Abstract
The avian eggshell is a critical physical barrier, which permits extra-uterine development of the embryo. Its formation involves the fastest known biomineralization process in vertebrates. The eggshell consists of proteins and proteoglycans that interact with the mineral phase to impart its specific microstructure and mechanical properties. In this study, we investigated the role of epidermal growth factor (EGF)-like repeats and discoidin-like domains 3 (EDIL3) and milk fat globule–EGF factor 8 (MFGE8), two glycoproteins that are consistently detected in eggshell proteomes. We verified their common evolutionary history and identified the timing of the duplication event giving rise to these two distinct proteins. Edil3/mfge8 chromosomal locations revealed a nested syntenous relationship with other genes (hapln1/hapln3 and vcan/acan) that are also involved in vertebrate calcification. EDIL3 and MFGE8 proteins possess EGF-like and coagulation factor 5/8 (F5/8C) domains, and their 3D structures predicted that they bind calcium and extracellular vesicles. In chicken, we confirmed the presence of EDIL3 and MFGE8 proteins in eggshell, uterine fluid, and uterus. We observed that only edil3 is overexpressed in tissues in which eggshell mineralization takes place and that this overexpression occurs only at the onset of shell calcification. We therefore propose a model in which EDIL3 and, to a lesser extent, MFGE8 proteins guide vesicles containing amorphous calcium carbonate to the mineralization site. This model was supported by the observation that extracellular vesicles accumulate in uterine fluid during eggshell calcification and that they contain high levels of calcium, carbon, and oxygen that correspond to calcium carbonate.
Keywords: biomineralization, calcium transport, extracellular vesicles, protein evolution, glycoprotein, avian eggshell, EGF-like repeats and discoidin-like domains 3 (EDIL3), milk fat globule-EGF factor 8 (MFGE8), uterine fluid, vesicular transport
Introduction
The avian eggshell is a physical barrier against mechanical and microbial stresses that allows extra-uterine development of the embryo in a closed chamber. The eggshell consists of 95% calcium carbonate (CaCO3, calcite), 3.5% organic matrix (OM2; proteins, polysaccharides, and proteoglycans), and 1.5% water (1, 2). This biomineral is composed of five layers: inner and outer eggshell membranes, two contiguous calcified layers (mammillary knobs and palisades), and the cuticle (3). Shell formation is extracellular and is the fastest biomineralization process in vertebrates (4). Egg formation occurs in the hen reproductive tract (1, 5). In chickens, the forming egg reaches the red isthmus (RI) at 4.5 h post-ovulation (p.o.), where shell biomineralization is initiated on organic cores. The egg then enters the uterus (Ut), where it will remain for 18–19 h during shell calcification. Calcium carbonate is initially deposited as an amorphous phase (ACC), which progressively transforms into calcite (6).
During its formation, the shell is bathed in a uterine fluid (UF) secreted by uterine cells that contains the organic and mineral precursors necessary for shell calcification (7, 8). Matrix proteins stabilize ACC, promote crystal nucleation, select the calcite polymorph, and regulate the evolution of crystal size and morphology (4, 7, 9, 10). These matrix–mineral interactions determine the orientation of calcite crystals, which results in the complex ultrastructure of the eggshell, its texture, and consequently its mechanical properties. Over the last couple of decades, numerous studies have defined the chicken eggshell matrix proteome (11–18). Currently, the entire dataset of reported proteins contains around 900 nonredundant proteins for the chicken eggshell and more than 600 for the UF (19, 20). Among them, EDIL3 (EGF)-like repeats and discoidin-like domains 3) and MFGE8 (milk fat globule–EGF factor 8) are reported in the majority of transcriptomics and proteomics studies associated with chicken eggshell biomineralization (11, 13–18, 21, 22). These two proteins were also identified in other avian eggshell proteomes (quail, turkey, and guinea fowl) (23–25).
MFGE8 and EDIL3 are two secreted glycoproteins that have been well-described in mammals and exhibit sequence similarity to each other. In avian eggshell, it was suggested that both EDIL3 and MFGE8 could bind Ca2+, due to an EGF-like calcium-binding domain, and could favor crystal nucleation or change crystal morphology by selective interaction with calcium ion-rich crystallographic faces (19, 21).
Nevertheless, the function of EDIL3 and MFGE8 is poorly understood. In this study, we have used bioinformatics tools to decipher the common evolutionary history of edil3 and mfge8 genes. We also explored the role of EDIL3 and MFGE8 in chicken eggshell mineralization by predicting their molecular features, quantifying their gene expression, and measuring protein levels in tissues involved in eggshell formation. Our study suggests that EDIL3 and MFGE8 would bind to vesicles and calcium carbonate to guide vesicular transport providing mineral cargo during avian eggshell biomineralization.
Results
Evolutionary history of EDIL3 and MFGE8: synteny and phylogenetic analysis
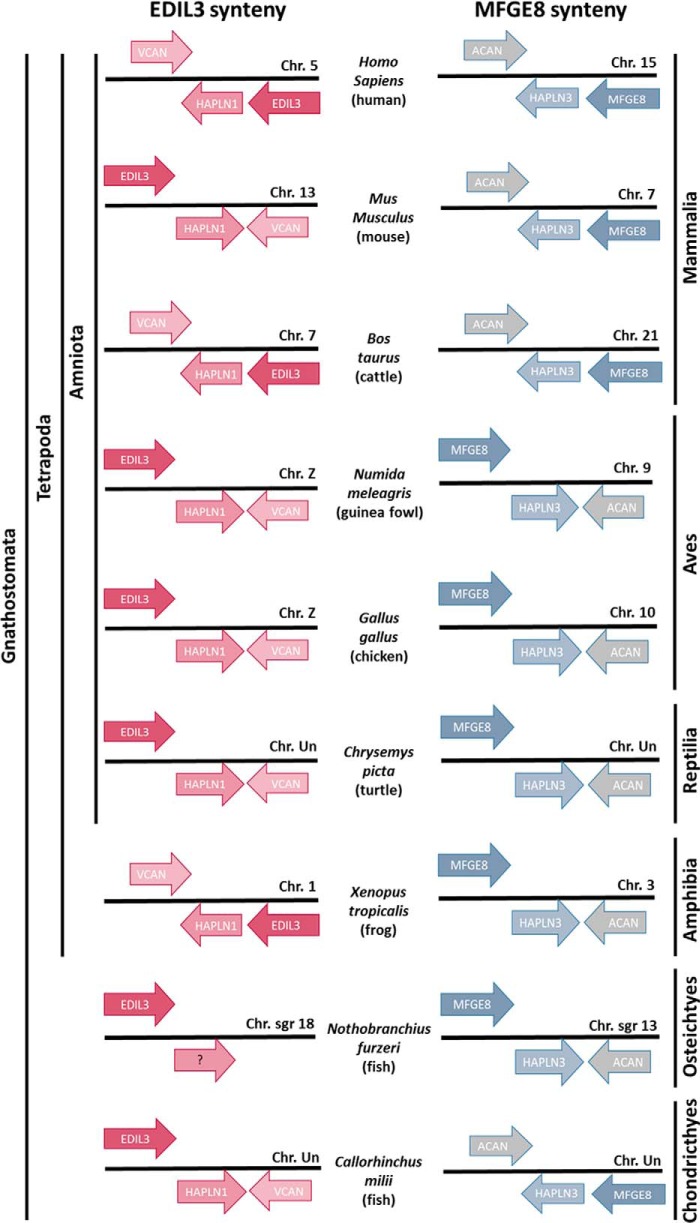
EDIL3 and MFGE8 are two related proteins that share 54% identity and 69% similarity in Gallus gallus (EDIL3, XP_424906.3; MFGE8, NP_001264039.1; Table S1). They are encoded by two distinct genes, edil3 and mfge8, which are localized on chromosome Z and chromosome 10, respectively. We compared their gene loci in different vertebrate classes (fishes, amphibians, reptiles, birds, and mammals; Fig. 1). Edil3 and mfge8 were observed to be nested in syntenous clusters containing hapln1 (hyaluronan and proteoglycan link protein 1) and vcan (versican) adjacent to edil3, and hapln3 (hyaluronan and proteoglycan link protein 3) and acan (aggrecan) adjacent to mfge8 (Fig. 1). HAPLN1 and HAPLN3 belong to hyaluronan and proteoglycan link protein family, and VCAN and ACAN belong to the lectican family. In chicken, HAPLN1 is related to HAPLN3 with 48% identity/66% similarity for conservative substitutions, whereas VCAN and ACAN displayed 18% identity/27% similarity (Table S1). The direction of mfge8 gene cluster transcription is unchanged from Chondrichthyes (Callorhinchus milii) to mammalia (Homo sapiens) (Fig. 1). This observation is also true for the edil3 cluster during vertebrate evolution (Fig. 1). Therefore, the edil3 and mfge8 clusters are highly conserved in Gnathostoma. In birds, the edil3 gene cluster is consistently observed on chromosome Z, and the mfge8 cluster is found on a nonsexual chromosome (Table S2). These results confirm that the edil3 and mfge8 gene loci contain paralogous genes resulting from a duplication event.
Figure 1.
Chromosomal arrangements of edil3 and mfge8 gene clusters for several vertebrate classes. Gene organization was investigated on the NCBI genome explorer. Arrows indicate the direction of gene transcription.
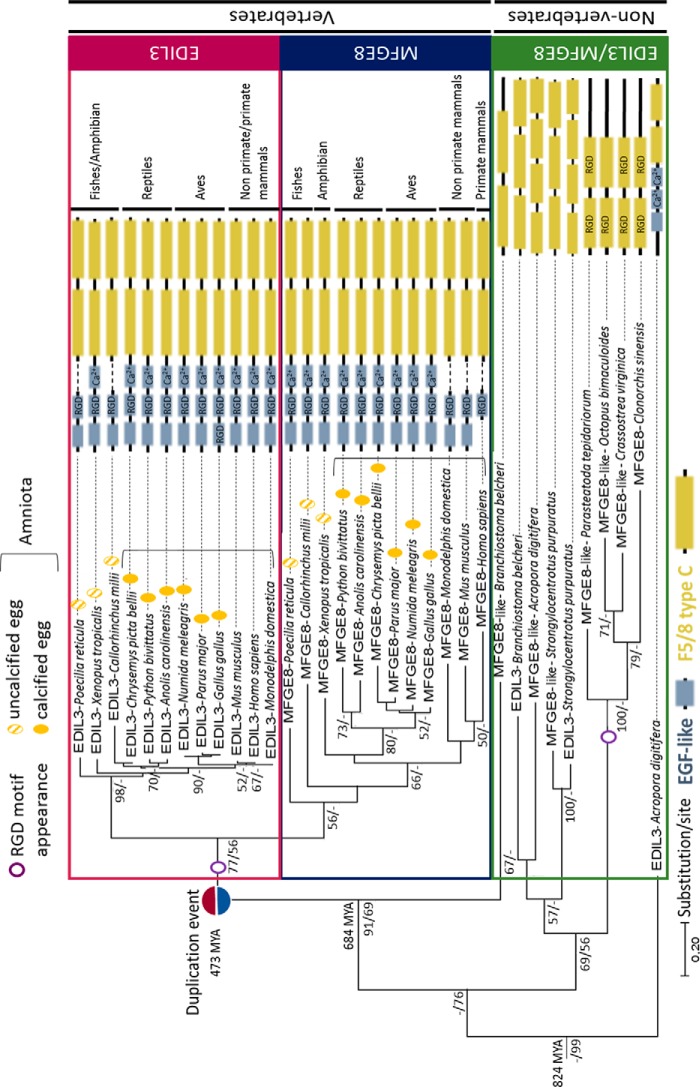
To determine the timing of divergence between these two genes, we performed phylogenetic tree analysis of the protein sequences to investigate the evolutionary history of EDIL3 and MFGE8 in metazoans (Fig. 2). We first investigated the presence of the two proteins in eukaryotes. Blast analysis revealed that EDIL3 and/or MFGE8 were absent from protozoans to sponges. Consequently, the phylogenetic reconstruction was performed with EDIL3/MFGE8 protein sequences from eumetazoan species (Fig. 2). The NCBI database contains a large number of EDIL3/MFGE8 orthologs. To perform this analysis, we loaded 34 EDIL3/MFGE8 sequences from 19 different species representing different taxa. These sequences belong to nonvertebrates (one cnidarian, one platyhelminthe, two molluscs, one arthropod, one echinoderm, and one cephalochordate) and vertebrates (two fishes, one amphibian, three reptiles, three birds, and three mammals; Table S3). MFGE8 sequences were often annotated as lactadherin in various databases (Table S3). For consistency during our analysis, all lactadherin sequences were renamed MFGE8 (Fig. 2). Sequences were aligned using Clustal Omega multiple alignment. A phylogenetic tree was constructed using Maximum Likelihood and Bayesian inference methods, which both conducted to similar tree topology, and Maximum Likelihood result was displayed (Fig. 2). According to the topology of the tree, with coral Acropora digitifera EDIL3 protein as a rooted sequence, two distinct groups emerged within the phylogenetic tree. The first one contained nonvertebrate EDIL3/MFGE8 sequences, and the second one contained vertebrate EDIL3/MFGE8 sequences. The latter group had a cephalochordate MFGE8 sequence as a root and is then divided into two clusters: vertebrate EDIL3 and vertebrate MFGE8 sequences, respectively. Inside both clusters, we noticed that species were also grouped according to their class (actinopterygii, amphibian, reptiles, aves, and mammals).
Figure 2.
Phylogenetic tree reconstruction of EDIL3 and MFGE8 common evolutionary histories in Eumetazoa. Multiple alignment of protein sequences (LG substitution model) was performed using Clustal Omega, and the phylogenetic tree was reconstructed using Maximum Likelihood (in MEGA7) and Bayesian inference (in BEAST version 1.10.4) methods. The topology of the tree is that resulting from Maximum Likelihood. Node values correspond to the percentage of Maximum Likelihood bootstrap values/Bayesian posterior probabilities, respectively. Only values above 50 are indicated. The divergence times were analyzed on Timetree. The schematic representation of EDIL3 and MFGE8 protein domains were acquired with HomoloGene of NCBI and several databases (ExPASy-PROSITE, Ensembl, and SMART). RGD, integrin-binding site; Ca2+, calcium-binding site. NCBI accession numbers for each protein are listed in Table S3.
In Fig. 2, the number of substitutions by site indicated that EDIL3 had undergone fewer changes during vertebrate evolution compared with MFGE8. Indeed, MFGE8 branches were longer than EDIL3 branches, and the EDIL3 cluster bootstrap value (node value, 98) was higher than the value for the MFGE8 cluster (node value, 56). This result indicated that vertebrate sequences of MFGE8 were more divergent from each other than within the EDIL3 sequences. In nonvertebrates, EDIL3 and MFGE8 sequences did not form two distinct clusters. Moreover, we did not consistently observe both EDIL3 and MFGE8 protein sequences; in several nonvertebrates species, only one sequence is present. These are mostly annotated as lactadherin-like (Table S3). The presence of both genes in the same nonvertebrate species, and their proximity in the tree, suggested that another duplication event affecting the ancestral edil3/mfge8-like gene appeared in the nonvertebrate lineages, as observed for the coral A. digitifera, the cephalochordate Branchiostoma belcheri and the sea urchin Strongylocentrotus purpuratus. We also observed during the course of vertebrate evolution a distinction between edil3 and mfge8 emerging around 480 million years ago (MYA), which led to further diversification/divergence of their sequences and gave rise to vertebrate edil3 and mfge8. This event occurred after the split of nonvertebrate/vertebrate lineages and before the split between fishes/amphibian phyla. Therefore, the edil3/mfge8 divergence appeared between 684 and 473 million years ago.
A comparison of protein domains was performed throughout eumetazoan evolution (Fig. 2). In nonvertebrates, only EDIL3 of the coral A. digitifera possesses EGF-like domains, although this domain was present in all EDIL3/MFGE8 vertebrate species. The most primitive vertebrate of the study (C. milii) possessed three EGF-like domains in MFGE8 but only two in its EDIL3. With the notable exception of fishes, all EDIL3 vertebrate sequences contain three EGF-like domains. Three EGF-like domains are also present in nonmammalian MFGE8 sequences, whereas two and one are present in nonprimate mammals and primates, respectively. A Ca2+-binding motif is also present in A. digitifera EDIL3 and in all other sequences except in fishes EDIL3, mammalian MFGE8, and nonvertebrates. An RGD motif (integrin (ITG)-binding) was also detected in EGF-like domains of all vertebrate MFGE8s and EDIL3s. In nonvertebrates, this motif is observed in F5/8C domains, when present. This latter domain was observed in all vertebrate and nonvertebrate sequences. A double RGD motif was also detected in three nonvertebrate sequences and in G. gallus EDIL3. However, the vertebrate RGD sequences are found in the EGF-like domains, whereas the invertebrate RGDs are located within the F5/8C regions.
Edil3 and mfge8 expression in G. gallus tissues at various stages of shell mineralization
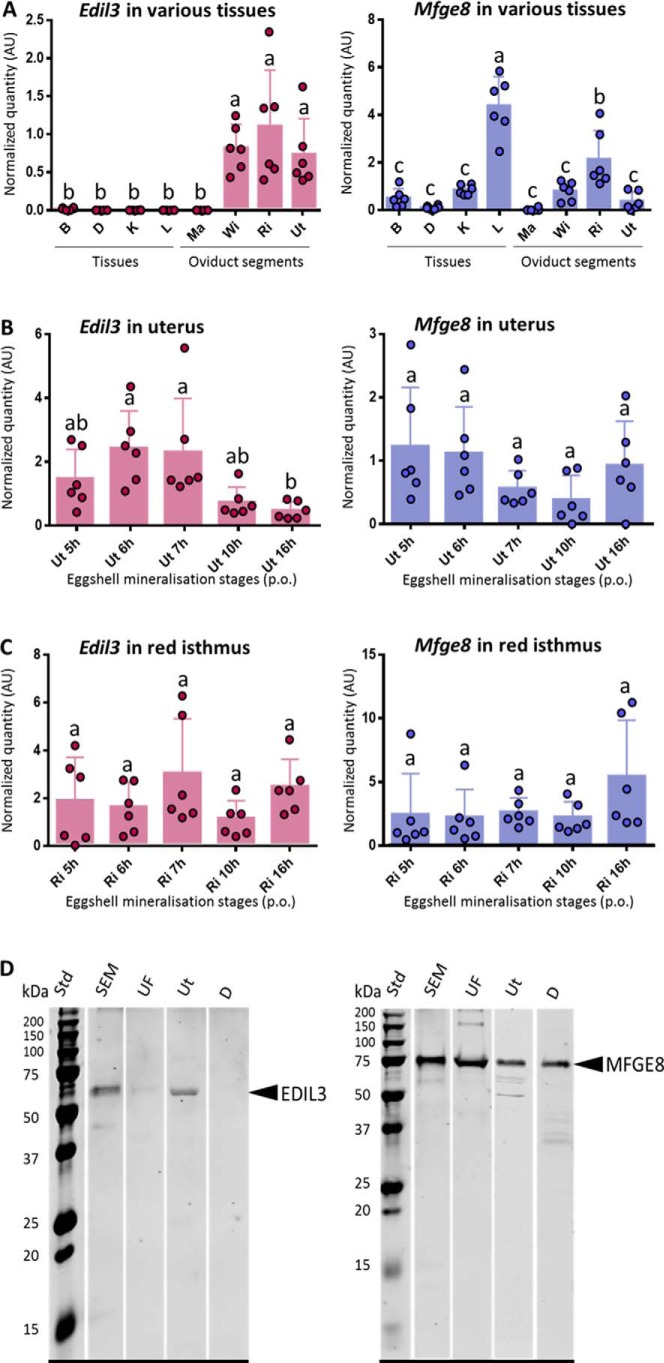
We investigated the level of expression of both genes in oviduct tissues involved in egg white and eggshell formation (Fig. 3A). These are the magnum (Ma), where egg white proteins are synthesized, and the white isthmus (WI), where the shell membranes are formed. In addition, the red isthmus (RI) and uterus (Ut) are involved in the onset and development of shell mineralization, respectively. Gene expression in these oviduct segments was compared with samples from mid-shaft tibial bone (B) (mineralized tissue), duodenum (D) (high calcium intake), kidney (K) (involved in ion exchange), and finally liver (L) as an important tissue of general metabolism. Levels of expression were measured in tissues harvested during the onset of the rapid growth phase of shell mineralization (10 h p.o., except for B collected at 18 h p.o. when shell mineralization and bone demineralization are completed).
Figure 3.
A–C, gene expression of edil3 and mfge8 in oviduct regions and four tissues of G. gallus at key events of eggshell biomineralization. Each tissue and stage were determined on six different laying hens (n = 6). A, Edil3 and mfge8 expression in oviduct regions and four tissues of G. gallus. All tissues were harvested at 10 h post-ovulation from 40-week-old hens, except for tibial bone (B), which is from 18-h post-ovulation in 90-week-old hens. B, Edil3 and mfge8 expression in uterus at five key stages of eggshell biomineralization (5–7, 10, and 16 h post-ovulation). C, Edil3 and mfge8 expression in red isthmus at five key stages of eggshell biomineralization (5–7, 10, and 16 h post-ovulation). Relative expression levels of genes were normalized with eight housekeeping genes (see under “Experimental procedures”). Error bars indicate standard deviations. Different letters indicate significant differences in levels of expression (p value <0.05), based on one-way ANOVA and pairwise Tukey test. D, Western blot analysis of EDIL3 and MFGE8 levels in SEM, UF, Ut, and D. For each sample, 15 μg of protein was subjected to electrophoretic separation and electro-transfer. The membranes were probed with anti-EDIL3 (left) or anti-MFGE8 (right) antibodies. AlexaFluor® 680-labeled secondary antibody was used for the detection. Std,: molecular mass standard.
Edil3 expression was low in D, L, Ma, B, and K and not significantly different from background levels (Fig. 3A). Edil3 was significantly expressed in tissues involved in shell formation in contrast to the five other tissues. They were, however, not significantly different from each other. RI/B and Ut/B expression ratios were 58.6 and 39.2, respectively, and 846.9 and 566.9, respectively, for RI/K and Ut/K ratios. Mfge8 was expressed in all tissues (Fig. 3A). Liver exhibited the highest level of mfge8 expression followed by RI. Mfge8 expression levels were significantly different in these two tissues, which were both significantly different from the six other tissues. Ratios of RI expression were 73, 18.3, 4.1, 2.5, and 2.6 with Ma, D, B, K, and WI, respectively.
Levels of edil3 and mfge8 expression in oviduct regions that are active during eggshell mineralization (Ut and RI) were measured at five key stages of shell calcification. Edil3 was expressed at all stages in Ut and RI and was significantly higher in Ut for stages 6 h and 7 h p.o. compared with 16 h p.o. (Fig. 3, B and C), when ACC is massively deposited with the first aggregates of calcite forming around mammillary knobs. Edil3 expression compared with 16-h p.o. exhibited a fold change of 5.6 and 4.9 at 6 h and 7 h p.o., respectively. In contrast, from 5 h to 16 h p.o., no significant difference was observed for edil3 in RI and for mfge8 in Ut and RI (Fig. 3, B and C).
EDIL3 and MFGE8 protein levels during eggshell formation
The presence of EDIL3 and MFGE8 proteins was investigated in soluble eggshell matrix (SEM), UF, Ut, and D (which is involved in ion transfer without mineralization). Using anti-EDIL3 antibodies, a single immunoband was observed around 60 kDa (expected size 54 kDa) in SEM, Ut, and to a lesser extent in UF (Fig. 3D). No immunoreactive band was observed in D. MFGE8 exhibited an intense immunoreactive band in all tissue samples, including D (Fig. 3D) at 75 kDa (expected 53 kDa). The higher than predicted molecular mass observed for MFGE8 could be due to post-translational modification such as glycosylation, which has been reported for this protein (26, 27). These results are in accordance with expression measurements, and they confirmed that EDIL3 is specific to the oviduct tissue involved in shell mineralization, whereas MFGE8 is present in a variety of tissues or organs, including those responsible for eggshell calcification (Fig. 3).
Characterization of G. gallus EDIL3 and MFGE8 functional domains
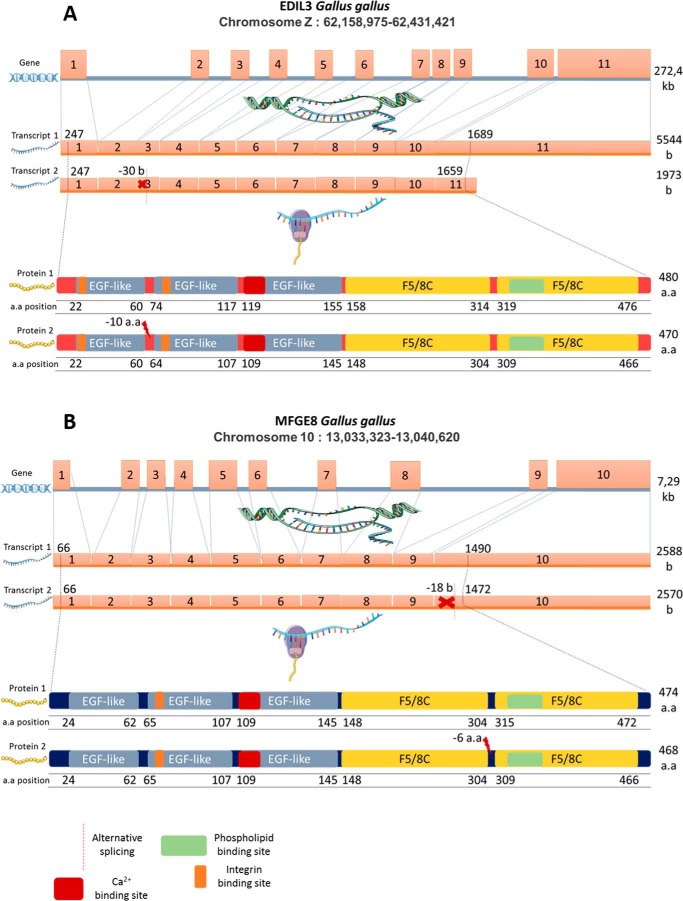
We explored gene and protein sequences of EDIL3 and MFGE8 using bioinformatics tools. G. gallus edil3 is made of 11 exons and is located on sex chromosome Z (Fig. 4). Its transcription leads to two different transcripts of 5544 and 1973 nucleotides coding for 480 and 470 amino acid proteins, respectively (NCBI). Differences in transcript size are the consequence of a 30-nucleotide deletion in exon 3 and alternative splicing of exon 11. This latter alternative splicing does not affect the translated protein because it modifies the 3′-UTR region in the mRNA. Stop codons are located at positions 1657–1659 and 1687–1689 in the short and long mRNA variants, respectively. The 30-nucleotide in-frame deletion of exon 3 resulted in a loss of 10 amino acids between two EGF-like domains. The G. gallus mfge8 gene is located on chromosome 10 and is made of 10 exons (Fig. 4). As is the case for edil3, mfge8 also exhibited two transcripts of 2588 and 2570 bases, which encode proteins of 474 and 468 amino acids, respectively (NCBI). The shorter transcript is the result of an 18-nucleotide deletion at the onset of the last exon, which induces a loss of six amino acids between two F5/8C domains of the corresponding protein. The remaining sequence is not affected by this deletion.
Figure 4.
Gene, mRNA variants, and protein isoforms for G. gallus EDIL3 (A) and MFGE8 (B). The domain positions and sequence lengths were determined using the NCBI database. Edil3 Gene ID, 427326; transcript 1, XM_424906.6; transcript 2, XM_004949448.3; protein isoform 1, XP_424906.3; protein isoform 2, XP_004949505.1. Mfge8 Gene ID, 415494; transcript 1, NM_001277110.1; transcript 2, NM_001277111.1; protein isoform 1, NP_001264039.1; protein isoform 2, NP_001264040.1. Elements were from Servier Medical Art (https://smart.servier.com/),3 licensed under a Creative Commons Attribution 3.0 Unported License.
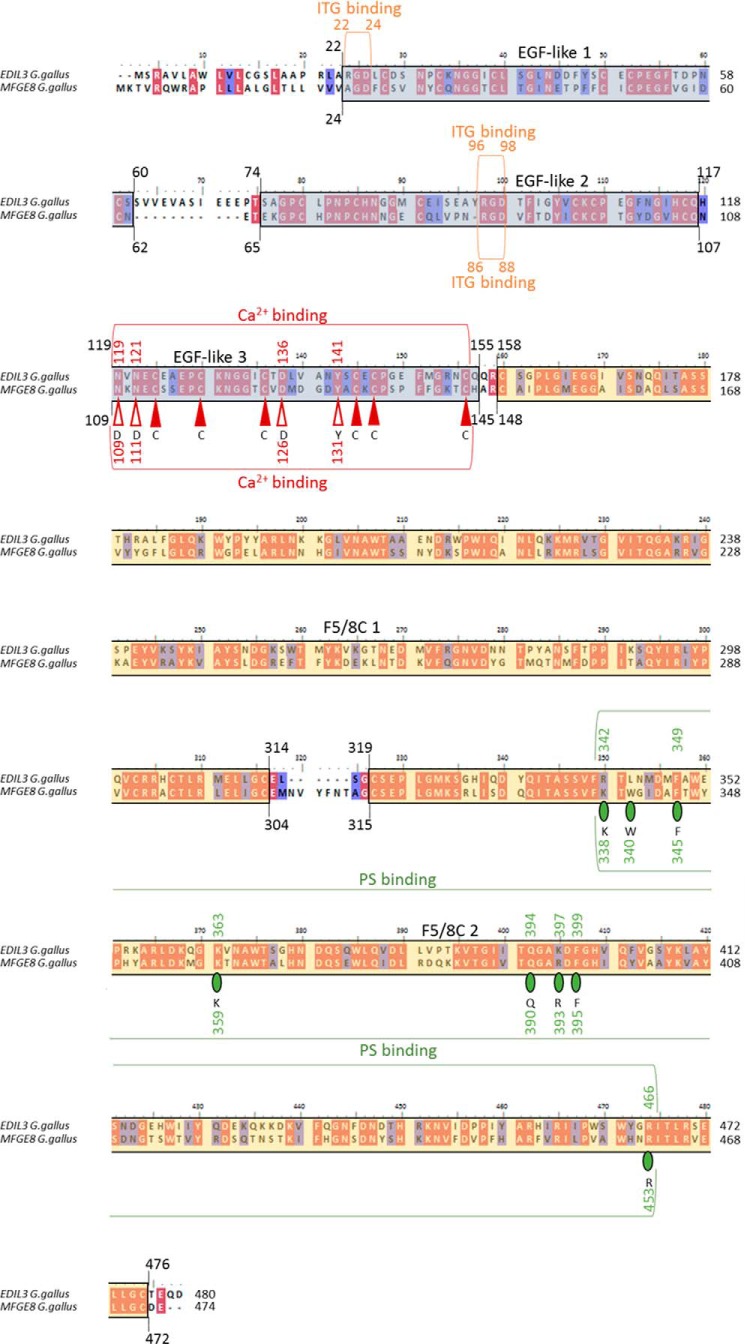
EDIL3 and MFGE8 G. gallus proteins were aligned with MEGA7 software, and protein domains were investigated using PROSITE (Fig. 5). Both proteins exhibited three EGF-like domains at the N terminus (Fig. 5, protein 1 in Fig. 4). Residues 22–60 and 24–62 correspond to EDIL3 and MFGE8 EGF-like 1 domains, respectively. Other EGF-like domains were located at positions 74–117, 65–107 and 119–155, 109–145 for domains 2 and 3 of EDIL3 and MFGE8, respectively. EDIL3 EGF-like 1 and 2 domains possess an ITG-binding site (RGD motif in positions 22–24 and 96–98), whereas this motif was only present in the EGF-like 2 domain for MFGE8 (position 86–88). Both proteins also possess a Ca2+-binding motif in the EGF-like 3 domain. This motif was composed of about 40 residues with six cysteine residues that form three disulfide bonds with the following consensus pattern: nXnnC-X(3,14)-C-X(3,7)-CXXbXXXXAXC-X(1,6)-C-X(8,13)-CX. This Ca2+-binding motif was found in positions 119–143 and 109–133 for EDIL3 and MFGE8, respectively. Alignments of G. gallus EDIL3 and MFGE8 EGF-like 3 domains with H. sapiens coagulation factors IX/X EGF-like domains are represented on Fig. S1A. The Ca2+ consensus sequence of human coagulation factors IX/X are depicted in Fig. 5. The Ca2+-binding motifs of EGF-like 3 domains translated from the longer transcripts of G. gallus EDIL3 (Asn-119, Asn-121, Asp-136, and Tyr-141) and MFGE8 (Asn-109, Asn-111, Asp-126, and Tyr-131) were highly conserved when compared with the EGF-like Ca2+-binding consensus sequences of the human protein (Asp-1, Asp-3, Asp-18, and Tyr-23; Fig. 5 and Fig. S1A). However both Asps of the human coagulation factors IX/X Ca2+-binding motif were replaced by Asn (residue with a similar polar property to Asp) in each G. gallus protein. Cysteines were strictly conserved between both proteins at the Ca2+-binding site.
Figure 5.
Domain prediction for G. gallus EDIL3 and MFGE8 protein sequences. GenBankTM accession numbers used are EDIL3 XP424906.3 and MFGE8 NP_001264039.1. Alignment was performed using Clustal Omega method in MEGA7 and then annotated with BioEdit. Red- and blue-colored residues indicate identity and similarity, respectively. Blue and yellow backgrounds correspond to EGF-like and F5/8C domains, respectively. Red vertical arrows (filled for cysteines) and green circles correspond to amino acid residues involved in Ca2+ binding and PS binding, respectively. The letters below arrows and circles indicate the consensus pattern for Ca2+-binding and PS-binding residues of human coagulation factors IX/X and mouse MFGE8, respectively. Indicated positions correspond to the residue numbering for the full-length sequences of G. gallus EDIL3 and MFGE8.
We also observed two F5/8C domains in both EDIL3 and MFGE8 C-terminal regions (positions 158–314, 319–476 and 148–304, 315–472, respectively, Fig. 5). Finally, phosphatidylserine-binding (PS-binding) motifs have been reported in the mammalian sequences, and we investigated the presence of this motif in both EDIL3 and MFGE8 from G. gallus. The following residues (Lys-24, Trp-26, Phe-31, Lys-45, Gln-76, Arg-79, Phe-81, and Arg-146) were described in Mus musculus protein as involved in PS binding (28). The same residues were observed in positions 338, 340, 345, 359, 390, 393, 395, and 453 of the F5/8C two domains of G. gallus MFGE8, whereas EDIL3 showed identical and similar residues (R-FKQKFR) in positions 342, 349, 363, 394, 397, 399, and 466 (Fig. 5 and Fig. S1B). Trp is not present in the EDIL3 sequence, and conservative substitutions (Lys/Arg and Arg/Lys) are observed in positions 342 and 397, respectively. However, an absence of Trp at this position did not affect the function of the PS-binding site (28); consequently, these data suggest potential functional PS-binding activity for both G. gallus EDIL3 and MFGE8 proteins.
We performed computational predictions of secondary structure for chicken EDIL3 and MFGE8 using PSIPRED version 3.3 and the I-TASSER program server. Twenty two β-sheets and two α-helices were predicted for EDIL3, and 25 β-sheets and two α-helices were predicted for MFGE8 proteins, with good correspondence between the 22 β-sheets of EDIL3 and those in MFGE8. We also observed that EGF-like domains are made up of two β-sheets in each protein. The first F5/8C contains 11 β-sheets in MFGE8 and nine in EDIL3, whereas the second is similar in both proteins with seven β-sheets.
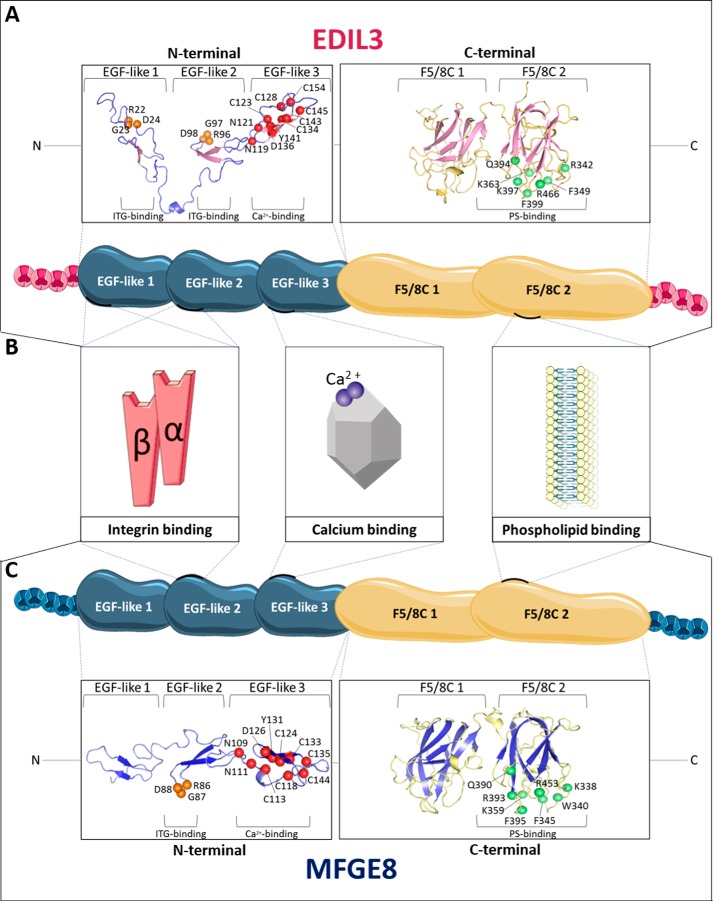
The 3D structures of EDIL3 and MFGE8 were also predicted with the I-TASSER program server (molecular homology modeling), and the spatial arrangement of amino acids involved in each binding function (ITG, Ca2+, and PS) was highlighted on the 3D structure models using PyMOL Molecular Graphics system (Fig. 6, A and C). A close spatial proximity among residues was observed in accordance with their ability to interact with their substrates, and consequently, we predicted that these domains are functional. The conformation of the Ca2+-binding domain was composed, as expected, of three disulfide bridges necessary to maintain the protein conformation for interaction with Ca2+ (Fig. 6, A and C). The spatial arrangement of residues in the second F5/8C domains associated with the PS-binding motif in G. gallus is highlighted (Fig. 6, A and C). This motif involves seven and eight amino acids for EDIL3 and MFGE8, respectively. Residues are located in three different loops as observed for the mammalian sequences (Fig. 6, A and C, green balls). Only Gln-390 was observed in a β-sheet in the MFGE8 F5/8C 2 domain. Both EDIL3 and MFGE8 RGD motifs involved in ITG binding of the second EGF-like domains were localized to a loop between two β-sheets (Fig. 6, orange balls). Structural modeling also indicates that the major difference between EDIL3 and MFGE8 is associated with the first EGF-like domain (Fig. 6), for which an additional specific RGD motif was observed in EDIL3. This additional RGD motif is not located in a loop between β-sheets.
Figure 6.
In silico prediction and structure modeling of EDIL3 and MFGE8 protein domains and binding sites. A, tertiary structure prediction of EGF-like and F5/8C regions of G. gallus EDIL3 (XP_424906.3). B, schematic representation of G. gallus EDIL3 and MFGE8 protein domains, including their binding sites. C, tertiary structure prediction of EGF-like and F5/8C regions of G. gallus MFGE8 (NP_001264039.1). The tertiary structure prediction models were generated by EGF-like and F5/8C domain regions splitting using the I-TASSER template-based modeling. Following prediction modeling, the generated tertiary structures were edited using PyMOL. Orange balls indicate RGD residues of the ITG-binding sites; red balls illustrate residues of the Ca2+-binding sites, and green balls display PS-binding sites. The position of the residues is indicated according to their location in the entire protein. Elements were from Servier Medical Art, licensed under a Creative Commons Attribution 3.0 Unported License.
Discussion
EDIL3 and MFGE8 are two proteins widely detected in proteomic studies associated with chicken eggshell biomineralization (11, 13–18, 21). In chicken these two proteins exhibited a high abundance in partially mineralized eggshell at the initiation of biomineralization, and consequently, they are thought to be potential major actors in the calcification process (21).
MFGE8 and EDIL3 are homologously-secreted glycoproteins that have been well-described in mammals. The mfge8 transcript was first identified in mouse mammary epithelial cells (29). MFGE8 is known to participate in a wide variety of cellular interactions such as macrophage and apoptotic cell bridging, adhesion between sperm and the zona pellucida of the egg, or exosome function (30–34). EDIL3 was first identified in mouse as an extracellular matrix protein involved in embryonic vascular development and demonstrating an angiogenic role (35–37). In mammals, EDIL3 is physiologically expressed in various tissues such as brain, intestine, heart, and kidney (38). EDIL3 was also detected during cancer development and is expressed by primary tumors from breast and colon cancers (39). In breast cancer, EDIL3 was identified on extracellular vesicles (EV) promoting cellular invasion and was suggested to be a potential biomarker for monitoring this metastatic cancer (40). In developing chicken embryos, experiments have been performed on chorioallantoic membrane (CAM) using mammalian EDIL3 (DEL1). DEL1 was found to be involved in embryonic vascular remodeling, in mediating the breakdown of existing vascular blood, and inducing restructuring of the CAM vasculature (35). In contrast, a potent pro-angiogenic activity was reported for DEL1 in chicken CAM, which was mediated by integrin (αvβ3) activation via the RGD motif (36).
Our phylogenetic analysis demonstrated that vertebrate edil3 and mfge8 are derived from a duplication event of an ancestral common gene. The divergence point between these two paralogs was estimated to be about 480 MYA. Gene duplication processes are crucial to generate genes with novel or altered functions (41). The duplication events giving rise to edil3 and mfge8 in early vertebrates emerged prior to the appearance of the amniotic egg at the split between amphibians (uncalcified egg) and reptiles (calcified egg, amniota) that occurred around 350 MYA (42, 43). According to our study, the function of EDIL3 and MFGE8 was recruited to diverse biological processes, including eggshell calcification.
In nonprimate mammals, two homologous domains of epidermal growth factor from Drosophila NOTCH (EGF-like) at the N terminus and one tandem of discoidin-like domains similar to those of coagulation factors V and VIII (F5/8C) at the C terminus were described in MFGE8 (29, 44). Our evolutionary study confirmed the presence of these domains in mammals and also explored additional eumetazoan sequences used in this study. The F5/8C domains appear to be present in all species sequences indicating an origin of this domain in the common ancestor of eumetazoans. The number of F5/8C domains is variable in nonvertebrates (from two to four), whereas it is constant in vertebrates, in which two F5/8C were reported. The PS-binding site of MFGE8 orthologs was previously described in cow (Bos taurus) and mouse (M. musculus) (28, 45–48). The following residues (Lys-24, Trp-26, Phe-31, Lys-45, Gln-76, Arg-79, Phe-81, and Arg-146) are major residues contributing to the interaction between the F5/8C 2 domain of MFGE8 and PS in the mouse (28). Several studies reported the mechanism of mammalian MFGE8 F5/8C binding to PS (47, 49). PS binding allows MFGE8 to complex with vesicles or cell membranes through their second F5/8C domain to perform physiological functions such as cellular interactions and exosome transport (30, 31, 33). Thus, mammalian MFGE8 binds to PS-containing vesicles in a two-step mechanism that explains the strong enrichment of MFGE8 in exosomes from dendritic cells (30, 47). We identified this motif in the G. gallus EDIL3/MFGE8 sequences suggesting that chicken EDIL3 and MFGE8 proteins can also bind to membrane phospholipids.
EGF-like domains are present in all vertebrates but absent in nonvertebrates with the notable exception of EDIL3 of the Cnidaria A. digitifera, which is the most primitive eumetazoan species used in this study. In vertebrates, three EGF-like domains were present, except for EDIL3 in fishes (two domains) and MFGE8 in mammals (one and two domains in primate and nonprimates, respectively). This result confirms previous findings on mammalian MFGE8 (27, 50). The highest-conserved EGF-like domain in vertebrate species includes an RGD motif (arginine, glycine, and aspartic acid) in both MFGE8 and EDIL3 proteins. This motif displays a strong affinity for αvβ3/αvβ5 ITGs, which are expressed at the cell surface and widely present in vertebrates, underlying its major role in cellular interactions (30, 31, 33, 34). It is present in the second and third EGF-like domains in vertebrates. In nonvertebrates, the RGD site is found in the F5/8C domain of protostomian species (arthropods, molluscs, and trematodes) and is completely absent in others (Fig. 5). These results suggest the independent acquisition of the RGD motif in protostomians and in vertebrates. We also predict a Ca2+-binding motif in the third EGF-like domain of EDIL3 and MFGE8 in the majority of the vertebrate sequences analyzed. However, this motif was not observed in mammalian MFGE8 or in EDIL3 of fishes selected for this study. The Ca2+-binding motif was therefore lost in MFGE8 at the split of oviparous/viviparous vertebrates. Interestingly, MFGE8 was reported as the major component of the milk fat globule membrane, which is a calcium-rich milieu (51). In nonvertebrate species, Ca2+-binding sites were only found in the EDIL3 sequence of the staghorn coral (A. digitifera), which precipitates a calcium carbonate exoskeleton. However, we did not identify Ca2+-binding motifs in protein sequences of the purple sea urchin (S. purpuratus) or the eastern oyster (Crassostrea virginica), which also mineralize a CaCO3 skeleton. The cephalochordate B. belcheri, which is a “fish-like” species without a calcareous skeleton, did not exhibit this motif.
The question of the role(s) of these two proteins in avian eggshell formation is the main focus of this study. EDIL3 and MFGE8 were initially proposed to be important molecular actors in chicken eggshell mineralization (21). They were quantified at the four key steps of shell mineralization and classified according to their abundance (21). Among the 216 eggshell proteins quantified in this study, EDIL3 abundance was among the top 10 proteins at the two stages corresponding to the transformation of ACC into calcite crystals. The abundance of MFGE8 was intermediate in the shell, whatever the stage of shell mineralization (21). A comparative study revealed that MFGE8 is overabundant in the proteome of strong eggshells compared with weaker shells (15). In addition to their detection in numerous investigations of the chicken eggshell proteome, both proteins were identified in other avian eggshell proteomes (quail, turkey, and guinea fowl) (23–25). The experiments performed in this study support our hypothesis that EDIL3 plays a key role during eggshell formation. Edil3 was overexpressed in the oviduct regions where eggshell formation takes place (WI, RI, and Ut), and EDIL3 protein was present in SEM, UF, and Ut. We also compared edil3 expression levels with those of other uterine genes during eggshell calcification. We have previously observed by RNA-Seq that the edil3 counting per million (CPM) value was about three times higher than the median CPM value of genes coding for eggshell matrix proteins (52, 53). The median emPAI of proteins quantified in the shell proteome is 5.92, whereas the maximum emPAI was obtained for lysozyme with a value of 13537. The second most abundant protein was Ovocleidin-116 (OC-116, MEPE) with an emPAI value of 7168. EDIL3 exhibited a value of 3523 and was the sixth most abundant eggshell protein. MFGE8 was the 34th most abundant protein with an emPAI value of 37.5. The role of MFGE8 in chicken eggshell biomineralization is not so clear. Indeed, mfge8 was significantly overexpressed in RI where shell mineralization is initiated, but its expression is not specific to the oviduct segment where shell mineralization occurs; moreover, it is ubiquitous in all tested tissues, with the highest levels in liver. Nevertheless, its levels in uterus determined by RNA-Seq were about two times higher than the median CPM value for genes encoding matrix proteins (52, 53).
In mammals, MFGE8 was reported to be involved in spermatozoid-egg zona pellucida-binding (54). In bird eggs, the vitelline membranes that surround the yolk contain zona pellucida proteins that originate from the liver (55). Therefore, MFGE8 could play a role in spermatozoid adhesion. Western blotting confirmed the presence of MFGE8 protein in shell, uterine fluid, and uterus but also in duodenum. In birds, the duodenum is not involved in shell calcification but plays a key role in the uptake of dietary calcium to provide the ions necessary for bone and eggshell formation (56).
Our exploration of edil3 and mfge8 chromosomal arrangements throughout vertebrate evolution highlighted a strongly conserved synteny for both genes (Fig. 1). In avian species, we observed that edil3 and mfge8 are localized on chromosome Z and on a nonsexual chromosome, respectively. Interestingly, quantitative trait loci affecting chicken eggshell strength are present on the Z chromosome (57, 58). Moreover, we observed that the adjacent genes at the edil3 and mfge8 loci are hapln1/vcan and hapln3/acan, respectively. HAPLN3 is one of the most abundant proteins in the chicken eggshell, and HAPLN1 was identified in the eggshell membranes of fertilized eggs (21, 59). Proteoglycans play an important role in eggshell biomineralization; for example, the dermatan sulfate proteoglycan ovocleidin-116 (MEPE) is abundant in eggshells (21, 60–63). Proteoglycan macromolecules combine a protein core with a negatively charged complex polysaccharide and strongly interact with calcium (64). In mammals, HAPLN1, ACAN, and VCAN are cartilage matrix proteins, and they participate in multimolecular aggregates (65–67).
Numerous studies on mammals revealed the participation of vesicles in bone and cartilage mineralization (68–73). A noncrystalline calcium phosphate phase was shown within intracellular vesicles of bone-lining cells (74). Membrane-bound mineral particles have been identified in osteoblasts (75), and numerous vesicles containing calcium phosphate are also present in the circulatory system or in the extracellular space adjacent to osteoblasts during embryonic chicken long-bone development (76). Additionally, MFGE8 is an abundant protein in mineralizing vesicles from chicken embryonic femurs and was also present with EDIL3 in mammalian cartilage vesicles (77–79). Furthermore, HAPLN1, ACAN, and VCAN were also identified in these vesicles (77, 78, 80). Recently, an RNA-Seq study reported that several vesicular genes are highly expressed in hen uterus (52). For example, pdcd6ip and sdcbp encode proteins (PDCD6IP and Syntenin-1) found in matrix vesicles isolated from bone or cartilage (79). In addition, EM studies of hen utero-vaginal junction detected a large number of vesicles released into the lumen (81). Spherical voids can be visualized by scanning EM in the calcified eggshell (82, 83), which we hypothesize are the remnants of mineralization-associated matrix vesicles. Spherical structures (≈300 nm) are observed within the organic matrix of decalcified eggshell (82); they are observed throughout the entire palisade layer and upper mammillary knobs, and they are immunoreactive for the eggshell proteoglycan ovocleidin-116 (84–87).
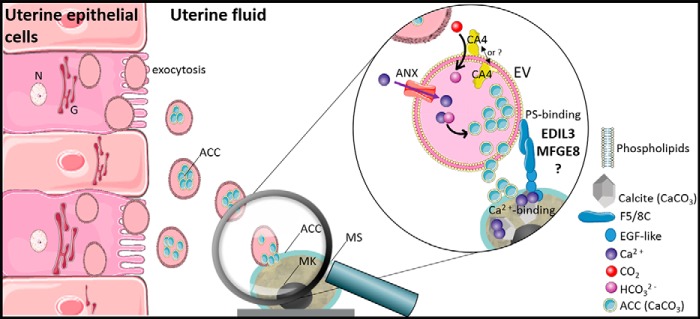
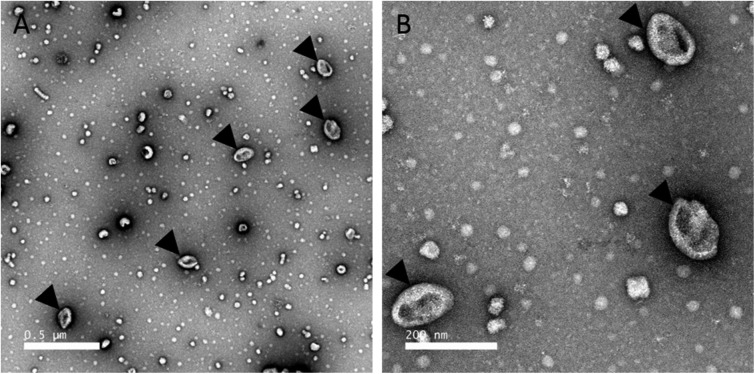
We hypothesize that extracellular vesicles are involved in eggshell formation. Chicken eggshell biomineralization requires ACC to supply ions for rapid mineralization (88, 89). Indeed, ACC was reported as an important highly-reactive translational phase for the formation of sea urchin spines or molluscan shells (89–91). In chicken eggshell, ACC is first massively deposited on the entire surface of the shell membranes, and then it accumulates at specific nucleation sites to supply ions to form calcite (6). ACC is highly unstable under physiological conditions, and consequently, the vesicles observed in eggshell could stabilize and transport ACC during mineralization. In vitro studies have shown that liposome vesicles can stabilize ACC and prevent its premature crystallization (92). Indeed, uterine fluid is a supersaturated solution (93); therefore, transport of ACC via vesicles through the uterine fluid would avoid ectopic calcification, as proposed for bone mineralization by calcium phosphate (75). We also reported in this study that G. gallus EDIL3 and MFGE8 have Ca2+-binding and PS-binding sites. PS binding would allow their attachment to vesicle membranes, and the Ca2+-binding domain could interact with the mineral phase. We therefore propose a model in which EDIL3 and MFGE8 would guide vesicles containing ACC cargo to the site of mineralization (Fig. 7). This hypothesis is supported by the description in dendritic cell exosomes of MFGE8 targeting effector cells ITG as a guiding protein (30). The presence of vesicles in CaCO3 mineralization has been described in sea urchin embryos (94). In this study, EDIL3 and MFGE8 were observed in Ut and UF and would be present in vesicles originating from uterine epithelial cells, transiting through the UF and targeting Ca2+ to the mineralization site to deliver the mineral precursor (Fig. 7). We used transmission electronic microscopy (TEM) to analyze uterine fluid, and we confirmed the presence of EV in the 100–300-nm size range in this milieu (Fig. 8). We also performed energy-dispersive X-ray spectroscopy (EDS) on vesicles to detect elements present in vesicles (Fig. 9). Several mapping measurements were obtained across a vesicle (Fig. 9B) or mapped point by point at different sites where vesicles were present or absent (Fig. 9C). The spectra showed that calcium, carbon, and oxygen were present in vesicles in agreement with calcium carbonate. However, phosphorus and chlorine, representing alternative counterions for calcium (phosphate and chloride), were not detected at these levels. This result is robust evidence for calcium carbonate inside UF vesicles and highlights their contribution to the transport of calcium necessary for calcification.
Figure 7.
Schematic representation of proposed vesicular transport during eggshell mineralisation. EVs originate from the plasma membrane of the uterine epithelial cells and accumulate in the UF. ANX and CA4 provide Ca2+ and HCO3− ions, respectively, for the accumulation of ACC inside the EV. This mechanism implies ACC stabilization by additional proteins. EDIL3 and MFGE8 are present at the EV surface due to their PS-binding motif. EVs target the mineralization site (MS) through Ca2+ binding of EDIL3/MFGE8 and deliver the ACC required for mineral formation. MK, mammillary knobs; N, nucleus; G, Golgi. Elements were from Servier Medical Art, licensed under a Creative Commons Attribution 3.0 Unported License.
Figure 8.
Transmission EM of EV fractions purified from uterine fluid harvested at 7 h post-ovulation. A, low magnification view. B, higher magnification TEM micrograph of the EVs. EVs were negatively stained with 2% uranyl acetate (see “Experimental procedures”). Black arrowheads indicate extracellular EVs. Scale bars, A, 0.5 μm; B, 200 nm.
Figure 9.
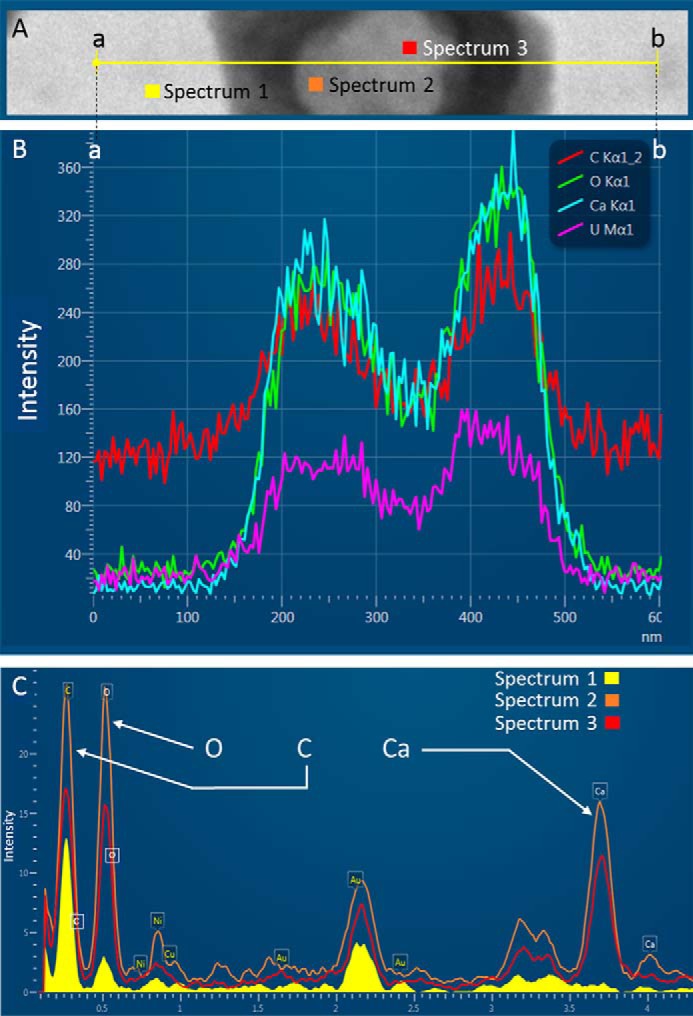
TEM and EDS on EVs from UF collected at 16 h p.o. A, transmission electron microphotography of an extracellular vesicle observed in uterine fluid. The yellow line (from points a to b) indicates the mapping zone across which EDS analysis was performed to obtain elemental spectra along the line for the indicated elements (B). C, spectrum 1 corresponds to EDS analysis of background in the absence of vesicles, and spectrum 2 and spectrum 3 correspond to EDS analysis at two different locations within a vesicle, as indicated in A. UFs were negatively stained with uranyl acetate 2%. Intensities are given in arbitrary units.
Moreover, we confirmed that both proteins are incorporated into the shell matrix during mineralization. Additionally, our results highlighted edil3 and mfge8 expression throughout eggshell formation and notably at the early stages for edil3 when ACC deposition is massive (6). This scenario would take place during the entire process of shell formation, as ACC is required at the mineralization front (6).
We surveyed previous chicken eggshell and uterine fluid proteomic studies (11, 15, 19, 21) to identify additional proteins potentially involved in our proposed vesicular transport model (Table 1). Annexins (ANX) should be involved in vesicular transport as shown in vitro for cartilage or bone (95–97). Indeed, ANXs can act as a Ca2+ channel in vesicle membranes (Fig. 7) (98–100). ANXA1, ANXA2, ANXA5, and ANXA8 are detected in the chicken eggshell, and their encoding genes are highly expressed in uterine cells (11, 15, 21, 52). ANXA2 is also identified in other bird eggshell proteomes such as duck, quail, turkey, and guinea fowl, suggesting a common role in shell biomineralization (25, 101). Eggshell CaCO3 requires bicarbonate ions in addition to calcium to form ACC. This role of the HCO3− supplier inside the vesicles could be ensured by carbonic anhydrases (CAs), which transform soluble CO2 into bicarbonate ions (Fig. 7). Indeed, the UF has a high partial pressure of CO2 (100 mm Hg), which is elevated 2-fold compared with plasma (93, 102). Among the members of the carbonic anhydrase family, CA4 is present in the eggshell proteome of chicken, duck, quail, turkey, and guinea fowl (25, 101). CA4 is a glycosylphosphatidylinositol-anchored protein that is attached to plasma membrane, and consequently, we predict that it is present in the extracellular vesicles. Ovocleidin-17 (OC-17), ovalbumin (OVAL), and lysozyme (LYZ) are three chicken eggshell proteins described as ACC stabilizers (103–105). Finally, we used the Vesiclepedia database (106) to explore vesicular markers among chicken shell proteins already identified (Table 1). We found seven groups of EV marker proteins according to their function: chaperone proteins (HSP), biogenesis factors (PDCD6IP, EZR, and syntenin-1), cell adhesion (FN1), intracellular trafficking proteins (Rab10), membrane organizer (CD9), signaling proteins (YWHAZ and YWHAE), and other cargo (ALB, GSN, VCP, etc.). Moreover, EZR, FN1, GSN, HSPA5, PDCD6IP, and syntenin-1 are present in several avian eggshell proteomes (chicken, duck, quail, and turkey) (101). Their presence in several proteomes highlighted their central role in the shell mineralization process. These proteins could play functional roles in the vesicular transport model proposed herein.
Table 1.
Proteins involved in the proposed vesicular transport
All proteins have been reported in proteomic studies of chicken eggshell (ES) or uterine fluid (UF) (11, 15, 19, 21). ANX and CA can provide Ca2+ or HCO3−, respectively. ACC can be stabilized by LYZ (103), OC17 (105), or OVAL (104). EV markers are from the Vesiclepedia database (106).
| Mineralizing activity |
EV markers found in ES or UF |
|||
|---|---|---|---|---|
| Targeting | Ion accumulation (Ca2+ and HCO3−) | ACC stabilization | Function | Protein symbol |
| EDIL3 | ANXA1 | LYZ | Chaperone | HSP90AA1 |
| MFGE8 | ANXA2 | OC17 | HSP90B1 | |
| ANXA5 | OVAL | HSPA5 | ||
| ANXA8 | HSPA8 | |||
| CA2 | Biogenesis factor | PDCD6IP | ||
| CA4 | Syntenin-1 | |||
| EZR | ||||
| TSG101 | ||||
| Cell adhesion | FN1 | |||
| Intracellular trafficking | RAB10 | |||
| Membrane organiszer | CD9 | |||
| Signaling protein | YWHAE | |||
| YWHAZ | ||||
| Other (cargo) | ALB | |||
| VCP | ||||
| TPI1 | ||||
| GSN | ||||
| LDHB | ||||
| ENO1 | ||||
| GDI2 | ||||
In this study, we used a bioinformatics approach to delineate the evolutionary history of edil3/mfge8 in Eumetazoa, and we suggest that EDIL3 and MFGE8 proteins in vertebrates are derived from a duplication event, as supported by observed synteny in these and adjacent genes. Our gene expression and immunochemistry results allow us to propose a coherent model for EDIL3 and MFGE8 function in vesicles that deliver stabilized ACC for shell mineralization. This scenario involves the vesicular transport of both Ca2+ and HCO3− in the uterine fluid and predicts the involvement of additional vesicular genes, such as annexins. We confirmed the presence of extracellular vesicles in the uterine fluid and detected the elements of calcium, carbon, and oxygen (calcium carbonate) within them.
Experimental procedures
Ethical statement animal handling and housing
The present experiments, including animal handling protocols, were performed in accordance with European Communities Council Directives concerning the practice for the care and use of animals for scientific purposes and the French Ministry of Agriculture on animal experimentation under the supervision of authorized scientists (authorization no. 7323, delivered by the “Direction Départementale de la Protection des Populations d'Indre et Loire-France,” DDPP). Experimental unit (UE-PEAT 1295), where the birds were housed, has an agreement to rear birds and to euthanize experimental animals (decree no. C31-175-1 of August 28, 2012, delivered by the “Préfecture d'Indre et Loire”). The experimental protocol was accepted by the CEEA VdL Ethical Committee (French National Ethics Committee for Animal Experimentation no. 19) and the French Ministry under no. 16099-015902.
Uterine fluid, egg, and tissue collections
Thirty six mature brown laying hens (ISA-Hendrix, 40 and 90 weeks old) were placed in individual furnished cages with automatic devices for recording of the oviposition times. Animals were fed ad libitum (layer mash) using a commercial feed for layers. Laying hens were subjected to a cycle of 14 h of light/10 h of darkness. Following collection and opening of eggs, the eggshells were thoroughly washed with water, air-dried, and stored at −20 °C.
Uterine fluids were collected as described previously (7). Briefly, egg expulsion was induced by an intravenous injection of 50 μg of Dinolytic® prostaglandin-F2α (Zoetis, Malakoff, France) at 7–9 and 16 h p.o., which correspond to the initiation and active phase of shell mineralization. Immediately after egg expulsion, UFs were collected by gravity in a tube placed at the entrance of the everted vagina. UFs were directly frozen in liquid nitrogen in the form of beads or diluted into PBS. UF samples were stored at −20 °C until use. An additional UF aliquot was also diluted 5× for SDS-PAGE into Laemmli sample buffer, for a final composition of 0.0415 m Tris-HCl, pH 6.8, 1% SDS, 10% glycerol, 1.66% β-mercaptoethanol, and 0.01% bromphenol blue.
Tissues were collected from 30 40-week-old and six 90-week-old laying hens. Animals were euthanized with Dolethal® (Vetoquinol, Magny-vernois, France) at the initial phase of eggshell mineralization (5, 6, and 7 h p.o., when the nucleation sites appear and early mineralization starts), at the beginning of the active calcification phase (10 h p.o.) or during the linear growth phase of rapid calcification (16 h p.o.). Various tissues (D, K, and L) and oviduct regions (Ma, WI, RI, and Ut) were collected from birds at each stage of shell calcification. Mid-shaft tibial bone (B) from 90-week-old hens (18 h p.o., at the end of bone resorption) were also harvested. Tissues were directly immersed in liquid nitrogen and were kept at −80 °C.
Soluble eggshell matrix and tissue protein extraction
Eggshell matrix proteins from eggs were extracted as detailed previously (107). Briefly, eggshell pieces with eggshell membranes were immersed in 154 mm NaCl solution containing protease inhibitors (2.5 mm benzamidine-HCl, 50 mm ϵ-amino-n-caproic acid, 0.5 mm N-ethylmaleimide, and 1 mm phenylmethylsulfonyl fluoride) and ground into a fine powder. Eggshell powders were fully demineralized by immersion in 20% acetic acid with overnight stirring at 4 °C. The resulting suspensions were dialyzed (cutoff at 3500 Da) against demineralized water during 24 h at 4 °C and lyophilized. Samples were then incubated overnight at 4 °C in 4 m guanidine HCl, 5 mm benzamidine HCl, 0.1 m ϵ-amino-n-caproic acid, 10 mm EDTA, 50 mm sodium acetate, and 1 mm phenylmethylsulfonyl fluoride. Samples were then dialyzed (cutoff at 3500 Da) against 0.5 m sodium acetate, pH 7.4, solution during 24 h at 4 °C and then centrifuged for 10 min at 2000 × g at 4 °C. The resulting supernatants were stored at −20 °C.
Harvested tissues were homogenized with ULTRA-TURRAX® (IKA, Staufen, Germany) in Tris-buffered saline (TBS: 50 mm Tris-HCl, 77 mm NaCl, pH 7.4, and protease inhibitors), for 0.2 g/ml tissue in each extraction solution. Samples were then centrifuged for 30 min at 10,000 × g, and supernatants were stored at −20 °C.
Electrophoresis and Western blot analysis
Protein concentrations were determined in various samples (SEM: Ut 10 h, UF 9 h, and D 10 h), using Bio-Rad DC Protein assay kit II (Bio-Rad, Marnes-la Coquette, France) in accordance with the manufacturer's instructions and using BSA as a standard. Fifteen micrograms of each sample (SEM: UF, Ut, and D) were diluted into Laemmli sample buffer (5:1), as described previously, and boiled for 5 min. Protein samples were then separated on 12% polyacrylamide gels (Mini-Protean II electrophoresis cell, Bio-Rad, Marnes-la-Coquette, France) and transferred onto 0.2-μm nitrocellulose blotting membrane (GE Healthcare, Little Chalfont, UK) for Western blot analysis. Membranes were washed 5 min in TBS (50 mm Tris-HCl, 150 mm NaCl, pH 7.4) and incubated for 2 h with the Odyssey® blocking buffer (LI-COR, Bad Homburg, Germany) in TBS (1:1). Membranes were then incubated for 3 h in the blocking solution, containing 0.1% Tween 20, with rabbit polyclonal anti-EDIL3 (1: 1000; SAB2105802, Sigma-Aldrich, Saint-Quentin Fallavier, France) or rabbit anti-MFGE8 peptide (GKAEYVRAYKVAYS) (1:1000; ProteoGenix, Schiltigheim, France), and sequentially washed 5 min in TBS, 0.1% Tween 20. The membranes were then incubated 1 h in the blocking solution, 0.1% Tween 20 and AlexaFluor® 680 goat anti-rabbit secondary antibody (1: 20,000, Invitrogen), washed three times in TBS, 0.1% Tween 20, and two times in TBS. Immunobands were revealed with Odyssey® imaging system (LI-COR, Bad Homburg, Germany) with the 700 nm channel.
Gene expression analysis
Total RNA was extracted from frozen samples (B, D, K, L, Ma, WI, RI, and Ut). RNAs from K, D, Ma, WI, RI, and Ut were extracted using NucleoSpin RNA® commercial kit (Macherey-Nagel, Düren, Germany), and the B total RNA was extracted using RNA Now® (Ozyme, Montigny-le-Bretonneux, France) and tandem TRI Reagent®–TRI Reagent LS® commercial kits (Sigma-Aldrich, Saint-Quentin Fallavier, France). Tissues were extracted at 10 h p.o., except for tibial bone, as this sample was solely collected at 18 h p.o. Additionally, Ut and RI samples were extracted at all stages (5- 7, 10, and 16 h p.o.). Total RNAs from each sample were treated with TURBO DNA-free kit (Invitrogen). RNA concentration was measured at 260 nm, and their integrity was assessed on 1.5% agarose gels. Total RNA samples (1 μg) were reverse-transcribed using RNase H–Moloney murine leukemia virus reverse transcriptase (Superscript II, Invitrogen) and Oligo(dT)TM primers (Invitrogen).
Primers to detect expression of G. gallus edil3, mfge8, and eight housekeeping genes (b2m, eif3i, gapdh, gusb, stag2, tbp, sdha, and ppia) were designed using Primer-BLAST (NCBI, https://www.ncbi.nlm.nih.gov/) (Table S4) and synthesized (Eurogentec, Lièges, Belgium). Their efficiencies were evaluated by real-time quantitative RT-PCR using LightCycler® 480 SYBR Green I Master and LightCycler® 480 Instrument II (Roche Applied Science, Bâle, Switzerland). Edil3, mfge8, and housekeeping gene expressions were quantified using the Biomark microfluidic system from Fluidigm, in which every sample–gene combination was quantified using a 96.96 Dynamic ArrayTM IFCs (BMK-M-96.96, Fluidigm, San Francisco) at the Get-Genotoul platform (Toulouse, France). After specific pre-amplification and according to the manufacturer's specifications, the PCR was performed using the following thermal protocol: Thermal Mix at 50 °C, 2 min; 70 °C, 30 min; 25 °C, 10 min; Hot Start at 50 °C, 2 min; 95 °C, 10 min; PCR cycle of 35 cycles of 95 °C, 15 s; 60 °C, 60 s; and melting analysis of 60 °C, 30 s; 95 °C, 1 °C/3 s. RT-qPCR results were analyzed using Fluidigm real-time PCR analysis software version 4.1.3. Six biological replicates and two technical replicates were performed for each sample. GenNorm software was used for validation of housekeeping gene stabilities. Edil3 and mfge8 normalized quantities were calculated using the following formula: gene efficiency (Ctcalibrateur − Ctsample)/geometric average quantity of housekeeping genes. Normalized quantities of edil3 and mfge8 gene transcripts were compared between various measured tissues and oviduct parts using one-way ANOVA and Tukey pairwise test analysis on Minitab® 18 software with p value<0.05 to detect significance.
Synteny and phylogeny of EDIL3 and MFGE8
Synteny of edil3 and mfge8 genes were analyzed using the Genome data viewer of NCBI for eight vertebrate species (three mammals, two birds, one nonavian reptile, one amphibian, and two fishes).
EDIL3 and MFGE8 protein sequences of 12 vertebrate and 7 nonvertebrate species (Table S3) were aligned using Clustal Omega multiple alignment in MEGA7 (108). The appropriate substitution model, determined using MEGA7 substitution model, was the LG model. The phylogenetic tree was built with MEGA7 using Maximum Likelihood method (LG substitution model) with 1000 repetitions and the bootstrap method to evaluate branch robustness. Additionally, phylogenetic analysis using Bayesian inference was performed using BEAST version 1.10.4 (109) with 10,000,000 generations and a sampling of tree each 1000 steps. Trees were then summarized with TreeAnnotator (distributed with BEAST version 1.10.4) and displayed using FigTree version 1.4.4 (115) (http://tree.bio.ed.ac.uk/software/figtree/).3 Divergence times between species were determined using Timetree (www.timetree.org/)3 (113) and are reported on the generated phylogenetic tree.
Distinct protein domain conservation throughout the evolution of eumetazoans was investigated with HomoloGene (NCBI) and domain sequence analysis in several databases and genome browsers (ExPASy-PROSITE, https://prosite.expasy.org/; Ensembl, https://www.ensembl.org/index.html; and SMART, http://smart.embl-heidelberg.de/).3
Analysis of gene and protein sequences of EDIL3 and MFGE8 in G. gallus
G. gallus edil3 and mfge8 gene, mRNA, and protein sequences were collected from the NCBI database (edil3 Gene ID, 427326; edil3 transcripts, XM_424906.6 and XM_004949448.3; EDIL3 proteins, XP_424906.3 and XP_004949505.1; and mfge8 Gene ID, 415494; mfge8 transcripts, NM_001277110.1 and NM_001277111.1; and MFGE8 proteins, NP_001264039.1 and NP_001264040.1). NCBI Genome data viewer was used to identify the exon/intron boundaries and alternative splicing of each gene. EDIL3/MFGE8 (XP_424906.3/NP_001264039.1), HAPLN1/HAPLN3 (XP_015136083.1/XP_413868.3), and VCAN/ACAN (XP_015136071.1/XP_015147464.1) pairwise alignments were performed using Clustal Omega in MEGA7. BioEdit software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html)3 was used to determine identity and similarity percentages between sequences. Protein domains were characterized using PROSITE (https://prosite.expasy.org/)3 and NCBI graphical view. G. gallus EDIL3 and MFGE8 residues involved in Ca2+-binding site and PS binding were identified by homology with H. sapiens coagulation factor IX and X EGF domain residues and M. musculus MFGE8 F5/8C domain residues (28, 110). Alignment of G. gallus EDIL3 (XP_424906.3) and MFGE8 (NP_001264039.1) F5/8C 2 with M. musculus MFGE8 (NP_001038954.1) F5/8C 2 was performed using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) (114).3
G. gallus EDIL3 and MFGE8 secondary and tertiary structure homology
Secondary structure of G. gallus EDIL3 (XP_424906.3) and MFGE8 (NP_001264039.1) peptide signal regions was predicted using PSIPRED version 3.3 (http://bioinf.cs.ucl.ac.uk/psipred/).3 EGF-like and F5/8C sequence regions were then loaded on the I-TASSER program server (111), to generate secondary and tertiary structural predictions. This template-based modeling program computed protein 3D structure predictions by molecular homology for each region of EDIL3 and MFGE8. The 3D structure prediction models were edited on PyMOL Molecular Graphics system.
Purification of extracellular vesicles from uterine fluid, examination by transmission EM, and energy-dispersive X-ray analysis
Immediately after dilution, EVs were isolated from UF harvested at 7 h p.o., using previously described methodology (112). Briefly, diluted UFs were centrifuged at 100 × g for 15 min and then at 12,000 × g for 15 min to remove cell debris (Sorvall ST40 R, Thermo Fisher Scientific, Waltham, MA). Two successive ultracentrifugation steps at 100,000 × g (Beckman Coulter L8–70M, Beckman, Fullerton) were then performed during 90 min to pellet the EVs. The pellet was suspended in 50 μl of PBS and stored at −20 °C until TEM analysis.
Ten microliters of EV fraction were deposited on an electron microscope grid (Copper + Carbon) and then dried for 1 min. Excess liquid was absorbed with filter paper, and EVs were stained with 2% uranyl acetate. The grid was then immediately observed with TEM-1400 Plus electron microscope (120 kV; JEOL, Tokyo, Japan). Additionally, frozen beads (20 μl) of uterine fluid harvested at 16 h p.o. and negatively stained with 2% uranyl acetate were deposited on electron microscope grids, and EDS (Oxford 65-mm2 detector, Aztec software) measurements were performed to detect calcium, oxygen, and carbon.
Author contributions
L. S., N. L. R., M. T. H., and J. G. conceptualization; L. S., N. L. R., M. T. H., and J. G. validation; L. S. and N. L. R. investigation; L. S. and N. L. R. methodology; L. S. writing-original draft; L. S. and J. G. project administration; N. L. R. resources; N. L. R. formal analysis; N. L. R., M. T. H., and J. G. writing-review and editing; J. G. supervision; J. G. funding acquisition.
Supplementary Material
Acknowledgments
We are grateful to Jacky Ezagal for technical skills and help in molecular biology experiments. We also thank the experimental unit (UE-PEAT) for the care of birds, the GeT-GenoToul Platform Toulouse for performing the real time quantitative PCR with the Biomark Fluidigm system, and Pierre-Ivan Raynal and Sonia Georgeault from Electronic Microscopy Platform of the Université François Rabelais de Tours for the microscopic observations.
This work was supported in part by Canadian Natural Sciences and Engineering Research Council (NSERC) Discovery Program RGPIN-2016-04410 (to M. H.) and Université François Rabelais de Tours and the Région Centre Val de Loire (to L. S.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Fig. S1 and Tables S1–S4.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- OM
- organic matrix
- ACAN
- aggrecan
- ACC
- amorphous calcium carbonate
- ANX
- annexin
- B
- tibial bone
- DEL1
- developmental endothelial locus-1
- EDIL3
- EGF-like repeats and discoidin I-like domains 3
- EGF-like
- epidermal growth factor-like
- EV
- extracellular vesicle
- F5/8C
- coagulation factor 5/8 domain
- HCO3−
- bicarbonate ion
- ITG
- integrin
- K
- kidney
- L
- liver
- Ma
- magnum
- MFGE8
- milk fat globule-EGF factor 8
- MYA
- million years ago
- p.o.
- post-ovulation
- PS
- phosphatidylserine
- RI
- red isthmus
- SEM
- soluble eggshell matrix
- UF
- uterine fluid
- Ut
- uterus
- VCAN
- versican
- WI
- white isthmus
- TEM
- transmission EM
- ANOVA
- analysis of variance
- CAM
- chorioallantoic membrane
- EDS
- energy dispersive X-ray spectroscopy
- RNA-Seq
- RNA sequencing
- emPAI
- exponentially modified protein abundance index.
References
- 1. Nys Y., Gautron J., Garcia-Ruiz J. M., and Hincke M. T. (2004) Avian eggshell mineralization: biochemical and functional characterization of matrix proteins. Cr. Palevol. 3, 549–562 [Google Scholar]
- 2. Hincke M. T., Nys Y., Gautron J., Mann K., Rodriguez-Navarro A. B., and McKee M. D. (2012) The eggshell: structure, composition and mineralization. Front. Biosci. 17, 1266–1280 10.2741/3985 [DOI] [PubMed] [Google Scholar]
- 3. Hamilton R. M. G. (1986) The Microstructure of the hens eggshell- a short review. Food Microstruct. 5, 99–110 [Google Scholar]
- 4. Arias J. L., Fink D. J., Xiao S. Q., Heuer A. H., and Caplan A. I. (1993) Biomineralization and eggshells: cell-mediated acellular compartments of mineralized extracellular matrix. Int. Rev. Cytol. 145, 217–250 10.1016/S0074-7696(08)60428-3 [DOI] [PubMed] [Google Scholar]
- 5. Nys Y., Hincke M. T., Arias J. L., Garcia-Ruiz J. M., and Solomon S. E. (1999) Avian eggshell mineralization. Poult. Avian Biol. Rev. 10, 143–166 [Google Scholar]
- 6. Rodríguez-Navarro A. B., Marie P., Nys Y., Hincke M. T., and Gautron J. (2015) Amorphous calcium carbonate controls avian eggshell mineralization: a new paradigm for understanding rapid eggshell calcification. J. Struct. Biol. 190, 291–303 10.1016/j.jsb.2015.04.014 [DOI] [PubMed] [Google Scholar]
- 7. Gautron J., Hincke M. T., and Nys Y. (1997) Precursor matrix proteins in the uterine fluid change with stages of eggshell formation in hens. Connect. Tissue Res. 36, 195–210 10.3109/03008209709160220 [DOI] [PubMed] [Google Scholar]
- 8. Jonchère V., Brionne A., Gautron J., and Nys Y. (2012) Identification of uterine ion transporters for mineralisation precursors of the avian eggshell. BMC Physiol. 12, 10 10.1186/1472-6793-12-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dominguez-Vera J. M., Gautron J., Garcia-Ruiz J. M., and Nys Y. (2000) The effect of avian uterine fluid on the growth behavior of calcite crystals. Poult. Sci. 79, 901–907 10.1093/ps/79.6.901 [DOI] [PubMed] [Google Scholar]
- 10. Hernandez-Hernandez A., Gomez-Morales J., Rodriguez-Navarro A. B., Gautron J., Nys Y., and Garcia-Ruiz J. M. (2008) Identification of some active proteins in the process of hen eggshell formation. Cryst. Growth Des. 8, 4330–4339 10.1021/cg800786s [DOI] [Google Scholar]
- 11. Mann K., Macek B., and Olsen J. V. (2006) Proteomic analysis of the acid-soluble organic matrix of the chicken calcified eggshell layer. Proteomics 6, 3801–3810 10.1002/pmic.200600120 [DOI] [PubMed] [Google Scholar]
- 12. Mann K., Olsen J. V., Macek B., Gnad F., and Mann M. (2007) Phosphoproteins of the chicken eggshell calcified layer. Proteomics 7, 106–115 10.1002/pmic.200600635 [DOI] [PubMed] [Google Scholar]
- 13. Mikšík I., Sedláková P., Lacinová K., Pataridis S., and Eckhardt A. (2010) Determination of insoluble avian eggshell matrix proteins. Anal. Bioanal. Chem. 397, 205–214 10.1007/s00216-009-3326-3 [DOI] [PubMed] [Google Scholar]
- 14. Rose-Martel M., Du J., and Hincke M. T. (2012) Proteomic analysis provides new insight into the chicken eggshell cuticle. J. Proteomics 75, 2697–2706 10.1016/j.jprot.2012.03.019 [DOI] [PubMed] [Google Scholar]
- 15. Sun C., Xu G., and Yang N. (2013) Differential label-free quantitative proteomic analysis of avian eggshell matrix and uterine fluid proteins associated with eggshell mechanical property. Proteomics 13, 3523–3536 10.1002/pmic.201300286 [DOI] [PubMed] [Google Scholar]
- 16. Kaweewong K., Garnjanagoonchorn W., Jirapakkul W., and Roytrakul S. (2013) Solubilization and identification of hen eggshell membrane proteins during different times of chicken embryo development using the proteomic approach. Protein J. 32, 297–308 10.1007/s10930-013-9487-0 [DOI] [PubMed] [Google Scholar]
- 17. Mikšík I., Ergang P., and Pácha J. (2014) Proteomic analysis of chicken eggshell cuticle membrane layer. Anal. Bioanal. Chem. 406, 7633–7640 10.1007/s00216-014-8213-x [DOI] [PubMed] [Google Scholar]
- 18. Ahmed T. A., Suso H. P., and Hincke M. T. (2017) In-depth comparative analysis of the chicken eggshell membrane proteome. J. Proteomics 155, 49–62 10.1016/j.jprot.2017.01.002 [DOI] [PubMed] [Google Scholar]
- 19. Marie P., Labas V., Brionne A., Harichaux G., Hennequet-Antier C., Nys Y., and Gautron J. (2015) Quantitative proteomics and bioinformatic analysis provide new insight into protein function during avian eggshell biomineralization. J. Proteomics 113, 178–193 10.1016/j.jprot.2014.09.024 [DOI] [PubMed] [Google Scholar]
- 20. Gautron J., Guyot N., Brionne A., and Réhault-Godbert S. (2019) in Eggs as Functional Foods and Nutraceuticals for Human Health (Wu J., ed) pp. 259–284, The Royal Society Of Chemistry, London, UK. [Google Scholar]
- 21. Marie P., Labas V., Brionne A., Harichaux G., Hennequet-Antier C., Rodriguez-Navarro A. B., Nys Y., and Gautron J. (2015) Quantitative proteomics provides new insights into chicken eggshell matrix protein functions during the primary events of mineralisation and the active calcification phase. J. Proteomics 126, 140–154 10.1016/j.jprot.2015.05.034 [DOI] [PubMed] [Google Scholar]
- 22. Brionne A., Nys Y., Hennequet-Antier C., and Gautron J. (2014) Hen uterine gene expression profiling during eggshell formation reveals putative proteins involved in the supply of minerals or in the shell mineralization process. BMC Genomics 15, 220 10.1186/1471-2164-15-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mann K., and Mann M. (2013) The proteome of the calcified layer organic matrix of turkey (Meleagris gallopavo) eggshell. Proteome Sci. 11, 40 10.1186/1477-5956-11-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mann K., and Mann M. (2015) Proteomic analysis of quail calcified eggshell matrix: a comparison to chicken and turkey eggshell proteomes. Proteome Sci. 13, 22 10.1186/s12953-015-0078-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deleted in proof.
- 26. Oshima K., Aoki N., Negi M., Kishi M., Kitajima K., and Matsuda T. (1999) Lactation-dependent expression of an mRNA splice variant with an exon for a multiply O-glycosylated domain of mouse milk fat globule glycoprotein MFG-E8. Biochem. Biophys. Res. Commun. 254, 522–528 10.1006/bbrc.1998.0107 [DOI] [PubMed] [Google Scholar]
- 27. Oshima K., Yasueda T., Nishio S., and Matsuda T. (2014) in MFG-E8 and Inflammation (Wang P., ed) pp. 1–31, Springer, Dordrecht, The Netherlands. [Google Scholar]
- 28. Ye H., Li B., Subramanian V., Choi B. H., Liang Y., Harikishore A., Chakraborty G., Baek K., and Yoon H. S. (2013) NMR solution structure of C2 domain of MFG-E8 and insights into its molecular recognition with phosphatidylserine. Biochim. Biophys. Acta 1828, 1083–1093 10.1016/j.bbamem.2012.12.009 [DOI] [PubMed] [Google Scholar]
- 29. Stubbs J. D., Lekutis C., Singer K. L., Bui A., Yuzuki D., Srinivasan U., and Parry G. (1990) Cdna cloning of a mouse mammary epithelial-cell surface protein reveals the existence of epidermal growth factor-like domains linked to factor-Viii-like sequences. Proc. Natl. Acad. Sci. U.S.A. 87, 8417–8421 10.1073/pnas.87.21.8417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Théry C., Regnault A., Garin J., Wolfers J., Zitvogel L., Ricciardi-Castagnoli P., Raposo G., and Amigorena S. (1999) Molecular characterization of dendritic cell-derived exosomes: Selective accumulation of the heat shock protein hsc73. J. Cell Biol. 147, 599–610 10.1083/jcb.147.3.599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hanayama R., Tanaka M., Miwa K., Shinohara A., Iwamatsu A., and Nagata S. (2002) Identification of a factor that links apoptotic cells to phagocytes. Nature 417, 182–187 10.1038/417182a [DOI] [PubMed] [Google Scholar]
- 32. Ensslin M. A., and Shur B. D. (2003) Identification of mouse sperm SED1, a bimotif EGF repeat and discoidin-domain protein involved in sperm-egg binding. Cell 114, 405–417 10.1016/S0092-8674(03)00643-3 [DOI] [PubMed] [Google Scholar]
- 33. Gatti J. L., Métayer S., Belghazi M., Dacheux F., and Dacheux J. L. (2005) Identification, proteomic profiling, and origin of ram epididymal fluid exosome-like vesicles. Biol. Reprod. 72, 1452–1465 10.1095/biolreprod.104.036426 [DOI] [PubMed] [Google Scholar]
- 34. Ait-Oufella H., Kinugawa K., Zoll J., Simon T., Boddaert J., Heeneman S., Blanc-Brude O., Barateau V., Potteaux S., Merval R., Esposito B., Teissier E., Daemen M. J., Lesèche G., Boulanger C., et al. (2007) Lactadherin deficiency leads to apoptotic cell accumulation and accelerated atherosclerosis in mice. Circulation 115, 2168–2177 10.1161/CIRCULATIONAHA.106.662080 [DOI] [PubMed] [Google Scholar]
- 35. Hidai C., Zupancic T., Penta K., Mikhail A., Kawana M., Quertermous E. E., Aoka Y., Fukagawa M., Matsui Y., Platika D., Auerbach R., Hogan B. L., Snodgrass R., and Quertermous T. (1998) Cloning and characterization of developmental endothelial locus-1: An embryonic endothelial cell protein that binds the αvβ3 integrin receptor. Gene Dev. 12, 21–33 10.1101/gad.12.1.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Penta K., Varner J. A., Liaw L., Hidai C., Schatzman R., and Quertermous T. (1999) Del1 induces integrin signaling and angiogenesis by ligation of αVβ3. J. Biol. Chem. 274, 11101–11109 10.1074/jbc.274.16.11101 [DOI] [PubMed] [Google Scholar]
- 37. Zhong J., Eliceiri B., Stupack D., Penta K., Sakamoto G., Quertermous T., Coleman M., Boudreau N., and Varner J. A. (2003) Neovascularization of ischemic tissues by gene delivery of the extracellular matrix protein Del-1. J. Clin. Invest. 112, 30–41 10.1172/JCI17034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zou X., Qiao H., Jiang X., Dong X., Jiang H., and Sun X. (2009) Downregulation of developmentally regulated endothelial cell locus-1 inhibits the growth of colon cancer. J. Biomed. Sci. 16, 33 10.1186/1423-0127-16-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Aoka Y., Johnson F. L., Penta K., Hirata Ki K., Hidai C., Schatzman R., Varner J. A., and Quertermous T. (2002) The embryonic angiogenic factor Del1 accelerates tumor growth by enhancing vascular formation. Microvasc. Res. 64, 148–161 10.1006/mvre.2002.2414 [DOI] [PubMed] [Google Scholar]
- 40. Lee J. E., Moon P. G., Cho Y. E., Kim Y. B., Kim I. S., Park H., and Baek M. C. (2016) Identification of EDIL3 on extracellular vesicles involved in breast cancer cell invasion. J. Proteomics 131, 17–28 10.1016/j.jprot.2015.10.005 [DOI] [PubMed] [Google Scholar]
- 41. Pavlopoulou A., Pampalakis G., Michalopoulos I., and Sotiropoulou G. (2010) Evolutionary history of tissue kallikreins. PLoS ONE 5, e13781 10.1371/journal.pone.0013781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Packard M. J., and Seymour R. S. (1997) in Amniote Origins (Sumida S.S., and Martin K. L.M., eds) pp. 265–290, Academic Press, London [Google Scholar]
- 43. Romer A. S. (1957) Origin of the amniote egg. Scientific Monthly 85, 57–63 [Google Scholar]
- 44. Larocca D., Peterson J. A., Urrea R., Kuniyoshi J., Bistrain A. M., and Ceriani R. L. (1991) A Mr 46,000 human-milk fat globule protein that is highly expressed in human breast tumors contains factor Viii-like domains. Cancer Res. 51, 4994–4998 [PubMed] [Google Scholar]
- 45. Andersen M. H., Berglund L., Rasmussen J. T., and Petersen T. E. (1997) Bovine PAS-6/7 binds α(V)β(5) integrin and anionic phospholipids through two domains. Biochemistry 36, 5441–5446 10.1021/bi963119m [DOI] [PubMed] [Google Scholar]
- 46. Shi J., Heegaard C. W., Rasmussen J. T., and Gilbert G. E. (2004) Lactadherin binds selectively to membranes containing phosphatidyl-l-serine and increased curvature. Biochim. Biophys. Acta 1667, 82–90 10.1016/j.bbamem.2004.09.006 [DOI] [PubMed] [Google Scholar]
- 47. Otzen D. E., Blans K., Wang H., Gilbert G. E., and Rasmussen J. T. (2012) Lactadherin binds to phosphatidylserine-containing vesicles in a two-step mechanism sensitive to vesicle size and composition. Biochim. Biophys. Acta 1818, 1019–1027 10.1016/j.bbamem.2011.08.032 [DOI] [PubMed] [Google Scholar]
- 48. Shao C., Novakovic V. A., Head J. F., Seaton B. A., and Gilbert G. E. (2008) Crystal structure of lactadherin C2 domain at 1.7A resolution with mutational and computational analyses of its membrane-binding motif. J. Biol. Chem. 283, 7230–7241 10.1074/jbc.M705195200 [DOI] [PubMed] [Google Scholar]
- 49. Oshima K., Aoki N., Kato T., Kitajima K., and Matsuda T. (2002) Secretion of a peripheral membrane protein, MFG-E8, as a complex with membrane vesicles. Eur. J. Biochem. 269, 1209–1218 10.1046/j.1432-1033.2002.02758.x [DOI] [PubMed] [Google Scholar]
- 50. Podlaha O., Webb D. M., and Zhang J. (2006) Accelerated evolution and loss of a domain of the sperm–egg-binding protein SED1 in ancestral primates. Mol. Biol. Evol. 23, 1828–1831 10.1093/molbev/msl066 [DOI] [PubMed] [Google Scholar]
- 51. Hvarregaard J., Andersen M. H., Berglund L., Rasmussen J. T., and Petersen T. E. (1996) Characterization of glycoprotein PAS-6/7 from membranes of bovine milk fat globules. Eur. J. Biochem. 240, 628–636 10.1111/j.1432-1033.1996.0628h.x [DOI] [PubMed] [Google Scholar]
- 52. Gautron J. (2018) ANR IMPACT report, Agence National de la Recherche, Paris, France. [Google Scholar]
- 53. Deleted in proof.
- 54. Raymond A., Ensslin M. A., and Shur B. D. (2009) SED1/MFG-E8: a Bi-Motif protein that orchestrates diverse cellular interactions. J. Cell Biochem. 106, 957–966 10.1002/jcb.22076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nys Y., and Guyot N. (2011) in Improving the Safety and Quality of Eggs and Egg Products (Yves N., Maureen B., Filip V.I.,) pp. 83–132, Wooding head publishing, Cambridge, UK. [Google Scholar]
- 56. Nys Y., Parkes C. O., and Thomasset M. (1986) Effects of suppression and resumption of shell formation and parathyroid hormone on uterine calcium-binding protein, carbonic anhydrase activity, and intestinal calcium absorption in hens. Gen. Comp. Endocrinol. 64, 293–299 10.1016/0016-6480(86)90016-X [DOI] [PubMed] [Google Scholar]
- 57. Ankra-Badu G., and Aggrey S. (2005) Identification of candidate genes at quantitative trait loci on chicken chromosome Z using orthologous comparison of chicken, mouse, and human genomes. In Silico Biol. 5, 593–604 [PubMed] [Google Scholar]
- 58. Tuiskula-Haavisto M., Honkatukia M., Vilkki J., de Koning D. J., Schulman N. F., and Maki-Tanila A. (2002) Mapping of quantitative trait loci affecting quality and production traits in egg layers. Poult. Sci. 81, 919–927 10.1093/ps/81.7.919 [DOI] [PubMed] [Google Scholar]
- 59. Cordeiro C. M., and Hincke M. T. (2016) Quantitative proteomics analysis of eggshell membrane proteins during chick embryonic development. J. Proteomics 130, 11–25 10.1016/j.jprot.2015.08.014 [DOI] [PubMed] [Google Scholar]
- 60. Carrino D. A., Dennis J. E., Wu T. M., Arias J. L., Fernandez M. S., Rodriguez J. P., Fink D. J., Heuer A. H., and Caplan A. I. (1996) The avian eggshell extracellular matrix as a model for biomineralization. Connect. Tissue Res. 35, 325–329 10.3109/03008209609029207 [DOI] [PubMed] [Google Scholar]
- 61. Fernandez M. S., Araya M., and Arias J. L. (1997) Eggshells are shaped by a precise spatio-temporal arrangement of sequentially deposited macromolecules. Matrix Biol. 16, 13–20 10.1016/S0945-053X(97)90112-8 [DOI] [PubMed] [Google Scholar]
- 62. Nakano T., Ikawa N., and Ozimek L. (2001) Extraction of glycosaminoglycans from chicken eggshell. Poult. Sci. 80, 681–684 10.1093/ps/80.5.681 [DOI] [PubMed] [Google Scholar]
- 63. Nakano T., Ikawa N., and Ozimek L. (2002) Galactosaminoglycan composition in chicken eggshell. Poult. Sci. 81, 709–714 10.1093/ps/81.5.709 [DOI] [PubMed] [Google Scholar]
- 64. Hunter G. K., Wong K. S., and Kim J. J. (1988) Binding of calcium to glycosaminoglycans: an equilibrium dialysis study. Arch. Biochem. Biophys. 260, 161–167 10.1016/0003-9861(88)90437-7 [DOI] [PubMed] [Google Scholar]
- 65. Kiani C., Chen L., Wu Y. J., Yee A. J., and Yang B. B. (2002) Structure and function of aggrecan. Cell Res. 12, 19–32 10.1038/sj.cr.7290106 [DOI] [PubMed] [Google Scholar]
- 66. Shibata S., Fukada K., Imai H., Abe T., and Yamashita Y. (2003) In situ hybridization and immunohistochemistry of versican, aggrecan and link protein, and histochemistry of hyaluronan in the developing mouse limb bud cartilage. J. Anat. 203, 425–432 10.1046/j.1469-7580.2003.00226.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Matsumoto K., Kamiya N., Suwan K., Atsumi F., Shimizu K., Shinomura T., Yamada Y., Kimata K., and Watanabe H. (2006) Identification and characterization of versican/PG-M aggregates in cartilage. J. Biol. Chem. 281, 18257–18263 10.1074/jbc.M510330200 [DOI] [PubMed] [Google Scholar]
- 68. Anderson H. C. (1969) Vesicles associated with calcification in the matrix of epiphyseal cartilage. J. Cell Biol. 41, 59–72 10.1083/jcb.41.1.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Xiao Z., Camalier C. E., Nagashima K., Chan K. C., Lucas D. A., de la Cruz M. J., Gignac M., Lockett S., Issaq H. J., Veenstra T. D., Conrads T. P., and Beck G. R. Jr. (2007) Analysis of the extracellular matrix vesicle proteome in mineralizing osteoblasts. J. Cell Physiol. 210, 325–335 10.1002/jcp.20826 [DOI] [PubMed] [Google Scholar]
- 70. Golub E. E. (2009) Role of matrix vesicles in biomineralization. Biochim. Biophys. Acta 1790, 1592–1598 10.1016/j.bbagen.2009.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Morhayim J., Baroncelli M., and van Leeuwen J. P. (2014) Extracellular vesicles: specialized bone messengers. Arch. Biochem. Biophys. 561, 38–45 10.1016/j.abb.2014.05.011 [DOI] [PubMed] [Google Scholar]
- 72. Bottini M., Mebarek S., Anderson K. L., Strzelecka-Kiliszek A., Bozycki L., Simão A. M. S., Bolean M., Ciancaglini P., Pikula J. B., Pikula S., Magne D., Volkmann N., Hanein D., Millán J. L., and Buchet R. (2018) Matrix vesicles from chondrocytes and osteoblasts: their biogenesis, properties, functions and biomimetic models. Biochim. Biophys. Acta Gen. Subj. 1862, 532–546 10.1016/j.bbagen.2017.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hasegawa T., Yamamoto T., Tsuchiya E., Hongo H., Tsuboi K., Kudo A., Abe M., Yoshida T., Nagai T., Khadiza N., Yokoyama A., Oda K., Ozawa H., de Freitas P. H. L., Li M., and Amizuka N. (2017) Ultrastructural and biochemical aspects of matrix vesicle-mediated mineralization. Jpn. Dent. Sci. Rev. 53, 34–45 10.1016/j.jdsr.2016.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mahamid J., Sharir A., Gur D., Zelzer E., Addadi L., and Weiner S. (2011) Bone mineralization proceeds through intracellular calcium phosphate loaded vesicles: a cryo-electron microscopy study. J. Struct. Biol. 174, 527–535 10.1016/j.jsb.2011.03.014 [DOI] [PubMed] [Google Scholar]
- 75. Kerschnitzki M., Akiva A., Ben Shoham A., Asscher Y., Wagermaier W., Fratzl P., Addadi L., and Weiner S. (2016) Bone mineralization pathways during the rapid growth of embryonic chicken long bones. J. Struct. Biol. 195, 82–92 10.1016/j.jsb.2016.04.011 [DOI] [PubMed] [Google Scholar]
- 76. Kerschnitzki M., Akiva A., Shoham A. B., Koifman N., Shimoni E., Rechav K., Arraf A. A., Schultheiss T. M., Talmon Y., Zelzer E., Weiner S., and Addadi L. (2016) Transport of membrane-bound mineral particles in blood vessels during chicken embryonic bone development. Bone 83, 65–72 10.1016/j.bone.2015.10.009 [DOI] [PubMed] [Google Scholar]
- 77. Balcerzak M., Malinowska A., Thouverey C., Sekrecka A., Dadlez M., Buchet R., and Pikula S. (2008) Proteome analysis of matrix vesicles isolated from femurs of chicken embryo. Proteomics 8, 192–205 10.1002/pmic.200700612 [DOI] [PubMed] [Google Scholar]
- 78. Rosenthal A. K., Gohr C. M., Ninomiya J., and Wakim B. T. (2011) Proteomic analysis of articular cartilage vesicles from normal and osteoarthritic cartilage. Arthritis Rheum. 63, 401–411 10.1002/art.30120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Shapiro I. M., Landis W. J., and Risbud M. V. (2015) Matrix vesicles: are they anchored exosomes? Bone 79, 29–36 10.1016/j.bone.2015.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Thouverey C., Malinowska A., Balcerzak M., Strzelecka-Kiliszek A., Buchet R., Dadlez M., and Pikula S. (2011) Proteomic characterization of biogenesis and functions of matrix vesicles released from mineralizing human osteoblast-like cells. J. Proteomics 74, 1123–1134 10.1016/j.jprot.2011.04.005 [DOI] [PubMed] [Google Scholar]
- 81. Waqas M. Y., Yang P., Ahmed N., Zhang Q., Liu T., Li Q., Hu L., Hong C., and Chen Q. (2016) Characterization of the ultrastructure in the uterovaginal junction of the hen. Poult. Sci. 95, 2112–2119 10.3382/ps/pew141 [DOI] [PubMed] [Google Scholar]
- 82. Hincke M. T., Chien Y. C., Gerstenfeld L. C., and McKee M. D. (2008) Colloidal-gold immunocytochemical localization of osteopontin in avian eggshell gland and eggshell. J. Histochem. Cytochem. 56, 467–476 10.1369/jhc.2008.950576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chien Y. C., Hincke M. T., and McKee M. D. (2009) Ultrastructure of avian eggshell during resorption following egg fertilization. J. Struct. Biol. 168, 527–538 10.1016/j.jsb.2009.07.005 [DOI] [PubMed] [Google Scholar]
- 84. Wyburn G. M., Johnston H. S., Draper M. H., and Davidson M. F. (1973) The ultrastructure of the shell forming region of the oviduct and the development of the shell of Gallus domesticus. Q. J. Exp. Physiol. Cogn. Med. Sci. 58, 143–151 [PubMed] [Google Scholar]
- 85. Dennis J. E., Xiao S. Q., Agarwal M., Fink D. J., Heuer A. H., and Caplan A. I. (1996) Microstructure of matrix and mineral components of eggshells from White Leghorn chickens (G. gallus). J. Morphol. 228, 287–306 [DOI] [PubMed] [Google Scholar]
- 86. Fraser A. C., Bain M. M., and Solomon S. E. (1998) Organic matrix morphology and distribution in the palisade layer of eggshells sampled at selected periods during lay. Br. Poult. Sci. 39, 225–228 10.1080/00071669889169 [DOI] [PubMed] [Google Scholar]
- 87. Hincke M. T., Gautron J., Tsang C. P., McKee M. D., and Nys Y. (1999) Molecular cloning and ultrastructural localization of the core protein of an eggshell matrix proteoglycan, ovocleidin-116. J. Biol. Chem. 274, 32915–32923 10.1074/jbc.274.46.32915 [DOI] [PubMed] [Google Scholar]
- 88. Brecevic L., and Nielsen A. E. (1989) Solubility of amorphous calcium-carbonate. J. Cryst. Growth 98, 504–510 10.1016/0022-0248(89)90168-1 [DOI] [Google Scholar]
- 89. Addadi L., Raz S., and Weiner S. (2003) Taking advantage of disorder: amorphous calcium carbonate and its roles in biomineralization. Adv. Mater. 15, 959–970 10.1002/adma.200300381 [DOI] [Google Scholar]
- 90. Weiss I. M., Tuross N., Addadi L., and Weiner S. (2002) Mollusc larval shell formation: amorphous calcium carbonate is a precursor phase for aragonite. J. Exp. Zool. 293, 478–491 10.1002/jez.90004 [DOI] [PubMed] [Google Scholar]
- 91. Luquet G. (2012) Biomineralizations: insights and prospects from crustaceans. Zookeys 2012, 103–121 10.3897/zookeys.176.2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Tester C. C., Brock R. E., Wu C. H., Krejci M. R., Weigand S., and Joester D. (2011) In vitro synthesis and stabilization of amorphous calcium carbonate (ACC) nanoparticles within liposomes. CrystEngComm 13, 3975–3978 10.1039/c1ce05153a [DOI] [Google Scholar]
- 93. Nys Y., Zawadzki J., Gautron J., and Mills A. D. (1991) Whitening of brown-shelled eggs: mineral composition of uterine fluid and rate of protoporphyrin deposition. Poult. Sci. 70, 1236–1245 10.3382/ps.0701236 [DOI] [PubMed] [Google Scholar]
- 94. Vidavsky N., Masic A., Schertel A., Weiner S., and Addadi L. (2015) Mineral-bearing vesicle transport in sea urchin embryos. J. Struct. Biol. 192, 358–365 10.1016/j.jsb.2015.09.017 [DOI] [PubMed] [Google Scholar]
- 95. Kirsch T., Harrison G., Golub E. E., and Nah H. D. (2000) The roles of annexins and types II and X collagen in matrix vesicle-mediated mineralization of growth plate cartilage. J. Biol. Chem. 275, 35577–35583 10.1074/jbc.M005648200 [DOI] [PubMed] [Google Scholar]
- 96. Genge B. R., Wu L. N., and Wuthier R. E. (2007) In vitro modeling of matrix vesicle nucleation: synergistic stimulation of mineral formation by annexin A5 and phosphatidylserine. J. Biol. Chem. 282, 26035–26045 10.1074/jbc.M701057200 [DOI] [PubMed] [Google Scholar]
- 97. Merolli A., and Santin M. (2009) Role of phosphatidyl-serine in bone repair and its technological exploitation. Molecules 14, 5367–5381 10.3390/molecules14125367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Rojas E., Arispe N., Haigler H. T., Burns A. L., and Pollard H. B. (1992) Identification of annexins as calcium channels in biological-membranes. Bone Miner. 17, 214–218 10.1016/0169-6009(92)90739-Z [DOI] [PubMed] [Google Scholar]
- 99. Wuthier R. E., Wu L. N., Sauer G. R., Genge B. R., Yoshimori T., and Ishikawa Y. (1992) Mechanism of matrix vesicle calcification–characterization of ion channels and the nucleational core of growth plate vesicles. Bone Miner. 17, 290–295 10.1016/0169-6009(92)90753-Z [DOI] [PubMed] [Google Scholar]
- 100. Kirsch T., Nah H. D., Demuth D. R., Harrison G., Golub E. E., Adams S. L., and Pacifici M. (1997) Annexin V-mediated calcium flux across membranes is dependent on the lipid composition: implications for cartilage mineralization. Biochemistry 36, 3359–3367 10.1021/bi9626867 [DOI] [PubMed] [Google Scholar]
- 101. Zhu F., Zhang F., Hincke M., Yin Z. T., Chen S. R., Yang N., and Hou Z. C. (2019) iTRAQ-based quantitative proteomic analysis of duck eggshell during biomineralization. Proteomics 19, e1900011 10.1002/pmic.201900011 [DOI] [PubMed] [Google Scholar]
- 102. Arad Z., Eylath U., Ginsburg M., and Eyal-Giladi H. (1989) Changes in uterine fluid composition and acid-base status during shell formation in the chicken. Am. J. Physiol. 257, R732–R737 10.1152/ajpregu.1989.257.4.R732 [DOI] [PubMed] [Google Scholar]
- 103. Voinescu A. E., Touraud D., Lecker A., Pfitzner A., Kunz W., and Ninham B. W. (2007) Mineralization of CaCO3 in the presence of egg white lysozyme. Langmuir 23, 12269–12274 10.1021/la701892v [DOI] [PubMed] [Google Scholar]
- 104. Pipich V., Balz M., Wolf S. E., Tremel W., and Schwahn D. (2008) Nucleation and growth of CaCO3 mediated by the egg-white protein ovalbumin: a time-resolved in situ study using small-angle neutron scattering. J. Am. Chem. Soc. 130, 6879–6892 10.1021/ja801798h [DOI] [PubMed] [Google Scholar]
- 105. Freeman C. L., Harding J. H., Quigley D., and Rodger P. M. (2010) Structural control of crystal nuclei by an eggshell protein. Angew. Chem. Int. Ed. Engl. 49, 5135–5137 10.1002/anie.201000679 [DOI] [PubMed] [Google Scholar]
- 106. Pathan M., Fonseka P., Chitti S. V., Kang T., Sanwlani R., Van Deun J., Hendrix A., and Mathivanan S. (2019) Vesiclepedia 2019: a compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res. 47, D516–D519 10.1093/nar/gky1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Gautron J., Hincke M. T., Panheleux M., Garcia-Ruiz J. M., Boldicke T., and Nys Y. (2001) Ovotransferrin is a matrix protein of the hen eggshell membranes and basal calcified layer. Connect. Tissue Res. 42, 255–267 10.3109/03008200109016840 [DOI] [PubMed] [Google Scholar]
- 108. Kumar S., Stecher G., and Tamura K. (2016) MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Drummond A. J., and Rambaut A. (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Handford P. A., Baron M., Mayhew M., Willis A., Beesley T., Brownlee G. G., and Campbell I. D. (1990) The first EGF-like domain from human factor IX contains a high-affinity calcium binding site. EMBO J. 9, 475–480 10.1002/j.1460-2075.1990.tb08133.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Yang J., and Zhang Y. (2015) I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 43, W174–W181 10.1093/nar/gkv342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Thery C., Amigorena S., Raposo G., and Clayton A. (2006) Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, Chapter 3, Unit 3.22 10.1002/0471143030.cb0322s30 [DOI] [PubMed] [Google Scholar]
- 113. Kumar S., Stecher G., Suleski M., and Hedges S. B. (2017) TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819 10.1093/molbev/msx116 [DOI] [PubMed] [Google Scholar]
- 114. Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S., Park Y. M., Buso N., and Lopez R. (2015) The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 43, W580–W584 10.1093/nar/gkv279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Rambaut A. (2009) FigTree, Version 1.4.4 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.