Abstract
The three-dimensional cell culture system is an increasingly important technique for discovering new biological aspects of cancer cells. In the present study it was demonstrated that bladder cancer cell lines, RT4 and 5637, spontaneously formed round multicellular spheroids (MCSs) in suspension by the aggregation method. MCSs consisted of cells differentially expressing luminal/basal markers. Western blotting showed that PPARγ and forkhead box A1 (FOXA1)of luminal markers were expressed to a lesser extent in MCSs than in parental cells grown in two-dimensional (2D) adherent culture. Cells in MCSs in suspension proliferated less efficiently, and were more resistant to cisplatin (CDDP) and gemcitabine than parental cells grown in 2D culture. Culturing cell lines as MCSs in suspension is a notable platform to decipher alternative biological aspects of bladder cancer cells, which could not be unraveled by the conventional 2D adherent culture.
Keywords: bladder cancer, cell line, multicellular spheroid, chemosensitivity, molecular subtype, cell culture
Introduction
Three-dimensional (3D) cell culture has emerged in the field of cancer research as a tool for capturing critical biological properties of cancer, especially for solid tumors. Multicellular spheroids (MCSs) developed in 3D culture are believed to more closely mimic solid tumors with respect to cell-cell interactions, hypoxia, drug penetration, and nutrition gradients, which are irreproducible in conventional two-dimensional (2D) cell culture (1,2). MCSs generated from cancer cell lines alter their molecular phenotypes in various types of cancer, including ovarian, colon, prostate, head and neck, endometrial, and lung (3–11). Along with the molecular phenotypic changes, functional properties in 3D culture, e.g. cell proliferation and sensitivity to drugs, also change compared to their parental cells grown in 2D adherent culture (3–7,12). Evidence for MCSs derived from bladder cancer cell lines is limited. Our group found that MCSs generated from bladder cancer cell lines changed protein levels of delta N p63 alpha, E-cadherin, and N-cadherin, implicating epithelial-mesenchymal transition between MCSs in suspension and parental cells grown in 2D adherent culture (13). Intriguingly, CHIR99021, a GSK3 inhibitor and Wnt pathway activator, promoted cell proliferation only in 3D culture but not in 2D culture (14). These data further corroborate the importance of 3D cell culture to discover new biological aspects of cancer cells.
Methodologies preparing MCSs from cell lines vary among researches, including the U-bottom ultra-low adherence plates method (6,7,9), the hanging drop method (8), the microgravity system using a rotating bioreactor (10), culture within extracellular matrix (4,5), and nano-imprinted plates based on scaffold-based technologies (12). The difference among these methodologies can cause biological differences among MCSs prepared by each method. Particularly, the presence of extracellular matrix impacts structural, molecular, and functional characteristics of MCSs (13,15,16). We have adopted a modified aggregation-based method using plates coated in-house with poly-2-hydroxyethyl methacrylate (poly-HEMA), which prevents cells from attaching to plates, enhances cell aggregation, and consequently yields MCSs in suspension (3,14). This method enables rapid, easy, and cost-effective preparation of MCSs from conventional cell lines.
In this study, we described the process of spontaneous formation of MCSs in suspension from human bladder cancer cell lines by the aggregation-based method. Histological evaluation for these MCSs was carried out using immunohistochemistry with antibodies against luminal and basal markers for urothelium. We also compared molecular characteristics of bladder cancer cells between MCSs in suspension and parental cells grown in conventional 2D culture. Cellular proliferation and chemosensitivity to cisplatin (CDDP) and gemcitabine were also assessed in 3D and 2D cultures for comparison.
Materials and methods
Cell culture
RT4 and 5637 were selected to examine as representative luminal and basal human bladder cancer cell lines, respectively (17). RT4 and 5637 were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) in November 2015, and cultured as previously described (14). Briefly, cells were cultured at 37°C under 5% CO2 in RPMI 1640 containing 10% fetal bovine serum. MCSs from cell lines were prepared by the aggregation-based method. 2×105 or 1,000 cells (respectively) were seeded in 6- or 96-well U-bottom plates coated with poly-HEMA (Sigma, St. Louis, MO, USA). MCSs were formed 24 h after seeding and used in further experiments. Images of cultured cells were taken by EVOS Cell Imaging Systems (Thermo Fischer Scientific, Waltham, MA, USA).
Histological evaluation
Histological evaluation was performed as previously described (15). MCSs were embedded in iPGell (Diagnocine, Hackensack, NJ, USA) and fixed in 10% buffered formalin. Sections of 4 µm were stained with hematoxylin-eosin for histological evaluation. Immunohistochemistry was performed using primary antibodies; Ki 67 (ab16667, Abcam, Cambridge, MA, USA), cleaved caspase 3 (9661, Cell Signaling Technology, Danvers, MA, USA), cytokeratin 14 (CK14) (ab181595, Abcam), cytokeratin 20 (CK20) (ab76126, Abcam), cytokeratin 5 (CK5) (PRB-160P, Covance, Princeton, NJ, USA), peroxisome proliferator-activated receptor gamma (PPARγ) (2435, Cell Signaling Technology), forkhead box A1 (FOXA1) (ab170933, Abcam), and p63 (CM163A, Biocare Medical, Pacheco, CA, USA).
Western blotting
Western blotting was performed as previously described (14). Primary antibodies used included; β-actin (A2228, Sigma), FOXA1 (ab170933, Abcam), CK20 (ab76126, Abcam), CK5 (PRB-160P, Covance), CK14 (ab181595, Abcam), and PPARγ (2435, Cell Signaling Technology).
Immunocytochemistry
Immunocytochemistry was performed as previously described (15). Briefly, Cells were seeded on Lab-Tek II Chamber Slide (Thermo Fischer Scientific) 24 h before fixation. Cells were fixed with 4% paraformaldehyde/PBS for 15 min. After permeabilization and blocking with Dako Protein Block Seum-Free (Dako) containing 1% Triton X-100 for 15 min, cells were incubated at 4°C with anti-CK14 (Abcam) or CK20 antibody (Abcam) overnight followed by incubation at room temperature with Alexa-488 conjugated secondary antibody (Thermo Fischer Sicentific) for 1 h. After counterstaining with Hoechst 33342 (Thermo Fischer Sicentific), images of fluorescent cells were obtained using EVOS Cell Imaging Systems (Thermo Fischer Sicentific).
Quantitative real-time polymerase chain reaction (qRT-PCR)
qRT-PCR analysis was performed as previously described (14). Briefly, total RNA was extracted from cells using the RNeasy system (Qiagen, Hilden, Germany) after RT4 and 5637 cells were grown in 2D adherent culture and in 3D suspension culture. qRT-PCR was performed using the StepOnePlus system (Applied Biosystems, Foster City, CA, USA). TaqMan gene expression assays for KRT20 (CK20), KRT18 (CK18), KRT14 (CK14), KRT5 (CK5), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were used. All primers were purchased from Thermo Fischer Scientific. GAPDH was used for normalization.
Viability assay and growth assay
Viability assay and growth assay were performed as previously described (14). Briefly, for viability assay in 2D culture, 3,000 and 1,500 cells of RT4 and 5637 were seeded in a 96-well tissue-treated plate in triplicate, respectively. For viability assay in 3D culture, MCSs were prepared in 96-well U-bottom plates as described above. Twenty-four hours after seeding, cells were treated with PBS or CDDP (Sigma) or gemcitabine (Sigma) for 72 h. To measure cell viability, CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI, USA) and FLUOstar OPTIMA (BMG Labtech, Ortenberg, Germany) were used according to manufacture's protocols. Relative cell proliferation was calculated by dividing the viability of the indicated day by that of day 1. Following this, dose-response curves of CDDP and gemcitabine were depicted and IC50 was calculated using GraphPad Prism 7 (GraphPad Software, San Diego, CA, USA) with non-linear (curve fit) regression algorithms. For growth assay in 3D cultures, Images of MCSs were collected at days 1 and 6 by the EVOS Cell Imaging Systems, and the areas occupied by the MCSs were measured using Image J software (National Institutes of Health, Bethesda, MD, USA). Relative growth rates were calculated by dividing the area of MCSs at day 6 by that at day 1.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 7 (GraphPad Software). Data are presented as mean ± standard deviation (SD). Two-group analysis was performed by the unpaired t-test for the results of RT-qPCR, and results were considered statistically significant at P≤0.05.
Results
Bladder cancer cell lines RT4 and 5637 spontaneously form MCSs in suspension by the aggregation-based method
Twenty-four hours after RT4 and 5637 cells were seeded in a 6-well plate coated with poly-HEMA, multiple MSCs were found floating (Fig. 1A). 5637-derived MCSs were apparently darker than RT4-derived MCSs. Time-lapse images showed that both cells aggregated with each other over time, eventually forming a round-shaped MCS in suspension (Fig. 1B). Hematoxylin-eosin staining of MCSs demonstrated that MCSs derived from RT4 and 5637 were packed with cells (Fig. 1C). Cells in RT4-derived MCSs had larger cytoplasm and lower nuclear/cytoplasmic ratio than cells in 5637-derived MCSs (Fig 1C). These data demonstrated rapid generation of MCSs in suspension from bladder cancer cell lines RT4 and 5637 by the aggregation-based method.
Figure 1.
Preparation of MCSs from bladder cancer cell lines RT4 and 5637. (A) Bright field images show the morphology of RT4 and 5637 cells grown in 2D adherent culture and as MCSs in 3D suspension culture. Scale bar, 400 µm. (B) Time-course images of RT4 and 5637 cultured in a poly-HEMA coated U-bottom culture well. (C) H&E staining of RT4- and 5637-derived MCSs. Scale bar, 100 µm. MCSs, multicellular spheroids; H&E, hematoxylin and eosin staining.
RT4- and 5637-derived MCSs consist of cells expressing different levels of luminal/basal differentiation markers
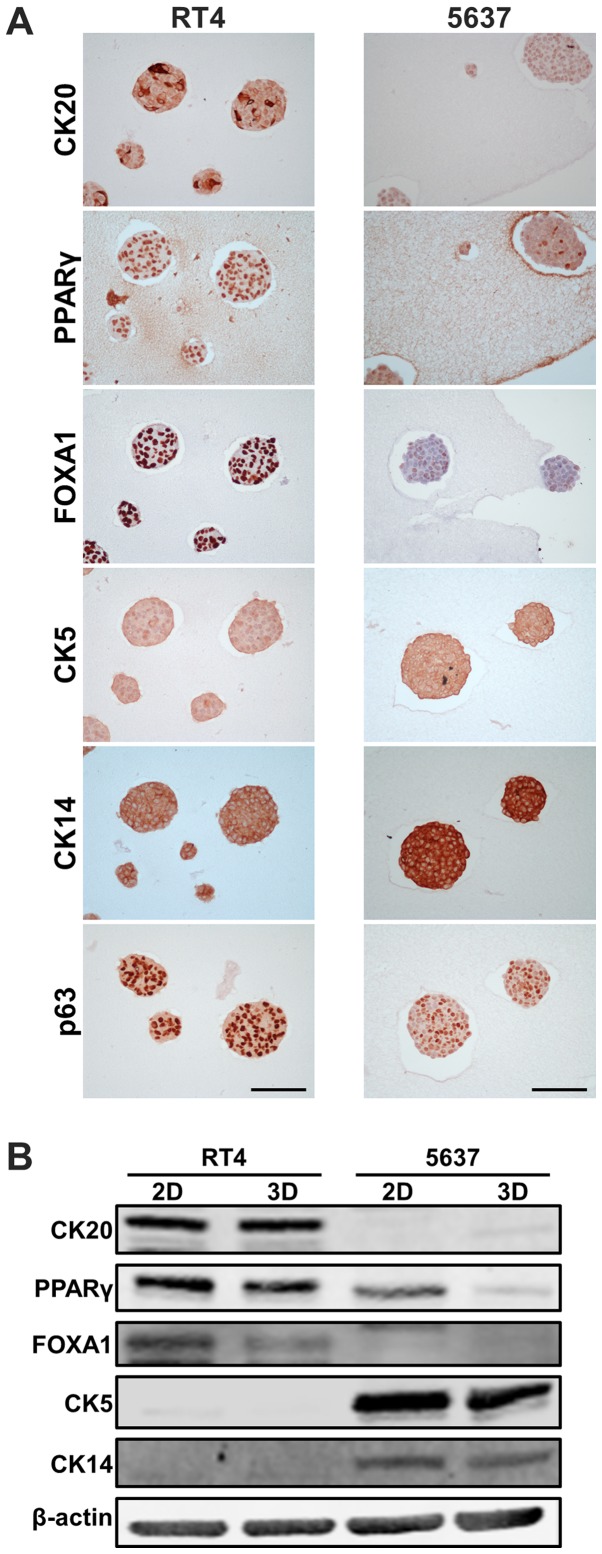
Molecular subtype membership has been established in bladder cancer, exhibiting a broad spectrum of bladder cancer biology. Subtypes are clustered into luminal and basal at the highest level (18). We sought to characterize RT4- and 5637-derived MCSs with the expression levels of luminal/basal markers. Immunohistochemistry showed that luminal markers, including CK20, PPARγ, and FOXA1, were expressed more in RT4-derived MCSs than in 5637-derived MCSs, while basal markers of CK5 and CK14 were expressed more in 5637-derived MCSs than in RT4-derived MCSs (Fig. 2A). These data were consistent with the findings that RT4 and 5637 were luminal and basal subtypes according to transcriptome analysis in conventional 2D adherent culture (17). Heterogeneous expression of CK20 in RT4-derived MCSs, and that of PPARγ and p63 in 5637-derived MCSs were observed within a single MCS (Fig. 2A), demonstrating that one MCS consisted of cells heterogeneously expressing luminal/basal markers. Heterogeneous expression of CK20 was also observed among RT4 cells grown in 2D adherent culture (Fig. S1).
Figure 2.
Luminal/basal markers in RT4- and 5637-derived MCSs. (A) Immunohistochemical staining of RT4- and 5637-deried MCSs using antibodies against luminal markers (CK20, PPARγ and FOXA1) and basal markers (CK5, CK14 and p63). Scale bar, 100 µm. (B) Western blotting shows protein levels of luminal and basal markers in RT4 and 5637 cells cultured in 2D adherent culture and as MCSs in 3D suspension culture. β-actin was used as a loading control. MCSs, multicellular spheroids; MCSs, multicellular spheroids; CK, cytokeratin; PPARγ, peroxisome proliferator-activated receptor γ; FOXA1, forkhead box A1.
Next, expression levels of luminal/basal markers were compared between MCSs in 3D suspension culture and parental cells grown in 2D adherent culture. Western blotting showed that luminal markers, including CK20, PPARγ, and FOXA1, were expressed more in RT4-derived MCSs than in 5637-derived MCSs, while basal markers of CK5 and CK14 were expressed more in 5637-derived MCSs than in RT4-derived MCSs (Fig. 2B), which was in line with the immunohistochemical findings. PPARγ and FOXA1 were expressed less in MCSs than in parental cells grown in 2D culture, while other markers examined were equally expressed in cells in both culture conditions (Fig. 2B). Quantitative real-time polymerase chain reaction analysis demonstrated significant difference in mRNA expression levels of KRT20, KRT18, and KRT14 between MCSs in 3D suspension culture and parental cells grown in 2D adherent culture (Fig. S2).
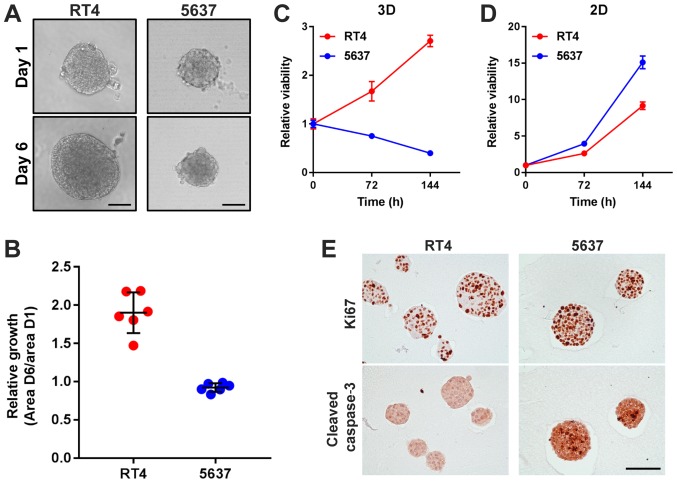
RT4-derived MCSs grow more rapidly than 5637-derived MCSs, but RT4 proliferates less than 5637 in 2D adherent culture. Next, cellular ability of proliferation was examined both in 3D suspension culture and in 2D adherent culture. RT4-derived MCSs apparently grew over time under microscopic evaluation, while 5637-derived MCSs slightly shrunk over 120 h of culture (Fig. 3A and B). Consistent with the microscopic findings, cellular viability of RT4-derived MCSs increased and that of 5637-derived MCSs decreased with time (Fig 3C). In 2D adherent culture, cellular viability of both RT4 and 5637 increased with time (Fig. 3D). Of interest, cell viability of 5637 decreased with time when cultured as MCSs in suspension but increased more rapidly than that of RT4 in 2D culture (Fig. 3C and D). Immunohistochemistry revealed that Ki67 staining was expressed in almost all of the cells in both RT4- and 5637-derived MCSs, while cleaved caspase 3 was profoundly found in 5637-derived MCSs but not in RT4-derived MCSs (Fig. 3E). These data suggested that 5637 cells were more susceptible to apoptosis than RT4 cells in MCSs in suspension, resulting in more efficient growth of RT4-derived MCSs than 5637-derived MCSs.
Figure 3.
RT4-derived MCSs grow more rapidly than 5637-derived MCSs, but RT4 proliferates less than 5637 in 2D adherent culture. (A) Representative images of RT4- and 5637-derived MCSs at day 1 and day 6. Scale bar, 100 µm. (B) Relative growth of RT4- and 5637-derived MCSs was calculated by dividing area at day 6 by that at day 1, and depicted in the plot. Each dot represents relative growth of one MCS. Relative proliferation of RT4 and 5637 cells cultured (C) in 3D suspension culture and (D) in 2D adherent culture. Relative proliferation at 72 and 144 h were measured by dividing the amount of ATP at 72 and 144 h by that at day 1, and depicted in the plot. Values represent the mean ± SD from 3 independent experiments. (E) Immunohistochemical staining of RT4- and 5637-derived MCSs using antibodies against Ki67 and cleaved caspase 3. Scale bar, 100 µm. MCSs, multicellular spheroids.
Cells in RT4- and 5637-derived MCSs are more resistant to CDDP and gemcitabine than parental cells grown in 2D adherent culture
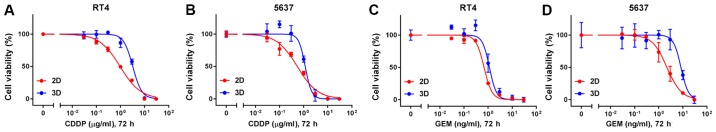
Lastly, chemosensitivity was compared between MCSs in 3D suspension culture and cells grown in 2D adherent culture. CDDP and gemcitabine, which are the main chemotherapeutic agents of the current standard regimen to treat advanced urothelial cancer, were selected to test. IC50 of CDDP of both RT4- and 5637-derived MCSs were higher than those of parental cells grown in 2D adherent culture (Fig. 4A and B and Table I). Similarly, IC50 of gemcitabine of both RT4- and 5637-derived MCSs were higher than those of parental cells in 2D adherent culture (Fig. 4C and D and Table I). RT4 cells were more resistant to CDDP than 5637 cells both in 2D and in 3D cultures (Fig. 4A and B and Table I). Likewise, RT4 cells were more sensitive to gemcitabine than 5637 cells both in 2D and in 3D cultures (Fig. 4C and D and Table I). The difference in sensitivity to CDDP and gemcitabine between RT4 and 5637 cells were more conspicuous in 3D culture than 2D culture (Table I).
Figure 4.
RT4- and 5637-derived MCSs are more resistant to CDDP and gemcitabine than parental cells grown in 2D adherent culture. Dose-response curves of (A and B) CDDP and (C and D) gemcitabine on proliferation of RT4 and 5637 cells grown as MCSs in 3D suspension culture and in 2D adherent culture. Cells were treated with the indicated dose of CDDP or gemcitabine for 72 h. After treatment, relative viability was calculated and dose-response curves were depicted. Values represent the mean ± SD from 3 independent experiments. CDDP, cisplatin; GEM, gemcitabine; MCSs, multicellular spheroids.
Table I.
IC50 of CDDP and gemcitabine on proliferation of RT4 and 5637 cells grown as MCSs in 3D suspension culture and in 2D adherent culture.
| CDDP, µg/ml | Gemcitabine, ng/ml | ||||
|---|---|---|---|---|---|
| Cell line | Culture type | IC50 | 95% CI | IC50 | 95% CI |
| RT4 | 2D | 0.9601 | 0.8388–1.098 | 0.6785 | 0.6159–0.7458 |
| 3D | 3.245 | 2.984–3.521 | 1.134 | 0.9862–1.362 | |
| 5637 | 2D | 0.5424 | 0.4442–0.6599 | 2.2 | 1.852–2.615 |
| 3D | 1.158 | NA-1.317 | 8.309 | 6.662–9.9 | |
CI, confidence interval; MCSs, multicellular spheroidsl; CDDP, cisplatin.
Discussion
In the present study, we demonstrated that human bladder cancer cell lines RT4 and 5637 spontaneously aggregated with each other over time and formed round MCSs by being prevented from attaching to culture plates and kept in suspension. MCSs were composed of cells differentially expressing luminal/basal markers. PPARγ and FOXA1 of luminal markers were less expressed in MCSs than in parental cells grown in 2D adherent culture. Cells in MCSs in suspension proliferated less efficiently, and were more resistant to CDDP and gemcitabine than parental cells grown in 2D culture.
Morphology of MCSs varies among cell lines, from a round sphere with a smooth surface to a loose aggregate with irregular structure (3–7,9,12). Formation of the round contour of MCSs was apparently associated with the presence of E-cadherin, which is an adhesion molecule tightening cell-cell contact, in cell lines of ovarian, colon, and head and neck cancer (3,6,9). Cell lines lacking E-cadherin expression tended to form loose aggregates in 3D culture (3,6,9). As for bladder cancer, RT4 and 5637 have E-cadherin expression (13,17), and were able to form round MCSs (Fig. 1). Functional blocking of E-cadherin disrupted round MCSs into loose aggregates of 5637 cells in 3D suspension culture (data not shown). Moreover, J82, which is one of the mesenchymal cell lines of bladder cancer and lacks a detectable level of E-cadherin (17), also formed loose aggregates in 3D suspension culture (13). These data suggest that adhesion molecules like E-cadherin would be indispensable to form a round sphere with a smooth surface in 3D suspension culture.
Cells cultured as MCSs are generally less proliferative than parental cells grown in 2D adherent culture irrespectively of methodologies to produce MCSs (3,5,6,9), which was shown to be true of RT4 and 5637 cells (Fig. 3C and D). Less proliferative ability of MCSs in suspension could be attributable to the absence of cellular contact with the extracellular matrix, as the cellular adhesion to the extracellular matrix like basement membrane activates intracellular signaling of proliferation (19). Another reason for the less proliferation of MCSs could be explained by anoikis, a particular form of apoptosis in epithelial cells induced by the lack of cellular contact with the extracellular matrix (20). Interestingly, 5637 cells could proliferate more rapidly than RT4 cells in 2D adherent culture but were less proliferative as MCSs in 3D suspension culture (Fig. 3C and D). Cleaved caspase 3, an apoptotic marker, were found only in 5637-derived MCSs but not in RT4-derived MCSs (Fig. 3E). Therefore, 5637 cells might be more vulnerable to anoikis than RT4 cells, leading to the dynamic difference of cellular proliferation between culture conditions.
IC50 of CDDP and gemcitabine of RT4 and 5637 as MCSs were consistently higher than those of parental cells in 2D adherent culture (Fig. 4 and Table 1). These results were expected from the abovementioned less proliferative characteristics of MCSs because cytotoxic agents typically target rapidly dividing cells. Intriguingly, in vitro screening experiments identified cytotoxic drugs especially to slow-proliferating cancer cells, which are supposedly responsible for resistance to typical chemotherapeutic agents and late relapse (21). These drugs targeting slow-proliferating cells might be more effective to RT4 and 5637 cells in 3D culture than in 2D culture, which would indicate the impact of culture conditions on drug sensitivity of cancer cells.
We showed that PPARγ and FOXA1 of luminal markers were expressed less in RT4- and 5637-derived MCSs than in parental cells grown in 2D culture, while delta N p63 alpha, a transcriptional factor positively controlling basal cell gene signature in bladder cancer cells (22), was previously found reversibly upregulated in MCSs in suspension (14). These findings lead to the hypothesis that cellular plasticity of bladder cancer cells within the luminal/basal spectrum could occur depending on culture conditions (23). Of note, divergent luminal/basal subtypes were found within the same tumor of human bladder cancer despite sharing identical genomic alterations, implicating subtype-specific plasticity of bladder cancer cells (24). We are currently planning to perform transcriptome analysis based on our hypothesis to comprehensively characterize their phenotypic alterations between culture conditions.
In summary, we described the process of developing MCSs from two bladder cancer cell lines, RT4 and 5637, using the aggregation-based method. Molecular and functional characteristics of RT4 and 5637 cells were found remarkably different between MCSs in 3D suspension culture and parental cells grown in conventional 2D adherent culture. Culturing cell lines as MCSs in suspension is a notable platform to decipher alternative aspects of biology of bladder cancer cells, which could not be unraveled by the conventional 2D adherent culture.
Supplementary Material
Acknowledgements
Not applicable.
Funding
This work was supported by the grant from the Greenberg Bladder Cancer Institute.
Availability of data and materials
The datasets used analyzed in the present study are available from the corresponding author on reasonable request.
Authors' contributions
TY and TJB conceived and designed the study. TY and XL performed the experiments. TY performed the statistical analysis. NAS, MK, GJ and DJM contributed to the analysis and interpretation of the data. TY and TJB wrote the paper. NAS, MK, GJ and DJM provided discussion and critically revised the paper for important intellectual content and gave final approval of the paper to be published. DJM and TJB supervised the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Sharma SV, Haber DA, Settleman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer. 2010;10:241–253. doi: 10.1038/nrc2820. [DOI] [PubMed] [Google Scholar]
- 2.Yamada KM, Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell. 2007;130:601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Lee JM, Mhawech-Fauceglia P, Lee N, Parsanian LC, Lin YG, Gayther SA, Lawrenson K. A three-dimensional microenvironment alters protein expression and chemosensitivity of epithelial ovarian cancer cells in vitro. Lab Invest. 2013;93:528–542. doi: 10.1038/labinvest.2013.41. [DOI] [PubMed] [Google Scholar]
- 4.Härmä V, Virtanen J, Mäkelä R, Happonen A, Mpindi JP, Knuuttila M, Kohonen P, Lötjönen J, Kallioniemi O, Nees M. A comprehensive panel of three-dimensional models for studies of prostate cancer growth, invasion and drug responses. PLoS One. 2010;5:e10431. doi: 10.1371/journal.pone.0010431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luca AC, Mersch S, Deenen R, Schmidt S, Messner I, Schäfer KL, Baldus SE, Huckenbeck W, Piekorz RP, Knoefel WT, et al. Impact of the 3D microenvironment on phenotype, gene expression, and EGFR inhibition of colorectal cancer cell lines. PLoS One. 2013;8:e59689. doi: 10.1371/journal.pone.0059689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riedl A, Schlederer M, Pudelko K, Stadler M, Walter S, Unterleuthner D, Unger C, Kramer N, Hengstschläger M, Kenner L, et al. Comparison of cancer cells in 2D vs 3D culture reveals differences in AKT-mTOR-S6K signaling and drug responses. J Cell Sci. 2017;130:203–218. doi: 10.1242/jcs.188102. [DOI] [PubMed] [Google Scholar]
- 7.Ekert JE, Johnson K, Strake B, Pardinas J, Jarantow S, Perkinson R, Colter DC. Three-dimensional lung tumor microenvironment modulates therapeutic compound responsiveness in vitro-implication for drug development. PLoS One. 2014;9:e92248. doi: 10.1371/journal.pone.0092248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paullin T, Powell C, Menzie C, Hill R, Cheng F, Martyniuk CJ, Westerheide SD. Spheroid growth in ovarian cancer alters transcriptome responses for stress pathways and epigenetic responses. PLoS One. 2017;12:e0182930. doi: 10.1371/journal.pone.0182930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt M, Scholz CJ, Polednik C, Roller J. Spheroid-based 3-dimensional culture models: Gene expression and functionality in head and neck cancer. Oncol Rep. 2016;35:2431–2440. doi: 10.3892/or.2016.4581. [DOI] [PubMed] [Google Scholar]
- 10.Grun B, Benjamin E, Sinclair J, Timms JF, Jacobs IJ, Gayther SA, Dafou D. Three-dimensional in vitro cell biology models of ovarian and endometrial cancer. Cell Prolif. 2009;42:219–228. doi: 10.1111/j.1365-2184.2008.00579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadletz L, Heiduschka G, Domayer J, Schmid R, Enzenhofer E, Thurnher D. Evaluation of spheroid head and neck squamous cell carcinoma cell models in comparison to monolayer cultures. Oncol Lett. 2015;10:1281–1286. doi: 10.3892/ol.2015.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imamura Y, Mukohara T, Shimono Y, Funakoshi Y, Chayahara N, Toyoda M, Kiyota N, Takao S, Kono S, Nakatsura T, Minami H. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol Rep. 2015;33:1837–1843. doi: 10.3892/or.2015.3767. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida T, Okuyama H, Nakayama M, Endo H, Tomita Y, Nonomura N, Nishimura K, Inoue M. Dynamic change in p63 protein expression during implantation of urothelial cancer clusters. Neoplasia. 2015;17:574–585. doi: 10.1016/j.neo.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshida T, Sopko NA, Kates M, Liu X, Joice G, McConkey DJ, Bivalacqua TJ. Three-dimensional organoid culture reveals involvement of Wnt/β-catenin pathway in proliferation of bladder cancer cells. Oncotarget. 2018;9:11060–11070. doi: 10.18632/oncotarget.24308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida T, Kates M, Sopko NA, Liu X, Singh AK, Bishai WR, Joice G, McConkey DJ, Bivalacqua TJ. Ex vivo culture of tumor cells from N-methyl-N-nitrosourea-induced bladder cancer in rats: Development of organoids and an immortalized cell line. Urol Oncol. 2018;36:160.e23–160.e32. doi: 10.1016/j.urolonc.2017.11.024. [DOI] [PubMed] [Google Scholar]
- 16.Okuyama H, Kondo J, Sato Y, Endo H, Nakajima A, Piulats JM, Tomita Y, Fujiwara T, Itoh Y, Mizoguchi A, et al. Dynamic change of polarity in primary cultured spheroids of human colorectal adenocarcinoma and its role in metastasis. Am J Pathol. 2016;186:899–911. doi: 10.1016/j.ajpath.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 17.Warrick JI, Walter V, Yamashita H, Chung E, Shuman L, Amponsa VO, Zheng Z, Chan W, Whitcomb TL, Yue F, et al. Cooperate to drive luminal subtype in bladder cancer: A molecular analysis of established human cell lines. Sci Rep. 2016;6:38531. doi: 10.1038/srep38531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, Hinoue T, Laird PW, Hoadley KA, Akbani R, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2018;174:1033. doi: 10.1016/j.cell.2018.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pickup MW, Mouw JK, Weaver VM. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014;15:1243–1253. doi: 10.15252/embr.201439246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frisch SM, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kondoh E, Mori S, Yamaguchi K, Baba T, Matsumura N, Cory Barnett J, Whitaker RS, Konishi I, Fujii S, Berchuck A, Murphy SK. Targeting slow-proliferating ovarian cancer cells. Int J Cancer. 2010;126:2448–2456. doi: 10.1002/ijc.24919. [DOI] [PubMed] [Google Scholar]
- 22.Choi W, Porten S, Kim S, Willis D, Plimack ER, Hoffman-Censits J, Roth B, Cheng T, Tran M, Lee IL, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25:152–165. doi: 10.1016/j.ccr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee SH, Hu W, Matulay JT, Silva MV, Owczarek TB, Kim K, Chua CW, Barlow LJ, Kandoth C, Williams AB, et al. Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell. 2018;173:515–528. doi: 10.1016/j.cell.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hovelson DH, Udager AM, McDaniel AS, Grivas P, Palmbos P, Tamura S, Lazo de la Vega L, Palapattu G, Veeneman B, El-Sawy L, et al. Targeted DNA and RNA sequencing of paired urothelial and squamous bladder cancers reveals discordant genomic and transcriptomic events and unique therapeutic implications. Eur Urol. 2018;74:741–753. doi: 10.1016/j.eururo.2018.06.047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used analyzed in the present study are available from the corresponding author on reasonable request.