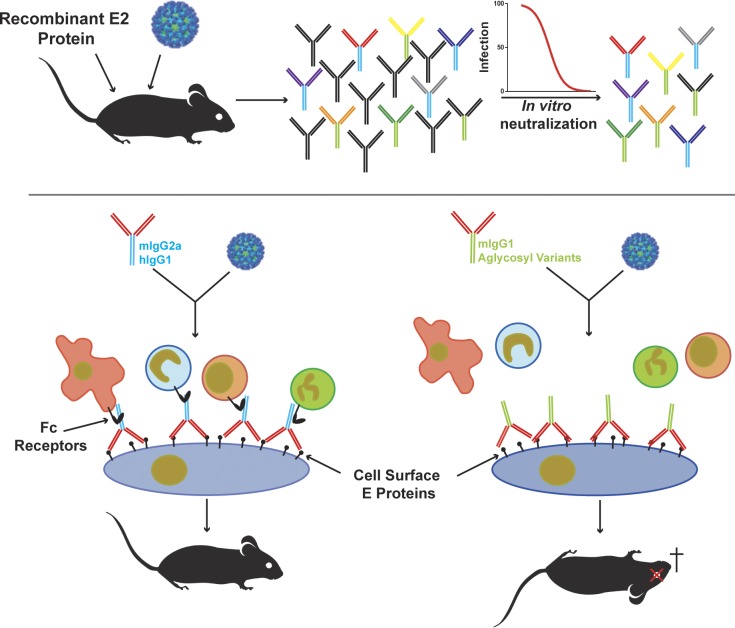
The authors generate and characterize a panel of highly neutralizing antibodies against Mayaro virus, an emerging alphavirus. They find that neutralization activity alone is not sufficient for antibodies to protect mice from disease and that Fc effector functions also are required.
Abstract
Despite causing outbreaks of fever and arthritis in multiple countries, no countermeasures exist against Mayaro virus (MAYV), an emerging mosquito-transmitted alphavirus. We generated 18 neutralizing mAbs against MAYV, 11 of which had “elite” activity that inhibited infection with EC50 values of <10 ng/ml. Antibodies with the greatest inhibitory capacity in cell culture mapped to epitopes near the fusion peptide of E1 and in domain B of the E2 glycoproteins. Unexpectedly, many of the elite neutralizing mAbs failed to prevent MAYV infection and disease in vivo. Instead, the most protective mAbs bound viral antigen on the cell surface with high avidity and promoted specific Fc effector functions, including phagocytosis by neutrophils and monocytes. In subclass switching studies, murine IgG2a and humanized IgG1 mAb variants controlled infection better than murine IgG1 and humanized IgG1-N297Q variants. An optimally protective antibody response to MAYV and possibly other alphaviruses may require tandem virus neutralization by the Fab moiety and effector functions of the Fc region.
Graphical Abstract
Introduction
Mayaro virus (MAYV) is a mosquito-transmitted arthritogenic alphavirus in the Togaviridae family of positive-stranded RNA viruses. MAYV was first described in 1954 in Trinidad (Causey and Maroja, 1957) and now circulates in the Caribbean Islands and South America (Causey and Maroja, 1957; Pinheiro et al., 1981; Azevedo et al., 2009). MAYV infection causes an acute febrile illness that initially is indistinguishable from other arboviruses, including chikungunya virus (CHIKV), dengue, and Zika viruses. A study of a MAYV outbreak in Brazil showed a high incidence of severe arthralgia (55%) and myalgia (49%), which can persist for months to years (Mourão et al., 2012). There are no vaccines or therapeutics available for the prevention or treatment of MAYV infection.
MAYV is transmitted principally by Haemagogus species mosquitoes among primates in a sylvatic cycle, with intermittent spillover to human populations. However, there is concern that even single mutations in MAYV could lead to changes in vector competence, as occurred with CHIKV in La Reunion Island in 2006. In that case, CHIKV acquired the ability to infect Aedes albopictus mosquitoes more efficiently, which resulted in an urban transmission cycle (Tsetsarkin et al., 2007). Aedes mosquitoes can be infected by MAYV, although the levels of virus in blood required for transmission from viremic humans are considered insufficient for epidemic transmission (Brustolin et al., 2018). In comparison, four different Anopheles mosquitoes can transmit MAYV (Brustolin et al., 2018). As these species are native to distinct geographic regions (Africa, Asia, and North America), they could facilitate the global spread of MAYV.
The MAYV genome encodes for four nonstructural proteins (nsp1–nsp4) and five structural proteins (capsid, E3, E2, 6K, and E1). Alphavirus envelope glycoproteins associate in the endoplasmic reticulum and form a heterotrimer comprised of the E3, E2, and E1 proteins (Carleton et al., 1997). E3 is cleaved by furin-like proteases during the maturation process in the trans-Golgi network (Heidner et al., 1996). The mature alphavirus virion is ∼700 Å in diameter and contains a lipid bilayer with 240 E2–E1 heterodimers assembled into 80 trimeric spikes with T = 4 icosahedral symmetry (Paredes et al., 1993; Cheng et al., 1995; Kostyuchenko et al., 2011) and a nucleocapsid containing a single copy of genomic RNA. As the E1 and E2 glycoproteins are displayed prominently on the viral spike, they are targets of antibody responses.
Although mAbs that specifically bind the envelope glycoproteins of MAYV have not yet been described, others generated in the context of immunization with CHIKV antigens have shown cross-reactivity and cross-neutralizing activity with MAYV (Fox et al., 2015). In the context of human polyclonal antibody responses to MAYV, antibody responses to E1 and E2 proteins have been detected, with greater cross-reactivity among those recognizing E2 (Smith et al., 2018). However, these studies have not fully elucidated the epitopes and mechanisms by which antibodies against MAYV are protective in vivo.
Here, we produced and characterized a panel of 18 neutralizing anti-MAYV mAbs after immunizing mice with infectious virus and recombinant MAYV E2 protein. 11 of these anti-MAYV mAbs showed exceptional potency in cell culture with half-maximal effective inhibitory concentration (EC50) of <10 ng/ml. All but two of these antibodies were capable of neutralizing 11 different strains of MAYV that we tested. Several anti-MAYV mAbs cross-neutralized related arthritogenic alphaviruses, including CHIKV, Una virus (UNAV), Ross River virus (RRV), and O’nyong’nyong virus (ONNV), whereas others were MAYV specific. Our neutralizing anti-MAYV mAbs generally blocked infection at a postattachment step, preventing fusion of virus–host membranes and impairing viral egress. Epitope mapping analyses revealed that 16 of the mAbs bound epitopes in the B domain of the E2 glycoprotein, with two others recognizing a site in the E1 glycoprotein proximal to the fusion loop (FL) peptide. A subset of the strongly neutralizing mAbs protected mice from lethal MAYV challenge and virus-induced joint swelling and dissemination. Despite the potent neutralizing activity observed in cell culture, protection required fragment crystallizable (Fc) effector functions, since isotype-switched or aglycosyl variants with less or no capacity to interact with the complement component C1q or activating Fc-γ receptors (FcγR) lost protective activity in vivo. Our results in mice suggest that strategies to induce highly protective antibodies against MAYV may require designs that optimize both neutralizing activity and Fc effector functions.
Results
Generation of anti-MAYV mAbs
We inoculated BALB/c mice with MAYV strain CH and boosted twice with recombinant, bacterially produced MAYV E2 ectodomain (amino acids 1–340) in Freund’s incomplete adjuvant and then one additional time with MAYV-CH (see Materials and methods), as we did to generate anti-CHIKV mAbs (Pal et al., 2013). Serum had neutralization titers in cell culture of >1/10,000 against MAYV-CH. After a final intravenous boost with MAYV-CH, we performed splenocyte-myeloma fusions from four mice. Subsequently, 151 anti-MAYV mAbs were subcloned and isolated. Supernatant from 18 of these hybridomas contained antibodies that reduced MAYV infection by >80% in Vero cells. These mAbs were isotyped (Table 1) and purified by protein A affinity chromatography.
Table 1. Anti-MAYV mAbs.
| Antibody | Isotype | KD, equilibrium | CH | BeH407 | FSB0311 | IQU2950 | OBS2209 | Uruma | BeH343155 | BeH506151 | TRVL15537 | BeH473130 | BeH428890 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAY-115 | IgG2a | n.b. | 2 (1–4) | 2 (1–3) | 5 (3–9) | 7 (4–14) | 9 (5–17) | 1 (1–3) | 166 (93–102) | 33 (14–67) | 32 (17–61) | 64 (36–115) | 92 (54–146) |
| MAY-116 | IgG1 | 20 ± 2 | 11 (9–16) | 37 (19–76) | 54 (23–105) | 11 (5–15) | 3 (1–5) | 11 (11–15) | 15 (8–32) | 65 (32–135) | 10 (6–20) | 23 (11–49) | 19 (4–96) |
| MAY-117 | IgG1 | 11 ± 3 | 5 (3–9) | 4 (2–9) | 5 (3–11) | 28 (16–51) | 3 (2–6) | 57 (26–113) | 8 (3–18) | 18 (10–33) | 60 (27–120) | 9 (6–17) | 39 (18–85) |
| MAY-118 | IgG1 | 13 ± 4 | 2 (1–3) | 6 (2–13) | 3 (2–5) | 24 (13–45) | 33 (16–71) | 48 (29–95) | 16 (7–34) | 8 (4–15) | 12 (10–33) | 36 (17–69) | 11 (7–18) |
| MAY-119 | IgG1 | 14 ± 4 | 2 (1–4) | 16 (9–31) | 1 (1–3) | 25 (14–55) | 2 (1–4) | 19 (10–36) | 8 (5–17) | 20 (13–39) | 50 (28–102) | 23 (10–46) | 25 (13–47) |
| MAY-120 | IgG1 | 18 ± 1 | 3 (1–6) | 10 (5–20) | 6 (3–11) | 5 (2–10) | 1 (1–2) | 41 (25–72) | 3 (1–7) | 68 (32–140) | 26 (12–55) | 13 (5–28) | 22 (14–36) |
| MAY-121 | IgG1 | 47 ± 13 | 4 (2–6) | 20 (11–36) | 7 (4–11) | 33 (23–49) | 3 (2–8) | 5 (4–7) | 280 (175–450) | 34 (23–49) | 34 (15–83) | 28 (19–50) | 45 (22–92) |
| MAY-122 | IgG2a | 4 ± 1 | 2 (1–6) | 3 (2–6) | 6 (3–10) | 11 (4–32) | 6 (3–13) | 5 (4–8) | 10 (6–16) | 7 (6–8) | 7 (3–13) | 5 (3–11) | 6 (4–10) |
| MAY-125 | IgG2a | 58 ± 19 | 5 (3–10) | 22 (14–42) | 5 (2–12) | 10 (6–19) | 10 (5–21) | 61 (28–122) | 30 (21–43) | 9 (3–17) | 103 (52–213) | 25 (17–58) | 35 (24–53) |
| MAY-130 | IgG1 | 3 ± 1 | 1 (1–2) | 5 (3–10) | 1 (1–2) | 4 (1–10) | 3 (1–10) | 2 (1–4) | 2 (1–3) | 25 (14–41) | 41 (27–62) | 5 (2–13) | 3 (2–4) |
| MAY-131 | IgG2a | n.b. | 7 (3–12) | 8 (2–14) | 3 (1–5) | 14 (11–17) | 12 (4–29) | 22 (15–30) | 64 (22–104) | 42 (36–47) | 22 (13–32) | 61 (45–78) | 43 (32–55) |
| MAY-133 | IgG2a | 243 ± 64 | 38 (14–106) | 25 (14–44) | 34 (25–32) | 50 (28–91) | 3 (2–5) | >10,000 | 31 (22–46) | 42 (30–57) | 1,911 (940–4,429) | 30 (18–51) | 39 (21–75) |
| MAY-134 | IgG2a | 152 ± 69 | 4 (2–7) | 9 (4–13) | 27 (19–39) | 50 (37–90) | 3 (2–4) | 17 (12–23) | 33 (21–53) | 34 (27–44) | 26 (18–38) | 41 (27–62) | 44 (22–90) |
| MAY-139 | IgG2a | 22 ± 11 | 36 (17–77) | 246 (133–342) | 22 (8–65) | 38 (15–103) | 3 (1–15) | 62 (38–105) | 27 (17–42) | 26 (16–42) | 33 (8–159) | 27 (15–29) | 39 (21–75) |
| MAY-140 | IgG2a | 170 ± 29 | 1,315 (408–6,040) | 18 (11–30) | >10,000 | 4 (2–8) | >10,000 | 69 (19–345) | 5 (2–6) | 3,788 (732–6,403) | 3 (1–15) | >10,000 | 2 (1–3) |
| MAY-146 | IgG1 | 18 ± 5 | 30 (26–34) | 63 (55–72) | 42 (37–47) | 22 (14–30) | 47 (22–89) | 33 (19–345) | 74 (65–83) | 29 (24–36) | 57 (30–84) | 85 (64–106) | 73 (65–82) |
| MAY-147 | IgG1 | 38 ± 8 | 23 (15–30) | 16 (11–22) | 98 (58–169) | 233 (96–630) | 11 (4–27) | 18 (9–25) | 88 (37–229) | 108 (28–151) | 105 (43–269) | 41 (22–77) | 112 (71–178) |
| MAY-151 | IgG1 | 29 ± 7 | 66 (48–55) | 52 (47–57) | 6 (2–18) | 59 (29–87) | 52 (42–83) | 27 (20–35) | 7 (2–13) | 78 (66–91) | 21 (17–26) | 8 (4–14) | 5 (1–11) |
IgG subclasses, KD, equilibrium values, and neutralization values of the 18 inhibitory anti-MAYV mAbs. IgG subclasses were determined by ELISA. KD, equilibrium values (nanomoles) were measured by BLI using recombinant MAYV E2 protein. EC50 values (ng/ml) along with 95% confidence intervals (in parentheses) were calculated from FRNT. Data are the mean and 95% confidence of two experiments performed in triplicate. Bold font indicates antibodies with elite neutralizing activity (<10 ng/ml) against the indicated MAYV strain. n.b., nonbinding.
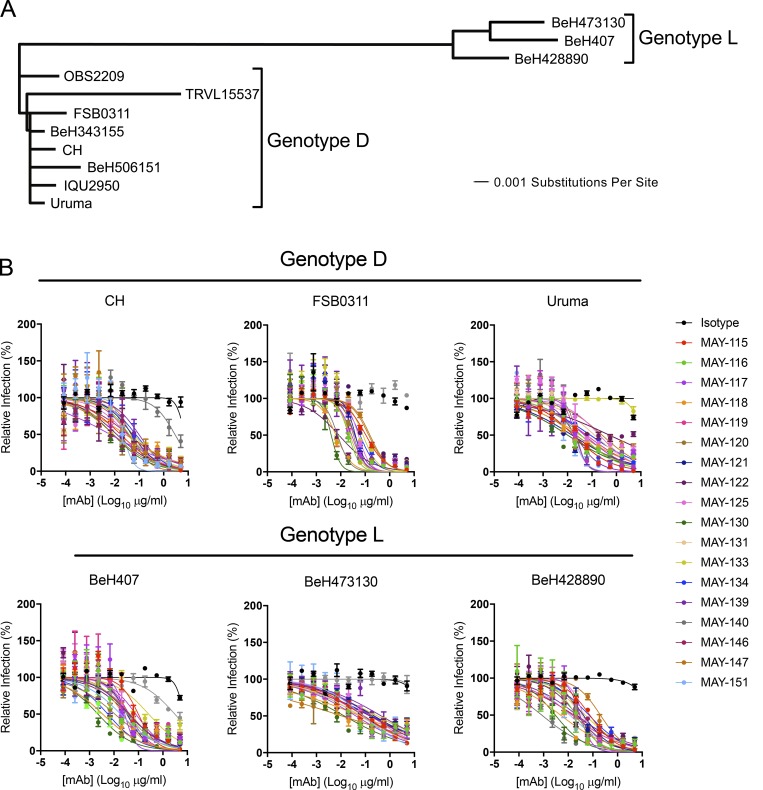
The purified mAbs were evaluated by focus reduction neutralization test (FRNT) for inhibition of infection of the immunizing MAYV-CH strain as well as 10 other MAYV strains (described in Powers et al., 2006) encompassing the predominant D and L genotypes (Figs. 1 A and S1), isolated between 1955 and 2002 (Table S1). These strains exhibit a high degree of amino acid sequence identity in the E1 (94–100%; Fig. S2) and E2 (96–100%; Fig. S3) glycoproteins. Only a single strain in the N genotype exists (Auguste et al., 2015), and it was not available to us for evaluation. 11 of the 18 mAbs potently inhibited infection of MAYV-CH (Table 1 and Fig. 1 B) and the majority neutralized a range of MAYV strains from the D and L genotypes. Only two mAbs showed a complete loss of neutralization activity against one or more MAYV strains (MAY-133: Uruma strain; and MAY-140: FSB0311, OBS6443, and BeH473130 strains; Table 1). For others, a >10-fold decrease in inhibitory activity against different MAYV strains compared with MAYV-CH was observed with 13 of the 18 mAbs. Only mAbs MAY-116, MAY-122, MAY-131, MAY-146, and MAY-151 maintained potency (<10-fold reduction in EC50 values compared with CH strain) across all tested MAYV strains (Table 1). We also performed neutralization assays with MAYV-BeH407 in C2C12 myoblast and embryonic fibroblast cells and observed similar inhibitory activity (Fig. S1). Because our vaccination strategy included boosting with recombinant E2 protein, we measured antibody affinity to E2 protein using biolayer interferometry (BLI; Tables 1 and S2). The KD, equilibrium (equilibrium dissociation constant) rate for the mAbs ranged from 3 to 243 nM, with eight demonstrating high-affinity monovalent binding of <20 nM. MAY-115 and MAY-131 had no detectable binding to E2, suggesting they recognized a different structural protein (i.e., E1) or a quaternary epitope on E2 displayed exclusively on the virion. MAY-122, likely the most consistently potent neutralizing antibody across all MAYV strains, had the second highest affinity, with a KD of 4 nM.
Figure 1.
Anti-MAYV mAbs neutralize strains from both D and L genotypes. (A) Viral strains were subjected to next-generation sequencing. A phylogenetic tree was generated using a Jukes–Cantor genetic distance model by aligning structural gene nucleotide sequences for each MAYV strain. (B) Serial dilutions of mAbs were incubated with 102 FFU of the indicated MAYV strain, representing either genotypes D (top) or L (bottom), before inoculation of Vero cells. Cells were overlaid with methylcellulose and incubated for 18 h. Viral foci were stained, counted, and plotted relative to a no-antibody control. Data are representative of two experiments performed in triplicate. Error bars represent SD within one experiment.
Breadth of alphavirus neutralization of anti-MAYV mAbs
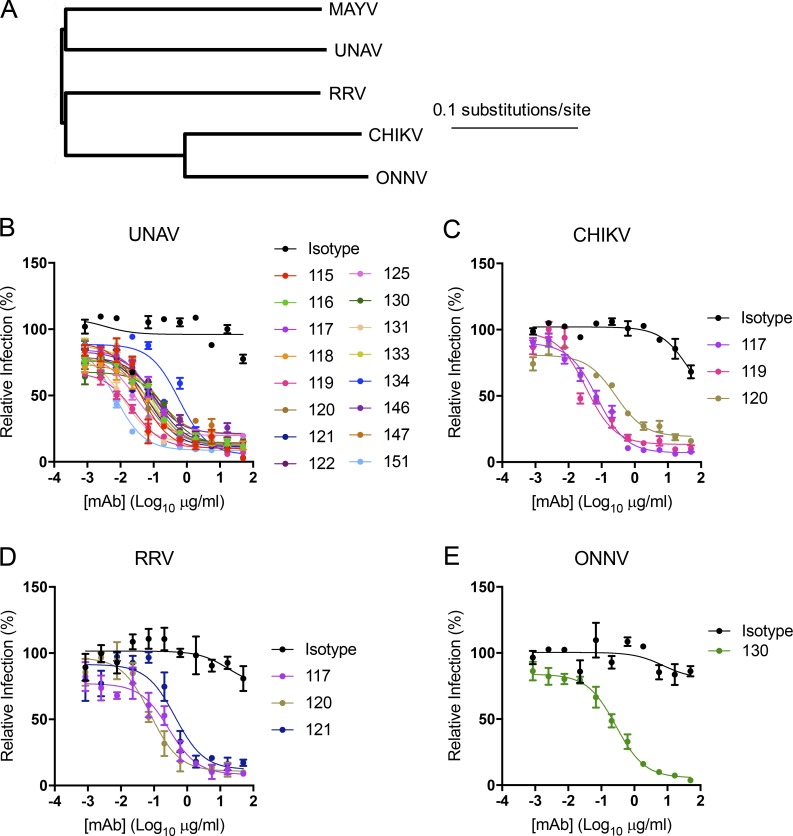
To assess whether the anti-MAYV mAbs inhibited related arthritogenic alphaviruses, we first assessed cross-reactive binding by flow cytometry to permeabilized cells infected with UNAV, CHIKV, RRV, and ONNV (Fig. 2 A). Of the 18 mAbs tested, 16, 10, 3, and 1 cross-reacted with UNAV, CHIKV, RRV, and ONNV, respectively (Table 2). We next tested the cross-reactive mAbs for their ability to cross-neutralize infection. All 16 mAbs that bound UNAV-infected cells neutralized this virus (Fig. 2 B). Similar results were observed with RRV and ONNV, with all mAbs that bound infected cells neutralizing the respective virus (Fig. 2, D and E). For CHIKV, only 3 of the 10 mAbs that bound infected cells substantially inhibited CHIKV infection in cells (Fig. 2 C). Although MAY-117 and MAY-120 neutralized UNAV, CHIKV, and RRV, none of our anti-MAYV mAbs inhibited infection of all of the arthritogenic alphaviruses that we tested (Fig. 2).
Figure 2.
Cross-reactivity and cross-neutralization of related arthritogenic alphaviruses. (A) Dendrogram showing the phylogenetic relationship of the structural genes of related alphaviruses used for cross-neutralization testing. (B–E) Neutralization assays were performed with anti-MAYV mAbs that bound to cells infected by UNAV (B), CHIKV (C), RRV (D), or ONNV (E). Serial dilutions of the indicated mAbs were incubated with 102 FFU of the indicated alphavirus before inoculation of Vero cells as described in Fig. 1. Data are representative of two experiments performed in triplicate. Error bars represent SD within one experiment.
Table 2. Cross-reactivity of anti-MAYV mAbs with other alphaviruses.
| Antibody | MAYV | UNAV | CHIKV | RRV | ONNV |
|---|---|---|---|---|---|
| MAY-115 | + | + | − | − | − |
| MAY-116 | + | + | + | − | − |
| MAY-117 | + | + | + | + | − |
| MAY-118 | + | + | + | − | − |
| MAY-119 | + | + | + | − | − |
| MAY-120 | + | + | + | + | − |
| MAY-121 | + | + | + | + | − |
| MAY-122 | + | + | − | − | − |
| MAY-125 | + | + | + | − | − |
| MAY-130 | + | + | + | − | + |
| MAY-131 | + | + | − | − | − |
| MAY-133 | + | + | − | − | − |
| MAY-134 | + | + | − | − | − |
| MAY-139 | + | − | − | − | − |
| MAY-140 | + | − | − | − | − |
| MAY-146 | + | + | + | − | − |
| MAY-147 | + | + | − | − | − |
| MAY-151 | + | + | + | − | − |
Vero cells infected with the indicated alphavirus were incubated with anti-MAYV mAbs. Binding to infected cells was determined by flow cytometry. Data are representative of two experiments performed in triplicate. + indicates cross-reactive and − indicates non–cross-reactive.
Mechanism of neutralization by anti-MAYV mAbs
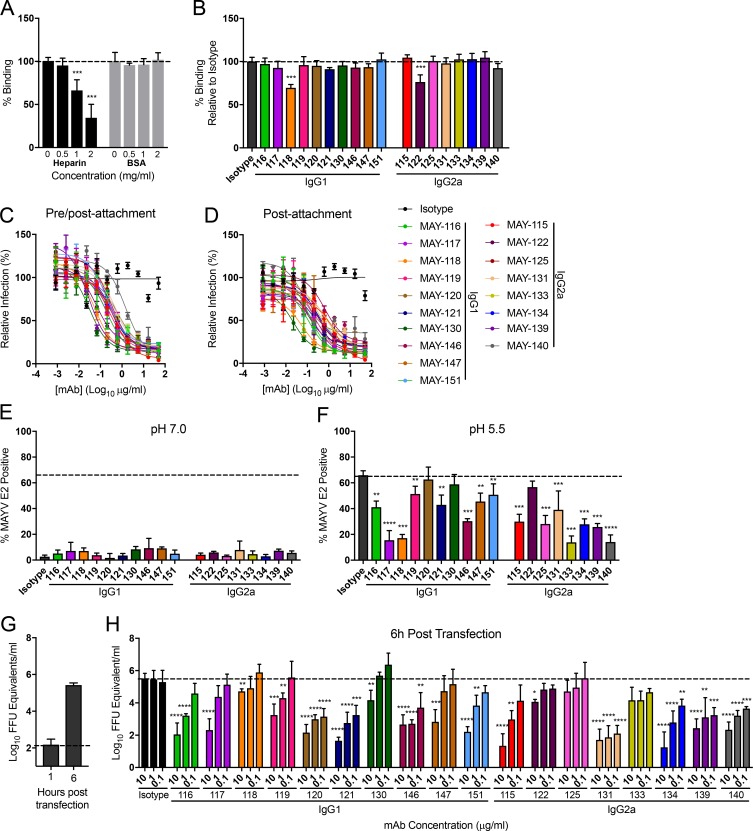
Antibodies against alphaviruses can inhibit attachment, internalization, fusion, or budding and egress (Fox et al., 2015; Jin et al., 2015, 2018; Kim et al., 2019). To assess how the panel of neutralizing anti-MAYV mAbs blocked infection, we first performed attachment inhibition assays in which mAbs and virus were premixed at 4°C before addition to Vero cells. After extensive washing, viral RNA adsorbed to cells was measured by quantitative RT-PCR (qRT-PCR). As a positive control, pretreatment of MAYV with soluble heparin, a molecule that closely mimics the attachment factor heparan sulfate (Klimstra et al., 1998), blocked MAYV attachment dose dependently (Fig. 3 A). Only MAY-118 and MAY-122 modestly inhibited virus binding, with 28% and 24% reductions, respectively (Fig. 3 B).
Figure 3.
Neutralizing mAbs block MAYV infection at postattachment steps. (A and B) Attachment inhibition assay. MAYV-CH was incubated with soluble heparin (0.5 to 2 mg/ml; A), BSA (0.5–2 mg/ml; A), or anti-MAYV mAbs (10 µg/ml; B) for 1 h before addition to Vero cells at 4°C. After unbound virus was removed by extensive rinsing, cell-adsorbed viral RNA was quantified by qRT-PCR, standardized to GAPDH levels, and plotted relative to an untreated (A) or isotype mAb-treated (B) control. Data are the mean and SD of three experiments performed in triplicate (one-way ANOVA with Dunnett’s post-test compared with the isotype control mAb). (C) Pre/postattachment neutralization assays. Serial dilutions of anti-MAYV mAbs were incubated with 102 FFU of MAYV and added to Vero cells. Infection proceeded for 18 h before foci were stained, counted, and plotted relative to a no-antibody control. Data are the mean and SD of two experiments performed in triplicate. (D) Postattachment neutralization assay. 102 FFU of MAYV was adsorbed to Vero cells at 4°C. Unbound virus was removed by extensive washing, and serial dilutions of anti-MAYV mAbs were added. Infection proceeded for 18 h at 37°C. Viral foci were stained, counted, and plotted relative to a no-mAb control. (E and F) FFWO assay. Cells were incubated with virus at 4°C. After removing unbound virus by rinsing, cells were treated with 1 µg/ml of the indicated mAbs and then pulsed for 2 min with medium at pH 7.6 (E) or pH 5.5 (F) at 37°C. After pH neutralization, cells were cultured in medium supplemented with 20 mM NH4Cl to prevent viral fusion via canonical endosomal pathways. Fusion inhibition was measured by flow cytometry by staining cells for MAYV E2 antigen 18 h later. Data are the mean and SD of three experiments performed in duplicate (one-way ANOVA with Dunnett’s post-test compared with the isotype mAb control). (G and H) Egress inhibition assay. RNA isolated from MAYV-infected cells was transfected into Vero cells. Medium was added with the indicated concentrations of anti-MAYV mAbs. RNase A–resistant encapsidated viral RNA in the supernatant was quantified by qRT-PCR at 1 h (G) or 6 h (H) after transfection. Data are the mean and SD of three experiments performed in duplicate (one-way ANOVA with Dunnett’s post-test compared with the isotype mAb control). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
We next determined whether our panel of neutralizing mAbs inhibited at a postattachment step. Neutralization assays were performed by adsorbing virus to cells at 4°C before adding mAbs. All anti-MAYV mAbs tested potently inhibited infection when added before virus attachment to cells (Fig. 3 C), as expected. The mAbs also had similar EC50 values when added after virus attachment (Fig. 3 D). As these results suggested that mAb inhibition occurred after virus binding to cells, we assessed their capacity to block viral fusion. We performed fusion-from-without (FFWO) assays (Edwards and Brown, 1986). MAYV was bound to target cells at 4°C before addition of mAbs. After removing unbound antibody and virus, membrane fusion was induced by a pulse exposure to acidic medium (pH 5.5). After pH neutralization, cells were incubated in medium supplemented with 20 mM NH4Cl to prevent MAYV fusion via canonical endosomal pathways. After 18 h, cells were stained for intracellular MAYV E2 proteins. As expected, neutral pH medium did not result in expression of MAYV E2 antigen, as plasma membrane fusion did not occur (Fig. 3 E). However, a short exposure of cell surface–adsorbed virus to acidic pH resulted in viral fusion and MAYV E2 antigen expression, and 16 of the 18 mAbs blocked this, whereas the isotype control mAbs did not (Fig. 3 F).
We evaluated whether anti-MAYV neutralizing mAbs could block viral egress, presumably by binding to structural glycoproteins on the plasma membrane and inhibiting assembly or budding (Fox et al., 2015; Jin et al., 2015, 2018). Vero cells were transfected with RNA isolated from MAYV-infected cells. The cells were treated with anti-MAYV mAbs, and the amount of RNase A–resistant, encapsidated viral RNA released into the supernatant was monitored. At 1 h, the levels of viral RNA in the supernatant were at the limit of detection (Fig. 3 G), indicating an absence of de novo synthesized and secreted virions. By 6 h, however, a 10,000-fold increase in encapsidated viral RNA was observed in the supernatant of the isotype control mAb–treated cells (Fig. 3 G). All of the mAbs tested, except MAY-125 and MAY-133, diminished the amount of viral RNA in the supernatant at doses of 10 µg/ml, suggesting a capacity to block MAYV egress. The potency of this inhibition was variable among the mAbs tested, as 11 of the 18 mAbs lost the ability to inhibit egress when a concentration of 0.1 µg/ml was used (Fig. 3 H).
Epitope mapping of anti-MAYV mAbs
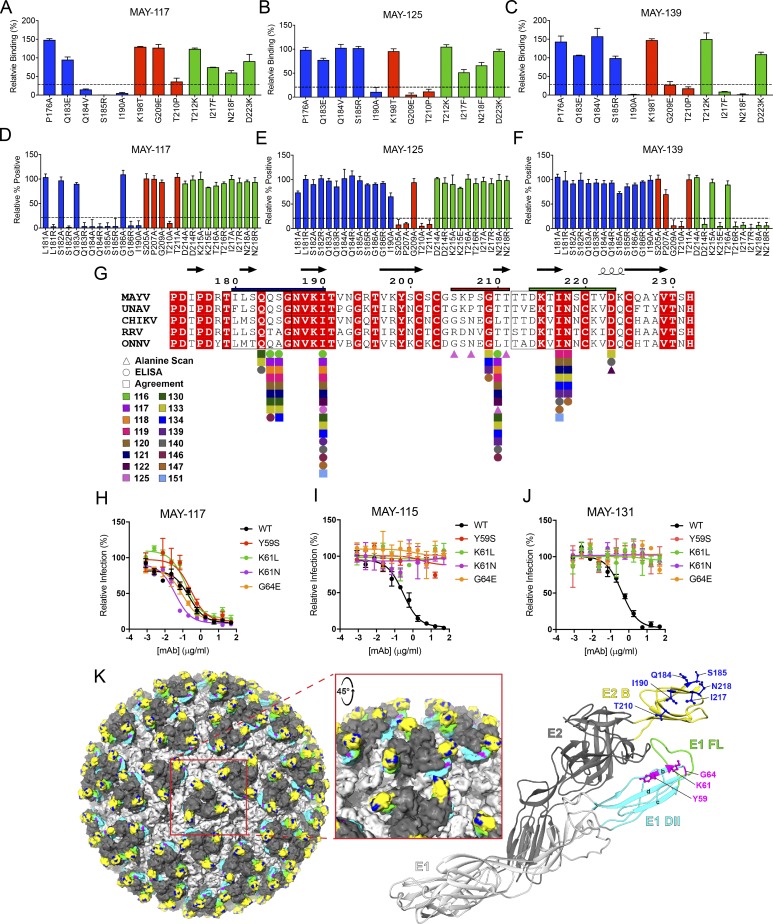
Our BLI data indicated that the majority of mAbs bound to the E2 protein (Tables 1 and S2). We next tested whether these mAbs recognized the B domain of E2, a target of neutralizing antibodies against other alphaviruses (Fox et al., 2015; Weger-Lucarelli et al., 2015). Of the 18 mAbs tested, 16 bound to the E2 B domain by ELISA (Fig. S4, A and B). We used two methods to map these B domain–specific mAbs at the amino acid level: (1) mAb binding to recombinant E2 proteins produced with substitutions in predicted solvent-exposed amino acids in the B domain (based on the CHIKV pE2-E1 structure; PDB: 3N42) from nonconserved residues present in other alphaviruses (Fig. 4, A–C; and Fig. S4 C); and (2) alanine scanning mutagenesis of the E2 B domain in the context of the full-length structural polyprotein (C-E3-E2-6K-E1). 293T cells were transfected with plasmids containing a single alanine substitution in residues 173–231 of the E2 protein, except at position 227, when the alanine was substituted to serine (Fig. 4, D–F; and Fig. S4 D). The 16 mAbs mapped to three regions within the E2 B domain: residues 181–190 (Fig. 4, A, D, and G; blue), 205–211 (Fig. 4, B, E, and G; red), and 214–218 (Fig. 4, C, F, and G; green). A majority of the mAbs bound to one of two loops (amino acids 179–186 and 212–218) that are predicted to protrude from the E2–E1 heterodimer. We made additional charge substitutions in these loops to determine if secondary structure was important for mAb binding. Whereas mutations to residues T212 and T213 impacted protein expression and could not be evaluated, the remaining mutations in these loops led to decreased binding of the mAbs mapping to these regions (Fig. 4, D and F; and Fig. S4 D). In several instances, charge mutations led to loss of mAb binding when alanine mutations did not. Likely, the mAbs do not recognize these residues directly but rather the chemical properties of the amino acid side chains maintain the architecture of the loop and preserve the epitopes.
Figure 4.
Mapping of neutralizing anti-MAYV mAbs to sites within E1 and E2 proteins. (A–C) Solvent-exposed residues on the MAYV E2 B domain were changed to the indicated amino acids. WT and mutant MAYV B domain proteins were purified, and binding to anti-MAYV mAbs was tested by ELISA. Regions were divided into groups A (blue), B (red), and C (green) based on patterns of mutations that resulted in loss of binding of mAbs. Representative mAbs from each group are shown (MAY-117, group A [A]; MAY-125, group B [B]; and MAY-139, group C [C]), with the remainder of the data in Fig. S4. (D–F) 293T cells were transfected with a C-E3-E2-6K-E1 plasmid containing alanine mutations in the B domain of E2 and tested for binding with anti-MAYV mAbs by flow cytometry. Additional arginine or glutamic acid changes were made to residues in two loops in the B domain (residues 179–186 and 212–218). Representative mAbs from three binding groups are shown (MAY-117, group A [D]; MAY-125, group B [E]; and MAY-139, group C [F]), with the remainder of the data in Fig. S4. Critical residues were defined as those with ≤25% binding to an individual mAb but ≥75% binding to an oligoclonal pool of anti-MAYV mAbs. Data are from three experiments performed in triplicate. Error bars represent SD within one experiment. (G) Alignment of the B domain of E2 of MAYV, UNAV, CHIKV, RRV, and ONNV with the critical interaction residues identified for each mAb marked. Residues mapped by structure-guided mutagenesis and ELISA (circles), alanine scanning mutagenesis and flow cytometry (triangles), or both (squares) are marked. Boxes around amino acids 179–186 and 212–218 indicate loops in the B domain. Bars above the B domain indicate regions used to define binding groups A (blue), B (red), and C (green). (H–J) Neutralization escape mutants were generated by serial passage of MAYV (strain CH) in the presence of MAY-115 or MAY-131. Sequence-confirmed mutations were introduced into an infectious cDNA clone of the parental MAYV strain and tested for neutralization by MAY-117 (H), MAY-115 (I), or MAY-131 (J). Data are representative of two experiments performed in triplicate. Error bars represent SD within on experiment. (K) Key residues necessary for mAb engagement are highlighted on the surface representation of CHIKV (Basore et al., 2019; left; PDB: 6NK5) and depicted as balls and sticks on a ribbon diagram of the predicted structure of MAYV E2-E1 monomer generated using Phyre2 (right). Inset: Zoomed-in view of a trimeric spike. The E1 glycoprotein is in light gray, with the FL in green and the B-, C-, and D-strands of domain II in cyan. The E2 glycoprotein is in dark gray, with the B domain in yellow. Epitope-mapped residues in E2 domain B and E1 domain II are colored in blue and magenta, respectively.
Two mAbs, MAY-115 and MAY-131, did not bind to recombinant E2 proteins (Fig. S4, A and B). To determine their epitopes, we selected for neutralization escape viruses. MAYV CH was propagated in Vero cells in the presence of MAY-115 or MAY-131 for at least six passages until neutralization activity was lost. The structural genes of the escape mutants were sequenced. Mutations in the E1 gene were identified for both MAY-115 (Y59S and K61L) and MAY-131 (K61N and G64E) escape mutants. These changes were introduced into the MAYV-CH infectious cDNA plasmid (Weise et al., 2014), and mutant viruses were produced. As expected, these mutant viruses were neutralized by the anti-E2 MAY-117 (Fig. 4 H). In comparison, both MAY-115 (Fig. 4 I) and MAY-131 (Fig. 4 J) showed reduced neutralizing activity against viruses containing any of the single mutations. All of the neutralizing mAbs we mapped localized to sites proximal to the FL in the E1 protein of the heterodimer (Fig. 4 K; E2 B domain mAb epitopes in blue, E1 domain II mAb epitopes in magenta, and FL in green).
Protection against lethal MAYV challenge by mAbs
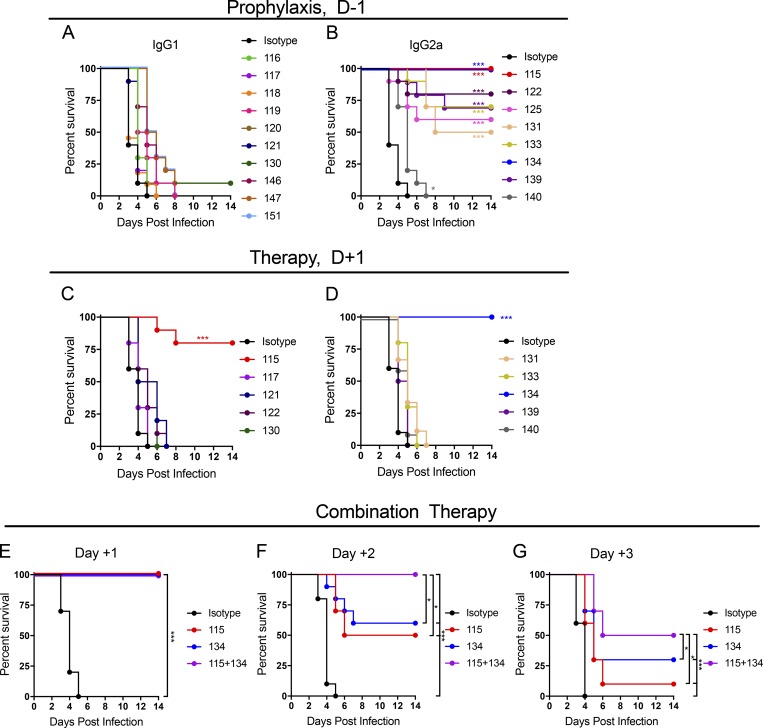
To determine if mAbs could protect in vivo, we developed a challenge model of MAYV infection in mice that resulted in uniform mortality. 4-wk-old C57BL/6 mice were administered a single intraperitoneal injection of 100 µg of anti-interferon alpha and beta receptor subunit 1 (anti-Ifnar1) mAb (MAR-5A3; Sheehan et al., 2006) 1 d before subcutaneous inoculation with MAYV strain BeH407 in the footpad. Although MAYV is not lethal in immunocompetent mice, infected animals treated with anti-Ifnar1 mAbs began to succumb at 3 d post infection (dpi) and experienced 100% mortality by 6 dpi (Fig. 5, A and B). To test whether anti-MAYV mAbs could protect in this model, we administered a single 100-µg mAb dose as prophylaxis 1 d before virus inoculation. Even though many mAbs had strong neutralizing activity (11 had EC50 values against MAYV BeH407 of <10 ng/ml, and 17 had values <100 ng/ml), only 9 of 18 protected mice against lethal challenge (Fig. 5, A and B), with only two mAbs (MAY-115 and MAY-134) preventing mortality completely. Remarkably, all of the mAbs that protected mice against lethal MAYV infection were of the IgG2a subclass (Fig. 5 B). Despite similar EC50 values to their IgG2a counterparts (Table 1), strongly neutralizing anti-MAYV IgG1 mAbs failed to protect against virus-induced mortality (Fig. 5 A).
Figure 5.
Antibody protection against lethal MAYV challenge. A lethal MAYV challenge model was developed by treating 4 wk-old C57BL/6J male mice with a single 100-µg dose of anti-Ifnar1 mAb 1 d before subcutaneous virus inoculation. (A and B) Prophylaxis studies. A single dose of anti-MAYV mAbs (100 µg/mouse; ∼6/mg/kg) was administered 1 d before inoculation with MAYV-BeH407, and survival was monitored. Antibodies are presented in IgG subclass groups: mouse IgG1 (A) or mouse IgG2a (B). Data are from two experiments. (C and D) Therapeutic studies. Indicated mAbs (100 µg/mouse) were given 1 d after virus inoculation, and survival was monitored. Data are from two experiments. (E–G) Combination mAb therapy. 200 total µg of MAY-115, MAY-134, or a combination (100 µg each) was given to mice beginning at 1, 2, or 3 dpi, and survival was monitored. Data are from two experiments. In this figure, n = 10, log-rank test with Bonferroni correction compared with the isotype mAb control treated group. *, P < 0.05; ***, P < 0.001.
For the protective IgG2a mAbs, we determined their therapeutic activity in a postexposure treatment design. We administered MAY-115, MAY-122, MAY-131, MAY-133, MAY-134, MAY-139, or MAY-140 as well as the slightly protective (MAY-121) and nonprotective (MAY-117) mAbs 24 h after virus inoculation of anti-Ifnar1 mAb–treated mice. Only MAY-115 and MAY-134 conferred substantial protection, with 80% and 100% survival rates, respectively (Fig. 5, C and D). Because MAY-115 and MAY-134 bound distinct epitopes (Fig. 4 B), we tested them in combination. We treated mice with 200 µg of MAY-115, MAY-134, or a combination of 100 µg each of MAY-115 and MAY-134 at 1, 2, or 3 dpi in the lethal challenge model. The higher dose of antibody conferred protection in all mice treated 1 dpi (Fig. 5 E). MAY-115 and MAY-134 protected 50% and 60% of mice when given individually at 2 dpi, whereas combination therapy protected all mice (Fig. 5 F). When treatment was begun at 3 dpi, MAY-115 or MAY-134 protected 10% and 30%, respectively, whereas combination therapy protected 50% of mice (Fig. 5 G). Thus, a combination of anti-E1 and E2 mAbs provided superior therapeutic activity compared with the individual mAbs against MAYV in the lethal challenge model.
Protection against MAYV-induced musculoskeletal disease by mAbs
As MAYV causes arthritis and musculoskeletal disease, we assessed whether our protective mAbs also mitigated joint-associated infection and swelling. 4-wk-old C57BL/6 mice were inoculated subcutaneously in the foot with MAYV-BeH407, and ipsilateral and contralateral ankle joint swelling was measured. In the ipsilateral ankle, MAYV caused swelling beginning at 2 dpi that persisted until ∼12 dpi (Fig. 6, A and B); in the contralateral ankle, swelling was observed at 7 dpi (Fig. 6 A). We tested mAbs with therapeutic activity in the lethal challenge model for their ability to limit MAYV-associated joint swelling along with the nonprotective MAY-117 (IgG1) and an isotype control mAb. For each mAb, 100 µg was administered 1 d before subcutaneous inoculation with MAYV-BeH407, and ankle swelling was measured using digital calipers (Fig. 6, B–G). Compared with an isotype control mAb, all tested anti-MAYV mAbs reduced swelling in both the ipsilateral and contralateral feet. MAY-115, MAY-130, MAY-131, and MAY-133 blocked swelling in both feet. Mice treated with MAY-117, which failed to protect against lethal challenge in anti-Ifnar1 mAb–treated mice, showed swelling of the contralateral foot, although less than the isotype control mAb (Fig. 6 C). Animals treated with MAY-122 exhibited a partial reduction in swelling of the ipsilateral foot and delayed swelling in the contralateral foot (Fig. 6, D and E).
Figure 6.
Antibody protection of MAYV-induced musculoskeletal disease. (A) Swelling observed in the ipsilateral (4 dpi, left) and contralateral (7 dpi, right) ankle following infection with MAYV-BeH407. (B–G) Protective mAbs in the lethal challenge model (Fig. 5) were tested for activity against MAYV-induced musculoskeletal disease. 4-wk-old C57BL/6J male mice were given 100 µg of indicated mAbs via intraperitoneal route 1 d before subcutaneous inoculation of 103 FFU MAYV-BeH407 in the foot. Swelling was measured in the ipsilateral (B, D, and F) and contralateral (C, E, and G) ankles using digital calipers. Data are the mean and SEM of two experiments (n = 10 mice, two-way ANOVA with Tukey’s post-test). (H–M) Virus titers in the ipsilateral (H and I) or contralateral (J and K) feet or draining inguinal lymph node (L and M) at 1 (H, J, and L) and 7 (I, K, and M) dpi after prophylaxis of mice with 100 µg of the indicated mAbs. Animals were perfused with PBS before tissue collection. Viral titers were determined by qRT-PCR. Data are from two experiments (n = 10 mice, one-way ANOVA with Dunnett’s post-test). *, P < 0.05; **, P < 0.01; ***, P < 0.001, ****, P < 0.0001. The experiments in B, D, and F or C, E, and G were performed concurrently, and thus a single isotype control mAbs was used and included in each graph.
To determine the effect of anti-MAYV mAbs on viral dissemination, we treated mice in the musculoskeletal disease model with either highly (MAY-115 or MAY-134) or partially (MAY-117) protective mAbs. Mice treated with MAY-115 or MAY-134 had less viral RNA in the ipsilateral foot at 1 dpi compared with the isotype control, and viral RNA was cleared at 7 dpi (Fig. 6, H and I). Mice treated with MAY-117 had less viral RNA in the ipsilateral foot than animals given the isotype control mAb but had higher levels at both 1 and 7 dpi than animals treated with MAY-115 or MAY-134. Consistent with these data, MAY-115 and MAY-134 prevented viral dissemination to the contralateral foot and the draining lymph node, whereas MAY-117 did not (Fig. 6, J–M).
Antibody effector functions are required for anti-MAYV mAb–mediated protection
We observed marked differences in mAb protection against MAYV challenge (Fig. 5) despite similarities in neutralization potency and epitope localization (Table 1 and Fig. 4). Because the protective IgG2a mAbs against MAYV bound avidly to viral antigens on the surface of infected cells (Table 3), we hypothesized that optimal in vivo activity required Fc effector functions of the antibody and recognition of viral proteins on the plasma membrane. To begin to evaluate the contribution of Fc effector functions to protection, we tested the ability of anti-MAYV mAbs to promote neutrophil and monocyte-dependent phagocytosis of beads coated with recombinant MAYV E2 protein; we were unable to express MAYV E1 protein and thus could not interrogate anti-E1 mAbs for these studies. Several of the protective anti-MAYV E2 mAbs (e.g., MAY-122, MAY-133, and MAY-134) promoted phagocytosis of MAYV antigen–coated beads in murine neutrophils and monocytes (Fig. S5, A and B). Capillary electrophoresis analysis revealed heterogeneity in the abundance of specific glycans on anti-MAYV E2 mAbs (Fig. S5 C). Dominant afucosylated, digalactosylated, and sialylated (G2S1 and GS2S) glycans appeared to associate positively with protection, whereas fucosylated (G2S1F) and highly inflammatory agalactosylated (G0) glycoforms were negatively associated (see Table 3).
Table 3. Summary of properties of anti-MAYV IgG2a mAbs.
| Antibody | Binding group | Protection (%) | EC50 binding | EC50 neutralization | Glycans | ADNP | ADCP |
|---|---|---|---|---|---|---|---|
| MAY-134 | E2: A, C | 100 | 31 (24–47) | 9 (4–13) | G2S2, G2S1, G2S1F, | ++ | ++ |
| G0F, G1F, G2F | |||||||
| MAY-122 | E2: B | 80 | 7 (5–11) | 3 (2–6) | G2S2, G2S1F, G0F, | ++ | + |
| G1F-G1FB, G2F | |||||||
| MAY-133 | E2: A, C | 70 | 27 (21–38) | 25 (14–44) | G2S2, G2S1, G0F, | ++ | ++ |
| G1F-G1FB, G2F | |||||||
| MAY-139 | E2: C | 70 | 18 (9–55) | 246 (133–342) | G2S2, G2S1, G2S1F, G0, | − | − |
| G0F, G1F-G1FB, GIFB-G2, G2F | |||||||
| MAY-125 | E2: B | 60 | 3 (2–8) | 22 (14–42) | G0F, G1F-G1FB, | − | − |
| G1FB-G2, G2F | |||||||
| MAY-140 | E2: A | 0 | 16 (12–22) | 18 (11–30) | G0F, G1F | − | − |
| MAY-115 | E1 | 100 | 2 (1–7) | 2 (1–3) | N/A | N/A | N/A |
| MAY-131 | E1 | 50 | 2 (1–4) | 8 (2–14) | N/A | N/A | N/A |
IgG2a mAbs that bind to MAYV-E2 (top) or E1 (bottom) are listed along with the mapped binding group, percentage of mice protected from lethal challenge, EC50 values for binding to MAYV-BeH407 infected Vero cells (EC50 binding) at 4°C, neutralization of MAYV-BeH407 (EC50 neutralization), glycan profile, and antibody-dependent neutrophil phagocytosis and antibody-dependent cellular phagocytosis activity. Antibodies are listed in order from most protective to least protective in each class. EC50 values are nanograms per milliliter. ADCP, antibody-dependent cellular phagocytosis; ADNP, antibody-dependent neutrophil phagocytosis; N/A, not applicable.
We next evaluated the effect of mAb isotype on protective anti-MAYV mAb activity in anti-Ifnar1 mAb-treated C57BL/6 mice lacking the common signaling γ chain and activating FcγRs (FcγR−/−). We performed prophylaxis studies with MAY-115 and MAYV-134 by administering 100 µg of mAb 1 d before virus inoculation. Isotype control mAb–treated FcγR−/− mice sustained 100% mortality by day 4 (Fig. 7 A), whereas animals treated with MAY-115 or MAY-134 were protected only partially, with a 30–40% survival rate, respectively. This result contrasts with the ability of the mAbs to confer complete protection as prophylaxis in anti-Ifnar1 mAb–treated WT mice (see Fig. 5). As expected, MAY-117, a neutralizing mAb of the IgG1 isotype, failed to protect against MAYV in FcγR−/− mice.
Figure 7.
Antibody protection against MAYV depends on the IgG subclass and Fc effector functions. (A) 4-wk-old C57BL/6 male and female FcγR−/− mice were administered 100 µg of the indicated anti-MAYV mAbs and 100 µg of anti-Ifnar1 mAb 1 d before subcutaneous inoculation with 103 FFU of MAYV-BeH407. Data are from two experiments. (B) Recombinant, isotype-switched mAbs (MAY-115 and MAY-134) were tested by ELISA for binding to soluble mouse and human FcγRs. Binding is presented relative to the mouse IgG2a isotype control of MAY-115 (left) or MAY-134 (right). Data are the mean and SD of two experiments performed in triplicate. (C and D) Neutralizing activity of isotype-switched MAY-115 (C) or MAY-134 (D) was measured by FRNT against MAYV-BeH407. Data are representative of three experiments performed in triplicate. (E) 4-wk-old FcγR−/− mice were administered 100 µg of the indicated MAY-115 IgG and 100 µg of anti-Ifnar1 mAb 1 d before subcutaneous inoculation with 103 FFU of MAYV-BeH407. Data are from two experiments. (F–I) Protection by isotype-switched mouse IgG2a mAbs. MAY-115 (F and G) and MAY-134 (H and I) isotype-switched mAbs were tested as prophylaxis against lethal challenge (F and H) or musculoskeletal disease (G and I) as described in Figs. 5 and 6. Survival data are from two experiments. Swelling data are the mean and SD from two experiments (n = 10 mice, two-way ANOVA with Sidak’s post-test). (J–L) Protection by isotype-switched mouse IgG1 mAbs. MAY-117 (J) and MAY-130 (K) isotype-switched mAbs were tested as prophylaxis against lethal challenge (L). Survival data are from two experiments (A, E, F, H, and L; n = 10 mice, log-rank test with Bonferroni correction compared with the isotype control or indicated treatment groups). The experiments in F and H or G and I were performed concurrently, and thus a single isotype control mAbs was used and included in each graph. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To further define the role of mAb subclass in MAYV protection, we performed isotype-switching studies. Antibody isotypes bind FcγRs with different affinities: mouse IgG2a binds strongly to mouse FcγR I and FcγR IV, whereas mouse IgG1 does not (Mancardi et al., 2008). We cloned the variable regions of MAY-115 and MAY-134 and inserted them into mouse IgG1 and IgG2a antibody heavy chain expression vectors. We also cloned the variable regions of MAY-115 and MAY-134 as human IgG1 antibodies, which have comparable binding to mouse FcγR I and FcγR IV as mIgG2a (Dekkers et al., 2017). Finally, we generated an aglycosyl variant of human IgG1 (N297Q) that abrogates binding to FcγR and C1q (Tao and Morrison, 1989). We first tested the binding of the isotype-switched mAbs to mouse and human FcγRs by ELISA; we observed little or no binding of mouse IgG1 and human IgG1-N297Q antibodies to mouse FcγR I, mouse FcγR III, mouse FcγR IV, human FcγR I, and human FcγR IIIa (Fig. 7 B). Human IgG1 mAbs bound to mouse FcγR I and FcγR IV similarly to mouse IgG2a but showed less binding to mouse FcγR III. FRNT assays confirmed that isotype-switched mAbs retained neutralizing activity (Fig. 7, C and D). As expected, all isotype-switched variants of MAY-115 had similar defects in protection against MAYV challenge in FcγR−/− mice (Fig. 7 E).
We administered the isotype-switched mAbs to mice in both the lethal challenge and musculoskeletal disease models. When switched from mouse IgG2a to IgG1, both MAY-115 and MAY-134 lost their ability to fully protect mice, with 60% of animals succumbing in each case (Fig. 7, F and H). Human IgG1 isotype-switched variants of MAY-115 and MAYV-134 exhibited slightly reduced activity in vivo compared with mouse IgG2a but were more protective than mouse IgG1 or human IgG1-N297Q mAbs. Combination therapy experiments showed decreased efficacy when one of the two mAbs was a human IgG1-N297Q variant, suggesting that protection depends on effector functions of both mAbs (Fig. S5 D). In the musculoskeletal disease model, all isotype-switched mAbs reduced joint swelling in the ipsilateral foot compared with isotype control mAbs, although differences were apparent (Fig. 7, G and I). Whereas mouse IgG2a and human IgG1 forms of MAY-115 and MAY-134 mAbs prevented swelling completely, mouse IgG1 and human IgG1-N297Q variants showed only partial decreases in swelling. We observed roughly equivalent half-lives of all of the isotype-switched mAbs we tested (Fig. S5, E–J), which indicated that the decreased protection observed with particular mAb variants was not due to more rapid clearance in vivo.
Even though many of our IgG1 mAbs had similarly potent neutralizing activity against MAYV compared with IgG2a mAbs, they failed to protect against lethal challenge. To corroborate a key role for IgG subclass in protection against MAYV, we switched the isotype of MAY-117 and MAY-130, two nonprotective neutralizing mAbs, from mouse IgG1 to human IgG1, which engage FcγRs weakly and strongly, respectively. The isotype-switched mAbs had similar neutralization activity against MAYV BeH407 as the parental mAbs (Fig. 7, J and K). As expected, uniform mortality was seen in MAYV-infected mice treated with the mouse IgG1 forms of MAY-117 or MAY-130 (Fig. 7 L). In contrast, we observed significant protection against lethal MAYV challenge when mAbs MAY-117 or MAY-130 of the human IgG1 isotype was administered. These experiments confirm that effector functions contribute to optimal antibody-mediated protection of mice from lethal MAYV infection.
Discussion
Our goal was to produce type-specific and broadly neutralizing mAbs that have therapeutic utility and can shed light on the requisite components of a protective humoral immune response against MAYV. We generated 151 hybridomas and identified 18 that produced mAbs with potent neutralizing activity. Of these, 11 had “elite” activity, with EC50 values against the immunizing MAYV strain of <10 ng/ml. 16 of these mAbs neutralized the closely related UNAV, with a smaller subset showing a breadth of inhibitory activity against the more distant CHIKV, RRV, and ONNV.
Although many mAbs neutralized a broad spectrum of MAYV isolates from both the D and L genotypes, there were exceptions. MAY-133 had no neutralizing activity against MAYV-Uruma, and MAY-140 lacked activity against MAYV-FSB0311, MAYV-OBS6443, and MAYV-BeH473130. This observation was unexpected, because both mAbs engage residues (for MAY-133: Q183, Q184, I190, G209, T210, I217, N218, and D223; for MAY-140: Q183, I190, G209, T210, I217, and D223) on the E2 protein that are shared by other mAbs that neutralized all MAYV strains and even some other alphaviruses. Indeed, these interaction residues are conserved across all MAYV strains that we tested. Although further studies are warranted, we hypothesize that the display of particular residues on E2 varies in a strain-dependent manner, which could affect neutralization by a given antibody. Thus, substitutions in the envelope proteins of Uruma, FSB0311, OBS6443, and BeH473130 strains of MAYV could affect epitope exposure, as has been described for flaviviruses (Austin et al., 2012; Goo et al., 2017; Fernandez et al., 2018). Analysis of the virion structures of these different MAYV strains and the display of their structural glycoproteins may elucidate why certain antibodies fail to neutralize individual viruses even though their binding residues appear conserved.
The neutralizing mAbs that we characterized all blocked virus entry at postattachment steps. Only mAbs MAY-117 and MAY-122 showed nominal blocking of viral binding to cells, and these mAbs also blocked postattachment entry steps. The majority of the mAbs, with the exception of MAY-120 and MAY-122, inhibited plasma membrane fusion. These results are similar to studies with other alphaviruses, including CHIKV (Fox et al., 2015; Jin et al., 2015, 2018), Venezuelan equine encephalitis (Porta et al., 2014), and eastern equine encephalitis virus (Kim et al., 2019), indicating that potently neutralizing mAbs may share a common mechanism of action. Indeed, all of the strongly inhibitory mAbs we analyzed bound to regions in E2 and E1 that are proximal to the FL peptide. While entry blockade likely contributed to the potency of virus neutralization, 16 of the 18 mAbs also inhibited viral egress. This activity may occur as a result of cross-linking of adjacent E protein complexes at the cell surface and/or conformational shifts induced by anti-B domain antibodies (Jin et al., 2015, 2018) that prevent budding from infected cells. As the regions in E1 and in the B domain of E2 to which our mAbs mapped are conserved among MAYV strains, they could be targeted by vaccines to induce broadly inhibitory humoral responses against MAYV and closely related viruses.
Although all of the neutralizing E2-specific anti-MAYV mAbs that we mapped localized to the B domain, it is likely that that neutralizing epitopes also exist in the A domain. Neutralizing mAbs that bind to the A domain of E2 of CHIKV (Fox et al., 2015; Jin et al., 2015, 2018; Smith et al., 2015) and eastern equine encephalitis virus (Kim et al., 2019) have been described. Furthermore, vaccine-generated anti-MAYV-neutralizing antibodies preferentially bound to epitopes within the A domain of E2 (Choi et al., 2019). Our isolation of B rather than A domain mAbs may be a product of our vaccination strategy. Although the initial immunization and the final boosts were performed with infectious virus, we boosted with recombinant E2 protein, which may have skewed the immune response toward B domain antibodies. Analysis of the antibody responses of different mouse strains or naturally infected humans will be necessary to establish the immunodominance of neutralizing epitopes in the MAYV envelope proteins.
The neutralizing mAbs showed disparate levels of protection in mice. Although many had exceptional neutralizing potency in cell culture, only 8 of the 18 mAbs conferred significant protection in vivo. In this subset, there were differing levels of protection ranging from partial to complete protection (Table 3), and there was no correlation with the shape of the neutralization curve or presence of a resistant fraction. The most protective anti-E2 mAbs bound viral antigen on the surface of infected cells with high avidity and promoted phagocytosis of E2 protein–coated beads by both neutrophils and monocytes. These results are consistent with prior studies with the related Sindbis and Semliki Forest viruses, which showed that nonneutralizing mAbs can protect against lethal infection (Schmaljohn et al., 1982; Wust et al., 1987). We observed a preliminary correlation with protection in vivo and dominant afucosylated, digalactosylated, and sialylated glycan specifies (sialic acid: G2S2 and G2S1). The cytotoxic function of afucosylated glycans, which enhance Fc-effector function, coupled to enhanced galactosylation/sialylation, which dampen inflammation (Kaneko et al., 2006; Anthony et al., 2008), may promote optimal viral clearance and minimize pathology. Further glycan analysis and protection data with a larger panel of anti-MAYV mAbs is needed to corroborate these results, as was performed with anti-Ebola virus mAbs (Saphire et al., 2018).
Antibody protection against lethal challenge and musculoskeletal disease required Fc effector functions, albeit the latter to a lesser degree. Our 18 neutralizing mAbs were composed of two IgG subclasses: 10 IgG1 and 8 IgG2a. Remarkably, only the IgG2a anti-MAYV mAbs conferred substantial protection against lethal challenge. Mouse IgG2a antibodies not only interact with more FcγRs on immune cells but also bind specific receptors (e.g., FcγR I) with higher affinity than IgG1 (Mancardi et al., 2008; Dekkers et al., 2017). The capacity for an IgG subclass to engage FcγRs correlated with in vivo activity, as MAY-115 and MAY-134 were more protective as IgG2a than IgG1 antibodies even though their neutralizing activity in vitro and half-lives in vivo were essentially equivalent. The dependence of protection on effector function was corroborated by comparing isotype-switched anti-MAYV human IgG1 variants with and without (N297Q) heavy-chain N-linked glycosylation. Moreover, nonprotective mouse IgG1 mAbs (MAY-117 and MAY-130) became protective when they were switched to a human IgG1 subclass, which engages activating FcγRs more efficiently. The contribution of FcγRs to anti-MAYV-mediated protection was confirmed through challenge studies in mice lacking all activating FcγRs. Antibody recognition of MAYV or other alphavirus antigens on the cell surface may be an important mechanism for clearance of infected cells through Fc effector–function dependent phagocytosis or cytolysis by specific myeloid cell subsets (Fox et al., 2019). Studies are planned to define the dominant effector functions and individual FcγRs that contribute to mAb protection in our MAYV models, as has been performed to varying degrees with antibodies against influenza (DiLillo et al., 2014), hepatitis B (Li et al., 2017), human immunodeficiency virus (Hessell et al., 2007), West Nile virus (Vogt et al., 2011), and CHIKV (Fox et al., 2019). Experiments are needed to determine if other IgG subclasses (e.g., mouse IgG2b, human IgG2, and human IgG3) confer protection against MAYV in these models and whether there is an impact of C1q, which differentially binds IgG subclasses, on protection. Finally, our combination therapy studies with MAY-115 and MAY-134, which bind discrete epitopes on E1 and E2 proteins, suggest that inclusion of multiple mAbs in a cocktail likely will confer greater protection than either constituent alone. This effect could be due to synergy of neutralization, effector functions, or the control of resistance against either of the two individual mAbs (Pal et al., 2013, 2014).
Antibodies with similar neutralization profiles and mechanisms of action did not protect mice from MAYV infection equivalently. Thus, the importance of IgG subclass and effector function activity likely should be optimized for vaccines or therapeutic antibodies against MAYV and other alphavirus infections. Three candidate MAYV vaccines have been described, including a chemically inactivated virion (Robinson et al., 1976), a live-attenuated virus (Weise et al., 2014), and a DNA plasmid vaccine (Choi et al., 2019). Although all protected in mice, the antibody repertoires were not fully analyzed. Whereas the DNA plasmid vaccine produced IgG2a antibodies in mice, the majority were against epitopes on E1 and E2 not identified in our screens. Our results in mice suggest than immunization strategies that use protein scaffolds (Correia et al., 2014) and specific TH1-skewing adjuvants could target regions in the B domain of E2, the FL of E1, and possibly other sites to generate neutralizing antibodies that block viral fusion and egress and optimize Fc effector functions.
Materials and methods
Animal ethics statement
All animal experiments and procedures were performed in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, Bethesda, MD. The protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (assurance number A3381-01). Injections were performed under anesthesia that was induced and maintained with ketamine hydrochloride and xylazine, and all efforts were made to minimize animal suffering.
Cell lines
Vero, HEK-293T, C2C12, and MEF cells were cultured in DMEM supplemented with 10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, 1× MEM nonessential amino acids, 1 mM sodium pyruvate, 2 mM L-glutamine, and 10 mM Hepes, pH 7.3. Hybridomas were cultured in IMDM supplemented with 20% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, and 1 mM sodium pyruvate. Expi293 cells were maintained in Expi293 medium (Gibco).
Viruses
MAYV strains (Beh428890, BeH473130, BeH343155, BeH506151, FSB0311, IQU2950, OBS6443, TRVL15537, BeH407, and Uruma), CHIKV (La Reunion OPY1), UNAV (CoAr2380), RRV (T48), and ONNV (M30) all were obtained from the World Reference Center for Emerging Viruses and Arboviruses (R. Tesh, K. Plante, and S. Weaver, University of Texas Medical Branch, Galveston, TX) and passaged in Vero cells from lyophilized stocks. Recombinant viruses were produced after linearization of a prS2 vector containing cDNA from MAYV strain CH (Weise et al., 2014) that was generously provided by S. Weaver (University of Texas Medical Branch, Galveston, TX). After in vitro transcription with mMESSAGE mMACHINE SP6 transcription kit (Invitrogen) and transfection into BHK-21 cells, p0 virus stocks were harvested and passaged once (p1) in Vero cells. Virus titers were determined by focus forming assay (Fox et al., 2015).
Mouse studies
4-wk-old WT C57BL/6J male mice were purchased from the Jackson Laboratory. 4-wk-old common γ-chain–deficient (FcRγ−/−) C57BL/6 mice were obtained commercially (Taconic) and bred at the Washington University Animal Facility. Anti-MAYV mAbs were administered by intraperitoneal injection at specified times before or after inoculation in the left footpad with 103 focus-forming units (FFU) MAYV in HBSS supplemented with 1% heat-inactivated FBS. Foot swelling was monitored via measurements (width × height) using digital calipers. Tissues were harvested after perfusion with 40 ml PBS and titered by qRT-PCR using RNA isolated from viral stocks as a standard curve to determine FFU equivalents. For lethal challenge experiments, mice were administered via intraperitoneal injection a single 100-µg dose of anti-Ifnar1 mAb MAR1-5A3 (Sheehan et al., 2006; Bio X Cell) 1 d before infection.
mAb generation
10-wk-old female BALB/c mice were inoculated with 103 FFU of MAYV-CH. Mice were boosted with 100 µg of recombinant MAYV E2 protein mixed 1:1 with Freund’s incomplete adjuvant at 14, 28, and 42 d after initial infection. At 56 dpi, mice were boosted with 106 FFU of MAYV-CH. 3 d before splenocyte harvest, at 70 dpi, mice were inoculated with 1010 FFU of MAYV-CH that was produced by centrifuging MAYV through a 20% sucrose cushion at ∼175,000 ×g for 2 h. Spleens were harvested at 73 dpi and fused with P3X63 Ag.8.6.5.3 mouse myeloma cells as described previously (Pal et al., 2013). Hybridoma supernatants were screened for antibodies that bound to recombinant MAYV E2 in an ELISA and/or to MAYV-infected cells by flow cytometry. Neat hybridoma supernatants were screened for neutralization of MAYV-CH using an FRNT (described below). Selected mAbs were isotyped by ELISA and purified by protein A affinity columns using a commercial vendor (Bio X Cell).
FRNTs
Anti-MAYV mAbs were diluted serially and incubated with 102 FFU of the specified MAYV strain for 1 h at 37°C in triplicate wells. Virus-mAb mixtures were incubated on Vero cells for 60 min at 37°C before being overlaid with 1% methylcellulose in MEM supplemented with 10 mM Hepes, pH 7.3, 100 U/ml of penicillin, 100 µg/ml of streptomycin, 2 mM L-glutamine, and 2% FBS. 18 h after virus inoculation, cells were fixed with 1% paraformaldehyde (PFA) in PBS. Cells were then washed and incubated with 1 µg/ml CHK-48 (Fox et al., 2015), a cross-reactive anti-CHIKV mAb, for 2 h. Cells were washed and incubated with 500 ng/ml HRP-conjugated goat anti-mouse IgG (Sigma-Aldrich) for 1 h. Foci of infection were detected using TrueBlue substrate (KPL) and counted using a Biospot plate reader (Cellular Technology). Wells containing virus incubated with mAbs were compared with wells treated with virus containing no mAb. The EC50 value was calculated using nonlinear regression analysis after constraining the bottom to 0 and the top to 100.
Attachment inhibition assay
104 FFU of MAYV was incubated with mAbs, soluble heparin (H3393; Sigma), or BSA (Sigma) at the specified concentration for 1 h at 37°C. The mixture was chilled on ice and added to Vero cells and incubated at 4°C for 1 h. Cells were washed six times with chilled PBS before addition of lysis buffer and extraction of RNA using an RNAeasy Mini Kit (QIAGEN). MAYV RNA was quantified using a Taqman RNA-to-Ct 1-step kit (Thermo Fisher Scientific) and a 5′ untranslated region and nsp1 specific primer/probe set (Waggoner et al., 2018). MAYV RNA levels were normalized to GAPDH, and the relative fold change was compared with cells treated with an isotype control mAb.
Pre- and postattachment assays
Pre- and postattachment neutralization assays were performed by first incubating serially diluted anti-MAYV mAbs with 102 FFU of MAYV for 1 h at 37°C. The mAb–virus complexes were then added to Vero cells for 1 h at 37°C. Cells were overlaid with 1% (wt/vol) methylcellulose in modified Eagle medium supplemented with 2% FBS. Postattachment neutralization assays were performed by first incubating Vero cells with 102 FFU of MAYV for 1 h at 4°C. Cells were washed extensively with cold DMEM to remove unbound virus. Diluted anti-MAYV mAbs were added to virus-adsorbed cells and incubated for 1 h at 4°C. After a 15-min incubation at 37°C to allow virus internalization, cells were overlaid with methylcellulose. Pre- and postattachment neutralization assays were processed similarly using the FRNT described above.
Fusion inhibition assays
FFWO assays were performed after allowing virus adsorption to BHK-21 cells (multiplicity of infection [MOI] of 25) for 1 h at 4°C. Unbound virus was removed by rinsing cells with chilled PBS. Diluted anti-MAYV mAbs (1 µg/ml) were added to virus-adsorbed cells for 30 min at 4°C. Cells were washed with chilled PBS. FFWO was induced by pulsing with fusion medium (RPMI 1640, 10 mM Hepes, 0.2% BSA, and 30 mM succinic acid, pH 5.5) for 2 min at 37°C. A nonfusion control was included using control media (RPMI 1640, 10 mM Hepes, pH 7.6, and 0.2% BSA). After the pulse, cells were washed twice with chilled PBS and incubated in DMEM supplemented with 5% FBS, 10 mM Hepes, pH 7.3, 100 U/ml of penicillin, 100 µg/ml of streptomycin, and 20 mM NH4Cl to prevent de novo infection via canonical endocytosis pathways. Infection was allowed to proceed for 6 h, and cells were detached and fixed with 4% PFA in PBS. Cells were stained with MAY-118 (0.5 µg/ml) in permeabilization buffer (PBS + 1% BSA + 0.1% saponin) and incubated for 1 h at 4°C. After two washes with permeabilization buffer, MAYV E2 antigen was detected with Alexa Fluor 647–conjugated goat anti-mouse IgG (1:1,000 dilution; Thermo Fisher Scientific). After two washes with permeabilization buffer, cells were resuspended in 100 µl and analyzed on a MACSQuant Analyzer (Miltenyi Biotec).
Egress inhibition assays
Vero cells were inoculated with MAYV at an MOI of 0.1. After 24 h, cells were washed and lysed, and total RNA was collected using an RNAeasy Mini kit (QIAGEN). The amount of viral RNA present was measured by qRT-PCR, and the stocks were aliquoted and stored at −80°C. Vero cells were transfected with viral RNA (equivalent to an MOI of 10) and incubated for 3 h at 37°C. The cell culture media was replaced with 50 µl of medium containing dilutions of anti-MAYV mAbs. The cells then were incubated for 3 h at 37°C before supernatant was collected. The samples were incubated with PBS containing 50 µg/ml RNase A (Sigma) to remove nonencapsidated RNA. RNA was isolated from the supernatant using an RNAeasy Mini kit, and MAYV RNA was measured using qRT-PCR as described above.
Protein expression and purification
The MAYV E2 ectodomain (residues 1–340) was cloned into the pET21a expression vector and expressed in BL21 (DE3) Escherichia coli cells. Protein production was induced using 1 mM IPTG, where E2 partitioned into the inclusion body fraction and was refolded using an oxidative refolding protocol (Nelson et al., 2014). Briefly, 10 ml solubilized inclusion body was injected at 1 ml/h into a 1-liter volume of arginine refolding buffer (400 mM L-arginine, 100 mM Tris, pH 8.5, 5 mM reduced glutathione, 0.5 mM oxidized glutathione, and 0.2 mM PMSF) and then allowed to stir slowly overnight at 4°C. The refolded protein was filtered, concentrated using a 30-kD-cutoff stirred cell concentrator (EMD Millipore), and purified by HiLoad 16/600 Superdex 75 size exclusion chromatography (GE Healthcare). B domain mutants were generated using a QuikChange II XL Site-Directed Mutagenesis Kit (Agilent) and verified by DNA sequencing. Mutants were confirmed by direct sequencing of plasmid DNA and expressed and purified as described above.
MAYV E2-E1 structure prediction and substitution design
The predicted structure of the MAYV E2-E1 heterodimer was generated for strain TRVL4675 (GenBank accession no. AAO33335.1) using the intensive modeling mode of Phyre2 (Kelley et al., 2015). Cutoffs of residues to display were based on the crystal structure of CHIKV p62E1 (PDB: 3N42; Voss et al., 2010). Substitution mutations in the B domain of E2 were designed by first estimating the solvent accessibility of each residue using Naccess (Hubbard and Thornton, 1993). A residue was chosen to undergo substitution if it was predicted to be solvent accessible and conserved among arthritogenic alphaviruses, but not encephalitic alphaviruses.
BLI binding assays
The binding affinity of purified recombinant MAYV E2 ectodomain protein to MAYV mAbs was monitored at 25°C using an Octet-Red96 device (Pall ForteBio). 100 µg of each mAb was mixed with biotin (EZ-Link-NHS-PEG4-Biotin; Thermo Fisher Scientific) at a molar ratio of 20:1 biotin/protein and incubated at room temperature for 30 min. The unreacted biotin was removed by passage through a desalting column (5 ml Zeba Spin 7 kD molecular weight cutoff; Thermo Fisher Scientific). The biotinylated mAbs were loaded onto streptavidin biosensor pins (ForteBio) until saturation, typically 10 µg/ml for 2 min, in 10 mM Hepes, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% P20 surfactant, and 1% BSA. The pins were equilibrated in binding buffer alone before being plunged into wells containing various concentrations of MAYV E2 and then placed back into binding buffer to allow for dissociation. Real-time data were analyzed using BIAevaluation 3.1 (GE Healthcare). Kinetic profiles and steady-state equilibrium concentration curves were fitted using a global 1:1 binding algorithm with drifting baseline.
MAYV mAb mapping by ELISA
Purified recombinant MAYV E2 ectodomain WT or mutant proteins (100 ng/well) were adsorbed overnight at 4°C on Maxisorp immunocapture ELISA plates (Thermo Fisher Scientific) in a sodium bicarbonate buffer (pH 9.3). Plates were washed three times with PBS and 0.05% Tween 20 for 1 h at 37°C followed by incubation of 0.5 µg/ml of indicated mAbs for 1 h at room temperature. Plates were washed again and then sequentially incubated with 2 µg/ml of HRP-conjugated goat anti-mouse IgG and tetramethylbenzidine substrate (Invitrogen). The reaction was stopped by the addition of 1 N H2SO4, and emission at 450 nm was read using an iMark microplate reader (Bio-Rad). Critical residues were defined as those with ≤25% binding to an individual mAb but ≥75% binding to an oligoclonal pool of anti-MAYV mAbs.
Alanine scanning mutagenesis
A pcDNA3.1(+) plasmid expressing a codon-optimized MAYV-TRVL4675 structural polyprotein (C, E3, E2, 6K, and E1 genes) was synthesized and mutated by Genewiz. Alanine scanning mutagenesis was performed on amino acids in the B domain of the E2 protein (residues 189–231), whereas charge mutations to arginine were made on residues 179–186 and 212–218, with the exception of 215, which was changed to glutamic acid. Plasmids were transfected into HEK-293T cells using Lipofectamine 3000 (Thermo Fisher Scientific). 18 h later, cells were chilled to 4°C, washed with PBS, and incubated with anti-MAYV mAbs (10 µg/ml) in PBS with 2% FBS for 1 h at 4°C. An oligoclonal mixture of all the anti-MAYV mAbs was used as a control for mutant E2 protein expression. Anti-MAYV mAb binding was detected using Alexa Fluor 647–conjugated goat anti-mouse IgG (Thermo Fisher Scientific) diluted 1:1,000. After 1 h, cells were washed, fixed with 1% PFA in PBS and analyzed by flow cytometry using a MACSQuant Analyzer (Miltenyi Biotec). Using previously described criteria (Smith et al., 2015), critical residues were defined as those with ≤25% binding to an individual mAb but ≥75% binding to an oligoclonal pool of anti-MAYV mAbs when detected by flow cytometry.
Generation of neutralization escape mutants
105 FFU of MAYV-CH was incubated with 1 µg/ml of MAY-115 or MAY-131 for 1 h at 37°C. The virus–mAb mixtures were added to Vero cells. After 24 h, half of the virus supernatant was frozen at −80°C, whereas the other half was incubated with 1 µg/ml of MAY-115 or MAY-131 for 1 h at 37°C. This virus–mAb mixture was added to Vero cells and harvested after 24 h. This protocol was performed a total of six passages before escape mutants were confirmed by FRNT. Viral RNA was isolated from supernatant pools using a MagMAX Viral Isolation kit (Applied Biosystems), and cDNA was generated using SuperScript IV Reverse transcription using Oligo(dT)20 primers (Thermo Fisher Scientific). Viral structural genes were amplified using the forward primer 5′-GGTCCTAAATAGGTGCTCTACACG-3′ and reverse primer 5′-ACTTTGAGAAGGTRAATCAWAAGTACCG-3′. The amplicons were sequenced by Genewiz, and mutants were produced using a Phusion Site-Directed Mutagenesis Kit (Thermo Fisher Scientific) in the MAYV-CH infectious cDNA clone described above.
Isotype switching of mAbs
MAY-115 and MAY-134 variable regions were sequenced and cloned using previously described methods (Ho et al., 2016). Total RNA was isolated from hybridomas, and cDNA was produced using random hexamers and Oligo(dT)20 using a SuperScript IV First Strand Synthesis kit (Invitrogen). Heavy and light chain variable regions were amplified and sequenced using mouse-specific primer sets (Ho et al., 2016). Allele-specific primers were used to amplify variable regions and append Gibson assembly sequences to the 5′ and 3′ ends. The variable regions then were cloned into plasmids containing the constant regions of human IgG1 (pAbVec-hIgG1) or mouse IgG1 (pAbVec-mIgG1) or the appropriate kappa chain (pAbVec-hIgKappa or pAbVect-mIgKappa) using NEBuilder (New England Biolabs). The human IgG1-N297Q vector was produced by site-directed mutagenesis of the human IgG1 vector using a Phusion site-directed mutagenesis kit. Antibodies were produced by cotransfecting Expi293 cells with an appropriate heavy and kappa chain plasmid using Hype5 transfection reagent (Oz Biosciences). 4 d after transfection, supernatant was collected and mAbs were purified on a Protein A Agarose column (Pierce).
Antibody half-life measurement
4-wk-old WT C57BL/6J male mice were administered 200 µg of anti-MAYV mAbs by intraperitoneal injection. At specified time points, mice were euthanized and perfused with 40 ml PBS, and feet were collected. The supernatant from foot homogenates was analyzed for anti-MAYV mAbs by whole-virus capture ELISA using a standard curve of MAY-115 mIgG2a. Briefly, ELISA plates were coated with 104 FFU/well MAYV-CH at 4°C overnight. Plates were blocked using 5% BSA in PBS for 2 h at 37°C and then incubated for 2 h with serial dilutions of foot homogenate supernatant in parallel with a serial dilution of a known quantity of MAY-115 mIgG2a. After washing, bound antibody was detected using goat anti-mouse or anti-human HRP-conjugated antibodies (1:1,000; Southern Biotech). Plates were developed using tetramethylbenzidine substrate (Invitrogen), and the reaction was stopped with H2SO4. ELISA plates were read using a TriBar LB941 plate reader (Berthold Technologies). The optical density values from the known quantity of MAY-115-mIgG2a were fitted to a standard curve and compared with the optical density values of the foot homogenate supernatant to determine amount of MAYV-specific antibody present in the tissue.
FcγR ELISA
ELISA plates were coated with 200 ng/well of soluble FcγR proteins (R&D Systems) at 4°C overnight. Plates were blocked using 5% BSA in PBS for 2 h at 37°C. Anti-MAYV mAbs were added (1 µg/well) and incubated at room temperature for 2 h. Unbound antibody was washed from the plate, and bound antibody was detected as described above.
Next-generation sequencing
Following vRNA extraction of MAYV strains, samples were amplified using a random RT-PCR protocol. The resulting amplicons were used to generate a library using the NEBNext (NEB) kit with indexing as described (Zhao et al., 2017). Libraries from multiple indexed samples were pooled and sequenced on the 2 × 250 bp Illumina MiSeq platform. Consensus sequences were generated using Geneious v10.2.2 by mapping short reads onto the genome of MAYV-BeH742930.
Antibody binding to infected cells
Vero cells were inoculated with MAYV-BeH407 at an MOI of 1. 18 h later, cells were detached with TrypleE (Thermo Fisher Scientific) and washed with PBS supplemented with 2% BSA at 4°C. Cells were incubated with fivefold dilutions (1 µg/ml to 20.5 pg/ml) of anti-MAYV mAbs for 30 min at 4°C. Bound mAbs were detected using an Alexa Fluor 647–conjugated goat anti-mouse IgG antibody (Thermo Fisher Scientific) before fixation with 2% PFA for 10 min. Antibody-bound cells were detected by flow cytometry using a MACSQuant analyzer (Miltenyi Biotec). The percentage of cells positive for a given mAb was compared with cells stained with a saturating amount of (100 µg/ml) an oligoclonal mixture of anti-MAYV mAbs. Binding EC50 values were determined by plotting the percentage of positive cells relative to the oligoclonal mAb control and fitting the data to a nonlinear regression curve.
Antibody-dependent neutrophil phagocytosis
Recombinant MAYV E2 protein was biotinylated and conjugated to streptavidin-coated Alexa Fluor 488 beads. MAYV E2–coated beads were incubated with fivefold dilutions of antibodies (mAbs: 5–0.0016 µg/ml) in cell culture medium for 2 h at 37°C. Bone marrow cells were harvested from C57BL/6 mice. Cells were washed with PBS, and 5.0 × 104 cells per well were added to bead-antibody immune complexes and incubated for 1 h at 37°C. Cells were stained with the following antibodies: CD11b APC (clone M1/70; BioLegend), CD11c APC/Cy7 (clone N418; BioLegend), Ly6G Pacific Blue (clone 1A8; BioLegend), Ly-6C BV605 (clone HK1.4; BioLegend), and CD3 PE/Cy7 (clone 17A2; BioLegend). Cells were fixed with 4% PFA and analyzed on an IntelliCyt iQue Screener Plus flow cytometer. Neutrophils were defined as CD3− and CD11c− cells that were Ly6C−, CD11b+, and Ly6G+. The phagocytic score was determined using the following calculation: (percentage of Alexa Fluor 488+ cells) × (Alexa Fluor 488 geometric mean fluorescent intensity of Alexa Fluor 488+ cells)/10,000.
Antibody-dependent cellular phagocytosis
Recombinant MAYV E2 protein was biotinylated and conjugated to streptavidin-coated Alexa Fluor 488 beads. MAYV E2–coated beads were incubated with fivefold dilutions of antibodies (mAbs: 5–0.0016 µg/ml) in cell culture medium for 2 h at 37°C. J774A.1 (TIB-67; ATCC) murine monocyte cells were added to bead–antibody immune complexes (5.0 × 104 cells per well) and incubated for 1 h at 37°C. Cells were washed in 5 mM EDTA PBS, fixed with 4% PFA, and analyzed on an IntelliCyt iQue Screener Plus flow cytometer. The phagocytic score was determined using the following calculation: (percentage of Alexa Fluor 488+ cells) × (Alexa Fluor 488 geometric mean fluorescent intensity of Alexa Fluor 488+cells)/10,000.
Glycan analysis
The relative abundance of antibody glycan structures was quantified by capillary electrophoresis, as previously described (Mahan et al., 2015). Briefly, mAbs were purified using protein G magnetic beads (New England Biolabs) and then treated with IdeZ protease (New England Biolabs) to release the antibody Fab portion. The Fc-attached beads were washed, and N-glycans were removed from the Fc domains and labeled with 8-aminopyrene-1,3,6-trisulfonic acid (APTS) using the GlycanAssure APTS Kit (Thermo Fisher Scientific), as described in the manufacturer’s protocol. Labeled glycans were loaded onto the 3500 Genetic Analyzer (Thermo Fisher Scientific). Peaks for differentially galactosylated, fucosylated, bisected (GlcNAc), and sialylated structures were identified (13 peaks were reproducibly observed). The relative abundance of each glycan structure was determined by calculating the area under the curve, normalized for equal amounts of loaded APTS dye, of each peak divided by the total area of all peaks.
Statistical analysis
Statistical significance was assigned with P values <0.05 using GraphPad Prism version 7.0. The specific test for each dataset is indicated in respective figure legends and was selected based on the number of comparison groups and variance of the data. For foot swelling analysis, significance was determined by a two-way ANOVA with Tukey’s post-test (more than two groups) or Sidak’s post-test (between two groups). Viral burden data were analyzed by a one-way ANOVA with a Dunnett’s post-test. Survival curve analysis was analyzed by the log rank test. A Bonferroni correction was used depending on the number of comparison groups.
Data availability
Next-generation sequencing data have been deposited in the National Center for Biotechnology Information Trace and Short-Read Archive accession number PRJNA525867 and consensus sequences were deposited with GenBank accession numbers MK573238–MK573246. No proprietary software was used in the data analysis.
Online supplemental material
Fig. S1 shows the neutralization curves for each mAb with each h strain of MAYV. Figs. S2 and S3 show an amino acid alignment and relative identity of MAYV E1 and E2 proteins, respectively, of the strains used in this study. Fig. S4 shows binding data of the mAbs to E2 and E2 B domain in ELISA as well as the full dataset of antibody mapping data for each mAb. Fig. S5 shows the glycan profile and antibody effector activity for the anti-MAYV E2 mAbs as well as the half-lives of the isotype switched mAbs in vivo. Table S1 provides strain information for the strains used in this study. Table S2 provides kinetic and equilibrium binding values for the mAbs for MAYV E2 protein measured by BLI.
Supplementary Material
Acknowledgments
The authors thank S. Vernon and J. Blatter for assistance with next-generation sequencing.
This work was supported by the National Institutes of Health (grants R01 AI089591, R01 AI114816, and U19AI142790).
M.S. Diamond is a consultant for Inbios and Atreca and is on the Scientific Advisory Board of Moderna. The authors declare no further competing financial interests.
Author contributions: Conceptualization: J.T. Earnest, K. Basore, V. Roy, G. Alter, D.H. Fremont, and M.S. Diamond; Methodology: J.T. Earnest, K. Basore, D. Wang, G. Alter, D.H. Fremont, and M.S. Diamond; Investigation: J.T. Earnest, K. Basore, V. Roy, A.L. Bailey, and D. Wang; Writing (original draft): J.T. Earnest, K. Basore, and M.S. Diamond; Writing (review and editing): J.T. Earnest, K. Basore, V. Roy, A.L. Bailey, D. Wang, G. Alter, D.H. Fremont, and M.S. Diamond; Mentorship: G. Alter, D.H. Fremont, and M.S. Diamond; Funding acquisition: D.H. Fremont and M.S. Diamond.
References
- Anthony R.M., Nimmerjahn F., Ashline D.J., Reinhold V.N., Paulson J.C., and Ravetch J.V.. 2008. Recapitulation of IVIG anti-inflammatory activity with a recombinant IgG Fc. Science. 320:373–376. 10.1126/science.1154315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auguste A.J., Liria J., Forrester N.L., Giambalvo D., Moncada M., Long K.C., Morón D., de Manzione N., Tesh R.B., Halsey E.S., et al. 2015. Evolutionary and ecological characterization of Mayaro virus strains isolated during an outbreak, Venezuela, 2010. Emerg. Infect. Dis. 21:1742–1750. 10.3201/eid2110.141660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin S.K., Dowd K.A., Shrestha B., Nelson C.A., Edeling M.A., Johnson S., Pierson T.C., Diamond M.S., and Fremont D.H.. 2012. Structural basis of differential neutralization of DENV-1 genotypes by an antibody that recognizes a cryptic epitope. PLoS Pathog. 8:e1002930 10.1371/journal.ppat.1002930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo R.S., Silva E.V., Carvalho V.L., Rodrigues S.G., Nunes-Neto J.P., Monteiro H., Peixoto V.S., Chiang J.O., Nunes M.R., and Vasconcelos P.F.. 2009. Mayaro fever virus, Brazilian Amazon. Emerg. Infect. Dis. 15:1830–1832. 10.3201/eid1511.090461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basore K., Kim A.S., Nelson C.A., Zhang R., Smith B.K., Uranga C., Vang L., Cheng M., Gross M.L., Smith J., et al. 2019. Cryo-EM structure of Chikungunya virus in complex with Mxra8 receptor. Cell. 177:1725–1737.e16. 10.1016/j.cell.2019.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brustolin M., Pujhari S., Henderson C.A., and Rasgon J.L.. 2018. Anopheles mosquitoes may drive invasion and transmission of Mayaro virus across geographically diverse regions. PLoS Negl. Trop. Dis. 12:e0006895 10.1371/journal.pntd.0006895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton M., Lee H., Mulvey M., and Brown D.T.. 1997. Role of glycoprotein PE2 in formation and maturation of the Sindbis virus spike. J. Virol. 71:1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causey O.R., and Maroja O.M.. 1957. Mayaro virus: a new human disease agent. III. Investigation of an epidemic of acute febrile illness on the river Guama in Pará, Brazil, and isolation of Mayaro virus as causative agent. Am. J. Trop. Med. Hyg. 6:1017–1023. 10.4269/ajtmh.1957.6.1017 [DOI] [PubMed] [Google Scholar]
- Cheng R.H., Kuhn R.J., Olson N.H., Rossmann M.G., Choi H.K., Smith T.J., and Baker T.S.. 1995. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell. 80:621–630. 10.1016/0092-8674(95)90516-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H., Kudchodkar S.B., Reuschel E.L., Asija K., Borole P., Ho M., Wojtak K., Reed C., Ramos S., Bopp N.E., et al. 2019. Protective immunity by an engineered DNA vaccine for Mayaro virus. PLoS Negl. Trop. Dis. 13:e0007042 10.1371/journal.pntd.0007042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia B.E., Bates J.T., Loomis R.J., Baneyx G., Carrico C., Jardine J.G., Rupert P., Correnti C., Kalyuzhniy O., Vittal V., et al. 2014. Proof of principle for epitope-focused vaccine design. Nature. 507:201–206. 10.1038/nature12966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers G., Bentlage A.E.H., Stegmann T.C., Howie H.L., Lissenberg-Thunnissen S., Zimring J., Rispens T., and Vidarsson G.. 2017. Affinity of human IgG subclasses to mouse Fc gamma receptors. MAbs. 9:767–773. 10.1080/19420862.2017.1323159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo D.J., Tan G.S., Palese P., and Ravetch J.V.. 2014. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat. Med. 20:143–151. 10.1038/nm.3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J., and Brown D.T.. 1986. Sindbis virus-mediated cell fusion from without is a two-step event. J. Gen. Virol. 67:377–380. 10.1099/0022-1317-67-2-377 [DOI] [PubMed] [Google Scholar]
- Fernandez E., Kose N., Edeling M.A., Adhikari J., Sapparapu G., Lazarte S.M., Nelson C.A., Govero J., Gross M.L., Fremont D.H., et al. 2018. Mouse and Human Monoclonal Antibodies Protect against Infection by Multiple Genotypes of Japanese Encephalitis Virus. MBio. 9:e00008–e00018. 10.1128/mBio.00008-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J.M., Long F., Edeling M.A., Lin H., van Duijl-Richter M.K.S., Fong R.H., Kahle K.M., Smit J.M., Jin J., Simmons G., et al. 2015. Broadly Neutralizing Alphavirus Antibodies Bind an Epitope on E2 and Inhibit Entry and Egress. Cell. 163:1095–1107. 10.1016/j.cell.2015.10.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J.M., Roy V., Gunn B.M., Huang L., Edeling M.A., Mack M., Fremont D.H., Doranz B.J., Johnson S., Alter G., and Diamond M.S.. 2019. Optimal therapeutic activity of monoclonal antibodies against chikungunya virus requires Fc-FcγR interaction on monocytes. Sci. Immunol. 4:eaav5062 10.1126/sciimmunol.aav5062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goo L., VanBlargan L.A., Dowd K.A., Diamond M.S., and Pierson T.C.. 2017. A single mutation in the envelope protein modulates flavivirus antigenicity, stability, and pathogenesis. PLoS Pathog. 13:e1006178 10.1371/journal.ppat.1006178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidner H.W., Knott T.A., and Johnston R.E.. 1996. Differential processing of sindbis virus glycoprotein PE2 in cultured vertebrate and arthropod cells. J. Virol. 70:2069–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessell A.J., Hangartner L., Hunter M., Havenith C.E., Beurskens F.J., Bakker J.M., Lanigan C.M., Landucci G., Forthal D.N., Parren P.W., et al. 2007. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 449:101–104. 10.1038/nature06106 [DOI] [PubMed] [Google Scholar]
- Ho I.Y., Bunker J.J., Erickson S.A., Neu K.E., Huang M., Cortese M., Pulendran B., and Wilson P.C.. 2016. Refined protocol for generating monoclonal antibodies from single human and murine B cells. J. Immunol. Methods. 438:67–70. 10.1016/j.jim.2016.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard S., and Thornton J.. 1993. NACCESS Computer Program. Department of Biochemistry and Molecular Biology, University College London, London. [Google Scholar]
- Jin J., Liss N.M., Chen D.H., Liao M., Fox J.M., Shimak R.M., Fong R.H., Chafets D., Bakkour S., Keating S., et al. 2015. Neutralizing Monoclonal Antibodies Block Chikungunya Virus Entry and Release by Targeting an Epitope Critical to Viral Pathogenesis. Cell Reports. 13:2553–2564. 10.1016/j.celrep.2015.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Galaz-Montoya J.G., Sherman M.B., Sun S.Y., Goldsmith C.S., O’Toole E.T., Ackerman L., Carlson L.A., Weaver S.C., Chiu W., and Simmons G.. 2018. Neutralizing Antibodies Inhibit Chikungunya Virus Budding at the Plasma Membrane. Cell Host Microbe. 24:417–428.e5. 10.1016/j.chom.2018.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y., Nimmerjahn F., and Ravetch J.V.. 2006. Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science. 313:670–673. 10.1126/science.1129594 [DOI] [PubMed] [Google Scholar]
- Kelley L.A., Mezulis S., Yates C.M., Wass M.N., and Sternberg M.J.. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10:845–858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A.S., Austin S.K., Gardner C.L., Zuiani A., Reed D.S., Trobaugh D.W., Sun C., Basore K., Williamson L.E., Crowe J.E. Jr., et al. 2019. Protective antibodies against Eastern equine encephalitis virus bind to epitopes in domains A and B of the E2 glycoprotein. Nat. Microbiol. 4:187–197. 10.1038/s41564-018-0286-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimstra W.B., Ryman K.D., and Johnston R.E.. 1998. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J. Virol. 72:7357–7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyuchenko V.A., Jakana J., Liu X., Haddow A.D., Aung M., Weaver S.C., Chiu W., and Lok S.M.. 2011. The structure of barmah forest virus as revealed by cryo-electron microscopy at a 6-angstrom resolution has detailed transmembrane protein architecture and interactions. J. Virol. 85:9327–9333. 10.1128/JVI.05015-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., He W., Liu X., Zheng S., Qi Y., Li H., Mao F., Liu J., Sun Y., Pan L., et al. 2017. A potent human neutralizing antibody Fc-dependently reduces established HBV infections. eLife. 6:e26738 10.7554/eLife.26738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahan A.E., Tedesco J., Dionne K., Baruah K., Cheng H.D., De Jager P.L., Barouch D.H., Suscovich T., Ackerman M., Crispin M., and Alter G.. 2015. A method for high-throughput, sensitive analysis of IgG Fc and Fab glycosylation by capillary electrophoresis. J. Immunol. Methods. 417:34–44. 10.1016/j.jim.2014.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancardi D.A., Iannascoli B., Hoos S., England P., Daëron M., and Bruhns P.. 2008. FcgammaRIV is a mouse IgE receptor that resembles macrophage FcepsilonRI in humans and promotes IgE-induced lung inflammation. J. Clin. Invest. 118:3738–3750. 10.1172/JCI36452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourão M.P.G., Bastos Mde.S., de Figueiredo R.P., Gimaque J.B.L., Galusso Edos.S., Kramer V.M., de Oliveira C.M.C., Naveca F.G., and Figueiredo L.T.M.. 2012. Mayaro fever in the city of Manaus, Brazil, 2007-2008. Vector Borne Zoonotic Dis. 12:42–46. 10.1089/vbz.2011.0669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C.A., Lee C.A., and Fremont D.H.. 2014. Oxidative refolding from inclusion bodies. Methods Mol. Biol. 1140:145–157. 10.1007/978-1-4939-0354-2_11 [DOI] [PubMed] [Google Scholar]
- Pal P., Dowd K.A., Brien J.D., Edeling M.A., Gorlatov S., Johnson S., Lee I., Akahata W., Nabel G.J., Richter M.K., et al. 2013. Development of a highly protective combination monoclonal antibody therapy against Chikungunya virus. PLoS Pathog. 9:e1003312 10.1371/journal.ppat.1003312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal P., Fox J.M., Hawman D.W., Huang Y.J., Messaoudi I., Kreklywich C., Denton M., Legasse A.W., Smith P.P., Johnson S., et al. 2014. Chikungunya viruses that escape monoclonal antibody therapy are clinically attenuated, stable, and not purified in mosquitoes. J. Virol. 88:8213–8226. 10.1128/JVI.01032-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes A.M., Brown D.T., Rothnagel R., Chiu W., Schoepp R.J., Johnston R.E., and Prasad B.V.. 1993. Three-dimensional structure of a membrane-containing virus. Proc. Natl. Acad. Sci. USA. 90:9095–9099. 10.1073/pnas.90.19.9095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro F.P., Freitas R.B., Travassos da Rosa J.F., Gabbay Y.B., Mello W.A., and LeDuc J.W.. 1981. An outbreak of Mayaro virus disease in Belterra, Brazil. I. Clinical and virological findings. Am. J. Trop. Med. Hyg. 30:674–681. 10.4269/ajtmh.1981.30.674 [DOI] [PubMed] [Google Scholar]
- Porta J., Jose J., Roehrig J.T., Blair C.D., Kuhn R.J., and Rossmann M.G.. 2014. Locking and blocking the viral landscape of an alphavirus with neutralizing antibodies. J. Virol. 88:9616–9623. 10.1128/JVI.01286-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers A.M., Aguilar P.V., Chandler L.J., Brault A.C., Meakins T.A., Watts D., Russell K.L., Olson J., Vasconcelos P.F., Da Rosa A.T., et al. 2006. Genetic relationships among Mayaro and Una viruses suggest distinct patterns of transmission. Am. J. Trop. Med. Hyg. 75:461–469. 10.4269/ajtmh.2006.75.461 [DOI] [PubMed] [Google Scholar]
- Robinson D.M., Cole F.E. Jr., McManus A.T., and Pedersen C.E. Jr. 1976. Inactivated Mayaro vaccine produced in human diploid cell cultures. Mil. Med. 141:163–166. 10.1093/milmed/141.3.163 [DOI] [PubMed] [Google Scholar]
- Saphire E.O., Schendel S.L., Fusco M.L., Gangavarapu K., Gunn B.M., Wec A.Z., Halfmann P.J., Brannan J.M., Herbert A.S., Qiu X., et al. Viral Hemorrhagic Fever Immunotherapeutic Consortium . 2018. Systematic Analysis of Monoclonal Antibodies against Ebola Virus GP Defines Features that Contribute to Protection. Cell. 174:938–952.e13. 10.1016/j.cell.2018.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaljohn A.L., Johnson E.D., Dalrymple J.M., and Cole G.A.. 1982. Non-neutralizing monoclonal antibodies can prevent lethal alphavirus encephalitis. Nature. 297:70–72. 10.1038/297070a0 [DOI] [PubMed] [Google Scholar]
- Sheehan K.C., Lai K.S., Dunn G.P., Bruce A.T., Diamond M.S., Heutel J.D., Dungo-Arthur C., Carrero J.A., White J.M., Hertzog P.J., and Schreiber R.D.. 2006. Blocking monoclonal antibodies specific for mouse IFN-alpha/beta receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection. J. Interferon Cytokine Res. 26:804–819. [DOI] [PubMed] [Google Scholar]
- Smith J.L., Pugh C.L., Cisney E.D., Keasey S.L., Guevara C., Ampuero J.S., Comach G., Gomez D., Ochoa-Diaz M., Hontz R.D., and Ulrich R.G.. 2018. Human Antibody Responses to Emerging Mayaro Virus and Cocirculating Alphavirus Infections Examined by Using Structural Proteins from Nine New and Old World Lineages. MSphere. 3:e00003–e00018. 10.1128/mSphere.00003-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.A., Silva L.A., Fox J.M., Flyak A.I., Kose N., Sapparapu G., Khomandiak S., Ashbrook A.W., Kahle K.M., Fong R.H., et al. 2015. Isolation and Characterization of Broad and Ultrapotent Human Monoclonal Antibodies with Therapeutic Activity against Chikungunya Virus. Cell Host Microbe. 18:86–95. 10.1016/j.chom.2015.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao M.H., and Morrison S.L.. 1989. Studies of aglycosylated chimeric mouse-human IgG. Role of carbohydrate in the structure and effector functions mediated by the human IgG constant region. J. Immunol. 143:2595–2601. [PubMed] [Google Scholar]
- Tsetsarkin K.A., Vanlandingham D.L., McGee C.E., and Higgs S.. 2007. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 3:e201 10.1371/journal.ppat.0030201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt M.R., Dowd K.A., Engle M., Tesh R.B., Johnson S., Pierson T.C., and Diamond M.S.. 2011. Poorly neutralizing cross-reactive antibodies against the fusion loop of West Nile virus envelope protein protect in vivo via Fcgamma receptor and complement-dependent effector mechanisms. J. Virol. 85:11567–11580. 10.1128/JVI.05859-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss J.E., Vaney M.C., Duquerroy S., Vonrhein C., Girard-Blanc C., Crublet E., Thompson A., Bricogne G., and Rey F.A.. 2010. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature. 468:709–712. 10.1038/nature09555 [DOI] [PubMed] [Google Scholar]
- Waggoner J.J., Rojas A., Mohamed-Hadley A., de Guillén Y.A., and Pinsky B.A.. 2018. Real-time RT-PCR for Mayaro virus detection in plasma and urine. J. Clin. Virol. 98:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weger-Lucarelli J., Aliota M.T., Kamlangdee A., and Osorio J.E.. 2015. Identifying the Role of E2 Domains on Alphavirus Neutralization and Protective Immune Responses. PLoS Negl. Trop. Dis. 9:e0004163–e0004163. 10.1371/journal.pntd.0004163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise W.J., Hermance M.E., Forrester N., Adams A.P., Langsjoen R., Gorchakov R., Wang E., Alcorn M.D.H., Tsetsarkin K., and Weaver S.C.. 2014. A novel live-attenuated vaccine candidate for mayaro Fever. PLoS Negl. Trop. Dis. 8:e2969–e2969. 10.1371/journal.pntd.0002969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wust C.J., Crombie R., and Brown A.. 1987. Passive protection across subgroups of alphaviruses by hyperimmune non-cross-neutralizing anti-Sindbis serum. Proc. Soc. Exp. Biol. Med. 184:56–63. [DOI] [PubMed] [Google Scholar]
- Zhao G., Vatanen T., Droit L., Park A., Kostic A.D., Poon T.W., Vlamakis H., Siljander H., Härkönen T., Hämäläinen A.M., et al. 2017. Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children. Proc. Natl. Acad. Sci. USA. 114:E6166–E6175. 10.1073/pnas.1706359114 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Next-generation sequencing data have been deposited in the National Center for Biotechnology Information Trace and Short-Read Archive accession number PRJNA525867 and consensus sequences were deposited with GenBank accession numbers MK573238–MK573246. No proprietary software was used in the data analysis.