haddick PC, Bush K, Dilly GA, Niere F, Zemelman BV, Raab-Graham KF, 2015. Rapid antidepressants stimulate the decoupling of GABA receptors from GIRK/Kir3 channels through increased protein stability of 14–3-3eta. Mol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman ER, Niere F, Raab-Graham KF, 2013. mTORC1-dependent protein synthesis underlying rapid antidepressant effect requires GABABR signaling. Neuropharmacology 73, 192–203. [DOI] [PubMed] [Google Scholar]
- Zarate CA Jr., Singh JB, Carlson PJ, Brutsche NE, Ameli R, Luckenbaugh DA, Charney DS, Manji HK, 2006. A randomized trial of an N-methyl-D-aspartate antagonist in treatment-resistant major depression. Arch Gen Psychiatry 63, 856–864. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Ren H, Dwyer DS, Peoples RW, 2015. Different sites of alcohol action in the NMDA receptor GluN2A and GluN2B subunits. Neuropharmacology 97, 240–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Yuan Q, Mash DC, Goldman D, 2011. Substance-specific and shared transcription and epigenetic changes in the human hippocampus chronically exposed to cocaine and alcohol. Proc Natl Acad Sci U S A 108, 6626–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Stein RA, Yoshioka C, Lee CH, Goehring A, McHaourab HS, Gouaux E, 2016. Mechanism of NMDA Receptor Inhibition and Activation. Cell 165, 704–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Stroebel D, Yao CA, Taly A, Paoletti P, 2013. Allosteric signaling and dynamics of the clamshell-like NMDA receptor GluN1 N-terminal domain. Nat Struct Mol Biol 20, 477–485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data is publicly available at SRA Accession numbers TBD.