Abstract
Red deer and wild boar are two major game species whose populations are managed and live in areas impacted by human activities. Measuring and understanding the impact of landscape features on individual movements and spatial patterns of genetic variability in these species is thus of importance for managers. A large number of individuals sampled across Wallonia (Belgium) for both species have been genotyped using microsatellite markers (respectively > 1700 and > 1200 genotyped individuals) and some individuals have also been followed using a capture-mark-recapture (CMR) protocol. The combined data set represents an unprecedented opportunity to study and compare the environmental factors impacting the interconnectivity of these large mammals. The present study describes and uses a landscape genetic workflow to compare spatial patterns of genetic variability and the impact of environmental factors on genetic differentiation. For the latter analyses, we investigate the correlation between genetic and environmental distances (pairwise approach) and also between local genetic dissimilarity and environmental conditions (point approach). Preliminary analyses of CMR data confirm that motorways act as significant barriers to dispersal. However, analyses performed with the pairwise approach do not highlight any evidence of an impact of motorways on genetic differentiation, which is presumably due to their recent establishment. Complementary analyses performed with the point approach reveal that low altitude tends to be associated with higher genetic dissimilarity. From a methodological point of view, the present workflow illustrates the complementary application of both pairwise and point approaches, as well as univariate and multivariate analyses.
Subject terms: Population genetics, Population genetics
Introduction
Landscape connectivity is a key aspect in species conservation (e.g., Correa Ayram et al. 2016) and wildlife management (e.g., Dobson et al. 1997). It is important to identify and measure the impact of environmental factors on connectivity among individuals. Measures of connectivity can either come from direct records of individual dispersal events (e.g., based on GPS collar or capture-mark-recapture [CMR] data) or estimates of genetic differentiation informed by molecular data analysis (e.g., DNA sequences, microsatellites; Broquet and Petit 2009). Although the first approach provides direct measures of individual movements, the second approach, addressed by landscape genetics, is rather a measure of “efficient connectivity”, providing additional information on the reproductive success of migrants (Baguette et al. 2013). Combining CMR data and landscape genetic analyses can thus provide different but potentially complementary insights into intra-specific connectivity (Lowe and Allendorf 2010).
Originating a decade ago, landscape genetics is a rapidly evolving field that aims to analyse the interaction between landscape features and microevolutionary processes, such as gene flow, genetic drift and selection (Manel et al. 2003; Storfer et al. 2010). This field now includes an increasing number of methods to relate matrices of genetic and environmental distances, in order to investigate landscape effects on genetic differentiation or similarity (Balkenhol et al. 2009; Manel and Holderegger 2013). An environmental distance can be defined as a measure of separation between sampling locations that incorporates the effects of differing environmental permeabilities (Spear et al. 2010). Environmental distances among sampled individuals can, for instance, be estimated using circuit theory (McRae 2006) and then compared with inter-individual measures of genetic distances.
As pointed out by Richardson et al. (2016), to date, most landscape genetic studies on plants or animals have focused on a single species. From a conservation perspective, moving towards comparative analyses of multiple species on the same landscape can improve the evaluation of candidate areas as corridors for movement. Furthermore, despite some recent methodological developments, landscape genetics still faces several methodological challenges. For instance, although circuit theory allows estimating environmental distances based on realistic path models, pairwise distances computed on distinct environmental layers (rasters) will all take the spatial distance into account. When resulting distance matrices are analysed in a multivariate framework, the same spatial distance variable is then automatically included several times in the statistical analysis. This aspect further highlights the necessity to properly deal with the collinearity among tested environmental distances. Prunier et al. (2015) have proposed to solve multicollinearity issues by coupling multiple regressions with commonality analyses (CAs). Although this procedure allows identifying the unique and common contributions of each environmental variable to the variance in measures of genetic differentiation, biological interpretations are not always straightforward as spatial distance remains included in each variable identified as important. As detailed in the present study, we propose to circumvent this problem by (i) applying and comparing both univariate and multivariate analyses of pairwise environmental and genetic distances, the univariate procedure being precisely focused on the comparison between environmental and pure spatial distances to help identifying factors explaining genetic differentiation better than spatial distance alone, but also by (ii) directly comparing local environmental values with local estimates of genetic (dis)similarity.
We here study two major game species co-distributed in Wallonia (Belgium): red deer (Cervus elaphus) and wild boar (Sus scrofa), two ungulate species living in the absence of natural predators and whose populations are controlled and managed according to hunting schedules. Human activity and land use in general are known to potentially impact these species and can, for instance, reduce inter-populations connectivity as a result of habitat fragmentation (e.g., Kuehn et al. 2003; Hartl et al. 2005). Due to their distinct ecology, the two species may be differently impacted by the same environmental factors. Frantz et al. (2012) suggested the impact of a Belgian motorway on the genetic population structure of C. elaphus but not for S. scrofa. Yet, their study focused on a geographically limited sampling, and their conclusions were solely based on the visual comparison between the motorway position and inferred genetic clusters. Given these previous results and because these species may be directly impacted by human activities and landscape management, a particular emphasis is given here to the analysis of anthropogenic factors such as main roads and agricultural areas.
The overall goals of the present study are to analyse and compare the spatial patterns of genetic variability of both species, as well as the impact of environmental factors on their intra-specific genetic differentiation. In addition to the preliminary analysis of available CMR data, we have built genetic microsatellites data sets (i) to infer genetic clusters, (ii) to map the genetic variability with a novel approach allowing the inclusion of all the inter-individual distances, i.e., even for pair of individuals that are not adjacent on a connectivity network, (iii) to use both univariate and multivariate procedures to assess the correlation between genetic differentiation and environmental factors. The latter analyses were performed following two complementary but distinct approaches: first, a pairwise approach in which genetic distances are compared with environmental distances computed with circuit theory, and second, a newly introduced point approach in which local genetic dissimilarity measures are compared with local environmental conditions. The analytical workflow used in the present study is in Fig. 1.
Fig. 1.
Analytical workflow of comparative landscape genetics used in the present study
Material and methods
Analysing CMR data
CMR data sets were composed of 105 and 1673 CMR movement records for C. elaphus and S. scrofa, respectively (Fig. S1; Prévot and Licoppe 2013). Each CMR movement record was considered to be an independent movement vector with a starting location, dispersal duration and end location. As a preliminary step to landscape genetic analyses, we used these CMR data to study the impact of motorways (Fig. S2) on the dispersal frequency of both species. For this purpose, we implemented a randomisation procedure to obtain a null distribution for the number of motorway-crossing events. As detailed in Appendix S1, this procedure consisted of randomly rotating CMR movement vectors around their starting locations and re-counting the number of crossing events. Observed numbers of motorway-crossing events were then compared with their null distribution to assess their level of significance with a one-sided probability test. In this study, we performed 1000 randomisation steps to assess, for both species, the significance level associated with the number of observed motorways crossing events. The automated procedure is implemented in an R script detailed in a tutorial (Appendix S1).
Sampling and genotyping
A total of 1733 individual samples for C. elaphus (Frantz et al. 2012) and 1253 individual samples for S. scrofa were collected from Wallonia, the southern part of Belgium, between 2003 and 2009 for C. elaphus and between 2005 and 2013 for S. scrofa (Fig. S1). Samples were collected from harvested animals during legal hunts. DNA was extracted from frozen tissue using a chloroform-based extraction method (Doyle and Doyle 1990). Samples were gathered from spleen or ear pieces for S. scrofa and from muscle or ear pieces for C. elaphus. All the PCRs were performed using the Qiagen Multiplex Kit (Qiagen, Hilden, Germany) in a total volume of 5 µl and approximately 15 ng of DNA, using a Verity Thermocycler (Applied Biosystems, Warrington, UK). For C. elaphus genotyping, we used the same 13 microsatellite loci as Frantz et al. (2006), organised in three multiplex PCRs. Detailed information on the PCR composition and reaction times can be found in Dellicour et al. (2011). For S. scrofa, we used the same 14 microsatellite markers as Frantz et al. (2009) and microsatellite genotyping was performed in two multiplex PCRs. The first multiplex contained loci S0002, S0026, S0097, Sw857, Sw911 and Sw122, and the second multiplex loci S0005, S0090, S0155, S0226, Sw240, Sw632 and Sw936. PCRs were performed with 1X Qiagen Multiplex Master Mix and 0.5X Q-solution. The final concentration of each primer was 0.05 µM, except for S0097 at 0.08 µM, SW122 at 0.07 µM and S0090 at 0.1 µM. Thermal cycling conditions consisted of initial denaturation at 95 °C for 15 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 90 s and extension at 72 °C for 1 min, with a final extension at 60 °C for 30 min. Amplification products were detected using an ABI 3130xl Genetic Analyser (Applied Biosystems) and the data were analysed using GeneMapper version 4.0 (Applied Biosystems). Properties of the microsatellite loci used in this study are summarised in Table S1. Smaller versions of these data sets, i.e., 876 C. elaphus and 325 S. scrofa individuals, were previously used by Frantz et al. (2012) to perform preliminary investigations of the impact of a motorway using a clustering approach. More recently, Frantz et al. (2017) also used the entire C. elaphus data set to discuss the identification and presence of non-autochthonous individuals (see below). As previously mentioned in another study presenting a first subset of the present C. elaphus data set (412 individuals, Frantz et al. 2006), all the microsatellite data were cross-read and double-checked to correct potential errors that had occurred during data entry. In the context of this first study (Frantz et al. 2006), 30 samples were also chosen randomly, re-extracted and re-genotyped, to check for genotyping repeatability. They identified one allelic dropout, which corresponds to a genotyping error rate of 0.0013 per allele.
Removing potentially illegally translocated individuals
Frantz et al. (2017) recently highlighted the presence of illegally translocated red deer in Wallonia. To avoid any bias in our landscape genetic analyses, we attempted to discard the corresponding genetic profiles from our data sets. Using a leave-one-out procedure, we calculated exclusion probabilities for each individual sampled in the present study area with the Monte Carlo method of Paetkau et al. (2004) available in the program GENECLASS 2.0 (Piry et al. 2004). We performed exclusions based on 104 simulated multi-locus genotypes and set the threshold for exclusion of individuals to 0.01 (Paetkau et al. 2004). Individuals identified as illegally-translocated (26 C. elaphus and 22 S. scrofa individuals) were removed from the original matrices, resulting in final matrices made of 1707 C. elaphus and 1231 S. scrofa individuals. This approach was deliberately conservative to ensure that illegally introduced, non-local individuals were removed from data sets before further analyses.
Inferring genetic clusters
Similar to Frantz et al. (2012), we analysed the genetic population structure of both species using two different clustering algorithms: methods implemented in STRUCTURE 2.3 (Pritchard et al. 2000) and GENELAND 4.5.0 (Guillot et al. 2005). STRUCTURE uses a Bayesian algorithm to assign individuals to clusters by minimising deviations from Hardy–Weinberg equilibrium and linkage equilibria. With this first method, we estimated the number of clusters (K) with 10 independent runs of K = 1–10 carried out with 106 Markov chain Monte Carlo (MCMC) iterations after a burn-in period of 105 iterations, using the model with correlated allele frequencies and assuming admixture. The most probable number of clusters was identified based on the log-likelihood values (and their convergence) associated with each K (hereafter referenced as the “log(P(K)) method”), as well as on the ΔK method of Evanno et al. (2005; hereafter referenced as the “Evanno method”). The first method elucidates the general (non-hierarchical) structure whereas the Evanno method identifies the highest hierarchical level of population structure (Evanno et al. 2005). Finally, we used the “Greedy” algorithm implemented in the software CLUMPP (Jakobsson and Rosenberg 2007) to calculate individual Q ancestry values (one Q-value per cluster) averaged over 10 runs, indicating the percentage of membership of each individual to each inferred cluster.
We also analysed both data sets using GENELAND, which employs a Bayesian clustering model that additionally considers the sampling coordinates of each individual when inferring genetic population structure. The number of genetic clusters was determined by running the algorithm 10 times, allowing K to vary from 1 to 10, with the following parameters: 106 MCMC iterations with a thinning of 1000, maximum rate of the Poisson process fixed to 100, and maximum number of nuclei in the Poisson-Voronoi tessellation fixed to 300. After inferring the number of clusters, the algorithm was run a further 100 times with K fixed to the inferred optimal number of clusters, with 250,000 MCMC iterations, a thinning of 250 and the other parameters left unchanged. Excluding the first 100 values as a burn-in, the mean logarithm of the posterior probability was calculated for each of the 100 runs and the posterior probability of cluster membership for each pixel of the spatial domain was then computed for the 10 runs with the highest values. For each individual, the probability of cluster membership was averaged across these 10 runs.
In addition to identifying the cluster associated with the highest percentage of membership for each individual, we generated interpolation maps for each inferred cluster. These maps were based on percentages of membership inferred for each individual and were generated with an inverse distance interpolation implemented in the R function GDivPAL (Dellicour and Mardulyn 2014). The interpolation was restricted to a minimum convex hull defined by all the sampling locations.
Computing inter-individual genetic distances
Three different but complementary inter-individual genetic distance metrics were computed in this study: the Bray–Curtis dissimilarity (“BCD”, Bray and Curtis 1957), the Rousset’s â distance (“aR”, Rousset 2000) and the Loiselle’s kinship coefficient (“LKC”, Loiselle et al. 1995). Inter-individual BCDs were estimated with the R package “gstudio” (github.com/dyerlab/gstudio) and can be defined as the proportions of alleles that are different between pairs of individuals. We used the program SPAGeDi 1.5 (Hardy and Vekemans 2002) to compute inter-individual aR distances, which correspond to FST/(1 − FST) ratios, but are estimated between pairs of individuals instead of populations. Although BCD and aR are measures of inter-individual genetic differentiation, a kinship coefficient as LKC between two individuals A and B is commonly defined as the probability of identity-by-descent between a random gene from A and a random gene from B (Hardy 2003). LKC values were also estimated with SPAGeDi 1.5 (Hardy and Vekemans 2002). We used three different metrics of genetic distances in an attempt to capture the various possible characteristics of genetic data sets.
Working with inter-individual rather than inter-population distances makes it possible to avoid having to arbitrarily define “populations” or “groups” among sampled sequences (see, for instance, Manel et al. 2003; Prunier et al. 2013; Lecocq et al. 2016). Having to define such groups is associated with several limitations: the partition is often completely arbitrary, hides intra-group differentiation and decreases the number of available pairwise distance estimates (Troupin et al. 2017). As preliminary analyses, isolation-by-distance was tested by regressing pairwise genetic distances against pairwise log-transformed geographic distances (i.e., great-circle distances estimated with the R package “fields”; Nychka et al. 2017). Correlation between genetic and log-transformed geographic distances were tested with Mantel tests (Mantel 1967) based on 1000 permutations and performed with the R package “vegan” (Oksanen et al. 2011).
Mapping inter-individual genetic distances
Mapping spatial patterns of inter-individual distances represents an alternative, but still descriptive, approach to clustering methods. In the present study, inter-individual genetic distances were mapped using the software MAPI (Piry et al. 2016). MAPI implements a recent method based on a spatial network in which samples are linked by ellipses and grids of hexagonal cells encompassing the study area. Pairwise metric values, attributed to ellipses, are averaged and assigned to cells they intersect following the principle that the larger the ellipse, the smaller its contribution to cells below it (Piry et al. 2016). This method thus allows generating maps reporting the degree of genetic dissimilarity associated with each cell of the grid. Cells with higher dissimilarity values reveal that geographically close individuals tend to be more genetically different. One of the advantages of this method is that it allows inclusion of all the inter-individual distances, i.e., even those estimated for pairs of individuals that are not adjacent on a connectivity network. We used the randomisation procedure implemented in the program to test whether distance values associated with the ellipses are independent from sampling locations. In this procedure, sample locations are permuted and at each permutation, new distance values are computed to create a null distribution for each grid cell. Once a false discovery rate correction is applied, cells where observed values are smaller (larger) than the 5% lower (resp. 95% higher) permuted values can then be aggregated to define areas where genetic dissimilarity is significantly smaller (resp. higher) than expected by chance (for further details, see Fig. 2 in Piry et al. 2016).
Fig. 2.
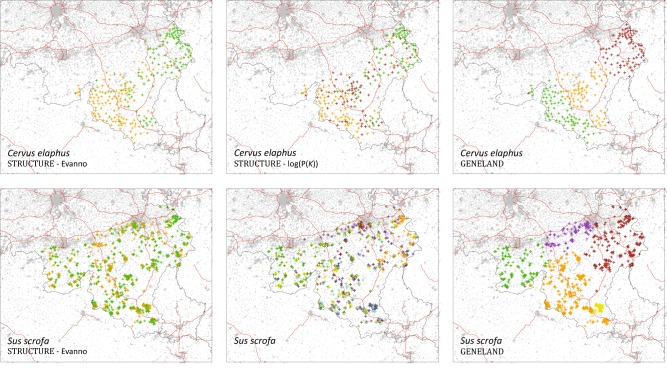
Results of the clustering analyses performed with STRUCTURE and GENELAND. One specific colour has been assigned to each individual and refers to the inferred cluster for which the individual has the highest percentage of membership. Grey areas and red lines respectively correspond to artificial areas and motorways (see also Figure S2)
MAPI also allows for specification of a sampling precision radius (error circle). As S. scrofa samples are associated with relatively precise sampling coordinates compared to study area, we specified an arbitrary but small sampling precision radius of 2 km. Sampling locations associated with C. elaphus samples are less precise as we only know the hunting administrative area of origin (Figs. S3-4) and centroid points of these polygons thus correspond to the most precise sampling coordinates that we were able to obtain. To estimate the average sampling precision radius associated with these geographic coordinates, we generated potential sampling points randomly distributed within sampled administrative areas. The number of sampling locations simulated per administrative area was proportional to the area of the corresponding polygon (with a maximum of 10,000 locations simulated within the largest sampled administrative area) and simulated locations falling outside the forest coverage were eventually discarded. Remaining simulated sampling points were then used to estimate the distribution of geographic distances between potential sampling locations and the centroid point of the corresponding hunting administrative area. The estimated distribution was then used to determine a relevant radius that could realistically define the uncertainty related to the sampling precision of C. elaphus. Based on the 0.95 quantile of this distribution (4.966 km), we set the sampling precision radius for C. elaphus to 5 km (see Figure S3 and its legend for further details). Other MAPI parameters for all computations were set as follows: an eccentricity of 0.975 (default), a minimal distance of 100 m to exclude pairwise relations between two samples from the same location, 1000 permutations (default) and an α value set to 0.05, corresponding to the 5% and 95% thresholds cited above. Hexagonal grids were built using a halfwidth of 2 km for C. elaphus and 1 km for S. scrofa.
Investigating the impact of environmental factors
To assess the impact of environmental factors on effective dispersal, we first tested the correlation between pairwise genetic and environmental distances. Environmental distances were computed by analysing several environmental rasters with the program CIRCUITSCAPE 4.0.5 (McRae 2006; McRae et al. 2008) that implements a method based on circuit theory. In CIRCUITSCAPE, environmental rasters were treated by the algorithm as resistance and/or conductance factors, i.e., factors impeding or facilitating movements. We investigated the influence of several environmental variables (Fig. S2): elevation (tested both as a conductance and a resistance factor), motorways, primary roads, railways, rivers and streams (tested as resistance factors), and the most important land cover variables for the study area, i.e., agricultural and artificial areas (tested as resistance factors), as well as broad leaved, coniferous and mixed forests (tested as conductance factors). Note that a more global “forest areas” variable combining the three kinds of forest cover (broad leaved, coniferous and mixed) had also been tested (results not shown). The elevation raster came from the Consortium for Spatial Information (CGIAR-CSI; srtm.csi.cgiar.org; resolution: 0.05 arcmin), the original motorways, primary roads, railways, rivers and streams shapefiles from the OpenStreetMap database (download.geofabrik.de) and the land cover rasters were retrieved from the Corine Land Cover 2012 raster (CLC; www.eea.europa.eu; resolution: ∼100 m). We generated distinct land cover rasters from the original CLC raster by creating lower resolution rasters (∼1000 m) whose cell values equalled the number of ~100 m pixels of each land cover category within the broader ∼1000 m pixels. The motorways, primary roads, railways, rivers and streams rasters were generated by rasterising the original shapefiles on an empty raster with a resolution of 0.5 arcmin and by assigning a resistance value equal to (1 + k) to each cell crossed by a linear feature. We tested three different values for the parameter k: 10, 100 and 1000. As the raster cells that are not crossed by a linear feature were assigned a uniform value of “1”, k thus defines the additional resistance when the cell does contain such a potential landscape barrier (see Laenen et al. [2016] for a similar approach). By varying the value of the parameter k, we thus explore the impact of different linear transformations of these original grids, i.e., a binary raster only indicating the absence (raster cell = 0) or the presence (raster cell = 1) of the infrastructure at a given cell. More parametric transformations are in theory possible (see, e.g., Peterman et al. 2014) but are a priori not relevant in the context of binary environmental variables like rasterised roads or water flows.
In a second step, we tested the correlation between mapped genetic distances and environmental values. Mapped genetic distance values were those estimated by MAPI for each cell of a hexagonal grid covering the study area in the case of C. elaphus and for a subset of cells in the case of S. scrofa. In the latter case, we subsampled the hexagonal grid using the “sample.grid” function from the R package “GSIF” (Hengl et al. 2017) to obtain a cell density of 0.101 cell/km², as in the case of C. elaphus. To obtain environmental values corresponding to these MAPI values, we averaged environmental raster values falling in each of these hexagonal cells. In the end, we obtained a vector of values for each genetic metric and each environmental variable, with the position of each value corresponding to a specific hexagonal cell. Environmental variables with skewed distributions were log-transformed to achieve normality. Furthermore, given that the present approach does not allow the analysis of linear environmental features such as roads, railways and water flows, we focused on the analysis of elevation and land cover variables. Although this approach also compares genetic distance estimates with environmental variables, it tests a different type of correlation. Indeed, in the first approach based on the comparison between genetic and environmental distances, we explicitly test whether inter-individual genetic differentiation can be explained by the environmental costs of travelling across the study area (hereafter referenced as the “pairwise approach”). As with this second approach based on the comparison between local MAPI estimates and environmental values, we rather test whether specific environmental conditions can be related to globally smaller or higher values of genetic differentiation (hereafter referenced as the “point approach”). The two approaches are thus slightly different but together allow investigating the impact of environmental heterogeneity measured both by pairwise spatial distances and local environmental measures.
As data used in the point approach were gathered from a grid, we expected spatial autocorrelation to affect output from multivariate linear models. To suppress spatial autocorrelation in residuals, we modelled spatial connectivity among data points using a Delaunay triangulation. We then completed our linear models with a subset of spatial eigenvectors (or Moran’s Eigenvector Maps “MEM”) selected from all possible MEMs using the “mem.select” function from the R package “adespatial” (Dray et al. 2018). Specifically, we used the minimisation of Moran’s I in the residuals (MIRs) approach (Bauman et al. 2018) to identify the set of MEMs removing spatial autocorrelation in the residuals so that the Moran’s I statistic was not significant at a threshold α = 0.1. Any environmental predictor acting as a cross-over suppressor in presence of retained spatial predictors was discarded (see below for details about statistical suppression) and the MIR optimisation procedure was repeated.
We used multiple regressions on distance matrices (MRDMs) for the pairwise approach and multiple linear regressions (LRs) for the point approach, both coupled with CAs (Newton and Spurrell 1967), hereafter respectively referenced as “MRDM-CA” and “LR-CA”. CA is a detailed variance-partitioning procedure that can be used to deal with dependence among spatial predictors (Prunier et al. 2015; Ray-Mukherjee et al. 2014). This approach computes both the “unique” and “common” contributions of predictors to the variance in the response variable. Specifically, unique (U) and common (C) effects respectively represent the amount of variance in the response variable that is accounted for by a single predictor and that can be jointly explained by several predictors together. MRDM-CA and LR-CA were performed using R packages “ecodist” (Goslee and Urban 2007) and “yhat” (Nimon et al. 2008). MRDM were based on 1000 permutations. After the first MRDM/LR-CA analyses, total suppressors were identified and discarded in a series of successive MRDM/LR-CA analyses, until all the suppressors were removed (Dellicour et al. 2017). A predictor may be considered a total suppressor when its unique contribution is counterbalanced by its (negative) common contribution (classical suppression) or when its regression coefficient and its correlation coefficient are of opposite signs (cross-over suppression; Paulhus et al. 2004; Prunier et al. 2017): it shares no or little variance with the response variable but is responsible for artefactual relationships among variables due to the removal of the irrelevant variance in other (suppressed) predictors. Discarding such suppressor variables can potentially purify the relationship between remaining predictors and the response variable (Prunier et al. 2017). For the pairwise approach, we also included the so-called “null” raster, i.e., a raster with uniform cell values equal to “1”, as a negative control. The purpose of analysing such a raster with CIRCUITSCAPE is to create a variable corresponding to geographical distance alone. We argue that this is a relevant procedure to integrate geographical distance as a negative control and avoid erroneous (false positive) conclusions in the interpretation of such univariate/multivariate procedures. For the point approach, MEMs were not included in the CAs because of the large number of spatial eigenvectors retained by the MIR optimisation procedure.
For the pairwise approach, we also performed univariate tests. Mantel tests based on 1000 permutations were performed between each matrix of pairwise genetic distances and each matrix of environmental distances. The purpose of including these univariate tests in the analytical workflow was to directly compare R2 values with the R2 value estimated from the regression between genetic distances and environmental distances computed on a “null” raster. In this raster, a unique value of “1” is assigned to all the cells and distances computed from it are thus a proxy for pairwise geographic distances but here computed with the same circuit theory algorithm used to compute pairwise distances from environmental rasters. Comparing R2 values estimated from the regressions with distances computed on an environmental raster (R2env) and on the “null” raster (R2null) makes it possible to investigate whether considering environmental heterogeneity improves the correlation with genetic distances (see, e.g., Dellicour et al. 2016, Dellicour et al. 2018).
Results
Analysing CMR data
Of the 105 CMR movement records available for C. elaphus, only one crossed a motorway segment. For that species, the randomisation test returned a p-value of 0.076, which is close to but still higher than the commonly used threshold value of 5%. As for S. scrofa, the CMR data set was larger and, of the 1673 available CMR records, 22 crossed a motorway segment. In that case, randomisation indicated that this number was highly significant (p-value < 0.001), meaning that the proportion of CMR vectors crossing motorways was lower than expected by chance (see Appendix S1 for more details). Preliminary analyses of CMR data thus formally confirm that motorways limit dispersal for at least one of the two species.
Inferring genetic clusters
Results of the clustering analyses are reported in Fig. 2, where, for each individual, the inferred cluster associated with the highest percentage of membership is displayed (see also Figure S5 for a summary of STRUCTURE results). In addition, we also generated interpolation maps for each cluster taken separately. These alternative representations give a more detailed overview of the clustering results (Figs. S6-S11). Globally, clusters inferred by GENELAND appear to be clearly more spatially distinct than those inferred by STRUCTURE (Fig. 2). In the case of C. elaphus, STRUCTURE inferred two or three clusters, respectively with the Evanno and log(P(K)) methods (Fig. S5). For S. scrofa, STRUCTURE inferred two and eight clusters, respectively with the Evanno and log(P(K)) methods (Fig. S5).
Mapping inter-individual genetic distances
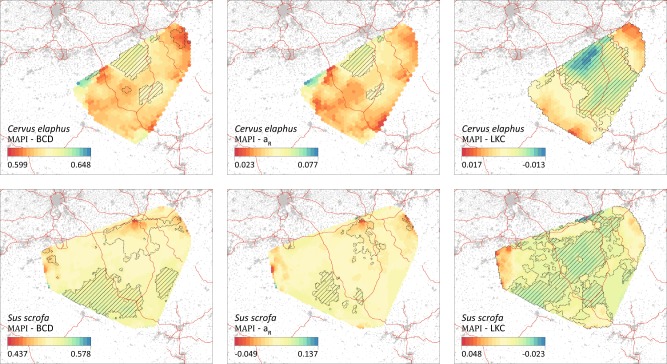
In this study, the three different pairwise genetic distances (BCD, aR and LKC) were mapped with the method implemented in MAPI (Fig. 3; Piry et al. 2016). For C. elaphus, the BCD and aR surfaces revealed two central areas associated with significantly higher genetic dissimilarity (Fig. 3). The surface obtained for the same species but based on LKC, which is a kinship rather than distance metric, and is quite different. In that case, there is a broad central band of significantly higher genetic dissimilarity (low kinship values) surrounded by areas of significantly lower genetic dissimilarity (Fig. 3). For S. scrofa, BCD and aR surfaces again appear similar. The BCD surface displays two main continuous areas of significantly higher and one main area of significantly lower genetic dissimilarity. The aR surface only reveals narrower areas of significantly lower/higher dissimilarity but those are located at the same position. As for the LKC surface obtained for S. scrofa, the resulting pattern is more complex and the map is again mainly divided into areas of significantly lower or higher dissimilarity (Fig. 3).
Fig. 3.
MAPI graphs based on the three inter-individual genetic distances (BCD, aR and LKC; see the text for further details). Polygons with dashed contours and with embedded hatching respectively correspond to areas with significantly lower and higher inter-individual dissimilarity than expected by chance. Grey areas outside the MAPI surfaces and red lines respectively correspond to artificial areas and motorways (see also Figure S2). In these graphs, levels of genetic dissimilarity are indicated by a colour scale ranging from red (lower genetic dissimilarity) to blue (higher genetic dissimilarity). Contrary to the genetic distances BCD and aR, the kinship coefficient of Loiselle (LKC) is a measure of genetic similarity. In a comparison purpose, the order of LKC values has been inverted in the colour scale to correspond to the visualisations obtained for BCD and aR, i.e. colour scales uniformly ranging from lower to higher dissimilarity measures
The ellipses-based method implemented in MAPI already includes the effect of geographical distance: genetic distances estimated between individuals sampled from distant locations will contribute relatively less to genetic distance estimates attributed to underneath grid cells (Piry et al. 2016). These results thus provide a relevant visualisation of the information contained in inter-individual genetic distances. As displayed in Fig. 3, identified areas associated with significantly higher genetic dissimilarity do not necessarily tend to correspond to areas divided/crossed by a motorway segment. Although only based on a preliminary visual comparison, this approach does not highlight any obvious patterns related to this particular landscape feature.
Investigating the impact of environmental factors
Results of univariate regressions on distance matrices (pairwise approach) revealed that almost all the tested environmental factors are associated with a low but significant determination coefficient R2 (Table S2). Yet, only two environmental rasters lead to a determination coefficient R2 higher than the one obtained when environmental distances have been computed with the “null” raster. These two factors were both identified in the context of the analysis of S. scrofa using the BCD metric and are the elevation and artificial area rasters treated as resistance factors (Table S2). None of the other tested factors led to an increase in R2 compared with the R2 value obtained from the analysis of environmental distances computed from the “null” raster. In other words, environmental distances between sampling locations do not explain differences in pairwise genetic distances better than the corresponding proxy measure of pairwise spatial distance computed with circuit theory.
Results of multivariate analyses between genetic and environmental distances (pairwise approach) are all reported in Table 1. These MRDM-CA results indicate that (i) significant global MRDM R2 associated with these analyses are all very small (< 2.5%), and that (ii) none of the tested environmental factors showed a unique contribution to the variance in the dependant variable higher than 1%. These results are coherent between the three different genetic distance metrics used in this study but also with the univariate results based on the same distance matrices. On the opposite, multivariate analyses based on MAPI estimates and environmental values (point approach) revealed that for C. elaphus the elevation is associated with a significant regression coefficient, as well as CA unique contributions equal to ~0.17 and ~0.08 for tests based on genetic distances BCD and aR, respectively (Table 2). Note that for both these genetic distances, the regression coefficient associated with the elevation factor is negative, meaning that low-elevated areas tend to be associated with higher genetic dissimilarity. For S. scrofa and considering BCD distances, successive LR-CA analyses performed after removing identified suppressors did not lead to the identification of any environmental factor associated with a unique contribution to the variance in the dependant variable higher than 5%.
Table 1.
Results of the multivariate analyses performed with the pairwise approach
| Species | Genetic distance | Environmental factors | r | β | U | C | |
|---|---|---|---|---|---|---|---|
| C. elaphus |
BCD R2 = 0.016* |
Null raster (R) | 0.120 | 0.105* | 0.0087 | 0.0058 | |
| Artificial areas (R) | 0.059 | 0.019* | 0.0003 | 0.0031 | |||
| Coniferous forests (C) | 0.072 | 0.022* | 0.0004 | 0.0048 | |||
| Primary roads (R; k = 1000) | 0.034 | 0.008* | 0.0001 | 0.0011 | |||
| Rivers (R; k = 10) | 0.014 | 0.004* | 0.0000 | 0.0002 | |||
|
a R R2 = 0.013* |
Null raster (R) | 0.111 | 0.103* | 0.0095 | 0.0027 | ||
| Artificial areas (R) | 0.051 | 0.021* | 0.0004 | 0.0022 | |||
| Primary roads (R; k = 1000) | 0.028 | 0.004* | 0.0000 | 0.0008 | |||
|
LKC R2 = 0.015* |
Null raster (R) | − 0.120 | − 0.117* | 0.0108 | 0.0037 | ||
| Agricultural areas (R) | − 0.054 | − 0.005* | 0.0000 | 0.0029 | |||
| Coniferous forests (C) | − 0.053 | − 0.004* | 0.0000 | 0.0028 | |||
| Motorways (R; k = 1000) | − 0.009 | − 0.004* | 0.0000 | 0.0001 | |||
| Railways (R; k = 10) | − 0.013 | − 0.000 | 0.0000 | 0.0002 | |||
| S. scrofa |
BCD R2 = 0.024* |
Null raster (R) | 0.093 | 0.105* | 0.0007 | 0.0080 | |
| Elevation (C) | − 0.030 | − 0.040* | 0.0001 | 0.0008 | |||
| Elevation (R) | 0.116 | 0.012* | 0.0000 | 0.0135 | |||
| Agricultural areas (R) | − 0.115 | − 0.092* | 0.0036 | 0.0096 | |||
| Artificial areas (R) | 0.039 | 0.019* | 0.0002 | 0.0013 | |||
|
a R R2 = 0.012* |
Null raster (R) | 0.096 | 0.029* | 0.0002 | 0.0089 | ||
| Elevation (R) | 0.065 | 0.076* | 0.0013 | 0.0029 | |||
| Agricultural areas (R) | 0.061 | 0.027* | 0.0004 | 0.0033 | |||
| Coniferous forests (C) | 0.037 | 0.046* | 0.0005 | 0.0009 | |||
| Motorways (R; k = 1000) | 0.026 | 0.018* | 0.0003 | 0.0003 | |||
|
LKC R2 = 0.011* |
0 negative | Null raster (R) | − 0.102 | − 0.085* | 0.0018 | 0.0086 | |
| Elevation (R) | − 0.049 | − 0.021* | 0.0001 | 0.0023 | |||
| Artificial areas (R) | − 0.013 | − 0.011* | 0.0001 | 0.0001 | |||
| Coniferous forests (C) | − 0.044 | − 0.017* | 0.0001 | 0.0018 | |||
| Railways (R; k = 10) | − 0.007 | − 0.002 | 0.0000 | 0.0000 | |||
For each species and each genetic distance, the table provides the results of a MRDM (multiple regressions on distance matrices) analysis and additional parameters derived from CA (commonality analysis) after having successively removed identified suppressors. For each environmental factor, are provided: Pearson’s correlation coefficient (r), β weights (β), as well as unique and common contributions (U, C) of environmental distances to the variance in the dependent variable. MRDM-CAs were each time performed between one genetic distances matrix and several matrices of environmental distances. (*) refers to significant determination coefficients R2 or β-values (p-values < 0.05 after Benjamini–Hochberg correction), “C”/“R” indicate if the considered environmental raster was respectively treated as a conductance or resistance factor, and k corresponds to the parameter used to transform the initial raster file (see the text for further details)
Table 2.
Results of the multivariate analyses performed with the point approach
| Species | Genetic distance | Spatial predictors (MEM) | Environmental factors | r (with MEMs) | β (with MEMs) | U (without MEMs) | C (without MEMs) |
|---|---|---|---|---|---|---|---|
| C. elaphus |
BCD R2 = 0.264* |
84 positive | Elevation | − 0.488 | − 0.461* | 0.1739 | 0.0640 |
| 0 negative | Agricultural areas | 0.214 | 0.175* | 0.0199 | 0.0257 | ||
| Broad leaved forests | 0.068 | 0.024 | 0.0004 | 0.0043 | |||
|
a R R2 = 0.211* |
83 positive | Elevation | − 0.437 | − 0.362* | 0.0828 | 0.1080 | |
| 0 negative | Artificial areas | 0.263 | 0.126* | 0.0131 | 0.0560 | ||
| Coniferous forests | − 0.331 | − 0.060 | 0.0021 | 0.1075 | |||
|
LKC R2 = 0.160* |
83 positive | Elevation | 0.138 | 0.163* | 0.0218 | − 0.0029 | |
| 0 negative | Agricultural areas | − 0.362 | − 0.250* | 0.0402 | 0.0907 | ||
| Broad leaved forests | 0.264 | 0.182* | 0.0200 | 0.0497 | |||
| S. scrofa |
BCD R2 = 0.193* |
98 positive | Elevation | 0.288 | 0.233* | 0.0371 | 0.0458 |
| 0 negative | Artificial areas | − 0.350 | − 0.111* | 0.0068 | 0.1157 | ||
| Broad leaved forests | 0.268 | 0.255* | 0.0487 | 0.0228 | |||
| Mixed forests | 0.177 | 0.107* | 0.0100 | 0.0212 | |||
|
a R R2 = 0.034* |
117 positive | Artificial areas | − 0.086 | − 0.022 | 0.0004 | 0.0069 | |
| 0 negative | Broad leaved forests | 0.183 | 0.175* | 0.0266 | 0.0069 | ||
| LKC | 124 positive | Mixed forests | − 0.179 | − 0.241* | – | – | |
| R2 = 0.043* | 0 negative |
For each species and each genetic distance, the table first provides the number of retained positive and negative spatial predictors (MEM) and the list of retained environmental factors. For each environmental factor, the table also reports: the Pearson’s correlation coefficient (r), the β weights (β) and the corresponding p-value with Benjamini–Hochberg correction after suppression of spatial autocorrelation in model residuals, as well as the unique and common contributions (U and C) of environmental factors to the variance in the dependent variable. Unique and common contribution of each factor were derived from CA (commonality analysis) performed in absence of spatial predictors (MEM). Genetic distance values were estimated with MAPI and environmental values were obtained by averaging environmental raster values falling in each corresponding hexagonal cell. Spatial predictors were computed as Eigenvector maps from a spatial connectivity model based on a Delaunay triangulation. (*) refers to significant determination coefficients R2 or β-values (p-values < 0.05 after Benjamini–Hochberg correction)
Discussion
Univariate and multivariate analyses performed in this study converge towards the same conclusion, i.e., the absence of a significant and global impact of most of the tested environmental factors on inter-individual measures of pairwise genetic differentiation. Correlated with lower genetic dissimilarity for C. elaphus, only the elevation factor is associated with a notable unique contribution when estimated with the point approach. Regarding the small altitude range of the study area (0–694 m), the trend related to the elevation is not necessarily easy to interpret and such correlation could be due to an indirect, unidentified factor. For instance, higher altitude areas could be correlated with more landscape connectivity within the forest network and/or less pressure related to human presence and activities.
As mentioned in the introduction, Frantz et al. (2012) previously identified a likely impact of motorways on C. elaphus population structure. Yet, their study was based on a smaller sample encompassing a fraction of the present study area. Indeed, only a western area of Wallonia was considered and their sampling included individuals separated by only one motorway segment (the E411; Fig. S2). Furthermore, their conclusions were driven by clustering results, i.e. by making visual comparisons between inferred clusters and motorway segment positions. In the present study, clusters inferred by STRUCTURE do not seem to correspond to areas delineated by main motorways. Only the GENELAND analysis identifies, for C. elaphus, two out of three clusters that tend to visually match with the position of the E411 (extended by the E25 in the South; Fig. S2), although some individuals from the second cluster (coloured in orange in Fig. 2) are located on both sides of a motorway. Yet, it is noteworthy that the study area is now broader, and that significant isolation-by-distance signals are identified for both species and whatever the inter-individual genetic distance considered in this study (Fig. S12). It has been shown that Bayesian clustering methods such as STRUCTURE and GENELAND can be impacted by isolation-by-distance by overestimating the number of real genetic clusters (Frantz et al. 2009). It is thus important to complement the interpretation of such clustering results with the results obtained from alternative methods. Such alternative methods can also be based on a visualisation approach (e.g., MAPI, Piry et al. 2016), or on statistical analyses of genetic and environmental distances (e.g., MRDM-CA, Prunier et al. 2015).
Here we do not question the fact that these important roads actually have a role of barriers to dispersal. In that context, our preliminary analyses of CMR records confirms a significant impact of the motorways on the dispersal frequency of S. scrofa, whereas the size of the C. elaphus CMR data is probably too small to detect any impact. The specific question addressed in the present study is rather to what extent these barriers have already impacted the genetic differentiation of each species across the study area and whether the still existing gene flow is sufficient to alleviate differentiation. When based on the entire C. elaphus or S. scrofa data sets, analyses of inter-individual distances do not reveal any obvious evidence of an impact of motorways on inter-individual differentiation. As they were mainly built during the 70s and 80s, Belgian Motorways crossing the study areas are still relatively recent (Fig. S13). Furthermore, the E411 is not necessarily older than other motorways such as the E25, depending on the considered local segments. One potential explanation of the lack of global impact detected on inter-individual differentiation could be that there were too few generations of individuals separated by these motorways to allow the identification of such a trend, despite the use of an individual-based approach (Landguth et al. 2010, Prunier et al. 2013). In addition to motorway bridges already crossing over rivers or small roads, several wildlife crossings were also installed on the motorway network (Fig. S13). Although it is difficult to quantify their influence, they could altogether have a role by increasing the permeability of motorways to the effective dispersal in C. elaphus and S. scrofa.
In this study, we perform correlation tests based on the comparison of pairwise genetic and environmental distance matrices but also between synthetic vectors of local measures of genetic differentiation and environmental variables. The latter “point approach” is based on the genetic distance estimates provided by MAPI and assigned to each cell of a grid covering the study area. Estimates assigned to these cells can be used to perform a direct comparison with associated environmental values. As illustrated in Table 2, this approach can allow the detection of significant trends that may not be evident with the pairwise approach based on distance matrices. For instance, the comparison between MAPI estimates and environmental distances highlight that for C. elaphus, elevation is significantly associated with lower genetic dissimilarity. These results illustrate that both approaches can be complementary and used within the same study to perform an extensive analysis of the impact of environmental factors on genetic differentiation. In addition, we also perform all these correlation tests using univariate and multivariate procedures. Univariate tests were particularly relevant for the correlations among distance matrices because each regression between genetic and environmental distances could be compared with the regression based on distances computed from the “null” raster. When analysing the entire data set, resulting differences among determination coefficients already indicate that, although significant, none of the environmental factors explain the genetic differentiation better than the spatial distance alone. Importantly, univariate and multivariate results are coherent and we argue that performing both procedures can help for the overall interpretation of the results. Indeed, in this case, the absence of positive MRDM-CA results is easily explained by the comparison of univariate determination coefficients. The analytical workflow used here (Fig. 1) allowed us to describe, analyse and compare co-distributed data in an extensive landscape genetic study. Yet, one interesting further perspective could be to perform a simulation-based study to evaluate exactly to what extent clustering and CA methods are able to detect the impact of more or less recent barriers to dispersal.
In conclusion, we further promote the use of complementary approaches when studying the impact of environmental factors on genetic differentiation (Lowe and Allendorf 2010). For instance, CMR data can be analysed in addition to genetic data to assess the impact of barriers on current dispersal frequency, which is an aspect that can be undetected when analysing population structure or inter-individual differentiation. Indeed, although metrics such as inter-individual genetic distances will be treated as measures of effective gene flow, movement-based data will rather inform on actual migration events. As illustrated in the present study, CMR data can detect an impact of barrier to migration in a situation where relative isolation is probably too recent for already having an impact on inter-individual differentiation. In such context, movement and genetic data are thus complementary, because investigating two different aspects, i.e., current versus long-term impact of barriers to migration.
Furthermore, visualisation approaches, e.g., MAPI or mapping genetic clusters, are complementary to formal statistical analyses of the impact of environmental factors on genetic differentiation. Visualisation approaches are useful to represent genetic differentiation or population structure and to visually compare genetic (dis)similarity or genetic clusters with some landscape features. However, although such approaches represent valuable representation tools, it is a reasonable analytical strategy to complement these visualisations with more formal statistical analyses. When analysing large data sets and multiple environmental factors, some trends could not be detected when solely performing visualisation analyses. Apparent trends, for instance the impact of barriers such as motorways, should always be formally tested.
Formal statistical analyses can also be performed with complementary approaches investigating distinct aspects of the same data set. As discussed above, we here used both so-called pairwise and point approaches, comparing genetic and environmental distance matrices or vectors of environmental and genetic dissimilarity values. Although the first traditional approach specifically tests the correlation between genetic differentiation between areas and the environmental cost to move between these areas, the second approach rather tests the association between local genetic dissimilarity and environmental conditions. Again, the complementarity of these two approaches lies in the fact that they do not test the same aspect. As outlined above, a significant correlation can be identified with one approach and not with the other: although elevation is not identified as a significant factor correlated to inter-individual differentiation by the pairwise approach, the point approach here reveals that low altitude tends to be associated with higher genetic dissimilarity in C. elaphus.
Finally, some complementary statistical analyses can also be performed in a univariate or multivariate framework. On one hand, univariate analyses can help explore the data sets and exclude the potential effect of a series of environmental factors. For instance, univariate analyses can be used to compare (i) the correlation between genetic distances and environmental distances computed on a specific environmental raster and (ii) the correlation between the same genetic distances and the distances computed on a corresponding “null” raster (i.e., with cell values uniformly equal to “1”). Pairwise distances computed on this uniform raster will constitute a proxy for geographic distance and a negative control. If the correlation computed in (i) is not higher than the correlation computed in (ii), it is already a clear indication that the tested environmental factor does not seem to better explain inter-individual genetic differentiation than geographic distance alone (see Table S2 for an example of such comparisons). On the other hand, multivariate analyses represent a more integrated approach. However, failure to deal with multicollinearity among environmental variables can represent a major flaw in landscape genetics (Prunier et al. 2015), hence the importance of approaches to properly account for it, such as the CAs used in the present study.
Data accessibility
Example files related to the Appendix S1, microsatellite genotypes and CMR data were deposited in the Dryad Digital Repository (10.5061/dryad.c6t0470).
Supplementary information
Acknowledgements
We are grateful to three anonymous reviewers for their constructive comments, as well as to Mandev Gill for his comments and proofreading of the final version of our manuscript. This work was supported by grants from the Public Service of Wallonia (PSW), General Directorate for Agriculture, Natural Resources and Environment. We are indebted to the local Services of the Nature and Forest Department (General Directorate for Agriculture, Natural Resources and Environment of the Public Service of Wallonia) for providing us with samples. We are also grateful to Olivier Hardy, Paul Bastide, Benoit Manet and Romain Candaele for their useful advices on this study. This research used resources of the Plateforme Technologique de Calcul Intensif (PTCI) located at the University of Namur, Belgium, which is supported by the F.R.S.-FNRS under the convention No. 2.4520.11. The PTCI is member of the Consortium des Équipements de Calcul Intensif (CÉCI). SD was supported by the Wiener-Anspach Foundation and the Fonds Wetenschappelijk Onderzoek (FWO, Flanders, Belgium) and is currently funded by the Fonds National de la Recherche Scientifique (FNRS, Belgium).
Author contributions
SD designed the study. M-CE and M-CF generated the raw data in the laboratory. AL prepared the CMR data. SD, JGP, SP and ACF performed the data analysis. SD wrote the first draft of the manuscript. All authors discussed the results, edited and approved the contents of the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41437-019-0183-5) contains supplementary material, which is available to authorized users.
References
- Baguette M, Blanchet S, Legrand D, Stevens VM, Turlure C. Individual dispersal, landscape connectivity and ecological networks. Biol Rev. 2013;88:310–326. doi: 10.1111/brv.12000. [DOI] [PubMed] [Google Scholar]
- Balkenhol N, Waits LP, Dezzani RJ. Statistical approaches in landscape genetics: an evaluation of methods for linking landscape and genetic data. Ecography. 2009;32:818–830. doi: 10.1111/j.1600-0587.2009.05807.x. [DOI] [Google Scholar]
- Bauman David, Drouet Thomas, Fortin Marie-Josée, Dray Stéphane. Optimizing the choice of a spatial weighting matrix in eigenvector-based methods. Ecology. 2018;99(10):2159–2166. doi: 10.1002/ecy.2469. [DOI] [PubMed] [Google Scholar]
- Bray JR, Curtis JT. An ordination of the upland forestcommunities of Southern Wisconsin. Ecol Monogr. 1957;27:326–349. doi: 10.2307/1942268. [DOI] [Google Scholar]
- Broquet T, Petit EJ. Molecular estimation of dispersal for ecology and population genetics. Annu Rev Ecol Evol Syst. 2009;40:193–216. doi: 10.1146/annurev.ecolsys.110308.120324. [DOI] [Google Scholar]
- Correa Ayram CA, Mendoza ME, Etter A, Salicrup DRP. Habitat connectivity in biodiversity conservation: A review of recent studies and applications. Progress Phys Geogr. 2016;40:7–37. doi: 10.1177/0309133315598713. [DOI] [Google Scholar]
- Dellicour S, Frantz AC, Colyn M, Bertouille S, Chaumont F, Flamand MC. Population structure and genetic diversity of red deer (Cervus elaphus) in forest fragments in north-western France. Conserv Genet. 2011;12:1287–1297. doi: 10.1007/s10592-011-0230-0. [DOI] [Google Scholar]
- Dellicour S, Mardulyn P. SPADS 1.0: a toolbox to perform spatial analyses on DNA sequence data sets. Mol Ecol Resour. 2014;14:647–651. doi: 10.1111/1755-0998.12200. [DOI] [PubMed] [Google Scholar]
- Dellicour S, Vrancken B, Trovão NS, Fargette D, Lemey P. On the importance of negative controls in viral landscape phylogeography. Virus Evol. 2018;4:vey023. doi: 10.1093/ve/vey023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellicour S, Rose R, Pybus OG. Explaining the geographic spread of emerging epidemics: a framework for comparing viral phylogenies and environmental landscape data. BMC Bioinformatics. 2016;17:1–12. doi: 10.1186/s12859-016-0924-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellicour S, Gerard M, Prunier JG, Dewulf A, Kuhlmann M, Michez D. Distribution and predictors of wing shape and size variability in three sister species of solitary bees. PLoS ONE. 2017;12:e0173109. doi: 10.1371/journal.pone.0173109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson AP, Bradshaw AD, Baker AJM. Hopes for the future: Restoration ecology and conservation biology. Science. 1997;277:515–522. doi: 10.1126/science.277.5325.515. [DOI] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Dray, S, Blanchet, G, Borcard, D, Clappe, S, Guenard, G, Jombart, T, Larocque, G, Legendre, P, Madi, N & Wagner HW (2018) adespatial: Multivariate multiscale spatial analysis. R package version 0.3-0. https://CRAN.R-project.org/package=adespatial
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Frantz AC, Bertouille S, Eloy MC, Licoppe A, Chaumon TF, Flamand MC. Comparative landscape genetic analyses show a Belgian motorway to be a gene flow barrier for red deer (Cervus elaphus), but not wild boars (Sus scrofa) Mol Ecol. 2012;21:3445–3457. doi: 10.1111/j.1365-294X.2012.05623.x. [DOI] [PubMed] [Google Scholar]
- Frantz AC, Cellina S, Krier A, Schley L, Burke T. Using spatial Bayesian methods to determine the genetic structure of a continuously distributed population: clusters or isolation by distance? J Appl Ecol. 2009;46:493–505. doi: 10.1111/j.1365-2664.2008.01606.x. [DOI] [Google Scholar]
- Frantz AC, Pourtois JT, Heuertz M, Schley L, Flamand MC, Krier A, Bertouille S, Chaumont F, Burke T. Genetic structure and assignment tests demonstrate illegal translocation of red deer (Cervus elaphus) into a continuous population. Mol Ecol. 2006;15:3191–3203. doi: 10.1111/j.1365-294X.2006.03022.x. [DOI] [PubMed] [Google Scholar]
- Frantz AC, Zachos FE, Bertouille S, Eloy MC, Colyn M, Flamand MC. Using genetic tools to estimate the prevalence of non-native red deer (Cervus elaphus) in a Western European population. Ecol Evol. 2017;7:7650–7660. doi: 10.1002/ece3.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goslee SC, Urban DL. The “ecodist” package for dissimilarity-based analysis of ecological data. J Stat Softw. 2007;22:19. doi: 10.18637/jss.v022.i07. [DOI] [Google Scholar]
- Guillot G, Mortier F, Estoup A. GENELAND: a computer package for landscape genetics. Mol Ecol Notes. 2005;5:712–715. doi: 10.1111/j.1471-8286.2005.01031.x. [DOI] [Google Scholar]
- Hardy OJ. Estimation of pairwise relatedness between individuals and characterization of isolation-by-distance processes using dominant genetic markers. Mol Ecol. 2003;12:1577–1588. doi: 10.1046/j.1365-294X.2003.01835.x. [DOI] [PubMed] [Google Scholar]
- Hardy OJ, Vekemans X. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes. 2002;2:618–620. doi: 10.1046/j.1471-8286.2002.00305.x. [DOI] [Google Scholar]
- Hartl GB, Zachos FE, Nadlinger K, Ratkiewicz M, Klein F, Lang G. Allozyme and mitochondrial DNA analysis of French red deer (Cervus elaphus) populations: genetic structure and its implications for management and conservation. Mamm Biol. 2005;70:24–34. doi: 10.1078/1616-5047-00173. [DOI] [Google Scholar]
- Hengl, T, Kempen, B, Heuvelink, G, Malone, B (2017) GSIF: Global Soil Information Facilities – tools (standards and functions) and sample datasets for global soil mapping. R package version 0.5-4. https://CRAN.R-project.org/package=GSIF
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Kuehn R, Schroeder W, Pirchner F, Rottmann O. Genetic diversity, gene flow and drift in Bavarian red deer populations (Cervus elaphus) Conserv Genet. 2003;4:157–166. doi: 10.1023/A:1023394707884. [DOI] [Google Scholar]
- Laenen L, Dellicour S, Vergote V, Nauwelaers I, De Coster S, Verbeeck I, Vanmechelen B, Lemey P, Maes P. Spatio-temporal analysis of Nova virus, a divergent hantavirus circulating in the European mole in Belgium. Mol Ecol. 2016;25:5994–6008. doi: 10.1111/mec.13887. [DOI] [PubMed] [Google Scholar]
- Landguth EL, Cushman SA, Schwartz MK, McKelvey KS, Murphy M, Luikart G. Quantifying the lag time to detect barriers in landscape genetics. Mol Ecol. 2010;19:4179–4191. doi: 10.1111/j.1365-294X.2010.04808.x. [DOI] [PubMed] [Google Scholar]
- Lecocq T, Gérard M, Michez D, Dellicour S. Conservation genetics of European bees: new insights from the continental scale. Conserv Genet. 2016;18:585–596. doi: 10.1007/s10592-016-0917-3. [DOI] [Google Scholar]
- Loiselle BA, Sork VL, Nason J, Graham C. Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae) Am J Bot. 1995;82:1420–1425. doi: 10.1002/j.1537-2197.1995.tb12679.x. [DOI] [Google Scholar]
- Lowe WH, Allendorf FW. What can genetics tell us about population connectivity? Mol Ecol. 2010;19:3038–3051. doi: 10.1111/j.1365-294X.2010.04688.x. [DOI] [PubMed] [Google Scholar]
- Manel S, Holderegger R. Ten years of landscape genetics. Trends Ecol Evol. 2013;28:614–621. doi: 10.1016/j.tree.2013.05.012. [DOI] [PubMed] [Google Scholar]
- Manel S, Schwartz MK, Luikart G, Taberlet P. Landscape genetics: Combining landscape ecology and population genetics. Trends Ecol Evol. 2003;18:189–197. doi: 10.1016/S0169-5347(03)00008-9. [DOI] [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- McRae BH. Isolation by resistance. Evolution. 2006;60:1551–1561. doi: 10.1111/j.0014-3820.2006.tb00500.x. [DOI] [PubMed] [Google Scholar]
- McRae BH, Dickson BG, Keitt TH, Shah VB. Using circuit theory to model connectivity in ecology, evolution, and conservation. Ecology. 2008;89:2712–2724. doi: 10.1890/07-1861.1. [DOI] [PubMed] [Google Scholar]
- Newton RG, Spurrell DJ. A development of multiple regression for the analysis of routine data. J R Stat Soc Appl Stat. 1967;16:51–64. [Google Scholar]
- Nimon K, Lewis M, Kane R, Haynes RM. An R package to compute commonality coefficients in the multiple regression case: an introduction to the package and a practical example. Behav Res Methods. 2008;40:457–466. doi: 10.3758/BRM.40.2.457. [DOI] [PubMed] [Google Scholar]
- Nychka D, Furrer R, Paige J, Sain S (2017) fields: Tools for spatial data. University Corporation for Atmospheric Research, Boulder, CO, USA, 2017. www.image.ucar.edu/~nychka/Fields
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2011) Vegan: community ecology package. R package version 2.4-5. https://CRAN.R-project.org/package=vegan
- Paetkau D, Slade R, Burden M, Estoup A. Genetic assignment methods for the direct, real-time estimation of migration rate: a simulation-based exploration of accuracy and power. Mol Ecol. 2004;13:55–65. doi: 10.1046/j.1365-294X.2004.02008.x. [DOI] [PubMed] [Google Scholar]
- Paulhus DL, Robins RW, Trzesniewski KH, Tracy JL. Two replicable suppressor situations in personality research. Multivar Behav Res. 2004;39:303–328. doi: 10.1207/s15327906mbr3902_7. [DOI] [PubMed] [Google Scholar]
- Peterman WE, Connette GM, Semlitsch RD, Eggert LS. Ecological resistance surfaces predict fine-scale genetic differentiation in a terrestrial woodland salamander. Mol Ecol. 2014;23:2402–2413. doi: 10.1111/mec.12747. [DOI] [PubMed] [Google Scholar]
- Piry S, Alapetite A, Cornuet JM, Paetkau D, Baudouin L, Estoup A. GENECLASS2: a software for genetic assignment and first-generation migrant detection. J Hered. 2004;95:536–539. doi: 10.1093/jhered/esh074. [DOI] [PubMed] [Google Scholar]
- Piry S, Chapuis MP, Gauffre B, Papaïx J, Cruaud A, Berthier K. Mapping Averaged Pairwise Information (MAPI): a new exploratory tool to uncover spatial structure. Methods Ecol Evol. 2016;7:1463–1475. doi: 10.1111/2041-210X.12616. [DOI] [Google Scholar]
- Prévot C, Licoppe A. Comparing red deer (Cervus elaphus L.) and wild boar (Sus scrofa L.) dispersal patterns in southern Belgium. Eur J Wildl Res. 2013;59:795–803. doi: 10.1007/s10344-013-0732-9. [DOI] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunier JG, Kaufmann B, Fenet S, Picard D, Pompanon F, Joly P, Lena JP. Optimizing the trade-off between spatial and genetic sampling efforts in patchy populations: towards a better assessment of functional connectivity using an individual-based sampling scheme. Mol Ecol. 2013;22:5516–5530. doi: 10.1111/mec.12499. [DOI] [PubMed] [Google Scholar]
- Prunier JG, Colyn M, Legendre X, Nimon KF, Flamand MC. Multicollinearity in spatial genetics: separating the wheat from the chaff using commonality analyses. Mol Ecol. 2015;24:263–283. doi: 10.1111/mec.13029. [DOI] [PubMed] [Google Scholar]
- Prunier Jérôme G., Dubut Vincent, Loot Géraldine, Tudesque Loïc, Blanchet Simon. The relative contribution of river network structure and anthropogenic stressors to spatial patterns of genetic diversity in two freshwater fishes: A multiple-stressors approach. Freshwater Biology. 2017;63(1):6–21. doi: 10.1111/fwb.13034. [DOI] [Google Scholar]
- Ray-Mukherjee J, Nimon K, Mukherjee S, Morris DW, Slotow R, Hamer M. Using commonality analysis in multiple regressions: a tool to decompose regression effects in the face of multicollinearity. Methods Ecol Evol. 2014;5:320–328. doi: 10.1111/2041-210X.12166. [DOI] [Google Scholar]
- Richardson JL, Brady SP, Wang IJ, Spear SF. Navigating the pitfalls and promise of landscape genetics. Mol Ecol. 2016;25:849–863. doi: 10.1111/mec.13527. [DOI] [PubMed] [Google Scholar]
- Rousset F. Genetic differentiation between individuals. J Evolut Biol. 2000;13:58–62. doi: 10.1046/j.1420-9101.2000.00137.x. [DOI] [Google Scholar]
- Spear SF, Balkenhol N, Fortin MJ, McRae BH, Scribner KIM. Use of resistance surfaces for landscape genetic studies: considerations for parameterization and analysis. Mol Ecol. 2010;19:3576–3591. doi: 10.1111/j.1365-294X.2010.04657.x. [DOI] [PubMed] [Google Scholar]
- Storfer A, Murphy MA, Spear SF, Holderegger R, Waits LP. Landscape genetics: where are we now? Mol Ecol. 2010;19:3496–3514. doi: 10.1111/j.1365-294X.2010.04691.x. [DOI] [PubMed] [Google Scholar]
- Troupin Cécile, Picard-Meyer Evelyne, Dellicour Simon, Casademont Isabelle, Kergoat Lauriane, Lepelletier Anthony, Dacheux Laurent, Baele Guy, Monchâtre-Leroy Elodie, Cliquet Florence, Lemey Philippe, Bourhy Hervé. Host Genetic Variation Does Not Determine Spatio-Temporal Patterns of European Bat 1 Lyssavirus. Genome Biology and Evolution. 2017;9(11):3202–3213. doi: 10.1093/gbe/evx236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Example files related to the Appendix S1, microsatellite genotypes and CMR data were deposited in the Dryad Digital Repository (10.5061/dryad.c6t0470).