ABSTRACT
Prokaryotic CRISPR-Cas adaptive immune systems rely on small non-coding RNAs derived from CRISPR loci to recognize and destroy complementary nucleic acids. However, the mechanism of Type IV CRISPR RNA (crRNA) biogenesis is poorly understood. To dissect the mechanism of Type IV CRISPR RNA biogenesis, we determined the x-ray crystal structure of the putative Type IV CRISPR associated endoribonuclease Cas6 from Mahella australiensis (Ma Cas6-IV) and characterized its enzymatic activity with RNA cleavage assays. We show that Ma Cas6-IV specifically cleaves Type IV crRNA repeats at the 3ʹ side of a predicted stem loop, with a metal-independent, single-turnover mechanism that relies on a histidine and a tyrosine located within the putative endonuclease active site. Structure and sequence alignments with Cas6 orthologs reveal that although Ma Cas6-IV shares little sequence homology with other Cas6 proteins, all share common structural features that bind distinct crRNA repeat sequences. This analysis of Type IV crRNA biogenesis provides a structural and biochemical framework for understanding the similarities and differences of crRNA biogenesis across multi-subunit Class 1 CRISPR immune systems.
KEYWORDS: CRISPR, Cas6, Csf5, Type IV CRISPR, X-ray crystallography, RNA, endonuclease, prokaryotic immunity, Mahella australiensis
Introduction
Bacteria and archaea use small RNAs derived from CRISPR (Clustered Regularly Interspaced Short Palindromic Repeat) loci to guide CRISPR associated (Cas) proteins to complementary targets such as invasive phage and plasmid DNA [1–8]. Once bound, crRNA-guided ribonucleoprotein complexes undergo conformational rearrangements that activate trans- or cis-acting nucleases to degrade bound targets [9]. Thus, a critical step of prokaryotic crRNA-mediated immunity is crRNA biogenesis, where long, CRISPR-derived transcripts are processed into small crRNA guides.
Although crRNA biogenesis is a fundamental process of all known CRISPR adaptive immune systems, the proteins and mechanisms involved vary across the two CRISPR system classes (1 and 2), six types (I, II, III, IV, V and VI), and more than thirty-three distinct subtypes [10–13]. For example, some single-subunit Class 2 proteins rely on RNase III to cleave duplexed RNAs formed from the base pairing of crRNA repeats with a second trans-activating CRISPR RNA (e.g. Type II-A, and V-B) [14,15], while other Class 2 systems process crRNAs with endonuclease activities that reside within single subunit effector proteins (types V-A, and VI) [16,17].
Most multi-subunit Class 1 systems process crRNAs with CRISPR associated endonucleases, called Cas6, which share conserved structural motifs that bind crRNAs [11]. In general, Cas6 enzymes use a metal-ion-independent mechanism to cleave crRNAs on the 3ʹ-side of stem-loops formed within the palindromic CRISPR repeat [1,18]. Cleavage is generally catalyzed by stabilizing nucleophilic attack from the 2ʹ hydroxyl located upstream from the scissile phosphate [19–22]. In spite of these similarities, Cas6 amino acid sequences are remarkably diverse [23], and several structural and mechanistic differences have been observed [11]. For example, often a histidine is used to catalyze cleavage [20,23–27], but other residues, such as lysine, have been shown to catalyze the reaction when histidine is not present (e.g. subtype I-A) [28,29]. Additionally, distinct Cas6 proteins associate differently with processed crRNAs after cleavage. In subtypes I-B, I-E, I-D and I-F, Cas6 makes structural and base specific interactions with the stable stem-loop formed by the palindromic CRISPR repeat and typically stays bound after cleavage to form a component of the multi-subunit interference complex [19,21,22,24,25,30–33]. In contrast, the repeats of subtypes I-A, III-A, and III-B are less stable, allowing Cas6 to dissociate from the processed crRNA and to perform multi-turnover crRNA cleavage [26,28,29,34].
Type IV CRISPR systems are categorized as Class 1 as they are predicted to form multi-subunit crRNA-guided complexes [6,10,13]. However, the enzymatic functions of Type IV systems remain largely unknown, including the mechanisms of crRNA biogenesis. All Type IV systems contain genes predicted to encode a multi-subunit complex consisting of a large-subunit (csf1, cas8-like), backbone (csf2, cas7-like), and tail (csf3, cas5-like) [10]. Type IV-A systems contain additional genes that encode for a putative helicase (dinG) and a cas6 endonuclease, while Type IV-B systems lack these accessory genes, and interestingly, often lack a CRISPR locus. Distinct Type IV-A systems contain diverse cas6 gene sequences, including genes designated as cas6e and cas6f (cas6 sequences observed in subtypes I-E and I-F), and a Type IV-specific cas6-like gene called csf5 [13].
The presence of cas6 homologs suggests that Type IV-A systems process their own crRNAs through a Cas6-mediated mechanism. Supporting this prediction, it was recently shown that in vivo assembly of a Type IV-A crRNA-protein complex required the Cas6-homolog Csf5 [35]. Independently purified Csf5 remained bound to a processed crRNA, suggesting Csf5 mediates single-turnover crRNA cleavage. However, Csf5 was unstable without a co-expressed crRNA, therefore, it could not be purified in its apo form and a mechanistic analysis of Csf5 catalytic activity was not assayed. Thus, a need remains for a mechanistic description of Type IV crRNA processing.
To better understand the mechanism of crRNA biogenesis in Type IV CRISPR systems we recombinantly expressed and purified the apo Type IV Cas6 protein from the thermophilic anaerobe Mahella australiensis (Ma Cas6-IV) [36]. The genome of M. australiensis harbors putative Type I-B and Type IV-A CRISPR systems at two distinct loci (Figure 1(a)) each with distinct CRISPR repeat sequences which we have designated CRISPR I and CRISPR IV, respectively. We analyzed the Ma Cas6-IV-mediated processing of Type IV crRNA repeats with cleavage assays and identified a putative nuclease active site by determining the structure of Ma Cas6-IV. Additionally we show that the crRNA is cleaved on the 3ʹ side of the predicted stem-loop structure, with nucleophilic attack on the scissile phosphate coming from the 2ʹ hydroxyl of base G22 of the repeat. These results provide a biochemical analysis of Type IV crRNA biogenesis and suggest that although various mechanisms exist, Cas6-mediated metal-independent processing of crRNA is a conserved process across diverse Class 1 systems.
Figure 1.
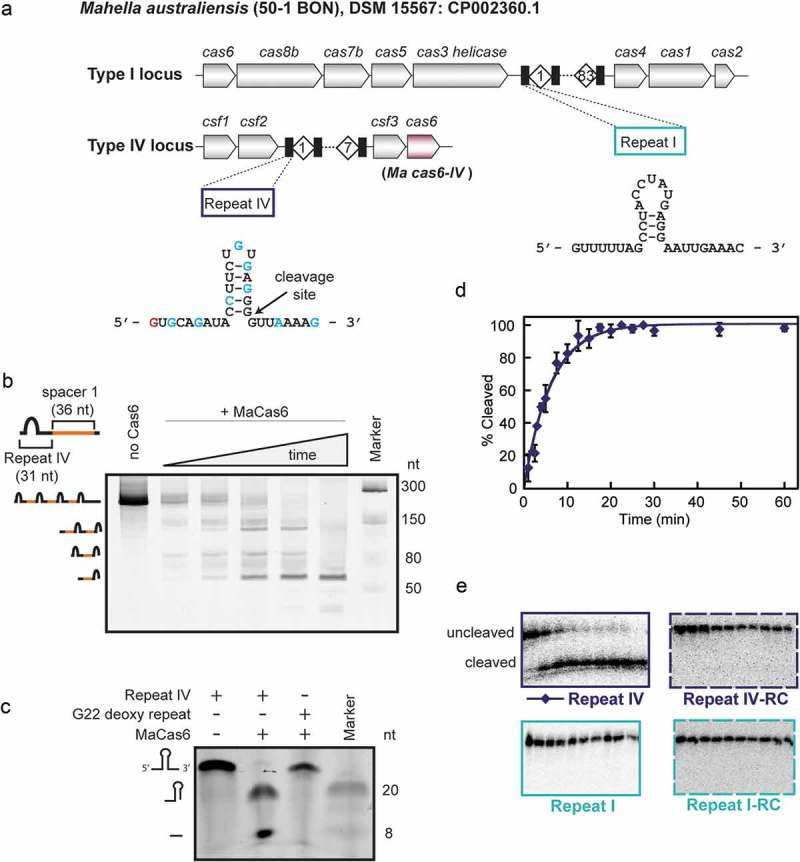
The Type IV Cas6 from M. australiensis (Ma Cas6-IV) cleaves the CRISPR repeat of the Type IV associated CRISPR. (a) M. australiensis contains a Type I CRISPR system and a Type IV CRISPR system, each with a distinct repeat sequence designated as Repeat I and Repeat IV. Nucleotides highlighted in blue indicate a purine-purine or pyrimidine-pyrimidine shift in sequence. Bases colored red indicate broader mutations. (b) Ma Cas6-IV-mediated processing of a pre-crRNA composed of four Repeat IV sequences interspersed with the first spacer sequence from the M. australiensis Type IV CRISPR. (c) SYBR Gold stained gel of the 30 nt long Repeat IV and the G22 2ʹ-deoxy repeat incubated with Ma Cas6-IV. The G22 2ʹ-deoxy repeat remains uncleaved in the presence of Ma Cas6-IV (d) Fit to cleavage data of a single radiolabeled repeat by Ma Cas6-IV. Data for the Repeat IV cleavage were fit as described in [24]. Error bars denote standard deviation between two or more experiments. (e) Ma Cas6-IV-mediated cleavage trials of 5ʹ-32P-end labeled Repeat IV, Repeat I, and their reverse complements (Repeat IV-RC, Repeat I-RC). Data were not fit for Repeat I or the RC sequences because no cleavage was observed.
Results
Ma Cas6-IV processes pre-crRNAs
To investigate the mechanism of Type IV crRNA biogenesis, we recombinantly expressed and purified N-terminally His-tagged Cas6 protein from the M. australiensis Type IV CRISPR system (Ma Cas6-IV) (Figure 1(a) and Supplemental Figure S1). To determine if Ma Cas6-IV processes crRNAs, we performed an RNA cleavage assay with a pre-crRNA transcribed in vitro with the same directionality as the Type IV cas genes. The pre-crRNA was designed to contain four Type IV repeat sequences, each 31 nucleotides in length, and three 36 nucleotide spacer sequences, each identical to the first spacer observed in the native CRISPR IV (Supplemental Figure S2). The first three of eight repeats observed in the Mahella CRISPR are identical, and this sequence was used in the pre-crRNA design (Supplemental Figure S2). Switches in purine or pyrimidine identity occur within the repeat at positions along the 5ʹ and 3ʹ arms and stem of the repeat hairpin. We note that the base switches at the base of the stem would not disrupt base pairing if both Watson Crick and G-U wobble base pairs are acceptable for binding, suggesting the stem of the repeat may be recognized primarily through shape rather than base-specific interactions (Figure 1(a) and Supplemental Figure S2). Other switches in the arms and loop of the hairpin likewise suggest that those positions are recognized through shape, or are not necessary at all for binding.
Our initial nuclease assay showed that Ma Cas6-IV cleaves the pre-crRNA at regular intervals, producing several species of smaller RNAs with differences in length of ~ 67 nucleotides (the length of the repeat + spacer) (Figure 1(b)). This result indicated that Ma Cas6-IV processes the pre-crRNA into smaller crRNAs with a mechanism similar to other Cas6 nucleases that bind and cleave conserved features of the palindromic CRISPR repeat [11]. To evaluate this activity in more detail, we showed that Ma Cas6-IV cleaves a radio- or fluorescein-labeled 30-nucleotide RNA identical in sequence to the first crRNA repeat of CRISPR IV (Figure 1(c–e)). As M. australiensis is a thermophile [36], we were not surprised to observe that optimal Ma Cas6-IV cleavage occurs at temperatures around 50°C, indicating the cleavage mechanism is thermostable (Supplemental Figure S3). Additionally, Ma Cas6-IV was able to cleave crRNAs in the presence of high concentrations of the metal chelator EDTA (see Methods section), indicating that the cleavage mechanism is metal-ion-independent, consistent with other Class 1 crRNA processing mechanisms [1,18,21,28].
To determine where on the CRISPR repeat Ma Cas6-IV cuts, a small RNA marker was run alongside a cleaved repeat stained with SYBR gold (Figure 1(c)). We observed that the smaller of two distinct cleavage products ran next to the 8 nt band, while the larger ran next to a 20 nt band, indicating Ma Cas6-IV cleaves the repeat asymmetrically on the 3ʹ-side of the repeat near position 22. To confirm the location of cleavage we incubated Ma Cas6-IV with a repeat containing a 2ʹ-deoxy sugar at nucleotide position G22 (G22 2ʹ-deoxy repeat). Ma Cas6-IV is unable to cleave the G22 2ʹ-deoxy repeat indicating the scissile phosphate resides on nucleotide 23 of the repeat, and cleavage is mediated through nucleophilic attack of the 2ʹ hydroxyl of position G22.
Previous analysis of Cas6 endonucleases from the thermophile Thermus thermophilus (TtCas6A and TtCas6B) showed that some Cas6 endonucleases encoded in organisms with multiple CRISPR loci are capable of cleaving more than one CRISPR repeat sequence [24]. As M. australiensis contains two different CRISPR loci in its genome, each with a distinct CRISPR repeat sequence, Ma Cas6-IV could potentially cleave both Type IV and Type I repeats (Figure 1(a)). To measure the sequence specificity of Ma Cas6-IV, in addition to the Type IV repeat, we tested for nuclease activity against the Type I repeat, the Type I repeat-reverse complement, and the Type IV repeat-reverse complement (Figure 1(c)). Unlike the Cas6 enzymes from Thermus thermophilus that cleave multiple repeat sequences, Ma Cas6-IV cleaves only the Type IV crRNA, indicating Ma Cas6-IV is specific for the Type IV repeat sequence.
Structure of Ma Cas6-IV
Our initial analysis showed that Ma Cas6-IV specifically cleaves Type IV crRNA repeats, but mechanistic details, such as the turnover kinetics for this enzyme and the features important for binding and cleaving the crRNA, remained unknown. Sequence alignments of Ma Cas6-IV with Type I and Type III Cas6 orthologs, as well as the recently investigated Type IV-specific Csf5 protein revealed low sequence identity (~ 16%) (Supplemental Table 1), including in the region of the putative active site. Thus, the mechanistic details of Ma Cas6-IV could not be deduced through simple amino acid sequence alignments with previously characterized proteins.
To better understand Ma Cas6-IV function, we determined the x-ray crystal structure of apo Ma Cas6-IV at 1.76 Å resolution (Figure 2 and Table 1). Crystals of N-terminally His-tagged Ma Cas6-IV were grown using sitting-drop vapor diffusion and formed in a hexagonal space group (P61) (Supplemental Figure S4). Our initial attempts to solve the crystal structure using existing Cas6 structures as molecular replacement models failed. We hypothesized that this was due to structural differences between Ma Cas6-IV and the available Cas6 models, so we instead solved the structure using Single-wavelength Anomalous Dispersion methods with crystals soaked overnight in Potassium Tetrachloroplatinate (See Methods and Table 1).
Figure 2.
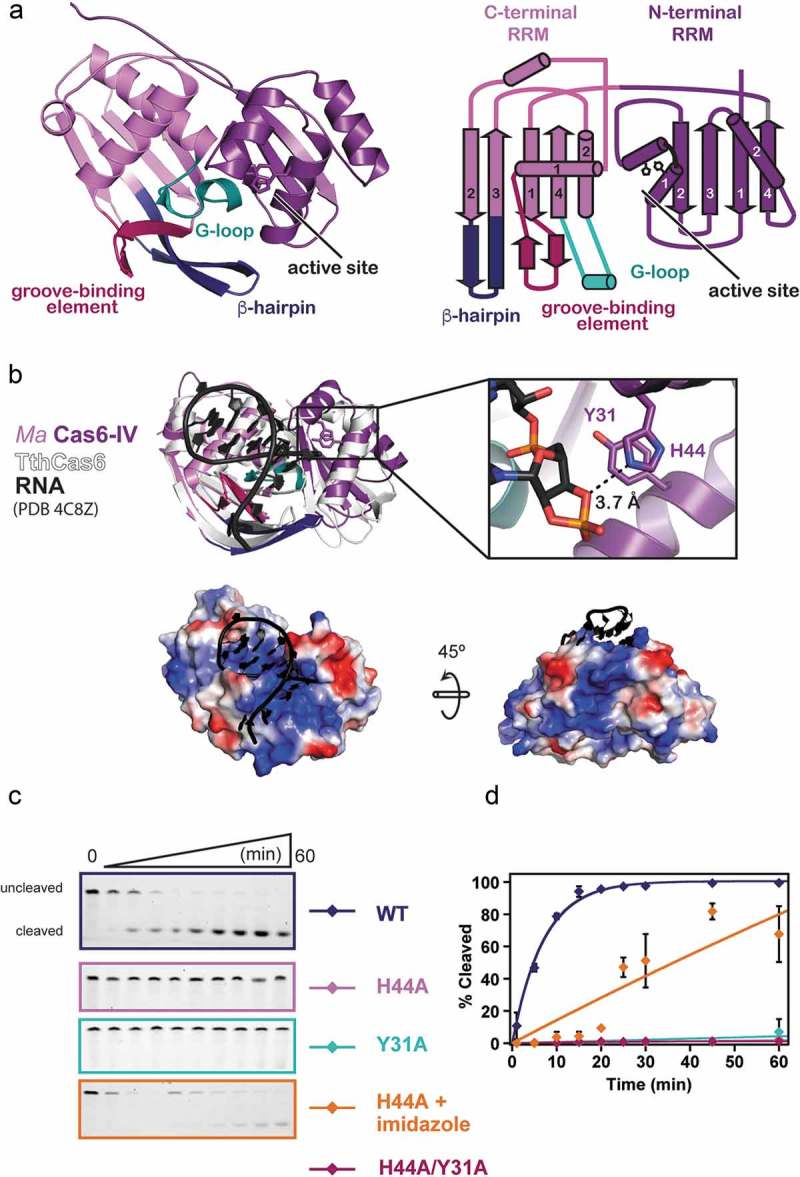
Structure of apo Ma Cas6-IV and identification of the active site residues, His44 and Tyr31. (a) Ribbon model (left) and topology diagram (right) of the Ma Cas6-IV structure. The two RRM domains, C-terminal structural motifs involved in binding the crRNA, and putative active site are indicated. (b) Alignment of apo Ma Cas6-IV to RNA-bound TthCas6 (PDB:4C8Z) with a zoomed in view of the Ma Cas6-IV active site containing His44 and Tyr31 residues. RMSD of the alignment is 1.26 Å over 95 out of 257 Cα carbons. The bottom panels show surface renditions of Ma Cas6-IV aligned with the RNA of PDB 4C8Z. Predicted positively charged surface is colored blue and negatively charged surface is colored red. (c) Cleavage assays of Repeat IV by predicted active site mutants of Ma Cas6-IV (left) with cleavage percentages fit to a pseudo-first order rate equation (right). Cleavage activity of H44A Ma Cas6-IV in the presence of 500 mM imidazole is also shown.
Table 1.
Data collection and refinement statistics.
| Dataset | Native | SAD (K2• Pt Cl4 – soak) |
|---|---|---|
| Data collection | ||
| Beamline Space group |
SSRL 12–2 P61 |
SSRL 12–2 P61 |
| Cell dimensions | ||
| a, b, c (Å) | 85.5, 85.5, 142.6 | 85.9, 85.9, 144.5 |
| α, β, γ (deg) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Wavelength (Å) | 1.55 | 1.0718 |
| Resolution (Å) | 40–1.76 (1.82–1.76)* | 40–2.95 (3.06–2.95) |
| Rmerge(%) | 5.3 (72.7) | 6.5 (22.3) |
| CC1/2 | 0.995 (0.736) | 0.994 (0.941) |
| I/σI | 25.3 (2.2) | 15.4 (3.5) |
| Observations | 350,790 (23,595) | 95,034 (5484) |
| Unique reflections | 58,149 (5618) | 24,773 (2359) |
| Multiplicity | 6.0 (4.2) | 3.8 (2.3) |
| Completeness (%) | 99.6 (96.3) | 99.3 (94.9) |
| Refinement | ||
| Resolution† (Å) | 40–1.76 (1.82–1.76) | |
| No. reflections | 148,794 | |
| Rwork/Rfree (%) | 17.1/19.6 | |
| No. atoms | 4202 | |
| Protein Water Ligands |
3861 246 95 |
|
| B-factors | ||
| mean | 43.84 | |
| Protein | 42.84 | |
| Water | 50.51 | |
| Ligands | 78.20 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.008 | |
| Bond angles (deg) Ramachandran Favored (%) Allowed (%) Outliers (%) Clashscore |
1.20 98.72 1.28 0 9.30 |
|
*Values in parentheses are for highest-resolution shell.
†Resolution limits use the criterion of I/σI > 2.0
The structure of Ma Cas6-IV reveals that two protein chains reside in the asymmetric unit. The electron density is well resolved and continuous, allowing all 229 amino acids of both proteins to be modeled along with 7 additional nucleotides of the histidine affinity tag (Supplemental Figure S4). The monomers are arranged with two-fold symmetry and are superimposable with an R.M.S.D. < 0.29 Å over 227 of 237 Cα carbons. Although the structure indicates Ma Cas6-IV is capable of forming a dimer, size exclusion profiles indicate it purifies as a monomer (Supplemental Figure 1). Additionally, analysis with the PDBePISA server reveals the dimer interface is 1067 Å2 with a Complex Formation Significance Score (CSS) of 0.066 [37] (Supplemental Figure S5). The low CSS score (scores range from 0 to 1) implies the interface is not significant for complex formation. These results, along with the observation that the dimer conformation is fundamentally different from other Cas6 dimers shown to be functionally relevant [24,29], suggest the observed dimer may solely be the result of crystal packing.
Each monomer is composed of two modified RRM (RNA recognition motif) domains, also known as ferredoxin-like folds (Figure 2(a)). Canonical RRM domains adopt a β1, α1, β2, β3, α2, β4 fold, where the two helices pack against the concave face of an anti-parallel beta-sheet formed by the four beta-strands [38]. The two RRM domains of Ma Cas6-IV express this secondary structure, but in each domain the first alpha-helix has been modified into a helix-turn-helix. Additionally, the C-terminal RRM contains extended features between β1 and α1, β2 and β3, and α2 and β4. To identify the function of these motifs we aligned our structure with the available Cas6 structures in the PDB using the secondary structure matching (SSM) tool in the molecular modeling program Coot (Figure 2, Supplemental Table 1, and Supplemental Figure S6) [39]. Our structural alignments reveal that the C-terminal RRM extensions align with conserved features important for binding the crRNA and positioning the scissile phosphate into the nuclease active site. These motifs have been previously described as the groove-binding element (GBE), the β-hairpin, and the glycine-rich loop (G-loop) (Figure 2(a)) [11]. In Ma Cas6-IV the GBE forms a β-hairpin. In RNA-bound structures of other Cas6 proteins, the GBE typically makes sequence and shape specific contacts within the major groove of the crRNA stem-loop [21,24,25,40]. An alignment of 184 Cα carbons with the RNA-bound TthCas6A protein from Thermus thermophilus (PDB 4C8Z, RMSD 2.45 Å) positions the GBE from Ma Cas6-IV into the major groove of the bound RNA, suggesting that the GBE of Ma Cas6-IV binds the major groove of the crRNA stem-loop (Figure 2(b) and Supplemental Figure S7). The β-hairpin motif occurs in the majority of Cas6 enzymes, and typically contacts the base of the crRNA stem-loop and positions the scissile phosphate into the active site [21,25,28]. Our alignment with the RNA-bound TthCas6A shows the tip of the Ma Cas6-IV β-hairpin pointing away from the scissile phosphate, suggesting this feature may undergo a conformational change upon binding the crRNA [25]. The G-loop, in the Ma Cas6-IV structure resides between α2 and β4 and forms a small loop-helix-loop structure. In other Cas6 enzymes, this motif is involved in binding the crRNA through ionic interactions along the phosphate backbone [23–25,41]. The G-loop in Ma Cas6-IV contains two lysine residues that help form a large positively charged patch on the surface of the protein (Figure 2(b)). In our alignment, the crRNA of the RNA-bound structure is positioned on top of this positively charged surface, suggesting the lysines within the G-loop of Ma Cas6-IV may form ionic interactions with the negatively charged backbone of the CRISPR IV crRNA. Interestingly, this positive patch extends towards the back of the C-terminal RRM, suggesting a possible trajectory of the crRNA that wraps around the protein as seen in Type III-A Cas6 enzymes (Figure 2(b) bottom and Supplemental Figure S7) [26,28,34].
His44 and Tyr31 are catalytic residues of the Ma Cas6-IV active site
Cas6 nuclease active sites are typically located in the cleft between the two RRM folds [11] but, without a substrate bound, our structure of apo Ma Cas6-IV was not sufficient to determine the location of the Ma Cas6-IV active site. In our alignment with TthCas6A bound to RNA, Ma Cas6-IV residues His44 and Tyr31 are located within 4 Å of the aligned scissile phosphate, suggesting they could be involved in catalysis. Indeed, in the TthCas6A protein, a histidine in this region was shown to catalyze cleavage [24], and histidine residues in this region have been shown to have a role in catalysis of several other Cas6 enzymes [20,23,25–27]. To determine if the Ma Cas6-IV residues His44 and Tyr31 are responsible for catalysis of RNA cleavage, we created alanine mutants of each residue, alone and in tandem, and assayed their ability to cleave Repeat IV. Mutation of either residue to alanine severely reduced cleavage (Figure 2(c,d)). Previous analyses of Cas6 catalytic activity showed that, in some cases, nuclease activity lost upon histidine mutation could be restored with the addition of imidazole [24]. To better understand how the histidine catalyzes cleavage, we added 500 mM imidazole to our H44A Ma Cas6-IV mutant cleavage reaction. Under these conditions, H44A Ma Cas6-IV partially regained its cleavage activity (Figure 2(c,d)), suggesting the imidazole ring compensates for the histidine mutant, consistent with a model where the active site histidine plays a role in catalysis [20,42].
Ma Cas6-IV is a single-turnover enzyme to the CRISPR repeat RNA substrate
Many Cas6 enzymes exhibit single-turnover characteristics, remaining bound to their cleaved crRNA products, while others dissociate from cleaved crRNAs allowing for multi-turnover activity [11]. To determine the turnover number of the Ma Cas6-IV enzyme, we performed nuclease assays with constant concentrations of Repeat IV and varying concentrations of Ma Cas6-IV corresponding to the following ratios of Cas6 to RNA substrate; 2:1, 1:1, 1:2, 1:4, and 1:8. In both the 2:1 and 1:1 conditions nearly all of the RNA substrate was cleaved. In each successive condition, a cleavage amplitude was reached where less total RNA was cleaved than in the previous condition (Figure 3). When excess substrate is present, cleavage does not go to completion, but rather corresponds to the cleavage of one RNA molecule per Cas6 active site. Thus, we conclude that Ma Cas6-IV is a single-turnover enzyme.
Figure 3.
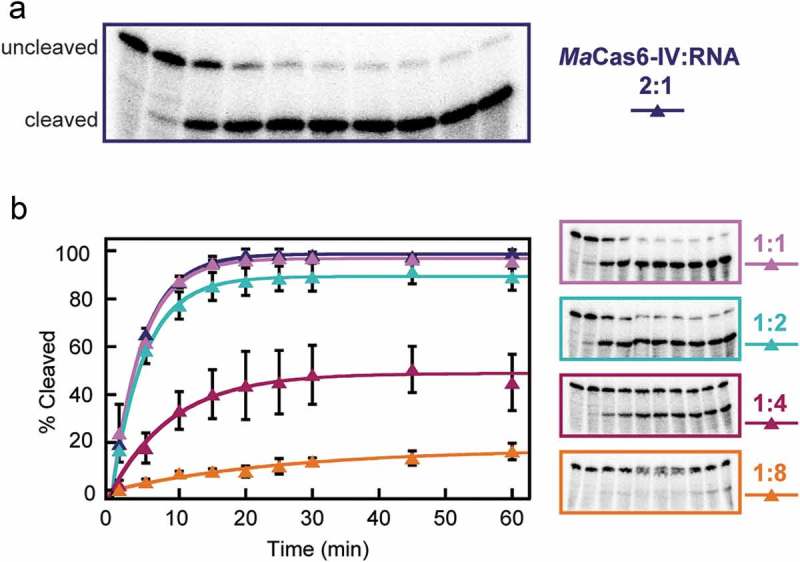
Ma Cas6-IV exhibits single-turnover characteristics for Repeat IV. (a) Cleavage of Repeat 4 by Ma Cas6-IV with a Cas6:Repeat IV ratio of 2:1. (b) Isotherms and electrophoresis gels of additional ratios of Cas6:Repeat are shown. The data were fit as described in [24]. Error bars denote standard deviation between three experiments.
Structural comparison of Ma Cas6-IV to Csf5
Although Type IV systems are similar in gene arrangement and identity, Type IV Cas6 proteins are unexpectedly diverse in sequence (Figure 4 and Supplemental Table 1). Type IV systems contain several Cas6 variants, including sequences that resemble Cas6e and Cas6f, and others that encode a Type IV-specific Cas6 homolog called Csf5 [13]. Structural comparisons of Ma Cas6-IV with Csf5 from Aromatoleum aromaticum reveal they both contain the dual RRM domain scaffold generally observed in Cas6 proteins (Figure 4(a)). The C-terminal RRM domains of both enzymes contain the motifs that bind crRNA (GBE, β-hairpin, and G-loop), but the C-terminal domain of Csf5 differs from Ma Cas6-IV in that the second alpha helix (α2) of the canonical RRM fold is absent (Supplemental Figure S6). In both Csf5 and Ma Cas6-IV the α1 helices of the N-terminal RRM domains have been replaced with helix-turn-helix motifs that house putative active-site residues (Figure 4(b)). However, instead of the small loop sequence observed in Ma Cas6-IV that connects the helix-loop-helix to β2, Csf5 has an insertion of ~40 amino acids called the α-helical finger domain that contains two additional helices. One of these helices interacts with the minor groove of the crRNA stem-loop, providing additional contacts for binding the crRNA that may provide additional specificity toward Type IV crRNA repeats.
Figure 4.
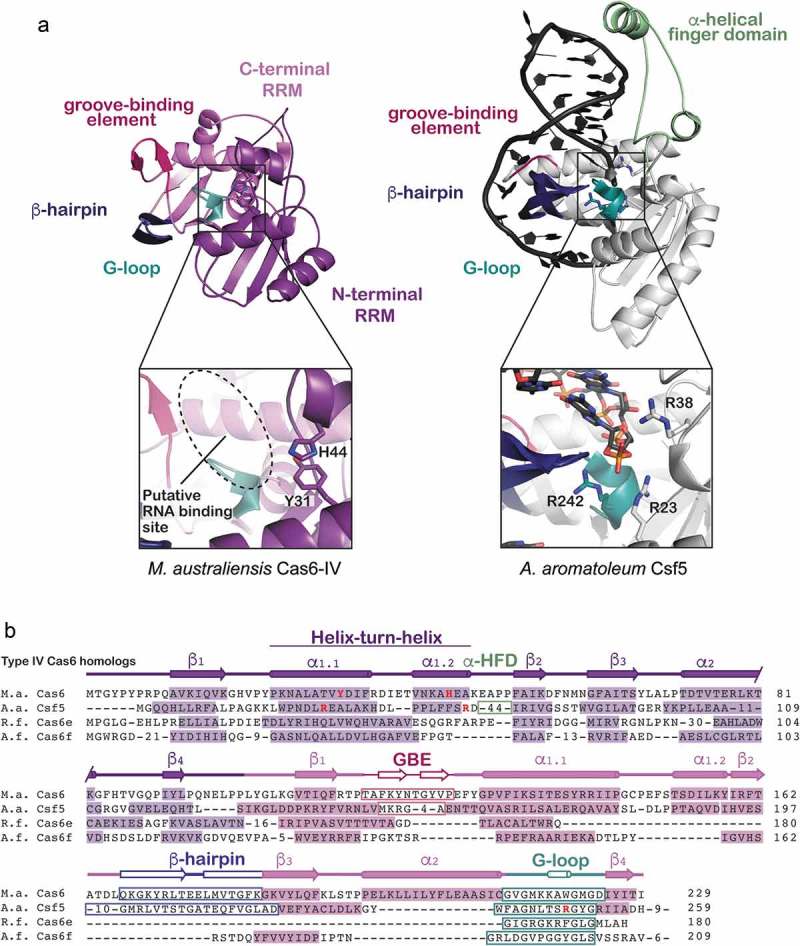
Structure and sequence alignments of Ma Cas6-IV with other Type IV RNA endonucleases. (a) A structural comparison of Ma Cas6-IV with the Cas6-homolog Csf5 from Aromatoleum aromaticum (PDB 6H9I). Features involved in binding crRNAs are indicated. The Csf5 protein contains a large insert called the alpha-helical finger domain (light green) that is not observed in Ma Cas6-IV. Residues predicted to activate cleavage of the crRNA are indicated in the inset below. (b) Sequence alignment of Ma Cas6-IV with other RNA endonucleases observed in Type IV systems. The N- and C-terminal RRM secondary structure elements are indicated, as well as features which bind crRNA, including the groove-binding element (GBE), beta-hairpin, and glycine-rich loop (G-loop). The alpha-helical finger domain (α-HFD) insert of Csf5 is also indicated. Active site residues of Ma Cas6-IV and Csf5 are bolded in red. Cas6e and Cas6f sequences are noticeably shorter than Ma Cas6-IV and Csf5, lacking large portions of the C-terminal RRM.
Interestingly, despite structural similarity the mechanisms of cleavage utilized by these protein homologs are diverse. Both Csf5 and Ma Cas6-IV contain a histidine in the N-terminal RRM at the same sequence position (H44). In Ma Cas6-IV the histidine resides in the helix-turn-helix and is within H-bonding distance of the scissile phosphate (Supplemental Figure S7). However, in Csf5 this histidine resides within the 40 amino acid insert several Ångstroms away from the scissile phosphate, suggesting it does not participate in nuclease activity. In support of this hypothesis, an H44A mutant of Csf5 did not impair cleavage [35]. Rather, mutation of arginine residues located on the helix-turn-helix and the G-loop (R23A, R38A, R242A) impaired cleavage (Figure 4(a)). Notably several of these arginines are located in similar positions to the active site residues of Ma Cas6-IV (His44 and Tyr31) (Figure 4(b) and Supplemental Figure S7), revealing that although these Type IV crRNA processing enzymes are diverse in sequence, they rely on similar structural themes to bind and cleave crRNA substrates.
Discussion
Because Type IV CRISPR systems have just recently been discovered, the biological function of these systems remains largely unknown. We hypothesized that pre-crRNA transcripts from adjacent CRISPR loci would be processed into small crRNA-guides to direct a biological function. To understand the feasibility and mechanism of Type IV crRNA biogenesis, we characterized the structure and activity of a Type IV-A Cas6 crRNA endonuclease from the microbe Mahella australiensis (Ma Cas6-IV). We observed that Ma Cas6-IV processes pre-crRNAs in vitro by cleaving within the CRISPR repeat sequence, through a mechanism that is metal-independent and facilitated by residues His44 and Tyr31. Analysis of repeat cleavage products suggests that the repeat is cleaved asymmetrically on the 3ʹ side of the predicted stem loop with the 5ʹ-phosphate of the 23rd nucleotide as the scissile phosphate. We show that Ma Cas6-IV cleaves crRNAs with single-turnover kinetics, suggesting the enzyme stays bound after cleavage to one side of the repeat. Binding assays and crystal structures show that in several Type I systems Cas6 remains bound to its crRNA product, forming part of the interference complex that uses the crRNA as a guide to bind complementary targets [30–33]. We hypothesize that the single-turnover kinetics we observe are due to Ma Cas6-IV remaining bound to the 5ʹ side of its cleaved crRNA product, suggesting Ma Cas6-IV may be a component of a multi-subunit complex that assembles on the processed crRNA. Alternatively Ma Cas6-IV could undergo a conformational change upon release of both sides of the crRNA that does not allow for additional repeats to be cleaved. Further work is needed to determine how Ma Cas6-IV interacts with the repeat cleavage products and whether it forms a part of the predicted multi-subunit complex.
Phylogenetic work previously predicted that multi-subunit complexes would form in Type IV-A systems and would be composed of Csf2 (Cas7-like backbone protein), Csf3 (Cas5-like tail protein) and the Type IV signature protein Csf1 [6,10,13]. Supporting this prediction, Özcan and colleagues recombinantly expressed and isolated a Type IV-A complex containing these exact subunits and the Cas6-homolog Csf5 [35]. This recent work, coupled with our observations, suggest that in Type IV-A systems Cas6 mediates cleavage of the pre-crRNA, creating small crRNA guides that are then bound by Csf1, Csf2, and Csf3 to form a multi-subunit crRNA-guided complex. It appears that in some cases Cas6 forms a part of the multi-subunit complex, but it remains unknown if this is a conserved feature of all Type IV-A systems.
The function of Type IV systems also remains unknown. A recent phylogenetic analysis reported that many Type IV system spacers are complementary to known phage sequences [6], suggesting the system acts as an immune system. However, analysis of the spacers of Mahella australiensis using the CRISPRTarget tool [43] reported no plausible matches, with only three spacer sequences retaining some complementarity to phage sequences, but with multiple mismatches (Supplemental Figure S8). Even if it were clear that Type IV systems target phage or plasmid sequences the function of such targeting would remain unknown, as no nuclease domains have yet been clearly identified in any of the Type IV systems. Thus, a more detailed analysis of the structure and function of these systems is needed.
In contrast to Type IV-A systems, Type IV-B systems do not encode Cas6 enzymes and are not associated with CRISPR loci. However, Type IV-B systems do encode the Csf1, Csf2, and Csf3 proteins, suggesting they may be capable of forming a complex on processed crRNA guides. It has been proposed that Type IV-B systems may utilize crRNAs generated from other CRISPR systems to form complexes and may have a non-defense function [44]. However, it has yet to be shown that Type IV-B proteins can form a complex on a crRNA.
Cas6-like RNA endonucleases associated with Type IV-A systems are quite diverse in amino acid sequence. A small group of systems encode for the Type IV-specific Csf5 protein, but others encode variants of Cas6 that are dissimilar in sequence but retain the dual RRM architecture observed in Cas6 proteins (Figure 4(b)). The wide range of Cas6 sequences across Type IV-A systems suggests that perhaps Cas6 enzymes were acquired by Type IV-B systems at different times in evolutionary history. We postulate that such fusions also included a CRISPR locus, providing a crRNA-guide upon which the Csf1, Csf2, and Csf3 proteins could assemble. Interestingly, the protein for which Ma Cas6-IV is most similar in structure and sequence is an orphaned Cas6-homolog from Thermus thermophilus (TthCas6A) (Figure 2, Supplemental Figure S2, and Supplemental Table 1). TthCas6A is not found within a CRISPR system but is adjacent to a CRISPR locus, suggesting it could combine with other systems in tandem with its CRISPR locus. We speculate that perhaps a similar orphaned Cas6 sequence combined with a Type IV-B system to produce the Type IV-A system of Mahella australiensis.
This work and a recent study on the Type IV system from A. aromaticum [35] have displayed that Type IV-A systems process crRNA guides through a Cas6-mediated mechanism. Additionally it has been shown that multi-protein complexes assemble onto these crRNAs [35]. However, the biological function of both Type IV-A and Type IV-B systems remains unknown and awaits additional biochemical, in vivo, and structural studies.
Methods
Expression and purification of Ma Cas6-IV
The full-length M. australiensis Cas6 gene sequence (Uniprot AEE97687.1) was obtained from IDT (Integrated DNA Technologies) as a gBlock. The sequence was PCR amplified with primers containing LIC (Ligation Independent Cloning) overhangs, and was subcloned, using ligation independent cloning, into the 2B-T transfer vector. Ma Cas6-IV mutants were created using the Q5 Site-Directed Mutagenesis kit (New England BioLabs (NEB)).
Recombinant protein expression was induced in BL21 DE3 cells with 0.1 mM of IPTG (isopropyl β-D-1-thiogalactopyranoside) at an optical cell density (O. D. 600 nM) of ~0.3–0.4. After induction, cells were grown at 16°C for 24 hours and then pelleted via centrifugation. Pelleted cells were added at a ratio of 1:8 (g of cells: mL of buffer) to 25mM NaPO4 (pH 7.5), 500mM NaCl, 25mM imidazole, 5% glycerol, 0.01% Triton X-100, 1 mM Tris [2-carboxyethyl] phosphine hydrochloride (TCEP), 0.5 mM phenylmethylsulphonyl fluoride (PMSF), and 0.1 mM lysozyme. Cells were lysed by sonication. Sonicated lysate was clarified via centrifugation. Clarified lysate was placed over a HisTrap FF column (GE Healthcare) and the bound protein was eluted with a high imidazole buffer (25mM NaPO4 (pH 7.5), 500 mM NaCl, 700 mM imidazole, 5% glycerol, 1 mM TCEP) then desalted into 25 mM NaPO4 (pH 7.5), 150 mM NaCl, 1mM TCEP, 5% glycerol with a HiPrep 26/10 desalting column (GE healthcare). Desalted protein was placed over a HiTrap SP FF column (GE healthcare) and eluted with a NaCl gradient. The protein was further purified with a Superdex 200 pg 26/600 column (GE Healthcare) and eluted in 100 mM HEPES (pH 7.5), 150 KCl, 5% glycerol, 1 mM TCEP. The protein was then concentrated to 6–8 mg/ml using Vivaspin centrifugation concentrators. Wild type and mutant protein constructs were expressed and purified with the same protocol.
Generation of RNA substrates
The Ma pre-crRNA was generated using in vitro transcription with the HiScribe T7 Quick kit (NEB) and a plasmid linearized immediately after the encoded Ma CRISPR under control of a T7 promoter. All small RNA oligonucleotides were synthesized by IDT with or without a 3ʹ fluorescein label. Radiolabeled substrates were 5ʹ-end labeled with (γ-32P)-ATP (Perkin Elmer) and T4 polynucleotide kinase (NEB). Labeled RNAs were separated from excess ATP with a MicroSpin G-25 column (GE Healthcare), then gel purified on a 12% denaturing (7M urea) polyacrylamide gel, ethanol precipitated, and recovered in water.
Pre-crRNA cleavage assay
15 µM Cas6 and 0.25 µM Ma pre-crRNA were incubated at 50°C in 20 mM HEPES (pH 7.5), 100 mM KCl, 1 mM β-Mercaptoethanol, and 50 mM EDTA. 10 µl aliquots were removed at indicated timepoints and quenched with 50 µl acid phenol chloroform and briefly centrifuged. Eight µl of the aqueous layer was mixed with formamide RNA loading buffer and resolved on a 12% denaturing (7M urea) polyacrylamide gel with a Low Range ssRNA ladder (NEB). The gel was stained with SYBR Gold Nucleic Acid Gel stain (Invitrogen) and imaged with a ChemiDoc MP Imaging system (Bio-Rad).
Repeat cleavage and turnover assays
2.5 µM Cas6 and 5 nM 5ʹ-end 32P or 1μM 3ʹ-end fluorescein labeled RNA substrate were incubated at 50°C in 20 mM HEPES (pH 7.5), 100 mM KCl, 1 mM β-Mercaptoethanol, and 50 mM EDTA. Time points were collected and resolved on a gel as described for the pre-crRNA cleavage assay. Fluorescein labeled substrates were imaged and quantified with a ChemiDoc MP Imaging system (Bio-Rad). Gels containing 32P labeled substrates were dried, exposed to a phosphor storage screen, and scanned with a Typhoon (GE Healthcare) phosphorimager. Cleaved and uncleaved fractions were quantified using ImageQuant (GE Healthcare) software. All data were fit as described by Niewoehner and colleagues [24]. Briefly, using Kaleidagraph (Synergy Software), the cleavage assay data were fit to the equation:
Reported data is the average of three experiments and error bars represent standard deviations. To recover the cleavage activity of the Ma Cas6-IV H63A mutant, 500 mM imidazole was added to the reaction buffer.
The turnover kinetics of Ma Cas6-IV were tested by performing cleavage assays as described above, except with 1000 nM substrate (5 nM of which was radiolabeled) and varying concentrations of Ma Cas6-IV (2000, 1000, 500, 250, and 125 nM).
Crystal growth and structure determination
Ma Cas6-IV was crystallized at 22°C using the sitting drop vapor diffusion method. Drops were made by mixing equal volumes of protein (6–8 mg/ml in 100 mM HEPES (pH 7.5), 150 KCl, 5% glycerol, 1 mM TCEP) and reservoir solution (0.1 M sodium acetate trihydrate (pH 4.8–5.1) and 1.60–1.75 M ammonium sulphate). Most crystals grew to a size large enough for data collection in about a week.
As initial attempts at solving the crystal structure using molecular replacement with existing models failed, and because we were unable to grow crystals of appreciable size with selenomethionine derivatized protein, we solved the structure by soaking in heavy metals. Crystals were soaked in tetrachloroplatinate overnight and then back soaked into cryo-protectant containing the mother liquor solution and 30% dextrose followed by flash freezing. Native and anomalous diffraction data were collected remotely at SSRL beamline 12–2 with high redundancy and completeness (Table 1). The Phenix tool autosol was used to identify an initial substructure, phase the data, density modify initial maps, and autobuild an initial model [45–49]. Although incomplete, the initial autobuild model was sufficient for molecular replacement for phasing the native data [50]. Iterative rounds of model building and refinement using Coot and Phenix.refine tools produced the final model [39,51].
Structure and sequence alignments
Ma Cas6-IV was aligned with Cas6, and Cas5d models available in the PDB using the SSM (Secondary Structure Matching) tool in Coot [39]. After alignment Coot produces statistics that list the number of Cα carbons aligned, the R.M.S.D. of the alignment, and the percentage of residues aligned that are identical (see Supplemental Table 1.). Alignment of diverse Type IV Cas6 sequences that had structures available (Ma Cas6-IV and Aa Csf5) was done in Coot using SSM. However the Cas6e and Cas6f sequences were aligned manually using secondary structure predicted by the PSIPRED tool as a guide [52].
Funding Statement
This work was supported by the Foundation for the National Institutes of Health [P41GM103393];U.S. Department of Energy [DE-AC02-76SF00515].
Acknowledgments
Research in the Jackson Lab is supported by Utah State University New Faculty Start-up funding from the Department of Chemistry and Biochemistry, the Research and Graduate Studies Office, and the College of Science. The Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory is supported by the U.S. Department of Energy, Contract No. DE-AC02-76SF00515, and the National Institutes of Health NIGMS (P41GM103393).
Disclosure statement
No potential conflict of interest was reported by the authors.
Data deposition
The model coordinates and structure factors for the apo Ma Cas6-IV structure have been deposited in the Protein Data Bank under PDB code: 6NJY.
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Brouns SJJ, Jore MM, Lundgren M, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Barrangou R. Diversity of CRISPR-Cas immune systems and molecular machines. Genome Biol. 2015;16:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wright AV, Nuñez JK, Doudna JA. Biology and applications of CRISPR systems: harnessing Nature’s toolbox for genome engineering. Cell. 2016;164:29–44. [DOI] [PubMed] [Google Scholar]
- [4].Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. [DOI] [PubMed] [Google Scholar]
- [5].Mohanraju P, Makarova KS, Zetsche B, et al. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016;353:aad5147. [DOI] [PubMed] [Google Scholar]
- [6].Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science, New Series 2007;315:1709–1712. [DOI] [PubMed] [Google Scholar]
- [8].Gasiunas G, Sinkunas T, Siksnys V. Molecular mechanisms of CRISPR-mediated microbial immunity. Cell Mol Life Sci. 2014;71:449–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jackson RN, van Erp PB, Sternberg SH, et al. Conformational regulation of CRISPR-associated nucleases. Curr Opin Microbiol. 2017;37:110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Makarova KS, Wolf YI, Koonin EV. Classification and Nomenclature of CRISPR-Cas systems: where from here? The CRISPR J 2018;1:325–336.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hochstrasser ML, Doudna JA. Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends Biochem Sci. 2015;40:58–66. [DOI] [PubMed] [Google Scholar]
- [12].Charpentier E, Richter H, van der Oost J, et al. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity. FEMS Microbiol Rev. 2015;39:428–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Makarova KS, Wolf YI, Alkhnbashi OS, et al. An updated evolutionary classification of CRISPR–cas systems. Nature Rev Microbiol. 2015;13:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Deltcheva E, Chylinski K, Sharma CM, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Shmakov S, Abudayyeh OO, Makarova KS, et al. Discovery and functional characterization of diverse Class 2 CRISPR-Cas systems. Mol Cell. 2015;60:385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Fonfara I, Richter H, Bratovič M, et al. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016;532:517–521. [DOI] [PubMed] [Google Scholar]
- [17].East-Seletsky A, O’Connell MR, Knight SC, et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538:270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Carte J, Wang R, Li H, et al. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Carte J, Pfister NT, Compton MM, et al. Binding and cleavage of CRISPR RNA by Cas6. RNA. 2010;16:2181–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Haurwitz RE, Sternberg SH, Doudna JA. Csy4 relies on an unusual catalytic dyad to position and cleave CRISPR RNA: mechanism of CRISPR RNA cleavage. Embo J. 2012;31:2824–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Haurwitz RE, Jinek M, Wiedenheft B, et al. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sternberg SH, Haurwitz RE, Doudna JA. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA. 2012;18:661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jesser R, Behler J, Benda C, et al. Biochemical analysis of the Cas6-1 RNA endonuclease associated with the subtype I-D CRISPR-Cas system in Synechocystis sp. PCC 6803. RNA Biol. 2018;1–11. DOI: 10.1080/15476286.2018.1447742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Niewoehner O, Jinek M, Doudna JA. Evolution of CRISPR RNA recognition and processing by Cas6 endonucleases. Nucleic Acids Res. 2014;42:1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sashital DG, Jinek M, Doudna JA. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat Struct Mol Biol. 2011;18:680–687. [DOI] [PubMed] [Google Scholar]
- [26].Wang R, Preamplume G, Terns MP, et al. Interaction of the Cas6 Riboendonuclease with CRISPR RNAs: recognition and Cleavage. Structure. 2011;19:257–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lee M, Tseng S, Yang J, et al. Expression, purification, crystallization, and X-ray structural analysis of CRISPR-Associated protein Cas6 from methanocaldococcus jannaschii. Crystals. 2017;7:344. [Google Scholar]
- [28].Shao Y, Recognition LH. Cleavage of a nonstructured CRISPR RNA by its processing endoribonuclease Cas6. Structure. 2013;21:385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Reeks J, Sokolowski RD, Graham S, et al. Structure of a dimeric crenarchaeal Cas6 enzyme with an atypical active site for CRISPR RNA processing. Biochem J. 2013;452:223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jackson RN, Golden SM, van EPBG, et al. Crystal structure of the CRISPR RNA–guided surveillance complex from Escherichia coli. Science. 2014;345:1473–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhao H, Sheng G, Wang J, et al. Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature. 2014;515:147–150. [DOI] [PubMed] [Google Scholar]
- [32].Mulepati S, Héroux A, Bailey S. Crystal structure of a CRISPR RNA–guided surveillance complex bound to a ssDNA target. Science. 2014;345:1479–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pausch P, Müller-Esparza H, Gleditzsch D, et al. Structural variation of Type I-F CRISPR RNA guided DNA surveillance. Mol Cell. 2017;67(622–632.e4). DOI: 10.1016/j.molcel.2017.06.036 [DOI] [PubMed] [Google Scholar]
- [34].Shao Y, Richter H, Sun S, et al. A non-stem-loop CRISPR RNA is processed by dual binding Cas6. Structure. 2016;24:547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Özcan A, Pausch P, Linden A, et al. Type IV CRISPR RNA processing and effector complex formation in Aromatoleum aromaticum. Nat Microbiol. 2018. DOI: 10.1038/s41564-018-0274-8. [DOI] [PubMed] [Google Scholar]
- [36].Salinas MB. Mahella australiensis gen. nov., sp. nov., a moderately thermophilic anaerobic bacterium isolated from an Australian oil well. Int J Syst Evol Microbiol. 2004;54:2169–2173. [DOI] [PubMed] [Google Scholar]
- [37].Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. [DOI] [PubMed] [Google Scholar]
- [38].Maris C, Dominguez C, Allain FH-T. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression: the RRM domain, a plastic RNA-binding platform. Febs J. 2005;272:2118–2131. [DOI] [PubMed] [Google Scholar]
- [39].Emsley P, Lohkamp B, Scott WG, et al. Features and development of Coot. Acta Crystallogr Sect D: Biol Crystallogr 2010;66:486–501.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gesner EM, Schellenberg MJ, Garside EL, et al. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat Struct Mol Biol. 2011;18:688–692. [DOI] [PubMed] [Google Scholar]
- [41].Wei W, Zhang S, Fleming J, et al. Mycobacterium tuberculosis type III-A CRISPR/Cas system crRNA and its maturation have atypical features. Faseb J. 2018. fj.201800557RR DOI: 10.1096/fj.201800557RR. [DOI] [PubMed] [Google Scholar]
- [42].Lee HY, Haurwitz RE, Apffel A, et al. RNA-protein analysis using a conditional CRISPR nuclease. Proc Nat Acad Sci. 2013;110:5416–5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Biswas A, Gagnon JN, Brouns SJJ, et al. CRISPRTarget. RNA Biol. 2013;10:817–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Faure G, Makarova KS, Koonin EV. CRISPR–cas: complex Functional Networks and Multiple Roles beyond Adaptive Immunity. J Mol Biol. 2018. DOI: 10.1016/j.jmb.2018.08.030 [DOI] [PubMed] [Google Scholar]
- [45].Terwilliger TC, Adams PD, Read RJ, et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr Sect D: Biol Crystallogr 2009;65:582–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Grosse-Kunstleve RW, Adams PD. Substructure search procedures for macromolecular structures. Acta Crystallogr Sect D: Biol Crystallogr. 2003;59:1966–1973.. [DOI] [PubMed] [Google Scholar]
- [47].Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr Sect D: Biol Crystallogr. 2000;56:965–972.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].McCoy AJ, Storoni LC, Read RJ. Simple algorithm for a maximum-likelihood SAD function. Acta Crystallogr Sect D: Biol Crystallogr. 2004;60:1220–1228.. [DOI] [PubMed] [Google Scholar]
- [49].Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr Sect D: Biol Crystallogr. 2003;59:38–44.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bunkóczi G, Echols N, McCoy AJ, et al. Phaser.MRage : automated molecular replacement. Acta Crystallogr Sect D: Biol Crystallogr. 2013;69:2276–2286.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Afonine PV, Grosse-Kunstleve RW, Echols N, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr Sect D: Biol Crystallogr. 2012;68:352–367.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Buchan DWA, Minneci F, Nugent TCO, et al. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013;41:W349–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


