Abstract
Anthropogenic disturbances can have strong impacts on lichen communities, as well as on individual species of lichenized fungi. Traditionally, lichen monitoring studies are based on the presence and abundance of fungal morphospecies. However, the photobionts, as well photobiont mycobiont interactions also contribute to the structure, composition, and resilience of lichen communities. Here we assess the genetic diversity and interaction patterns of algal and fungal partners in lichen communities along an anthropogenic disturbance gradient in Białowieża Forest (Poland). We sampled a total of 224 lichen thalli in a protected, a managed, and a disturbed area of the forest, and sequenced internal transcribed spacer (ITS) ribosomal DNA (rDNA) of both, fungal and algal partners. Sequence clustering using a 97% similarity threshold resulted in 46 fungal and 23 green algal operational taxonomic units (OTUs). Most of the recovered photobiont OTUs (14 out of 23) had no similar hit in the NCBI-BLAST search, suggesting that even in well studied regions, such as central Europe, a lot of photobiont diversity is yet undiscovered. If a mycobiont was present at more than one site, it was typically associated with the same photobiont OTU(s). Generalist species, i.e., taxa that associate with multiple symbiont partners, occurred in all three disturbance regimes, suggesting that such taxa have few limitations in colonizing or persisting in disturbed areas. Trebouxia jamesii associated with 53% of the fungal OTUs, and was generally the most common photobiont OTU in all areas, implying that lichens that associate with this symbiont are not limited by the availability of compatible photobionts in Central European forests, regardless of land use intensity.
Keywords: barcoding, biological indicators, ITS, managed forests, species interaction network, photobiont
1. Introduction
Lichens are frequently used as bioindicators for air pollution, climatic changes, and other anthropogenic disturbances [1,2]. The symbiotic association consisting of a fungal partner (mycobiont) and a photosynthetic partner (photobiont) is particularly sensitive to changes in substrate properties and microclimatic conditions. Anthropogenic activities in forests change host tree properties (e.g., stand age, and tree species composition), which has an effect, for instance, on the availability of bark with a particular texture or pH, as well as the microclimatic conditions, and can ultimately change the lichen community patterns of the regions under management [3,4]. Thus, lichens have been used as indicators of anthropogenic disturbance and forest health for decades [5,6,7].
Environmental assessments using lichens as indicators typically rely on presence and abundance data, based on morphological identification of the lichen-forming fungus [2,8,9,10]. However, morphological identification of specimens could bias diversity estimates, because cryptic (= molecular) diversity is not regarded. Furthermore, this approach neglects an assessment of the associated microalgae, which may also contribute to geographic distribution, and community composition of lichenized fungi [11,12]. For example, climatic factors can influence the altitudinal distribution of symbiotic green algae [13,14], or specific interaction patterns between mycobionts and photobionts [15,16]. Presently, we know much more about the diversity and distribution of fungal partners of lichens in forests [2,5,9,17,18]. However, a more comprehensive understanding of lichen communities requires an assessment of the diversities, distributions, and interactions of both partners since the availability of compatible algae is a prerequisite for the establishment and maintenance of lichen populations.
Molecular species delimitation has been effectively implemented in the last decades to uncover previously unknown lineages and to estimate species boundaries in both, lichenized fungi and their symbiotic algae [19,20,21,22,23,24]. In this regard, internal transcribed spacer (ITS) ribosomal DNA (rDNA) has been widely used as a barcode for both mycobionts [25,26,27], and their associated photobionts [24,26,28,29,30]. The availability of universal primers specific for the photobiont ITS have facilitated the identification of photobionts, often across different genera [31,32,33,34,35]. Nevertheless, photobionts of many lichens remain unidentified, hindering our understanding of patterns of symbiont interactions. Generating a molecular barcode of both partners in natural lichen communities will help us narrow down this knowledge gap and provide a more concise perspective on symbiont interaction patterns in lichens.
Symbiont interactions in lichens can be described in terms of specificity and selectivity. Specificity refers to the phylogenetic range of possible partners [36,37,38]. Selectivity refers to preferential association with a specific partner when more than one partner is available. Both specialist and generalist interactions have been reported for lichens [19,39,40,41]. It is expected that lichen communities harboring organisms with low selectivity and flexible partner choice may be more resilient to anthropogenic disturbances than communities consisting of specialized lichens. This is because loss of the photobiont species may not result in the immediate loss of the lichen due to flexible partner choice. The identification of both symbionts allows us to assess the interaction patterns present in a community. A better understanding of symbiont interaction patterns could help devise forestry practices that minimize the impact on lichen communities.
The objective of this study is to assess the impact of land use intensity on mycobiont and photobiont diversity and interaction patterns in lichen communities in a central European forest. For this, we generated ITS sequences of both symbiotic partners from lichens collected in a protected, a managed, and a disturbed area of Białowieża Forest, Poland.
2. Materials and Methods
2.1. Study Area and Sampling Design
The specimens for the present study were collected by AŁ and MK, in 2016 from trees located in three different areas in Białowieża Forest, which differ in the degree of land use intensity (Table S1). Białowieża Forest in Central-Eastern Europe is one of the largest remaining parts of an extensive temperate forest system that once constituted the natural vegetation of the European lowlands. The first study area (“protected”) is present in forest section No. 256 (52°45′45″ N 23°52′42″ E) and is the best-preserved forest ecosystem of Białowieża Forest comprising trees of different age, and dead wood present in the form of abundant logs and snags [10,42]. Some of the oldest trees in this area are 100 to 200 years old (oaks and hornbeam respectively). The second study area (“managed”) is located west of Budy village in forest section No. 335D (52°44′03″ N 23°42′59″ E) outside the national park. This area contains trees of different age, but no logs and snags, and only a few stumps of cut trees, indicating management activity. The oldest trees in this area are about 100 years old (Picea abies). The dominant trees are mostly young and mature spruce (Picea abies) and hornbeam trees (Carpinus betulus), a few mature oaks (Quercus robur) and aspen (Populus tremula), and very few lime trees (Tilia cordata). The third study area (“disturbed”) is located south of Czerlonka village in forest section No. 469C (52°41′16″ N 23°43′02″ E). This area is the most disturbed, located close to a road and buildings, without dead wood in the form of decaying logs or snags. Presence of several stumps with cut marks indicates past logging activities. The oldest trees are rare (pines and spruces, about 170 years old), and most of the trees in this area (e.g., hornbeam) are younger (about 100 years or less). A few apple trees, possibly dispersed from a neighboring orchard, are also present in this area.
Samples in the three areas were collected from about 60 living trees of different species from near the ground to about 2 m up the stem. From each tree 1 to 3 samples were collected. Detailed specimen information is given in Supplementary S1 (Supplementary Materials). Specimens are deposited in the following herbaria: The Jan Kochanowski University in Kielce (KTC) and the University of Gdańsk (UGDA).
2.2. DNA Sequencing and Alignments
Total genomic DNA was extracted from lichen thalli using the cetyl-trimethyl ammonium bromide (CTAB) method [43]. We amplified fungal and algal internal transcribed spacer (ITS) ribosomal DNA (rDNA) using general primers or taxon-specific primers [27,30,44,45] (Table S2). PCRs were carried out in a volume of 25 µL. Each reaction mix contained 2.5 µL buffer, 0.13 µL (0.65 U) TaKaRa ExTaq (Takara Bio Europe, SAS, Saint-Germain-en-Laye, France), 2.0 µL dNTP mix (2.5 mM each), 1.0 µL each (10 mM) of the primer set (forward and reverse), c. 20 ng of template, and 16 µL H2O. DNA concentration was measured with a nanophotometer (Implen) and by gel images. Reactions were performed under the following cycling conditions: Initial denaturation at 95 °C for 4 min, followed by 35 cycles of 95 °C for 30 s, 50 °C/60 °C for 30 s (annealing temperature for fungal ITS and Trebouxia ITS: 50 °C, for the Trentepohlia ITS: 60 °C) and 72 °C for 1 min, and final elongation at 72 °C for 5 min. PCR products were checked for amplification on 1% agarose gels and sequenced using the Big Dye Terminator v.3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA) as follows: 1 min at 95 °C, and 30 cycles of 10 s at 96 °C, 10 s at 50 °C, and 2 min at 60 °C. Products were purified and then detected on an ABI PRISM 3730 DNA Analyzer (Applied Biosystems). Sequences were assembled using GENEIOUS v.5.4 [46]. Gaps were treated as missing data and ambiguously aligned regions were excluded. The sequences are deposited in GenBank (accession numbers- fungal ITS: MN387000-MN387223; photobiont ITS: MN396904-MN397127).
Bands of expected size were extracted using the peqGOLD Gel Extraction Kit (PEQLAB Biotechnologie GmbH, Erlangen, Germany) and labeled for cycle sequencing using the Big Dye Terminator v.3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA) and sequenced as follows: 1 min at 96 °C, and 26 cycles of 20 s at 96 °C, 5 s at 50 °C, and 2 min at 60 °C. Products were purified using the Big Dye XTerminator Purification kit (Life Technologies, Foster City, CA, USA) and then detected on an ABI PRISM 3730 DNA Analyzer (Applied Biosystems). Sequences were assembled using GENEIOUS v.5.4 [46]. Gaps were treated as missing data and ambiguously aligned regions were excluded.
2.3. Species Identification, OTU Analysis, and Association Network
Fungal and algal OTUs were inferred using a 97% similarity threshold using vsearch v2.8.4, which uses a fast heuristic based on nucleotides shared by the sequences in order to identify similar sequences [47]. The delimited fungal OTUs were assigned a species name based on morphological identification, and NCBI BLAST hits of ≥97% similarity to sequences present in GenBank (Table S1). We implemented the following scheme for identifying fungal species: If the first NCBI BLAST hit was the same as morphological identification (based on >97% sequence similarity), the morphological identification was confirmed. If the ITS sequence of the identified species was not present in GenBank, and no significantly similar BLAST hits were retrieved, the morphological identification was retained. If the first BLAST hit corresponded to the morphological identification but the sequence similarity was <97%, the species was given the morphological name along with the suffix sp. 1 and so on.
Assigning a taxon name to the ITS sequence of a lichen photobiont using NCBI BLAST searches is impaired by the incompleteness of the data base, the lack of reliably identified algal species, and the presently unresolved phylogeny and systematics of the large and variable genus Trebouxia. Blast searches often result in several similar hits, and different putative species names for a sequence. To reliably allocate OTUs to an algal species we decided to use only the reference ITS sequences derived from algal cultures. We aligned our algal ITS dataset to the ITS sequences of the reference ITS sequences of Trebouxia and Trentepohlia from the (1) SAG culture collection, Göttingen, Germany (Sammlung von Algenkulturen der Universität Göttingen, University of Goettingen, Germany), which maintains the largest and most comprehensive culture collection of microalgae, consisting of about 1400 species belonging to 500 genera, representing almost all classes and phyla of eukaryotic algae and cyanobacteria, and (2) UTEX culture collection, Texas, USA (University of Texas, USA), which maintains cultures of about 3000 strains of algae. Overall, 40 ITS reference sequences of Trebouxia, and Trentepohlia, representing 26 species, were aligned with our algal ITS data set of 224 sequences representing 23 OTUs.
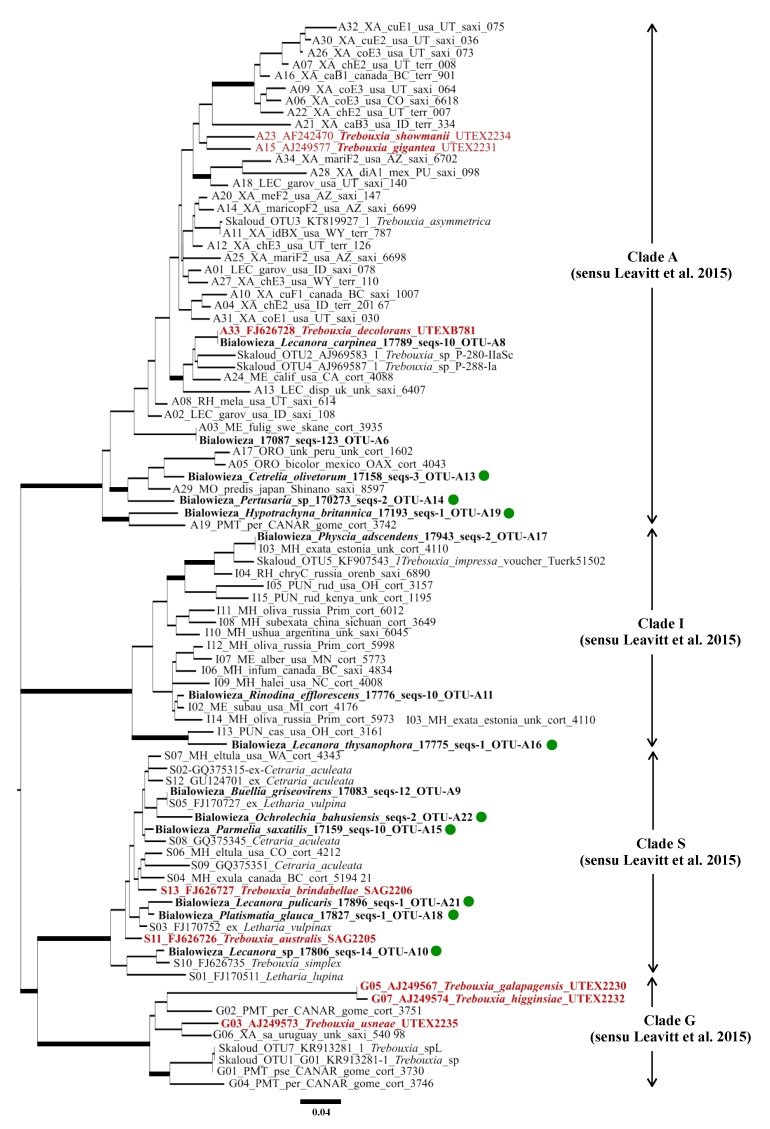
In order to link the photobiont OTUs recovered in our study to the photobionts found in other studies, we performed a sequence-similarity based search in the NCBI database with 97% similarity as the threshold for the same photobiont OTU. This demonstrates, if the photobiont was reported from other lichens elsewhere. Additionally, to infer the other potential fungal hosts of the photobionts reported in our study, we generated a maximum likelihood tree with 1000 bootstraps from an alignment of the 14 representative Trebouxia OTUs from our study, eight representative Trebouxia OTUs from Škaloud et al. [48] and 69 representative Trebouxia OTUs from Leavitt et al. [19] (Figure 1).
Figure 1.
Internal transcribed spacer (ITS) gene tree constructed with the representative Trebouxia operational taxonomic units (OTUs) from this study, and 69 and seven representative OTUs from Leavitt et al. [19], and Škaloud et al. [48] respectively. Four major clades from Leavitt et al. [19] are indicated. ITS sequences from this study are in bold, and those from the SAG and UTEX culture collection are in red and bold. For the sake of reference to the already published dataset, the names of the representative OTUs from Leavitt et al. [19] and Škaloud et al. [48] are retained. The reference OTUs from this study are named according to the following scheme: Locality, DNA/sample ID, number of sequences of that OTU in the dataset, and lastly, Trebouxia OTU number. Novel OTUs are indicated with a green dot in front of the name of the representative OTU sequence.
The association network of the lichen-forming fungi and photobionts was inferred using the function plotweb v1.4.4 in the bipartite package in R [49,50].
3. Results
3.1. Genetic Diversity and Identification of the Lichen Symbionts
We detected 46 mycobiont and 23 photobiont OTUs in 224 lichen samples collected from the three areas in Białowieża Forest (Table 1). Between 25–32 fungal, and 11–17 algal OTUs were retrieved from each area in the Białowieża Forest (Table 1). We did not find high cryptic diversity in the fungal partners (Table S1 and Table 2). Only five fungal OTUs had no significant hit in GenBank, i.e., >97% sequence similarity (Table S1). In contrast, we found high cryptic diversity in the algal partners (Table S1 and Table 3). Only seven OTUs (out of 23) could be assigned a name based on clustering with the reference ITS sequences derived from the algal cultures deposited in SAG or the UTEX culture collection (Table 3), two algal OTUs had previously been reported from other lichens, but are not formally described, and 14 algal OTUs (10 Trebouxia and four Trentepohlia) had no significant hit in GenBank.
Table 1.
OTUs recovered from the forest lichen community.
| Site | # Samples | # Fungal OTUs | # Algal OTUs | Fungal OTUs Shared | Algal OTUs Shared | % Unique F OTUs | % Unique A OTUs |
|---|---|---|---|---|---|---|---|
| protected | 78 | 32 | 17 (6) | NA | NA | 34.3% | 35.3% |
| managed | 75 | 25 | 13 (2) | NA | NA | 4% | 15.4% |
| disturbed | 71 | 27 | 11 | NA | NA | 27% | 27.3% |
| overall | 224 | 46 | 23 | 12 | 7 | NA | NA |
Table 2.
Fungal–algal associations of lichens in a protected, a managed, and a disturbed area of Białowieża Forest.
| Fungal OTU | Overall Frequency Fungal OTU | Protected | Managed | Disturbed | |||
|---|---|---|---|---|---|---|---|
| Frequency Fungal OTU | Associated Algal OTU(s) | Frequency of Fungal OTU | Associated Algal OTU(s) | Frequency Fungal OTU | Associated Algal OTU(s) | ||
| Biatora mendax | 1 | 1 | T. jamesii | 0 | -- | 0 | -- |
| Biatora sp. 1 | 2 | 2 | Apatococcus sp. | 0 | -- | 0 | -- |
| Buellia griseovirens | 14 | 2 | T. suecica, T. simplex | 4 | T. simplex | 8 | T. suecica, T. simplex, Trebouxia (A22) |
| Cetrelia olivetorum | 2 | 2 | Trebouxia (A13) | 0 | -- | 0 | -- |
| Cladonia chlorophaea | 1 | 0 | -- | 0 | -- | 1 | Asterochloris phycobiontica |
| Cladonia coniocraea | 3 | 0 | -- | 2 | Asterochloris phycobiontica | 1 | Asterochloris phycobiontica |
| Cladonia digitata | 2 | 1 | Asterochloris phycobiontica | 1 | Asterochloris phycobiontica | 0 | -- |
| Cladonia fimbriata | 3 | 0 | -- | 0 | -- | 3 | Asterochloris phycobiontica |
| Evernia prunastri | 4 | 0 | -- | 0 | -- | 4 | T. jamesii |
| Fellhanera gyrophorica | 5 | 2 | Parachloroidium sp. | 3 | Parachloroidium sp. | 0 | -- |
| Graphis betulina | 1 | 1 | Trentepohlia (A4) | 0 | -- | 0 | -- |
| Graphis sp. 1 | 3 | 2 | Trebouxia (A11) | 1 | Trentepohlia sp SAG11880 (A1) | 0 | -- |
| Graphis sp. 2 | 3 | 0 | -- | 2 | Trentepohlia (A4) | 1 | Trentepohlia (A4) |
| Hypocenomyce scalaris | 2 | 0 | -- | 0 | -- | 2 | T. suecica |
| Hypogymnia physodes | 4 | 0 | -- | 1 | T. suecica | 3 | T. suecica |
| Hypotrachyna revoluta | 1 | 1 | Trebouxia (A18) | 0 | -- | 0 | -- |
| Lecanora argentata | 20 | 4 | T. jamesii | 12 | T. jamesii | 4 | T. jamesii |
| Lecanora carpinea | 7 | 1 | T. decolorans | 3 | T. decolorans | 3 | T. decolorans |
| Lecanora chlarotera | 1 | 0 | -- | 0 | -- | 1 | T. jamesii |
| Lecanora glabrata | 14 | 7 | T. jamesii | 7 | T. jamesii | 0 | -- |
| Lecanora pulicaris | 1 | 0 | -- | 0 | -- | 1 | Trebouxia (A20) |
| Lecanora stanislai | 2 | 0 | -- | 1 | T. simplex | 1 | T. simplex |
| Lecanora thysanophora | 2 | 1 | T. jamesii | 1 | Trebouxia (A15) | 0 | -- |
| Lecidella elaeochroma | 9 | 3 | T. jamesii | 3 | T. jamesii | 3 | T. jamesii, T. decolorans |
| Loxospora elatina | 2 | 1 | T. jamesii | 1 | Asterochloris phycobiontica | 0 | -- |
| Melanelixia glabratula | 20 | 5 | T. jamesii | 6 | T. jamesii | 9 | T. jamesii |
| Menegazzia terebrata | 1 | 1 | Trebouxia (A13) | 0 | -- | 0 | -- |
| Ochrolechia bahusiensis | 3 | 0 | -- | 2 | T. suecica, T. simplex | 1 | Trebouxia (A22) |
| Parmelia saxatilis | 3 | 1 | T. suecica | 0 | -- | 2 | T. suecica |
| Parmelia sulcata | 6 | 2 | Trebouxia (A11) | 2 | Trebouxia (A11) | 2 | Trebouxia (A11) |
| Parmelia sulcata s.l. | 2 | 1 | Trebouxia (A11) | 0 | -- | 1 | Trebouxia (A11) |
| Pertusaria amara | 19 | 8 | T. jamesii | 7 | T. jamesii | 4 | T. jamesii |
| Pertusaria coccodes | 9 | 5 | T. jamesii | 3 | T. jamesii | 1 | T. jamesii |
| Pertusaria leioplaca | 9 | 3 | T. jamesii | 5 | T. jamesii | 1 | T. jamesii |
| Pertusaria sp. 1 | 4 | 3 | T. jamesii, Trebouxia (A14) | 1 | Trebouxia (A14) | 0 | -- |
| Phlyctis argena | 9 | 1 | T. jamesii | 4 | Dictyochloropsis sp. | 4 | T. jamesii, Dictyochloropsis sp. |
| Physcia adscendens | 1 | 0 | -- | 0 | -- | 1 | T. flava |
| Platismatia glauca | 5 | 1 | T. suecica | 1 | Trebouxia (A17) | 3 | Asterochloris phycobiontica, T. suecica |
| Pseudevernia furfuracea | 2 | 0 | -- | 0 | -- | 2 | T. suecica |
| Pyrenula nitida | 4 | 4 | Trentepohlia (A2) | 0 | -- | 0 | -- |
| Pyrenula nitidella | 1 | 1 | Trentepohlia (A2) | 0 | -- | 0 | -- |
| Ramalina farinacea | 7 | 2 | T. jamesii | 1 | T. jamesii | 4 | T. jamesii |
| Ramalina pollinaria | 3 | 3 | T. jamesii | 0 | -- | 0 | -- |
| Rinodina sp. | 1 | 0 | -- | 1 | Trebouxia (A11) | 0 | -- |
| Ropalospora viridis | 1 | 1 | Trebouxia (A19) | 0 | -- | 0 | -- |
| Thelotrema lepadinum | 5 | 5 | Trentepohlia (A3), Trentepohlia (A5) | 0 | -- | 0 | -- |
Table 3.
Algal OTUs: Summary of the sampled photobiont sequences, including their frequency and fungal hosts.
| OTUs | Photobiont sp. | Overall Occurrence | Associated # Fungal OTU(s) | Fungal Hosts (This Study) | Clade/OTU sensu Leavitt et al. [19] | Top NCBI Hits/Other Studies |
|---|---|---|---|---|---|---|
| A0 | Asterochloris phycobiontica strain SAG 26.81 | 11 | 7 | Cladonia gracilis, C. coniocraea, C. digitata, C. chlorophaea, C. fimbriata, Loxospora elatina, Platismatia glauca | NA | Cladonia symphycarpa, Lepraria caesioalba, Varicellaria carneonivea |
| A1 | Trentepohlia sp SAG11880 | 3 | 1 | Graphis pulverulenta, Graphis scripta | NA | Graphis scripta |
| A2 | Trentepohlia (A2) | 4 | 1 | Pyrenula nitida, Pyrenula nitidella | NA | None |
| A3 | Trentepohlia (A3) | 4 | 1 | Thelotrema lepadinum | NA | Trentepohlia annulata strain SAG 20.94 (96.5%) |
| A4 | Trentepohlia (A4) | 4 | 2 | Graphis betulina, Graphis sp. 2 | NA | None |
| A5 | Trentepohlia (A5) | 1 | 1 | Thelotrema sp. | NA | None |
| A6 | Trebouxia jamesii | 123 | 16 | Biatora sp. 2, Evernia prunastri, Lecanora argentata, L. chlarotera, L. glabrata, L. thysanophora, Lecidella elaeochroma, Loxospora elatina, Melanelixia glabratula, Pertusaria amara, P. coccodes, P. leioplaca, P. sp. 1, Phlyctis argena, Ramalina farinacea, R. pollinaria | Clade A, OTU A03 |
Lecanora bicincta, L. rupicola, Protoparmelia badia, P. montagnei, Tephromela atra |
| A7 | Parachloroidium sp. | 5 | 1 | Fellhanera sp. | NA | None |
| A8 | Trebouxia decolorans UTEXB781 | 9 | 2 | Lecidella elaeochroma, Lecanora carpinea | Clade A, OTU A33 |
Xanthoria candelaria, X. elegans, X. parietina |
| A9 | Trebouxia suecica/ T. australis | 19 | 7 | Buellia griseovirens, Ochrolechia bahusiensis, Hypocenomyce scalaris, Hypogymnia physodes, Parmelia saxatilis, Platismatia glauca, Pseudevernia furfuracea | Clade S, OTU S05 |
Cetraria islandica |
| A10 | Trebouxia simplex | 12 | 3 | Buellia griseovirens, Lecanora stanislai, Ochrolechia bahusiensis | Clade S, OTU S10 |
Bryoria furcellata, Boreoplaca ultrafrigida, Imshaugia aleurites, Psora decipiens, Protoparmelia ochrococca, Thamnolia vermicularis, Tuckermannopsis americana, Umbilicaria esculenta, Pseudevernia consocians |
| A11 | Trebouxia (A11) | 9 | 2 | Parmelia sulcata, Rinodina sp. | Clade I, OTU 102 |
Melanelixia subaurifera |
| A12 | Dictyochloropsis sp. | 7 | 1 | Phlyctis argena | NA | None |
| A13 | Trebouxia (A13) | 3 | 2 | Cetrelia olivetorum, Menegazzia terebrata | Clade A, NONE | None |
| A14 | Trebouxia (A14) | 2 | 1 | Pertusaria sp. 1 | Clade A, NONE | None |
| A15 | Trebouxia (A15) | 1 | 1 | Lecanora thysanophora | Clade S, OTU S08 |
None |
| A16 | Trebouxia flava UTEX181 | 1 | 1 | Physcia adscendens | Clade I, NONE |
Physconia grisea, P. distorta, P. enteroxantha, Tephromela atra |
| A17 | Trebouxia (A17) | 1 | 1 | Platismatia glauca | Clade I, OTU I03 |
None |
| A18 | Trebouxia (A18) | 1 | 1 | Hypotrachyna britannica | Clade S, NONE | None |
| A19 | Trebouxia (A19) | 1 | 1 | Ropalospora sp. | Clade A, NONE | None |
| A20 | Trebouxia (A20) | 1 | 1 | Lecanora pulicaris | Clade S, NONE | None |
| A21 | Apatococcus | 2 | 1 | Biatora sp. 1 | NA | None |
| A22 | Trebouxia (A22) | 2 | 2 | Buellia griseovirens, Ochrolechia androgyna | Clade S/NONE | None |
Out of 46 fungal OTUs, 11 belong to Parmeliaceae (nine genera), eight to Lecanoraceae (all to the genus Lecanora), four each to Pertusariaceae (all Pertusaria), Cladoniaceae (all Cladonia), Ramalineaceae (two each to the genera Ramalina and Biatora), and Graphidaceae (three Graphis and Thelotrema lepadinum), two each to Physciaceae (Physcia adscendens and Rinodina efflorescens) and Pyrenulaceae (both to the genus Pyrenula), and one each to Caliciaceae (Buellia griseovirens), Ochrolechiaceae (Ochrolechia bahusiensis), Ophioparmaceae (Hypocenomyce scalaris), Pilocarpaceae (Fellhanera gyrophorica), Phlyctidaceae (Phlyctis argena), Ropalosporaceae (Ropalospora viridis), and Sarrameanaceae (Loxospora elatina). As for the algal partners, out of 23 OTUs, 14 belonged to Trebouxia, five to Trentepohlia, and one each to Apatococcus, Asterochloris, Dictyochloropsis, and Parachloroidium (Table 3). Trebouxia jamesii was the most common photobiont associating with 16 mycobiont OTUs (out of 46) from six different families, followed by Asterochloris phycobiontica and Trebouxia suecica, each of which associated with seven mycobiont OTUs from three different families (Table 3).
The ITS phylogeny of the green algal genus Trebouxia generated from a combined dataset comprising representative OTUs from this study, Leavitt et al. [19] and Škaloud et al. [48] consisted of four supported clades sensu Leavitt et al. [19], namely Clade A (T. arboricola/T. gigantea clade), clade G (T. galapagensis/T. usneae), clade I (T. impressa/T. gelatinosa), and clade S (T. simplex/T. letharii/T. jamesii) Figure 1). From our study, six Trebouxia OTUs grouped within clade S, three within clade I, and five within clade A. None of the Trebouxia OTUs retrieved in our study clustered with Clade G (Figure 1).
3.2. Lichen Community Composition in the Three Regions
About 26% of the fungal and 30% of the algal OTUs respectively were shared among the three areas (Table 1). We found that 12 lichens were common to the three regions: Buellia griseovirens, Lecanora argentata, L. carpinea, Lecidella elaeochroma, Melanelixia glabratula, Parmelia sulcata, Pertusaria amara, P. coccodes, P. leioplaca, Phlyctis argena, Platismatia glauca, and Ramalina farinacea (Table 2). Lichens reported only from the protected area are Pyrenula nitida, and Thelotrema lepadinum. As for the photobionts, Trebouxia jamesii (OTU A6) was the most common photobiont in all the regions, overall associating with 53% of the sampled lichens (Table 3). Mycobiont species associating with Trentepohlia algae were more common in the protected area (Table 2).
3.3. Symbiont Interaction Patterns
Several lichenized fungi and green algae associated with more than one partner, indicating the presence of many generalized lichen associations in Białowieża Forest (Figure 2). The bipartite network of symbiotic interactions between fungi and algae was asymmetric in species richness and included 46 fungal and 23 algal OTUs; resulting in a mean of two algae interacting per mycobiont (Figure 2). Among the mycobiont–photobiont associations, seven were one-to-one associations, and the rest involved more than two symbionts (Figure 2). Out of the seven one-to-one associations, three were unique to the protected region, two to the protected and managed region, and two were found only in the disturbed region. About 20% of the mycobionts (nine fungal OTUs) and 43% of the photobionts (10 photobiont OTUs) associated with more than one partner (Table 2 and Table 3).
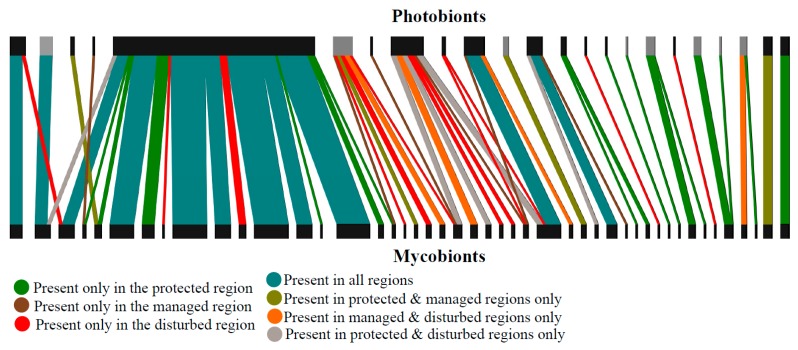
Figure 2.
Association network based on fungal and algal ITS sequences, given a 97% similarity BLASTn threshold. Black and grey horizontal bars on the top represent Trebouxia and Trentepohlia OTUs respectively. Colored vertical bars represent fungal algal associations.
Overall, we found 58 fungal–algal pairs, of which only 11 pairs were common to all regions (Figure 2, represented by blue colored vertical bars). Four fungal–algal pairs were present in the protected and managed regions and absent from the disturbed region (Figure 2, green colored vertical bars). The number of unique pairs was 16 in the protected, six in the managed, and 12 in the disturbed region.
4. Discussion
4.1. Genetic Diversity of Symbionts
Accurate recognition of fungal and algal lineages is important to understand the diversity patterns, specificity, and selectivity of symbionts [51]. In the case of the mycobionts, for most of the samples there were no conflicts between the morphological identification and the significant hits of the ITS sequence in GenBank database. For photobionts, only seven algal OTUs could be assigned to described species (Table 3). Fourteen photobiont OTUs had no significant similarity to ITS sequences in GenBank (Supplementary S1). Thus, our study highlights the presence a surprisingly large fraction of unknown diversity in the green algal genera Trebouxia and Trentepohlia, even in well studied ecosystems and geographic regions, such as forests in Central Europe.
Members of the genus Trebouxia constitute the most common lichenized algae associating with more than 20% of all lichenized fungi [52]. However, due to the paucity of taxonomically relevant morphological characters, only about 36 Trebouxia species have been formally described and deposited in the UTEX and SAG culture collections. Studies based on molecular data, however, indicate that the diversity of lichen-associated Trebouxia is high, with vastly undiscovered species-level diversity [19,30,53]. For instance, only 30% of the Trebouxia lineages which associated with 10 genera in Parmeliaceae could be allotted to described species [19]. Similarly, only 25% of the Trebouxia species which associated with the genus Protoparmelia (Parmeliaceae) could be assigned to previously described species [40]. Our study is in concordance with these studies suggesting the presence of vastly undiscovered species level diversity in Trebouxia.
4.2. Lichen Community Composition in the Three Regions
We found that 24% of the fungal and 30% of the algal OTUs were shared among all studied areas, suggesting that anthropogenic disturbance does not strongly affect these taxa (Table 2 and Table 3). Particularly those mycobionts associating with Trebouxia jamesii are not limited by the presence of a compatible photobiont in either disturbance regime, since this algal OTU was the most common photobiont in all studied areas. The protected area was characterized by the highest numbers of mycobiont and photobiont OTUs that were exclusively found in that area. One reason for this observation could be long-term ecological continuity and the presence of old trees in this habitat. Old trees provide variable microhabitat (e.g., bark with deep cracks; [7,54], stable substrate conditions over decades or centuries, i.e., longer periods of time available for colonization and growth [55], and stable microclimate, which can all lead to the development of a diverse lichen community. Some of the taxa we found in the protected area have been reported to preferentially occur on old trees (older than 180 years), such as Lecanora glabrata, Pyrenula nitida, and Thelotrema lepadinum and might be excellent indicators of long-term ecological continuity in forests [56]. Some of the taxa that are known to occur on younger trees (50–120 years old) we found in all three studied areas, i.e., Buellia griseovirens, Lecidella elaeochroma, Parmelia sulcata, and Pertusaria amara [56]. Overall, our study corroborates the importance of stand age for lichens [4,54,55,56].
Another reason for the higher number of unique fungal and algal OTUs in the protected region could be changes in the microclimatic conditions following anthropogenic disturbance. Epiphytic lichens are sensitive to microclimatic conditions, such as local humidity, and may change their vertical position on the trees [57,58]. As sampling was conducted only up to a maximum of 200 cm tree height in all sampling sites, taxa may have been missed that shifted higher up on the stem in response to anthropogenic activity.
The susceptibility of lichens to stand age-related properties renders the stand rotation length a crucial determinant of how severely forestry practices affect diversity loss. In fact, shorter rotation lengths have been associated with increased biodiversity loss of certain other organisms such as bryophytes [56,59]. Implementing the information on lichen substrate-age preference in decisions about rotation lengths, i.e., the duration between the plantation and final felling, may help mitigate biodiversity loss. Furthermore, apart from depleting the forest of old trees, many forestry practices lead to simplified forest structure with reduced diversity regarding trees species and age. Encouraging practices, which promote habitat diversity, for instance, retention practices, whereby living and dead trees are retained at final harvest, could be a way to minimize excessive habitat uniformity. Retention practices have also been shown to support higher biodiversity and a greater abundance of several species [60,61,62], including lichens [63,64,65]. Sustainable forestry practices encompassing various aspects of diversity and life-style traits of organisms could help avoiding future bottlenecks in habitat availability for lichens.
4.3. Symbiont Interaction Pattern
Our results indicate that generalist mycobionts and photobionts are present in all three areas of Białowieża forest. This could be attributed to the fact that generalist taxa sharing a common symbiont have a higher probability of finding a compatible partner and re-establishing after disturbance, provided the suitable microclimatic and substrate conditions are available. Identifying generalist taxa is simpler than identifying specialists, and often even low sampling might be sufficient to reveal low symbiont specificity. For instance, in our study we found that Lecidella elaeochroma, Pertusaria leioplaca, Lecanora thysanophora, Loxospora elatina, Phlyctis argena, Platismatia glauca, Ochrolechia bahusiensis, and Thelotrema lepadinum, associate with two or more than two photosynthetic partners (Table 2). The generalist interaction pattern of these mycobionts was revealed even if the number of samples per lichen was lower than 10. Furthermore, generalist taxa are more likely to be picked up under a random sampling scheme, because they are more common in the community. On the other hand, identifying highly specific taxa, i.e., one-to-one interactions, requires a larger sampling than that performed in the present study. A restricted sampling design may be misleading and may erroneously indicate a generalist as a specialist. For instance, although we found that Lecanora argentata (20 samples), L. glabrata (14 samples), Melanelixia glabratula (20 samples), and Pertusaria amara (19 samples) associate with only one photobiont species (Table 2), it is possible that they associate with a different photobiont species in more distant regions. The pattern observed in our study could be an artifact of photobiont-species availability. Samples from different geographic regions would be required to affirm if these lichens have high photobiont specificity and always associate with the same photobiont species as found in our study. Thus, identifying a highly specific mycobiont is more complex and is often obscured by sampling limitations. Interpretation on mycobiont specificity therefore must be ideally based on systematic, range wide sampling as the same lichen might be associated with a different photobiont species and vice-versa in different geographic regions.
Photobiont sharing is a common phenomenon in lichens [39,41,66,67]. Consequently, photobiont interaction patterns can also be inferred via by sequence-similarity based search to investigate if the same photobiont species was reported from other lichens. For instance, although Trebouxia flava (OTU A19) was recovered only once in our dataset (associated with Physcia adscendens; Table 3), sequence-similarity search revealed that Trebouxia flava associates with Physconia grisea, P. distorta, P. enteroxantha, and Tephromela atra as well. This suggests that, in spite of its low occurrence in the Białowieża Forest, Trebouxia flava is a generalist, associating with multiple mycobionts across its range. Similarly, based on our dataset, we suggest that Trebouxia OTU A13 displays high specificity (although not exclusive one-to-one pattern) as it associates with only two mycobionts, Cetrelia olivetorum and Menegazzia terebrata (Table 3) and has not been reported to be associated with any other lichen so far.
Overall, mycobionts display high partner selectivity, whereas photobionts display low partner selectivity. For instance, Trebouxia jamesii associates with 16 different fungal OTUs, corresponding to 53% of the sampled lichens (Table 3). Most of the fungal partners of T. jamesii, however, associate only with T. jamesii. Lack of reciprocal high selectivity is a common phenomenon in lichens [19,38,40]. Low selectivity enhances the chances of the fungal partner to find compatible algae. For instance, T. jamesii-associating lichen-forming fungi might find it easier to retrieve the compatible partner from the photobiont pool and consequently their colonization in the disturbed region may not be limited by photobiont availability. This is particularly important for lichens, which have a phase in their reproductive cycle, during which fungi and algae disperse separately, e.g., in sexual reproduction after fungal spore release [31,68,69]. Thus, the processes of relichenization (fungal and algal partners joining to form a new lichen thallus) and recolonization of novel or disturbed habitats, might be comparatively easier for mycobionts associating with generalist algae. This might explain the presence of T. jamesii-associated lichens under all three disturbance regimes.
Out of 58 fungal-algal pairs, only 11 pairs were common to all regions (Figure 2, represented by blue colored vertical bars) whereas four others were present in the protected and managed regions only and absent from the disturbed region (Figure 2, green colored vertical bars). Each region harbored certain number of unique pairs, not present in the other regions. These findings are similar to those reported in other studies suggesting that distinct communities are present in the forest regions with different management types [64,70]. This could be attributed to the altered stand structure, habitat and microclimatic conditions, and/or differences in the photobiont pool among communities. Monitoring studies targeting these lichens are essential to infer the status of these fungal–algal pairs in the managed/disturbed regions.
Our study gives a preliminary account of the community structure of lichen mycobionts and photobionts in a temperate forest ecosystem. It provides baseline data on diversities and interactions of lichen symbionts, which can be informative for designing future studies involving lichen communities. More than half of the photobiont diversity presented here was sequenced for the first time, and many fungal–algal interaction pairs were shown for the first time. Initial assumptions that forest management practices could act as filters for particular photobionts, or that the same mycobiont associates with different algae in differentially disturbed areas could not be confirmed.
4.4. Impact of Forest Disturbances on Organisms and Communities
The magnitude of the impact of anthropogenic activities varies from organism to organism. This might depend on many factors including the life-cycle traits of a species. In this regard, a recent meta-analysis surveying lichen restoration after anthropogenic activities suggested that it might take up to 150–180 years for epiphytic lichen communities to reach a diversity similar to that of old-growth forests [67,71]. This is much longer than the average restoration time for other organisms, such as amphibians (<30 years; [72]), reptiles (<20 years; [72]), ectomycorrhizal fungi (90 years), epiphytic plants (100 years), and some common forest animals (average 20–40 years for bats, birds, and invertebrates; [73]). In general, restoring lichen communities might take longer because of multiple factors; (a) symbiotic lifestyle, (b) high partner specificity, (c) substrate or host tree specificity, (d) forest age (old forests harbor different microhabitats, light, and moisture conditions than managed/disturbed regions), and (e) slow growth rate.
5. Conclusions
Our study provided a glimpse into the lichen community composition, shared taxa among disturbance regimes and symbiont interaction patterns present in the different disturbance regimes of the Białowieża Forest. Furthermore, it provides an account of the photobiont partners of several lichens for the first time. This is valuable information when interpreting the symbiont interaction patterns of lichens from other communities. We suggest that the colonization and reestablishment of generalist taxa may be simpler after disturbance of due to higher chances of obtaining the compatible partner from the photobiont pool of the community.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/2076-2607/7/9/335/s1.
Author Contributions
Conceptualization: I.S. and M.K.; methodology, I.S., M.K., J.O.; validation, I.S., M.K, A.Ł., F.D.G. and G.S.; formal analysis, G.S., F.D.G., J.O.; investigation, G.S., J.O.; resources, I.S., M.K., A.Ł.; data curation, M.K., A.Ł., G.S.; writing—original draft preparation, G.S., I.S.; writing—review and editing, G.S., I.S., F.D.G.; visualization, G.S.; supervision, I.S.; project administration, I.S.; funding acquisition, I.S.
Funding
This study was funded by Senckenberg BiK-F and a European Commission SYNTHESYS grant to M.K.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Aragón G., Martínez I., Izquierdo P., Belinchón R., Escudero A. Effects of forest management on epiphytic lichen diversity in Mediterranean forests. Appl. Veg. Sci. 2010;13:183–194. doi: 10.1111/j.1654-109X.2009.01060.x. [DOI] [Google Scholar]
- 2.Ardelean I.V., Keller C., Scheidegger C. Effects of management on lichen species richness, ecological traits and community structure in the Rodnei mountains national park (Romania) PLoS ONE. 2015;10:e0145808. doi: 10.1371/journal.pone.0145808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellis C.J., Coppins B.J. Contrasting functional traits maintain lichen epiphyte diversity in response to climate and autogenic succession. J. Biogeogr. 2006;33:1643–1656. doi: 10.1111/j.1365-2699.2006.01522.x. [DOI] [Google Scholar]
- 4.Boch S., Prati D., Hessenmöller D., Schulze E.-D., Fischer M. Richness of lichen species, especially of threatened ones, is promoted by management methods furthering stand continuity. PLoS ONE. 2013;8:e55461. doi: 10.1371/journal.pone.0055461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giordani P., Brunialti G., Bacaro G., Nascimbene J. Functional traits of epiphytic lichens as potential indicators of environmental conditions in forest ecosystems. Ecol. Indic. 2012;18:413–420. doi: 10.1016/j.ecolind.2011.12.006. [DOI] [Google Scholar]
- 6.Zedda L. The epiphytic lichens on Quercus in Sardinia (Italy) and their value as ecological indicators. Englera. 2002;24:5–416. doi: 10.2307/3776783. [DOI] [Google Scholar]
- 7.Jovan S. Lichen Bioindication of Biodiversity, Air Quality, and Climate: Baseline Results From Monitoring in Washington, Oregon, and California. For. Sci. 2008;115:737. [Google Scholar]
- 8.Aragón G., Rico V.J., Belinchón R. Lichen diversity from Cazorla, Segura and Las Villas Biosphere Reserve (SE Spain) Nov. Hedwigia. 2006;82:31–50. doi: 10.1127/0029-5035/2006/0082-0031. [DOI] [Google Scholar]
- 9.Nascimbene J., Marini L., Nimis P.L. Influence of forest management on epiphytic lichens in a temperate beech forest of northern Italy. For. Ecol. Manage. 2007;247:43–47. doi: 10.1016/j.foreco.2007.04.011. [DOI] [Google Scholar]
- 10.Łubek A., Kukwa M., Jaroszewicz B., Czortek P. Changes in the epiphytic lichen biota of Białowieża Primeval Forest are not explained by climate warming. Sci. Total Environ. 2018;643:468–478. doi: 10.1016/j.scitotenv.2018.06.222. [DOI] [PubMed] [Google Scholar]
- 11.Peksa O., Skaloud P. Do photobionts influence the ecology of lichens? A case study of environmental preferences in symbiotic green alga Asterochloris (Trebouxiophyceae) Mol. Ecol. 2011;20:3936–3948. doi: 10.1111/j.1365-294X.2011.05168.x. [DOI] [PubMed] [Google Scholar]
- 12.Yahr R., Vilgalys R., DePriest P.T. Geographic variation in algal partners of Cladonia subtenuis (Cladoniaceae) highlights the dynamic nature of a lichen symbiosis. New Phytol. 2006;171:847–860. doi: 10.1111/j.1469-8137.2006.01792.x. [DOI] [PubMed] [Google Scholar]
- 13.Vargas Castillo R., Beck A. Photobiont selectivity and specificity in Caloplaca species in a fog-induced community in the Atacama Desert, northern Chile. Fungal Biol. 2012;116:665–676. doi: 10.1016/j.funbio.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Dal Grande F., Rolshausen G., Divakar P.K., Crespo A., Otte J., Schleuning M., Schmitt I. Environment and host identity structure communities of green algal symbionts in lichens. New Phytol. 2018;217:277–289. doi: 10.1111/nph.14770. [DOI] [PubMed] [Google Scholar]
- 15.Rolshausen G., Dal Grande F., Sadowska-Deś A.D., Otte J., Schmitt I. Quantifying the climatic niche of symbiont partners in a lichen symbiosis indicates mutualist-mediated niche expansions. Ecography (Cop.). 2018;41:1380–1392. doi: 10.1111/ecog.03457. [DOI] [Google Scholar]
- 16.Fernández-Mendoza F., Domaschke S., García M.A., Jordan P., Martín M.P., Printzen C. Population structure of mycobionts and photobionts of the widespread lichen Cetraria aculeata. Mol. Ecol. 2011;20:1208–1232. doi: 10.1111/j.1365-294X.2010.04993.x. [DOI] [PubMed] [Google Scholar]
- 17.Kalwij J.M., Wagner H.H., Scheidegger C. Effects of stand-level disturbances on the spatial distribution of a lichen indicator. Ecol. Appl. 2005;15:2015–2024. doi: 10.1890/04-1912. [DOI] [Google Scholar]
- 18.Singh G., Dal Grande F., Werth S., Scheidegger C. Long-term consequences of disturbances on reproductive strategies of the rare epiphytic lichen Lobaria pulmonaria: Clonality a gift and a curse. FEMS Microbiol. Ecol. 2015;91:1–11. doi: 10.1093/femsec/fiu009. [DOI] [PubMed] [Google Scholar]
- 19.Leavitt S.D., Kraichak E., Nelsen M.P., Altermann S., Divakar P.K., Alors D., Esslinger T.L., Crespo A., Lumbsch T. Fungal specificity and selectivity for algae play a major role in determining lichen partnerships across diverse ecogeographic regions in the lichen-forming family Parmeliaceae (Ascomycota) Mol. Ecol. 2015;24:3779–3797. doi: 10.1111/mec.13271. [DOI] [PubMed] [Google Scholar]
- 20.Lumbsch H.T., Leavitt S.D. Goodbye morphology? A paradigm shift in the delimitation of species in lichenized fungi. Fungal Divers. 2011;50:59–72. doi: 10.1007/s13225-011-0123-z. [DOI] [Google Scholar]
- 21.Taylor J.W., Jacobson D.J., Kroken S., Kasuga T., Geiser D.M., Hibbett D.S., Fisher M.C. Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 2000;31:21–32. doi: 10.1006/fgbi.2000.1228. [DOI] [PubMed] [Google Scholar]
- 22.Grube M., Kroken S. Molecular approaches and the concept of species and species complexes in lichenized fungi. Mycol. Res. 2000;104:1284–1294. doi: 10.1017/S0953756200003476. [DOI] [Google Scholar]
- 23.Škaloud P., Friedl T., Hallmann C., Beck A., Dal Grande F. Taxonomic revision and species delimitation of coccoid green algae currently assigned to the genus Dictyochloropsis (Trebouxiophyceae, Chlorophyta) J. Phycol. 2016;52:599–617. doi: 10.1111/jpy.12422. [DOI] [PubMed] [Google Scholar]
- 24.Sadowska-Deś A.D., Dal Grande F., Lumbsch H.T., Beck A., Otte J., Hur J.-S., Kim J.A., Schmitt I. Integrating coalescent and phylogenetic approaches to delimit species in the lichen photobiont Trebouxia. Mol. Phylogenet. Evol. 2014;76:202–210. doi: 10.1016/j.ympev.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 25.Kelly L.J., Hollingsworth P.M., Coppins B.J., Ellis C.J., Harrold P., Tosh J., Yahr R. DNA barcoding of lichenized fungi demonstrates high identification success in a floristic context. New Phytol. 2011;191:288–300. doi: 10.1111/j.1469-8137.2011.03677.x. [DOI] [PubMed] [Google Scholar]
- 26.Schoch C.L., Seifert K.A., Huhndorf S., Robert V., Spouge J.L., Levesque C.A., Chen W. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardes M., Bruns T.D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 28.Leliaert F., Verbruggen H., Vanormelingen P., Steen F., López-Bautista J.M., Zuccarello G.C., De Clerck O. DNA-based species delimitation in algae. Eur. J. Phycol. 2014;49:179–196. doi: 10.1080/09670262.2014.904524. [DOI] [Google Scholar]
- 29.Piercey-Normore M.D. The lichen-forming ascomycete Evernia mesomorpha associates with multiple genotypes of Trebouxia jamesii. New Phytol. 2006;169:331–344. doi: 10.1111/j.1469-8137.2005.01576.x. [DOI] [PubMed] [Google Scholar]
- 30.Kroken S., Taylor J.W. Phylogenetic species, reproductive mode, and specificity of the green alga Trebouxia forming lichens with the fungal genus Letharia. Bryologist. 2000;103:645–660. doi: 10.1639/0007-2745(2000)103[0645:PSRMAS]2.0.CO;2. [DOI] [Google Scholar]
- 31.Romeike J., Friedl T., Helms G., Ott S. Genetic diversity of algal and fungal partners in four species of Umbilicaria (Lichenized Ascomycetes) along a transect of the Antarctic peninsula. Mol. Biol. Evol. 2002;19:1209–1217. doi: 10.1093/oxfordjournals.molbev.a004181. [DOI] [PubMed] [Google Scholar]
- 32.Skaloud P., Peksa O. Evolutionary inferences based on ITS rDNA and actin sequences reveal extensive diversity of the common lichen alga Asterochloris (Trebouxiophyceae, Chlorophyta) Mol. Phylogenet. Evol. 2010;54:36–46. doi: 10.1016/j.ympev.2009.09.035. [DOI] [PubMed] [Google Scholar]
- 33.Muggia L., Baloch E., Stabentheiner E., Grube M., Wedin M. Photobiont association and genetic diversity of the optionally lichenized fungus Schizoxylon albescens. FEMS Microbiol. Ecol. 2011;75:255–272. doi: 10.1111/j.1574-6941.2010.01002.x. [DOI] [PubMed] [Google Scholar]
- 34.Rikkinen J. Molecular studies on cyanobacterial diversity in lichen symbioses. MycoKeys. 2013;6:3–32. doi: 10.3897/mycokeys.6.3869. [DOI] [Google Scholar]
- 35.Leavitt S.D., Esslinger T.L., Spribille T., Divakar P.K., Thorsten Lumbsch H. Multilocus phylogeny of the lichen-forming fungal genus Melanohalea (Parmeliaceae, Ascomycota): Insights on diversity, distributions, and a comparison of species tree and concatenated topologies. Mol. Phylogenet. Evol. 2013;66:138–152. doi: 10.1016/j.ympev.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 36.Galun M., Bubrick P. Physiological interactions between the partners of the lichen symbiosis. In: Linskens H.F., Heslop-Harrison J., editors. Cellular Interactions. Springer; Berlin/Heidelberg, Germany: 1984. pp. 362–401. [Google Scholar]
- 37.Beck A., Friedl T., Rambold G. Selectivity of photobiont choice in a defined lichen community: Inferences from cultural and molecular studies. New Phytol. 1998;139:709–720. doi: 10.1046/j.1469-8137.1998.00231.x. [DOI] [Google Scholar]
- 38.Yahr R., Vilgalys R., Depriest P. Strong fungal specifity and selectivity for algal symbionts in Florida scrub Cladonia lichens. Mol. Ecol. 2004;13:3367–3378. doi: 10.1111/j.1365-294X.2004.02350.x. [DOI] [PubMed] [Google Scholar]
- 39.Dal Grande F., Beck A., Cornejo C., Singh G., Cheenacharoen S., Nelsen M.P., Scheidegger C. Molecular phylogeny and symbiotic selectivity of the green algal genus Dictyochloropsis s.l. (Trebouxiophyceae): A polyphyletic and widespread group forming photobiont-mediated guilds in the lichen family Lobariaceae. New Phytol. 2014;202:455–5470. doi: 10.1111/nph.12678. [DOI] [PubMed] [Google Scholar]
- 40.Singh G., Dal Grande F., Divakar P.K., Otte J., Crespo A., Schmitt I. Fungal–algal association patterns in lichen symbiosis linked to macroclimate. New Phytol. 2017;214:317–329. doi: 10.1111/nph.14366. [DOI] [PubMed] [Google Scholar]
- 41.Jüriado I., Kaasalainen U., Jylhä M., Rikkinen J. Relationships between mycobiont identity, photobiont specificity and ecological preferences in the lichen genus Peltigera (Ascomycota) in Estonia (northeastern Europe) Fungal Ecol. 2019;39:45–54. doi: 10.1016/j.funeco.2018.11.005. [DOI] [Google Scholar]
- 42.Sabatini F.M., Burrascano S., Keeton W.S., Levers C., Lindner M., Pötzschner F., Verkerk P.J., Bauhus J., Buchwald E., Chaskovsky O., et al. Where are Europe’s last primary forests? Divers. Distrib. 2018;24:1426–1439. doi: 10.1111/ddi.12778. [DOI] [Google Scholar]
- 43.Cubero O.F., Crespo A. Isolation of nucleic acids from lichens. In: Kranner I., Beckett R., Varma A., editors. Protocols in Lichenology. Springer; Berlin, Germany: 2002. pp. 381–391. [Google Scholar]
- 44.White T.J., Bruns T.D., Lee S.B., Taylor J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M., Gelfand H., Sninsky J., White T., editors. PCR Protocols: A Guide to Methods and Applications. Academic Press; Cambridge, MA, USA: 1990. pp. 315–322. [Google Scholar]
- 45.Hametner C., Stocker-Wörgötter E., Grube M. New insights into diversity and selectivity of trentepohlialean lichen photobionts from the extratropics. Symbiosis. 2014;63:31–40. doi: 10.1007/s13199-014-0285-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Drummond A.J., Ashton B., Buxton S., Cheung M., Cooper A., Duran C., Field M., Heled J., Kearse M., Markowitz S., et al. Geneious. [(accessed on 11 April 2019)];2011 Available online: https://www.geneious.com/
- 47.Rognes T., Flouri T., Nichols B., Quince C., Mahé F. VSEARCH: A versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.ŠKALOUD P., MOYA P., MOLINS A., PEKSA O., SANTOS-GUERRA A., BARRENO E. Untangling the hidden intrathalline microalgal diversity in Parmotrema pseudotinctorum: Trebouxia crespoana sp. nov. Lichenologist. 2018;50:357–369. doi: 10.1017/S0024282918000208. [DOI] [Google Scholar]
- 49.Paradis E., Claude J., Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 50.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2017. [Google Scholar]
- 51.de Vienne D.M., Giraud T., Shykoff J.A. When can host shifts produce congruent host and parasite phylogenies? A simulation approach. J. Evol. Biol. 2007;20:1428–1438. doi: 10.1111/j.1420-9101.2007.01340.x. [DOI] [PubMed] [Google Scholar]
- 52.Rambold G., Triebel D. The Inter-lecanoralean Associations. Bibl. Lichenol. 1992;48:58027–58028. [Google Scholar]
- 53.Nyati S., Scherrer S., Werth S., Honegger R. Green-algal photobiont diversity (Trebouxia spp.) in representatives of Teloschistaceae (Lecanoromycetes, lichen-forming ascomycetes) Lichenol. 2014;46:189–212. doi: 10.1017/S0024282913000819. [DOI] [Google Scholar]
- 54.Hedenås H., Ericson L. Epiphytic macrolichens as conservation indicators: Successional sequence in Populus tremula stands. Biol. Conserv. 2000;93:43–53. doi: 10.1016/S0006-3207(99)00113-5. [DOI] [Google Scholar]
- 55.Ranius T., Johansson P., Berg N., Niklasson M. The influence of tree age and microhabitat quality on the occurrence of crustose lichens associated with old oaks. J. Veg. Sci. 2008;19:653–662. doi: 10.3170/2008-8-18433. [DOI] [Google Scholar]
- 56.Fritz Ö., Niklasson M., Churski M. Tree age is a key factor for the conservation of epiphytic lichens and bryophytes in beech forests. Appl. Veg. Sci. 2009;12:93–106. doi: 10.1111/j.1654-109X.2009.01007.x. [DOI] [Google Scholar]
- 57.McCune B. Gradients in epiphyte biomass in three Pseudotsuga-Tsuga forests of different ages in Western Oregon and Washington. Bryologist. 1993;96:405. doi: 10.2307/3243870. [DOI] [Google Scholar]
- 58.Rambo T.B., Harris R.H., Larson B.W., Majors S., Peterson E.T., Widmer M., Shaw D.C., Rosso A., Proctor J., Camacho F.J., et al. Vertical Profile of Epiphytes in a Pacific Northwest Old-growth Forest. [(accessed on 24 June 2019)];1997 Available online: https://www.semanticscholar.org/paper/Vertical-Profile-of-Epiphytes-in-a-Pacific-Forest-Rambo-Harris/a29a6936685890d2fdb33ad87f92ad63a8598120.
- 59.Felton A., Sonesson J., Nilsson U., Lämås T., Lundmark T., Nordin A., Ranius T., Roberge J.-M. Varying rotation lengths in northern production forests: Implications for habitats provided by retention and production trees. Ambio. 2017;46:324–334. doi: 10.1007/s13280-017-0909-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fedrowitz K., Koricheva J., Baker S.C., Lindenmayer D.B., Palik B., Rosenvald R., Beese W., Franklin J.F., Kouki J., Macdonald E., et al. REVIEW: Can retention forestry help conserve biodiversity? A meta-analysis. J. Appl. Ecol. 2014;51:1669–1679. doi: 10.1111/1365-2664.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gustafsson L., Baker S.C., Bauhus J., Beese W.J., Brodie A., Kouki J., Lindenmayer D.B., Lõhmus A., Pastur G.M., Messier C., et al. Retention Forestry to Maintain Multifunctional Forests: A World Perspective. Bioscience. 2012;62:633–645. doi: 10.1525/bio.2012.62.7.6. [DOI] [Google Scholar]
- 62.Gustafsson L., Kouki J., Sverdrup-Thygeson A. Tree retention as a conservation measure in clear-cut forests of northern Europe: A review of ecological consequences. Scand. J. For. Res. 2010;25:295–308. doi: 10.1080/02827581.2010.497495. [DOI] [Google Scholar]
- 63.Lõhmus A., Lõhmus P. Epiphyte communities on the trunks of retention trees stabilise in 5 years after timber harvesting, but remain threatened due to tree loss. Biol. Conserv. 2010;143:891–898. doi: 10.1016/j.biocon.2009.12.036. [DOI] [Google Scholar]
- 64.Lundström J., Jonsson F., Perhans K., Gustafsson L. Lichen species richness on retained aspens increases with time since clear-cutting. For. Ecol. Manage. 2013;293:49–56. doi: 10.1016/j.foreco.2012.12.027. [DOI] [Google Scholar]
- 65.Hedenås H., Hedström P. Conservation of epiphytic lichens: Significance of remnant aspen (Populus tremula) trees in clear-cuts. Biol. Conserv. 2007;135:388–395. doi: 10.1016/j.biocon.2006.10.011. [DOI] [Google Scholar]
- 66.Rikkinen J., Oksanen I., Lohtander K. Lichen guilds share related cyanobacterial symbionts. Science. 2002;297:357. doi: 10.1126/science.1072961. [DOI] [PubMed] [Google Scholar]
- 67.Gjerde I., Blom H.H., Lindblom L., Saetersdal M., Schei F.H. Community assembly in epiphytic lichens in early stages of colonization. Ecology. 2012;93:749–759. doi: 10.1890/11-1018.1. [DOI] [PubMed] [Google Scholar]
- 68.Sanders W.B., Lucking R. Reproductive strategies, relichenization and thallus development observed in situ in leaf-dwelling lichen communities. New Phytol. 2002;155:425–435. doi: 10.1046/j.1469-8137.2002.00472.x. [DOI] [PubMed] [Google Scholar]
- 69.Insarova I.D., Blagoveshchenskaya E.Y. Lichen symbiosis: Search and recognition of partners. Biol. Bull. 2016;43:408–418. doi: 10.1134/S1062359016040038. [DOI] [PubMed] [Google Scholar]
- 70.Goldmann K., Schöning I., Buscot F., Wubet T. Forest management type influences diversity and community composition of soil fungi across temperate forest ecosystems. Front. Microbiol. 2015;6:1300. doi: 10.3389/fmicb.2015.01300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Spake R., Ezard T.H.G., Martin P.A., Newton A.C., Doncaster C.P. A meta-analysis of functional group responses to forest recovery outside of the tropics. Conserv. Biol. 2015;29:1695–1703. doi: 10.1111/cobi.12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thompson M.E., Donnelly M.A. Effects of secondary forest succession on amphibians and reptiles: A review and meta-analysis. Copeia. 2018;106:10–19. doi: 10.1643/CH-17-654. [DOI] [Google Scholar]
- 73.Dunn R.R. Recovery of faunal communities during tropical forest regeneration. Conserv. Biol. 2004;18:302–309. doi: 10.1111/j.1523-1739.2004.00151.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.