Abstract
Background
Human papillomaviruses (HPV) cause over 500 000 cervical cancers each year, most of which occur in low-resource settings. Human papillomavirus genotyping is important to study natural history and vaccine efficacy. We evaluated TypeSeq, a novel, next-generation, sequencing-based assay that detects 51 HPV genotypes, in 2 large international epidemiologic studies.
Methods
TypeSeq was evaluated in 2804 cervical specimens from the Study to Understand Cervical Cancer Endpoints and Early Determinants (SUCCEED) and in 2357 specimens from the Costa Rica Vaccine Trial (CVT). Positive agreement and risks of precancer for individual genotypes were calculated for TypeSeq in comparison to Linear Array (SUCCEED). In CVT, positive agreement and vaccine efficacy were calculated for TypeSeq and SPF10-LiPA.
Results
We observed high overall and positive agreement for most genotypes between TypeSeq and Linear Array in SUCCEED and SPF10-LiPA in CVT. There was no significant difference in risk of precancer between TypeSeq and Linear Array in SUCCEED or in estimates of vaccine efficacy between TypeSeq and SPF10-LiPA in CVT.
Conclusions
The agreement of TypeSeq with Linear Array and SPF10-LiPA, 2 well established standards for HPV genotyping, demonstrates its high accuracy. TypeSeq provides high-throughput, affordable HPV genotyping for world-wide studies of cervical precancer risk and of HPV vaccine efficacy.
Keywords: HPV, TypeSeq, vaccine efficacy, genotyping, screening
Human papillomaviruses (HPV) are a major cause of invasive cancers world-wide, with up to 600 000 cancers related to HPV occurring in 2012, 500 000 of which were cervical cancers. Less common HPV-related cancer sites include the vulva, vagina, anus, penis, and oral cavity [1]. Over 200 HPV genotypes have been identified so far that vary by tissue tropism and carcinogenic potential. Only a small group of 12–13 types is responsible for most HPV-related cancers (referred to as high risk [HR] or carcinogenic types). Risk of precancer and cancer varies substantially within this group. HPV16 is by far the most carcinogenic type, causing over 60% of invasive cervical cancers and most HPV-related noncervical cancers, followed by HPV18 (15% of cervical cancers). At the cervix, both HPV16 and HPV18 increase in relative prevalence from HPV infections to precancers and cancers [2].
Detection of individual HPV genotypes associated with infections is important for research and, to some degree, for clinical use [3–5]. Epidemiologic research regarding the natural history of HPV depends on typing, because transmission, clearance, transformation, and invasive potential may differ by genotype. In a clinical setting, HPV testing is more efficient than the Pap test for primary cervical cancer screening [4, 6–9]. Evaluation of type-specific risk for precancer and cancer is necessary to decide which genotypes should be included in HPV assays and which genotypes should be detected individually. Recently approved assays use partial genotyping for HPV16 and HPV18 (and sometimes HPV45) for additional risk stratification [10]; more extended genotyping is under consideration.
Typing is important to estimate HPV vaccine efficacy (VE) [11]. Human papillomavirus vaccine trials and postvaccination surveillance programs require accurate and reliable HPV genotyping assays that detect a wide range of genotypes that are either (1) directly targeted by the vaccine, (2) targets of cross-protection, or (3) not directly or indirectly affected by vaccines [11]. There is a growing public health need for reliable, low-cost HPV genotyping methods applicable to large populations.
Few commercial assays are currently available for HPV genotyping, and most are laborious, low-throughput, and expensive. Next-generation sequencing (NGS) technology provides massively parallel sequencing capacity that can be used for detection of viral deoxyribonucleic acid (DNA) in clinical specimens. We recently developed TypeSeq, a novel NGS-based HPV genotyping assay that allows for highly automated, high-throughput testing at low cost [12]. In this study, we present the validation of TypeSeq in 2 large studies with a focus on detection of cervical precancers and on estimating VE.
METHODS
Study Descriptions
Human papillomavirus genotyping was performed in 2804 samples from women enrolled in the Study to Understand Cervical Cancer Early Endpoints and Determinants (SUCCEED) and in 2357 samples from women enrolled into the Costa Rica HPV Vaccine Trial (CVT). Of note, 1000 samples were randomly selected from each arm in CVT, and the remaining 357 samples were enriched for SPF-10 HPV16/18-positive samples.
This study is registered with Clinicaltrials.gov (NCT00128661). GlaxoSmithKline Biologicals provided vaccine and support for aspects of the trial associated with regulatory submission needs of the company under a Clinical Trials Agreement (FDA BB-IND 7920) during the 4-year, randomized, blinded phase of our study.
SUCCEED Population, DNA Extraction, and Linear Array Genotyping
SUCCEED was a cross-sectional study of women 18 years of age or older with an abnormal Pap smear who were referred to colposcopy or treatment at the University of Oklahoma (OUHSC) between 2003 and 2011. Written informed consent was obtained from all women enrolled in the study, and Institutional Review Board approval was provided by OUHSC and the US National Cancer Institute (NCI) [13].
The DNA isolation method used for SUCCEED has been described previously [14]. Linear Array HPV Genotyping System (Roche Molecular Diagnostics) genotyping was performed as described previously [14–16]. Up to 80 patient specimens, 3 HPV16-positive controls, and 1 HPV-negative control were amplified in each batch using the Linear Array (LA). Detection of both β-globin concentration control probes was required to report genotyping results. A hybridization signal was called “positive” when an unambiguous, continuous band was observed on the array.
CVT Population, DNA Extraction, and SPF10-LiPA Genotyping
The CVT was a community-based, double-blind, randomized, controlled phase III trial of the bivalent vaccine (Cervarix). As previously described, 18- to 25-year-old women residing in the provinces of Guanacaste and Puntarenas, Costa Rica, identified via a population census specifically conducted for the study, were invited to participate by attending a study clinic [17]. The trial was approved by human subjects review committees of the NCI and Instituto Costarricense de Investigación y Enseñanza en Nutrición y Salud ([INCIENSA] Costa Rica).
Extracted DNA from cervical specimens was polymerase chain reaction (PCR)-amplified and hybridized to HPV-specific probes using the SPF10 HPV DNA enzyme immunoassay (DEIA) system and the LiPA25, version 1, line detection system (SPF10 DEIA/LiPA25/TS16/18 system) [17, 18], following the manufacturer’s instructions. In addition, all specimens positive for HPV DNA using SPF10 DEIA but negative for HPV16 or HPV18 by LiPA25 were also tested with type-specific primers/probes for the presence of HPV16 and HPV18 DNA [18, 19].
TypeSeq Genotyping
TypeSeq is able to detect the following 51 types: HPV3, 6, 11, 13, 16, 18, 26, 28, 30, 31, 32, 33, 34, 35, 39, 40, 42, 43, 44, 45, 51, 52, 53, 54, 56, 58, 59, 61, 62, 66, 67, 68, 69, 70, 71, 72, 73, 74, 76, 81, 82, 83, 84, 85, 86, 87, 89, 90, 91, 97, 114 (see Supplementary Table 1). Up to 950 patient specimens, 12 HPV-positive control pools, 2 HPV-negative human-positive controls, and 2 no-template controls were processed per batch, with 1 additional no-template control randomly located on each 96-well plate of specimens. Human beta-2-microglobulin (B2M) gene (GenBank accession number NG_012920) served as the internal positive control in each reaction.
In brief, the stage 1 (S1) type-specific multiplex amplification and copy number standardization PCR was performed in a final reaction volume of 12 µL, containing 5 µL purified genomic DNA. The S1 primer pool contained 127 RNase H2-dependent primers (Integrated DNA Technologies, Coralville, IA), targeting 1 human gene (B2M) and a region of the L1 gene. The S2 primer pool contained nested B2M primers and 170 nested HPV unmodified primers (Integrated DNA Technologies). After cycling, unincorporated primers were degraded with Exonuclease I (Lucigen, Middleton, WI), then 2 µL was used as template for the 10-µL stage 3 (S3) PCR. During the S3 PCR, Ion sequencing adapters and dual barcodes were incorporated into amplicons via the universal priming sites. The PCR products were pooled and purified, then sequenced on the Ion S5 platform (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s instructions. Dual barcode demultiplexing, quality filtering and HPV genotyping detection were performed using a custom plugin developed in-house, run within the Torrent Suite software (Thermo Fisher Scientific). A minimum of 850 total HPV reads or 300 B2M reads per sample was required to report genotyping results, otherwise the sample was reported as “failed to amplify”. Positive HPV type calls required a minimum of 127–212 reads, depending on type.
Five hundred fifty-eight CVT specimens were tested in 2 independent batches on different days to assess TypeSeq interbatch reproducibility. Batch 1 was performed using manual pipetting, and batch 2 was done by automated pipetting using a JANUS liquid handler (PerkinElmer, Waltham, MA).
Statistical Analysis
We calculated overall agreement and positive agreement between TypeSeq and LA genotyping in SUCCEED, and between TypeSeq and SPF10-LiPA genotyping in CVT, and we calculated McNemar P values. We further evaluated agreement between hierarchical categories of HPV positivity between the paired tests, combining any carcinogenic type (HR+), other HPV types, and types only detected by TypeSeq but not by the paired assay. We compared the observed frequencies of 2-type combinations for the 37 genotypes detected by LA and for the 51 genotypes detected by TypeSeq with expected frequencies in all women with at least 2 concurrent HPV infections. To obtain expected frequencies for a 2-type combination, the observed genotype frequencies for both types were multiplied and the result was multiplied with the total number of subjects.
In SUCCEED, we compared the risk of precancer and cancer (cervical intraepithelial neoplasia [CIN]3+) for each genotype as determined by TypeSeq and LA. For testing statistical significance, we compared the proportion of TS+/LA− and TS−/LA+ samples that were positive for CIN3+ using Fisher’s exact test. To evaluate the performance of the assay for HPV vaccination studies, we recomputed VE at the 4-year study visit in CVT using HPV results generated by TypeSeq and compared these estimates to those obtained in the initial round of testing using the SPF10 DEIA/LiPA25/TS16/18 system. Vaccine efficacy against one-time detection of HPV16/18, HPV31/33/45, and other carcinogenic HPV types (excluding HPV16, 18, 31, 33, and 45) based on the TypeSeq and SPF10 tests in the intention-to-treat cohort (all vaccinated women) was computed. For each arm, we defined the prevalence as the proportion of the number of events among the number of women at the 4-year study visit. The complement of the ratios of the HPV prevalence in the HPV arm and the control arm are the VE estimates. We calculated these VE estimates within the randomly sampled population and weighted back to the entire CVT. Exact confidence intervals (CIs) [20] for VE were calculated based on the binomial distribution of the number of events in the HPV arm among the total number of events in the HPV and control arms [21]. Vaccine efficacy obtained by the SPF10-LiPA system was predefined as the comparator. The prespecified goal of the analysis was that the 95% CI of the TypeSeq VE estimate included the SPF10-LiPA/DEIA VE point estimate, to assure comparability of the VE estimates between both tests.
RESULTS
Prevalence of 51 Human Papillomavirus Genotypes Detected by TypeSeq in SUCCEED and CVT
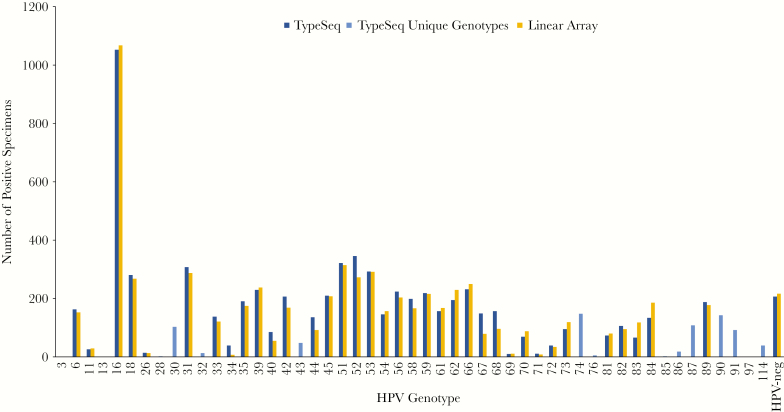
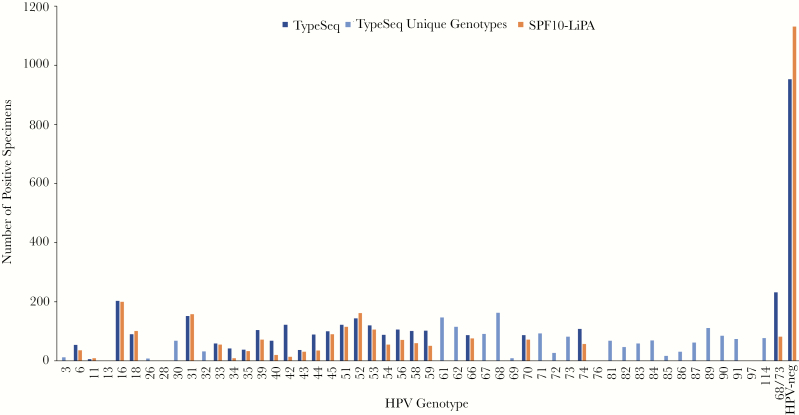
We evaluated TypeSeq results of cervical samples from 2804 women enrolled in SUCCEED and from 2357 women enrolled in CVT, respectively. In SUCCEED, a colposcopy referral population enriched for precancers and cancers, most women were HPV positive and HPV16 was by far the most common type (Figure 1). Among the 37 HPV genotypes detected by both TypeSeq and LA, the number of infections detected was very similar for both assays. The most common types detected by TypeSeq not included in LA were HPV30, 43, 74, 87, 90, 91, and 114; none has known strong disease associations. In CVT, an HPV vaccination trial of young women, a large proportion of women were negative for all 51 types detected using TypeSeq. For most of the 26 types detected by both TypeSeq and SPF10-LiPA, the number of infections was similar for both assays (Figure 2). To evaluate possible cross-reactivity between HPV genotypes, we compared observed versus expected numbers of 2‐genotype combinations detected by LA and TypeSeq in SUCCEED. With LA, we observed substantially more combinations of HPV56 and HPV66, which are closely related and have been previously described to cross-react (see Supplementary Figure 1A) [15]. In contrast, there was no type combination observed substantially more frequently than expected for TypeSeq, suggesting no cross-reactivity (see Supplementary Figure 1B).
Figure 1.
TypeSeq versus Linear Array (LA) positives by human papillomavirus (HPV) genotype for 2804 clinical specimens. The number of positive specimens are shown for LA and TypeSeq for the 37 genotypes detectable by both assays. The genotypes uniquely detectable by TypeSeq are displayed as “TypeSeq Unique Genotypes”. Linear Array’s HPV55 positives are show as HPV44 according to the current PAVE classification. HPV68 represents results for the HPV68 lineages C to F (previously “68b”) detectable by LA. HPV82 represents a combined result for HPV82 and 82v (IS39), which are detected individually by both assays.
Figure 2.
TypeSeq versus SPF10-LiPA positives by human papillomavirus (HPV) genotype for 2357 clinical specimens. The number of positive specimens are shown for SPF10-LiPA and TypeSeq for the 27 genotypes detectable by both assays. The genotypes uniquely detectable by TypeSeq are displayed as “TypeSeq Unique Genotypes”.
To assess TypeSeq reproducibility on clinical specimens, we tested 558 CVT specimens in duplicate using manual and automated processing (Table 1). Agreements for testing positive were 93.1% for any HR type, 93.2% for HPV16/18 combined, and ranged from 71.4% (HPV59) to 100% (HPV58) for individual HR types. Positive agreements were 60% or higher for 33 of the 35 LR types with positive specimens in either batch. None of the discrepancies were statistically significant for any type (P < .05, McNemar). Five types (HPV3, 13, 28, 76, and 97) were not detected in either batch.
Table 1.
TypeSeq Reproducibility Testing on 558 CVT Clinical Specimens in Duplicatea
| Number of Specimens | %Agreement | ||||||
|---|---|---|---|---|---|---|---|
| HPV Genotype | R1−/R2− | R1+/R2− | R1−/R2+ | R1+/R2+ | Total | Positive | McNemar P Value |
| Any HPV | 175 | 21 | 14 | 348 | 93.7 | 90.9 | .31 |
| Any HR-HPVb | 313 | 12 | 5 | 228 | 97.0 | 93.1 | .15 |
| 16/18 | 514 | 1 | 2 | 41 | 99.5 | 93.2 | 1 |
| 3 | 558 | 0 | 0 | 0 | 100.0 | NE | NE |
| 6 | 544 | 2 | 0 | 12 | 99.6 | 85.7 | .48 |
| 11 | 557 | 0 | 0 | 1 | 100.0 | 100.0 | NE |
| 13 | 558 | 0 | 0 | 0 | 100.0 | NE | NE |
| 16 | 522 | 1 | 1 | 34 | 99.6 | 94.4 | .48 |
| 18 | 548 | 0 | 1 | 9 | 99.8 | 90.0 | 1 |
| 26 | 555 | 0 | 0 | 3 | 100.0 | 100.0 | NE |
| 28 | 558 | 0 | 0 | 0 | 100.0 | NE | NE |
| 30 | 535 | 3 | 3 | 17 | 98.9 | 73.9 | .68 |
| 31 | 531 | 1 | 2 | 24 | 99.5 | 88.9 | 1 |
| 32 | 548 | 2 | 0 | 8 | 99.6 | 80.0 | .48 |
| 33 | 546 | 1 | 0 | 11 | 99.8 | 91.7 | 1 |
| 34 | 547 | 1 | 2 | 8 | 99.5 | 72.7 | 1 |
| 35 | 543 | 0 | 1 | 14 | 99.8 | 93.3 | 1 |
| 39 | 528 | 2 | 1 | 27 | 99.5 | 90.0 | 1 |
| 40 | 546 | 2 | 3 | 7 | 99.1 | 58.3 | 1 |
| 42 | 536 | 6 | 0 | 16 | 98.9 | 72.7 | .041 |
| 43 | 550 | 3 | 2 | 3 | 99.1 | 37.5 | 1 |
| 44 | 533 | 3 | 5 | 17 | 98.6 | 68.0 | .72 |
| 45 | 541 | 2 | 0 | 15 | 99.6 | 88.2 | .48 |
| 51 | 533 | 2 | 5 | 18 | 98.7 | 72.0 | .45 |
| 52 | 510 | 2 | 1 | 45 | 99.5 | 93.8 | 1 |
| 53 | 529 | 4 | 3 | 22 | 98.7 | 75.9 | 1 |
| 54 | 541 | 2 | 4 | 11 | 98.9 | 64.7 | .68 |
| 56 | 532 | 1 | 0 | 25 | 99.8 | 96.2 | 1 |
| 58 | 525 | 0 | 0 | 33 | 100.0 | 100.0 | NE |
| 59 | 530 | 7 | 1 | 20 | 98.6 | 71.4 | .077 |
| 61 | 534 | 0 | 3 | 21 | 99.5 | 87.5 | .25 |
| 62 | 534 | 1 | 3 | 20 | 99.3 | 83.3 | .62 |
| 66 | 528 | 1 | 0 | 29 | 99.8 | 96.7 | 1 |
| 67 | 536 | 0 | 4 | 18 | 99.3 | 81.8 | .13 |
| 68a | 527 | 4 | 4 | 23 | 98.6 | 74.2 | .72 |
| 68b | 549 | 1 | 0 | 8 | 99.8 | 88.9 | 1 |
| 69 | 556 | 0 | 0 | 2 | 100.0 | 100.0 | NE |
| 70 | 533 | 2 | 3 | 20 | 99.1 | 80.0 | 1 |
| 71 | 540 | 0 | 1 | 17 | 99.8 | 94.4 | 1 |
| 72 | 551 | 0 | 2 | 5 | 99.6 | 71.4 | .48 |
| 73 | 546 | 2 | 2 | 8 | 99.3 | 66.7 | .62 |
| 74 | 528 | 6 | 2 | 22 | 98.6 | 73.3 | .28 |
| 76 | 558 | 0 | 0 | 0 | 100.0 | NE | NE |
| 81 | 540 | 3 | 0 | 15 | 99.5 | 83.3 | .25 |
| 82 | 555 | 1 | 0 | 2 | 99.8 | 66.7 | 1 |
| 82v | 547 | 1 | 1 | 9 | 99.6 | 81.8 | .48 |
| 83 | 545 | 2 | 1 | 10 | 99.5 | 76.9 | 1 |
| 84 | 543 | 0 | 1 | 14 | 99.8 | 93.3 | 1 |
| 85 | 552 | 0 | 1 | 5 | 99.8 | 83.3 | 1 |
| 86 | 549 | 1 | 1 | 7 | 99.6 | 77.8 | .48 |
| 87 | 535 | 3 | 2 | 18 | 99.1 | 78.3 | 1 |
| 89 | 538 | 4 | 1 | 15 | 99.1 | 75.0 | .37 |
| 90 | 543 | 1 | 3 | 11 | 99.3 | 73.3 | .62 |
| 91 | 543 | 0 | 0 | 15 | 100.0 | 100.0 | NE |
| 97 | 558 | 0 | 0 | 0 | 100.0 | NE | NE |
| 114 | 543 | 3 | 3 | 9 | 98.9 | 60.0 | .68 |
Abbreviations: CVT, Costa Rica Vaccine Trial; HPV, human papillomavirus; HR-HPV, high-risk HPV; NE, not evaluable; R1, replicate 1; R2, replicate 2.
aClinical specimens were tested in duplicate by manual (R1) or automated (R2) processing.
bHR-HPV represents HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68.
Agreement of TypeSeq With Linear Array and SPF10-LiPA for Individual Human Papillomavirus Types
We evaluated the agreement of TypeSeq with LA in SUCCEED and SPF10-LiPA in CVT. Comparing TypeSeq with LA in SUCCEED, 10 of 13 carcinogenic types had at least 80% positive agreement and HPV16 had 94% positive agreement (Table 2). The 3 types with lower agreement were HPV51, 52, and 56; all showed higher detection with TypeSeq. Six types (HPV31, 33, 35, 52, 56, and 58) showed a significantly higher positivity for TypeSeq among the discrepant results (McNemar, P < .05). There was high agreement for detecting overall HPV positivity, and positive agreement for detecting any HR type reached 95% between TypeSeq and LA (see Supplementary Table 2).
Table 2.
Agreement Between TypeSeq and Linear Array for 13 Carcinogenic Types Among 2804 Women in SUCCEED
| HPV Type | N LA | % LA | N TypeSeq | % TypeSeq | N Both+ | % Both+ | N LA+/TypeSeq− | % LA+/TypeSeq− | N LA−/TypeSeq+ | % LA−/TypeSeq+ | N Neither+ | % Neither+ | % Agreement | % Positive Agreement | McNemar P Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16 | 1067 | 38.1 | 1052 | 37.6 | 1025 | 36.6 | 42 | 1.5 | 27 | 1.0 | 1707 | 60.9 | 97.5 | 93.7 | .09 |
| 18 | 268 | 9.6 | 280 | 10.0 | 250 | 8.9 | 18 | 0.6 | 30 | 1.1 | 2503 | 89.4 | 98.3 | 83.9 | .11 |
| 31 | 287 | 10.3 | 307 | 11.0 | 275 | 9.8 | 12 | 0.4 | 32 | 1.1 | 2482 | 88.6 | 98.4 | 86.2 | .003 |
| 33 | 121 | 4.3 | 138 | 4.9 | 120 | 4.3 | 1 | 0.04 | 18 | 0.6 | 2662 | 95.0 | 99.3 | 86.3 | .00004 |
| 35 | 175 | 6.3 | 191 | 6.8 | 167 | 6.0 | 8 | 0.3 | 24 | 0.9 | 2602 | 92.9 | 98.9 | 83.9 | .004 |
| 39 | 238 | 8.5 | 230 | 8.2 | 216 | 7.71 | 22 | 0.8 | 14 | 0.5 | 2549 | 91.0 | 98.7 | 85.7 | .24 |
| 45 | 208 | 7.4 | 210 | 7.5 | 189 | 6.75 | 19 | 0.7 | 21 | 0.8 | 2572 | 91.8 | 98.6 | 82.5 | .87 |
| 51 | 314 | 11.2 | 321 | 11.5 | 266 | 9.5 | 48 | 1.7 | 55 | 2.0 | 2432 | 86.8 | 96.3 | 72.1 | .55 |
| 52 | 273 | 9.7 | 345 | 12.3 | 250 | 8.92 | 23 | 0.8 | 95 | 3.4 | 2436 | 86.9 | 95.8 | 67.9 | .0002 |
| 56 | 204 | 7.3 | 224 | 8.0 | 184 | 6.57 | 20 | 0.7 | 40 | 1.4 | 2557 | 91.3 | 97.9 | 75.4 | .01 |
| 58 | 167 | 6.0 | 199 | 7.1 | 163 | 5.82 | 4 | 0.1 | 36 | 1.3 | 2598 | 92.8 | 98.6 | 80.3 | <.00001 |
| 59 | 216 | 7.7 | 219 | 7.8 | 197 | 7.03 | 19 | 0.7 | 22 | 0.8 | 2563 | 91.5 | 98.5 | 82.8 | .76 |
| 68a | 96 | 3.4 | 103 | 3.7 | 90 | 3.21 | 6 | 0.2 | 13 | 0.5 | 2692 | 96.1 | 99.3 | 82.6 | .17 |
Abbreviations: HPV, human papillomavirus; LA, Linear Array; SUCCEED, Study to Understand Cervical Cancer Endpoints and Early Determinants.
aHPV68 represents results for the lineages detectable by LA (C to F, formerly “68b”). TypeSeq results for HPV68 lineages A and B (formerly “68a”), which are uniquely detectable by TypeSeq, were excluded from this analysis.
Positive agreement between TypeSeq and SPF10-LiPA was somewhat lower, with 4 types having at least 70% and 9 types having at least 60% positive agreement (Table 3). The positive agreement for HPV16 reached 80%. The types with lower agreement were HPV35, 58, 59, and 68/73; all showed higher detection by TypeSeq. Five types (HPV39, 56, 58, 59, and 68) showed a significantly higher positivity by TypeSeq among the discrepant results (P < .00001, McNemar), whereas HPV52 was significantly more positive in SPF10-LiPA (P = .03). The positive agreement between TypeSeq and SPF10-LiPA in the random draw of 1998 samples from both study arms was slightly lower for the types that were oversampled (see Supplementary Table 3). Positive agreement for detecting any HR type reached 77% between TypeSeq and SPF10-LiPA (see Supplementary Table 4).
Table 3.
Agreement Between TypeSeq and SPF10-LiPA for 13 Carcinogenic Types Among 2357 Women in CVT
| HPV Type | N SPF10 | % SPF10 | N TypeSeq | % TypeSeq | N Both+ | % Both+ | N SPF10+/TypeSeq− | % SPF10+/TypeSeq− | N SPF10− /TypeSeq+ | % SPF10− /TypeSeq+ | N Neither+ | % Neither+ | % Agreement | % Positive Agreement | McNemar P Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16 | 200 | 8.5 | 203 | 8.6 | 179 | 7.6 | 21 | 0.89 | 24 | 1.0 | 2133 | 90.5 | 98.1 | 79.9 | .38 |
| 18 | 101 | 4.3 | 90 | 3.8 | 84 | 3.6 | 17 | 0.72 | 6 | 0.3 | 2250 | 95.5 | 99.0 | 78.5 | .43 |
| 31 | 159 | 6.8 | 152 | 6.5 | 125 | 5.3 | 34 | 1.44 | 27 | 1.2 | 2171 | 92.1 | 97.4 | 67.2 | .44 |
| 33 | 55 | 2.3 | 59 | 2.5 | 46 | 2.0 | 9 | 0.38 | 13 | 0.6 | 2289 | 97.1 | 99.1 | 67.7 | .39 |
| 35 | 33 | 1.4 | 38 | 1.6 | 25 | 1.1 | 8 | 0.34 | 13 | 0.6 | 2311 | 98.1 | 99.1 | 54.4 | .28 |
| 39 | 72 | 3.1 | 104 | 4.4 | 66 | 2.8 | 6 | 0.25 | 38 | 1.6 | 2247 | 95.3 | 98.1 | 60.0 | <.00001 |
| 45 | 90 | 3.8 | 100 | 4.2 | 80 | 3.4 | 10 | 0.42 | 20 | 0.9 | 2247 | 95.3 | 98.7 | 72.7 | .07 |
| 51 | 115 | 4.9 | 122 | 5.2 | 96 | 4.1 | 19 | 0.81 | 26 | 1.1 | 2216 | 94.0 | 98.1 | 68.1 | .30 |
| 52 | 163 | 6.9 | 144 | 6.1 | 120 | 5.1 | 43 | 1.82 | 24 | 1.0 | 2170 | 92.1 | 97.2 | 64.2 | .03 |
| 56 | 71 | 3.0 | 106 | 4.5 | 69 | 2.9 | 2 | 0.08 | 37 | 1.6 | 2249 | 95.4 | 98.4 | 63.9 | <.00001 |
| 58 | 60 | 2.6 | 101 | 4.3 | 58 | 2.5 | 2 | 0.08 | 43 | 1.8 | 2254 | 95.6 | 98.1 | 56.3 | <.00001 |
| 59 | 51 | 2.2 | 102 | 4.3 | 49 | 2.1 | 2 | 0.08 | 53 | 2.3 | 2253 | 95.6 | 97.7 | 47.1 | <.00001 |
| 68/73 | 83 | 3.5 | 232 | 9.8 | 78 | 3.3 | 5 | 0.21 | 154 | 6.5 | 2120 | 89.9 | 93.3 | 32.9 | <.00001 |
Abbreviations: CVT, Costa Rica Vaccine Trial; HPV, human papillomavirus.
Human Papillomavirus Genotype Prevalence by TypeSeq and Linear Array in CIN3+ From SUCCEED
We compared the genotype prevalence in cervical precancer and cancer (CIN3+) by TypeSeq and LA (Table 4). Overall, there was high concordance between both assays for all genotypes. Cases with HPV39, 52, 56, and 58 showed a difference of at least 5 CIN3+ cases between both assays; TypeSeq detected more CIN3+ for all these types except for HPV39-positive cases, in which it detected fewer cases of CIN3+.
Table 4.
Prevalence of HPV Genotypes in CIN3+ by TypeSeq and Linear Array
| HPV Type | TS+ | TS+ CIN3+ | Risk TS (%) | LA+ | LA+ CIN3+ | Risk LA (%) | TS−/LA− | TS−/LA+ | TS−/LA+ CIN3+ | Risk TS−/LA+a (%) | TS+/LA− | TS+/LA− CIN3+ | Risk TS+/LA−a (%) | TS+/LA+ | TS+/LA+ CIN3+ | Risk TS+/LA+ (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16 | 1052 | 474 | 45.1 | 1067 | 476 | 44.6 | 1707 | 42 | 5 | 11.9 | 27 | 3 | 11.1 | 1025 | 471 | 46.0 |
| 18 | 280 | 75 | 26.8 | 268 | 74 | 27.6 | 2503 | 18 | 6 | 33.3 | 30 | 7 | 23.3 | 250 | 68 | 27.2 |
| 31 | 307 | 81 | 26.4 | 287 | 78 | 27.2 | 2482 | 12 | 2 | 16.7 | 32 | 5 | 15.6 | 275 | 76 | 27.6 |
| 33 | 138 | 39 | 28.3 | 121 | 35 | 28.9 | 2662 | 1 | 0 | 0.0 | 18 | 4 | 22.2 | 120 | 35 | 29.2 |
| 35 | 191 | 37 | 19.4 | 175 | 33 | 18.9 | 2602 | 8 | 2 | 25.0 | 24 | 6 | 25.0 | 167 | 31 | 18.6 |
| 39 | 230 | 27 | 11.7 | 238 | 33 | 13.9 | 2549 | 22 | 7 | 31.8 | 14 | 1 | 7.1 | 216 | 26 | 12.0 |
| 45 | 210 | 56 | 26.7 | 208 | 60 | 28.8 | 2572 | 19 | 9 | 47.4 | 21 | 5 | 23.8 | 189 | 51 | 27.0 |
| 51 | 321 | 43 | 13.4 | 314 | 40 | 12.7 | 2432 | 48 | 8 | 16.7 | 55 | 11 | 20.0 | 266 | 32 | 12.0 |
| 52 | 345 | 63 | 18.3 | 273 | 54 | 19.8 | 2436 | 23 | 7 | 30.3 | 95 | 16 | 16.8 | 250 | 47 | 18.8 |
| 56 | 224 | 28 | 12.5 | 204 | 21 | 10.3 | 2557 | 20 | 2 | 10.0 | 40 | 9 | 22.5 | 184 | 19 | 10.3 |
| 58 | 199 | 42 | 21.1 | 167 | 34 | 20.4 | 2598 | 4 | 0 | 0.0 | 36 | 8 | 22.2 | 163 | 34 | 20.9 |
| 59 | 219 | 38 | 17.4 | 216 | 39 | 18.1 | 2563 | 19 | 4 | 21.1 | 22 | 3 | 13.6 | 197 | 35 | 17.8 |
| 68 | 103 | 21 | 20.4 | 96 | 19 | 19.8 | 2692 | 6 | 0 | 0.0 | 13 | 2 | 15.4 | 90 | 19 | 21.1 |
Abbreviations: CIN, cervical intraepithelial neoplasia; HPV, human papillomavirus; LA, Linear Array; TS, TypeSeq.
aThe difference in risk between TS−/LA and TS+/LA− was not significant for any of the types (P > .5).
When comparing all TypeSeq-positive and all LA-positive women, the risk of CIN3+ was very similar for all genotypes. There was no significant difference in the risk of CIN3+ between the discrepant categories (TS−/LA+ vs TS+/LA−) for any genotype (P > .05).
Estimation of Vaccine Efficacy Based on TypeSeq Compared With SPF10-LiPA Test Results
To evaluate the performance of TypeSeq for detecting viral endpoints in vaccine trials and population surveillance studies, we calculated vaccine efficiency of the bivalent HPV vaccine based on HPV genotyping with TypeSeq compared with the reference standard in CVT, SPF10-LiPA (Table 5). Vaccine efficiency estimates were computed in a population naive to HPV infections at the time of vaccination as well as the full analytic cohort for HPV16/18 (the types included in the bivalent vaccine), HPV31, 33, 45 (3 types that show cross-protection for the bivalent vaccine), and 7 other types that are typically not affected by the vaccine. The number of infections detected by both assays was similar. In all comparisons, there was no statistically significant difference in VE as determined by TypeSeq compared with SPF10-LiPA, and the prespecified threshold for assay comparability was achieved in all population and HPV genotype groups.
Table 5.
Bivalent Vaccine Efficacy Determined by TypeSeq and SPF10-LiPA
| HPV16/18 | HPV31, 33, 45 | Other (HPV35, 39, 51, 52, 56, 58, 59) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ATP | ITT | ATP | ITT | ATP | ITT | ||||||||
| Assay | Trial Arm | Events/ Women | VE (%) (95% CI) | Events/ Women | VE (%) (95% CI) | Events/ Women | VE (%) (95% CI) | Events/ Women | VE (%) (95% CI) | Events/ Women | VE (%) (95% CI) | Events/ Women | VE (%) (95% CI) |
| All Women Tested by TypeSeq | |||||||||||||
| SPF10-LiPA | HPV | 49/820 | 66.6 | 62/1104 | 69.2 | 69/823 | 45.7 | 99/1104 | 39.4 | 153/824 | 12.7 | 208/1104 | 9.8 |
| HAV | 164/916 | (54.3–75.9) | 228/1250 | (59.4–76.9) | 142/920 | (27.8–59.4) | 185/1250 | (22.8–52.7) | 196/922 | (−7.9 to 29.4) | 261/1250 | (−8.2 to 24.8) | |
| TypeSeq | HPV | 43/820 | 70.2 | 60/1104 | 69.5 | 66/823 | 52.7 | 90/1104 | 49.6 | 182/824 | 13.7 | 246/1104 | 7.8 |
| HAV | 161/916 | (58.5–78.9) | 223/1250 | (59.7–77.2) | 156/920 | (37.1–64.7) | 202/1250 | (35.5–60.8) | 236/922 | (−4.7 to 28.9) | 302/1250 | (−9.1 to 22.1 | |
| Random Sample From Both Arms | |||||||||||||
| SPF10-LiPA | HPV | 16/741 | 72.6 | 20/998 | 74.0 | 20/742 | 64.4 | 32/998 | 53.5 | 127/743 | 1.2 | 175/998 | −6.3 |
| HAV | 58/735 | (53.2–84.7) | 77/1000 | (58.0–84.4) | 56/739 | (41.4–79.1) | 69/1000 | (29.7–69.8) | 128/740 | (−26.4 to 22.7) | 165/1000 | (−31.5 to 14.1) | |
| TypeSeq | HPV | 17/741 | 73.2 | 26/998 | 69.0 | 23/742 | 67.3 | 35/998 | 58.2 | 153/743 | 4.2 | 212/998 | −8.4 |
| HAV | 63/735 | (55.0–84.7) | 84/1000 | (52.3–80.3) | 70/739 | (48.1–79.9) | 84/1000 | (38.4–72.1) | 159/740 | (−19.7 to 23.3) | 196/1000 | (−31.7 to 10.8) | |
| Random Sample Weighted Back to Full Study | |||||||||||||
| SPF10-LiPA | HPV | 49/2379 | 70.4 | 62/3175 | 72.7 | 69/2384 | 51.8 | 100/3175 | 46.3 | 385/2387 | 2.7 | 516/3175 | 0.3 |
| HAV | 164/2340 | (61.6–77.2) | 228/3167 | (66.3–77.9) | 142/2352 | (39.7–61.4) | 185/3167 | (37.0–54.2) | 390/2354 | (−19.9 to 21.1) | 516/3167 | (−19.5 to 16.8) | |
| TypeSeq | HPV | 55/2379 | 67.9 | 82/3175 | 65.7 | 82/2384 | 55.9 | 112/3175 | 52.6 | 460/2387 | 6.0 | 623/3175 | −2.8 |
| HAV | 169/2340 | (54.6–77.4) | 239/3167 | (54.0–74.5) | 184/2352 | (40.9–67.1) | 236/3167 | (39.5–62.8) | 483/2354 | (−13.2 to 22.1) | 605/3167 | (−21.0 to 12.7) | |
Abbreviations: ATP, according-to-protocol; CI, confidence interval; HAV, control arm; HPV, vaccine arm; ITT, intention-to-treat; VE, vaccine efficacy.
DISCUSSION
TypeSeq is a novel, affordable, high-throughput NGS assay for detection of 51 HPV genotypes. We validated TypeSeq in over 5100 cervical specimens from 2 large epidemiological studies. TypeSeq showed high agreement with 2 widely established HPV genotyping assays, LA and SPF10-LiPA. In a large study of cervical precancers, no difference in risk of CIN3+ associated with individual genotypes was observed between TypeSeq and LA. In a randomized controlled HPV vaccine trial, no difference in VE for vaccine types and cross-protective types was observed between TypeSeq and SPF10-LiPA.
Human papillomavirus genotyping is central to understanding natural histories of HPV-related diseases and to HPV-based prevention efforts [1, 22]. Natural history differs between genotypes with respect to HPV acquisition and progression to precancer [23, 24]. Most importantly, the risk of cancer differs substantially between individual genotypes [2]. However, currently, very few HPV genotyping assays are commercially available, and most established assays are costly and laborious, which has limited HPV genotyping particularly in large longitudinal population-based studies with multiple rounds of specimen sampling. More important, the performance of HPV assays needs to strike a delicate balance between sensitivity and specificity, so that all clinically important infections, but not irrelevant minor infections or viral depositions, are detected.
We are now entering a new era of HPV-based prevention of cervical cancers, with 3 highly efficacious HPV vaccines available that cover different HPV types. Evaluation of VE is shifting to viral endpoints, which requires accurate and reproducible HPV genotyping [11]. A new focus of HPV vaccine studies evaluates efficacy and effectiveness of vaccination with fewer doses, particularly 1-dose vaccination [25]. Likewise, postvaccination surveillance requires affordable and reliable HPV genotyping of specimens from large populations.
At the same time, HPV genotyping is important to determine which types to include in HPV assays and which types to detect individually for additional risk stratification [1]. Given its comparable performance to LA, an assay that performs similarly to HPV assays approved for HPV screening [26], TypeSeq could also be evaluated as an assay for screening and triage.
TypeSeq can process up to 950 samples per batch. The turnaround time for this batch size is 3 days, with a hands-on time of 12 hours. Human papillomavirus genotype calling is fully automated, and results are immediately exported into a spreadsheet. All genotypes are reported individually, but the software can mask and group genotypes, allowing customization of the assay to specific needs.
To improve detection of HPV genotypes in multiple infections, the assay normalizes the input viral DNA over a wide range of concentrations. Therefore, the assay does not provide information about viral load. However, this is a minor limitation since the clinical relevance of viral load is limited [27].
The unique features of TypeSeq, particularly the high throughput, low cost, and high accuracy, make it an attractive assay for many applications. It can provide reliable HPV genotyping for large natural history studies with multiple sampling. We successfully evaluated TypeSeq in a randomized controlled vaccine trial, and, based on its performance, we plan to use it in future HPV vaccination studies. The performance of TypeSeq is similar to LA, a widely evaluated assay that has high agreement with clinical HPV tests like Cobas and Onclarity [28]. Further evaluation of TypeSeq as a clinical test for primary screening with partial genotyping is supported by our findings. Evaluation of other specimen types, such as anal and oral swab samples, as well as tissue specimens, using the assay is currently underway.
TypeSeq requires an infrastructure suitable for NGS technology and investment in sequencing equipment. However, these technologies are advancing quickly, and new robust equipment can now be made available in all regions of the world. Furthermore, smaller and more achievable NGS platforms are now available that reduce up-front costs. Due to the highly integrated and automated workflow of the assay, technology transfer is feasible and is currently happening in other laboratories.
CONCLUSIONS
In summary, we have validated a novel NGS-based HPV genotyping assay that addresses a widespread need for high-throughput, affordable HPV genotyping for research, surveillance, and clinical management.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We extend a special thanks to the women of Guanacaste and Puntarenas, Costa Rica, who gave of themselves in participating in this effort. In Costa Rica, we acknowledge the tremendous effort and dedication of the staff involved in this project; we would like to specifically acknowledge the meaningful contributions by Carlos Avila, Loretto Carvajal, Rebecca Ocampo, Cristian Montero, Diego Guillen, Jorge Morales, and Mario Alfaro. In the United States, we extend our appreciation to the team from Information Management Services responsible for the development and maintenance of the data system used in the trial and who serve as the data management center for this effort, especially Jean Cyr, Julie Buckland, John Schussler, and Brian Befano. We thank Dr. Diane Solomon (Costa Rica Vaccine Trial [CVT]: medical monitor and quality control pathologist) for invaluable contributions during the randomized blinded phase of the trial and the design of the long-term follow-up (LTFU) and Nora Macklin (CVT) and Kate Torres (LTFU) for the expertise in coordinating the study. We thank the members of the Data and Safety Monitoring Board charged with protecting the safety and interest of participants during the randomized, blinded phase of our study (Steve Self, Chair, Adriana Benavides, Luis Diego Calzada, Ruth Karron, Ritu Nayar, and Nancy Roach) and members of the external Scientific HPV Working Group who have contributed to the success of our efforts over the years (Joanna Cain and Elizabeth Fontham, Co-Chairs, Diane Davey, David DeMets, Anne Gershon, Elizabeth Holly, Silvia Lara, Henriette Raventós, Wasima Rida, Luis Rosero-Bixby, Gypsyamber D’Souza, and Richard Roden).
Author contributions. N. W., M. S., S. W., D. R., J. B., A. R. K., M. Y., M. C., L. M., and R. H. conceived the project. A. R. K., A. H., B. C., C. P., and A. C. R. provided the CVT specimens. W. Q. and L.-J. V. D. performed the SPF10-LiPA genotyping. T. D., J. W., and R. Z. provided the Study to Understand Cervical Cancer Endpoints and Early Determinants (SUCCEED) specimens and performed the Linear Array genotyping. S. W. performed the TypeSeq genotyping. D. R. performed the TypeSeq bioinformatics analysis. J. S. performed the CVT vaccine efficacy analysis. N. W. performed the assay concordance and statistical analyses. The manuscript was drafted by N. W. and reviewed by all coauthors. The National Cancer Institute and Costa Rica investigators are responsible for the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation of the manuscript.
Financial support. This work was funded in whole or in part with federal funds from the National Cancer Institute at the National Institutes of Health (HHSN261200800001E). The Costa Rica HPV Vaccine Trial is a long-standing collaboration between investigators in Costa Rica and the National Cancer Institute at the National Institutes of Health. The trial is sponsored and funded by the National Cancer Institute at the National Institutes of Health (Contract N01-CP-11005), with funding support from the National Institutes of Health Office of Research on Women’s Health.
Potential conflicts of interest. John T. Schiller and Douglas R. Lowy report that they are named inventors on US Government-owned HPV vaccine patents that are licensed to GlaxoSmithKline and Merck and for which the National Cancer Institute receives licensing fees. They are entitled to limited royalties as specified by federal law. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Members of the Costa Rica HPV Vaccine Trial (CVT) Group. Costa Rican Agency for Biomedical Research (formerly Proyecto Epidemiológico Guanacaste, Fundación INCIENSA), San José, Costa Rica—Bernal Cortés (specimen and repository manager), Paula González (long-term follow-up study [LTFU]; co-principal investigator), Rolando Herrero (CVT: co-principal investigator), Silvia E. Jiménez (trial coordinator), Carolina Porras (co-investigator), Ana Cecilia Rodríguez (co-investigator). United States National Cancer Institute (NCI), Bethesda, MD—Allan Hildesheim (co-principal investigator and NCI co-project officer), Aimée R. Kreimer (LTFU; co-principal investigator and NCI co-project officer), Douglas R. Lowy (HPV virologist), Mark Schiffman (CVT; medical monitor and NCI co-project officer), John T. Schiller (HPV virologist), Mark Sherman (CVT; quality control pathologist), Sholom Wacholder (statistician). Leidos Biomedical Research, Inc., Frederick National Laboratory for Cancer Research, Frederick, MD (HPV Immunology Laboratory)—Ligia A. Pinto, Troy J. Kemp, Georgetown University, Washington, DC, and Mary K. Sidawy (CVT; histopathologist). DDL Diagnostic Laboratory, Netherlands (HPV DNA Testing)—Wim Quint, Leen-Jan van Doorn, and Linda Struijk. University of California, San Francisco, CA—Joel M. Palefsky, Teresa M. Darragh, University of Virginia, Charlottesville, VA, and Mark H. Stoler.
Presented in part: 31st International Papillomavirus Conference, March 2017, Cape Town, South Africa.
Contributor Information
The CVT Group:
San José, Paula González, Rolando Herrero, Silvia E Jiménez, Carolina Porras, Ana Cecilia Rodríguez, Allan Hildesheim, Aimée R Kreimer, Douglas R Lowy, Mark Schiffman, John T Schiller, Mark Sherman, Sholom Wacholder, Ligia A Pinto, Troy J Kemp, Mary K Sidawy, Wim Quint, Leen-Jan van Doorn, Linda Struijk, Joel M Palefsky, Teresa M Darragh, and Mark H Stoler
References
- 1. Schiffman M, Hyun N, Raine-Bennett TR, et al. A cohort study of cervical screening using partial HPV typing and cytology triage. Int J Cancer 2016; 139:2606–15. [DOI] [PubMed] [Google Scholar]
- 2. Guan P, Howell-Jones R, Li N, et al. Human papillomavirus types in 115,789 HPV-positive women: a meta-analysis from cervical infection to cancer. Int J Cancer 2012; 131:2349–59. [DOI] [PubMed] [Google Scholar]
- 3. Wentzensen N, Schiffman M, Palmer T, Arbyn M. Triage of HPV positive women in cervical cancer screening. J Clin Virol 2016; 76(Suppl 1):49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huh WK, Ault KA, Chelmow D, et al. Use of primary high-risk human papillomavirus testing for cervical cancer screening: interim clinical guidance. Gynecol Oncol 2015; 136:178–82. [DOI] [PubMed] [Google Scholar]
- 5. Del Mistro A, Adcock R, Carozzi F, et al. Human papilloma virus genotyping for the cross-sectional and longitudinal probability of developing cervical intraepithelial neoplasia grade 2 or more. Int J Cancer 2018; 143:333–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wentzensen N, Arbyn M, Berkhof J, et al. Eurogin 2016 Roadmap: how HPV knowledge is changing screening practice. Int J Cancer 2017; 140:2192–200. [DOI] [PubMed] [Google Scholar]
- 7. Ronco G, Dillner J, Elfström KM, et al. Efficacy of HPV-based screening for prevention of invasive cervical cancer: follow-up of four European randomised controlled trials. Lancet 2014; 383:524–32. [DOI] [PubMed] [Google Scholar]
- 8. Dillner J, Rebolj M, Birembaut P, et al. Long term predictive values of cytology and human papillomavirus testing in cervical cancer screening: joint European cohort study. BMJ 2008; 337:a1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gage JC, Schiffman M, Katki HA, et al. Reassurance against future risk of precancer and cancer conferred by a negative human papillomavirus test. J Natl Cancer Inst 2014; 106: dju153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wright TC, Stoler MH, Behrens CM, Sharma A, Zhang G, Wright TL. Primary cervical cancer screening with human papillomavirus: end of study results from the ATHENA study using HPV as the first-line screening test. Gynecol Oncol 2015; 136:189–97. [DOI] [PubMed] [Google Scholar]
- 11. Lowy DR, Herrero R, Hildesheim A; Participants in the IARC/NCI workshop on Primary Endpoints for Prophylactic HPV Vaccine Trials Primary endpoints for future prophylactic human papillomavirus vaccine trials: towards infection and immunobridging. Lancet Oncol 2015; 16:e226–33. [DOI] [PubMed] [Google Scholar]
- 12. Wagner S, Roberson D, Boland J, et al. Development of the TypeSeq assay for detection of 51 HPV genotypes by next generation sequencing. J Clin Microbiol 2019; 57: e01794–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang SS, Zuna RE, Wentzensen N, et al. Human papillomavirus cofactors by disease progression and human papillomavirus types in the study to understand cervical cancer early endpoints and determinants. Cancer Epidemiol Biomarkers Prev 2009; 18:113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dunn ST, Allen RA, Wang S, Walker J, Schiffman M. DNA extraction: an understudied and important aspect of HPV genotyping using PCR-based methods. J Virol Methods 2007; 143:45–54. [DOI] [PubMed] [Google Scholar]
- 15. Wentzensen N, Schiffman M, Dunn T, et al. Multiple human papillomavirus genotype infections in cervical cancer progression in the study to understand cervical cancer early endpoints and determinants. Int J Cancer 2009; 125:2151–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wentzensen N, Schiffman M, Dunn ST, et al. Grading the severity of cervical neoplasia based on combined histopathology, cytopathology, and HPV genotype distribution among 1,700 women referred to colposcopy in Oklahoma. Int J Cancer 2009; 124:964–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Herrero R, Hildesheim A, Rodríguez AC, et al. Rationale and design of a community-based double-blind randomized clinical trial of an HPV 16 and 18 vaccine in Guanacaste, Costa Rica. Vaccine 2008; 26:4795–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kleter B, van Doorn LJ, ter Schegget J, et al. Novel short-fragment PCR assay for highly sensitive broad-spectrum detection of anogenital human papillomaviruses. Am J Pathol 1998; 153:1731–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kleter B, van Doorn LJ, Schrauwen L, et al. Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J Clin Microbiol 1999; 37:2508–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Agresti A. Categorical data analysis. 2nd ed Hoboken, NJ: John Wiley and Sons Inc, 2002. [Google Scholar]
- 21. Rothman KJ, Boice HD, Austin H.. Epidemiologic analysis with a programmable calculator. New ed Boston, MA: Epidemiology Resources, 1982. [Google Scholar]
- 22. Schiffman M, Wentzensen N, Wacholder S, Kinney W, Gage JC, Castle PE. Human papillomavirus testing in the prevention of cervical cancer. J Natl Cancer Inst 2011; 103:368–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rodríguez AC, Schiffman M, Herrero R, et al. Rapid clearance of human papillomavirus and implications for clinical focus on persistent infections. J Natl Cancer Inst 2008; 100:513–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rodríguez AC, Schiffman M, Herrero R, et al. Longitudinal study of human papillomavirus persistence and cervical intraepithelial neoplasia grade 2/3: critical role of duration of infection. J Natl Cancer Inst 2010; 102:315–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kreimer AR, Herrero R, Sampson JN, et al. Evidence for single-dose protection by the bivalent HPV vaccine-review of the Costa Rica HPV vaccine trial and future research studies. Vaccine 2018; 36:4774–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schiffman M, Boyle S, Raine-Bennett T, et al. The role of human papillomavirus genotyping in cervical cancer screening: a large-scale evaluation of the cobas HPV test. Cancer Epidemiol Biomarkers Prev 2015; 24:1304–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schiffman M, Doorbar J, Wentzensen N, et al. Carcinogenic human papillomavirus infection. Nat Rev Dis Primers 2016; 2:16086. [DOI] [PubMed] [Google Scholar]
- 28. Demarco M, Carter-Pokras O, Hyun N, et al. Validation of a human papillomavirus (HPV) DNA cervical screening test that provides expanded HPV typing. J Clin Microbiol 2018; 56:e01910–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.