Kindlin-3 regulates podosome stability by recruiting leupaxin to podosomes, which in turn controls PTP-PEST activity and paxillin phosphorylation. Kindlin-3 deficiency allows formation of initial adhesion patches containing talin, vinculin, and paxillin, whereas paxillin family proteins are dispensable for podosome formation.
Abstract
Binding of kindlins to integrins is required for integrin activation, stable ligand binding, and subsequent intracellular signaling. How hematopoietic kindlin-3 contributes to the assembly and stability of the adhesion complex is not known. Here we report that kindlin-3 recruits leupaxin into podosomes and thereby regulates paxillin phosphorylation and podosome turnover. We demonstrate that the activity of the protein tyrosine phosphatase PTP-PEST, which controls paxillin phosphorylation, requires leupaxin. In contrast, despite sharing the same binding mode with leupaxin, paxillin recruitment into podosomes is kindlin-3 independent. Instead, we found paxillin together with talin and vinculin in initial adhesion patches of kindlin-3–null cells. Surprisingly, despite its presence in these early adhesion patches, podosomes can form in the absence of paxillin or any paxillin member. In conclusion, our findings show that kindlin-3 not only activates and clusters integrins into podosomes but also regulates their lifetime by recruiting leupaxin, which controls PTP-PEST activity and thereby paxillin phosphorylation and downstream signaling.
Introduction
Integrin-mediated cell-matrix adhesions anchor cells on or within extracellular matrices, sense the physical properties of the environment, and translate them into biochemical signals (Hynes, 2002). These signals are combined with those from other signaling pathways to regulate a variety of cellular functions such as cell proliferation, differentiation, and survival (Legate et al., 2009; Humphries et al., 2019). The ability of integrins to efficiently and stably bind to their extracellular ligands is regulated by changes in their affinity to ligands and the concentration of activated integrins into clusters (Iwamoto and Calderwood, 2015). Both aspects of integrin regulation require the binding of two intracellular adapter proteins, talin and kindlin, to the integrin β subunit cytoplasmic domain (Moser et al., 2009; Sun et al., 2019). These initial events are followed by a complex assembly of a multitude of adapter and signaling molecules, which define the biochemical and physical properties of the adhesion complex (Harburger and Calderwood, 2009).
Different integrin-mediated cell adhesion complexes exist depending on the cell type, integrin expression, or matrix composition (Block et al., 2008). Here we focus on podosomes, which are adhesion structures that show distinct morphological characteristics compared with focal adhesions, although they are composed of almost the same components (Marchisio et al., 1988; Destaing et al., 2003; Calle et al., 2006). Podosomes have been observed in myeloid, endothelial, and some c-Src–transformed tumor cells. Each podosome is organized into two domains: a dense actin core, which contains actin-regulatory proteins, and a ring of signaling and adaptor proteins embedded in an actin cloud, which surrounds the actin core and is connected to the extracellular matrix via integrins (Linder and Kopp, 2005; Murphy and Courtneidge, 2011). Podosomes are more dynamic than focal adhesions with lifetimes within the minute scale and are involved in matrix degradation and cell invasion (Block et al., 2008). Talin-1 and kindlin-3 are both required for proper podosome assembly (Schmidt et al., 2011; Zou et al., 2013), most likely due to their essential role in integrin activation, clustering, and linkage to the actin cytoskeleton. But how these two proteins orchestrate the complex assembly of podosomes and regulate their stability and turnover, which are important for processes such as cell adhesion, migration, matrix degradation, transmigration, and tumor invasion, has not been investigated at the molecular level.
The adapter protein paxillin is one of the earliest proteins to be detected in nascent adhesions at the leading edge of the cell (Digman et al., 2008), and accumulation of paxillin is the first visible step in podosome assembly (Luxenburg et al., 2012). In addition to its role in the assembly of adhesion structures, paxillin regulates focal adhesion and podosome disassembly. Phosphorylation of paxillin at Y31 and Y118 is involved in focal adhesion turnover and dissolving podosomal belts (Laukaitis et al., 2001; Ballestrem et al., 2006; Badowski et al., 2008). Mutation of both tyrosines into non-phosphorylated amino acids impairs the disassembly of adhesion structures (Webb et al., 2004). Recently, a direct interaction between kindlin-2 and paxillin has been reported, which is crucial for paxillin recruitment to newly formed adhesion sites and induction of cell spreading in fibroblasts (Theodosiou et al., 2016; Böttcher et al., 2017). Moreover, interaction of paxillin with kindlin-3, the hematopoietic member of the kindlin family, was shown to promote platelet integrin activation (Gao et al., 2017).
In the present study, we aimed to elucidate a signaling pathway downstream of kindlin-3, which is involved in podosome assembly and stability regulation and identified leupaxin, a member of the paxillin gene family, as a new kindlin-3 interactor. Overall our study shows that kindlin-3 has a dual function in podosome regulation: besides its essential role in integrin regulation and proper podosome assembly, it also regulates podosome turnover by recruiting leupaxin into podosomes to control phospho-paxillin levels and thereby increasing the podosome lifetime.
Results
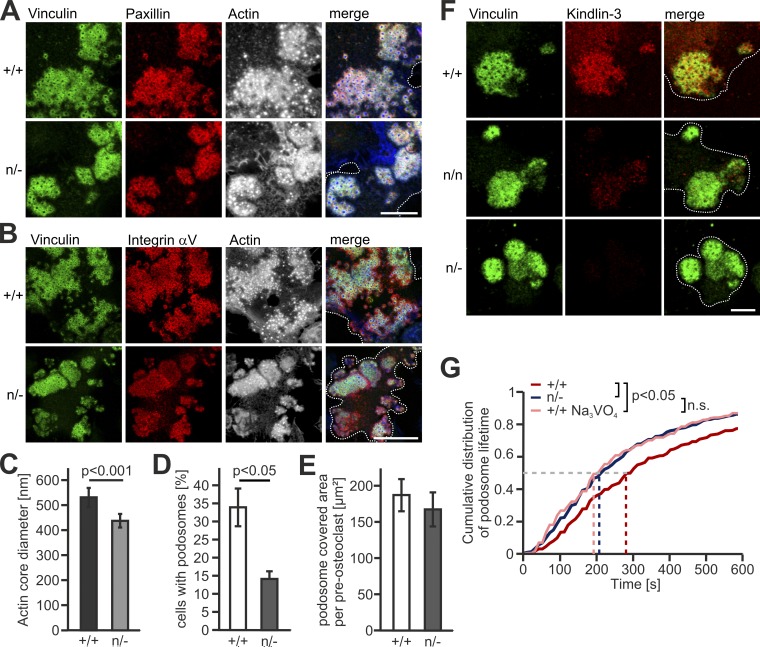
Low kindlin-3 expression results in reduced podosome lifetime
We have previously shown that kindlin-3–deficient cells fail to assemble definitive podosomes because of their inability to activate, cluster, and recruit integrins (Schmidt et al., 2011). Since the adhesion structures are not properly formed in the absence of kindlin-3, the role of kindlin-3 in integrin-mediated outside-in signaling could not be studied yet. We overcame this problem by strongly reducing kindlin-3 levels rather than eliminating it. We achieved this by generating preosteoclasts from kindlin-3 hypomorphic mice (K3n/−), which express only 5% of normal kindlin-3 levels due to the presence of a neomycin resistance cassette within the kindlin-3 gene locus (Klapproth et al., 2015). Despite the very low kindlin-3 expression, K3n/− cells formed podosomal clusters, in which plaque proteins such as vinculin and paxillin, as well as αV integrins, are correctly targeted to the podosomal ring (Fig. 1, A and B). Their actin core diameter, however, was reduced (Fig. 1 C). Although fewer K3n/− cells formed podosomal clusters (Fig. 1 D), their cluster size was similar to that of control preosteoclasts (Fig. 1 E). Importantly and consistent with its strongly reduced expression, kindlin-3 is hardly detectable by immunofluorescence (IF) in podosomal clusters of K3n/n (10% kindlin-3) and below detection level in K3n/− preosteoclasts (Fig. 1 F). To further characterize the podosomes of K3n/− cells, we determined their turnover by tracking individual podosomes of control and K3n/− cells by life cell imaging. We transfected the cells with LifeAct-GFP, which binds to F-actin and labels the central podosomal actin core (Riedl et al., 2008). Whereas 50% of WT podosomes could be imaged for 260 s or longer, half of the podosomes in K3n/− cells were already disassembled after 200 s (Fig. 1 G and Videos 1 and 2). These data suggest two distinct functions of kindlin-3: (1) the assembly of podosomes, which requires just a small amount of kindlin-3 and is presumably due to kindlin-3–mediated integrin activation (inside-out signaling), and (2) stabilizing podosomes, which requires high kindlin-3 levels and is probably controlled by kindlin-3–mediated integrin outside-in signaling. The second mechanism is unexplored and was the focus of the following studies.
Figure 1.
Low kindlin-3 expression results in reduced podosome lifetime. (A) IF stainings of WT (+/+) and K3n/− (n/−) preosteoclasts for vinculin (green), paxillin (red) and actin (white/blue in merge). Scale bar, 10 µm. (B) IF stainings for vinculin (green), integrin αV (red), and actin (white/blue in merge) in podosomes of +/+ and n/− preosteoclasts. Scale bar, 20 µm. (C) Diameter of the podosomal actin cores in +/+ and n/− cells. 10 actin cores in two regions of three cells were measured per experiment. n = 5. (D) Percentage of podosome forming +/+ and n/− preosteoclasts. n = 4. (E) Quantification of the basal cell surface area that podosomal clusters cover in +/+ and n/− preosteoclasts. n = 48/50 from five different preparations. (F) Vinculin (green) and kindlin-3 (red) IF stainings of +/+ and n/− preosteoclasts. Scale bar, 5 µm. (G) Cumulative distribution of podosome lifetimes in +/+ untreated or treated with orthovanadate and n/− preosteoclasts. 15–20 podosomes were measured per cell. Two to five cells were analyzed in each of six different dishes per genotype. See also Videos 1 and 2. Dotted white lines mark cell borders.
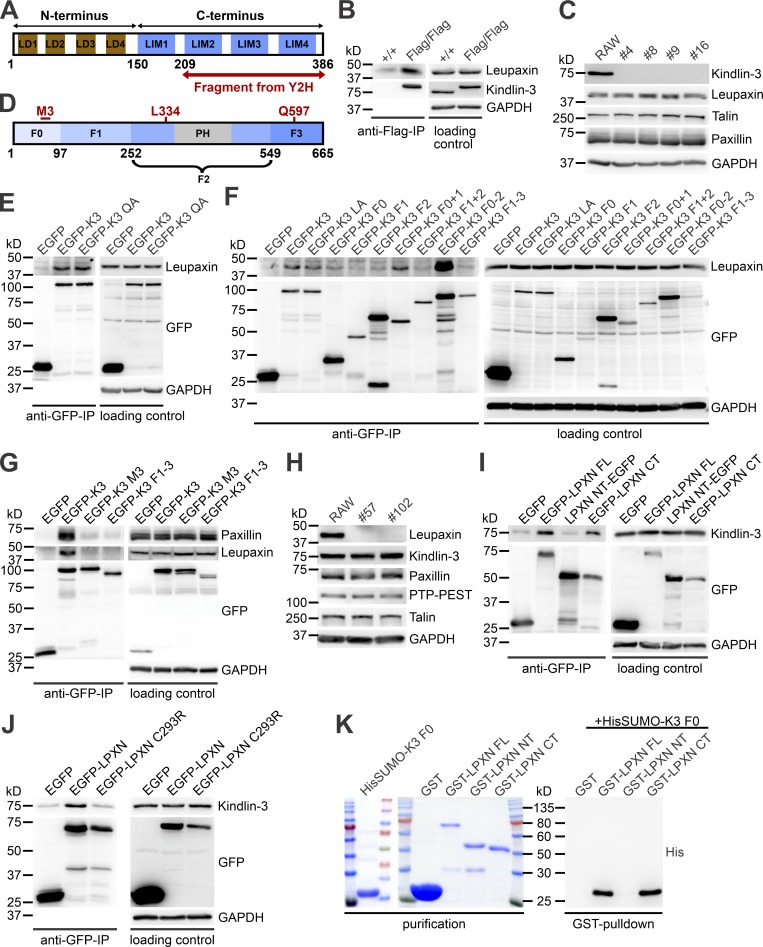
Identification of leupaxin as a new kindlin-3 interactor
To elucidate the molecular mechanism of kindlin-3–mediated integrin outside-in signaling, we performed yeast-two-hybrid assays with kindlin-3 as bait using a mouse spleen cDNA library. In two screens, five cDNAs were isolated. One encoded the C terminus (CT) of leupaxin, a cytoskeleton adapter protein belonging to the paxillin protein family (Brown and Turner, 2004). Leupaxin is preferentially expressed in hematopoietic cells and consists of four leucine-aspartic acid (LD)–rich motifs in the N terminus (NT) and four Lin11, Isl-1, and Mec-3 (LIM) domains in the CT (Fig. 2 A).
Figure 2.
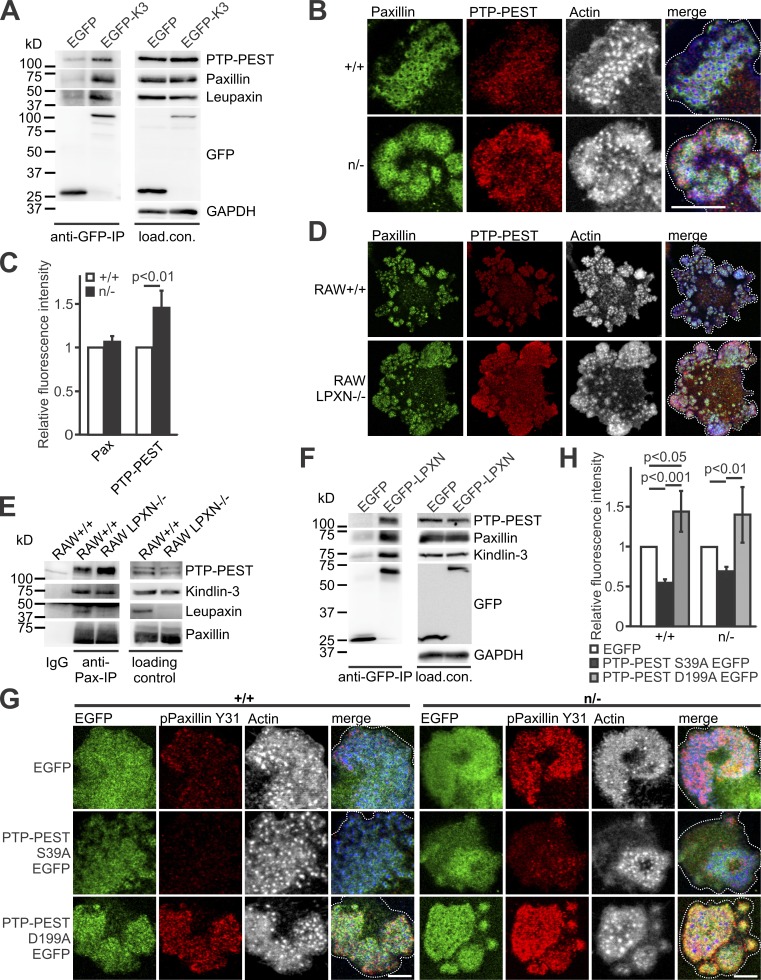
Identification and characterization of leupaxin as new kindlin-3 interactor. (A) Domain structure of leupaxin. A C-terminal leupaxin fragment bound to kindlin-3 in a yeast-two-hybrid screen. (B) Flag-immunoprecipitation (IP) from lysates of +/+ and Flag-tagged kindlin-3 expressing (Flag/Flag) bone marrow–derived macrophages to verify interaction with endogenous leupaxin. (C) Western blot analyses of leupaxin, talin, and paxillin expression in +/+ RAW cells and four different K3−/− RAW cell clones. (D) Domain structure of kindlin-3. (E) GFP-IP from lysates of K3−/− RAW cells expressing EGFP, EGFP-K3, or the EGFP-tagged K3 QA mutant analyzed for leupaxin. (F) GFP-IP from lysates of K3−/− RAW cells expressing different EGFP-K3 fragments to identify the leupaxin-interacting domain. (G) GFP-IP from lysates of K3−/− RAW cells expressing EGFP, EGFP-K3, EGFP-K3 M3 mutant, or EGFP-K3 F1–3 to investigate the interaction with leupaxin and paxillin. (H) Western blot analyses of +/+ RAW cells and two leupaxin−/− RAW cell clones for their expression of kindlin-3, paxillin, PTP-PEST, and talin. (I) GFP-IP from lysates of leupaxin−/− RAW cells reconstituted with EGFP-tagged FL leupaxin (LPXN), a N-terminal fragment (NT), or its CT to determine interaction with kindlin-3. (J) GFP-IP from lysates of leupaxin−/− RAW cells reconstituted either with EGFP-tagged WT leupaxin or a leupaxin mutant (C293R) analyzed for kindlin-3 binding. (K) GST-pulldown with GST, GST-leupaxin FL, GST-leupaxin NT, and GST-leupaxin CT incubated with His-Sumo-tagged K3 F0.
To verify the interaction between kindlin-3 and leupaxin in hematopoietic cells, we used Flag–kindlin-3 knockin mice, in which a triple Flag sequence was inserted after the start codon of the kindlin-3 gene (Fig. S1, A and B). Flag-tagged kindlin-3 was expressed at normal levels (Fig. S1 C) and localized indistinguishably from kindlin-3 in the podosomal ring structure of macrophages (Fig. S1 D; Ussar et al., 2006). We differentiated macrophages from bone marrow of WT and Flag–kindlin-3 knockin mice and immunoprecipitated Flag-tagged kindlin-3 (Fig. 2 B). These immunoprecipitates contained leupaxin, supporting the direct binding between the two proteins suggested by the yeast-two-hybrid interaction.
To further map the leupaxin binding site within kindlin-3 we used RAW 246.7 (RAW) cells, a murine macrophage/monocyte-like cell line, in which we deleted the kindlin-3 kursiv gene by CRISPR/Cas9 technology. Kindlin-3–deficient RAW cells expressed normal levels of leupaxin, talin, and paxillin in all analyzed clones (Fig. 2 C) and were used for reexpression of various kindlin-3 mutants and domains (Fig. 2 D). We then transfected EGFP-tagged WT kindlin-3 and an integrin-binding mutant kindlin-3 (K3 QA; Moser et al., 2008) into these cells, performed a GFP immunoprecipitation, and again confirmed the interaction between WT kindlin-3 and leupaxin. Since the kindlin-3 integrin-binding mutant precipitated similar amounts of leupaxin as the WT kindlin-3 (Fig. 2 E), this indicates that association of kindlin-3 with leupaxin does not require kindlin-3 to be bound to the integrin tail. Furthermore, the interaction between kindlin-3 and integrin-linked kinase (ILK), which is mediated via the F2 domain (Fukuda et al., 2014; Huet-Calderwood et al., 2014), had no impact on kindlin-3/leupaxin interaction, as ILK-binding mutant kindlin-3 (K3 LA) showed normal leupaxin binding (Fig. 2 F). Additionally, we conducted immunoprecipitation experiments with truncated kindlin-3 constructs, which mapped the leupaxin binding site to the F0 domain. Binding of the F0 domain to leupaxin was rather weak but was enhanced in the presence of the F1 and F2 domains, indicating that these domains stabilize either the F0 domain or its interaction with leupaxin. On the contrary, all kindlin-3 constructs that lacked the F0 domain did not bind (Fig. 2 F). The strong binding of a kindlin-3 construct lacking the integrin-binding F3 domain confirmed that association of kindlin-3 with integrins was not required for leupaxin binding.
It has been shown recently that an exposed loop within the N-terminal F0 domain of kindlins mediates direct interaction with paxillin (Gao et al., 2017). To investigate whether the same motif (called the M3 cluster) conveys leupaxin binding, we substituted the M3 residues of kindlin-3 for alanines (K3 M3) and compared leupaxin and paxillin binding to WT kindlin-3, the kindlin-3 M3 mutant, and kindlin-3 lacking the F0 domain (K3 F1–3). After retroviral transduction into kindlin-3–deficient RAW cells, immunoprecipitation experiments revealed that when the F0 domain of kindlin-3 is mutated or absent, binding of both leupaxin and paxillin is strongly diminished (Fig. 2 G).
To map the kindlin-3 binding site within leupaxin, we generated leupaxin-deficient RAW cells using the CRISPR/Cas9 system. Loss of leupaxin had no effect on kindlin-3, paxillin, Hic-5, protein tyrosine phosphatase PEST (PTP-PEST), and talin expression (Fig. 2 H). Reconstitution of these cells with EGFP-tagged full-length (FL) leupaxin, its NT or CT, followed by GFP-immunoprecipitations revealed that endogenous kindlin-3 was precipitated with the FL and the C-terminal LIM domain–containing region of leupaxin, confirming the result of the yeast-two-hybrid assay (Fig. 2 I). Moreover, a specific mutation within the LIM3 domain of leupaxin (C293R), which disrupts the Zn finger motif and thereby the structural folding of this domain (Chen and Kroog, 2010; Robertson and Ostergaard, 2011), abolished kindlin-3/leupaxin interaction (Fig. 2 J). Finally, we proved direct kindlin-3/leupaxin interaction by GST-pulldown experiments with bacterially expressed HisSumo-tagged kindlin-3 F0 domain incubated with GST-FL leupaxin or the N- and C-terminal leupaxin LD and LIM domains, respectively (Fig. 2 K). Altogether, these results indicate that the M3 loop in the F0 domain of kindlin-3 interacts with the LIM domains of leupaxin, and this interaction is impaired by disrupting LIM3.
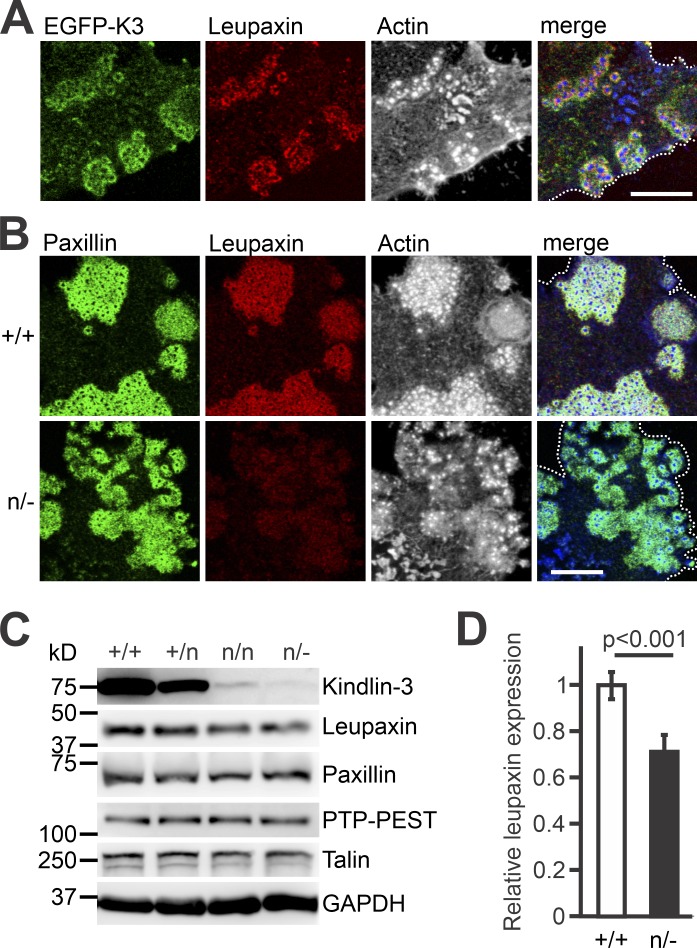
Kindlin-3–mediated leupaxin recruitment into podosomes reduces paxillin phosphorylation and increases podosome stability
Leupaxin is a cytoskeleton adapter protein, which localizes in focal adhesions of cancer cells, podosomes of macrophages, and sealing zones of osteoclasts (Gupta et al., 2003; Chen and Kroog, 2010; Tanaka et al., 2010). IF staining of preosteoclasts, differentiated from bone marrow cells and retrovirally transduced with EGFP-K3, revealed colocalization of leupaxin and kindlin-3 in the podosomal ring (Fig. 3 A). We then analyzed whether low kindlin-3 expression impacts on leupaxin localization and found strongly reduced leupaxin levels in podosomes of K3n/− cells. In contrast, paxillin was found at comparable levels, indicating that leupaxin but not its paralog paxillin requires kindlin-3 for podosomal targeting (Fig. 3 B). In support of this observation, we detected a slight reduction in total cellular leupaxin levels to ∼70% in K3n/− cells, suggesting that the interaction with kindlin-3 stabilizes cellular leupaxin, whereas paxillin levels remained unchanged (Fig. 3, C and D).
Figure 3.
Reduced kindlin-3 expression impairs leupaxin recruitment to podosomes. (A) IF staining of a K3−/− preosteoclast retrovirally transduced with EGFP-K3 (green), for leupaxin (red) and F-actin (white/blue in merge). (B) IF stainings of K3+/+ and K3n/− preosteoclasts for paxillin (green), leupaxin (red), and actin (white/blue in merge). (C) Western blot analyses on lysates of K3+/+, K3+/n, K3n/n, and K3n/− macrophages for leupaxin, paxillin, PTP-PEST, and talin expression. (D) Densitometric quantification of leupaxin expression in K3n/− macrophages relative to WT cells. All values were normalized to the corresponding GAPDH signal. n = 6. Scale bars, 10 µm. Dotted white lines mark cell borders.
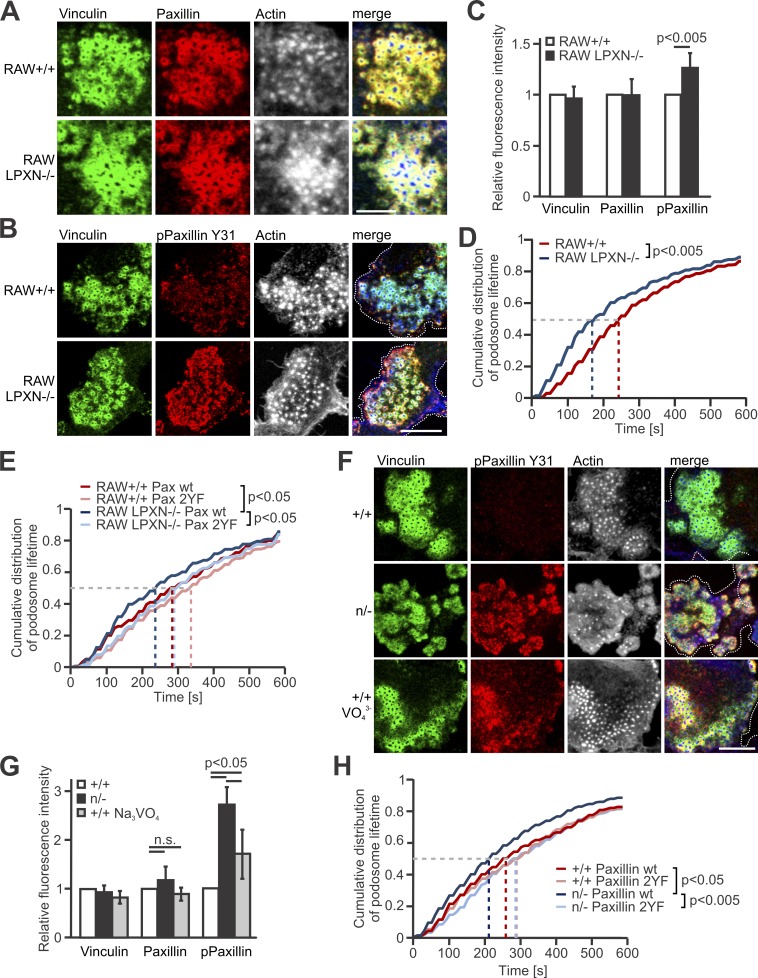
Next, we investigated whether the interaction between kindlin-3 and leupaxin is involved in podosome lifetime regulation. Previous studies suggested that leupaxin is a central component of the osteoclast podosomal signaling complex and functions as a paxillin counterpart by suppressing the tyrosine phosphorylation of paxillin in focal adhesions (Lipsky et al., 1998; Gupta et al., 2003; Sahu et al., 2007a; Tanaka et al., 2010). Nevertheless, leupaxin-null RAW cells formed podosome clusters, which were comparable to control cells (Fig. 4 A). We quantified the fluorescence intensity of vinculin, paxillin, and phospho-paxillin Y31 and Y118 within podosomal units of control and leupaxin-null RAW cells by confocal microscopy and analyzed paxillin phosphorylation upon adhesion by Western blotting. Whereas vinculin and paxillin showed normal expression in podosomes, we measured an increase in phospho-paxillin Y31 and Y118 signal in leupaxin-null cells (Fig. 4, B and C; and Fig. S2, A and B). As paxillin phosphorylation at Y31 and Y118 is known to promote adhesion turnover, podosome disassembly, and reorganization (Badowski et al., 2008), we also determined the podosome lifetime in leupaxin-null and control cells like we did for K3n/− cells. Whereas 50% of the podosomes in WT RAW cells can be imaged for ∼250 s or longer, the mean lifetime of podosomes in leupaxin-null RAW cells was reduced to ∼180 s (Fig. 4 D). To confirm that this reduction is due to elevated paxillin phosphorylation on tyrosines Y31 and Y118, we transfected the cells with mCherry-tagged WT or nonphosphorylatable paxillin (paxillin-2YF) and indeed measured an extension of the podosomal lifetime of paxillin-2YF expressing WT and leupaxin-null RAW cells compared with cells transfected with WT paxillin (Fig. 4 E).
Figure 4.
Loss of leupaxin and reduced kindlin-3 expression result in increased podosomal paxillin phosphorylation and decreased podosome lifetime. (A) IF stainings of +/+ RAW cells and leupaxin−/− RAW cells for vinculin, (green), paxillin (red), and actin (white/blue in merge) Scale bar, 10 µm. (B) IF stainings of +/+ and leupaxin−/− RAW cells for vinculin (green), phospho-paxillin Y31 (red), and actin (white/blue in merge). Scale bar, 5 µm. (C) Quantification of vinculin, total paxillin, and phospho-paxillin Y31 recruitment to podosome clusters assessed by measuring fluorescence intensity (MFI) of confocal images. MFIs of +/+ cells were set to 1. In each independent experiment, five podosome regions in each of at least 10 cells were measured. n ≥ 6. (D) Cumulative distribution of podosome lifetime in +/+ and leupaxin−/− RAW cells. 10–30 podosomes were measured per cell. At least two cells were analyzed in each of eight independent experiments. (E) Control RAW cells and different clones of leupaxin−/− RAW cells were cotransfected with WT paxillin-Cherry or a non-phosphorylatable mutant paxillin-Cherry (paxillin 2YF) and LifeAct-GFP. Podosome lifetime was assessed and blotted as cumulative distribution. 10–30 podosomes were measured per cell. At least 2 cells were analyzed in each of 5 different dishes. (F) IF stainings of +/+ preosteoclasts untreated or treated with Na3VO4 and K3n/− preosteoclasts for phosphorylated paxillin (red). Vinculin (green) and actin (white/blue in merge) served as control stainings. Scale bar, 10 µm. (G) Quantification of the recruitment of vinculin, paxillin and phospho-paxillin Y31 to podosome clusters of +/+ preosteoclasts untreated or treated with Na3VO4 and K3n/− preosteoclasts by measuring MFI of confocal images. MFIs of untreated +/+ cells were set to 1. At least five podosome regions in each of at least 10 cells were measured per experiment. n ≥ 5. (H) Control and K3n/− preosteoclasts were transfected with LifeAct-GFP, left untreated or treated with Na3VO4, or cotransfected with WT paxillin-Cherry or a nonphosphorylatable mutant paxillin-Cherry (paxillin 2YF). The cells were imaged for 10 min with a 15-s time interval. Podosome lifetime was assessed and blotted as cumulative distribution. 10–30 podosomes were measured per cell. Two to five cells were analyzed per condition in each of six different dishes. Dotted white lines mark cell borders.
We then investigated whether the reduced podosomal lifetime of K3n/− cells is also due to increased paxillin phosphorylation. In fact, we measured strongly increased phospho-paxillin levels in K3n/− podosomes of preosteoclasts (Fig. 4, F and G; and Fig. S2 C), which could be stabilized again by expression of a phospho-dead paxillin mutant (paxillin 2YF; Fig. 4 H). To further confirm our hypothesis that adhesion-mediated paxillin de-phosphorylation stabilizes podosomes, which is impaired at low kindlin-3 levels, we directly interfered with protein phosphorylation experimentally. We treated WT cells with orthovanadate, a general tyrosine phosphatase inhibitor. Orthovanadate treatment of WT preosteoclasts resulted in an increase in paxillin phosphorylation (Fig. 4, F and G) and a decrease in podosome lifetime to a similar extent as in K3n/− cells (Fig. 1 G).
Finally, we tested whether a retroviral overexpression of leupaxin rescues paxillin phosphorylation in K3n/− cells. Notably, although overexpressed leupaxin localized to podosomes of K3n/− cells, it failed to reduce paxillin phosphorylation, suggesting that kindlin-3 not only recruits leupaxin into podosomes but also facilitates leupaxin-mediated paxillin dephosphorylation (Fig. S2, D and E). In sum, these experiments indicate that low kindlin-3 expression results in impaired leupaxin podosomal targeting, increased paxillin phosphorylation, and high podosomal turnover.
Paxillin dephosphorylation by PTP-PEST depends on kindlin-3–mediated recruitment of leupaxin into podosomes
How does the reduced recruitment of leupaxin in kindlin-3 hypomorphic cells control paxillin dephosphorylation? We speculated that the protein tyrosine phosphatase PTP-PEST, which interacts with both paxillin and leupaxin (Shen et al., 1998; Gupta et al., 2003), might be involved in this process. In support of this hypothesis, an anti-GFP immunoprecipitation with lysates from kindlin-3–deficient RAW cells expressing GFP-tagged kindlin-3 showed PTP-PEST in association with kindlin-3, leupaxin, and paxillin (Fig. 5 A). Unexpectedly, we detected more PTP-PEST in podosomal clusters of K3n/− preosteoclasts and leupaxin-null RAW cells (Fig. 5, B–D), probably due to the increased binding of PTP-PEST to phosphorylated paxillin. This hypothesis was supported by anti-paxillin coimmunoprecipitation experiments showing more PTP-PEST associated with paxillin in leupaxin-null cells compared with control cells (Fig. 5 E). This experiment and an EGFP-leupaxin immunoprecipitation also showed leupaxin and kindlin-3 in association with paxillin and PTP-PEST (Fig. 5, E and F). The finding that both phospho-paxillin Y31 and PTP-PEST levels were elevated in K3n/− and leupaxin-null podosomal clusters suggests that either PTP-PEST does not dephosphorylate paxillin or its enzymatic activity requires leupaxin. To address these possibilities, we lentivirally transduced WT and K3n/− preosteoclasts with constructs expressing EGFP as a control, an EGFP-tagged constitutive active PTP-PEST mutant (S39A), or a dominant-negative form of the phosphatase (D199A; Garton et al., 1996; Motohashi et al., 2014) and measured paxillin Y31 phosphorylation levels by confocal microscopy. Constitutive active PTP-PEST reduced Y31 phosphorylation in podosome clusters of WT and K3n/− cells, and dominant-negative PTP-PEST increased phosphorylation (Fig. 5, G and H). These data suggest that in the absence of leupaxin, PTP-PEST can still bind phospho-paxillin but does not dephosphorylate it.
Figure 5.
Dephosphorylation of paxillin by PTP-PEST depends on kindlin-3–mediated recruitment of leupaxin into podosomes. (A) GFP-IP from K3−/− RAW cells, retrovirally transduced with GFP alone or a N-terminally GFP-tagged kindlin-3 analyzed for leupaxin, paxillin, and PTP-PEST. (B) IF staining of +/+ and n/− preosteoclasts for paxillin (green), PTP-PEST (red), and actin (white/blue in merge). Scale bar, 10 µm. (C) IF stainings shown in B were quantified by measuring fluorescence intensity. Values from WT cells were set to 1. In each independent experiment, five podosome regions in each of at least 10 cells were measured. n = 5. (D) Control and leupaxin−/− RAW cells stained for paxillin (green), PTP-PEST (red), and actin (white/blue in merge). Scale bar, 20 µm. (E) IP using a mouse–anti-paxillin antibody or an IgG control with lysates from +/+ and leupaxin−/− RAW cells. Binding of PTP-PEST, kindlin-3, and leupaxin was tested. (F) GFP-IP from lysates of leupaxin−/− RAW cells, retrovirally transduced with GFP alone or a N-terminally GFP-tagged leupaxin analyzed for kindlin-3, paxillin, and PTP-PEST. (G) Control and n/− preosteoclasts lentivirally transduced with EGFP, PTP-PEST S39A EGFP, or PTP-PEST D199A EGFP and stained for paxillin phosphorylated at Y31 (red) and actin (white/blue in merge). Scale bars, 5 µm. (H) Quantification of paxillin phosphorylation levels observed in G. n = 5. Dotted white lines mark cell borders.
Paxillin recruitment to primitive adhesion patches of actin cores precedes podosome maturation and is kindlin-3–independent
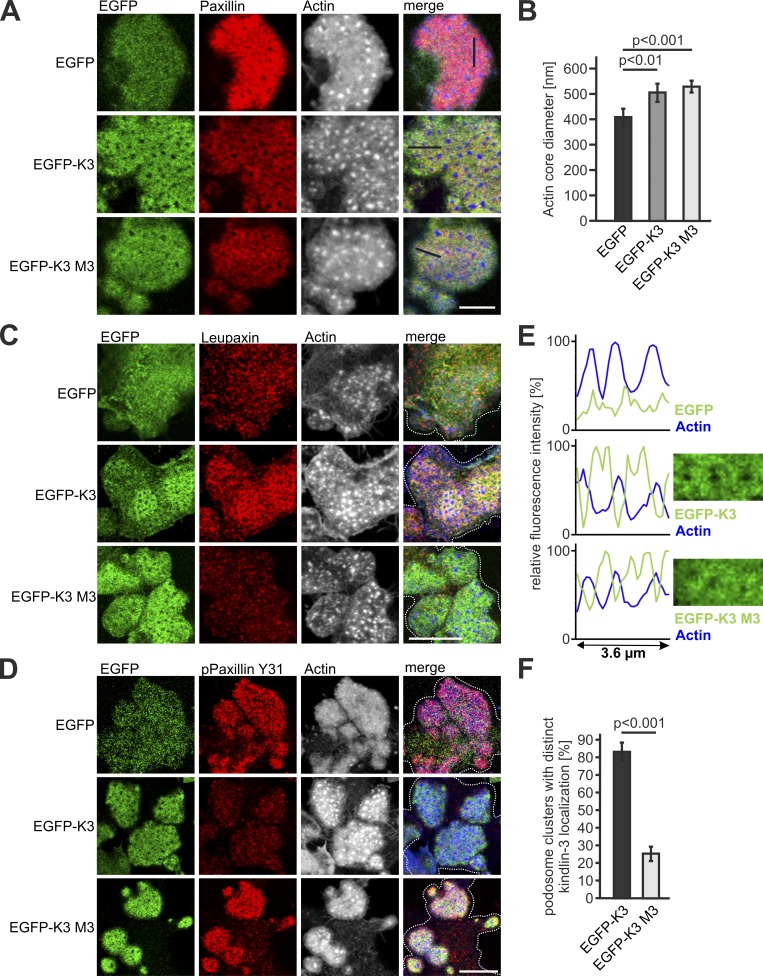
A striking observation of our study was that in contrast to leupaxin, paxillin is normally targeted to podosomes at very low kindlin-3 levels. This suggests that either paxillin recruitment to podosomes is kindlin-3 independent, or very low kindlin-3 levels are sufficient for paxillin podosomal targeting. To address this point, we differentiated preosteoclasts from the fetal liver of kindlin-3 knockout embryos and transduced them with WT and kindlin-3 M3 retroviral constructs. Both WT kindlin-3 and kindlin-3 M3 mutant rescued podosome formation, which is also indicated by the restored podosomal actin core diameter (Fig. 6, A and B). Moreover, they showed colocalization with paxillin in the podosomal ring structure, demonstrating that paxillin binding to the kindlin-3 F0 domain is not needed for podosome targeting (Fig. 6 A). In contrast, leupaxin recruitment depends on this interaction, and hence kindlin-3 M3 transduced cells exhibited increased phospho-paxillin levels (Fig. 6, C and D). However, their podosomes appeared more diffuse, with kindlin-3 and paxillin aberrantly extending into the actin cores, suggesting that the individual podosomes are less organized (Fig. 6, E and F). Moreover, consistent with a kindlin-3–independent paxillin localization to podosomes, we found paxillin already strongly accumulated in actin-rich adhesion patches of kindlin-3–null cells (Fig. 6 A). These adhesion patches were also enriched for vinculin and talin, but lack α4 and β1 integrins, indicating that paxillin is not recruited via binding to α4 integrin (Fig. S3, A–D). The lack of kindlin-3 probably explains the absence of leupaxin within these adhesion patches and the higher phospho-paxillin levels compared with that within podosomal clusters of cells expressing WT kindlin-3 (Fig. 6 D). These data on the one hand show that paxillin recruitment to podosomes does not depend on interaction with kindlin-3 and on the other hand imply that during podosome formation, initial, actin-rich adhesion patches, which contain actin cores surrounded by the adapter proteins talin, vinculin, and paxillin, are formed independently of kindlin-3. Kindlin-3 is subsequently needed to recruit and cluster integrins as well as to induce integrin-mediated signaling leading to leupaxin-dependent regulation of phospho-paxillin levels.
Figure 6.
Leupaxin-binding mutant kindlin-3 fails to recruit leupaxin to podosomes and to reduce paxillin phosphorylation. (A, C, and D) IF staining and confocal imaging of K3−/− preosteoclasts retrovirally transduced with EGFP, WT EGFP-K3, or the EGFP-K3 M3 mutant (green) for paxillin (red, A), leupaxin (red, C), and phospho-paxillin Y31 (red, D) and actin (white/blue in merge). Scale bars, 5 µm (A) and 10 µm (C and D). (B) Actin core diameter of K3−/− preosteoclasts expressing EGFP, EGPF-K3, or the EGFP-K3 M3 mutant. 10 actin cores in two regions of three to five cells were measured per experiment. n = 5. (E) Fluorescence intensity profile through three actin-cores (indicated by the black lines in A) of K3−/− preosteoclasts expressing EGFP, WT EGPF-K3, or the EGFP-K3 M3 mutant. (F) Percentage of cells with podosome clusters, which reveal discrete localization of EGFP-K3 and EGFP-K3 M3 in the podosomal ring. 50 cells were evaluated per condition in each of three independent experiments. Dotted white lines mark cell borders.
Paxillin family proteins are not crucial for podosome formation but for their signaling
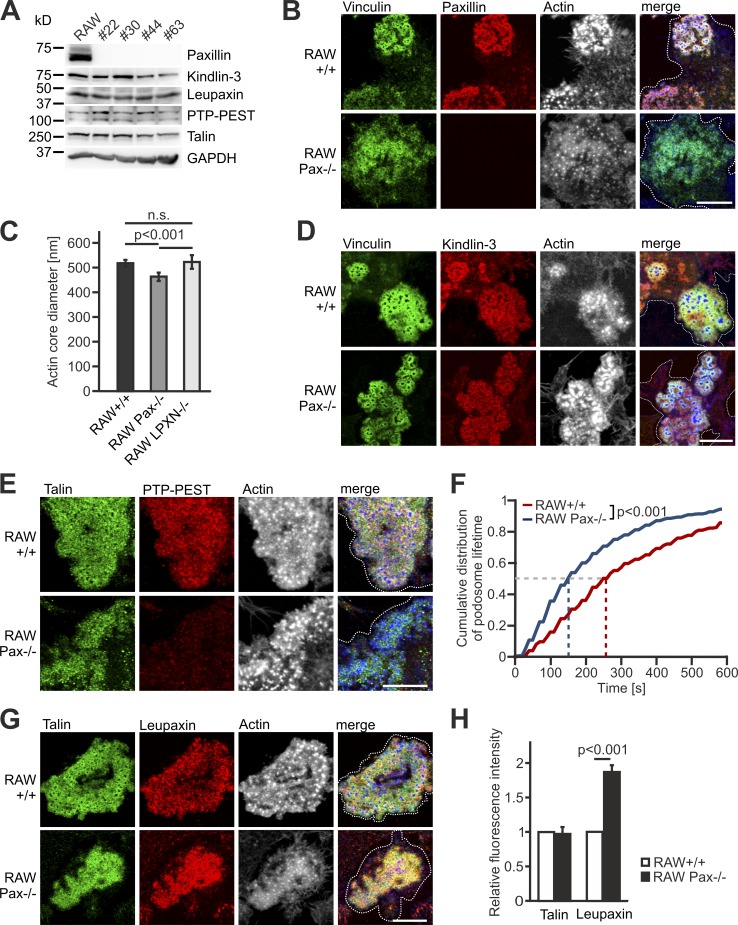
Based on the described interaction of paxillin with all members of the kindlin family and its early presence in forming podosomes (Luxenburg et al., 2012; Gao et al., 2017), we wondered whether paxillin instead recruits kindlin-3 to podosomes and how its deficiency affects podosome formation and dynamics. To this end, we mutated the paxillin gene by CRISPR/Cas9 in RAW cells. Paxillin-deficient RAW cells expressed normal levels of kindlin-3, leupaxin, and PTP-PEST (Fig. 7 A), and showed no induction of the third paxillin family member, Hic-5 (Fig. S4 A). Surprisingly, paxillin-null RAW cells did form podosomes, which were organized into kindlin-3–, talin-, and vinculin-positive rings surrounding an ∼10% smaller actin core compared with control podosomes (Fig. 7, B–E). We often noticed a more diffuse staining of the plaque proteins, indicating a mild defect in podosomal organization similar to cells expressing the kindlin-3 M3 mutant. In addition, we measured a strongly reduced podosomal lifetime in paxillin-null cells; while 50% of control podosomes dissolved after 260 s, paxillin-null cells had a half-life of only 150 s (Fig. 7 F). Of note, PTP-PEST was virtually absent from the adhesion patches of paxillin-null cells, consistent with the coelevation of PTP-PEST and phospho-paxillin when kindlin-3 activity is reduced, and suggesting that phospho-paxillin is responsible for recruiting PTP-PEST to podosomes (Fig. 7 E). These data indicate that paxillin is dispensable for podosome formation, but podosomal organization and stability depend on the adapter protein paxillin.
Figure 7.
Paxillin-deficient cells form podosomes with smaller actin cores and strongly reduced lifetime. (A) Kindlin-3, leupaxin, PTP-PEST, and talin expression in +/+ RAW cells and four different paxillin−/− RAW cell clones analyzed by Western blotting. (B) IF stainings of +/+ and paxillin−/− RAW cells for vinculin (green), paxillin (red), and actin (white/blue in merge). (C) Diameter of the podosome actin cores in +/+, paxillin−/−, and leupaxin−/− RAW cells. 10 actin cores in two regions of five to eight cells were measured in each experiment. n = 10/6/7. (D) IF stainings for vinculin (green), kindlin-3 (red), and actin (white/blue in merge) on +/+ and paxillin−/− RAW cells. (E) IF stainings for talin (green), PTP-PEST (red) and actin (white/blue in merge) on +/+ and paxillin−/− RAW cells. (F) Control RAW cells and different clones of paxillin−/− RAW cells were transfected with LifeAct-GFP and imaged at a spinning disc microscope for 10 min with a 15-s time interval. The cumulative distribution of these measurements is shown. 10–30 podosomes were measured per cell. At least four cells were analyzed in each of six independent experiments. (G) Confocal images of talin (green), leupaxin (red), and actin (white/blue in merge) IF stainings of +/+ and paxillin−/− RAW cells. (H) IF stainings shown in G were quantified by measuring fluorescence intensity. Values from WT cells were set to 1. In each independent experiment, five podosome regions in each of at least 10 cells were measured. n = 4. Scale bars, 10 µm. Dotted white lines mark cell borders.
The rather mild podosome defect in paxillin-null cells suggested partial functional compensation by leupaxin, which was indeed found at higher levels in podosomal rings of paxillin-null cells (Fig. 7, G and H). To further investigate the relationship between kindlin-3, leupaxin, and paxillin, we retrovirally transduced paxillin and leupaxin-deficient RAW cells with EGFP-K3 and performed EGFP-coimmunoprecipitation experiments. In both experiments, loss of one paxillin family member did not result in increased binding of the other member to kindlin-3, suggesting a lack of competitive binding (Fig. S4, B and C). This may be explained by an excess of kindlin-3 relative to the levels of paxillin and leupaxin, which we confirmed by a whole proteome analysis of WT RAW cells (Fig. S4 D).
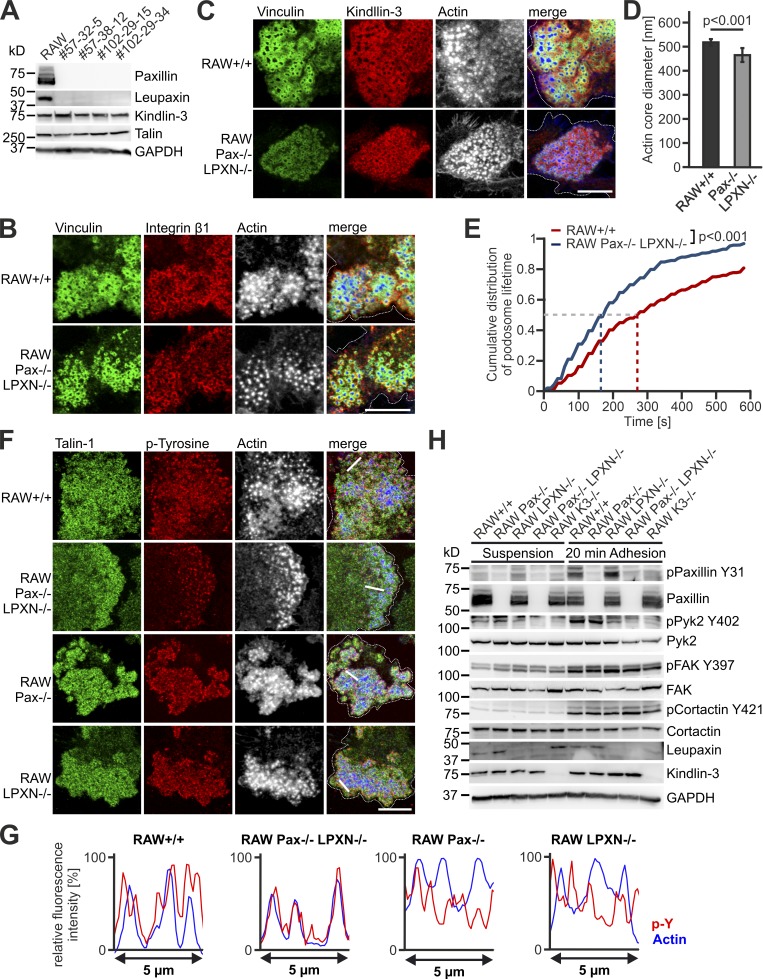
To directly address the potential compensatory effects between paxillin and leupaxin, we generated paxillin and leupaxin double knockout (dKO) cells, which showed normal kindlin-3 and talin expression (Fig. 8 A). To our surprise, the dKO cells were also able to form podosomes and podosome clusters, in which kindlin-3, talin, vinculin, and β1-integrin localized to the podosomal ring surrounding an actin core with a similarly reduced size like in paxillin-null cells (Fig. 8, B–D and F). Podosome lifetime was decreased to a similar extent as in paxillin−/− cells (Fig. 8 E). Notably, these cells showed no induction of Hic-5 expression at either the mRNA or the protein level (Fig. S4, E and F). We then expressed GFP–Hic-5 in dKO cells, which showed correct localization in the podosomal ring, however, it failed to rescue the reduced actin core size of dKO cells, again showing specific functions for the different paxillin family members (Pignatelli et al., 2012; Petropoulos et al., 2016; Fig. S4, G and H). Thus, recruitment of kindlin-3 and talin into podosomal rings as well as general organization of podosomes can occur in the absence of paxillin family members.
Figure 8.
Characterization of podosomes from paxillin/leupaxin double deficient RAW cells. (A) Western blot analyses of +/+ RAW cells and four different clones of paxillin/leupaxin dKO RAW cells for their expression of kindlin-3 and talin. (B and C) IF stainings of +/+ and paxillin/leupaxin dKO RAW cells for vinculin (green), integrin β1 (red, B), kindin-3 (red, C) and actin (white/blue in merge). (D) Diameter of the podosome actin cores in +/+ and paxillin/leupaxin dKO RAW cells. 10 actin cores in two regions of five to eight cells were measured in each experiment. n = 9/7. (E) Control RAW cells and different clones of paxillin/leupaxin dKO RAW cells were transfected with LifeAct-GFP and imaged at a spinning disc microscope for 10 min with a 15-s time interval. The cumulative distribution of these measurements is shown. 20 podosomes were measured per cell. At least four cells were analyzed in each of six dishes. (F) IF stainings for talin (green), tyrosine-phosphorylated proteins (red) and actin (white/blue in merge) on +/+, paxillin/leupaxin dKO, paxillin−/− and leupaxin−/− RAW cells. (G) Fluorescence intensity profiles of actin (blue) and phospho-tyrosine (p-Y; red) through three actin cores (indicated by the white lines in E) of +/+, paxillin/leupaxin dKO, paxillin−/−, and leupaxin−/− RAW cells. (H) Western blot analyses of the phosphorylation status of paxillin, Pyk2, FAK and cortactin in +/+, paxillin−/−, leupaxin−/−, K3−/− and paxillin/leupaxin dKO RAW cells kept in suspension or adherent to fibronectin. Scale bars, 10 µm. Dotted white lines mark cell borders.
We then studied whether paxillin family members are critically involved in podosome/integrin signaling and function. Consistent with previous studies (Luxenburg et al., 2006, 2007), strong phospho-tyrosine signals were detected in the podosomal ring of control cells, and also present in paxillin- and leupaxin-null cells (Fig. 8 F). In contrast, dKO cells revealed a dotted distribution of phospho-tyrosine signals colocalizing with the actin core, which was also illustrated by fluorescence intensity profiles across actin cores (Fig. 8, F and G). In addition, we investigated protein phosphorylation upon adhesion-mediated signaling to fibronectin by Western blot analyses (Fig. 8 H). Consistent with the IF stainings, we measured strong induction of paxillin phosphorylation at Y31 in leupaxin-null cells and only a weak increase in kindlin-3 knockout cells, which was probably due to the low number of adhesion complexes formed by kindlin-3–null cells. Phosphorylation of the actin core marker cortactin was comparable between the different cell lines. We detected a reduction in Pyk2 phosphorylation in leupaxin-null cells and virtually no Pyk2 phosphorylation at Y402 in dKO and kindlin-3–null cells, while FAK phosphorylation at Y397 was not reduced.
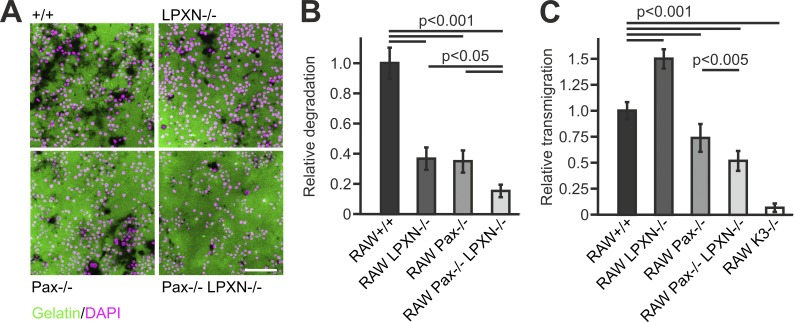
Finally, we tested whether these changes in podosome dynamics and adhesion signaling affect cell migration and matrix degradation and found reduced gelatin degradation of paxillin−/− and leupaxin−/− cells, which was further exacerbated in dKO cells (Fig. 9, A and B). Cell migration was assessed by Transwell assays showing reduced transmigration in paxillin−/− and dKO cells, whereas loss of leupaxin increased migration (Fig. 9 C).
Figure 9.
Loss of leupaxin and/or paxillin affect matrix degradation and cell migration. (A) IF images of DAPI-stained (magenta) +/+, leupaxin−/−, paxillin−/−, and paxillin/leupaxin dKO RAW cells seeded on Oregon Green 488–labeled gelatin (green) for 24 h. Scale bar, 100 µm. (B) Relative degradation capacity (area of degraded collagen normalized by number of nuclei) quantified from images shown in A. (C) Migration of these and K3−/− RAW cells in relation to +/+ cells assessed by Transwell assays.
Discussion
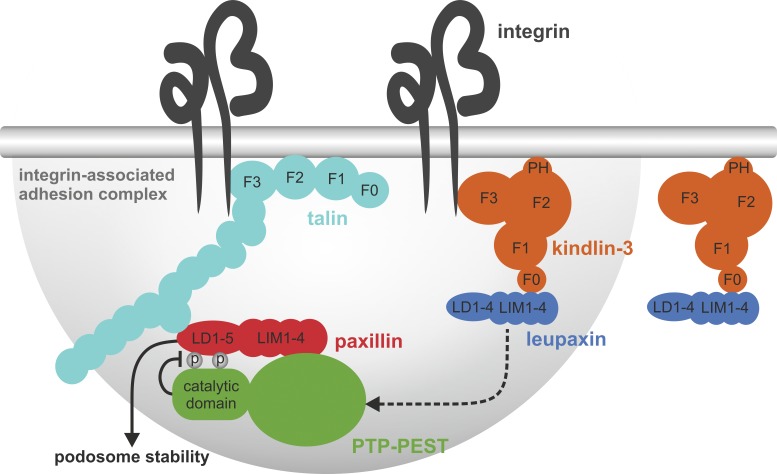
The role of kindlin-3 in regulating integrin activity in cooperation with talin is well established in hematopoietic cells. The central aim of this study was to investigate how kindlin-3 regulates integrin signaling and adhesion structure formation in hematopoietic cells, which is far less understood. To this end, we performed a Y2H screen and identified leupaxin as a new kindlin-3 interactor. Leupaxin is primarily expressed in hematopoietic cells and belongs to the paxillin gene family of adapter proteins, which also includes paxillin and Hic-5 (Lipsky et al., 1998). We show that recruitment of leupaxin into podosomes depends on the interaction between a loop within the F0 domain of kindlin-3 and leupaxin LIM domains. The same loop also mediates binding to paxillin, indicating a conserved binding mode between kindlin and paxillin members (Gao et al., 2017). The finding that leupaxin binding to kindlin-3 is enhanced by removal of the F3 domain of kindlin-3 suggests that the F3 domain exerts an inhibitory effect via steric hindrance. Such a model is consistent with the fact that in the cloverleaf domain structure of kindlins, the C-terminal F3 domain and the N-terminal F0 domain are in proximity with each other (Li et al., 2017). Notably, leupaxin binds equally well to WT kindlin-3 and an integrin binding-defective mutant of kindlin-3 (QA), suggesting that their interaction occurs outside of the adhesion site and thus that kindlin-3/leupaxin interaction may also have functions other than regulating adhesion stability (Fig. 10). Which signals modulate their interaction or whether this interaction is of constitutive nature is unclear yet and requires further studies.
Figure 10.
A kindlin-3/leupaxin complex regulates paxillin tyrosine phosphorylation and podosome stability. Kindlin-3/leupaxin interaction is independent of integrin binding. However, kindlin-3 targets leupaxin into the podosome adhesion complex. In contrast, paxillin recruitment to podosomes occurs independent of kindlin-3. The recruitment of leupaxin into podosomes enables the tyrosine phosphatase PTP-PEST to dephosphorylate paxillin at Y31 and Y118, resulting in increased podosome lifetime and stability. PH, pleckstrin homology domain.
An important finding of our study is that although leupaxin and paxillin bind kindlin-3 in a similar manner and have multiple binding partners such as FAK, Pyk2, and PTP-PEST in common (Brown et al., 1996; Lipsky et al., 1998; Gupta et al., 2003; Brown and Turner, 2004; Vanarotti et al., 2016), only leupaxin recruitment to podosomes is kindlin-3 dependent. While both paxillin and leupaxin act as scaffolds within adhesion complexes, leupaxin also regulates paxillin activity by suppressing its tyrosine phosphorylation (Tanaka et al., 2010). Phosphorylation of paxillin on Y31 and Y118, which occurs in response to integrin-mediated adhesion signaling, creates new SH2 docking sites for other signaling proteins such as FAK, Crk, and p120RasGAP and thereby functions as a molecular switch, which changes the adhesive and signaling properties of the adhesion complex (Zaidel-Bar et al., 2007; Deakin and Turner, 2008). One important function of paxillin tyrosine phosphorylation is to enhance adhesion disassembly (Nakamura et al., 2000; Webb et al., 2004; Badowski et al., 2008). So kindlin-3 not only is essential for the assembly of podosomes by activating and clustering integrins within podosomes (Schmidt et al., 2011) but also stabilizes and prolongs the lifetime of these adhesions through recruitment of leupaxin.
How leupaxin regulates paxillin phosphorylation is not clear. Based on the reported interaction between the phosphatase PTP-PEST with leupaxin and paxillin (Shen et al., 1998; Sahu et al., 2007b) and the higher phospho-paxillin levels in PTP-PEST–null cells (Angers-Loustau et al., 1999), we hypothesized that PTP-PEST might be critically involved in that process. Several findings supported this hypothesis. First, we found more PTP-PEST in podosomes of kindlin-3 hypomorphic and leupaxin-null cells, which have higher levels of phospho-paxillin, and PTP-PEST does not localize to podosomes in paxillin-null cells. Second, immunoprecipitation experiments revealed more PTP-PEST in association with paxillin in leupaxin-null cells compared with WT cells, indicating that PTP-PEST primarily associates with phosphorylated paxillin. Third, overexpression of a constitutive active form of PTP-PEST reduced paxillin phosphorylation. In sum, these findings suggest an impaired tyrosine phosphatase activity of PTP-PEST in cells where leupaxin is not efficiently recruited to podosomes. However, the molecular details of how leupaxin regulates PTP-PEST activity, e.g., by direct interaction or by recruiting a kinase or phosphatase that acts on PTP-PEST, require further investigations.
Another interesting aspect of our study concerns the role of paxillin family members in podosome formation and signaling. The first evidence for a direct interaction between paxillin and kindlin family members came from recent studies in fibroblasts, which showed that recruitment of paxillin by kindlin-2 to small, peripheral forming adhesion sites, also known as nascent adhesions, precedes focal adhesion maturation and is required for lamellipodia formation and cell spreading (Theodosiou et al., 2016; Böttcher et al., 2017). Another study reported that paxillin binding to both kindlin and talin stabilizes talin binding to the integrin cytoplasmic domain, thereby promoting integrin activation in CHO cells and platelets (Gao et al., 2017). These studies suggested that interactions between kindlins and paxillin are particularly important during the initial phases of adhesion formation by stabilizing the activation state of talin-bound integrin and establishing signaling platforms that further promote cell adhesion and spreading. Consistent with this idea is the observation of very early local accumulation of paxillin at sites where podosomes assemble (Luxenburg et al., 2012). However, in contrast to the kindlin-2–dependent paxillin recruitment to focal complexes, paxillin accumulation at sites where podosomes form is kindlin-3 independent, as shown by normal paxillin localization in podosomes of cells expressing a paxillin-binding mutant kindlin-3 or in adhesion patches of kindlin-3–deficient cells. Notably, paxillin together with talin and vinculin are already present within these initial adhesion sites of kindlin-3–null cells and surround small, less interconnected actin cores that lack a ring of integrins. It is important to note here that hematopoietic cells only express kindlin-3 (Ussar et al., 2006; Moser et al., 2008), which was also confirmed by the whole proteome analysis of RAW cells. Thus, paxillin in complex with vinculin and talin may already form a preassembled complex within immature adhesion patches before kindlin-3 and integrins are recruited to finally assemble podosomes (Deakin and Turner, 2008). The kindlin-independent mechanism of paxillin targeting to podosomes is also consistent with the recent finding that paxillin recruitment to invadosomes, which share many similarities to podosomes, is mediated via the N-terminal LD domains, whereas targeting to focal adhesions is mediated via the LIM2 and LIM3 domains (Brown et al., 1996). Thus, targeting paxillin family members during formation of podosomes and nascent adhesions occurs via different mechanisms.
Finally, our study shows that podosomes can be formed in the absence of any paxillin family member. Although paxillin-deficient podosomes appear less organized and higher leupaxin levels suggest compensation, the overall organization of podosomes does not further deteriorate when both paxillin and leupaxin are absent. Since Hic-5 is not expressed in RAW cells, these cells obviously find a way by which other adapter proteins help out to organize the adhesion structure. Nevertheless, podosomes from dKO cells may exhibit more subtle defects such as an altered equilibrium between the podosome actin core and the actin cloud, which may impact on the mechanical properties of the podosome. These unapparent defects become evident in functional assays such as cell migration and matrix degradation, which are affected when one or more paxillin family members are absent, or at the molecular level when specific signaling pathways such as Pyk2 phosphorylation are investigated. In this context, the role of paxillin family members in the formation and function of invadosomes, which are similar to podosomes, has recently been investigated in src-transformed fibroblasts (Petropoulos et al., 2016). While paxillin deficiency also affected invadosome assembly, loss of both paxillin and Hic-5 abolished invadosome formation. This contrasts with our study and might be explained by differences in cell type or the src-driven process of invadosome assembly, which depends on the presence of either paxillin or Hic-5. Unfortunately, the role of leupaxin was not addressed in this study (Petropoulos et al., 2016).
In sum, our study shows that kindlin-3 regulates the turnover and lifetime of podosomes in myeloid cells by recruiting leupaxin to the adhesion complex, which in turn controls PTP-PEST enzymatic activity and paxillin phosphorylation (Fig. 10); that initial adhesion patches containing talin, vinculin, and paxillin form in the absence of kindlin-3; and that podosomes can form independent of paxillin family proteins.
Materials and methods
Mice
Kindlin-3–deficient (K3−/−) and kindlin-3 hypomorphic (K3n/+; K3n/n; K3n/−) mice were described previously (Moser et al., 2008; Klapproth et al., 2015). To generate Flag-tagged kindlin-3 knockin mice, a targeting vector was cloned in which a triple Flag sequence was inserted into exon 2 after the codon for methionine at position 4. In addition, we introduced a loxP-flanked neomycin resistance cassette into intron 2 of the Kindlin-3 gene. Correctly targeted R1 embryonic stem cells were injected into C57BL/6 blastocysts and the resulting chimeric mice were crossed with deleter-Cre mice to remove the loxP-flanked neomycin cassette (Betz et al., 1996).
Mouse experiments were performed with the approval of the District Government of Bavaria.
Reagents
Recombinant murine macrophage colony-stimulating factor (M-CSF) and receptor activator of NF-κB ligand (RANKL) were obtained from Peprotech. Bovine plasma fibronectin was purchased from Merck Millipore and murine TNF-α from R&D Systems. A list of key resources is shown in Table S1.
Antibodies
The following antibodies were used for immunostaining of cells: mouse anti-vinculin antibody, mouse anti-Flag M2-Cy3 antibody (Sigma-Aldrich); rat anti-integrin αV-PE (Millipore); integrin α4-PE (BD PharMingen); mouse anti-cathepsin K (Calbiochem); rabbit anti-paxillin and mouse anti-leupaxin (Abcam); rabbit anti-paxillin Y31, rabbit anti-paxillin Y118, and mouse anti-PTP-PEST (Thermo Fisher Scientific); rabbit anti–kindlin-3 antibody (homemade [Ussar et al., 2006]); rabbit anti-integrin β1 (homemade [Azimifar et al., 2012]); rabbit anti-talin-1 (Abcam); and mouse anti p-tyrosine (pY99, Santa Cruz Biotechnology). Phalloidin dyes and secondary antibodies were obtained from Invitrogen (Thermo Fisher Scientific).
The following antibodies were used for Western blotting: mouse anti-GAPDH (Merck); mouse anti-talin (Sigma-Aldrich); mouse anti-paxillin, rabbit anti-paxillin Y118 (Thermo Fisher Scientific); mouse anti-leupaxin (Abcam); mouse anti-PTP-PEST, rabbit anti-paxillin Y31 (Thermo Fisher Scientific); mouse anti-Hic-5 (BD Biosciences); rabbit anti-Pyk2, rabbit anti-phospho-Pyk2 Y402, rabbit anti-FAK, rabbit anit-phospho-FAK Y397, rabbit anti-cortactin, rabbit anti-phospho-cortactin Y421, rabbit anti-His-tag (all from Cell Signaling Technology); mouse anti-GFP (homemade cell culture supernatant); and rabbit anti–kindlin-3 antibody (homemade [Ussar et al., 2006]). HRP-labeled secondary antibodies were purchased from Jackson ImmunoResearch Laboratories.
Cell culture
Preosteoclasts were differentiated from bone marrow (K3+/+ and K3n/−) or from embryonic day 14.5 fetal liver cells (K3−/−). Cells were kept in complete α-MEM supplemented with 20 ng/ml M-CSF overnight. Non-adherent cells were collected after 24 h. Leukocytes were isolated from the interface after centrifugation at 1,000 g for 20 min in leukocyte separation medium (Laboratories Eurobio), and then washed with α-MEM medium and seeded at a concentration of 2,000–2,500 cells/mm2 in osteoclast differentiation medium (α-MEM [Gibco] containing 10% FCS, 100 U/ml penicillin, 100 µg/ml streptomycin, 60 ng/ml M-CSF, and 40 ng/ml RANKL). Cells were cultured at 37°C in 5% CO2 for 3–5 d, and medium was changed every second day. Preosteoclasts were defined as adherent cells that were treated with osteoclast differentiation medium for at least 3 d but had not yet rearranged their cytoskeleton and translocated the nuclei to the cell periphery.
RAW cells (RAW 264.7) were cultured in D10 medium (DMEM containing 10% FCS, 100 U/ml penicillin, 100 µg/ml streptomycin, 2 mM L-Glu, and nonessential amino acids; all from Gibco/Thermo Fisher Scientific). For differentiation into preosteoclasts, RAW cells were treated with osteoclast differentiation medium for ∼5 d.
Bone marrow–derived macrophages were generated by treating bone marrow cells with R10 medium (RPMI 1640 [Gibco] containing 10% FCS, 25 mM Hepes, 100 U/ml penicillin, 100 µg/ml streptomycin, 2 mM L-Glu, nonessential amino acids, and 50 µM β-mercaptoethanol) supplemented with M-CSF for 6 d.
For transient transfection of RAW cells, Lipofectamine 3000 (Invitrogen) was used according to the manufacturer’s protocol. Primary preosteoclasts were transfected using the Mouse Macrophage Nucleofector Kit from Lonza.
Plasmids and viral infections
All kindlin-3 constructs were cloned into pEGFP-C1 to express EGFP-tagged proteins. The subdomains were defined according to Huet-Calderwood et al. (2014): the F0 domain from aa 1–97, the F1 domain from aa 98–252, the F2 from aa 253–549, and the F3 domain from aa 550–665. Point mutations (Q597 to A, L334A, Q57A, D58A, W59A, S60A, and D61A) were generated by site-directed mutagenesis.
EGFP, EGFP–kindlin-3, EGFP–kindlin-3 M3 mutant, and EGFP-leupaxin were directionally cloned into the pCLMFG retroviral vector (Naviaux et al., 1996) using the XhoI and NotI restriction sites. Vesicular stomatitis virus G–pseudotyped retroviral vectors were produced in 293T (human embryonic kidney) cells. Viral particles were concentrated from cell culture supernatant as described previously (Pfeifer et al., 2000) and used for infection of preosteoclasts at day three of differentiation.
Murine FL leupaxin and the C-terminal part (aa 150–386) were cloned into pEGFP-C1, and the N-terminal part (1–149) into pEGFP-N1. The C293R mutation was introduced by site-directed mutagenesis. Murine Hic-5 was cloned into pEGFP-C1.
LifeAct-GFP vector (Riedl et al., 2008) was provided by M. Sixt (Institute of Science and Technology, Klosterneuburg, Austria). Mouse paxillin cDNA was cloned into pCherry-C1. Mutations (Y31 to F and Y118 to F) were introduced by site-directed mutagenesis (pCherry-C1 paxillin 2YF).
Human PTP-PEST cDNA was subcloned into pJet to introduce mutations (S39 to A and D199 to A) and then cloned into a lentiviral vector (rrl-CMV-GFP; provided by A. Pfeifer, University of Bonn, Bonn, Germany).
CRISPR/Cas9 mediated gene ablation
The CRISPR/Cas9 guiding sequence was designed with the help of the CRISPR design webpage of the Zhang laboratory. The sequence for the single guide RNA was cloned into the GFP-expressing vector pX458 (for kindlin-3 targeting: 5′-GGTGGCACCCACTTTATTCAG-3′, for leupaxin targeting: 5′-GTCCAAAGACCTTGTCATCGC-3′), and RAW cells were transfected using Lipofectamine LTX.
For paxillin ablation, gRNA was generated with a GeneArt Precision gRNA Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions (paxillin target sequence: 5′-TGAACTTGACCGGCTGTTAC-3′). 10 µg Cas9 protein (Thermo Fisher Scientific) were combined with 2.5 µg gRNA and incubated for 5 min. The complexes together with a GFP-encoding plasmid were electroporated into 106 RAW cells using the NEON transfection system (Thermo Fisher Scientific; conditions: resuspension buffer R, 1,680 V, 20 ms, 1 pulse, 107 cells/ml).
After 2 d, GFP-positive cells were FACS-sorted and seeded at very low concentrations to allow picking of single cell clones. Clones were checked by Western blot for the presence or absence of kindlin-3, leupaxin, and paxillin, respectively.
Immunoprecipitation
For immunoprecipitations, cells were washed twice with PBS, and 0.5 mM dithiobis(succinimidyl propionate) (Thermo Fisher Scientific) dissolved in PBS was added for 30 min at RT to cross-link proximal proteins. The reaction was stopped by incubation with 50 mM Tris, pH 7.5, for 10 min. Cells were washed with cold PBS and lysed in mammalian protein extraction reagent (Thermo Fisher Scientific) supplemented with protease inhibitors (Roche Diagnostics) and phosphatase inhibitor cocktails (Sigma-Aldrich). µMACS GFP and DYKDDDDK (to recognize the Flag-tag) Isolation kits (Miltenyi) were used according to the manufacturer’s protocol using 1–2 mg protein lysate. Endogenous paxillin immunoprecipitation was performed using 5 µg mouse anti-paxillin antibody (BD Biosciences) and Pierce Protein A/G Magnetic Beads following the user’s guide. Mouse IgG1 (Sigma-Aldrich) was used as control. Immunoprecipitations and 40 µg per loading control were subjected to 10% SDS-PAGE and subsequent Western blotting.
IF microscopy
Preosteoclasts were stained as previously described (Schmidt et al., 2011). For most antibody stainings, cells were fixed with 4% PFA for 10 min. For kindlin-3 labeling, cells were fixed with 1% PFA for 10 min followed by a 10-min incubation with ice-cold aceton. Leupaxin was stained in cells fixed with 1.5% PFA for 12 min. Cells were imaged at RT with a Leica TCS SP5 X confocal microscope (Leica Microsystems) using 63× NA 1.40 oil objective lenses and Leica Confocal Software (LAS AF). Single channels were imaged sequentially. All pictures were processed with Photoshop (Adobe Systems).
Protein recruitment to podosome clusters was quantified using ImageJ software (US National Institutes of Health). Fluorescence intensities were assessed in five randomly chosen areas (∼10 µm2) within podosome clusters per cell. All values were corrected by background fluorescence. Intensities of control stainings were measured at the identical areas in the corresponding channels (vinculin/paxillin, vinculin/phospho-paxillin Y31, and paxillin/PTP-PEST).
To assess podosome lifetime, preosteoclasts transfected with LifeAct-GFP (Riedl et al., 2008) were imaged with a custom-made spinning disc confocal microscope (Visitron System) based on a Zeiss Observer Z1 and a Yokogawa spinning disk, equipped with a 100× NA 1.45 oil objective and an EVOLVE EM512 digital camera (Photometrics). Where stated, the cells were additionally transfected with pCherry-C1 paxillin WT or pCherry-C1 paxillin 2YF. The cells were either left untreated or treated with 5 mM Na3VO4 and imaged at the spinning disc microscope for 10 min with a 15-s time interval. The lifetime of a single podosome was analyzed from these time-lapse videos by measuring the time until a podosome present at time point 0 disappeared. 10–30 podosomes were measured per cell. 2–10 cells were analyzed per condition in each of two to eight independent experiments. Podosome lifetime was analyzed using ImageJ software and the MTrackJ plugin (National Institutes of Health).
Actin core size was measured upon phalloidin staining. Two podosome containing regions were chosen per cell, and 10 podosome core diameters were measured per area. 4–10 cells were analyzed in each of five experiments by a person blinded to the genotype of the cells.
Adhesion signaling
RAW cells were trypsinized and kept in serum-free DMEM in suspension for 3 h. Then cells were either kept in suspension or plated on a 6-cm cell culture dish coated with 5 µg/ml fibronectin for 20 min. Cells were carefully washed with ice-cold PBS and lysed in RIPA buffer, containing protease inhibitors and phosphatase inhibitor cocktails (Sigma-Aldrich). 40 µg of lysates was subjected to 10% SDS-PAGE and subsequent Western blot analyses.
Sample preparation and in-solution digest
Proteins were extracted from pelleted and snap-frozen cells with 4% SDS, heated for 5 min to 95°C, and afterward precipitated with ice-cold acetone at −20°C for 2.5 h. The proteins were dissolved in 300 µl of 8 M urea buffer, and protein concentrations were determined using the Pierce 660 nm Protein Assay (Thermo Fisher Scientific). Per sample, 50 µg of protein was reduced with 5 mM dithiothreitol for 60 min, followed by carbamidomethylation with 40 mM iodoacetamide for 45 min in the dark at RT. A predigest with the protease LysC (Wako Pure Chemicals Industries) was performed for 3 h, followed by a digest with trypsin (Promega) for 16 h. The digest was terminated by the acidification of the sample to 1% formic acid, and the resulting peptides were separated from salts and detergents using the C18 based Stop and Go extraction tips (Rappsilber et al., 2003).
Liquid chromatography–tandem mass spectrometry analysis and data processing
An Easy nLC 1200 ultra-high-performance liquid chromatography coupled to a QExactive HF-X Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific) was used for the proteomic analysis with the following settings. Peptides were fractionated using in-house–made 50-cm columns packed with 1.7-µm C18 beads using a binary buffer system, consisting of Buffer A (0.1% formic acid) and Buffer B (80% acetonitrile in 0.1% formic acid). All samples were analyzed over a 90-min gradient, raising the content of Buffer B from 4% to 23% over 65 min, then from 23% to 55% over 13 min, followed by washing with 95% Buffer B. Spectra for full MS were acquired at a resolution of 60,000 at 200 m/z, and the automated gain control target was set to 3 × 106 with a maximum injection time of 20 ms. The dynamic exclusion was set to 20 s. The MS2 measurements were acquired using a resolution of 15,000 at 200 m/z with a top 22 data-dependent mode. Here the automated gain control target was set to 1 × 105 with an injection time of 22 ms. The normalized collision energy in the higher-energy collisional dissociation cell was 25.
The raw data were analyzed using the Andromeda search engine implemented in the MaxQuant software 1.5.3.8 (Cox et al., 2011). Parameters in MaxQuant were set to default with trypsin selected as protease for digestion. A mouse database from Uniprot (16.06.17) with contaminants was used for peptide and protein identification.
Transwell assay
Cell migration was tested in Transwell cell culture inserts (24-well, 8-µm pore, polyester membrane; Merck Millipore) coated with 1 mg/ml fibrinogen in TBS for 2 h at 37°C. 105 cells were stimulated with 2 µg/ml TNF-α (R&D Systems) in DMEM/F-12 containing 0.1% FCS and added to the upper reservoir. 1 ml DMEM/F-12 with 0.1% FCS was filled in the lower reservoir. The experiment was started by adding 1 µg/ml LPS to the lower reservoir. After 6 h, the cells were fixed for 15 min with 4% PFA and stained with DAPI. Not-transmigrated cells were removed with a cotton swab, and six pictures were taken per Transwell with an Evos FL cell imaging system (Thermo Fisher Scientific) with a 10× objective to assess the number of transmigrated cells.
Gelatin degradation assay
12-mm glass coverslips were coated with 10 µg/ml Oregon Green 488–conjugated gelatin (Invitrogen) in PBS for 30 min. Slides were washed with PBS and fixed with 4% PFA for 15 min. Residual PFA was removed by extensive washing in PBS and incubation with full medium for 30 min. 2 × 105 cells that were cultured in osteoclast differentiation medium for 24 h were seeded per coverslip. Cells were fixed with 4% PFA the following day and stained with DAPI. Five pictures were taken per slide with a Leica TCS SP5 X confocal microscope using a 40× NA 1.25–0.75 oil objective. Gelatin degradation was quantified by measuring the degraded area using ImageJ software and correlated to the number of nuclei.
Quantitative real-time PCR
mRNA expression level of Hic-5 was assessed by quantitative real-time PCR. An RNeasy Mini Kit (Qiagen) was used to isolate RNA, and reverse transcription was performed using a iScript cDNA Synthesis Kit (Bio-Rad Laboratories). Quantitative PCR was done with iQ SYBR Green Supermix (Bio-Rad Laboratories) in LightCycler 480 Multiwell Plate 96, white (Roche) on a LightCycler 480 II (Roche Molecular Systems) under standard conditions. The following primers were used: Hic-5 forward: 5′-GTAACCAACCCATCCGACAC-3′, Hic-5 reverse: 5′-GCTGAGCATGGAAATGGTTT-3′ (as published by Rashid et al., 2017); GAPDH forward: 5′-TCGTGGATCTGACGTGCCGCCTG-3′, and GAPDH reverse: 5′-CACCACCCTGTTGCTGTAGCCGT-3′. All samples were measured in quadruplicate. Data were analyzed using the 2−ΔΔCT-method (Livak and Schmittgen, 2001).
Recombinant protein expression, purification, and pulldown
FL leupaxin, leupaxin NT, or leupaxin CT cDNAs were cloned into the bacterial expression vector pGEX-2T, kindlin-3 F0 domain into pCoofy17 (Scholz et al., 2013). GST, N-terminally GST-tagged leupaxin FL, NT, or CT and HisSUMO-tagged kindlin-3 F0 domain were expressed in Rosetta (DE3)–competent cells. Bacteria were cultured in 50–100 ml Luria-Bertani medium at 37°C to an OD600 of 0.6 to 0.7. Proteins were expressed at 18°C overnight upon induction with 0.2 mM IPTG. For leupaxin expression, cultures were supplemented with 500 µM ZnCl2. Protein purification, pulldowns, and wash steps were performed in 25 mM Tris buffer, pH 7.5, containing 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.01% NP-40 Substitute (Sigma-Aldrich), protease inhibitors, and 10 µM ZnCl2 (for leupaxin only). Bacteria pellets were lysed by incubation with lysozyme (10 µg/ml) and sonication. Lysates were cleared by centrifugation. HisSUMO–kindlin-3 F0 was batch-purified using PureCube 100 INDIGO Ni-Agarose beads (Cube Biotech). GST and GST-tagged leupaxin FL, NT, or CT were bound to 40 µl Glutathione Magnetic Agarose Beads (Jena Bioscience) for 1 h at RT. After washing extensively, 10 µl of loaded beads per condition were blocked with 5% BSA for 1.5 h at 4°C, and, subsequently, purified kindlin-3 F0 domain was added for 1 h at 4°C in the presence of 0.5% BSA. After washing, bound proteins were eluted by incubation for 5 min at 95°C in Lämmli buffer. Samples were subjected to SDS-PAGE and Western blot.
Statistical analysis
Data are presented as means ± 95% confidence interval. Statistical analyses were performed using the software GraphPad Prism. Fluorescence intensities were log-transformed before statistical analyses, as upon transformation, normal distribution can be assumed. For statistical evaluation of podosome lifetime measurements, we determined the time at which 50% of the observed podosomes had disappeared for each cell culture dish. These values were subjected to statistical analyses, assuming normal distribution.
Paired or unpaired Student's t tests were used to compare two different datasets if normal distribution could be assumed. A Mann–Whitney test was performed to compare two datasets if normal distribution could not be implied. To evaluate three or more datasets, one-way ANOVA was performed followed by a Tukey’s or a Sidak’s multiple comparison test, as recommended by GraphPad Prism.
Data were tested for significance as follows: paired Student’s t test for fluorescence intensity measurements; unpaired Student's t test for podosome lifetime measurements, actin core diameter, and leupaxin expression; Mann–Whitney test for cells that form podosomes; one-way ANOVA with Tukey’s multiple comparison test for fluorescence intensity measurements, actin core diameter, and gelatin degradation assay; and one-way ANOVA with Sidak’s multiple comparison test for podosome lifetime measurements and Transwell assay.
A difference between datasets was considered to be significant if P < 0.05.
Online supplemental material
Fig. S1 describes the generation of Flag-tagged kindin-3 knockin mice, Flag-tagged kindlin-3 protein expression, and its localization to podosomes. Fig. S2 shows by IF stainings and Western blotting that loss of leupaxin or a reduced kindlin-3 expression result in increased phosphorylation of paxillin at Y118. Overexpression of leupaxin in K3n/− cells is not sufficient to lower paxillin phosphorylation at Y31. Fig. S3 shows confocal images of IF stainings of various podosomal marker proteins in control and K3n/− cells. Fig. S4 shows that Hic-5 expression is not induced in paxillin/leupaxin dKO cells, and retroviral overexpression of Hic-5 cannot rescue the podosome defect of paxillin/leupaxin dKO RAW cells. Furthermore, immunoprecipitation and quantitative proteomics experiments reveal that leupaxin and paxillin do not compete for kindlin-3 binding. Table S1 shows a list of the main reagents and resources used in the study. Videos 1 and 2 show +/+ and K3n/− preosteoclasts expressing LifeAct-GFP, revealing shorter podosome lifetime at low kindlin-3 expression levels.
Supplementary Material
Acknowledgments
The authors thank Soo Jin Min-Weiβenhorn and the people from the transgenic facility of the Max Planck Institute of Biochemistry for help with generating the Flag–kindlin-3 mice. We thank Reinhard Fässler for his generous support and Arnoud Sonnenberg, Stefan Linder, and Nick Brown for critically reading the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB914 TP A01) and the Max Planck Society.
The authors declare no competing financial interests.
Author contributions: S. Klapproth, T. Bromberger, and M. Moser designed and performed the experiments and analyzed data. C. Türk and M. Krüger performed mass spectrometry and analyzed the data. M. Moser supervised the work, and S. Klapproth and M. Moser wrote the paper with input from all other authors.
References
- Angers-Loustau A., Côté J.F., Charest A., Dowbenko D., Spencer S., Lasky L.A., and Tremblay M.L.. 1999. Protein tyrosine phosphatase-PEST regulates focal adhesion disassembly, migration, and cytokinesis in fibroblasts. J. Cell Biol. 144:1019–1031. 10.1083/jcb.144.5.1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimifar S.B., Böttcher R.T., Zanivan S., Grashoff C., Krüger M., Legate K.R., Mann M., and Fässler R.. 2012. Induction of membrane circular dorsal ruffles requires co-signalling of integrin-ILK-complex and EGF receptor. J. Cell Sci. 125:435–448. 10.1242/jcs.091652 [DOI] [PubMed] [Google Scholar]
- Badowski C., Pawlak G., Grichine A., Chabadel A., Oddou C., Jurdic P., Pfaff M., Albigès-Rizo C., and Block M.R.. 2008. Paxillin phosphorylation controls invadopodia/podosomes spatiotemporal organization. Mol. Biol. Cell. 19:633–645. 10.1091/mbc.e06-01-0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballestrem C., Erez N., Kirchner J., Kam Z., Bershadsky A., and Geiger B.. 2006. Molecular mapping of tyrosine-phosphorylated proteins in focal adhesions using fluorescence resonance energy transfer. J. Cell Sci. 119:866–875. 10.1242/jcs.02794 [DOI] [PubMed] [Google Scholar]
- Betz U.A., Vosshenrich C.A., Rajewsky K., and Müller W.. 1996. Bypass of lethality with mosaic mice generated by Cre-loxP-mediated recombination. Curr. Biol. 6:1307–1316. 10.1016/S0960-9822(02)70717-3 [DOI] [PubMed] [Google Scholar]
- Block M.R., Badowski C., Millon-Fremillon A., Bouvard D., Bouin A.P., Faurobert E., Gerber-Scokaert D., Planus E., and Albiges-Rizo C.. 2008. Podosome-type adhesions and focal adhesions, so alike yet so different. Eur. J. Cell Biol. 87:491–506. 10.1016/j.ejcb.2008.02.012 [DOI] [PubMed] [Google Scholar]
- Böttcher R.T., Veelders M., Rombaut P., Faix J., Theodosiou M., Stradal T.E., Rottner K., Zent R., Herzog F., and Fässler R.. 2017. Kindlin-2 recruits paxillin and Arp2/3 to promote membrane protrusions during initial cell spreading. J. Cell Biol. 216:3785–3798. 10.1083/jcb.201701176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M.C., and Turner C.E.. 2004. Paxillin: adapting to change. Physiol. Rev. 84:1315–1339. 10.1152/physrev.00002.2004 [DOI] [PubMed] [Google Scholar]
- Brown M.C., Perrotta J.A., and Turner C.E.. 1996. Identification of LIM3 as the principal determinant of paxillin focal adhesion localization and characterization of a novel motif on paxillin directing vinculin and focal adhesion kinase binding. J. Cell Biol. 135:1109–1123. 10.1083/jcb.135.4.1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calle Y., Burns S., Thrasher A.J., and Jones G.E.. 2006. The leukocyte podosome. Eur. J. Cell Biol. 85:151–157. 10.1016/j.ejcb.2005.09.003 [DOI] [PubMed] [Google Scholar]
- Chen P.W., and Kroog G.S.. 2010. Leupaxin is similar to paxillin in focal adhesion targeting and tyrosine phosphorylation but has distinct roles in cell adhesion and spreading. Cell Adhes. Migr. 4:527–540. 10.4161/cam.4.4.12399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., and Mann M.. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10:1794–1805. 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- Deakin N.O., and Turner C.E.. 2008. Paxillin comes of age. J. Cell Sci. 121:2435–2444. 10.1242/jcs.018044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destaing O., Saltel F., Géminard J.C., Jurdic P., and Bard F.. 2003. Podosomes display actin turnover and dynamic self-organization in osteoclasts expressing actin-green fluorescent protein. Mol. Biol. Cell. 14:407–416. 10.1091/mbc.e02-07-0389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digman M.A., Brown C.M., Horwitz A.R., Mantulin W.W., and Gratton E.. 2008. Paxillin dynamics measured during adhesion assembly and disassembly by correlation spectroscopy. Biophys. J. 94:2819–2831. 10.1529/biophysj.107.104984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda K., Bledzka K., Yang J., Perera H.D., Plow E.F., and Qin J.. 2014. Molecular basis of kindlin-2 binding to integrin-linked kinase pseudokinase for regulating cell adhesion. J. Biol. Chem. 289:28363–28375. 10.1074/jbc.M114.596692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Huang M., Lai J., Mao K., Sun P., Cao Z., Hu Y., Zhang Y., Schulte M.L., Jin C., et al. . 2017. Kindlin supports platelet integrin αIIbβ3 activation by interacting with paxillin. J. Cell Sci. 130:3764–3775. 10.1242/jcs.205641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garton A.J., Flint A.J., and Tonks N.K.. 1996. Identification of p130(cas) as a substrate for the cytosolic protein tyrosine phosphatase PTP-PEST. Mol. Cell. Biol. 16:6408–6418. 10.1128/MCB.16.11.6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Lee B.S., Khadeer M.A., Tang Z., Chellaiah M., Abu-Amer Y., Goldknopf J., and Hruska K.A.. 2003. Leupaxin is a critical adaptor protein in the adhesion zone of the osteoclast. J. Bone Miner. Res. 18:669–685. 10.1359/jbmr.2003.18.4.669 [DOI] [PubMed] [Google Scholar]
- Harburger D.S., and Calderwood D.A.. 2009. Integrin signalling at a glance. J. Cell Sci. 122:159–163. 10.1242/jcs.018093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet-Calderwood C., Brahme N.N., Kumar N., Stiegler A.L., Raghavan S., Boggon T.J., and Calderwood D.A.. 2014. Differences in binding to the ILK complex determines kindlin isoform adhesion localization and integrin activation. J. Cell Sci. 127:4308–4321. 10.1242/jcs.155879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries J.D., Chastney M.R., Askari J.A., and Humphries M.J.. 2019. Signal transduction via integrin adhesion complexes. Curr. Opin. Cell Biol. 56:14–21. 10.1016/j.ceb.2018.08.004 [DOI] [PubMed] [Google Scholar]
- Hynes R.O. 2002. Integrins: bidirectional, allosteric signaling machines. Cell. 110:673–687. 10.1016/S0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- Iwamoto D.V., and Calderwood D.A.. 2015. Regulation of integrin-mediated adhesions. Curr. Opin. Cell Biol. 36:41–47. 10.1016/j.ceb.2015.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapproth S., Moretti F.A., Zeiler M., Ruppert R., Breithaupt U., Mueller S., Haas R., Mann M., Sperandio M., Fässler R., and Moser M.. 2015. Minimal amounts of kindlin-3 suffice for basal platelet and leukocyte functions in mice. Blood. 126:2592–2600. 10.1182/blood-2015-04-639310 [DOI] [PubMed] [Google Scholar]
- Laukaitis C.M., Webb D.J., Donais K., and Horwitz A.F.. 2001. Differential dynamics of alpha 5 integrin, paxillin, and alpha-actinin during formation and disassembly of adhesions in migrating cells. J. Cell Biol. 153:1427–1440. 10.1083/jcb.153.7.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legate K.R., Wickström S.A., and Fässler R.. 2009. Genetic and cell biological analysis of integrin outside-in signaling. Genes Dev. 23:397–418. 10.1101/gad.1758709 [DOI] [PubMed] [Google Scholar]
- Li H., Deng Y., Sun K., Yang H., Liu J., Wang M., Zhang Z., Lin J., Wu C., Wei Z., and Yu C.. 2017. Structural basis of kindlin-mediated integrin recognition and activation. Proc. Natl. Acad. Sci. USA. 114:9349–9354. 10.1073/pnas.1703064114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder S., and Kopp P.. 2005. Podosomes at a glance. J. Cell Sci. 118:2079–2082. 10.1242/jcs.02390 [DOI] [PubMed] [Google Scholar]
- Lipsky B.P., Beals C.R., and Staunton D.E.. 1998. Leupaxin is a novel LIM domain protein that forms a complex with PYK2. J. Biol. Chem. 273:11709–11713. 10.1074/jbc.273.19.11709 [DOI] [PubMed] [Google Scholar]
- Livak K.J., and Schmittgen T.D.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Luxenburg C., Parsons J.T., Addadi L., and Geiger B.. 2006. Involvement of the Src-cortactin pathway in podosome formation and turnover during polarization of cultured osteoclasts. J. Cell Sci. 119:4878–4888. 10.1242/jcs.03271 [DOI] [PubMed] [Google Scholar]
- Luxenburg C., Geblinger D., Klein E., Anderson K., Hanein D., Geiger B., and Addadi L.. 2007. The architecture of the adhesive apparatus of cultured osteoclasts: from podosome formation to sealing zone assembly. PLoS One. 2:e179 10.1371/journal.pone.0000179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxenburg C., Winograd-Katz S., Addadi L., and Geiger B.. 2012. Involvement of actin polymerization in podosome dynamics. J. Cell Sci. 125:1666–1672. 10.1242/jcs.075903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchisio P.C., Bergui L., Corbascio G.C., Cremona O., D’Urso N., Schena M., Tesio L., and Caligaris-Cappio F.. 1988. Vinculin, talin, and integrins are localized at specific adhesion sites of malignant B lymphocytes. Blood. 72:830–833. [PubMed] [Google Scholar]
- Moser M., Nieswandt B., Ussar S., Pozgajova M., and Fässler R.. 2008. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat. Med. 14:325–330. 10.1038/nm1722 [DOI] [PubMed] [Google Scholar]
- Moser M., Legate K.R., Zent R., and Fässler R.. 2009. The tail of integrins, talin, and kindlins. Science. 324:895–899. 10.1126/science.1163865 [DOI] [PubMed] [Google Scholar]
- Motohashi S., Koizumi K., Honda R., Maruyama A., Palmer H.E., and Mashima K.. 2014. Protein tyrosine phosphatase-PEST (PTP-PEST) regulates mast cell-activating signals in PTP activity-dependent and -independent manners. Cell. Immunol. 289:128–134. 10.1016/j.cellimm.2014.04.003 [DOI] [PubMed] [Google Scholar]
- Murphy D.A., and Courtneidge S.A.. 2011. The ‘ins’ and ‘outs’ of podosomes and invadopodia: characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 12:413–426. 10.1038/nrm3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K., Yano H., Uchida H., Hashimoto S., Schaefer E., and Sabe H.. 2000. Tyrosine phosphorylation of paxillin alpha is involved in temporospatial regulation of paxillin-containing focal adhesion formation and F-actin organization in motile cells. J. Biol. Chem. 275:27155–27164. [DOI] [PubMed] [Google Scholar]
- Naviaux R.K., Costanzi E., Haas M., and Verma I.M.. 1996. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 70:5701–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropoulos C., Oddou C., Emadali A., Hiriart-Bryant E., Boyault C., Faurobert E., Vande Pol S., Kim-Kaneyama J.R., Kraut A., Coute Y., et al. . 2016. Roles of paxillin family members in adhesion and ECM degradation coupling at invadosomes. J. Cell Biol. 213:585–599. 10.1083/jcb.201510036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer A., Kessler T., Silletti S., Cheresh D.A., and Verma I.M.. 2000. Suppression of angiogenesis by lentiviral delivery of PEX, a noncatalytic fragment of matrix metalloproteinase 2. Proc. Natl. Acad. Sci. USA. 97:12227–12232. 10.1073/pnas.220399597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatelli J., LaLonde S.E., LaLonde D.P., Clarke D., and Turner C.E.. 2012. Actopaxin (α-parvin) phosphorylation is required for matrix degradation and cancer cell invasion. J. Biol. Chem. 287:37309–37320. 10.1074/jbc.M112.385229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Ishihama Y., and Mann M.. 2003. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75:663–670. 10.1021/ac026117i [DOI] [PubMed] [Google Scholar]
- Rashid M., Belmont J., Carpenter D., Turner C.E., and Olson E.C.. 2017. Neural-specific deletion of the focal adhesion adaptor protein paxillin slows migration speed and delays cortical layer formation. Development. 144:4002–4014. 10.1242/dev.147934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z., et al. . 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 5:605–607. 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson L.K., and Ostergaard H.L.. 2011. Paxillin associates with the microtubule cytoskeleton and the immunological synapse of CTL through its leucine-aspartic acid domains and contributes to microtubule organizing center reorientation. J. Immunol. 187:5824–5833. 10.4049/jimmunol.1003690 [DOI] [PubMed] [Google Scholar]
- Sahu S.N., Khadeer M.A., Robertson B.W., Núñez S.M., Bai G., and Gupta A.. 2007a Association of leupaxin with Src in osteoclasts. Am. J. Physiol. Cell Physiol. 292:C581–C590. 10.1152/ajpcell.00636.2005 [DOI] [PubMed] [Google Scholar]
- Sahu S.N., Nunez S., Bai G., and Gupta A.. 2007b Interaction of Pyk2 and PTP-PEST with leupaxin in prostate cancer cells. Am. J. Physiol. Cell Physiol. 292:C2288–C2296. 10.1152/ajpcell.00503.2006 [DOI] [PubMed] [Google Scholar]
- Schmidt S., Nakchbandi I., Ruppert R., Kawelke N., Hess M.W., Pfaller K., Jurdic P., Fässler R., and Moser M.. 2011. Kindlin-3-mediated signaling from multiple integrin classes is required for osteoclast-mediated bone resorption. J. Cell Biol. 192:883–897. 10.1083/jcb.201007141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz J., Besir H., Strasser C., and Suppmann S.. 2013. A new method to customize protein expression vectors for fast, efficient and background free parallel cloning. BMC Biotechnol. 13:12 10.1186/1472-6750-13-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Schneider G., Cloutier J.F., Veillette A., and Schaller M.D.. 1998. Direct association of protein-tyrosine phosphatase PTP-PEST with paxillin. J. Biol. Chem. 273:6474–6481. 10.1074/jbc.273.11.6474 [DOI] [PubMed] [Google Scholar]
- Sun Z., Costell M., and Fässler R.. 2019. Integrin activation by talin, kindlin and mechanical forces. Nat. Cell Biol. 21:25–31. 10.1038/s41556-018-0234-9 [DOI] [PubMed] [Google Scholar]
- Tanaka T., Moriwaki K., Murata S., and Miyasaka M.. 2010. LIM domain-containing adaptor, leupaxin, localizes in focal adhesion and suppresses the integrin-induced tyrosine phosphorylation of paxillin. Cancer Sci. 101:363–368. 10.1111/j.1349-7006.2009.01398.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodosiou M., Widmaier M., Böttcher R.T., Rognoni E., Veelders M., Bharadwaj M., Lambacher A., Austen K., Müller D.J., Zent R., and Fässler R.. 2016. Kindlin-2 cooperates with talin to activate integrins and induces cell spreading by directly binding paxillin. eLife. 5:e10130 10.7554/eLife.10130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ussar S., Wang H.V., Linder S., Fässler R., and Moser M.. 2006. The Kindlins: subcellular localization and expression during murine development. Exp. Cell Res. 312:3142–3151. 10.1016/j.yexcr.2006.06.030 [DOI] [PubMed] [Google Scholar]
- Vanarotti M.S., Finkelstein D.B., Guibao C.D., Nourse A., Miller D.J., and Zheng J.J.. 2016. Structural Basis for the Interaction between Pyk2-FAT Domain and Leupaxin LD Repeats. Biochemistry. 55:1332–1345. 10.1021/acs.biochem.5b01274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb D.J., Donais K., Whitmore L.A., Thomas S.M., Turner C.E., Parsons J.T., and Horwitz A.F.. 2004. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 6:154–161. 10.1038/ncb1094 [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar R., Itzkovitz S., Ma’ayan A., Iyengar R., and Geiger B.. 2007. Functional atlas of the integrin adhesome. Nat. Cell Biol. 9:858–867. 10.1038/ncb0807-858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W., Izawa T., Zhu T., Chappel J., Otero K., Monkley S.J., Critchley D.R., Petrich B.G., Morozov A., Ginsberg M.H., and Teitelbaum S.L.. 2013. Talin1 and Rap1 are critical for osteoclast function. Mol. Cell. Biol. 33:830–844. 10.1128/MCB.00790-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.