RRAS2 is involved in setting the threshold for negative selection of T cells in the thymus. In its absence, most autoreactive T cells are eliminated, and, consequently, mice become resistant to development of autoimmune diseases in experimental models.
Abstract
Signal strength controls the outcome of αβ T cell selection in the thymus, resulting in death if the affinity of the rearranged TCR is below the threshold for positive selection, or if the affinity of the TCR is above the threshold for negative selection. Here we show that deletion of the GTPase RRAS2 results in exacerbated negative selection and above-normal expression of positive selection markers. Furthermore, Rras2−/− mice are resistant to autoimmunity both in a model of inflammatory bowel disease (IBD) and in a model of myelin oligodendrocyte glycoprotein (MOG)–induced experimental autoimmune encephalomyelitis (EAE). We show that MOG-specific T cells in Rras2−/− mice have reduced affinity for MOG/I-Ab tetramers, suggesting that enhanced negative selection leads to selection of TCRs with lower affinity for the self-MOG peptide. An analysis of the TCR repertoire shows alterations that mostly affect the TCRα variable (TRAV) locus with specific VJ combinations and CDR3α sequences that are absent in Rras2−/− mice, suggesting their involvement in autoimmunity.
Graphical Abstract
Introduction
T cell development in the thymus is an active process that implies different intracellular signaling events regulating cell differentiation, proliferation, and survival. This process generates an anticipatory peripheral T cell repertoire able to promote a proper adaptive defense to future antigens derived from pathogens. Thymic development follows well-defined maturation steps according to the expression of CD4 and CD8 coreceptors: CD4−CD8− (double negative [DN]), CD4+CD8+ (double positive [DP]), and CD4+ or CD8+ (single positive [SP]; SP4 or SP8). At the DN stage, the expression of a correctly rearranged TCRβ chain together with the invariant pTα chain allows thymocytes to mature to the DP stage and rearrange the TCRα chain to express a mature α/β TCR (Falk et al., 2001). To promote only the survival of functional self-tolerant T cells, DP thymocytes are subjected to a stringent selection process based on the affinity of their randomly recombined TCR for self-peptides presented by MHC molecules (self-pMHC). Thymocytes with TCRs that fail to interact, and those with TCRs that interact with too much affinity with self-pMHCs, enter into an apoptotic process called “death by neglect” or “negative selection,” respectively. Only those thymocytes with TCRs that interact with low but sufficient affinity are “positively selected” and progress toward SP4 or SP8 stages. Hence, TCR signaling is essential during development of thymocytes and controls their fate (death, survival, and differentiation; Starr et al., 2003; Gascoigne et al., 2016).
Engagement of the TCR with cognate pMHC promotes a conformational change and the initiation of downstream signaling cascades, beginning with the recruitment of Nck and phosphorylation of CD3 immunoreceptor tyrosine activation motifs (ITAMs) by the Src-kinase, Lck. ITAM phosphorylation generates docking sites for the Syk-tyrosine kinase ZAP-70, which in turn phosphorylates the adaptor LAT and promotes the generation of the early TCR scaffold complex together with SLP76 and PLCγ1, activating multiple biochemical pathways (Cantrell, 2015; Alcover et al., 2018). In vivo studies have shown that thymocyte selection is dependent on the activation of early TCR signaling molecules, such as the kinases and adaptors Lck, ZAP70, Nck, LAT, and SLP76 (Zamoyska et al., 2003; Singer et al., 2008; Borroto et al., 2013). In addition to those TCR signal transducers that play a role at all stages of a T cell’s life, new molecular players are being discovered, such as Themis and Tespa1, which regulate TCR signaling during negative and positive selection (Wang et al., 2012; Gascoigne et al., 2016; Choi et al., 2017).
Activation of the TCR downstream pathways PI3K-Akt and ERK via Ras proteins is of great importance during thymocyte development. Thus, PI3K-Akt has been described to control pre-TCR-dependent differentiation, positive and negative selection signals, CD4/CD8 ratio, differentiation, and thymic exit (Na et al., 2003; Rodríguez-Borlado et al., 2003; Barbee and Alberola-Ila, 2005; Fayard et al., 2010). Likewise, Ras-ERK signals have been shown to be required for pre-TCR signaling, as well as to determine positive or negative selection, depending on the kinetics of ERK activation (Fischer et al., 2005; Daniels et al., 2006). The Ras family of small GTPases consists of 39 genes in humans (Rojas et al., 2012). Although some of their members are frequently mutated in human cancer, their role in physiological processes is not as well defined. Using germline knockout mouse lines of classical RAS subfamily members Nras and Hras, it was shown that these genes mediate neither positive nor negative selection (Iborra et al., 2011). In contrast, the implication of several guanosine nucleotide exchange factor proteins such as RasGRP and Sos in thymocyte selection processes is remarkable, suggesting that Ras GTPases are indeed involved (Dower et al., 2000; Iborra et al., 2011; Kortum et al., 2013). In our previous work, we described that RRAS2, a small GTPase of the RRAS subfamily, interacts with ITAMs of the TCR and BCR and that it plays a fundamental role in the survival and homeostatic proliferation of mature T and B cells, presumably through the activation of the PI3K-Akt pathway (Delgado et al., 2009; Martínez-Martín et al., 2009). More recently, we have shown that RRAS2 has a nonredundant, B cell–intrinsic essential role during the germinal center response by controlling B cell metabolism (Mendoza et al., 2018).
In the present study, we describe the relevance of RRAS2 during T cell development. We found that Rras2─/─ mice have reduced DP thymocyte numbers and show an activated thymocyte phenotype with increased up-regulation of Nur77 and Bim pro-apoptotic molecules. Moreover, the absence of RRAS2 in DP OT-I TCR transgenic thymocytes makes them more sensitive to apoptosis induced by OVA antigen-variant peptides of low affinity, indicating an enhanced negative selection process. Consequently, Rras2─/─ mice have an altered peripheral TCR repertoire and show less susceptibility to undergo autoimmune disorders, such as inflammatory bowel disease (IBD) or experimental autoimmune encephalomyelitis (EAE). We hypothesize that RRAS2 could be an important molecule in the TCR signaling cascade, setting the threshold for negative selection in thymocytes by activating the pro-survival PI3K-Akt pathway.
Results
Reduced numbers of DP thymocytes in Rras2−/− mice
To investigate the role of RRAS2 during T cell maturation in the thymus, we first studied the distribution of thymocytes within the four major thymic populations in Rras2−/− mice and WT (Rras2+/+ WT) controls. We found that thymuses of Rras2−/− mice were slightly bigger than those of their WT counterparts and had increased numbers of thymocytes of the DN (CD4–CD8–), CD4SP (CD4+CD8–), and CD8SP (CD4–CD8+) populations, although those differences were only significant for CD4SP cells (Fig. 1 A). However, analysis of RRAS2 deficiency in terms of the relative distribution into the four populations showed that Rras2−/− mice presented a small, but significant, reduction in the percentage of DP (CD4+CD8+) thymocytes, accompanied by a small increase in the percentage of CD4SP and CD8SP (Fig. 1 B). The effects on the relative distribution of thymic populations were accompanied by a slight reduction of TCR expression in Rras2−/− mice compared with WT in the DP, CD4SP, and CD8SP populations (Fig. 1 C). This reduction of TCR expression was concomitant with a small but significant reduction of TCR and CD4 or CD8 coreceptor expression in mature T cells of peripheral lymphoid organs (Fig. S1 A). The reduced expression of the TCR and coreceptors may be indicative of a defect in positive or negative selection at the DP stage in Rras2−/− mice. Another piece of information that indicated a defect in positive or negative selection at the DP stage was that the number of DP thymocytes negative or with low TCR (or CD3) expression was not smaller in Rras2−/− mice than in WT controls (Fig. 1 D). In fact, the number of DP CD3low thymocytes was a bit higher, in line with the general tendency to have bigger thymuses in the absence of RRAS2 (Fig. 1 A). The result was very different when the number of DP thymocytes positive for CD3 was analyzed; Rras2−/− mice have reduced number of DP positive thymocytes once they express the TCR (Fig. 1 D). This result shows that defects in Rras2−/− mice during thymic selection occur after the TCR is expressed.
Figure 1.
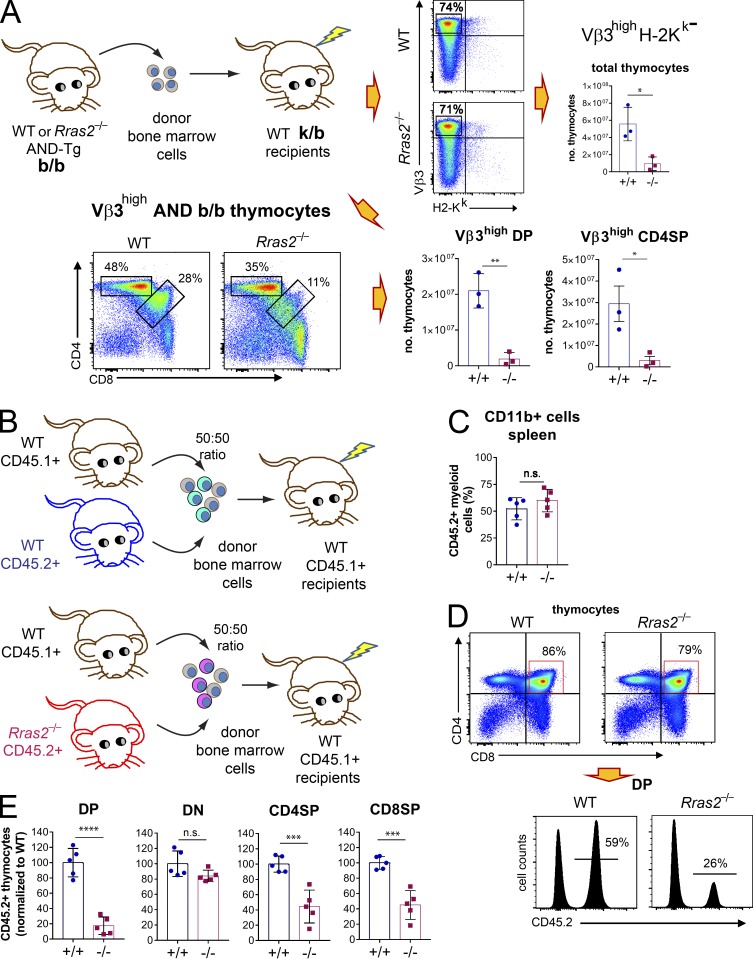
Reduced percentage of DP thymocytes in Rras2-deficient mice. (A) Flow cytometry analysis of WT and Rras2-deficient thymocytes in 6-wk-old C57BL/6 mice according to the expression of CD4 and CD8α markers. Bar plots to the right represent the number of CD4ˉCD8ˉ (DN), CD4+CD8+ (DP), CD4+CD8ˉ (CD4SP), CD4ˉCD8+ (CD8SP), and total thymocytes in both genotypes (n = 8 mice per group). Blue circles, Rras2+/+; red squares, Rras2−/−. (B) Percentages of DN, DP, SP4, and DP8 thymocytes in mice analyzed as in A. n = 11 mice per group. (C) Flow cytometry analysis of CD3 expression in the indicated thymocyte populations. An overlay plot of Rras2+/+ (continuous line) and Rras2−/− (broken lines) is shown on the left, and quantification of CD3 expression is shown in the bar blots on the right. n = 4 mice per group. MFI, mean fluorescence intensity. (D) Number of DP thymocytes according to the expression of CD3. Two-color plots show CD3 expression versus DP thymocyte size. Bar plots below represent the number of DP thymocytes expressing or not expressing CD3 for both genotypes Rras2+/+ (blue circles) and Rras2−/− (red squares). n = 9 mice per group. FS, forward scatter. (E) Flow cytometry analysis of WT and Rras2-deficient thymocytes in ANDTg mice in a pure b/b background and in a mixed k/b background after gating within the transgenic Vβ3high thymocytes. Bar plots to the right represent the percentage of thymocyte subpopulations for both genotypes. Blue circles, Rras2+/+; red squares, Rras2−/−. n = 4–7 mice per group. (F) Number of DN, DP, SP4, and DP8 thymocytes in AND TCR mice in a pure b/b background and in a mixed k/b background. Blue circles, Rras2+/+; red squares, Rras2−/−. n = 4–6 mice per group. Quantitative data in all panels are means ± SEM. *, P < 0.05; **, P < 0.005; ****, P < 0.0005; n.s., not significant (unpaired two-tailed Student’s t test). All phenotyping experiments in this figure were repeated at least three times.
We did not detect major alterations in the percentage of natural regulatory T cells (nT reg cells) within the CD4SP population in the thymus. This is to be expected given that nT reg cell differentiation is believed to occur for thymocytes with TCR affinities for self-pMHC that lie between the affinities that drive positive and negative selection (Hsieh et al., 2012; Klein et al., 2014). Instead, we did detect a small reduction of the percentage of nT reg cells in spleen, but not in lymph nodes, of Rras2−/− compared with WT mice (Fig. S1 B).
To determine if RRAS2 deficiency leads to defective selection in the thymus, we crossed Rras2−/− mice and their WT counterparts with mice transgenic for the AND TCR (ANDTg). The AND TCR is specific for a moth cytochrome c peptide presented by I-Ek (Kaye et al., 1989). The number of ANDTg thymocytes in mice homozygous for the I-Ek allele is reduced in comparison with I-Ek heterozygous mice (Kaye et al., 1992). Importantly, this suggests that the I-Ek allele induces a certain extent of negative selection, possibly because of a higher affinity of the ANDTg receptor for this allele compared with the also positively selecting I-Ab allele (Kaye et al., 1992). To determine whether RRAS2 affected negative selection, we backcrossed ANDTg Rras2−/− mice in mixed k/b background with C57BL/6 mice for eight generations to create a line of mice homozygous for the H-2b haplotype (b/b) or bearing one H-2k haplotype (k/b). ANDTg WT DP thymocytes are selected toward CD4SP mature thymocytes in both the k/b and b/b haplotype (Fig. 1 E). Compared with WT, mice deficient in RRAS2 in a k/b background had a lower percentage of DP thymocytes and slightly higher percentage of CD4SP (Fig. 1 E). The analysis of the number of thymocytes, however, showed a strong and significant reduction of the DP and the CD4SP populations (Fig. 1 F). By contrast, there were no significant differences in the distribution or size of thymic populations between WT and Rras2−/− mice in the absence of the I-Ek haplotype (b/b background; Fig. 1, E and F). AND TCR expression was not significantly different in DP and CD4SP thymocytes of WT or Rras2−/− mice in a k/b background (Fig. S1 D). Therefore, the reduced number of DP and CD4SP thymocytes in the presence of the I-Ek haplotype suggests that the absence of RRAS2 enhances negative selection.
To investigate if the effect of RRAS2 deficiency on thymic selection was T cell–intrinsic, we adoptively transferred bone marrow precursors from ANDTg Rras2−/− or WT mice of the b/b background into lethally irradiated WT nontransgenic k/b recipient mice. Thymuses of reconstituted mice were analyzed for major subset distribution within the donor-derived, ANDTg-specific cells (Vβ3high, H-2k–negative). The comparison between donor cells of WT and Rras2−/− genotypes showed a strong reduction in the number of DP and CD4SP thymocytes in the absence of RRAS2 (Fig. 2 A). This result shows that the effect of RRAS2 deficiency on thymic selection is indeed T cell–intrinsic.
Figure 2.
Reduced numbers of DP thymocytes in Rras2−/− mice result from a T cell–intrinsic defect. (A) WT adult mice of mixed k/b background were lethally irradiated and reconstituted with bone marrow cells from either Rras2+/+ or Rras2−/− AND TCR transgenic mice of b/b background. 2 mo after reconstitution, thymocyte count and DP and CD4SP subset distribution were analyzed within the donor H-2k negative Vβ3+ (AND b/b) population. Quantitative data are means ± SEM (n = 3). *, P < 0.05; **, P < 0.005 (unpaired two-tailed Student’s t test). This experiment was repeated twice. (B) WT adult mice of b/b background expressing the CD45.1 allele were lethally irradiated and reconstituted with an equal mixture of bone marrow cells from WT CD45.1+ mice and either Rras2+/+ or Rras2−/− mice expressing the CD45.2 allele. (C) 2 mo after reconstitution, chimerism was analyzed by measuring the distribution of the CD45.2 allele within myeloid CD11b+ spleen cells. (D) The effect of RRAS2 deficiency on the distribution of thymocyte subpopulations was analyzed according to the expression of the CD45.2 allele. An example of CD45.2 expression within the DP thymocyte population of chimeric mice is shown in the plots below. (E) Relative contribution of the donor CD45.2+ cells to the different thymocyte populations. Data points from two different experiments are integrated by normalizing the percentage of CD45.2+ cells to those of the mean values in the WT controls within each experiment. Quantitative data in all panels are means ± SEM (n = 5 mice per group). ****, P < 0.00005; ***, P < 0.0005; (unpaired two-tailed Student’s t test). The experiment of B–E was repeated twice. n.s., not significant.
At this point, we examined whether the relative reduction of DP thymocytes detected in Rras2−/− mice with a polyclonal TCR repertoire (Fig. 1 A) was also T cell intrinsic. To enhance our possibility of observing the potential effect of RRAS2 deficiency, we performed this experiment under competitive conditions. Donor bone marrow cells of WT or Rras2−/− genotype bearing the CD45.2 allele were mixed with an equal proportion (50:50) of donor WT bone marrow cells bearing the CD45.1 allele. The mixture was subsequently inoculated into lethally irradiated CD45.1+ WT recipient mice (Fig. 2 B). 2 mo after bone marrow reconstitution, the expression of the CD45.2 allele within the myeloid CD11b+ compartment in the spleen was close to 50%, regardless of the Rras2 genotype (Fig. 2 C). The analysis of thymic populations showed, by contrast, a very different distribution. The percentage of thymocytes bearing the CD45.2 allele within the DP, CD4SP, and CD8SP subsets was strongly reduced in Rras2−/− donor thymocytes, compared with their WT controls (Fig. 2, D and E). The percentage of CD45.2+ thymocytes within the DN subset did not differ significantly between the two genotypes (Fig. 2 E). Furthermore, the distribution of DN thymocytes into the four major subpopulations defined by the expression of CD44 and CD25 markers was not significantly different between WT and Rras2−/− mice in the bone marrow reconstitution experiment (Fig. S1 E), either in nontransgenic mice (Fig. S1 F) or in ANDTg mice (Fig. S1 G). These data indicate that RRAS2 deficiency induces a defect in thymic selection beyond the DN stage in both TCR transgenic and nontransgenic mice bearing a polyclonal TCR repertoire.
Enhanced negative selection in Rras2−/− mice
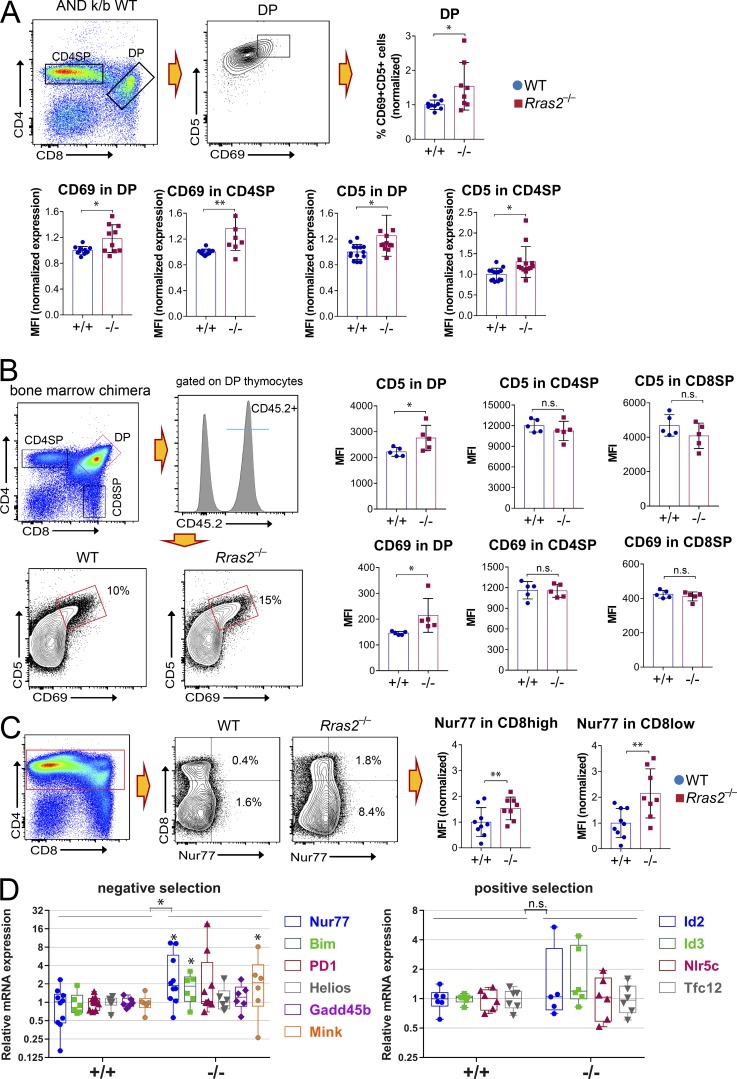
The differential effect of RRAS2 deficiency on selection of ANDTg thymocytes in the presence and absence of I-Ek (Fig. 1, E and F) suggested that reduced DP thymocyte number was due to enhanced negative selection. To further investigate if RRAS2 deficiency enhanced negative selection or was, in contrast, the consequence of deficient positive selection, we analyzed the expression of positive and negative selection markers in DP, transitional, and CD4SP ANDTg thymocytes in the k/b background. DP thymocytes that receive a TCR signal up-regulate CD69 and CD5, which are considered markers of TCR signaling strength (Bendelac et al., 1992; Brändle et al., 1994; Azzam et al., 1998). In fact, CD69 expression correlates with the nature of positive or negative selecting peptides and is higher in the latter (Merkenschlager et al., 1997). When comparing the expression of CD5 and CD69 in DP and CD4SP thymocytes of WT and Rras2−/−mice, we found that RRAS2 deficiency did not decrease the expression of those markers. Rather, it had the opposite effect, such that CD5 and CD69 were significantly more up-regulated than in WT thymocytes (Fig. 3 A). This increased up-regulation of CD5 and CD69 markers suggests that ANDTg Rras2−/− thymocytes are not deficient in positive selection. To test if the same effect held true for non-TCR transgenic thymocytes, we studied the expression of CD5 and CD69 in thymic Rras2−/− and WT populations under the competitive conditions described for the experiments in Fig. 2. We found that both markers were significantly more up-regulated in Rras2−/− DP thymocytes than in their WT counterparts and that Rras2−/− and WT SP thymocytes showed no differences in expression level (Fig. 3 B). These data suggest that positive selection was not impaired in the absence of RRAS2 and that Rras2−/− DP thymocytes actually received stronger signals via their TCR.
Figure 3.
Enhanced expression of positive selection and negative selection markers in thymocytes from Rras2−/− mice. (A) Thymocytes from AND TCR transgenic mice in mixed k/b background were analyzed for expression of positive selection markers CD5 and CD69 within the DP and CD4SP subsets. Bar plots show quantitative data from a total of three experiments. Percentage and mean fluorescence intensity (MFI) values are represented normalized to those of the mean values in the WT controls within each experiment. Mean percentage and MFI in WT thymocytes are set to 1. Quantitative data are means ± SEM (n = 7–12 mice per group). *, P < 0.05; **, P < 0.005 (unpaired two-tailed Student’s t test). This experiment was repeated three times. (B) Expression of positive selection markers CD5 and CD69 in the thymic subsets of CD45.2+ thymocytes from the bone marrow competition experiments of Fig. 2, B–E. An example of the gating strategy is shown for the DP population. Quantitative MFI data for the entire DP, CD4SP, and CD8SP populations are means ± SEM (n = 5 mice per group). *, P < 0.05 (unpaired two-tailed Student’s t test). This experiment was repeated twice. (C) Thymocytes from AND TCR transgenic mice in mixed k/b background were analyzed for expression of the negative selection marker Nur77 within CD4+ subsets, as indicated. Bar plots show the normalized expression of Nur77 in the CD8high (DP) and CD8low (transitional and CD4SP) subpopulations in a combination of three experiments. MFI values are represented normalized to those of the mean values in the WT controls within each experiment. Quantitative data are means ± SEM (n = 8–9 mice per group). **, P < 0.005 (unpaired two-tailed Student’s t test). This experiment was repeated three times. (D) RT-qPCR analysis of gene expression in sorted DP thymocytes from Rras2+/+ and Rras2−/− mice. Genes are grouped according to their differential expression during positive and negative selection (Mingueneau et al., 2013). An interleaved box and whiskers plot showing all data points and the maximum and minimum has been chosen for data representation. Values are normalized to Rras2+/+. *, P < 0.05. An unpaired two-tailed Student’s t test was used to compare significance of individual genes; a two-way ANOVA test was used to compare gene sets. This experiment was repeated four times. n.s., not significant.
Another marker of TCR signaling strength is the orphan nuclear hormone receptor Nur77, which is considered a marker of negative selection in vivo (Moran et al., 2011). Nur77 expression was up-regulated in ANDTg DP (CD8high in Fig. 3 C) and CD4SP thymocytes going through intermediate CD8low stages (CD8low, Fig. 3 C) in Rras2−/− mice compared with WT. To determine if additional negative selection markers were up-regulated in the absence of RRAS2 in non-TCR transgenic thymocytes, we performed a quantitative RT-PCR (RT-qPCR) analysis of gene expression on FACS-sorted DP thymocytes using a panel of genes previously characterized as differentially regulated during positive or negative selection (Mingueneau et al., 2013). The comparison between Rras2−/− and WT genotypes showed a significant transcriptional up-regulation of genes associated with negative selection, whereas expression of genes associated with positive selection was not significantly altered in the absence of RRAS2 (Fig. 3 D). Altogether, the results shown in Fig. 3 indicate that positive selection is not deficient in Rras2−/− mice and that stronger negative selection occurs according to the specific marker expression.
Another model to study the effect of RRAS2 deficiency on positive versus negative selection is that of OT1 TCR-transgenic mice. The OT1 TCR recognizes a peptide derived from OVA (OVAp) presented by H2-Kb and drives differentiation of OT1 T cells into the CD8+ lineage. A phenotypic analysis of adult Rras2−/− and WT mice showed that the former had relatively fewer DP thymocytes (CD4highCD8high, Fig. 4 A) and more transitional CD4lowCD8low and CD4+CD8low thymocytes. Unlike in mice with a polyclonal TCR repertoire (Fig. S1), OT1 CD8 T cells showed no differences in the expression of CD8β in peripheral T cells from Rras2−/− mice compared with WT (Fig. 4 B).
Figure 4.
Enhanced activation of Rras2-deficient DP thymocytes in response to low- and high-affinity TCR ligands. (A) Rras2+/+ or Rras2−/− OT1 TCR transgenic mice were analyzed for the distribution of thymocytes among DP (CD4highCD8high), CD8SP (CD4ˉCD8+), and intermediate maturation subsets (CD4lowCD8low; CD4+CD8low; n = 5 mice per group). (B) Expression of CD8α and CD8β subunits within gated CD8α+ T cells in lymph nodes of OT1 TCR transgenic mice. The percentage of DP CD8α+CD8β+ T cells and the MFI for both markers is shown in the bar plots to the right (n = 3 mice per group). (C) CD69 and Annexin V expression was analyzed in Rras2+/+ and Rras2−/− DP OT1 TCR transgenic thymocytes after a 24-h incubation with the indicated concentrations of OVA peptide derivatives of different affinity. Annexin V values were adjusted using a semilog line function. Quantitative data are means ± SEM (n = 5). **, P < 0.005 (unpaired two-tailed Student’s t test). All experiments in this figure were repeated twice. n.s., not significant.
A sharp affinity threshold between positively selecting and negatively selecting peptides for the OT1 TCR has been defined based on the induction of CD69 expression (Daniels et al., 2006). We incubated total thymocytes from Rras2−/− and WT OT1Tg mice for 24 h with splenic antigen-presenting cells loaded with OVAp variants with characterized affinity for the OT1 TCR. In this system, the Q4H7 peptide variant is positive selecting, whereas the Q4R7 peptide is just above the threshold of affinity necessary for negative selection (Daniels et al., 2006). We found that all OVAp derivatives induced more CD69 and annexin V expression (a marker of apoptosis) in RRAS2-deficient DP thymocytes than in their WT counterparts regardless of their concentrations and affinities (Fig. 4 C). These results are in agreement with the observation that induction of the TCR engagement marker CD69 in Rras2−/− DP thymocytes is not weaker but rather stronger than in WT DP ones and that induction of apoptosis is consistently stronger. These results suggest that RRAS2 deficiency does not lead to defective TCR signaling in response to low-affinity antigens in OT1 mice and reinforce the idea that RRAS2 deficiency enhances negative selection.
Defective PI3K-Akt pathway activation in thymocytes from Rras2−/− mice
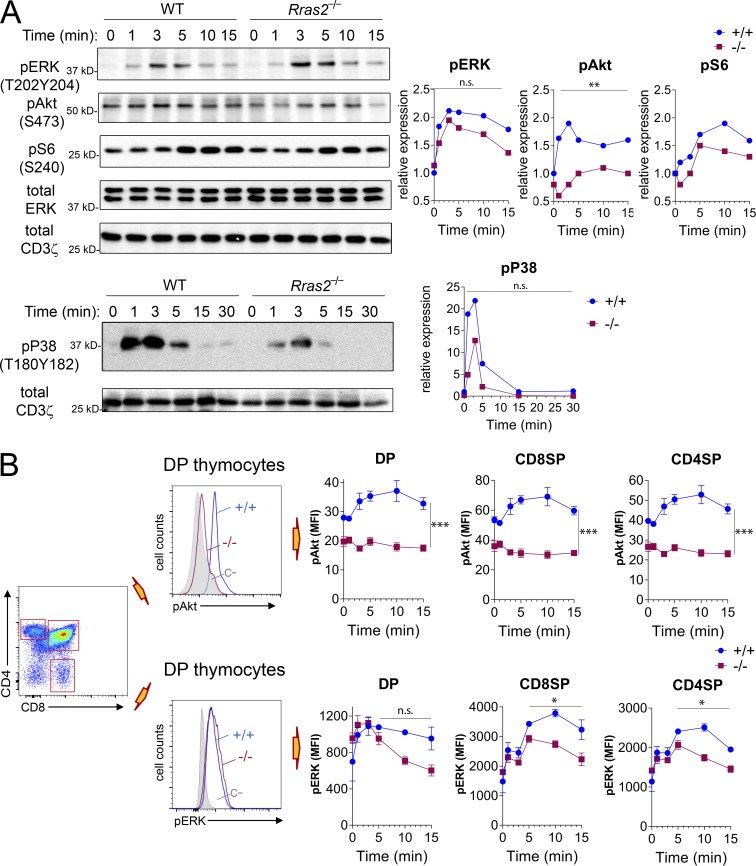
Differences in the intensity and kinetics of activation of the Raf-ERK pathway after TCR triggering have been correlated with the effect of positive-selecting versus negative-selecting ligands (Teixeiro and Daniels, 2010). Thus, negative selection correlates with an intense but brief phosphorylation of ERK, whereas positive selection correlates with weaker but more sustained ERK phosphorylation. On the other hand, although classic Ras GTPases activate the Raf-ERK pathway, RRAS2 is rather an activator of the PI3K-Akt pathway. We therefore investigated induction of both pathways after a short-term stimulation with anti-CD3, a stimulus likely mimicking negative selection. We first analyzed the induction of phosphorylation of ERK and Akt by Western blotting of post-nuclear lysates of total thymocytes stimulated with anti-CD3 in vitro. We found that dual ERK phosphorylation by MEK-1 at Thr202 and Tyr204 peaked at 3 min of stimulation with similar kinetics in thymocytes from WT and Rras2−/− mice (Fig. 5 A). However, phosphorylation of Akt at residue Ser473 by mTORC2 and phosphorylation of S6 by p70S6K, which is downstream of Akt, in residue Ser240 were reduced at all time points in the absence of RRAS2 compared with the WT. These results suggested that ERK phosphorylation is not affected by RRAS2 deficiency, whereas activation of the PI3K-Akt pathway is strongly inhibited.
Figure 5.
Deficient activation of the PI3K-Akt and Ras-ERK pathways in thymocytes from Rras2−/− mice. (A) Western blot of total thymocyte lysates stimulated with anti-CD3 antibody for the indicated times. Immunoblots were performed with the indicated phospho-specific antibodies. Immunoblots with antibodies to total ERK and/or CD3ζ were used as loading controls. A densitometric quantification of the intensity of the protein bands relative to the loading controls is shown to the right. A representative experiment out of four has been chosen for display. (B) Phosphoflow analysis of phospho-Akt (Ser473) and phospho-ERK (Thr202Tyr204) expression in thymocyte subpopulations after stimulation with anti-CD3 for the indicated time points. An overlay plot is shown in the middle panels illustrating changes in phospho-protein expression. Staining with an irrelevant rabbit antibody was used as negative control (C–; gray histograms). Quantitative data in the right panels are means ± SEM of triplicate values. *, P < 0.05; **, P < 0.005; ***, P < 0.0005 (paired two-tailed Student’s t test). Significance was assessed for the entire dataset except for pERK in B, where it was assessed for time points 5–15 min. All experiments in this figure were repeated three times. n.s., not significant.
We also examined if RRAS2 deficiency altered the phosphorylation of MKK6 (p38), since higher activation of this MAP kinase has been correlated with stronger negative selection (Sugawara et al., 1998). We detected reduced phosphorylation of p38 in thymocytes of Rras2−/− mice (Fig. 5 A), suggesting that enhanced negative selection in RRAS2-deficient mice is not due to increased p38 activity.
Since Western blotting was performed on total thymocyte lysates, we also measured ERK and Akt phosphorylation in thymic subsets by phospho-flow cytometry to study their regulation in thymic subpopulations. Induction of Akt phosphorylation by stimulation with anti-CD3 was not detectable in DP, CD4SP, or CD8SP thymocytes of Rras2−/− mice, whereas clear induction was observed in WT controls (Fig. 5 B). The effect of RRAS2 deficiency on ERK phosphorylation was, however, more complex: whereas ERK phosphorylation peaked between 3–5 min in RRAS2-deficient DP and SP thymocytes as in the WT controls, ERK phosphorylation was less sustained after 5 min of stimulation (Fig. 5 B). In summary, the Western blot and phospho-flow data indicate that thymocytes from Rras2−/− mice are deficient in the activation of the PI3K-Akt pathway upon stimulation with anti-CD3 and that ERK phosphorylation might be less sustained than in WT thymocytes.
Resistance to autoimmunity in Rras2−/− mice
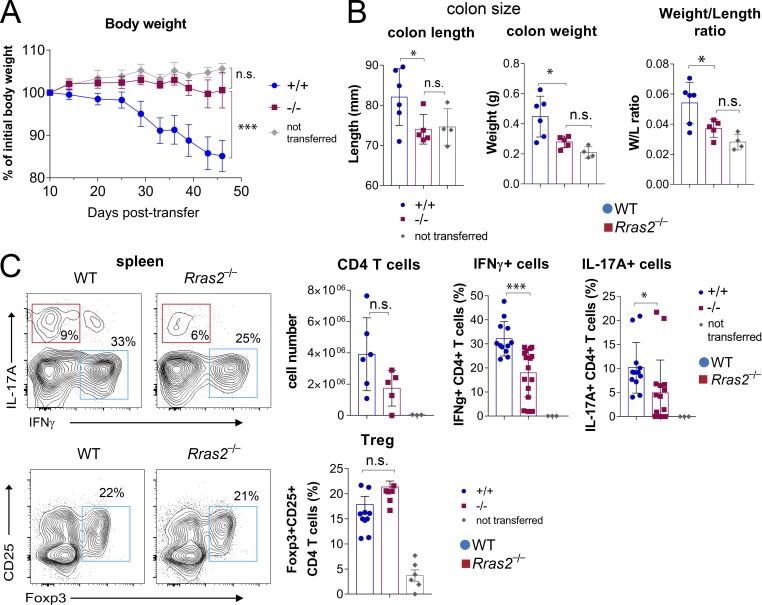
A consequence of enhanced negative selection in the thymus should be reduced autoreactivity of the mature T cells exiting the thymus and a reduction in the intensity or the propensity to develop autoimmune diseases in which T cells are a major driver. To test this hypothesis, we first selected a model of IBD, in which CD4+ T cells depleted of nT reg cells are adoptively transferred into lymphopenic mice. The homeostatic expansion of the transferred T cells is supposed to benefit the expansion of autoreactive T cells, which infiltrate the intestinal mucosa and cause symptoms similar to those observed in Crohn’s disease and ulcerative colitis. The adoptive transfer of CD4+CD25− T cells from WT donors to RAG1−/− lymphopenic mice caused a progressive loss of body weight that was not detected when transferred T cells originated from Rras2−/− mice (Fig. 6 A). Body weight loss in mice receiving WT T cells was accompanied by an increase in the length and weight of the colon, as well as the colon weight/length ratio, compared with those of the RAG1−/− control mice not inoculated with T cells (Fig. 6 B). In contrast, RAG1−/− mice that received T cells from Rras2−/− mice were indistinguishable from noninoculated mice. The absence of IBD symptoms in mice transferred with RRAS2-deficient T cells was not due to a defective expansion of the cells, because CD4+ T cell numbers detected in spleens of adoptively transferred mice were not significantly different from those of mice transferred with WT cells (Fig. 6 C). Although the number of total CD4+ T cells was not significantly different, the presence of pro-inflammatory effector Th cells producing IFNγ or IL-17A in spleen was significantly reduced in mice that received RRAS2-deficient versus WT T cells (Fig. 6 C). By contrast, no differences were observed for the Foxp3+CD25+–induced T reg cell population. These results indicate that conventional CD4+ T cells selected in the absence of RRAS2 are unable to promote IBD.
Figure 6.
Adoptive transfer of CD25ˉ CD4+ T cells from Rras2−/− mice to recipient Rag1−/− mice does not induce IBD symptoms. (A) Body weight loss of Rag1−/− mice transferred with Rras2+/+ (blue circles) or Rras2−/− (red squares) naive CD4 T cells or not transferred (gray circles). (B) Colon length, colon weight, and colon weight/length ratio of mice shown in A, sacrificed at day 46. (C) Total CD4+ T cell counts and expression of intracellular IFNγ, IL-17A, and Foxp3 by gated CD4+ T cells of spleens of adoptively transferred mice in A and B after 6 h of incubation with PMA plus ionomycin. Quantitative data in all panels are means ± SEM (n = 4–6 mice per group). *, P < 0.05; ***, P < 0.0005 (unpaired two-tailed Student’s t test). All experiments in this figure were repeated twice. n.s., not significant.
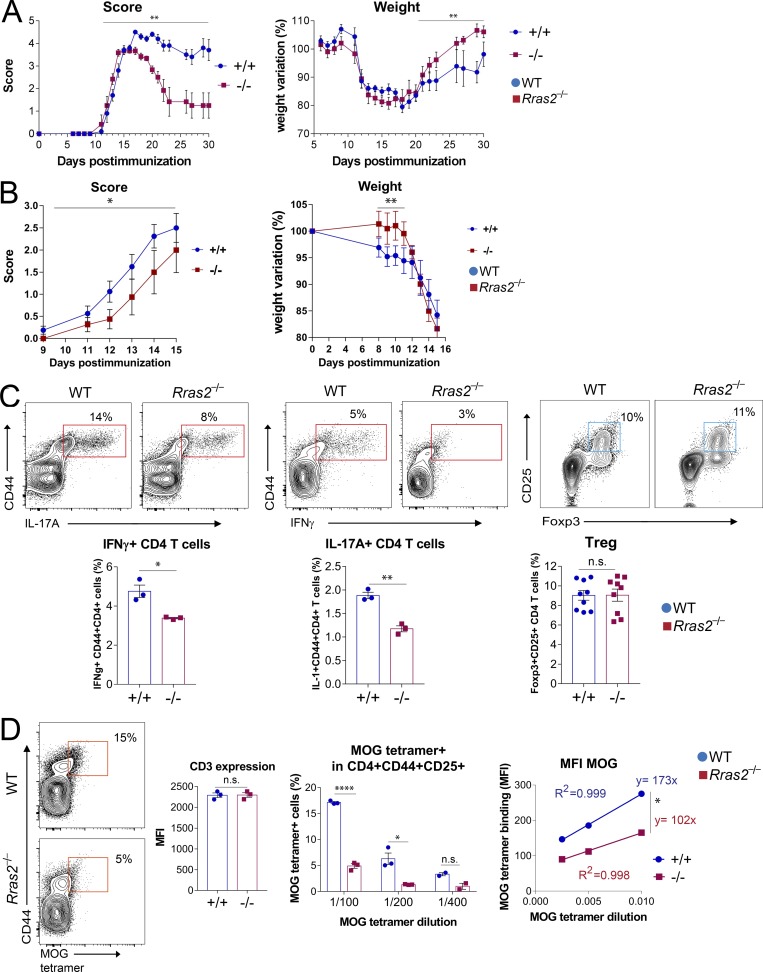
We used a second model of autoimmunity, EAE, in which tolerance to self-antigens is broken by immunization with a self-peptide in the presence of a strong adjuvant. This model allows detection of T cells reactive with the self-peptide used for immunization. EAE is induced upon immunization of female C57BL/6 mice with a peptide derived from myelin oligodendrocyte glycoprotein (MOG peptide). Immunization of WT mice led to a progressive loss of neurological capabilities that led to a complete paralysis of the hind limbs (Fig. 7 A). Immunization of Rras2−/− mice led to a milder disease with faster recovery from symptoms and less body weight loss than immunized WT mice (Fig. 7 A). We next tested if the milder form of EAE in Rras2−/− mice compared with WT was T cell intrinsic. To this aim, we reconstituted lethally irradiated WT mice with bone marrow of either WT or Rras2−/− mice and immunized with MOG 2 mo after reconstitution. RRAS2-deficient mice developed EAE symptoms at a significantly lower rate than WT mice and began to lose weight with a delay of 4 d (Fig. 7 B). These data indicate that the resistance to EAE of Rras2−/− mice is at least in part T cell intrinsic.
Figure 7.
Rras2−/− mice are resistant to EAE in a MOG immunization-induced model. (A) Score and body weight evolution in Rras2+/+ (blue) and Rras2−/− (red) mice immunized with MOG. Scoring was as follows: 0, normal behavior; 1, weakness at the distal portion of the tail; 1.5, complete flaccidity of the tail; 2, moderate hind limb weakness; 2.5, severe hind limb weakness; 3, ataxia; 3.5, partial hind limb paralysis; 4, complete hind limb paralysis; 4.5, complete hind limb paralysis accompanied by muscle stiffness; 5, moribund state and hence sacrificed for humanitarian reasons. Quantitative data are means ± SEM (n = 5–6). **, P < 0.005. Score difference significance was assessed using a nonparametric Wilcoxon matched-pair signed rank test. Weight variations were compared using an unpaired two-tailed Student’s t test. This experiment was repeated three times. (B) Score and body weight evolution in lethally irradiated WT C57BL/6 mice reconstituted with bone marrow from either Rras2+/+ (blue) or Rras2−/− (red) mice and immunized with MOG. Quantitative data are means ± SEM (n = 8). *, P < 0.05; **, P < 0.005. Score difference significance was assessed using a nonparametric Wilcoxon matched-pair signed rank test; weight variations were compared using an unpaired two-tailed Student’s t test. This experiment was repeated twice. (C) Detection of inflammatory cytokine-producing T cells and T reg cells in spleen cells 7 d after immunization with MOG. For IFNγ and IL-17A production, spleen cells were stimulated overnight with PMA plus ionomycin and treated with brefeldin A before intracellular staining with anti-IFNγ and anti-IL17A antibodies. Quantitative data are means ± SEM (n = 3). In a different experiment, the presence of T reg cells was evaluated in spleen cells of MOG-immunized mice by staining with CD25 and intracellular staining with Foxp3. Quantitative data are means ± SEM (n = 9). *, P < 0.05; **, P < 0.005 (unpaired two-tailed Student’s t test). (D) Detection of MOG tetramer+ within the CD4+CD44+CD25+ cell population of splenocytes was evaluated 7 d after immunization and 48 h of in vitro stimulation with MOG peptide. The bar plots show the percentage of MOG tetramer+ cells within the CD4+ population and CD3 expression in CD4+ T cells. The scatter plot to the right shows the MFI of MOG tetramer binding within the positive population. A linear regression fit is shown as well as the slope of the line plots. Quantitative data are means ± SEM (n = 3). *, P < 0.05; ****, P < 0.00005 (unpaired two-tailed Student’s t test). Experiments in C and D were repeated three times. n.s., not significant.
We next searched for the presence of MOG-responsive pro-inflammatory T cells in lymphoid organs of immunized mice. We found a reduced percentage of IFNγ- and IL-17A–producing CD4+ T cells in draining lymph nodes of Rras2−/− mice compared with WT (Fig. 7 C), suggesting that a weaker reactivity with MOG leads to less generation of pro-inflammatory effector CD4 T cells in RRAS2-deficient mice. By contrast, we did not detect significant differences in the percentage of Foxp3+CD25+CD4+ T reg cells, suggesting that resistance to EAE is not due to a higher abundance of T reg cells (Fig. 7 C).
The mice in Fig. 7 A were sacrificed at day 30, and the presence of MOG-reactive CD4+ T cells was evaluated in spleens by flow cytometry, gating on the CD4+TCRβ+ T cell population bearing the CD44 marker of activation. Staining with different dilutions of an I-Ab tetramer loaded with the immunizing MOG peptide allowed us to detect 8% MOG tetramer+ CD4+CD44+ T cells in WT mice and 6% in Rras2−/− mice (Fig. S2 A). The poorer expansion of MOG tetramer+ CD4 T cells in Rras2−/− mice could result from an underlying defect of RRAS2 deficient mature T cells in responding to TCR triggering. However, we found that the response of Rras2−/−CD4+ and CD8+ T cells to anti-CD3 stimulation in terms of CD69 expression and cytokine production (IL-2 and IFNγ) were unaffected (Fig. S3, A and B). The proliferative response to anti-CD3 plus anti-CD28 stimulation was not impaired in Rras2−/− T cells; in fact, RRAS2-deficient CD4 T cells proliferated faster than their WT counterparts (Fig. S3 C). These data indicate that the response to polyclonal stimulation of mature T cells is not defective in the absence of RRAS2. Furthermore, the proliferative response of OT1 CD8+ T cells to stimulation with its agonist peptide in vivo was indistinguishable from those of WT cells (Fig. S3 D). We therefore reasoned that the dissimilar percentages of MOG tetramer+ CD4 T cells in the EAE experiment in vivo were due to a different responsiveness derived from an exacerbated negative selection in the thymus. Along these lines, staining with the MOG tetramer was weaker in Rras2−/− mice at all tetramer dilutions in spite of the fact that TCR expression was identical to WT MOG tetramer+ T cells, as shown by co-staining with an anti-TCRβ antibody (Fig. S2 A). Interestingly, we did not detect reduced reactivity of Rras2−/− mice to immunization with a nonself OVAp compared with their WT counterparts (Fig. S2 B), suggesting that the response to strong, foreign antigens is not diminished in the thymus. The reduction of the percentage of MOG tetramer+ CD4+ T cells (Fig. S2 A) in Rras2−/− mice data suggested a weaker reactivity of MOG tetramer with MOG-specific CD4+ T cells in immunized Rras2−/− mice than in WT controls. To confirm this idea, we analyzed the MOG-responsive CD4+ T cell population within shorter times after immunization. In this case, we collected the popliteal and inguinal draining lymph nodes and examined the percentage of MOG tetramer+ CD4 T cells after a brief MOG antigen-mediated expansion in vitro. The results showed a 3.5-fold reduction in the percentage of MOG tetramer+ cells within the CD4+CD44+ population (Fig. 7 D, center) in Rras2−/− mice compared with WT. Of note, the adjustment to a line plot of MOG tetramer concentrations versus fluorescence intensity reflected a smaller slope for staining of Rras2−/− compared with WT CD4 T cells (Fig. 7 D, right). Since TCR expression assessed with an anti-CD3 antibody in MOG tetramer+ CD4+ T cells was equal in WT and Rras2−/− mice (Fig. 7 D, center), the smaller slope suggests that MOG-reactive CD4+ T cells in Rras2−/− mice have less affinity for their antigen than in WT mice.
Analysis of the TCR repertoire in Rras2−/− mice points to TRAV4N-3 and TRAV4D-3 as Vα sequences biased toward autoimmunity
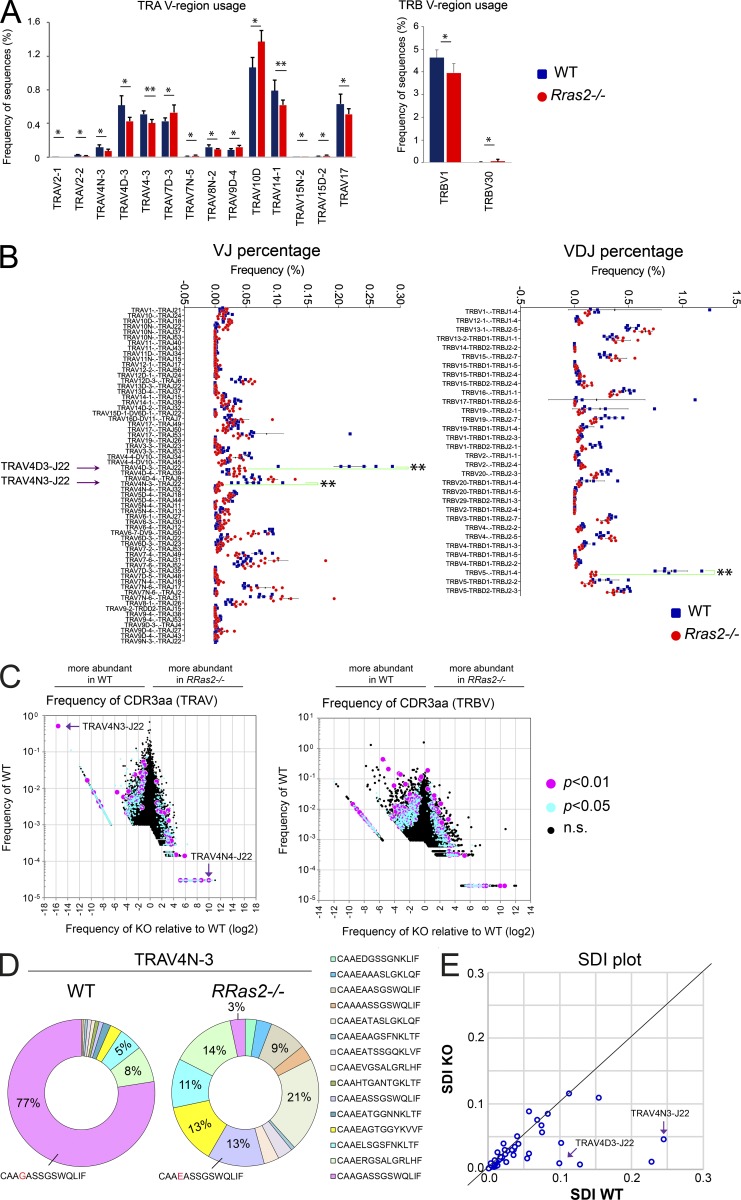
A consequence of enhanced negative selection in Rras2−/− mice might be manifested in a skewed TCR repertoire resulting from a lower frequency of the most autoreactive T cell clones. To study changes in the TCR repertoire, we purified total CD4+ T cells from nonimmunized WT and Rras2−/− mice and sequenced TRAV- and TRBV-encoding mRNAs after unbiased PCR expansion. TRAV is the gene locus encoding Vα variable regions, and TRBV is the gene locus encoding Vβ variable regions. An average of 357,785 ± 47,449 (mean ± SD; n = 12) reads/mouse for TRAV and of 365,643 ± 43,269 TRBV reads/mouse were obtained from WT and Rras2−/− mice (Datasets 1 and 2). Sequence frequency comparison showed significant differences in the use of two Vβ regions and fourteen Vα regions, with a generally lower representation in Rras2−/− mice (Fig. 8 A). The analysis of the usage of VJ combinations (TRA) and VDJ combinations (TRB) showed prominent significant differences for certain combinations (Fig. 8 B). Two VJ (TRAV4D3-J22 and TRAV4N3-J22) and one VDJ combination (TRBV5-J1-4) that are relatively abundant in WT T cells were detected at greatly reduced frequencies in Rras2−/− T cells (fivefold, eightfold, and fivefold, respectively). Interestingly, the two VJ combinations have been associated with MOG reactivity in the EAE model (Kieback et al., 2016), and therefore their diminished representation within the Rras2−/− T cell repertoire could explain the reduced susceptibility of Rras2−/− mice to MOG-induced EAE (Fig. 7 A). Furthermore, assuming they represent the high-affinity MOG clones, their scarce appearance could also explain the lower binding affinity for MOG tetramer within the remaining MOG-specific repertoire in these mice (Fig. 7 C).
Figure 8.
Rras2−/−mice have a biased TCR repertoire in peripheral CD4+ T cells. (A) Bar graphs show significant differences in TCRα and TCRβ V region usage between WT (blue) and Rras2−/− (red) mice. Vα and Vβ families are ordered according to the P value (most significant to the left). Data are means ± SD (n = 6 mice per group). *, P < 0.05; **, P < 0.005 (unpaired two-tailed Student’s t test). (B) Analysis of TCR clone diversity based on TCRα VJ rearrangement (left panel) and TCRβ VDJ rearrangement (right panel). The graph shows the expression frequency of clones showing significant differences between WT and Rras2−/− mice (P < 0.05). Arrows emphasize two TCRα VJ rearrangements and one TCRβ VDJ rearrangement for which representation in the repertoire differs strongly between WT and Rras2−/− CD4+ T cells. (C) Clonotype diversity graphs of TCRα (left) and TCRβ (right) based on the expression frequency of CDR3 sequences. Purple and blue dots illustrate all CDR3 clones with an expression that is significantly different between both groups of animals (P < 0.05 and 0.01, respectively). To calculate log2 of the KO/WT frequency ratio (x axis), the 0 values for Rras2−/− and WT sequences were arbitrarily set as 0.00001 and 0.00003, respectively. (D) Frequency of specific CDR3α sequences for CD4 T cell clones bearing the TRAV4N-3 Vα region. (E) Representation of the Simpson Diversity Index calculated for all Vα families with a number of reads >100 in WT and Rras2−/− CD4+ T cells. A line has been hand drawn to indicate a diagonal of equal index values. Data in this figure result from the aggregation of two experiments (three mice of each genotype per group and per experiment) performed independently. n.s., not significant.
In addition to frequencies of VJ and VDJ recombinations, we analyzed the frequencies of CDR3 loop amino acid sequences in WT and Rras2−/− mice, which in TCRα reside at the VJ junction and in TCRβ at the VDJ junction (Glusman et al., 2001). The CDR3α and CDR3β loops are especially relevant during thymic selection because they interact with self-peptides in the MHC-I and MHC-II grooves (Garcia and Adams, 2005). This analysis showed that various CDR3 sequences present in the WT repertoire at a wide range of frequencies were strongly underrepresented, or even absent, in the RRAS2-deficient T cell repertoire. These sequences might therefore belong to clones that were more strongly negatively selected in the absence of RRAS2 (Fig. 8 C). The TRAV sequence with the clearest overrepresentation in WT mice versus Rras2−/− mice was a clone with the CDR3α sequence (CAAGASSGSWQLIF, from here on termed “GASS”) encoded by the VJ combination TRAV4N3-TRAJ22, which was present with the mean frequency of 0.5% in WT T cells and absent in Rras2−/− T cells (Fig. 8 C, arrow). Of note, the CDR3 sequences most frequently found in Rras2−/− T cells compared with their WT counterparts corresponded to clones of no or very low abundance in WT T cells (<0.001%; Fig. 8 C). This suggests that those TCRs that are rare in WT mice could emerge in the absence of RRAS2 because of enhanced positive selection.
The comparison of preselection and post-selection TCR sequences has shown a shortening of CDR3β length as evidence of TCR repertoire shaping during positive and negative selection (Matsutani et al., 2007). We found no significant differences in CDR3α and CDR3β length between the post-selection WT and RRAS2-deficient T cell repertoires, except for very short or very long ones, when all sequences were globally considered, regardless of their V region usage (Fig. S4 A). However, we checked the TRAV4N-3 and TRAV4D-3 Vα regions in more detail, given the strong reduction of these regions in the repertoire of Rras2-deficient animals. There was a strong significant effect on the CDR3α length of these Vα regions (Fig. S4 B). Namely, the frequency of T cell clones bearing the CDR3α sequence of 42 nucleotides (14 amino acids) of TRAV4N-3 and TRAV4D-3 Vα was reduced by two-thirds in Rras2−/− mice, whereas the frequency of T cell clones bearing the 13-amino acid CDR3α associated to those Vα, was not significantly different from WT mice. The analysis of the CDR3 sequence frequencies bearing any of the two Vα regions showed that the sequence GASS, already detected in Fig. 8 C, was present in 77% of the TRAV4N-3 T cell clones found in WT mice (Fig. 8 D), as well as in 70% of the TRAV4D-3 T cell clones (Fig. S4 C). This sequence was virtually absent in T cells bearing any of the two Vα regions of Rras2−/− mice. At the same time, a very similar sequence (CAAEASSGSWQLIF) was found in T cells bearing TRAV4N-3 in Rras2−/− mice at relatively high frequency (13%; Fig. 8 D), suggesting a selection against T cells bearing a glycine residue in position 4 of the CDR3α and a contribution of the 14–amino acid sequence with glycine in position 4 to self-peptide recognition. Glycine in position 4 was found encoded by three different codons (gga, ggg, ggt; Dataset 1), indicating that the sequence was present in more than one TCR clone. Interestingly, the GASS CDR3α sequence was found in T cells of Rras2−/− mice, but not in those of the WT, associated with another Vα sequence (TRAV4N4; Fig. 8 C). This might indicate that deletion of TCRα sequences in the absence of RRAS2 is dictated by both the CDR3α sequence and the germline Vα-encoded CDR1α and CDR2α sequences and that the latter CDRs also contribute to autoreactivity.
To determine if the overabundance of T cell clones bearing a single CDR3α sequence in WT versus Rras2−/− CD4+ T cells was restricted to the TRAV4N-3 and TRAV4D-3 Vα regions or was more widespread, we studied the diversity of CDR3 sequences generated by each Vα. When comparing the number of different sequences generated by each Vα, we did not observe statistically significant differences between WT and Rras2−/− cells (Fig. S4 D). A different issue is how homogenously a given sequence is distributed within each Vα family. For this, we calculated the Simpson index (Simpson, 1949), which measures the degree of concentration when individuals are classified into types, obtaining a value of 1 when there is no diversity (all the sequences in the group are equal) and a value of 0 when the diversity is maximal (all the sequences inside the group are different). We obtained an average Simpson index of 0.0441 (± 0.1124) for WT sequences, and of 0.0326 (± 0.1069) for Rras2−/− sequences. This small difference was statistically significant (P = 0.016; paired t test), suggesting that there is in general less diversity in the TCRα sequences, classified by their Vα, in WT than in Rras2−/− mice. In other words, in WT mice there is overrepresentation of particular TCRα sequences that is not found, or found to a lower extent, in Rras2−/− mice (Fig. 8 E, Fig. S4 E, and Dataset 3). This indicates that the effect of RRAS2 deficiency on the overrepresentation of GASS-encoding clones (Fig. 8 D and Fig S4 C) is extended to other clones expressing other CDR3α and other Vα regions.
Finally, we aimed to determine if the differences detected by RNA sequencing (RNaseq) in peripheral CD4 T cells of WT versus Rras2−/− mice regarding expression of TRAV4N-3 and TRAV4D-3 Vα sequences were already present in the thymus. To this end, we developed a qPCR strategy to detect the TRAV4N-3 and TRAV4D-3 sequences based on primers that partially anneal to the CDR1α and CDR3α regions (Fig. S5 A). With that set of primers, we found a significant difference of expression within the same sets of samples used for RNaseq (Fig. S5 B). Once we confirmed that the qPCR strategy could distinguish differences in expression of the TRAV4N-3 and TRAV4D-3 sequences, we ran new samples of mRNA extracted from purified peripheral CD4+ T cells and from thymic CD4SP. We found that the TRAV4N-3 and TRAV4D-3 sequences were already underrepresented in the thymus of Rras2−/− mice compared with the frequency in WT mice (Fig. S5 C).
Discussion
We have found that RRAS2 plays an important role in shaping the T cell repertoire in the thymus. In the absence of RRAS2, DP thymocytes undergo a more stringent negative selection, resulting in peripheral resistance to autoimmunity manifested in two models of disease. While autoantigens in the adoptive transfer IBD model are unknown, in the EAE model the disease is unleashed by breaking self-tolerance through immunization with a peptide derived from MOG protein in the presence of strong adjuvants. Interestingly, we found that a faster improvement from neurological impairment symptoms in Rras2−/−mice was correlated with a lower affinity of MOG-reactive CD4+ T cells for MOG tetramers in mice. Furthermore, the comparative analysis of TRAV and TRBV repertoires in peripheral T cells from Rras2−/− and WT mice has allowed us to identify two Vα regions (TRAV4D-3 and TRAV4N-3), one Jα region (J22), and the 14–amino acid CDR3α GASS sequence as TCRα combinations that are strongly disfavored in the absence of RRAS2. TRAV4N-3 and TRAV4D-3 sequences have been previously linked to MOG recognition (Kieback et al., 2016), although they are not always linked to J22 or to the GASS sequence. Interestingly, one of the MOG-reactive T cell clones bears a TRAV4ND-3 Vα sequence associated to J22 with a CDR3α sequence (CAADSSGSWQLIF, sequence N5-575; Fillatreau et al., 2012) similar to GASS, suggesting that the latter might be involved in MOG recognition, as well. Another interesting finding, in our opinion, is that the GASS sequence, highly abundant in WT mice but absent in Rras2−/− mice when associated to TRAV4N-3-J22, is present in Rras2−/− but not in WT mice when associated to TRAV4N-4-J22. These data suggest that T cells bearing the GASS sequence are negatively selected when this is associated to TRAV4N-3 and TRAV4D-3 in Rras2−/− mice, but positively selected when associated to TRAV4N-4-J22. Therefore, it is possible that the GASS sequence favors either negative selection or positive selection depending on the context of the germline-encoded CDR1α and CDR2α sequences. The subsequent corollary would be that positive and negative selections are the result of a global effect of the three CDRs on interaction with self-pMHC in the thymus. The selection processes are hence not necessarily the result of a stepwise contribution first of germline CDR1 and CDR2 interacting with positions in MHC α-helices for positive selection and then of the peptide-interacting CDR3 for negative selection (Huseby et al., 2005; Vrisekoop et al., 2014).
The absence of particular VJ and VDJ combinations and CDR3 sequences in Rras2−/− mice, which on the other hand are highly abundant in WT mice, reinforces the idea that RRAS2 helps to set the threshold for negative selection in the thymus. RRAS2 could do so by providing a pro-survival signal for those DP thymocytes that bear a TCR affinity just above this threshold. This idea is sustained by the enhanced deletion of AND TCR-bearing thymocytes in the presence of the H-2k haplotype, by the overexpression of Nur77 and other markers of negative selection, and by the lack of inhibition of transcription of genes associated with positive selection. In this regard, the analysis of sequence diversity does not show impoverishment or enrichment in sequence diversity for TCRα in Rras2−/− versus WT CD4+ T cells, suggesting that the overall effect of RRAS2 deficiency is not that of reducing or increasing positive selection. By contrast, the Simpson analysis of diversity shows that there is overrepresentation of certain CD4 T cell clones in WT mice and not in Rras2−/− mice, of which those that express the GASS sequence in the TRAV4N-3 and TRAV4D-3 Vα families are just examples. In our opinion, the overrepresentation of independent clones bearing certain TCRα sequences in WT mice is due to the enhanced signaling fitness of thymocytes bearing those sequences. During immune responses in mice (Malherbe et al., 2004) and humans (Price et al., 2005), the T cell repertoire becomes restricted to a few dominant clones that have higher affinity for their ligand. Skewing of the preselection repertoire, based on quantitative parameters of the TCR self-pMHC interactions, also occurs in the thymus (Savage and Davis, 2001). This indicates that the strength of the TCR–MHC interaction is a driving force in shaping the immature and mature T cell repertoire. Our observation that abundant clones in WT mice, which include previously identified autoreactive clones, disappear in the thymus of Rras2−/− mice and the finding that these latter mice are more resistant to experimental autoimmune diseases support the hypothesis that strongly represented clones in the thymus of nonmanipulated WT mice are enriched for autoreactive T cells. As such, those clones are deleted in the absence of RRAS2 due to increased negative selection.
The reduced autoimmune capacity of the Rras2−/− T cell repertoire could in principle also be due to an impairment of positive selection of low-affinity autoreactive clones. The lower TCR-mediated signaling capacity of the Rras2−/− T cell precursors might prohibit them, as well as self-tolerant low-affinity clones, from reaching the minimally required positive selection signaling threshold. We consider this scenario less likely for three reasons. First, the MOG-specific T cell repertoire of Rras2−/− mice has a lower affinity for this antigen compared with the T cell repertoire in WT mice, showing that signaling in absence of RRAS2 permits inclusion of such low-affinity clones in the mature T cell repertoire. Second, the two TCR-transgenic models of positive selection we tested (OT-1; AND on a pure H-2b background) show no obvious defects in this process in Rras2−/− mice. Finally, the enhanced functional response of Rras2−/− OT-1 thymocytes to positively selecting ligands compared with WT thymocytes indicates that it is not the ability to respond to weak ligands that is inhibited but that it may actually change the fate of the thymocytes from positive selection to negative selection.
The nature of the signaling pathways activated by RRAS2 involved in protection against negative selection is not fully understood, although there are some clues derived from previous knowledge. RRAS2 is an activator of the PI3K-Akt pathway rather than the Raf-ERK pathway, and it has been previously described to deliver pro-survival signals to mature T and B cells (Delgado et al., 2009). The role of the PI3K-Akt pathway during thymic differentiation of T cells has been investigated mainly on the pre-TCR–driven transition from DN to DP thymocytes, where the PI3K-Akt pathway is required to maintain thymocyte survival during β-selection rather than thymocyte proliferation (Juntilla and Koretzky, 2008). In contrast, we have not detected a significant effect on the DN populations in Rras2−/− mice, suggesting that RRAS2 does not contribute in an important manner to pre-TCR signaling. The role of the PI3K-Akt pathway beyond the DP stage has not been studied in detail. An early work described that deletion of Pten, a negative regulator of PI3K activity, leads to increased thymic cellularity due to a possible negative selection defect (Suzuki et al., 2001). Interestingly, the overexpression of Pten beyond the β-selection checkpoint resulted in reduced DP and SP thymic populations, which was interpreted as a defective maturation to DP of a transitional intermediate SP population (Xue et al., 2008). However, the idea that reduced DP and SP thymocytes was due to enhanced negative selection was not discarded. Likewise, a double deletion of the p110δ and p110γ catalytic subunits of PI3Kδ and PI3Kγ, respectively, resulted in massive death at the DP stage, although pre-TCR function and DP generation remained unperturbed (Swat et al., 2006). These data point to a role for the PI3K pathway in the survival of thymocytes beyond the DP stage. Mechanistically, the anti-apoptotic role of the PI3K-Akt pathway in negative selection could be driven by Akt-mediated phosphorylation and degradation of the pro-apoptotic transcription factor involved in negative selection, Nur77 (Masuyama et al., 2001; Huo et al., 2010). Therefore, RRAS2 deficiency would lead to low PI3K-Akt activity and enhanced susceptibility to apoptosis induced by relatively high-affinity TCR interactions with self-pMHC complexes. The pronounced defect in Akt phosphorylation detected in Rras2−/− thymocytes demonstrates that RRAS2 is required for the activation of the PI3K-Akt pathway by TCR triggering in thymocytes. However, although less evident, we have also found that Rras2−/− thymocytes are deficient in maintaining a sustained ERK phosphorylation in response to TCR triggering. Interestingly, positive selection was associated with weak but prolonged ERK phosphorylation, whereas negative selection was associated with a rapid, transitory peak of ERK phosphorylation (reviewed in Teixeiro and Daniels, 2010). Therefore, it could be that enhanced negative selection in the absence of RRAS2 is due to defective sustained ERK phosphorylation rather than to defective activation of the PI3K-Akt pathway. Nonetheless, it is also known that PI3K activates ERK and that both Ras-ERK and PI3K-Akt pathways are intensely interconnected (Mendoza et al., 2011). At this stage, it is not possible to ascribe the protective effect of RRAS2 during negative selection to either the Raf-ERK or the PI3K-Akt pathway, although we favor a model in which it is the integration of the global effect on both pathways that determines the final outcome. Of note, if such balance determines positive and negative selection, the closely related small GTPases HRAS and NRAS do not seem to significantly contribute to this equilibrium, since simultaneous germline deletion of both genes did not cause detectable effects on positive and negative selections (Iborra et al., 2011). Accordingly, we have identified a new type of regulator of selection thresholds in the thymus, different in class from other proteins identified as key players, such as Themis, a possible adaptor for tyrosine phosphatases (Fu et al., 2013; Paster et al., 2015); Bim, a pro-apoptotic protein acting through the mitochondrial pathway; and Nur77, a transcriptional regulator of negative selection. Since RRAS2 directly binds the TCR (Delgado et al., 2009), one could speculate that it is upstream of the other players. Further work will help to determine the existence of such functional connections.
Materials and methods
Mice
Rras2−/− mice were generated as previously described (Delgado et al., 2009). These mice were crossed with AND TCRTg mice expressing I-Ab or I-Ek MHC class II (Kaye et al., 1989) in order to generate Rras2−/− ANDTg I-Ab/I-Ek. Rras2−/− mice were also crossed with mice transgenic for the OT2 TCR specific for a peptide corresponding to residues 323–339 of chicken OVA presented by I-Ab (Barnden et al., 1998). C57BL/6 mice bearing the pan-leukocyte marker allele CD45.1 were kindly provided by Dr. Carlos Ardavín (Centro Nacional de Biotecnología, Madrid, Spain). Cd3e−/− mice, deficient in the expression of CD3ε were obtained from Jackson Laboratories (DeJarnette et al., 1998). Mice homozygous for the Rag1tm1Mom mutation lack both T and B cells (Mombaerts et al., 1992) and were kindly provided by Dr. César Cobaleda (Centro de Biología Molecular Severo Ochoa, Madrid, Spain). All animals were backcrossed to the C57BL/6J background (Envigo) for ≥10 generations. For all in vivo experiments, age (5–8 wk) and sex were matched between the Rras2+/+ (WT) and Rras2−/− (KO) mice. Mice were maintained under specific pathogen-free conditions in the animal facility of the Centro de Biología Molecular Severo Ochoa in accordance with applicable national and European guidelines. All animal procedures were approved by the ethical committee of the Centro de Biología Molecular Severo Ochoa.
Antibodies and reagents
The following antibodies were used: anti-mouse CD45R-V450 -biotin -APC (RA3-6B2), CD4-PerCP -647 -FITC (RM4-5), CD8-biotin -V405 -PerCP -647 (53–6.7), CD11b-biotin (M1/70), purified CD16/32 (2,4G2), CD19–PE-Cy7 (1D3), CD25-APC (3C7), CD43-biotin (S7), CD45.1–APC-Cy7 (A20), CD45.2-APC (104), Gr1-biotin (RB6-8C5), CD44-PE (IM7), TCRβ-FITC (H57-597), CD3ε-PerCP (2C11), Vβ3-biotin -PE (KJ25), CD5-PE -biotin (53–7.3), IL-17a–PerCP (TC11-18H10), CD69-FITC -PerCP (H1.2F3), and NK1.1-biotin from BD Pharmigen; anti-mouse F4/80–biotin (BM8), H2-Kk–V405 (AF3-12.1.3), Foxp3-PE (NRRF-30), and Nur77-PE (12.14) from eBioscience; anti-mouse IFNγ–APC from Milteny; anti-mouse Vα2–PerCP (B20.1) from BioLegend; rabbit anti-mouse pERK (T202 Y204), pAKT (S473), pS6 (S240), total ERK, and total CD3ε from Cell Signaling.
I-Ab MOG3555 Tetramer-PE was acquired from MBL. I-Ab OVA329-337 Tetramer-APC was a generous donation of the National Institutes of Health Tetramer Core Facility. OVAp (SIINFEKL), Q4R7, Q4H7 (SIIQFEHL), E1 (EIINFEKL), and classII-OVA (AAHAEINEA) peptides were from Anaspec. CellTrace Violet (CTV; C34557) was from Invitrogen. OVA (A5503) and LPS (L2630) were from Sigma-Aldrich.
Cell preparation
The thymus, lymph nodes, and spleen from 5–8-wk-old mice were homogenized with 40 µm strainers and washed in PBS containing 2% (vol/vol) FBS. Spleen cells were resuspended for 3 min in AcK buffer (0.15 M NH4Cl, 10 mM KHCO3, 0.1 mM EDTA, pH 7.2–7.4) to lyse the erythrocytes and then washed in PBS 2% FBS.
For in vitro cultures, cells were maintained in RPMI with 10% FBS supplemented with 2 mM L-glutamine, 100 U/ml penicillin, 100 U/ml streptomycin, 20 µM β-mercaptoethanol, and 10 mM sodium pyruvate.
Flow cytometry
Mouse single-cell suspensions were incubated with fluorescence-labeled antibodies for 30 min at 4°C after blocking Fc receptors using anti-CD16/32 antibody. Afterwards, cells were washed in PBS+1%BSA, and data were collected on a FACS Canto II. Analyses were performed using the FlowJo program.
Bone marrow reconstitution
ANDTg TCR expressing I-Ek MHC class II mice were irradiated lethally with 1,000 rads the day before reconstitution. Bone marrow cells from Rras2+/+ or Rras2−/− ANDTg TCR mice expressing only I-Ab were harvested and injected in the irradiated receptor mice. Thymuses of reconstituted animals were analyzed by flow cytometry 6–8 wk after reconstitution.
For mixed bone marrow competition experiments, bone marrow cells from Rras2+/+ (CD45.2+) or Rras2−/− (CD45.2+) were mixed with bone marrow cells in a 50:50 ratio of C57BL/6 CD45.1+ mice. Mixed bone marrow cells were used to reconstitute lethally irradiated (10 Gy) C57BL/6 CD45.1+ receptor mice. Thymuses of reconstituted mice were collected and analyzed by flow cytometry 8 wk after reconstitution.
Analysis of T cell activation
Thymocytes were resuspended in RPMI plus 20 mM Hepes and left in starving conditions for 1 h. Cells were stimulated at different time points with soluble anti-CD3 (2C11) antibody (0, 1, 5, 15, and 30 min) at 37°C.
When T cell stimulation was analyzed by immunoblotting, after stimulation, cells were lysed in Brij96 lysis buffer containing protease and phosphatase inhibitors (1% Brij96, 140 mM NaCl, 10 mM Tris–HCl [pH 7.8], 10 mM iodoacetamide, 1 mM PMSF, 1 µg/ml leupeptin, 1 µg/ml aprotinin, 1 mM sodium orthovanadate, 20 mM sodium fluoride, and 5 mM MgCl2). After removing the nuclei by low-speed centrifugation, cell lysates were resolved by SDS-PAGE and immunoblotted using standard protocols. The membranes were probed with anti-phospho AKT, anti-phospho ERK, anti-phospho p70S6, or anti-total ERK antibodies (Cell Signaling), visualizing the protein bands by ECL (Pierce).
When T cell stimulation was analyzed by flow cytometry, after stimulation, cells were fixed in 4% paraformaldehyde. First, cells were stained extracellularly with anti-CD4 and anti-CD8 antibodies in order to differentiate the four different thymocyte subpopulations, and afterwards permeabilized with 0.1% NP-40 and incubated with rabbit anti-phosho ERK and anti-phosho AKT (Cell Signaling) followed by an anti-rabbit Ig Alexa 488 antibody (Jackson ImmunoReasearch). Phosphorylation levels were quantified by flow cytometry.
In vitro apoptosis assay
Splenocytes from Cd3e−/− mice were incubated for 2 h with different concentrations of OVAp, Q4R7, Q4H7, and E1 peptides based on Ref (Daniels et al., 2006). Afterwards, cells were washed, resuspended in RPMI medium, and plated in a 96-well plate together with 0.2 × 106 thymocytes from Rras2+/+ or Rras2−/− OTI mice. 20 h later, cells were stained for CD4, CD8, Vα2, and CD69. Annexin V labeling on the membrane of thymocytes was measured by staining with an anti-Annexin V (BD) following the manufacter’s instructions.
In vivo T cell proliferation
CD4 T cells from lymph nodes and spleens of Rras2+/+ and Rras2−/− OT2tg mice, expressing the CD45.2 allele, were purified by negative selection using a mix of biotinylated antibodies: anti-B220, anti-CD8, anti-NK1.1, anti-CD11b, anti-Gr1, and anti-F4/80. Afterwards, stained cells were incubated with streptavidin beads (Dynabeads Invitrogen) for 30 min and separated using a Dynal Invitrogen Beads Separator. Proliferation of OT2 cells was assessed using CTV labeling as specified by the manufacturer (Thermo Fisher Scientific). A total of 106 CTV-labeled OT2 T cells were inoculated intravenously into CD45.1+ receptor mice, and 24 h later they were immunized intraperitoneally with 200 µg OVA plus 50 µg LPS in saline buffer. 5 d after immunization, splenocytes were analyzed by flow cytometry for CTV dilution within the transferred OT2 T cells.
In vitro T cell activation
A total of 0.5 × 106 splenocytes from Rras2+/+ and Rras2−/− mice were plated in 96-well plates that had been preincubated with anti-CD3 antibody (20 µg/ml) with or without 1 µg/ml soluble anti-CD28. 24 h later, CD69 expression was analyzed by flow cytometry.
RT-PCR
DP thymocytes were sorted (using a FACSAria Fusion BSC II) from Rras2+/+ and Rras2−/− mice, and their RNA was isolated using the RNAeasy Plus Mini Kit (QIAGEN). cDNA was synthesized with SuperScript III (Invitrogen) using Oligo-dT primers. RT-qPCR was performed in triplicate using the reverse transcription reaction with SYBR Green PCR Master Mix and gene-specific primers in an ABI 7300 Real Time PCR System. Obtained cycle threshold (Ct) values were used to calculate mRNA levels relative to hypoxanthine-guanine phosphoribosyltransferase and GAPDH expression using the 2-ΔCt method.
IBD
A total of 0.5 × 106 sorted CD4+ CD25− naive T cells from spleens of Rras2+/+ and Rras2−/− mice were inoculated intraperitoneally into age-matched Rag1tm1Mom mice. Total body weight was monitored one to two times per week, and mice were sacrificed 7 wk after initiation of the experiment. The colon was removed and flushed with PBS, and the length and weight was measured from the rectum to the cecum. Spleens were removed, and cells were counted and analyzed by flow cytometry to detect the percentage of IL-17– and IFNγ-producing CD4+ T cells after treatment with PMA (50 ng/ml) plus ionomycin (750 µg/ml) for 12 h in the presence of 10 µg/ml brefeldin A for the final 4 h.
EAE
EAE was induced in female Rras2+/+ and Rras2−/− C57BL/6 mice (6–8 wk old; 20 g body weight) by subcutaneously injecting a total of 150 µg MOG35–55 (Espikem) emulsified in Freund’s complete adjuvant (Sigma-Aldrich) supplemented with Mycobacterium tuberculosis (1 mg/ml; H37Ra strain from Difco) into both femoral regions. 200 ng pertussis toxin (Sigma-Aldrich) per mouse were immediately injected intraperitoneally. A second dose of pertussis toxin was administered 48 h after immunization. The animals were weighted and inspected for clinical signs of disease. Disease severity of EAE was assessed according to the EAE severity scale: 0, normal behavior; 1, weakness at the distal portion of the tail; 1.5, complete flaccidity of the tail; 2, moderate hind limb weakness; 2.5, severe hind limb weakness; 3, ataxia; 3.5, partial hind limb paralysis; 4, complete hind limb paralysis; 4.5, complete hind limb paralysis accompanied by muscle stiffness; 5, moribund state and hence sacrificed for humanitarian reasons.
Alternatively, mice were sacrificed 7 d after immunization, before clinical disease signals appeared, in order to analyze the presence of MOG+ activated T cells in the popliteal lymph nodes by staining the cells with I-Ab MOG35-55 tetramer (National Institutes of Health tetramer core facility) at different dilutions for 30 min. Mice were also analyzed 30 d after immunization, at which time activated MOG+ T cells were also assessed in the spleen by flow cytometry.
To test the T cell–intrinsic dependency of EAE resistance, 8-wk-old female C57BL/6 mice were lethally irradiated (10 Gy) and reconstituted with 6 × 106 of a bone marrow mixture of cells containing 80% bone marrow cells from female Cd3e−/− mice and 20% bone marrow cells from either female Rras2+/+ or Rras2−/− mice. At 8 wk after reconstitution, mice were immunized and scored as above.
Immunization with class II-restricted OVA peptide
8-wk-old female Rras2+/+ and Rras2−/− mice were immunized by inoculation of 25 µl of a PBS solution containing 100 µg OVA 329–337 peptide (AAHAEINEA) and 15 µg LPS (Sigma-Aldrich) into each hind limb footpad. Mice were euthanized 7 d after immunization.
Analysis of TCR repertoire
CD4+ T cells from the spleens of six Rras2+/+ and six Rras2−/− 8-wk-old female and male mice (in equal proportion in both groups) were purified by negative selection using a mix of biotinylated antibodies: anti-B220, anti-CD8, anti-NK1.1, anti-CD11b, anti-Gr1, and anti-F4/80. Afterwards, stained cells were incubated with streptavidin beads (Dynabeads Invitrogen) for 30 min and separated using the Dynal Invitrogen Beads Separator. mRNA was extracted from purified CD4+ T cells using the RNAeasy Plus Mini Kit (QIAGEN). cDNA libraries were prepared using the 5′RACE-based protocol (i.e., onе read covers CDR3 and its surroundings, and another one covers the 5′UTR and downstream sequence of the V gene) with the SMARTer Mouse TCR α/β Profiling Kit (Takara) following the manufacturer’s instructions. Briefly, the reverse transcription reaction adds nontemplate nucleotides to the first strand of the cDNA that are used as template nucleotides for posterior PCR oligonucelotides. Afterwards, a semi-nested PCR with the first PCR amplification of the full-length variable region of the TCR is performed, followed by a subsequent round of PCR to further amplify full-length sequences of V(D)J variable regions of TCRα and/or TCRβ subunits and to incorporate adapter sequences for Illumina sequencing.
The cDNA libraries were sequenced on an Illumina MiSeq (2 × 300 nt). The quality of sequencing reads was assessed using FastQC. To extract CDR3 sequences; to determine V, D, and J genes; and to assemble clonotypes by CDR3 sequences, the raw sequencing reads were processed using the MiXCR v2.1.9 software (Bolotin et al., 2015) programs Align (with the parameters -OvParameters.geneFeatureToAlign = VTranscript --species mmu --chain TRA,TRB), Assemble (with the parameters -OseparateByJ = true -OseparateByV = true), and exportClones (with the parameters --filter-out-of-frames --filter-stops). The clones present (containing at least one read) in less than three samples of either six Rras2+/+ or six Rras2−/− mice were filtered out. Further analysis of clones was conducted using in-house written R-scripts and VDJtools v1.1.7 (Shugay et al., 2015).
Determination of TRAV4N3-J22 and TRAV4D3-J22 expression by qPCR analysis
Purified CD4+ T cells and CD44SP from lymph nodes and thymus, respectively, were obtained by cell sorting. RNA and cDNA extraction from these cells was performed according to the manufacturers’ instructions (Qiagen kit #74136 and Invitrogen kit #11752-050, respectively). qPCR was performed at 63.6°C using specific primers for TRAV4N3 and TRAVD3 sequences (Fw: 5′-AGATGCAATTTTTCTATCGCTGC-3′; Rv: 5′-TCCAAAGATGAGTTGCCAGCT-3′) and GAPDH. Relative gene expression levels of the sequences were obtained by normalizing the obtained Ct to GAPDH.
Diversity analysis
Sequence diversity among the clones generated by each V type gene were measured using the Simpson index (Simpson, 1949). Sequences from Rras2+/+ and Rras2−/− samples were classified independently according to their V type, and the Simpson index was calculated, with an in-house perl script, for each group, whenever there were >100 sequences for the WT and the Rras2−/− for the group.
Statistical analysis
Statistical parameters including the exact value of n and the means ± SD or SEM are described in the figures and figure legends. The nonparametric Wilcoxon–Mann–Whitney U test and the parametric Student’s t test were used as indicated to assess the difference in means.
Data availability
Raw sequencing data are available in the National Center for Biotechnology Information Sequence Read Archive under the accession number PRJNA488012.
Online supplemental material
Fig. S1 shows the expression of TCR and coreceptors as well as the presence of nT reg cells in WT and Rras2−/− mice. Fig. S2 shows the response of WT and Rras2−/− T cells to TCR-triggering stimuli. Fig. S3 shows the distribution of CDR3 lengths in the TCRα and TCRα chains. Fig. S4 shows the representation of T cell clones bearing particular TCRα sequences in WT and Rras2−/− mice. Fig S5 shows the sequences of annealing primers and qPCR data showing expression of particular TCRα sequences. Dataset 1 shows TRA sequences per mouse. Dataset 2 shows TRB sequences per mouse. Dataset 3 shows an analysis of TRA sequence diversity within Vα families.
Supplementary Material
Acknowledgments
We are indebted to Cristina Prieto, Valentina Blanco, and Tania Gómez for their expert technical assistance, the National Institutes of Health (Bethesda, MA) tetramer core facility for provision of MHC–peptide complexes, the Centre for Genomic Regulation Genomics Unit for sequencing T-cell repertoire data, and the Centre or Genomic Regulation Bioinformatics Unit for providing support in sequencing data analysis.
This work was funded by grants from Comisión Interministerial de Ciencia y Tecnología (SAF2016-76394-R to B. Alarcon), the “Comunidad de Madrid” (S2017/BMD-3671 to M. Fresno and B. Alarcon), and the European Research Council (ERC 2013-Advanced Grant 334763 “NOVARIPP” to B. Alarcon). The Centre for Genomic Regulation acknowledges support of the Spanish Ministry of Economy and Competitiveness, “Centro de Excelencia Severo Ochoa,” the Centres de Recerca de Catalunya Program/Generalitat de Catalunya, and the Centro de Biología Molecular Severo Ochoa to the Fundación Ramón Areces.
The authors declare no competing financial interests.
Author contributions: A. Martínez-Riaño designed and performed the research, analyzed the data, and wrote the manuscript. E.R. Bovolenta and V.L. Boccasavia assisted with analysis of TCR sequence expression in the thymus. C.L. Oeste edited the manuscript. J. Ponomarenko analyzed TCR repertoire sequencing data, prepared related figures, and edited the manuscript. D. Abia contributed to the figures presenting the sequencing data analysis. M. Fresno and H.M. van Santen helped to write the manuscript and supervised research. B. Alarcon supervised and designed the research, analyzed the data, and wrote the manuscript.
References
- Alcover A., Alarcon B., and Di Bartolo V.. 2018. Cell biology of T cell receptor expression and regulation. Annu. Rev. Immunol. 36:103–125. [DOI] [PubMed] [Google Scholar]
- Azzam H.S., Grinberg A., Lui K., Shen H., Shores E.W., and Love P.E.. 1998. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J. Exp. Med. 188:2301–2311. 10.1084/jem.188.12.2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbee S.D., and Alberola-Ila J.. 2005. Phosphatidylinositol 3-kinase regulates thymic exit. J. Immunol. 174:1230–1238. 10.4049/jimmunol.174.3.1230 [DOI] [PubMed] [Google Scholar]
- Barnden M.J., Allison J., Heath W.R., and Carbone F.R.. 1998. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 76:34–40. 10.1046/j.1440-1711.1998.00709.x [DOI] [PubMed] [Google Scholar]
- Bendelac A., Matzinger P., Seder R.A., Paul W.E., and Schwartz R.H.. 1992. Activation events during thymic selection. J. Exp. Med. 175:731–742. 10.1084/jem.175.3.731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin D.A., Poslavsky S., Mitrophanov I., Shugay M., Mamedov I.Z., Putintseva E.V., and Chudakov D.M.. 2015. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods. 12:380–381. 10.1038/nmeth.3364 [DOI] [PubMed] [Google Scholar]
- Borroto A., Arellano I., Dopfer E.P., Prouza M., Suchànek M., Fuentes M., Orfao A., Schamel W.W., and Alarcón B.. 2013. Nck recruitment to the TCR required for ZAP70 activation during thymic development. J. Immunol. 190:1103–1112. 10.4049/jimmunol.1202055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brändle D., Müller S., Müller C., Hengartner H., and Pircher H.. 1994. Regulation of RAG-1 and CD69 expression in the thymus during positive and negative selection. Eur. J. Immunol. 24:145–151. 10.1002/eji.1830240122 [DOI] [PubMed] [Google Scholar]
- Cantrell D. 2015. Signaling in lymphocyte activation. Cold Spring Harb. Perspect. Biol. 7:a018788 10.1101/cshperspect.a018788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S., Cornall R., Lesourne R., and Love P.E.. 2017. THEMIS: two models, different thresholds. Trends Immunol. 38:622–632. 10.1016/j.it.2017.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels M.A., Teixeiro E., Gill J., Hausmann B., Roubaty D., Holmberg K., Werlen G., Holländer G.A., Gascoigne N.R., and Palmer E.. 2006. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature. 444:724–729. 10.1038/nature05269 [DOI] [PubMed] [Google Scholar]
- DeJarnette J.B., Sommers C.L., Huang K., Woodside K.J., Emmons R., Katz K., Shores E.W., and Love P.E.. 1998. Specific requirement for CD3epsilon in T cell development. Proc. Natl. Acad. Sci. USA. 95:14909–14914. 10.1073/pnas.95.25.14909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado P., Cubelos B., Calleja E., Martínez-Martín N., Ciprés A., Mérida I., Bellas C., Bustelo X.R., and Alarcón B.. 2009. Essential function for the GTPase TC21 in homeostatic antigen receptor signaling. Nat. Immunol. 10:880–888. 10.1038/ni.1749 [DOI] [PubMed] [Google Scholar]
- Dower N.A., Stang S.L., Bottorff D.A., Ebinu J.O., Dickie P., Ostergaard H.L., and Stone J.C.. 2000. RasGRP is essential for mouse thymocyte differentiation and TCR signaling. Nat. Immunol. 1:317–321. 10.1038/79766 [DOI] [PubMed] [Google Scholar]
- Falk I., Nerz G., Haidl I., Krotkova A., and Eichmann K.. 2001. Immature thymocytes that fail to express TCRbeta and/or TCRgamma delta proteins die by apoptotic cell death in the CD44(-)CD25(-) (DN4) subset. Eur. J. Immunol. 31:3308–3317. [DOI] [PubMed] [Google Scholar]
- Fayard E., Moncayo G., Hemmings B.A., and Holländer G.A.. 2010. Phosphatidylinositol 3-kinase signaling in thymocytes: the need for stringent control. Sci. Signal. 3:re5 10.1126/scisignal.3135re5 [DOI] [PubMed] [Google Scholar]
- Fillatreau S., Uckert W., Kieback E., Anderton S., Lampropoulou V., Stervbo U., and Bunse M.. 2012. TCR transgenic mouse model for immune disease. European Patent Application 12175113.5, priority date July 5, 2012.
- Fischer A.M., Katayama C.D., Pagès G., Pouysségur J., and Hedrick S.M.. 2005. The role of erk1 and erk2 in multiple stages of T cell development. Immunity. 23:431–443. 10.1016/j.immuni.2005.08.013 [DOI] [PubMed] [Google Scholar]
- Fu G., Casas J., Rigaud S., Rybakin V., Lambolez F., Brzostek J., Hoerter J.A., Paster W., Acuto O., Cheroutre H., et al. . 2013. Themis sets the signal threshold for positive and negative selection in T-cell development. Nature. 504:441–445. 10.1038/nature12718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia K.C., and Adams E.J.. 2005. How the T cell receptor sees antigen--a structural view. Cell. 122:333–336. 10.1016/j.cell.2005.07.015 [DOI] [PubMed] [Google Scholar]
- Gascoigne N.R., Rybakin V., Acuto O., and Brzostek J.. 2016. TCR Signal Strength and T Cell Development. Annu. Rev. Cell Dev. Biol. 32:327–348. 10.1146/annurev-cellbio-111315-125324 [DOI] [PubMed] [Google Scholar]
- Glusman G., Rowen L., Lee I., Boysen C., Roach J.C., Smit A.F., Wang K., Koop B.F., and Hood L.. 2001. Comparative genomics of the human and mouse T cell receptor loci. Immunity. 15:337–349. 10.1016/S1074-7613(01)00200-X [DOI] [PubMed] [Google Scholar]
- Hsieh C.S., Lee H.M., and Lio C.W.. 2012. Selection of regulatory T cells in the thymus. Nat. Rev. Immunol. 12:157–167. 10.1038/nri3155 [DOI] [PubMed] [Google Scholar]
- Huo J., Xu S., and Lam K.P.. 2010. Fas apoptosis inhibitory molecule regulates T cell receptor-mediated apoptosis of thymocytes by modulating Akt activation and Nur77 expression. J. Biol. Chem. 285:11827–11835. 10.1074/jbc.M109.072744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huseby E.S., White J., Crawford F., Vass T., Becker D., Pinilla C., Marrack P., and Kappler J.W.. 2005. How the T cell repertoire becomes peptide and MHC specific. Cell. 122:247–260. 10.1016/j.cell.2005.05.013 [DOI] [PubMed] [Google Scholar]
- Iborra S., Soto M., Stark-Aroeira L., Castellano E., Alarcón B., Alonso C., Santos E., and Fernández-Malavé E.. 2011. H-ras and N-ras are dispensable for T-cell development and activation but critical for protective Th1 immunity. Blood. 117:5102–5111. 10.1182/blood-2010-10-315770 [DOI] [PubMed] [Google Scholar]
- Juntilla M.M., and Koretzky G.A.. 2008. Critical roles of the PI3K/Akt signaling pathway in T cell development. Immunol. Lett. 116:104–110. 10.1016/j.imlet.2007.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye J., Hsu M.L., Sauron M.E., Jameson S.C., Gascoigne N.R., and Hedrick S.M.. 1989. Selective development of CD4+ T cells in transgenic mice expressing a class II MHC-restricted antigen receptor. Nature. 341:746–749. 10.1038/341746a0 [DOI] [PubMed] [Google Scholar]
- Kaye J., Vasquez N.J., and Hedrick S.M.. 1992. Involvement of the same region of the T cell antigen receptor in thymic selection and foreign peptide recognition. J. Immunol. 148:3342–3353. [PubMed] [Google Scholar]
- Kieback E., Hilgenberg E., Stervbo U., Lampropoulou V., Shen P., Bunse M., Jaimes Y., Boudinot P., Radbruch A., Klemm U., et al. . 2016. Thymus-Derived Regulatory T Cells Are Positively Selected on Natural Self-Antigen through Cognate Interactions of High Functional Avidity. Immunity. 44:1114–1126. 10.1016/j.immuni.2016.04.018 [DOI] [PubMed] [Google Scholar]
- Klein L., Kyewski B., Allen P.M., and Hogquist K.A.. 2014. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat. Rev. Immunol. 14:377–391. 10.1038/nri3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortum R.L., Rouquette-Jazdanian A.K., and Samelson L.E.. 2013. Ras and extracellular signal-regulated kinase signaling in thymocytes and T cells. Trends Immunol. 34:259–268. 10.1016/j.it.2013.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malherbe L., Hausl C., Teyton L., and McHeyzer-Williams M.G.. 2004. Clonal selection of helper T cells is determined by an affinity threshold with no further skewing of TCR binding properties. Immunity. 21:669–679. 10.1016/j.immuni.2004.09.008 [DOI] [PubMed] [Google Scholar]
- Martínez-Martín N., Risueño R.M., Morreale A., Zaldívar I., Fernández-Arenas E., Herranz F., Ortiz A.R., and Alarcón B.. 2009. Cooperativity between T cell receptor complexes revealed by conformational mutants of CD3epsilon. Sci. Signal. 2:ra43 10.1126/scisignal.2000402 [DOI] [PubMed] [Google Scholar]
- Masuyama N., Oishi K., Mori Y., Ueno T., Takahama Y., and Gotoh Y.. 2001. Akt inhibits the orphan nuclear receptor Nur77 and T-cell apoptosis. J. Biol. Chem. 276:32799–32805. 10.1074/jbc.M105431200 [DOI] [PubMed] [Google Scholar]
- Matsutani T., Ohmori T., Ogata M., Soga H., Kasahara S., Yoshioka T., Suzuki R., and Itoh T.. 2007. Comparison of CDR3 length among thymocyte subpopulations: impacts of MHC and BV segment on the CDR3 shortening. Mol. Immunol. 44:2378–2387. 10.1016/j.molimm.2006.10.026 [DOI] [PubMed] [Google Scholar]
- Mendoza M.C., Er E.E., and Blenis J.. 2011. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem. Sci. 36:320–328. 10.1016/j.tibs.2011.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza P., Martínez-Martín N., Bovolenta E.R., Reyes-Garau D., Hernansanz-Agustín P., Delgado P., Diaz-Muñoz M.D., Oeste C.L., Fernández-Pisonero I., Castellano E., et al. . 2018. R-Ras2 is required for germinal center formation to aid B cells during energetically demanding processes. Sci. Signal. 11:11 10.1126/scisignal.aal1506 [DOI] [PubMed] [Google Scholar]
- Merkenschlager M., Graf D., Lovatt M., Bommhardt U., Zamoyska R., and Fisher A.G.. 1997. How many thymocytes audition for selection? J. Exp. Med. 186:1149–1158. 10.1084/jem.186.7.1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingueneau M., Kreslavsky T., Gray D., Heng T., Cruse R., Ericson J., Bendall S., Spitzer M.H., Nolan G.P., Kobayashi K., et al. Immunological Genome Consortium . 2013. The transcriptional landscape of αβ T cell differentiation. Nat. Immunol. 14:619–632. 10.1038/ni.2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P., Iacomini J., Johnson R.S., Herrup K., Tonegawa S., and Papaioannou V.E.. 1992. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 68:869–877. 10.1016/0092-8674(92)90030-G [DOI] [PubMed] [Google Scholar]
- Moran A.E., Holzapfel K.L., Xing Y., Cunningham N.R., Maltzman J.S., Punt J., and Hogquist K.A.. 2011. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 208:1279–1289. 10.1084/jem.20110308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na S.Y., Patra A., Scheuring Y., Marx A., Tolaini M., Kioussis D., Hemmings B.A., Hünig T., and Bommhardt U.. 2003. Constitutively active protein kinase B enhances Lck and Erk activities and influences thymocyte selection and activation. J. Immunol. 171:1285–1296. 10.4049/jimmunol.171.3.1285 [DOI] [PubMed] [Google Scholar]
- Paster W., Bruger A.M., Katsch K., Grégoire C., Roncagalli R., Fu G., Gascoigne N.R., Nika K., Cohnen A., Feller S.M., et al. . 2015. A THEMIS:SHP1 complex promotes T-cell survival. EMBO J. 34:393–409. 10.15252/embj.201387725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price D.A., Brenchley J.M., Ruff L.E., Betts M.R., Hill B.J., Roederer M., Koup R.A., Migueles S.A., Gostick E., Wooldridge L., et al. . 2005. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J. Exp. Med. 202:1349–1361. 10.1084/jem.20051357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Borlado L., Barber D.F., Hernández C., Rodríguez-Marcos M.A., Sánchez A., Hirsch E., Wymann M., Martínez-A C., and Carrera A.C.. 2003. Phosphatidylinositol 3-kinase regulates the CD4/CD8 T cell differentiation ratio. J. Immunol. 170:4475–4482. 10.4049/jimmunol.170.9.4475 [DOI] [PubMed] [Google Scholar]
- Rojas A.M., Fuentes G., Rausell A., and Valencia A.. 2012. The Ras protein superfamily: evolutionary tree and role of conserved amino acids. J. Cell Biol. 196:189–201. 10.1083/jcb.201103008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage P.A., and Davis M.M.. 2001. A kinetic window constricts the T cell receptor repertoire in the thymus. Immunity. 14:243–252. 10.1016/S1074-7613(01)00106-6 [DOI] [PubMed] [Google Scholar]
- Shugay M., Bagaev D.V., Turchaninova M.A., Bolotin D.A., Britanova O.V., Putintseva E.V., Pogorelyy M.V., Nazarov V.I., Zvyagin I.V., Kirgizova V.I., et al. . 2015. VDJtools: unifying post-analysis of T cell receptor repertoires. PLOS Comput. Biol. 11:e1004503 10.1371/journal.pcbi.1004503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson E.H. 1949. Measurement of diversity. Nature. 163:688 10.1038/163688a0 [DOI] [Google Scholar]
- Singer A., Adoro S., and Park J.H.. 2008. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat. Rev. Immunol. 8:788–801. 10.1038/nri2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.K., Jameson S.C., and Hogquist K.A.. 2003. Positive and negative selection of T cells. Annu. Rev. Immunol. 21:139–176. 10.1146/annurev.immunol.21.120601.141107 [DOI] [PubMed] [Google Scholar]
- Sugawara T., Moriguchi T., Nishida E., and Takahama Y.. 1998. Differential roles of ERK and p38 MAP kinase pathways in positive and negative selection of T lymphocytes. Immunity. 9:565–574. 10.1016/S1074-7613(00)80639-1 [DOI] [PubMed] [Google Scholar]
- Suzuki A., Yamaguchi M.T., Ohteki T., Sasaki T., Kaisho T., Kimura Y., Yoshida R., Wakeham A., Higuchi T., Fukumoto M., et al. . 2001. T cell-specific loss of Pten leads to defects in central and peripheral tolerance. Immunity. 14:523–534. 10.1016/S1074-7613(01)00134-0 [DOI] [PubMed] [Google Scholar]
- Swat W., Montgrain V., Doggett T.A., Douangpanya J., Puri K., Vermi W., and Diacovo T.G.. 2006. Essential role of PI3Kdelta and PI3Kgamma in thymocyte survival. Blood. 107:2415–2422. 10.1182/blood-2005-08-3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeiro E., and Daniels M.A.. 2010. ERK and cell death: ERK location and T cell selection. FEBS J. 277:30–38. 10.1111/j.1742-4658.2009.07368.x [DOI] [PubMed] [Google Scholar]
- Vrisekoop N., Monteiro J.P., Mandl J.N., and Germain R.N.. 2014. Revisiting thymic positive selection and the mature T cell repertoire for antigen. Immunity. 41:181–190. 10.1016/j.immuni.2014.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Zheng M., Lei L., Ji J., Yao Y., Qiu Y., Ma L., Lou J., Ouyang C., Zhang X., et al. . 2012. Tespa1 is involved in late thymocyte development through the regulation of TCR-mediated signaling. Nat. Immunol. 13:560–568. 10.1038/ni.2301 [DOI] [PubMed] [Google Scholar]
- Xue L., Chiang L., Kang C., and Winoto A.. 2008. The role of the PI3K-AKT kinase pathway in T-cell development beyond the beta checkpoint. Eur. J. Immunol. 38:3200–3207. 10.1002/eji.200838614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamoyska R., Basson A., Filby A., Legname G., Lovatt M., and Seddon B.. 2003. The influence of the src-family kinases, Lck and Fyn, on T cell differentiation, survival and activation. Immunol. Rev. 191:107–118. 10.1034/j.1600-065X.2003.00015.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data are available in the National Center for Biotechnology Information Sequence Read Archive under the accession number PRJNA488012.