An anti-idiotype recognizing the germline version of a broadly neutralizing antibody was utilized to identify human B cells expressing similar germline antibodies and was converted to an immunogen to stimulate transgenic murine B cells in vivo despite the presence of anergy.
Abstract
Many tested vaccines fail to provide protection against disease despite the induction of antibodies that bind the pathogen of interest. In light of this, there is much interest in rationally designed subunit vaccines that direct the antibody response to protective epitopes. Here, we produced a panel of anti-idiotype antibodies able to specifically recognize the inferred germline version of the human immunodeficiency virus 1 (HIV-1) broadly neutralizing antibody b12 (iglb12). We determined the crystal structure of two anti-idiotypes in complex with iglb12 and used these anti-idiotypes to identify rare naive human B cells expressing B cell receptors with similarity to iglb12. Immunization with a multimerized version of this anti-idiotype induced the proliferation of transgenic murine B cells expressing the iglb12 heavy chain in vivo, despite the presence of deletion and anergy within this population. Together, our data indicate that anti-idiotypes are a valuable tool for the study and induction of potentially protective antibodies.
Introduction
There are many pathogens, such as HIV-1, respiratory syncytial virus, and influenza virus, for which the development of a protective vaccine has been elusive despite decades of effort. In many cases, the failure does not appear to be an absolute inability of the immune system to produce protective antibodies, since they have been characterized in some infected individuals (Beeler and van Wyke Coelingh, 1989; Burton et al., 1991; Okuno et al., 1993; Johnson et al., 1997; Karron et al., 1997; Scanlan et al., 2002; Kashyap et al., 2008; Throsby et al., 2008; Ekiert et al., 2009, 2011; Scheid et al., 2009, 2011; Sui et al., 2009; Walker et al., 2009, 2011; Wu et al., 2010; Corti et al., 2011; Dreyfus et al., 2012, 2013; Huang et al., 2012, 2014; Magro et al., 2012; Mouquet et al., 2012; Liao et al., 2013; Ngwuta et al., 2015). It is not entirely clear why traditional vaccine development approaches have failed to induce similar protective antibodies, but mounting evidence suggests that protective vaccines for these pathogens will likely need to stimulate specific lineages of antibodies targeting defined epitopes (Pantaleo and Koup, 2004; Rappuoli et al., 2016; Lang et al., 2017; Robbiani et al., 2017; Goodwin et al., 2018; Kwong and Mascola, 2018). In contrast, traditional vaccines generally stimulate polyclonal antibody responses against whole pathogens, or large subunits from pathogens, rather than focus the immune response toward known protective epitopes.
The observation that a subset of HIV-1–infected individuals develop broadly neutralizing antibodies (bNAbs) targeting the HIV-1 envelope protein (Env) that are capable of neutralizing diverse HIV-1 viral isolates over the course of infection demonstrates that the human immune system is capable of generating bNAb responses provided it receives the appropriate antigenic stimulation. Indeed, numerous bNAbs have been isolated from infected individuals that define the sites of vulnerability and mechanisms of neutralization of HIV-1 (West et al., 2014; Burton and Hangartner, 2016; Kwong and Mascola, 2018). Importantly, bNAbs have been shown to protect from experimental infection in animal models, suggesting that they will be an important component of an effective vaccine against HIV-1 (Mascola et al., 1999; Shibata et al., 1999; Parren et al., 2001; Hessell et al., 2009; Moldt et al., 2012; Pietzsch et al., 2012; Gruell et al., 2013; Balazs et al., 2014; Shingai et al., 2014; Gautam et al., 2016; Liu et al., 2016).
Unfortunately, bNAbs have not been elicited by vaccination in humans, despite the use of diverse recombinant Env-derived immunogens and immunization schemes (Burton et al., 2004; Cohen and Dolin, 2013; Schiffner et al., 2013). Recent advances in the amplification of antibody genes from individual B cells have revealed that bNAbs often require extensive somatic mutation to achieve broad and potent neutralizing activity (Scheid et al., 2009; Dimitrov, 2010; Klein et al., 2013a; Kwong et al., 2013; Mascola and Haynes, 2013; West et al., 2014). Although mutated bNAbs are capable of broad Env recognition and virus neutralization, most inferred germline bNAb variants do not display reactivity with any Env variant tested, which led us and others to propose that one reason for the lack of bNAb elicitation through immunization is the lack of stimulation of B cells expressing bNAb progenitor B cell receptors (BCRs) by Env-based immunogens (Xiao et al., 2009; Hoot et al., 2013; Jardine et al., 2013; McGuire et al., 2013; Andrabi et al., 2015; Zhou et al., 2015; Gorman et al., 2016).
Env-based immunogens capable of binding to certain germline bNAbs have been recently developed that effectively activate B cells in knock-in mice expressing the desired BCRs in vivo (Dosenovic et al., 2015; Escolano et al., 2016; Jardine et al., 2016; McGuire et al., 2016; Steichen et al., 2016; Abbott et al., 2018). For many bNAbs, however, antigens capable of being bound by the corresponding germline forms have not yet been identified or designed. Here, we demonstrate the use of anti-idiotypes as an alternative approach to structure-based immunogen design to target the inferred germline version of the HIV-1 bNAb b12. We focused upon b12 because despite being one of the first HIV-1 bNAbs isolated, Env-based immunogens have not been identified or developed to recognize its ancestral germline form (Xiao et al., 2009; Hoot et al., 2013). The mature form of b12 was isolated through phage display (Barbas et al., 1992), and it used a heavy chain derived from VH1-3*02/DH2-21/JH6*03 and light chain derived from VK3-20*01/JK2 (Xiao et al., 2009; Hoot et al., 2013). Using anti-idiotypes specific for the inferred germline version of b12 (iglb12) as baits for single B cell sorting, we identified a subset of human germline BCRs using VH1-3 with some heavy chain CDRH3 similarity to iglb12. While crystal structures indicated that one of these anti-iglb12 idiotypes made extensive contacts with the iglb12 CDRH3 region encoded by DH2-21, this gene segment was not enriched in sorted B cells. Moreover, we could not identify any CDRH3 regions with >50% amino acid sequence identity to iglb12 within a dataset including hundreds of BCRs. In light of this, we hypothesized that B cells expressing BCRs containing CDRH3s with high similarity to iglb12 may be deleted from the repertoire as a result of autoreactivity with self-antigens. In line with this notion, we demonstrate that the iglb12 antibody stains HEp-2 cells, a feature of other autoreactive HIV-1–specific antibodies (Haynes et al., 2005; Verkoczy et al., 2010; Ota et al., 2013). To evaluate the consequence of this autoreactivity in vivo, we generated mice with the iglb12 heavy chain knocked into the endogenous murine heavy chain locus. The iglb12 heavy chain knock-in mice exhibited B cell deletion during development, with the surviving B cells exhibiting BCR down-regulation and anergy. Despite the presence of anergy, immunization with a multimerized version of an anti-iglb12 idiotype stimulated the proliferation and differentiation of transgenic B cells expressing the iglb12 heavy chain. In summary, our results establish a proof of concept that anti-idiotypes can be used as immunogens to identify and stimulate the ancestral germline version of protective antibodies.
Results
Generation and characterization of anti-iglb12 idiotypes
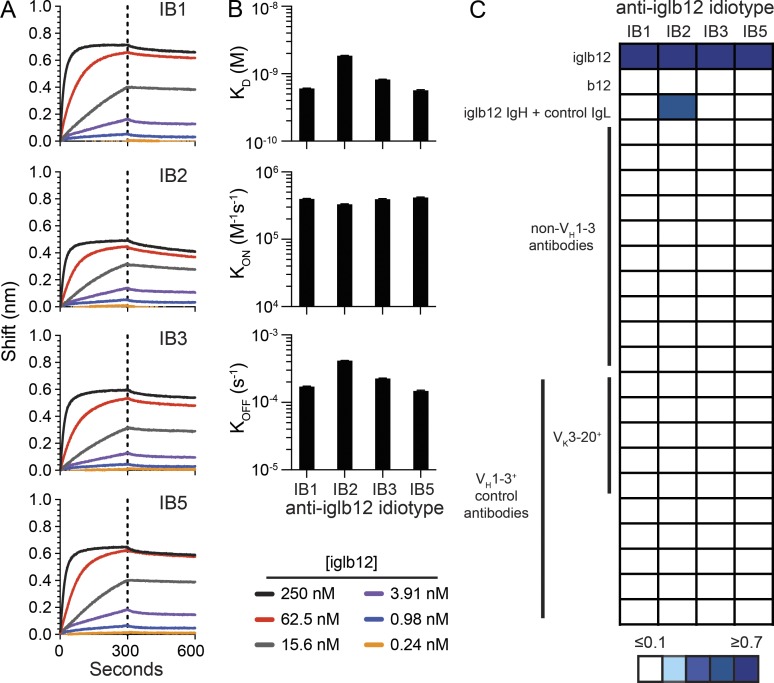
To generate anti-idiotype antibodies to iglb12 (Hoot et al., 2013), we immunized and boosted mice intraperitoneally with iglb12 in adjuvant. 3 d after the final immunization, splenocytes were isolated and fused to myeloma cells, resulting in the generation of hundreds of hybridomas producing antibodies able to bind iglb12. Nearly all of the clones were excluded because they bound to a panel of control germline antibodies as well as iglb12, suggesting specificity for the heavy or light chain constant regions of human Ig (data not shown). Importantly, four clones (IB1, IB2, IB3, and IB5) exhibited binding to iglb12 with similar apparent affinities (dissociation constants [KD] of ∼10−9) but did not bind a panel of control antibodies derived from germline BCR sequences of anti–HIV-1 antibodies (Fig. 1, A–C; and Table S1). The anti-iglb12 idiotypes also failed to bind the fully mature version of b12, indicating a preference for the unmutated sequence (Fig. 1 C). One clone, IB2, bound a chimera expressing the iglb12 heavy chain and a noncognate control light chain, while the other three clones, IB1, IB3, and IB5, did not bind this chimera (Fig. 1 C), suggesting different modes of recognition between IB2 and IB1, IB3, and IB5. We next assessed whether binding of these clones was specific for the iglb12 heavy chain CDRH3 region as opposed to a dependence on VH1-3, the variable gene segment used by iglb12. Consistent with our clones specifically recognizing features unique to iglb12, all four clones failed to bind naturally paired control antibodies using VH1-3 (DeKosky et al., 2016) whether or not they were paired with a Vk3-20 light chain, from which the b12 light chain is derived (Fig. 1 C). Together, these data suggest that these four clones preferentially recognized the iglb12 idiotype.
Figure 1.
Identification and characterization of anti-iglb12 idiotypes. (A) BLI analysis of the purified products of four hybridomas named IB1, IB2, IB3, and IB5 binding to different concentrations of iglb12. (B) The affinity (KD) of the anti-idiotype for iglb12 was calculated based upon the on rate (KON) and off rate (KOFF) determined by BLI. (C) The heat map displays the maximum shift (nm) measured by BLI when IB1, IB2, IB3, or IB5 were incubated with the listed antibodies and antibody groups. Detailed information on sequences of the non–VH1-3 and VH1-3+ control antibodies can be found in Table S1. Data are representative of two to three similar experiments.
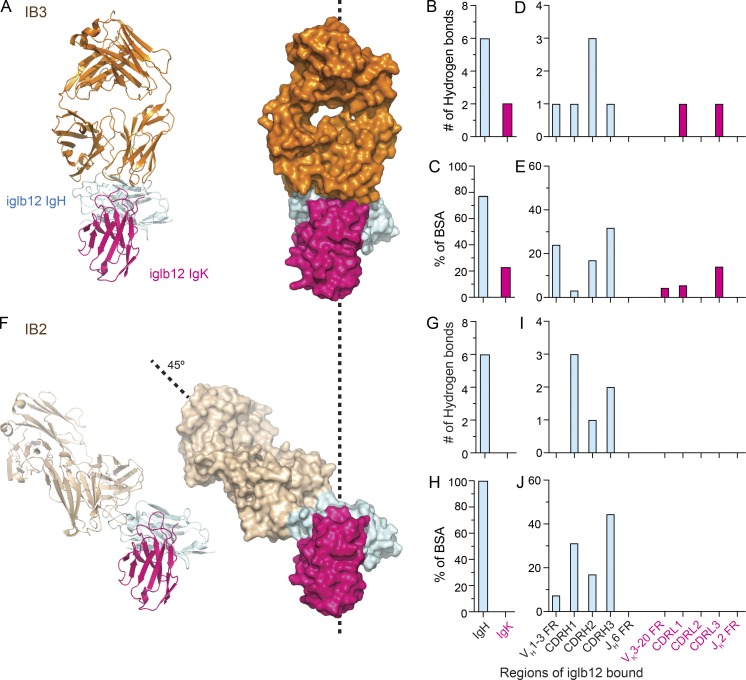
Among the four anti-iglb12 idiotypes identified by our screen, we focused upon IB2 and IB3 because we solved their crystal structures in complex with iglb12 at a resolution of 2.6 Å and 3.0 Å, respectively. The crystal structures revealed that IB2 and IB3 displayed distinct modes of interaction with iglb12 (Fig. 2). IB3 interacted with both the heavy and light chains of iglb12 (Fig. 2, A–C), consistent with biolayer interferometry (BLI) data (Fig. 1 C). Most interactions occurred within the heavy chain of iglb12 (Fig. 2, B and C), which contributed 76% of the buried surface area (BSA) and six of the eight hydrogen bonds. One hydrogen bond occurred in each of the CDRH3 and CDRL3 regions of iglb12, with the remaining six hydrogen bonds and BSA occurring in the CDR1, CDR2, and framework regions of iglb12, encoded by VH1-3 or VK3-20 (Fig. 2, D and E). In contrast, IB2 approached iglb12 at a 45° angle relative to IB3 and bound exclusively to the iglb12 heavy chain (Fig. 2, A and F). The CDRH3 region of iglb12 contributed nearly half of the BSA and two of the six hydrogen bonds, with the remaining BSA and hydrogen bonds largely focused upon CDRH1 and CDRH2, which are encoded by VH1-3 (Fig. 2, G–J). The extensive interactions with the iglb12 heavy chain CDRH3 likely explain why IB2 and IB3 do not bind other antibodies using VH1-3 (Fig. 1 C).
Figure 2.
Crystal structure analysis of anti-idiotypes bound to iglb12. (A and F) Ribbon diagrams and surface representation of Fab fragments of anti-iglb12 idiotypes IB3 bound to a Fab fragment of iglb12 (A) and IB2 bound to an iglb12 c/scFv (F) at a resolution of 3.0 Å and 2.6 Å, respectively. (B–E and G–J) Quantitation of the number of hydrogen bonds and the percentage of total BSA within the listed regions of iglb12 bound by IB3 (B–E) and IB2 (G–J), as determined from the crystal structures.
Identification of iglb12-like BCRs using anti-iglb12 idiotypes
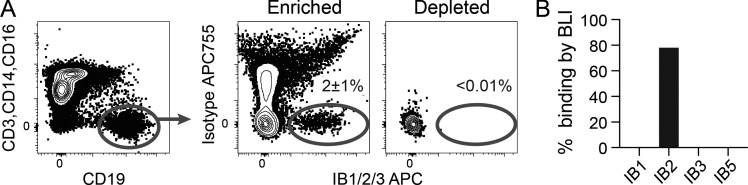
We next assessed whether the anti-iglb12 idiotypes could be used to identify B cells expressing iglb12-like BCRs from within the polyclonal human B cell repertoire of HIV-uninfected individuals using single B cell sorting followed by RT-PCR amplification and sequencing of heavy and light chain genes. In an initial set of experiments, IB1, IB2, and IB3 were biotinylated and tetramerized using streptavidin conjugated to allophycocyanin (APC) and incubated together with 100 million human peripheral blood mononuclear cells (PBMCs) before enrichment using anti-APC microbeads. Using this approach ∼2% of B cells in the APC-enriched fraction bound to IB1/2/3-APC, but not APC-Dylight755 (APC755)–conjugated isotype control tetramers (Fig. 3 A). Highlighting the efficiency of this enrichment approach, few IB1/2/3-APC+ B cells could be detected in the APC-depleted fraction (Fig. 3 A). Using this approach, single B cells binding IB1/2/3 tetramers were sorted into individual wells of a 96-well plate, and the heavy and light chains expressed by these cells were sequenced using nested RT-PCR (Tiller et al., 2008). In total, we FACS-purified 91 single IB1/2/3-binding B cells from an individual. This yielded 66 heavy chain and 38 light chain sequences, with 31 heavy and light chain pairs (Table S2). To confirm the specificity of our flow cytometry approach, antibodies were produced from paired heavy and light chains from 10 cells. 80% of these antibodies bound IB2, but not IB1, IB3, or IB5, when assessed by BLI (Fig. 3 B). Interestingly, the eight antibodies that bound IB2 all used the same heavy chain variable region as igbl12, VH1-3, while the two antibodies that failed to bind any of the anti-idiotypes used other VH alleles (Table S2). In two of the cloned antibodies, the VH1-3+ heavy chains were paired with VK3-20+ light chains, suggesting that the lack of binding to IB1, IB3, and IB5 was not due to the absence of this segment (Fig. 1 C). These results highlight the high specificity of our flow cytometry approach and suggest that IB2 is bound by a higher number of cells compared with other anti-idiotypes.
Figure 3.
Identification of human B cells able to bind anti-iglb12 idiotypes. (A) Detection of live CD19+ CD3− CD14− CD16− B cells from human PBMCs that bound a cocktail containing IB1-APC, IB2-APC, and IB3-APC tetramers in fractions enriched or depleted of APC+ cells using anti-APC microbeads before flow cytometry. APC755 tetramers containing isotype control antibodies were included in these experiments to exclude B cells specific for the APC, streptavidin, and conserved portions of the anti-idiotypes. The displayed plots were derived from ∼100,000 APC-depleted or ∼400,000 APC-enriched cells derived from 100 million PBMCs and representative of three similar experiments. The mean percentage ± SD of IB1/2/3-APC+ APC755− B cells in the enriched and depleted fractions from three individuals is shown on the plots. (B) The frequency of ten antibodies cloned from IB1/IB2/IB3-APC+ APC755− B cells from an individual were assayed for binding to IB1, IB2, IB3, or IB5 by BLI. Each antibody was assessed for binding in two to three independent experiments.
To gain a more in-depth understanding of the BCRs recognized by individual anti-idiotypic antibodies, we next conducted sorts with IB2 and IB3 labeled with different fluorophores. We focused our analysis on these anti-idiotypes because the high-resolution crystal structures of IB2 and IB3 complexed with iglb12 allowed us to make predictions about the BCRs they may preferentially recognize. From the crystal structures, we predicted that IB2 would be highly selective for B cells expressing BCRs using VH1-3 with CDRH3 regions similar to iglb12. In contrast, we predicted that IB3 would be selective for B cells expressing BCRs using VH1-3 with more diverse CDRH3s paired with Vk3-20 light chains with some CDRL3 similarity to iglb12. Due to the random nature of V(D)J recombination that gives rise to CDR3 regions, we did not expect to find B cells expressing CDRH3s or CDRL3s identical to those of iglb12.
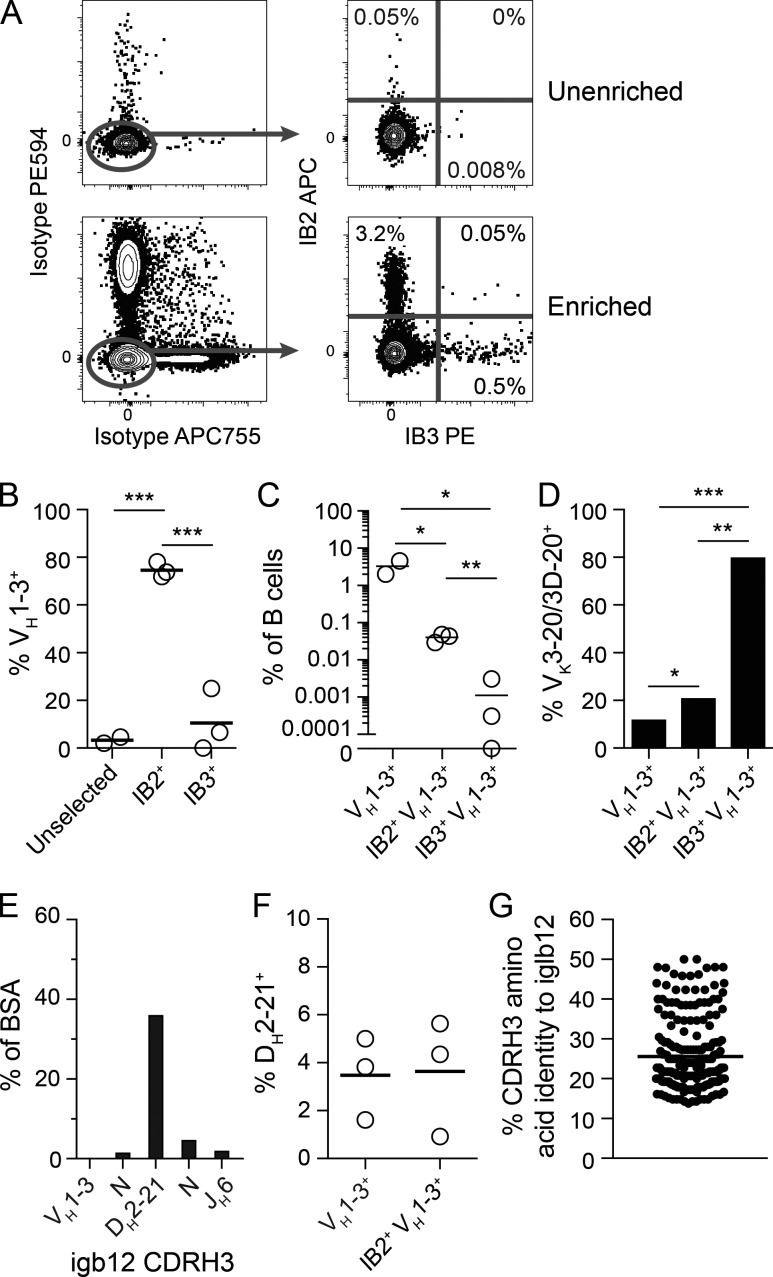
To test these predictions, IB2 and IB3 were biotinylated and tetramerized using streptavidin conjugated to APC or R-phycoerythrin (PE). Staining PBMCs with anti-idiotype tetramers revealed that <0.05% and 0.008% of the live B cell population from HIV-uninfected individuals bound IB2-APC or IB3-PE, respectively, but not APC755- or PE-Dylight594 (PE594)–conjugated isotype control tetramers (Fig. 4 A). To more easily identify rare B cells binding IB2 and/or IB3, tetramer-binding cells were enriched using APC-specific and PE-specific magnetic microbeads from samples of 200 million PBMCs before flow cytometry (Fig. 4 A). Using this approach, single B cells binding IB2 and/or IB3 tetramers were sorted into individual wells of a 96-well plate, and the heavy and kappa light chains expressed by these cells were sequenced using nested RT-PCR (Tiller et al., 2008). In total, we FACS-purified 576 single IB2-binding B cells and 150 single IB3-binding B cells from three individuals. From IB2-binding B cells, we obtained 302 heavy chain and 174 kappa light chain sequences, with 136 heavy and light chain pairs (Table S3). IB3-binding B cells yielded 84 heavy chain and 44 kappa light chain sequences, with 27 heavy and light chain pairs (Table S4). We were unable to recover heavy or light chain sequences from the small number of sorted cells binding both IB2 and IB3 tetramers.
Figure 4.
Identification and analysis of human B cells able to bind anti-iglb12 idiotypes. (A) Detection of live CD19+ CD3− CD14− CD16− B cells from human PBMCs that bound IB2-PE and IB3-APC tetramers with or without enrichment using anti-PE and anti-APC microbeads before flow cytometry. PE594 and APC755 tetramers containing isotype control antibodies were included in these experiments to exclude B cells specific for the PE, APC, streptavidin, and conserved portions of the anti-idiotypes. The displayed plots were derived from ∼400,000 unfractionated PBMCs or ∼400,000 PE- and APC-enriched cells derived from 200 million PBMCs and representative of three individuals in three independent experiments. The percentages of single and double positive B cells in the enriched and unenriched fractions from one individual are shown on the plots. (B) The frequency of heavy chains using VH1-3 within the population of IB2+ B cells, IB3+ B cells, and a population of control B cells that was not selected based upon antigen binding are displayed for three individuals. (C) The frequency of total VH1-3+, IB2+ VH1-3+, and IB3+ VH1-3+ within the entire B cell repertoire is displayed for three individuals. (D) The frequency of VK3-20/VK3D-20 usage among 120 IB2+ VH1-3+ BCRs and 10 IB3+ VH1-3+ BCRs using kappa light chains pooled from three individuals are compared with 259 VH1-3+ BCRs using kappa light chains from a control dataset derived from naive B cells (DeKosky et al., 2016). (E) Quantitation of the percent BSA between IB2 and the segments derived from VH1-3, DH2-21, JH6, and N nucleotide additions within iglb12 heavy chain CDRH3 from the crystal structures displayed in Fig. 2. (F) DH2-21 usage among 228 IB2+ VH1-3+ BCRs pooled from three individuals are compared with 467 VH1-3+ BCRs from a control dataset derived from naive B cells (DeKosky et al., 2016). (G) Comparison of CDRH3 similarity from 228 IB2+ VH1-3+ BCRs compared with the CDRH3 of iglb12 using pairwise alignment. The P values (*, P < 0.05; **, P < 0.01; ***, P < 0.001) in B and C were determined using an unpaired two-tailed Student’s t test, and the P values in D were determined by Fisher’s exact test.
The BCR sequences from B cells binding to IB2 or IB3 were next compared with iglb12. On average, over 71% of the IB2-binding B cells from all three individuals used the same VH gene as iglb12, VH1-3 (Fig. 4 B). In contrast, only 4.6% of individually sorted B cells expressed VH1-3 when neither anti-idiotype was used to select cells for sorting (Fig. 4 B), consistent with previous studies (DeKosky et al., 2016). In terms of the entire B cell repertoire, an average of 0.04% of the B cells bound IB2 and used VH1-3 (Fig. 4 C). In contrast, only 11% of the IB3-binding B cells used VH1-3, which amounted to ∼0.001% of the B cell repertoire (Fig. 4, B and C). Consistent with the crystal structure revealing that IB3 interacts with both the heavy and light chains of iglb12 (Fig. 2, B and C), four out of the five IB3+ VH1-3+ BCRs in which light chain sequences were recovered used the same light chain as igbl12, VK3-20, or the closely related VK3D-20 (Fig. 4 D). Surprisingly, IB2+ VH1-3+ BCRs exhibited a slight preference for VK3-20 (Fig. 4 D), which was not predicted by the heavy chain–only mode of binding indicated by the crystal structure (Fig. 2, F–J).
We next examined the unique heavy chain CDRH3 regions used by IB2+ VH1-3+ BCRs in search of further similarity to iglb12. Since we were only able to detect 10 IB3+ VH1-3+ BCRs in this dataset, these BCRs were not included in this analysis. The crystal structure of IB2 bound to iglb12 revealed that 44% of the BSA was contributed by the CDRH3 of iglb12 (Fig. 2 J). Nearly all of the CDRH3 BSA, 82% (i.e., 36% of the total BSA), is focused on the portion encoded by DH2-21 (Fig. 4 E). However, only 3.6% of IB2+ VH1-3+ BCRs used DH2-21, which was not different from control VH1-3+ BCRs (Fig. 4 F). Of the CDRH3 regions in IB2+ VH1-3+ BCRs, the two most similar to iglb12 CDRH3 only displayed 50% amino acid identity (Fig. 4 G).
Analysis of iglb12 autoreactivity
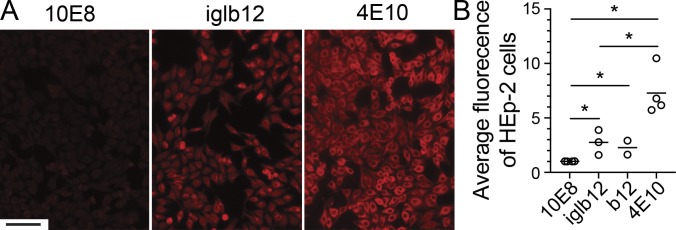
The moderate similarity of IB2+ BCR sequences to iglb12 led us to consider whether there were biological mechanisms selecting against CDRH3s that more closely matched iglb12. Specifically, we hypothesized that the iglb12 heavy chain was autoreactive, resulting in deletion of similar sequences from the B cell repertoire. Autoreactivity of iglb12 has not been reported, and conflicting reports of autoreactivity have been reported for the mature form of b12. One study reported that mature b12 exhibited autoreactivity using an in vitro assay (Haynes et al., 2005; Ota et al., 2013), however B cell development in transgenic mice expressing this antibody was normal (Ota et al., 2013). As a preliminary assessment of potential autoreactivity, we measured iglb12 binding to human HEp-2 cells using immunofluorescence. This approach revealed that iglb12 bound moderately to HEp-2 cells compared with 4E10 (Fig. 5, A and B), an HIV-specific antibody previously shown to exhibit binding in this assay (Haynes et al., 2005; Verkoczy et al., 2010). The iglb12 staining was similar to the mature form of b12, both of which were brighter than that of 10E8 (Fig. 5, A and B), an antibody that was previously shown to be nonreactive in this assay (Haynes et al., 2005; Verkoczy et al., 2010). Together, these data suggest that iglb12 is autoreactive.
Figure 5.
Assessment of iglb12 autoreactivity. (A and B) Representative immunofluorescence images (A; scale bar, 100 µm) and quantitation of iglb12 and the mature form of b12 bound to human HEp-2 cells compared with a positive control antibody shown previously to bind HEp-2 cells, 4E10, and a negative control antibody, 10E8 (B). Data points in B were combined from four independent experiments and represent the average Alexa Fluor 594 fluorescence per HEp-2 cell. The P values (*, P < 0.04) were determined using an unpaired two-tailed Student’s t test, and the bar indicates the mean (n = 2–4).
Deletion and anergy in transgenic mice expressing iglb12 heavy chain
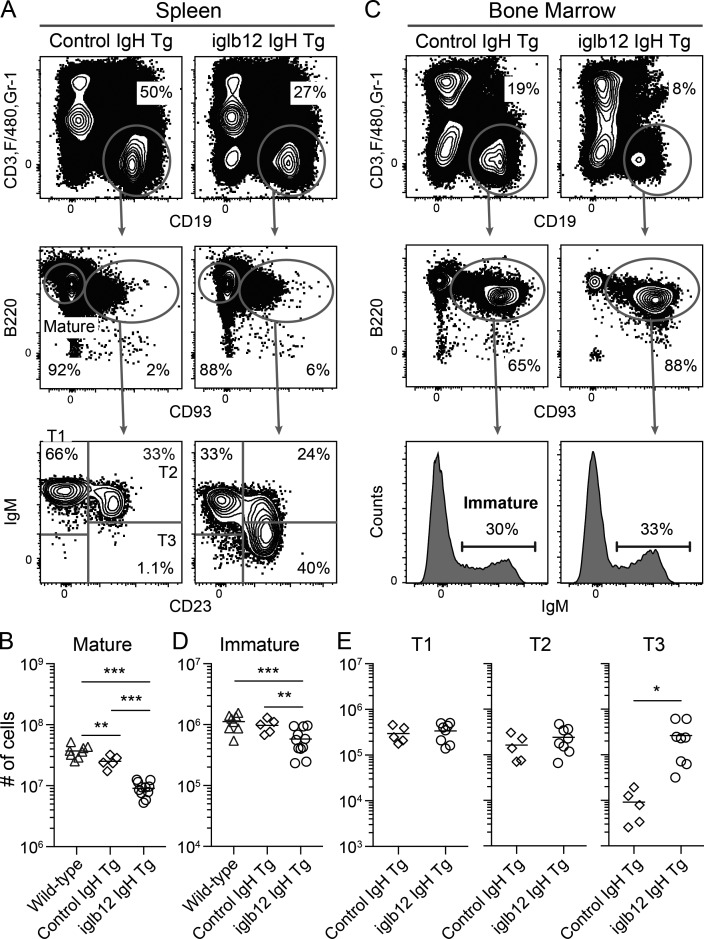
To assess whether the autoreactivity detected in the HEp-2 assay resulted in functional consequences in vivo, we generated a transgenic mouse in which all B cells expressed the iglb12 heavy chain. Previous work has demonstrated that B cells from BCR knock-in mice expressing 4E10, but not mature b12, are deleted from the repertoire as a consequence of autoreactivity (Doyle-Cooper et al., 2013; Ota et al., 2013). We found that iglb12 heavy chain transgenic mice displayed a three- to fourfold reduction in the number of CD19+ B220+ CD93− CD3− F4/80− Gr-1− mature B cells in the spleen compared with WT mice or mice expressing a control inferred germline heavy chain generated using the same knock-in strategy (Jardine et al., 2015; Fig. 6, A and B). Deletion could be traced back to the immature stage in the bone marrow, where iglb12 heavy chain transgenic mice contained approximately twofold fewer CD19+ B220+ CD93+ IgM+ CD3− F4/80− Gr-1− mature B cells compared with both WT and control transgenic animals (Fig. 6, C and D). Together, the data indicate that a significant portion of murine B cells expressing an iglb12 heavy chain are deleted in the bone marrow.
Figure 6.
Analysis of the development and subset distribution of transgenic B cells expressing the iglb12 heavy chain. (A and C) Representative flow cytometric gating of B cell (CD19+ CD3− Gr-1− F4/80−) subsets in the (A) spleen and (C) bone marrow of transgenic mice expressing the heavy chain from iglb12 (iglb12 IgH transgenic [Tg]) or a control population expressing the inferred germline heavy chain of VRC01 (control IgH transgenic). The percentages of cells within each gate are from representative animals. (B, D, and E) Combined data from four experiments showing the total number of (B) mature B cells in the spleen, (D) immature B cells in the bone marrow, and (E) the transitional T1, T2, and T3 B cell subsets in the spleen of individual transgenic mice (n = 5–11). The bar indicates the mean, and the P values (*, P < 0.04; **, P < 0.01; ***, P < 0.001) were determined using an unpaired two-tailed Student’s t test.
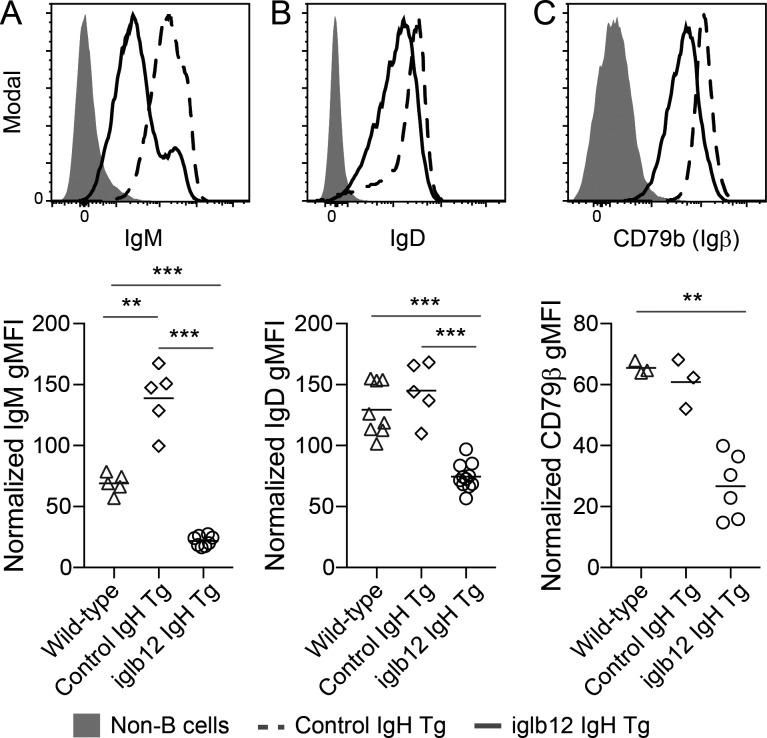
Previous studies have shown that autoreactive B cells that escape deletion are often rendered functionally unresponsive to antigenic stimulation, a state referred to as anergy (Goodnow et al., 1988, 2010; Cyster et al., 1994; Fulcher and Basten, 1994; Klinman, 1996; Nemazee, 2006, 2017). Given this, we next considered whether the B cells remaining in iglb12 heavy chain transgenic mice were anergic. Previously, several phenotypes of anergic B cells have been described in mice, including CD93+ CD23+ IgMLOW “T3” transitional phenotype, as well as mature B cells expressing low levels of IgM (Allman et al., 2001; Merrell et al., 2006; Taylor et al., 2012; Agrawal et al., 2013; Sabouri et al., 2016). In agreement with the hypothesis that circulating iglb12 heavy chain transgenic B cells were anergic, the number of T3 cells was increased 29-fold in these animals compared with control transgenic animals (Fig. 6, C and E). In addition, IgM expression was 6.4-fold lower on mature B cells from iglb12 heavy chain transgenic mice compared with control heavy chain transgenic animals (Fig. 7 A). IgD expression was also reduced 1.9-fold on mature B cells from iglb12 heavy chain transgenic mice compared with control transgenic animals (Fig. 7 B), which suggested a global down-regulation of surface BCR on these cells. Down-regulated surface BCR expression by iglb12 heavy chain transgenic mature B cells was confirmed through the assessment of CD79β (Igβ; Fig. 7 C), a component of the BCR signaling complex required for surface BCR expression.
Figure 7.
Reduced BCR expression by mature transgenic B cells expressing the iglb12 heavy chain. (A–C) Representative flow cytometric analysis and quantitation of (A) IgM, (B) IgD, and (C) CD79β expression by mature CD93− B220+ CD19+ CD3− Gr-1− F4/80− B cells in the spleen of iglb12 heavy chain transgenic (Tg), control heavy chain transgenic, and WT mice. Data are pooled from four independent experiments, which were normalized to account for experiment-to-experiment variability by displaying the data as a geometric mean fluorescence intensity (gMFI) fold increase over background fluorescence in CD19− B220− non-B cells. The bar indicates the mean, and P values (**, P < 0.001; ***, P < 0.001) were determined using an unpaired two-tailed Student’s t test (n = 3–11).
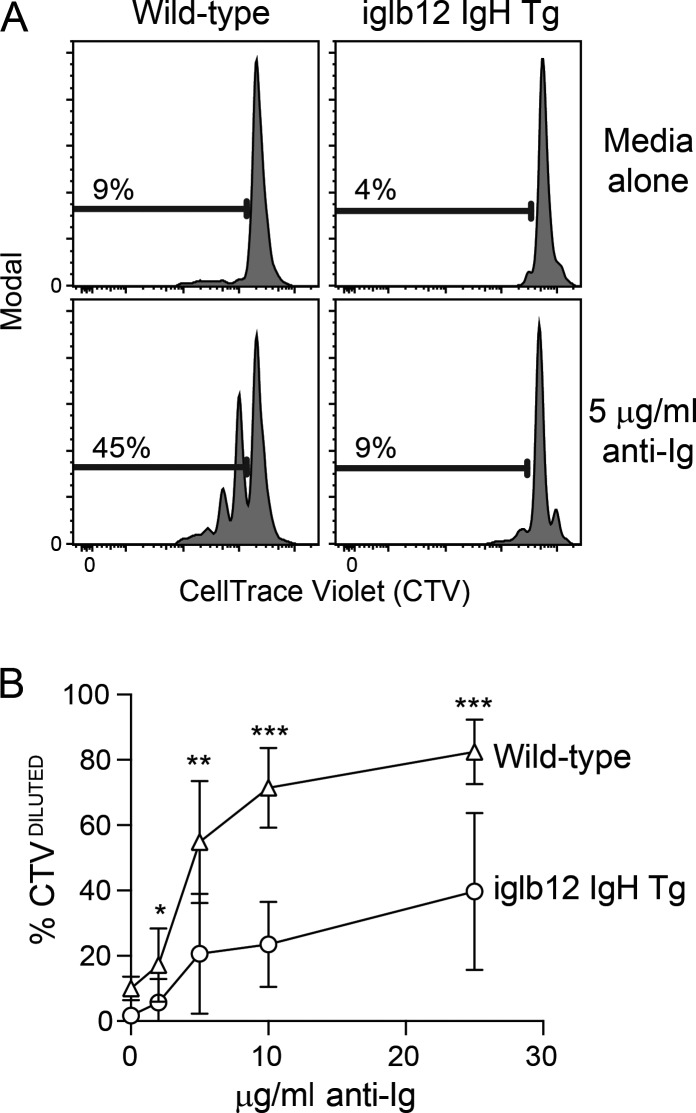
The increased frequency of iglb12 heavy chain transgenic B cells exhibiting an anergic phenotype in combination with decreased BCR expression suggested that these cells would respond poorly to BCR stimulation. To test this, we labeled purified mature CD93− B cells from WT and iglb12 heavy chain transgenic mice with CellTrace Violet (CTV) and incubated them with varying concentrations of anti-Ig for 72 h in vitro before analysis. At all concentrations of anti-Ig, fewer iglb12 heavy chain transgenic B cells had diluted CTV compared with B cells from WT mice (Fig. 8, A and B). These data suggest that the iglb12 heavy chain transgenic B cells that escape deletion are functionally anergic. Combined with the HEp-2 staining data, our murine data suggest that the iglb12 heavy chain is autoreactive, supporting the hypothesis that deletion and anergy of iglb12-like BCRs occurs in humans.
Figure 8.
In vitro response of transgenic B cells expressing the iglb12 heavy chain. (A and B) Representative flow cytometric analysis (A) and quantitation of the percentage of CTVDILUTED B cells (B) from individual iglb12 heavy chain transgenic (Tg) or WT control mice were labeled with CTV following in vitro culture in the presence of 2, 5, 10, or 25 µg/ml polyclonal goat anti-mouse Ig or media alone for 72 h. The percentages of B cells in the gates are shown in A. (B) Mean percentage ± SD of CTVDILUTED cells found in samples from seven individual mice from three independent experiments. The P values (*, P < 0.05; **, P < 0.01; ***, P < 0.001) were determined using an unpaired two-tailed Student’s t test.
Activation and differentiation of iglb12 heavy chain transgenic B cells in vivo following anti-iglb12 injection
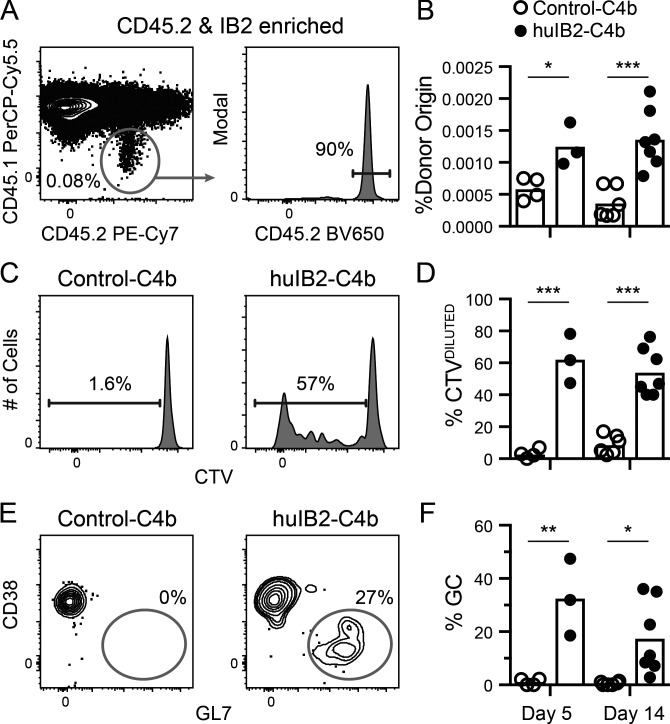
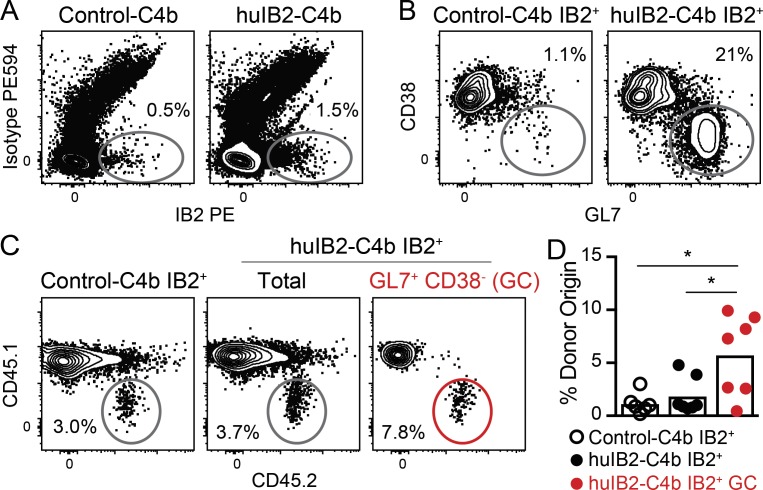
The presence of anergy within iglb12 heavy chain transgenic B cells suggests that immunogens targeting this population would need to overcome these functional defects. Before performing immunization experiments, we modified IB2 to ensure optimal murine responses. Since IB2 was generated in a mouse, it would likely generate a poor response if injected unmodified, since tolerance mechanisms would be expected to eliminate most sources of T cell help targeting it. To increase possible CD4+ T cell help, the heavy and light chain constant regions used by this anti-idiotype were humanized (Tiller et al., 2009). Since multimerization can overcome anergic responses (Cooke et al., 1994; Gautam et al., 2016; McGuire et al., 2016), humanized IB2 antigen-binding fragment (Fab) was fused to a modified multimerization domain of the chicken C4b-binding protein (huIB2-C4b), which self-assembles into a multimer containing up to seven humanized IB2 Fab domains (Ogun et al., 2008; Hofmeyer et al., 2013). To further garner T cell help, the 2W epitope (Rees et al., 1999) was added to the C-terminus of C4b separated by a cathepsin consensus sequence in order to promote endosomal processing and MHC presentation (Schneider et al., 2000). For immunization experiments, 4 × 105 B cells from CD45.2+ iglb12 heavy chain transgenic mice were labeled with CTV and transferred into CD45.1+ WT recipient mice. 1 d after transfer, the mice were immunized with huIB2-C4b or an irrelevant humanized isotype control-C4b fusion (control-C4b). 5 or 14 d following the injection of the control-C4b, CD45.2+ donor B cells accounted for an average of ∼0.0006% of total B cells in control recipient animals, which we detected by enriching for CD45.2+ cells before analysis (Fig. 9, A and B). As expected of resting cells that had not encountered antigen and are therefore not proliferating, these cells largely retained high levels of CTV (Fig. 9, C and D). In contrast, 5 d following injection of huIB2-C4b, donor B cells increased twofold to an average of ∼0.001% of total B cells (Fig. 9, B–D). Approximately half of the donor B cells had low levels of CTV, indicating that proliferation accounted for the increased frequency of donor cells (Fig. 9, B–D). In addition to expansion, ∼33% of transgenic B cells in huIB2-C4b–immunized mice bound GL7 and down-regulated CD38 (Fig. 9, E and F), a phenotype of germinal center B cells (Allen et al., 2007; Kurosaki et al., 2010; Victora et al., 2010; Vinuesa et al., 2010; Kitano et al., 2011). In these experiments, IB2-binding germinal center B cells derived from both the transgenic donor and recipient animals could be detected using IB2 tetramers (Fig. 10, A–C). This analysis revealed that transgenic CD45.2+ iglb12 B cells accounted for 5.8% of the IB2-binding germinal center B cells (Fig. 10 D). This frequency represented an increase from the 1–2% of transgenic cells found in cells outside the germinal center in both huIB2-C4b and control-C4b injected animals (Fig. 10 D). Together, these results indicate that anti-iglb12 idiotype antibodies can be used as immunogens that can overcome anergy and induce B cell activation and entry into germinal centers.
Figure 9.
In vivo response of transgenic B cells expressing the iglb12 heavy chain following the injection of a multimerized anti-iglb12 idiotype. Data from three experiments in which 4 × 105 purified B cells from CD45.2+ iglb12 heavy chain transgenic mice were labeled with CTV and adoptively transferred retro-orbitally into WT CD45.1+ recipients 1 d before intraperitoneal immunization with 20 µg huIB2-C4b or control-C4b and 25 µg of Sigma Adjuvant System. 5 or 14 d later, spleen and lymph nodes from individual mice were pooled and enriched for CD45.2-biotin+ iglb12 heavy chain transgenic donor cells using anti-biotin microbeads before analysis by flow cytometry. (A and B) Representative gating (A) and quantitation (B) of CD45.2+ CD45.1− CD19+ CD3− Gr-1− F4/80− donor iglb12 heavy chain transgenic B cells from the CD45.2-enriched fractions from individual recipient mice. The percentage on the plot in A represents the frequency of donor cells among only the B cells in the enriched fraction, while the percentages in B represent the frequency of donor B cells when B cells from the depleted fraction are included in the calculation. (C and D) Representative gating (C) and quantitation (D) of donor iglb12 heavy chain transgenic B cells with diluted CTV. The percentages of cells within gates in C are from representative animals. (E and F) Representative gating (E) and quantitation (F) of donor GL7+ CD38− iglb12 heavy chain transgenic germinal center (GC) B cells. The percentages of cells within gates in E are from representative animals. The bars in B, D, and F plot the mean from individual recipient mice, and P values (*, P < 0.05; **, P < 0.01; ***, P < 0.001) were determined using an unpaired two-tailed Student’s t test (n = 3–7).
Figure 10.
Transgenic B cells expressing the iglb12 heavy chain enter germinal centers despite competition from WT cells. Data from two experiments in which 4 × 105 purified B cells from CD45.2+ iglb12 heavy chain transgenic mice were labeled with CTV and adoptively transferred retro-orbitally into WT CD45.1+ recipients 1 d before intraperitoneal immunization with 20 µg huIB2-C4b or control-C4b and 25 µg Sigma Adjuvant System. 14 d later, spleen and lymph nodes from individual mice were pooled and enriched for IB2-PE+ and CD45.2-biotin+ iglb12 heavy chain transgenic donor cells using anti-PE and anti-biotin microbeads before analysis by flow cytometry. (A) Representative flow cytometric analysis of live CD19+ CD3− Gr-1− F4/80− B cells binding IB2-PE tetramers, but not isotype control PE594 tetramers. (B) Representative gating of GL7+ CD38− germinal center B cells in the IB2+ population described in A. (C and D) Representative gating (C) and quantitation(D) of the frequency of CD45.2+ CD45.1− donor B cells within the total IB2+ and IB2+ GL7+ CD38− germinal center populations described in A and B. The percentages of cells within gates in A–C are from representative animals. The bars in D represent the mean from individual recipient mice (n = 6–7), and P values (*, P < 0.05) were determined using an unpaired two-tailed Student’s t test.
Discussion
Our results demonstrate that anti-idiotype antibodies can identify and activate bNAb precursor B cells in vivo and represent a potential alternative or complement to structure-based immunogen design. Anti-idiotypes present several advantages. First, antigens derived from pathogens are generally difficult to produce and are often highly unstable or difficult to maintain in the desired conformation, especially after injection. From a manufacturing standpoint, anti-idiotypes are easily produced once identified and would be expected to be highly stable in vivo, as evidenced by the licensure of several therapeutic antibodies. Another advantage to this approach is that one could simultaneously use numerous anti-idiotypes to stimulate multiple potentially protective lineages. However, it must be recognized that it is unlikely that the injection of a single anti-idiotype containing immunogen would induce the production of a protective antibody in situations where high levels of somatic mutation are required (Scheid et al., 2009; Dimitrov, 2010; Pancera et al., 2010; Zhou et al., 2010; Haynes et al., 2012; Mouquet et al., 2012; Jardine et al., 2013; Klein et al., 2013a,b; McGuire et al., 2013; Escolano et al., 2016). In these situations, successive injections of immunogens containing anti-idiotypes of increasingly mature antibodies may guide antibody maturation toward highly protective antibodies. Alternatively, anti-idiotypes may be better suited for activation of protective antibodies that require less somatic mutation, such as those that can neutralize Middle East respiratory syndrome (Agrawal et al., 2016) and respiratory syncytial virus (Goodwin et al., 2018), or as the first step in a maturation pathway that includes pathogen-derived antigen in subsequent boosts.
To identify four clones that produced anti-iglb12 idiotypes, we used multiplexed screening approaches to evaluate thousands of hybridomas. In future studies the screening burden can be further reduced by enrichment or sorting of B cells expressing BCRs with desirable binding properties before and/or after hybridoma fusion (Taylor et al., 2012; Spanier et al., 2016).
To our knowledge, this study represents the first time anti-idiotypes have been used to identify and analyze naive B cells expressing BCRs with similarities to the inferred germline sequence of a broadly neutralizing HIV-1–specific antibody. An important result within these analyses is the finding that different anti-idiotypic antibodies can have different modes of interaction with BCRs similar to the one of interest. The binding profile analysis (Fig. 1 C) and crystal structure data (Fig. 2) suggested that IB2 would select only for BCRs with iglb12-like VH1-3+ heavy chains, while IB3 would select for BCRs with iglb12-like VH1-3+ heavy and VK3-20+ light chains, perhaps as a subset of the IB2-binding population. Sorting and sequencing BCRs from human B cells was therefore important and revealed that IB2 and IB3 selected for completely different populations of cells, with IB2 selecting far more stringently for VH1-3+ BCRs compared with IB3. Given these findings, we are exploring ways to better predict which anti-idiotypes will be better at identifying BCRs of interest. Nevertheless, our results indicate that probing the preimmune repertoire with anti-idiotypes or other potential immunogens is an informative step in vaccine design.
In the course of our sequencing experiments, we screened through the equivalent of 30 million B cells and only identified a few CDRH3 sequences recognized by the anti-idiotypes that showed >50% amino acid identity to iglb12. It is conceivable that the anti-idiotypes were able to recognize rare CDRH3 sequences with higher identity to iglb12 but that we were unable to identify them due to limitations in the efficiency of cell sorting and BCR sequencing. The ability to screen a larger number of cells currently exceeds our capacity but is obtainable using enrichment and cell sorting in combination with droplet-based BCR sequencing approaches.
Nevertheless, the absence of CDRH3 sequences with high similarity to iglb12 led us to hypothesize that these sequences may be purged from the repertoire due to autoreactivity. Support for this hypothesis came from iglb12 staining of HEp-2 cells and experiments in which B cell deletion and anergy were observed in transgenic mice expressing the iglb12 heavy chain. These tolerance effects may explain why b12-like bNAbs have not been commonly identified from HIV-infected individuals in several studies. In contrast, bNAbs with other specificities are present and are identified in a large fraction of HIV-1 subjects who develop bNAb responses, such as those targeting the N332 supersite (Pancera et al., 2014; Landais et al., 2016; Rusert et al., 2016; Ditse et al., 2018).
The existence of deletion and anergy in mice suggested that immunization strategies able to overcome tolerance may be required to stimulate the production of b12-like bNAbs in humans. The need to overcome tolerance is not unique to iglb12 knock-in mice, since deletion and anergy have also been detected in mice expressing the inferred germline forms of the HIV-1 bNAbs 4E10 and 3BNC60 (McGuire et al., 2016; Zhang et al., 2016). Autoreactivity is not a unique feature of some HIV-specific lineages and has been detected in assessments of influenza bNAbs (Andrews et al., 2015; Bajic et al., 2019). Notably, it remains possible that the tolerance exhibited in transgenic mice expressing the iglb12 heavy chain is an artifact of the inability to accurately infer the true unmutated heavy chain CDRH3 from the mature b12 antibody. For example, the CDRH3 in this sequence contains two N regions that cannot be inferred and are instead carried over from the mature sequence. It is possible that the true germline heavy chain CDRH3 had different sequences in these locations that would not have resulted in deletion and anergy.
Importantly, activation and germinal center entry of iglb12-expressing B cells is just the first step of the maturation process. This lineage would need to be guided toward broad Env binding and neutralization, which will likely require many rounds of mutation and selection. Future work will explore whether prime boost experiments with IB2/IB3 and Env can be used to guide the maturation of bNAb responses.
Overall, our study demonstrates the utility of using anti-idiotypes to identify and stimulate B cells expressing desirable BCRs. Future work is aimed at including anti-idiotypes in the vaccine design process, both as tools to identify cells of interest and as immunogens themselves.
Materials and methods
Human PBMC collection and storage
Blood was obtained by venipuncture from healthy, HIV-seronegative adult volunteers enrolled in the General Quality Control study in Seattle, WA, which was approved by the Fred Hutchinson Cancer Research Center institutional review board. Informed consent was obtained from all study participants before enrollment into the parent protocols. PBMCs were isolated from whole blood using Accuspin System Histopaque-1077 (Sigma-Aldrich) resuspended in 10% dimethylsulfoxide in heat-inactivated fetal bovine serum and cryopreserved in liquid nitrogen before use.
Animals
All experiments were performed in accordance with relevant institutional and national guidelines and were approved by Fred Hutchinson Cancer Research Center Institutional Animal Care and Use Committee. 6–14-wk-old male and female WT C57BL/6J and B6.SJL-Ptprca Pepcb/BoyJ (CD45.1+) mice were purchased from the Jackson Laboratory or bred in house. Generation of iglb12 heavy chain transgenic mice and control heavy chain transgenic mice was performed at The Scripps Research Institute Mouse Genetics Core Facility similarly to what was described previously for the mature b12 heavy chain (Ota et al., 2013). Briefly, the DQ52-JH cluster was replaced in a C57BL/6-derived C2 embryonic stem cell line with the iglb12 heavy chain VDJ exon or inferred germline VRC01 heavy chain VDJ exon. Mice derived from these modified embryonic cells carrying the transgenic heavy chain were interbred and homozygous transgenic heavy chain+/+ male or female mice were used for experiments. Control transgenic animals expressing the iglVRC01 heavy chain were generated in an identical manner (Jardine et al., 2015).
Anti-idiotype hybridoma generation and purification
The sequence of iglb12 has been previously reported (Hoot et al., 2013, and the CDRH3 and CDRL3 sequences are listed in Table S2. Anti-idiotypic antibodies were generated by the Fred Hutchinson Cancer Research Center Antibody Technology Core. Briefly, 8–12-wk-old female BALB/c mice (Jackson Laboratory) were immunized and boosted intraperitoneally with iglb12 in adjuvant five times over a 5-mo period. 3 d following the final boost, splenocytes were electrofused to myeloma cells, and thousands of hybridoma colonies were screened for binding to iglb12 and a panel of control germline antibodies. Hybridomas producing antibodies specific for iglb12 were subcloned from single cells and expanded before anti-idiotype purification. Supernatant from these expanded hybridomas was harvested and purified over protein G resin (Pierce) following the manufacturer’s recommendations. To produce recombinant anti-idiotypes, RNA was extracted from 1 × 106 cells using the RNeasy kit (Qiagen) and the heavy and light chain sequences of the murine hybridomas were by obtained using the mouse Ig-primer set (69831; EMD Millipore) using the protocol developed by Seigel et al. (Siegel, 2009). Sequences were codon optimized and cloned into pTT3-based IgG expression vectors with human constant regions (Snijder et al., 2018) using In-Fusion cloning (Clontech).
Tetramer production
Purified anti-idiotype antibodies were biotinylated using an EZ-link Sulfo-NHS-LC-Biotinylation kit (Thermo Fisher Scientific) using a 1:1 molar ratio of biotin to anti-idiotype. Unconjugated biotin was removed by centrifugation using a 50-kD Amicon Ultra size exclusion column (EMD Millipore). To determine the average number of biotin molecules bound to each anti-idiotype molecule, streptavidin-PE (ProZyme) was titrated into a fixed amount of biotinylated anti-idiotype in increasing concentrations and incubated at room temperature for 30 min. Samples were run on an SDS-PAGE gel (Bio-Rad Laboratories) and transferred to nitrocellulose and incubated with streptavidin–Alexa Fluor 680 (1:10,000; Thermo Fisher Scientific) to determine the point at which there was excess biotin available for the streptavidin–Alexa Fluor 680 reagent to bind. Biotinylated anti-idiotypes were mixed with streptavidin-PE or streptavidin-APC at the ratio determined above to fully saturate streptavidin and incubated for 30 min at room temperature. Unconjugated anti-idiotypes were removed by several rounds of dilution and concentration using a 300K Nanosep centrifugal device (Pall Corporation). Control PE594 and APC755 tetramers were created by mixing isotype control antibodies with streptavidin-PE preconjugated with Dylight 594 (PE594) or streptavidin-APC preconjugated with Dylight 755 (APC755; both from Thermo Fisher Scientific) following the manufacturer’s instructions. On average, PE594 and APC755 contained 4–8 Dylight molecules per PE/APC. The concentration of each anti-idiotype tetramer was calculated by measuring the absorbance of PE (565 nm, extinction coefficient = 1.96 µM−1 cm−1) or APC (650 nm, extinction coefficient = 0.7 µM−1 cm−1) and the lot-specific ratio of streptavidin-PE or streptavidin-APC.
Tetramer enrichment
For human PBMC analysis, 200 × 106 frozen cells were thawed into RPMI with 10% fetal bovine serum (Thermo Fisher Scientific), 100 U/ml penicillin (Thermo Fisher Scientific), 100 µg/ml streptomycin (Thermo Fisher Scientific), and 0.0275 mM 2-mercaptoethanol (Sigma-Aldrich). Cells were centrifuged and resuspended to 0.2 ml in ice-cold FACS buffer composed of 1× Dulbecco's PBS (DPBS) with 1% newborn calf serum (Thermo Fisher Scientific) containing 2% rat and mouse serums. PE594- and APC755-conjugated control anti-idiotype tetramers were added at a final concentration of 5–15 nM and incubated on ice for 5 min to allow binding to B cells of unwanted specificities. IB2-PE and IB3-APC or IB1-APC, IB2-APC, and IB3-APC tetramers were added at a final concentration of 5 nM and incubated on ice for 25 min, followed by a 10-ml wash with ice-cold FACS buffer. 25 μl of both anti-PE- and/or anti-APC-conjugated microbeads (Miltenyi Biotec) were added and incubated on ice for 30 min. 3 ml of FACS buffer was then added to the cell mixture and passed over a magnetized LS column (Miltenyi Biotec). The column was washed once with 5 ml ice-cold FACS buffer and then removed from the magnetic field. 5 ml ice-cold FACS buffer was pushed through the unmagnetized column twice using a plunger to elute the bound cell fraction.
Adoptive transfer experiments and immunizations
Purified naive mature B cells were prepared from spleens of iglb12 heavy chain transgenic mice using a B cell negative selection kit (Miltenyi Biotec) supplemented with 1.25 µg/ml biotinylated anti-CD93 (AA4.1; eBioscience) to remove transitional B cells. Purified B cells were washed in 1× DPBS, adjusted to a concentration of 5 × 107 cells/ml in warm 1× DPBS and incubated with 5 µg CTV (Thermo Fisher Scientific) for 12 min at 37°C before being washed with media containing 10% fetal bovine serum (Thermo Fisher Scientific). Cells were resuspended in warm 1× DPBS and 4 × 105 cells per mouse were injected retro-orbitally into CD45.1+ recipients. 1 d following the transfer, recipient mice were injected intraperitoneally with 0.2 ml of the following solution: 0.1 ml (25 µg) diluted Sigma Adjuvant System (Sigma-Aldrich) and 20 µg of a humanized version of IB2 containing Fabs fused to the 2W epitope (EAWGALANWAVDSA) heptamerized on the C4B nanoparticle as described previously (Hofmeyer et al., 2013) in 0.1 ml 1× DPBS. A humanized isotype control Fab-C4b in diluted Sigma Adjuvant System was used as a control. 5 d following injection, the spleen, inguinal, brachial, cervical, and mesenteric lymph nodes were harvested into 1× DPBS, minced, and then digested as described previously (Taylor et al., 2015) in dispase (0.8 mg/ml; Thermo Fisher Scientific), collagenase (0.2 mg/ml; Roche), and DNase (0.1 mg/ml; Roche) for 20 min at 37°C. Enzymes were inactivated with 5 mM EDTA, and single-cell suspensions were obtained. Samples were centrifuged and cell pellets resuspended to 0.2 ml in 1× DPBS containing 1% newborn calf serum, 5 µg/ml of anti-CD16/CD3 (2.4G2, BioXCell), and 1.25 µg/ml anti-CD45.2 biotin (104; eBioscience) and enriched using 25 µl anti-biotin microbeads (Miltenyi Biotec) as described for tetramer enrichment.
Flow cytometry
After centrifugation, supernatant was discarded and cell pellets were resuspended in FACS buffer. The entire enriched fraction and 1/40th of the flow-through or unenriched fractions and were incubated with antibodies against surface markers in a total volume of 50 µl for 25 min on ice and washed with FACS buffer. Human PBMCs were labeled with 6.24 µg/ml anti-IgM FITC (G20-127; BD Biosciences), 4 µg/ml anti-IgD PerCP-Cy5.5 (IA6-2; BD Biosciences), 2 µg/ml anti-CD20 eFluor 450 (2H7; eBioscience), 1 µl anti-CD27 BV650 or BV480 (L128; BD Biosciences), 1 µl anti-CD3 BV711 (UCT1; BD Biosciences), 1 µl anti-CD14 BV711 (MϕP9; BD Biosciences), 2.8 µg/ml anti-CD16 BV711 (3G8; BioLegend), and 1 µl anti-CD19 PE-Cy7 (HIB19; BD Biosciences).
In adoptive transfer experiments, mouse spleen and lymph node cells were labeled with 2.5 µg/ml GL7 FITC (BD Biosciences), 2 µg/ml anti-CD45.1 PerCP-Cy5.5 (A20; BioLegend), 2 µg/ml anti-CD38 Alexa Fluor 700 (90; eBioscience), 2 µg/ml anti-CD45.2 PE-Cy7 (104; BioLegend), 2 µg/ml anti-CD3 BV510 (145-2C11; BD Biosciences), 2 µg/ml anti-F4/80 BV510 (BM8; BioLegend), 2 µg/ml anti–Gr-1 BV510 (RB6-8C5; BD Biosciences), 2 µg/ml anti-B220 BV786 (RA3-6B2; BD Biosciences), and 2 µg/ml anti-CD19 BUV395 (1D3; BD Biosciences). 1 µg/ml streptavidin-BV650 (BD Biosciences) was also included to detect donor cells that were enriched using anti-CD45.2 biotin.
For B cell subset analysis, cells were labeled with 2.5 µg/ml anti-CD79β FITC (HM79-12; BD Biosciences), 2 µg/ml anti-IgD PerCP-Cy5.5 (11-26c.2a; BD Biosciences), 2 µg/ml anti-CD21 PE-Cy7 (eBio8D9; eBioscience), 2 µg/ml anti-CD93 BV421 (AA4.1; BD Biosciences), 2 µg/ml anti-CD3 BV510 (145-2C11; BD Biosciences), 2 µg/ml anti-F4/80 BV510 (BM8; BioLegend), 2 µg/ml anti-Gr-1 BV510 (RB6-8C5; BD Biosciences), 2 µg/ml anti-IgMb BV650 (AF6-78; BD Biosciences), 2 µg/ml anti-CD23 BV786 (B3B4; BD Biosciences), 2 µg/ml anti-B220 BUV395 (RA3-6B2; BD Biosciences), and 2 µg/ml anti-CD19 BUV737 (1D3; BD Biosciences).
For in vitro proliferation experiments, cells were labeled with 2 µg/ml anti-CD3 BV510 (145-2C11; BD Biosciences), 2 µg/ml anti-F4/80 BV510 (BM8; BioLegend), 2 µg/ml anti-Gr-1 BV510 (RB6-8C5; BD Biosciences), and 2 µg/ml anti-CD19 BUV395 (1D3; BD Biosciences).
For all experiments, 0.25 µl Fixable Viability Dye eFluor 506 or eFluor 780 (eBioscience) or Fixable Viability Stain 700 (BD Biosciences) was included in the surface antibody cocktail to distinguish live from dead cells. Flow cytometry was performed on a five-laser (355 nm, 405 nm, 488 nm, 561 nm, and 640 nm) LSR II, LSRFortessa, FACSARIA II, or FACSymphony device (BD Biosciences) and analyzed with FlowJo 10 software (Tree Star). 2 × 104 fluorescent AccuCheck counting beads (Thermo Fisher Scientific) were added to the samples before flow cytometry and used to calculate total numbers of cell subtypes in the column-bound and flow-through suspensions, as previously described (Taylor et al., 2015).
In vitro proliferation assays
Naive mature B cells were purified and labeled with CTV as described for adoptive transfer experiments. Cells were adjusted to a concentration of 2 × 106 cells/ml in RPMI (Thermo Fisher Scientific) containing 10% fetal bovine serum (Thermo Fisher Scientific), 100 U/ml penicillin (Thermo Fisher Scientific), 0.1 mg/ml streptomycin, 2 mM L-glutamine (Thermo Fisher Scientific), 0.02 mg/ml gentamicin (Thermo Fisher Scientific), and 0.027.5 mM 2-mercaptoethanol (Sigma-Aldrich). 2 × 105 cells from each sample were added per well of a 96-well flat bottomed plate, and cells were cultured in the presence or absence of 2, 5, 10, or 25 µg/ml F(ab′)2 goat anti-mouse Ig (Jackson ImmunoResearch) in a final volume of 0.2 ml and incubated for 72 h at 37°C before analysis by flow cytometry.
Single-cell sorting
Single human B cells were isolated using a FACSARIA II cell sorter with Diva configuration (BD Biosciences) following tetramer enrichment and cell surface marker staining using human PBMCs as described above. Single B cells from defined subpopulations were sorted according to surface marker expression patterns. Specifically, IB2- or IB3-specific B cells were sorted using side scatter–based doublet discrimination, followed by gating on FVD− CD19+ CD20+ CD3− CD14− CD16− B cells that bound IB2-PE or IB3-APC tetramers but not PE650 or APC755 tetramers containing isotype control antibodies. We also sorted 576 control FVD− CD19+ CD20+ CD3− CD14− CD16− CD27− IgM/D+ B cells from two individuals that were not stained with an anti-idiotype. Single cells were sorted into individual wells of 96-well PCR plates (Eppendorf) containing 10 µl/well ice-cold lysis buffer containing 0.25 µl (12.5 U) RNaseOUT (Thermo Fisher Scientific), 2.5 µl 5× SuperScript IV First Strand Buffer (Thermo Fisher Scientific), 0.625 µl 0.1 M dithiothreitol (Thermo Fisher Scientific), 0.3125 µl 10% Igepal detergent (Sigma-Aldrich), and 6.625 µl diethyl pyrocarbonate–treated water. Plates were sealed with adhesive PCR plate seals (Thermo Fisher Scientific), centrifuged briefly, and immediately frozen on dry ice before storage at −80°C.
Nested RT-PCR BCR sequencing and analysis
RT was performed using SuperScript IV (Thermo Fisher Scientific) as previously described (Tiller et al., 2009; Wu et al., 2010). Briefly, 3 µl RT reaction mix consisting of 1.5 µl 50 µM random hexamers (Thermo Fisher Scientific), 0.4 µl 25 mM deoxyribonucleotide triphosphates (dNTPs; Thermo Fisher Scientific), 0.5 µl (10 U) SuperScript IV RT, and 0.6 µl water was added to each well containing a single sorted B cell in 10 µl lysis buffer and incubated at 50°C for 1 h. Following RT, 2 µl cDNA was added to 19 µl PCR reaction mix so that the final reaction contained 0.2 µl (0.5 U) HotStarTaq Polymerase (Qiagen), 0.075 µl 50 µM 3′ reverse primers, 0.115 µl 50 µM 5′ forward primers, 0.24 µl 25 mM dNTPs, 1.9 µl 10× buffer (Qiagen), and 16.5 µl water. The PCR program was 94°C 30 s, 57°C 30 s, 72°C 55 s, 50 cycles, 72°C 10 min for IgM/IgG/kappa light chains and 94°C 30 s, 60°C 30 s, 72°C 55 s, 50 cycles, 72°C 10 min for lambda light chains. After the first round of PCR, 2 µl of the PCR product was added to 19 µl of the second-round PCR reaction so that the final reaction contained 0.2 µl (0.5 U) HotStarTaq Polymerase, 0.075 µl 50 µM 3′ reverse primers, 0.075 µl 50 µM 5′ forward primers, 0.24 µl 25 mM dNTPs, 1.9 µl 10× buffer, and 16.5 µl water. PCR programs were the same as the first round of PCR. 4 μl of the PCR product was run on an agarose gel to confirm the presence of a ∼500-bp heavy chain band or 450-bp light chain band. 5 μl from PCR reactions showing the presence of heavy or light chain amplicons was mixed with 2 µl of ExoSAP-IT (Thermo Fisher Scientific) and incubated at 37°C 15 min, followed by 80°C for 15 min to hydrolyze excess primers and nucleotides. Hydrolyzed second-round PCR products were sequenced by Genewiz with the respective reverse primer, and sequences were analyzed using IMGT/V-Quest to identify V, D, and J gene segments.
Cloning and expression of human BCRs
Non-VH1-3–derived control antibodies were previous described (Hoot et al., 2013; Liao et al., 2013; McGuire et al., 2014a,b). Paired heavy chain VDJ and light chain VJ sequences from BCR sequencing experiments or naive VH1-3+ B cells identified previously (DeKosky et al., 2016) were codon optimized and cloned into pTT3-derived expression vectors containing the human IgG1, IgK, or IgL constant regions using In-Fusion cloning (Clontech).
BLI analysis
BLI assays were performed on the Octet.Red instrument (ForteBio). For anti-idiotype binding screens, anti-mouse IgG capture sensors (ForteBio) were immersed in kinetics buffer (1× PBS, 0.01% BSA, 0.02% Tween 20, and 0.005% NaN3, pH 7.4) containing 5 µg/ml purified anti-idiotypic antibody for 100 s. After loading, the baseline signal was then recorded for 60 s in kinetics buffer. The sensors were then immersed in kinetics buffer containing 20 µg/ml purified human antibody for a 100 s association step. The maximum response was determined by averaging the nanometer shift over the last 5 s of the association step after subtracting the background signal from each analyte-containing well using empty anti-mouse IgG Fc capture sensors at each time point.
Kinetic analyses were performed at room temperature with shaking at 500 rpm. Anti-mouse Ig capture sensors were immersed in kinetics buffer containing 20 µg/ml purified anti-idiotypic antibody for 240 s. After loading, the baseline signal was then recorded for 60 s in kinetics buffer. The sensors were next immersed into wells containing serial dilutions of purified iglb12 Fab in kinetics buffer for a 300-s association phase, followed by immersion in kinetics buffer for an additional 600-s dissociation phase. The background was signal measured from each analyte-containing well using sensors loaded with a negative control antibody and subtracted from the signal obtained with each corresponding ligand-coupled sensor at each time point. Kinetic analyses were performed at least twice with an independently prepared analyte dilution series. Curve fitting was performed using a 1:1 binding model and ForteBio data analysis software. The mean on rate and off rate values were determined by averaging all binding curves in each dilution series that matched the theoretical fit with an R2 value of ≥0.95.
HEp-2 binding assay
The ZEUS IFA ANA HEp-2 Test System was used as recommended by the manufacturer (Zeus Scientific) with minor changes. Briefly, HEp-2–coated slides were incubated with 25 µl antibody at 0.1 mg/ml for 30 min at room temperature. Slides were then washed three times in 1× DPBS followed by the incubation of 25 µl of 0.01 mg/ml goat anti-human IgG Alexa Fluor 594 (Thermo Fisher Scientific) in 1× DPBS for 30 min at room temperature in the dark. After washing three times with 1× DPBS, slides incubated with ZEUS IFA ANA HEp-2 Test System fixative, and coverslips were applied. Images were acquired using the EVOS Cell Imaging System (Thermo Fisher Scientific) and analyzed using ImageJ to determine the average Alexa Fluor 594 fluorescence per HEp-2 cell for each image.
Fab and cleavable single-chain variable fragment (c/scFv) expression, purification, and complex preparation
IB2 and IB3 IgG purified from hybridomas were digested over activated Ficin on Agarose Resin (Thermo Fisher Scientific) for 48 h to obtain Fabs. Fabs for iglb12 were generated by digesting with 1 µg Lys C (Roche) for 10 mg of IgG overnight at 37°C. The Fabs were purified from Fc regions by collecting the fraction that did not bind to a Protein A column (Thermo Fisher Scientific) and by size exclusion chromatography using Superdex 200 (GE Healthcare Life Sciences) in 5 mM Hepes and 150 mM NaCl.
A c/scFv iglb12 was cloned and expressed using lentiviral-based stable transduction of human embryonic kidney suspension adapted freestyle 293F cells (Thermo Fisher Scientific) as described previously (Bandaranayake et al., 2011). Briefly, plasmids containing appropriate constructs were incorporated into replication-incompetent lentiviral particles and transduced into human embryonic kidney suspension adapted freestyle 293F cells plated in fresh media at 106/ml in 10 ml and incubated at 37°C with 8% CO2 and 80% humidity. Cells were supplemented with 25 ml fresh freestyle media 8–16 h after transduction. 3 d following transduction, cells were replated at a cell density of 0.5 × 106/ml in cultures as large as 2 liters. Culture supernatants were harvested by centrifugation at 8,000 rpm to remove cells and clarified by passing through 0.22-μm filter (Thermo Fisher Scientific). Clarified supernatants were supplemented with 150 mM NaCl and concentrated using a 10,000 molecular weight cutoff Amicon Stirred Cell concentrator (EMD Millipore). Recombinant proteins were further purified by size exclusion chromatography using a Superdex 75 and/or Superdex 200 column (GE Healthcare) running in 25 mM piperazine-N,N′-bis(2-ethanesylfonic acid) pH 7.0, 150 mM NaCl, 1 mM EDTA, and 0.02% NaN3. Eluted fractions were analyzed using SDS-PAGE to assess purity. Protein concentrations were determined using molar extinction coefficients and absorbance at 280 nm.
Crystallization and data collection
Complexes of IB2 Fab + iglb12 c/scFv and IB3 Fab + iglb12 Fab were formed by mixing both components at a 1:1 molar ratio and incubating overnight at 4°C. Complexes were purified by size exclusion chromatography using Superdex 200 in 5 mM Hepes and 150 mM NaCl at pH 7.4. Crystals of IB2 Fab + iglb12 c/scFv and IB3 Fab + iglb12 Fab were obtained using a mosquito and an NT8 dispensing robots, and screening was done using commercially available screens (Rigaku Wizard Precipitant Synergy block #2, Molecular Dimensions Proplex screen HT-96, Hampton Research Crystal Screen HT) by mixing 0.1 µl/0.1 µl (protein/reservoir) by the vapor diffusion method. Crystals used for diffraction data were grown in the following conditions: IB3 Fab + iglb12 Fab crystals were grown in solution containing 0.2 M ammonium phosphate monobasic, 0.1 M Tris, pH 8.5, and 50%(+/−) 2-methyl-2,4-pentanediol. IB2 Fab + iglb12 c/scFv crystals were grown in solution containing 13.4% polyethylene glycol 8000, 13.4 PEG 400, 0.1 M MgCl2, and 0.1 M Tris, pH 8.5, using the vapor diffusion method. Crystals were cryoprotected in solutions containing 30% molar excess of their original reagents and 20% glycerol for IB3 Fab + iglb12 Fab and 20% 2R-3R Butanediol for IB2 Fab + iglb12 c/scFv. Crystal diffracted to 3 Å for IB3 Fab + iglb12 Fab and 2.6Å for IB2 Fab + iglb12 c/scFv. Data were collected with advanced light source 5.1 and 5.2 and processed using HKL2000 (Otwinowski and Minor, 1997).
Structure solution and refinement
The structures were solved by molecular replacement using Phaser in CCP4 (Collaborative Computational Project, Number 4, 1994) and PDB accession number 2NY7 (Zhou et al., 2007) for the mature b12 portion as a search model. Iterating rounds of structure building and refinement was performed in COOT (Emsley and Cowtan, 2004) and Phenix (Adams et al., 2010). The refinement statistics are summarized in Table S5, and the structural figures were made with Pymol (DeLano, 2002).
Accession numbers
Atomic coordinates and structure factors of the IB2/iglb12 (6OL6) and IB3/iglb12 (6OL5) complex x-ray crystal structures have been deposited in the Protein Data Bank. The heavy and light chain sequences of IB1 (MK775682 and MK775686), IB2 (MK775683 and MK775687), IB3 (MK775684 and MK775688), and IB5 (MK775685 and MK775689) have been deposited in GenBank.
Online supplemental material
Table S1 contains sequence information for the control antibodies used in Fig. 1 C. Table S2 contains sequence information for the heavy and light chain sequences derived from B cells binding the IB1/2/3 tetramer cocktail. Table S3 contains sequence information for the heavy and light chain sequences derived from human B cells binding the IB2 tetramers. Table S4 contains sequence information for the heavy and light chain sequences derived from human B cells binding the IB3 tetramers. Table S5 contains data collection and refinement statistics for the crystal structures shown in Fig. 2.
Supplementary Material
Acknowledgments
We thank A. Macy, M. Baker, D. Alwan, and the Fred Hutchinson Cancer Research Center Flow Cytometry Shared Resource for technical assistance and S. Voght for careful reading of the manuscript. We thank the James B. Pendleton Charitable Trust for generous support of Formulatrix robotic instruments. We also thank J. McElrath for PBMC (Fred Hutchinson Cancer Research Center, Seattle, Washington).
X-ray diffraction data were collected at the Berkeley Center for Structural Biology beamlines 5.0.1 and 5.0.2, which are supported in part by the National Institutes of Health, National Institute of General Medical Sciences. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences of the US Department of Energy under contract DE-AC02-05CH11231. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award numbers R01AI122912 (J.J. Taylor), R01AI081625 (L. Stamatatos), and R21AI127249 (A.T. McGuire). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare no competing financial interests.
Author contributions: L. Stamatatos, A.T. McGuire, and J.J. Taylor designed the study, analyzed results, and wrote the manuscript; T. Bancroft conducted flow cytometry, cell sorting, adoptive transfer, and BCR sequencing experiments, analyzed results, and wrote the manuscript; B.L. DeBuysscher conducted and analyzed HEp-2 binding experiments; H.R. Steach assisted with cell sorting experiments; K.S. Fitzpatrick conducted and analyzed flow cytometry experiments; C.E. Weidle and M. Pancera determined crystal structures; A. Schwartz conducted adoptive transfer and in vitro stimulation experiments; A. Wall, M.D. Gray, and J. Feng, performed binding screens, cloned and purified recombinant antibodies or antigens, and conducted BLI experiments; M.M. Gewe and R.K. Strong provided the iglb12 c/scFv; and P.D. Skog, C. Doyle-Cooper, T. Ota, and D. Nemazee generated iglb12 IgH transgenics.
References
- Abbott R.K., Lee J.H., Menis S., Skog P., Rossi M., Ota T., Kulp D.W., Bhullar D., Kalyuzhniy O., Havenar-Daughton C., et al. 2018. Precursor Frequency and Affinity Determine B Cell Competitive Fitness in Germinal Centers, Tested with Germline-Targeting HIV Vaccine Immunogens. Immunity. 48:133–146.e6. 10.1016/j.immuni.2017.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A.S., Ying T., Tao X., Garron T., Algaissi A., Wang Y., Wang L., Peng B.H., Jiang S., Dimitrov D.S., and Tseng C.T.. 2016. Passive Transfer of A Germline-like Neutralizing Human Monoclonal Antibody Protects Transgenic Mice Against Lethal Middle East Respiratory Syndrome Coronavirus Infection. Sci. Rep. 6:31629 10.1038/srep31629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal S., Smith S.A., Tangye S.G., and Sewell W.A.. 2013. Transitional B cell subsets in human bone marrow. Clin. Exp. Immunol. 174:53–59. 10.1111/cei.12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen C.D., Okada T., and Cyster J.G.. 2007. Germinal-center organization and cellular dynamics. Immunity. 27:190–202. 10.1016/j.immuni.2007.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allman D., Lindsley R.C., DeMuth W., Rudd K., Shinton S.A., and Hardy R.R.. 2001. Resolution of three nonproliferative immature splenic B cell subsets reveals multiple selection points during peripheral B cell maturation. J. Immunol. 167:6834–6840. 10.4049/jimmunol.167.12.6834 [DOI] [PubMed] [Google Scholar]
- Andrabi R., Voss J.E., Liang C.H., Briney B., McCoy L.E., Wu C.Y., Wong C.H., Poignard P., and Burton D.R.. 2015. Identification of Common Features in Prototype Broadly Neutralizing Antibodies to HIV Envelope V2 Apex to Facilitate Vaccine Design. Immunity. 43:959–973. 10.1016/j.immuni.2015.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S.F., Huang Y., Kaur K., Popova L.I., Ho I.Y., Pauli N.T., Henry Dunand C.J., Taylor W.M., Lim S., Huang M., et al. 2015. Immune history profoundly affects broadly protective B cell responses to influenza. Sci. Transl. Med. 7:316ra192 10.1126/scitranslmed.aad0522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic G., van der Poel C.E., Kuraoka M., Schmidt A.G., Carroll M.C., Kelsoe G., and Harrison S.C.. 2019. Autoreactivity profiles of influenza hemagglutinin broadly neutralizing antibodies. Sci. Rep. 9:3492 10.1038/s41598-019-40175-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazs A.B., Ouyang Y., Hong C.M., Chen J., Nguyen S.M., Rao D.S., An D.S., and Baltimore D.. 2014. Vectored immunoprophylaxis protects humanized mice from mucosal HIV transmission. Nat. Med. 20:296–300. 10.1038/nm.3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandaranayake A.D., Correnti C., Ryu B.Y., Brault M., Strong R.K., and Rawlings D.J.. 2011. Daedalus: a robust, turnkey platform for rapid production of decigram quantities of active recombinant proteins in human cell lines using novel lentiviral vectors. Nucleic Acids Res. 39:e143 10.1093/nar/gkr706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbas C.F. III, Björling E., Chiodi F., Dunlop N., Cababa D., Jones T.M., Zebedee S.L., Persson M.A., Nara P.L., Norrby E., et al. 1992. Recombinant human Fab fragments neutralize human type 1 immunodeficiency virus in vitro. Proc. Natl. Acad. Sci. USA. 89:9339–9343. 10.1073/pnas.89.19.9339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeler J.A., and van Wyke Coelingh K.. 1989. Neutralization epitopes of the F glycoprotein of respiratory syncytial virus: effect of mutation upon fusion function. J. Virol. 63:2941–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton D.R., and Hangartner L.. 2016. Broadly Neutralizing Antibodies to HIV and Their Role in Vaccine Design. Annu. Rev. Immunol. 34:635–659. 10.1146/annurev-immunol-041015-055515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton D.R., Barbas C.F. III, Persson M.A., Koenig S., Chanock R.M., and Lerner R.A.. 1991. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc. Natl. Acad. Sci. USA. 88:10134–10137. 10.1073/pnas.88.22.10134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton D.R., Desrosiers R.C., Doms R.W., Koff W.C., Kwong P.D., Moore J.P., Nabel G.J., Sodroski J., Wilson I.A., and Wyatt R.T.. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5:233–236. 10.1038/ni0304-233 [DOI] [PubMed] [Google Scholar]
- Cohen Y.Z., and Dolin R.. 2013. Novel HIV vaccine strategies: overview and perspective. Ther. Adv. Vaccines. 1:99–112. 10.1177/2051013613494535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763. 10.1107/S0907444994003112 [DOI] [PubMed] [Google Scholar]
- Cooke M.P., Heath A.W., Shokat K.M., Zeng Y., Finkelman F.D., Linsley P.S., Howard M., and Goodnow C.C.. 1994. Immunoglobulin signal transduction guides the specificity of B cell-T cell interactions and is blocked in tolerant self-reactive B cells. J. Exp. Med. 179:425–438. 10.1084/jem.179.2.425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Voss J., Gamblin S.J., Codoni G., Macagno A., Jarrossay D., Vachieri S.G., Pinna D., Minola A., Vanzetta F., et al. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 333:850–856. 10.1126/science.1205669 [DOI] [PubMed] [Google Scholar]
- Cyster J.G., Hartley S.B., and Goodnow C.C.. 1994. Competition for follicular niches excludes self-reactive cells from the recirculating B-cell repertoire. Nature. 371:389–395. 10.1038/371389a0 [DOI] [PubMed] [Google Scholar]
- DeKosky B.J., Lungu O.I., Park D., Johnson E.L., Charab W., Chrysostomou C., Kuroda D., Ellington A.D., Ippolito G.C., Gray J.J., and Georgiou G.. 2016. Large-scale sequence and structural comparisons of human naive and antigen-experienced antibody repertoires. Proc. Natl. Acad. Sci. USA. 113:E2636–E2645. 10.1073/pnas.1525510113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W.L. 2002. Unraveling hot spots in binding interfaces: progress and challenges. Curr. Opin. Struct. Biol. 12:14–20. 10.1016/S0959-440X(02)00283-X [DOI] [PubMed] [Google Scholar]
- Dimitrov D.S. 2010. Therapeutic antibodies, vaccines and antibodyomes. MAbs. 2:347–356. 10.4161/mabs.2.3.11779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditse Z., Muenchhoff M., Adland E., Jooste P., Goulder P., Moore P.L., and Morris L.. 2018. HIV-1 Subtype C-Infected Children with Exceptional Neutralization Breadth Exhibit Polyclonal Responses Targeting Known Epitopes. J. Virol. 92:92 10.1128/JVI.00878-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenovic P., von Boehmer L., Escolano A., Jardine J., Freund N.T., Gitlin A.D., McGuire A.T., Kulp D.W., Oliveira T., Scharf L., et al. 2015. Immunization for HIV-1 Broadly Neutralizing Antibodies in Human Ig Knockin Mice. Cell. 161:1505–1515. 10.1016/j.cell.2015.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle-Cooper C., Hudson K.E., Cooper A.B., Ota T., Skog P., Dawson P.E., Zwick M.B., Schief W.R., Burton D.R., and Nemazee D.. 2013. Immune tolerance negatively regulates B cells in knock-in mice expressing broadly neutralizing HIV antibody 4E10. J. Immunol. 191:3186–3191. 10.4049/jimmunol.1301285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus C., Laursen N.S., Kwaks T., Zuijdgeest D., Khayat R., Ekiert D.C., Lee J.H., Metlagel Z., Bujny M.V., Jongeneelen M., et al. 2012. Highly conserved protective epitopes on influenza B viruses. Science. 337:1343–1348. 10.1126/science.1222908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus C., Ekiert D.C., and Wilson I.A.. 2013. Structure of a classical broadly neutralizing stem antibody in complex with a pandemic H2 influenza virus hemagglutinin. J. Virol. 87:7149–7154. 10.1128/JVI.02975-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert D.C., Bhabha G., Elsliger M.A., Friesen R.H., Jongeneelen M., Throsby M., Goudsmit J., and Wilson I.A.. 2009. Antibody recognition of a highly conserved influenza virus epitope. Science. 324:246–251. 10.1126/science.1171491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert D.C., Friesen R.H., Bhabha G., Kwaks T., Jongeneelen M., Yu W., Ophorst C., Cox F., Korse H.J., Brandenburg B., et al. 2011. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 333:843–850. 10.1126/science.1204839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., and Cowtan K.. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Escolano A., Steichen J.M., Dosenovic P., Kulp D.W., Golijanin J., Sok D., Freund N.T., Gitlin A.D., Oliveira T., Araki T., et al. 2016. Sequential Immunization Elicits Broadly Neutralizing Anti-HIV-1 Antibodies in Ig Knockin Mice. Cell. 166:1445–1458.e12. 10.1016/j.cell.2016.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher D.A., and Basten A.. 1994. Reduced life span of anergic self-reactive B cells in a double-transgenic model. J. Exp. Med. 179:125–134. 10.1084/jem.179.1.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam R., Nishimura Y., Pegu A., Nason M.C., Klein F., Gazumyan A., Golijanin J., Buckler-White A., Sadjadpour R., Wang K., et al. 2016. A single injection of anti-HIV-1 antibodies protects against repeated SHIV challenges. Nature. 533:105–109. 10.1038/nature17677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodnow C.C.J., Crosbie J., Adelstein S., Lavoie T.B., Smith-Gill S.J., Brink R.A., Pritchard-Briscoe H., Wotherspoon J.S., Loblay R.H., Raphael K., et al. 1988. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature. 334:676–682. 10.1038/334676a0 [DOI] [PubMed] [Google Scholar]
- Goodnow C.C., Vinuesa C.G., Randall K.L., Mackay F., and Brink R.. 2010. Control systems and decision making for antibody production. Nat. Immunol. 11:681–688. 10.1038/ni.1900 [DOI] [PubMed] [Google Scholar]
- Goodwin E., Gilman M.S.A., Wrapp D., Chen M., Ngwuta J.O., Moin S.M., Bai P., Sivasubramanian A., Connor R.I., Wright P.F., et al. 2018. Infants Infected with Respiratory Syncytial Virus Generate Potent Neutralizing Antibodies that Lack Somatic Hypermutation. Immunity. 48:339–349.e5. 10.1016/j.immuni.2018.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J., Soto C., Yang M.M., Davenport T.M., Guttman M., Bailer R.T., Chambers M., Chuang G.Y., DeKosky B.J., Doria-Rose N.A., et al. NISC Comparative Sequencing Program . 2016. Structures of HIV-1 Env V1V2 with broadly neutralizing antibodies reveal commonalities that enable vaccine design. Nat. Struct. Mol. Biol. 23:81–90. 10.1038/nsmb.3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruell H., Bournazos S., Ravetch J.V., Ploss A., Nussenzweig M.C., and Pietzsch J.. 2013. Antibody and antiretroviral preexposure prophylaxis prevent cervicovaginal HIV-1 infection in a transgenic mouse model. J. Virol. 87:8535–8544. 10.1128/JVI.00868-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes B.F., Fleming J., St Clair E.W., Katinger H., Stiegler G., Kunert R., Robinson J., Scearce R.M., Plonk K., Staats H.F., et al. 2005. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 308:1906–1908. 10.1126/science.1111781 [DOI] [PubMed] [Google Scholar]
- Haynes B.F., Kelsoe G., Harrison S.C., and Kepler T.B.. 2012. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat. Biotechnol. 30:423–433. 10.1038/nbt.2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessell A.J., Rakasz E.G., Poignard P., Hangartner L., Landucci G., Forthal D.N., Koff W.C., Watkins D.I., and Burton D.R.. 2009. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 5:e1000433 10.1371/journal.ppat.1000433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmeyer T., Schmelz S., Degiacomi M.T., Dal Peraro M., Daneschdar M., Scrima A., van den Heuvel J., Heinz D.W., and Kolmar H.. 2013. Arranged sevenfold: structural insights into the C-terminal oligomerization domain of human C4b-binding protein. J. Mol. Biol. 425:1302–1317. 10.1016/j.jmb.2012.12.017 [DOI] [PubMed] [Google Scholar]
- Hoot S., McGuire A.T., Cohen K.W., Strong R.K., Hangartner L., Klein F., Diskin R., Scheid J.F., Sather D.N., Burton D.R., and Stamatatos L.. 2013. Recombinant HIV envelope proteins fail to engage germline versions of anti-CD4bs bNAbs. PLoS Pathog. 9:e1003106 10.1371/journal.ppat.1003106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Ofek G., Laub L., Louder M.K., Doria-Rose N.A., Longo N.S., Imamichi H., Bailer R.T., Chakrabarti B., Sharma S.K., et al. 2012. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 491:406–412. 10.1038/nature11544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Kang B.H., Pancera M., Lee J.H., Tong T., Feng Y., Imamichi H., Georgiev I.S., Chuang G.Y., Druz A., et al. 2014. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature. 515:138–142. 10.1038/nature13601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J., Julien J.P., Menis S., Ota T., Kalyuzhniy O., McGuire A., Sok D., Huang P.S., MacPherson S., Jones M., et al. 2013. Rational HIV immunogen design to target specific germline B cell receptors. Science. 340:711–716. 10.1126/science.1234150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J.G., Ota T., Sok D., Pauthner M., Kulp D.W., Kalyuzhniy O., Skog P.D., Thinnes T.C., Bhullar D., Briney B., et al. 2015. HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science. 349:156–161. 10.1126/science.aac5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J.G., Kulp D.W., Havenar-Daughton C., Sarkar A., Briney B., Sok D., Sesterhenn F., Ereño-Orbea J., Kalyuzhniy O., Deresa I., et al. 2016. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science. 351:1458–1463. 10.1126/science.aad9195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S., Oliver C., Prince G.A., Hemming V.G., Pfarr D.S., Wang S.C., Dormitzer M., O’Grady J., Koenig S., Tamura J.K., et al. 1997. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J. Infect. Dis. 176:1215–1224. 10.1086/514115 [DOI] [PubMed] [Google Scholar]
- Karron R.A., Buonagurio D.A., Georgiu A.F., Whitehead S.S., Adamus J.E., Clements-Mann M.L., Harris D.O., Randolph V.B., Udem S.A., Murphy B.R., and Sidhu M.S.. 1997. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc. Natl. Acad. Sci. USA. 94:13961–13966. 10.1073/pnas.94.25.13961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap A.K., Steel J., Oner A.F., Dillon M.A., Swale R.E., Wall K.M., Perry K.J., Faynboym A., Ilhan M., Horowitz M., et al. 2008. Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc. Natl. Acad. Sci. USA. 105:5986–5991. 10.1073/pnas.0801367105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano M., Moriyama S., Ando Y., Hikida M., Mori Y., Kurosaki T., and Okada T.. 2011. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity. 34:961–972. 10.1016/j.immuni.2011.03.025 [DOI] [PubMed] [Google Scholar]
- Klein F., Diskin R., Scheid J.F., Gaebler C., Mouquet H., Georgiev I.S., Pancera M., Zhou T., Incesu R.B., Fu B.Z., et al. 2013a Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 153:126–138. 10.1016/j.cell.2013.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F., Mouquet H., Dosenovic P., Scheid J.F., Scharf L., and Nussenzweig M.C.. 2013b Antibodies in HIV-1 vaccine development and therapy. Science. 341:1199–1204. 10.1126/science.1241144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinman N.R. 1996. The “clonal selection hypothesis” and current concepts of B cell tolerance. Immunity. 5:189–195. 10.1016/S1074-7613(00)80314-3 [DOI] [PubMed] [Google Scholar]
- Kurosaki T., Aiba Y., Kometani K., Moriyama S., and Takahashi Y.. 2010. Unique properties of memory B cells of different isotypes. Immunol. Rev. 237:104–116. 10.1111/j.1600-065X.2010.00939.x [DOI] [PubMed] [Google Scholar]
- Kwong P.D., and Mascola J.R.. 2018. HIV-1 Vaccines Based on Antibody Identification, B Cell Ontogeny, and Epitope Structure. Immunity. 48:855–871. 10.1016/j.immuni.2018.04.029 [DOI] [PubMed] [Google Scholar]
- Kwong P.D., Mascola J.R., and Nabel G.J.. 2013. Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nat. Rev. Immunol. 13:693–701. 10.1038/nri3516 [DOI] [PubMed] [Google Scholar]
- Landais E., Huang X., Havenar-Daughton C., Murrell B., Price M.A., Wickramasinghe L., Ramos A., Bian C.B., Simek M., Allen S., et al. 2016. Broadly Neutralizing Antibody Responses in a Large Longitudinal Sub-Saharan HIV Primary Infection Cohort. PLoS Pathog. 12:e1005369 10.1371/journal.ppat.1005369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang S., Xie J., Zhu X., Wu N.C., Lerner R.A., and Wilson I.A.. 2017. Antibody 27F3 Broadly Targets Influenza A Group 1 and 2 Hemagglutinins through a Further Variation in VH1-69 Antibody Orientation on the HA Stem. Cell Reports. 20:2935–2943. 10.1016/j.celrep.2017.08.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H.X., Lynch R., Zhou T., Gao F., Alam S.M., Boyd S.D., Fire A.Z., Roskin K.M., Schramm C.A., Zhang Z., et al. NISC Comparative Sequencing Program . 2013. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 496:469–476. 10.1038/nature12053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Ghneim K., Sok D., Bosche W.J., Li Y., Chipriano E., Berkemeier B., Oswald K., Borducchi E., Cabral C., et al. 2016. Antibody-mediated protection against SHIV challenge includes systemic clearance of distal virus. Science. 353:1045–1049. 10.1126/science.aag0491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magro M., Mas V., Chappell K., Vázquez M., Cano O., Luque D., Terrón M.C., Melero J.A., and Palomo C.. 2012. Neutralizing antibodies against the preactive form of respiratory syncytial virus fusion protein offer unique possibilities for clinical intervention. Proc. Natl. Acad. Sci. USA. 109:3089–3094. 10.1073/pnas.1115941109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola J.R., and Haynes B.F.. 2013. HIV-1 neutralizing antibodies: understanding nature’s pathways. Immunol. Rev. 254:225–244. 10.1111/imr.12075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola J.R., Lewis M.G., Stiegler G., Harris D., VanCott T.C., Hayes D., Louder M.K., Brown C.R., Sapan C.V., Frankel S.S., et al. 1999. Protection of Macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J. Virol. 73:4009–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire A.T., Hoot S., Dreyer A.M., Lippy A., Stuart A., Cohen K.W., Jardine J., Menis S., Scheid J.F., West A.P., et al. 2013. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J. Exp. Med. 210:655–663. 10.1084/jem.20122824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire A.T., Dreyer A.M., Carbonetti S., Lippy A., Glenn J., Scheid J.F., Mouquet H., and Stamatatos L.. 2014a HIV antibodies. Antigen modification regulates competition of broad and narrow neutralizing HIV antibodies. Science. 346:1380–1383. 10.1126/science.1259206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire A.T., Glenn J.A., Lippy A., and Stamatatos L.. 2014b Diverse recombinant HIV-1 Envs fail to activate B cells expressing the germline B cell receptors of the broadly neutralizing anti-HIV-1 antibodies PG9 and 447-52D. J. Virol. 88:2645–2657. 10.1128/JVI.03228-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire A.T., Gray M.D., Dosenovic P., Gitlin A.D., Freund N.T., Petersen J., Correnti C., Johnsen W., Kegel R., Stuart A.B., et al. 2016. Specifically modified Env immunogens activate B-cell precursors of broadly neutralizing HIV-1 antibodies in transgenic mice. Nat. Commun. 7:10618 10.1038/ncomms10618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrell K.T., Benschop R.J., Gauld S.B., Aviszus K., Decote-Ricardo D., Wysocki L.J., and Cambier J.C.. 2006. Identification of anergic B cells within a wild-type repertoire. Immunity. 25:953–962. 10.1016/j.immuni.2006.10.017 [DOI] [PubMed] [Google Scholar]
- Moldt B., Rakasz E.G., Schultz N., Chan-Hui P.Y., Swiderek K., Weisgrau K.L., Piaskowski S.M., Bergman Z., Watkins D.I., Poignard P., and Burton D.R.. 2012. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc. Natl. Acad. Sci. USA. 109:18921–18925. 10.1073/pnas.1214785109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet H., Scharf L., Euler Z., Liu Y., Eden C., Scheid J.F., Halper-Stromberg A., Gnanapragasam P.N., Spencer D.I., Seaman M.S., et al. 2012. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc. Natl. Acad. Sci. USA. 109:E3268–E3277. 10.1073/pnas.1217207109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemazee D. 2006. Receptor editing in lymphocyte development and central tolerance. Nat. Rev. Immunol. 6:728–740. 10.1038/nri1939 [DOI] [PubMed] [Google Scholar]
- Nemazee D. 2017. Mechanisms of central tolerance for B cells. Nat. Rev. Immunol. 17:281–294. 10.1038/nri.2017.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngwuta J.O., Chen M., Modjarrad K., Joyce M.G., Kanekiyo M., Kumar A., Yassine H.M., Moin S.M., Killikelly A.M., Chuang G.Y., et al. 2015. Prefusion F-specific antibodies determine the magnitude of RSV neutralizing activity in human sera. Sci. Transl. Med. 7:309ra162 10.1126/scitranslmed.aac4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogun S.A., Dumon-Seignovert L., Marchand J.B., Holder A.A., and Hill F.. 2008. The oligomerization domain of C4-binding protein (C4bp) acts as an adjuvant, and the fusion protein comprised of the 19-kilodalton merozoite surface protein 1 fused with the murine C4bp domain protects mice against malaria. Infect. Immun. 76:3817–3823. 10.1128/IAI.01369-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y., Isegawa Y., Sasao F., and Ueda S.. 1993. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J. Virol. 67:2552–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T., Doyle-Cooper C., Cooper A.B., Doores K.J., Aoki-Ota M., Le K., Schief W.R., Wyatt R.T., Burton D.R., and Nemazee D.. 2013. B cells from knock-in mice expressing broadly neutralizing HIV antibody b12 carry an innocuous B cell receptor responsive to HIV vaccine candidates. J. Immunol. 191:3179–3185. 10.4049/jimmunol.1301283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., and Minor W.. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326. 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- Pancera M., McLellan J.S., Wu X., Zhu J., Changela A., Schmidt S.D., Yang Y., Zhou T., Phogat S., Mascola J.R., and Kwong P.D.. 2010. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J. Virol. 84:8098–8110. 10.1128/JVI.00966-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M., Zhou T., Druz A., Georgiev I.S., Soto C., Gorman J., Huang J., Acharya P., Chuang G.Y., Ofek G., et al. 2014. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 514:455–461. 10.1038/nature13808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaleo G., and Koup R.A.. 2004. Correlates of immune protection in HIV-1 infection: what we know, what we don’t know, what we should know. Nat. Med. 10:806–810. 10.1038/nm0804-806 [DOI] [PubMed] [Google Scholar]
- Parren P.W., Marx P.A., Hessell A.J., Luckay A., Harouse J., Cheng-Mayer C., Moore J.P., and Burton D.R.. 2001. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 75:8340–8347. 10.1128/JVI.75.17.8340-8347.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietzsch J., Gruell H., Bournazos S., Donovan B.M., Klein F., Diskin R., Seaman M.S., Bjorkman P.J., Ravetch J.V., Ploss A., and Nussenzweig M.C.. 2012. A mouse model for HIV-1 entry. Proc. Natl. Acad. Sci. USA. 109:15859–15864. 10.1073/pnas.1213409109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappuoli R., Bottomley M.J., D’Oro U., Finco O., and De Gregorio E.. 2016. Reverse vaccinology 2.0: Human immunology instructs vaccine antigen design. J. Exp. Med. 213:469–481. 10.1084/jem.20151960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees W., Bender J., Teague T.K., Kedl R.M., Crawford F., Marrack P., and Kappler J.. 1999. An inverse relationship between T cell receptor affinity and antigen dose during CD4(+) T cell responses in vivo and in vitro. Proc. Natl. Acad. Sci. USA. 96:9781–9786. 10.1073/pnas.96.17.9781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Bozzacco L., Keeffe J.R., Khouri R., Olsen P.C., Gazumyan A., Schaefer-Babajew D., Avila-Rios S., Nogueira L., Patel R., et al. 2017. Recurrent Potent Human Neutralizing Antibodies to Zika Virus in Brazil and Mexico. Cell. 169:597–609.e11. 10.1016/j.cell.2017.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusert P., Kouyos R.D., Kadelka C., Ebner H., Schanz M., Huber M., Braun D.L., Hozé N., Scherrer A., Magnus C., et al. Swiss HIV Cohort Study . 2016. Determinants of HIV-1 broadly neutralizing antibody induction. Nat. Med. 22:1260–1267. 10.1038/nm.4187 [DOI] [PubMed] [Google Scholar]
- Sabouri Z., Perotti S., Spierings E., Humburg P., Yabas M., Bergmann H., Horikawa K., Roots C., Lambe S., Young C., et al. 2016. IgD attenuates the IgM-induced anergy response in transitional and mature B cells. Nat. Commun. 7:13381 10.1038/ncomms13381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan C.N., Pantophlet R., Wormald M.R., Ollmann Saphire E., Stanfield R., Wilson I.A., Katinger H., Dwek R.A., Rudd P.M., and Burton D.R.. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1-->2 mannose residues on the outer face of gp120. J. Virol. 76:7306–7321. 10.1128/JVI.76.14.7306-7321.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid J.F., Mouquet H., Feldhahn N., Seaman M.S., Velinzon K., Pietzsch J., Ott R.G., Anthony R.M., Zebroski H., Hurley A., et al. 2009. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 458:636–640. 10.1038/nature07930 [DOI] [PubMed] [Google Scholar]
- Scheid J.F., Mouquet H., Ueberheide B., Diskin R., Klein F., Oliveira T.Y., Pietzsch J., Fenyo D., Abadir A., Velinzon K., et al. 2011. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 333:1633–1637. 10.1126/science.1207227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffner T., Sattentau Q.J., and Dorrell L.. 2013. Development of prophylactic vaccines against HIV-1. Retrovirology. 10:72 10.1186/1742-4690-10-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S.C., Ohmen J., Fosdick L., Gladstone B., Guo J., Ametani A., Sercarz E.E., and Deng H.. 2000. Cutting edge: introduction of an endopeptidase cleavage motif into a determinant flanking region of hen egg lysozyme results in enhanced T cell determinant display. J. Immunol. 165:20–23. 10.4049/jimmunol.165.1.20 [DOI] [PubMed] [Google Scholar]
- Shibata R., Igarashi T., Haigwood N., Buckler-White A., Ogert R., Ross W., Willey R., Cho M.W., and Martin M.A.. 1999. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat. Med. 5:204–210. 10.1038/5568 [DOI] [PubMed] [Google Scholar]
- Shingai M., Donau O.K., Plishka R.J., Buckler-White A., Mascola J.R., Nabel G.J., Nason M.C., Montefiori D., Moldt B., Poignard P., et al. 2014. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J. Exp. Med. 211:2061–2074. 10.1084/jem.20132494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R.W. 2009. Antibody affinity optimization using yeast cell surface display. Methods Mol. Biol. 504:351–383. 10.1007/978-1-60327-569-9_20 [DOI] [PubMed] [Google Scholar]
- Snijder J., Ortego M.S., Weidle C., Stuart A.B., Gray M.D., McElrath M.J., Pancera M., Veesler D., and McGuire A.T.. 2018. An Antibody Targeting the Fusion Machinery Neutralizes Dual-Tropic Infection and Defines a Site of Vulnerability on Epstein-Barr Virus. Immunity. 48:799–811.e9. 10.1016/j.immuni.2018.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanier J.A., Frederick D.R., Taylor J.J., Heffernan J.R., Kotov D.I., Martinov T., Osum K.C., Ruggiero J.L., Rust B.J., Landry S.J., et al. 2016. Efficient generation of monoclonal antibodies against peptide in the context of MHCII using magnetic enrichment. Nat. Commun. 7:11804 10.1038/ncomms11804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steichen J.M., Kulp D.W., Tokatlian T., Escolano A., Dosenovic P., Stanfield R.L., McCoy L.E., Ozorowski G., Hu X., Kalyuzhniy O., et al. 2016. HIV Vaccine Design to Target Germline Precursors of Glycan-Dependent Broadly Neutralizing Antibodies. Immunity. 45:483–496. 10.1016/j.immuni.2016.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J., Hwang W.C., Perez S., Wei G., Aird D., Chen L.M., Santelli E., Stec B., Cadwell G., Ali M., et al. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 16:265–273. 10.1038/nsmb.1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.J., Martinez R.J., Titcombe P.J., Barsness L.O., Thomas S.R., Zhang N., Katzman S.D., Jenkins M.K., and Mueller D.L.. 2012. Deletion and anergy of polyclonal B cells specific for ubiquitous membrane-bound self-antigen. J. Exp. Med. 209:2065–2077. 10.1084/jem.20112272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.J., Pape K.A., Steach H.R., and Jenkins M.K.. 2015. Humoral immunity. Apoptosis and antigen affinity limit effector cell differentiation of a single naïve B cell. Science. 347:784–787. 10.1126/science.aaa1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throsby M., van den Brink E., Jongeneelen M., Poon L.L., Alard P., Cornelissen L., Bakker A., Cox F., van Deventer E., Guan Y., et al. 2008. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One. 3:e3942 10.1371/journal.pone.0003942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., and Wardemann H.. 2008. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 329:112–124. 10.1016/j.jim.2007.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T., Busse C.E., and Wardemann H.. 2009. Cloning and expression of murine Ig genes from single B cells. J. Immunol. Methods. 350:183–193. 10.1016/j.jim.2009.08.009 [DOI] [PubMed] [Google Scholar]
- Verkoczy L., Diaz M., Holl T.M., Ouyang Y.B., Bouton-Verville H., Alam S.M., Liao H.X., Kelsoe G., and Haynes B.F.. 2010. Autoreactivity in an HIV-1 broadly reactive neutralizing antibody variable region heavy chain induces immunologic tolerance. Proc. Natl. Acad. Sci. USA. 107:181–186. 10.1073/pnas.0912914107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora G.D., Schwickert T.A., Fooksman D.R., Kamphorst A.O., Meyer-Hermann M., Dustin M.L., and Nussenzweig M.C.. 2010. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 143:592–605. 10.1016/j.cell.2010.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuesa C.G., Linterman M.A., Goodnow C.C., and Randall K.L.. 2010. T cells and follicular dendritic cells in germinal center B-cell formation and selection. Immunol. Rev. 237:72–89. 10.1111/j.1600-065X.2010.00937.x [DOI] [PubMed] [Google Scholar]
- Walker L.M., Phogat S.K., Chan-Hui P.Y., Wagner D., Phung P., Goss J.L., Wrin T., Simek M.D., Fling S., Mitcham J.L., et al. Protocol G Principal Investigators . 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 326:285–289. 10.1126/science.1178746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.M., Huber M., Doores K.J., Falkowska E., Pejchal R., Julien J.P., Wang S.K., Ramos A., Chan-Hui P.Y., Moyle M., et al. Protocol G Principal Investigators . 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 477:466–470. 10.1038/nature10373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.P. Jr., Scharf L., Scheid J.F., Klein F., Bjorkman P.J., and Nussenzweig M.C.. 2014. Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell. 156:633–648. 10.1016/j.cell.2014.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Yang Z.Y., Li Y., Hogerkorp C.M., Schief W.R., Seaman M.S., Zhou T., Schmidt S.D., Wu L., Xu L., et al. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 329:856–861. 10.1126/science.1187659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Chen W., Feng Y., Zhu Z., Prabakaran P., Wang Y., Zhang M.Y., Longo N.S., and Dimitrov D.S.. 2009. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem. Biophys. Res. Commun. 390:404–409. 10.1016/j.bbrc.2009.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Verkoczy L., Wiehe K., Munir Alam S., Nicely N.I., Santra S., Bradley T., Pemble C.W. IV, Zhang J., Gao F., et al. 2016. Initiation of immune tolerance-controlled HIV gp41 neutralizing B cell lineages. Sci. Transl. Med. 8:336ra62 10.1126/scitranslmed.aaf0618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Xu L., Dey B., Hessell A.J., Van Ryk D., Xiang S.H., Yang X., Zhang M.Y., Zwick M.B., Arthos J., et al. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 445:732–737. 10.1038/nature05580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Georgiev I., Wu X., Yang Z.Y., Dai K., Finzi A., Kwon Y.D., Scheid J.F., Shi W., Xu L., et al. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 329:811–817. 10.1126/science.1192819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Lynch R.M., Chen L., Acharya P., Wu X., Doria-Rose N.A., Joyce M.G., Lingwood D., Soto C., Bailer R.T., et al. NISC Comparative Sequencing Program . 2015. Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors. Cell. 161:1280–1292. 10.1016/j.cell.2015.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.