Abstract
Background and Objectives: Antimicrobial resistance (AMR) is increasing worldwide and imposes significant life-threatening risks to several different populations, especially to those in intensive care units (ICU). The most commonly isolated organisms in ICU comprise gram-negative bacilli (GNB), and these represent a leading cause of serious infections. This study was conducted to describe the prevalence of resistance in GNB isolated from patients in adults, pediatric, and neonatal ICU in a tertiary-care hospital in Mérida, Mexico. Materials and Methods: A retrospective study was done on samples collected in Neonatal (NICU), Pediatric (PICU) and Adult (AICU) ICU of Unidad Médica de Alta Especialidad, Instituto Mexicano del Seguro Social in Mérida, México. The identification of isolates and antimicrobial susceptibility testing were performed using an automated system. Results: A total of 517 GNB strains were isolated. The most common positive culture was bronchial secretions. Pseudomonas aeruginosa was the prevalent pathogen in NICU and PICU, whereas Escherichia coli was common in the AICU. Overall, GNB exhibited a high resistance rates for Ampicillin (95.85%), Cefuroxime (84.17%), Piperacillin (82.93%), Cefotaxime (78.07%), Ceftriaxone (77.41%), Aztreonam (75.23%), Cefazolin (75.00%), and Ceftazidime (73.19%). There are significant differences in the resistance rates of GNB from different ICUs for penicillins, cephalosporins, carbapenems and fluoroquinolones drugs. Escherichia coli (multidrug-resistant [MDR] = 91.57%, highly resistant microorganisms [HRMO] = 90.36%) and Acinetobacter baumannii (MDR = 86.79%, HRMO = 83.02%) exhibited the highest percentage of MDR and HRMO profiles. The prevalence of the extended-spectrum beta-lactamases (ESBL)-producing isolates was 83.13% in E. coli, 78.84% in Klebsiella pneumoniae, and 66.67% in Proteus mirabilis, respectively. Conclusions: The high resistance rates to drugs were exhibited by our GNB isolates. Continuous surveillance and control of the use of antimicrobials are urgently needed to reduce the emergence and spreading of MDR, HRMO, and/or ESBL-producing bacilli.
Keywords: antimicrobial resistance, Gram-negative bacilli, enterobacteriaceae, non-fermenting Gram-negative bacilli, ESBL, MDR, HRMO
1. Introduction
According to the World Health Organization (WHO), antimicrobial resistance (AMR) is a threat to the prevention of and therapy against infectious diseases. It is a global problem caused mainly by poor administration and inadequate therapy, and the use of antimicrobials in an abusive manner without medical supervision. AMR leads to longer hospital stays, higher medical costs, and increased mortality [1]. The cost annual for AMR in the U.S. healthcare system has been estimated as ranging from US $21–$34 billion dollars, accompanied by more than 8 million additional days in the hospital [2]. Additionally, in the U.S., more than 2 million persons were infected with resistant bacteria and 23,000 died as a result of these infections [3]. In countries of the European Union, the costs for infections caused by resistant bacteria exceeded 1.6 million €, with 2.5 million additional days of hospital stays, and these infections caused 25,000 deaths [4].
AMR increases by 700,000, the number of deaths annually, and for 2050, the number is estimated to be 10,000,000, overtaking that of cancer [5]. AMR is increasing global [6]. In recent years, a great number of resistant strains have emerged in different pathogens, and multidrug-resistant bacteria (MDR) gram-negative pathogens are also becoming increasingly prevalent in the community [2]. The specific definition of MDR is labelled as such because of their in vitro resistance to three or more antimicrobial classes of drugs [7]. In a recent report, the WHO published a list of antibiotic-resistant priority pathogens, in which bacteria with critical priority included Acinetobacter spp. and Pseudomonas spp. with carbapenem resistance, and Enterobacteriaceae family members that produced the extended-spectrum beta-lactamases (ESBL) and carbapenemases [8].
The mechanisms of drug resistance fall into several broad categories including drug degradation/alteration (such as ESBL, aminoglycoside-modifying enzymes, or Chloramphenicol acetyltransferases); the modification of drug binding sites/target; the changes in cell permeability and efflux pump expression, resulting in reduced intracellular drug accumulation [9].
Recent studies of the prevalence of AMR have shown a high increase in resistant infections, especially in intensive care unit (ICU) areas [10,11,12,13]. The patients in ICU are vulnerable as they are exposed to different invasive procedures, such as intubation, mechanical ventilation, and vascular access. In addition, many of the drugs employed can give rise to inhibition of their immune response [14]. The different studies have reported that the most commonly isolated organisms in patients in ICU comprise gram-negative bacilli (GNB), mainly Enterobacteriaceae species, and non-fermenting gram-negative bacilli (NF-GNB), such as Pseudomonas and Acinetobacter species [13,15,16]. GNB were responsible for 45–70% of ventilator-associated pneumonias, for 20–30% of vascular pathway infections (catheter), and for commonly caused infections in surgical wounds and in the urinary tract that led to sepsis [17]. In addition, in the last several years, the occurrence of highly resistant microorganisms (HRMO) is a major threat to patients in ICU, leading to worse outcomes, the need for isolation measures, and the demand for second-line or rescue antibiotics [18].
AMR varies from country to country and from to region to region depending on policies for antimicrobial use and the knowledge of its prevalence, and local resistance patterns may aid in better management of patients and the development of better antimicrobial stewardship programs. The aim of this study was to determine the prevalence of AMR in the clinical isolates of GNB from adult, pediatric, and neonatal patients in these ICU at a tertiary-care hospital in Mexico.
2. Materials and Methods
2.1. Site and Period of Study
This retrospective study involved all bacteriological samples collected in neonatal (NICU), pediatric (PICU), and adult (AICU) patients of these ICU at the Unidad Médica de Alta Especialidad (UMAE), Instituto Mexicano del Seguro Social (IMSS), in Mérida, state of Yucatán, Mexico. The protocol was approved by the Local Committees on Health Research 3203 and Ethics 32038 of the UMAE with registration number R-2018-3203-021. The data of the culture reports during the period from 1 August, 2016 to 31 December, 2018 were analyzed.
2.2. Microbiology
2.2.1. Culture and Identification
The clinical samples were collected appropriately at the bed-side and then transported to the Microbiology Laboratory. For this purpose, this study employed sterile bottles, sterile dry cotton swabs, versaTREK REDOX bottles (Thermo Fisher Scientific, Inc., Waltham, MA, USA), and sterile tubes with a Brain Heart Infusion medium depending on the sample type. Each specimen was plated onto MacConkey, Blood, Mannitol Salt, and Chocolate agars (Becton Dickinson & Co., Franklin Lakes, NJ, USA). The inoculated agars were incubated aerobically at 36 °C for 24 h. The bacterial species were classified by morphological-colony characteristics, Gram staining, and by routine rapid tests, such as catalase and oxidase. The organism identification was carried out using the automated MICROSCAN WalkAway® system (Beckman Coulter, Inc., Brea, CA, USA). MICROSCAN panels utilized modified conventional and chromogenic tests for the identification of Enterobacteriaceae species and NF-GNB. The identification was based on the detection of pH changes, substrate utilization, biochemical overlays, and growth in the presence of antimicrobial agents after 12–48 h of incubation at 35 °C [19].
2.2.2. Test Drugs
A total of eight types of drugs were tested for all microorganisms. The drugs included the following: (a) Penicillins, such as Ampicillin and Piperacillin; (b) penicillin-beta-lactamase inhibitor combination, such as Ampicillin/Sulbactam, Piperacillin/Tazobactam, and Ticarcillin/Clavulanic Acid; (c) cephalosporins, such as Cefepime, Cefotaxime, Cefuroxime, Cefazolin, Cefotetan, Ceftriaxone, and Ceftazidime; (d) aminoglycosides, such as Amikacin, Gentamicin, and Tobramycin; (e) carbapenems, such as Imipenem and Meropenem; (f) fluoroquinolones, such as Ciprofloxacin, Levofloxacin, and Moxifloxacin; (g) trimethoprims, such as Trimethoprim/Sulfamethoxazole, and (h) monobactams, such as Aztreonam.
2.2.3. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility was carried out with the help of the MICROSCAN WalkAway® system using Negative/urine combo 55 and Negative combo 68 panels. The tests carried out on MICROSCAN were miniaturizations of broth microdilution susceptibility tests that were dehydrated. The various antimicrobial agents were diluted in Mueller-Hilton broth supplemented with calcium and magnesium to concentrations bridging the range-of-clinical-interest after inoculation and rehydration with a standardized suspension of the organism and incubation at 35 °C for a minimum of 16 h [20,21]. The minimal inhibitory concentration (MIC) for the test organism was determined by observing the lowest antimicrobial concentration exhibiting growth inhibition, and the quality antimicrobial susceptibility pattern (S: Susceptible, I: Intermediate, and R: Resistant) was determined per Clinical and Laboratory Standards Institute (CLSI) guidelines [20].
2.2.4. Detection of Extended-Spectrum Beta-Lactamases (ESBL)
The screening for ESBL detection was performed on the MICROSCAN WalkAway® system employing antimicrobial susceptibility tests using Ceftazidime and Cefotaxime, with or without Clavulanic Acid [20].
2.3. Statistical Analysis
The data were entered and analyzed using Microsoft Excel. The distribution of the specimens and bacterial isolates was expressed as percentages. AMR, ESBL-producing, MDR, and HRMO profiles of clinical isolates were analyzed and expressed as percentages.
3. Results
3.1. Bacterial Isolates
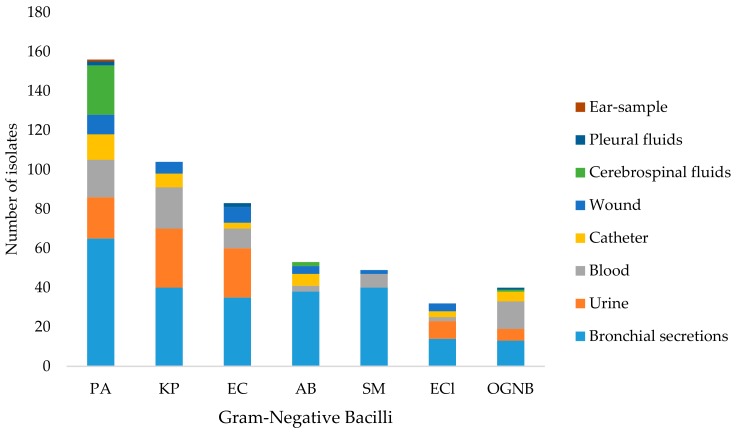
A total of 2,711 clinical samples were analyzed and 517 GNB strains were isolated. The distribution of GNB recovered from various clinical specimens of ICU patients is summarized in Figure 1 and Table 1. The majority of isolates were recovered from the clinical samples of bronchial secretions (n = 245), urinary samples (n = 91), and blood samples (n = 76). Pseudomonas aeruginosa was the predominant isolate (n = 156, 30.17%), followed by K. pneumoniae (n = 104, 20.12%), and E. coli (n = 83, 16.05%).
Figure 1.
Distribution of Gram-negative bacilli (GNB) recovered from clinical samples of patients in intensive care units (ICU). PA: P. aeruginosa, KP: K. pneumoniae, EC: E. coli, AB: A. baumannii; SM: Stenotrophomonas maltophilia; ECl: Enterobacter cloacae; OGNB: Other GNB. OGNB included P. mirabilis (n = 9), Serratia marcescens (n = 9), Burkholderia spp. (n = 9), A. lwoffi (n = 8), and Klebsiella oxytoca (n = 5).
Table 1.
Distribution of GNB recovered from clinical samples of patients in ICU.
| Sample | Total Number of Isolates | PA | KP | EC | AB | SM | ECl | OGNB |
|---|---|---|---|---|---|---|---|---|
| Bronchial secretions | 245 | 65 | 40 | 35 | 38 | 40 | 14 | 13 |
| Urine | 91 | 21 | 30 | 25 | 0 | 0 | 9 | 6 |
| Blood | 76 | 19 | 21 | 10 | 3 | 7 | 2 | 14 |
| Catheter | 37 | 13 | 7 | 3 | 6 | 0 | 3 | 5 |
| Wound | 34 | 10 | 6 | 8 | 4 | 2 | 4 | 0 |
| Cerebrospinal fluids | 28 | 25 | 0 | 0 | 2 | 0 | 0 | 1 |
| Pleural fluids | 5 | 2 | 0 | 2 | 0 | 0 | 0 | 1 |
| Ear-sample | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total (%) |
517 | 156 (30.17) | 104 (20.12) | 83 (16.05) | 53 (10.25) | 49 (9.48) |
32
(6.19) |
40
(7.74) |
GNB: Gram-Negative Bacilli, ICU: intensive care units, PA: P. aeruginosa, KP: K. pneumoniae, EC: E. coli, AB: A. baumannii, SM: S. maltophilia, ECl: E. cloacae, OGNB: Other GNB, including P. mirabilis (n = 9), S. marcescens (n = 9), Burkholderia spp. (n = 9), A. lwoffi (n = 8), and K. oxytoca (n = 5).
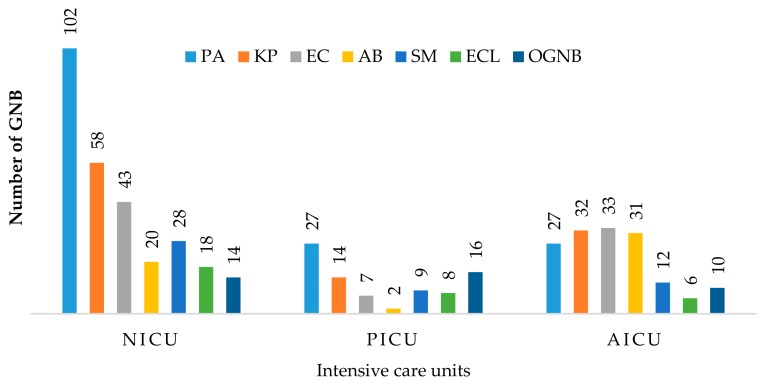
According to the distribution of isolates by ICU type, P. aeruginosa and K. pneumoniae were the most prevalent pathogens in NICU and PICU. Moreover, E. coli, K. pneumoniae, A. baumannii, and P. aeruginosa were the most frequent isolates from the AICU (Figure 2).
Figure 2.
Frequency of GNB among three intensive care units (Neonatal, NICU, Pediatric, PICU, and Adult, AICU) PA: P. aeruginosa, KP: K. pneumoniae, EC: E. coli, AB: A. baumannii; SM: S. maltophilia; ECl: E. cloacae; OGNB: Other GNB. OGNB including P. mirabilis (n = 9), S. marcescens (n = 9), Burkholderia spp. (n = 9), A. lwoffi (n = 8). and K. oxytoca (n = 5).
3.2. AMR of the Clinical Isolates
The drug resistance for GNB clinical isolates is depicted in Table 2. Overall, the highest resistant rate was found for Ampicillin (95.85%), followed by Cefuroxime (84.17%), Piperacillin (82.93%), Cefotaxime (78.07%), Ceftriaxone (77.41%), Aztreonam (75.23%), Cefazolin (75.00%), and Ceftazidime (73.19%). The lowest resistance rate was observed for Cefotetan (10.96%), and carbapenems (27.42–33.41%).
Table 2.
Proportions and percentage of drug-resistant GNB recovered from clinical samples of patients in ICU.
| Antimicrobial Drugs |
PA | KN | EC | AB | SM | ECl | OGNB | Overall Isolates |
|---|---|---|---|---|---|---|---|---|
| Ampicillin | 101/104 (97.12) |
78/83 (93.98) | 30/32 (93.75) | 22/22 (100) |
231/241 (95.85) |
|||
| Ampicillin/ Sulbactam |
71/104 (68.27) | 64/83 (77.11) | 28/53 (52.83) | 24/32 (75.00) | 13/29 (44.83) |
200/301 (66.67) |
||
| Piperacillin | 12/17 (70.59) | 30/30 (100) |
20/25 (80.00) |
6/8 (75.00) |
0/2 (0.00) |
68/82 (82.93) |
||
| Piperacillin/ Tazobactam |
71/155 (45.80) | 28/103 (27.18) | 28/81 (34.57) | 8/32 (25.00) | 4/22 (18.18) |
139/393 (35.36) |
||
| Ticarcillin/ Clavulanic acid |
26/40 (65.00) | 11/28 (39.29) |
16/29 (55.17) |
16/28 (57.14) | 10/28 (35.71) | 4/7 (57.14) |
2/14 (14.29) |
85/174 (48.85) |
| Cefazolin | 25/41 (60.97) | 34/411 (82.93) | 8/8 (100) | 8/10 (80.00) |
75/100 (75.00) |
|||
| Cefotetan | 7/62 (11.29) | 1/57 (1.75) |
6/16 (37.50) | 2/11 (18.18) |
16/146 (10.96) |
|||
| Cefuroxime | 84/103 (81.55) | 73/83 (87.95) | 27/32 (84.98) | 18/22 (81.82) |
202/240 (84.17) |
|||
| Cefotaxime | 84/104 (80.76) | 72/83 (86.75) | 40/53 (75.47) | 22/32 (68.75) | 17/29 (58.62) |
235/301 (78.07) |
||
| Ceftazidime | 100/154 (64.93) | 84/104 (80.76) | 73/83 (87.95) | 43/53 (81.13) | 28/49 (57.14) | 22/32 (68.75) | 24/36 (66.67) |
374/511 (73.19) |
| Ceftriaxone | 84/104 (80.76) | 73/83 (87.95) | 36/53 (67.92) | 23/32 (71.88) | 17/29 (58.62) |
233/301 (77.41) |
||
| Cefepime | 83/155 (55.13) | 83/104 (79.80) | 71/83 (85.54) | 40/53 (75.47) | 19/32 (59.37) | 17/29 (58.62) |
313/456 (68.64) |
|
| Aztreonam | 24/57 (42.11) | 65/74 (87.84) | 56/63 (88.89) | 14/16 (87.50) | 8/12 (66.67) |
167/222 (75.23) |
||
| Imipenem | 108/155 (69.68) | 10/104 (9.62) | 3/83 (3.61) | 7/32 (21.88) | 2/15 (13.33) |
130/389 (33.41) |
||
| Meropenem | 84/155 (54.19) | 7/104 (6.73) | 2/83 (2.41) | 30/53 (56.60) | 1/32 (3.13) | 3/36 (8.33) |
127/463 (27.42) |
|
| Ciprofloxacin | 65/155 (41.93) | 32/104 (30.77) | 65/83 (78.31) | 42/53 (79.25) | 16/32 (50.00) | 8/29 (27.59) |
228/456 (49.34) |
|
| Levofloxacin | 61/155 (39.35) | 16/104 (15.38) | 63/83 (75.90) | 32/53 (60.38) | 3/49 (6.12) | 4/32 (12.50) | 7/36 (19.44) |
186/512 (36.32) |
| Moxifloxacin | 5/28 (17.86) | 21/30 (70.00) | 1/8 (12.50) | 1/9 (11.11) |
28/75 (37.33) |
|||
| Amikacin | 90/155 (58.06) | 14/104 (13.46) | 8/83 (9.64) | 31/53 (58.49) | 5/32 (15.63) | 10/29 (34.48) |
158/456 (34.65) |
|
| Gentamicin | 80/155 (51.61) |
48/104 (46.15) | 52/83 (62.65) | 45/53 (84.91) | 9/32 (28.13) | 8/29 (27.59) |
242/456 (53.07) |
|
| Tobramycin | 66/155 (42.58) | 63/102 (61.76) | 64/81 (79.01) | 34/53 (64.15) | 15/31 (48.39) | 18/29 (62.07) |
260/451 (57.65) |
|
| Trimethoprim/ Sulfamethoxazole |
66/102 (64.71) | 52/82 (63.41) | 41/53 (77.36) | 4/49 (8.16) | 15/31 (48.39) | 15/35 (42.86) |
193/352 (54.83) |
Data that were available in Microbiology Laboratory reports were informed. PA: P. aeruginosa; KP: K. pneumoniae; EC: E. coli; AB: A. baumannii; SM: S. maltophilia; ECl: E. cloacae; OGNB: Other GNB. OGNB include: P. mirabilis (n = 9). S. marcescens (n = 9), Burkholderia spp. (n = 9), A. lwoffi (n = 8), and K. oxytoca (n = 5).
The enterobacterial isolates of K. pneumoniae, E. coli, and E. cloacae revealed high resistance rates to penicillins (97.12–75.00%), which was reversed by the addition of an ESBL inhibitor, such as Sulbactam and Tazobactam, cephalosporins (100–59.37%) except for Cefotetan, Aztreonam (88.89–87.50%), and Tobramycin (79.01–48.39%). Additionally, clinical isolates of E. coli displayed high resistance rates to fluoroquinolones (78.31–70.00%).
The clinical isolates of A. baumannii exhibited high resistance rates to third- and fourth-generation cephalosporins (81.13–67.92%), Ciprofloxacin (79.25%), Gentamicin (84.91%), and Trimethoprim/Sulfamethoxazole (77.36%). The clinical isolates of P. aeruginosa revealed high resistance rates to Piperacillin (70.59%) and Imipenem (69.68%). Other GNB (OGNB) that included P. mirabilis, S. marcescens, Burkholderia spp., A. lwoffi, and K. oxytoca revealed high resistance rates to Ampicillin (100%), Cefazolin (80.00%), Cefuroxime (81.82%), Ceftazidime (66.67%), and Aztreonam (66.67%).
3.3. MDR, HRMO Profiles, and ESBL-Producing
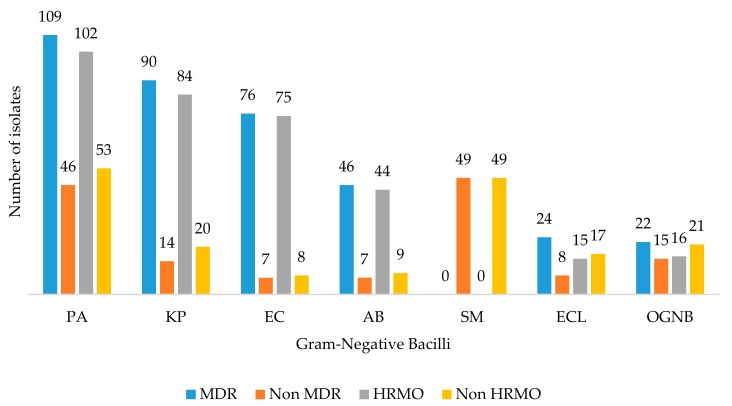
The frequency of MDR and HRMO rates is illustrated in Figure 3. Escherichia coli (MDR = 91.57%, HRMO = 90.36%), A. baumannii (MDR = 86.79%, HRMO = 83.02%), and K. pneumoniae (MDR = 83.65%, HRMO = 80.77%) exhibited the highest percentage of MDR and HRMO profiles in clinical isolates and, overall, 71.65% and 65.50% of total GNB exhibited MDR and HRMO profiles, respectively.
Figure 3.
Frequency of multidrug-resistant (MDR) and highly resistant microorganisms (HRMO) GNB recovered from patients in the three ICU. PA: P. aeruginosa; KP: K. pneumoniae; EC: E. coli; AB: A. baumannii; SM: S. maltophilia; ECl: E. cloacae; OGNB: Other GNB. OGNB include P. mirabilis (n = 9), S. marcescens (n = 9), Burkholderia spp. (n = 9), A. lwoffi (n = 8). and K. oxytoca (n = 5).
Moreover, the prevalence of the ESBL-producing was 83.13% in E. coli, 78.84% in K. pneumonia, and 66.67% in P. mirabilis.
4. Discussion
In hospitals ICU, due to their characteristics, allow for concentration factors that lead to AMR. These factors include the frequent use of broad-spectrum antimicrobial drugs, invasive procedures and devices, patients with a high frequency of comorbidity, and prolonged hospital stays, among others [13]. The present study investigated the spectrum of the GNB clinical isolates of patients hospitalized in ICU and their AMR pattern in a tertiary-care level hospital in the state of Yucatán, Mexico.
4.1. Bacterial Isolates
A total of 517 GNB were isolated from 2,711 clinical samples analyzed. In this study, the most common organism isolated from patients in an ICU was P. aeruginosa (n = 156; 30.41%). Our findings were in agreement with another study conducted in an ICU of a tertiary-care hospital located in India [11].
Klebsiella pneumoniae (n = 104; 20.27%) and E. coli (n = 83; 16.17%) were the most common enterobacterial clinical isolates, similar to those previously reported by Moolchandani et al. [11], in clinical isolates of patients in an ICU in India.
In the case of NF-GNB, the most prevalent species isolated were P. aeruginosa (n = 156; 30.41%) and A. baumannii (n = 53; 10.33%). Similar data were reported in studies conducted in three hospitals in Mexico. The most prevalent species found was P. aeruginosa (24%) followed by A. baumannii (12.5%) [22].
The most common culture-positive clinical samples received during the present study were bronchial secretions (40.64%). Similar findings were obtained by Moolchandani et al. [11] in a study conducted in an ICU of a tertiary-care level hospital in Southern India [10]. This is probably due to ventilator-associated pneumonia, which is one of principal infections in ICU and which occurs 48–72 h after the initiation of mechanical ventilation [23]. Interestingly, the most prevalent GNB isolate for bronchial secretions was the NF-GNB P. aeruginosa (42.30%). In the ICU of a hospital in Southern India, NF-GNB Acinetobacter spp. (36.00%) and Pseudomonas spp. (27.6%) were those most prevalent in bronchial secretions. This suggests that this is due to an increase in the colonization of NF-GNB in the respiratory tract of patients on prolonged mechanical ventilation [11].
4.2. AMR of the Clinical Isolates
AMR in hospitals is driven by the failures of hospitals in terms of hygiene, selective pressures created by the overuse of antimicrobial drugs, and mobile genetic elements that can encode mechanisms of bacterial resistance [24]. The increasing drug resistance of K. pneumoniae and E. coli in the last few decades has been a problem worldwide [25], and these GNB were the most prevalent among our enterobacterial clinical isolates. The clinical isolates of K. pneumoniae and E. coli exhibited high resistance rates to various groups of antimicrobial drugs such as penicillins (100–80.00%), cephalosporins (87.95–60.97%), Aztreonam (88.89–87.84%), and Tobramycin (79.01–61.76%). The high resistance rates to penicillins and cephalosporins were also detected in clinical isolates of patients in ICU from Vietnam [13], India [11], and Nepal [10]. Our results of drug resistance rates to Aztreonam and Tobramycin were higher than those reported for clinical isolates of patients in a hospital ICU in Saudi Arabia, where the resistance rates reported for these antimicrobials were within the range of 65.2–30.6% [26].
In addition, the clinical isolates of E. coli exhibited high resistance rates to all quinolone drugs evaluated (78.31–70.00%). In a Latin-American country such as Venezuela, since 2011, the resistance rates of E. coli to quinolones reached 46.51% and, for 2012, the tendency for Levofloxacin and Moxifloxacin was of 100% [27]. On the other hand, in a study conducted at three hospitals in Mexico, the resistance rates of E. coli to Ciprofloxacin was 55.56% [22]. Regarding another part of the world, such as ICU in Nepal and India, the number of strains resistant to Ciprofloxacin is greater, up to 90.1%, and to Levofloxacin, 77.4% [10,11].
NF-GNB have high intrinsic resistance to antimicrobial drugs due to the low outer-membrane permeability, together with secondary resistance mechanisms, such as inducible cephalosporinase or antibiotic efflux pumps. Therefore, the number of antimicrobial drugs is reduced [28]. Among the NF-GNB isolates of our patients, A. baumannii exhibited high resistance rates to a large number of antimicrobial drugs tested, such as cephalosporins, Ciprofloxacin, Gentamicin, and Trimethoprim/Sulfamethoxazole. Cephalosporin drugs play an important role in antimicrobial treatment. The A. baumannii isolates demonstrated high resistance rates, between 79.25 and 75.47%. The high resistance rates to Cephalosporin were also detected in a multicenter study conducted in three hospitals in Mexico. The resistance rates reported for these groups of antimicrobials was in the range of 100–55.56% [22]. In an ICU from hospitals located in Argentina, Chile, and Venezuela, in terms of A. baumannii, the number of resistant isolates was above 60% between the years 2004 and 2010 [27]. On the other hand, recent reports in hospitals in countries in Africa and South Asia revealed higher results than this study: Cephalosporin resistance rates in clinical isolates from patients in ICU of hospitals in Morocco and Vietnam ranged from 94.66–95.8% and from 93–95%, respectively [12,13]. In a hospital in Nepal, entire isolates were resistant to Ceftazidime and Cefepime [10]. In the case of Ciprofloxacin, Gentamicin, and Trimethoprim/Sulfamethoxazole, our results revealed the prevalence of resistance in more than 79.25% of A. baumannii clinical isolates. Our resistance results were lower than those reported for the clinical isolates of patients in hospital ICU in India [11], Vietnam [13], and Saudi Arabia [26]. Parajuli et al. [10] suggested that these high resistance levels of Acinetobacter spp. may be due to the elevated change of the acquisition of resistant genes and their ability to persist and multiply in a hospital environment [10].
The clinical isolates of P. aeruginosa exhibited high resistance rates to Imipenem (69.68%) and Piperacillin (70.59%). The effect of Piperacillin was improved with a β-lactamase inhibitor such as Tazobactam. Our findings on Imipenem are disturbing. P. aeruginosa was the most common organism isolated and 69.68% of these clinical isolates were resistant to Imipenem. Imipenem is the best reserve drug employed in the treatment of infections caused by MDR strains. Imipenem-resistant strains are often resistant to other antimicrobial drugs, and the prognosis is worse yet in terms of mortality and morbidity [29]. Imipenem resistance may be mediated by diminished outer membrane permeability, efflux pump over-expression, and carbapenemase enzymes. This later type of resistance is the most important clinically because these enzymes hydrolyze all or nearly all beta-lactams drugs. Carbapenemases are encoded by genes that are horizontally transferable by plasmids or transposons and commonly associated with genes encoding for other resistance determinants [30]. Carbapenemases, such as IMP- (Imipenem-resistant Pseudomonas), GES- (Guiana extended-spectrum beta-lactamase), VIM- (Veron integron-encoded metallo-beta-lactamase), and OXA-type (oxacillin-hydrolyzing), have been reported in clinical isolates of P. aeruginosa from patients in a hospital in Mexico [31,32]. On the other hand, our Imipenem resistance rates were higher than those reported for the clinical isolates from patients in ICU of a hospital in Saudi Arabia (38.2%) [26]. Similar findings were reported in isolates from patients in ICU in hospitals in Vietnam (79.3%) [13] and Nepal (62.5%) [10].
4.3. MDR, HRMO Profiles, and ESBL-Producing
This study found that 70.96% of GNB isolates demonstrated an MDR profile. This is high compared with that previously reported by Moolchandani et al. [11] in a study conducted in patients of ICU of hospitals in India (55.7%) [11]. Specifically, the clinical isolates of E. coli (91.57%), A. baumannii (86.79%), and K. pneumoniae (83.65%) comprised the GNB species with the greatest number of MDR strains. Our findings were in accordance with those of Moolchandani et al. [11] with respect to A. baumannii (82.1%). However, these were higher for E. coli (26.3%) and Klebsiella spp. (52.4%) [11]. In addition, the prevalence of the MDR profile of clinical isolates from this research was higher than that reported for patients at the Hospital Universitario of Monterrey, Nuevo León, Mexico, where the global prevalence of GNB with an MDR profile was 46.2% and included A. baumannii (74%), E. coli (40%), P. aeruginosa (34%), K. pneumoniae (22%), E. cloacae (9%), and Serratia spp. (7%) [33]. The prevalence of MDR-GNB is high even in developed countries. During 2013 in the U.S., 63% of isolates of A. baumannii presented an MDR profile, and it has been estimated that 7,300 infections were caused by these isolates [34].
HRMO are microorganisms that 1) are known to cause disease, 2) have acquired an AMR pattern that hampers therapy, and 3) have the potential to spread if, in addition to standard precautions, no transmission-based precautions are taken [35]. Kluytmans-VandenBergh et al. [35] defined HRMO as (1) Enterobacteriaceae that are ESBL-producing and/or resistant to carbapenems and/or resistant to fluoroquinolones and aminoglycosides (F and A), (2) Acinetobacter spp. that are resistant to carbapenems and/or resistant to F and A, (3) S. maltophilia resistant to Co-trimoxazole, and (4) P. aeruginosa resistant to at least three of following antimicrobial or antimicrobial groups: Piperacillin, Ceftazidime, F and A, and/or carbapenems [35,36]. This study demonstrated a high number of clinical isolates with an HRMO profile. A total of 65.50% of GNB exhibited the HRMO profile. The highest percentages of HRMO were recovered in clinical isolates of E. coli (90.36%), A. baumannii (83.02%), and K. pneumoniae (80.77%).
One of type of drug resistance comprises ESBL enzymes in the Enterobacteriaceae family, which permit the bacteria to resist the effect of penicillins and cephalosporins of first-, second-, and third-generation penicillins and cephalphorins, as well as Aztreonam [37]. In this study, 83.13% (69/83) of the clinical isolates of E. coli, 78.84% (82/104), of K. pneumoniae, and 66.67% (6/9) of P. mirabilis were ESBL-producing. The data in this study were higher than those reported in studies conducted in a hospital ICU from Nepal and India. With respect to research developed at a hospital in Nepal, ESBL isolates of E. coli and Klebsiella spp. exhibited 70.90% and 59.40%, respectively [10]. In case of a hospital in India, the prevalence of ESBL-producing was 47.6% for E. coli, 15.4% for Klebsiella spp., and 0% for Proteus spp. [38]. On the other hand, from 2009–2011, 13.7% and 16.6% of clinical isolates of E. coli were ESBL-producing in the U.S. and in European countries, respectively. The ESBL-producing isolates of Klebsiella spp. were reported to have undergone an increase in the prevalence of 27.5–42.8% in Europe during the same years [39]. Even so, the data of prevalence of ESBL-producing isolates of E. coli (83.13%) were higher than those reported by the SMART (Study for Monitoring Antimicrobial Resistance Trends) group with 41% [40]. Moreover, the SENTRY Antimicrobial Surveillance Program of Mexico reported a prevalence of 33% of ESBL-producing K. pneumoniae and at our hospital, 78.84% was found. This study’s data were also higher than those reported for Argentina (60%), Brazil (50%), and Chile (59%) [40]. The research studies on prevalence in Monterrey, N.L., Mexico, informed ESBL-producing in 35.9% of clinical isolates of K. pneumoniae and 35.6% of E. coli [41]. Morfín-Otero et al. [42] reported lower data for E. coli (16.3%) and K. pneumoniae (26.9%) in isolates of patients in Guadalajara Jalisco, Mexico [42]. In Mexico, CTX-M-15, and SHV-12 are the most commonly detected ESBL from patients in hospitals. In addition, CTX-M-9, SHV-5, GES-1, GES-19, TEM-1 and TLA-1 have also been found [43,44,45,46]. ESBL-producing in K. pneumoniae and E. coli isolates support the high level of resistance to Ampicillin (97.12–93.98%), Cephalosporin (91.18–60.97%), and Aztreonam (88.89–87.84%) in this research.
4.4. Study Limitations
First, MICROSCAN WalkAway® system are not able to differentiate among the species of the Burkholderia cepacia complex. Second, the data of MIC testing in/of clinical isolates were taken from the Microbiology Laboratory reports and sometimes these were incomplete. Third, the identification of MDR and HRMO profiles were based on the number of antimicrobial drug classes that were evaluated in the Microbiology Laboratory.
5. Conclusions
It can be concluded from this study that the most commonly isolated GNB were P. aeruginosa, K. pneumoniae, E. coli, and A. baumannii. The enterobacterial isolates revealed high resistance rates to Ampicillin and Aztreonam. Additionally, E. coli demonstrated high resistance rates to quinolones. Generally speaking, A. baumannii isolates were highly resistant to a large number of drugs tested. The clinical isolates of P. aeruginosa exhibited high resistance rates to Imipenem, the best reserve drug employed in the treatment of MDR infections. Finally, this study showed a higher rate of MDR and HRMO profiles and ESBL-producing in GNB clinical isolates. This finding herein emphasizes the need for continuous surveillance and rational treatment strategies to reduce the emergence and spreading of MDR- and HMRO-GNB.
Acknowledgments
JG-JG is a Medical Physician student who received a Scientific Summer Fellowship in 2017 from Universidad Juárez Autónoma de Tabasco. The authors are grateful to Maggie Brunner, M.A., for the English language review of this article.
Author Contributions
Conceptualization, U.-C.A.H. and M.-S.G.M.; methodology, U.-C.A.H., G.-O.C., M.-S.G.M.; software, U.-C.A.H., G.-O.C. and J.-G.J.G.; validation, U.-C.A.H., L.-C.I.G. and M.-S.G.M.; data curation, L.-C.I.G. and J.-G.J.G.; formal analysis, G.-O.C., M.-S.G.M.; investigation, U.-C.A.H. and M.-S.G.M.; writing—original draft preparation, U.-C.A.H. and M.-S.G.M.; writing—review and editing, U.-C.A.H., G.-O.C., L.-C.I.G., J.-G.J.G. and M.-S.G.M.; supervision, U.-C.A.H. and M.-S.G.M.; project administration, U.-C.A.H. and M.-S.G.M.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.World Health Organization . WHO Antimicrobial Resistance. WHO; Geneva, Switzerland: 2018. [(accessed on 12 July 2019)]. Available online: http://www.who.int/en/news-room/fact-heets/detail/antimicrobial-resistance. [Google Scholar]
- 2.Ventola C.L. The antibiotic resistance crisis Part 1: Causes and threats. Pharm. Ther. 2015;40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 3.Blair J.M., Webber M.A., Baylay A.J., Ogbolu D.O., Piddock L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015;13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 4.Fair R.J., Tor Y. Antibiotics and bacterial resistance in the 21st century. Perspect. Med. Chem. 2014;28:25–64. doi: 10.4137/PMC.S14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Kraker M.E.A., Stewardson A.J., Harbarth S. Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med. 2016;13:e1002184. doi: 10.1371/journal.pmed.1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson D.J., Jenkins T.C., Evans S.R., Harris A.D., Weinstein R.A., Tamma P.D., Han J.H., Banerjee R., Patel R., Zaoutis T., et al. The role of stewardship in addressing antibacterial resistance: Stewardship and infection control committee of the antibacterial resistance leadership Group. Clin. Infect. Dis. 2017;64:S36–S40. doi: 10.1093/cid/ciw830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magiorakos A.P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbarth S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization . WHO Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. WHO; Geneva, Switzerland: 2017. [(accessed on 12 July 2019)]. Available online: https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf. [Google Scholar]
- 9.Santajit S., Indrawattana N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed. Res. Int. 2016;2016:1–8. doi: 10.1155/2016/2475067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parajuli N.P., Acharya S.P., Mishra S.K., Parajuli K., Rijal B.P., Pokhrel B.M. High burden of antimicrobial resistance among Gram negative bacteria causing healthcare associated infections in a critical care unit of Nepal. Antimicrob. Resist. Infect. Control. 2017;6:1–9. doi: 10.1186/s13756-017-0222-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moolchandani K., Sastry A.S., Deepashree R., Sistla S., Harish B.N., Mandal J. Antimicrobial resistance surveillance among intensive care units of a tertiary care hospital in South India. J. Clin. Diagn. Res. 2017;11:DC01–DC07. doi: 10.7860/JCDR/2017/23717.9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uwingabiye J., Frikh M., Lemnouer A., Bssaibis F., Belefquih B., Maleb A., Dahraoui S., Belyamani L., Bait A., Haimeur C., et al. Acinetobacter infections prevalence and frequency of the antibiotics resistance: Comparative study of intensive care units versus other hospital units. Pan Afr. Med. J. 2016;23:1–10. doi: 10.11604/pamj.2016.23.191.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tran G.M., Ho-Le T.P., Ha D.T., Tran-Nguyen C.H., Nguyen T.S.M., Pham T.T.N., Nguyen T.A., Nguyen D.A., Hoang H.Q., Tran N.V., et al. Patterns of antimicrobial resistance in intensive care unit patients: A study in Vietnam. BMC Infect. Dis. 2017;17:429. doi: 10.1186/s12879-017-2529-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Álvarez C., Cortés J., Arango Á., Correa C., Leal A. Resistencia antimicrobiana en unidades de cuidado intensivo de Bogotá, Colombia, 2001–2003. Revista de Salud Pública. 2006;8:86–101. doi: 10.1590/S0124-00642006000400008. [DOI] [PubMed] [Google Scholar]
- 15.Hamishehkar H., Shadmehr P., Mahmoodpoor A., Mashayekhi S.O., Entezari-Maleki T. Antimicrobial susceptibility patterns among bacteria isolated from intensive care units of the largest teaching hospital at the northwest of Iran. Braz. J. Pharm. Sci. 2016;52:403–412. doi: 10.1590/s1984-82502016000300006. [DOI] [Google Scholar]
- 16.Esfahani B.N., Basiri R., Mirhosseini S.M.M., Moghim S., Dolatkhah S. Nosocomial infections in intensive care unit: Pattern of antibiotic-resistance in Iranian community. Adv. Biomed. Res. 2017;6:1–5. doi: 10.4103/2277-9175.205527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruppé É., Woerther P.L., Barbier F. Mechanisms of antimicrobial resistance in Gram-negative bacilli. Ann. Intens. Care. 2015;5:1–15. doi: 10.1186/s13613-015-0061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aardema H., Arends J.P., de Smet A.M., Zijlstra J.G. Burden of highly resistant microorganisms in a Dutch intensive care unit. Neth. J. Med. 2015;73:169–174. [PubMed] [Google Scholar]
- 19.MacFaddin J.F. Biochemical Tests for Identification of Medical Bacteria. 3rd ed. Williams and Wilkins; Baltimore, MD, USA: 2000. [Google Scholar]
- 20.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing 2013. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2013. Twenty third informational supplement M100-S23. [Google Scholar]
- 21.Tilton R.C., Lieberman L., Gerlach E.H. Microdilution antibiotic susceptibility test: Examination of certain variables. Appl. Microbiol. 1973;26:658–665. doi: 10.1128/am.26.5.658-665.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutiérrez-Muñoz J., Ramírez-Corona A., Martínez-Bustamante M.E., Coria-Lorenzo J.J., Armenta-Gallegos L., Ayala-Franco J.R., Bernal-Granillo S.M., Flores-Zaleta F.J., García-Pérez F.E., Monjardín-Rochín J.A., et al. Estudio multicéntrico de resistencias bacterianas nosocomiales en México. Rev. Latin. Infect. Pediatr. 2017;30:68–75. [Google Scholar]
- 23.Charles M.P., Kali A., Easow J.M., Joseph N.M., Ravishankar M., Srinivasan S., Kumar S., Umadevi S. Ventilator-associated pneumonia. Australas. Med. J. 2014;7:334–344. doi: 10.4066/AMJ.2014.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinstein R.A. Controlling antimicrobial resistance in hospitals: Infection control and use of antibiotics. Emerg. Infect. Dis. 2001;7:188–192. doi: 10.3201/eid0702.010206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durdu B., Meric-Koc M., Hakyemez I.N., Akkoyunlu Y., Daskaya H., Sumbul-Gultepe B., Aslan T. Risk Factors affecting patterns of antibiotic resistance and treatment efficacy in extreme drug resistance in intensive care unit-acquired Klebsiella pneumoniae infections: A 5-year analysis. Med. Sci. Monit. 2019;7:174–183. doi: 10.12659/MSM.911338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ibrahim M.E. High antimicrobial resistant rates among Gram-negative pathogens in intensive care units. A retrospective study at a tertiary care hospital in Southwest Saudi Arabia. Saudi Med. J. 2018;39:1035–1043. doi: 10.15537/smj.2018.10.22944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luna C.M., Rodríguez-Noriega E., Bavestrello L., Guzmán-Blanco M. Gram-negative infections in adult intensive care units of Latin America and the Caribbean. Crit. Care Res. Pract. 2014;2014:1–12. doi: 10.1155/2014/480463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hancock R.E. Resistance mechanisms in Pseudomonas aeruginosa and other nonfermentative Gram-Negative bacteria. Clin. Infect. Dis. 1998;1:S93–S99. doi: 10.1086/514909. [DOI] [PubMed] [Google Scholar]
- 29.Ameen N., Memon Z., Shaheen S., Fatima G., Ahmed F. Imipenem resistant Pseudomonas aeruginosa: The fall of the final quarterback. Pak. J. Med. Sci. 2015;31:561–565. doi: 10.12669/pjms.313.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meletis G. Carbapenem resistance: Overview of the problem and future perspectives. Ther. Adv. Infect. Dis. 2016;3:15–21. doi: 10.1177/2049936115621709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.López-García A., Rocha-Gracia C., Bello-López E., Juárez-Zelocualtecalt C., Sáenz Y., Castañeda-Lucio M., López-Pliego L., González-Vázquez M.C., Torres C., Ayala-Núñez T., et al. Characterization of antimicrobial resistance mechanisms in carbapenem-resistant Pseudomonas aeruginosa carrying IMP variants recovered from a Mexican hospital. Inf. Drug Resist. 2018;11:1523–1536. doi: 10.2147/IDR.S173455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garza-Ramos U., Barrios H., Reyna-Flores F., Tamayo-Legorreta E., Catalán-Nájera J.C., Morfín-Otero R., Rodríguez-Noriega E., Volkow P., Cornejo-Juárez P., González A., et al. Widespread of ESBL- and carbapenemase GES-type genes on carbapenem-resistant Pseudomonas aeruginosa clinical isolates: A multicenter study in Mexican hospitals. Diagn. Microbiol. Infect. Dis. 2015;81:135–137. doi: 10.1016/j.diagmicrobio.2014.09.029. [DOI] [PubMed] [Google Scholar]
- 33.Garza-González E., Llaca-Díaz J.M., Bosques-Padilla F.J., González G.M. Prevalence of multidrug-resistant bacteria at a tertiary-care teaching hospital in Mexico: Special focus on Acinetobacter baumannii. Chemotherapy. 2010;56:275–279. doi: 10.1159/000319903. [DOI] [PubMed] [Google Scholar]
- 34.US Centers for Disease Control and Prevention Antibiotic Resistance Threats in the United States. [(accessed on 12 July 2018)];2013 Available online: https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf.
- 35.Kluytmans-Vandenbergh M.F., Kluytmans J.A., Voss A. Dutch guideline for preventing nosocomial transmission of Highly Resistant Microorganisms (HRMO) Infection. 2005;33:309–313. doi: 10.1007/s15010-005-5079-z. [DOI] [PubMed] [Google Scholar]
- 36.Souverein D., Euser S.M., Herpers B.L., Kluytmans J., Rossen J.W.A., Den-Boer J.W. Association between rectal colonization with Highly Resistant Gram-negative Rods (HR-GNRs) and subsequent infection with HR-GNRs in clinical patients: A one-year historical cohort study. PLoS ONE. 2017;14:e0211016. doi: 10.1371/journal.pone.0211016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrew B., Kagirita A., Bazira J. Prevalence of Extended-Spectrum Beta-Lactamases-Producing microorganisms in patients admitted at KRRH, southwestern Uganda. Int. J. Microb. 2017;2017:1–5. doi: 10.1155/2017/3183076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arora A., Jain C., Saxena S., Kaur R. Profile of drug resistant Gram negative bacteria from ICU at a tertiary care center of India. Asian J. Med. Health. 2017;3:1–7. doi: 10.9734/AJMAH/2017/31434. [DOI] [Google Scholar]
- 39.Sader H., Farrell D., Flamm R., Jones R. Antimicrobial susceptibility of Gram-negative organisms isolated from patients hospitalized in intensive care units in United States and European hospitals (2009–2011) Diagn. Microbiol. Infect. Dis. 2014;78:443–448. doi: 10.1016/j.diagmicrobio.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 40.Guzmán-Blanco M., Labarca J.A., Villegas M.V., Gotuzzo E. Extended spectrum β-lactamase producers among nosocomial Enterobacteriaceae in Latin America. Braz. J. Infect. Dis. 2014;18:421–433. doi: 10.1016/j.bjid.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garza-González E., Ibarra S.I.M., Llaca-Díaz J.M., González G.M. Molecular characterization and antimicrobial susceptibility of extended-spectrum β-lactamase-producing Enterobacteriaceae isolates at a tertiary-care centre in Monterrey, Mexico. J. Med. Microbiol. 2011;60:84–90. doi: 10.1099/jmm.0.022970-0. [DOI] [PubMed] [Google Scholar]
- 42.Morfín-Otero R., Mendoza-Olazarán S., Silva-Sánchez J., Rodríguez-Noriega E., Laca-Díaz J., Tinoco-Carrillo P., Petersen L., López P., Reyna-Flores F., Alcántar-Curiel D., et al. Characterization of Enterobacteriaceae isolates obtained from a tertiary care hospital in Mexico, which produces extended-spectrum beta-lactamase. Microb. Drug Resist. 2013;19:378–383. doi: 10.1089/mdr.2012.0263. [DOI] [PubMed] [Google Scholar]
- 43.Aquino-Andrade A., Mérida-Vieyra J., Arias de la Garza E., Arzate-Barbosa P., De Colsa-Ranero A.E. Carbapenemase-producing Enterobacteriaceae in Mexico: Report of seven non-clonal cases in a pediatric hospital. BMC Microbiol. 2018;18:38. doi: 10.1186/s12866-018-1166-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barrios H., Garza-Ramos U., Mejía-Miranda I., Reyna-Flores F., Sánchez-Pérez A., Mosqueda-García D., Silva-Sánchez J. ESBL-producing Escherichia coli and Klebsiella pneumoniae: The most prevalent clinical isolates obtained between 2005 and 2012 in Mexico. J. Glob. Antimicrob. Resist. 2017;10:243–246. doi: 10.1016/j.jgar.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 45.Alcántar-Curiel M.D., Alpuche-Aranda C.M., Varona-Bobadilla H.J., Gayosso-Vázquez C., Jarillo-Quijada M.D., Frías-Mendívil M., Sanjuan-Padrón L., Santos-Preciado J.I. Risk factors for extended-spectrum β-lactamases-producing Escherichia coli urinary tract infections in a tertiary hospital. Salud Pública de México. 2015;57:412–418. doi: 10.21149/spm.v57i5.7621. [DOI] [PubMed] [Google Scholar]
- 46.Mérida-Vieyra J., De Colsa A., Calderón-Castañeda Y., Arzate-Barbosa P., Aquino-Andrade A. First report of group CTX-M-9 Extended spectrum beta- lactamases in Escherichia coli isolates from pediatric patients in Mexico. PLoS ONE. 2016;11:e0168608. doi: 10.1371/journal.pone.0168608. [DOI] [PMC free article] [PubMed] [Google Scholar]