This study documents the involvement of ETF in anaerobic toluene metabolism as the physiological electron acceptor for benzylsuccinyl-CoA dehydrogenase. While toluene-degrading denitrifying proteobacteria use a common ETF species, which is also used for other β-oxidation pathways, obligately anaerobic sulfate- or ferric-iron-reducing bacteria use specialized ETF paralogues for toluene degradation. Based on the structure and sequence conservation of these ETFs, they form a new clade that is only remotely related to the previously characterized members of the ETF family. An exhaustive analysis of the available sequences indicated that the protein family consists of several closely related clades of proven or potential electron-bifurcating ETF species and many deeply branching nonbifurcating clades, which either follow the host phylogeny or are affiliated according to functional criteria.
KEYWORDS: Geobacter, X-ray structure, anaerobic toluene degradation, benzylsuccinyl-CoA dehydrogenase, electron transfer, flavin
ABSTRACT
(R)-Benzylsuccinate is the characteristic initial intermediate of anaerobic toluene metabolism, which is formed by a radical-type addition of toluene to fumarate. Its further degradation proceeds by activation to the coenzyme A (CoA)-thioester and β-oxidation involving a specific (R)-2-benzylsuccinyl-CoA dehydrogenase (BbsG) affiliated with the family of acyl-CoA dehydrogenases. In this report, we present the biochemical properties of electron transfer flavoproteins (ETFs) from the strictly anaerobic toluene-degrading species Geobacter metallireducens and Desulfobacula toluolica and the facultatively anaerobic bacterium Aromatoleum aromaticum. We determined the X-ray structure of the ETF paralogue involved in toluene metabolism of G. metallireducens, revealing strong overall similarities to previously characterized ETF variants but significantly different structural properties in the hinge regions mediating conformational changes. We also show that all strictly anaerobic toluene degraders utilize one of multiple genome-encoded related ETF paralogues, which constitute a distinct clade of similar sequences in the ETF family, for β-oxidation of benzylsuccinate. In contrast, facultatively anaerobic toluene degraders contain only one ETF species, which is utilized in all β-oxidation pathways. Our phylogenetic analysis of the known sequences of the ETF family suggests that at least 36 different clades can be differentiated, which are defined either by the taxonomic group of the respective host species (e.g., clade P for Proteobacteria) or by functional specialization (e.g., clade T for anaerobic toluene degradation).
IMPORTANCE This study documents the involvement of ETF in anaerobic toluene metabolism as the physiological electron acceptor for benzylsuccinyl-CoA dehydrogenase. While toluene-degrading denitrifying proteobacteria use a common ETF species, which is also used for other β-oxidation pathways, obligately anaerobic sulfate- or ferric-iron-reducing bacteria use specialized ETF paralogues for toluene degradation. Based on the structure and sequence conservation of these ETFs, they form a new clade that is only remotely related to the previously characterized members of the ETF family. An exhaustive analysis of the available sequences indicated that the protein family consists of several closely related clades of proven or potential electron-bifurcating ETF species and many deeply branching nonbifurcating clades, which either follow the host phylogeny or are affiliated according to functional criteria.
INTRODUCTION
Alkylbenzenes are an important class of aromatic hydrocarbons that occur naturally in crude oil, coal, and mineral oil-related products or are produced by incomplete combustion events. Their relatively high water solubility makes them amenable to transport with surface or groundwater flows. Therefore, they are widespread in the environment and occur at low concentrations even in noncontaminated subsurface systems such as groundwater, sediments, or interstitial rock deposits (1–3). The small amounts of oxygen present in these habitats are rapidly consumed by microbial activity and cannot be replenished from the surface, leading to anoxic conditions. Therefore, anaerobic alkylbenzene metabolism plays a major role in subsurface environments and should be taken into account for bioremediation efforts for fuel-contaminated aquifers or other subsurface environments. Typical bacterial species accounting for these processes are either facultatively anaerobic denitrifiers or strictly anaerobic sulfate- or metal-ion-reducing bacteria (1). The latter group consists especially of species affiliated with the metal ion-reducing genera Geobacter (Deltaproteobacteria) and Georgfuchsia (Betaproteobacteria) or various genera of sulfate-reducing bacteria affiliated with either the Gram-negative Deltaproteobacteria or the Gram-positive Firmicutes.
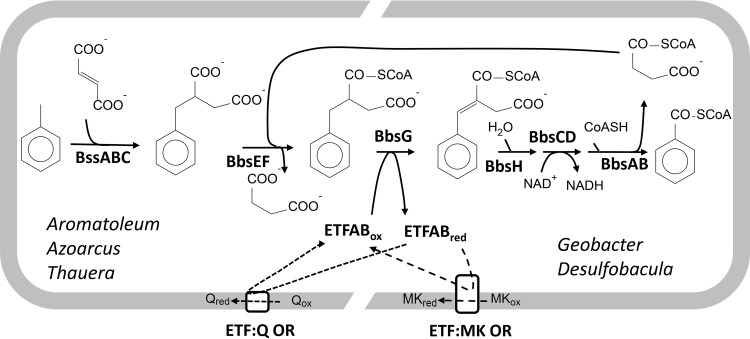
Toluene was one of the first hydrocarbons shown to be degraded anaerobically and has since been widely used as a model compound for studying the pathways involved (1, 4, 5). The most critical step in toluene degradation is the initial attack on the hydrocarbon, which is exclusively catalyzed by oxygen-dependent mono- or dioxygenases in aerobic degradation pathways. In contrast, all currently investigated anaerobic toluene degraders initiate the pathway through formation of (R)-benzylsuccinate from toluene and fumarate in a radical addition reaction catalyzed by the glycyl radical enzyme benzylsuccinate synthase (BSS) (Fig. 1) (6–8). The same pathway was found in many bacteria of different physiological groups that couple to various electron acceptors in anaerobic respiration, such as nitrate, sulfate, iron(III), manganese(IV), or carbonate, or degrade toluene phototrophically (1, 8). While most studies were performed with facultatively anaerobic denitrifying betaproteobacteria (9, 10), the best studied strictly anaerobic toluene-degrading species are iron(III)-reducing Geobacter metallireducens and sulfate-reducing Desulfobacula toluolica (11, 12).
FIG 1.
Pathway of anaerobic toluene degradation via benzylsuccinate. The left half shows the proposed pathway of facultative anaerobic bacteria and the right half shows that of strictly anaerobic bacteria, transferring the electrons from benzylsuccinyl-CoA oxidation to ubiquinone or menaquinone, respectively. BssABC, benzylsuccinate synthase; BbsEF, benzylsuccinate CoA-transferase; BbsG, benzylsuccinyl-CoA dehydrogenase; BbsH, enoyl-CoA hydratase; BbsCD, alcohol dehydrogenase; BbsAB, benzoylsuccinyl-CoA thiolase; ETF, electron transfer flavoprotein; ETF:Q OR, ETF-quinone oxidoreductase; ETF:MK OR, hypothetical ETF-menaquinone oxidoreductase.
Degradation of toluene continues via a pathway of β-oxidation of (R)-benzylsuccinate to 2-benzoylsuccinyl coenzyme A (2-benzoylsuccinyl-CoA), which is subsequently cleaved to succinyl-CoA and benzoyl-CoA (Fig. 1), the common intermediate for further degradation of aromatic compounds (13). The activation of (R)-benzylsuccinate to (R)-2-benzylsuccinyl-CoA by the benzylsuccinate CoA-transferase BbsEF (14, 15) initiates the pathway. The next step is the specific oxidation of (R)-2-benzylsuccinyl-CoA to (E)-2-benzylidenesuccinyl-CoA by benzylsuccinyl-CoA dehydrogenase (BbsG), which belongs to the family of flavin adenine dinucleotide (FAD)-containing acyl-CoA dehydrogenases (16). The subsequent reactions include hydration of the double bond of (E)-2-benzylidenesuccinyl-CoA by the enoyl-CoA hydratase BbsH, oxidation of the alcohol intermediate to benzoylsuccinyl-CoA by the alcohol dehydrogenase BbsCD, and thiolytic cleavage by benzoylsuccinyl-CoA thiolase BbsAB (13). These enzymes are encoded in the bss (for BSS) and bbs (for β-oxidation of benzylsuccinate) operons and are highly conserved in anaerobic toluene degraders (Fig. 1). However, the facultatively and strictly anaerobic species appear to differ in the physiological electron acceptor of BbsG. Enzymes of the acyl-CoA dehydrogenase family usually couple to soluble electron transfer flavoproteins (ETFs), which then transfer the electron further to the quinone pool by reacting with membrane-bound oxidoreductases (Fig. 1) (17–19). Facultatively anaerobic toluene-degrading bacteria (e.g., Aromatoleum aromaticum) contain single genes coding for the subunits of ETF and the corresponding quinone oxidoreductase, which are not linked to the bss or bbs genes, whereas all known strictly anaerobic toluene degraders contain multiple genes for ETF paralogues, including a copy within their bbs operons (8, 20, 21). In addition, the various etfAB genes of the latter group are mostly adjacent to genes for paralogous membrane proteins that may be involved in electron transfer from ETF to menaquinone (Fig. 1) (8, 20). In particular, G. metallireducens contains genes for nine ETF paralogues, while D. toluolica contains five (20, 21). Remarkably, D. toluolica synthesizes two very similar ETF paralogues (ETFDt1 and ETFDt2) when degrading either toluene or p-cresol, which are metabolized via fumarate addition but by separately induced isoenzymes (20). In this work, we set out to characterize the structures and biochemical properties of the ETF paralogues involved in toluene degradation, to compare them with previously characterized members of the ETF family, and to investigate the diversity of ETF variants among different organisms.
RESULTS
Purification of ETF proteins and their biochemical properties.
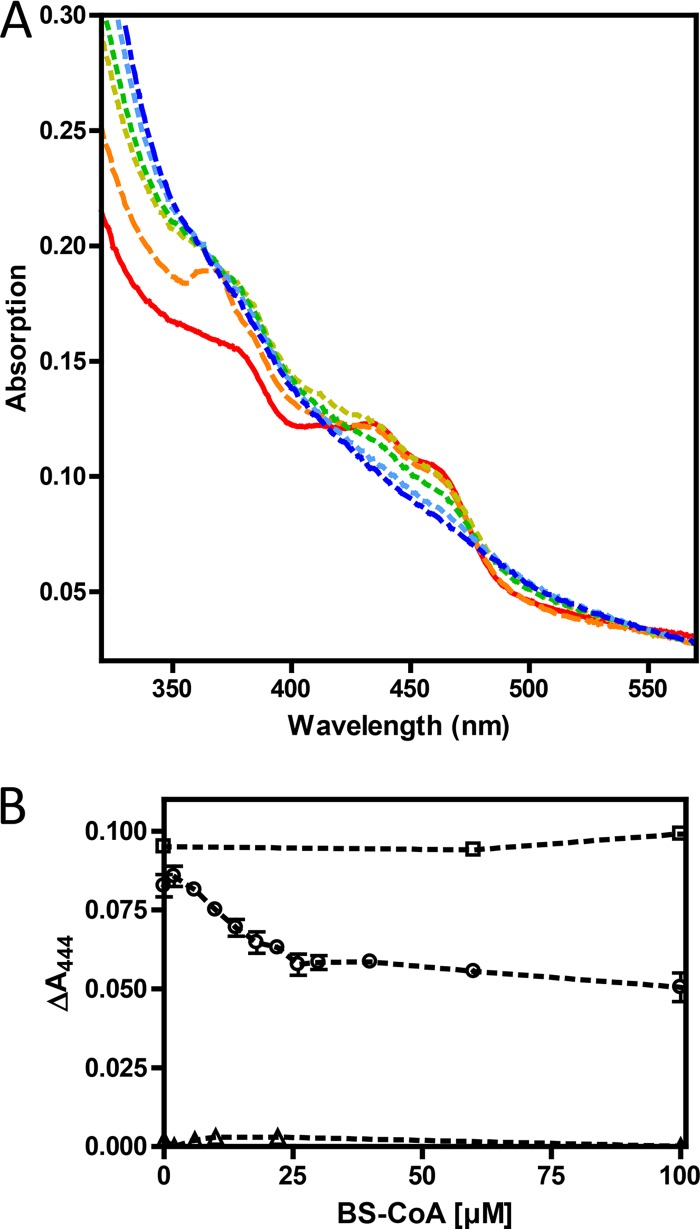
The genes coding for the two subunits of ETFs from three phylogenetically divergent bacterial species that are capable of anaerobic toluene degradation were cloned and overexpressed in Escherichia coli as His tag or Strep-tag fusion proteins. The genes consisted of the single copy of etfAB in the facultative (aerobic/denitrifying) species A. aromaticum (coding for ETFAa) (GenBank accession numbers WP_011239497 and WP_011239498) and the genes for the ETF paralogues in the respective bbs operons of the obligately anaerobic species G. metallireducens and D. toluolica (ETFGm5, GenBank accession numbers WP_004511541 and WP_004511542; ETFDt1, GenBank accession numbers WP_014955682 and WP_014955683). All three proteins were produced in soluble forms in recombinant E. coli cultures and were purified at reasonable yields in one step via affinity chromatography (see Materials and Methods for details). All three ETF variants exhibited the characteristic UV-visible (UV-Vis) and fluorescence spectra of flavoproteins (Fig. 2). In the case of ETFGm5, the flavin cofactor was extracted by heat denaturation of the protein and was confirmed to be FAD by thin-layer chromatography (TLC) analysis (data not shown). The relative FAD contents of the three different ETFs were calculated as 0.62 to 0.73 mol/mol, based on their absorption at 450 nm (εFAD = 11,300 M−1 cm−1) and the respective protein concentrations (for details, see Table 1). In the case of ETFGm5, reconstitution experiments were performed by incubation with FAD and passage of the protein over a desalting column, resulting in an increase in the FAD content of the protein from 0.73 to 0.98 mol/mol (Table 1). All three ETF species exhibited low activities in reducing iodonitrotetrazolium chloride (INT) with NADH as the electron donor, which were used to follow the proteins during their purification procedures. This activity was observed previously for several electron-bifurcating ETFs (22, 23) but also for a nonbifurcating ETF from Syntrophus aciditrophicus (24). The electron-bifurcating ETFAf1 was characterized and measured in parallel with the three ETFs of the toluene-degrading bacteria (22) and showed 75- to 350-fold higher NADH-INT redox activity levels under the same experimental conditions (Table 1).
FIG 2.
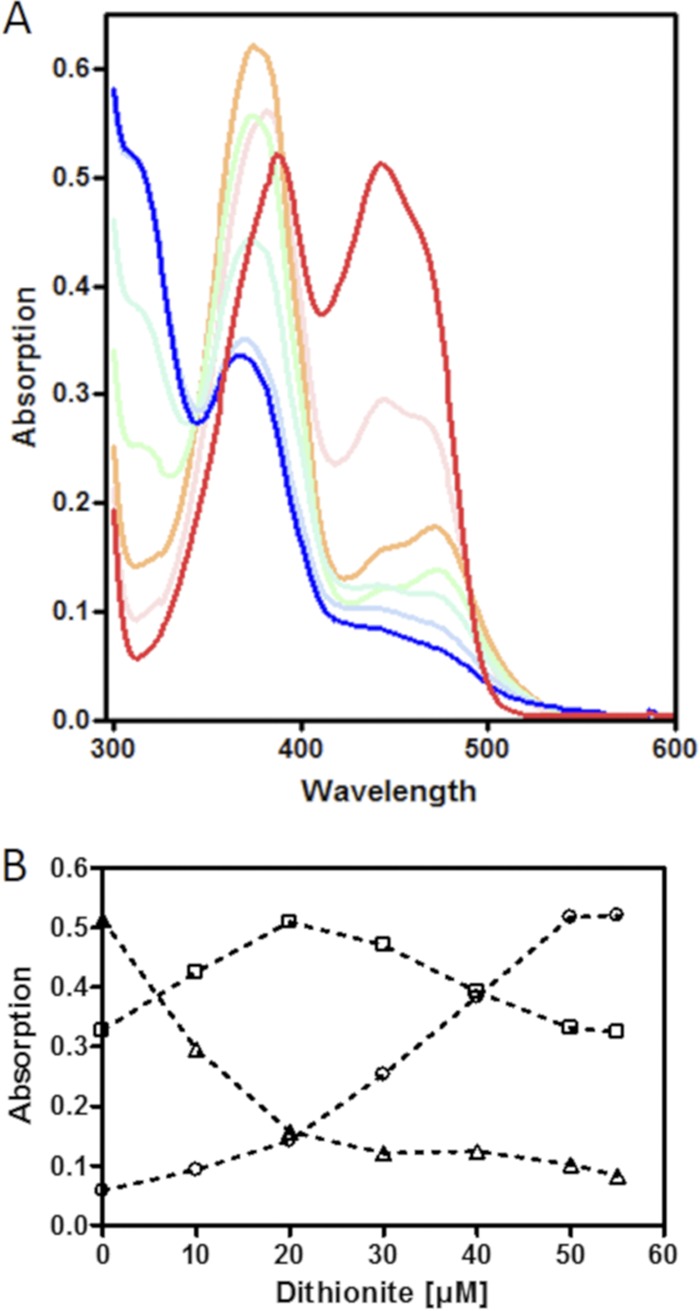
UV-Vis spectroscopic characterization of ETFGm5. (A) UV-Vis spectral changes of purified ETFGm5 (60 μM) during reduction with dithionite from the fully oxidized form (red line) to the fully reduced form (blue line). Dithionite concentrations were increased by 10 μM between each measurement. (B) Tracings of the different redox stages of the protein by their maximum absorption values at representative wavelengths, as follows: oxidized FAD, 444 nm (triangles); semiquinone anion, 360 nm (squares); reduced FADH−, 311 nm (circles).
TABLE 1.
Properties of ETF species
| ETF species | Mass (Da) | FAD content (mol/mol) | NADH-INT activity (U/mg) | Reference(s) |
|---|---|---|---|---|
| ETFGm5 | 60,558 | 0.73 | 0.022 | This study |
| ETFGm5 after reconstruction | 0.98 | This study | ||
| ETFDt1 | 59,752 | 0.62 | 0.007 | This study |
| ETFAa | 60,917 | 0.66 | 0.033 | This study |
| ETFAf1 | 65,451 | 0.7 | 2.5 | This study, 26 |
| ETFAf1 after reconstruction | 2.0 | This study |
The redox properties of ETFGm5 were determined by reducing the protein gradually with dithionite (Fig. 2). This revealed an almost complete initial conversion of oxidized FAD to the anionic FAD semiquinone form, followed by its further reduction to the fully reduced FADH− form, as indicated by the changes of the respective UV-Vis spectra (25). No absorption at wavelengths longer than 500 nm was observed under any conditions, indicating that the neutral semiquinone radical form of ETF was not present (Fig. 2). We also tried to obtain information on the midpoint redox potentials of ETFGm5 by mixing the protein with the redox dyes thionine, methylene blue, and Nile blue and titrating the mixture with increasing concentrations of dithionite. Unfortunately, the absorption peaks of the semiquinone anion radical (maximum at 365 nm) and the reduced FADH− form (maxima at 360 and 310 nm) showed too much interference with the absorption of the redox dyes, so that only the conversion of oxidized FAD (maximum at 444 nm) to the semiquinone could be evaluated. The absorption values at the respective absorption maxima of FAD and the redox dyes were recorded and normalized to 0 to 100%. After these values were plotted against the added dithionite concentrations and curve fitting was performed, a midpoint potential of about 0 ± 10 mV was interpolated for the FAD/FAD*− couple of ETFGm5, relative to the known midpoint redox values of the dyes at pH 7 (data not shown).
Purification of benzylsuccinyl-CoA dehydrogenases and their properties.
The bbsG genes coding for the benzylsuccinyl-CoA dehydrogenases of G. metallireducens (BbsGGm) (GenBank accession number WP_004511544) and Thauera aromatica (BbsGTa) (GenBank accession number AAF89842) were cloned and overexpressed in E. coli. BbsGTa from Thauera aromatica was characterized previously (16) and was used as proxy for the highly similar A. aromaticum BbsG (92% sequence identity). Like the ETFs, both BbsG orthologues were obtained in good yields as soluble proteins. They exhibited typical flavoprotein UV-Vis spectra, indicating the presence of a FAD cofactor, as reported previously (16). In contrast to ETF, reduction of BbsG with dithionite led directly to the fully reduced form without transient semiquinone formation (see also reference 16). The purified proteins contained relative FAD contents of 0.92 to 0.99 mol/mol, as calculated from the absorption values at 450 nm (εFAD = 11.300 M−1 cm−1) (Table 2). Catalytic parameters for the BbsG orthologues were compared by determining their apparent kinetic parameters, which were quite similar [apparent Vmax values of 0.70 and 0.73 U/mg and Km values of 98 and 140 μM for (R)-benzylsuccinyl-CoA for BbsGGm and BbsGTa, respectively] (Table 2). We also checked whether BbsG was able to catalyze the reverse reaction, namely, reducing benzylidenesuccinyl-CoA to benzylsuccinyl-CoA, in the presence of reduced methyl viologen as a thermodynamically appropriate electron donor. Both BbsG orthologues were found to be highly active in catalyzing this reaction under strictly anoxic conditions. The reaction was dependent on the presence of the enzyme and yielded specific activities of 7.0 and 4.4 U/mg for BbsGTa and BbsGGm, respectively, exceeding the respective ferricenium-dependent benzylsuccinyl-CoA-oxidizing activities by factors of 6 to 10 (Table 2). The other catalytic parameters of the two BbsG orthologues showed only one major deviation, namely, in their pH dependences. BbsGGm activity was optimal at pH 6.4 and decreased sharply at pH values lower than 6.2 or higher than 7.0 (data not shown). In contrast, BbsGTa exhibited its pH optimum at pH 8.0 (16), where BbsGGm retained only 10% of its activity.
TABLE 2.
Properties of BbsG orthologues (benzylsuccinyl-CoA dehydrogenases)
| BbsG species | Subunit size (kDa) | FAD content (mol/mol) | Forward reaction |
Reverse reaction sp act (U/mg) | pH optimum | Temp optimum (°C) | |
|---|---|---|---|---|---|---|---|
| Vmax (U/mg) | Km (μM) | ||||||
| BbsGGm | 46.36 | 0.99 | 0.70 ± 0.04 | 98 ± 15 | 4.4 | 6.4 | 50 |
| BbsGTa | 45.58 | 0.92 | 0.73 ± 0.06 | 140 ± 30 | 7.0 | 8.0 | 50 |
Coupling of benzylsuccinyl-CoA dehydrogenase to ETFGm5.
To test for its expected function as a physiological electron acceptor for the oxidation of benzylsuccinyl-CoA, ETFGm5 was mixed with a small amount of BbsG and titrated with increasing concentrations of chemically synthesized benzylsuccinyl-CoA (Fig. 3). To obtain clear data, interference of oxygen as a potential oxidant for the reduced ETF had to be excluded by performing the experiments under anaerobic conditions in a glove box. Because the experiment was impaired by increasing protein precipitation with increasing substrate concentrations, we recorded the absorption differences between the observed maxima and isosbestic points at 444 and 478 nm, respectively. Because the effects of light scattering by protein precipitation should be very similar at these close wavelengths, the absorption differences should correlate with increased FAD reduction in ETFGm5 with the added substrate concentrations. Control assays without added BbsG or ETF showed that BbsG needed to be present to catalyze benzylsuccinyl-CoA oxidation and that the amount of added BbsG was negligible to cause changes in FAD absorbance (Fig. 3B). A similar observation was reported previously for the electron transfer from glutaryl-CoA to the paralogous ETFGm4 of G. metallireducens (26).
FIG 3.
Reduction of ETFGm5 from enzymatic oxidation of benzylsuccinyl-CoA. Purified ETFGm5 (12.5 μM) was mixed with catalytic amounts of BbsG (1 μM) and titrated with chemically synthesized benzylsuccinyl-CoA (BS-CoA), which was initially added at 2 μM and then increased in increments as indicated. The substrate consisted of both regioisomers from racemic benzylsuccinate and contained ∼35% 2-(R)-benzylsuccinyl-CoA (16). (A) Recorded spectra ranging from the fully oxidized form (red) to the fully reduced form (blue). (B) Concentration-dependent reduction of ETF, including standard deviations from three experiments. Circles represent the full assay as shown in panel A, squares represent the controls without BbsG, and triangles represent the controls without ETF. To correct for the background increases due to precipitation, the absorption differences between the maximum at 444 nm and the isosbestic point at 478 nm were plotted.
Electron-bifurcating abilities of benzylsuccinyl-CoA dehydrogenase with various ETF species.
A particularly interesting aspect in characterizing ETF isoforms from strictly anaerobic bacteria is the question of whether they may be involved in electron bifurcation reactions like those recently characterized for the butyrate or ethanol/acetate fermentation pathways of Clostridium species, Acidaminococcus fermentans, or related species (22, 23, 27, 28). The reaction proceeds by coupling the exergonic NADH-dependent reduction of crotonyl-CoA to butyryl-CoA with simultaneous endergonic reduction of ferredoxin or flavodoxin, leading to bifurcation of the electrons from NADH to crotonyl-CoA and oxidized ferredoxin (27, 28). Accordingly, the reverse reaction may be running with electron confurcation from butyryl-CoA plus reduced ferredoxin to NAD. Electron bifurcation is thought to be restricted to special ETF species that contain two FAD cofactors per ETFAB dimer, rather than the usual single FAD and AMP cofactors per ETFAB (29). Here we analyzed the extent to which electron bifurcation reactions are possible with the redox couple benzylsuccinyl-CoA/benzylidenesuccinyl-CoA, using either purified ETFGm5 or the bifurcation-competent ETFAf1 from A. fermentans (22).
To test for electron bifurcation, either BbsGGm or BbsGTa was added to an assay mixture containing NADH, ferredoxin, and hydrogenase from Clostridium pasteurianum plus either ETFGm5 or ETFAf1, which supports electron bifurcation (26). The reaction was started by adding benzylidenesuccinyl-CoA, and the reaction was followed photometrically by recording the oxidation of NADH. As a positive control, BbsG was replaced by butyryl-CoA dehydrogenase (BDH) of A. fermentans (BDHAf), and the reaction was started with crotonyl-CoA (Table 3). These assays revealed no activity with ETFGm5 in any combination, indicating its inability to engage in electron bifurcation. However, low electron-bifurcating activity, amounting to ∼3% of the value recorded for the positive control, was recorded for a combination of BbsGGm with ETFAf1 when started with benzylidenesuccinyl-CoA (Table 3). Although the recorded activity level was very low, it indicates that electron bifurcation is possible only with an appropriate ETF species such as ETFAf1. The low level of activity may be caused by insufficient interactions between benzylsuccinyl-CoA dehydrogenase and ETFAf1.
TABLE 3.
Electron bifurcation capabilities of different ETF species
| Enzyme | Starting substrate | Activity (mU/mg) |
|
|---|---|---|---|
| ETFGm5 | ETFAf1 | ||
| BbsGGm | Benzylidenesuccinyl-CoA | <1 | 7 ± 1 |
| BbsGTa | Benzylidenesuccinyl-CoA | <1 | 6 ± 1 |
| BDHAf | Crotonyl-CoA | <1 | 220 ± 40 |
| None | Benzylidenesuccinyl-CoA | <1 | <1 |
Structure of ETFGm5.
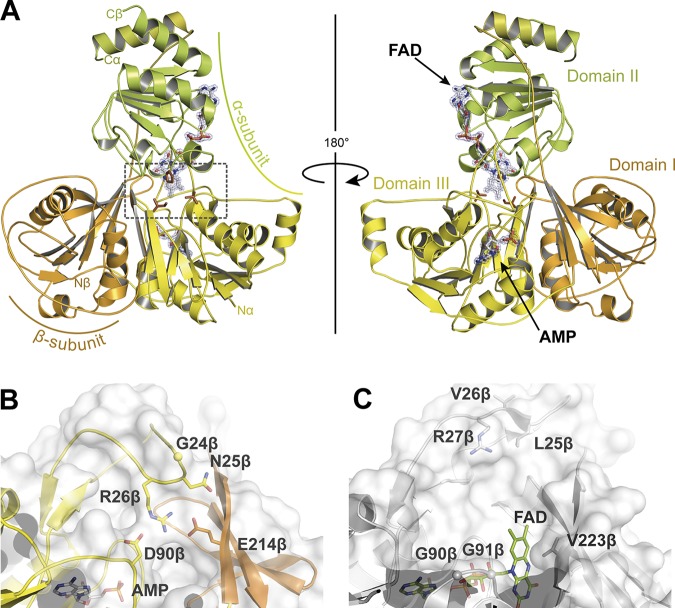
We obtained crystals of native and selenomethionine-substituted ETFGm5 and used these to determine the crystal structure at 1.7-Å resolution, with final Rwork and Rfree values of 13.2% and 16.7%, respectively. For further details on data collection and refinement, see Table 4. ETFGm5 crystallized in orthorhombic space group P212121 and revealed a heterodimer consisting of the α and β subunits within the asymmetric unit (Fig. 4A). The entire mature protein is structurally defined by continuous electron density. The ETFGm5 heterodimer shares the three-domain architecture that is known from other ETF proteins (22, 29–32). Domains I and III have similar overall folds that belong to the adenine nucleotide α-hydrolase (AANH) superfamily. In domain I, the typical α/β/α-fold is composed of six parallel β-strands with a rudimentary seventh strand and three α-helices, while the α/β/α-fold of domain III consists of seven parallel β-strands sandwiched by four α-helices. Furthermore, domain III has one additional α-helix and two additional β-strands, compared to other ETF structures. The domains are flanked on both sides by an antiparallel β-sheet consisting of four strands, with three strands from one domain and one strand from the other domain. Despite their common folds, domains I and III lack sequence similarity, which was also observed in ETFs from humans (30) and other organisms (32, 33). As in other ETF structures, domain I is built by the N-terminal portion of the α subunit of ETFGm5, while domain III consists mainly of the β subunit. In contrast, both subunits contribute to domain II, which is composed of the C-terminal α-helix of the β subunit and the C-terminal part of the α subunit. In this domain, five parallel β-strands are sandwiched by six surrounding α-helices.
TABLE 4.
Data collection and refinement statistics
| Parameter | Result(s) for selenomethionine-substituted ETFGma |
|---|---|
| Beamline | ID29 at ESRF (Grenoble, France) |
| Detector | Pilatus 6M |
| Detection wavelength (Å) | 0.97939 |
| Resolution range (Å) | 43.74–1.7 (1.761–1.700) |
| Space group | P212121 |
| Unit cell | |
| Distances (Å) | 54.73, 72.75, 133.88 |
| Angles (°) | 90, 90, 90 |
| Total no. of reflections | 744,382 (71,000) |
| No. of unique reflections | 59,428 (5,846) |
| Multiplicity | 12.5 (12.1) |
| Completeness (%) | 99.65 (99.40) |
| Mean I/sigma(I) ratio | 19.85 (4.25) |
| Wilson B factor (Å2) | 17.48 |
| Rmerge | 0.09647 (0.5937) |
| Rmeas | 0.1006 |
| CC1/2 | 0.999 (0.926) |
| CC* | 1.000 (0.981) |
| Rwork | 0.1319 (0.1728) |
| Rfree | 0.1667 (0.2273) |
| No. of nonhydrogen atoms | 4,921 |
| Macromolecules | 4,149 |
| Ligands | 53 |
| Water | 719 |
| No. of protein residues | 545 |
| Root mean square | |
| Bond (Å) | 0.015 |
| Angle (°) | 1.57 |
| Ramachandran favored (%) | 99 |
| Ramachandran outliers (%) | 0 |
| Average B factor (Å2) | 23.70 |
| Macromolecules | 21.60 |
| Ligands | 12.70 |
| Water | 36.70 |
Statistics for the highest-resolution shell are shown in parentheses.
FIG 4.
Structural model of ETFGm5. (A) Cartoon representation of front and rear views of ETFGm5 (domain I, orange; domain II, green; domain III, yellow). The ligands AMP and FAD are indicated by arrows and depicted as stick models with white carbon atoms and their associated F0F1 electron densities at sigma level 2.0. The residues constituting the hydrophobic hinge cluster, P220α, I157β, and V178β, are shown as atom-colored-stick models with carbon atoms in brown. The dashed box in the left model indicates the area of the hinge region, which is shown in further detail in Fig. 5. (B) Surface of domain III of ETFGm5. Amino acids protruding into the funnel-like structure are indicated. (C) Surface of domain III of bifurcating ETFAf, indicating the open funnel-like structure allowing access to the second FAD cofactor. Amino acids lining the respective funnel walls are indicated.
The crystal structure of ETFGm5 confirms the expected presence of one FAD and one AMP bound in the heterodimer (Fig. 4A). The AMP cofactor is deeply buried within domain III in an anti conformation and is well shielded from the FAD cofactor by loop 9, which forms the interface between domains II and III. This arrangement of the bound AMP is consistent with that of other nonbifurcating ETF proteins, in which a structural function is attributed to the AMP nucleotide, rather than it being involved in electron transfer. The ability of ETF species to bind AMP might be a legacy of ancestral ETF-type proteins, which might have contained a second bound FAD within domain III, as still observed in electron-bifurcating ETF (22). When the structure of ETFGm5 was compared with that of either the electron-bifurcating ETFAf1 from A. fermentans or the nonbifurcating ETF from human mitochondria, Pseudomonas denitrificans, or Methylophilus methylotrophus, a funnel-like opening was found to exist on the surface of domain III in ETFAf1, which is lined by residues L25β to R27β, G90β and G91β, and V223β and thus may allow access to the second FAD cofactor (Fig. 4C). In contrast, all structurally characterized nonbifurcating ETFs, including ETFGm5, contain different structural elements that block access to the AMP-binding site at this location. For example, human ETF contains two loops (K23β to D32β and P93β to R98β) that extend into the funnel, whereas several other amino acid side chains of ETFGm5 (e.g., N25β, R26β, D90β, and E214β) protrude into the funnel-like domain, prohibiting access to the interior (Fig. 4B).
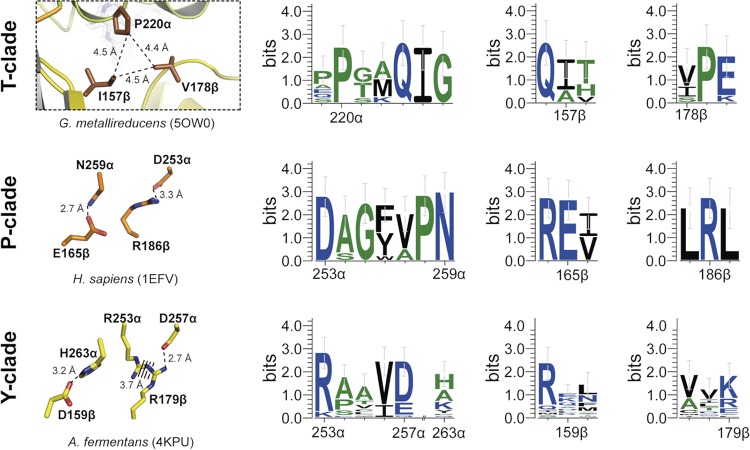
The additional electron density in domain II of ETFGm5 was unambiguously assigned to its single FAD cofactor (Fig. 4A). It is noncovalently bound in a stretched manner, primarily by polar contacts, including a network of 13 water molecules. The ADP portion of the dinucleotide is surface exposed, while the isoalloxazine ring is embedded in a small pocket. This structure resembles the electron-transfer-incompetent closed conformation of other known ETF species. For ETFs from human mitochondria or the closely related alphaproteobacterium P. denitrificans, an essential role of E165β, as part of a hinge mechanism, in maintaining the closed conformation by stabilizing the nonproductive state of domain II was proposed (34). An E165βA mutation led to enhanced occupancy of the open active state and a 100-fold increase of the kcat/Km value of the mutant, compared to that of the wild-type protein. However, the glutamate and its hydrogen-bond-forming partner residue N259α (Fig. 5) are apparently conserved only in the standard group 1 ETF from proteobacteria (suggested to be renamed clade P; see below) and not in structurally characterized ETF species from other clades. For example, the trimethylamine dehydrogenase-linked ETF from M. methylotrophus (the only characterized member of clade J) contains the conserved glutamate (E163β), but it interacts with R236β instead of an asparagine. Analogously, the bifurcating ETFAf1 of A. fermentans (clade Y) contains an aspartate (D159β) in place of the conserved glutamate, which is hydrogen bonded to H263α but is not conserved in the other known bifurcating ETF species (Fig. 5). The structural model of ETFGm5 presented here, as the first member of clade T, reveals an even more notable difference in the composition of its hinge region. Here, the glutamate is substituted by an isoleucine (I157β), which forms a small hydrophobic cluster between domain II and domain III, together with residues V178β and P220α (Fig. 5). This set of hydrophobic residues is highly conserved among all ETFs of clade T and seems to be unique for this clade, corroborating the suggested clade affiliation of the members of the ETF family (Fig. 5 and 6). Therefore, the particular ways of forming the interface between the mobile domains of ETF, which need to switch back and forth between the conformations that are active versus inactive in electron transfer, appear to be highly divergent among the different ETF clades.
FIG 5.
Comparison of the hinge regions between domains II and III of ETF clades T, P, and Y. Structural details are shown in the left panels, and conservation of the relevant interacting amino acids within members of the respective clade is indicated in the right panels. Clade J was omitted because it contains too few members to produce statistically significant results. Broken lines indicate distances. The color scheme is as in Fig. 4. Sequence conservation patterns of relevant locations are based on the sequences from Fig. 6 using the numbering of the respective ETFs.
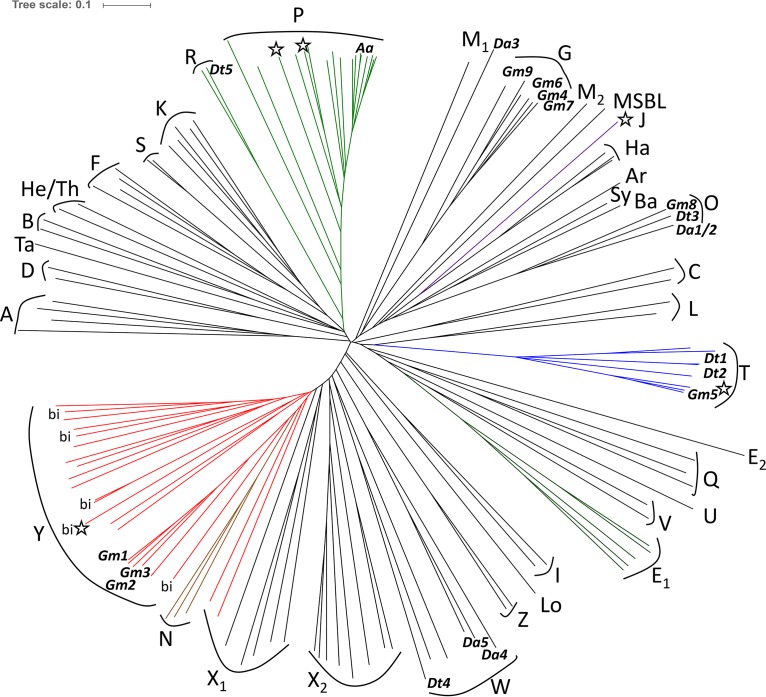
FIG 6.
Unrooted phylogenetic tree representing evolutionary branches of the ETFAB family proteins. Structurally characterized proteins are labeled with stars, known bifurcating ETFs are labeled with “bi,” and the positions of ETFAa (Aa) and all of the paralogues from G. metallireducens (Gm1 to Gm9), D. toluolica (Dt1 to Dt5), and D. alkenivorans (Da1 to Da5) are indicated. Color-labeled clades are as follows: P, green; T, blue; Y, red; N, brown. Bacterial groups are as follows: A, aerobic Actinobacteria; B, aerobic Bacteroidetes; C, Chlorobi; D, Deinococcus/Thermus; E1 and E2, Enterobacteriales; F, aerobic Firmicutes; I, Chloroflexi; K, anaerobic Firmicutes; L, Limnochorda; M1 and M2, paralogues of Thermomicrobium; P, aerobic Proteobacteria; S, Syntrophus; U, Propionibacterium; V, Sporomusa/Syntrophobacter; W, Thermodesulfobacterium/Desulfobacterales. Archaeal groups are as follows: Ar, Archaeoglobus; Ba, Bathyarchaeota; Ha, Haloarchaea; He/Th, Heimdallarchaeota/Thaumarchaeota; Lo, Lokiarchaeota; MSBL, archaeal MSBL-1 phylum; Q, ETF paralogues of acidophilic aerobic archaea; Sy, Syntrophoarchaeum; Ta, Thalassoarchaea. Functionally defined groups are as follows: G, glutaryl-CoA dehydrogenase coupled; J, trimethylamine dehydrogenase coupled; N, nitrogen fixation related; O, β-oxidation related; R, benzoyl-CoA reductase related in sulfate reducers; T, toluene related; X1 and X2, potentially bifurcating; Y, bifurcating; Z, nonbifurcating paralogues. GenBank accession numbers of the respective ETFA sequences (archaea in bold) are as follows: A, EMY33520, ACS29836, NP_217544, and EFD70844; B, ABQ04505 and WP_018342142; C, ACD89420 and WP_011358480; D, EED10602 and WP_010887615; E1, EFJ87054, EFJ86261, EDP99951, KKJ88755, and EEI47268; E2, EFJ85051; F, WP_012685393, BAB06818, ALS22938, and KIX83781; G, WP_035589839, WP_012529901, WP_004513053, WP_011366019, WP_004514142, and WP_004512525; I, YP_001637120 and ABX04479; J, WP_019880640; K, WP_027363721, ABZ83674, WP_011344236, BAF59734, and WP_018212273; L, BAS26084 and WP_006904367; M1 and M2, ACM05675 and ACM04988; N, WP_011389921, ACH83433, and ANL06469; O, WP_011366021, WP_014956641, ACL05007, and ACL04905; P, CAI09844, WP_011766221, WP_053421827, WP_011748831, WP_027458991, WP_011286977, WP_014237359, WP_011046469, WP_021247815, WP_059038764, WP_011555870, WP_010918612, CEI09958, NP_000117, WP_021248657, Q12480, and WP_011390826; Q, CAC11358, WP_011177083, and AAY79707; R, WP_014955896 and KGO33720; S, ABC78522 and SEL95069; T, WP_004511541, WP_012647507, WP_014955682, WP_014956022, KGO32932, WP_027716136, ADB04301, and WP_054702711; U, CBL55769; V, SDF02418 and WP_011700714; W, WP_014958616, WP_049674793, ACL06852, ACL05721, WP_035218962, and AEH23377; X1, WP_014355959, ABZ85146, EDK35507, WP_077744614, WP_015043044, KXT39316, and ADA67362; X2, CAC11474, AAY79731, WP_048060312, WP_013604503, WP_013266183, ACB40911, WP_013679410, and ABO09547; Y, AJY69063, WP_004514075, WP_004514036, WP_011366012, KPK44661, WP_014355266, ADB46955, WP_014017062, WP_044503462, EDK32511, WP_066047471, WP_003428573, WP_003437556, WP_074183331, WP_057979362, WP_011391607, EFW36455, WP_076488762, BAR51057, KXH77764, and KPQ43842; Z, ADB47358 and CCC74206; Ar, WP_015591020; Ba, KPV62836; Ha, WP_008848967, WP_074792744, and ELY25127; He, OLS26127; Lo, KKK41863; MBSL, KXA89304; Sy, OFV66880; Ta, ANV79625; Th, AIF20582.
Clade partition of the ETF family.
After realizing that the ETF species related to toluene metabolism in strictly anaerobic bacteria belong to a specialized ETF subgroup (clade T) with distinctive differences from the previously known ETFs, we continued to investigate the diversity of the naturally occurring ETFs from bacteria, archaea, and eukaryotic mitochondria. We included ETF proteins encoded by single gene copies per species, as well as selections of paralogues from species with multiple gene copies, and used artificially concatenated sequences of the two subunits derived from directly neighboring etfAB-like genes for the analysis. In particular, we included all known paralogous etfAB gene products from the genomes of the toluene-degrading species G. metallireducens (nine sequences) and D. toluolica (five sequences), as well as those of the anaerobic alkane-degrading species Desulfatibacillum alkenivorans (five sequences). The resulting phylogenetic tree indicates that ETF species exhibit enormous diversity, with 37 clades (Fig. 6), rather than the previously suggested 3 clades (33) or 5 clades (35). This diversity reflects both intensive coevolution with the phylogenetic lineages of the host organisms (16 clades representing bacterial phyla and 9 representing archaeal phyla or subphyla) and functional specialization of paralogues (12 clades), which apparently evolved for metabolic purposes, particularly in anaerobic bacteria or archaea, and were probably spread by lateral gene transfer.
Almost all experimentally proven electron-bifurcating ETF proteins (23, 28) are affiliated with a single clade (clade Y, symbolizing bifurcation), which is one of the few clades containing proteins from organisms of widely different phylogenetic origins, including strictly anaerobic bacteria and archaea. All members of this clade appear to share electron-bifurcating functions and to contain a second FAD cofactor instead of the AMP found otherwise, because they contain the amino acids involved in binding the second FAD as highly conserved residues (27). Some of these proteins, including the G. metallireducens paralogues ETFGm1 to ETFGm3, even contain an additional ferredoxin domain fused to the N terminus of ETFA, which contains two predicted [Fe4S4] clusters. One of these G. metallireducens clade Y paralogues has recently been suggested to be involved in an electron bifurcation process necessary for benzoyl-CoA reduction (36). ETF sequences of clade Y are also present in the bacterial genera Spirochaeta, Treponema, Tannerella, and Nitrospira, as well as in archaea affiliated with the candidate phylum Thorarchaeota or the anaerobic methane-oxidizing “Candidatus Methanoperedens” group (Fig. 6). The closely related clade N consists of a second group of proven electron-bifurcating ETF paralogues, namely, the FixAB proteins, which are present in several nitrogen-fixing proteobacterial species and provide nitrogenase with low-potential electrons (37). Clades X1 (Clostridium, Heliobacterium, Bacteroides, Thermotoga, and Dehalobacter species), X2 (several Crenarchaeota species), W (sulfate-reducing Deltaproteobacteria or Thermodesulfobacteriaceae species), and Z (ETF paralogues from Acidaminococcus or Megasphaera species) also are closely associated with the known electron-bifurcating ETF clades Y and N (Fig. 6), but their electron-bifurcating capabilities are still questionable. One of the members of clade Z, the second ETF paralogue of A. fermentans (ETFAf2), has been found to be incapable of electron bifurcation (W. Buckel and N. P. Chowdhury, unpublished data), suggesting that they may be evolutionary intermediates for the transition between bifurcating ETFs and nonbifurcating ETFs. The large phylogenetic diversity of the ETF species affiliated with clade Y suggests that this clade represents the evolutionary origin of the ETF family. Starting from electron-bifurcating proteins with two FAD cofactors, the proteins might have lost the second FAD cofactor and evolved further to the proteins of the other clades of the family.
Some of the clades of nonbifurcating ETFs seem to have coevolved with defined phylogenetic microbial groups, particularly those representing aerobic or facultatively anaerobic species, which usually have only a single copy of the ETF genes in their genomes. Separate ETF clades can be assigned to the respective (mostly aerobic) members of the Proteobacteria (including the mitochondrial ETFs of eukaryotes, which are derived from Alphaproteobacteria) (clade P), Firmicutes (clades F and K), Actinobacteria, Bacteroidetes, Chlorobi, the Thermus/Deinococcus group, and Chloroflexi (clades A, B, C, D, and I, respectively). Separate clades are also represented by the ETFs from aerobic or denitrifying archaeal Halobacteria (clade Ha) and ETF paralogues of the acidophilic Euryarchaeota or Crenarchaeota (clade Q), while several other archaeal clades are represented by only a few known sequences and represent the Archaeoglobales (clade Ar), as well as candidate groups Thalassoarchaea (clade Ta), Heimdallarchaeota (clade He), Thaumarchaeota (clade Th), Syntrophoarchaeum (clade Sy), Bathyarchaeota (clade Ba), Lokiarchaeota (clade Lo), and the archaeal MSBL-1 phylum (Fig. 6). The sequences of clades Ha and MSBL-1 exhibit branch points close to that of a trimethylamine dehydrogenase-associated ETF (clade J). Many strictly anaerobic members of bacterial or archaeal phyla are either lacking recognizable genes for ETF (examples include most lactic acid bacteria, archaeal thermococci, and methanogens) or contain single or multiple ETF species belonging to different clusters than those of the aerobic members. For example, the ETFs of the proteobacterial Enterobacteriales/Vibrionales (clades E1 and E2) and those of anaerobic members of the Bacteroidetes or Actinobacteria (e.g., Bacteroides or Propionibacterium) belong to separate evolutionary branches (clades Y and U, respectively), compared to those of the aerobic members of the respective phyla (Fig. 6). It appears that especially the ETFs of anaerobic microbes are more likely to be affiliated with clades defined by common functionality, e.g., the proteins involved in anaerobic toluene (clade T) or glutarate (clade G) degradation. In particular, many species of the strictly anaerobic Firmicutes or Deltaproteobacteria contain multiple ETF paralogues that are affiliated with several unrelated sequence clades (Fig. 6). For example, the nine ETF paralogues of G. metallireducens belong to clades G, O, T, and Y, whereas the five paralogues of D. toluolica belong to clades O, R, T, and W, and the five of D. alkenivorans belong to clades G, O, and W (Fig. 6).
DISCUSSION
In this study, we investigate the function and structure of the ETF paralogue ETFGm5 from the strictly anaerobic toluene-degrading species G. metallireducens and compare it to the related proteins ETFDt1 from D. toluolica and ETFAa from A. aromaticum. ETFGm5 and ETFDt1 are encoded within the respective bbs operons coding for the enzymes of β-oxidation of benzylsuccinate and therefore are specifically linked to anaerobic toluene metabolism. Moreover, they belong to the common clade T (for toluene) of the ETF family, which contains exclusively ETFs from strictly anaerobic toluene degraders that are linked to toluene metabolism. They are apparently used for channeling the electrons from benzylsuccinyl-CoA oxidation to menaquinone, probably via a special membrane-bound oxidoreductase that is encoded next to the genes of the ETF subunits (Fig. 1). Like a large number of other strictly anaerobic bacteria, G. metallireducens and D. toluolica contain multiple copies of ETF-encoding genes in their genomes. Their genomic contexts and phylogenetic affiliations suggest that some of these paralogues are involved in electron bifurcation processes and others in specific β-oxidation-like metabolic modules. For example, the paralogous ETFGm4 from G. metallireducens (Fig. 6) has been shown to be induced upon growth on benzoate and to serve as an electron acceptor for glutaryl-CoA dehydrogenase in the benzoyl-CoA degradation pathway (26). In contrast, denitrifying toluene-degrading proteobacteria, such as Thauera or Aromatoleum species, apparently utilize only one standard ETF protein for toluene degradation, as well as for all other β-oxidation-like pathways. The corresponding proteins belong to clade P, which contains most of the ETF sequences of proteobacteria and mitochondria (Fig. 6).
A fundamental difference among these ETF species of different clades appears to be how the electrons are transferred to the respective quinones. For the ETF species of clade P (Fig. 6), a well-characterized membrane-bound ETF:(ubi)quinone oxidoreductase of the FixC superfamily (pfam05187) transfers the electrons further to ubiquinone (Fig. 1), the standard electron carrier in Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria species (17, 38). Genes coding for this type of enzyme are absent in the genomes of Geobacter and Desulfobacula species. Therefore, it is assumed that the electron transfer from ETF to menaquinone (the quinone type of Deltaproteobacteria [38]) is taken over by a different enzyme (Fig. 1), most likely the product of a directly adjacent gene linked to most of the etfAB paralogues (26). This protein is predicted to be an integral membrane redox protein of 71.8 kDa, which exhibits a cytochrome b-like domain (pfam02665) in its N-terminal half, consisting of six membrane-spanning helices and two heme-binding sites, followed by a domain containing two [Fe4S4] clusters in its midsection and a heterodisulfide reductase-like Cys-rich domain in its C-terminal half.
The biochemical properties of the purified proteins ETFGm5 and ETFDt1 are similar to those of ETFAa (this study) but also to those of the previously characterized paralogue ETFGm4 (26) and many other ETF species from aerobic proteobacteria or mitochondria (33). All of these proteins contain only one FAD cofactor and an additional bound AMP as a second cofactor per heterodimer. Moreover, the biochemical and spectroscopic properties of ETFGm5 and ETFDt1 were highly similar to those of other characterized ETF species. In particular, the UV-Vis spectra of ETFGm5 and ETFGm4 exhibited a sequential one-electron conversion to FADH− via a half-reduced anionic flavin semiquinone upon stepwise chemical reduction with dithionite (Fig. 2B), whereas both proteins were simultaneously reduced by two electrons (at least predominantly, with only traces of visible semiquinones) by the natural reductants benzylsuccinyl-CoA or glutaryl-CoA with the respective dehydrogenases (Fig. 3) (26, 33). In contrast, the FAD cofactor of BbsG is reduced or reoxidized without a detectable semiquinone intermediate with either benzylsuccinyl-CoA or dithionite (16). This confirms the general observation that electron transfer from the FAD cofactor of acyl-CoA dehydrogenases to the FAD of ETF occurs usually in the form of a hydride equivalent, generating a two-electron-reduced ETF.
The overall structure of ETFGm5 is very similar to those of the previously solved ETFs, especially for domains II and III, whereas domain I contains several structural differences, which made it necessary to use selenomethionine substitution and single-wavelength anomalous diffraction (SAD) phasing to determine the structure. A distinct feature is the hydrophobic nature of its hinge region with the aliphatic side chains of I157β, V178β, and P220α. This hinge clearly differs from that of the structurally characterized ETFs of clades P, J, and Y, in which hydrogen bonds or salt bridges between polar residues prevail. It remains to be seen how these differences affect the closed-open transition of clade T ETFs.
Phylogenetic analysis of ETF sequences from organisms of all domains has shown that they are distributed in many different similarity clades, which often coincide with the taxonomic groups of the respective bacteria and archaea. In this respect, it is important to note that the eukaryotic ETF sequences are not grouped with the sequences of the host organisms but with those of their mitochondria (derived from Alphaproteobacteria). Apparently, the types of ETF proteins vary fundamentally between the major taxonomic phyla. In addition to the phylum-associated ETF clades, at least 12 additional clades are often present in strictly anaerobic bacteria or archaea with multiple etfAB gene copies. These proteins seem to be specialized for specific functions and are coinduced with certain degradation pathways. They are represented by clades G and T, which are involved in anaerobic glutaryl-CoA and toluene metabolism, respectively, but also by clades Y, N, X1, X2, and W, which include the proven (22, 23, 37, 39, 40) or potential electron-bifurcating members of the family.
Our analysis indicates that there are many more variations in ETF-mediated electron transfer reactions to be detected than currently known from the previously proposed ETF clades (three clades were proposed in reference 33 and five clades in reference 35). These previously considered ETF clades correspond to clade P (previously class 1 ETFs), the ETFs of Enterobacteriales contained in clades E1 and E2 (class 3 ETFs) and clade N, the clade J ETFs associated with trimethylamine dehydrogenase (included in class 2), and the few characterized bifurcating ETFs of clade Y (included in class 2). The recently proposed classes 4 and 5 (35) correspond to clade F from Firmicutes and clade B from aerobic Bacteroidetes. Moreover, the proposed substructure within the predicted bifurcating ETFs in reference 35 resembles a combination of clades Y, N, X1, X2, and W in our study. We think that we provide evidence supporting the proposed ETF clade partition by establishing the distinctive structural properties of ETFGm5 as a member of the new clade T, and we expect that there is much more variability to be detected among ETF species of other clades in various microorganisms (Fig. 6).
MATERIALS AND METHODS
Chemicals.
The chemicals used in this study were of the highest available purity (generally ≥99%). Unless otherwise specified, the chemicals were purchased from AppliChem (Darmstadt, Germany), Fluka (Steinheim, Germany), Merck (Darmstadt, Germany), Roth (Karlsruhe, Germany), or Sigma-Aldrich (Taufkirchen, Germany). Benzylsuccinyl-CoA and benzylidenesuccinyl-CoA were chemically synthesized as described (16).
Growth of bacteria and preparation of cell extracts.
Thauera aromatica K172 (41) (DSM 6984), Desulfobacula toluolica (42) (DSM 7467/Tol2), and Geobacter metallireducens GS-15 (43) (DSM 7210) were obtained from the Leibniz Institute German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany) and grown as described previously (41–44). Recombinant Escherichia coli cells were grown aerobically in the presence of the appropriate antibiotics in Luria-Bertani (LB) medium.
Extracts of recombinant E. coli cells were prepared at 25°C either aerobically or under strictly anoxic conditions (except for centrifugation steps in airtight beakers) in a glove box with a N2/H2 (95:5 [vol/vol]) atmosphere (Coy Laboratory Products Inc., USA). Cultures were harvested in the exponential growth phase by centrifugation at 10,000 × g for 10 min at 4°C (Sorvall RC5B Plus centrifuge with SLA 3000 rotor; Du Pont Instruments) and suspended in 2 to 3 volumes (wt/vol) of 10 mM triethanolamine/NaOH buffer (pH 7.5) with 10% (vol/vol) glycerol, containing 0.05 mg/ml DNase I. The suspension was then transferred to a French pressure cell (American Instruments Company), and the cells were lysed by one passage at 137 MPa. The lysate was centrifuged at 100,000 × g for 60 min at 4°C (L-60 ultracentrifuge and type 90 Ti fixed-angle rotor; Beckmann, Munich, Germany). Protein concentrations of the supernatant (cell extract) were determined by the Bradford method (48), using bovine serum albumin as the standard. The cell extracts prepared in this way were used immediately for assays or further purification steps.
Cloning of genes and protein overproduction.
The genes encoding the two subunits of ETFAa from Aromatoleum aromaticum EbN1 (genes ebA6510 and ebA6511; forward primer, GGCCGGCATATGAAGATCCTCGTACCCGTTAAGCGCGTGG; reverse primer, GGCCGGAGATCTGCCTACCGCGTTGGTCAGCTCGG) and ETFGm5 from G. metallireducens (genes Gmet1525 and Gmet1526; forward primer, GGCCGGCATAGCAGATAGTCGTCTTGGC; reverse primer, GCCATAGCTGATATTCCAG) were cloned in vector pET16b (Novagen; Merck), which adds a 5′ His tag sequence to the respective etfB genes (ebA6511 and Gm_1525). Overproduction was performed in E. coli Rosetta (DE3)pLys in LB medium after induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The genes encoding ETFDt1 from D. toluolica (TOL2_RS00890 and TOL2_RS00895; forward primer, AAGCTCTTCAATGCATATTGCGGTATTAGCAAAG; reverse primer, AAGCTCTTCACCCCTTACCCTCCATAATCGCCT) were cloned in pAsg-IBA3 (IBA Lifesciences, Göttingen, Germany), fusing a C-terminal Strep-tag sequence to the etfA gene (TOL2_RS00895). Overproduction was performed in E. coli DH5α in LB medium after induction with anhydrotetracycline. The bbsG gene from T. aromatica (coding for BbsGTa) was cloned without a tag in the expression plasmid pTrc99A (16). Overexpression was performed in E. coli DH5α in LB medium after induction with IPTG. The gene for BbsGGm (Gmet1523) was cloned with a fused 5′ His tag sequence in pET16b (forward primer, GGCCGGCATATGGATTTCAGCATACCCG; reverse primer, GGCCGGGGATCCTCACTCCTCGTCGGC) and expressed in E. coli Rosetta (DE3)pLys.
Purification of proteins.
Strep-tagged proteins were purified via affinity chromatography on a Strep-Tactin column (5 ml; IBA Lifesciences) equilibrated with buffer A (100 mM Tris-HCl buffer, 150 mM KCl [pH 7.5]). After the column was loaded with cell extract and washed with 20 column volumes of buffer A, the bound proteins were eluted with 4 volumes of buffer A containing 2.5 mM desthiobiotin. Proteins with a His tag fusion were purified on an IMAC Fast Flow column (2 ml; GE Healthcare). The column was equilibrated with buffer B (20 mM potassium phosphate buffer [pH 7.4], 0.5 M sodium chloride). Cell extract was applied and bound protein was eluted with a linear gradient of 0 to 250 mM imidazole in buffer B (15 column volumes). Untagged BbsGTa was purified as described previously (16). Fractions containing ETF or BbsG, as identified by their respective enzyme activities, were pooled, concentrated by centrifugation (4,500 × g at 4°C in 10-kDa-cutoff Centricon Vivaspin 20 units; Sartorius), and desalted (PD-10 desalting columns; GE Healthcare). After addition of 10% glycerol, proteins were stored at –20°C.
Cofactor extraction, TLC, and UV-Vis spectroscopy.
The flavin cofactor was extracted by heat denaturation (10 min at 95°C) of the respective protein and centrifugation (15 min at 16,000 × g). The supernatant was used for UV-Vis spectroscopy (200 to 800 nm) or TLC on silica gel plates (TLC silica gel 60 F254 plates; Merck), with a solvent of n-butanol–acetic acid–5% (wt/vol) Na2HPO4 in H2O (4:1:5). Flavin-containing spots after chromatography were analyzed with UV light. The proteins were characterized by UV spectroscopy after stepwise reduction with dithionite. For ETFGm5, the approximate midpoint redox potential was determined by mixing the protein with a redox dye (methylene blue, Nile blue, or thionine) and titrating the added spectra with dithionite.
Preparation of FAD-free apoprotein and cofactor reconstitution experiments.
To obtain FAD-free apoprotein, the enzyme was incubated in 100 mM Tris-HCl (pH 8.0) with 2 M potassium bromide and washed by centrifugation (4,500 × g at 4°C) in membrane centrifugation units (10-kDa-cutoff Centricon Vivaspin 20 units; Sartorius) until all traces of FAD were removed from the protein fraction (as determined by UV-Vis spectroscopy). The apoprotein was than reconstituted by incubation (12 h at 4°C) in 100 mM Tris-HCl (pH 8.0) with 1 mM FAD or flavin mononucleotide (FMN). Unbound cofactor was removed with a desalting column (PD-10; GE Healthcare).
Activity assays.
Oxidation of benzylsuccinyl-CoA by BbsG was monitored photometrically with ferricenium hexafluorophosphate as an artificial electron acceptor, as described previously (16). The reverse reaction, reduction of benzylidenesuccinyl-CoA (phenylitaconyl-CoA), was monitored photometrically at 600 nm with reduced methylviologen (0.1 mM) as an artificial electron donor, in 100 mM Tris-HCl buffer (pH 7.5), under strictly anoxic conditions. The same conditions were used with other CoA-thioesters for specificity determination. The activity of ETF for mediating the electron transfer from NADH to the artificial electron acceptor INT was measured in 100 mM Tris-HCl buffer (pH 7.5) containing 1 mM INT and 0.1 mM NADH, as described previously (22). The role of ETFGm5 as an electron acceptor for BbsG was tested by mixing a catalytic amount of BbsGGm or BbsGTa (0.1 mg/ml) with 10-fold higher concentrations of ETFGm5 in 100 mM Tris-HCl buffer (pH 7.5) under anoxic conditions, adding increasing amounts of (R)-benzylsuccinyl-CoA, and recording the changes in the UV-Vis spectra. The potential electron-bifurcating activity of ETFs was determined photometrically; ETF and BbsG species of different organisms were combined to study their potential interactions. We monitored the reduction of benzylidenesuccinyl-CoA via the respective BbsG variant in the presence of NADH and ferredoxin (a second final electron acceptor). Continuous reoxidation of ferredoxin was achieved with a hydrogenase, as described previously (22, 23). The reaction was performed under anoxic conditions at 30°C, started with the addition of benzylidenesuccinyl-CoA, and monitored for the oxidation of NADH at 340 nm (ε = 6.3 cm−1 mM−1) (45).
Phylogenetic analysis.
Amino acid sequences of ETF species from the investigated organisms and from reference strains of various phylogenetic groups were aligned using the Clustal Omega algorithm (www.ebi.ac.uk/Tools/msa/clustalo). Based on this alignment, a neighbor-joining tree was constructed using the Program iTOL (itol.embl.de).
Crystallization and structure determination.
l-Selenomethionine-labeled ETFGm5 was concentrated to a final concentration of 10 mg/ml in 300 mM NaCl-20 mM HEPES (pH 7.5). Sitting drops were set with 500 nl protein and 500 nl mother liquor solution (15% [wt/vol] polyethylene glycol 3350, 0.1 M HEPES [pH 7.5]) and incubated at 4°C. The crystals were flash frozen in liquid nitrogen, using 20% (vol/vol) glycerol as a cryoprotectant, and the data set was collected at beamline ID29 at the European Synchrotron Radiation Facility (ESRF) (Grenoble, France). The structure was solved at 1.7-Å resolution by SAD phasing using Autosol from the PHENIX suite (46). Automatic refinement and manual model building were performed with Phenix.refine and Coot, respectively (46, 47). The model analysis and figure generation were performed with PyMOL, v1.8.3.2.
Data availability.
The structure data for ETFGm5 have been deposited in the Protein Data Bank under accession number 5OW0.
ACKNOWLEDGMENTS
This work was supported by the German Research Foundation via Priority Program 1319 and CRC 987 and by the Center for Synthetic Microbiology (Marburg, Germany).
We acknowledge Andrea Schmidt for technical support, Ralf Poeschke for assistance in the MarXtal crystallization laboratory, and Stephan Kiontke for support in the first crystallization experiments.
REFERENCES
- 1.Heider J, Schühle K. 2013. Anaerobic biodegradation of hydrocarbons including methane, p 601–630. In Rosenberg E, DeLong EF, Thompson F, Lory S, Stackebrandt E (ed), The prokaryotes: prokaryotic physiology and biochemistry, 4th ed Springer-Verlag, Heidelberg, Germany. [Google Scholar]
- 2.Jobelius C, Ruth B, Griebler C, Meckenstock RU, Hollender J, Reineke A, Frimmel FH, Zwiener C. 2011. Metabolites indicate hot spots of biodegradation and biogeochemical gradients in a high-resolution monitoring well. Environ Sci Technol 45:474–481. doi: 10.1021/es1030867. [DOI] [PubMed] [Google Scholar]
- 3.Parisi VA, Brubaker GR, Zenker MJ, Prince RC, Gieg LM, da Silva ML, Alvarez PJ, Suflita JM. 2009. Field metabolomics and laboratory assessments of anaerobic intrinsic bioremediation of hydrocarbons at a petroleum‐contaminated site. Microb Biotechnol 2:202–212. doi: 10.1111/j.1751-7915.2009.00077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boll M, Heider J. 2010. Anaerobic degradation of hydrocarbons: mechanisms of C-H-bond activation in the absence of oxygen, p 1011–1024. In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology. Springer-Verlag, Heidelberg, Germany. [Google Scholar]
- 5.Fuchs G, Boll M, Heider J. 2011. Microbial degradation of aromatic compounds: from one strategy to four. Nat Rev Microbiol 9:803–816. doi: 10.1038/nrmicro2652. [DOI] [PubMed] [Google Scholar]
- 6.Biegert T, Fuchs G, Heider J. 1996. Evidence that anaerobic oxidation of toluene in the denitrifying bacterium Thauera aromatica is initiated by formation of benzylsuccinate from toluene and fumarate. Eur J Biochem 238:661–668. doi: 10.1111/j.1432-1033.1996.0661w.x. [DOI] [PubMed] [Google Scholar]
- 7.Szaleniec M, Heider J. 2016. Modeling of the reaction mechanism of enzymatic radical C-C coupling by benzylsuccinate synthase. Int J Mol Sci 17:514–535. doi: 10.3390/ijms17040514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heider J, Szaleniec M, Martins BM, Seyhan D, Buckel W, Golding BT. 2016. Structure and function of benzylsuccinate synthase and related fumarate-adding glycyl radical enzymes. J Mol Microbiol Biotechnol 26:29–44. doi: 10.1159/000441656. [DOI] [PubMed] [Google Scholar]
- 9.Leuthner B, Leutwein C, Schulz H, Hörth P, Haehnel W, Schiltz E, Schägger H, Heider J. 1998. Biochemical and genetic characterization of benzylsuccinate synthase from Thauera aromatica: a new glycyl radical enzyme catalysing the first step in anaerobic toluene metabolism. Mol Microbiol 28:615–628. doi: 10.1046/j.1365-2958.1998.00826.x. [DOI] [PubMed] [Google Scholar]
- 10.Achong GR, Rodriguez AM, Spormann AM. 2001. Benzylsuccinate synthase of Azoarcus sp. strain T: cloning, sequencing, transcriptional organization, and its role in anaerobic toluene and m-xylene mineralization. J Bacteriol 183:6763–6770. doi: 10.1128/JB.183.23.6763-6770.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rabus R, Heider J. 1998. Initial reactions of anaerobic metabolism of alkylbenzenes in denitrifying and sulfate-reducing bacteria. Arch Microbiol 170:377–384. doi: 10.1007/s002030050656. [DOI] [Google Scholar]
- 12.Kane SR, Beller HR, Legler TC, Anderson RT. 2002. Biochemical and genetic evidence of benzylsuccinate synthase in toluene-degrading, ferric iron-reducing Geobacter metallireducens. Biodegradation 13:149–154. doi: 10.1023/A:1020454831407. [DOI] [PubMed] [Google Scholar]
- 13.Leuthner B, Heider J. 2000. Anaerobic toluene catabolism of Thauera aromatica: the bbs operon codes for enzymes of β oxidation of the intermediate benzylsuccinate. J Bacteriol 182:272–277. doi: 10.1128/jb.182.2.272-277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leutwein C, Heider J. 1999. Anaerobic toluene-catabolic pathway in denitrifying Thauera aromatica: activation and β-oxidation of the first intermediate, (R)-(+)-benzylsuccinate. Microbiology 145:3265–3271. doi: 10.1099/00221287-145-11-3265. [DOI] [PubMed] [Google Scholar]
- 15.Leutwein C, Heider J. 2001. Succinyl-CoA:(R)-benzylsuccinate CoA-transferase: an enzyme of the anaerobic toluene catabolic pathway in denitrifying bacteria. J Bacteriol 183:4288–4295. doi: 10.1128/JB.183.14.4288-4295.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leutwein C, Heider J. 2002. (R)-Benzylsuccinyl-CoA dehydrogenase of Thauera aromatica, an enzyme of the anaerobic toluene catabolic pathway. Arch Microbiol 178:517–524. doi: 10.1007/s00203-002-0484-5. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Frerman FE, Kim J-J. 2006. Structure of electron transfer flavoprotein-ubiquinone oxidoreductase and electron transfer to the mitochondrial ubiquinone pool. Proc Natl Acad Sci U S A 103:16212–16217. doi: 10.1073/pnas.0604567103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watmough NJ, Frerman FE. 2010. The electron transfer flavoprotein: ubiquinone oxidoreductases. Biochim Biophys Acta 1797:1910–1916. doi: 10.1016/j.bbabio.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 19.Ramsay RR, Steenkamp DJ, Husain M. 1987. Reactions of electron-transfer flavoprotein and electron-transfer flavoprotein: ubiquinone oxidoreductase. Biochem J 241:883–892. doi: 10.1042/bj2410883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wöhlbrand L, Jacob JH, Kube M, Mussmann M, Jarling R, Beck A, Amann R, Wilkes H, Reinhardt R, Rabus R. 2013. Complete genome, catabolic sub-proteomes and key-metabolites of Desulfobacula toluolica Tol2, a marine, aromatic compound-degrading, sulfate-reducing bacterium. Environ Microbiol 15:1334–1355. doi: 10.1111/j.1462-2920.2012.02885.x. [DOI] [PubMed] [Google Scholar]
- 21.Aklujkar M, Krushkal J, DiBartolo G, Lapidus A, Land ML, Lovley DR. 2009. The genome sequence of Geobacter metallireducens: features of metabolism, physiology and regulation common and dissimilar to Geobacter sulfurreducens. BMC Microbiol 9:109–130. doi: 10.1186/1471-2180-9-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chowdhury NP, Mowafy AM, Demmer JK, Upadhyay V, Koelzer S, Jayamani E, Kahnt J, Hornung M, Demmer U, Ermler U, Buckel W. 2014. Studies on the mechanism of electron bifurcation catalyzed by electron transferring flavoprotein (Etf) and butyryl-CoA dehydrogenase (Bcd) of Acidaminococcus fermentans. J Biol Chem 289:5145–5157. doi: 10.1074/jbc.M113.521013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li F, Hinderberger J, Seedorf H, Zhang J, Buckel W, Thauer RK. 2008. Coupled ferredoxin and crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri. J Bacteriol 190:843–850. doi: 10.1128/JB.01417-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Djurjevic I. 2011. Production of glutaconic acid in recombinant Escherichia coli. PhD dissertation. Philipps University Marburg, Marburg, Germany. [Google Scholar]
- 25.Liu B, Liu H, Zhong D, Lin C. 2010. Searching for a photocycle of the cryptochrome photoreceptors. Curr Opin Plant Biol 13:578–586. doi: 10.1016/j.pbi.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Estelmann S, Boll M. 2014. Glutaryl‐coenzyme A dehydrogenase from Geobacter metallireducens: interaction with electron transferring flavoprotein and kinetic basis of unidirectional catalysis. FEBS J 281:5120–5131. doi: 10.1111/febs.13051. [DOI] [PubMed] [Google Scholar]
- 27.Buckel W, Thauer RK. 2013. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim Biophys Acta 1827:94–113. doi: 10.1016/j.bbabio.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 28.Herrmann G, Jayamani E, Mai G, Buckel W. 2008. Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J Bacteriol 190:784–791. doi: 10.1128/JB.01422-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frerman FE. 1988. Acyl-CoA dehydrogenases, electron transfer flavoprotein and electron transfer flavoprotein dehydrogenase. Biochem Soc Trans 16:416–418. doi: 10.1042/bst0160416. [DOI] [PubMed] [Google Scholar]
- 30.Roberts DL, Frerman FE, Kim J-J. 1996. Three-dimensional structure of human electron transfer flavoprotein to 2.1-Å resolution. Proc Natl Acad Sci U S A 93:14355–14360. doi: 10.1073/pnas.93.25.14355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts DL, Salazar D, Fulmer JP, Frerman FE, Kim J-J. 1999. Crystal structure of Paracoccus denitrificans electron transfer flavoprotein: structural and electrostatic analysis of a conserved flavin binding domain. Biochemistry 38:1977–1989. doi: 10.1021/bi9820917. [DOI] [PubMed] [Google Scholar]
- 32.Leys D, Basran J, Talfournier F, Sutcliffe MJ, Scrutton NS. 2003. Extensive conformational sampling in a ternary electron transfer complex. Nat Struct Mol Biol 10:219–225. doi: 10.1038/nsb894. [DOI] [PubMed] [Google Scholar]
- 33.Toogood HS, Leys D, Scrutton NS. 2007. Dynamics driving function: new insights from electron transferring flavoproteins and partner complexes. FEBS J 274:5481–5504. doi: 10.1111/j.1742-4658.2007.06107.x. [DOI] [PubMed] [Google Scholar]
- 34.Toogood HS, van Thiel A, Scrutton NS, Leys D. 2005. Stabilization of non-productive conformations underpins rapid electron transfer to electron-transferring flavoprotein. J Biol Chem 280:30361–30366. doi: 10.1074/jbc.M505562200. [DOI] [PubMed] [Google Scholar]
- 35.Garcia Costas AM, Poudel S, Miller A-F, Schut GJ, Ledbetter RN, Fixen KR, Seefeldt LC, Adams MWW, Harwood CS, Boyd ES, Peters JW. 2017. Defining electron bifurcation in the electron-transferring flavoprotein family. J Bacteriol 199:e00440-17. doi: 10.1128/JB.00440-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huwiler SG, Löffler C, Anselmann SEL, Stärk H-J, von Bergen M, Flechsler J, Rachel R, Boll M. 2019. One-megadalton metalloenzyme complex in Geobacter metallireducens involved in benzene ring reduction beyond the biological redox window. Proc Natl Acad Sci U S A 116:2259–2264. doi: 10.1073/pnas.1819636116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ledbetter RN, Garcia Costas AM, Lubner CE, Mulder DW, Tokmina-Lukaszewska M, Artz JH, Patterson A, Magnuson TS, Jay ZJ, Duan HD, Miller J, Plunkett MH, Hoben JP, Barney BM, Carlson RP, Miller A-F, Bothner B, King PW, Peters JW, Seefeldt LC. 2017. The electron bifurcating FixABCX protein complex from Azotobacter vinelandii: generation of low-potential reducing equivalents for nitrogenase catalysis. Biochemistry 56:4177–4190. doi: 10.1021/acs.biochem.7b00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pelosi L, Ducluzeau A-L, Loiseau L, Barras F, Schneider D, Junier I, Pierrel F. 2016. Evolution of ubiquinone biosynthesis: multiple proteobacterial enzymes with various regioselectivities to catalyze three contiguous aromatic hydroxylation reactions. mSystems 1:e00091-16. doi: 10.1128/mSystems.00091-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bertsch J, Parthasarathy A, Buckel W, Müller V. 2013. An electron-bifurcating caffeyl-CoA reductase. J Biol Chem 288:11304–11311. doi: 10.1074/jbc.M112.444919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuchmann K, Müller V. 2016. Energetics and application of heterotrophy in acetogenic bacteria. Appl Environ Microbiol 82:4056–4069. doi: 10.1128/AEM.00882-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anders HJ, Kaetzke A, Kämpfer P, Ludwig W, Fuchs G. 1995. Taxonomic position of aromatic-degrading denitrifying pseudomonad strains K 172 and KB 740 and their description as new members of the genera Thauera, as Thauera aromatica sp. nov., and Azoarcus, as Azoarcus evansii sp. nov., respectively, members of the beta subclass of the Proteobacteria. Int J Syst Bacteriol 45:327–333. doi: 10.1099/00207713-45-2-327. [DOI] [PubMed] [Google Scholar]
- 42.Rabus R, Nordhaus R, Ludwig W, Widdel F. 1993. Complete oxidation of toluene under strictly anoxic conditions by a new sulfate-reducing bacterium. Appl Environ Microbiol 59:1444–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lovley DR, Giovannoni SJ, White DC, Champine JE, Phillips EJ, Gorby YA, Goodwin S. 1993. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch Microbiol 159:336–344. doi: 10.1007/BF00290916. [DOI] [PubMed] [Google Scholar]
- 44.Tschech A, Fuchs G. 1987. Anaerobic degradation of phenol by pure cultures of newly isolated denitrifying pseudomonads. Arch Microbiol 148:213–217. doi: 10.1007/BF00414814. [DOI] [PubMed] [Google Scholar]
- 45.Ziegenhorn J, Senn M, Bücher T. 1976. Molar absorptivities of beta-NADH and beta-NADPH. Clin Chem 22:151–160. [PubMed] [Google Scholar]
- 46.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bradford MM. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The structure data for ETFGm5 have been deposited in the Protein Data Bank under accession number 5OW0.