Abstract
A number of studies from different countries have characterized mcr-1-harboring plasmids isolated from food; however, nothing has been reported about it in South Korea. In this study, we report the characterization of mcr-1 plasmids from pan drug-resistant (PDR) Escherichia coli strains isolated from retail food in the country. Colistin-resistant E. coli strains were isolated from retail raw chicken, and PCR was carried out to detect the mcr-1 gene. Whole genome sequencing of the mcr-1-positive strains was performed for further characterization. The results of whole genome sequencing revealed that all mcr-1 plasmids belonged to the IncI2 type. In addition to the mcr-1 plasmids, all of the isolates also carried additional plasmids possessing multiple antibiotic resistance genes, and the PDR was mediated by resistant plasmids except for fluoroquinolone resistance resulting from mutations in gyrA and parC. Interestingly, the mcr-1 plasmids were transferred by conjugation to other pathogenic strains including enterohemorrhagic E. coli (EHEC), enterotoxigenic E. coli (ETEC), enteroaggregative E. coli (EAEC), Salmonella, and Klebsiella at the frequencies of 10−3−10−6, 10−2−10−5, 10−4−10−5, 10−4−10−6, and 10−5−10−6, respectively. The results showed that mcr-1 plasmids can be easily transmitted to pathogenic bacteria by conjugation.
Keywords: Escherichia coli, mcr-1, food, retail raw chicken
1. Introduction
The emergence of multidrug-resistant (MDR) pathogens, such as ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumonia, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp., or recently Enterobacteriaceae) and the lack of effective antimicrobials are a serious issue in public health [1]. Colistin is one of the last-resort antibiotics to treat MDR Gram-negative bacteria, and its binding to the lipid A moiety of lipopolysaccharide (LPS) destabilizes the outer membrane and results in cell death [2]. Colistin resistance is mainly associated with the modification of LPS, such as phosphoethanolamine modification of lipid A [3,4,5], and causes serous clinical problems in the control of Gram-negative pathogens in ESKAPE [6,7,8].
Since the first discovery of the mobilized colistin resistance (mcr)-1 gene on plasmids in Escherichia coli by Liu et al. in China, 2016 [4], E. coli strains harboring mcr-1 have been reported in many countries throughout America, Asia, and Europe [4,9,10] and have been isolated from various sources, such as animals, humans, environmental samples, and food [10,11,12]. In South Korea, mcr-1-positive E. coli strains have been isolated in livestock and humans [13,14]. However, there have been no studies about mcr-1-positive E. coli from retail food in the country.
Several recent reports also suggested that the spread of mcr-1 to multidrug-resistant bacteria can contribute to the development of the pan drug-resistant (PDR) phenotype, since mcr-1 on plasmids can be easily disseminated [3,15]. The increasing number of PDR bacteria, including E. coli, is considered a threat to public health [16]. E. coli is a major cause of human diseases, such as urinary tract infections, sepsis, and pneumonia [17]. Therefore, the emergence of PDR E. coli isolates, particularly those harboring extended-spectrum β-lactamase (ESBL) and plasmid-mediated quinolone resistance (PMQR) genes, have aggravated the public health burdens of antibiotic resistance.
A number of studies have shown that retail chicken is a major reservoir of disseminating antibiotic-resistant E. coli to humans [9,18,19]. In this study, we aimed at isolating mcr-1-positive E. coli strains from retail chicken in South Korea, characterizing the DNA sequence of the mcr-1 plasmids, and determining the frequencies of conjugational transfer of mcr-1 plasmids to other Gram-negative pathogens.
2. Materials and Methods
2.1. Bacterial Strains and Culture Methods
Three mcr-1-positive E. coli strains (JSMCR1, FORC81 and FORC82) were isolated from retail raw chicken in South Korea in our previous study [20]. The mcr-1-positive E. coli strains, E. coli ATCC 43889 (Enterohemorrhagic E. coli; EHEC), E. coli NCCP 14039 (Enteroaggregative E. coli; EAEC), enterotoxigenic E. coli (ETEC, a laboratory collection), Salmonella enetrica serovar Typhimurium SL1344, and K. pneumoniae (a laboratory collection) were cultured on Luria–Bertani (LB) media at 37 °C. The pathogenic E. coli strains (EHEC, ETEC, EAEC), Salmonella Typhimurium, and Klebsiella were used as recipient strains in the conjugation assay.
2.2. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was performed with a broth dilution method as described previously [21,22] with ten antibiotics, including ampicillin, cephalothin, tetracycline, chloramphenicol, ciprofloxacin, kanamycin, gentamicin, streptomycin, polymyxin B, and colistin. E. coli ATCC 25922 was used as the quality control strain.
2.3. Conjugation Assay
For the selection of transconjugants, we first obtained spontaneous mutants of streptomycin-resistant recipient strains by culturing them on LB media supplemented with streptomycin. Donor and recipient cells were prepared by transferring 1% inoculum from overnight cultures into fresh LB broth, followed by incubation at 37 °C for 4 h with constant shaking. E. coli was conjugated with recipient cells at a ratio of 1:1. Cells were pelleted by centrifugation, washed twice with 10 mM MgSO4, and resuspended in 50 µL of MgSO4. The mixture of donor and recipient cells were spread on LB agars supplemented with streptomycin (2 µg/mL) and colistin (4 µg/mL). Transconjugants were confirmed with PCR using mcr-1-specific primers [4] and the recipient strains. Conjugation frequencies were calculated as the number of transconjugants per recipient cell.
2.4. Whole-Genome Sequencing
Whole-genome sequencing and assembly were performed commercially at ChunLab Inc. (Seoul, South Korea). The whole genome of E. coli JSMCR1, FORC81 and FORC82 was sequenced using PacBio RS II (Pacific Biosciences, Menlo Park, CA, USA). The genome sequences were annotated using the online Rapid Annotation Subsequencing Technology (RAST) and CLC Main Workbench 3.6.1 (CLC bio, Aarhus, Denmark), and deposited in the GenBank database with accession numbers CP030152-CP030157 (JSMCR1), CP029057-CP029061 (FORC81), and CP026641-CP026644 (FORC82).
3. Results and Discussion
3.1. Whole-Genome Sequencing of mcr-1-Positive E. coli Strains
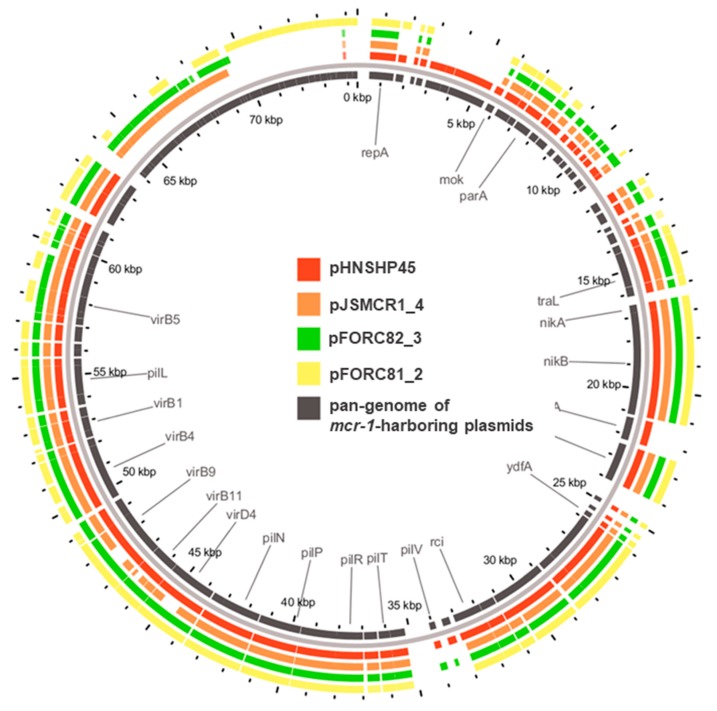
Three mcr-1-positive E. coli strains (JSMCR1, FORC81 and FORC82) were isolated from retail chicken in South Korea. The results of whole genome sequencing revealed that the three mcr-1-postive E. coli strains possessed multiple plasmids and some of the plasmids harbored a number of antibiotic resistance genes (Table 1 and Figure S1). The three mcr-1-harboring plasmids belonged to the IncI2 type and possessed the genetic elements for bacterial conjugation (Table 1). The three mcr-1-harboring plasmids were similar to pHNSHP45 (accession no. KP347127), the first mcr-1-harboring plasmid isolated in China; pJSMCR1_4 (96% query coverage, 100% max nucleotide identity), pFORC81_2 (89% query coverage, 99% max nucleotide identity), and pFORC82_3 (97% query coverage, 99% max nucleotide identity). Unlike pHNSHP45, however, insertion sequences were not found in the plasmids (Figure 1). The sequences of the mcr-1-harboring plasmids were similar to the IncI2-type mcr-1-harboring plasmids, which were isolated from livestock and humans in Korea [13,14]. This is also consistent with a previous extensive analysis revealing that IncI2 is predominant in mcr-1-harboring plasmids in Asia, whereas IncHI2 plasmids are predominant in Europe [23].
Table 1.
Plasmids present in E. coli isolates harboring mcr-1.
| E. coli Strain | Plasmid | Size (bp) | GenBank Accession No. | Inc Group | Resistance Genes |
|---|---|---|---|---|---|
| JSMCR1 | pJSMCR1_1 | 152,677 | CP030153 | IncFIB, IncFII | aph(3′)-Ia, aac(3)-IId, blaCTX-M-65, fosA3 |
| pJSMCR1_2 | 134,064 | CP030154 | p0111 | aadA1, blaOXA-10, qnrS1, floR, cmlA1, arr-2, tet(A), dfrA14 | |
| pJSMCR1_3 | 109,689 | CP030155 | IncI1 | - | |
| pJSMCR1_4 | 61,828 | CP030156 | IncI2 | mcr-1 | |
| pJSMCR1_5 | 29,589 | CP030157 | IncX4 | - | |
| FORC81 | pFORC81_1 | 253,947 | CP029058 | IncI1, IncFII |
aadA1, aadA2, aac(3)-IId, blaTEM-1B,
qnrS1, floR, cmlA1, sul3, tet(A), dfrA12 |
| pFORC81_2 | 61,917 | CP029059 | IncI2 | mcr-1 | |
| pFORC81_3 | 38,749 | CP029060 | IncX1 | - | |
| pFORC81_4 | 32,945 | CP029061 | IncI1 | blaTEM-1B, floR | |
| FORC82 | pFORC82_1 | 250,778 | CP026642 | IncHI2A, IncHI2, IncN |
aadA1, aph(3′’)-Ib, aph(6)-Id, blaCTX-M-65, blaOXA-10, qnrS1, mph(A), floR, cmlA1,
arr-2, sul2, tet(M), tet(A), dfrA14 |
| pFORC82_2 | 101,404 | CP026643 | IncFIC, IncFIB | - | |
| pFORC82_3 | 65,206 | CP026644 | IncI2 | mcr-1 |
Figure 1.
Sequence comparison of mcr-1-harboring plasmids. pHNSHP45 was used as a reference. Black inner ring indicated the pan-genome of the mcr-1-harboring plasmid.
3.2. Antimicrobial Susceptibility Profiles and Other Resistance Genes
All of the mcr-1-positive E. coli strains were highly resistant to most of the tested antibiotics belonging to different classes (Table 2). In particular, E. coli JSMCR1 was resistant to all the antibiotics tested in this study. Based on the results of whole genome sequencing, all isolates carried a few plasmids with different replicon types and multiple other antibiotic resistance genes conferring resistance to several different antibiotic classes (Table 1 and Figure S1). Whole-genome sequencing discovered point mutations in gyrA and parC, which confer resistance to fluoroquinolones; however, other antibiotic resistance genes were not found in the chromosome of the three strains, suggesting that pan drug resistance is primarily mediated by resistance plasmids in the strains.
Table 2.
Minimum inhibitory concentrations (MICs) of mcr-1-positive E. coli strains and their transconjugants.
| Strain | Origin a | mcr-1 Gene b | MIC c (μg/mL) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | CEF | TET | CHL | CIP | KAN | GEN | STR | POL | COL | |||
| E. coli JSMCR1 | WT | + | >64 | >64 | 128 | >64 | >8 | >32 | >64 | 64 | 8 | 8 |
| E. coli FORC81 | WT | + | >64 | 64 | >128 | >64 | >8 | 8 | >64 | 8 | 8 | 8 |
| E. coli FORC82 | WT | + | >64 | >64 | 128 | 64 | 2 | 4 | 2 | 2 | 8 | 8 |
| EHEC (E. coli ATCC 43889) | WT | - | ≤0.5 | ≤0.5 | ≤0.0628 | ≤0.5 | ≤0.0039 | ≤0.25 | ≤0.5 | >128 | ≤0.25 | ≤0.25 |
| pJSMCR1/EHEC | TC | + | ≤0.5 | ≤0.5 | ≤0.0628 | ≤0.5 | ≤0.0039 | ≤0.25 | ≤0.5 | >128 | 4 | 4 |
| pFORC81/EHEC | TC | + | ≤0.5 | ≤0.5 | ≤0.0628 | ≤0.5 | ≤0.0039 | ≤0.25 | ≤0.5 | >128 | 4 | 4 |
| pFORC82/EHEC | TC | + | ≤0.5 | ≤0.5 | ≤0.0628 | ≤0.5 | ≤0.0039 | ≤0.25 | ≤0.5 | >128 | 4 | 4 |
| ETEC (isolate) | WT | - | >64 | 8 | 32 | 64 | 0.0312 | >32 | >64 | >128 | 4 | 2 |
| pJSMCR1/ETEC | TC | + | >64 | 8 | 32 | 64 | 0.0312 | >32 | >64 | >128 | 8 | 8 |
| pFORC81/ETEC | TC | + | >64 | 8 | 32 | 64 | 0.0312 | >32 | >64 | >128 | 8 | 8 |
| pFORC82/ETEC | TC | + | >64 | 8 | 32 | 64 | 0.0312 | >32 | >64 | >128 | 8 | 8 |
| EAEC (E. coli NCCP 14039) | WT | - | >64 | 16 | >128 | 4 | 0.0312 | 8 | 2 | >128 | 4 | 2 |
| pJSMCR1/EAEC | TC | + | >64 | 16 | >128 | 4 | 0.0312 | 8 | 2 | >128 | 8 | 4 |
| pFORC81/EAEC | TC | + | >64 | 16 | >128 | 4 | 0.0312 | 8 | 2 | >128 | 8 | 8 |
| pFORC82/EAEC | TC | + | >64 | 16 | >128 | 4 | 0.0312 | 8 | 2 | >128 | 8 | 8 |
| S. Typhimurium SL1344 | WT | - | 2 | 2 | 0.5 | 4 | 0.0156 | 4 | 2 | >128 | 4 | 4 |
| pJSMCR1/SL1344 | TC | + | 2 | 2 | 0.5 | 4 | 0.0156 | 4 | 2 | >128 | 8 | 8 |
| pFORC81/SL1344 | TC | + | 2 | 2 | 0.5 | 4 | 0.0156 | 4 | 2 | >128 | 8 | 8 |
| pFORC82/SL1344 | TC | + | 2 | 2 | 0.5 | 4 | 0.0156 | 4 | 2 | >128 | 8 | 8 |
| Klebsiella (isolate) | WT | - | >64 | 16 | >128 | >64 | >16 | >32 | 1 | >128 | 4 | 4 |
| pJSMCR1/Klebsiella | TC | + | >64 | 16 | >128 | >64 | >16 | >32 | 1 | >128 | 8 | 16 |
| pFORC81/Klebsiella | TC | + | >64 | 16 | >128 | >64 | >16 | >32 | 1 | >128 | 8 | 16 |
| pFORC82/Klebsiella | TC | + | >64 | 16 | >128 | >64 | >16 | >32 | 1 | >128 | 32 | 16 |
a WT: wild type, TC: Transconjugant. b Presence (+) or absence (-) of mcr-1, based on PCR and confirmed by sequencing. c AMP: ampicillin, CEF: cephalothin, TET: tetracycline, CHL: chloramphenicol, CIP: ciprofloxacin, KAN: Kanamycin; GEN: gentamicin; STR: streptomycin; POL: polymyxin B; COL: colistin.
3.3. Conjugation Assay
The conjugation experiments were performed with pathogenic E. coli strains (EHEC, ETEC, EAEC), Salmonella Typhimurium, and Klebsiella as the recipient strains. The results of the antimicrobial susceptibility test showed that only the minimum inhibitory concentrations (MICs) of polymyxin B and colistin were increased in transconjugants from two-fold to 16-fold compared to their parental strains (Table 2). However, other plasmids harboring antibiotic resistance genes, which were simultaneously present in the mcr-1-postive strains, were not transferred to the recipient strains under the experimental settings based on PCR testing with primers specific to the plasmids. These results indicated that mcr-1 plasmids are highly transmissible compared to other resistance plasmids; this presumably enables mcr-1 plasmids to be disseminated from E. coli to Gram-negative bacteria [4,24].
To confirm this, we determined the conjugational frequency of mcr-1 plasmids and found that the transmission rates of the mcr-1 plasmids were 10−3–10−6 in EHEC, 10−2–10−5 in ETEC, 10−4–10−5 in EAEC, 10−4–10−6 in Salmonella, and 10−5–10−6 in Klebsiella (Figure 2). Conjugation frequencies varied depending on the recipient strain. For instance, the conjugation frequencies of pFORC81-2 were as high as 3 × 10−3 in ETEC and as low as 2 × 10−6 in EHEC (Figure 2). The frequencies of conjugational transfer of the mcr-1 plasmids to E. coli strains were similar to previous studies [25]. Furthermore, in this study, we demonstrated that mcr-1 plasmids were transmitted to other Gram-negative bacteria, such as Salmonella and Klebsiella, at the frequencies comparable to those observed in E. coli (Figure 2).
Figure 2.
Conjugation frequencies of three mcr-1 plasmids from E. coli isolates from retail chicken. The data represent the means and standard deviations of the results from three independent experiments.
4. Conclusions
In this study, we isolated and characterized three mcr-1 plasmids from PDR E. coli strains from retail raw chicken in South Korea. Although the number of isolated plasmids was small, the findings of this study are important because this is the first report about mcr-1 plasmids originating from retail food in the country. The whole genome sequencing of the PDR E. coli strains showed that all the genetic determinants for antibiotic resistance were associated with plasmids, except for fluoroquinolone resistance caused by point mutations in gyrA and parC. The mcr-1 plasmids were highly transferrable to pathogenic E. coli strains, Salmonella, and Klebsiella. This may allow colistin resistance to easily spread in the food supply system.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/2076-2607/7/9/344/s1. Figure S1: Circular map of plasmid pJSMCR1_1 (A), pJSMCR1_2 (B), pFORC81_1 (C), and pFORC82_1 (D).
Author Contributions
S.R. and B.J. designed the study; J.K., B.K.H., and H.C. performed the experiments. J.K. and B.J. analyzed the data; J.K. and B.J. wrote the manuscript; J.K., Y.W., S.H.C., S.R., and B.J. critically reviewed the manuscript.
Funding
This research was funded by a grant (14162MFDS972, 19162MFDS037) from the Ministry of Food and Drug Safety in 2019. J.K. was supported by the BK21 Plus Program of Department of Agricultural Biotechnology, Seoul National University, Seoul, Korea.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Gajdács M. The concept of an ideal antibiotic: Implications for drug design. Molecules. 2019;24:892. doi: 10.3390/molecules24050892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rhouma M., Beaudry F., Theriault W., Letellier A. Colistin in pig production: Chemistry, mechanism of antibacterial action, microbial resistance emergence, and one health perspectives. Front. Microbiol. 2016;7:1789. doi: 10.3389/fmicb.2016.01789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao R., Hu Y., Li Z., Sun J., Wang Q., Lin J., Ye H., Liu F., Srinivas S., Li D. Dissemination and mechanism for the mcr-1 colistin resistance. PLoS Pathog. 2016;12:e1005957. doi: 10.1371/journal.ppat.1005957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y.-Y., Wang Y., Walsh T.R., Yi L.-X., Zhang R., Spencer J., Doi Y., Tian G., Dong B., Huang X. Emergence of plasmid-mediated colistin resistance mechanism mcr-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 5.Carroll L.M., Gaballa A., Guldimann C., Sullivan G., Henderson L.O., Wiedmann M. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. mBio. 2019;10:e00853-19. doi: 10.1128/mBio.00853-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El Chakhtoura N.G., Saade E., Iovleva A., Yasmin M., Wilson B., Perez F., Bonomo R.A. Therapies for multidrug resistant and extensively drug-resistant non-fermenting Gram-negative bacteria causing nosocomial infections: a perilous journey toward ‘molecularly targeted’therapy. Expert Rev. Anti-Infect. Ther. 2018;16:89–110. doi: 10.1080/14787210.2018.1425139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gajdács M., Urbán E. Epidemiological trends and resistance associated with Stenotrophomonas maltophilia bacteremia: a 10-year retrospective cohort study in a tertiary-care hospital in hungary. Diseases. 2019;7:41. doi: 10.3390/diseases7020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boucher H.W., Talbot G.H., Bradley J.S., Edwards J.E., Gilbert D., Rice L.B., Scheld M., Spellberg B., Bartlett J. Bad bugs, no drugs: No ESKAPE! an update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 9.do Monte D.F., Fernandes M.R., Cerdeira L., Esposito F., Galvão J.A., Franco B.D., Lincopan N., Landgraf M. Chicken meat as reservoir of colistin-resistant Escherichia coli carrying mcr-1 genes in South America. Antimicrob. Agents Chemother. 2017;61:e02718-16. doi: 10.1128/AAC.02718-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perrin-Guyomard A., Bruneau M., Houée P., Deleurme K., Legrandois P., Poirier C., Soumet C., Sanders P. Prevalence of mcr-1 in commensal Escherichia coli from French livestock, 2007 to 2014. Eurosurveillance. 2016;21:1–3. doi: 10.2807/1560-7917.ES.2016.21.6.30135. [DOI] [PubMed] [Google Scholar]
- 11.Guenther S., Falgenhauer L., Semmler T., Imirzalioglu C., Chakraborty T., Roesler U., Roschanski N. Environmental emission of multiresistant Escherichia coli carrying the colistin resistance gene mcr-1 from German swine farms. J. Antimicrob. Chemother. 2017;72:1289–1292. doi: 10.1093/jac/dkw585. [DOI] [PubMed] [Google Scholar]
- 12.Donà V., Bernasconi O.J., Pires J., Collaud A., Overesch G., Ramette A., Perreten V., Endimiani A. Heterogeneous genetic location of mcr-1 in colistin-resistant Escherichia coli isolated from humans and retail chicken meat in Switzerland: emergence of mcr-1-carrying IncK2 plasmids. Antimicrob. Agents Chemother. 2017;61:e01245-17. doi: 10.1128/AAC.01245-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim S.-K., Kang H.Y., Lee K., Moon D.-C., Lee H.-S., Jung S.-C. First detection of the mcr-1 gene in Escherichia coli isolated from livestock between 2013 and 2015 in South Korea. Antimicrob. Agents Chemother. 2016;AAC:01472-16. doi: 10.1128/AAC.01472-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim E.S., Chong Y.P., Park S.-J., Kim M.-N., Kim S.-H., Lee S.-O., Choi S.-H., Woo J.H., Jeong J.-Y., Kim Y.S. Detection and genetic features of mcr-1-producing plasmid in human Escherichia coli infection in South korea. Diagn. Microbiol. Infect. Dis. 2017;89:158–160. doi: 10.1016/j.diagmicrobio.2017.06.020. [DOI] [PubMed] [Google Scholar]
- 15.Du H., Chen L., Tang Y.-W., Kreiswirth B.N. Emergence of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet Infect. Dis. 2016;16:287–288. doi: 10.1016/S1473-3099(16)00056-6. [DOI] [PubMed] [Google Scholar]
- 16.Abat C., Fournier P.-E., Jimeno M.-T., Rolain J.-M., Raoult D. Extremely and pandrug-resistant bacteria extra-deaths: myth or reality? Eur. J. Clin. Microbiol. Infect. Dis. 2018;37:1687–1697. doi: 10.1007/s10096-018-3300-0. [DOI] [PubMed] [Google Scholar]
- 17.Smith J.L., Fratamico P.M., Gunther N.W. Extraintestinal pathogenic Escherichia coli. Foodborne Pathog. Dis. 2007;4:134–163. doi: 10.1089/fpd.2007.0087. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y., Zhang R., Li J., Wu Z., Yin W., Schwarz S., Tyrrell J.M., Zheng Y., Wang S., Shen Z. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat. Microbiol. 2017;2:16260. doi: 10.1038/nmicrobiol.2016.260. [DOI] [PubMed] [Google Scholar]
- 19.Schrauwen E.J., Huizinga P., van Spreuwel N., Verhulst C., Kluytmans-van den Bergh M.F., Kluytmans J.A. High prevalence of the mcr-1 gene in retail chicken meat in the Netherlands in 2015. Antimicrob. Resist. Infect. Control. 2017;6:83. doi: 10.1186/s13756-017-0242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park H., Kim J., Ryu S., Jeon B. The predominance of blaCTX-M-65 and blaCTX-M-55 in extended-spectrum β-lactamase-producing Escherichia coli from retail raw chicken in South korea. J. Glob. Antimicrob. Resist. 2019;17:216–220. doi: 10.1016/j.jgar.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 21.Clinical and Laboratory Standards Institute Performance standards for antimicrobial susceptibility testing: M100–S27. [(accessed on 1 January 2017)];2017 Available online: https://clsi.org/standards-development/document-correction-notices/
- 22.Bell S., Gatus B., Pham J., Rafferty D. Antibiotic susceptibility testing by the CDS method. [(accessed on 12 July 2019)];1999 A concise laboratory manual. Available online: http://cdstest.net/manual/
- 23.Matamoros S., Hattem J.M., Arcilla M.S., Willemse N., Melles D.C., Penders J., Vinh T.N., Hoa N., Jong M.D., Schultsz C. Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci. Rep. 2017;7:15364. doi: 10.1038/s41598-017-15539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang C., Feng Y., Liu F., Jiang H., Qu Z., Lei M., Wang J., Zhang B., Hu Y., Ding J. A phage-like IncY plasmid carrying the mcr-1 gene in Escherichia coli from a pig farm in China. Antimicrob. Agents Chemother. 2017;61:e02035-16. doi: 10.1128/AAC.02035-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee J.-Y., Lim S.-K., Choi Y., Moon D.-C., Shin J., Ko K.S. Whole sequences and characteristics of mcr-1-harboring plasmids of Escherichia coli strains isolated from livestock in South Korea. Microb. Drug Resist. 2018;24:489–492. doi: 10.1089/mdr.2017.0369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.