Abstract
Neuropilin-1 (NRP-1), a member of the NRP-family, has been reported to be vital for tumor angiogenesis, growth and metastasis. As a co-receptor of vascular endothelial growth factor (VEGF), NRP-1 can bind to VEGF and meditate vascular development through the VEGF-VEGF receptor 2 (VEGFR2) signaling pathway. Furthermore, NRP-1 is capable of binding with platelet-derived growth factor (PDGF) to regulate the PDGF-PDGF receptor (PDGR) signaling pathway in tumor angiogenesis. In the present study, The DNA was obtained from the paraffin-embedded tissues of patients with advanced gastric cancer (AGC), amplified using PCR and subsequently sequenced to determine the polymorphisms within NRP-1, VEGFR2 [kinase insert domain receptor (KDR)] and PDGF. The effect of the functional polymorphism of the aforementioned genes on the overall survival (OS) and progression-free survival (PFS) of 81 patients with advanced gastric cancer was examined. Three single nucleotide polymorphisms (SNPs) of KDR were significantly associated with clinical outcomes. The rs1870377 TT genotype was positively associated with longer OS and PFS times compared with the AA+AT genotype (PFS, P=0.012; OS, P=0.038), the rs7692791 wild-type TT genotype was positively associated with longer PFS time and the rs2034965 AA+GA genotype was associated with shorter OS time (P=0.034). With regards to the SNPs of NRP-1, the rs2065364 AA genotype was significantly associated with improved OS and PFS times (PFS, P=0.023; OS, P=0.045). Following multivariate analysis using Cox proportional hazards regression models, patients with the KDR rs7692791 TT genotype experienced a longer PFS time compared with those with the CT genotype (P=0.016), and patients with the NRP-1 rs2065364 variant-type AA genotype still experienced a longer PFS time compared with those patients with the AG+GG genotypes (P=0.006). Regarding OS, the results demonstrated that the KDR rs2034965 AG+GG genotypes presented with a significant reduction in OS time (P=0.029), and that the KDR rs1870377 AT+AA genotypes had worse OS times compared with the wild-type TT genotype (P=0.021). In addition, increased mortality risk and AGC progression were significantly associated with the number of adverse alleles for combinations of NRP-1 rs2065364 and KDR rs1870377. In conclusion, the data from the present study demonstrated that the selected KDR and NRP-1 gene polymorphisms may be potential prognostic biomarkers in AGC.
Keywords: gastric cancer, polymorphism, NRP-1, KDR, PDGF
Introduction
Gastric cancer is one of the most common cancer types worldwide and remains the third leading cause of cancer-associated mortality, accounting for >783,000 deaths worldwide in 2018 (1). The diagnostic rate of early gastric cancer is only 10% in China, and most patients are at an advanced stage when clinically diagnosed, which confers a poor prognosis (2).
Systematic chemotherapy is the major treatment for patients with advanced gastric cancer (AGC). Platinum-fluoropyrimidine- and paclitaxel-fluoropyrimidine-based chemotherapy regimens are recommended as the first-line treatments in line with the Chinese Society of Clinical Oncology guidelines (3).
However, patients with the same tumor stage and receiving similar treatment can exhibit different clinical outcomes, and gastric cancer is a complex disease and its prognosis and progression are significantly affected by genetic and environmental factors (4). Identifying predictive genetic biomarkers could therefore contribute to the development of individualized therapy and follow-up strategies (5).
Neuropilin-1 (NRP-1) is a type I transmembrane glycoprotein distributed at the surface of cells that has been reported to affect neuronal axon guidance and embryonic angiogenesis (6), and to serve as a co-receptor regulating tumorigenesis in the vascular endothelial growth factor (VEGF)-VEGF receptor 2 (VEGFR2) [kinase insert domain receptor (KDR)] or platelet-derived growth factor (PDGF)-PDGF receptor (PDGFR) signaling pathways (7,8). VEGFRs are a type of tyrosine kinase receptor, and include VEGFR1 and VEGFR2 (KDR), which can be activated by binding with VEGF ligands (9). The VEGF-VEGFR2 signaling pathway is the leading pathway that activates the proliferation and migration of endothelial cells, therefore promoting angiogenesis and stimulating tumor growth and invasion (10,11). Furthermore, PDGF isoforms can transduce signals via binding to structurally similar α- and β-tyrosine kinase receptors, known as PDGFRα and PDGFRβ, respectively. The PDGF-PDGFR signaling pathway serves critical roles in regulating proliferation and survival of certain cell types (e.g. hematopoietic stem cell, vascular endothelial cell and vascular smooth muscle cell) during embryogenesis, and overexpression or mutation of the PDGF-PDGFR pathway can stimulate tumor cell proliferation (12,13). Previous studies have reported that polymorphisms within VEGF and KDR impacted their expression at the gene level (14,15). Thus, polymorphisms of these two signaling pathways may affect AGC prognosis by regulating the expression of the aforementioned genes and therefore affect the survival of patients with AGC. The present study investigated the association between polymorphisms of the NRP-1, KDR, PDGFβ, PDGFRβ and PDGFRα genes and the prognosis of patients with AGC.
Materials and methods
Study population
A total of 100 patients with AGC from the Second Affiliated Hospital of Dalian Medical University (Dalian, Liaoning, China) were recruited between January 2011 and June 2016. The inclusion criteria were as follows: i) Patients were histopathologically diagnosed with gastric adenocarcinoma; ii) patients had inoperable locally advanced, metastatic or recurrent gastric cancer (AGC); iii) patients had an Eastern Cooperative Oncology Group performance status (ECOG-PS) ≤2 (16); and iv) patients underwent at least 2 cycles of chemotherapy at the Second Affiliated Hospital when diagnosed with postoperative recurrence or inoperable advanced gastric cancer. The exclusion criteria were as follows: i) Patients who received chemotherapy, radiotherapy and/or biological treatment previously; ii) patients with an ECOG-PS >2; and iii) patients with multiple primary malignant neoplasms. The 100 patients were followed up by clinic visits and phone calls every 2 months, and clinical outcomes were recorded until October 2018. Genotype information was not available for 8 patients, 5 cases were lost to follow-up and 6 patients failed to receive the protocol treatment. Therefore, 81 patients were analyzed in the present study. Unresectable patients were staged according to imaging and gastroscopy when histopathologically diagnosed by biospy, and the postoperative recurrence patients were staged according to postoperative pathology. Tumors were staged using the 7th edition of the Tumor-Node-Metastasis (TNM) staging system of the International Union Against Cancer/American Joint Committee on Cancer (17). Chemotherapy was given prior to the present study, and regimens included platinum and fluoropyrimidine [cisplatin (D) 80 mg/m2 on day 1 and fluorouracil (F) 750 mg/m2 from day 1–4; D 80 mg/m2 on day 1 and capecitabine (X) 1,000 mg/m2 from day 1–14], and paclitaxel (P) and fluoropyrimidine [P, 150 mg/m2 on day 1 and F 750 mg/m2 from day 1–5; P 150 mg/m2 on day 1 and X 1,000 mg/m2 from day 1–14]. The regimens were repeated every 21 days. Chemotherapy was stopped in case of disease progression, patient refusal or grade 3–4 toxicity according to National Cancer Institute Common Terminology Criteria for Adverse Events version 4.03 (18).
SNP selection
The SNP loci of the target genes were selected from the public SNP database of the 1,000 Genome Project in the National Center for Biotechnology Information (NCBI) using minor allele frequency (MAF) >0.1 in the Chinese Han population and the Hardy-Weinberg equilibrium with a P-value of >0.1, then tagSNPs with a cut-off value of R2>0.8, and covering the gene and flanking 3 kb either side of the gene regions were chosen by the Genome Variation Server (https://gvs.gs.washington.edu). In total, 66 tagSNPs (27 from KDR gene, 32 from NRP-1 gene and 7 from PDGFβ) were selected, however due to financial constraints, 10 SNPs (rs7692791, rs6838752, rs2034965, rs1531290, rs13109660 from KDR, rs2070296, rs2804495, rs2065364 from NRP-1, and rs4821877, and rs9622978 from PDGFβ) were randomly selected from the tagSNPs. In addition, five disease-associated SNPs (rs1870377 and rs2305948 from KDR, rs6554162 and rs1800812 from PDGFRα, and rs2302273 from PDGFRβ) were selected according to their use in previous literature (19–24). Finally, the 15 SNPs (Table SI) of KDR rs7692791, rs2305948, rs6838752, rs2034965, rs1531290, rs13109660 and rs1870377, of NRP-1 rs2070296, rs2804495 and rs2065364, of PDGFβ rs4821877 and rs9622978, of PDGFRα rs6554162 and rs1800812, and of PDGFRβ rs2302273, were obtained from the SNP database of the NCBI (http://www.ncbi.nlm.nih.gov/SNP).
SNP genotyping
The tissues from patients with AGC were obtained via biopsy or surgery, fixed with 10% neutral buffer formalin for 24 h at room temperature, immersed in 60°C paraffin, embedded in a paraffin block and stored at 4°C. Genomic DNA was extracted from paraffin-embedded tissues of patients with AGC using the QIAamp DNA FFPE Tissue kit (Qiagen GmbH) according to the manufacturer's instructions. The polymerase chain reaction (PCR) primers for SNPs were designed using Sequenom Assay Design 3.1 software (Sequenom) and are listed in Table SII. A thermocycler (PTC-100PCR; MJ Research) and KAPA Taq HotStart DNA polymerase (Kapa Biosystems; Roche Diagnostics) were used for PCR amplification, the thermal cycling program employed was as follows: 94°C for 5 min, followed by 35 cycles of 30 sec at 94°C, then 30 sec of annealing at 60°C, 30 sec of extension at 72°C, and a final elongation step at 72°C for 10 min. The PCR products were sequenced using a 3730XL DNA Analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Statistical analysis
In the present study, the genetic model was divided into 3 types, namely, general, dominant and recessive models, as follows: Dominant model, MW+MM vs. WW; recessive model, WW+WM vs. MM; and general model, MM vs. WM vs. WW, where W indicates the wild-type allele and M the mutant allele). Before analysis, the Hardy-Weinberg equation for the equilibrium of allele distributions was tested by the χ2 test (Table I) and the SNPs with a P-value of <0.05 were excluded. The progression-free survival (PFS) and overall survival (OS) probabilities were estimated using the Kaplan-Meier method. The association between SNPs and PFS and OS were analyzed by log-rank tests and Cox regression analyses. Hazard ratios (HR) and 95% confidence intervals (CIs) were estimated for the uivariate and multivariate analyses usingy Cox regression analyses. Bonferroni's correction was applied for multiple comparisons (with the significance level set at P<0.025). Statistical analyses were conducted using SPSS v.21.0 (IBM Corp.). All tests were two-sided, and P<0.05 was considered to indicate a statistically significant difference.
Table I.
Hardy-Weinberg equilibrium test results of selected SNPs.
| Gene | SNP | χ2 | P-value |
|---|---|---|---|
| KDR | rs7692791 | 0.11 | 0.739 |
| rs2305948 | 0.02 | 0.902 | |
| rs6838752 | 0.65 | 0.418 | |
| rs2034965 | 1.66 | 0.197 | |
| rs13109660 | 0.03 | 0.860 | |
| rs1870377 | 0.58 | 0.455 | |
| rs1531290 | 3.37 | 0.067 | |
| NRP-1 | rs2070296 | 3.46 | 0.062 |
| rs2804495 | 0.08 | 0.775 | |
| rs2065364 | 1.00 | 0.317 | |
| PDGFβ | rs9622978 | 11.54 | 0.007a |
| rs4821877 | 0.00 | 0.998 | |
| PDGFRα | rs6554162 | 0.00 | 0.951 |
| rs1800812 | 30.3 | <0.001a | |
| PDGFRβ | rs2302273 | 7.04 | 0.007a |
P<0.05. KDR, kinase insert domain receptor; NRP-1, neuropilin-1; PDGF, platelet-derived growth factor; PDGFR, PDGF receptor; SNP, single nucleotide polymorphism.
Results
Patient clinical characteristics
A total of 81 patients were recruited in the present study, including 56 men (69.1%) and 25 women (30.9%). The age of the patients ranged from 30 to 83 years, and the mean age was 60.7±10.1 years. By October 2018, 79 patients were deceased, 2 had been lost during follow-up, and the median PFS and OS times were 5.5 and 11.0 months, respectively. The association between clinical pathological features and survival time are listed in Table II. The results demonstrated that TNM stage analyzed by Kaplan-Meier analysis was significantly associated with longer OS time (log-rank, P=0.047), and the platinum-based chemotherapy regimen was significantly associated with longer PFS time (log-rank, P=0.025). Associations between survival time and other clinical characteristics were not identified.
Table II.
Association between characteristics and prognosis of patients with advanced gastric cancer.
| Log-rank P-value | |||||
|---|---|---|---|---|---|
| Variables | n | mPFS (95% CI) | mOS (95% CI) | PFS | OS |
| Sex | 0.433 | 0.703 | |||
| Male | 56 | 5.0 (3.3–6.7) | 11 (9.6–12.4) | ||
| Female | 25 | 6.0 (3.6–8.4) | 12 (9.4–14.5) | ||
| Age, years | 0.773 | 0.898 | |||
| >60 | 33 | 5.5 (2.8–8.2) | 10.2 (6.0–14.4) | ||
| ≥60 | 48 | 6.0 (4.8–7.2) | 11.0 (10.0–12.0) | ||
| N stage | 0.590 | 0.081 | |||
| N1+N2 | 49 | 5.0 (3.6–6.4) | 11.6 (10.4–12.8) | ||
| N3 | 32 | 5.0 (2.8–7.2) | 10.2 (8.6–11.8) | ||
| TNM stage | 0.080 | 0.047a | |||
| I, II and III | 26 | 6.8 (5.9–7.7) | 12.0 (8.5–15.5) | ||
| IV | 55 | 4.5 (2.8–6.2) | 10.5 (8.5–12.5) | ||
| Tumor size, cm | 0.803 | 0.916 | |||
| >5 | 31 | 5.0 (2.9–7.1) | 11.0 (9.0–13.0) | ||
| ≥5 | 50 | 6.0 (4.4–7.6) | 11.0 (9.5–12.5) | ||
| Differentiation | 0.415 | 0.079 | |||
| Well to moderate | 27 | 6.0 (4.5–7.5) | 14.8 (8.0–21.6) | ||
| Poor | 54 | 4.5 (3.5–5.5) | 10.2 (8.2–12.2) | ||
| Platinum chemotherapy regimen | 0.025a | 0.359 | |||
| Platinum included | 38 | 6 (4.6–7.4) | 11.6 (9.3–13.9) | ||
| Non-platinum included | 43 | 4.5 (3.3–5.7) | 10.5 (9.0–12.0) | ||
| Paclitaxel chemotherapy regimen | 0.393 | 0.484 | |||
| Paclitaxel included | 39 | 4.4 (2.9–5.9) | 11.0 (7.1–14.9) | ||
| Non-paclitaxel included | 42 | 6 (4.6–7.4) | 11.0 (10.2–11.8) | ||
P<0.05. CI, confidence interval; OS, overall survival (months); mOS, median overall survival; PFS, progression-free survival (months); mPFS, median progression-free survival; TNM, Tumor-Node-Metastasis.
Associations between genotype and survival time
Associations between genotype and prognosis were estimated by Kaplan-Meier analysis, statistical significance was determined by the log-rank test and the genotype information are listed in Table SIII. The associations between the three types of genetic models (general, dominant and recessive) and survival time were analyzed (Table III). The results demonstrated that of all the selected SNPs, five SNPs (KDR rs7692791, KDR rs1870377, KDR rs2034965, NRP-1 rs2065364 and NRP-1 rs2804495) were significantly associated with PFS or OS; however, SNPs from PDGF and PDGFR genes were not associated with clinical outcomes.
Table III.
Effect of SNPs in selected genes on the prognosis in patients with advanced gastric cancer.
| Log-rank P-value for PFS | Log-rank P-value for OS | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene | SNP | Allelic change | General | Dominant | Recessive | General | Dominant | Recessive |
| KDR | rs7692791 | T/C | 0.032a | 0.009a | 0.281 | 0.227 | 0.093 | 0.364 |
| rs2305948 | C/T | 0.619 | 0.329 | 0.871 | 0.277 | 0.109 | 0.748 | |
| rs6838752 | T/C | 0.097 | 0.137 | 0.053 | 0.203 | 0.254 | 0.095 | |
| rs2034965 | G/A | 0.155 | 0.065 | 0.240 | 0.065 | 0.031 | 0.883 | |
| rs13109660 | G/A | 0.795 | 0.522 | 0.687 | 0.365 | 0.376 | 0.481 | |
| rs1870377 | T/A | 0.030a | 0.008a | 0.256 | 0.091 | 0.032a | 0.250 | |
| rs1531290 | A/G | 0.236 | 0.128 | 0.313 | 0.451 | 0.845 | 0.207 | |
| NRP-1 | rs2070296 | G/A | 0.498 | 0.417 | 0.486 | 0.993 | 0.964 | 0.909 |
| rs2804495 | G/T | 0.064 | 0.150 | 0.028a | 0.085 | 0.308 | 0.029 | |
| rs2065364 | G/A | 0.052 | 0.300 | 0.015a | 0.113 | 0.587 | 0.037a | |
| PDGFβ | rs4821877 | C/T | 0.712 | 0.490 | 0.862 | 0.949 | 0.933 | 0.747 |
| PDGFRα | rs6554162 | G/A | 0.513 | 0.322 | 0.751 | 0.501 | 0.413 | 0.561 |
P<0.05. KDR, kinase insert domain receptor; NRP-1, neuropilin-1; PDGF, platelet-derived growth factor; PDGFR, PDGF receptor; OS, overall survival; PFS, progression-free survival; SNP, single nucleotide polymorphism.
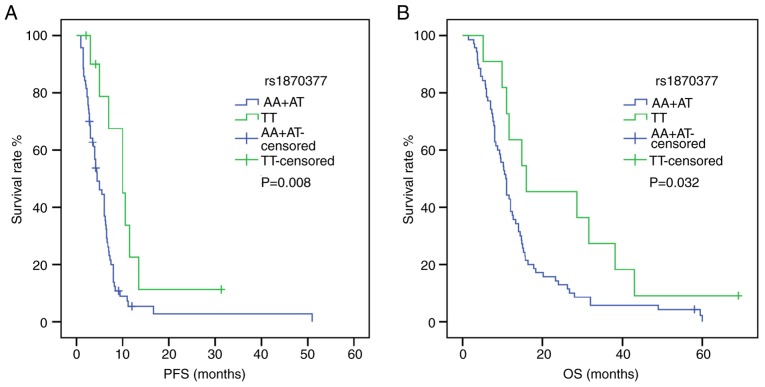
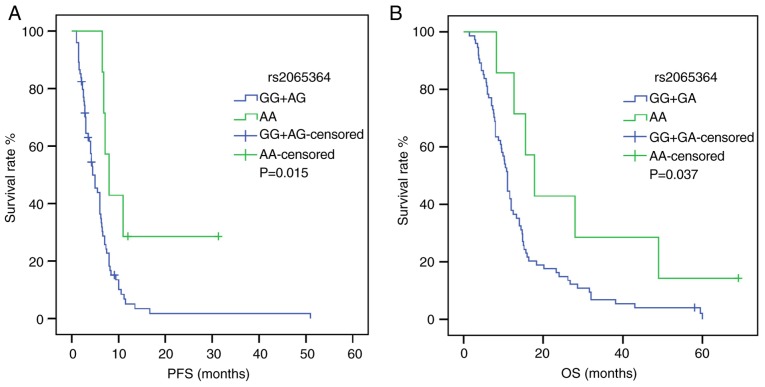
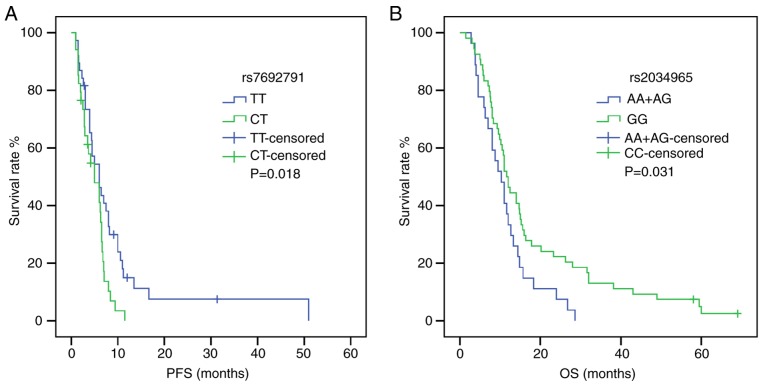
Following univariate analysis (Tables IV and V), the dominant model of KDR rs1870377 indicated that AA+AT carriers were associated with shorter PFS and OS times compared with TT carriers [PFS: HR, 2.618; 95% CI, 1.235–5.550; P=0.012; OS: HR, 2.041; 95% CI, 1.042–3.999; P=0.038) (Fig. 1). Furthermore, in a recessive model of NRP-1 rs2065364, AA genotype carriers exhibited more favorable PFS and OS times compared with the GG+AG genotypes (PFS: HR, 2.896; 95% CI, 1.159–7.237; P=0.023; OS: HR, 2.367; 95% CI, 1.019–5.496; P=0.045) (Fig. 2). However, KDR rs1870377 variant AA and AT genotype were significantly associated with poor PFS times compared with wild-type TT (AA vs. TT: HR, 3.221; 95% CI, 1.356–7.651; P=0.008; AT vs. TT: HR, 2.545, 95% CI, 1.159–5.589; P=0.020; Table IV) (Fig. S1). Furthermore, the KDR rs7692791 CT genotype was associated with lower PFS times compared with the wild-type TT genotype (HR, 1.829, 95% CI, 1.091–3.066, P=0.022; Table IV) (Fig. 3). In addition, in the dominant model of KDR rs2034965, AA+GA genotypes were significantly associated with reduced OS times (HR, 1.687; 95% CI, 1.039–2.738; P=0.034; Table IV) (Fig. 3). Statistical significance between SNPs and survival time in other polymorphisms was not found.
Table IV.
Associations of SNPs in selected genes and PFS in patients with advanced gastric cancer.
| Univariate analysis | Multivariate analysis | |||||||
|---|---|---|---|---|---|---|---|---|
| SNP | Outcome | mPFS, months | Model | Log-rank P-value | HR (95% CI) | P-value | HR (95% CI)a | P-valuea |
| KDR rs7692791 | PFS | General | 0.032 | 0.020 | 0.012 | |||
| 4.2 | CC | 0.099b | 1.926 (0.859–4.319) | 0.112 | 2.053 (0.855–4.929) | 0.107 | ||
| 5.0 | CT | 0.018b | 1.829 (1.091–3.066) | 0.022 | 1.969 (1.150–3.369) | 0.013 | ||
| 6.0 | TT | Reference | Reference | |||||
| Dominant | 0.009 | 0.010 | 0.006 | |||||
| 6.0 | TT | Reference | Reference | |||||
| 5.0 | CC+CT | 1.892 (1.156–3.098) | 0.011 | 1.982 (1.196–3.284) | 0.008 | |||
| KDR rs1870377 | PFS | General | 0.030 | 0.017 | 0.127 | |||
| 4.0 | AA | 0.005b | 3.221 (1.356–7.651) | 0.008 | 2.892 (0.987–8.474) | 0.053 | ||
| 5.5 | AT | 0.015b | 2.545 (1.159–5.589) | 0.020 | 1.778 (0.724–4.366) | 0.209 | ||
| 10.0 | TT | Reference | Reference | |||||
| Dominant | 0.008 | 0.009 | 0.051 | |||||
| 10.0 | TT | Reference | Reference | |||||
| 4.5 | AA+AT | 2.618 (1.235–5.550) | 0.012 | 1.970 (0.861–4.503) | 0.108 | |||
| NRP-1 rs2065364 | PFS | Recessive | 0.015 | 0.017 | 0.004 | |||
| 8.0 | AA | Reference | Reference | |||||
| 4.5 | AG+GG | 2.896 (1.159–7.237) | 0.023 | 3.905 (1.485–10.268) | 0.006 | |||
Adjusted for age, sex, N stage, TNM stage, platinum included or not and differentiation.
Bonferroni-adjusted P-value=0.05/2, so P<0.025 was considered statistically significant. CI, confidence interval; HR, hazard ratio; KDR, kinase insert domain receptor; NRP-1, neuropilin-1; PFS, progression-free survival; mPFS, median progression-free survival; SNP, single nucleotide polymorphism.
Table V.
Associations of SNPs in selected genes and OS in patients with advanced gastric cancer.
| Univariate analysis | Multivariate analysis | |||||||
|---|---|---|---|---|---|---|---|---|
| SNP | Outcome | mOS, months | Model | Log-rank P-value | HR (95% CI) | P-value | HR (95% CI)a | P-value |
| KDR rs2034965 | OS | Dominant | 0.031 | 0.032 | 0.029 | |||
| 11.6 | GG | Reference | Reference | |||||
| 10.3 | AA+GA | 1.687 (1.039–2.738) | 0.034 | 1.978 (1.193–3.280) | 0.008 | |||
| NRP1 rs2065364 | OS | Recessive | 0.037 | 0.039 | 0.105 | |||
| 17.8 | AA | Reference | Reference | |||||
| 11.0 | AG+GG | 2.367 (1.019–5.496) | 0.045 | 2.048 (0.847–4.952) | 0.112 | |||
| KDR rs1870377 | OS | Dominant | 0.032 | 0.034 | 0.035 | |||
| 16.0 | TT | Reference | Reference | |||||
| 10.8 | AA+AT | 2.041 (1.042–3.999) | 0.038 | 2.264 (1.130–4.536) | 0.021 | |||
| NRP1 rs2804495 | OS | Recessive | 0.029 | 0.031 | 0.084 | |||
| 8.8 | TT | Reference | Reference | |||||
| 12.0 | GT+GG | 1.710 (1.046–2.796) | 0.033 | 1.570 (0.924–2.667) | 0.095 | |||
Adjusted for age, gender, N stage, TNM stage and differentiation. CI, confidence interval; HR, hazard ratio; KDR, kinase insert domain receptor; NRP-1, neuropilin-1; OS, overall survival; mOS, median overall survival; SNP, single nucleotide polymorphism.
Figure 1.
Effect of kinase insert domain receptor rs1870377 on survival time in patients carrying the AA+AT and TT genotypes. (A) PFS curve. (B) OS curve. P-values were obtained by log-rank tests. OS, overall survival; PFS, progression-free survival.
Figure 2.
Effect of neuropilin-1 rs2065364 on survival time in patients carrying GG+AG and AA genotypes. (A) PFS curve. (B) OS curve. P-values were obtained by log-rank tests. OS, overall survival; PFS, progression-free survival.
Figure 3.
Effect of kinase insert domain receptor rs7692791 and rs2034965 on survival time. (A) PFS curve of rs7692791 in patients with TT and CT genotypes. (B) OS curve of rs2034965 in patients with AA+AG and GG genotypes. P-values were obtained by log-rank tests. OS, overall survival; PFS, progression-free survival.
For multivariate analysis (Tables IV and V), adjustments were performed for different variables in PFS and OS. Variables that were considered clinically relevant, such as age and TNM stage, or that presented an association with survival time following univariate analysis as listed in Table II were entered into a multivariate Cox proportional-hazards regression model. KDR rs7692791 remained significantly associated with PFS, and the TT genotype was associated with better prognosis compared with the CT genotype (HR, 1.969; 95% CI, 1.150–3.369; P=0.013). Furthermore, the association between NRP-1 rs2065364 AG+GG genotypes and shorter PFS remained significant following adjustment (HR, 3.905; 95% CI, 1.485–10.268; P=0.006). Furthermore, KDR rs2034965 AA+GA genotypes remained significantly associated with worse OS following adjustment (HR, 1.978; 95% CI, 1.193–3.280, P=0.008), and the KDR rs1870377 AA+AT genotypes were also associated with shorter OS compared with the wild-type TT genotype following adjustment (HR, 2.264; 95% CI, 1.130–4.536; P=0.021).
Effect of risk allele combinations on PFS and OS
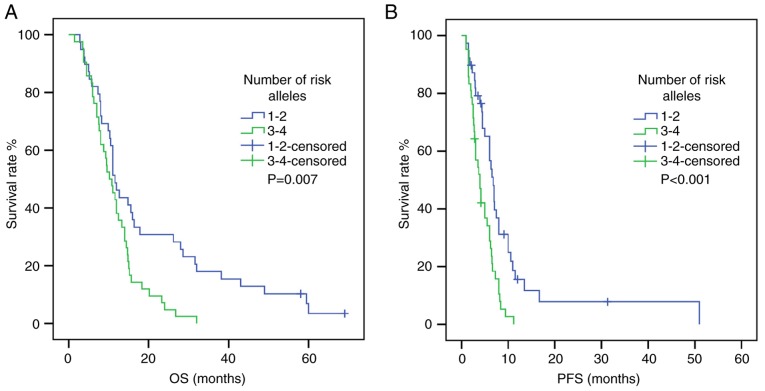
To study the combined effects of polymorphisms on survival time, risk alleles were selected according to the aforementioned results. The NRP-1 rs2065364G allele and the KDR rs1870377 A allele were found to be unfavorable for PFS and OS. Subsequently, the NRP-1 rs2065364/KDR rs1870377 combination was tested for its association with survival time and numbers of ‘risk alleles’ (Tables VI and VII). The results suggested that patients carrying >2 risk alleles were more likely to have shorter PFS and OS times compared with carriers with 1–2 risk alleles (PFS: HR, 0.427; 95% CI, 0.260–0.701; P=0.008; OS: HR, 0.523; 95% CI, 0.323–0.845; P=0.008; Tables VI and VII) (Fig. 4). Following adjustment, this association was also significant (PFS: HR, 0.427; 95% CI, 0.257–0.709; P=0.001; OS: HR, 0.511; 95% CI, 0.314–0.833; P=0.007).
Table VI.
Association between number of risk alleles and overall survival in patients with advanced gastric cancer.
| Univariate analysis | Multivariate analysisa | ||||
|---|---|---|---|---|---|
| Alleles combination | n | HR (95% CI) | P-value | HR (95% CI) | P-value |
| rs2065364/rs1870377 | |||||
| 1-2 risk alleles | 39 | 0.523 (0.323–0.845) | 0.008 | 0.511 (0.314–0.833) | 0.007 |
| 3-4 risk alleles | 42 | Reference | Reference | ||
Adjusted for age, sex, N stage, TNM stage, platinum included or not and differentiation. CI, confidence interval; HR, hazard ratio; OS, overall survival.
Table VII.
Association between number of risk alleles and progression-free survival in patients with advanced gastric cancer.
| Univariate analysis | Multivariate analysisa | ||||
|---|---|---|---|---|---|
| Alleles combination | n | HR (95% CI) | P-value | HR (95% CI) | P-value |
| rs2065364/rs1870377 | |||||
| 1-2 risk alleles | 39 | 0.427 (0.260–0.701) | 0.008 | 0.427 (0.257–0.709) | 0.001 |
| 3-4 risk alleles | 42 | Reference | Reference | ||
Adjusted for age, sex, N stage, TNM stage, platinum included or not and differentiation. CI, confidence interval; HR, hazard ratio; PFS, progression-free survival.
Figure 4.
Association between number of risk alleles (rs1870377/rs2065364 combination) and survival time. (A) PFS curve. (B) OS curve. P-values were obtained by log-rank tests. OS, overall survival; PFS, progression-free survival.
Discussion
The results from the present study demonstrated that polymorphisms of NRP-1 and KDR genes were associated with clinical outcome in patients with AGC. Following univariate analysis, KDR rs1870377 AA+AT genotypes were found to be associated with shorter PFS and OS times compared with the wild-type TT genotype, and the KDR rs1870377 variant AA and AT genotypes were significantly associated with poor PFS time compared with wild-type TT genotype. Furthermore, the NRP-1 rs2065364 homozygous mutant AA genotype was significantly associated with higher PFS and OS times compared with the GG+AG genotypes. The genotypes of KDR rs7692971 and KDR rs2034965 were also significantly associated with higher PFS and OS times, respectively. Following adjustment, the KDR rs7692791 TT genotype was associated with increased PFS time compared with the CT genotype, and the NRP-1 rs2065364 AG+GG genotypes were associated with shorter PFS times compared with the AA genotype. The KDR rs2034965 AA+GA genotypes were associated with worse OS times compared with the GG genotype. The KDR rs1870377 AA+AT genotypes were associated with shorter OS times compared with the TT genotype. Additionally, increasing number of risk alleles with the NRP-1 rs2065364/KDR rs1870377 combination was significantly associated with shorter OS and PFS times. These results demonstrated that NRP-1 rs2065364, KDR rs7692791, KDR rs2034965 and KDR rs1870377 may be considered as independent indicators of prognosis in patients with AGC.
NRP-1 was originally found to be crucial for neuronal axon guidance and embryonic angiogenesis, and was identified as a novel receptor involved in angiogenesis (6–8). Previous studies reported that the NRP-1 gene is associated with tumorigenesis and progression. One study reported that NRP-1 overexpression is associated with the promotion of gastric cancer migration, invasion and growth (25). Lin et al (26) demonstrated that NRP-1 is a novel TEA domain transcription factor target that serves a crucial role in hepatocellular carcinoma tumorigenesis. A previous demonstrated that NRP-1 is abnormally highly expressed in non-small cell lung tumor tissue, and is associated with patient prognosis (27). Another study reported that NRP-1 affects the chemosensitivity of cancer cells (28), Wey et al (28) demonstrated that NRP-1 overexpression in pancreatic cancer cell lines is associated with increased chemoresistance to gemcitabine in vitro. Yue et al (29) reported that NRP-1 overexpression increases osteosarcoma cell survival following exposure to doxorubicin. To the best of our knowledge, no study has demonstrated the association between NRP-1 SNPs and cancer. The present study confirmed that the NRP-1 rs2065364 AA genotype was associated with increased PFS time compared with the AG+GG genotypes. Further molecular investigation is required to reveal the underlying mechanisms involved.
KDR (VEGFR-2) is a tyrosine kinase receptor that can regulate signal transduction by binding to VEGF via its extracellular domain (9). VEGF/VEGFR2 is an important signaling pathway that can promote proliferation, survival and migration of vascular endothelial cells and increase vascular permeability (9,10). The cellular processes mediated by the VEGF-VEGFR2 signaling cascade can lead to angiogenesis and therefore regulate tumor growth and invasion, and therapeutic resistance (10,11). Previous studies reported that KDR gene polymorphisms are associated with clinical outcomes in various types of cancer, including colorectal cancer, glioma, hepatocellular carcinoma and gastric cancer. Torben et al (30) reported that VEGFR2 1192C>T and −604T>C polymorphisms were associated with increased microvessel density in colorectal cancer. A previous study of glioma in the Chinese population demonstrated that three SNPs of VEGFR2 (rs7667298, rs2305948 and rs1870377) are correlated with an increased risk of a glioma when homozygous (31). Another study described that the VEGFR-2 rs2305948 T polymorphism frequency is higher in patients with gastroenteropancreatic neuroendocrine neoplasms compared with that in the healthy population (19). In the present study, among the genetic variations of the VEGFR2 gene, the KDR rs1870377 and KDR rs7692791 TT genotypes were found to be associated with a better prognosis, and the KDR rs2034965 GG genotype was associated with increased OS time. Zhu et al (20) demonstrated that the VEGFR2 rs1870377 TT genotype confers a favorable prognosis in gastric cancer. Furthermore, Wang et al (21) investigated the correlation between polymorphisms of four genes from the epidermal growth factor receptor (EGFR) pathway and the clinical outcome of 363 patients with hepatocellular carcinoma, and reported that EGFR rs2034965 with the AA genotype is negatively correlated with disease-free survival. These results were consistent with the results from the present study; however, inconsistent results were reported in other types of cancer, and Kim et al (22) reported that the VEGFR2 rs1870377 TT genotype is associated with shorter OS time in patients with diffuse large B cell lymphoma. Furthermore, it was reported that rs7692791 C allele is significantly correlated with increased OS and DFS in hepatocellular carcinoma (21). These discordances may be partly attributed to the different types of cancer, the different clinical characteristics of the patients and the study sizes. The rs1870377 mutation is located in the coding region of KDR and is a missense mutation. The functional role of this gene polymorphism remains unclear.
Proteins from the PDGF family are crucial to stimulate the proliferation, survival and migration of mesenchymal cells (32), This family consists of 5 different isoforms, named disulphide-bonded homodimers of A-, B-, C- and D-polypeptide chains and the heterodimer PDGF-AB. PDGFR is classified as a receptor tyrosine kinase, and the 5 PDGF isoforms can activate cellular responses via PDGFRα and PDGFRβ (32,33). Overactivation of the PDGF-PDGFR signaling pathway has been reported to be associated with tumorigenesis (34). PDGFR gene mutations have been found in malignancies. Point mutations in PDGFRα were found in ~5% of gastrointestinal stroma tumors, which led to amino acid residue changes, therefore activating PDGFR kinase activity (35). In addition, a study reported that rs1800812 T allele and rs6554162 G allele in PDGFRα were related to decreased frequency in patients with papillary thyroid cancer compared with that in the healthy population (23). A previous study demonstrated that two SNPs in PDGFβ (rs5757573 T>C and rs6001516 C>T) were associated with an increased risk of pancreatic cancer (36). Furthermore, Volz et al (24) found that the SNP (rs2302273 C>T) in PDGFRβ gene was associated with a significantly longer PFS time in patients with metastatic colorectal cancer. However, in the present study, no relevance was found between SNPs and prognosis.
The present study had some limitations. Firstly, only 81 patients with AGC were eligible for statistical analysis. Since the sample size was relatively small, the results from this study should be considered as preliminary data and for generation of a hypothesis for subsequent investigation. Secondly, since the patients studied had AGC, it is not known whether the results could be applicable to patients with other types of gastric cancer. Further investigation should therefore be conducted to validate the results.
In conclusion, the results from the present study demonstrated that KDR rs7692791 and NRP-1 rs2065364 were positively associated with PFS. Furthermore, KDR rs2034965 and KDR rs1870377 significantly negatively correlated with OS time following multivariate analysis in patients with AGC. In addition, the numbers of ‘risk alleles’ of NRP-1 rs2065364/KDR rs1870377 combination were significantly associated with survival time. These results suggested that genetic variants in NRP-1 and KDR genes may affect the biological features and prognosis of patients with AGC. Due to limited funding, the underlying mechanisms were not explored, and further investigation is required to verify these results.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Dan Lv (Department of Medical Oncology, The Second Hospital Affiliated to Dalian Medical University) and Professor Na Gao (Department of Obstetrics and Gynecology, First Affiliated Hospital of Dalian Medical University) for their technical assistance.
Funding
No funding was recieved.
Availability of data and materials
All data and materials generated and/or used during the study are available from the corresponding author upon reasonable request.
Authors' contributions
TW and YS designed the study. YS collected the patients' clinical data. YJZ and YS performed the experiments. YJZ and YS analyzed the data and wrote the manuscript. TW contributed to the revision of the manuscript. All the authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Clinical Research Ethics Committee of The Second Hospital Affiliated to Dalian Medical University and was conducted in accordance with The Declaration of Helsinki. Participants were fully informed of the procedures and provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Li Z, Shan F, Miao R, Xue K, Li Z, Gao C, Chen N, Gao X, Li S, Ji J. Current status of diagnosis and treatment of early gastric cancer in China-Data from China gastrointestinal cancer surgery union. Zhonghua Wei Chang Wai Ke Za Zhi. 2018;21:168–174. (In Chinese) [PubMed] [Google Scholar]
- 3.Wang FH, Shen L, Li J, Zhou ZW, Liang H, Zhang XT, Tang L, Xin Y, Jin J, Zhang YJ, et al. The Chinese Society of Clinical Oncology (CSCO): Clinical Guidelines for the Diagnosis and Treatment of Gastric Cancer. Cancer Commun (Lond) 2019;39:10. doi: 10.1186/s40880-019-0349-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654–2664. doi: 10.1016/S0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 5.Chen K, Yang D, Li X, Sun B, Song F, Cao W, Brat DJ, Gao Z, Li H, Liang H, et al. Mutational landscape of gastric adenocarcinoma in Chinese: Implications for prognosis and therapy. Proc Natl Acad Sci USA. 2015;112:1107–1112. doi: 10.1073/pnas.1422640112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujisawa H, Takagi S, Hirata T. Growth-associated expression of a membrane protein, neuropilin, in Xenopus optic nerve fibers. Dev Neurosci. 1995;17:343–349. doi: 10.1159/000111304. [DOI] [PubMed] [Google Scholar]
- 7.Muhl L, Folestad EB, Gladh H, Wang Y, Moessinger C, Jakobsson L, Eriksson U. Neuropilin 1 binds PDGF-D and is a co-receptor in PDGF-D-PDGFRβ signaling. J Cell Sci. 2017;130:1365–1378. doi: 10.1242/jcs.200493. [DOI] [PubMed] [Google Scholar]
- 8.Raimondi C. Neuropilin-1 enforces extracellular matrix signalling via ABL1 to promote angiogenesis. Biochem Soc Trans. 2014;42:1429–1434. doi: 10.1042/BST20140141. [DOI] [PubMed] [Google Scholar]
- 9.Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 10.Karaman S, Leppänen VM, Alitalo K. Vascular endothelial growth factor signaling in development and disease. Development. 2018;145(pii):dev151019. doi: 10.1242/dev.151019. [DOI] [PubMed] [Google Scholar]
- 11.Shibuya M, Claesson-Welsh L. Signal transduction by VEGF receptors in regulation of angiogenesis and lymphangiogenesis. Exp Cell Res. 2006;312:549–560. doi: 10.1016/j.yexcr.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 12.Farooqi AA, Siddik ZH. Platelet-derived growth factor (PDGF) signalling in cancer: Rapidly emerging signalling landscape. Cell Biochem Funct. 2015;33:257–265. doi: 10.1002/cbf.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heldin CH, Lennartsson J, Westermark B. Involvement of platelet-derived growth factor ligands and receptors in tumorigenesis. J Intern Med. 2018;283:16–44. doi: 10.1111/joim.12690. [DOI] [PubMed] [Google Scholar]
- 14.Sa-Nguanraksa D, Kooptiwut S, Chuangsuwanich T, Pongpruttipan T, Malasit P, O-Charoenrat P. Vascular endothelial growth factor polymorphisms affect gene expression and tumor aggressiveness in patients with breast cancer. Mol Med Rep. 2014;9:1044–1048. doi: 10.3892/mmr.2014.1890. [DOI] [PubMed] [Google Scholar]
- 15.Glubb DM, Cerri E, Giese A, Zhang W, Mirza O, Thompson EE, Chen P, Das S, Jassem J, Rzyman W, et al. Novel functional germline variants in the VEGF receptor 2 gene and their effect on gene expression and microvessel density in lung cancer. Clin Cancer Res. 2011;17:5257–5267. doi: 10.1158/1078-0432.CCR-11-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oken M, Creech R, Tormey D, Horton J, Davis TE, McFadden ET, Carbone PP. Toxicity and response criteria of the Eastern Cooperative Oncology Group. Am J Clin Oncol. 1982;5:649–655. doi: 10.1097/00000421-198212000-00014. [DOI] [PubMed] [Google Scholar]
- 17.Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A., III . AJCC Cancer Staging Manual. 7th. Springer; New York, NY: 2010. [Google Scholar]
- 18.National Cancer Institute (NCI), corp-author Common Terminology Criteria for Adverse Events CTCAE) 2010 Version 4.03. [Google Scholar]
- 19.Berardi R, Torniai M, Partelli S, Rubini C, Pagliaretta S, Savini A, Polenta V, Santoni M, Giampieri R, Onorati S, et al. Impact of vascular endothelial growth factor (VEGF) and vascular endothelial growth factor receptor (VEGFR) single nucleotide polymorphisms on outcome in gastroenteropancreatic neuroendocrine neoplasms. PLoS One. 2018;13:e0197035. doi: 10.1371/journal.pone.0197035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu X, Wang Y, Xue W, Wang R, Wang L, Zhu ML, Zheng L. The VEGFR-2 protein and the VEGFR-2 rs1870377 A>T genetic polymorphism are prognostic factors for gastric cancer. Cancer Biol Ther. 2019;20:497–504. doi: 10.1080/15384047.2018.1537575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W, Ma XP, Shi Z, Zhang P, Ding DL, Huang HX, Saiyin HG, Chen TY, Lu PX, Wang NJ, et al. Epidermal growth factor receptor pathway polymorphisms and the prognosis of hepatocellular carcinoma. Am J Cancer Res. 2015;5:396–410. [PMC free article] [PubMed] [Google Scholar]
- 22.Kim MK, Suh C, Chi HS, Cho HS, Bae YK, Lee KH, Lee GW, Kim IS, Eom HS, Kong SY, et al. VEGFA and VEGFR2 genetic polymorphisms and survival in patients with diffuse large B cell lymphoma. Cancer Sci. 2012;103:497–503. doi: 10.1111/j.1349-7006.2011.02168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim MJ, Kim SK, Park HJ, Chung DH, Park HK, Lee JS, Kwon KH, Chung JH. PDGFRA promoter polymorphisms are associated with the risk of papillary thyroid cancer. Mol Med Rep. 2012;5:1267–1270. doi: 10.3892/mmr.2012.784. [DOI] [PubMed] [Google Scholar]
- 24.Volz NB, Stintzing S, Zhang W, Yang D, Ning Y, Wakatsuki T, El-Khoueiry RE, Li JE, Kardosh A, Loupakis F, et al. Genes involved in pericyte-driven tumor maturation predict treatment benefit of first-line FOLFIRI plus bevacizumab in patients with metastatic colorectal cancer. Pharmacogenomics J. 2015;15:69–76. doi: 10.1038/tpj.2014.40. [DOI] [PubMed] [Google Scholar]
- 25.Peng Y, Liu YM, Li LC, Wang LL, Wu XL. MicroRNA-338 inhibits growth, invasion and metastasis of gastric cancer by targeting NRP1 expression. PLoS One. 2014;9:e94422. doi: 10.1371/journal.pone.0094422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin J, Zhang Y, Wu J, Li L, Chen N, Ni P, Song L, Liu X. Neuropilin 1 (NRP1) is a novel tumor marker in hepatocellular carcinoma. Clin Chim Acta. 2018;485:158–165. doi: 10.1016/j.cca.2018.06.046. [DOI] [PubMed] [Google Scholar]
- 27.Ding M, Liu L, Hu C, Liu Y, Qiao Y, Jiang X. Expression of VEGFR2 and NRP-1 in non-small cell lung cancer and their clinical significance. Chin J Cancer Res. 2014;26:669–677. doi: 10.3978/j.issn.1000-9604.2014.12.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wey JS, Gray MJ, Fan F, Belcheva A, McCarty MF, Stoeltzing O, Somcio R, Liu W, Evans DB, Klagsbrun M, et al. Overexpression of neuropilin-1 promotes constitutive MAPK signalling and chemoresistance in pancreatic cancer cells. Br J Cancer. 2005;93:233–241. doi: 10.1038/sj.bjc.6602663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yue B, Ma JF, Yao G, Yang MD, Cheng H, Liu GY. Knockdown of neuropilin-1 suppresses invasion, angiogenesis and increases the chemosensitivity to doxorubicin in osteosarcoma cells-an in vitro study. Eur Rev Med Pharmacol Sci. 2014;18:1735–1741. [PubMed] [Google Scholar]
- 30.Hansen TF, Sørensen FB, Spindler KL, Olsen DA, Andersen RF, Lindebjerg J, Brandslund I, Jakobsen A. Microvessel density and the association with single nucleotide polymorphisms of the vascular endothelial growth factor receptor 2 in patients with colorectal cancer. Virchows Arch. 2010;456:251–260. doi: 10.1007/s00428-009-0878-8. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, Yang J, Chen Y, Mao Q, Li S, Xiong W, Lin Y, Chen J, Ge J. Genetic variants of VEGF (rs201963 and rs3025039) and KDR (rs7667298, rs2305948, and rs1870377) are associated with glioma risk in a Han Chinese population: A case-control study. Mol Neurobiol. 2016;53:2610–2618. doi: 10.1007/s12035-015-9240-0. [DOI] [PubMed] [Google Scholar]
- 32.Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22:1276–1312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heldin CH, Westermark B. Mechanism of action and in vivo role of platelet-derived growth factor. Physiol Rev. 1999;79:1283–1316. doi: 10.1152/physrev.1999.79.4.1283. [DOI] [PubMed] [Google Scholar]
- 34.Pietras K, Sjöblom T, Rubin K, Heldin CH, Ostman A. PDGF receptors as cancer drug targets. Cancer Cell. 2003;3:439–443. doi: 10.1016/S1535-6108(03)00089-8. [DOI] [PubMed] [Google Scholar]
- 35.Heinrich MC, Corless CL, Duensing A, McGreevey L, Chen CJ, Joseph N, Singer S, Griffith DJ, Haley A, Town A, et al. PDGFRA activating mutations, in gastrointestinal stromal tumors. Science. 2003;299:708–710. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- 36.Duan B, Hu J, Liu H, Wang Y, Li H, Liu S, Xie J, Owzar K, Abbruzzese J, Hurwitz H, et al. Genetic variants in the platelet-derived growth factor subunit B gene associated with pancreatic cancer risk. Int J Cancer. 2018;142:1322–1331. doi: 10.1002/ijc.31171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and materials generated and/or used during the study are available from the corresponding author upon reasonable request.